Abstract
Background
The aim of this study was to investigate the genotypic differences between different sequence type MRSA isolates, especially focusing on silent rpoB474 mutations and the relationship between such mutations and ciprofloxacin resistance.
Methods
Seventy-nine MRSA isolates were obtained for antibiotic susceptibility tests and molecular study.
Results
Among these isolates, we found that the MIC50, MIC90, and minimum inhibitory concentration (MIC) range of ciprofloxacin were much higher for the isolates without the rpoB474 mutation than for isolates with the rpoB474 mutation. A total of 87.5% of the isolates with the rpoB474 mutation were susceptible to ciprofloxacin, but none of the isolates without the rpoB474 mutation were susceptible to ciprofloxacin. For 27 MRSA isolates without rpo474 silent mutation but with gyrA86/126 silent mutation, all of them belonged to SCCmec III, and had high ciprofloxacin MIC levels. For another 44 MRSA isolates with rpo474 silent mutation but without gyrA86/126 silent mutation, all of them showed low ciprofloxacin MIC levels, all of them belonged to either SCCmec IV or V. Furthermore, MRSA ciprofloxacin resistance was found to be associated with the mutations gyrA S84L/parC S80F or gyrA S84L, and S85P/parC S80Y.
Conclusion
Most occurrences of this rpoB474 silent mutation were found in community acquired-MRSA (CA-MRSA) isolates with susceptibility to most antibiotics, especially for ciprofloxacin and vice versa. Thus, this mutation may help to differentiate the different microbiologic characteristics of MRSA clinical isolates.
Keywords: MRSA, silent mutation, rpoB474, ciprofloxacin, resistance
Introduction
The emergence of methicillin-resistant Staphylococcus aureus (MRSA) infections has become a substantial treatment challenge in hospital-associated settings and in community settings around the world.1–3 In addition to methicillin, MRSA can develop antimicrobial resistance against several different antibiotic classes, including β-lactams, quinolones, trimethoprim-sulfamethoxazole, erythromycin, clindamycin, linezolid, and daptomycin, through various resistance gene mutations, including blaZ, mecA, parC, gyrA, gyrB, sulA, dfrB, erm, cfr, and mprF.4–8 There is no exception for the rpoB gene, which encodes the β-subunit of the bacterial RNA polymerase. The mutation of this gene following rifampicin therapy can often lead to the emergence of rifampicin resistance.9 Therefore, rifampicin resistance can be detected by sequencing the rpoB gene. Hellmark et al demonstrated that rpoB sequencing could be an accurate method of species identification in staphylococci,9 and Marty et al reported that the highly discriminatory rpoB species-specific PCR-RFLP analysis allows for fast and simple molecular identification of Staphylococcus and other bacteria.10 This finding suggests that the rpoB gene may have the potential role as one of the core gene candidates for phylogenetic analyses and bacterial identification. Our recent study on the rpoB gene found that a point mutation in codon 474 (AAC → AAT) located in cluster I region11 was present in 60% of our clinical MRSA isolates, and this mutation was not associated with any induction of rifampicin resistance and was even associated with lower antibiotic resistance rates, especially for ciprofloxacin.12 Therefore, this study was conducted to investigate the genotypic differences among different sequence type MRSA isolates, especially focusing on silent mutations and the relationship between such mutations and ciprofloxacin resistance.
Materials and methods
Clinical isolates
Seventy-nine MRSA isolates were obtained from the Tigecycline In-vitro Surveillance in Taiwan (TIST) study at 22 hospitals from 2006 to 2010.13 Staphylococci were identified by colony morphology, Gram stain, and coagulase test results. MRSA was further confirmed by the tube coagulase test and growth on 6 μg/mL oxacillin salt agar screening plates. The mecA gene was confirmed using a PCR method. Isolates were stored at −70°C in Protect Bacterial Preservers (Technical Service Consultants Limited, Heywood, UK) until use.
