Abstract
A variety of data models have been developed to provide a standardized data interface that supports organizing clinical research data into a standard structure for building the integrated data repositories. HL7 Fast Healthcare Interoperability Resources (FHIR) is emerging as a next generation standards framework for facilitating health care and electronic health records-based data exchange. The objective of the study is to design and assess a consensus-based approach for harmonizing the OHDSI CDM with HL7 FHIR. We leverage a FHIR W5 (Who, What, When, Where, and Why) Classification System for designing the harmonization approaches and assess their utility in achieving the consensus among curators using a standard inter-rater agreement measure. Moderate agreement was achieved for the model-level harmonization (kappa=0.50) whereas only fair agreement was achieved for the property-level harmonization (kappa=0.21). FHIR W5 is a useful tool in designing the harmonization approaches between data models and FHIR, and facilitating the consensus achievement.
Keywords: Reference Standards, Observational Study, Vocabulary, Controlled
Introduction
Integrated Data Repositories (IDRs)(1, 2) are needed to combine molecular and phenotypic data, making data available with analytic tools. This is especially important for clinical research investigators with limited computing resources. In a 2010 survey conducted by the Clinical and Translational Science Award (CTSA) consortium(3), IDR was defined as a data warehouse integrating various sources of clinical data to support queries for a range of research-like functions. Survey results suggest that individual organizations are progressing in their approaches to the development, management, and use of IDRs as a means to support a broad array of research. A variety of data models have been developed to provide a standardized data interface that supports organizing clinical research data into a standard structure in such IDRs. These data models include the Observational Medical Outcomes Partnership (OMOP) Common Data Model (CDM)(4, 5), the National Patient-Centered Research Networks (PCORnet) CDM(6), and the Informatics for Integrating Biology and the Bedside (i2b2) Star Schema(7).
The OMOP was a public-private partnership established to inform the appropriate use of observational healthcare databases for studying the effects of medical products. The OMOP community is actively using the OMOP CDM(4, 5) for their various research purposes. Observational Health Data Sciences and Informatics (OHDSI) has been established as a multi-stakeholder, interdisciplinary collaborative to create open-source solutions that bring out the value of observational health data through large-scale analytics. The OHDSI collaborative includes all of the original OMOP research investigators and develops its tools using the OMOP CDM, which will continue to be an open-source, community standard for observational healthcare data.
The PCORnet CDM(6) is based on the Mini-Sentinel CDM and has been informed by other distributed initiatives such as the HMO Research network, the Vaccine Safety Datalink, and the ONC Standards & Interoperability Framework Query Health Initiative. The CDM leverages standard terminologies and coding systems for healthcare to enable interoperability and ensure responsiveness to evolving data standards.
The i2b2 is an open-source clinical data analytics platform that provides a component-based architecture and a flexible analytical database design(7-9). The i2b2 Star Schema was developed as a CDM that enables conformant transformation of patient data to a common data structure and representation of meaning. i2b2-based solutions have been widely used in clinical research communities such as the Shared Health Research Information Networks (SHRINE)(10), and the PCOR-net(11). Building on the i2b2 framework, the tranSMART platform(12-14) is an analytic platform that also incorporates the ability to load molecular datatypes, including those derived from next generation sequencing (NGS).
These data models serve well as a layer of standardization for clinical research data within their own research network; however, if the investigators want to reuse and integrate these research datasets and applications in broader clinical research communities across different research networks they still face huge challenges. This situation demands a global data model as a reference standard to facilitate data model harmonization and data integration.
HL7 Fast Healthcare Interoperability Resources (FHIR) is emerging as a next generation standards framework for facilitating health care and electronic health records-based data exchange(15). However, it has been a chellenging issue for the standards and research communities to build a consistent and measurable approach for enabling the consensus achieveing in terms of data model hamronization efforts. The objective of the study is to design and assess a consensus-based approach for harmonizing the OHDSI CDM with HL7 FHIR. We leverage the FHIR W5 (Who, What, When, Where, and Why) Classification System(16, 17) for designing the harmonization approach and assess its utility in achieving the consensus among curators using a standard inter-rater agreement measure. The outcome of this study would provide guidance to harmonize different data models with FHIR in future.
