Abstract
Major depression is a highly complex disabling psychiatric disorder affecting millions of people worldwide. Despite the availability of several classes of antidepressants, a substantial percentage of patients are unresponsive to these medications. A better understanding of the neurobiology of depression and the mechanisms underlying antidepressant response is thus critically needed. We previously reported that mice lacking CREB-regulated transcription coactivator 1 (CRTC1) exhibit a depressive-like phenotype and a blunted antidepressant response to the selective serotonin reuptake inhibitor fluoxetine. In this study, we similarly show that Crtc1‒/‒ mice are resistant to the antidepressant effect of chronic desipramine in a behavioral despair paradigm. Supporting the blunted response to this tricyclic antidepressant, we found that desipramine does not significantly increase the expression of Bdnf and Nr4a1-3 in the hippocampus and prefrontal cortex of Crtc1‒/‒ mice. Epigenetic regulation of neuroplasticity gene expression has been associated with depression and antidepressant response, and histone deacetylase (HDAC) inhibitors have been shown to have antidepressant-like properties. Here, we show that unlike conventional antidepressants, chronic systemic administration of the HDAC inhibitor SAHA partially rescues the depressive-like behavior of Crtc1‒/‒ mice. This behavioral effect is accompanied by an increased expression of Bdnf, but not Nr4a1-3, in the prefrontal cortex of these mice, suggesting that this epigenetic intervention restores the expression of a subset of genes by acting downstream of CRTC1. These findings suggest that CRTC1 alterations may be associated with treatment-resistant depression, and support the interesting possibility that targeting HDACs may be a useful therapeutic strategy in antidepressant development.
Keywords: mood disorders, antidepressants, animal model of depression, CREB coactivator, BDNF, HDAC inhibitor, SAHA, epigenetics
1. Introduction
Mood disorders (including major depressive disorders and bipolar disorders) are a major cause of disability worldwide, with an estimated lifetime prevalence of 16% (Kessler et al., 2005). Although the underlying etiological mechanisms are complex and still unclear, studies over the past decades have highlighted the importance of the transcription factor cyclic adenosine monophosphate (cAMP) response element-binding protein (CREB) and one of its target genes brain-derived neurotrophic factor (Bdnf) (Blendy, 2006; Krishnan and Nestler, 2008; Martinowich et al., 2007). CREB is a pleiotropic transcription factor involved, in particular, in neuronal growth and survival, neurogenesis, synaptic plasticity and long-term memory (Blendy, 2006; Carlezon et al., 2005; Lonze and Ginty, 2002). BDNF is also implicated in these processes, partly through the induction of its expression by CREB (Martinowich and Lu, 2008; Martinowich et al., 2007). CREB and BDNF have both been involved in depression and antidepressant treatment. Indeed, overexpression of CREB and BDNF in the hippocampus (HIP) results in antidepressant effects (Chen et al., 2001; Shirayama et al., 2002) and inversely, chronic stress reduces Bdnf expression in the same structure (Smith et al., 1995). Of note, antidepressants such as fluoxetine and desipramine were also shown to upregulate Bdnf expression in the prefrontal cortex (PFC) and HIP of rodents in a CREB-dependent manner (Conti et al., 2002).
CREB-regulated transcription coactivator 1 (CRTC1) is a potent CREB coactivator activated by cAMP and Ca2+ signaling that trigger its dephosphorylation and translocation to the nucleus, where it binds to CREB, helps to recruit CREB-binding protein (CBP) and RNA polymerase II, and thus activates CREB-dependent gene transcription (Altarejos and Montminy, 2011; Ch'ng et al., 2012; Conkright et al., 2003; Kovacs et al., 2007; Ravnskjaer et al., 2007; Screaton et al., 2004). CRTC1 is involved in activity-induced dendritic growth and late phase long-term potentiation (L-LTP) (Kovacs et al., 2007; Li et al., 2009; Zhou et al., 2006). Moreover, Bdnf expression as well as BDNF-dependent dendritic growth require CRTC1 (Finsterwald et al., 2010; Kovacs et al., 2007; Zhou et al., 2006). Region-specific CRTC1’s activation is critical for activity-dependent immediate early genes regulation and long-term fear memory (Nonaka et al., 2014a; Sekeres et al., 2012). CRTC1-dependent transcription of Bdnf and orphan nuclear receptors 4a (Nr4a) is also involved in cognitive impairments related to Alzheimer’s disease (Parra-Damas et al., 2014).
To further investigate the role of CRTC1 in the brain, we generated a CRTC1-deficient mouse line that exhibits several endophenotypes related to mood disorders and a blunted response to the antidepressant fluoxetine in behavioral despair-related paradigms (Breuillaud et al., 2009; Breuillaud et al., 2012). At the molecular level, Crtc1‒/‒ mice have a decreased HIP and PFC expression of several CREB-regulated neuroplasticity genes, including notably Bdnf and Nr4a1-3 (Breuillaud et al., 2012). Altogether, this suggests that CRTC1, as a key regulator of neuroplasticity-related genes, is critical for mood regulation and antidepressant response. However, the direct or indirect mechanisms linking CRTC1’s deficiency and behavioral consequences are still unclear. As CRTC1 promotes CBP recruitment, which has an intrinsic histone acetyltransferase (HAT) activity, these mechanisms might include CRTC1-related epigenetic gene regulation. Noteworthily, epigenetic mechanisms have been widely associated with depressive disorders and with CREB and Bdnf regulation in the frame of stress and antidepressant response (Lin et al., 2012; Tsankova et al., 2007; Tsankova et al., 2006; Vecsey et al., 2007; Vialou et al., 2013).
In this study, we were interested in further investigating the involvement of CRTC1 in antidepressant response and the underlying molecular mechanisms. We first tested the effects of chronic desipramine on the behavior of Crtc1‒/‒ mice, which revealed a blunted behavioral response to this antidepressant. We also observed that desipramine failed to induce Bdnf and Nr4a1-3 expression in the HIP and PFC of Crtc1‒/‒ mice. By acting at the epigenetic levels and treating the animals with a HDAC inhibitor, we were able to partially rescue the depressive-like behavior of Crtc1‒/‒ mice. This was paralleled by an increased expression of Bdnf, but not Nr4a1-3, in the PFC of these animals.
2. Materials and Methods
2.1. Animals
Crtc1‒/‒ mice and wild-type (WT) littermates were generated and genotyped as previously described (Breuillaud et al., 2009). Mice were housed under a 12-h light-dark cycle with ad libitum access to water and standard rodent chow diet. Male mice were weaned at 21 days and group-housed until being isolated at 5 weeks of age in order to avoid wounding of cage mates by aggressive Crtc1‒/‒ males (Breuillaud et al., 2012). All animal experiments were conducted in accordance with the Swiss Federal Veterinary Office’s guidelines and were approved by the Cantonal Veterinary Service. Behavioral procedures began when mice reached the age of 8 weeks.
2.2. Drugs and treatment
Desipramine hydrochloride was purchased from Sigma (St-Louis, MO, USA). Mice received desipramine in the drinking water at a concentration of 100 mg/l, which corresponds to approximately 20 mg/kg of body weight/day. Desipramine solution was changed every week and concentration was adjusted depending on the weight gain and average water consumption of the mice. Suberoylanilide hydroxamic acid (SAHA, also known as vorinostat) was purchased from Selleck Chemicals (Houston, TX, USA). Mice received SAHA through daily intra-peritoneal injections. SAHA was first dissolved in DMSO at a dose of 50 mg/ml. This stock solution was diluted 1:10 every day in saline solution (final concentration: 5 mg/ml) prior to injections at a dose of 25 mg/kg of body weight. Vehicle groups were injected with a solution of 10% DMSO in saline.
2.3. Repeated open-space forced swim procedure (OSFS)
The repeated OSFS procedure was performed as previously described (Breuillaud et al., 2012; Stone and Lin, 2011). Briefly, swimming was carried out in rat tub cages (24x43x23 cm) filled with ~14 cm of lukewarm tap water (34±0.5°C) and colored with ~10 ml of milk. Mice undertook individual daily swim of 15 minutes during 4 consecutive days (Days -4 to -1). On day 0, desipramine or SAHA treatment started. Swimming sessions were repeated at 3 or 4 days of intervals during 3 weeks (Days 2, 5, 9, 12, 16, 19, 23) under continuous treatment (Fig. 1A). Water was changed after 4 mice had swum, in order to maintain water temperature. Swimming sessions were videotaped from above. Time spent immobile (drifting with no observable movement of limbs or tail) was manually recorded.
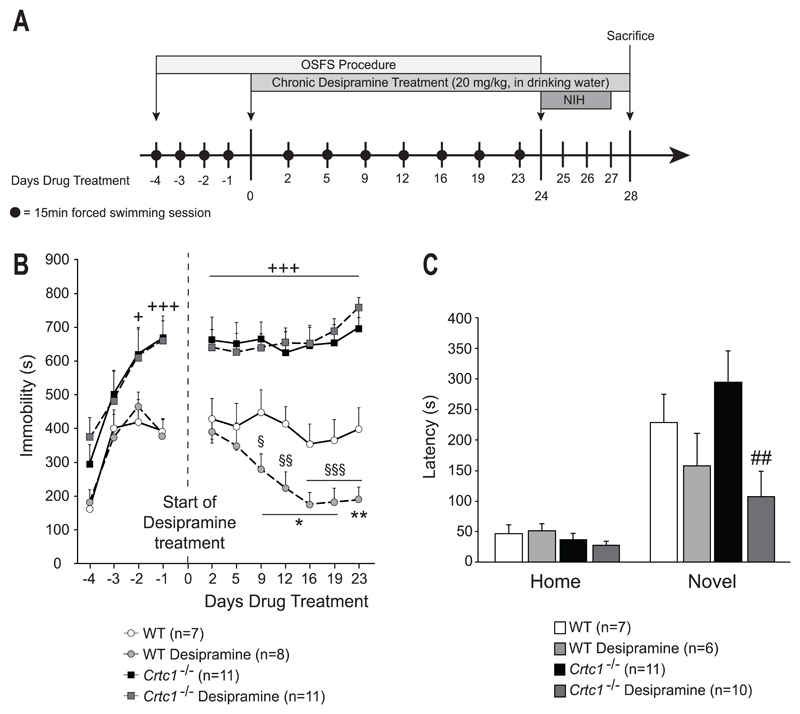
Fig. 1. Behavioral response of Crtc1‒/‒ mice to desipramine.