Antibiotic susceptibility test
Antibiotics tested included chloramphenicol, erythromycin, gentamicin, minocycline, oxacillin, rifampin, and vancomycin (Sigma-Aldrich Co., St Louis, MO, USA), fosfomycin (Ercros, Barcelona, Spain), linezolid and tigecycline (Pfizer, Inc., New York, NY, USA), fusidic acid (Leo Pharma, Ballerup, Denmark), teicoplanin (Sanofi-Aventis, Bridgewater, NJ, USA), ciprofloxacin (Bayer AG, Leverkusen, Germany), daptomycin (Merck & Co., Inc., Kenilworth, NJ, USA). Interpretation criteria for the susceptibility test and the minimum inhibitory concentration (MIC) determined by the agar dilution tests were based on the recommendations of the Clinical and Laboratory Standards Institute (CLSI) or the British Society for Antimicrobial Chemotherapy.14–16 For the fosfomycin susceptibility test, glucose-6-phosphate (25 mg/L) was added to the agar plate. The daptomycin susceptibility test was performed in Mueller–Hinton broth adjusted to 50 mg/L of calcium per standard methodology. Mueller–Hinton agar (Thermo Fisher Scientific, Waltham, MA, USA) was employed for S. aureus MIC determination. Inocula were prepared by suspending growth from overnight cultures in saline to a turbidity of a 0.5 McFarland standard. Inoculated plates were then incubated in ambient air at 37°C for 24 h. S. aureus ATCC 29213 was included as the control strain in each of the MIC measurements.
Determination of the mecA, PVL, rpoB, gyrA and parC gene mutation
PCR for the mecA gene was performed according to the protocol described by Vannuffel et al.17 S. aureus ATCC BAA-1707, USA400 was used as the positive control. A 433-bp nucleotide fragment located in the lukS-PV and lukF-PV operons was amplified by PCR using the primers and conditions described by Lina et al.18 The primer sequence was lukPV-forward (ATC ATT AGG TAA AAT GTC TGG ACA TGA TCC A) and reverse (GCA TCA AAT GTA TTG GAT AGC AAA AGC). The PCR method was used to amplify these genes including rpoB, gyrA, and parC with gene mutations using primers and previously described cycling conditions.19, 20 The following primers were used: (a) rpoB-forward (CCG TCG TTT ACG TTC TGT AGG) and reverse (AAA GCC GAA TTC ATT TAC ACG); (b) gyrA-forward (AAT GAA CAA GGT ATG ACA CC) and reverse (TAC GCG CTT CAG TAT AAC GC); (c) parC-forward (ACT TGA AGA TGT TTT AGG TGA T) and reverse (TTA GGA AAT CTT GAT GGC AA). Template DNA for PCR was prepared using InstaGene™ Matrix as recommended by the manufacturer (Bio-Rad Laboratories Inc., Hercules, CA, USA). After amplification, PCR products were purified from excess primers and nucleotides using a QIAquick PCR Purification Kit (QIAGEN, Valencia, CA, USA) and sequenced with the same primers by the dideoxy chain termination method in an ABI PRISM 3730 sequence analyzer (Thermo Fisher Scientific, Waltham, MA, USA).
