Abstract
The pace of discovery in eScience is increasingly dependent on a scientist’s ability to acquire, curate, integrate, analyze, and share large and diverse collections of data. It is all too common for investigators to spend inordinate amounts of time developing ad hoc procedures to manage their data. In previous work, we presented Deriva, a Scientific Asset Management System, designed to accelerate data driven discovery. In this paper, we report on the use of Deriva in a number of substantial and diverse eScience applications. We describe the lessons we have learned, both from the perspective of the Deriva technology, as well as the ability and willingness of scientists to incorporate Scientific Asset Management into their daily workflows.
I. Introduction
While much attention has been given to the process of publication, citation, and access of curated scientific data in shared data repositories [1], little attention has been paid to the data management needs that show up in daily practice of data-rich scientific collaborations that lead to sharable data. In current practice, scientists organize and share data via ad hoc directory structures, coding critical metadata via a combination of file naming conventions, text files, or spreadsheets. As a result, data generated during the course of an investigation may be lost, mislabeled, regenerated or misused because it cannot be found or correctly identified. Frequently data are generated by scientists who are not experts in information systems, and they are overwhelmed by the complexity of data management tasks. Taken together, the lack of robust tools and mechanisms for data management contribute to significant investigator overheads [2] and unreproducible data [3].
Efficient creation of high-quality, reusable and sharable data would clearly benefit from tools and processes that reduce the complexity and overhead of collecting, organizing and preparing data for data-driven scientific investigations. We have argued that a data management approach based on scientific digital asset management [4] can significantly streamline the process of managing complex and evolving data collections, and in doing so, accelerate “knowledge turns [5]” for scientific discovery. Digital asset management systems (DAMS) enable the management tasks and decisions surrounding the “ingestion, annotation, cataloging, storage, retrieval and distribution of digital assets [6].” Our hypothesis is that a DAMS platform tailored for eScience could have a similar impact as commercial DAMS products have had in professional media and creative industries — transforming how scientists interact with their data on a daily basis, making them more efficient and effective.
To test this hypothesis, we have developed Discovery Environment for Relational Information and Versioned Assets (Deriva), a platform which provides an end-to-end ecosystem for managing scientific data from acquisition through analysis and publication. Using Deriva, we have conducted a multi-year study on the impact of these techniques within the context of diverse data-driven scientific collaborations at scales spanning from small basic research investigations to international data sharing consortia. To evaluate the effectiveness of the scientific asset management based approach, we examine its use in detail in two representative use cases.
This paper makes the following contributions. We:
summarize the features of Deriva, a platform designed to increase the productivity of data driven scientific discovery via digital asset management concepts,
show how a single platform for asset management can be configured and applied to distinct scientific discovery domains, and
describe how daily workflows of diverse domain scientists have been improved by incorporation of scientific asset management systems.
The rest of this paper is organized as follows. In Section II we discuss key requirements for scientific asset management. In Section III, we describe Deriva, our platform for scientific asset management. In Section IV, we discuss Deriva and the FAIR guidelines. In Section V, we describe two Deriva use cases in detail. We then summarize key lessons learned from our experiences in Section VI. We review related work in Section VII and conclusions in Section VIII.
II. Scientific Asset Management Requirements
Based on an analysis of a broad set of potential use cases, we propose that an eScience DAMS ecosystem should provide the following capabilities:
Acquisition of diverse scientific assets
Data and metadata may be sourced from legacy data sets, outputs of computations, specialized instruments and instrument control software, existing databases and lab information management systems, or sources like spreadsheets, text files, and manual data entry.
Model-driven organization and discovery of assets
A scientific DAMS must support diverse models that evolve over time. DAMS systems in the consumer space provide users with intuitive and interactive ways of organizing and discovering assets structured by an underlying model. Unlike music, for example, where there is a well understood domain model (e.g. albums, artists, composers, tracks), the models for scientific investigations may vary radically from instance to instance and change over time as the discovery process unfolds.
Storage and retrieval of eScience data assets
Assets may be very large, and may be physically distributed in local, enterprise, and cloud based storage systems.
Rights management and access control
Scientific data sharing may involve data use agreements, access to proprietary data, time driven data embargoes, and different user roles within and across collaborations. Access control and associated policy may be required at levels of granularity that go from an entire data set, to a single data element, both for data assets and metadata.
Integration into analytics ecosystem
Data driven discovery is the result of repeated knowledge turns, which consume existing data and produce derived data and/or metadata which may be ingested as new assets. Users must be able to identify the data that is required for a knowledge turn, assemble and export relevant data for consumption by diverse computational tools and re-ingest the results.
III. The Deriva Platform
To address the requirements detailed above, we have created a collaborative scientific asset management platform called Deriva. The platform is designed to support collaboration through the full life-cycle of scientific data including initial experiment design; prototype and production data acquisition; ad hoc and routine analyses; and publication.
Core principles underlying the design of Deriva are:
loosely coupled web services architecture with well defined public interfaces for every component,
use of Entity Relationship Models (ERM) that leverage standardized vocabularies, with adaptive components that can automatically respond to evolving ER models,
model-driven user interfaces to enable navigation and discovery as the data model evolves,
data-oriented protocols where distributed components coordinate complex activities via data state changes.
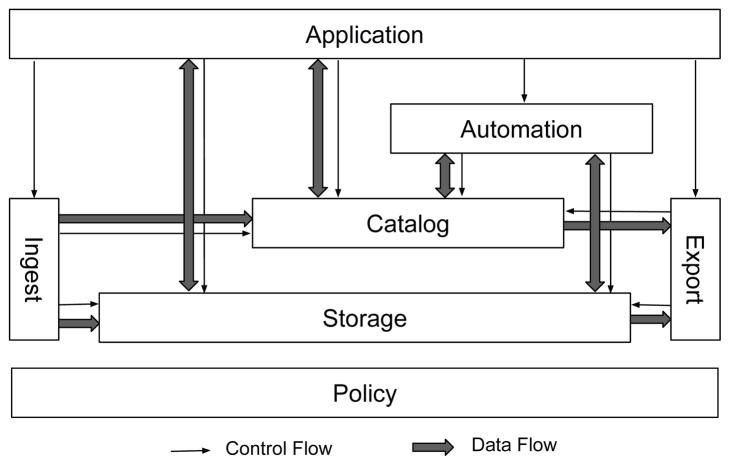
Deriva uses an entity-relationship data model to catalog and organize assets which may be digital objects (i.e. files) or references to physical objects, such as proteins or mice. Assets are characterized by contextualizing metadata which places an asset into the model by relating it to a specific entity. Additional descriptive metadata are used to describe additional attributes and relationships between assets. Figure 1 illustrates the Deriva architecture. We summarize each component below. More details are provided in [4].
