Abstract
Motivation: Fluorescence Correlation Spectroscopy (FCS) is a popular tool for measuring molecular mobility and how mobility relates to molecular interaction dynamics and bioactivity in living cells. The FCS technique has been significantly advanced by its combination with super-resolution STED microscopy (STED-FCS). Specifically, the use of gated detection has shown great potential for enhancing STED-FCS, but has also created a demand for software which is efficient and also implements the latest algorithms. Prior to this study, no open software has been available which would allow practical time-gating and correlation of point data derived from STED-FCS experiments.
Results: The product of this study is a piece of stand-alone software called FoCuS-point. FoCuS-point utilizes advanced time-correlated single-photon counting (TCSPC) correlation algorithms along with time-gated filtering and innovative data visualization. The software has been designed to be highly user-friendly and is tailored to handle batches of data with tools designed to process files in bulk. FoCuS-point also includes advanced fitting algorithms which allow the parameters of the correlation curves and thus the kinetics of diffusion to be established quickly and efficiently.
Availability and implementation: FoCuS-point is written in python and is available through the github repository: https://github.com/dwaithe/FCS_point_correlator. Furthermore, compiled versions of the code are available as executables which can be run directly in Linux, Windows and Mac OSX operating systems.
Contact: dominic.waithe@imm.ox.ac.uk
1 Introduction
Fluorescence Correlation Spectroscopy (FCS) is a well established statistical technique for characterizing diffusion and kinetics of fluorescently labelled molecules in solution (Ehrenberg and Rigler, 1974; Magde et al., 1972). Through the advent of confocal microscopes, which allow detection to within small femtoliter volumes, it was then possible to apply FCS techniques to study kinetics of a broad range of biological molecules at the surface or within the cytoplasm of biological cells. (Schwille et al., 1999; Widengren and Rigler, 1998). More recently, super-resolution techniques have opened up a new vista in biological imaging, and one of these techniques known as STimulated Emission Depletion (STED) microscopy (Hell and Wichmann, 1994) has been combined with FCS to study diffusion of molecules below the diffraction limit of light (Kastrup etal., 2005). Truly capable of studying membranes at the nanoscale <200 nm, the application of STED-FCS to biological imaging has illuminated the kinetics of proteins and lipids whose mechanisms have until now remained elusive (Eggeling etal., 2009). Until recently, the STED-FCS technique has only been applied to study a few select mechanisms due to the technically demanding and specialist nature of the STED-FCS acquisition systems. Commercial systems are now available which are both reliable and have the sensitivity required to perform STED-FCS on cells (Clausen etal., 2015). The use of continuous wave (CW) STED lasers which, although highly functional and suitable for deployment in commercial systems, has the caveat that the output fluorescence is contaminated with a non-negligible amount of light derived from outside the confined effective fluorescence observation volume. Fortunately, this artefact can be removed through employing a time-gating filtering scheme (Moffitt et al., 2011; Vicidomini etal., 2011; 2015). Time-gating is a technique made possible through Time-Correlated Single Photon Counting (TCSPC) and allows individual photons to be filtered based on their arrival times at the detector. Time-gating of TCSPC data generated during STED laser illumination allows photons which are derived from outside the confined effective fluorescence observation to be filtered out, restoring the effective observation volume size when using CW lasers. Furthermore, TCSPC derived data is rich in information and many of its properties can be utilized for characterization of the system under investigation, such as fluorescence lifetime information (Enderlein and Gregor, 2005).
Given the recent availability of commercial STED microscopes and applicability of FCS techniques for studying biological systems there is a real demand for software solutions which can harness the power of these techniques in a way which is user-friendly and practical. Some generalizable software solutions do exist which enable robust and practical analysis of FCS data from a variety of source microscopes (Müller etal., 2014; Wachsmuth etal., 2015), but no software solution yet exists which allow full utilization of STED-FCS derived data. FoCuS-point is a tool which successfully addresses the requirements of STED-FCS and uniquely facilitates the application of this technique in the field of cell biology.
