Abstract
In many cases of familial amyotrophic lateral sclerosis (ALS), mutant forms of the Cu,Zn superoxide dismutase protein (SOD1) misfold and aggregate in motor neurons. Monomers of the normally homodimeric SOD1 have been found in patient tissue, presymptomatic mouse models of ALS, and in vitro misfolding assays which suggests that monomerization might be an early step in the pathological SOD1 misfolding pathway. In this study, we targeted the dimer interface with small molecules that might act as chemical chaperones to stabilize the native dimer and prevent downstream misfolding and aggregation. We performed a computational screen with a library of ~4400 drugs and natural compounds that were docked to two pockets around the SOD1 dimer interface. Of the resultant hits, seven were tested for misfolding and aggregation inhibition activity with A4V mutant SOD1. Quercitrin, quercetin-3-β-d-glucoside (Q3BDG), and, to a markedly lesser extent, epigallocatechin gallate (EGCG) were found to combat misfolding and aggregation induced by hydrogen peroxide, a physiologically relevant stress, as assessed by a gel-based assay and 8-anilinonaphthalene-1-suflonic acid (ANS) fluorescence. Isothermal titration calorimetry (ITC) and a colourimetric assay determined that these molecules directly bind A4V SOD1. Based on these findings, we speculate that quercitrin and Q3BDG may be potential therapeutic inhibitors of misfolding and aggregation in SOD1-associated ALS.
Keywords: amyotrophic lateral sclerosis; chemical chaperones; Cu,Zn superoxide dismutase; native-state stabilizers; protein misfolding
Introduction
Amyotrophic lateral sclerosis (ALS) is a late-onset neurodegenerative disease in which motor neurons die, leading to progressive paralysis and death. Although the large majority of cases are sporadic (sALS) with no known cause, ~10% are familial (fALS) where mutations in specific proteins are associated with the disease (Majoor-Krakauer et al., 2003). Of these familial cases, ~10–20% of them involve mutations in the Cu,Zn superoxide dismutase protein (SOD1) (Rosen et al., 1993). In SOD1 ALS, the SOD1 protein misfolds and aggregates primarily in motor neurons (Kato et al., 2001). The disease, however, does not seem to be caused by the loss of SOD1 function. ALS patients have mutant protein present from birth, thus one would expect the disease to start earlier in life if mutation caused a loss of function. In fact, SOD1 knock-out mice do not develop ALS-like symptoms (Reaume et al., 1996). On the contrary, it is sufficient to overexpress the ALS mutant protein in mice alongside their endogenous wild-type SOD1 for them to model the disease (Gurney et al., 1994; Wong et al., 1995). Considering that many of the ALS mutants retain activity similar to wild-type (Borchelt et al., 1994), it is well established that SOD1 mutants cause ALS through a gain of toxic function rather than a loss of its antioxidant activity.
It is not clear, however, what this toxic function is nor what conformations of the protein cause disease. While investigating this, our lab previously identified that monomerization of the native homodimer is an early and key step in SOD1 oxidation-induced misfolding and aggregation (Rakhit et al., 2004). This led to our development of a conformation-specific antibody that would only bind the SOD1 exposed dimer interface of the misfolded protein (SEDI antibody) which was used to confirm the presence of this monomeric species in SOD1 ALS patient spinal cords (Rakhit et al., 2007). If monomerization is an early step in the misfolding process, one would expect that it would be present prior to symptom onset and, indeed, in SOD1 ALS mouse model spinal cords stained with SEDI, SOD1 monomers begin to appear at 63 days of age and continue to be present when the initial limb weakness appears at 100 days (Rakhit et al., 2007).
In an effort to further map the SOD1 misfolding pathway, our lab developed a battery of assays to kinetically monitor the release of copper and zinc ions, dimer dissociation, and changes in the beta-barrel conformation while the protein was unfolded in chemical denaturant. Although monomerization was still found to be an early unfolding step, an increased tendency to release copper ions was common among ALS-causing mutant SOD1. The apparent effect, then, of the ALS mutations tested is that, if a mutant SOD1 molecule were to misfold, it would have a significantly higher probability to populate a monomeric, copper-deficient intermediate (Mulligan et al., 2008; Ip et al., 2011). This is curious because fully folded and metalated SOD1 is an extraordinarily stable protein with a Tm of 93°C (Stathopulos et al., 2003), activity in 10 M urea or 4% SDS, and protease resistance (Forman and Fridovich, 1973). Many SOD1 mutants retain similarly high stability while others are seemingly more stable (Rodriguez et al., 2002; Shaw and Valentine, 2007). Combined with the fact that most ALS cases occur over the age of 40, this raises the possibility that SOD1 mutations make the protein vulnerable to some additional stress that causes misfolding.
One candidate that might induce misfolding is oxidative stress. ALS patient tissue and animal disease models both show increased oxidative damage of proteins, lipids, and DNA in disease-specific sites, namely motor neurons in the spinal cord (Ferrante et al., 1997; Andrus et al., 1998). SOD1 disease-causing mutants have been found to be more susceptible to oxidative damage which causes monomerization, misfolding, and aggregation (Rakhit et al., 2002). Motor neurons are also more active, thus their mitochondria produce more reactive oxygen species (ROS) than other cell types. In fact, a positive feedback loop might occur where misfolded SOD1 associates with mitochondrial outer membranes in diseased motor neurons causing dysfunction and increased ROS generation (Sasaki and Iwata, 1996; Velde et al., 2008). SOD1 itself faces an occupational hazard since it interacts with ROS as both the reactant (superoxide) and product (H2O2) of its catalytic activity. Combined with the higher concentration and half-life of SOD1 in motor neurons (Pardo et al., 1995), there is a greater possibility of SOD1 accumulating oxidative damage which causes misfolding uniquely in motor neurons rather than in any other cell type.
With these insights into the SOD1 misfolding pathway and possible damaging agents, this current study is an attempt to rationally identify small molecules that might stabilize the SOD1 dimer interface to prevent oxidation-induced monomerization and downstream misfolding and aggregation. Currently, there are no effective treatments for any form of ALS: the only drug in use is riluzole which extends patient lives by ~3–6 months with little increase in quality of life (Lacomblez et al., 1996). In an effort to remedy this situation, we investigated potential chemical chaperones by computationally docking a library of ~4400 compounds to two regions around the SOD1 dimer interface. Of the resultant hits, seven were selected for in vitro testing on the SOD1 dimer interface mutant A4V, the most common ALS-causing SOD1 mutation found in North America (Juneja et al., 1997), treated with hydrogen peroxide (H2O2), and their abilities to inhibit aggregation and misfolding were assessed using gel and fluorescence-based assays. Two compounds, quercitrin and quercetin 3-β-d-glucoside (Q3BDG), were found to possess these qualities while another related molecule, epigallocatechin gallate (EGCG), inhibited misfolding in a limited fashion. The binding strengths of these molecules with SOD1 were measured which also confirmed that these drugs were directly interacting with the protein.
Materials and methods
Reagents
All small molecules experimentally tested were purchased from Santa Cruz Biotechnology Inc. (CA, USA) with the exceptions of chenodeoxycholic acid which was purchased from Sigma-Aldrich (MO, USA). Stock solutions of each compound were prepared by dissolving each small molecule in 100% anhydrous ethanol and storing solutions at −20°C. Protein concentrations were measured using the bicinchoninic acid (BCA) assay (using bovine serum albumin as a standard), and the reagents were purchased from Thermo Scientific (MA, USA). All other reagents were purchased from Sigma-Aldrich (MO, USA) unless otherwise specified.