Fig. 1.
Deriva architecture consisting of metadata catalog (ERMrest), object storage (Hatrac), web applications (Chaise), ingest/export and automation agents (IObox), and policy enforcement and authentication.
A. ERMrest: Model-Neutral Relational Metadata Store
Entity Relationship Model via Representational State Transfer (ERMrest) provides a relational metadata store as a web service, allowing general entity relationship modeling and manipulation of data resources via web protocols. ERMrest enables the evolving and dynamic data models needed for describing, contextualizing, and linking scientific assets. ERMrest is designed to organize and contextualize large-scale data assets that are stored in the Hatrac object store or other cloud based storage systems (e.g., Amazon S3).
ERMrest is a multi-tenant system where catalogs may be provisioned for separate tenants, each with its own access-control lists (ACLs), ERM, and content following that model. Unlike conventional data management solutions which are designed or deployed with a priori knowledge of the data model, ERMrest continuously adapts, providing service interfaces to initiate and immediately reflect ERM changes.
To enable concurrent activities and controlled collaboration, ERMrest enforces fine-grained access and update policy on models and content. Cloud-based authentication and group management is used to assign users to roles [7]. Different policies for model visibility, query, insert, update and delete can be set on an entity type (table) or on specific attributes (columns). Dynamic policies can also consult data values, further differentiating access decisions for individual entities (rows) or entity classes (rows sharing a common coded value). Since dynamic policies consult data, sufficiently trusted users can alter policy by editing the associated data values, allowing for complex social scenarios such as rights delegation.
B. Hatrac: Version-Tracking Object Store
While assets may be stored in file systems or general cloud storage, scientific data benefits from additional assurances such as data immutability and integrity. Hatrac is a specialized object store that provides a service interface to create and access immutable objects. Objects are organized into hierarchical namespaces and access can be granted to entire namespaces or to individual objects. End-to-end checksumming in the protocol ensures integrity. Hatrac can be deployed on a conventional Linux server or cloud hosted as an interface to Amazon S3 or compatible object stores.
C. Chaise: Model-Driven Web Applications
Chaise is a suite of dynamic, model-driven, Web applications for searching, browsing, importing, editing, and exporting digital assets and metadata. Chaise introspects the current ERM stored in an ERMrest catalog and dynamically generates complex user interface applications for various modes of interaction. Current Chaise applications are:
RecordSet generates a faceted search interface, enabling users to find entities via criteria such as attribute values or relationships;
Record generates a detailed presentation of the selected entity in the model along with data connected to the entity via its relationships in the model; and
RecordEdit generates a multi-record entry and edit application for creating and curating metadata records.
Relationships in the data model can be traversed as navigable links, enabling the user to interactively explore the data, for example, going from a biosample to individual observations that have been made on the biosample. Chaise has integrated support for vocabulary terms, promoting reuse of existing terms or controlled vocabulary, while providing a means for dynamically adding new terms as needed.
Chaise is highly extensible via its integrated Markdown renderer and Mustache [8] template engine with built-in plugins we developed for including iframe elements. Through this feature, Chaise applications can integrate virtually any standard Web components to customize a Deriva deployment to its use case requirements. For example, we routinely add data visualizations such as scatter plots, histograms, as well as specialized data viewers such as volume renderers.
D. IObox: Data-Driven Workflow Agent
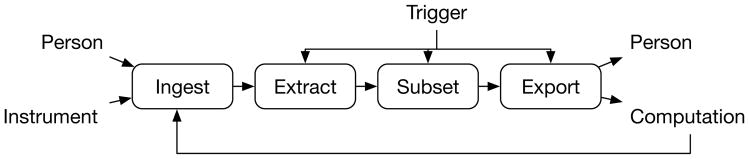
Deriva automates data-driven workflows via its IObox framework as depicted in Figure 2. It supports three different usage scenarios for ingest and extract operations. 1) Data are generated by a scientific instrument such as a microscope or sequencer. An agent monitors a directory for incoming files and automatically uploads them to Deriva, much like new photos are imported into a photo album from a smartphone without user intervention. 2) Data are generated by an external data system such as a laboratory information system. An agent extracts, transforms and loads (ETL) relational data to Deriva based on a predefined configuration, e.g. nightly or when data entries are of a certain status. 3) Data are generated by arbitrary means and are located on a file system. For bulk assets, e.g. a batch of files, an interactive graphical client can be used to examine the assets, extract necessary metadata and ingest the assets, notifying the user immediately if it fails. For infrequently generated assets, users use the Chaise RecordEdit tool to identify where in the data model the assets should be tracked, manually input appropriate metadata, and upload the files.
Fig. 2.
IObox enabled workflow depiction. Boxes indicate key operations performed by IObox while interactions and data flow are indicated by arrows.
IObox provides two general mechanisms for interfacing with external computational agents via its subset and export operations. 1) Condition-Action (CA) processing asynchronously initiates agent execution based on the catalog state. Agents may be co-located with ERMrest or distributed to remote servers or the cloud. And 2) BDBag [9] asset export allows the user to bundle data collections for use in other analysis tools, such as Python, R, or platforms such as Galaxy.
A key aspect of these tools is that they “contextualize” the asset within the research protocol that produced it. Our general strategy is to link and annotate an asset as accurately as possible when it is initially acquired. For example, a BAM file containing RNA-Seq data should be associated with the specific replicate, sample, and experiment. The general steps are to introspect the data model, and then infer additional details from the asset contents such as its file name, who/what produced the file, a barcoded label, and/or embedded file metadata to properly contextualize the asset. Finally, to ensure immutability, the asset itself is typically copied into the Hatrac object store. Given the model agnostic approach of Deriva, the IObox framework is readily adapted and extended through configuration parameters or Python scripting.
