Abstract
To understand the biosynthesis and functional role of gibberellins (GAs) in developing seeds, we isolated Cv20ox, a cDNA clone from watermelon (Citrullus lanatus) that shows significant amino acid homology with GA 20-oxidases. The complementary DNA clone was expressed in Escherichia coli as a fusion protein, which oxidized GA12 at C-20 to the C19 compound GA9, a precursor of bioactive GAs. RNA-blot analysis showed that the Cv20ox gene was expressed specifically in developing seeds. The gene was strongly expressed in the integument tissues, and it was also expressed weakly in inner seed tissues. In parthenocarpic fruits induced by 1-(2-chloro-4-pyridyl)-3-phenylurea treatment, the expression pattern of Cv20ox did not change, indicating that the GA 20-oxidase gene is expressed primarily in the maternal cells of developing seeds. The promoter of Cv20ox was isolated and fused to the β-glucuronidase (GUS) gene. In a transient expression system, β-glucuronidase staining was detectable only in the integument tissues of developing watermelon seeds.
GAs have been characterized as important hormones that control many aspects of plant development, including seed germination, shoot elongation, flower formation and development, fruit-setting, seed development, and anthocyanin biosynthesis (Weiss et al., 1992; Hooley, 1994). GAs are synthesized from the tetracyclic diterpene ent-kaurene by two types of enzymes (Hedden, 1997; Hedden and Kamiya, 1997). Monooxygenases synthesize GA12 and GA53 in the early reactions. These GAs are converted to physiologically active C19-GAs by GA 20-oxidases and 3β-hydroxylases, two principal dioxygenases. The GA 20-oxidases form a C19 skeleton by the sequential oxidation of C-20 of GA12 and GA53. The 3β-hydroxylases catalyze 3β-hydroxylation of the C19 skeleton, resulting in the formation of GAs that are biologically active. Genomic and cDNA clones encoding GA 20-oxidase and 3β-hydroxylase have been isolated from several plant species, and their enzymatic properties have been identified with recombinant enzymes produced in Escherichia coli (Lange et al., 1994; Chiang et al., 1995; Phillips et al., 1995; Xu et al., 1995; Wu et al., 1996; Garcia-Martinez et al., 1997; Hedden and Kamiya, 1997; Lange, 1997; Lester et al., 1997; MacMillan et al., 1997; Toyomasu et al., 1998).
It has been shown that the expression of the two dioxygenases is repressed in feedback regulation by GA treatment (Chiang et al., 1995; Phillips et al., 1995; Xu et al., 1995; Martin et al., 1996). It has also been demonstrated that the expression of the GA 20-oxidases in Arabidopsis (Xu et al., 1995) and spinach (Wu et al., 1996) is enhanced under long-day conditions. Studies on GA 20-oxidases in Arabidopsis, bean, pea, and tomato plants have demonstrated that plants contain multiple GA 20-oxidase genes that are regulated spatially and temporally during development. These genes appear to be involved in different GA-regulated processes, such as stem elongation, flower formation, and fruit growth (Phillips et al., 1995; Garcia-Martinez et al., 1997; Rebers et al., 1999).
Developing seeds contain more abundant amounts of GAs than any other plant organ, and have therefore been used frequently to elucidate GA biosynthetic pathways in plants (Graebe, 1987; Lange et al., 1997; MacMillan et al., 1997; Rodrigo et al., 1997). However, little is known about the roles of GAs and the exact locations of synthesis in developing seeds. It has recently been suggested that GAs are involved in the early and late stages of seed development in some plant species (Eeuwens and Schwabe et al., 1975; Graebe, 1987; Phillips et al., 1995; Lange, 1997; MacMillan et al., 1997; Swain et al., 1997). Several GA-deficient mutants that have altered seed development have been reported in the pea plant. The lh-2 mutation dramatically reduces GA levels in developing seeds and increases seed abortion (Swain et al., 1993, 1995).
It has been reported that some of the GA 20-oxidases are expressed specifically in seeds or fruit. YAP169 of Arabidopsis is expressed in siliques (Phillips et al., 1995), Pv85-26 of bean in developing seeds (Garcia-Martinez et al., 1997), and M3-8 of Marah macrocarpus in the embryos and endosperm of developing seeds (MacMillan et al., 1997). In present study, we report the isolation of the GA 20-oxidase gene from watermelon (Citrullus lanatus), which is expressed specifically in the maternal tissues of developing seeds.
MATERIALS AND METHODS
Plant Material
An F1 hybrid of watermelon (Citrullus lanatus [Thunb.] var Country Home) was used. Plants were cultivated in a greenhouse and maintained at 30°C to 35°C during the day and 15°C to 20°C at night. Parthenocarpic fruit development was induced with 100 mg/L N-(2-chloro-4-pyridyl)-N′-phenylurea (CPPU) in a 0.1% aqueous Tween 20 solution by application to the ovaries at the flowering stage. RNAs were extracted from the roots, hypocotyls, and cotyledons of seedlings grown in sterile vermiculite. Seed coats and the inner parts of seeds, including the embryo, endosperm, nucellus, and transfer cells, were separately collected after splitting seeds with a razor blade. Roots were washed thoroughly in tap water and blotted on a paper towel. All plant materials were quickly frozen in liquid N2 after harvest and stored at −70°C until use.
Construction of cDNA Library and Isolation of Full-Length ORF cDNA Clones
A cDNA library was constructed in a λ-ZAPII (Stratagene, La Jolla, CA) vector with poly(A+) RNAs extracted from watermelon seeds at 6 to 10 DAP using a cDNA synthesis kit (ZAP cDNA kit, Stratagene) and an in vitro packaging mix (Gigapack III Gold, Stratagene). A cDNA library of 4.0 × 106 independent recombinants was obtained and amplified in Escherichia coli XL1-Blue (Stratagene). Plaque hybridization experiments were performed with 4 × 105 plaques, which were lifted onto nitrocellulose membranes. The pBluescript plasmids containing cDNA inserts were in vivo rescued from the λ-bacteriophage using the f1 helper phage R408 (Stratagene).