In silico docking of small molecules to SOD1
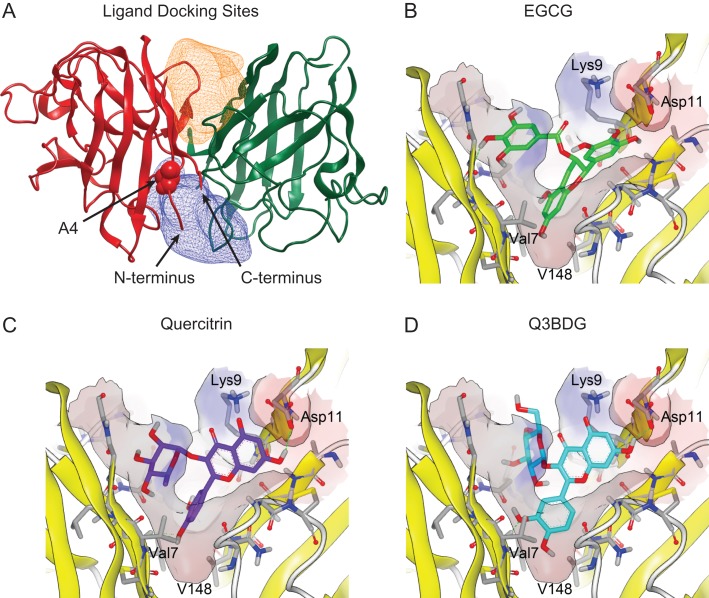
The in silico docking screen was done using OpenEye software (OpenEye Scientific Software Inc., Santa Fe, NM, USA, www.eyesopen.com). The receptors were generated from the SOD1 crystal structure (PDB ID: 1SPD) using the OpenEye GUI program ‘make_ receptor’. One receptor box of dimensions 16 Å × 21 Å × 16 Å encompassed Pocket 1 (Fig 1A, orange) and the other receptor box of dimensions 26 Å × 22 Å × 14 Å encompassed Pocket 2 (Fig 1A, blue). The ligand database was SWEETLEAD, a database of ~4400 chemical structures of approved drugs, herbal isolates, and regulated chemicals (Novick et al., 2013). Up to 200 three-dimensional conformers were generated for each chemical structure using OpenEye OMEGA (v2.4.6, 2013) (Hawkins et al., 2010). The conformers were then docked to the receptor using OpenEye FRED (v3.0.1, 2013) (McGann, 2011), which uses an exhaustive search algorithm to dock each rigid conformer into the rigid receptor. Docking poses were scored using Chemgauss4, a Gaussian function that sums the pairwise interactions between the docked ligand and protein receptor (McGann et al., 2003). The top-scoring pose for each of the top 50 ligands was examined using OpenEye VIDA (v4.2.1, 2013).
Fig. 1.
Computer docking of drugs into the SOD1 structure. The structure of wild-type SOD1 (PDB ID: 1SPD) is shown with the simulated regions highlighted. (A) The constructed binding pockets are displayed as meshes that encompass the volume that drugs were docked into. ‘Pocket 1’ is on top of the depicted dimer interface while ‘Pocket 2’ is on the bottom. These two regions around the dimer interface were isolated from this structure using the OpenEye GUI program ‘make_receptor’. The alanine at residue four for the subunit on the left is shown as space-filling balls and labeled. The N- and C-termini of this left subunit are indicated with arrows. (B–D) Small molecules from the SWEETLEAD database were allowed to dock into these regions using OpenEye FRED, and their binding energies were evaluated using Chemgauss4 scoring. The predicted binding of EGCG, quercitrin, and Q3BDG to Pocket 1 are shown in a similar orientation as the overall SOD1 molecule depicted in panel (A). Structures were rendered using VIDA 4.3.0.4. The predicted binding of Nice, BPP, CDCA, and HupA are shown in Supplementary Figure 5.
Cloning and construction of Escherichia coli expression vector for A4V SOD1
Our lab had previously cloned a (His)6-tagged Venus yellow fluorescent protein (vYFP) into pET23d vector DNA (Arslan and Chakrabartty, 2009). We have used the same techniques to clone (His)6-vYFP and a TEV cleavage site into other DNA vectors including pET30a. The cDNA encoding the unaltered wild-type human SOD1 protein (a gift from Dr Janice Robertson) was amplified by PCR using two primers that included the restriction sites BamHI and XhoI:
SOD1 BamHI (+)(5′-CGCGGATCCGCGACGAAGGC-3′)
SOD1 XhoI (–)(5′-GTAATTGGGATCGCCCAAACTCGAGCGGT-3′).
The SOD1 PCR product was purified using a PCR purification kit (Qiagen, Limburg, Netherlands) and digested with BamHI and XhoI. This product was further purified and ligated into our pET30a-(His)6-(vYFP)-(TEV cleavage site) construct that was similarly digested with BamHI and XhoI restriction enzymes and purified. The resulting pET30a-(His)6-(vYFP)-(TEV cleavage site)-SOD1 sequence was verified by DNA sequencing from ACGT Corporation (Ontario, Canada). All DNA restriction enzymes were purchased from New England Biolabs (MA, USA).
The A4V SOD1 mutation was created using mutagenic primers to introduce the desired mutation ((+) 5′-TCCGCGACAAGGTCGTGTGCGTCCTGAAG-3′ and (–) 5′-CTTCAGGACGCACACGACCTTCGTCGCGGA-3′). PCR was performed on our pET30a construct to introduce the mutation using Pfu polymerase and following the manufacturer's protocol (Thermo Scientific, MA, USA). The mutagenesis was confirmed by complete sequencing of the SOD1 insert. The final sequence codes for the expression of a (His)6-(vYFP)-(TEV cleavage site)-A4V SOD1 fusion protein, referred to as YFP-A4V SOD1.
A4V SOD1 protein expression and purification
YFP-A4V SOD1 was expressed in Escherichia coli strain BL21. Bacteria were grown in Luria broth (LB – Novagen, WI, USA) and protein expression was induced at an OD600nm reading of 0.6 with 0.5 mM isopropyl β-d-thiogalactopyranoside (IPTG – Invitrogen Life Technologies, CA, USA). At the time of induction, the culture was supplemented with CuCl2 (150 μM) and ZnCl2 (150 μM) and shaken overnight at 15°C. Bacteria were harvested by centrifugation (5000g for 15 min at 4°C) and resuspended in lysis buffer containing 300 mM NaCl, 100 mM KCl, 50 mM Tris (pH 7.4), 2 mM β-mercaptoethanol, and EDTA-free protease inhibitors (Roche, Basel, Switzerland). Cells were lysed using an EmulsiFlex-C5 homogenizer (Avestin Inc., Ottawa, Canada), and the sample was centrifuged for 45 min at 35 000 rpm using an L8-70M ultracentrifuge (Beckman Coulter Canada LP, Ontario, Canada). The supernatant was passed through a gravity column of Ni-NTA agarose (Qiagen, Limburg, Netherlands), and the column was washed three times with lysis buffer containing 40 mM imidazole. The fusion protein was eluted off of the column with lysis buffer containing 250 mM imidazole and dialyzed overnight against 20 mM HEPES (pH 7.5). The fusion protein sample was cleaved using TEV protease (20:1 fusion protein:TEV) at room temperature overnight and the sample was again passed through a Ni-NTA column. The purified cleaved SOD1 was collected in the flow-through, spin-concentrated, quantified using the BCA assay, and then metalated.