(A) Experimental design and timeline. The OSFS protocol started when mice reached the age of 8 weeks. Mice underwent forced swimming session during the first four days (day -4 to -1). On day 0, desipramine treatment started and continued until the end of the experiment. Swimming session were regularly repeated until day 24. The NIH test was then applied from day 24 until day 27. On day 28, animals were sacrificed. (B) Effects of chronic desipramine in the OSFS model of depression on Crtc1‒/‒ mice and WT littermates. During the four consecutive daily swimming sessions prior to treatment (Days -4 to -1), all groups increased their immobility time. Crtc1‒/‒ mice were significantly more immobile than WT mice during the two last sessions (+p<0.05, +++p<0.001 vs. WT untreated). From day 0 to day 23, WT and Crtc1‒/‒ mice received either water (n=7 and n=11 respectively) or chronic desipramine treatment (20 mg/kg in drinking water, n=8 and n=11 respectively). Crtc1‒/‒ mice were significantly more immobile than WT mice, regardless of the treatment, during the whole procedure (+++p<0.001, vs. WT untreated). Desipramine-treated WT mice showed a progressive decrease in immobility time starting from day 9 of treatment (§p<0.05, §§p<0.01, §§§p<0.001, vs. themselves on day 2). Their immobility time was also significantly lower that untreated WT mice (*p<0.05, **p<0.01, vs. WT untreated). No effect of desipramine was seen in Crtc1‒/‒ mice. (C) Effects of chronic desipramine on the NIH paradigm in Crtc1‒/‒ and WT mice. Latencies to drink sweetened condensed milk are shown in the homecage and in the novel environment. Two mice were removed for having latency scores > 2 SD from the mean. One mouse was removed for never having drunk the milk during the habituation and test phases. No significant difference of latency between the different groups was observed in the homecage conditions. In the novel environment, desipramine-treated WT mice (n=7) showed a non-significant trend to a decreased latency as compared to untreated WT mice (n=6). Desipramine significantly reduced the latency of Crtc1‒/‒ mice (n=10) compared to untreated Crtc1‒/‒ mice (n=11) (##p<0.05 vs. Crtc1‒/‒ mice). Data are mean ±SEM.
2.4. Novelty-induced hypophagia (NIH)
NIH procedure started on day 24, one day after the last swimming session (Fig. 1A). Mice were trained to drink sweetened condensed milk (1:3, condensed milk: water) for two consecutive days (day 24 and 25), 2x1h each day. Milk was presented in homemade drinking tubes made of 10 ml conical tubes (Sarsedt) with a hole at the bottom closed by a glass bead (5 mm diameter) to make a sipper. Tubes were placed through wire cage lids. On the third day (day 26), mice were tested in homecage conditions (light intensity: ~27 lux). Mice were videotaped during 15 min from the side and latency to drink was measured. On the fourth day (day 27), mice were placed in a novel cage without bedding, under bright light (~1200 lux), and the same measurements were made. The cage was cleaned with water and 70% ethanol between each mouse and before the first mouse.
2.5. Brain microdissection and micropunching
On day 28, one day after the end of the NIH procedure, all mice were sacrificed by cervical dislocation and decapitation. The brain was rapidly placed in a stainless steel adult mouse brain slicer matrix with 1 mm coronal section slice intervals. Six coronal slices were made from the second frontal slice channel of the matrix. Brain slices were placed on microscope slides and immediately frozen in dry ice, and then stored at -80°C. Medial PFC and dorsal HIP were collected with a micropunch (ø 1 mm, Stoelting, Wood Dale, IL, USA) in corresponding brain slices. PFC and HIP samples were kept at -80°C.
2.6. Gene expression analysis
Total RNA was extracted and purified from the PFC and HIP micropunches, using the RNAeasy Plus Minikit (Qiagen, Venlo, Netherland) according to the manufacturer’s instructions. RNA concentrations were measured by UV spectrophotometry with a NanoDrop Lite (Thermo Scientific, Wilmington, DE, USA). cDNA was prepared in a 50 μl reaction by reverse transcription, using 100 ng of RNA with Taqman Reagents and random hexamers (Applied Biosystems, Foster City, CA, USA). 0.8 μl of cDNA was amplified on a 96-well plate using the SYBR Green PCR Master Mix (Applied Biosystem). Amplification was performed with an ABIPRISM 7500 real-time PCR system (Applied Biosystem). The program was 2 min at 50°C, 10 min at 95°C, followed by 45 cycles of 15 s at 95°C and 1 min at 60°C. Relative gene expression was quantified using the comparative ΔΔ Ct method and normalized with β-actin transcript levels.
The following primers were used, at a concentration of 250 nM: β-actin forward 5’-GCTTCTTTGCAGCTCCTTCGT-3’, β-actin reverse 5’-ATATCGTCATCCATGGCGAAC-3’, Crtc1 forward 5’-CAGGACTTGGGCCTGGAA-3’, Crtc1 reverse 5’-AGACAGACAAGACCCTTTCTAAGCA-3’; Bdnf forward 5’-AAAACCATAAGGACGC GGACTT-3’, Bdnf reverse 5’-GAGGCTCCAAAGGCACTTGA-3’; Bdnf exIV forward 5’-GTAAG AGTCTAGAACCTTGGGGACC-3’, Bdnf exIV reverse 5’-GGATGGTCATCACTCTTCTCACCT-3’; Nr4a1 forward 5’-AAAATCCCTGGCTTCATTGAG-3’, Nr4a1 reverse 5’-TTAGA TCGGTATGCCAGGCG-3’; Nr4a2 forward 5’-CGGTTTCAGAAGTGCCTAGC-3’, Nr4a2 reverse 5’-TTGCCTGGAACCTGGAATAG-3’; Nr4a3 forward 5’-TGGCTCGACTCCATTAAAGAC-3’, Nr4a3 reverse 5’-TGCATAGCTCCTCCACTCTCT-3’; Hdac1 forward 5’-TTCCAACATGACCAACCAGA-3’, Hdac1 reverse 5’-GGCAGCATCCTCAAGTTCTC-3’, Hdac2 forward 5’-GGGACAGGCTTGGTTGTTTC-3’, Hdac2 reverse 5’-GAGCATCAGCAATGGCAAGT-3’, Hdac3 forward 5’-AGAGAGGTCCCGAGGAGAAC-3’, Hdac3 reverse 5’-ACTCTTGGGGACACAGCATC-3’, Hdac4 forward 5’-CAATCCCACAGTCTCCGTGT-3’, Hdac4 reverse 5’-CAGCACCCCACTAAGGTTCA-3’, Hdac5 forward 5’-TGTCACCGCCAGATGTTTTG-3’, Hdac5 reverse 5’-TGAGCAGAGCCGAGACACAG-3’, Hdac7 forward 5’-GGTGGACCCCCTTTCAGAAG-3’, Hdac7 reverse 5’-TGGGTAGCCAGGAGTCTGGA-3’, Hdac9 forward 5’-GCGAGACACAGATGCTCAGAC-3’, Hdac9 reverse 5’-TGGGTTTTCCTTCCATTGCT-3’.
2.7. Effects of SAHA on histone acetylation
Mice were weaned and housed according to 2.1. At the age of 8 weeks, Crtc1‒/‒ mice and WT littermates received a single injection of either SAHA or vehicle as in 2.2. Animals were sacrificed 2h after the injection, with a similar procedure as in 2.5. Histone acetylation was measured by Western blotting.
2.8. Western Blot
Histones were extracted from HIP and PFC micropunches using the Histone Purification Minikit (Active Motif, Carlsbad, CA, USA) according to the manufacturer’s instructions. Samples with poor histone yield were excluded. 1 μg of extracted histones were diluted 1:1 with sample buffer [65.8 mM Tris-HCl pH 6.8, 26.3% glycerol, 2.1% SDS, 100mM DTT, 0.01% bromophenol blue], separated on a 15% SDS-polyacrylamide gel, and proteins were transferred to polyvinylidene difluoride (PVDF) membranes with a Transblot Turbo Transfer System (BioRad, Hercules, CA, USA). Blots were blocked for 1 hour at room temperature (RT) in TBST [10 mM Tris-HCl (pH 7.4), 150 mM NaCl, 0.1% Tween-20], supplemented with 5% skim milk powder. Blots were subsequently incubated with a primary antibody in TBST plus 5% bovine serum albumin (BSA) overnight at 4°C. Finally, PVDF membranes were incubated for 1 hour at RT with horseradish peroxidase (HRP)-conjugated secondary antibodies in TBST plus 5% skim milk powder, and developed using a Pierce ECL Western chemiluminescence detection kit (Thermo Scientific). The following antibodies and dilutions were used: rabbit α-Acetyl-Histone H3, 1:2500 (EMD Millipore, Temecula, CA, USA), rabbit α-Histone H3, 1:2000 (Abcam, Cambridge, UK), rabbit α-Acetyl-Histone H4, 1:2000 (EMD Millipore), rabbit α-Histone H4, 1:2000 (EMD Millipore), donkey HRP-α-rabbit 1:2000 (GE Healthcare, Little Chalfont, UK). Quantification of band intensity was performed with Image J software (National Institute of Health, Bethesda, MD, USA). Acetyl-H3 and acetyl-H4 band intensities were normalized with total H3 and H4 signals, respectively.
