Abstract
Fungal xylanases from Trichoderma spp. are potent elicitors of defense responses in various plants. To determine whether enzymatic activity is necessary for elicitor activity, we used site-directed mutagenesis to reduce the catalytic activity of xylanase II from Trichoderma reesei. For this, the glutamic acid residue at position 210, which is part of the active center in this family of enzymes, was changed to either aspartic acid (E210D) or serine (E210S). Wild-type and mutated forms of xylanase II were expressed in yeast cells and purified to homogeneity. Compared with the wild-type form of xylanase II, E210D had >100-fold and E210S 1,000-fold lower enzymatic activity. In contrast, these mutated forms showed no comparable drop in elicitor activity. They fully stimulated medium alkalinization and ethylene biosynthesis in suspension-cultured tomato (Lycopersicon esculentum) cells, as well as hypersensitive necrosis in leaves of tomato and tobacco (Nicotiana tabacum) plants. These results provide direct evidence that enzyme activity is not necessary for elicitor activity of fungal xylanase.
Many microorganisms that live as plant pathogens or as plant saprophytes dispose of an array of enzymes to degrade plant cell walls that include enzymes hydrolyzing cellulose, pectin, and xylan. Fungal endo-β-1,4-xylanases are well-known proteinaceous elicitors of defense response reactions in tobacco (Nicotiana tabacum) and tomato (Lycopersicon esculentum) plants (Bailey et al., 1990; Fluhr et al., 1991; Avni et al., 1994). When applied to tobacco or tomato leaves, these xylanases induce ethylene biosynthesis, the production of phytoalexins and pathogenesis-related proteins, and cause necrosis and hypersensitive cell death. In suspension-cultured cells of tobacco and tomato they induce rapid medium alkalinization, oxidative burst, and ethylene biosynthesis (Felix et al., 1991, 1993; Bailey et al., 1992; Yano et al., 1998). Endo-β-1,4-xylanases have been isolated and characterized from a variety of different plant-pathogenic and non-pathogenic fungi (Törrönen et al., 1992; Yagouchi et al., 1992; Wu et al., 1995; Apel-Birkhold and Walton, 1996; Luttig et al., 1997; Giesbert et al., 1998). Several of these β-1,4-xylanases are active elicitors, but the xylanase from Trichoderma viride has been used most extensively to study elicitor activity of fungal xylanases (Dean et al., 1989; Yano et al., 1998).
Due to the importance of endo-β-1,4-xylanases for industrial applications in the pulp and paper industry, this group of proteins has been characterized in detail (Wong and Saddler, 1992). Endo-β-1,4-xylanases (1,4-β-d-xylan xylanohydrolase; EC 3.2.1.8) belong to the class of glycosyl hydrolases, and within this class they have been assigned to two enzyme families: family 10 contains xylanases with a molecular mass over 30 kD and an acidic pI, and family 11 comprises xylanases with a molecular mass smaller than 30 kD and a basic pI (Henrissat and Bairoch, 1993; Biely et al., 1997). All endo-β-1,4-xylanases that have thus far been found to be elicitors of defense reactions in plants belong to the family 11 xylanases (Dean et al., 1989; Yano et al., 1998). Several endo-β-1,4-xylanases of bacterial and fungal origin that belong to family 11 have been crystallized and their three-dimensional structures determined (Campbell et al., 1993; Törrönen and Rouvinen, 1995; Krengel and Dijkstra, 1996). The overall three-dimensional structure of these proteins is very similar and has been described as the shape of a “right hand” with an active center formed by the thumb, palm, and fingers of the hand. Mutational analyses have led to the identification of two conserved Glu residues in the active center that are critical for enzymatic activity (Ko et al., 1992; Wakarchuk et al., 1994).
At present it is still uncertain how the xylanase elicitor is perceived by plant cells. Plant cells could perceive xylanase directly by a receptor for this protein (Hanania and Avni, 1997) or indirectly via plant cell wall fragments generated by its enzymatic activity (Bucheli et al., 1990). So far, elicitor activity and enzyme activity have not been clearly separated. Treatments that lead to losses in enzyme activity, such as treatment with protease or heat denaturation, also abolish elicitor activity. This means that the three-dimensional structure of xylanase is essential for elicitor activity and that the protein is not simply recognized by its primary structure (Fuchs et al., 1989; Lotan and Fluhr, 1990).
Indirect but cumulative evidence suggests that xylanase is recognized as a protein structure rather than via its enzymatic activity: (a) protoplasts that are lacking most of their cell walls react to xylanase (Sharon et al., 1993); (b) not all enzymes with endo-β-1,4-xylanase activity are active as elicitors (Yano et al., 1998); (c) efforts to isolate elicitor-active cell wall fragments released by endo-β-1,4-xylanase activity have not been successful (Dean et al., 1991); (d) a specific binding site for the endo-β-1,4-xylanase protein on tobacco protoplasts has been reported (Hanania and Avni, 1997). Even though these experiments suggest a direct recognition of xylanase, they do not exclude the involvement of endo-β-1,4-xylanase activity in the elicitation process. The goal of this study was to determine whether enzymatic activity of fungal β-1,4-xylanase is necessary for its elicitor activity. We addressed this question by creating enzymatically inactive forms of xylanase II cloned from Trichoderma reesei and testing them for their elicitor activity.
