Abstract
Mining, jewellery and metal-processing industries use cyanide for extracting gold and other valuable metals, generating large amounts of highly toxic wastewater. Biological treatments may be a clean alternative under the environmental point of view to the conventional physical or chemical processes used to remove cyanide and related compounds from these industrial effluents. Pseudomonas pseudoalcaligenes CECT5344 can grow under alkaline conditions using cyanide, cyanate or different nitriles as the sole nitrogen source, and is able to remove up to 12 mM total cyanide from a jewellery industry wastewater that contains cyanide free and complexed to metals. Complete genome sequencing of this bacterium has allowed the application of transcriptomic and proteomic techniques, providing a holistic view of the cyanide biodegradation process. The complex response to cyanide by the cyanotrophic bacterium P. pseudoalcaligenes CECT5344 and the potential biotechnological applications of this model organism in the bioremediation of cyanide-containing industrial residues are reviewed.
Keywords: biodegradation, bioplastics, cyanate, cyanide, jewellery wastewater, proteomics, Pseudomonas, transcriptomics
Cyanide assimilation by Pseudomonas pseudoalcaligenes.
INTRODUCTION: BIODEGRADATION OF CYANIDE AND RELATED COMPOUNDS
Cyanide is a powerful toxic chemical that shows high affinity for iron and other metals, blocking the function of different metalloproteins like the cytochrome c oxidase involved in the cellular respiratory process (Solomonson 1981; Jünemann 1997; Gupta, Balomajumder and Agarwal 2010; Luque-Almagro, Moreno-Vivián and Roldán 2016). Paradoxically, cyanide played a key role in the prebiotic synthesis of amino acids and nitrogenous bases (Oró 1961; Oró and Kamat 1961). Cyanide may be generated by many organisms, from bacteria, algae and fungi to higher plants and arthrophods. Plants produce cyanide during ethylene synthesis, and also generate cyanoglycosides and cyanolipids, whereas insects and other organisms use cyanide mainly for defensive or offensive purposes (Peiser et al.1984; Zagrobelny, Bak and Møller 2008). Some Pseudomonas strains and other bacteria contain a membrane-bound flavoenzyme, the HCN synthase, which oxidizes glycine to hydrogen cyanide and carbon dioxide under low oxygen levels at early stationary phase of growth (Laville et al.1998; Zdor 2014). However, environmental and health problems related to cyanide and its derivatives are caused by human activities, such as nitrile polymers production, plastics, paints, dyes and drugs synthesis, food processing, and particularly coal coking, steel handling, electroplating, jewellery industry and mining, which produce the largest amounts of liquid wastes containing cyanide (Akcil and Mudder 2003; Dash, Gaur and Balomajumder 2009; Luque-Almagro, Moreno-Vivián and Roldán 2016). These industrial wastes often contain metals, which increase the toxicity of the effluents that can be potentially released to the environment. Thus, the jewellery industry in Córdoba (Spain) generates an alkaline waste containing up to 1.5 M total cyanide, about 1 M in form of ion cyanide or weak metal–cyanide complexes and near 0.5 M as highly stable metal–cyanide complexes (Ibáñez et al.2017).
Different physico-chemical processes have been applied to remove cyanide, including oxidation by hydrogen peroxide, alkaline clorination, SO2/air mixture (INCO process) or ozonization, among other methods (Dash, Gaur and Balomajumder 2009). However, these conventional physical-chemical methods are expensive, require special equipment and maintenance, and are not effective to remove metal–cyanide complexes. Some microorganisms are capable of enzymatic cyanide detoxification, or even cyanide assimilation using this compound as a nitrogen source, so that the biodegradation technology may constitute a potential alternative for the treatment of cyanide-containing liquid industrial wastes (Raybuck 1992; Akcil and Mudder 2003; Ebbs 2004; Baxter and Cummings 2006; Huertas et al.2006; Dash, Gaur and Balomajumder 2009; Gupta, Balomajumder and Agarwal 2010; Kumar et al.2016; Luque-Almagro, Moreno-Vivián and Roldán 2016; Park, Sewell and Benedik 2017). Microbial cyanide degradation leads to the formation of non-toxic products, like ammonia and carbon dioxide, and takes place by different biochemical routes involving oxidative, reductive, hydrolytic or substitution/transfer reactions (Table 1).
Table 1.