2.9. Statistical analyses
Statistical analyses were performed using the Statistica 8.0 Software (StatSoft Inc., Tulsa, OK, USA). All data are presented as mean ± SEM. P-values of p < 0.05 were considered as statistically significant. A Shapiro-Wilk test was first performed to assess data normality. All results were found to follow normal distribution; therefore parametric tests could be used. For both behavioral and molecular analyses, a two-way ANOVA was performed to assess statistical differences for two factors (genotype and treatment). A Fisher LSD post-hoc test was carried out afterwards. For analyses of the OSFS data, a two-way ANOVA with repeated measures was performed as immobility time was repeatedly measured. A Fisher LSD post-hoc test was carried out afterwards.
3. Results
3.1. Crtc1‒/‒ mice have a blunted behavioral response to chronic desipramine treatment
Behavioral effects of desipramine were first assessed in the open-space forced swim (OSFS) model of depression (Fig. 1A and 1B). This protocol induces depressive-like symptoms that are reversed by chronic, but not acute, antidepressant treatments, and thus it has better face and construct validities than the conventional forced swim test (Breuillaud et al., 2012; Stone and Lin, 2011; Stone et al., 2008). All animals presented increased immobility time after the four days of consecutive swimming with a significant effect of time (F(1,37)=36.1, p<0.001) and genotype (F(1,111)=13.7, p<0.001). Crtc1‒/‒ mice had higher immobility time than WT mice during the last two sessions of the pre-test period (day -2 and -1) (p<0.001 for both days). This genotype effect remained significant after the beginning of the desipramine treatment (F(1,35)=71.34, p<0.001), indicating that Crtc1‒/‒ mice had higher immobility time that WT mice during the whole procedure. WT mice progressively responded to desipramine, as their immobility time significantly decreased over time. As compared to their own immobility time on day 2 of treatment, the effect of desipramine became significant starting from day 9 (p=0.029 on day 9, p=0.003 on day 12, p<0.001 on days 16, 19 and 23, vs. day 2). Their immobility time was also lower than untreated WT mice, starting from day 9 as well (p=0.032 on day 9, p=0.032 on day 12, p=0.023 on day 12, p=0.016 on day 19, p=0.009 on day 23, vs. WT untreated). Desipramine had no effect on the depressive-like behavior of Crtc1‒/‒ mice, indicating that these mice were resistant to desipramine in this paradigm. At the end of the procedure, mice were then tested in the novelty-induced hypophagia (NIH) test, a depression- and anxiety-related paradigm sensitive to chronic antidepressant treatment (Dulawa and Hen, 2005) (Fig. 1A and 1C). In homecage conditions, genotype or treatment had no effect on the latency to consume condensed milk, whereas a significant effect of desipramine was found in the novel environment (F(1,30)=6.52, p=0.016). Desipramine significantly decreased the latency of Crtc1‒/‒ mice (p=0.005, vs. untreated Crtc1‒/‒ mice), thus showing that these mice responded positively to desipramine in this paradigm. Comparably to their response to fluoxetine (Breuillaud et al., 2012), Crtc1‒/‒ mice show a blunted response to chronic desipramine in the OSFS test and a reduced latency to drink sweetened milk in the anxiogenic environment of the NIH, which suggest a differential involvement of CRTC1 in the behavioral response to antidepressants.
3.2. Desipramine fails to induce expression of neuroplasticity-related genes in Crtc1‒/‒ mice
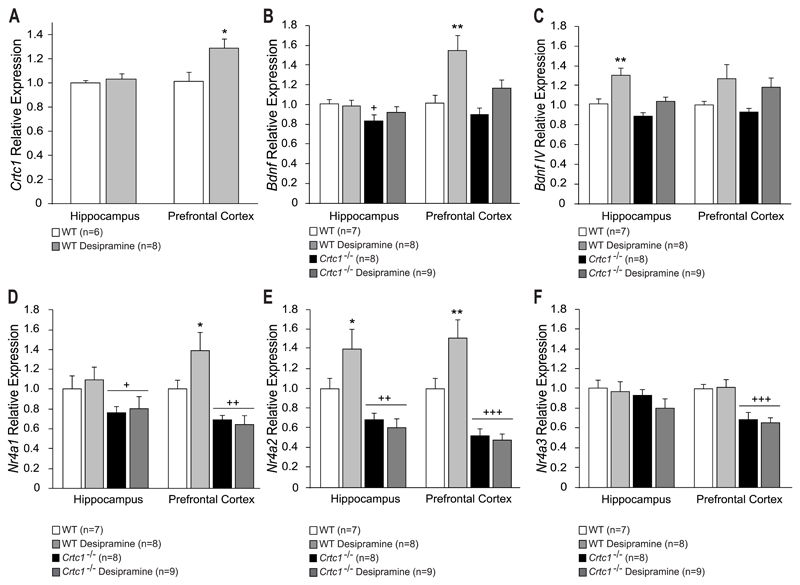
In order to investigate the molecular mechanisms underlying the blunted behavioral response of Crtc1‒/‒ mice to desipramine, gene expression analysis was performed in the HIP and PFC of these animals (Fig. 2). Crtc1‒/‒ mice have a reduced expression of several neuroplasticity-related genes in these brain regions involved in depressive disorders (Breuillaud et al., 2012). We were first interested in assessing a possible effect of desipramine on Crtc1 expression in WT mice (Fig. 2A). Desipramine upregulated Crtc1 mRNA in the PFC of WT mice (t=-2.35, df=12, p=0.037, vs. WT untreated), which further suggests a role for CRTC1 in antidepressant response. Total Bdnf expression was then measured in WT and Crtc1‒/‒ mice (Fig. 2B). In the HIP, genotype had a significant effect on Bdnf expression (F(1,27)=4.53, p=0.042), unlike desipramine that had no effect. Indeed, Crtc1‒/‒ mice had lower levels of Bdnf mRNA (p=0.044, vs. WT untreated) as previously observed in these animals (Breuillaud et al., 2012). In the PFC, a significant effect of treatment was found (F(1,22)=9.08, p=0.006), which revealed that desipramine significantly induced Bdnf expression in WT mice (p=0.003, vs. WT untreated), but failed to do so in Crtc1‒/‒ mice. Promoter IV-driven Bdnf expression (BdnfIV) analysis showed a significant effect of treatment in the HIP (F(1,22)=13.46, p=0.001), as desipramine induced an increased expression of BdnfIV mRNA in WT mice (p=0.002 vs. WT untreated), but not in Crtc1‒/‒ mice (Fig. 2C). No significant effect of genotype or treatment was observed in the PFC. We were then interested in measuring Nr4a1-3 gene expression, as these orphan nuclear receptors are downregulated in Crtc1‒/‒ mice (Breuillaud et al., 2012). Furthermore, NR4A1-3 have been shown to be involved in Bdnf regulation, in mood disorders and in neuroprotection (Schaffer et al., 2010; Volakakis et al., 2010; Volpicelli et al., 2007). Analysis of Nr4a1 expression (Fig. 2D) revealed a significant effect of genotype in the HIP (F(1,27)=6.71, p<0.015), showing that, independently of the treatment, Crtc1‒/‒ mice presented decreased levels of Nr4a1 (p=0.025 vs. WT untreated). In the PFC, genotype had a significant effect (F(1,26)=23.84, p<0.001). Desipramine significantly increased Nr4a1 expression in WT mice (p=0.026 vs. WT untreated), but not in Crtc1‒/‒ mice, which also presented decreased Nr4a1 levels (p=0.003 vs. WT untreated). Similar results were observed for Nr4a2 expression (Fig. 2E). A significant effects of genotype was observed in both structures (HIP: F(1,27)=22.79, p<0.001; PFC: F(1,27)=39.04, p<0.001), as Crtc1‒/‒ mice presented lower levels of Nr4a2 expression (HIP: p=0.002; PFC: p<0.001, vs. WT untreated). Furthermore, desipramine induced Nr4a2 expression in the HIP and PFC of WT mice (HIP: p=0.035; PFC: p=0.008, vs. WT untreated). A significant genotype*treatment interaction was found in the PFC (F(1, 27)=5.12, p=0.031) as desipramine had no effect in Crtc1‒/‒ mice. Finally, no effect of genotype or treatment was observed for Nr4a3 in the HIP (Fig. 2F), whereas only a genotype effect was found in the PFC, (F(1,27)=27.59, p<0.001), as Crtc1‒/‒ mice presented decreased Nr4a3 expression (p<0.001, vs. WT untreated). Altogether, these data show that desipramine fails to induce the expression of Bdnf, Bdnf IV, Nr4a1 and Nr4a2 in the PFC and HIP of Crtc1‒/‒ mice, therefore possibly explaining their abnormal behavioral response to this antidepressant.
Fig. 2. Molecular effects of desipramine in the HIP and PFC of Crtc1‒/‒ mice and WT littermates.