PCR Cloning
A pair of degenerate primers was designed to isolate the GA 20-oxidase gene. The following primers were synthesized: forward primer, 5′-ATGTGG(CT)(AC)NGA(AG)-GGNTT(CT)AC-3′; and reverse primer, 5′-GT(AG)TGNGC-NGCNAGNCCCAT-3′, where N is a mixture of A, C, G, and T. DNAs isolated from the 6 to 10 DAP seed cDNA library were used as templates. The PCR reaction was initiated by heating to 94°C for 5 min, then subjected to 40 cycles of 94°C for 1 min, 40°C or 50°C for 1 min, and 72°C for 1 min. The reaction was completed by a 10-min incubation at 72°C. The products were purified by agarose gel electrophoresis and cloned into pGEM-T Easy vector (Promega, Madison, WI).
DNA Sequence Analysis
The nucleotide sequence was determined using a DNA sequencing kit (Big Dye Terminator Cycle Sequencing Ready Reaction Kit, PE-Applied Biosystems, Foster City, CA) with a DNA sequencer (model ABI 373, PE-Applied Biosystems). Homology search within the databases was done using the BLAST program of the DNA databank of Japan (http//www.ddbj.nig.ac.jp/E-mail/homology.html).Alignments of amino acid sequences were performed using the Clustal W program (http//www.clustalw.genome.ad.jp/).
DNA and RNA Gel-Blot Analysis
The cetyltrimethylammonium bromide method was used to isolate total genomic DNA from the leaves of 2-week-old watermelon plants (Rogers and Bendich, 1988). Eight micrograms of DNA was digested with the appropriate restriction enzymes, separated on a 0.8% (w/v) agarose gel, blotted onto a nylon membrane, and hybridized with a 32P-labeled probe by the random-priming method (Sambrook et al., 1989). Total RNA was isolated from various organs using Tri Reagent (Molecular Research Center, Cincinnati, OH). Leaf and root samples were harvested from 2-week-old seedlings. Floral organ samples were obtained by dissecting mature flowers under a dissecting microscope. Ten to 25 μg of total RNA was used for the blot analysis, as described previously (Kang et al., 1997).
In Situ Hybridization
Watermelon seeds were fixed in FAA fixative solution (50% [v/v] ethanol, 0.9 m glacial acetic acid, and 3.7% [v/v] formaldehyde) for 15 h at 4°C, dehydrated with ethanol, infiltrated with xylene, and embedded in paraffin (Paraplast X-tra, Oxford Labware, St. Louis) (Mckhann and Hirsch, 1993). Eight to 10 μm of the sections were transferred on Vectabond-coated (Vector Laboratories, Burlingame, CA) slides and dried overnight at 45°C. The riboprobes were prepared using the DIG nucleic acid labeling kit (Roche Molecular Biochemicals, Mannheim, Germany) according to the manufacturer's protocol. The antisense and sense probes of the cDNA were synthesized using T7 and T3 RNA polymerase and hydrolyzed in bicarbonate at 60°C for 30 min. The sections were treated with a solution containing 20 μg mL−1 proteinase K at 37°C for 30 min. The samples were acetylated and hybridized in a solution containing 50% (v/v) formamide, 0.3 m NaCl, 20 mm Tris-HCl (pH 7.5), 5 mm EDTA, 5 mm Na2HPO4, 10% (w/v) dextran sulfate, 1× Denhardt's solution, 0.5 mg mL−1 yeast tRNA, 80 μg mL−1 salmon-sperm DNA, and 300 ng riboprobe mL−1 at 50°C for 18 h. After incubation, slides were treated with RNase (20 μg mL−1 at 37°C for 30 min) to remove free RNA probes, and then washed in 1× SSC at 55°C for 1 h and 0.1× SSC at 55°C for 1 h. Hybridized probes were visualized by an anti-digoxigenin-alkaline phosphatase conjugate and the color substrates nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (Roche Molecular Biochemicals).
Heterologous Expression in E. coli and Enzyme Assay
The GA 20-oxidase cDNA clone was inserted into the BamHI and PstI sites of the pMAL-c2 vector (New England Biolabs, Beverly, MA), generating a maltose binding protein-Cv20ox fusion. The fusion protein was isolated from the E. coli JM109 harboring the plasmid according to the manufacturer's instructions (Rebers et al., 1999). The enzyme preparations were assayed for enzyme activity by incubation with GAs (200 ng) at 30°C for 1 h under the conditions described previously (Toyomasu et al., 1997). GA12 and GA53 were a gift from Professor Takeshi Sassa (Yamagata University, Tsuruoka, Japan) and used as a substrate. After derivatization with methyl ester-trimethylsilyl ether, the product was subjected to full-scan analysis using a gas chromatograph-mass spectrometer (Finnigan MAT, San Jose, CA). GC-MS conditions were as described previously (Kawaide et al., 1995).
Cloning of the Promoter Region of the Cv20ox Gene
The promoter region of the Cv20ox gene was isolated with an in vitro PCR cloning kit (TaKaRa Biotechnology, Shiga, Japan). Ten micrograms of watermelon genomic DNA was digested with the HindIII restriction enzyme, ligated to the HindIII cassette, and amplified by PCR according to the manufacturer's instructions. PCR products were cloned into the pGEM-T Easy vector, and the resulting plasmid was named pGA2044.