IV. Deriva and “FAIR” Guidelines
The FAIR Data Principles are guidelines for scientific data designed to promote data reuse and to enhance “the ability of machines to automatically find and use the data, in addition to supporting its reuse by individuals.” [10]. These principles state that data should be Findable, Accessible, Interoperable, and Reusable. However, FAIR is not only desirable for published repository data, but for any data produced in an eScience investigation, at any point in time, regardless of the scope over which the data are shared or reused. Discovery will be accelerated, if data are FAIR at each step of a data driven eScience process. With this in mind, a goal of Deriva is to provide a foundation on which each data generation stage of a discovery process produces FAIR data.
Instead of storing assets as unstructured data, each community defines their digital assets, their associated properties and relationships (i.e. metadata) through an ER Model (‘F’, ‘R’) leveraging controlled vocabularies. All assets and key metadata entries are assigned unique accession numbers, are referenced by globally unique resource names and are explicitly linked (‘F’, ‘R’). Deriva exposes the ERM, provides web APIs and interfaces for users and user agents to accurately find those assets using faceted and basic text search (‘F’, ‘A’). All assets and data are accessible, subject to the community access control policy (‘A’). The metadata can be exported in standard CSV and BagIt [9] formats, while the digital assets are submitted and made available in standard formats defined by the community (‘I’, ‘R’).
V. Use Cases
Deriva has been used in scientific collaborations spanning from small teams engaged in data collection and analysis for basic science, to large, multi-national consortium producing data repositories. Table I provides some basic statistics across a number of of these deployments. Shown are the number of different types of key entities currently being used to represent the domain model, the number of records corresponding to those entities, the number of different asset types, and the total number and size of the assets currently being managed by the system.
TABLE I.
Summary of representative Deriva deployments.
| Name | Key Entities | Assets | |||
|---|---|---|---|---|---|
| Types | Count | Types | Count | TiB | |
| FaceBase | 6 | 6,598 | 13 | 2,773 | 2.6 |
| RBK | 10 | 501 | 4 | 763 | 0.58 |
| GUDMAP | 4 | 23,186 | 4 | 35,334 | 0.6 |
| GPCR | 7 | 180,481 | 17 | 232,078 | 0.06 |
| Synapse | 4 | 1,675 | 9 | 1,906 | 1.7 |
| CIRM | 4 | 28,740 | 1 | 5,429 | 17 |
In this section, we explore in more detail how Deriva has been used in practice in two specific use cases: FaceBase and GPCR. The FaceBase consortium represents a large-scale deployment involving a multi-site collaboration and community curation and data sharing endeavor. The GPCR represents complex, distributed collaboration and early-phase exploratory research. RBK and GUDMAP are projects related to molecular anatomy and are similar to FaceBase, while Synapse and CIRM are basic science projects similar in nature to GPCR.
A. FaceBase
1) Overview
FaceBase (www.facebase.org) is a craniofacial research consortium that produces and shares comprehensive craniofacial data and resources for the international research community. The consortium is organized around a “hub and spoke” model where 20 spokes create and contribute data while the Hub is responsible for integrating this data into a curated collection to be used by the broader craniofacial research community. Data in FaceBase are diverse as the spokes engage in a wide range of investigations including imaging, confocal microscopy, laser capture microdissection, DNA microarray, high-throughput sequencing, and enhancer reporter studies involving mouse, zebrafish, and human subjects. Spokes are expected to release data immediately to the public upon their production without embargoing data for their own research. As such, timely release of data is among the critical goals of FaceBase.
2) Challenges
Prior to adopting Deriva, FaceBase took a manual, centralized approach to curation using Drupal as a Content Management System (CMS) for storing and displaying metadata coupled with Apache Lucene for keyword search of metadata. Data were submitted to the hub zipped in bundles that typically contained 5–20 individual files each, which were stored on and served from a standard Web server. Hub curators entered free form text descriptions in the CMS with a set of “tags” and a link to the zip file. Tags covered information about the species, age stage, anatomy, phenotype, and other descriptive properties about the data set. Users of the FaceBase site could perform keyword searches over the content and some limited filtering on tags.
The initial approach to curation and publication of data suffered significant limitations. The key problems and user complaints included:
limited resources for centralized curation at the Hub resulted in long delays before data set release and the curation process was susceptible to transcription errors leading to additional delays;
lack of a detailed data model for describing data sets led to inconsistencies in the quality and level of detail for data set descriptions between different data producers, for example, the relationship between files and the bioassays that produced them or the biosamples to which they related could only be inferred from “meaningful” filenames with gene names, age stages, and other critical details embedded in them;
difficulty finding and narrowing search results due to: keyword search could return many “hits” but not allow users to narrow through precise filtering; hand coded lists of dataset properties that looked like but were not structured metadata tags in the CMS; inconsistent, nonstandard, or misspelled terms.
FaceBase was restructured to use a scientific asset management based approach to achieve the following goals:
streamline and accelerate the curation pipeline so that data sets should be made available almost as fast as they are produced;
simplify the sometimes cumbersome interactions between submitters and curators that often plagues large repositories;
reduce the effort and resource load on the Hub by distributing the curation responsibilities among the participating spoke projects rather than assuming all curation tasks at the Hub; and
increase the ability of users to find data of interest via intuitive data models coupled with faceted search and linked data navigation.
3) Implementation
The application of Deriva to Face-Base was complicated by the need to transition the data and processes from the existing Drupal based approach to ours. We took a staged approach:
An initial data model that mirrored the original data representation was created and a one off ETL (extract, transform, and load) process was developed to move all of the existing data from the CMS to an ERMrest catalog and Hatrac object store. Data was left in its initial format as a set of zip files.
An ensuing clean up of terms was performed as aided by Chaise to display alphabetized term lists directly from the catalog which revealed inconsistencies (e.g., “Msx1” vs “MSX1”).
A new more detailed data model was developed that was more representative of the structure of the actual and new data being provided.
As the transition was taking place, spokes used spreadsheet templates to describe metadata while the Hub took responsibility for updating the catalog using IObox.
A curation protocol has been established to streamline the process, and going forward the spokes will use IObox and Chaise to directly upload and curate newly contributed data following the protocol.
