Abstract
Background
Histone H2A deubiquitinase MYSM1 has recently been shown to be essential for hematopoiesis and hematopoietic stem cell (HSC) function in both mice and humans. However, conventional MYSM1 knockouts cause partial embryonic lethality and growth retardation, and it is difficult to convincingly remove the effects of environmental factors on HSC differentiation and function.
Material/Methods
MYSM1 conditional knockout (cKO) mice were efficiently induced by using the Vav1-cre transgenic system. The Vav-Cre MYSM1 cKO mice were then analyzed to verify the intrinsic role of MYSM1 in hematopoietic cells.
Results
MYSM1 cKO mice were viable and were born at normal litter sizes. At steady state, we observed a defect in hematopoiesis, including reduced bone marrow cellularity and abnormal HSC function. MYSM1 deletion drives HSCs from quiescence into rapid cycling, and MYSM1-deficient HSCs display impaired engraftment. In particular, the immature cycling cKO HSCs have elevated reactive oxygen species (ROS) levels and are prone to apoptosis, resulting in the exhaustion of the stem cell pool during stress response to 5-FU.
Conclusions
Our study using MYSM1 cKO mice confirms the important role of MYSM1 in maintaining HSC quiescence and survival.
MeSH Keywords: Apoptosis, Cell Survival, Hematopoietic Stem Cells, Histones, Ubiquitination
Background
Hematopoietic stem cells (HSCs) are responsible for sequential lineage commitment to generate all cell types in the blood, and this commitment is controlled by an interactive network of transcription factors and epigenetic regulators [1–3]. Components of the Polycomb Repressive Complex (PRC) 1, such as Bmi1, Mel18, and Ring1a/b, are important epigenetic regulators of HSC maintenance [4–7] and are critical for both normal and pathological hematopoiesis [8–10]. These Polycomb group (PcG) proteins repress gene expression, at least in part, via histone H2A ubiquitination (ubH2A) [7,11], which is thought to contribute to transcriptional repression by inhibiting transcription initiation [12] or restraining RNA pol II from elongation [13]. In the nucleus, PRC1 is the main H2A ubiquitin ligase, and the steady-state level of ubH2A is governed by the balance between PRC1 and H2A deubiquitinases, which are also involved in cell cycle progression, X chromosome inactivation, DNA damage repair, and cancer [14–16].
MYSM1 was recently identified as a histone H2A deubiquitinase, and its activity was initially described in the activation of several AR-regulated target genes in prostate cancer cells [17]. We and other independent groups have observed an essential role for MYSM1 in bone marrow hematopoietic development and lymphocyte generation in both MYSM1 knockout mice [18–23] and patients with MYSM1 mutations [24,25]. However, MYSM1−/− mice are generated from heterozygous pairings at less-than-expected Mendelian ratios. In addition to partial embryonic lethality, conventional MYSM1 knockouts result in multiple organ defects and growth retardation, which has limited the functional study of HSCs.
In the present study, we generated a MYSM1 conditional knockout (cKO) allele to study MYSY1 function in adult hematopoiesis using Vav1-cre. MYSM1 cKO mice were born at roughly Mendelian ratios. We found decreased bone marrow cellularity and abnormities in HSCs and their progenitors in cKO mice. We discovered that cKO mice could not maintain HSC quiescence, and we observed accelerated cell proliferation in cKO HSCs. Additionally, we discovered impaired engraftment, elevated ROS levels, and a high apoptosis rate in cKO HSCs. Our study shows that MYSM1 is required for maintaining HSC quiescence and survival.
Material and Methods
Mice
Floxed MYSM1 mice were generated as previously described [18,20]. (B6.Cg-Tg(Vav1-cre)A2Kio/J) mice were purchased from Jackson Laboratories (Bar Harbor, ME, USA) and were crossed with the floxed mice to create MYSM1 cKO mice. Mysm1-wild-type but Vav1-cre-positive animals were used as control mice. Mice were housed in specific-pathogen-free facilities, and all experiments were performed in accordance with the guidelines of the Fourth Military Medical University Institutional Animal Care and Use Committee.
Genotyping
Genomic DNA was isolated from mouse BM using the Qiagen DNeasy Blood and Tissue kit following the manufacturer’s instructions (cat#69504). Primer sequences for polymerase chain reaction (PCR) amplification of the MYSM1 floxed and cKO alleles were MYSM1 LoxP forward_1 5′-GCATGTTCCAGAAGAGGTATAAGG-3′, MYSM1 LoxP forward_2 5′-TGGATAGCACCATCAGTGATG-3′, and MYSM1 LoxP reverse 5′-CGCAGAATACGCAGTTTGTTG-3′.