Metalation of wild-type and A4V SOD1
Properly folded and metalated SOD1 binds one copper and zinc ion per subunit. Copper and zinc ions were added to purified SOD1 using the following procedures. SOD1 was dialyzed against three molar equivalents of CuCl2 in 10 mM sodium phosphate buffer (pH 3.6) for 48 h at 4°C. This low pH step prevents metal ions from occupying the zinc binding site, thus selectively placing copper ions in the copper site (Pantoliano et al., 1982; Lyons et al., 2000). Sample was dialysed into 20 mM HEPES (pH 7.5) containing a three-fold molar excess of ZnCl2 with repeated buffer changes to quickly remove any remaining CuCl2. Free copper ions had to be removed rapidly at pH 7.5 to prevent copper-catalyzed aggregation of SOD1 possibly through the formation of aberrant disulfide bonds. SOD1 was allowed to bind zinc ions for 48 h at 4°C. The excess free metal ions were removed by multiple cycles of dialysis in 20 mM HEPES (pH 7.5).
The amount of metal ions bound to SOD1 was measured using the metal indicator PAR as previously described (Crow et al., 1997). Briefly, 4.25 μM SOD1 was denatured in 6 M guanidine hydrochloride (GuHCl) in 20 mM HEPES (pH 7.5) for 30 min, and the metal status was detected using 100 μM PAR in the presence and absence of NTA. As PAR chelates divalent metal ions (both copper and zinc), its absorbance spectrum changes which can be monitored at 500 nm. NTA, however, chelates zinc ions with a stronger affinity than PAR, thus, in the presence of NTA, the remaining PAR absorbance will only reflect copper ion binding. The metal ion concentrations were interpolated from standard curves of 0–14 μM for each metal ion where a concentration of 8.5 μM was expected for both metal ions if 100% of the metal binding sites were populated. A4V SOD1 samples typically had >90% metal occupancy of both the zinc and copper sites as measured by PAR absorbance with a 1:1 ratio of zinc to copper.
Oxidation-induced aggregation of SOD1 & inhibition with small molecules
A4V SOD1 (30 μM) was misfolded using 5 mM H2O2 in the presence of 100 μM EDTA and 20 mM HEPES buffer, pH 7.5, at 37°C with or without 500 μM small molecules or 10 mM mannitol as an antioxidant control. Due to solubility issues, small molecules were solubilized in 99% ethanol, thus SOD1 was also exposed to 4% ethanol. As such, 4% ethanol was added to control samples.
Popular methods to quantify aggregate formation include right-angle light scattering and measuring turbidity. Unfortunately, many of these small molecules absorb light at wavelengths that interfere with these techniques, thus a gel-based assay was developed. A4V SOD1 was treated with H2O2 for 0–72 h and spun in a centrifuge for 30 min at 14 000 rpm (18 407 rcf). Supernatants were immediately added to Laemmli sample buffer containing 2% β-mercaptoethanol which were then boiled for 5 min. Samples were run on 15% SDS-PAGE tris-glycine gels and visualized using Coomassie Brilliant Blue. Gels were photographed and color images were converted to gray scale using Photoshop Creative Suite 5 (Adobe Systems Incorporated, CA, USA). ImageJ software (NIH, Maryland, USA) was used to subtract background and quantify the monomeric SOD1 bands. Band densities were normalized using Equation (1) where D is the density of the band, DNoH2O2 is the density of the untreated control, and DBlank is the density of an empty region of the gel. The general two exponent kinetic equation (2) was used in OriginPro 9.0 software (OriginLab, Massachusetts, USA) to fit the curves generated from the band densities where k1 and k2 are rate constants, A and B are the fractional contribution of each rate constant to the overall aggregation phenomenon, and t is time.
| 1 |
| 2 |
Oxidation-induced misfolding inhibition
A4V SOD1 was misfolded under similar conditions as in the aggregation inhibition assay except that 5 μM SOD1 was used, treatment was at room temperature for 24 h, and the chemical chaperone concentration varied between 0 and 100 μM. Mannitol, an antioxidant control, was also assayed with concentrations between 0 and 10 mM. After 24 h, 100 μM 8-anilinonapthalene-1-sulfonic acid (ANS) was added to each sample just prior to reading its fluorescence. Fluorescence was read using excitation 375 nm, emission 490 nm on a QM-1 fluorometer (Photon Technology International, New Jersey, USA). Average values of 60 reads within a minute were used for each sample of the misfolding curves. The quercitrin, Q3BDG, and EGCG curves were fit using OriginPro 9.0 to Equation (3) where Fmax and Fmin are the maximal and minimal fluorescence (no drug and no H2O2 samples, respectively), [D] is the drug concentration, EC50 is the effective concentration 50% value, p is a curve steepness coefficient, m is the slope of a line, and b is a constant. A linear component was required due to the absorbance properties of quercitrin, Q3BDG, and EGCG that interfered with the ANS fluorescence. This linear term was validated by adding increasing amounts of each molecule to ANS bound to 5 μM bovine serum albumin (BSA) which has natively exposed hydrophobic pockets. The curves were then normalized using these parameters.
| 3 |
Isothermal titration calorimetry
For ITC, 500 μM of small molecule dissolved in DMSO was titrated into 2.2 ml of 50 μM A4V SOD1 (100 μM monomeric SOD1) in 20 mM HEPES buffer, pH 7.5 at a rate of 15 μL every 3 min. A drug-into-buffer control was also run and subtracted from the protein data (Supplementary Fig. 1). Energy changes were recorded using a MicroCal VP-ITC (Malvern Instruments Inc., UK). ITC thermograms were fit to a model for a single set of identical binding sites using Equation (4) where V0 is the volume of the ITC chamber, i is the injection number, dVi is the change in volume per injection, Q(i) is the total heat content of the solution in V0 at injection i, and ΔQ(i) is the change in total heat of the solution between the i and i – 1 injections. Q(i) and Q(i – 1) were calculated using Equation (5) where i is the injection number, Q(i) is the total heat content of the solution in the ITC chamber at injection number i, n is the number of binding sites per protein molecule, Mi is the total protein concentration present in the ITC chamber, ΔH is the molar heat of ligand binding, V0 is the chamber volume, Xi is the total ligand concentration present in the ITC chamber, and Ka is the association equilibrium constant. Q(i) and Q(i – 1) calculated from Equation (5) were inserted into Equation (4) during the fitting process.
| 4 |
| 5 |
Further details of these equations can be found in the MicroCal ITC Data Analysis in Origin: Tutorial Guide (Version 5.0, 1998) and at reference (Freire et al., 1990).
The fitting of the ITC thermograms was done using nonlinear least squares regression in Origin software using proprietary macros provided by Malvern. Initial guesses for appropriate values of n, Ka, and ΔH were automatically generated, and iterative Levenberg–Marquadt nonlinear least squares fitting was used to modify the variables until there were no further improvements in the difference between the fit generated and empirically-derived curves. Kd was calculated by taking the inverse of Ka. The uncertainty of the fitted parameters was estimated by the fitting software.