Relevant enzymes involved in the degradation of cyano-compounds.
| Reaction type | Enzyme | Reaction |
|---|---|---|
| Oxidation | Cyanide dioxygenase | HCN + O2 + 2e− + 2H+ → NH3 + CO2 |
| Cyanide monooxygenase | HCN + O2 + 2e− + 2H+ → OCN− + H2O | |
| Cyanase | OCN− + HCO3− + 2H+ → NH3 + 2CO2 | |
| Reduction | Nitrogenase | HCN + 6e− + 6H+ → CH4 + NH3 |
| Hydrolysis | Cyanidase | HCN + 2H2O → HCOOH + NH3 |
| Cyanide hydratase | HCN + H2O → HCONH2 | |
| Formamidase | HCONH2 + H2O → HCOOH + NH3 | |
| Nitrilase | R−CN + 2H2O → R−COOH + NH3 | |
| Nitrile hidratase | R−CN + H2O → R−CONH2 | |
| Amidase | R−CONH2 + H2O → R−COOH + NH3 | |
| Substitution/transfer | 3−Cyanoalanine synthase | HCN + Cys → 3−cyanoalanine + H2S |
| Rhodanese | HCN + S2O3− → SCN− + SO32− |
Both free cyanide and nitriles may be degraded through hydrolytic reactions that produce ammonia and the corresponding carboxylic derivative either directly or forming an amide as intermediate. Cyanidase and cyanide hydratase act on HCN generating formic acid or formamide, respectively. Formamide is finally converted into ammonia and formic acid by a formamidase. On the other hand, nitrilase and nitrile hydratase use nitriles as substrates, but while nitrilase generates ammonia and the corresponding carboxylic acid in one-step reaction, nitrile hydratase forms an amide product that is transformed to ammonia and the carboxylic acid by an amidase (Table 1). The oxidative pathways transform cyanide into ammonia and carbon dioxide, and may also occur either in a single reaction catalyzed by the cyanide dioxygenase or in two steps generating cyanate as intermediate, by the action of the enzymes cyanide monooxygenase and cyanase, respectively. Nitrogenase is the enzyme involved in the reductive pathway, which generates ammonia and methane when a total of six electrons are transferred to cyanide. Finally, substitution/transfer reactions also allow cyanide transformation to less toxic products. Thus, cyanide and cysteine are converted into 3-cyanoalanine and sulfide by the 3-cyanoalanine synthase, whereas cyanide and thiosulfate are transformed into thiocyanate and sulfite through the enzyme rhodanese (Table 1).
Pseudomonas pseudoalcaligenes CECT5344, a model alkaliphilic cyanotroph
Several considerations could be regarded in order to carry out a successful biological treatment of industrial cyanide-containing wastewater. Obviously, cyanide biodegradation requires microbial strains that tolerate and degrade cyanide, which is converted into non-toxic products by a specific enzymatic pathway (Table 1). As cyanide inhibits cellular aerobic respiration, microorganisms growing with cyanide contain an alternative oxidase insensitive to cyanide like the quinol oxidase encoded by the cioAB genes, which belongs to the cytochrome bd family (Jünemann 1997; Arai et al.2014). In addition, removal of cyanide also demands an alkaline pH to prevent the formation of hydrogen cyanide (the pKa for HCN/CN− dissociation is 9.2). However, most cyanide-degrading microorganisms described up to date capable to grow at neutral pH, a condition in which almost all cyanide is found as the highly toxic HCN form that could evaporate causing environmental and human health risks. Finally, most cyanide-containing industrial effluents also contain other cyano-derivatives like cyanate and thiocyanate, and high amounts of metals and metal–cyanide compounds. Therefore, the biotreatment of these wastes of industrial origin requires the use of microorganisms capable to degrade all these different cyano-compounds and to tolerate metals, which must be incorporated into the cells in the concentrations just needed for the metabolic processes.
The bacterial strain P. pseudoalcaligenes CECT5344 was isolated from sludge of the Guadalquivir River (Córdoba, Spain) by enrichment techniques at pH 9.5 with 2 mM NaCN as nitrogen source (Luque-Almagro et al.2005a,b). This autochthonous, alkaliphilic bacterial strain can use cyanide, cyanate, some metal–cyanide complexes or different nitriles (cyanohydrins) as nitrogen source under alkaline pH, thus preventing volatile HCN formation (Luque-Almagro et al.2005a,b). This strain was able to tolerate a pH of 11.5 and up to 30 mM cyanide or 100 mM cyanate (Luque-Almagro et al.2005b, 2008).
The high cyanide tolerance of P. pseudoalcaligenes CECT5344 is mainly due to the presence of a quinol oxidase insensitive to cyanide encoded by the cioAB genes. A mutant strain harboring a disrupted cioA gene did not show cyanide-resistant respiration and was incapable of using cyanide as nitrogen source, demonstrating that this alternative quinol oxidase is required for cyanide tolerance (Quesada et al.2007). Interestingly, the cyanide-insensitive respiratory activity was connected to the cyanide assimilation route in the strain CECT5344. Thus, a malate:quinone oxidoreductase (Mqo) was associated with the alternative oxidase insensitive to cyanide (CioAB) in the membrane of the cells grown with cyanide. In the membrane electron transport chain, the Mqo activity directly transfers electrons from malate to the quinone pool. Then, oxaloacetate formed by the Mqo activity reacts chemically with cyanide to form the corresponding 2-hydroxynitrile. Finally, the nitrilase encoded by the nitC located in the nit1C gene cluster may degrade this nitrile, releasing ammonia used as nitrogen source (Luque-Almagro et al.2011b; Estepa et al.2012). The P. pseudoalcaligenes CECT5344 nit1C gene cluster includes the nitA gene encoding a putative Fis family σ54-dependent transcriptional activator that is transcribed divergently from the nitBCDEFGH genes. Mutant strains defective in nitA, nitB or nitC genes did not grow with cyanide or aliphatic nitriles, demonstrating that the nitrilase NitC is required for assimilation of both free cyanide and aliphatic nitriles (Estepa et al.2012).