Molecular typing methods
All isolates were analyzed by SCCmec typing, MLST typing, and pulsed-field gel electrophoresis (PFGE). The SCCmec types were determined by the multiplex PCR strategy developed by Kondo et al.21 The MLST was carried out as previously described.22 The sequences of the PCR products were compared with the existing sequences available on the MLST website (http://saureus.mlst.net) for S. aureus.23 DNA extraction and SmaI restriction were performed as previously described. The PFGE patterns were visually examined and interpreted according to the criteria developed by Tenover et al.24 The similarities of PFGE profiles of each strain were compared using a Dice coefficient at 1.0% of tolerance and 0.8% of optimization.
Results
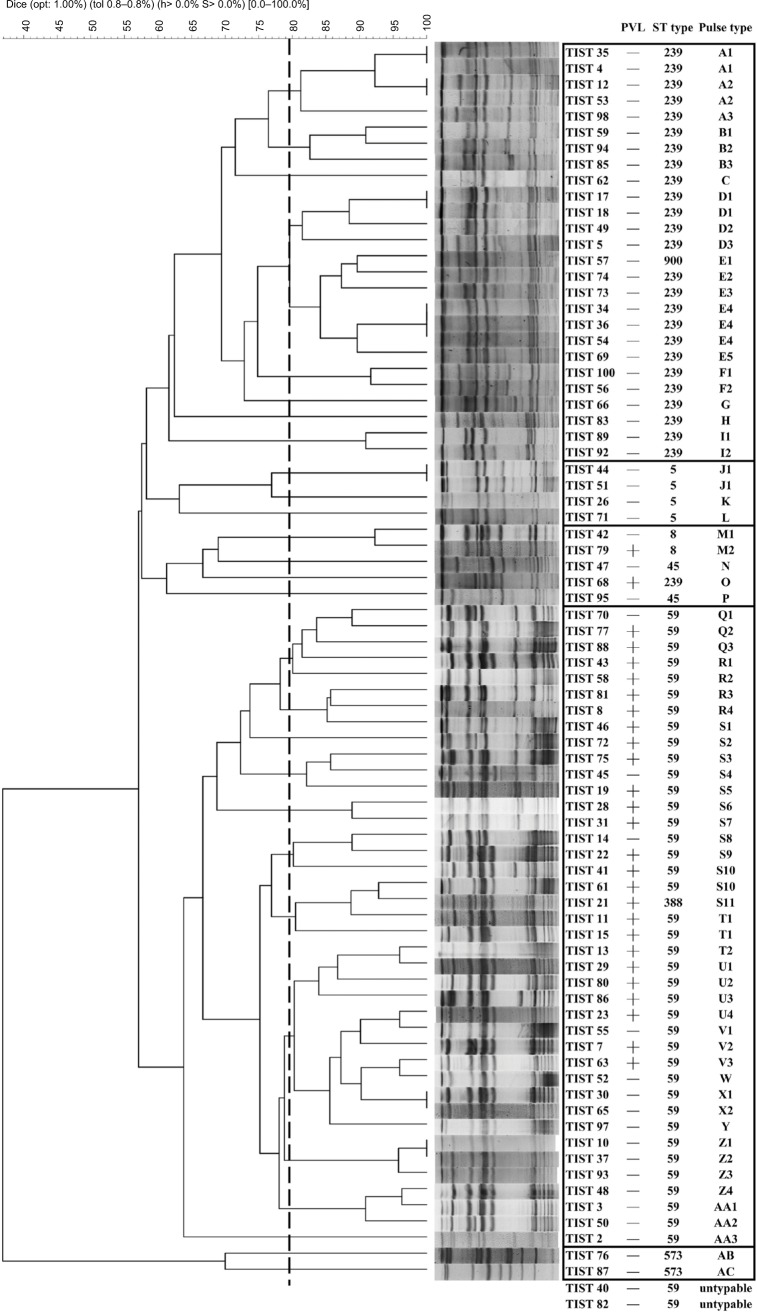
The PFGE patterns of all MRSA isolates were shown in Figure 1. Two isolates did not yield an interpretable result with PFGE analysis because of technical problems associated with degradation of the genomic DNA. This may lead to only very faint bands, degraded bands, or no banding patterns. Only 26 (32.9%) MRSA isolates were Panton–Valentine leukocidin (PVL) positive. The most common sequence type (ST) was ST59 (n=41, 51.8%), followed by ST239 (n=26, 32.9%), ST5 (n=4, 5.0%), ST8 (n=2, 2.5%), ST45 (n=2, 2.5%), ST573 (n=2, 2.5%), ST388 (n=1, 1.3%), and ST900 (n=1, 1.3%).
Figure 1.
PFGE and molecular patterns of MRSA isolates.
Abbreviations: PFGE, pulsed-field gel electrophoresis; MRSA, methicillin-resistant Staphylococcus aureus; opt, optimization; tol, tolerance; PVL, Panton–Valentine leucocidin; ST, sequence type; TIST, Tigecycline In-vitro Surveillance in Taiwan.
Table 1 shows the MICs and the susceptibility of 48 and 31 MRSA isolates with and without the rpoB474 silent mutation, respectively. All isolates were susceptible to vancomycin, teicoplanin, and daptomycin. For oxacillin, erythromycin, gentamicin, tigecycline, minocycline, fusidic acid, rifampicin, and fosfomycin, the susceptibility rates of 48 rpoB474 silent mutation isolates were higher than those of the 31 unmutated rpoB474 isolates. In contrast, the susceptibility rate against linezolid of the non-rpoB474 silent mutation isolates was 100% and higher than rpoB474 silent mutation isolates susceptibility rate of 89.6%. For ciprofloxacin, we found a significant difference regarding MICs and the susceptibility rate between isolates with and without the rpoB474 silent mutation. The MIC50, MIC90, and MIC range was much higher for the isolates without the rpoB474 silent mutation than with the rpoB474 silent mutation. A total of 87.5% of the isolates with the rpoB474 silent mutation were susceptible to ciprofloxacin, but none of the isolates without the rpoB474 silent mutation were susceptible to ciprofloxacin.
Table 1.