Colourimetric absorbance assay
Both quercitrin and Q3BDG change color when added to SOD1, going from a pale yellow to a markedly brighter shade. This phenomenon was used to quantify the binding strength of these compounds to SOD1. Absorbance spectra (Supplementary Fig. 2) were generated with 15 μM quercitrin or Q3BDG in the presence or absence of 50 μM SOD1. Absorbance was read using a SpectraMax M5 plate reader (Molecular Devices, California, USA). To determine the Kd of quercitrin and Q3BDG, 0–50 μM SOD1 was added to a constant 25 μM of drug. This was done to avoid changes in absorbance caused merely by drug concentration. Absorbance of each sample was read at 420 nm on the SpectraMax M5 plate reader. Resultant curves were fit and normalized using Equation (6) in OriginPro 9.0 software where B is the asymptotic maximum, Kd is the dissociation constant, [SOD1] is the concentration of SOD1, and C is the minimal absorbance.
| 6 |
Results
Computational docking of small molecules around the SOD1 dimer interface
To target the dimer interface for drug binding, we selected two regions around the interface that appeared to have space for small molecules to stably bind. These volumes can be seen in Fig. 1A as Pocket 1 (orange) and 2 (blue). The SWEETLEAD database of FDA-approved and natural molecules were docked into both of these pockets, and the predicted binding strengths were evaluated by Chemgauss4 score. Of the hits generated from this database, we selected seven small molecules to test in vitro for activity based on predicted binding strength and commercial availability. These small molecules were EGCG, quercitrin, quercetin 3-β-d-glucoside (Q3BDG), nicergoline (Nice), benzopurpurine 4B (BPP), chenodeoxycholic acid (CDCA), and huperzine A (HupA) (Chemgauss4 scores of these compounds in Supplementary Table I; Chemguass4 scores across the top 500 compounds in Supplementary Fig. 3; Chemical structures in Supplementary Fig. 4). EGCG, quercitrin, and Q3BDG were predicted to bind to Pocket 1 (Fig. 1B–D) whereas the other four bind to Pocket 2 (Supplementary Fig. 5).
Quercitrin and Q3BDG inhibit A4V SOD1 oxidation-induced aggregation
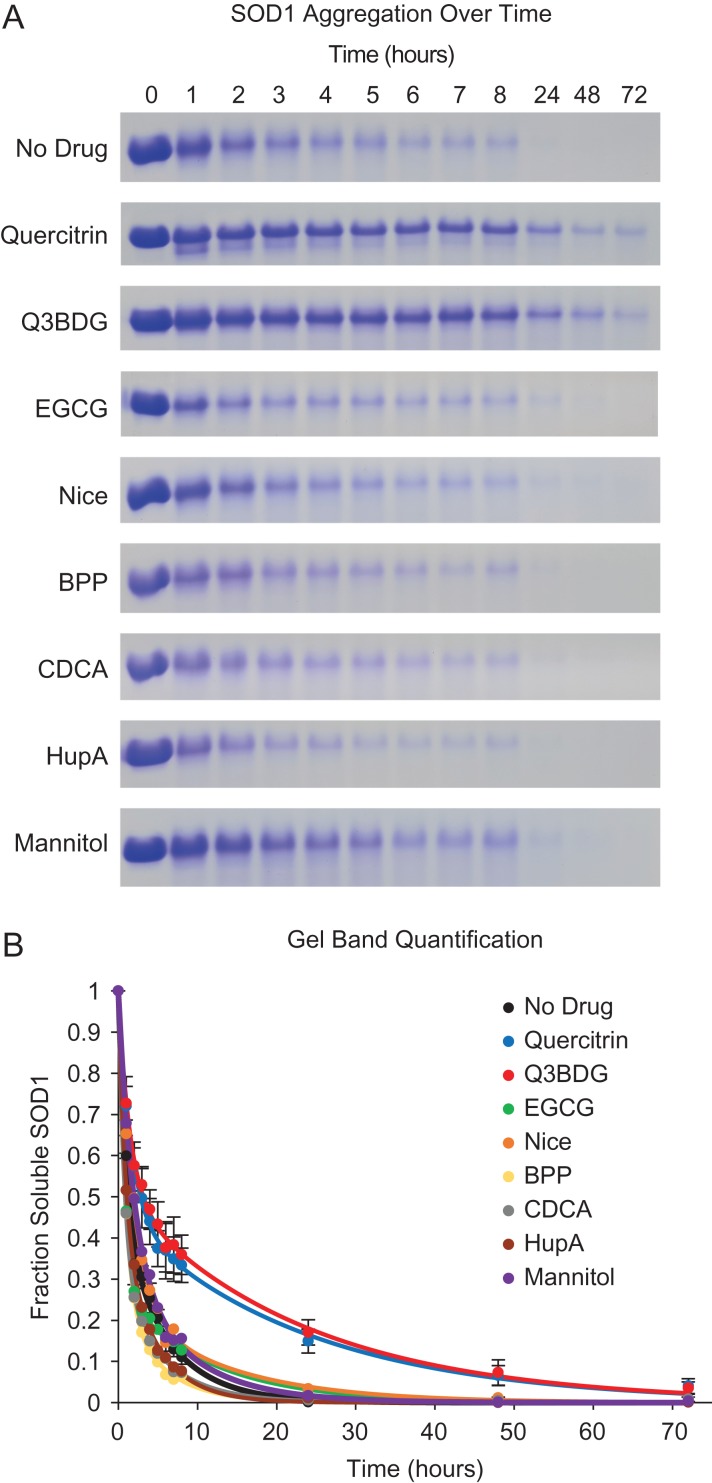
A4V SOD1 was treated with H2O2 at 37°C over a period of up to 3 days in the presence and absence of small molecules including mannitol as a general antioxidant control. Following the incubation time, samples were centrifuged, and the supernatants were loaded onto SDS-PAGE gels to detect the remaining soluble protein (Fig. 2; representative full gels in Supplementary Fig. 6). Aggregation was detected in this way instead of more typical absorbance-based assays because many of the small molecules absorb at UV wavelengths which would complicate the interpretation. Band quantities were normalized to the 0 h untreated control of their respective gels as well as any remaining background signal according to Equation (1). The amount of soluble SOD1 sharply decreased over the first 8 h with typically less than 5% remaining by 24 h of treatment. The noticeable exceptions to this were treatment with quercitrin or Q3BDG which retained significantly more soluble SOD1 over time. At 8 h, on average, quercitrin and Q3BDG had 33 ± 4% and 36 ± 5% soluble SOD1 remaining, respectively, compared to 11 ± 2% for the no drug control. By 24 h, quercitrin and Q3BDG had 15 ± 3% and 17 ± 3% soluble SOD1 left, respectively while the no drug control was under 1%. Mannitol, an effective neutralizer of radicals generated by H2O2 (Smirnoff and Cumbes, 1989), was tested as an antioxidant control. It had no effect on SOD1 aggregation. Curves were fit to a general two exponential kinetic equation as shown in Equation (2).
Fig. 2.