(A) Desipramine had no effect on Crtc1 expression in the HIP of WT mice. In the PFC, WT mice treated with desipramine (n=8) showed increased levels of Crtc1 expression as compared to untreated WT mice (n=6) (*p<0.05, vs. WT untreated). (B) Untreated Crtc1‒/‒ mice (n=8) displayed lower hippocampal Bdnf expression compared to WT mice (n=7) (+p<0.05, vs. WT untreated). Desipramine had no effect on Bdnf expression of WT and Crtc1‒/‒ mice (n=8 and n=9 respectively). In the PFC, desipramine significantly increased Bdnf levels of WT mice (*p<0.01 vs. WT untreated) but not Crtc1‒/‒ mice. (C) Hippocampal BdnfIV expression was increased in desipramine-treated WT mice (**p<0.01, vs. WT untreated), but not in Crtc1‒/‒ mice. In the PFC, desipramine did not have a significant effect on both genotypes. (D-F) Effects of desipramine on the expression of Nr4a1-3. (D) Crtc1‒/‒ mice displayed lower levels of Nr4a1 expression than WT mice in the HIP and PFC (+p<0.05, ++p<0.01, vs. WT untreated). Desipramine increased Nr4a1 levels of WT mice in the PFC but had no effect in the HIP (*p<0.05, vs. WT untreated). (E) Nr4a2 expression was found to be lower in the HIP and PFC of Crtc1‒/‒ mice compared to WT mice (+p<0.05, ++p<0.01, vs. WT untreated). Desipramine had no effect on Nr4a2 expression in Crtc1‒/‒ mice but increased its expression in the PFC of WT mice (*p<0.05, vs. WT untreated). (F) Desipramine increased Nr4a3 expression in the HIP of WT animal but had no effect on Crtc1‒/‒ mice (*p<0.05, vs. WT untreated). Crtc1‒/‒ mice displayed lower Nr4a3 expression than WT animals in the PFC (+p<0.05, vs. WT untreated). Data are mean ±SEM.
3.3. SAHA partially rescues the depressive-like phenotype of Crtc1‒/‒ mice
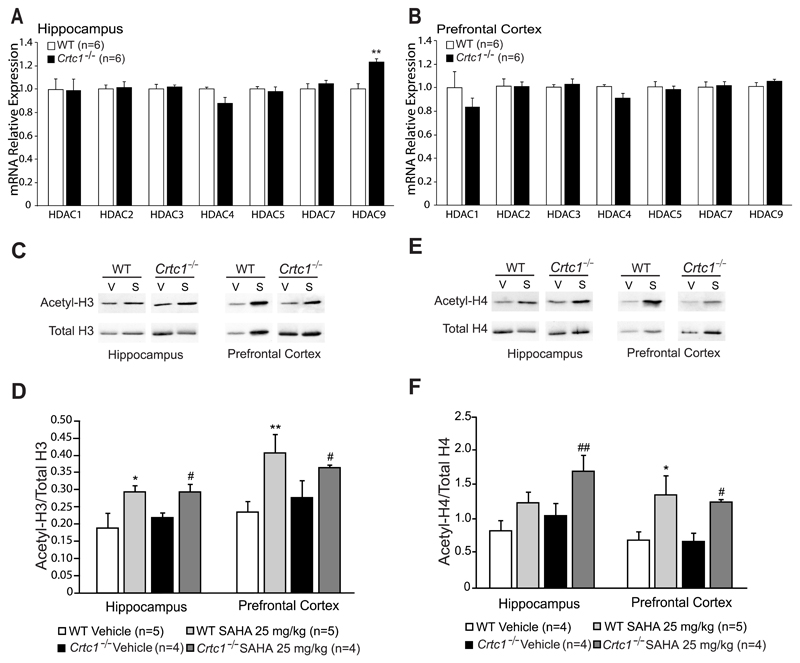
As CRTC1 is helping phosphorylated CREB to recruit the histone acetyltransferase CBP, we reasoned that HDAC inhibition might compensate for the possible deficit of CBP recruitment in Crtc1‒/‒ mice, and thus rescue the expression of neuroplasticity genes involved in mood regulation. Moreover, several lines of evidence both in humans and in animal models have suggested that pathological conditions may alter the expression of Hdac family members, mainly of class I and IIa (Covington et al., 2009; Gräff et al., 2012; Han et al., 2014; Morris and Monteggia, 2013; Tsankova et al., 2006; Uchida et al., 2011). Therefore, we first measured the expression of class I (Hdac1,2,3) and class IIa (Hdac4,5,7,9) members in the HIP and PFC of WT and Crtc1‒/‒ mice to determine whether mutant mice may present a possible altered epigenetic profile (Fig. 3A and 3B). We found that expression of most Hdac tested was globally unchanged in the HIP and PFC of Crtc1‒/‒ mice, except for Hdac9 that was slightly upregulated in the HIP of mutant mice (t=-4.29, df=10, p=0.002, vs. WT mice) (Fig. 3A). However, this increased expression of Hdac9 in the HIP of Crtc1‒/‒ mice did not influence global acetylation of histone H3 and H4, as revealed by Western blot analysis (Fig. 3C-F). This suggested that the behavioral and molecular phenotypes of Crtc1‒/‒ mice did not result from a major alteration of Hdac expression and histone acetylation.
Fig. 3. Expression of Hdac1,2,3,4,5,7,9 and effects of SAHA in the HIP and PFC of Crtc1‒/‒ and WT mice.
(A) Hippocampal mRNA expression of most Hdac class I (1,2,3) and IIa (4,5,7,9) was unchanged in Crtc1‒/‒ mice (n=6) with the exception of Hdac9 which was upregulated as compared to WT mice (n=6) (**p<0.01, vs. WT mice). (B) No difference of Hdac expression in the PFC of Crtc1‒/‒ mice. (C-F) Histone H3 and H4 acetylation 2h after a single SAHA injection (25 mg/kg) in Crtc1‒/‒ and WT mice. Immunoblot analysis of acetylated histones H3 and H4 (acetyl-H3 and acetyl-H4) and total H3 and H4 revealed single bands at the expected sizes of 15 kDa and 10kDa, respectively. Panel (C) shows a representative western blot for acetylated histone H3 (Acetyl-H3) and total histone H3 (total H3) in the HIP and PFC of WT and Crtc1‒/‒ mice. (D) SAHA-treated WT mice (n=5) presented higher levels of acetyl-H3 than vehicle-treated WT mice (n=5) in the hippocampus (*p<0.05, vs. WT Vehicle) and PFC (**p<0.01, vs. WT vehicle). SAHA-treated Crtc1‒/‒ mice (n=4) also presented increased histone H3 acetylation compared to vehicle-treated Crtc1‒/‒ mice (n=4) in the HIP and PFC (# p<0.05, vs. Crtc1‒/‒ Vehicle). Panel (E) shows a representative western blot for acetylated histone H4 (Acetyl-H4) and total histone H4 (total H4) in the HIP and PFC of WT and Crtc1‒/‒ mice. (F) SAHA-treated WT mice (n=4) presented a non-significant tendency to higher levels of acetyl-H4 than vehicle-treated WT mice (n=5) in the HIP. In the PFC, SAHA increased levels of H4 acetylation in WT mice (**p<0.01, vs. WT Vehicle). SAHA-treated Crtc1‒/‒ mice (n=4) presented increased histone H4 acetylation compared to vehicle-treated Crtc1‒/‒ mice (n=4) in the HIP and PFC (# p>0.05, ## p<0.01, vs. Crtc1‒/‒ Vehicle). Data are mean ±SEM.
HDAC inhibitors have been shown to have antidepressant effects in rodents and to regulate similar patterns of genes as antidepressants (Covington et al., 2009; Schroeder et al., 2007). SAHA is a hydroxamic acid compound that inhibits class I and II HDACs (Gräff and Tsai, 2013). Systemic administration of SAHA increases histone acetylation in the brain and has antidepressant effects in mice (Hockly et al., 2003; Uchida et al., 2011). Moreover, in primary neuronal cultures, SAHA induces a rapid increase in histone acetylation at Bdnf promoters I and IV, and an upregulation of Bdnf expression (Koppel and Timmusk, 2013). Therefore, SAHA was chosen to assess the behavioral response of WT and Crtc1‒/‒ mice to chronic HDAC inhibition. We first controlled that an intraperitoneal injection of SAHA indeed triggered an increase of histone acetylation in the brain of WT and Crtc1‒/‒ mice (Fig. 3C-F). Acetyl-H3 and Acetyl-H4 signals were quantified and normalized over total H3 and total H4 signals. Analysis of histone H3 acetylation (Fig. 3C-D) showed a significant effect of SAHA in the HIP (F(1, 14)=11.17, p=0.004) and in the PFC (F(1,14),=13.86, p=0.002) of WT (HIP: p=0.014, PFC: p=0.003, vs. WT vehicle) and Crtc1‒/‒ mice (HIP: p=0.017, PFC: p=0.032, vs. Crtc1‒/‒ vehicle). Similarly, SAHA had a significant effect on H4 acetylation (Fig. 3E-F) in the HIP (F(1, 14)=9.28, p=0.008) and in the PFC (F(1,13)=13.97, p=0.001) of WT (PFC: p=0.003, vs. WT vehicle) and Crtc1‒/‒ mice (HIP: p=0.003, PFC: p=0.034, vs. Crtc1‒/‒ vehicle).