Schema evolution
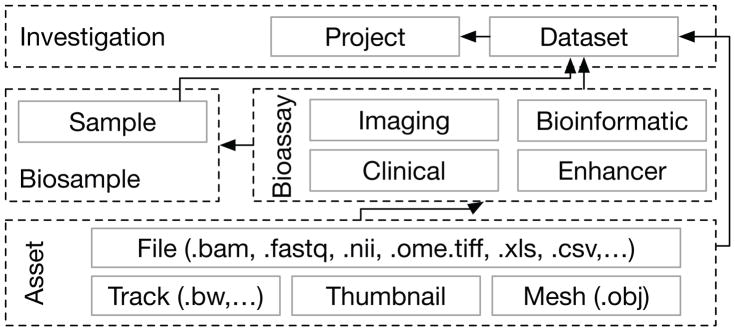
The Hub evolved the initial database schema to a structure that could represent the detailed information about the experiments, assays, samples, subjects and assets submitted by the spokes. The new model was informed by established conventions found in “Chado” [11] and “ISA” (Investigation Study Assay) [12] while accommodating the constraints imposed by the legacy database. The conceptual data model for FaceBase is depicted in Figure 3.
Fig. 3.
FaceBase ERM. Metadata are organized broadly as investigation, biosample, bioassay, and asset entities with relationships indicated by arrows.
Curation pipeline
To release data to the broader community as rapidly as possible, FaceBase has implemented a three stage pipeline, where datasets are either pending, released, or curated. Fine grain access control in ERMrest is used to control visibility, allowing spokes to upload data and edit metadata until data quality standards are met at which point the data or metadata are made visible and publicly available to users. Curation stages are explicitly represented in the data model.
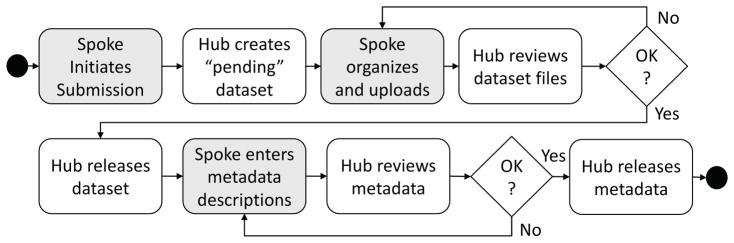
Dataset accessioning
The curation pipeline (see Figure 4) begins with investigators notifying the Hub that they wish to submit data. The spoke’s request must include basic metadata which cover essential details, similar to what may be found in Dublin Core or other data publication standards. The Hub then mints an accession number and creates a Dataset entity in the database which begins in a “pending” state. This pending dataset record serves the dual purpose of anchoring the curation process and all communication concerning it, and establishing a “landing page” for the dataset.
Fig. 4.
FaceBase Data Curation Pipeline. Shaded boxes indicate Spoke responsibilities versus clear boxes for the Hub’s activities.
Data acquisition
In the pending stage, data sets are submitted by the spokes using an IObox batch uploader or by transferring files to a hosted IObox operated by the Hub. IObox automatically extracts key information, such as file names and checksums, and registers the data files in the relevant fields in the data model. Thumbnails and 3D mesh files are stored in separate Hatrac namespaces with relaxed access control policies compared to general assets which FaceBase restricts to logged in end-users. In addition, genome tracks are stored under a namespace that serves as a virtual “trackhub” for the UCSC Genome Browser [13] and an embedded jBrowse Genome Browser [14]. Once ingested, the Hub reviews the data. If the consortium’s data quality standards are met, the Hub updates the data set’s status to “released” and the assets are made visible to the end-users.
Metadata curation
Metadata may be entered using Chaise or submitted in spreadsheets to the Hub for processing via the hosted IObox at any time in the process. In the interest of streamlining the release of data to the public, the process focuses on curating metadata after the data are released. The Hub currently uses Chaise for curation of the catalog and has begun training the spokes in its use so that they may directly enter metadata, with access control policies in ERMrest to control editing and visibility permissions. Once metadata are entered in the database, Hub staff review the entries to ensure that data quality standards have been satisfied and then upgrade the dataset to “curated” status. At this point, the complete dataset, its data and metadata, are fully available to the public.
4) Results
The implementation of Deriva has significantly impacted every aspect of how FaceBase curates and publishes data sets.
Structured, detailed data models now allow every individual asset to be represented in the database along with critical information about biosamples and bioassays;
Alignment with community ontologies (e.g., OBI, MP, HPO, ZFA, Theiler stages, etc.) and integration with the Monarch Initiative’s Phenogrid [15] have transformed FaceBase from a data silo to an interoperable data resource where data sets can be integrated with data produced outside of the consortium;
Powerful, dynamically generated search and browse interfaces now allow users to find and narrow searches using any attribute of any key entity in the database and then to navigate in a “linked data” style through the entire web of information in the database.
Quantifiable metrics indicate significant progress toward FaceBase goals:
FaceBase spokes have been able to publish data with detailed descriptions on 1,155 biosamples and 4,736 bioassays, up from essentially 0 using the prior approach;
FaceBase now measures its data curation cycle time in days rather than months meaning that users get access to new data sets rapidly;
Users now visit the new FaceBase data browser more than any other resource on the site accounting for 32.5% of unique page views with an extremely low bounce rate of 22.2%;
and most critically, in the past two years alone, FaceBase usage has grown from 4,699 unique users to 7,905 unique users with a two year total of 71,076 page views and 19,371 user sessions.
B. GPCR Consortium
1) Overview
G-protein-coupled receptors (GPCRs) play a critical role in a wide variety of human physiology and pathophysiological conditions. As a drug target, GPCRs are highly valuable with an estimated 30–40% of all drugs on the market, but mechanistically poorly understood. The best method to determine three dimensional GPCR structure is via X-Ray crystallography. However, the native form of a GPCR may not form a stable crystal, so many slight mutations, called constructs, are designed and evaluated. For each construct, the protein is synthesized using bacteria, and various tests (assays) using techniques such as flow cytometry, chromatography, and gel electrophoresis images are used to measure the quality, quantity and stability of the resulting proteins. Protein crystals are evaluated using high energy light sources, and its structure determined by analyzing the diffraction patterns.
The GPCR Consortium was formed to systematically evaluate a large number of GPCR structures. The consortium consists of three academic sites and five commercial partners. Given the data diversity, the complexity of the experimental process, and the scale and distribution of the GPCR Consortium effort, the conventional approach to data management would pose a significant obstacle to the goals of the consortium and so a data management solution based on Deriva was developed for organizing all consortium experiments and data.