A4V SOD1 oxidation-induced aggregation and its inhibition with quercitrin and Q3BDG. A4V SOD1 aggregation was induced with H2O2 at 37°C over various amounts of time in the presence and absence of small molecules. Samples were then centrifuged, and the supernatants were run on SDS-PAGE. (A) Representative SDS-PAGE gel cut-outs stained with Coomassie Brilliant Blue are shown. The bands displayed correspond to monomeric SOD1. (B) The band densities were quantified using ImageJ software. These values were normalized to the untreated (0 time point) and no drug controls as shown in Equation (1). These band density curves were fit to Equation (2) using Origin 9 software. The graph depicts the average of three replicates for each sample. For image clarity, error bars representing the standard error of the mean are only shown for the no drug, quercitrin, and Q3BDG samples.
Quercitrin, Q3BDG, and EGCG inhibit A4V SOD1 oxidation-induced misfolding
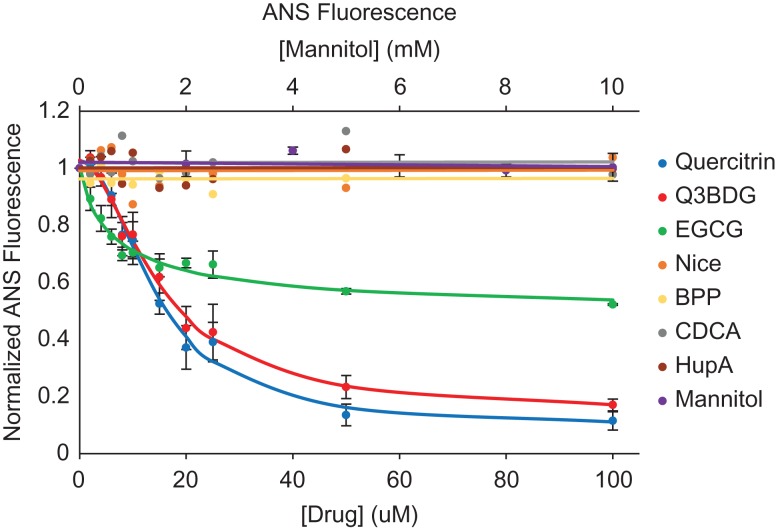
The dye anilinonaphthalene-8-sulfonic acid (ANS) binds to exposed hydrophobic pockets on misfolded proteins, which increases its fluorescence, allowing detection of misfolding intermediates and aggregates (Semisotnov et al., 1991; Kundu and Guptasarma, 2002). We used a lower concentration of SOD1 and lower incubation temperature to prevent the formation of large aggregates which could scatter light at the excitation wavelength of 375 nm and cause non-uniform excitation across samples. Quercitrin and Q3BDG almost completely prevent ANS-detectable H2O2-induced misfolding of A4V SOD1 in a dose-dependent manner while EGCG prevents about half (Fig. 3). To quantify this misfolding inhibition, the curves were fit to a Hill-like equation (Equation (3)) to derive effective concentration 50% (EC50) values (Table I). The EC50s of quercitrin, Q3BDG, and EGCG were 14.38 ± 1.11, 15.26 ± 1.25, and 7.25 ± 2.34 μM, respectively. Mannitol, an antioxidant control, had no effect on SOD1 misfolding.
Fig. 3.
A4V SOD1 oxidation-induced misfolding and its inhibition with quercitrin, Q3BDG, and EGCG. A4V SOD1 was misfolded using H2O2 at room temperature for 24 h in the presence and absence of increasing concentrations of the various drugs. Following this time period, ANS was added as a fluorescent dye that binds to exposed hydrophobic pockets where higher fluorescence indicates misfolding. The resultant fluorescence was normalized to the highest (no drug) and lowest (no H2O2) signals using Equation (3). The averages of three replicates are graphed here while error bars representing the standard error of the mean are shown for quercitrin, Q3BDG, EGCG, and mannitol. The EC50 values can be found in Table I.
Table I.
EC50 and Kd values determined for quercitrin, Q3BDG, and EGCG
| Compound | EC50 (μM) | Kd (ITC – μM) | Kd (colourimetric – μM) |
|---|---|---|---|
| Quercitrin | 14.38 ± 1.11 | 34 ± 23 | 27.70 ± 0.31 |
| Q3BDG | 15.26 ± 1.25 | 35 ± 20 | 23.65 ± 0.47 |
| EGCG | 7.25 ± 2.34 | 3 ± 1 |
The error indicated for EC50 and Kd (colourimetric) are the standard error of the mean of three replicates. The error for Kd (ITC) was estimated by the curve fitting software.
Quercitrin, Q3BDG, and EGCG directly bind A4V SOD1
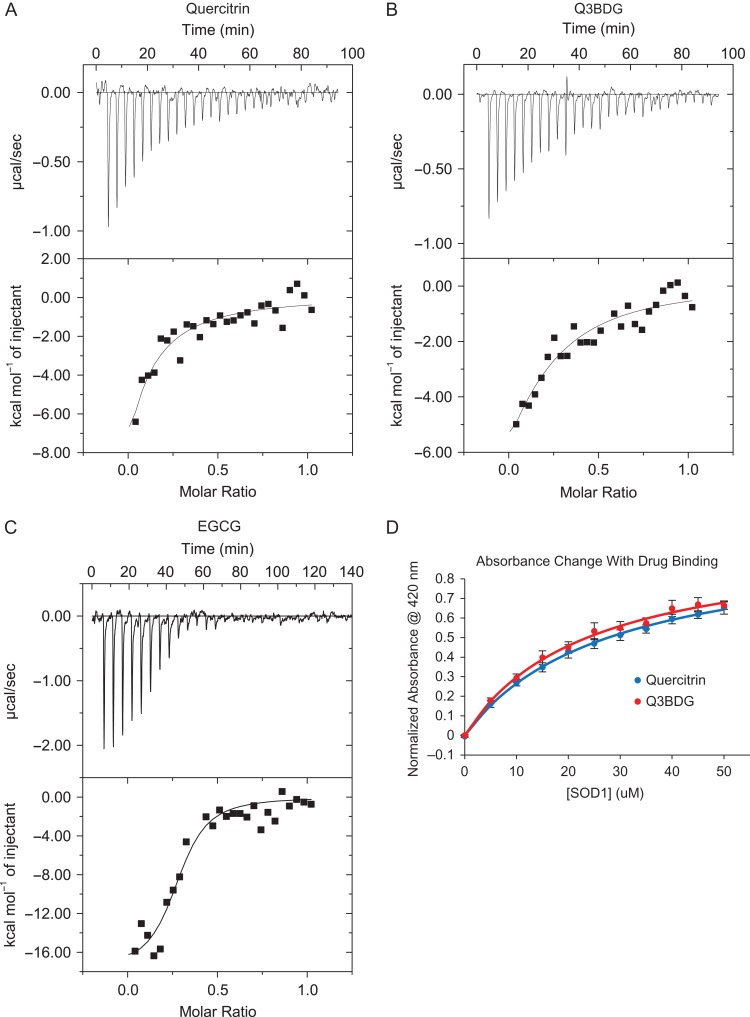
We used isothermal titration calorimetry (ITC) to demonstrate that quercitrin, Q3BDG, and EGCG bind directly to A4V SOD1. Quercitrin, Q3BDG, and EGCG were added by injection to A4V SOD1 using a MicroCal VP-ITC machine which recorded the changes in energy. The resultant titration and energy curves can be found in Fig. 4A–C. A drug-into-buffer control was subtracted from the curves of each drug (Supplementary Fig. 1). All three drug titration curves show an exothermic binding reaction and came to saturation, indicating a limited number of binding sites. In each case, saturation occurred at approximately a 1:2 drug:SOD1 molar ratio which suggests that the molecules are binding to the dimeric form of SOD1. The energy curves were fit to a model for a single set of identical binding sites to estimate the dissociation constants (Table I). For quercitrin, Q3BDG, and EGCG, the Kd values were 34 ± 23, 35 ± 20, and 3 ± 1 μM, respectively. Further parameters derived from the curve fits can be found in Supplementary Table II. The fits produced unrealistic values for the number of binding sites per molecule (n) and the molar heat of ligand binding (ΔH – see Equation (5)), and there were high estimated error values associated with the fit parameters. We believe this to be the case due to weak binding that is non-ideal for ITC analysis. Despite this, the derived Kd are similar to the ANS EC50 as well as the Kd determined in the following colourimetric experiments (Table I), thus, combined with the saturability of the curves, these ITC experiments demonstrate a quantifiable, direct interaction between these small molecules and A4V SOD1.