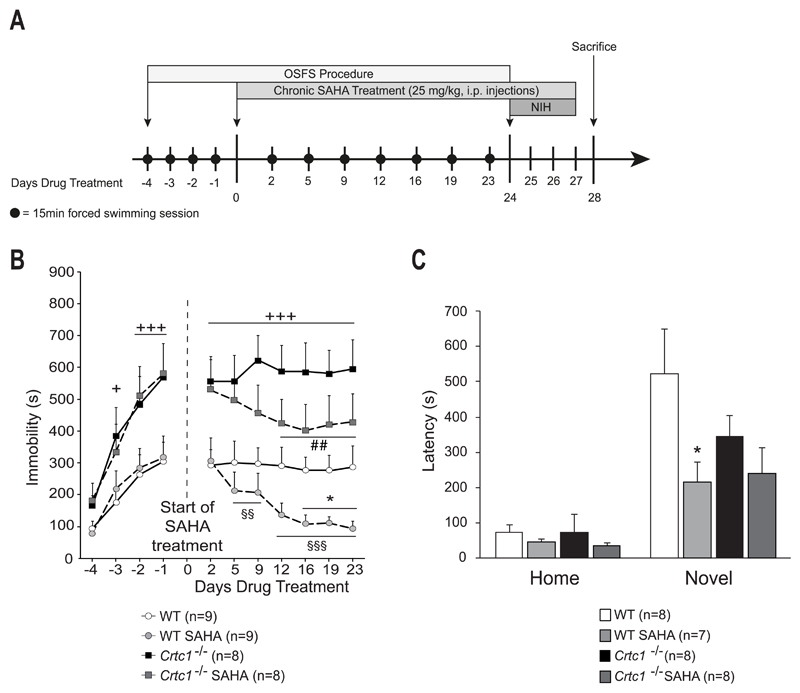
Having shown that SAHA crosses the blood-brain barrier and increases histone acetylation in the brain, we then tested the behavioral response of WT and Crtc1‒/‒ mice to chronic SAHA treatment. Animals were first tested in the OSFS protocol (Fig. 4A and 4B). During the initial four consecutive days (from day -4 to -1), all groups showed an increase in their immobility time, with a significant interaction of time and genotype (F(3,96)=3.89, p=0.011), and Crtc1‒/‒ mice progressively displayed increased immobility time as compared to WT mice (day -3: p=0.02, day-2: p=0.001, day-4: p<0.001, vs. WT mice). After the beginning of SAHA treatment, a significant effect of genotype (F(1,30)=20.21, p<0.001) and a significant interaction of time and treatment (F(6,180)=6.23, p<0.001) were observed. SAHA-treated mice rapidly decreased their immobility time, as compared to their score at the beginning of the treatment (day 5: p=0.01, day 9: p=0.007, day 12-23: p<0.001, vs. themselves on day 2). Their immobility time was also significantly lower than vehicle-treated WT mice on the last three sessions (day 16: p=0.037, day 19: p=0.049, day 23: p=0.046, vs. WT Vehicle). Vehicle-treated Crtc1‒/‒ mice displayed higher immobility time than WT mice during the whole procedure (day 2-23: p<0.001, vs. WT Vehicle). Crtc1‒/‒ mice responded to SAHA, as they progressively decreased their immobility time (day 12: p=0.007, day 16: p=0.001, day 19: p=0.005, day 23: p=0.009, vs. themselves on day 2). Animals were then tested in the NIH paradigm (Fig. 4A and 4C). In the homecage conditions, there was no effect of genotype or treatment, whereas a significant effect of genotype was observed in the novel environment (F(1,26)=4.55, p=0.042), where SAHA decreased the latency of WT mice (p=0.039, vs. WT Vehicle). There was not however a significant effect of SAHA on Crtc1‒/‒ mice, although SAHA-treated mutant mice presented a trend to a decreased latency. Taken together, these results show that SAHA improves the phenotype of Crtc1‒/‒ mice, as it partially rescued their depressive-like behavior in the OSFS paradigm.
Fig. 4. Behavioral response of Crtc1‒/‒ mice to SAHA.
(A) Experimental design and timeline. The OSFS protocol started when mice reached the age of 8 weeks. Mice underwent forced swimming session during the first four days (day -4 to -1). On day 0, SAHA treatment started and continued until the end of the experiment (last injection on day 27). Swimming session were regularly repeated until day 24. The NIH test was then applied from day 24 until day 27. On day 28, animals were sacrificed. (B) Effects of chronic SAHA in the OSFS model of depression on Crtc1‒/‒ mice and WT littermates. All groups increased their immobility time during the pre-treatment period (Day -4 to day -1). Crtc1‒/‒ mice presented higher immobility time than WT mice starting from day -2 (+p<0.05, +++p<0.001, vs. WT mice). From day 0 until day 23, WT and Crtc1‒/‒ mice were daily injected i.p. with either vehicle (n=9 and n=8 respectively) or 25 mg/kg SAHA (n=9 and n=8 respectively). Vehicle-treated Crtc1‒/‒ mice were significantly more immobile that vehicle-treated WT mice during the whole procedure (+++p<0.001, vs. WT Vehicle). SAHA-treated WT mice significantly decreased their immobility time starting from day 2 of treatment (§§p<0.01, §§§p<0.001, vs. themselves on day 2). Their immobility time was also significantly lower than vehicle-treated WT mice during the last three sessions (*p<0.05, vs. WT Vehicle). SAHA-treated Crtc1‒/‒ mice significantly decreased their immobility time starting from day 12 of treatment (##p<0.01, vs. themselves on day 2). (C) Effects of chronic SAHA on the NIH paradigm. Latencies to drink sweetened condensed milk are shown in the homecage and in the novel environment. Two mice were removed for having latency scores > 2 SD from the mean. One mouse was removed for never having drunk the milk during the habituation and test phases. In the homecage conditions, no effect of SAHA was observed in any groups. In the novel environment, SAHA-treated mice (n=7) presented significant shorter latencies to drink the milk, as compared to vehicle-treated WT mice (n=8) (*p<0.05, vs. WT Vehicle). SAHA-treated Crtc1‒/‒ mice showed a non-significant trend to a shorter latency than vehicle-treated Crtc1‒/‒ mice (n=8 for each group). Data are means ±SEM.
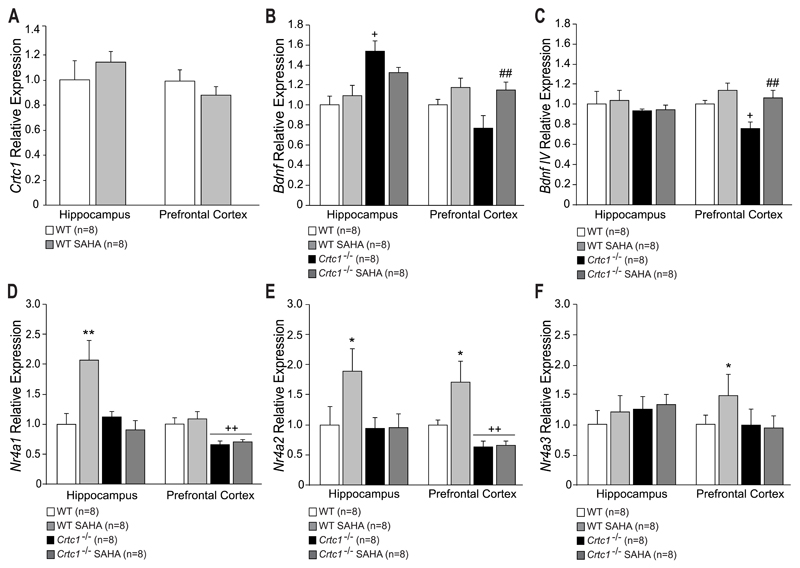
3.4. SAHA partially restores normal gene expression in Crtc1‒/‒ mice
Gene expression analysis of Crtc1, Bdnf, BdnfIV and Nr4a1-3 was performed in order to investigate the effects of chronic SAHA on these genes (Fig. 5). SAHA had no effect on Crtc1 expression in the HIP and PFC of WT mice (Fig. 5A). A significant effect of genotype on total Bdnf expression (Fig. 5B) was seen in the HIP (F(1,26)=18.06, p<0.001), as Crtc1‒/‒ mice surprisingly displayed higher Bdnf mRNA levels than WT mice (p=0.02, vs. WT Vehicle). In the PFC, a significant effect of treatment was observed (F(1,26)=8.52, p=0.007), as chronic SAHA increased Bdnf mRNA levels in Crtc1‒/‒ mice (p=0.009, vs. Crtc1‒/‒ Vehicle). Analysis of Bdnf promoter IV expression (Fig. 5C) did not reveal any effect of genotype or treatment in the HIP, whereas a significant genotype and treatment effect was observed in the PFC (genotype: F(1,25)=4.82, p=0.037; treatment: F(1,25)=10.11, p=0.003), where Crtc1‒/‒ mice displayed lower expression of BdnfIV (p=0.039, vs. WT Vehicle). SAHA treatment successfully restored BdnfIV levels in Crtc1‒/‒ mice (p=0.005, vs. Crtc1‒/‒ Vehicle). Measure of Nr4a1 mRNA expression (Fig. 5D) showed a significant interaction of genotype and treatment (F(1,25)=9.58, p=0.004), as SAHA increased Nr4a1 expression in the HIP of WT mice (p=0.001, vs. WT Vehicle), but had no effect in Crtc1‒/‒ mice. In the PFC, a significant effect of genotype was observed (F(1,27)=15.57, p<0.001), as Crtc1‒/‒ mice showed lower levels of Nr4a1 (p=0.005, vs. WT Vehicle). SAHA had no effect on both genotypes in this structure. Similar effects of SAHA on Nr4a2 expression were observed in the HIP (Fig. 5E), where SAHA increased Nr4a2 levels in WT mice (p=0.001, vs. WT Vehicle), but had no effect in Crtc1‒/‒ mice. SAHA also increased Nr4a2 levels in the PFC of WT mice (p=0.013, vs. WT mice). A significant effect of genotype was also visible in the same structure (F(1,24)=14.12, p<0.001), as Crtc1‒/‒ mice displayed decreased expression of Nr4a2 (p=0.005, vs. WT Vehicle). Genotype or treatment had no effect on Nr4a3 mRNA levels in the HIP (Fig. 5F). In the PFC, a significant interaction of genotype and treatment (F(1,25)=4.54, p<0.042) was observed, as SAHA increased Nr4a3 expression in WT mice only (p=0.012, vs. WT Vehicle).
Fig. 5. Molecular effects of SAHA in the HIP and PFC of Crtc1‒/‒ mice and WT littermates.