2) Challenges
The three academic sites, one in the US and two in China, are responsible for conducting GPCR research and generating the experiment data for the consortium. Within each academic site there are typically separate teams developing biomasses (bacteria that express the desired protein) and conducting the experiments. Each academic site maintains a local database system to keep track of construct design and biomass production conducted locally. Prior to using Deriva, all of the assays associated with the experimental process were managed in an ad hoc fashion, with each scientist copying all of her assays into an ad-hoc directory structure, recording the metadata in lab notebooks, etc. As a result, when examining previous experiments, we observed numerous instances in which a) assays for an experiment could not be located, b) experiment metadata associated with an assay could not be determined, c) construct designs were repeated, and d) experiment results were inconsistently noted. In addition, there was no mechanism to implement consortium-wide access control policies.
3) Implementation
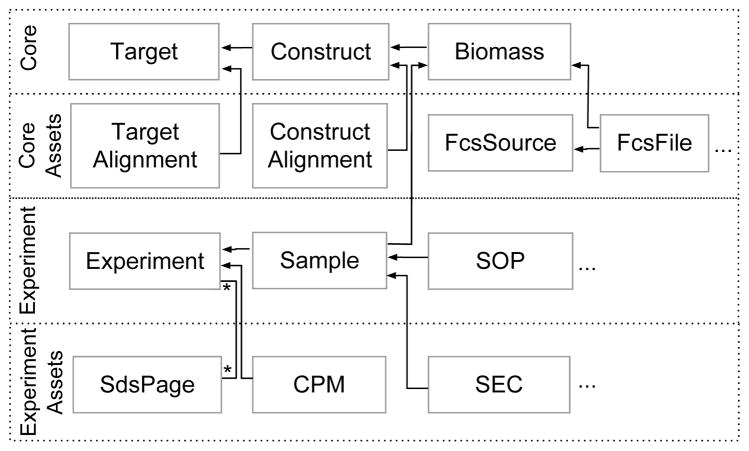
The GPCR data model was incrementally developed. Figure 5 illustrates essential parts of the catalog data model comprised of four tiers of entity types and relationships developed in phases: A core ERM captures the domain of GPCR targets, constructs, and biomasses; core asset metadata tracks alignment and flow cytometry data files; experiment metadata tracks experiment processes; and most recently, experiment asset metadata are beginning to track electrophoresis, stability, and chromatography data files. Concurrent with these major phases of model expansion, we also engaged the early users to review and refine the elements within each tier.
Fig. 5.
Select elements of GPCR catalog model. From top to bottom, four tiers of entities and relationships have been added in phases: core protein concepts; core assets including alignment and expression data; experiment metadata; and experiment assets capturing experimental results.
Construct design acquisition
The construct and biomass data are stored in the local database systems. We use IObox relational export and import agents to ingest these databases nightly to maintain a harmonized, multi-site record of core entities in the shared catalog.
Experiment design
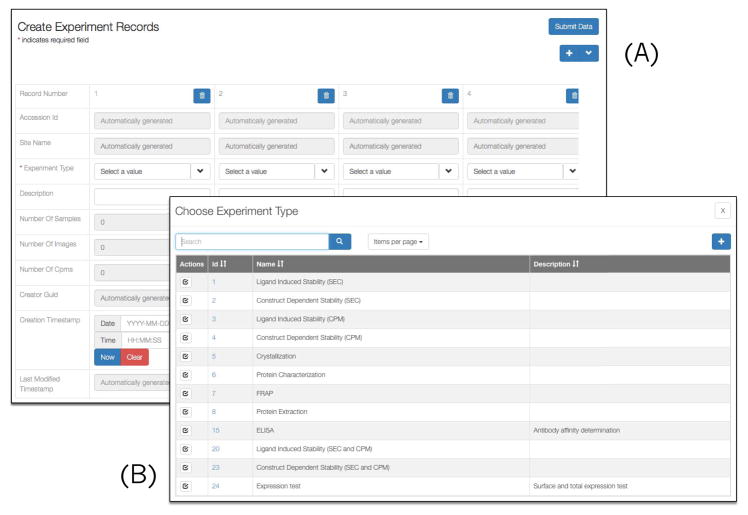
Contrary to the previous practice where the experiment details were noted in individuals’ spreadsheets, scientists were asked to use the Chaise RecordEdit tool to enter experiment related metadata such as experiment design, associated samples, purification protocols, or chemical composition (represented by the experiment entities). Figure 6 shows an example of Chaise screen-shots for creating multiple experiments simultaneously.
Fig. 6.
Use of RecordEdit application to create GPCR experiments.
Asset acquisition
IObox disk-monitoring agents are deployed at each site, for acquiring file-based experimental data such as flow cytometry ( .FCS), chromatography ( .CDF), gel images ( .JPG), and stability ( .CSV) data. The agents are configured with general ingest logic which examines file extension for asset type, and filename and directory patterns for additional metadata associated with the assets. Files are automatically added to the Hatrac object store, and metadata are added to the corresponding asset tables and cross-linked with other entities in ERMrest based on detected properties. For example, jsmith/20161013EXP1.jpg implies that the file is a gel image uploaded by ‘jsmith’ and it is linked to the experiment with ID ‘20161013EXP1’. Assets with unknown experimental context get uploaded with no experiment linkage and can be later cataloged using Chaise. For ad hoc assets such as publications or construct related documents, users can upload files along with their metadata using Chaise.
Condition-action processing
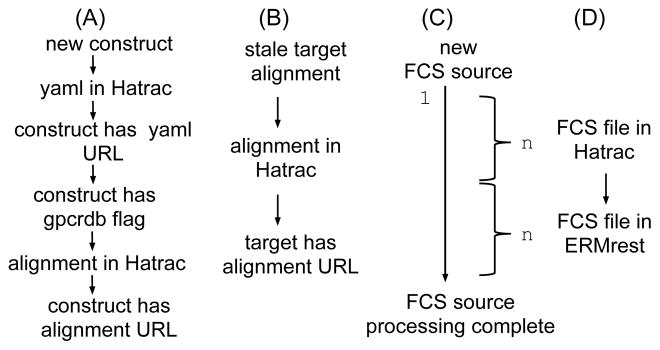
Raw assay data loaded into the system must be processed to make it useful to the scientist. Processing agents are triggered based on the existence or state of entities in the ERMrest catalog. Data are extracted from the catalog and object store using standard APIs and the results of the processing pipelines re-ingested into ERMrest and Hatrac as appropriate. Figure 7 depicts the main processing pipelines that are triggered on each type of asset. The flow cytometry (FCS) processing pipeline (Figure 7(c)) is illustrative of the process. As generated by the instrument, a FCS file contains results from many samples. The triggering condition is that the corresponding FCS source is “incomplete.” The processing pipeline expands the file into constituent single-sample FCS files, each of which is further processed and summarized, and the resulting new assets and associated entities are ingested into ERMrest and Hatrac (Figure 7(d)). The bulk action can restart multiple times, recognize already completed FCS file products, and continue working until completion.
