SUMMARY
Diet has a profound effect on tissue regeneration in diverse organisms, and low caloric states such as intermittent fasting have beneficial effects on organismal health and age-associated loss of tissue function. The role of adult stem and progenitor cells in responding to short-term fasting, and whether such responses improve regeneration, are not well-studied. Here, we show that a 24-hour fast augments intestinal stem cell (ISC) function in young and aged mice by inducing a fatty acid oxidation (FAO) program, and that pharmacological activation of this program mimics many effects of fasting. Acute genetic disruption of Cpt1a, the rate-limiting enzyme in FAO, abrogates ISC-enhancing effects of fasting but long-term Cpt1a deletion decreases ISC numbers and function, implicating a role for FAO in ISC maintenance. These findings highlight a role for FAO in mediating pro-regenerative effects of fasting in intestinal biology and may represent a viable strategy for enhancing intestinal regeneration.
TOC image
Mihaylova and Cheng et al. show that short-term fasting promotes intestinal stem and progenitor cell function in young and aged mice by inducing a robust fatty acid oxidation (FAO) program. PPARδ agonists emulate these effects, showing that fatty acid metabolism has positive effects on young and old ISCs.
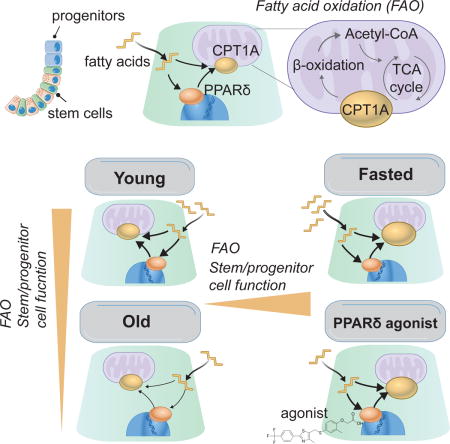
INTRODUCTION
Lgr5+ intestinal stem cells (ISCs) reside at the bottom of intestinal crypts adjacent to Paneth cells, which are a component of the stem cell niche, and remodel the intestinal epithelium in response to dietary signals (Barker et al., 2007; Sasaki et al., 2016; Sato et al., 2011; Yilmaz et al., 2012). Diet acts through cell autonomous and non-autonomous mechanisms to regulate the balance between ISC self-renewal and differentiation. For example, calorie restriction (CR) modulates ISC function through Paneth cell niche induced changes (Igarashi and Guarente, 2016; Yilmaz et al., 2012) whereas high fat diets (HFD) or cholesterol rich diets directly alter ISCs and progenitor function (Beyaz et al., 2016; Wang et al., 2018). Yet, to date, little is known about how ISCs and Paneth cells adapt to short-term fasts.
Acute fasting regimens have pro-longevity and regenerative effects in diverse species and may represent a dietary approach to enhance aged stem cell activity in tissues (Cheng et al., 2014; Longo and Mattson, 2014; Weindruch et al., 1986). Aging in lower organisms and mammals results in the attrition of stem cell numbers, function, or both in myriad tissues. Such age-related changes in stem cells are proposed to underlie some of the untoward consequences of organismal aging (Chandel et al., 2016; Jones and Rando, 2011; Li and Jasper, 2016; Lopez-Otin et al., 2013; Rafalski et al., 2012; Signer and Morrison, 2013). In Drosophila, for example, age-dependent dysregulation in intestinal stem cell homeostasis contributes to epithelial dysfunction (Biteau et al., 2008; Biteau et al., 2010; Choi et al., 2008; Haller et al., 2017; Resnik-Docampo et al., 2017). Similarly, the aging mammalian intestine undergoes diminishment of stem cell activity and impairment in its ability to repair itself after radiation-induced damage (Nalapareddy et al., 2017; Potten et al., 2001a) yet, little is understood about the mechanisms underlying these age-dependent changes. Finally, how old ISCs respond to a dietary changes and whether pharmacologic or dietary interventions can be leveraged to improve intestinal regeneration in old age requires investigation. Here, we interrogate how Lgr5+ ISCs in young and aged mice adapt to a short-term fast, a regimen in which no food is ingested for 24 hours (Goodrick et al., 1990).
RESULTS
Fasting increases crypt and ISC function
Short-term fasts improve adult stem cell function in diverse tissues (Cheng et al., 2014; Longo and Mattson, 2014; Mihaylova et al., 2014). To understand how fasting affects ISC function, we first assessed the potential of intestinal crypts to form multipotent, self-renewing mini-intestines (Beyaz et al., 2016; Sato et al., 2009). We assayed the organoid-forming capacities of crypts from young (3-4 months old) mice fed either standard chow ad libitum (AL) or fasted for 24 hours, an intervention where rodents lose 5-10% of their body mass (Figure S1A). Notably, fasting robustly boosted crypt organoid-forming capacity (Figure 1A) that correlated with the length of the fast (Figure S1B). To more precisely assess the effects of fasting on ISCs, we fasted the Lgr5-EGFP-IRES-CreERT2 reporter mice (hereinafter referred to as Lgr5-CreERT2), where the Lgr5+ ISCs are GFPhi and their more differentiated daughter progenitor cells are GFPlow and sorted ISCs for co-culture experiments with Paneth cells. Although fasting did not change the quantity of ISCs or progenitors (Figure S1C), fasting did enhance the organoid-forming capacity of ISCs alone or when co-cultured with AL Paneth cells, indicating that fasting intrinsically alters ISCs (Figure 1B). Interestingly, fasted Paneth cells co-cultured with AL ISCs (Figure S1D) did not augment the organoid-forming potential of ISCs, which is in contrast to what has been reported for CR Paneth cells (Igarashi and Guarente, 2016; Yilmaz et al., 2012). One possibility is that post-mitotic Paneth cells require prolonged exposure to low calorie states or mTOR inhibition to acquire the ability to boost ISC numbers, whereas proliferative ISCs dynamically adapt to acute fluctuations in nutrient availability. Overall, these findings demonstrate that a 24 hour fast has pro-regenerative effects on ISC function.
Figure 1. Fasting induces fatty acid oxidation and improves intestinal stem cell function.
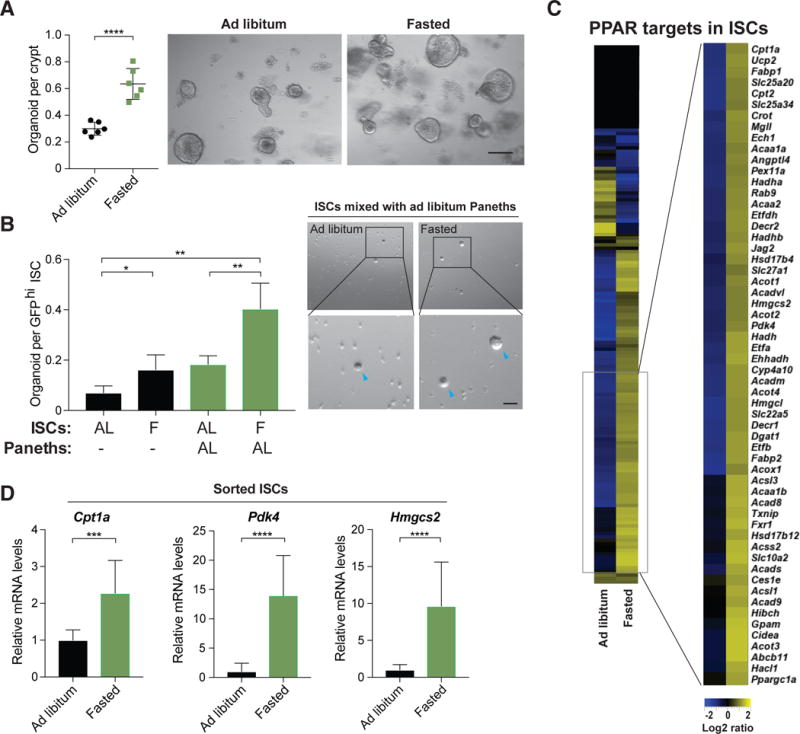
(A) Organoid frequency of crypts from 10-12 week old ad libitum or 24-hour fasted mice. Representative images of crypt culture from each condition. n=6 mice per group.
(B) ISCs from 24-hour fasted animals (F) had significantly enhanced organoid-forming capacity when mixed with age matched Paneth cells from ad libitum (AL) mice. Blue arrowheads indicate organoids. n=3 mice per group.
(C) Heat map of RNA-Seq analysis showing log2-transformed relative expression of PPAR target genes in ISCs (Lgr5-GFPhi) from AL or 24 hour fasted mice.