Cyanate is a less toxic cyanide derivative that may be formed by chemical or photochemical oxidation of cyanide (Nowakowska, Sterzel and Szczubialka 2006), and it is also a natural compound generated in cellular processes like urea and carbamoyl phosphate metabolism (Guilloton and Karst 1987). This fact could explain the widespread bacterial ability to degrade this compound, overcoming the toxicity of the cyanate-containing industrial wastes. Cyanase converts cyanate into carbon dioxide and ammonia (Table 1). Pseudomonas pseudoalcaligenes CECT5344 may also use cyanate as a nitrogen source, and contains a cynFABDS gene cluster organized into two transcriptional units: the cynF gene coding for a putative transcriptional regulator that belongs to the Fis family and the divergently transcribed cynABDS genes coding for the three components of a putative ABC-type transporter (CynABD) and the cyanase CynS (Luque-Almagro et al.2008). The cyanase activity of the strain CECT5344 was induced by both cyanate and cyanide. However, a cyanase-defective CynS− mutant strain assimilated cyanide at a similar rate to that observed in the wild type, indicating that cyanate is not generated in the cyanide assimilation pathway of the strain CECT5344 (Luque-Almagro et al.2008). Therefore, P. pseudoalcaligenes CECT5344 operates two separate and independent metabolic pathways for assimilation of cyanide compounds, with the nitrilase NitC or the cyanase CynS as key enzymes, respectively. The nitrilase NitC is essential for the growth with cyanide and nitriles, whereas the cyanase CynS is specifically required for cyanate assimilation.
Cells growing in the presence of cyanide must also solve additional problems, such as the decreased bioavailability of essential metals like iron and copper because of the formation of highly stable metal-cyanide compounds, and the potential cell damage caused by the oxidative stress generated by cyanide. Thus, in addition to cyanide-insensitive respiration, iron homeostasis and oxidative stress responses are essential for cyanide metabolism (Luque-Almagro et al.2007; Luque-Almagro, Moreno-Vivián and Roldán 2016). A proteomic analysis by two-dimensional gel electrophoresis revealed that P. pseudoalcaligenes CECT5344 displayed a response to cyanide that includes mechanisms against iron deprivation, oxidative stress and nitrogen starvation (Luque-Almagro et al.2007). Some proteins involved in iron homeostasis, such as the ferritin-like protein Dps and the dihydropicolinate synthase DapA, and oxidative stress resistance, like the alkyl-hydroperoxide reductase AhpC, were induced during cyanide assimilation. Therefore, induction of proteins related with iron acquisition and oxidative stress repairing and protection are also part of the resistance mechanisms that reduce the cyanide toxicity effects in P. pseudoalcaligenes CECT5344 (Luque-Almagro et al.2007).
The ferric uptake regulator (Fur) is a repressor that senses iron and controls the expression of genes related to iron uptake and siderophore biosynthesis, although it can also act as a positive regulator for the expression of specific genes through small regulatory RNAs (Escolar, Pérez-Martín and De Lorenzo 1999; Hantke 2001). In a Fur-defective mutant strain of P. pseudoalcaligenes CECT5344, the total cellular iron content was higher than that in the wild-type strain. In addition, the Fur mutant showed an increased susceptibility to iron-chelating agents and overexpressed the small regulatory RNA prrF gene in a constitutive manner, whereas in the wild-type strain CECT5344 expression of the prrF gene was repressed by iron availability through a Fur-dependent mechanism (Becerra et al.2014).
Although it has been discussed that the synthesis of siderophores for iron acquisition should be necessary for an efficient cyanide assimilation process (Luque-Almagro et al.2005b; Huertas et al.2006), it seems that P. pseudoalcaligenes CECT5344 does not produce siderophores and lacks putative genes involved in the synthesis of these ferric ion specific chelating biological agents. However, this cyanotrophic strain possesses iron uptake systems and it can use Prussian blue as a source of nitrogen and iron, generating decolored halos around the colonies when growing in agar plates (Luque-Almagro et al.2013; Becerra et al.2014).