Fig. 4.
Binding affinity of drugs to A4V SOD1 determined by ITC and absorbance. To confirm whether quercitrin, Q3BDG, and EGCG directly interact with A4V SOD1, their binding strengths were evaluated. (A–C) Drug was added by injection to A4V SOD1, and the changes in energy were recorded using a MicroCal VP-ITC as shown in the top graph of each pair. The energy required per mole of drug versus the molar ratio of drug to protein is depicted in the bottom graph of each pair. These curves were fit to a model for a single set of binding site using Origin software macros provided by Malvern Instruments to derive the dissociation constants which can be found in Table I. A drug-into-buffer control was subtracted from each sample (Supplementary Fig. 1). (D) It was observed that both quercitrin and Q3BDG undergo prominent color shifts from pale yellow to a bright yellow when added to A4V SOD1. These absorbance shifts (Supplementary Fig. 2) were used to characterize their binding affinities by adding increasing amounts of SOD1 to each drug and measuring the absorbance at 420 nm. Each point represents the average of three replicates, and error bars depict the standard error. Curves were fit and normalized using Equation (6) to determine the dissociation constants shown in Table I.
It was noticed that both quercitrin and Q3BDG go from a pale yellow to a bright yellow appearance when added to SOD1 (Supplementary Fig. 2). We used this phenomenon as another means to quantify their binding strength to SOD1 by monitoring the absorbance at 420 nm as we added increasing amounts of SOD1 to a constant amount of drug which is shown in Fig. 4D. Absorbance curves were fit and normalized to a binding equation (Equation (6)) which yielded the Kd values (Table I). The Kd values for quercitrin and Q3BDG were 27.70 ± 0.31 and 23.65 ± 0.47 μM, respectively.
Discussion
Our results demonstrate that treatment with H2O2 induces misfolding and aggregation of A4V SOD1 that can be inhibited by the presence of quercitrin, Q3BDG, and, to a limited extent, EGCG. Each of these polyphenolic compounds comes from the family of naturally occurring molecules known as flavonoids that are produced by plants as secondary metabolites (Manach et al., 2004). There has been increasing interest in flavonoids due to evidence that members of this family may have antioxidant, anti-inflammatory, and signaling effects which may be potentially useful for a variety of conditions such as heart disease (Jiang et al., 2015), diabetes (Babu et al., 2013), arthritis (Gardi et al., 2015), and neurodegenerative diseases like Alzheimer's (Francioso et al., 2015) and Parkinson's (Dutta and Mohanakumar, 2015).
Since quercitrin, Q3BDG, and EGCG have antioxidant properties (Rice-Evans, 1999; Wagner et al., 2006), we included an antioxidant control in the form of mannitol which is also known for its potent ROS scavenging, particularly of the hydroxyl radical that might form from H2O2 (Smirnoff and Cumbes, 1989). Mannitol was present up to 10 mM versus 5 mM H2O2 but had very little effect on both the aggregation and misfolding processes. EGCG, a strong antioxidant, also had a minimal effect on the aggregation process while inhibiting only half of the ANS-detectable misfolding. Given these antioxidant results, we can conclude that the ROS generation and oxidative damage being done to SOD1 happen in a highly specific manner to this protein rather than by injury to random sites throughout the protein's structure. This is consistent with our previous characterizations of oxidative damage to SOD1 where only histidines in the active site were modified by H2O2 or metal catalyzed oxidation (MCO) which were sufficient to induce metal ion release, large conformational changes, and aggregation (Rakhit et al., 2002; Mulligan et al., 2012). Other groups that studied bovine SOD1 had similar observations where H2O2 treatment modified active site histidines to 2-oxo-histidine facilitated by the active site copper ion through a reversal of the enzyme's activity to generate superoxide and through a Fenton-like reaction which produces hydroxyl radicals (Hodgson and Fridovich, 1975; Uchida and Kawakishi, 1994). These results from mannitol and EGCG are evidence that the inhibition of SOD1 misfolding and aggregation exhibited by quercitrin and Q3BDG are not through their antioxidant properties.
Our ITC and colourimetric results point to direct binding between the flavonoids and SOD1. The Kd values from these assays are in the μM range, which agree with the EC50 values from the ANS fluorescence assay. This suggests that quercitrin and Q3BDG are binding to and stabilizing the SOD1 native state. Although EGCG seems to bind tighter than the other flavonoids, it does not seem to have any effect on aggregation and only a moderate impact on misfolding. This is curious given the structural similarities between these three molecules. To gain some insights into why this effective difference exists, we can look at the predicted binding structures (Fig. 1B–D). Overall, quercitrin and Q3BDG share similar orientations with their fused rings pointing out of the dimer interface next to Lys9 and Asp11. In contrast, EGCG's fused ring is inserted between the SOD1 subunits which leads to closer contact with Val7 and Val148 on both subunits. Quercitrin and Q3BDG have their dihydroxy phenyl groups in this position which does not fill this pocket as fully as EGCG's fused ring. This difference in fit could account for the tighter binding of EGCG to SOD1 compared to quercitrin and Q3BDG.
In spite of this tighter binding, EGCG was far less effective at inhibiting aggregation and misfolding than both quercitrin and Q3BDG. The SOD1 used in this study was the A4V mutant which has a weakened dimer interface. While the majority of the A4V structure is conserved with the wild-type protein, there is a shift in monomer-monomer orientation and increased disorder around the N-terminus where the mutation resides (Hough et al., 2004). A main difference between the predicted binding of EGCG and that of quercitrin and Q3BDG is interaction with the N-terminus. Both quercitrin and Q3BDG are predicted to hydrogen bond with the carboxylic acid group of Asp11 of one subunit while their fused rings come into hydrophobic contact with the aliphatic portion of Lys9’s side chain on that same subunit. Q3BDG is also predicted to hydrogen bond with the amine group of Lys9’s side chain. On the other subunit of the dimer, quercitrin has a similar hydrophobic interaction between its rhamnose sugar and Lys9. In contrast, EGCG lacks these N-terminal interactions. The predicted binding structures were generated using the wild-type structure which suggests that quercitrin and Q3BDG might bind A4V mutant SOD1 and induce a native-like conformation in the N-terminus.