The MIC values of MRSA isolates according the presence of the rpoB474 silent mutation
| Antibiotics | No rpoB474 silent mutation (n=31) | rpoB474 silent mutation (n=48) | MIC interpretive criteria | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| MIC50, mg/L | MIC90, mg/L | MIC range, mg/L | Susceptibility, % | MIC50, mg/L | MIC90, mg/L | MIC range, mg/L | Susceptibility, % | S | I | R | |
| OXA | > 128 | > 128 | 4–> 128 | 0.0 | 16 | 128 | <1–> 128 | 14.6 | ≤2 | ≥ 4 | |
| ERY | > 128 | > 128 | 0.5–> 128 | 3.2 | > 128 | > 128 | 0.5–> 128 | 6.3 | ≤0.5 | 1–4 | ≥ 8 |
| GEN | > 128 | > 128 | <1–> 128 | 9.7 | <1 | > 128 | <1–> 128 | 50.0 | ≤4 | 8 | ≥ 16 |
| CM | 16 | 16 | 8–64 | 0.0 | 64 | 128 | 8–128 | 0.0 | ≤0.5 | 1–2 | ≥ 4 |
| VA | 2 | 2 | 1–2 | 100 | 1 | 2 | 1–2 | 100 | ≤2 | 4–8 | ≥ 16 |
| TGC | 0.5 | 1 | 0.25–1 | 83.9 | 0.5 | 0.5 | 0.25–1 | 95.8 | ≤0.5a | ||
| MNO | 8 | 8 | 0.125–8 | 45.2 | 0.25 | 0.5 | 0.125–8 | 95.8 | ≤4 | 8 | ≥ 16 |
| TEC | 2 | 2 | 0.5–2 | 100 | 1 | 1 | 0.5–2 | 100 | ≤8 | 16 | ≥ 32 |
| FA | 0.5 | 8 | 0.5–8 | 71.0 | 0.25 | 0.5 | 0.25–8 | 97.9 | ≤1b | ||
| LNZ | 4 | 4 | 2–4 | 100 | 4 | 8 | 2–8 | 89.6 | ≤4 | ≥ 8 | |
| CIP | > 64 | > 64 | 16–> 64 | 0.0 | 0.5 | 2 | 0.25–> 64 | 87.5 | ≤1 | 2 | ≥ 4 |
| RIF | 0.016 | 2 | 0.016–> 32 | 80.6 | 0.016 | 2 | 0.016–> 32 | 89.6 | ≤1 | 2 | ≥ 4 |
| FOS | 16 | 32 | 1–> 1024 | 90.3 | 4 | 8 | 1–> 1024 | 95.8 | ≤32b | ||
| DAP | 0.5 | 1 | 0.125–1 | 100 | 0.25 | 0.5 | 0.25–1 | 100 | ≤1 | ||
Notes:
MIC interpretive criteria were assessed according to the FDA guidelines.
MIC interpretive criteria were assessed according to the BSAC guidelines.
Abbreviations: MRSA, methicillin-resistant Staphylococcus aureus; MIC, minimum inhibitory concentration; OXA, oxacillin; ERY, erythromycin; GEN, gentamicin; CM, clindamycin; VA, vancomycin; TGC, tigecycline; MNO, minocycline; TEC, teicoplanin; FA, fusidic acid; LNZ, linezolid; CIP, ciprofloxacin; RIF, rifampicin; FOS, fosfomycin; DAP, daptomycin; S, susceptible; I, intermediate; R, resistant.
Table 2 shows the association between the ciprofloxacin MIC and the gene mutations. For 27 MRSA isolates without rpo474 silent mutation but with gyrA86/126 silent mutation (Group A), all of them belonged to SCCmec III and had the double mutations in the gyrA and parC genes, such as gyrA S84L/parC S80F or gyrA S84L. Moreover, all of Group A MRSA isolates showed high ciprofloxacin MIC levels. For four MRSA isolates with rpo474 silent mutation but without gyrA86/126 silent mutation (Group B), all of these presented with high ciprofloxacin MIC level, but only three of them had double mutations in the gyrA and parC genes. For four MRSA isolates without rpo474 and gyrA86/126 silent mutation (Group C), all of them had double mutations in the gyrA and parC genes, and high ciprofloxacin MIC levels. For 44 MRSA isolates with rpo474 silent mutation but without gyrA86/126 silent mutation (Group D), all of them showed low ciprofloxacin MIC level, but none of them had double mutations in the gyrA and parC genes. Among group D, MRSA isolates belonged to either SCCmec IV or V.
Table 2.