2 Implementation
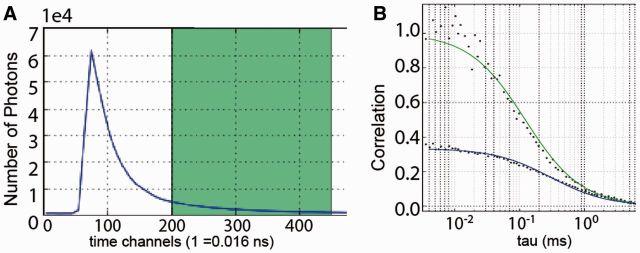
FoCuS-point is a compiled application written in python and makes use of the lmfit, numpy and matplotlib python libraries. When a file is processed using FoCuS-point, auto-correlation is performed on each input channel and cross-correlation is additionally performed in the case of two-channel data. FoCuS-point was intially designed to work with ‘.pt3’ files which are the output format from PicoQuant PicoHarp hardware, however correlation and time-gating can be applied to any file which contains TCSPC time-tag data if it is correctly parsed. In addition, FoCuS-point can also take already processed data in a variety of different formats. Correlation is a numerically intensive discipline and so algorithms have evolved which can efficiently perform this computation. For FoCuS-point an existing advanced correlation algorithm known as the Time-tag-to-correlation algorithm was implemented (Wahl etal., 2003) and was adapted to include normalization and to run efficiently in a python environment. From the TCSPC time-tag data, collected during an experiment, it is possible to visualize photons based on their macro and micro-scopic arrival times. Macro-scopic arrival times represent the arrival time of the photons from the beginning of the experiment, visualized as a time-series plot. Conversely, the micro-scopic arrival scheme represents the photon collection times subsequent to each pulse of the excitation laser. Micro-scopic arrival times for a population of photons are visualized as a life-time decay profile (Fig. 1A). Time-gating involves specifying a specific temporal window [tg1, tg2] on the micro time-scale and excluding from the subsequent analysis those photons which were outside of the gates. This process is performed in FoCuS-point by the user selecting the temporal window interactively, before applying the selected time-gate to a single file or multiple files and then correlating. FoCuS-point features a dedicated interface for the fitting of correlation functions which encompasses 2D and 3D diffusion, anomalous factor and also triplet states (Fig. 1B). Average transit times through the effective observation volume (and diffusion coefficients if diameters of the volume are supplied) as well as other parameters such as average particle numbers in the observation volume or anomaly factors in diffusion characteristics are extracted from FCS data through fitting of auto and cross-correlation functions. The user interface facility also encompasses data visualization using histogram and scatter plots for comparing parameters across large data collections. FoCuS-point facilitates data export through button-click copying to the clipboard and also the option of saving to a. csv file the fit parameters and correlated data. For further reference material relating to the software and the algorithms which it employs see the website: https://github.com/dwaithe/FCS_point_correlator.
Fig. 1.
(A) The fluorescence lifetime decay (blue line) is generated by the histogram over the micro-scopic arrival times of the detected photons (ns time scale), and a time gate is selected (green). (B) FCS data is calculated from the selected time-gated window and fitted by a selected model (green line), here compared to the FCS data generated from all (ungated) photons (blue line) indicating a decrease in decay (and thus transit time) as well as increase in amplitude upon gating, as expected (Clausen et al., 2015; Vicidomini et al., 2011)
3 Conclusion
FoCuS-point represents a computationally efficient user-friendly and easily deployable means of analyzing (gated) STED-FCS point source data.
Funding
Alfred Benzon Foundation, Medical Research Council (grant ref MC_UU_12010/unit programmes G0902418 and MC_UU_12025), MRC/BBSRC/EPSRC (grant ref MR/K01577X/1), Wolfson Foundation and Wellcome Trust (grant ref 104924/14/Z/14).
Conflict of Interest: none declared.
References
- Clausen M.P., et al. (2015) A straightforward approach for gated sted-fcs to investigate lipid membrane dynamics. Methods, 88, 67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggeling C., et al. (2009) Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature, 457, 1159–1162. [DOI] [PubMed] [Google Scholar]
- Ehrenberg M., Rigler R. (1974) Rotational Brownian motion and fluorescence intensify fluctuations. Chem. Phys., 4, 390–401. [Google Scholar]
- Enderlein J., Gregor I. (2005) Using fluorescence lifetime for discriminating detector afterpulsing in fluorescence-correlation spectroscopy. Rev. Sci. Instrum., 76, 033102. [Google Scholar]
- Hell S.W., Wichmann J. (1994) Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt. Lett., 19, 780–782. [DOI] [PubMed] [Google Scholar]
- Kastrup L., et al. (2005) Fluorescence fluctuation spectroscopy in subdiffraction focal volumes. Phys. Rev. Lett., 94, 178104. [DOI] [PubMed] [Google Scholar]
- Magde D., et al. (1972) Thermodynamic fluctuations in a reacting system—measurement by fluorescence correlation spectroscopy. Phys. Rev. Lett., 29, 705. [Google Scholar]
- Moffitt J.R., et al. (2011) Time-gating improves the spatial resolution of sted microscopy. Opt. Express, 19, 4242–4254. [DOI] [PubMed] [Google Scholar]
- Müller P., et al. (2014) Pycorrfit–generic data evaluation for fluorescence correlation spectroscopy. Bioinformatics, btu328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwille P., et al. (1999) Fluorescence correlation spectroscopy with single-molecule sensitivity on cell and model membranes. Cytometry, 36, 176–182. [DOI] [PubMed] [Google Scholar]
- Vicidomini G., et al. (2011) Sharper low-power sted nanoscopy by time gating. Nat. Methods, 8, 571–573. [DOI] [PubMed] [Google Scholar]
- Vicidomini G., et al. (2015) Sted-flcs: An advanced tool to reveal spatiotemporal heterogeneity of molecular membrane dynamics. Nano Lett., 15, 5912–5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachsmuth M., et al. (2015) High-throughput fluorescence correlation spectroscopy enables analysis of proteome dynamics in living cells. Nat. Biotechnol., 33, 384–389. [DOI] [PubMed] [Google Scholar]
- Wahl M., et al. (2003) Fast calculation of fluorescence correlation data with asynchronous time-correlated single-photon counting. Opt. Express, 11, 3583–3591. [DOI] [PubMed] [Google Scholar]
- Widengren J., Rigler R. (1998) Fluorescence correlation spectroscopy as a tool to investigate chemical reactions in solutions and on cell surfaces. Cell. Mol. Biol. (Noisy-le-Grand, France), 44, 857–879. [PubMed] [Google Scholar]