(A) SAHA had no effect on Crtc1 expression in the HIP and PFC of WT mice (n=8 for both group). (B) Untreated Crtc1‒/‒ mice (n=8) displayed higher hippocampal Bdnf expression compared to WT mice (+p<0.05, vs. WT mice). SAHA had no effect on Bdnf expression of WT and Crtc1‒/‒ mice (n=8 for each group). In the PFC, SAHA significantly increased Bdnf levels of Crtc1‒/‒ mice (##p<0.01 vs. Crtc1‒/‒ Vehicle). (C) Hippocampal BdnfIV expression was unchanged among the different groups. In the PFC, vehicle-treated Crtc1‒/‒ mice showed a decreased expression of BdnfIV (+p<0.05, vs. WT Vehicle). SAHA treatment significantly restored BdnfIV expression in Crtc1‒/‒ mice (##p<0.01, vs. Crtc1‒/‒ mice Vehicle). (D-F) Effects of SAHA on the expression of Nr4a1-3. (D) Crtc1‒/‒ mice displayed lower levels of Nr4a1 expression than WT mice in the PFC (++p<0.01, vs. WT Vehicle). Desipramine increased Nr4a1 levels of WT mice in the HIP but had no effect in the PFC (**p<0.01, vs. WT Vehicle). (E) Nr4a2 expression was found to be lower in the PFC of Crtc1‒/‒ mice compared to WT mice (++p<0.01, vs. WT Vehicle). SAHA had no effect on Nr4a2 expression in Crtc1‒/‒ mice but increased its expression in the HIP and PFC of WT mice (*p<0.05, vs. WT Vehicle). (F) SAHA had no effect on Nr4a3 expression in the HIP of all groups. In the PFC, SAHA increased Nr4a3 expression of WT animal but had no effect on Crtc1‒/‒ mice (*p<0.05, vs. WT Vehicle). Data are mean ±SEM.
Taken together, these results interestingly show that chronic SAHA treatment restored normal Bdnf and BdnfIV expression in the PFC of Crtc1‒/‒ mice, and that it increased Nr4a1-3 expression in WT mice, but not in Crtc1‒/‒ mice. However, it is worth noting that the relative gene expression levels of vehicle-injected Crtc1‒/‒ mice (Fig. 5, black bars) displayed several discrepancies, mostly in the HIP, as compared with those of mice that were exposed to the same behavioral paradigms, but not chronically injected (Fig. 2, black bars). The most striking difference was the surprising increased Bdnf expression in the HIP of vehicle-injected Crtc1‒/‒ mice (Fig. 5B). In addition, contrary to our previous observations and the results of Fig. 2, Nr4a1 and Nr4a2 levels were not reduced in the HIP of Crtc1‒/‒ mice. These inconsistencies prompted us to compare the expression levels of Bdnf, BdnfIV, and Nr4a1-3 in WT and Crtc1‒/‒ mice control groups of Figures 2 and 5 (supplementary Fig. S1). The apparent increase of Bdnf expression in the HIP of vehicle-injected Crtc1‒/‒ mice (Fig. 5B) appeared to be rather due to a decreased Bdnf expression in chronically injected WT mice (Fig. S1A). Similarly, hippocampal Nr4a1-3 levels seemed to be reduced mostly in WT mice chronically injected with vehicle (Fig. S1C-E). With a few exceptions, Crtc1‒/‒ mice appeared to be less affected by this chronic injection stress, which might explain the inconsistent data of Figure 5. In conclusion, the behavioral effect of chronic SAHA administration in Crtc1‒/‒ mice might be related to the restoration of normal Bdnf and BdnfIV expression in the PFC. However, unlike its effect in WT mice, chronic SAHA was not able to induce Nr4a1-3 expression in Crtc1‒/‒ mice, which suggests that the upregulation of these genes is not required to improve the behavioral despair of these animals in the OSFS paradigm.
4. Discussion
The generation and characterization of Crtc1‒/‒ mice highlighted a key role of CRTC1 as a mediator of neuroplasticity-related genes expression potentially involved in the pathophysiology of mood disorders (Breuillaud et al., 2012). Moreover, we showed that CRTC1 is required for proper behavioral response to chronic fluoxetine treatment. In the present study, we further investigated the role of CRTC1 in behavioral and molecular response to antidepressants. We were first interested to assess the effect of another class of antidepressants. The tricyclic antidepressant desipramine was selected as this type of compounds mainly acts on the noradrenergic system, while fluoxetine is a selective serotonin reuptake inhibitor. We observed that Crtc1‒/‒ mice responded similarly to desipramine and fluoxetine; i.e. desipramine had no effect on these animals in the OSFS protocol, whereas it had anxiolytic effects in the NIH paradigm. These results further suggest that CRTC1 is involved in behavioral response to antidepressants, mainly in behavioral despair-related paradigms, and that both noradrenergic and serotonergic pathways activated by antidepressants converge on CRTC1. We also observed that desipramine induced the expression of Crtc1 in the PFC of WT mice, which strengthens the hypothesis of a pivotal role of CRTC1 in antidepressant response.
Interestingly, desipramine, like fluoxetine in our previous study, still elicited behavioral response in Crtc1‒/‒ mice in the NIH paradigm. This test has the advantage to be sensitive to chronic, but not acute antidepressant treatment. However, hyponeophagic behavior can be related to depressive but also anxious behavior (Dulawa and Hen, 2005) and both fluoxetine and desipramine have anxiolytic effects (Zohar and Westenberg, 2000). Therefore, the behavioral response of Crtc1‒/‒ mice observed in the NIH might be due to the anxiolytic effects of these treatments. The fact that these drugs still have anxiolytic effect in the absence of CRTC1 suggests that these effects are either CRTC1-independent, or that alternative pathways can be used to counteract the absence of CRTC1.
Gene expression analyses revealed that desipramine induced the expression of Bdnf, BdnfIV, Nr4a1, and Nr4a2 in the HIP and PFC of WT mice, but failed to do so in Crtc1‒/‒ mice. While desipramine has been known for a long time to induce the expression of Bdnf (Nibuya et al., 1995), we report here that it also induces the expression of Nr4a1 and Nr4a2, which have been linked with mood disorders and Bdnf regulation (Buervenich et al., 2000; Volpicelli et al., 2007; Xing et al., 2006). Furthermore, we show that CRTC1 is required for the induction of Nr4a1 and Nr4a2 by chronic desipramine, thus suggesting a role for these nuclear receptors in the behavioral response to antidepressants.
Considering this hypothesis, we attempted to counteract CRTC1 deletion and restore normal gene expression in Crtc1‒/‒ mice by treating them with a HDAC inhibitor. Epigenetic gene regulation has been widely shown to be involved in mood disorders. Mechanisms such as DNA methylation and histone acetylation (mechanisms that respectively repress and activate gene transcription) were found to be altered in depressed patients and in response to chronic stress (Sun et al., 2013; Vialou et al., 2013). Antidepressants also induce epigenetic changes, such as histone H3 hyperacetylation (Tsankova et al., 2006). HDAC inhibitors were found to have antidepressant effects in rodents, and to regulate a subset of genes in a similar way as antidepressants drugs (Covington et al., 2009; Schroeder et al., 2007; Uchida et al., 2011). Of particular interest for this study, Bdnf and Nr4a1-3 have been shown to be induced by HDAC inhibitors (Hawk et al., 2012; Schroeder et al., 2007; Vecsey et al., 2007). In the light of these results, we injected intraperitoneally the HDAC inhibitor SAHA to WT and Crtc1‒/‒ mice, and confirmed that a systemic administration of SAHA increased histone acetylation in the brain. Mutant mice responded to chronic SAHA in the OSFS paradigm, as their immobility time decreased over time, indicating a successful antidepressant effect of SAHA in these animals. However, the effect was not complete, as treated Crtc1‒/‒ mice did not reach the level of WT animals. This suggests that SAHA was not able to completely rescue the depressive-like behavior of these animals. It is important to keep in mind that Crtc1‒/‒ mice are complete knock-out animal, therefore developmental effects of CRTC1 deficiency cannot be excluded. Thus, some behavioral and molecular impairment might not be completely reversible, and might explain the partial behavioral effects of SAHA. Nevertheless, this result indicates that the depressive-like behavior of Crtc1‒/‒ mice can be partially rescued by acting at the epigenetic level.
In the NIH, the effect of SAHA was less clear. While it reduced the latency of WT mice, it only produced a trend to a decreased latency in Crtc1‒/‒ mice. However, basal latency of vehicle-treated WT animals was abnormally high, which might indicate that the animals had higher basal anxiety, as compared with the NIH experiment of Fig. 1C. This might be explained by a possible effect of the three-week stressful OSFS procedure combined with chronic injections. All in all, NIH results suggest that SAHA had no effect on the anxiety of Crtc1‒/‒ mice, yet as WT animals seemed to present unusual higher anxiety, results of this test should be taken with caution.
Molecular investigation of SAHA effects allowed us to better understand the behavioral effects of this treatment on Crtc1‒/‒ mice. In the PFC, the decreased Bdnf and BdnfIV expression was successfully rescued by SAHA treatment. This rescue of Bdnf expression may be related to the decreased immobility of Crtc1‒/‒ mice in the OSFS, as BDNF alterations have often been linked with behavioral despair (Borsoi et al., 2014; Koponen et al., 2005; Shirayama et al., 2002; Siuciak et al., 1997). While no effect of SAHA was observed in the HIP, Crtc1‒/‒ mice elusively displayed higher Bdnf expression in this structure, instead of their usual lower levels. The comparison of Bdnf levels in control groups of WT and Crtc1‒/‒ mice of Figure 2 and 5 suggested that the stress generated by chronic daily vehicle injections decreased Bdnf expression only in WT and not in Crtc1‒/‒ mice (Fig. S1). While it is generally suggested that acute and chronic stress decrease Bdnf expression (Duman and Monteggia, 2006), several studies have shown opposite findings (Charrier et al., 2006; Marmigere et al., 2003; Nair et al., 2007), thus revealing a complex interplay between stress and BDNF. Crtc1‒/‒ mice might thus present an altered reactivity to stress, evidenced by differential Bdnf regulation. This highlights again the importance of CRTC1 in the regulation of Bdnf and stress response, and should be a focus for future investigations.