Methods
Materials
OMOP Common Data Model (CDM)
The OMOP CDM is “designed to support the conduct of research to identify and evaluate associations between interventions (e.g., drug exposure) and outcomes (e.g., adverse effects) caused by these interventions”(5). The design principles of the CDM include 1) suitable for purpose; 2) data protection; 3) design of domains; 4) rational for domains; 5) standardized vocabularies; 6) reuse of existing vocabularies; 7) maintaining source codes; 8) technology neutrality; 9) scalability; and 10) backwards compatibility. The CDM defines table schemas in a person-centric manner. As of September 18, 2016, version V5.0.1 of the CDM was released, which contains 39 tables in 6 categories: standardized clinical data, standardized health system data, standardized health economics, standardized metadata, standardized vocabularies and standardized derived elements. In fact, terminology normalization enabled by standard vocabularies with focus on SNOMED CT, LOINC and RxNorm is a strong characteristic of the OMOP CDM. Figure 1 shows a diagram highlighting the high-level relationships among the tables and categories.
Figure 1.
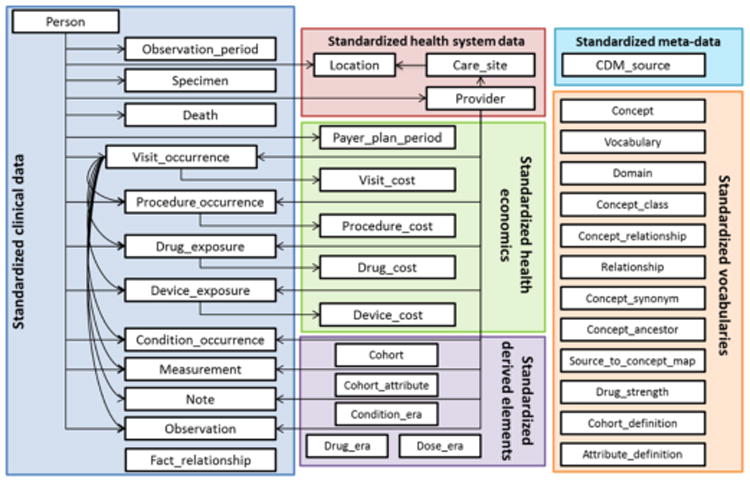
A diagram highlighting the high-level relationships among the tables and categories. (Source from the OMOP CDM document)
HL7 FHIR Core Resources
Detailed Clinical Models (DCMs) have been regarded as the basis for retaining computable meaning when data are exchanged between heterogeneous computer systems(18, 19). Amongst the emerging national and international initiatives on the standardization of DCM modeling are the Clinical Informatics Modeling Initiative (CIMI)(20) and FHIR)(14). The primary goal of CIMI is to “Improve the interoperability of healthcare systems through shared implementable clinical information models (A single curated collection)”. FHIR builds around the concept of “resources”. “Resources” here means small discrete concepts with clearly defined scope that can be maintained independently. Resources are the smallest units of a transaction, and each resource has a unique id that aligns with RESTful design philosophy. As of September 16, 2016, the version Draft Standard for Trials Use (DSTU) 2 has been released and the version of STU 3 was placed under ballot. Figure 2 shows a collection of FHIR core resources under the FHIR Clinical category in the released DSTU 2.
Figure 2.
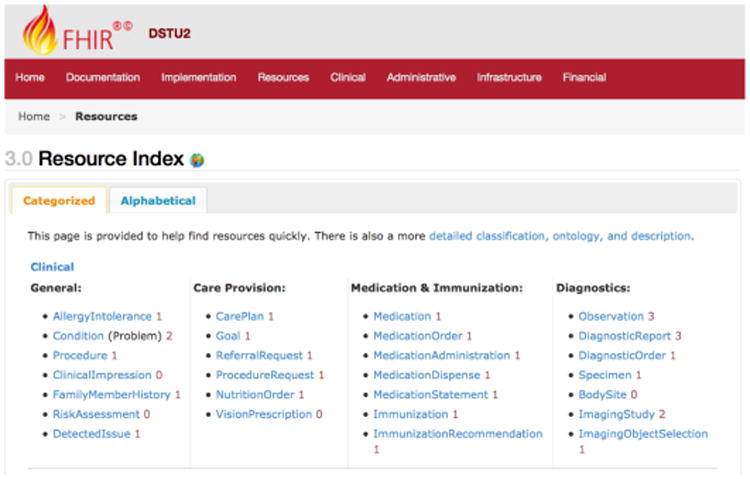
A screenshot showing a collection of FHIR core resources under the Clinical category in the released DSTU 2.