Fig. 7.
GPCR condition-action processing pipelines. Observable data states are depicted as labeled conditions, while processing actions are implied as arrows transitioning from one state to the next: A) a new construct is aligned using a third-party service, GPCRDB [16]; B) an aggregate alignment is maintained for each target, tracking its most recent construct alignments; C) a multi-sample FCS source file is processed in bulk, generating idempotent checkpoints for D) a single-sample FCS file.
Dashboards
The GPCR system consists of diverse data types, different ingest methods, and complex processing work-flows. To help different groups of stakeholders get a quick summary of the system, we have developed the following four dashboards. 1) Target dashboard summarizing individual target mean expressions and characterization based on its corresponding biomasses. Consortium members can easily identify active targets from this list. 2) Site dashboard summarizing the number of assets across asset types generated by different academic sites. This dashboard helps the PIs monitor the progress of each site. 3) Data submission dashboard summarizing the number of experiments, samples, and different assets created by individual users, as well as the number of orphaned assets that are yet to be cross-linked with an experiment or sample. This dashboard helps the PIs, lab managers and lab members get a quick summary of recent experiment activities. 4) Processing dashboard summarizing the real-time status of all condition-action processing pipelines in different time intervals (e.g. last 24 hours, last 7 days). System operators use this dashboard to detect processing problems and take prompt interventions.
Data exploration
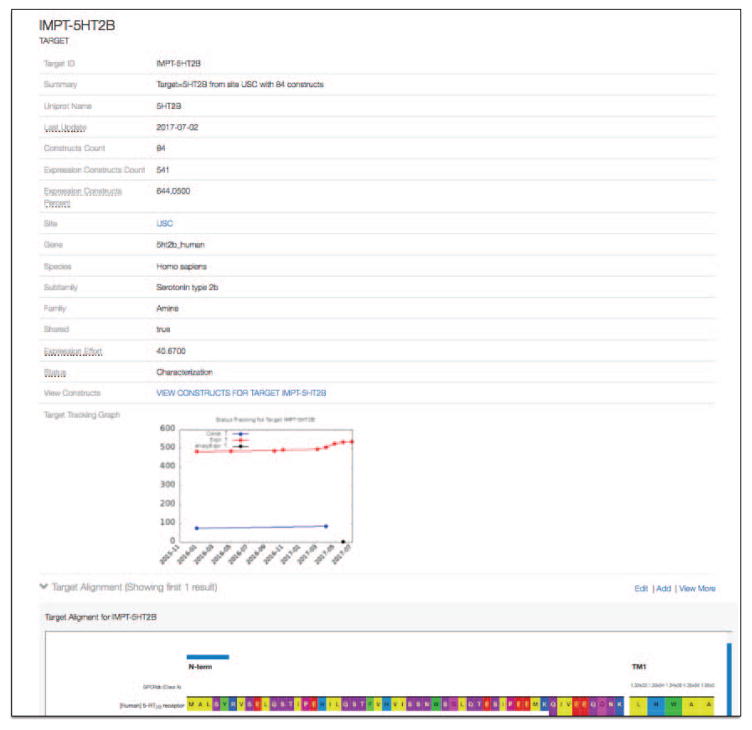
Chaise is the main user interface for exploring consortium data. We use a number of Chaise-supported “annotations” on the ER model to customize data presentation, such as embedding interactive visualization elements (e.g. FCS, CDF, and CPM plots) or thumbnail images. An overview of the different presentations generated by Chaise is shown in Figure 8.
Fig. 8.
Dynamically generated display of GPCR Target including metadata, activity tracking graph, and alignment.
Access control policy
GPCR data are subject to differentiated access controls, enforced by Hatrac and ERMrest to provide consistent policy enforcement for web browsers and any other networked clients. The academic members are able to access all data. Each site member can create experiment-related entries, but are only allowed to modify or delete entries associated within their site. Consortium members are allowed to access data associated with the consortium target list.
4) Results
The GPCR project (data.gpcrconsortium.org) has been in operation for 2 years. There are 122 users, 928 targets including non-GPCRs, 24,934 constructs, 87,422 expressions, and 97,739 FCS assets in the system. The experiment design process was deployed in March 2017. Since then there are 100 experiments, 375 samples, and 600 raw assets uploaded. We plan to expand the experiment data model tier to support the crystallization process and its assets in the future.
Prior to our involvement, industrial members had little insight on target status and had to wait for monthly updates. Now they can track progress in near real-time. Experimental results that were previously locked in scientists’ personal notebooks are now available across the consortium. Integrated acquisition, processing, and system state presentation GUIs have reduced the effort for users, increased visibility into workflow status, helped increase data quality, and freed up time for actual science. Finally, we are now in a position to start performing meta-analysis on experiments with the goal of helping guide experiment design.
The interaction between the technology platform and the daily practice of the data creators has been a critical element of GPCR. We rolled out an initial version of the experiment design model in December 2016. However, users were reluctant to enter data as it imposed additional step in their workflow. Upon further investigation, we found that scientists already entered needed information into other systems as part of their experiment workflow: biomass requests and setting up multi-sample chromatography assays. By slightly adjusting the investigators workflow, all the experiment-related metadata can be entered in the new GPCR system, and later exported by Chaise to use the data in downstream activities. The result was that we reduced the number of steps the investigator has to perform and increased data and metadata quality by eliminating manual double entry.
VI. Lessons Learned
Based on our experiences, a set of issues emerged across deployments. We summarize the issues and solutions here.