RNA extraction and real-time PCR
Total RNA was isolated from bone marrow with an RNeasy Mini kit (Qiagen, Alameda, CA, USA). Then, cDNA was synthesized with an iScript™ Advanced cDNA Synthesis kit (Bio-Rad, Hercules, CA, USA). PCR was performed in duplicate with a CFX96 PCR system and iTaq™ Universal SYBR® Green Supermix according to the manufacturer’s instructions (Bio-Rad, Hercules, CA, USA). Primer sequences for PCR were MYSM1_F 5′-CTGACGATGAAGATGTAG-3′, MYSM1_R 5′-GCATAATTCTCCTTGATTG-3′, GAPDH_F 5′-ACAATGAATA CGGCTACAG-3′, GAPDH_R 5′-GTCCAGGGTTTCTTACTC-3′.
Flow cytometric analyses
Sample preparation, cytometric analysis, and sorting were performed as described previously [26,27]. Cells were flushed out of bone marrow (BM) and were prepared and first stained for 20 minutes at 4°C with CD16/CD32 Fc-blocking antibody (2.4G2) in flow cytometry buffer, followed by incubation with a “cocktail” of antibody conjugated to fluorescein isothiocyanate (FITC), phycoerythrin (PE), peridinine chlorophyll protein complex–cyanine5.5 (PerCP-Cy5.5), phycoerythrin-indotricarbocyanine (PE-Cy7), allophycocyanin (APC), or allophycocyanin-indotricarbocyanine (APC-Cy7). For each staining, at least 100 000 events were collected for analysis. The following antibodies from BD Pharmingen (San Diego, CA, USA), eBioscience (San Diego, CA, USA) and BioLegend (San Diego, CA, USA) were used for flow cytometry: anti-mouse lineage cocktail (145-2C11, RB6-8C5, M1/70, RA3-6B2, Ter-119), anti-Sca1 (anti-Ly6A; D7), anti-CD117 (anti-c-Kit; 2B8), anti-CD127 (anti-IL-7Rα; A7R34), anti-CD150 (mShad150), anti-CD48 (HM48-1), anti-CD135 (Flt3;A2F10.1), anti-CD34 (RAM34), anti-B220 (RA3–6B2), anti-CD3e (145-2C11), anti-CD11b (M1/70), rat IgG2a k isotype (R35-95), rat IgG2b k isotype (A95-1), rat IgG1 k isotype (R3-34), and rat IgG1 λ isotype (A110-1). Data were collected using FACSCanto II (BD Pharmingen, San Diego, CA, USA) and were analyzed using FlowJo software (TreeStar, Ashland, OR, USA).
Cell proliferation studies
The in vivo incorporation of BrdU into LSK (Lin−Sca1+cKit+) cells was assessed using the FITC BrdU Flow kit (BD Pharmingen, San Diego, CA, USA). Mice were intraperitoneally injected with 2 mg/mL of BrdU for 5 days and then sacrificed. BM cells were prepared and stained with antibodies and analyzed by flow cytometry.
Pyronin and Hoechst staining
The quiescence of freshly isolated HSCs was determined by staining with Hoechst 33342 (Molecular Probes, Eugene, OR, USA) and Pyronin Y (Sigma-Aldrich, St. Louis, MO, USA). BM cells were resuspended in phosphate-buffered saline containing 2% (v/v) fetal calf serum and 10 mM Hoechst 33342. Cells were incubated for 30 minutes at 37°C and were then washed and subsequently resuspended in phosphate-buffered saline supplemented with 20 mM N-2-hydroxyethylpiperazine-N9-2-ethanesulfonic acid, pH 7.4, 1 mg/mL glucose, 10% (v/v) fetal calf serum, 10 mM Hoechst 33342, and 1 mg/mL Pyronin Y. Cells were incubated for an additional 15 minutes at 37°C and were then washed and stained for analysis by flow cytometry. Pyronin Y fluorescence was detected at 575 nm in the linear range.
Competitive repopulation and BM transplantation studies
For competitive-repopulation experiments, 1×103 of donor (CD45.2+) LSK cells were sorted from 8-week-old wild-type and cKO mice and mixed with 2×105 competitor BM (CD45.1+) cells, and the mixtures were transplanted intravenously into lethally irradiated (11 Gy) congenic (CD45.1+) recipients. Mice were sacrificed 8 weeks after transplantation. Blood and bone marrow cells were analyzed by FACS.
Treatment with 5-FU
Mice were given a single intraperitoneal dose of 5-FU (75 mg/kg body weight) on day 0. Mice were sacrificed on days 0, 2, 4, 6, and 10 after injection. BM cells were counted and analyzed by flow cytometry.
Statistical analysis
Statistical analysis was performed using SPSS 15.0 software (IBM, Armonk, NY, USA). Data are expressed as the mean ± standard deviation (s.d.) from at least 3 separate experiments. Differences were considered significant at * P<0.05 and ** P<0.01.