HL7 FHIR W5 Classification System
HL7 electronic health record (EHR) workgroup develops a FHIR Record Lifecycle Events Implementation Guide that “offers a methodology to support trusted EHR management using HL7 FHIR”(17). The fundamental assumption is that the EHR system captures Action taken and creates corresponding Record Entries. Actions have associated metadata (e.g., who, what, when, where, why, how, under what conditions, in what context). The corresponding Record Entry captures this metadata along with other Action and Record Entry related information. The FHIR W5 (Who, What, When, Where, and Why) Report(16) has been created for each release to establish consistent W5 metadata definitions across FHIR resources for tracking resource (instance) lifecycle events when content is created or updated, and/or when signature is applied. In the current implementation, the FHIR W5 Classification System plays two roles: 1) specifying a Resource Category system to classify the FHIR resources; and 2) specifying a Property Category system to classify the FHIR properties. For example, as shown in Figure 2, the FHIR resource Observation belongs to the category clinical.diagnostics. Table 1 and Table 2 show the categories specified in both systems respectively and examples from FHIR W5 Report for the properties of the FHIR resource Observation.
Table 1. The distribution of the FHIR core resources and OMOP CDM tables by the FHIR W5 resource category.
| FHIR W5 Resource Category | Number of FHIR Resources (DSTU2) | Number of OMOP CDM Tables |
|---|---|---|
| administrative | subtotal: 16(17%) | subtotal: 7 (18%) |
| administrative.device | 3 | 1 |
| admini strati ve. entity | 4 | 1 |
| administrative.group | 3 | 4 |
| administrative.individual | 6 | 1 |
| clinical | subtotal: 27 (28%) | subtotal: 13 (33%) |
| clinical.carepro vision | 6 | |
| clinical.diagnostics | 7 | 6 |
| clinical.general | 7 | 3 |
| clinical.medication | 7 | 4 |
| conformance | subtotal: 10(10%) | subtotal: 9 (23%) |
| conformance.behavior | 3 | |
| conformance.content | 2 | |
| conformance.misc | 2 | |
| conformance.terminology | 3 | 9 |
| financial | subtotal: 10(10%) | subtotal: 5 (13%) |
| financial.billing | 2 | 4 |
| financial.other | 1 | |
| flnancial.payment | 2 | 1 |
| financial.support | 5 | |
| infrastructure | subtotal: 16(17%) | subtotal: 3 (8%) |
| infrastructure.documents | 4 | 2 |
| infrastructure.exchange | 4 | 1 |
| infrastructure.information | 4 | |
| infrastructure.structure | 4 | |
| workflow | subtotal: 17(18%) | subtotal: 2 (5%) |
| workflow.encounter | 4 | 2 |
| workflow.order | 9 | |
| workflow. scheduling | 4 | |
| Total | 96 (100%) | 39 (100%) |
Table 2. The alignment results between the FHIR resource elements and the OMOP table fields.
| FHIRW5 Property Category | FHIR Observation | OMOP Observation | FHIR Drug Administration | OMOP Drug_Exposure |
|---|---|---|---|---|
| class | category | observation_type_concept_id | drug_type_concept_id | |
| context | encounter | visit_occurrence_id | encounter | visit_occurrence_id |
| grade | qualifier_concept_id | |||
| id | identifier | observation_id | identifier | drug_exposure_id |
| id. version | ||||
| status | status | status | ||
| what | code | observation_concept_id; value_as_number value_as_string value_as_concept_id unit_concept_id observation_source_value observation_source_concept_id unit_source_value qualifier_source_value |
drug_concept_id refills quantity days_supply sig route_concept_id effective_drug_dose dose_unit_concept_id lot_number drugs_source_value drug_source_concept_id drugs_source_value dose_unit_source_value |
|
| when.done | effective Time [x] | observation_date observation_time |
effect ivcTimc[x] | drug_exposure_end_date |
| when.ink | drug_exposure_start_date | |||
| when.planned | ||||
| when, recorded | issued | |||
| where | visit_occurrence_id | visit_occurrence_id | ||
| who | ||||
| who.actor | performer | provider_id | practitioner | provider_id |
| who.author | ||||
| who.cause | ||||
| who.focus | subject | person_id | patient | person_id |
| who.source | ||||
| who.witness | ||||
| why | stop_reason |
Methods
Measure the consensus in the model-level harmonization
The OMOP CDM consists of a collection of table schemas. Each table schema represents a particular OMOP domain, which is analogous to the resources as defined in FHIR. In the model level, we design the following two approaches to align the table schemas with the FHIR resources. The first approach is to categorize the OMOP table schemas using the FHIR W5 resource categories, and the second approach is to map the OMOP table schemas directly with the FHIR core resources.