A. Spreadsheets Are Poor Data Entry Tools
At the early stage of many of our deployments, new meta-data were imported into Deriva via spreadsheets based on predetermined templates that referenced controlled vocabulary in some cases. Spreadsheets are commonly used by domain scientists, and we expected that this would be a streamlined path towards data ingest. In practice, however, we found the use of spreadsheets to be idiosyncratic and subject to frequent human error. And, when we updated the data model or simply added new terms to the vocabulary, we had to redistribute the template to all data submitters which was prone to communication failures. Moreover, entity linkages rely on users supplying the right (foreign key) references and in general we found many users are not comfortable with creating proper references in multi-tab spreadsheets such as Microsoft Excel and will just type in the values directly or copy-and-paste whole rows. Based on this experience, we placed an increased focus on on-line tools for metadata entry that introspect the catalog schema, dynamically adapt to the evolving data models, and apply data integrity checking upon submission.
B. Detect Errors While You Have the User’s Attention
For asset submission, we initially created an upload agent that monitored shared directories, automatically harvesting metadata and ingesting that metadata along with the asset. We had good success with this approach in earlier deployments in which we were able to include sufficient metadata as part of the asset to ensure accurate placement into the data model, e.g. our CIRM use case used bar codes on microscope slides to indicate what experiment the resulting data belonged to. However, in use cases, such as GPCR we have less control over the content of an asset and are forced to rely on file naming protocols to cross-link the asset with existing metadata. Furthermore, in many bioscience use cases, data generating instruments are shared and users tend not to log in to their own account prior to acquiring data. This resulted in additional complexity to associate data with specific users when adding assets. In these cases, our agent based approach broke down, as the agent service running in the background reports errors asynchronously, after the user has turned their attention to something else. A minor violation of required naming conventions could delay data collection for an unnecessarily long period of time. Therefore, immediate feedback is required. These observations led us to structure our on-line tools to perform error checking synchronously when the user indicates the desire to ingest an asset, while executing transfers of assets asynchronously once the platform has determined that the contextualization of all the assets are correct.
C. Give Users Incentives to Change Their Ways
Most of our experiences to date have involved deployments of Deriva into an environment in which there was already an existing data management practice. While it is hard to find a user who disagrees with the concept of producing sharable and reusable data, and while there was broad agreement that current data management practices in our use cases were inadequate, it is also not surprising to observe that users were reluctant to change their daily practice.
Fortunately in many of our use cases, we were able to get users to alter their current practice by providing time savings in somewhat unexpected areas, hence incentivizing use. For example, in GPCR, we were able to use experiment design metadata and our data extraction routines to automatically generate configuration files for an instrument, eliminating a double manual data entry task that the researcher would have otherwise had to perform. Providing data and images as part of a paper preparation process has also been a strong motivator. In an image based use case, the ability to easily find and download JPEG images for publication was motivation enough to get users to adopt Deriva. By identifying and exploiting these small wins for a user, we can increase the uptake of our data management tools, which will in turn result in increased efficiencies that may be less directly observable to the domain scientist.
D. Users Want a Bird’s Eye View of Their Data
The domain models enabled by Deriva make it easier for users to find and access specific sets of data. However, we quickly learned that aggregate information about their assets were highly desired by users. Invariably, soon after users could see and explore their assets with Deriva, they would request various dashboards, summaries and roll ups. For example, program managers, lab managers, and principal investigators (PIs) often want to monitor the data submission progress.
In practice we see requirements for many different types of dashboards including counts of assets and entities, assets in a current state, roll ups over periods of time, etc. Fortunately, it is easy to create these dashboards in Deriva as database views over the underlying data model, which can be queried and displayed like any other normal model element. Support for creating views with web APIs in ERMrest is currently limited, and given the importance of dashboards and summaries, this is an area where the platform will be expanded.
E. Control Vocabulary But Not Too Much
We have experimented with different approaches to managing vocabulary. Initially, in CIRM our users wanted uncontrolled term lists to label specimens and experiments. This was soon plagued by numerous divergent terms and spelling errors. Fortunately, Deriva is extremely adept at helping users correct such issues. They eventually adopted a controlled terminology list that undergoes periodic review. Because they are a smaller more homogeneous group, their term list changes slowly and this approach works for them.
On the other hand, in GPCR and FaceBase we began with strictly controlled vocabulary. However, this presented a roadblock for users. In GPCR, users stopped entering data when they could not find a desired term for an annotation. In FaceBase, users entered their terms in non-standard locations of the metadata spreadsheet or added them to comments. This caused temporary data loss and lower quality of data annotations. We remedied these issues by taking a pragmatic approach to controlling the vocabulary. The data model still enforces the use of vocabulary terms but allows users to add new terms when appropriate.
Deriva aims to make it easier to reuse terms than to add new ones, yet at the same time is flexible to allow new terms. For example, investigators with appropriate access control can define new experiment types on the fly by adding elements to term tables which can then be used by other investigators. Other investigators see the complete list of possible experiment types before being given the option to add a new type.
F. Human Factors are Critical to Success
Human factors were a major driver in designing Deriva. We view this broadly from the perspective of how the technical components of Deriva interface into the daily workflow of the users, and the social structure in which research takes place. For example, an initial assumption was that users would be able to explore the data model rapidly by observing what links were available, and internalizing the model as they used the system. Unfortunately, this is not typically the case. While a computer scientist may think in terms of links and graph models for representing data, researchers in other eScience domains do not necessarily think this way. Fortunately, we have found that this barrier can be easily overcome by making users aware that there is an underlying model and providing a simple roadmap to the major model elements.
One important human factors metric is the number of manual steps required to complete common operations. For example, in biomedical investigations, experiments often consist of multiple samples that are prepared all at once (many instruments can handle 96 or more simultaneous data acquisitions). To help these users, we extended Chaise to support multi-record input mode, where the user can add multiple data-entry records to the same graphical form. Users also requested shortcuts to produce multiple records with only a few varying fields. As a result, we also added a mechanism to copy initial content from an existing or draft record into the newly created data-entry fields.
VII. Related Work
Digital repository systems, such as DSpace [17] and Globus Publish [18], provide object and data collection level metadata, similar to Deriva. Digital repositories are primarily concerned with publication, as opposed to the discovery process itself where one’s understanding of the domain model may evolve considerably and hence these systems have very simple metadata models (e.g. without relationships) and don’t support model evolution nor support easy creation of multiple catalogs.