(D) qRT-PCR analysis confirming PPAR target gene induction in ISCs (Lgr5-GFPhi) following a 24 hour fast. β actin was used as a housekeeping gene. n=7 mice.
Data are mean ± s.d. *p < 0.05, **p < 0.01, ***p < 0.001, ****p<0.0005 by Student’s t-test, unpaired. Scale bars, 20 μm (A) and 100 μm (B). See also Figure S1.
Fasting evokes a FAO program in ISCs
To gain mechanistic insight into how fasting mediates its effects in ISCs, bulk-population RNA sequencing analysis on ISCs from AL and fasted mice was performed (Figure 1C). Gene set enrichment analysis (GSEA) identified enrichment of the nuclear receptor peroxisome proliferator-activated receptor (PPAR) family targets, including genes involved in fatty acid oxidation (FAO) (Figure S1E)(Barish et al., 2006; Beyaz et al., 2016; Cangelosi and Yilmaz, 2016). We validated induction of select PPAR targets such as Pdk4, Cpt1a, Hmgcs2 at the mRNA and protein levels in ISCs and crypts (Figures 1D and S1G-S1I). In addition, fasting increased circulating free fatty acids (FFAs) such as palmitate (PA), which serve as FAO substrates and correspond with more organoid activity (Figures S1B and S1F). Interestingly, fasted crypt-formed organoids retained higher CPT1A protein levels (Figure S1J) after 48 hours in nutrient-rich culture, indicating that the fasting response endures in vitro. Given the strong induction of Cpt1a in fasted ISCs (Figures 1D, S1G and S1H) and coordinated elevation of circulating FFA (Figure S1F), we turned our attention to understanding whether CPT1A-mediated FAO drives the ISC response in fasting.
FAO mediates the effects of fasting in ISCs
We utilized etomoxir, an irreversible inhibitor of CPT1A, in the organoid assay to understand the functional requirement of FAO in crypts from AL and fasted mice (Ito et al., 2012). Indeed, while etomoxir treatment had minimal impact on baseline organoid initiation, it abrogated the organoid-enhancing effects of fasting (Figure 2A). Moreover, primary untreated organoids from fasted hosts have more self-renewal capacity (i.e. give rise to more daughter organoids) in secondary cultures than organoids from AL controls (Figure 2A), which illustrates that fasting elevates stem cell activity in a FAO-dependent manner.
Figure 2. Acute ablation of CPT1A abrogates the fasting-induced increase in crypt organoid formation.
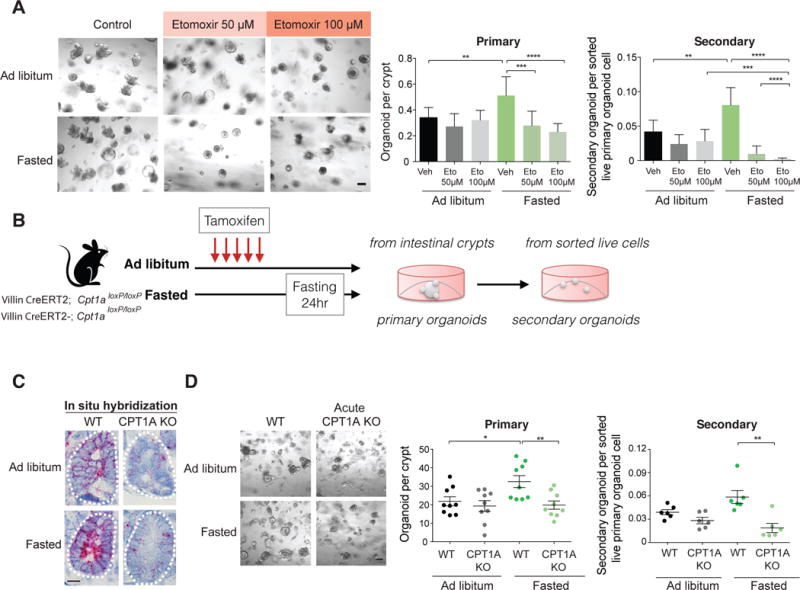
(A) Administration of etomoxir in culture blunted the effects of 24 hour fasting on organoid formation. Representative images: day 5 primary. n=3 mice.
(B) Schematic of acute CPT1A ablation in the intestine.
(C) Representative images of in situ hybridization of Cpt1a mRNA levels (red) in crypts of ad libitum or 24 hour fasted wild-type (WT) and CPT1A knockout (CPT1A KO) mice. See also Figures S2E-G. n=3 mice.
(D) Deletion of Cpt1a reduced primary and secondary organoid formation of crypts from 24 hour fasted mice. Representative images: day 3 primary organoids. n=3 mice.
Data are mean ± s.d. *p < 0.05, **p < 0.01, ***p < 0.001, ****p<0.0005 by Student’s t-test, unpaired. Scale bars, 20 μm (A, C and D). See also Figure S2.
Next, we sought to interrogate the in vivo requirement of FAO metabolism in AL and fasted ISCs in tamoxifen-inducible, intestinal-specific Villin-CreERT2; Cpt1aloxP/loxP conditional mice (Figure 2B) (el Marjou et al., 2004; Schoors et al., 2015). Acute ablation of Cpt1a in the intestine did not affect body mass and only slightly reduced small intestinal length and mass in fasted mice compared to AL controls (Figure S2B-D). Importantly, acute loss of Cpt1a (Figure 2B and C, Figure S2E-G) blocked the organoid-enhancing effects of fasting in primary and secondary cultures (Figure 2D). These observations highlight that fasting by shifting ISC metabolism towards more FAO stimulates regeneration.
To specifically decipher how Cpt1a influences Lgr5+ ISCs, we generated Lgr5-CreERT2; Rosa-LSL-LacZ; Cpt1aloxP/loxP mice. While tamoxifen treatment in these mice led to a reduction of CPT1A protein in Lgr5+ ISC (Figure S2H), this reduction did not perturb ISC numbers in vivo (Figure S2I). Similarly, in in vivo fate mapping experiments, Cpt1a-null ISCs had slightly less ability to generate LacZ+ progeny (Figure S2K) and had no impairment in initiating organoids (Figure S2J). Collectively, these data demonstrate that acute CPT1A loss is largely dispensable for Lgr5+ ISC maintenance but is necessary for fasting stimulated ISC activity.
ISCs depend on FAO for their long-term maintenance
We focused on the Villin-CreERT2; Cpt1aloxP/loxP model, where Cpt1a is strongly excised in all intestinal cells (Figure 3A) including ISCs, to distill the long-term consequences of Cpt1a loss. In contrast to 1 week of Cpt1a loss (Figure 2B-D, S2H-K), Cpt1a loss for 3 months (Figure 3A and B) reduced proliferation (Figure 3C), Lgr5+ ISC numbers (Figure 3D and Figure S3A) and crypt organoid-forming capacity (Figure 3E) that occurs independent of Cpt1a-null Paneth cells (Figure S3F) relative to controls. Also, there was a slight increase of crypt apoptotic bodies and caspase 3 positive cells in the knockout crypts (Figure S3B and S3C). Similar effects were noted when naïve organoids were passaged in the presence of etomoxir (Figure S2A), which did not further impair the organoid potential of Cpt1a-null crypts (Figure 3F). These changes occurred without influencing differentiation, intestinal length or body mass (Figure S3D and E). Lastly, chronic Cpt1a loss partially blunted the regenerative effects of fasting in crypts following ionizing radiation-induced damage (Figure S3G), indicating that fasting-stimulated FAO in ISCs and early progenitors contribute to repair after injury.
Figure 3. Long-term ablation of CPT1A diminishes ISC numbers and function.