As mentioned above, the toxicity of cyanide is based on its property for binding to the metal cofactors present in essential metalloproteins that are inhibited by cyanide (Gupta, Balomajumder and Agarwal 2010; Luque-Almagro et al.2011a), especially heme-containing proteins like cytochromes and iron–sulfur enzymes such as fumarase and aconitase. In this sense, the presence in P. pseudoalcaligenes CECT5344 of different isoforms of key iron-containing enzymes, such as the tricarboxylic acid cycle enzymes fumarase and aconitase, may also contribute to cyanide resistance (Igeño et al.2011).
Cyanomics: a holistic view of the cyanide biodegradation process
The genome of P. pseudoalcaligenes CECT5344 was first completely sequenced for a cyanide-degrading bacterium, becoming a functional instrument to develop transcriptomic and proteomic studies to analyze the global response to cyanide and industrial cyanide-containing residues. The application of these omic techniques to cyanide biodegradation has been termed ‘cyan-omics’ (Luque-Almagro, Moreno-Vivián and Roldán 2016).
The genome of P. pseudoalcaligenes CECT5344 was initially sequenced by combining genome-shotgun and 3k paired-end 454-pyrosequencing with PCR-based gap closure and sequencing of gap-spanning fosmids (Luque-Almagro et al.2013; Wibberg et al.2014). More recently, the genome was newly sequenced by using the PacBio single-molecule, real-time (SMRT®) sequencing (Wibberg et al.2016). The finished genome consists of a single 4696 984 bp chromosome with 65.34% GC content and 4514 genes, of which 4435 are protein-encoding genes. Sequence comparisons revealed that the strain CECT5344 is closely related to P. mendocina, sharing about 70% of total genes, although pathogenicity genes are not present in P. pseudoalcaligenes CECT5344, thus converting this strain in a safe candidate for biotechnological applications. The PacBio SMRT sequencing also allowed the establishment of the methyloms of the strain CECT5344, identifying the motifs with methylated adenine residues in the genome and the genes coding for different methyltransferases putatively involved in the methylation of these motifs (Wibberg et al.2016).
Genome sequencing of P. pseudoalcaligenes CECT5344 provides the basis for understanding the capability of this strain to survive in the presence of toxic cyano-derivatives. Thus, up to four nitrilase genes are present in the strain CECT5344, the largest number described for Pseudomonas species (Luque-Almagro et al.2013). One of these genes is nitC, which was found essential for degradation of cyanide and nitriles by its mutational analysis (Estepa et al.2012). The genome also contains the cynFABDS gene cluster required for cyanate assimilation, a putative rhodanese gene, three cio operons coding for cyanide-resistant alternative oxidases in addition to other gene clusters for different oxidases, and at least two copies of genes that code for key enzymes like malate:quinone oxidoreductase, fumarase and aconitase. Finally, the presence of a large number of genes for resistance and detoxification of metalloids and heavy metals, such as arsenate and mercury, also suggests that this strain could detoxify mining wastewaters that are frequently contaminated not only with cyanide but also with arsenic, mercury and other toxic metals (Luque-Almagro et al.2013).
Transcriptomic and proteomic techniques applied to a global analysis of the degradation of cyanide and cyanide-containing industrial residues have been only reported in the bacterium P. pseudoalcaligenes CECT5344. In order to characterize the transcriptomic response to cyanide in the strain CECT5344, DNA microarrays have been performed and compared with microarrays from cells grown with ammonium or in the absence of nitrogen source (nitrogen starvation) as control (Luque-Almagro et al.2015). This analysis revealed that cyanide causes significant changes in the gene expression. Induced by sodium cyanide or jewellery wastewater were the nitrilase nitC gene that is required for the assimilation of cyanide and also other nitrilase-encoding genes, the cyanase cynS gene, the cioAB genes essential for cyanide-insensitive respiration, the isc genes for Fe–S cluster assembly and the alkyl–hydroperoxide reductase ahpC gene involved in the response to oxidative stress, among other genes. Exclusively induced by the jewellery industrial residue were the malate:quinone oxidoreductase mqoB gene and other genes coding for metal pump extruders and their putative regulators (Luque-Almagro et al.2015).
A previous proteomic study by two-dimensional gel electrophoresis in P. pseudoalcaligenes CECT5344 revealed that cyanide induces a response to iron depletion, oxidative stress and nitrogen starvation (Luque-Almagro et al.2007). Recently, a second-generation gel-free quantitative proteomic technique has been used for the identification of P. pseudoalcaligenes CECT4344 proteins involved in cyanide and metals detoxification during bioremediation of the jewellery industry liquid residue (Ibáñez et al.2017). More than 150 proteins were significantly overproduced in cells degrading the jewellery wastewater. The genes coding for almost all these proteins were found induced by the industrial residue in the previous transcriptomic study (Luque-Almagro et al.2015), thus validating the DNA microarrays data and providing evidences that relevant transcribed genes are also translated to proteins. The jewellery wastewater has high concentrations of cyanide and the metals zinc, iron and copper. Accordingly, different proteins associated with oxidative stress, metal resistance/detoxification and iron–sulfur centers assembly/maturation were found induced by this residue in addition to proteins required for cyanide compounds assimilation and cyanide-resistant respiration. Also, several regulatory proteins of the GntR family were detected suggesting that GntR-like regulators are relevant in controlling assimilation of cyanide and metal–cyanide complexes (Ibáñez et al.2017).