Other research has explored topics of integrated metadata catalogs with key-value models [19]; distributed metadata catalogs with key-value models [20]; and distributed relational database access underpinning metadata catalogs [21]. However, these catalogs support a flat, per-asset description of data, and don’t support the structured models that ERMrest does, nor do they provide RESTful interfaces. Research on metadata catalogs has considered issues of flexible modeling [22], dynamic model generation and integration [23], and incorporating semantic representations [24] into metadata catalogs. We differ from this work in focusing on ER modeling as being more understandable by end users and integrating ER models into a RESTful web services architecture.
VIII. Conclusions and Future Work
In this paper, we have considered three questions: 1) can scientific asset management systems provide value to eScience investigations, 2) will users adjust their daily practices to use these systems, and 3) are there reusable platforms that can be readily adapted to diverse eScience applications. We answered these questions through a detailed description of two representative use cases: FaceBase and GPCR. We were also able to show that by focusing on human factors issues including user interface and daily workflows, we can provide tangible value to domain scientists (i.e. non-computer scientists) which results in them adapting their data creation and analysis activities to use these tools, as indicated by the quantifiable uptake in data curation and usage.
These use cases represent just two of the many real-world deployments of Deriva. Similar results were seen in our other deployments, further demonstrating that the asset management approach can improve the efficiency of the research process, and the quality of data produced by and used in experiments. In this paper we focused on biomedical applications, as they are complex and the users tend to be less computationally sophisticated. However, Deriva can be readily applied more broadly with, we believe, the same positive results.
Based on our experiences to date, we have identified a number of area for future work including providing web-based management of named views; streamlining external ontology integration processes; utilizing term relationships (e.g. part of) for more semantic search; and simplifying IObox software installation and update processes.
All Deriva tools (deriva.isrd.isi.edu) are open-source and are publicly available on github.
Acknowledgments
We would like to acknowledge our Deriva team, and GPCR collaborators Mike Hanson, Jeff Siu, and Raymond Stevens. The work presented in this paper was funded by the National Institutes of Health under awards 5U54EB020406, 1R01MH107238-01, 5U01DE024449 and 1U01DK107350, by the National Science Foundation under award 1446112, and the GPCR Consortium.
References
- 1.Borgman CL. The conundrum of sharing research data. Journal of the American Society for Information Science and Technology. 2012;63(6):1059–1078. [Google Scholar]
- 2.Kandel S, et al. Enterprise data analysis and visualization: An interview study. IEEE Transactions on Visualization and Computer Graphics. 2012;18(12):2917–2926. doi: 10.1109/TVCG.2012.219. [DOI] [PubMed] [Google Scholar]
- 3.Begley CG. Six red flags for suspect work. Nature. 2013 May;497(7450):433–4. doi: 10.1038/497433a. [DOI] [PubMed] [Google Scholar]
- 4.Schuler R, Kesselman C, Czjakowski K. Accelerating data-driven discovery with scientific asset management. IEEE 12th International Conference on eScience; IEEE; 2016. [Google Scholar]
- 5.Goble C, De Roure D, Bechhofer S. Accelerating scientists knowledge turns. International Joint Conference on Knowledge Discovery, Knowledge Engineering, and Knowledge Management; Springer; 2011. pp. 3–25. [Google Scholar]
- 6.van Niekerk A. The strategic management of media assets: A methodological approach. New Orleans Conference Proceedings: International Academy for Case Studies; Allied Academies; 2006. [Google Scholar]
- 7.Ananthakrishnan R, et al. Globus nexus: An identity, profile, and group management platform for science gateways and other collaborative science applications. Cluster Computing (CLUSTER), 2013 IEEE International Conference on; IEEE; 2013. pp. 1–3. [Google Scholar]
- 8.Mustache template engine. [Online]. Available: mustache.github.io.
- 9.Chard K, et al. I’ll take that to go: Big data bags and minimal identifiers for exchange of large, complex datasets. Big Data (Big Data), 2016 IEEE International Conference on; IEEE; 2016. pp. 319–328. [Google Scholar]
- 10.Wilkinson MD, et al. The FAIR guiding principles for scientific data management and stewardship. Scientific data. 2016;3:160018. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mungall CJ, Emmert DB. A chado case study: an ontology-based modular schema for representing genome-associated biological information. Bioinformatics. 2007;23(13):i337–i346. doi: 10.1093/bioinformatics/btm189. [DOI] [PubMed] [Google Scholar]
- 12.Sansone S-A, et al. Toward interoperable bioscience data. Nat Genet. 2012;44(2):121–126. doi: 10.1038/ng.1054. [Online]. Available: http://dx.doi.org/10.1038/ng.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The UCSC genome browser. [Online]. Available: genome.ucsc.edu.
- 14.The jBrowse genome browser. [Online]. Available: jbrowse.org.
- 15.Monarch initiative. [Online]. Available: monarchinitiative.org.
- 16.Horn F, et al. GPCRDB information system for g protein-coupled receptors. Nucleic acids research. 2003;31(1):294–297. doi: 10.1093/nar/gkg103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith M, et al. Dspace: An open source dynamic digital repository. Tech Rep. 2003 [Google Scholar]
- 18.Chard K, et al. Globus data publication as a service: Lowering barriers to reproducible science. e-Science (e-Science), 2015 IEEE 11th International Conference on; IEEE; 2015. pp. 401–410. [Google Scholar]
- 19.Rajasekar A, et al. iRODS primer: integrated rule-oriented data system. Synthesis Lectures on Information Concepts, Retrieval, and Services. 2010;2(1):1–143. [Google Scholar]
- 20.Koblitz B, Santos N, Pose V. The AMGA metadata service. Journal of Grid Computing. 2008;6(1):61–76. [Google Scholar]
- 21.Antonioletti M, et al. The design and implementation of Grid database services in OGSA-DAI. Concurrency and Computation: Practice and Experience. 2005;17(2–4):357–376. [Google Scholar]
- 22.Deelman E, et al. Grid-based metadata services. Scientific and Statistical Database Management, 2004. Proceedings. 16th International Conference on; IEEE; 2004. pp. 393–402. [Google Scholar]
- 23.Tuchinda R, et al. Artemis: Integrating scientific data on the grid. AAAI. 2004:892–899. [Google Scholar]
- 24.Wang X, et al. Semantic enabled metadata management in PetaShare. International Journal of Grid and Utility Computing. 2009;1(4):275–286. [Google Scholar]