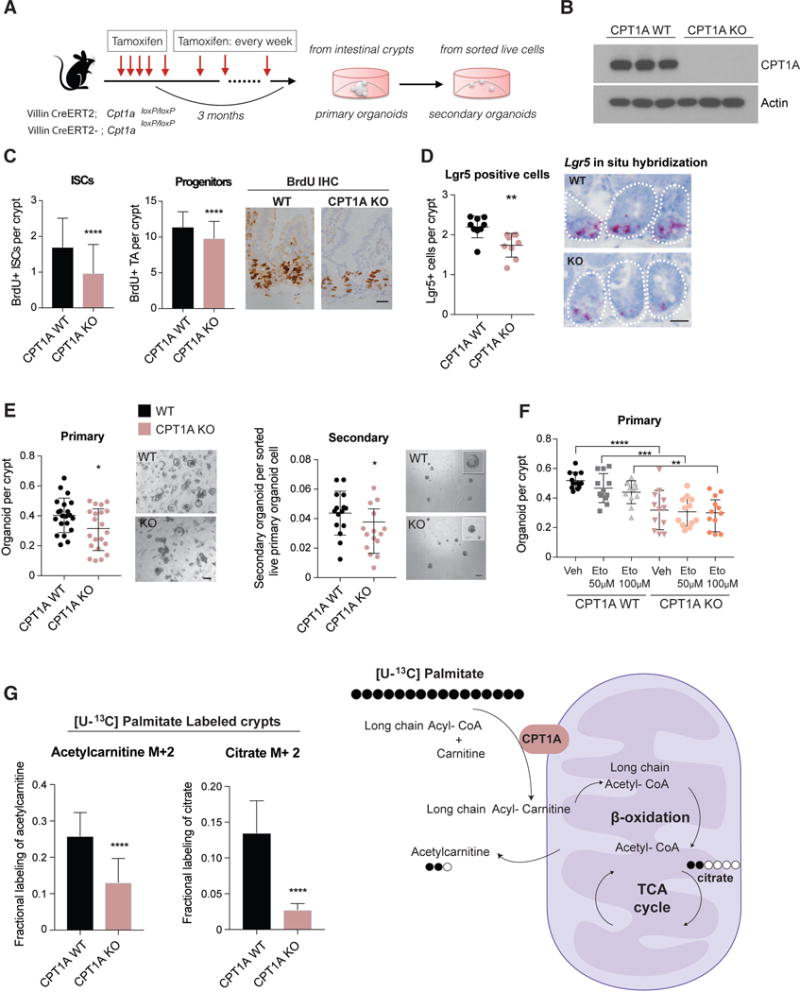
(A) Schematic of long-term (3 months) ablation of CPT1A in the intestine.
(B) Western blot analysis of CPT1A protein levels in intestinal crypts from WT and long-term deleted CPT1A KO mice.
(C) BrdU+ ISCs and progenitor cells in WT and CPT1A KO mice. (D) Long-term deletion of intestinal CPT1A decreased Lgr5+ stem cell numbers. n= 3 mice per group. Representative images of Lgr5+ cells by in situ hybridization (red).
(E) Long-term loss of CPT1A compromised organoid-forming capacity in primary and secondary cultures. n= 5 to 7 mice per group,
(F) In vitro etomoxir treatment did not further reduce the clonogenic ability of CPT1A KO crypts. n=4 mice.
(G) Long-term deletion of CPT1A significantly reduced the contribution of [U-13C] palmitate to acetylcarnitine and citrate. n=6 mice per group.
Data are mean ± s.d. *p < 0.05, **p < 0.01, ***p < 0.001, ****p<0.0005. Student’s t-test, unpaired. Scale bars, 50 μm (C and D), 20 μm primary and 100 μm secondary (E). Also see figure S3.
In labeled substrate metabolomics studies with [U-13C] palmitate, crypts lacking Cpt1a for 3 months had much lower fractional labeling of both M+2 acetylcarnitine (a proxy for acetyl-CoA derived from palmitate via FAO), (Kasumov et al., 2005; Li et al., 2015) and M+2 citrate compared to wild type crypts (Figure 3G and Figure S3H), while enrichment of [U-13C]-palmitate was similar between both (Figure S3I). Also, M+2 citrate labeling following tracing with [U-13C]-glucose was higher, indicating an increase in glucose oxidation in the absence of CPT1A (Figure S3J). As expected, a trend towards lower total acetylcarnitine levels and no change in total palmitate and citrate levels were noted (Figure S3K). Collectively, our data indicate that chronic disruption of FAO for 3 months through the loss of Cpt1a compromises intestinal stem and progenitor cell function and abrogates the pro-regenerative effects of fasting.
Age dependent decline of ISC numbers and function
Many recent studies have raised the possibility of exploiting fasting regimens to improve the function of aged tissues. To decipher whether a 24-hour fast can ameliorate some of the age-related decline in intestinal regeneration, we compared old mice of 17-24 months in age to young mice of 3-4 months in age, a range often used to study age-associated pathologies (Cerletti et al., 2012; Morrison et al., 1996). Aged mice have higher body mass (Figure S4A), a mild reduction in small intestinal length and no alteration in small intestinal mass compared to young controls (Figure S4B, C). Enumeration by flow cytometry revealed a significant, 62% reduction in Lgr5-GFPhi ISCs in aged mice and a trend towards fewer Lgr5-GFPlow progenitors (Figure 4A), while enteroendocrine or Goblet cell numbers within the crypt were unchanged and there was a slight regional increase in Paneth cell numbers (Figure S4D).
Figure 4. Physiological and pharmacological activation of PPAR delta and subsequent increase of FAO boost intestinal stem cell function in aged animals.
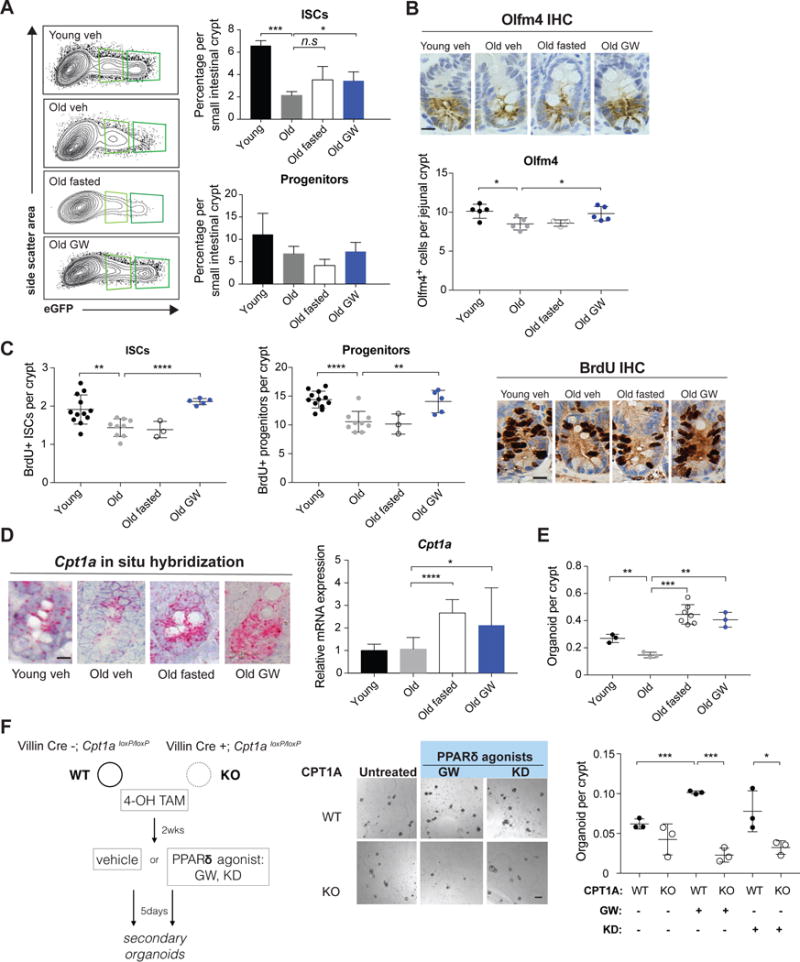
(A) FACS analysis of ISC (Lgr5-GFPhi) and progenitor cell (Lgr5-GFPlow) frequency in 4 month or 18-22 month old mice treated with either vehicle, GW501516 (GW, 3-4 weeks) or fasted for 24 hours. n=6 for young, n=7 for old, n=6 for old GW, n= 3 for 24 hour fasted mice.
(B) Quantification by IHC of Olfm4+ cells in young vehicle treated, old vehicle treated, old fasted and old GW treated animals. n=5 per group. (C) BrdU incorporation in ISCs and progenitors in young and aged animals following a 4-hour pulse . n=11 for young, n=9 for old, n= 3 for old fasted and n=5 for old GW treated mice.
(D) GW treatment and fasting strongly induce Cpt1a expression in aged intestinal stem and progenitor cells. Representative images of Cpt1a in situ hybridization (left) and qRT-PCR analysis of Cpt1a expression in sorted ISCs from corresponding treatments (right). n=12 for young, n=12 for old, n= 11 and n=8 for old GW treated mice.
(E) Intestinal crypts from aged animals have diminished capacity for organoid formation compared to young controls. GW treatment (3-4 weeks) and 24-hour fasting of old animals significantly boost organoid-forming capacity of crypts relative to controls. n= 3 independent experiments.
(F) Deletion of CPT1A prevented the organoid-enhancing effects of GW treatment. Cpt1aloxP/loxP (WT) and Villin-CreERT2; Cpt1aloxP/loxP (KO) organoids were treated with 4-OH tamoxifen in culture to delete CPT1A and subsequently treated with PPARδ agonists GW501516 (GW, 1 M) and KD3010 (KD, 3 M) or vehicle for 5 days. Equal number of live cells from the treated organoids were sorted to ascertain organoid initiation in secondary organoids.