Particle Bombardment and GUS Assay
The plasmid pGA2118 was constructed by inserting the 0.7-kb promoter fragment into the HindIII/PstI sites of pGA1230, a high-copy-number derivative of the GUS vector pBI101.2 (CLONTECH Laboratories, Palo Alto, CA). All target materials were bombarded using the Biolistic PDS-1000/He device (Bio-Rad Laboratories, Hercules, CA). The plasmid DNA was adsorbed to tungsten particles (M-10, Bio-Rad) according to the protocol recommended by the manufacturer. The target materials were positioned 8 cm below the macrocarrier stopping screen, which was positioned 2 cm below the 1100 PSI rupture disc. After bombardment, the samples were incubated for 30 h at 27°C in the dark. Histochemical GUS staining was performed according to the method of Jeon et al. (1999).
RESULTS
Isolation of a cDNA Homologous to GA 20-Oxidase
To isolate the genes responsible for GA biosynthesis, we first prepared a cDNA library from developing watermelon seeds. The initial number of plaque-forming units in the library was 4 × 106, which is sufficient to contain low-abundance clones. A DNA fragment of 300 bp was amplified from the watermelon seed cDNA library using degenerate PCR primers against conserved sequences in GA 20-oxidases. The PCR product was cloned in the pGEM-T Easy vector (Promega), and its sequence was determined to confirm the apparent GA 20-oxidase identity. Sequence comparison against genes in the database by the BLAST search program showed that its deduced amino acid sequence is highly homologous to the GA 20-oxidase from various plant species (Lange et al., 1994; Phillips et al., 1995; Xu et al., 1995; Garcia-Martinetz et al., 1997; Lange, 1997; MacMillan et al., 1997).
Since the clone was partial, a cDNA clone containing a full-length ORF of the putative GA 20-oxidase was isolated by screening the seed cDNA library. The clone, Cv20ox, consists of 1,420 nucleotides and encodes a putative protein of 379 amino acid residues (Fig. 1) (accession no. AF074709). The peptide has conserved regions typical of 2-oxoglutarate-dependent dioxygenases, including an Fe2+-binding motif consisting of His-239, Asp-241, and His-295 (Roach et al., 1995), and the putative 2-oxoglutarate-binding motif NYYPPCEKP (residues 222–230) (Xu et al., 1995). BLAST database searches showed that the Cv20ox peptide is highly homologous to the GA 20-oxidases (Fig. 1). The watermelon protein shares 76% and 63% identities with the GA 20-oxidases of M. macrocarpus (MacMillan et al., 1997) and pumpkin (Lange, 1997), respectively. The protein also shares >50% identity with the GA 20-oxidases of pea (Garcia-Martinez et al., 1997), bean (Garcia-Martinez et al., 1997), Arabidopsis (Phillips et al., 1995), spinach (Wu et al., 1996), and lettuce (Toyomasu et al., 1998).
Figure 1.
Alignment of the deduced amino acid sequence for the Cv20ox cDNA with GA 20-oxidases from different species. Regions identical to all GA 20-oxidases are boxed in black. The number symbols (#) indicates the putative Fe2+-binding motif typical for 2-oxoglutarate-dependent dioxygenases, and the asterisks (*) indicate the putative 2-oxoglutarate-binding motif. Dashes were introduced for the maximum sequence homology.
Functional Expression in E. coli
The high degree of similarity between the amino acid sequences of the watermelon Cv20ox protein and GA 20-oxidases suggested that Cv20ox encodes a GA 20-oxidase. This was confirmed by expressing the full-length ORF cDNA as a fusion protein in E. coli using the pMAL-c2 vector (New England Biolabs). The incubation of GA12 with the Cv20ox protein producedGA9. GA25 was also detected in the incubation in trace amounts (Table I). When GA53 was incubated with the recombinant protein, GA20 and GA44 were identified (data not shown).
Table I.
GC-MS data of methyl ester trimethylsilyl ether derivatives of metabolites from the substrate GA12 incubated with Cv20ox fusion protein
| Recombinant Protein | Substrate | Metabolite | KRIa | MS Data |
|---|---|---|---|---|
| m/z (% relative intensity) | ||||
| Cv20ox | GA12 | GA9 | 2,352 | 330 (4, [M+]), 298 (56), 270 (100), 243 (46), 226 (48) |
| GA25 | 2,476 | 404 (0.1, [M+]), 372 (20), 312 (90), 284 (100), 225 (47) |
KRI, Kovats retention index.
Genomic DNA-Blot Analysis
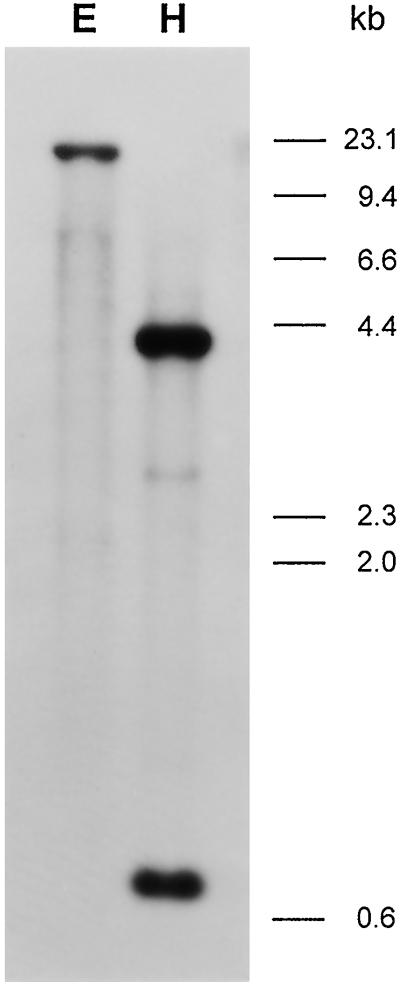
Genomic DNA-blot analysis was conducted to identify the copy number of the Cv20ox gene within the watermelon genome. The blot was hybridized with the cDNA probe and washed at low- or high-stringency conditions. Similar hybridization patterns were obtained from both stringency conditions. The result in Figure 2 shows that the clone hybridized to one or two major bands, indicating that there are one to two copies of the gene. The presence of weakly hybridizing bands suggests that there are additional genes that are related to Cv20ox.