Results
MYSM1 cKO mice show reduced BM cellularity
To overcome the embryonic lethality of MYSM1 germline knockout mice, we generated a MYSM1 conditional knockout allele. The mice with loxP sites flanking MYSM1 exon 3 have been described [18]. We bred floxed MYSM1 mice to Vav1-cre transgenic mice to generate MYSM1-floxed Vav1-cre mice, hereafter denoted as cKO mice, which were viable and fertile. Age-matched MYSM1-floxed Vav1-Cre mice, hereafter denoted as WT, were used as control. We observed highly efficient cre-mediated deletion of floxed MYSM1 in the bone marrow (BM) of cKO mice where there were no detectable floxed alleles (Figure 1A). Real-time PCR analysis showed that MYSM1 mRNA levels were greatly reduced in cKO BM cells (Figure 1B). Our analysis showed that MYSM1 cKO mice had a 3-fold decrease in total BM cell numbers and a more than 2-fold reduction in Lin− (CD11b−Gr-1−B220−CD3−Ter119−) cells, as well as a distinct reduction in the absolute numbers of LSK cells, which include HSCs and their earliest progenitors (Figure 1C–1E). The reduced BM cellularity in cKO mice was similar to the reduction observed in conventional MYSM1 knockouts [20].
Figure 1.
Loss of MYSM1 results in reduced BM cellularity (A) Agarose gel of polymerase chain reaction genotyping of bone marrow cells taken from WT (same as MYSM1fl/fl) and MYSM1 cKO (MYSM1fl/fl; Tg[Vav1-cre]) mice. (B) Real-time PCR analysis of MYSM1 in WT and cKO BM cells. The Ct values are normalized to GAPDH. Absolute number of (C) BM cells, (D) Lin− cells and (E) LSK cells per femur (hind leg) of 8- to 12-week-old WT and cKO mice; n=4 mice per group. ** P<0.01
MYSM1 deficiency results in a reduction of HSCs and progenitors
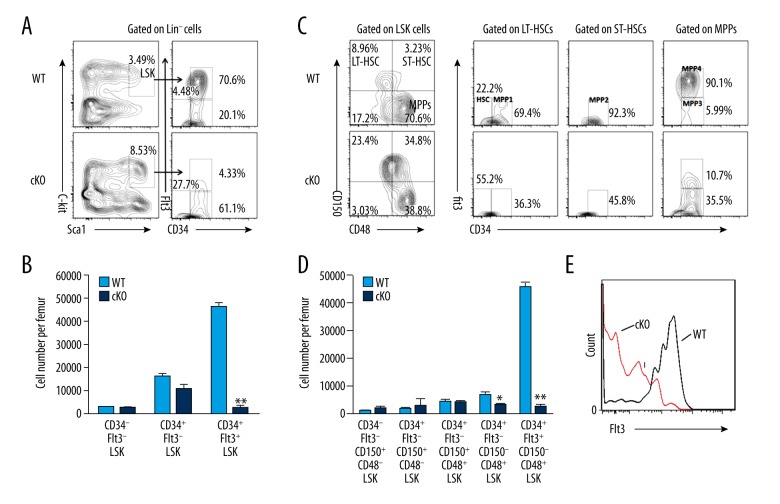
The LSK (Lin−Sca1+cKit+) fraction of BM cells is enriched for hematopoietic stem and progenitor cells (HSPCs). Although the LSK population is commonly studied, it is heterogeneous and includes multipotent progenitors and other committed progenitors that do not maintain their long-term reconstitution potential. To analyze the function of MYSM1 in each individual HSPC population, we further divided LSK cells based on CD34 and Flt3 expression. We discovered that the proportions of CD34−Flt3− and CD34+Flt3− LSK cells, which represent more primitive HSCs, were notably increased (Figure 2A). However, due to the reduced total number of cKO BM cells, the absolute numbers of CD34−Flt3− and CD34+Flt3− LSK cells in cKO mice were similar to those in WT (Figure 2B). On the other hand, the more mature multipotent progenitors, CD34+Flt3+LSK cells, were severely decreased both in proportion and in absolute number among cKO BM cells (Figure 2A, 2B). Similar results were obtained by CD150- and CD48-based immunophenotyping of LT-HSCs (CD150+CD48−LSK cells), ST-HSCs (CD150+CD48+LSK cells), and MPPs (CD150−CD48+LSK cells) (Figure 2C, left).
Figure 2.