Exploitation of P. pseudoalcaligenes genetic features for biotechnological applications
Sequencing of the genome of the strain CECT5344 has confirmed that this bacterium has a non-pathogenic lifestyle and is very well adapted to cyanide and metal-polluted habitats, being an ideal organism to be applied to biodegradation and bioremediation strategies (Wibberg et al.2016). The biotechnological potential of P. pseudoalcaligenes CECT5344 was first established by developing a method for spectrophotometric monitoring of cyanate in the bioremediation processes, using the enzyme cyanase partially purified from this strain immobilized in a packed reactor (Luque-Almagro et al.2003). Later, the capability of the strain CECT5344 to biodegrade cyanide at alkaline pH was demonstrated in a batch reactor using 2 mM NaCN as nitrogen source and 50 mM sodium acetate as carbon source (Huertas et al.2010). Cyanide degradation was found to be pH dependent, and took place at initial pH 10.0 without further control or at controlled constant pH 9.5 during all the degradation process. Under optimized conditions (10% dissolved oxygen saturation and constant pH 9.5), cyanide was totally depleted at a mean rate of 2.31 mg CN− L−1 OD−1 h−1 (Huertas et al.2010). More recently, P. pseudoalcaligenes CECT5344 was used in a reactor at controlled pH 9.0 and 10% oxygen saturation to degrade completely up to 12 mM total cyanide, including free cyanide and metal–cyanide complexes, from a jewellery industry wastewater (Ibáñez et al.2017). Alkaline biodegradation of such elevated cyanide concentration from an industrial residue also containing large amounts of metals demonstrates that the strain CECT5344 is an efficient model microorganism for biodegradation and bioremediation of industrial wastewaters. Chemical and biological processes could also be combined for treatment of industrial cyanide-containing wastes. Pseudomonas pseudoalcaligenes CECT5344 can grow with up to 100 mM cyanate, which is much less toxic than cyanide, and it could be expected that a chemical pre-treatment of the industrial residues with hydrogen peroxide or other oxidants to oxidize a significant amount of cyanide to cyanate will probably allow the biodegradation of even higher total cyanide concentrations.
Whole genome sequencing has also revealed the potential of P. pseudoalcaligenes CECT5344 for biodegradation of additional pollutants. This strain contains three atz gene clusters required for biodegradation of herbicides of the s-triazine family, such as atrazine, suggesting that the bioremediation of soils polluted with this type of herbicides may be successfully achieved. Putative genes for metabolic degradation of aromatic compounds like benzoate and naphthalene can be found in the genome of the strain CECT5344 (Luque-Almagro et al.2013). Furfural and furan derivatives are toxic by-products generated during treatment of lignocellulose-containing residues for bioethanol production. Genome of P. pseudoalcaligenes CECT5344 includes a hmfABCDE gene cluster that allows this strain to grow with furfural, leading to its possible application for the biodegradation of furanic compounds (Luque-Almagro et al.2013).
Of special interest is the presence in the genome of P. pseudoalcaligenes CECT5344 of three different pha gene regions probably required for the synthesis of polyhydroxyalkanoates (PHAs): two gene clusters (phaRP and phbRphaBAC) required for short chain length (scl) PHA metabolism and one gene cluster (phaIFDC2ZC1) involved in medium chain length (mcl) PHA production (Luque-Almagro et al.2013). The scl- and mcl-PHAs, which differ in their physical, thermal and plastic properties, have a valuable interest as renewable, biodegradable biomaterials (Luengo et al.2003). The functionality of the P. pseudoalcaligenes CECT5344 pha genes was recently confirmed, and mutant strains defective in the synthesis of scl-PHA or mcl-PHA have been obtained (Manso et al.2015). The wild-type strain accumulated both types of biopolymers (scl- and mcl-PHAs) as by-products during the cyanide-containing jewellery residue degradation process, demonstrating that the biodegradation process can be concomitantly used to produce these valuable materials (Manso et al.2015). In addition, a mutant strain deleted in the genes required for scl-PHA synthesis only accumulated mcl-PHAs, whereas a mutant lacking the synthase for mcl-PHAs only produced scl-PHAs. The double mutant strain was incapable of synthesizing PHAs (Manso et al.2015). Interestingly, the global transcriptomic and quantitative proteomic analyses revealed that genes and proteins involved in PHA production were overexpressed in cells grown with the jewellery residue (Luque-Almagro et al.2015; Ibáñez et al.2017). It is well known that PHAs are accumulated in the cytoplasm as carbon storage granules under conditions of nutrient imbalance or when environmental factors are not optimal (Luengo et al.2003). Therefore, the accumulation of PHAs in the cells growing with cyanide is probably a response against cyanide stress since this toxic compound is a poor nitrogen source for growth that causes a C/N imbalance. In fact, an involvement of the PII-type nitrogen regulatory protein GlnK in the cyanide-induced nitrogen deficiency response has been previously described (Luque-Almagro et al.2007).