Figure 2.
Genomic DNA-blot analysis of Cv20ox. Ten micrograms of watermelon genomic DNA was digested with EcoRI (E) or HindIII (H) and resolved on a 0.8% agarose gel. The 300-bp cDNA cloned by PCR was used as a probe. The positions of HindIII-digested λ-DNA size markers are shown on the right in kb.
Expression Pattern of the Cv20ox Gene
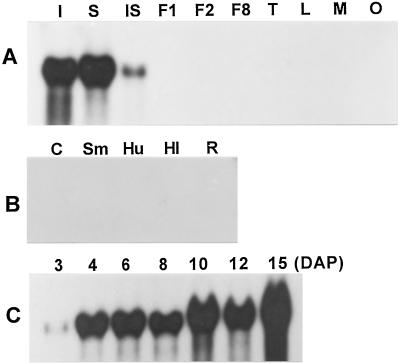
The spatial expression pattern of the GA 20-oxidase gene Cv20ox was studied by RNA-blot analysis using RNAs isolated from various reproductive and vegetative organs, including whole seeds, integuments, inner seed tissues, 1 DAP fruits, 2 to 3 DAP fruits, tendrils, young leaves, male flowers, and ovaries before pollination. The results show that a band of approximately 1.4 kb of the Cv20ox transcript was present in seeds, especially in the integument tissues, and was also present weakly in the inner part of the seeds (Fig. 3A). The transcript was not detectable in any other reproductive organs. RNA-blot analysis with mRNA isolated from cotyledons, hypocotyls, shoot meristems, and roots of seedlings grown 7 d after imbibition showed that the Cv20ox transcript was not present in any organ at this stage of development (Fig. 3B).
Figure 3.
Spatial and temporal expression of Cv20ox. A, The blot of the RNAs isolated from different parts of the mature plant was hybridized with the same probe as that used in genomic DNA-blot analysis. I, Integuments (8 DAP); S, whole seeds (8 DAP); IS, inner seed tissues (8 DAP); F1, fruits (1 DAP); F2, fruits (2–3 DAP); F8, fruits (8 DAP); T, tendrils; L, leaves; M, male flowers; O, ovaries before pollination. B, RNAs isolated from different organs of young plants grown for 15 d after imbibition were blotted and hybridized with the same probe as that described above. C, Cotyledons; Sm, shoot meristem including uppermost of hypocotyls; Hu, upper hypocotyls; Hl, lower hypocotyls; R, roots. Twenty-five micrograms of RNA was used for the blot analyses. C, Temporal expression pattern of Cv20ox during seed development. Twenty micrograms of RNAs isolated from whole seeds at seven different stages after pollination was resolved on a 0.8% agarose gel, transferred onto a nylon membrane, and hybridized with the Cv20ox probe.
The temporal expression pattern of the Cv20ox gene was studied during watermelon seed development. The transcript began to appear in seeds at 3 DAP, increased prominently at 4 DAP, and this high level was maintained until 15 DAP (Fig. 3C). The expression pattern of the gene during the late stage of fruit development could not be studied, because it was difficult to isolate RNAs from seeds after 20 DAP.
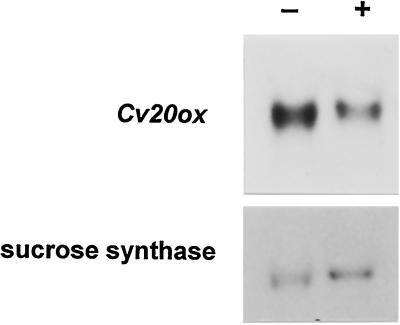
It has previously been shown that expression of GA 20-oxidase genes from several plants is feedback-regulated by active GAs (Phillips et al., 1995; Martin et al., 1996; Toyomasu et al., 1997). Therefore, we have examined whether the Cv20ox transcript level is altered by GA3 treatment in developing watermelon seeds. As a control, the level of Suc synthase transcript was measured. The result in Figure 4 shows that expression of Cv20ox was repressed by GA3 treatment, while the expression of the Suc synthase was not affected.
Figure 4.
Effects of GA3 treatment on Cv20ox gene expression. +, 3 DAP fruits injected with about 300 μL of 2 × 10−5 m GA3 dissolved in 0.1% Tween 20, and seeds were harvested 24 h after the treatment. −, Control RNA taken from seeds of fruits injected with 0.1% Tween 20 solution. The same blot was washed and rehybridized with the Suc synthase cDNA probe to confirm equal loading of RNAs.
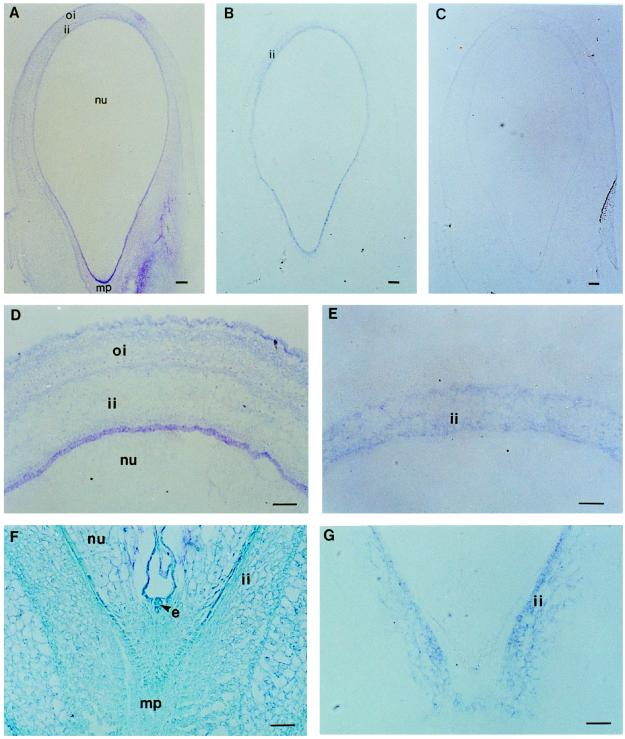
In situ hybridization experiments were conducted to localize the tissue types that express the Cv20ox gene in developing seeds (Fig. 5). It was revealed that the Cv20ox mRNAs were localized predominantly in the inner layer of the integument (Fig. 5, B, E, and G). This result is consistent with the RNA-blot experiment. However, the tissue type that expresses the Cv20ox gene in the inner parts of the watermelon seeds was not detectable by the method, probably due to a low-level expression.