Data are mean ± s.d. *p < 0.05, **p < 0.01, ***p < 0.001 by Student’s t-test, unpaired. Scale bars, 20 m (B, C and D) and 100 m (F). Also see figure S4.
Next, we asked whether ISC function declines in old age. To assess this, we co-cultured aged ISCs with either young or old Paneth cells and determined that aged ISCs have less clonogenic capacity compared to young ISCs (Figure S4E). Notably, aged Paneth cells had less capacity to boost aged ISC activity relative to young Paneth cells (Figure S4E), suggesting that aging has cell autonomous and non-autonomous (i.e. through Paneth cells) effects on ISCs. Consistent with a recent report (Nalapareddy et al., 2017), we found in in vivo fate mapping experiments that there was a significant decrease of labeled cells derived from Lgr5+ ISCs with aging, indicating slower turnover of the intestinal epithelium in elderly mice (Figure S4F). To further ascertain regenerative differences between young and aged ISCs, we lethally irradiated young and old mice and determined that at three days post irradiation there were a higher number of surviving crypts in young mice (Figure S4G). At this time point, there was no difference in the number of Ki67+ or cleaved caspase 3+ intestinal cells (Figure S4H). Together, these data demonstrate that aged ISCs have diminished regenerative capacity in the organoid assay, in in vivo fate mapping experiments, and in response to injury in vivo.
Fasting and agonist-enforced FAO improves ISC function in aged mice
Because fasting augments ISC function in young mice through a PPAR-FAO axis, we asked whether fasting or agonist-enforced FAO would also recapitulate these regenerative effects in aged ISCs (Barish et al., 2006; Beyaz et al., 2016. Fasting, as well as PPARδ activation with a small molecule agonist (GW501516, GW) that is a potent transcriptional PPARδ activator, induced intestinal FAO program that includes elevation in Cpt1a mRNA and protein levels (Figure 4D and Figure S4I and J) (Beyaz et al., 2016). GW treatment for 3-4 weeks, but not fasting, in old mice elevated Lgr5+ and Olfm4+ ISC/early progenitor numbers per crypt (Figure 4A and B) and proliferation (Figure 4C). However, both fasting and GW boosted aged crypt function in the organoid assay when compared to vehicle controls (Figure 4E). Finally, fasting in aged mice, compared to AL controls, augmented the number of labeled progeny in in vivo fate mapping experiments, indicating that fasting increases in vivo ISC and progenitor activity in elderly mice (Figure S4F). These results suggest that while both pharmacologic and fasting mediated FAO enhance the functional quality of ISCs, a short-term engagement of this PPARδ−CPT1a program with a 24 hour fast may not be a sufficient time period for ISCs to accumulate.
We next asked whether decreased basal FAO metabolism in aged ISCs correlates with their functional deficit in the organoid assay. To address this possibility, we labeled young and old crypts from mice treated with either vehicle or GW with [U-13C] palmitate for one hour and measured the generation of [U-13C]-labeled acetylcarnitine (M+2)(Kasumov et al., 2005; Kerner et al., 2014; Ventura et al., 1999). Interestingly, we consistently observed a mild but significant reduction in the fraction of [U13C]-labeled (M+2) acetylcarnitine in old crypts relative to young, indicating impairment of basal FAO in aged crypt cells (Figure S4K). GW treatment caused a ~30% increase in M+2 acetylcarnitine in young and old crypts. Finally, deletion of Cpt1a blocked the effects of GW or KD3010 (KD, an independent PPARδ agonist) treatment on organoids (Figure 4F), confirming that enforced PPARδ signaling enhances crypt function through FAO metabolism.
To decipher whether an exogenous substrate of FAO such as palmitic acid (PA) are affected by aging we tested the fasted plasma from young and old mice and determined that aged mice had lower fasting palmitate levels (Figure S4L). We next asked whether addition of exogenous PA to organoid cultures can recapitulate aspects of fasting on organoid self-renewal. We exposed young and old organoids to crypt media supplemented with PA for 1 week and consistent with fasting, PA dramatically improved primary organoid formation from both young and old mice compared to controls (Figure S4M). Moreover, primary organoids treated with PA gave rise to more secondary organoids, demonstrating that FAO substrates also boost ISC activity (Figure S4M). Thus, fasting through coupled intrinsic activation of an FAO stem cell program and an extrinsic increase in FAO substrates contributes to more ISC and progenitor activity.
DISCUSSION
It has long been appreciated that fasting has a profound impact on aging and tissue homeostasis (Mihaylova et al., 2014). Our data illustrate that a 24-hour fast augments ISC function through the activation of FAO, which subsequently improves ISC activity in young and aged mice. Fasting increases FAO in ISCs by driving both a robust PPAR-mediated FAO program in ISCs and by increasing circulating levels of triglycerides and free fatty acids (FFA) that can be then used by cells to generate acetyl-CoA for energy.
Although FAO is critical for tissues with high-energy needs like skeletal and cardiac muscle, little is known about the role of FAO in ISC biology. An important question is how does increased FAO boost ISC function. It was previously shown that FAO maintains the young hematopoietic stem cell pool through control of asymmetric divisions in vitro but this notion was not interrogated in vivo (Ito et al., 2012). Two recent studies demonstrated that limiting pyruvate oxidation enhances intestinal and hair follicle stem cells, raising the possibility that a switch to FAO may underlie some of these effects (Flores et al., 2017; Schell et al., 2017). Our data indicate that aged ISCs have a reduced capacity to utilize lipids for FAO (Figure S4K). Consistent with this notion, aging has been associated with impaired mitochondrial metabolism and FAO in a number of tissues (Houtkooper et al., 2011; Nguyen et al., 2013; Sengupta et al., 2010). Because addition of PA or induction of FAO with PPARδ agonists largely restores aged ISC function in the organoid assay, one possibility is that ISCs rely on FAO basally and a shortage in cellular energy hampers old ISC activity. For example, in contrast to acute Cpt1a loss, chronic Cpt1a loss reduces Lgr5+ ISCs numbers, function and proliferation (Figures 3C-F), highlighting a role for FAO metabolism in long-term ISC maintenance. A recent report (Richmond et al., 2015) raises the possibility that fasting, in a PTEN-dependent manner, permits dormant 4+ ISCs (i.e. Telomerase or Terthi cells) to contribute to intestinal turnover in the refed state. Fasting may enhance not only the function of Lgr5+ ISCs but also of dormant 4+ ISCs or the plasticity of differentiated progenitors, which collectively conspire to drive regeneration after injury (Asfaha et al., 2015; van Es et al., 2012; Yan et al., 2012).
We recently demonstrated that a chronic HFD, similar to fasting, augments ISC function through PPARδ signaling (Beyaz et al., 2016). Although fasting and HFD feeding are different in terms of overall calorie intake, both interventions actuate a PPARδ-mediated FAO program and increase circulating plasma FFAs. But, unlike in a chronic HFD, where the ISC are exposed to an overabundance of FFAs, in the fasted low energy state, ISCs need to scavenge energy from limiting amounts of FFAs for their maintenance. Whether FAO metabolism is necessary for the HFD ISC phenotype as it is in fasting remains to be determined.
Aging is a complex process comprised of a number of changes at the cellular and organismal level (Lopez-Otin et al., 2013). Our data indicate that aging alters the biology of Lgr5+ intestinal stem and niche cells, illustrating ISC aging reflects cell autonomous and non-autonomous alterations. Although previous studies have shown compromised regeneration and ISC function in old age (Kozar et al., 2013; Nalapareddy et al., 2017; Potten et al., 2001b), Kozar et al. through the use of an unbiased clonal labeling system reported that the number of functional stem cells does not decline with age. Given the extent of progenitor plasticity in the intestine (Buczacki et al., 2013; Tetteh et al., 2016; van Es et al., 2012), it is unclear whether the clonal activity noted by Kozar et al. (Kozar et al., 2013) in old age reflects not only Lgr5+ ISCs but also compensation from progenitors that may serve as facultative stem cells as Lgr5+ ISCs age. Here, we provide a dietary strategy that may improve baseline Lgr5+ ISC function, intestinal regeneration, as well as intestinal repair after injury in the elderly (Tinkum et al., 2015). Future studies will need to examine if supplementation of FAO substrates like lipids or FAO agonists may benefit patients afflicted with intestinal damage, particularly in old age.