The ciprofloxacin MIC values of MRSA isolates with respect to the rpoB474 and gyrA86/126 gene mutations
| rpoB 474/gyrA 86, 126 sm | Isolates | Ciprofloxacin MIC | gyrA | parC | SCCmec |
|---|---|---|---|---|---|
| Group A | |||||
| −/+ | TIST4 | > 64 | S84L | S80F | III |
| −/+ | TIST5 | > 64 | S84L | S80F | III |
| −/+ | TIST12 | > 64 | S84L | S80F | III |
| −/+ | TIST17 | > 64 | S84L | S80F | III |
| −/+ | TIST18 | > 64 | S84L | S80F | III |
| −/+ | TIST34 | > 64 | S84L | S80F | III |
| −/+ | TIST35 | > 64 | S84L | S80F | III |
| −/+ | TIST36 | > 64 | S84L/G108D | S80F | III |
| −/+ | TIST49 | > 64 | S84L | S80F | III |
| −/+ | TIST53 | > 64 | S84L | S80F | III |
| −/+ | TIST54 | > 64 | S84L | S80F | III |
| −/+ | TIST56 | > 64 | S84L | S80F | III |
| −/+ | TIST57 | 32 | S84L | S80F | III |
| −/+ | TIST59 | > 64 | S84L | S80F | III |
| −/+ | TIST62 | > 64 | S84L/S85P | S80F | III |
| −/+ | TIST66 | > 64 | S84L | S80F | III |
| −/+ | TIST68 | 64 | S84L/E88L | S80F/E84K | III |
| −/+ | TIST69 | 64 | S84L | S80F | III |
| −/+ | TIST73 | > 64 | S84L/S85P | S80F | III |
| −/+ | TIST74 | > 64 | S84L/S85P | S80F | III |
| −/+ | TIST83 | > 64 | S84L/S85P | S80F | III |
| −/+ | TIST85 | > 64 | S84L | S80F | III |
| −/+ | TIST89 | > 64 | S84L | S80F | III |
| −/+ | TIST92 | 64 | S84L/S85P | S80F | III |
| −/+ | TIST94 | > 64 | S84L | S80F/S81P | III |
| −/+ | TIST98 | > 64 | S84L | S80F | III |
| −/+ | TIST100 | > 64 | S84L | S80F | III |
| Group B | |||||
| +/− | TIST19 | 32 | x | x | V |
| +/− | TIST44 | > 64 | S84L | S80Y | II |
| +/− | TIST51 | > 64 | S84L | S80Y | II |
| +/− | TIST95 | 16 | S84L (110sm) | S80F (77sm) | IV |
| Group C | |||||
| −/− | TIST26 | > 64 | S84L/S85P | R61L/S80Y/S81P | II |
| −/− | TIST42 | > 64 | S84L | S80Y | IV |
| −/− | TIST47 | 16 | S84L (110sm) | S80F (77sm) | V |
| −/− | TIST71 | > 64 | S84L | S80Y | II |
| Group D | |||||
| +/− | TIST2 | 0.25 | x | x | IV |
| +/− | TIST3 | 0.5 | x | x | IV |
| +/− | TIST7 | 0.25 | x | x | IV |
| +/− | TIST8 | 0.25 | x | x | V |
| +/− | TIST10 | 0.25 | x | x | IV |
| +/− | TIST11 | 0.5 | x | x | V |
| +/− | TIST13 | 0.25 | x | x | V |
| +/− | TIST14 | 0.25 | x | x | V |
| +/− | TIST15 | 0.5 | x | x | V |
| +/− | TIST21 | 0.25 | x | x | V |
| +/− | TIST22 | 0.25 | x | x | V |
| +/− | TIST23 | 0.25 | x | x | IV |
| +/− | TIST28 | 0.25 | x | x | V |
| +/− | TIST29 | 0.25 | x | x | V |
| +/− | TIST30 | 0.25 | x | x | IV |
| +/− | TIST31 | 0.25 | x | x | V |
| +/− | TIST37 | 1 | x | x | IV |
| +/− | TIST40 | 1 | x | x (77/81/113sm) | V |
| +/− | TIST41 | 0.5 | x | x | V |
| +/− | TIST43 | 0.5 | x | x | V |
| +/− | TIST45 | 1 | x | x | V |
| +/− | TIST46 | 0.5 | x | x | V |
| +/− | TIST48 | 0.5 | x | x | IV |
| +/− | TIST50 | 1 | x | x | IV |
| +/− | TIST52 | 1 | x | x | IV |
| +/− | TIST55 | 1 | x | x | IV |
| +/− | TIST58 | 0.5 | x | x | V |
| +/− | TIST61 | 1 | x | x | V |
| +/− | TIST63 | 1 | x | x | IV |
| +/− | TIST65 | 0.25 | x | x | IV |
| +/− | TIST70 | 0.25 | x | x | V |
| +/− | TIST72 | 2 | x | E84K | V |
| +/− | TIST75 | 1 | x | x | IV |
| +/− | TIST76 | 1 | x | S80F/S123F | IV |
| +/− | TIST77 | 0.25 | x | x | V |
| +/− | TIST79 | 0.5 | x | x | IV |
| +/− | TIST80 | 0.5 | x | x | V |
| +/− | TIST81 | 0.25 | x | x | V |
| +/− | TIST82 | 1 | x | x (77/81/113sm) | V |
| +/− | TIST86 | 0.25 | x | x | V |
| +/− | TIST87 | 1 | x | S80F/S123F | IV |
| +/− | TIST88 | 0.25 | x | x | IV |
| +/− | TIST93 | 2 | x | S80F | IV |
| +/− | TIST97 | 0.25 | x | x | IV |
Notes: Group A: rpoB474 wild type/gyrA86 and126 mutation with high ciprofloxacin MIC. Group B: rpoB474 mutation/gyrA86 and126 wild type with high ciprofloxacin MIC. Group C: rpoB474 wild type/gyrA86 and126 wild type with high ciprofloxacin MIC. Group D: rpoB474 mutation/gyrA86 and126 wild type with low ciprofloxacin MIC.