For the first approach, we extracted all table names and their textual descriptions from the OMOP CDM documentation website(5). We designed a mapping application using an Excel spreadsheet that allows curators to assign a FHIR W5 resource category to each OMOP table name. As a pilot study, we asked the project team members to use the mapping application to assign the FHIR W5 resource category independently. The purposes of the pilot study are two-fold: 1) to examine the inter-rater agreement for categorizing the OMOP table schemas; and 2) to examine the domain coverage of the OMOP CDM by comparing with the FHIR core resources. Figure 3 shows the spreadsheet-based mapping For the second approach, we extracted the OMOP table names and their descriptions under the category of “standardized clinical data”(5) and the FHIR core resource names and their definitions from the DSTU2 version. We also designed the Excel spreadsheet-based mapping application. We used four SKOS mapping properties (exactMatch, closeMatch, broadMatch, and narrowMatch) as the mapping types. As a pilot study, we asked each project team member to use the mapping application to map the OMOP table schemas under the category of “standardized clinical data” to the FHIR core resources individually. The purpose of the pilot study is to examine the inter-rater agreement for creating the mappings.
Figure 3.
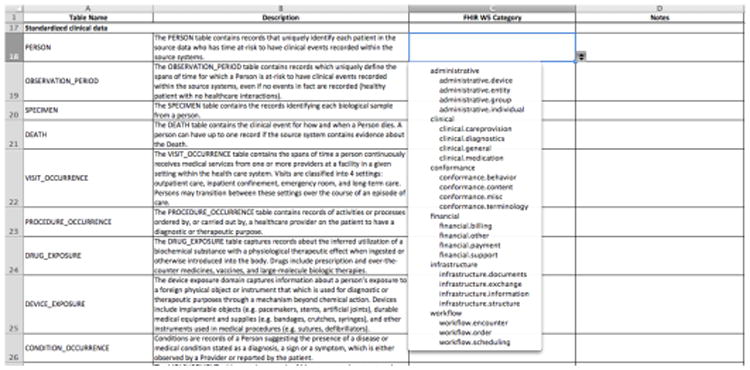
A screenshot of the spreadsheet-based mapping application for the FHIR W5 resource category.
Measure the consensus in the property-level harmonization
In the property level, we extracted the field names, data types and descriptions of a particular OMOP table from the OMOP CDM documentation website(5). Similarly, we designed a mapping application using an Excel spreadsheet (Figure 4) that allows curators to assign a property category to a particular OMOP table field. As a pilot study, we asked project team members to assign a FHIR W5 property category to the field names from two OMOP tables: Observation and Drug_Exposure. The purpose of this pilot study is to assess the utility of the FHIR W5 property category system in achieving consensus among curators and in serving as a proxy for aligning the table field names with the FHIR resource properties.
Figure 4.
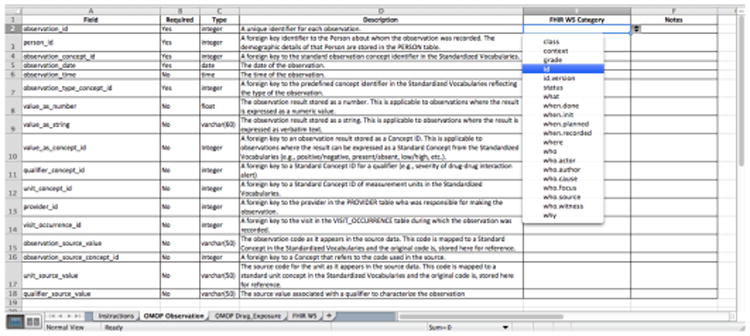
A screenshot of the spreadsheet-based mapping application for the FHIR W5 property category.