MYSM1 is vital in maintaining HSCs and progenitors. (A) Distribution of LSK cells and various LSK subsets in WT and cKO BM: Lin negative-gated cells were assessed based on their expression of Sca1 and c-Kit (left), whereas LSK-gated cells were further defined based on the expression of CD34 and Flt3 (right). Numbers adjacent to outlined areas indicate frequency. (B) Absolute numbers of LSK subsets per femur in WT and cKO mice based on the gates in a; n=4 mice per group. (C) Distribution of cells in LSK subsets in WT and cKO mice, assessed based on the expression of CD150 and CD48 (left), and expression of Flt3 and CD34 in WT and cKO BM LSK subsets (right). (D) Absolute numbers of LSK subsets per femur in WT and cKO mice based on the gates in c; n=4 mice per group. (E) Flt3 expression in WT and cKO LSK cells. * P<0.05, ** P<0.01
LSK cells can be further subcategorized on the basis of their expression of CD150, CD48, CD34, and Flt3 into the following 5 subsets: the most primitive HSC subset (CD34−Flt3−CD150+CD48−LSK) and the increasingly differentiated MPP1 (CD34+Flt3−CD150+CD48−LSK), MPP2 (CD34+Flt3−CD150+CD48+LSK), MPP3 (CD34+Flt3−CD150−CD48+LSK), and MPP4 subsets (CD34+Flt3+CD150−CD48+LSK) (Figure 2C, right) [28,29]. Our analysis showed that that the absolute numbers of MPP3 and MPP4 cells were dramatically decreased, although there was no statistically significant reduction in the most primitive HSC, MPP1, or MPP2 cells (Figure 2D). Specifically, the reduction was severe in the most mature MPP4 compartment, with a 9-fold reduction in the percentages and a 12-fold reduction in the number of cells in cKO mice compared with their WT controls (Figure 2C, 2D). We further noticed that the reduction of MPP4 resulted from a distinct decrease of the Flt3+LSK cells (Figure 2E). Together, these data demonstrate that MYSM1 is required for HSC differentiation, specifically during the transition to the MPP subsets.
MYSM1 is required to maintain HSC quiescence
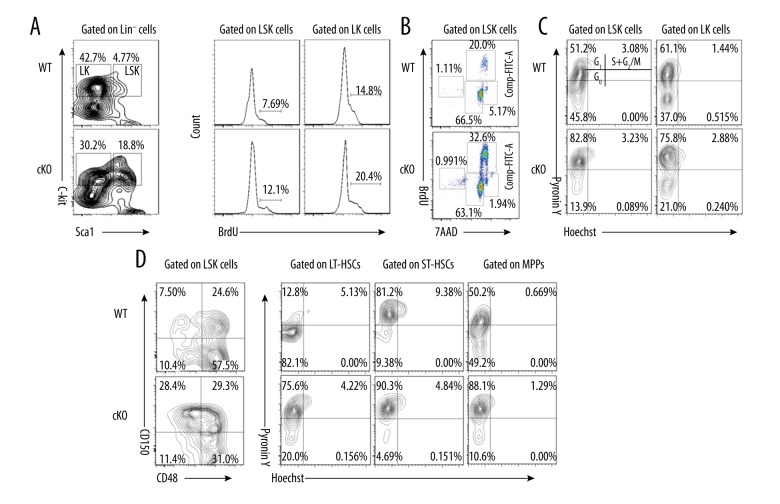
Adult HSCs rarely divide under steady-state conditions, and the majority of cells reside in the quiescent G0 phase of the cell cycle [28,30]. Quiescent HSCs possess remarkable proliferative capacity, allowing them to engage in massive and repetitive regenerative activities in response to hematopoietic damage. Bromodeoxyuridine (BrdU) is a thymidine analog that can be incorporated into DNA during the S phase of the cell cycle. As such, BrdU incorporation can be used to quantify the number of cells that are in S phase in the time period during which BrdU is available. We thus used an in vivo BrdU incorporation assay as a measure of cell proliferation in adult HSCs. At 5 days after BrdU injection, 12% of cKO LSK cells and 20% of cKO LK (Lin−Sca1−cKit+) cells were noted to have incorporated BrdU, whereas only approximately 8% of WT LSK and 15% of WT LK cells had incorporated BrdU (Figure 3A), indicating that MYSM1 cKO HSCs proliferate faster than WT HSCs. Cell cycle analysis showed that 32.6% of cKO LSK cells were in the S phase, whereas only 20.0% of WT LSK cells were in this phase (Figure 3B). We then measured the total RNA/DNA contents in the LSK cells and their subsets by Pyronin Y/Hoechst staining to evaluate HSC quiescence. We found that MYSM1 deficiency significantly reduced the proportion of LSK cells and LK cells in the G0 phase, whereas there was a significant increase in the proportion of LSK cells and LK cells in G1 phase (Figure 3C). When we used CD150 and CD48 to divide LSK cells into LT-HSCs, ST-HSCs, and MPPs, we observed drastically reduced proportions of G0 in all these subpopulations in cKO mice (Figure 3C), demonstrating that MYSM1 is required to maintain HSC quiescence and that deficiency in MYSM1 drives quiescent HSCs into rapid cycling.
Figure 3.
Loss of MYSM1 drives HSCs from quiescence to rapid proliferation. (A) WT and cKO mice received 2 mg BrdU intraperitoneally daily for 5 days. Incorporation of BrdU was analyzed by FACS in BM LSK and LK cells (n=4 per group). (B) Mice received 2 mg BrdU i.p. injection 1 hour before sacrifice. BM cells were isolated and stained for cell cycle analysis (n=4 per group). (C, D) WT and cKO BM cells were stained for HSC surface antigens followed by Hoechst 33258/Pyronin Y staining. Representative FACS plots of cells depicting G0 (bottom left quadrant), G1 (top left quadrant), and S/G2/M (top right quadrant) in (C) LSK and LK cells and (D) LSK subsets (n=4 per group).