Abbreviations: MRSA, methicillin-resistant Staphylococcus aureus; MIC, minimum inhibitory concentration; TIST, Tigecycline In-vitro Surveillance in Taiwan.
Table 3 summarizes the distribution of SCCmec types and silent mutations of rpoB474 and gyrA86/126 among all clinical isolates. For 27 SCCmec type III MRSA isolates, none had the rpoB474 silent mutation, but all had the gyrA86/126 silent mutation. In contrast, for most of the SCCmec type IV and V isolates, the rpoB474 silent mutation rate was 95.5% and 96.2%, respectively. None of the isolates had the gyrA86/126 silent mutation.
Table 3.
MRSA isolate SCCmec patterns
| SCCmec | Numbers | No. (%) of MRSA isolates with rpoB474 mutations | No. (%) of MRSA isolates with gyrA86/126 mutation |
|---|---|---|---|
| II | 4 | 2 (50) | 0 (0) |
| III | 27 | 0 (0) | 27 (100) |
| IV | 22 | 21 (95.5) | 0 (0) |
| V | 26 | 25 (96.2) | 0 (0) |
Notes: rpoB474: 474aac-aat silent mutation. gyrA86/126: 86att-atc/126gcg-gca silent mutation.
Abbreviation: MRSA, methicillin-resistant Staphylococcus aureus.
Discussion
This study had several interesting findings regarding the association between rpoB474 silent mutations and the different SCCmec and ST type MRSA isolates. Nearly all of the 48 MRSA isolates (Groups B and D) carried SCCmec type IV and V had the rpoB474 silent mutation. In contrast, none of them had the gyrA86/126 silent mutation and most of them do not have double mutations of gyrA/par C gene. Furthermore, these isolates exhibit high susceptibility to ciprofloxacin, and other antibiotics. All of these characteristics are consistent with microbiological features of community-acquired-MRSA (CA-MRSA).25,26 Additionally, most of the CA-MRSA isolates in this study were found to be ST59, which is the major type of CA-MRSA in Taiwan.27, 28 In contrast, for the group of 31 MRSA isolates without the rpoB474 silent mutation (Groups A and C), all of them had the double mutations in the gyrA and parC genes, and showed high ciprofloxacin MIC levels. Among them, SCCmec type III compromised most of the MRSA isolates (n=27, 87.1%). These findings indicate that MRSA without the rpoB474 silent mutation is associated with higher resistance to ciprofloxacin, and most of these isolates should be classified as hospital-acquired MRSA (HA-MRSA) carrying SCCmec type III. In summary, we found the different presentations between MRSA isolates with and without rpoB474 silent mutation and this mutation may help differentiate the different microbiologic characteristic of MRSA clinical isolates.
In this study, MRSA ciprofloxacin resistance was found to be associated with the mutations of gyrA S84L/parC S80F or gyrA S84L, S85P/parC S80Y. This is consistent with previous findings that double mutants in the parC and gyrA genes may cause high-level resistance to ciprofloxacin with an MIC value ≥ 64 mg/dL.29 Meanwhile, we also found a reciprocal relationship of rpoB474, gyrA86/126 silent mutation and gyrA/parC mutation that had never been reported before. Previous study suggested that rpoB mutation could be a “regulatory” mutation to decrease vancomycin susceptibility in heterogeneous vancomycin-intermediate S. aureus (hVISA) and VISA phenotype acquisition.30,31 In addition, the role of rpoB mutation in the ciprofloxacin resistance has been demonstrated in the study of Escherichia coli with ciprofloxacin-selected rpoB mutations.32 Pietsch et al found that the mutations in RNA polymerase can be served as novel contributors to the evolution of resistance to ciprofloxacin and also significantly increase the expression of mdtK, encoding a multidrug efflux transporter.32 However, rpoB474 and gyrA86/126 mutations in this study are silent mutations. In contrast to true mutation, they just have nucleotide change but do not result in new amino acid substitute. Therefore, whether the finding is incidental or significant among MRSA isolates remains unclear. Further study is warranted to investigate possible mechanisms.