CONCLUSIONS
The bacterium P. pseudoalcaligenes CECT5344 has the ability to grow in media containing high concentrations of cyano-compounds and metals, such as the effluents generated in mining, electroplating and jewellery industries, under alkaline pH that avoid hydrogen cyanide volatilization. This strain induces a great variety of metabolic changes that allow not only its tolerance and survival with these highly toxic compounds but also the utilization of cyanide and related compounds as nitrogen source for growth. This complex response includes a cyanide-resistant respiration, enzymatic pathways for assimilation of cyanide, nitriles and cyanate, defense mechanisms against oxidative stress and iron deprivation, and heavy metals and metalloids detoxification systems, among other processes (Fig. 1). Bioreactor experiments have confirmed that this cyanotrophic strain is a suitable model organism for treatment of cyanide- and metal-containing industrial effluents, being able to remove up to 12 mM total cyanide at pH 9.0. In addition, this strain is able to accumulate PHAs, a valuable by-product that is used for production of bioplastics and biomaterials, providing an additional value to the cyanide biodegradation process. Genome sequencing and transcriptomic and proteomic analyses have confirmed the great biotechnological potential of this cyanotrophic strain. Thus, the absence in the genome of P. pseudoalcaligenes CECT5344 of pathogenicity determinants, and the presence of genes required for degradation of other pollutants like aromatic compounds, s-triazine herbicides and furan derivatives, increases the biotechnological application spectrum of this strain. Future studies using P. pseudoalcaligenes CECT5344 in pure cultures or in co-cultures with other cyanotrophic bacteria or wastewater plant sludge, together with high-throughput comparative omic and meta-omic analyses, will enable the development and optimization of improved strategies for biodegradation and bioremediation of industrial cyanide- and metal-containing effluents.
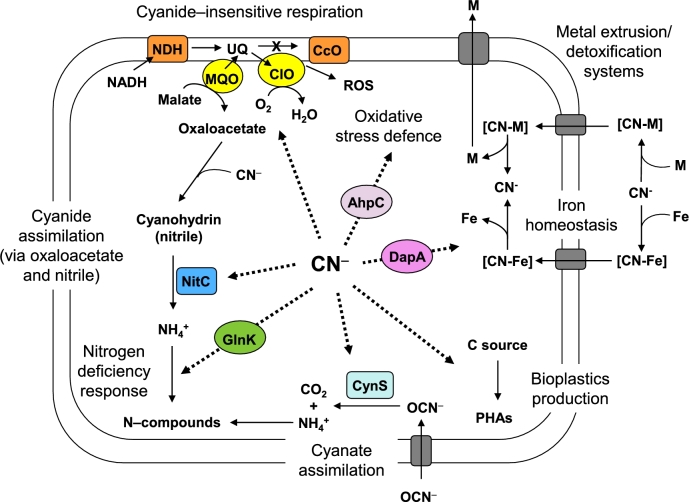
Figure 1.
Metabolic responses to cyanide and cyano-compounds in P. pseudoalcaligenes CECT5344. Dotted arrows indicate the different cyanide-induced responses. Some relevant cyanide-induced proteins like the nitrilase NitC, the cyanase CynS, the PII–type nitrogen regulator GlnK, the alkyl–hydroperoxide reductase AhpC and the dihydropicolinate synthase DapA are also shown. NDH, NADH dehydrogenase complex; UQ, ubiquinone; CcO, cytochrome c oxidase; MQO, malate:quinone oxidoreductase; CIO, cyanide–insensitive quinol oxidase; ROS, reactive oxygen species, PHAs, polyhydroxyalkanoates; M, metals; [CN–M], metal–cyanide complexes; [CN–Fe], iron–cyanide complexes.
Acknowledgements
Authors also thank FCC-Ámbito, SEVECO, AVENIR and MAGTEL for fruitful collaborations.
FUNDING
This work was funded by Grant BIO2015-64311-R (Ministerio de Economía, Industria y Competitividad, Spain, and FEDER, UE) Fondo Europeo de Desarrollo Regional Unión Europea and Grant CVI-7560 (Junta de Andalucía, Grupo BIO-117, Spain).
Conflict of interest. None declared.