MYSM1-deficient HSCs display impaired engraftment
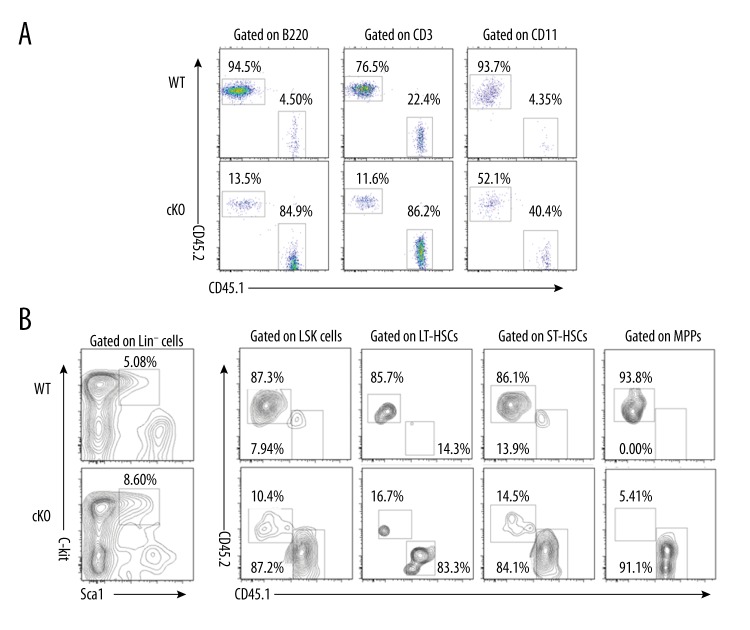
To test whether cKO HSCs were also affected in their functions, we next used competitive transplantation assays to compare the in vivo functions of cKO LSK cells. Each groups of lethally irradiated mice (CD45.1+) received 1×103 sorted LSK cells from WT or cKO mice (CD45.2+) along with 2×105 competitor BM cells (CD45.1+). The chimerism was followed in peripheral blood at 8 weeks post-transplantation. Loss of MYSM1 markedly affected LSK repopulating capacity. MYSM1-deficient LSK cells failed to reconstitute as many B cells, T cells, and myeloid cells as WT LSK cells did. Only 13.5% of B cells (B220+), 11.6% of T cells (CD3+), and 52.1% of myeloid cells (CD11+) were derived from the cKO donor, whereas 94.5% of B cells, 76.5% of T cells, and 93.7% of myeloid cells were derived from the WT donor (Figure 4A). Flow cytometry of BM cells revealed that in marked contrast to LSK and LSK subsets cells reconstitution by the WT LSK, engraftments of LSK cells from cKO mice were much lower (Figure 4B). These data demonstrate that normal HSC activity is critically dependent on MYSM1, and MYSM1-deficient HSCs display intrinsic functional defects and impaired engraftment.
Figure 4.
Cell-intrinsic defects of MYSM1-deficient HSCs in engraftment. (A, B) Donor chimerism in competitive repopulation assay. 1×103 LSK cells sorted from wild-type or cKO mice (CD45.2+) were transplanted into lethally irradiated recipients (CD45.1+) together with 2×105 competitor BM cells (CD45.1+). (A) Percentages of donor-derived cells (CD45.2+) in peripheral blood (PB) 8 weeks after transplantation. (B) Percentages of donor-derived cells in LSK and LSK subsets cells of BM 8 weeks after transplantation (n=4 per group).