Conclusion
We found the phenomenon about the relationship between rpoB474, gyrA86/126 silent mutation and gyrA/parC mutation with ciprofloxacin MIC and antibiotic resistance. Most occurrences of this rpoB474 silent mutation were found in CA-MRSA isolates with susceptibility to most antibiotics, especially for ciprofloxacin. In contrast, most of MRSA isolates without this mutation are HA-MRSA with high resistance to ciprofloxacin.
Acknowledgments
This study was supported by a research grant (nos. CMFHT10501, CMNCKU10509) from the Chi-Mei Medical Center Research Foundation, and MOST 105-2314-B-384-007-MY3 from Ministry of Science and Technology.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Boucher HW, Corey GR. Epidemiology of methicillin-resistant Staphylococcus aureus. Clin Infect Dis. 2008;46(Suppl 5):S344–S349. doi: 10.1086/533590. [DOI] [PubMed] [Google Scholar]
- 2.Chen CJ, Huang YC. Community-acquired methicillin-resistant Staphylococcus aureus in Taiwan. J Microbiol Immunol Infect. 2005;38(6):376–382. [PubMed] [Google Scholar]
- 3.Lee CM, Lai CC, Chiang HT, et al. Presence of multidrug-resistant organisms in the residents and environments of long-term care facilities in Taiwan. J Microbiol Immunol Infect. 2017;50(2):133–144. doi: 10.1016/j.jmii.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Mishra NN, Yang SJ, Sawa A, Rubio A, Nast CC, Yeaman MR, Bayer AS. Analysis of cell membrane characteristics of in vitro-selected daptomycin-resistant strains of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2009;53(6):2312–2318. doi: 10.1128/AAC.01682-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lowy FD. Antimicrobial resistance: the example of Staphylococcus aureus. J Clin Invest. 2003;111(9):1265–1273. doi: 10.1172/JCI18535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stryjewski ME, Corey GR. Methicillin-resistant Staphylococcus aureus: an evolving pathogen. Clin Infect Dis. 2014;58(Suppl 1):S10–S19. doi: 10.1093/cid/cit613. [DOI] [PubMed] [Google Scholar]
- 7.Malachowa N, DeLeo FR. Mobile genetic elements of Staphylococcus aureus. Cell Mol Life Sci. 2010;67(18):3057–3071. doi: 10.1007/s00018-010-0389-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu B, Kelesidis T, Tsiodras S, Hindler J, Humphries RM. The emerging problem of linezolid-resistant Staphylococcus. J Antimicrob Chemother. 2013;68(1):4–11. doi: 10.1093/jac/dks354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hellmark B, Soderquist B, Unemo M. Simultaneous species identification and detection of rifampicin resistance in staphylococci by sequencing of the rpoB gene. Eur J Clin Microbiol Infect Dis. 2009;28(2):183–190. doi: 10.1007/s10096-008-0604-5. [DOI] [PubMed] [Google Scholar]
- 10.Marty E, Buchs J, Eugster-Meier E, Lacroix C, Meile L. Identification of staphylococci and dominant lactic acid bacteria in spontaneously fermented Swiss meat products using PCR-RFLP. Food Microbiol. 2012;29(2):157–166. doi: 10.1016/j.fm.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 11.Aubry-Damon H, Galimand M, Gerbaud G, Courvalin P. rpoB mutation conferring rifampin resistance in Streptococcus pyogenes. Antimicrob Agents Chemother. 2002;46(5):1571–1573. doi: 10.1128/AAC.46.5.1571-1573.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang HJ, Lai CC, Hsueh PR, et al. RNA polymerase B subunit gene mutations in biofilm-embedded methicillin-resistant Staphylococcus aureus following rifampin treatment. J Microbiol Immunol Infect. 2016;49(3):394–401. doi: 10.1016/j.jmii.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 13.Hsueh PR. Tigecycline in-vitro surveillance in Taiwan (TIST) Int J Antimicrob Agents. 2008;32(Suppl 3):S173. doi: 10.1016/S0924-8579(08)70022-0. [DOI] [PubMed] [Google Scholar]
- 14.Andrews JM. Determination of minimum inhibitory concentrations. J Antimicrob Chemother. 2001;48(Suppl 1):5–16. doi: 10.1093/jac/48.suppl_1.5. [DOI] [PubMed] [Google Scholar]
- 15.CLSI . Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard. 9th ed. Wayne, PA: Clinical & Laboratory Standards Institute; 2006. pp. M7–A9. [Google Scholar]