Interestingly, SAHA was found to increase Nr4a1-3 expression in the HIP and/or PFC of WT mice, which could be correlated with its antidepressant effect in these mice. This is in line with previous studies showing that HDAC inhibitors increase Nr4a1-3 expression via the CREB-CBP pathway, which is a key mechanism for memory enhancement (Hawk et al., 2012; Vecsey et al., 2007). However, SAHA treatment was not able to increase Nr4a1-3 expression in Crtc1‒/‒ mice, and therefore these genes are apparently not involved in the partial rescue of their depressive-like behavior. These results suggest that CRTC1 is required for the CREB-CBP-mediated Nr4a1-3 transcription induced by HDAC inhibitors. As this pathway is involved in learning processes, these findings are in keeping with the recently characterized important role of CRTC1 in memory and cognitive behavior (Nonaka et al., 2014a; Nonaka et al., 2014b; Parra-Damas et al., 2014). Further characterization of cognitive and learning behaviors of Crtc1‒/‒ mice should better characterize CRTC1 function in these processes.
5. Conclusions
In this study, we provide evidence that CRTC1 is a key regulator of behavioral and molecular response to antidepressant. We show that CRTC1 is required for complete behavioral antidepressant response and for desipramine-induced expression of Bdnf and Nr4a1-2. By acting downstream of CRTC1, the HDAC inhibitor SAHA improves the depressive-like behavior of Crtc1‒/‒ mice. This partial rescue was paralleled by a restoration of Bdnf expression in the PFC, whereas SAHA was unable to increase Nr4a1-3 expression in these mice, thus suggesting that these orphan nuclear receptors do not play a major role in the improvement of their depressive-like behavior. In conclusion, several lines of evidence strongly suggest that CRTC1-deficient mice are a valuable animal model of depressive phenotypes and treatment resistance. As an important regulatory mediator of depression-related genes, CRTC1 provide a new focus for future research aiming at a better understanding of depression and new therapeutic approaches.
Supplementary Material
Acknowledgements
This work was funded by a grant from the Swiss National Science Foundation (grant number 31003A-135692), and partly supported by the National Centre of Competence in Research (NCCR) Synapsy.
Footnotes
Chemical compounds studied in this article
Desipramine hydrochloride (PubChem CID: 65327); Vorinostat (PubChem CID: 5311)
References
- Altarejos JY, Montminy M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat Rev Mol Cell Biol. 2011;12:141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blendy JA. The role of CREB in depression and antidepressant treatment. Biol Psychiatry. 2006;59:1144–1150. doi: 10.1016/j.biopsych.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Borsoi M, Antonio CB, Viana AF, Nardin P, Goncalves C, Rates SM. Immobility behavior during the forced swim test correlates with BNDF levels in the frontal cortex, but not with cognitive impairments. Physiol Behav. 2014;140C:79–88. doi: 10.1016/j.physbeh.2014.12.024. [DOI] [PubMed] [Google Scholar]
- Breuillaud L, Halfon O, Magistretti PJ, Pralong FP, Cardinaux JR. Mouse fertility is not dependent on the CREB coactivator Crtc1. Nat Med. 2009;15:989–990. doi: 10.1038/nm0909-989. author reply 991. [DOI] [PubMed] [Google Scholar]
- Breuillaud L, Rossetti C, Meylan EM, Merinat C, Halfon O, Magistretti PJ, Cardinaux JR. Deletion of CREB-regulated transcription coactivator 1 induces pathological aggression, depression-related behaviors, and neuroplasticity genes dysregulation in mice. Biol Psychiatry. 2012;72:528–536. doi: 10.1016/j.biopsych.2012.04.011. [DOI] [PubMed] [Google Scholar]
- Buervenich S, Carmine A, Arvidsson M, Xiang F, Zhang Z, Sydow O, Jonsson EG, Sedvall GC, Leonard S, Ross RG, Freedman R, et al. NURR1 mutations in cases of schizophrenia and manic-depressive disorder. Am J Med Genet. 2000;96:808–813. doi: 10.1002/1096-8628(20001204)96:6<808::aid-ajmg23>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Carlezon WA, Jr, Duman RS, Nestler EJ. The many faces of CREB. Trends Neurosci. 2005;28:436–445. doi: 10.1016/j.tins.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Ch'ng TH, Uzgil B, Lin P, Avliyakulov NK, O'Dell TJ, Martin KC. Activity-dependent transport of the transcriptional coactivator CRTC1 from synapse to nucleus. Cell. 2012;150:207–221. doi: 10.1016/j.cell.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier C, Chigr F, Tardivel C, Mahaut S, Jean A, Najimi M, Moyse E. BDNF regulation in the rat dorsal vagal complex during stress-induced anorexia. Brain Res. 2006;1107:52–57. doi: 10.1016/j.brainres.2006.05.099. [DOI] [PubMed] [Google Scholar]
- Chen AC, Shirayama Y, Shin KH, Neve RL, Duman RS. Expression of the cAMP response element binding protein (CREB) in hippocampus produces an antidepressant effect. Biol Psychiatry. 2001;49:753–762. doi: 10.1016/s0006-3223(00)01114-8. [DOI] [PubMed] [Google Scholar]
- Conkright MD, Canettieri G, Screaton R, Guzman E, Miraglia L, Hogenesch JB, Montminy M. TORCs: transducers of regulated CREB activity. Mol Cell. 2003;12:413–423. doi: 10.1016/j.molcel.2003.08.013. [DOI] [PubMed] [Google Scholar]
- Conti AC, Cryan JF, Dalvi A, Lucki I, Blendy JA. cAMP response element-binding protein is essential for the upregulation of brain-derived neurotrophic factor transcription, but not the behavioral or endocrine responses to antidepressant drugs. J Neurosci. 2002;22:3262–3268. doi: 10.1523/JNEUROSCI.22-08-03262.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington HE, 3rd, Maze I, LaPlant QC, Vialou VF, Ohnishi YN, Berton O, Fass DM, Renthal W, Rush AJ, 3rd, Wu EY, Ghose S, et al. Antidepressant actions of histone deacetylase inhibitors. J Neurosci. 2009;29:11451–11460. doi: 10.1523/JNEUROSCI.1758-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulawa SC, Hen R. Recent advances in animal models of chronic antidepressant effects: the novelty-induced hypophagia test. Neurosci Biobehav Rev. 2005;29:771–783. doi: 10.1016/j.neubiorev.2005.03.017. [DOI] [PubMed] [Google Scholar]
- Duman RS, Monteggia LM. A neurotrophic model for stress-related mood disorders. Biol Psychiatry. 2006;59:1116–1127. doi: 10.1016/j.biopsych.2006.02.013. [DOI] [PubMed] [Google Scholar]
- Finsterwald C, Fiumelli H, Cardinaux JR, Martin JL. Regulation of dendritic development by BDNF requires activation of CRTC1 by glutamate. J Biol Chem. 2010;285:28587–28595. doi: 10.1074/jbc.M110.125740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gräff J, Rei D, Guan JS, Wang WY, Seo J, Hennig KM, Nieland TJ, Fass DM, Kao PF, Kahn M, Su SC, et al. An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature. 2012;483:222–226. doi: 10.1038/nature10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gräff J, Tsai LH. Histone acetylation: molecular mnemonics on the chromatin. Nat Rev Neurosci. 2013;14:97–111. doi: 10.1038/nrn3427. [DOI] [PubMed] [Google Scholar]
- Han A, Sung YB, Chung SY, Kwon MS. Possible additional antidepressant-like mechanism of sodium butyrate: targeting the hippocampus. Neuropharmacology. 2014;81:292–302. doi: 10.1016/j.neuropharm.2014.02.017. [DOI] [PubMed] [Google Scholar]
- Hawk JD, Bookout AL, Poplawski SG, Bridi M, Rao AJ, Sulewski ME, Kroener BT, Manglesdorf DJ, Abel T. NR4A nuclear receptors support memory enhancement by histone deacetylase inhibitors. J Clin Invest. 2012;122:3593–3602. doi: 10.1172/JCI64145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockly E, Richon VM, Woodman B, Smith DL, Zhou X, Rosa E, Sathasivam K, Ghazi-Noori S, Mahal A, Lowden PA, Steffan JS, et al. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington's disease. Proc Natl Acad Sci U S A. 2003;100:2041–2046. doi: 10.1073/pnas.0437870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler RC, Berglund P, Demler O, Jin R, Merikangas KR, Walters EE. Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry. 2005;62:593–602. doi: 10.1001/archpsyc.62.6.593. [DOI] [PubMed] [Google Scholar]
- Koponen E, Rantamaki T, Voikar V, Saarelainen T, MacDonald E, Castren E. Enhanced BDNF signaling is associated with an antidepressant-like behavioral response and changes in brain monoamines. Cell Mol Neurobiol. 2005;25:973–980. doi: 10.1007/s10571-005-8468-z. [DOI] [PubMed] [Google Scholar]
- Koppel I, Timmusk T. Differential regulation of Bdnf expression in cortical neurons by class-selective histone deacetylase inhibitors. Neuropharmacology. 2013;75:106–115. doi: 10.1016/j.neuropharm.2013.07.015. [DOI] [PubMed] [Google Scholar]