Loss of MYSM1 results in elevated apoptosis and failure of recovery of HSC cells
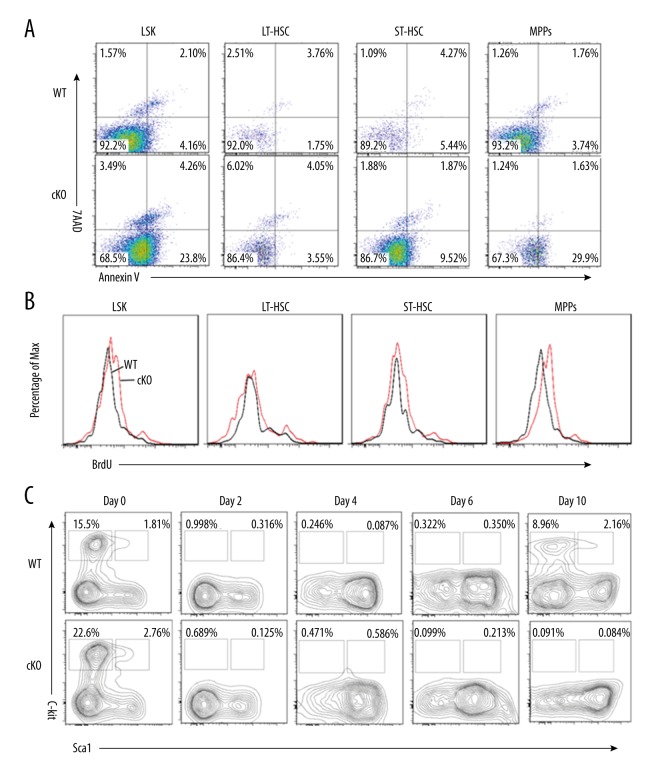
We then examined whether a reduction in the HSC pool size in MYSM1 cKO mice is caused by their increased apoptotic rate. Indeed, we observed an average of 23.8% apoptosis among MYSM1 cKO LSK cells, which was more than 5-fold higher than in WT mice (4.16%) (Figure 5A). Further analysis showed that although the apoptotic rate of LT-HSC was similar between WT and cKO BM cells, increased apoptosis was observed in cKO ST-HSC and MPPs cells as compared with WT controls (Figure 5A). Not surprisingly, we observed a high apoptosis rate in the more mature MPPs progenitors, which was consistent with the sharp reduction in both the frequencies and absolute cell numbers of these cells in the MYSM1 cKO BM (Figure 2A–2D). Our data further showed up-regulated levels of ROS in MYSM1 cKO LSK cells and MPPs cells (Figure 5B) and suggest that in addition to controlling HSC quiescence and driving them into cycling, MYSM1 balances the rate of apoptosis of HSCs and other progenitors to maintain the HSC pool size.
Figure 5.
MYSM1 is required to maintain HSC survival. (A) Analysis of apoptotic cells among LSK and LSK subsets cells from WT and cKO mice. Apoptotic cells were detected by staining with Annexin V and DAPI (n=3). (B) Reactive oxygen species (ROS) levels in LSK and LSK subsets cells from WT and cKO mice were measured using DCFDA, an ROS-sensitive fluorescent dye (n=3). (C) Eight-week-old WT and cKO mice were injected with a single dose of 5-FU (75 mg/kg, i.p.). Distribution of LSK and LK cells in WT and cKO BM were analyzed by FACS analysis (n=4 mice per group).
Because MYSM1 cKO HSCs are more likely to be actively cycling and are prone to apoptosis, we investigated whether these factors would result in exhaustion of the quiescent stem cell reservoir. Cycling cells are sensitive to 5-fluorouracil (5-FU) but quiescent HSCs are resistant. We administered a single dose of 5-FU (75 mg/kg, i.p.) to 8-week-old WT and cKO mice and monitored the LSK number and proportion. Examination by FACS analysis revealed that the LSK and LK cells in both groups were greatly decreased shortly after 5-FU administration. The LSK cells derived from WT mice had reverted back to the normal levels by day 10, whereas the cells derived from MYSM1 cKO mice were constantly proliferating and exhausting the stem cell reserves, leading to a steady reduction in LSK and more mature LK cell populations (Figure 5C).
Discussion
Histone ubiquitination is one of the major epigenetic modifications that occur on histone tails. Among the histone octamers, H2A is the most highly ubiquitinated protein. Monoubiquitination of histones H2A and H2B is reversible. PRC1 is responsible for monoubiquitination of H2A at lysine 119, whereas the removal of this ubiquitin modification requires ubiquitin-specific peptidases known as deubiquitinating enzymes (DUBs). Several DUBs, including USP16, MYSM1, USP21, and BAP1, have been identified as H2A-specific DUBs, whereas others, such as USP3, USP12, and USP46, display dual specificity toward both ubH2A and ubH2B [16]. Although initially described as a required protein of the activation of several target genes in prostate cancer cells, MYSM1 was later reported to be an important epigenetic regulator in hematopoiesis [18–22]. We and other groups found that MYSM1 deficiency in mice resulted in lymphopenia, anemia, and defects in B, NK, and dendritic cell development and function [18–22,31,32]. In human subjects, mutations in MYSM1 lead to defective hematopoiesis, causing anemia, thrombocytopenia, and lymphopenia, similar to the phenotypes in MYSM1−/− mice [24,25]. In terms of HSCs, our previous data showed that MYSM1 was required to maintain HSC self-renewal and differentiation, and MYSM1−/− mice failed to maintain HSC quiescence and pool size. In addition, studies have also described an essential role for MYSM1 in normal fetal liver hematopoiesis [23]. In the search for the mechanism by which MYSM1s function in HSC maintenance, we found that MYSM1 controls HSC homeostasis partly by regulating Gfi1 expression [20]. Interestingly, MYSM1−/−p53−/− mice show a full rescue of MYSM1−/− developmental and hematopoietic defects, and repression of the p53-target gene PUMA by MYSM1 is essential for the survival of hematopoietic multipotent progenitors, indicating that p53 activation is the driving mechanism for hematopoietic abnormalities in MYSM1 deficiency [33,34].