- 16.CLSI . Performance standards for antimicrobial susceptibility testing; Sixteenth Informational Supplement M100-S24. Wayne, PA: Clinical & Laboratory Standards Institute; 2014. [Google Scholar]
- 17.Vannuffel P, Gigi J, Ezzedine H, Vandercam B, Delmee M, Wauters G, Gala JL. Specific detection of methicillin-resistant Staphylococcus species by multiplex PCR. J Clin Microbiol. 1995;33(11):2864–2867. doi: 10.1128/jcm.33.11.2864-2867.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lina G, Piémont Y, Godail-Gamot F, et al. Involvement of Panton–Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999;29(5):1128–1132. doi: 10.1086/313461. [DOI] [PubMed] [Google Scholar]
- 19.Kwak YG, Truong-Bolduc QC, Bin Kim H, Song KH, Kim ES, Hooper DC. Association of norB overexpression and fluoroquinolone resistance in clinical isolates of Staphylococcus aureus from Korea. J Antimicrob Chemother. 2013;68(12):2766–2772. doi: 10.1093/jac/dkt286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wichelhaus TA, Schäfer V, Brade V, Böddinghaus B. Molecular characterization of rpoB mutations conferring cross-resistance to rifamycins on methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1999;43(11):2813–2816. doi: 10.1128/aac.43.11.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kondo Y, Ito T, Ma XX, Watanabe S, Kreiswirth BN, Etienne J, Hiramatsu K. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: Rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother. 2007;51(1):264–274. doi: 10.1128/AAC.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38(3):1008–1005. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ip M, Lyon DJ, Chio F, Enright MC, Cheng AF. Characterization of isolates of methicillin-resistant Staphylococcus aureus from Hong Kong by phage typing, pulsed-field gel electrophoresis, and fluorescent amplified-fragment length polymorphism analysis. J Clin Microbiol. 2003;41(11):4980–4985. doi: 10.1128/JCM.41.11.4980-4985.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: Criteria for bacterial strain typing. J Clin Microbiol. 1995;33(9):2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jung SI, Shin DH, Park KH, Shin JH. Antimicrobial susceptibility and clonal relatedness between community- and hospital-acquired methicillin-resistant Staphylococcus aureus from blood cultures. J Microbiol. 2006;44(3):336–343. [PubMed] [Google Scholar]
- 26.Gostev V, Kruglov A, Kalinogorskaya O, et al. Molecular epidemiology and antibiotic resistance of methicillin-resistant Staphylococcus aureus circulating in the Russian Federation. Infect Genet Evol. 2017;53:189–194. doi: 10.1016/j.meegid.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 27.Feng Y, Chen HL, Chen CJ, Chen CL, Chiu CH. Genome comparisons of two Taiwanese community-associated methicillin-resistant Staphylococcus aureus ST59 clones support the multi-origin theory of CA-MRSA. Infect Genet Evol. 2017;54:60–65. doi: 10.1016/j.meegid.2017.06.018. [DOI] [PubMed] [Google Scholar]
- 28.Huang YC, Chen CJ. Community-associated meticillin-resistant Staphylococcus aureus in children in Taiwan, 2000s. Int J Antimicrob Agents. 2011;38(1):2–8. doi: 10.1016/j.ijantimicag.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 29.Ferrero L, Cameron B, Crouzet J. Analysis of gyrA and grlA mutations in stepwise-selected ciprofloxacin-resistant mutants of Staphylococcus aureus. Antimicrob Agents Chemother. 1995;39(7):1554–1558. doi: 10.1128/aac.39.7.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe Y, Cui L, Katayama Y, Kozue K, Hiramatsu K. Impact of rpoB mutations on reduced vancomycin susceptibility in Staphylococcus aureus. J Clin Microbiol. 2011;49(7):2680–2684. doi: 10.1128/JCM.02144-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller N1, Rudoy RC. Vancomycin intermediate-resistant Staphylococcus aureus (VISA) Orthop Nurs. 2000;19(6):45–48. doi: 10.1097/00006416-200019060-00009. quiz 49–51. [DOI] [PubMed] [Google Scholar]
- 32.Pietsch F, Bergman JM, Brandis G, Marcusson LL, Zorzet A, Huseby DL, Hughes D. Ciprofloxacin selects for RNA polymerase mutations with pleiotropic antibiotic resistance effects. J Antimicrob Chemother. 2017;72(1):75–84. doi: 10.1093/jac/dkw364. [DOI] [PubMed] [Google Scholar]