STAR METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| rat anti-BrdU | Abcam | 6326 |
| rabbit monoclonal OLFM4 | gift from CST | clone PP7 |
| rabbit polyclonal lysozyme | Thermo | RB-372-A1 |
| rabbit chromogranin A | Abcam | 15160 |
| rabbit cleaved Caspase-3 | CST | 9664 |
| rabbit polyclonal Anti-RFP | Rockland | 600-401-379 |
| biotin-conjugated secondary donkey anti-rabbit | ImmunoResearch | 711-065-152 |
| biotin-conjugated secondary goat anti-rat | ImmunoResearch | 112-065-167 |
| anti-Cpt1a | Abcam | 128568 |
| anti-HMGCS2 | Sigma | AV41562 |
| anti- alpha tubulin | Santa Cruz | 8035 |
| anti- total S6 | CST | 2217 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Advanced DMEM | Gibco | 12491015 |
| DMEM/F12 | Gibco | 11320033 |
| SMEM | Life Technologies | 11380-037 |
| Murine EGF | PeproTech | 315-09 |
| Recombinant Murine Noggin | PeproTech | 250-38 |
| R-spondin | Sino Biological | 50316-M08S |
| N-acetyl-L-cysteine | Sigma-Aldrich | A9165 |
| N2 | Life Technologies | 17502001 |
| B27 | Life Technologies | 17504044 |
| Chir99021 | LC Laboratories | 04-0004 |
| Y-27632 | ApexBio | A3008 |
| JAG-1 protein | AnaSpec | AS-61298 |
| Palmitic acid | Cayman Chemical Company | 10006627 |
| Etomoxir | Sigma | E1905 |
| GW501516 | LC Laboratories | G-4789 |
| KD3010 | Dr. Albert R. La Spada | |
| Tamoxifen | Sigma-Aldrich | T5648-1G |
| 4-Hydroxytamoxifen | Calbiochem | 579002 |
| 13C-Palmitate | Sigma | 605573 |
| 13C-Glucose | Sigma | 389374 |
| Sunflower Seed Oil | Spectrum | S1929 |
| Deposited Data | ||
| RNA sequencing data | GEO repository | GEO: GSE89568 |
| Experimental Models: Organisms/Strains | ||
| Cpt1aloxP/loxP | Dr. Peter Carmeliet | |
| Villin-CreERT2 | Dr. Sylvie Robine | |
| Lgr5-EGFP-IRES-CreERT2;Rosa26-lacZ | The Jackson Laboratory | 008875 |
| Lgr5 -IRES-CreERT2 | Dr. Hans Clevers | |
| tdTomatoLSL | The Jackson Laboratory | 007909 |
| Oligonucleotides | ||
| See Table S1 for Primers for Real-time PCR | Integrated DNA Technologies (IDT) | N/A |
| Software and Algorithms | ||
| FlowJo v10 | FlowJo LLC. | https://www.flowjo.com/ |
| GraphPad Prism 7 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| Qlucore Omics Explorer 3.2 | Qlucore | https://www.qlucore.com/ |
| Other | ||
| In situ hybridization probe for mouse Cpt1a Mm-Cpt1a |
Advanced Cell Diagnostics, Inc. | 443071 |
| In situ hybridization probe for mouse Hmgcs2 Mm-Hmgcs2 |
Advanced Cell Diagnostics, Inc. | 437141 |
| In situ hybridization probe for mouse Lgr5 Mm-Lgr5 |
Advanced Cell Diagnostics, Inc. | 312171 |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ömer H. Yilmaz (ohyilmaz@mit.edu (Ö.H.Y).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Mice were under the husbandry care of Department of Comparative Medicine in both Whitehead Institute for Biomedical Research and Koch Institute for Integrative Cancer Research. The following strains were obtained from the Jackson Laboratory: Lgr5-EGFP-IRES-CreERT2 (strain name: B6.129P2-Lgr5tm1(cre/ERT2)Cle/J, stock number 008875), Rosa26-lacZ (strainname: B6.129S4-Gt(ROSA)26Sortm1Sor/J, Cpt1aloxP/loxP mouse strain was a gift from Dr. Peter Carmeliet and has been previously described24. Villin-CreERT2 was a gift from Sylvie Robine and previously described (el Marjou et al., 2004). For short-term Cpt1a deletion, Villin-CreERT2; Cpt1aloxP/loxP mice were administered 5 intraperetoneal (i.p.) tamoxifen injection (10mg per ml sunflower seed oil, 2mg per 25g of body weight) on alternate days before fasting. Tissue was harvested 24 hours post fasting. Long-term deletion for this model was achieved by the same initial tamoxifen administration protocol and mice were subsequently injected once per week thereafter until sacrifice. For the experiments using Lgr5-EGFP-IRES-CreERT2; Cpt1aloxP/loxP model, deletion of Cpt1a was achieved by the same initial tamoxifen administration protocol but tissue was harvested within 24hours post final tamoxifen injection. For lineage tracing experiments, i) Lgr5 -IRES-CreERT2 (Huch et al., 2013) (a gift from Dr. Hans Clevers) were crossed to tdTomatoLSL strain (Jackson labs #007909) and ii) Cpt1aloxP/loxP were crossed to Lgr5-EGFP-IRES-CreERT2; Rosa26-lacZ strain. Mice were injected with a single dose of tamoxifen and analyzed three days later. For aging experiments and profiling, young mice were 3 to 4 months of age at time of experiments. Lgr5-EGFP-IRES-CreERT2 were aged in both facilities and used between 17 and 24 months. C57BL/6 aged mice were ordered from the National Institute of Aging starting at age 19 months and housed in the same facility as the rest of the mice used in this study. Fasting was achieved through food removal from mice at 9 AM and mice were sacrificed approximately 24 hours later. Ad libitum controls were sex and age matched. GW501516 was purchased from Enzo and reconstituted in DMSO at 10mM concentration and further diluted 1:10 in a solution 5% Tween80, 5% PEG400, 90% sterile water. Mice were injected daily for 3-4 weeks. Both female and male mice were used in these studies.
Crypt Isolation and culturing
Isolated crypts were counted and embedded in Matrigel™ (Corning 356231 growth factor reduced) at 5-10 crypts per l and cultured in a modified form of crypt culture medium as described previously6. Standard (Gibco, Advanced DMEM) or lipid restricted (Gibco, DMEM/F12) medium was supplemented by EGF 40 ng ml−1 (Peprotech), Noggin 200 ng ml−1 (Peprotech), R-spondin 500 ng ml−1 (R&D or Sino Biological), N-acetyl-L-cysteine 1 M (Sigma-Aldrich), N2 1X (Life Technologies), B27 1X (Life Technologies), Chiron 10 M (Stemgent), Y-27632 dihydrochloride monohydrate 10 M (Sigma-Aldrich). Intestinal crypts were cultured in above mentioned media in 25-30 L droplets of Matrigel™ and plated onto a flat bottom 48-well plate (Corning 3548) and allowed to solidify for 20-30 minutes in a 37°C incubator. Three hundred microlit ers of crypt culture medium was then overlaid onto the Matrigel™, changed every three days, and maintained at 37°C in fully humidified ch ambers containing 5% CO2. Clonogenicity (colony-forming efficiency) was calculated by plating 50–300 crypts and assessing organoid formation 3-7 days or as specified after initiation of cultures. Palmitic acid (30 M; Cayman Chemical Company 10006627 conjugated to BSA), etomoxir (50 M and 100 M; Sigma, E1905) were added after overnight incubation to cultures in lipid-restricted media. GW501516 (1 M; LC Laboratories, G-4789) and KD3010 (3 M; from Dr. Albert R. La Spada’s laboratory) was added to cultures in standard media. Organoids from Cpt1aloxP/loxP (WT) and Villin-CreERT2; Cpt1aloxP/loxP (KO) were treated with to 4-OH Tamoxifen (TAM) for 2 passages (2 weeks) and then treated with PPARδ agonists GW501516 (GW, 1 μM) and KD3010 (KD, 3 μM) or vehicle for 5 days.
Isolated ISCs or progenitor cells were centrifuged at 250g for 5 minutes, re-suspended in the appropriate volume of crypt culture medium and seeded onto 25-30 l Matrigel™ (Corning 356231 growth factor reduced) containing 1 μM JAG-1 protein (AnaSpec, AS-61298) in a flat bottom 48-well plate (Corning 3548). Alternatively, ISCs and Paneth cells were mixed after sorting in a 1:1 ratio, centrifuged, and then seeded onto Matrigel™. The Matrigel™ and cells were allowed to solidify before adding 300 l of crypt culture medium. The crypt media was changed every second or third day. Organoids were quantified on days 3 unless otherwise specified.