Although functional studies of MYSM1 in hematopoiesis have been carried out, more detailed research is needed. In this study, we analyzed a conditional knockout of MYSM1 induced by Vav1-cre. At steady state, we observed reduced bone marrow cellularity and abnormal HSCs. Both the cell number and proportion of HSCs and multipotent progenitors in cKO mice differed from those in WT mice. In particular, the more mature multipotent progenitors (CD34+Flt3+LSK cells or MPP4 cells) in the cKO BM were more severely decreased in both proportion and absolute number, which is in accord with our previous studies in conventional MYSM1 knockout mice [20]. We also found that MYSM1 cKO HSCs had a loss of quiescence, unscheduled HSC proliferation, higher ROS levels, and increased apoptosis. Unlike c-Cbl-deficient mice [35] and Erg1-deficient mice [36], in which continuous and excessive HSC proliferation results in a larger HSC pool but without ‘stem cell exhaustion,’ MYSM1 cKO mice had a reduced pool of HSCs and failed to recover in response to stresses such as 5-FU due to premature proliferative exhaustion. In the competitive transplantation assays, cKO HSCs display impaired engraftment, although much less severe compared with MYSM1−/− mice, which we used in previous work [20]. We found that the overall phenotype we observed in the Vav1-cre MYSM1-floxed mouse line is similar to the phenotype seen in the full-MYSM1-knockout mice. However, due to the partial embryonic lethality, multiple organ defects, and growth retardation in conventional MYSM1 knockouts, cKO mice are more reliable in the functional study of HSCs.
Conclusions
In conclusion, together with our previous data, we clearly illustrate that MYSM1 is essential for maintaining HSC quiescence and survival. However, the detailed mechanisms of this phenomenon, and whether p53 signaling plays a key role in cKO HSCs, remains to be studied further.
Footnotes
Conflict of interest
None.
Source of support: This study was supported by the National Natural Science Foundation of China (81773003 and 81472633)
References
- 1.Sashida G, Iwama A. Epigenetic regulation of hematopoiesis. Int J Hematol. 2012;96:405–12. doi: 10.1007/s12185-012-1183-x. [DOI] [PubMed] [Google Scholar]
- 2.Rice KL, Hormaeche I, Licht JD. Epigenetic regulation of normal and malignant hematopoiesis. Oncogene. 2007;26:6697–714. doi: 10.1038/sj.onc.1210755. [DOI] [PubMed] [Google Scholar]
- 3.Cullen SM, Mayle A, Rossi L, Goodell MA. Hematopoietic stem cell development: An epigenetic journey. Curr Top Dev Biol. 2014;107:39–75. doi: 10.1016/B978-0-12-416022-4.00002-0. [DOI] [PubMed] [Google Scholar]
- 4.Park IK, Qian D, Kiel M, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–5. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 5.Akasaka T, Tsuji K, Kawahira H, et al. The role of mel-18, a mammalian Polycomb group gene, during IL-7-dependent proliferation of lymphocyte precursors. Immunity. 1997;7:135–46. doi: 10.1016/s1074-7613(00)80516-6. [DOI] [PubMed] [Google Scholar]
- 6.Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20:845–54. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 7.de Napoles M, Mermoud JE, Wakao R, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7:663–76. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 8.Martin-Perez D, Piris MA, Sanchez-Beato M. Polycomb proteins in hematologic malignancies. Blood. 2010;116:5465–75. doi: 10.1182/blood-2010-05-267096. [DOI] [PubMed] [Google Scholar]
- 9.Sauvageau M, Sauvageau G. Polycomb group proteins: Multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 2010;7:299–313. doi: 10.1016/j.stem.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwama A, Oguro H, Negishi M, et al. Epigenetic regulation of hematopoietic stem cell self-renewal by polycomb group genes. Int J Hematol. 2005;81:294–300. doi: 10.1532/IJH97.05011. [DOI] [PubMed] [Google Scholar]
- 11.Wang H, Wang L, Erdjument-Bromage H, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–78. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 12.Nakagawa T, Kajitani T, Togo S, et al. Deubiquitylation of histone H2A activates transcriptional initiation via trans-histone cross-talk with H3K4 di- and trimethylation. Genes Dev. 2008;22:37–49. doi: 10.1101/gad.1609708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stock JK, Giadrossi S, Casanova M, et al. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol. 2007;9:1428–35. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- 14.Joo HY, Zhai L, Yang C, et al. Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature. 2007;449:1068–72. doi: 10.1038/nature06256. [DOI] [PubMed] [Google Scholar]