- Kovacs KA, Steullet P, Steinmann M, Do KQ, Magistretti PJ, Halfon O, Cardinaux JR. TORC1 is a calcium- and cAMP-sensitive coincidence detector involved in hippocampal long-term synaptic plasticity. Proc Natl Acad Sci U S A. 2007;104:4700–4705. doi: 10.1073/pnas.0607524104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan V, Nestler EJ. The molecular neurobiology of depression. Nature. 2008;455:894–902. doi: 10.1038/nature07455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zhang C, Takemori H, Zhou Y, Xiong ZQ. TORC1 regulates activity-dependent CREB-target gene transcription and dendritic growth of developing cortical neurons. J Neurosci. 2009;29:2334–2343. doi: 10.1523/JNEUROSCI.2296-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Geng X, Dang W, Wu B, Dai Z, Li Y, Yang Y, Zhang H, Shi J. Molecular mechanisms associated with the antidepressant effects of the class I histone deacetylase inhibitor MS-275 in the rat ventrolateral orbital cortex. Brain Res. 2012;1447:119–125. doi: 10.1016/j.brainres.2012.01.053. [DOI] [PubMed] [Google Scholar]
- Lonze BE, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35:605–623. doi: 10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- Marmigere F, Givalois L, Rage F, Arancibia S, Tapia-Arancibia L. Rapid induction of BDNF expression in the hippocampus during immobilization stress challenge in adult rats. Hippocampus. 2003;13:646–655. doi: 10.1002/hipo.10109. [DOI] [PubMed] [Google Scholar]
- Martinowich K, Lu B. Interaction between BDNF and serotonin: role in mood disorders. Neuropsychopharmacology. 2008;33:73–83. doi: 10.1038/sj.npp.1301571. [DOI] [PubMed] [Google Scholar]
- Martinowich K, Manji H, Lu B. New insights into BDNF function in depression and anxiety. Nat Neurosci. 2007;10:1089–1093. doi: 10.1038/nn1971. [DOI] [PubMed] [Google Scholar]
- Morris MJ, Monteggia LM. Unique functional roles for class I and class II histone deacetylases in central nervous system development and function. Int J Dev Neurosci. 2013;31:370–381. doi: 10.1016/j.ijdevneu.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair A, Vadodaria KC, Banerjee SB, Benekareddy M, Dias BG, Duman RS, Vaidya VA. Stressor-specific regulation of distinct brain-derived neurotrophic factor transcripts and cyclic AMP response element-binding protein expression in the postnatal and adult rat hippocampus. Neuropsychopharmacology. 2007;32:1504–1519. doi: 10.1038/sj.npp.1301276. [DOI] [PubMed] [Google Scholar]
- Nibuya M, Morinobu S, Duman RS. Regulation of BDNF and trkB mRNA in rat brain by chronic electroconvulsive seizure and antidepressant drug treatments. J Neurosci. 1995;15:7539–7547. doi: 10.1523/JNEUROSCI.15-11-07539.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka M, Kim R, Fukushima H, Sasaki K, Suzuki K, Okamura M, Ishii Y, Kawashima T, Kamijo S, Takemoto-Kimura S, Okuno H, et al. Region-specific activation of CRTC1-CREB signaling mediates long-term fear memory. Neuron. 2014a;84:92–106. doi: 10.1016/j.neuron.2014.08.049. [DOI] [PubMed] [Google Scholar]
- Nonaka M, Kim R, Sharry S, Matsushima A, Takemoto-Kimura S, Bito H. Towards a better understanding of cognitive behaviors regulated by gene expression downstream of activity-dependent transcription factors. Neurobiol Learn Mem. 2014b;115C:21–29. doi: 10.1016/j.nlm.2014.08.010. [DOI] [PubMed] [Google Scholar]
- Parra-Damas A, Valero J, Chen M, Espana J, Martin E, Ferrer I, Rodriguez-Alvarez J, Saura CA. Crtc1 activates a transcriptional program deregulated at early Alzheimer's disease-related stages. J Neurosci. 2014;34:5776–5787. doi: 10.1523/JNEUROSCI.5288-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravnskjaer K, Kester H, Liu Y, Zhang X, Lee D, Yates JR, 3rd, Montminy M. Cooperative interactions between CBP and TORC2 confer selectivity to CREB target gene expression. EMBO J. 2007;26:2880–2889. doi: 10.1038/sj.emboj.7601715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer DJ, Tunc-Ozcan E, Shukla PK, Volenec A, Redei EE. Nuclear orphan receptor Nor-1 contributes to depressive behavior in the Wistar-Kyoto rat model of depression. Brain Res. 2010;1362:32–39. doi: 10.1016/j.brainres.2010.09.041. [DOI] [PubMed] [Google Scholar]
- Schroeder FA, Lin CL, Crusio WE, Akbarian S. Antidepressant-like effects of the histone deacetylase inhibitor, sodium butyrate, in the mouse. Biol Psychiatry. 2007;62:55–64. doi: 10.1016/j.biopsych.2006.06.036. [DOI] [PubMed] [Google Scholar]
- Screaton RA, Conkright MD, Katoh Y, Best JL, Canettieri G, Jeffries S, Guzman E, Niessen S, Yates JR, 3rd, Takemori H, Okamoto M, et al. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell. 2004;119:61–74. doi: 10.1016/j.cell.2004.09.015. [DOI] [PubMed] [Google Scholar]
- Sekeres MJ, Mercaldo V, Richards B, Sargin D, Mahadevan V, Woodin MA, Frankland PW, Josselyn SA. Increasing CRTC1 function in the dentate gyrus during memory formation or reactivation increases memory strength without compromising memory quality. J Neurosci. 2012;32:17857–17868. doi: 10.1523/JNEUROSCI.1419-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama Y, Chen AC, Nakagawa S, Russell DS, Duman RS. Brain-derived neurotrophic factor produces antidepressant effects in behavioral models of depression. J Neurosci. 2002;22:3251–3261. doi: 10.1523/JNEUROSCI.22-08-03251.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siuciak JA, Lewis DR, Wiegand SJ, Lindsay RM. Antidepressant-like effect of brain-derived neurotrophic factor (BDNF) Pharmacol Biochem Behav. 1997;56:131–137. doi: 10.1016/S0091-3057(96)00169-4. [DOI] [PubMed] [Google Scholar]
- Smith MA, Makino S, Kvetnansky R, Post RM. Stress and glucocorticoids affect the expression of brain-derived neurotrophic factor and neurotrophin-3 mRNAs in the hippocampus. J Neurosci. 1995;15:1768–1777. doi: 10.1523/JNEUROSCI.15-03-01768.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone EA, Lin Y. Open-space forced swim model of depression for mice. Curr Protoc Neurosci. 2011 doi: 10.1002/0471142301.ns0936s54. Chapter 9, Unit9 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone EA, Lin Y, Quartermain D. Evaluation of the repeated open-space swim model of depression in the mouse. Pharmacol Biochem Behav. 2008;91:190–195. doi: 10.1016/j.pbb.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Kennedy PJ, Nestler EJ. Epigenetics of the depressed brain: role of histone acetylation and methylation. Neuropsychopharmacology. 2013;38:124–137. doi: 10.1038/npp.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankova N, Renthal W, Kumar A, Nestler EJ. Epigenetic regulation in psychiatric disorders. Nat Rev Neurosci. 2007;8:355–367. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- Tsankova NM, Berton O, Renthal W, Kumar A, Neve RL, Nestler EJ. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci. 2006;9:519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- Uchida S, Hara K, Kobayashi A, Otsuki K, Yamagata H, Hobara T, Suzuki T, Miyata N, Watanabe Y. Epigenetic status of Gdnf in the ventral striatum determines susceptibility and adaptation to daily stressful events. Neuron. 2011;69:359–372. doi: 10.1016/j.neuron.2010.12.023. [DOI] [PubMed] [Google Scholar]
- Vecsey CG, Hawk JD, Lattal KM, Stein JM, Fabian SA, Attner MA, Cabrera SM, McDonough CB, Brindle PK, Abel T, Wood MA. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. J Neurosci. 2007;27:6128–6140. doi: 10.1523/JNEUROSCI.0296-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vialou V, Feng J, Robison AJ, Nestler EJ. Epigenetic mechanisms of depression and antidepressant action. Annu Rev Pharmacol Toxicol. 2013;53:59–87. doi: 10.1146/annurev-pharmtox-010611-134540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volakakis N, Kadkhodaei B, Joodmardi E, Wallis K, Panman L, Silvaggi J, Spiegelman BM, Perlmann T. NR4A orphan nuclear receptors as mediators of CREB-dependent neuroprotection. Proc Natl Acad Sci U S A. 2010;107:12317–12322. doi: 10.1073/pnas.1007088107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpicelli F, Caiazzo M, Greco D, Consales C, Leone L, Perrone-Capano C, Colucci D'Amato L, di Porzio U. Bdnf gene is a downstream target of Nurr1 transcription factor in rat midbrain neurons in vitro. J Neurochem. 2007;102:441–453. doi: 10.1111/j.1471-4159.2007.04494.x. [DOI] [PubMed] [Google Scholar]
- Xing G, Zhang L, Russell S, Post R. Reduction of dopamine-related transcription factors Nurr1 and NGFI-B in the prefrontal cortex in schizophrenia and bipolar disorders. Schizophr Res. 2006;84:36–56. doi: 10.1016/j.schres.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Wu H, Li S, Chen Q, Cheng XW, Zheng J, Takemori H, Xiong ZQ. Requirement of TORC1 for late-phase long-term potentiation in the hippocampus. PLoS One. 2006;1:e16. doi: 10.1371/journal.pone.0000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohar J, Westenberg HG. Anxiety disorders: a review of tricyclic antidepressants and selective serotonin reuptake inhibitors. Acta Psychiatr Scand Suppl. 2000;403:39–49. doi: 10.1111/j.1600-0447.2000.tb10947.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.