Inter-rater agreement analysis
As described previously, we asked project team members to complete the mappings independently. Fleiss' kappa statistics was calculated to assess inter-rater agreement(21). The measure calculates the degree of agreement in classification over that which would be expected by chance. The Kappa statistics value of 1 (k = 1) reflects complete agreement among raters and, a value of zero or less (k ≤ 0) shows no agreement.
Results
Model-level harmonization: Inter-rater agreement analysis for the FHIR W5 Resource Classification System
Five project team members were asked to complete the mapping spreadsheets and four valid responses were received. The average time used for the FHIR W5 Resource Category approach was 26 minutes (range from 24-30 minutes) whereas the average time used for the direct mapping approach was 29.25 minutes (range 17-35 minutes).
The overall Fleiss' kappa statistics for categorizing the OMOP CDM tables with the FHIR W5 Resource Classification System was calculated as 0.50 whereas the overall Fleiss' kappa statistics for directly mapping the OMOP CDM tables with the FHIR core resources was calculated as 0.48. The results indicated that for both harmonization approaches, the team achieved moderate agreement.
Model-level harmonization: Domain coverage between OMOP CDM and FHIR
After we calculated the Fleiss's kappa statistics, we had a team discussion to resolve the disagreement and achieved the consensus. Table 1 shows the distribution of the FHIR core resources and OMOP CDM tables by the FHIR W5 resource category.
The results indicated that the OMOP CDM covers the domains in all 6 categories as defined by FHIR W5. The distribution of the OMOP CDM tables in the category of clinical (33%), financial (13%) and administrative (18%) is comparable with FHIR. The percentage of the OMOP CDM tables in the category of infrastructure (8%) and workflow (5%) is lower than that in FHIR whereas the percentage of the OMOP CDM tables in the category of conformance (23%) is higher than that in FHIR. Interestingly, the OMOP CDM tables in the category conformance are all classified in the sub-category conformance.terminology, highlighting the OMOP CDM design principles emphasizing the use of standardized vocabularies.
Property-level harmonization: Inter-rater agreement analysis for the FHIR W5 Property Classification System
Similarly, five project team members were asked to complete the mapping spreadsheets and four valid responses were received. The average time used for the OMOP Observation table was 15.75 minutes (range 8-30 minutes) and the average time used for the OMOP Drug_Exposure was 16.75 minutes (range from 7-30 minutes).
The overall Fleiss' kappa statistics for categorizing the fields in the OMOP CDM tables “Observation” and “Drug_Exposure” with the FHIR W5 Property Classification System were both calculated as 0.21. The results indicated that for the property categorization approach, the team only achieved fair agreement. One of the main reasons is because that the FHIR W5 Property Classification System is new to most of the team members and only a limited documentation is available. We anticipate that more training orientation to curators would help improve the inter-rater agreement.
Property-level harmonization: Alignment between the FHIR resource elements and the OMOP table fields
We aligned the fields of the two OMOP tables Observation and Drug_Exposure with the elements from the FHIR resource Observation and DrugAdministration (based on the FHIR W5 Report(16)1 based on the FHIR W5 Property Classification System. The alignment results as shown in Table 2 indicated that a number of property categories including class, context, identifier, when.done, where, who.actor, who.focus are aligned reasonably well. A list of fields in both OMOP tables were categorized in the property category “what”, indicating this “what” category may need to be refined to have more specific subcategories.
Discussion
The use of data integration standards plays a critical role in the increasing adoption of the OHDSI CDM-based data repositories for clinical observational studies(22). While the OHDSI Vocabulary CDM(23) and its vocabulary services(24) have provided a solid foundation for enabling semantic interoperability across different clinical and research systems, the heterogenous data model use in these systems remains a major barrier for data integration and sharing. The emerging HL7 FHIR aims to serve as a global reference standard for exchanging healthcare and EHR data, and mappings to FHIR from different data models would greatly faciliate the secondary use of EHR data for clincial and translation research including the observational studies. Therefore, a number of independent efforts in harmonizing between FHIR and OHDSI CDM are currently underway. For example, Choi and Duke's team has developed a preliminary prototype known as the OHDSI on FHIR platform with OHDSI CDM mappings to FHIR resources(25). Actually, the FHIR Infrastructure Work Group is developing a Data Access Framework (DAF) FHIR Implementation Guide which includes the guidance on creating profiles and data element mappings between FHIR and OMOP CDM (mainly in the model level)(26). However, consensus building is a critical yet challenging factor in harmonizing and standardizing the mappings between different data models. Our consensus-based approach using the FHIR W5 category system is a preliminary effort in helping achieve the community-based agreement in a consistent and measurable manner.