- 15.Scheuermann JC, de Ayala Alonso AG, Oktaba K, et al. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465:243–47. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao J, Yan Q. Histone ubiquitination and deubiquitination in transcription, DNA damage response, and cancer. Front Oncol. 2012;2:26. doi: 10.3389/fonc.2012.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu P, Zhou W, Wang J, et al. A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Mol Cell. 2007;27:609–21. doi: 10.1016/j.molcel.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang XX, Nguyen Q, Chou Y, et al. Control of B cell development by the histone H2A deubiquitinase MYSM1. Immunity. 2011;35:883–96. doi: 10.1016/j.immuni.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nijnik A, Clare S, Hale C, et al. The critical role of histone H2A-deubiquitinase Mysm1 in hematopoiesis and lymphocyte differentiation. Blood. 2012;119:1370–79. doi: 10.1182/blood-2011-05-352666. [DOI] [PubMed] [Google Scholar]
- 20.Wang T, Nandakumar V, Jiang XX, et al. The control of hematopoietic stem cell maintenance, self-renewal, and differentiation by Mysm1-mediated epigenetic regulation. Blood. 2013;122:2812–22. doi: 10.1182/blood-2013-03-489641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Won H, Nandakumar V, Yates P, et al. Epigenetic control of dendritic cell development and fate determination of common myeloid progenitor by Mysm1. Blood. 2014;124:2647–56. doi: 10.1182/blood-2013-10-534313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nandakumar V, Chou Y, Zang L, et al. Epigenetic control of natural killer cell maturation by histone H2A deubiquitinase, MYSM1. Proc Natl Acad Sci USA. 2013;110(2013):E3927–36. doi: 10.1073/pnas.1308888110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forster M, Belle JI, Petrov JC, et al. Deubiquitinase MYSM1 is essential for normal fetal liver hematopoiesis and for the maintenance of hematopoietic stem cells in adult bone marrow. Stem Cells Dev. 2015;24:1865–77. doi: 10.1089/scd.2015.0058. [DOI] [PubMed] [Google Scholar]
- 24.Alsultan A, Shamseldin HE, Osman ME, et al. MYSM1 is mutated in a family with transient transfusion-dependent anemia, mild thrombocytopenia, and low NK- and B-cell counts. Blood. 2013;122:3844–45. doi: 10.1182/blood-2013-09-527127. [DOI] [PubMed] [Google Scholar]
- 25.Le Guen T, Touzot F, Andre-Schmutz I, et al. An in vivo genetic reversion highlights the crucial role of Myb-Like, SWIRM, and MPN domains 1 (MYSM1) in human hematopoiesis and lymphocyte differentiation. J Allergy Clin Immunol. 2015;136:1619–26. e1611–15. doi: 10.1016/j.jaci.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 26.Sharabi AB, Aldrich M, Sosic D, et al. Twist-2 controls myeloid lineage development and function. PLoS Biol. 2008;6:e316. doi: 10.1371/journal.pbio.0060316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song XT, Evel-Kabler K, Shen L, et al. A20 is an antigen presentation attenuator, and its inhibition overcomes regulatory T cell-mediated suppression. Nat Med. 2008;14:258–65. doi: 10.1038/nm1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson A, Laurenti E, Oser G, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135:1118–29. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- 29.Rathinam C, Matesic LE, Flavell RA. The E3 ligase Itch is a negative regulator of the homeostasis and function of hematopoietic stem cells. Nat Immunol. 2011;12:399–407. doi: 10.1038/ni.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takizawa H, Regoes RR, Boddupalli CS, et al. Dynamic variation in cycling of hematopoietic stem cells in steady state and inflammation. J Exp Med. 2011;208:273–84. doi: 10.1084/jem.20101643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang XX, Liu Y, Li H, et al. MYSM1/miR-150/FLT3 inhibits B1a cell proliferation. Oncotarget. 2016;7(42):68086–96. doi: 10.18632/oncotarget.11738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang XX, Chou Y, Jones L, et al. Epigenetic regulation of antibody responses by the histone H2A deubiquitinase MYSM1. Sci Rep. 2015;5:13755. doi: 10.1038/srep13755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Belle JI, Langlais D, Petrov JC, et al. p53 mediates loss of hematopoietic stem cell function and lymphopenia in Mysm1 deficiency. Blood. 2015;125:2344–48. doi: 10.1182/blood-2014-05-574111. [DOI] [PubMed] [Google Scholar]
- 34.Belle JI, Petrov JC, Langlais D, et al. Repression of p53-target gene Bbc3/PUMA by MYSM1 is essential for the survival of hematopoietic multipotent progenitors and contributes to stem cell maintenance. Cell Death Differ. 2016;23:759–75. doi: 10.1038/cdd.2015.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rathinam C, Thien CB, Langdon WY, et al. The E3 ubiquitin ligase c-Cbl restricts development and functions of hematopoietic stem cells. Genes Dev. 2008;22:992–97. doi: 10.1101/gad.1651408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Min IM, Pietramaggiori G, Kim FS, et al. The transcription factor EGR1 controls both the proliferation and localization of hematopoietic stem cells. Cell Stem Cell. 2008;2:380–91. doi: 10.1016/j.stem.2008.01.015. [DOI] [PubMed] [Google Scholar]