Figure 5.
Localization of the Cv20ox mRNA in watermelon seeds by in situ hybridization. All samples were sectioned longitudinally in parallel with the plane of seed. Micrographs show pollinated seeds at 5 DAP (A, B, and C), at 6 DAP (D and E), and at 4 DAP (F and G). The sections were hybridized to either an antisense (B, E, and G) or a sense (C) RNA probe of the entire cDNA that was labeled with digoxigenin. A, D, and F are toluidine-blue-stained samples. nu, Nucellus; ii, inner layer of integument; oi, outer layers of integument; mp, micropylar end; e, globular embryo. Bar = 100 μm.
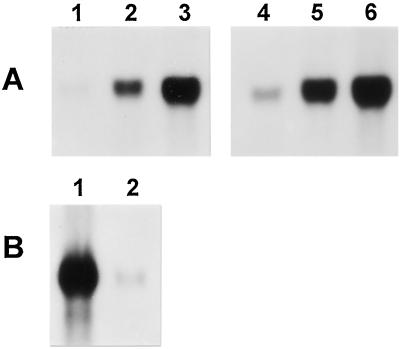
Expression Pattern of Cv20ox in Parthenocarpic Fruits Induced by CPPU Treatment
CPPU treatment resulted in earlier expression of the Cv20ox gene compared with pollinated controls (Fig. 6A). The earlier induction of the gene was probably due to promoted seed and fruit development in the CPPU-treated samples. The expression pattern of Cv20ox at 10 d after CPPU treatment showed that the transcript had accumulated primarily in the integument and also weakly in the inner parts of the seeds (Fig. 6B). Since the CPPU-treated seeds did not develop embryos or endosperm, the GA 20-oxidase transcript observed in the inner parts was probably present in nucellar or transfer cells.
Figure 6.
Effects of CPPU on Cv20ox gene expression. A, Comparative RNA-blot analysis of the Cv20ox gene between seeds harvested after normal pollination and after CPPU treatment. Lanes 1, 2, and 3, Seeds at 2, 3, and 4 DAP, respectively. Lanes 4, 5, and 6 are samples from 2, 3, and 4 d after CPPU treatment, respectively. B, The Cv20ox mRNA expression pattern in integuments (1) and inner seed tissues (2) of seeds at 10 d after CPPU treatment.
Isolation of the Cv20ox Promoter and Study of Promoter Activity Using Transient Expression System
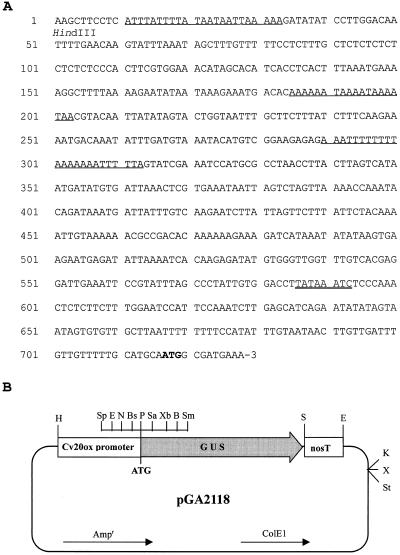
About 700 bp of the 5′-upstream region of the Cv20ox promoter was isolated from watermelon total genomic DNA. The sequence analysis showed that the putative TATA box (TATAAATC) is present 132 nucleotides upstream from the translation start codon (Fig. 7A). The 5′-upstream region was highly A/T rich, showing 72% of A/T content. The region contained three stretches of more than 19-bp A/T nucleotides. It has been reported that the 5′-upstream region of the ConA gene, which is one of the major seed-storage protein genes of Canavalia gladiata, is highly A/T-rich and contains several long A/T-rich sequences. It was suggested that the A/T-rich sequences of the 5′-upstream region play a role in transcriptional activation (Yamamoto et al., 1995).
Figure 7.
A, Nucleotide sequence of the 5′-upstream region of the Cv20ox gene. The putative TATA box is double-underlined and the translation start codon is in bold. Sequences consisting of 19 bp or more A/T are underlined. B, Plasmid map of pGA2118. The terminator region of the nopaline synthase gene (nosT) was placed after the GUS coding region. H, HindIII; Sp, SpeI; N, NotI; Bs, BstZI; Sa, SalI; Xb, XbaI; B, BamHI; Sm, SmaI; P, PstI; S, SacI; E, EcoRI; K, KpnI; X, XhoI; St, StuI.