Conclusions
In this study, we designed and assessed a consensus-based approach for harmonizing the OMOP CDM with HL7 FHIR. We leveraged a FHIR W5 (Who, What, When, Where, and Why) Classification System for designing the harmonization approaches and assessed their utility in achieving the consensus among curators using a standard inter-rater agreement measure. We demonstrated that the FHIR W5 classification system is a useful and promising tool for designing the harmonization approach between a data model and FHIR, and for facilitating the consensus achievement among curators. Future work includes: 1) building and refining the FHIR W5 Ontology using a community-based approach; 2) mapping the FHIR W5 ontology with other real-world data models (e.g., PCORnet CDM, i2b2); and 3) enhancing the mapping applications for effectively facilitating the alignment between various data models and FHIR.
Acknowledgments
This study is supported in part by NIH grants U01 HG009450, U01 CA180940, and R01 GM105688.
Footnotes
Note that not all properties of the FHIR resources are categorized in the FHIR W5 Report.
References
- 1.Huser V, Cimino JJ. Desiderata for healthcare integrated data repositories based on architectural comparison of three public repositories. AMIA Annual Symposium proceedings / AMIA Symposium AMIA Symposium. 2013;2013:648–56. Epub 2014/02/20. [PMC free article] [PubMed] [Google Scholar]
- 2.Wade TD, Zelarney PT, Hum RC, McGee S, Batson DH. Using patient lists to add value to integrated data repositories. Journal of biomedical informatics. 2014;52:72–7. doi: 10.1016/j.jbi.2014.02.010. Epub 2014/02/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacKenzie SL, Wyatt MC, Schuff R, Tenenbaum JD, Anderson N. Practices and perspectives on building integrated data repositories: results from a 2010 CTSA survey. Journal of the American Medical Informatics Association : JAMIA. 2012;19(e1):e119–24. doi: 10.1136/amiajnl-2011-000508. Epub 2012/03/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.The OMOP Common Data Model 2015. 2015 Sep 19; Available from: http://omop.org/CDM.
- 5.OMOP Common Data Model V5.0.1 2016. 2016 Sep 19; Available from: http://www.ohdsi.org/web/wiki/doku.php?id=documentation:cdm:single-page.
- 6.The PCORnet Common Data Model 2015. 2015 Sep 19; Available from: http://www.pcornet.org/pcornet-common-data-model/
- 7.i2b2: Informatics for Integrating Biology and the Bedside 2015. 2015 Apr 16; Available from: https://www.i2b2.org/
- 8.Klann JG, Murphy SN. Computing health quality measures using Informatics for Integrating Biology and the Bedside. Journal of medical Internet research. 2013;15(4):e75. doi: 10.2196/jmir.2493. Epub 2013/04/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klann JG, Buck MD, Brown J, Hadley M, Elmore R, Weber GM, Murphy SN. Query Health: standards-based, cross-platform population health surveillance. Journal of the American Medical Informatics Association : JAMIA. 2014;21(4):650–6. doi: 10.1136/amiajnl-2014-002707. Epub 2014/04/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weber GM, Murphy SN, McMurry AJ, Macfadden D, Nigrin DJ, Churchill S, Kohane IS. The Shared Health Research Information Network (SHRINE): a prototype federated query tool for clinical data repositories. Journal of the American Medical Informatics Association : JAMIA. 2009;16(5):624–30. doi: 10.1197/jamia.M3191. Epub 2009/07/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daugherty SE, Wahba S, Fleurence R. Patient-powered research networks: building capacity for conducting patient-centered clinical outcomes research. Journal of the American Medical Informatics Association : JAMIA. 2014;21(4):583–6. doi: 10.1136/amiajnl-2014-002758. Epub 2014/05/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Athey BD, Braxenthaler M, Haas M, Guo Y. tranSMART: An Open Source and Community-Driven Informatics and Data Sharing Platform for Clinical and Translational Research. AMIA Joint Summits on Translational Science proceedings AMIA Summit on Translational Science. 2013;2013:6–8. Epub 2013/12/05. [PMC free article] [PubMed] [Google Scholar]