For secondary organoid assays, primary organoids were dissociated TrypLE Express (5min at 32°C), resuspended with cold SMEM (Life Technologies,11380-037), centrifuged (5 minutes at 250g) and then resuspended in cold SMEM with the viability dye 7-AAD. Live cells were sorted and seeded onto Matrigel™ as previously described, and cultured in the standard crypt media without additional pharmacological or metabolites treatments. Secondary organoids were enumerated on day 3, unless otherwise specified.
RT-PCR and In Situ Hybridization
25,000 cells were sorted into Tri Reagent (Life Technologies) and total RNA was isolated according to the manufacturer's instructions with following modification: the aqueous phase containing total RNA was purified using RNeasy plus kit (Qiagen). RNA was converted to cDNA with cDNA synthesis kit (Bio-Rad). qRT-PCR was performed with diluted cDNA (1:5) in 3 wells for each primer and SYBR green master mix (Bio-Rad) on Bio-Rad iCycler RT-PCR detection system. Primers used are described in Key Resource Table. The in situ hybridization probes used in this study are as follows: mouse Cpt1a Mm-Cpt1a (REF 443071), mouse Hmgcs2 Mm-Hmgcs2 (REF 437141) and mouse Lgr5 Mm-Lgr5 (REF 312171). Both sense and antisense probes were generated to ensure specificity by in vitro transcription using DIG RNA labeling mix (Roche) according to the manufacturer’s instructions and to previously published detailed methods (Gregorieff and Clevers, 2010; van der Flier et al., 2009). Single-molecule in situ hybridization was performed using Advanced Cell Diagnostics RNAscope 2.0 HD Detection Kit.
Immunohistochemistry and Immunoblotting
As previously described (Beyaz et al., 2016), tissues were fixed in 10% formalin, paraffin embedded and sectioned. Antigen retrieval was performed with Borg Decloaker RTU solution (Biocare Medical) in a pressurized Decloaking Chamber (Biocare Medical) for 3 minutes. Antibodies used for immunohistochemistry: rat anti-BrdU (1:2000, Abcam 6326), rabbit monoclonal OLFM4 (1:10,000, gift from CST, clone PP7), rabbit polyclonal lysozyme (1:2000, Thermo RB-372-A1), rabbit chromogranin A (1:4000, Abcam 15160), . rabbit Cleaved Caspase-3 (1:500, CST #9664), Rabbit polyclonal Anti-RFP (1:500, Rockland 600-401-379). Biotin-conjugated secondary donkey anti-rabbit or anti-rat antibodies were used from Jackson ImmunoResearch. The Vectastain Elite ABC immunoperoxidase detection kit (Vector Labs PK-6101) followed by Dako Liquid DAB+ Substrate (Dako) were used for visualization. All antibody incubations involving tissue or sorted cells were performed with Common Antibody Diluent (Biogenex). The following antibodies were used for western blotting: anti-Cpt1a (1:1000, Abcam ab128568); anti-HMGCS2 (1:500, Sigma AV41562) and anti-alpha tubulin (1:3000, Santa Cruz sc- 8035), total S6 (1:1000, CST, 2217).
RNA-Seq data processing and differential expression analysis
Single-end, 40 base reads generated by Illumina HiSeq 2000 were subjected to quality control analysis using the FastQC program (Babraham Bioinformatics, UK), which revealed overall good quality of the sequencing libraries. The reads were then mapped to the genome of Mus musculus (mm10) using a RNA-Seq alignment software, STAR (Dobin et al., 2013), with default parameters and the mm10 gene annotation as splice junction database. Among all samples, on average 93.5% of the raw reads were able to be mapped to the genome, with 67.6% of the reads mapped uniquely. Reads aligning to individual genes were counted using htseq-count (Anders et al., 2015) in “union” mode. Read counts of different samples were normalized using the “geometric means” scaling method implemented in the DESeq R package (Anders and Huber, 2010). Significance of differential expression between two samples was determined using negative binomial test as implemented in the DESeq package. As recommended by developers of DESeq for samples without replicate, the following parameters were used in dispersion estimation: method="blind", fitType="local", sharingMode="fit-only". The p values were adjusted using Benjamini-Hochberg procedure. Differentially expressed genes were identified as genes meeting both of the following two criteria: 1) the absolute value of the log2 (FoldChange) needs to be greater than 1, and 2) the adjusted p value needs to be smaller than 0.05.
Clustering and heatmap of differentially expressed genes
Genes differentially expressed between two samples collected under different conditions were identified as described above. Lists of differentially expressed genes were clustered according to their log2 (Fold Change) value using a hierarchical clustering algorithm implemented in Qlucore Omics Explorer 3.2. The complete linkage clustering method was used. Data were first log-transformed and median centered for both genes and samples.
Gene set enrichment analysis (GSEA)
The GSEA tool developed by Broad Institute (Subramanian et al., 2005) was used to analyze potential enrichment of interesting gene sets affected by age, diet, etc. Genes were ranked according to their log2 (FoldChange) values and analyzed using the “pre-ranked” mode of the GSEA software using the following parameters: -mode Max_probe -norm meandiv -nperm 1000 -scoring_scheme weighted -set_max 500 -set_min 15. Several collections of gene sets, including the H, C2, C3, C5, C6, and C7 collections of the Molecular Signatures Database (MSigDB) (Subramanian et al., 2005) were analyzed.
13C-Palmitate and 13C-Glucose labeling and LC/MS Methods
Crypts were isolated from corresponding vehicle-treated or GW501516-treated mice and incubated in RPMI media containing above mentioned crypt components and 30mM 13C-Palmitate for 60 minutes. For glucose labeling, isolated crypts were incubated in 10mM 13C Glucose in RPMI. Crypts were then spun down and washed once with saline and resuspended in LC/MS grade 80% methanol solution containing internal standards (909 nM each of 17 isotopically labeled amino acids, Cambridge Isotope Laboratories product MSK-A2-1.2) and vortexed for 10 min. Samples were then spun down and dried in a vacuum dryer or dried under a stream of nitrogen. Samples were resuspended in 100 ul LC/MS grade water and analyzed by LC/MS as described (Birsoy et al., 2015). Briefly, 2 μL of each sample was injected onto a ZIC-pHILIC 2.1 × 150 mm (5 μm particle size) column (EMD Millipore). Buffer A was 20 mM ammonium carbonate, 0.1% ammonium hydroxide; buffer B was acetonitrile. The chromatographic gradient was run at a flow rate of 0.150 ml/min as follows: 0–20 min.: linear gradient from 80% to 20% B; 20–20.5 min.: linear gradient from 20% to 80% B; 20.5–28 min.: hold at 80% B. The mass spectrometer, a QExactive orbitrap instrument, was operated in full-scan, polarity switching mode with the spray voltage set to 3.0 kV, the heated capillary held at 275°C, and the HES I probe held at 350°C. The sheath gas flow was set to 40 units, the auxiliary gas flow was set to 15 units, and the sweep gas flow was set to 1 unit. The MS data acquisition was performed in a range of 70–1000 m/z, with the resolution set at 70,000, the AGC target at 10ˆ6, and the maximum injection time at 80 msec. Relative quantitation of polar metabolites was performed with XCalibur QuanBrowser 2.2 (Thermo Fisher Scientific) using a 5 ppm mass tolerance and referencing an in-house library of chemical standards. The fraction m+2 acetylcarnitine was calculated as the raw peak area of m+2 acetylcarnitine, divided by the sum of raw peak areas of unlabeled acetylcarnitine, m+1 acetylcarnitine, and m+2 acetylcarnitine. The same calculation was used for M+2 citrate.
IRRADIATION EXPERIMENTS
Mice were challenged by a lethal split dose of irradiation (7.5Gy x 2). Numbers of surviving crypts were enumerated after ionizing irradiation-induced (XRT) damage in young and aged mice by H and E.
QUANTIFICATION AND STATISTICAL ANALYSIS DATA
Unless otherwise specified in the figure legends, all experiments reported in this study were repeated at least three independent times. Unless otherwise specified in the main text or figure legends, all sample number (n) represent biological replicates. For murine organoid assays 2-4 wells per group with at least 3 different mice were analyzed. All center values shown in graphs refer to the mean. For analysis of the statistical significance of differences between two groups, we used two-tailed unpaired Student’s t-tests. No samples or animals were excluded from analysis, and sample size estimates were not used. Animals were randomly assigned to groups. Studies were not conducted blind with the exception of all histological analyses. All experiments involving mice were carried out with approval from the Committee for Animal Care at MIT and under supervision of the Department of Comparative Medicine at MIT.
DATA AVAILABILITY
RNA sequencing data that support the findings of this study have been deposited to GEO repository. GEO: GSE89568.
Supplementary Material
Highlight.