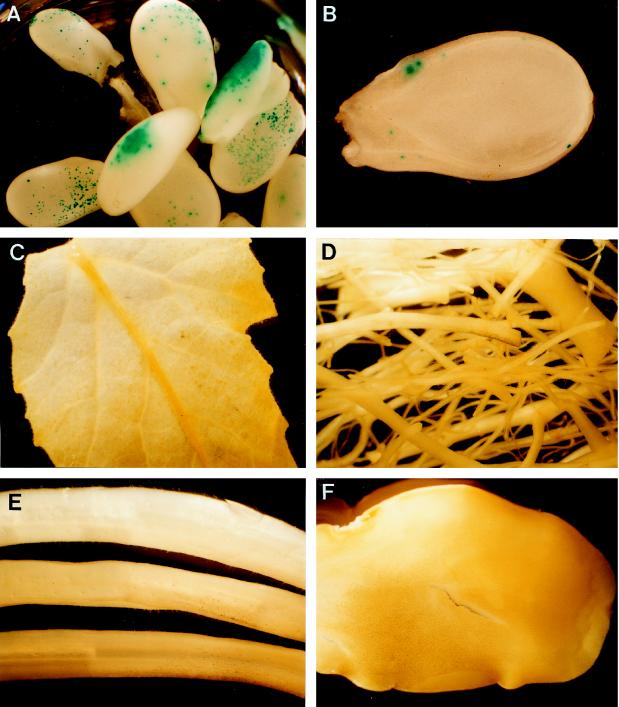
To study the tissue specificity of the Cv20ox promoter, we constructed a fusion between the Cv20ox promoter and the GUS coding region (Fig. 7B). The fusion molecule was introduced into various organs of watermelon plants by particle bombardment, and transient expression of the gene was measured by histochemical analysis of GUS activity. The results show that the Cv20ox promoter activity was detectable only in the integument tissues of the developing seeds, and very strongly in the layer close to nucellus (Fig. 8B). The promoter activity was not detectable in vegetative organs such as leaves (Fig. 8C), roots (Fig. 8D), hypocotyls (Fig. 8E), and cotyledons (Fig. 8F). The expression pattern coincided with that of the in situ localization experiment.
Figure 8.
Histochemical detection of GUS expression in different organs of watermelon plants after bombardment with tungsten particles coated with pGA2118. A, Developing seeds (8 DAP); B, inside of the seeds cut in parallel with the plane of the seeds (8 DAP); C to F, leaves, roots, hypocotyl, and cotyledons of seedlings, respectively.
DISCUSSION
We have isolated a cDNA clone, Cv20ox, from developing seeds of watermelon. The clone encodes a protein that is homologous to GA 20-oxidases, which are responsible for the formation of the C19 skeleton GAs. The Cv20ox protein has the conserved regions typical of 2-oxoglutarate-dependent dioxygenases, supporting the possibility that it is involved in the biosynthesis of GAs. Additional evidence that the clone is the Cv20ox gene was obtained by feedback regulation of the transcript level by application of GA3. Finally, the protein produced in E. coli catalyzed the conversion of GA12 to GA9 and GA25. Recombinant enzymes of the GA 20-oxidase cDNAs cloned from various plants have different catalytic properties, substrate affinities, and product distributions, which may indicate differences in GA metabolism between the species of origin (Garcia-Martinez et al., 1997). The watermelon GA 20-oxidase had the same activity as the recombinant YAP169 of Arabidopsis (Phillips et al., 1995) and M3-8 of M. macrocarpus (MacMillan et al., 1997), but seems to differ from Cm20ox of the pumpkin in its catalytic properties, which produced mainly GA25 from GA12 (Lange et al., 1994).
Among the various organs examined, the GA 20-oxidase gene is specifically expressed in developing seeds. Among the different tissues of the seeds, it was expressed very strongly in integument tissues and also weakly in inner parts of the seeds. This supports the fact that developing seeds are important in the synthesis of various GAs. An in situ localization experiment showed that the GA 20-oxidase transcript is present in the innermost tissues of the integument. However, the experiment failed to reveal the tissue types in the inner parts of the seeds that express the gene, probably since the gene is weakly expressed in the tissues. In parthenocarpic watermelon fruits induced by CPPU treatment, the transcript level of the GA 20-oxidase gene was not significantly altered, suggesting that the gene is expressed in maternal tissues, probably the nucellus or transfer cells. Transient expression analysis of the Cv20ox promoter using the GUS reporter also revealed that the gene is specifically expressed in the inner integument tissue. The promoter-reporter fusion may be used to determine cues mediating the regulation of Cv20ox during seed development.
There are several reports that the seed coat plays an important role during seed development. The role of the Suc-metabolizing enzymes in the seed coat has been studied in leguminous plants (Weber et al., 1997). A fava bean invertase gene, VfCWINV1, is specifically expressed in the veins and the thin-walled parenchyma, representing the unloading area of the seed coat during early seed development (Weber et al., 1995). The physiological and molecular features of Suc synthase in the pea seed coat suggest that the gene plays a central role in controlling the Suc concentration in that organ (Dejardin et al., 1997). The genetic analysis of the Arabidopsis banyuls (ban) mutant suggests that BANYULS functions as a negative regulator of flavonoid biosynthesis, preventing the accumulation of pigments in the seed coat during early embryogenesis (Albert et al., 1997). It is yet to be determined how expression of these genes is regulated and whether plant hormones are involved. Our observation that a GA 20-oxidase gene is specifically expressed in seed coats suggests that GAs may play an important role in controlling other genes in developing seed coats.
Our conclusion is consistent with previous results from Arabidopsis (Phillips et al., 1995), M. macrocarpus (MacMillan et al., 1997), and French bean (Garcia-Martinez et al., 1997) that a seed-specific GA 20-oxidase is present. However, the Cv20ox gene is unique in that it is expressed preferentially in integument tissues and not in embryos, endosperm, or cotyledons. This suggests that the GA 20-oxidase genes were differentiated diversely according to their biological roles. It is likely that there are additional GA 20-oxidase genes in watermelon that are active in vegetative or reproductive organs, as has been reported for Arabidopsis (Phillips et al., 1995) and French bean (Garcia-Martinez et al., 1997).