REFERENCES
- Akcil A, Mudder T. Microbial destruction of cyanide wastes in gold mining: process review. Biotechnol Lett 2003;25:445–50. [DOI] [PubMed] [Google Scholar]
- Arai H, Kawakami T, Osamura T et al. Enzymatic characterization and in vivo function of five terminal oxidases in Pseudomonas aeruginosa. J Bacteriol 2014;196:4206–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter J, Cummings SP. The current and future applications of microorganism in the bioremediation of cyanide contamination. Anton Leeuw 2006;90:1–17. [DOI] [PubMed] [Google Scholar]
- Becerra G, Merchán F, Blasco R et al. Characterization of a ferric uptake regulator (Fur)-mutant of the cyanotrophic bacterium Pseudomonas pseudoalcaligenes CECT5344. J Biotechnol 2014;190:2–10. [DOI] [PubMed] [Google Scholar]
- Dash RR, Gaur A, Balomajumder C. Cyanide in industrial wastewaters and its removal: a review on biotreatment. J Hazard Mater 2009;163:1–11. [DOI] [PubMed] [Google Scholar]
- Ebbs S. Biological degradation of cyanide compounds. Curr Opin Biotechnol 2004;15:231–6. [DOI] [PubMed] [Google Scholar]
- Escolar L, Pérez-Martín J, De Lorenzo V. Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriol 1999;181:6223–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estepa J, Luque-Almagro VM, Manso I et al. The nit1C gene cluster of Pseudomonas pseudoalcaligenes CECT5344 involved in assimilation of nitriles is essential for growth on cyanide. Environ Microbiol Rep 2012;4:326–34. [DOI] [PubMed] [Google Scholar]
- Guilloton M, Karst F. Isolation and characterization of E. coli mutants lacking inducible cyanase. J Gen Microbiol 1987;133:645–53. [DOI] [PubMed] [Google Scholar]
- Gupta N, Balomajumder C, Agarwal VK. Enzymatic mechanism and biochemistry for cyanide degradation: a review. J Hazard Mater 2010;176:1–13. [DOI] [PubMed] [Google Scholar]
- Hantke K. Iron and metal regulation in bacteria. Curr Opin Microbiol 2001;4:172–7. [DOI] [PubMed] [Google Scholar]
- Huertas MJ, Luque-Almagro VM, Martínez-Luque M et al. Cyanide metabolism of Pseudomonas pseudoalcaligenes CECT5344: role of siderophores. Biochem Soc Trans 2006;34:152–5. [DOI] [PubMed] [Google Scholar]
- Huertas MJ, Sáez LP, Roldán MD et al. Alkaline cyanide degradation by Pseudomonas pseudoalcaligenes CECT5344 in a batch reactor. Influence of pH. J Hazard Mater 2010;179:72–78. [DOI] [PubMed] [Google Scholar]
- Ibáñez MI, Cabello P, Luque-Almagro VM et al. Quantitative proteomic analysis of Pseudomonas pseudoalcaligenes CECT5344 in response to industrial cyanide-containing wastewaters using liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS). PLoS One 2017;12:e0172908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igeño MI, Becerra G, Guijo MI et al. Metabolic adaptation of Pseudomonas pseudoalcaligenes CECT5344 to cyanide: role of malate–quinone oxidoreductases, aconitase and fumarase isoenzymes. Biochm Soc Trans 2011;39:1849–53. [DOI] [PubMed] [Google Scholar]
- Jünemann S. Cytochrome bd terminal oxidase1All amino acid numbering refers to the E. coli enzyme.1. BBA-Bioenergetics 1997;1321:107–27. [DOI] [PubMed] [Google Scholar]
- Kumar R, Saha S, Dhaka S et al. Remediation of cyanide-contaminated environments through microbes and plants: a review of current knowledge and future perspectives. Geosystem Eng 2017;20:28–40. [Google Scholar]
- Laville J, Blumer C, von Schroetter C et al. Characterization of the hcnABC gene cluster encoding hydrogen cyanide synthase and anaerobic regulation by ANR in the strictly aerobic biocontrol agent Pseudomonas fluorescens CHA0. J Bacteriol 1998;180:3187–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luengo JM, García B, Sandoval A et al. Bioplastics from microorganisms. Curr Opin Microbiol 2003;6:251–60. [DOI] [PubMed] [Google Scholar]
- Luque-Almagro VM, Acera F, Igeño MI et al. Draft whole genome sequence of the cyanide-degrading bacterium Pseudomonas pseudoalcaligenes CECT5344. Environ Microbiol 2013;15:253–70. [DOI] [PubMed] [Google Scholar]
- Luque-Almagro VM, Blasco R, Fernández-Romero JM et al. Flow-injection spectrophotometric determination of cyanate in bioremediation processes by use of immobilised inducible cyanase. Anal Bioanal Chem 2003;377:1071–8. [DOI] [PubMed] [Google Scholar]