It is possible, however, that these molecules bind to SOD1 in a completely different fashion than predicted. Another group had followed a similar methodology of computationally targeting the dimer interface in a region similar to our ‘Pocket 1’ (Fig. 1A) for small molecule binding and experimentally testing hits for activity against metal chelation-induced aggregation and guanidinium chloride equilibrium unfolding (Ray et al., 2005). Many of their hits that were most effective at inhibiting aggregation and unfolding had a uracil or aza-uracil base with variable single ring attachments. A different group attempted to co-crystallize these (aza-)uracil-based compounds with L38V mutant SOD1 but were unsuccessful. In an attempt to get some binding information, this group co-crystallized L38V SOD1 with a related molecule, uridine 5′-monophosphate (UMP), and found that it bound between the ‘electrostatic’ and ‘zinc binding’ loops near the active site which is on the opposite side of the protein to the dimer interface (Antonyuk et al., 2010). As such, structural studies of quercitrin and Q3BDG bound to SOD1 are required to truly determine their binding modes and mechanisms of aggregation and misfolding inhibition which would be useful for chemically modifying these molecules to improve their binding and efficacy. It should be noted that our hits are structurally distinct from these (aza-)uracil-based molecules as quercitrin and Q3BDG have fused rings, do not contain any nitrogen atoms, and are significantly larger than these compounds that they selected to improve upon (Nowak et al., 2010). Our method of inducing aggregation using oxidative stress instead of EDTA chelation may also be a more physiologically relevant and distinct mode of misfolding and aggregation that would require different families of molecules to inhibit.
Other groups have already found some success using this flavonoid family of molecules with ALS models, particularly EGCG. Seong-Ho Koh et al. found that pre-treatment of motor neuron-like cell cultures expressing either wild-type or G93A mutant SOD1 with EGCG granted modest protection from H2O2 treatment (Koh et al., 2004). This same group treated a G93A SOD1 mouse model of ALS with EGCG and showed that this molecule delayed disease onset, progression, and death by inhibiting apoptosis in the spinal cord (Koh et al., 2006). Concurrently, Zhihao Xu et al. also treated G93A SOD1 mice with EGCG and found a significant delay in disease onset and death with reduced spinal cord inflammation and apoptosis (Xu et al., 2006). In an effort to determine the mechanism of EGCG action, Jixu Yu et al. showed that it protected rat spinal cord neuron cultures from threohydroxyaspartate(THA)-induced glutamate excitotoxicity (Yu et al., 2010).
Our results would predict that quercitrin and Q3BDG would be far more effective than EGCG. To our knowledge, however, no one to date has published any studies testing quercitrin and Q3BDG with ALS proteins or models. In fact, there are very few studies on quercitrin and Q3BDG with neurodegenerative disease models in general. Quercitrin protected rat primary hippocampal neuron cultures from Aβ25–35 peptide toxicity, a model of Alzheimer's disease, in a manner that reduced oxidative damage (Rattanajarasroj and Unchern, 2010). In a rat pheochromocytoma cell (PC12) oxidative stress model of Parkinson's disease, Q3BDG significantly increased cell viability by increasing the activities of a number of antioxidant enzymes. Interestingly, this included the superoxide dismutase family (Magalingam et al., 2014).
Given the limited number of studies on these drugs along with our own positive results, more research on these compounds with respect to neurodegenerative diseases is highly warranted, particularly for SOD1 ALS. Here, we showed that quercitrin and Q3BDG can inhibit oxidation-induced misfolding and aggregation of A4V SOD1 through a direct interaction that might stabilize the native state of the protein. Although these flavonoids could potentially be tested as a treatment for ALS due to their high bioavailability (Hollman et al., 1995), these quercetin glycoside derivatives might be metabolized into other forms once absorbed in vivo (Morand et al., 2000). Even if they remained intact, their weak binding and interactions with fully folded SOD1 (which is ubiquitously expressed in all cell types) would possibly necessitate a fairly high dose to be effective. As such, modifications to these molecules or synthesis of novel compounds based on predicted and co-crystal structures should be attempted to improve their binding strength, aggregation and misfolding inhibition efficacy, and resistance to metabolic processes. These flavonoids and their derivatives could then be tested for activity in ALS animal models.
Supplementary Material
Supplementary data
Supplementary data are available at Protein Engineering, Design & Selection online.
Funding
This work was supported by the ALS Society of Canada. Philbert Ip was also supported by a Canadian Institutes of Health Research (CIHR) Training Grant in Protein Folding.
References
- Andrus P.K., Fleck T.J., Gurney M.E. and Hall E.D. (1998) J. Neurochem., 71, 2041–2048. [DOI] [PubMed] [Google Scholar]
- Antonyuk S., Strange R.W. and Hasnain S.S. (2010) J. Med. Chem., 53, 1402–1406. [DOI] [PubMed] [Google Scholar]
- Arslan P.E. and Chakrabartty A. (2009) Biochem. Cell. Biol., 87, 631–639. [DOI] [PubMed] [Google Scholar]
- Babu P.V.A., Liu D. and Gilbert E.R. (2013) J. Nutr. Biochem., 24, 1777–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchelt D.R., Lee M.K., Slunt H.S. et al. (1994) Proc. Natl. Acad. Sci. USA, 91, 8292–8296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow J.P., Sampson J.B., Zhuang Y., Thompson J.A. and Beckman J.S. (1997) J. Neurochem., 69, 1936–1944. [DOI] [PubMed] [Google Scholar]
- Dutta D. and Mohanakumar K.P. (2015) Neurochem. Int., 89, 181–190. [DOI] [PubMed] [Google Scholar]
- Ferrante R.J., Browne S.E., Shinobu L.A., Bowling A.C., Baik M.J., MacGarvey U., Kowall N.W., Brown R.H. and Beal M.F. (1997) J. Neurochem., 69, 2064–2074. [DOI] [PubMed] [Google Scholar]
- Forman H.J. and Fridovich I. (1973) J. Biol. Chem., 248, 2645–2649. [PubMed] [Google Scholar]
- Francioso A., Punzi P., Boffi A., Lori C., Martire S., Giordano C., D'Erme M. and Mosca L. (2015) Bioorg. Med. Chem., 23, 1671–1683. [DOI] [PubMed] [Google Scholar]