The importance of GAs in seed development has been proposed by genetic and physiological studies. In the pea plant, GAs are required for embryo growth and seed development (Swain et al., 1997). The lh-2 (previously named lhi) mutation reduces GA levels remarkably in developing seeds and increases seed abortion (Swain et al., 1993, 1995). Physiological, biochemical, and genetic analysis of ls-1 and lh-2 suggest that GAs synthesized in the embryo and/or endosperm are required for seed development in the first few days after pollination, but not in older seeds (Swain et al., 1993, 1995; Ait-Ali et al., 1997). Moreover, the presence of seeds is very important for the developing pericarp, because of its need for phytohormones such GAs and auxins (van Huizen et al., 1997). In the tomato plant, GAs are also required for developing fruits and seeds, but only for a short time after fertilization (Groot et al., 1987). However, GAs are not necessary for seed or fruit development in Arabidopsis, although the seeds produce GAs (Barendse et al., 1986). We observed that in parthenocarpic fruits induced by CPPU treatment, the integument tissues developed normally at the early stages of seed development but failed to develop pigments at the later stage (H.G. Kang, S.H. Jun, J. Kim, and G. An, unpublished data). Therefore, GAs synthesized in zygotic tissues may be necessary for anthocyanin biosynthesis in the integuments.
ACKNOWLEDGMENTS
We thank Chahm An for critical reading of the manuscript, Masayo Sekimoto for her technical assistance with the enzyme assay of the recombinant protein, and Dr. Young-Yell Yang for useful discussions. We also thank Jeong-Hwan Choi and Yu-Chan Park for maintaining the watermelon plants. Watermelon seeds were kindly provided by Dongbu Hannong Seeds Co., Korea.
Footnotes
This work was funded in part by a grant from a special grant research program of the Ministry of Agriculture, Forestry and Fishery of Korea.
LITERATURE CITED
- Ait-Ali T, Swain SM, Reid JB, Sun T, Kamiya Y. The LS locus of pea encodes the gibberellin biosynthesis enzyme ent-kaurene synthase A. Plant J. 1997;11:443–454. doi: 10.1046/j.1365-313x.1997.11030443.x. [DOI] [PubMed] [Google Scholar]
- Albert S, Delseny M, Devic M. BANYULS, a novel negative regulator of flavonoid biosynthesis in the Arabidopsis seed coat. Plant J. 1997;11:289–299. doi: 10.1046/j.1365-313x.1997.11020289.x. [DOI] [PubMed] [Google Scholar]
- Barendse GWM, Kepczynski J, Karssen CM, Koornneef M. The role of endogenous gibberellins during fruit and seed development: studies on gibberellin-deficient genotypes of Arabidopsis thaliana. Physiol Plant. 1986;67:315–319. [Google Scholar]
- Chiang HH, Hwang I, Goodman HM. Isolation of the Arabidopsis GA4 locus. Plant Cell. 1995;7:195–201. doi: 10.1105/tpc.7.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejardin A, Rochat C, Maugenest S, Boutin J-P. Purification, characterization and physiological role of sucrose synthase in the pea seed coat (Pisum sativum L.) Planta. 1997;201:128–137. doi: 10.1007/BF01007697. [DOI] [PubMed] [Google Scholar]
- Eeuwens CJ, Schwabe WW. Seed and pod wall development in Pisum sativum L. in relation to extracted and applied hormones. J Exp Bot. 1975;26:1–14. [Google Scholar]
- Garcia-Martinez JL, Lopez-Diaz I, Sanchez-Beltran MJ, Phillips AL, Ward DA, Gaskin P, Hedden P. Isolation and transcript analysis of gibberellin 20-oxidase genes in pea and bean in relation to fruit development. Plant Mol Biol. 1997;33:1073–1084. doi: 10.1023/a:1005715722193. [DOI] [PubMed] [Google Scholar]
- Graebe JE. Gibberellin biosynthesis and control. Annu Rev Plant Physiol. 1987;38:419–465. [Google Scholar]
- Groot SPC, Bruinsma J, Karssen CM. The role of endogenous gibberellin in seed and fruit development of tomato: studies with a gibberellin-deficient mutant. Physiol Plant. 1987;71:184–190. [Google Scholar]
- Hedden P. The oxidases of gibberellin biosynthesis: their function and mechanism. Physiol Plant. 1997;101:709–719. [Google Scholar]
- Hedden P, Kamiya Y. Gibberellin biosynthesis: enzymes, genes and their regulation. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:431–460. doi: 10.1146/annurev.arplant.48.1.431. [DOI] [PubMed] [Google Scholar]
- Hooley R. Gibberellins: perception, transduction and responses. Plant Mol Biol. 1994;26:1529–1555. doi: 10.1007/BF00016489. [DOI] [PubMed] [Google Scholar]
- Jeon JS, Chung YY, Lee S, Yi GH, Oh BG, An G. Isolation and characterization of anther-specific gene, RA8, from rice (Oryza sativa L.) Plant Mol Biol. 1999;39:35–44. doi: 10.1023/a:1006157603096. [DOI] [PubMed] [Google Scholar]
- Kang HG, Jang S, Chung JE, Cho YG, An G. Characterization of two rice MADS box genes that control flowering time. Mol Cell. 1997;7:559–566. [PubMed] [Google Scholar]
- Kawaide H, Sassa T, Kamiya Y. Plant-like biosynthesis of gibberellin A1 in the fungus Phaeosphaeria sp. L487. Phytochemistry. 1995;39:305–310. [Google Scholar]