- Luque-Almagro VM, Blasco R, Huertas MJ et al. Alkaline cyanide biodegradation by Pseudomonas pseudoalcaligenes CECT5344: Figure 1. Biochem Soc Trans 2005a;33:168–9. [DOI] [PubMed] [Google Scholar]
- Luque-Almagro VM, Blasco R, Martínez-Luque M et al. Bacterial cyanide degradation is under review: Pseudomonas pseudoalcaligenes CECT5344, a case of an alkaliphilic cyanotroph. Biochem Soc Trans 2011a;39:269–74. [DOI] [PubMed] [Google Scholar]
- Luque-Almagro VM, Escribano MP, Manso I et al. DNA microarray analysis of the cyanotroph Pseudomonas pseudoalcaligenes CECT5344 in response to nitrogen starvation, cyanide and a jewelry wastewater. J Biotechnol 2015;214:171–81. [DOI] [PubMed] [Google Scholar]
- Luque-Almagro VM, Huertas MJ, Martínez-Luque M et al. Bacterial degradation of cyanide and its metal complexes under alkaline conditions. Appl Environ Microb 2005b;71:940–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luque-Almagro VM, Huertas MJ, Roldán MD et al. The cyanotrophic bacterium Pseudomonas pseudoalcaligenes CECT5344 responds to cyanide by defence mechanisms against iron deprivation, oxidative damage and nitrogen stress. Environ Microbiol 2007;9:1541–9. [DOI] [PubMed] [Google Scholar]
- Luque-Almagro VM, Huertas MJ, Sáez LP et al. Characterization of the Pseudomonas pseudoalcaligenes CECT5344 cyanase, an enzyme that is not essential for cyanide assimilation. Appl Environ Microb 2008;74:6280–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luque-Almagro VM, Merchán F, Blasco R et al. Cyanide degradation by Pseudomonas pseudoalcaligenes CECT5344 involves a malate : quinone oxidoreductase and an associated cyanide-insensitive electron transfer chain. Microbiology 2011b;157:739–46. [DOI] [PubMed] [Google Scholar]
- Luque-Almagro VM, Moreno-Vivián C, Roldán MD. Biodegradation of cyanide wastes from mining and jewellery industries. Curr Opin Biotechnol 2016;38:9–13. [DOI] [PubMed] [Google Scholar]
- Manso I, Ibáñez MI, de la Peña F et al. Pseudomonas pseudoalcaligenes CECT5344, a cyanide-degrading bacterium with by-product (polyhydroxyalkanoates) formation capacity. Microb Cell Fact 2015;14:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowska M, Sterzel M, Szczubialka K. Photosensitized oxidation of cyanide in aqueous solutions of photoactive modified hydroxyethylcellulose. J Polym Environ 2006;14:59–64. [Google Scholar]
- Oró J. Mechanism of synthesis of adenine from hydrogen cyanide under possible primitive Earth conditions. Nature 1961;191:1193–4. [DOI] [PubMed] [Google Scholar]
- Oró J, Kamat SS. Amino-acid synthesis from hydrogen cyanide under possible primitive earth conditions. Nature 1961;190:442–3. [DOI] [PubMed] [Google Scholar]
- Park JK, Sewell BT, Benedik MJ. Cyanide bioremediation: the potential of engineered nitrilases. Appl Microbiol Biot 2017;101:3029–42. [DOI] [PubMed] [Google Scholar]
- Peiser GD, Wang TT, Hoffman NE et al. Formation of cyanide from carbon 1 of 1-aminocyclopropane-1-carboxylic acid during its conversion to ethylene. P Natl Acad Sci USA 1984;81:3059–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada A, Guijo MI, Merchán F et al. Essential role of cytochrome bd-related oxidase in cyanide resistance of Pseudomonas pseudoalcaligenes CECT5344. Appl Environ Microb 2007;73:5118–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raybuck SA. Microbes and microbial enzymes for cyanide degradation. Biodegradation 1992;3:3–18. [DOI] [PubMed] [Google Scholar]
- Solomonson LP. Cyanide as a metabolic inhibitor. In: Vennesland B, Conn EE, Knowles CJ et al. (eds.) Cyanide in Biology. Academic Press, New York, 1981, 11–28. [Google Scholar]
- Wibberg D, Bremges A, Dammann-Kalinowski T et al. Finished genome sequence and methylome of the cyanide-degrading Pseudomonas pseudoalcaligenes strain CECT5344 as resolved by single-molecule real-time sequencing. J Biotechnol 2016;232:61–8. [DOI] [PubMed] [Google Scholar]
- Wibberg D, Luque-Almagro VM, Igeño MI et al. Complete genome sequence of the cyanide-degrading bacterium Pseudomonas pseudoalcaligenes CECT5344. J Biotechnol 2014;175:67–8. [DOI] [PubMed] [Google Scholar]
- Zagrobelny M, Bak S, Møller BL. Cyanogenesis in plants and arthropods. Phytochemistry 2008;69:1457–68. [DOI] [PubMed] [Google Scholar]
- Zdor RE. Bacterial cyanogenesis: impact on biotic interactions. J Appl Microbiol 2015;118:267–74. [DOI] [PubMed] [Google Scholar]