- Freire E., Obdulio L. and Straume M. (1990) Anal. Chem., 62, 950A–959A. [Google Scholar]
- Gardi C., Bauerova K., Stringa B. et al. (2015) Arch. Biochem. Biophys., 583, 150–157. [DOI] [PubMed] [Google Scholar]
- Gurney M.E., Pu H., Chiu A.Y. et al. (1994) Science, 264, 1772–1775. [DOI] [PubMed] [Google Scholar]
- Hawkins P.C.D., Skillman A.G., Warren G.L., Ellingson B.A. and Stahl M.T. (2010) J. Chem. Inf. Model., 50, 572–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson E.K. and Fridovich I. (1975) Biochemistry, 14, 5294–5299. [DOI] [PubMed] [Google Scholar]
- Hollman P.C., de Vries J.H., van Leeuwen S.D., Mengelers M.J. and Katan M.B. (1995) Am. J. Clin. Nutr., 62, 1276–1282. [DOI] [PubMed] [Google Scholar]
- Hough M.A., Grossmann J.G., Antonyuk S.V. et al. (2004) Proc. Natl. Acad. Sci. USA, 101, 5976–5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip P., Mulligan V.K. and Chakrabartty A. (2011) J. Mol. Biol., 409, 839–852. [DOI] [PubMed] [Google Scholar]
- Jiang W., Wei H. and He B. (2015) Thromb. Res., 135, 459–463. [DOI] [PubMed] [Google Scholar]
- Juneja T., Pericak-Vance M.A., Laing N.G., Dave S. and Siddique T. (1997) Neurology, 48, 55–57. [DOI] [PubMed] [Google Scholar]
- Kato S., Nakashima K., Horiuchi S. et al. (2001) Neuropathology, 21, 67–81. [DOI] [PubMed] [Google Scholar]
- Koh S.H., Kwon H., Kim K.S. et al. (2004) Toxicology, 202, 213–225. [DOI] [PubMed] [Google Scholar]
- Koh S.H., Lee S.M., Kim H.Y. et al. (2006) Neurosci. Lett., 395, 103–107. [DOI] [PubMed] [Google Scholar]
- Kundu B. and Guptasarma P. (2002) Biochem. Biophys. Res. Commun., 293, 572–577. [DOI] [PubMed] [Google Scholar]
- Lacomblez L., Bensimon G., Leigh P.N., Guillet P. and Meininger V. (1996) Lancet (London, England), 347, 1425–1431. [DOI] [PubMed] [Google Scholar]
- Lyons T.J., Nersissian A., Huang H., Yeom H., Nishida C.R., Graden J.A., Gralla E.B. and Valentine J.S. (2000) J. Biol. Inorg. Chem., 5, 189–203. [DOI] [PubMed] [Google Scholar]
- Magalingam K.B., Radhakrishnan A. and Haleagrahara N. (2014) BMC Res. Notes, 7, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majoor-Krakauer D., Willems P.J. and Hofman A. (2003) Clin. Genet., 63, 83–101. [DOI] [PubMed] [Google Scholar]
- Manach C., Scalbert A., Morand C., Rémésy C. and Jiménez L. (2004) Am. J. Clin. Nutr., 79, 727–747. [DOI] [PubMed] [Google Scholar]
- McGann M. (2011) J. Chem. Inf. Model, 51, 578–596. [DOI] [PubMed] [Google Scholar]
- McGann M.R., Almond H.R., Nicholls A., Grant J.A. and Brown F.K. (2003) Biopolymers, 68, 76–90. [DOI] [PubMed] [Google Scholar]
- Morand C., Manach C., Crespy V. and Remesy C. (2000) Free. Radic. Res., 33, 667–676. [DOI] [PubMed] [Google Scholar]
- Mulligan V.K., Kerman A., Ho S. and Chakrabartty A. (2008) J. Mol. Biol., 383, 424–436. [DOI] [PubMed] [Google Scholar]
- Mulligan V.K., Kerman A., Laister R.C., Sharda P.R., Arslan P.E. and Chakrabartty A. (2012) J. Mol. Biol., 421, 631–652. [DOI] [PubMed] [Google Scholar]
- Novick P.A., Ortiz O.F., Poelman J., Abdulhay A.Y. and Pande V.S. (2013) PLoS ONE, 8, e79568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak R.J., Cuny G.D., Choi S., Lansbury P.T. and Ray S.S. (2010) J. Med. Chem., 53, 2709–2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantoliano M.W., Valentine J.S., Mammone R.J. and Scholler D.M. (1982) J. Am. Chem. Soc., 104, 1717–1723. [Google Scholar]
- Pardo C.A., Xu Z., Borchelt D.R., Price D.L., Sisodia S.S. and Cleveland D.W. (1995) Proc. Natl. Acad. Sci. USA, 92, 954–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakhit R., Crow J.P., Lepock J.R., Kondejewski L.H., Cashman N.R. and Chakrabartty A. (2004) J. Biol. Chem., 279, 15499–15504. [DOI] [PubMed] [Google Scholar]
- Rakhit R., Cunningham P., Furtos-Matei A., Dahan S., Qi X.F., Crow J.P., Cashman N.R., Kondejewski L.H. and Chakrabartty A. (2002) J. Biol. Chem., 277, 47551–47556. [DOI] [PubMed] [Google Scholar]
- Rakhit R., Robertson J., Van de Velde C., Horne P., Ruth D.M., Griffin J., Cleveland D.W., Cashman N.R. and Chakrabartty A. (2007) Nat. Med., 13, 754–759. [DOI] [PubMed] [Google Scholar]
- Rattanajarasroj S. and Unchern S. (2010) Neurochem. Res., 35, 1196–1205. [DOI] [PubMed] [Google Scholar]
- Ray S.S., Nowak R.J., Brown R.H. and Lansbury P.T. (2005) Proc. Natl. Acad. Sci. USA, 102, 3639–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaume A.G., Elliott J.L., Hoffman E.K. et al. (1996) Nat. Genet., 13, 43–47. [DOI] [PubMed] [Google Scholar]
- Rice-Evans C. (1999) Proc. Soc. Exp. Biol. Med., 220, 262–266. [DOI] [PubMed] [Google Scholar]
- Rodriguez J.A., Valentine J.S., Eggers D.K., Roe J.A., Tiwari A., Brown R.H. and Hayward L.J. (2002) J. Biol. Chem., 277, 15932–15937. [DOI] [PubMed] [Google Scholar]
- Rosen D.R., Siddique T., Patterson D. et al. (1993) Nature, 362, 59–62. [DOI] [PubMed] [Google Scholar]
- Sasaki S. and Iwata M. (1996) Acta Neuropathol., 91, 278–283. [DOI] [PubMed] [Google Scholar]
- Semisotnov G.V., Rodionova N.A., Razgulyaev O.I., Uversky V.N., Gripas’ A.F. and Gilmanshin R.I. (1991) Biopolymers, 31, 119–128. [DOI] [PubMed] [Google Scholar]
- Shaw B.F. and Valentine J.S. (2007) Trends Biochem. Sci., 32, 78–85. [DOI] [PubMed] [Google Scholar]
- Smirnoff N. and Cumbes Q.J. (1989) Phytochemistry, 28, 1057–1060. [Google Scholar]
- Stathopulos P.B., Rumfeldt J.A.O., Scholz G.A., Irani R.A., Frey H.E., Hallewell R.A., Lepock J.R. and Meiering E.M. (2003) Proc. Natl. Acad. Sci. USA, 100, 7021–7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida K. and Kawakishi S. (1994) J. Biol. Chem., 269, 2405–2410. [PubMed] [Google Scholar]
- Velde V.C., Miller T.M., Cashman N.R. and Cleveland D.W. (2008) Proc. Natl. Acad. Sci. USA, 105, 4022–4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner C., Fachinetto R., Dalla Corte C.L., Brito V.B., Severo D., de Oliveira Costa Dias G., Morel A.F., Nogueira C.W. and Rocha J.B.T. (2006) Brain Res., 1107, 192–198. [DOI] [PubMed] [Google Scholar]
- Wong P.C., Pardo C.A., Borchelt D.R., Lee M.K., Copeland N.G., Jenkins N.A., Sisodia S.S., Cleveland D.W. and Price D.L. (1995) Neuron, 14, 1105–1116. [DOI] [PubMed] [Google Scholar]
- Xu Z., Chen S., Li X., Luo G., Li L. and Le W. (2006) Neurochem. Res., 31, 1263–1269. [DOI] [PubMed] [Google Scholar]
- Yu J., Jia Y., Guo Y., Chang G., Duan W., Sun M., Li B. and Li C. (2010) FEBS Lett., 584, 2921–2925. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.