Fasting induces fatty acid oxidation (FAO) in intestinal stem and progenitor cells
Aging reduces ISC numbers and function, correlating with decreased FAO
PPAR/CPT1a-mediated FAO augments ISC function in aging and during regeneration
PPARδ agonists boost and restore ISC and progenitor function
Acknowledgments
Ö.H.Y. is supported by the NIH R00 AG045144, R01CA211184, R01CA034992, V Foundation V Scholar Award, the Sidney Kimmel Scholar Award, the Pew-Stewart Trust Scholar Award, Kathy and Curt Marble Cancer Research Fund, and the American Federation of Aging Research (AFAR). D.M.S. is supported by NIH CA103866, Glenn/AFAR Breakthroughs in Gerontology Award and is an investigator of the Howard Hughes Medical Institute. M.M.M. is supported by the Damon Runyon Postdoctoral Fellowship, where she is a Robert Black Foundation Fellow, the Ludwig Postdoctoral Fellowship and NIH K99 Pathway to Independence award K99AG054760. C.W.C. is supported by Ludwig Postdoctoral Fellowship and Helen Hay Whitney Postdoctoral Fellowship. A.S.D. and A.R.L.S. are supported by the NIH R01 AG033082 and R01 NS065874. We thank members of The Hope Babette Tang (1983) Histology Facility at the Koch Institute. We also thank Sven Holder for histology support. We thank the Whitehead and Koch Institute Flow Cytometry Cores, and Stuart Levine of the MIT BioMicro Center. We thank Heather Keys for figure suggestions and members of the Sabatini and Yilmaz laboratories for discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONTRIBUTIONS:
M.M.M. and C.W.C. designed, preformed and interpreted all experiments with input from D.M.S. and Ö.H.Y. A.C.Q., S.T., L.C. participated in a number of experiments, in particular, immunohistochemistry, crypt isolations, RNA scope, palmitate tracing experiments, and in vitro treatments of crypt cultures. M.D.M. supplied protocols, young GW treated mice and FACS sorting. K.E.B.R. and A.E. participated in immunohistochemistry for BrdU and Olfm4 staining and enumeration. M.A.R. provided plasma of fasted mice. E.F. and C.L. developed 13C-palmitate tracing protocols and analyzed and interpreted all of the LC-MS tracing data. Y.H. and G.B. analyzed raw RNA-Seq data and prepared GSEA analysis. A.S.D and A.R.L.S supplied KD3010, and P.C. gifted CPT1A mice. P.K. contributed to aging data and interpretation. M.M.M. and C.W.C. wrote the manuscript with help from D.M.S. and Ö.H.Y.
DECLARATION OF INTERESTS
The authors M.M.M., D.M.S. and Ö.H.Y. have filed a patent related to these findings. The authors declare no competing interests.
References
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome biology. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asfaha S, Hayakawa Y, Muley A, Stokes S, Graham TA, Ericksen RE, Westphalen CB, von Burstin J, Mastracci TL, Worthley DL, et al. Krt19(+)/Lgr5(-) Cells Are Radioresistant Cancer-Initiating Stem Cells in the Colon and Intestine. Cell stem cell. 2015;16:627–638. doi: 10.1016/j.stem.2015.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barish GD, Narkar VA, Evans RM. PPAR delta: a dagger in the heart of the metabolic syndrome. The Journal of clinical investigation. 2006;116:590–597. doi: 10.1172/JCI27955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Beyaz S, Mana MD, Roper J, Kedrin D, Saadatpour A, Hong SJ, Bauer-Rowe KE, Xifaras ME, Akkad A, Arias E, et al. High-fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature. 2016;531:53–58. doi: 10.1038/nature17173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birsoy K, Wang T, Chen WW, Freinkman E, Abu-Remaileh M, Sabatini DM. An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell. 2015;162:540–551. doi: 10.1016/j.cell.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B, Hochmuth CE, Jasper H. JNK activity in somatic stem cells causes loss of tissue homeostasis in the aging Drosophila gut. Cell stem cell. 2008;3:442–455. doi: 10.1016/j.stem.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B, Karpac J, Supoyo S, Degennaro M, Lehmann R, Jasper H. Lifespan extension by preserving proliferative homeostasis in Drosophila. PLoS genetics. 2010;6:e1001159. doi: 10.1371/journal.pgen.1001159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buczacki SJ, Zecchini HI, Nicholson AM, Russell R, Vermeulen L, Kemp R, Winton DJ. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature. 2013;495:65–69. doi: 10.1038/nature11965. [DOI] [PubMed] [Google Scholar]
- Cangelosi AL, Yilmaz OH. High fat diet and stem cells: Linking diet to intestinal tumor formation. Cell cycle. 2016;15:1657–1658. doi: 10.1080/15384101.2016.1175275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerletti M, Jang YC, Finley LW, Haigis MC, Wagers AJ. Short-term calorie restriction enhances skeletal muscle stem cell function. Cell stem cell. 2012;10:515–519. doi: 10.1016/j.stem.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandel NS, Jasper H, Ho TT, Passegue E. Metabolic regulation of stem cell function in tissue homeostasis and organismal ageing. Nature cell biology. 2016;18:823–832. doi: 10.1038/ncb3385. [DOI] [PubMed] [Google Scholar]
- Cheng CW, Adams GB, Perin L, Wei M, Zhou X, Lam BS, Da Sacco S, Mirisola M, Quinn DI, Dorff TB, et al. Prolonged fasting reduces IGF-1/PKA to promote hematopoietic-stem-cell-based regeneration and reverse immunosuppression. Cell stem cell. 2014;14:810–823. doi: 10.1016/j.stem.2014.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi NH, Kim JG, Yang DJ, Kim YS, Yoo MA. Age-related changes in Drosophila midgut are associated with PVF2, a PDGF/VEGF-like growth factor. Aging cell. 2008;7:318–334. doi: 10.1111/j.1474-9726.2008.00380.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el Marjou F, Janssen KP, Chang BH, Li M, Hindie V, Chan L, Louvard D, Chambon P, Metzger D, Robine S. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39:186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- Flores A, Schell J, Krall AS, Jelinek D, Miranda M, Grigorian M, Braas D, White AC, Zhou JL, Graham NA, et al. Lactate dehydrogenase activity drives hair follicle stem cell activation. Nature cell biology. 2017;19:1017–1026. doi: 10.1038/ncb3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrick CL, Ingram DK, Reynolds MA, Freeman JR, Cider N. Effects of intermittent feeding upon body weight and lifespan in inbred mice: interaction of genotype and age. Mech Ageing Dev. 1990;55:69–87. doi: 10.1016/0047-6374(90)90107-q. [DOI] [PubMed] [Google Scholar]
- Gregorieff A, Clevers H. In situ hybridization to identify gut stem cells. Current protocols in stem cell biology. 2010 doi: 10.1002/9780470151808.sc02f01s12. Chapter 2, Unit 2F 1. [DOI] [PubMed] [Google Scholar]
- Haller S, Kapuria S, Riley RR, O'Leary MN, Schreiber KH, Andersen JK, Melov S, Que J, Rando TA, Rock J, et al. mTORC1 Activation during Repeated Regeneration Impairs Somatic Stem Cell Maintenance. Cell stem cell. 2017;21:806–818e805. doi: 10.1016/j.stem.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Argmann C, Houten SM, Canto C, Jeninga EH, Andreux PA, Thomas C, Doenlen R, Schoonjans K, Auwerx J. The metabolic footprint of aging in mice. Sci Rep. 2011;1:134. doi: 10.1038/srep00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M, Dorrell C, Boj SF, van Es JH, Li VS, van de Wetering M, Sato T, Hamer K, Sasaki N, Finegold MJ, et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013;494:247–250. doi: 10.1038/nature11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi M, Guarente L. mTORC1 and SIRT1 Cooperate to Foster Expansion of Gut Adult Stem Cells during Calorie Restriction. Cell. 2016;166:436–450. doi: 10.1016/j.cell.2016.05.044. [DOI] [PubMed] [Google Scholar]