- 13.Scheufele E, Aronzon D, Coopersmith R, McDuffie MT, Kapoor M, Uhrich CA, Avitabile JE, Liu J, Housman D, Palchuk MB. tranSMART: An Open Source Knowledge Management and High Content Data Analytics Platform. AMIA Joint Summits on Translational Science proceedings AMIA Summit on Translational Science. 2014;2014:96–101. Epub 2015/02/27. [PMC free article] [PubMed] [Google Scholar]
- 14.Novak AJ, Asmann YW, Maurer MJ, Wang C, Slager SL, Hodge LS, Manske M, Price-Troska T, Yang ZZ, Zimmermann MT, Nowakowski GS, Ansell SM, Witzig TE, McPhail E, Ketterling R, Feldman AL, Dogan A, Link BK, Habermann TM, Cerhan JR. Whole-exome analysis reveals novel somatic genomic alterations associated with outcome in immunochemotherapy-treated diffuse large B-cell lymphoma. Blood Cancer J. 2015;5:e346. doi: 10.1038/bcj.2015.69. Epub 2015/09/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang W, Wang C, Dawson DB, Thorland EC, Lundquist PA, Eckloff BW, Wu Y, Baheti S, Evans JM, Scherer SS, Dyck PJ, Klein CJ. Target-enrichment sequencing and copy number evaluation in inherited polyneuropathy. Neurology. 2016;86(19):1762–71. doi: 10.1212/WNL.0000000000002659. Epub 2016/05/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.FHIR W5 Report 2016. 2016 Sep 19; Available from: https://www.hl7.org/fhir/w5.html.
- 17.FHIR Record Lifecycle Events Implementation Guide 2016. 2016 Sep 19; Available from: http://hl7.org/fhir/2016Sep/ehrsrle/ehrsrle.html.
- 18.Coyle JF, Mori AR, Huff SM. Standards for detailed clinical models as the basis for medical data exchange and decision support. International journal of medical informatics. 2003;69(2-3):157–74. doi: 10.1016/s1386-5056(02)00103-x. Epub 2003/06/18. [DOI] [PubMed] [Google Scholar]
- 19.Jiang G, Evans J, Oniki TA, Coyle JF, Bain L, Huff SM, Kush RD, Chute CG. Harmonization of detailed clinical models with clinical study data standards. Methods of information in medicine. 2015;54(1):65–74. doi: 10.3414/ME13-02-0019. Epub 2014/11/27. [DOI] [PubMed] [Google Scholar]
- 20.Gamazon ER, Stranger BE. The impact of human copy number variation on gene expression. Brief Funct Genomics. 2015;14(5):352–7. doi: 10.1093/bfgp/elv017. Epub 2015/04/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fleiss' kappa 2016. 2016 Sep 22; Available from: https://en.wikipedia.org/wiki/Fleiss'_kappa.
- 22.Hripcsak G, Ryan PB, Duke JD, Shah NH, Park RW, Huser V, Suchard MA, Schuemie MJ, DeFalco FJ, Perotte A, Banda JM, Reich CG, Schilling LM, Matheny ME, Meeker D, Pratt N, Madigan D. Characterizing treatment pathways at scale using the OHDSI network. Proc Natl Acad Sci U S A. 2016;113(27):7329–36. doi: 10.1073/pnas.1510502113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.OHDSI Vocabulary CDM 2016. 2016 Dec 15; Available from: http://www.ohdsi.org/web/wiki/doku.php?id=documentation:cdm:standardized_vocabularies.
- 24.OHDSI Vocabulary Services 2016. 2016 Dec 15; Available from: http://athena.ohdsi.org/
- 25.Choi M, Starr R, Braunstein M, Duke JD. OHDSI on FHIR platform development with OMOP CDM mapping to FHIR resources 2016. 2016 Dec 15; Available from: http://www.ohdsi.org/web/wiki/lib/exe/fetch.php?media=resources:ohdsionfhir_gatech.pdf.
- 26.DAF-Research Profile List and Mappings from FHIR to PCORnet CDM and OMOP CDM 2017. 2017 Apr 6; Available from: http://hl7.org/fhir/us/daf-research/2017Jan/daf-research-profile.html.