- Lange T. Cloning gibberellin dioxygenase genes from pumpkin endosperm by heterologous expression of enzyme activities in Escherichia coli. Proc Natl Acad Sci USA. 1997;94:6553–6558. doi: 10.1073/pnas.94.12.6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange T, Hedden P, Graebe J. Expression cloning of a gibberellin 20-oxidase, a multifunctional enzyme involved in gibberellin biosynthesis. Proc Natl Acad Sci USA. 1994;91:8552–8556. doi: 10.1073/pnas.91.18.8552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange T, Robatzek S, Frisse A. Cloning and expression of a gibberellin 2β,3β-hydroxylase cDNA from pumpkin endosperm. Plant Cell. 1997;9:1459–1467. doi: 10.1105/tpc.9.8.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester DR, Ross JJ, Davies PJ, Reid JB. Mendel's stem length gene (Le) encodes a gibberellin 3β-hydroxylase. Plant Cell. 1997;9:1435–1443. doi: 10.1105/tpc.9.8.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMillan J, Ward DA, Phillips AL, Sanchez-Beltran MJ, Gaskin P, Lange T, Hedden P. Gibberellin biosynthesis from gibberellin A12-aldehyde in endosperm and embryos of Marah macrocarpus. Plant Physiol. 1997;113:1369–1377. doi: 10.1104/pp.113.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DN, Proebsting WM, Parks TD, Dougherty WG, Lange T, Lewis MJ, Gaskin P, Hedden P. Feed-back regulation of gibberellin biosynthesis and gene expression in Pisum sativum L. Planta. 1996;200:159–166. doi: 10.1007/BF00208304. [DOI] [PubMed] [Google Scholar]
- Mckhann HI, Hirsch AM. In situ localization of specific mRNAs in plant tissues. In: Glick BR, Thompson JE, editors. Methods in Plant Molecular Biology and Biotechnology. Boca Raton, FL: CRC Press; 1993. pp. 179–205. [Google Scholar]
- Phillips AL, Ward DA, Uknes S, Appleford NE, Lange T, Huttly AK, Gaskin P, Graebe JE, Hedden P. Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiol. 1995;108:1049–1057. doi: 10.1104/pp.108.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebers M, Kaneta T, Kawaide H, Yamaguchi S, Yang Y-Y, Imai R, Sekimoto H, Kamiya Y. Regulation of gibberellin biosynthesis genes during flower and early fruit developing of tomato. Plant J. 1999;17:241–250. doi: 10.1046/j.1365-313x.1999.00366.x. [DOI] [PubMed] [Google Scholar]
- Roach PL, Clifton IJ, Fulop V, Harlos K, Butron GJ, Hajdu J, Andersson I, Schofield CJ, Baldwin JE. Crystal structure of isopenicillin N synthase is the first from a new structural family of enzymes. Nature. 1995;375:700–704. doi: 10.1038/375700a0. [DOI] [PubMed] [Google Scholar]
- Rodrigo MJ, Hedden P, Garcia-Martinez JL, Santes CM, Gaskin P, Hedden P. The role of gibberellins A1 and A3 in fruit growth of Pisum sativum L. and the identification of gibberellins A4 and A7 in young seeds. Planta. 1997;20:446–453. [Google Scholar]
- Rogers SO, Bendich AJ. Extraction of DNA from plant tissues. In: Gelvin SB, Schilperoort RA, editors. Plant Molecular Manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1988. p. A6. :1–10. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Swain SM, Reid JB, Kamiya Y. Gibberellins are required for embryo growth and seed development in pea. Plant J. 1997;12:1329–1338. [Google Scholar]
- Swain SM, Reid JB, Ross JJ. Seed development in Pisum: the lhi allele reduces gibberellin levels in developing seeds, and increases seed abortion. Planta. 1993;191:482–488. [Google Scholar]
- Swain SM, Ross JJ, Reid JB, Kamiya Y. Gibberellins and pea seed development: expression of the lhi, ls and le mutations. Planta. 1995;195:426–433. [Google Scholar]
- Toyomasu T, Kawaide H, Mitsuhashi W, Inoue Y, Kamiya Y. Phytochrome regulates gibberellin biosynthesis during germination of photoblastic lettuce seeds. Plant Physiol. 1998;118:1517–1523. doi: 10.1104/pp.118.4.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyomasu T, Kawaide H, Sekimoto H, von Numers C, Phillips AL, Hedden P, Kamiya Y. Cloning and characterization of a cDNA encoding gibberellin 20-oxidase from rice (Oryza sativa) seedlings. Physiol Plant. 1997;99:111–118. [Google Scholar]
- van Huizen R, Ozga JA, Reinecke DM. Seed and hormonal regulation of gibberellin 20-oxidase expression in pea pericarp. Plant Physiol. 1997;115:123–128. doi: 10.1104/pp.115.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H, Borisjuk L, Heim U, Buchner P, Wobus U. Seed coat-associated invertases of fava bean control both unloading and storage functions: cloning of cDNAs and cell type-specific expression. Plant Cell. 1995;7:1835–1846. doi: 10.1105/tpc.7.11.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H, Borisjuk L, Wobus U. Sugar import and metabolism during seed development. Trends Plant Sci. 1997;2:169–174. [Google Scholar]
- Weiss D, van Blokland R, Kooter JM, Mol JNM, van Tunen AJ. Gibberellic acid regulates chalcone synthase gene transcription in the corolla of Petunia hybrida. Plant Physiol. 1992;98:191–197. doi: 10.1104/pp.98.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Li L, Gage DA, Zeevaart JAD. Molecular cloning and photoperiod-regulated expression of gibberellin 20-oxidase from the long-day plant spinach. Plant Physiol. 1996;110:547–554. doi: 10.1104/pp.110.2.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YL, Li L, Wu K, Peeters AJ, Gage DA, Zeevaart JAD. The GA5 locus of Arabidopsis thaliana encodes a multifunctional gibberellin 20-oxidase: molecular cloning and functional expression. Proc Natl Acad Sci USA. 1995;92:6640–6644. doi: 10.1073/pnas.92.14.6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Nishihara M, Morikawa H, Yamauchi D, Minamikawa T. Promoter analysis of seed storage protein genes from Canavalia gladiata D.C. Plant Mol Biol. 1995;27:729–741. doi: 10.1007/BF00020226. [DOI] [PubMed] [Google Scholar]