- Ito K, Carracedo A, Weiss D, Arai F, Ala U, Avigan DE, Schafer ZT, Evans RM, Suda T, Lee CH, et al. A PML-PPAR-delta pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nature medicine. 2012;18:1350–1358. doi: 10.1038/nm.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DL, Rando TA. Emerging models and paradigms for stem cell ageing. Nature cell biology. 2011;13:506–512. doi: 10.1038/ncb0511-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasumov T, Adams JE, Bian F, David F, Thomas KR, Jobbins KA, Minkler PE, Hoppel CL, Brunengraber H. Probing peroxisomal beta-oxidation and the labelling of acetyl-CoA proxies with [1-(13C)]octanoate and [3-(13C)]octanoate in the perfused rat liver. The Biochemical journal. 2005;389:397–401. doi: 10.1042/BJ20050144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerner J, Minkler PE, Lesnefsky EJ, Hoppel CL. Fatty acid chain elongation in palmitate-perfused working rat heart: mitochondrial acetyl-CoA is the source of two-carbon units for chain elongation. The Journal of biological chemistry. 2014;289:10223–10234. doi: 10.1074/jbc.M113.524314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozar S, Morrissey E, Nicholson AM, van der Heijden M, Zecchini HI, Kemp R, Tavare S, Vermeulen L, Winton DJ. Continuous clonal labeling reveals small numbers of functional stem cells in intestinal crypts and adenomas. Cell stem cell. 2013;13:626–633. doi: 10.1016/j.stem.2013.08.001. [DOI] [PubMed] [Google Scholar]
- Li H, Jasper H. Gastrointestinal stem cells in health and disease: from flies to humans. Dis Model Mech. 2016;9:487–499. doi: 10.1242/dmm.024232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Deng S, Ibarra RA, Anderson VE, Brunengraber H, Zhang GF. Multiple mass isotopomer tracing of acetyl-CoA metabolism in Langendorff-perfused rat hearts: channeling of acetyl-CoA from pyruvate dehydrogenase to carnitine acetyltransferase. The Journal of biological chemistry. 2015;290:8121–8132. doi: 10.1074/jbc.M114.631549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo VD, Mattson MP. Fasting: molecular mechanisms and clinical applications. Cell metabolism. 2014;19:181–192. doi: 10.1016/j.cmet.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylova MM, Sabatini DM, Yilmaz OH. Dietary and metabolic control of stem cell function in physiology and cancer. Cell stem cell. 2014;14:292–305. doi: 10.1016/j.stem.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Wandycz AM, Akashi K, Globerson A, Weissman IL. The aging of hematopoietic stem cells. Nature medicine. 1996;2:1011–1016. doi: 10.1038/nm0996-1011. [DOI] [PubMed] [Google Scholar]
- Nalapareddy K, Nattamai KJ, Kumar RS, Karns R, Wikenheiser-Brokamp KA, Sampson LL, Mahe MM, Sundaram N, Yacyshyn MB, Yacyshyn B, et al. Canonical Wnt Signaling Ameliorates Aging of Intestinal Stem Cells. Cell reports. 2017;18:2608–2621. doi: 10.1016/j.celrep.2017.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D, Samson SL, Reddy VT, Gonzalez EV, Sekhar RV. Impaired mitochondrial fatty acid oxidation and insulin resistance in aging: novel protective role of glutathione. Aging cell. 2013;12:415–425. doi: 10.1111/acel.12073. [DOI] [PubMed] [Google Scholar]
- Potten CS, Martin K, Kirkwood TB. Ageing of murine small intestinal stem cells. Novartis Foundation symposium. 2001a;235:66–79. doi: 10.1002/0470868694.ch7. discussion 79-84, 101-104. [DOI] [PubMed] [Google Scholar]
- Potten CS, Martin K, Kirkwood TB. Ageing of murine small intestinal stem cells. Novartis Found Symp. 2001b;235:66–79. doi: 10.1002/0470868694.ch7. discussion 79-84, 101-104. [DOI] [PubMed] [Google Scholar]
- Rafalski VA, Mancini E, Brunet A. Energy metabolism and energy-sensing pathways in mammalian embryonic and adult stem cell fate. Journal of cell science. 2012;125:5597–5608. doi: 10.1242/jcs.114827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnik-Docampo M, Koehler CL, Clark RI, Schinaman JM, Sauer V, Wong DM, Lewis S, D'Alterio C, Walker DW, Jones DL. Tricellular junctions regulate intestinal stem cell behaviour to maintain homeostasis. Nature cell biology. 2017;19:52–59. doi: 10.1038/ncb3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond CA, Shah MS, Deary LT, Trotier DC, Thomas H, Ambruzs DM, Jiang L, Whiles BB, Rickner HD, Montgomery RK, et al. Dormant Intestinal Stem Cells Are Regulated by PTEN and Nutritional Status. Cell reports. 2015;13:2403–2411. doi: 10.1016/j.celrep.2015.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki N, Sachs N, Wiebrands K, Ellenbroek SI, Fumagalli A, Lyubimova A, Begthel H, van den Born M, van Es JH, Karthaus WR, et al. Reg4+ deep crypt secretory cells function as epithelial niche for Lgr5+ stem cells in colon. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:E5399–5407. doi: 10.1073/pnas.1607327113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M, Clevers H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009 doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Schell JC, Wisidagama DR, Bensard C, Zhao H, Wei P, Tanner J, Flores A, Mohlman J, Sorensen LK, Earl CS, et al. Control of intestinal stem cell function and proliferation by mitochondrial pyruvate metabolism. Nature cell biology. 2017;19:1027–1036. doi: 10.1038/ncb3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoors S, Bruning U, Missiaen R, Queiroz KC, Borgers G, Elia I, Zecchin A, Cantelmo AR, Christen S, Goveia J, et al. Fatty acid carbon is essential for dNTP synthesis in endothelial cells. Nature. 2015;520:192–197. doi: 10.1038/nature14362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Peterson TR, Laplante M, Oh S, Sabatini DM. mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature. 2010;468:1100–1104. doi: 10.1038/nature09584. [DOI] [PubMed] [Google Scholar]
- Signer RA, Morrison SJ. Mechanisms that regulate stem cell aging and life span. Cell stem cell. 2013;12:152–165. doi: 10.1016/j.stem.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetteh PW, Basak O, Farin HF, Wiebrands K, Kretzschmar K, Begthel H, van den Born M, Korving J, de Sauvage F, van Es JH, et al. Replacement of Lost Lgr5-Positive Stem Cells through Plasticity of Their Enterocyte-Lineage Daughters. Cell stem cell. 2016;18:203–213. doi: 10.1016/j.stem.2016.01.001. [DOI] [PubMed] [Google Scholar]
- Tinkum KL, Stemler KM, White LS, Loza AJ, Jeter-Jones S, Michalski BM, Kuzmicki C, Pless R, Stappenbeck TS, Piwnica-Worms D, et al. Fasting protects mice from lethal DNA damage by promoting small intestinal epithelial stem cell survival. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E7148–7154. doi: 10.1073/pnas.1509249112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Flier LG, van Gijn ME, Hatzis P, Kujala P, Haegebarth A, Stange DE, Begthel H, van den Born M, Guryev V, Oving I, et al. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell. 2009;136:903–912. doi: 10.1016/j.cell.2009.01.031. [DOI] [PubMed] [Google Scholar]
- van Es JH, Sato T, van de Wetering M, Lyubimova A, Nee AN, Gregorieff A, Sasaki N, Zeinstra L, van den Born M, Korving J, et al. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nature cell biology. 2012;14:1099–1104. doi: 10.1038/ncb2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura FV, Costa CG, Struys EA, Ruiter J, Allers P, Ijlst L, Tavares de Almeida I, Duran M, Jakobs C, Wanders RJ. Quantitative acylcarnitine profiling in fibroblasts using [U-13C] palmitic acid: an improved tool for the diagnosis of fatty acid oxidation defects. Clin Chim Acta. 1999;281:1–17. doi: 10.1016/s0009-8981(98)00188-0. [DOI] [PubMed] [Google Scholar]
- Wang B, Rong X, Palladino END, Wang J, Fogelman AM, Martin MG, Alrefai WA, Ford DA, Tontonoz P. Phospholipid Remodeling and Cholesterol Availability Regulate Intestinal Stemness and Tumorigenesis. Cell stem cell. 2018;22:206–220e204. doi: 10.1016/j.stem.2017.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weindruch R, Walford RL, Fligiel S, Guthrie D. The retardation of aging in mice by dietary restriction: longevity, cancer, immunity and lifetime energy intake. J Nutr. 1986;116:641–654. doi: 10.1093/jn/116.4.641. [DOI] [PubMed] [Google Scholar]
- Yan KS, Chia LA, Li X, Ootani A, Su J, Lee JY, Su N, Luo Y, Heilshorn SC, Amieva MR, et al. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:466–471. doi: 10.1073/pnas.1118857109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz OH, Katajisto P, Lamming DW, Gultekin Y, Bauer-Rowe KE, Sengupta S, Birsoy K, Dursun A, Yilmaz VO, Selig M, et al. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature. 2012;486:490–495. doi: 10.1038/nature11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data that support the findings of this study have been deposited to GEO repository. GEO: GSE89568.