MATERIALS AND METHODS
Strains, Plant Material, and Growth Conditions
Saccharomyces cerevisiae strain Y294 (MATα, leu2-3, 112, ura3-52, his3, and trp1-289) (La Grange et al., 1996) was used as a host to express wild-type and mutated forms of the xylanase II gene (XYN2) from Trichoderma reesei QM 6a. Cultures were grown and maintained in complex (yeast peptone dextrose) or synthetic medium at 30°C (La Grange et al., 1996). Recombinant plasmids were constructed and amplified in Escherichia coli XL1 Blue (Stratagene, La Jolla, CA). The tomato (Lycopersicon esculentum) cell line Msk8 was maintained and subcultured as described earlier (Felix et al., 1991). Cells were used for experiments 5 to 8 d after subculturing. Tomato (cv Moneymaker) and tobacco (Nicotiana tabacum cv Havana 425) plants were grown in soil in a greenhouse.
DNA Manipulations and Plasmid Construction
Standard molecular techniques were used throughout (Sambrook et al., 1989). The yeast expression clone pDLG5 harboring the XYN2 gene of T. reesei under the control of the inducible ADH2 promoter was kindly provided by Dr. W.H. van Zyl (Department of Microbiology, University of Stellenbosch, South Africa).
For site-directed mutagenesis by inverse PCR (Clackson et al., 1991) the XYN2 gene was first subcloned into pBluescript II SK− (Stratagene). For this, a BglII restriction site was introduced into the multiple cloning site of pBluescript II SK by inverse PCR using the two phosphorylated back-to-back primers AGATCTATCGATACCGTCGAC and AAGCTTGATATCGAATTCCTG. The reaction conditions were as follows: 100 ng of template DNA, 0.25 μm of each of the primers, 0.2 mm dNTPs, and 5 units of Pfu DNA polymerase (Stratagene) in Pfu reaction buffer. After an initial denaturation step of 1 min at 95°C, 25 cycles of denaturation, annealing, and polymerization were carried out for 45 s at 95°C, 45 s at 50°C, and 6 min at 72°C, respectively. The amplified DNA fragment was purified and self-ligated, resulting in plasmid pBSBlgII. pDLG5 was digested with BglII and EcoRI, and the XYN2 fragment was isolated. XYN2 was ligated into pBSBglII, resulting in plasmid pXyn2. Mutations of amino acid E210 to S and D were introduced in pXyn2 using inverse PCR amplification with back-to-back primer pairs containing a mutation in the forward primer (E–S forward primer GCCGTGTCGGGTTACTTTAGCT; E–D forward primer GCCGTGGACGGTTACTTTAGCT). The reverse primer for both amplifications was GGTTGCGGGACCAGCCGTAC.
The reaction conditions were as described above except that the elongation time was 8 min. Amplified fragments were isolated and self-ligated, resulting in plasmids pXynES (E–S) and pXynED (E–D). Mutations were confirmed by sequencing the inserts of pXynES and pXynED. pXynES and pXynED were digested with BglII and EcoRI, the mutated XYN2-containing fragments were isolated and ligated into the expression vector pDLG1 (La Grange et al., 1996). The resulting plasmids were termed pE210S and pE210D, respectively. Plasmids were transformed into S. cerevisiae Y294 using the lithium acetate method (Gietz et al., 1992) and selected on synthetic medium minus uracil plates.
Protein Expression and Purification
Recombinant S. cerevisiae (200-mL cultures) were grown for 4 d in yeast-peptone-Gal medium (1% [w/v] yeast extract, 2% [w/v] peptone, and 0.8% [w/v] Gal) at 30°C on a rotary shaker at 250 rpm. Cells were removed by centrifugation (10 min at 8,000g) and the supernatants were dialyzed against water in tubing with a molecular cutoff of 6,000 to 8,000 (Spectra/Por, The Spectrum Companies, Gardena, CA). Dialyzed supernatants were pre-purified by passing over cation-exchange resin (SP Trisacryl M, Sepracor-IBF Biotechnics, Villeneuve-la-Garenne, France) at pH 4.5, and eluted with 500 mm NaCl. After dialysis and lyophilization, the pre-purified xylanase preparations were dissolved in 10 mm citrate buffer, pH 4.5, and separated on a MonoS FPLC column (Pharmacia Biotech, Uppsala) using a gradient of 0 to 500 mm NaCl for elution. Fractions used for further experiments were dialyzed as described above.
Enzyme Activity Assay
Endo-β-1,4-xylanase activity was measured using the modified dinitrosalicylic acid method (Bailey et al., 1992). 4-O-Methyl glucuronoxylan (product 7500, Roth, Karlsruhe, Germany) was used as a substrate at a concentration of 1% (w/v) and increases in reducing sugars were determined after 5 min of incubation. Samples were measured in triplicate at different dilutions and activity was derived from linear regression analysis.
Assays for Elicitor Activity
The extracellular alkalinization response was measured in suspension-cultured tomato cells using a small, combined pH electrode (Metrohm, Herisau, Switzerland) as described previously (Felix et al., 1993). Alternatively, extracellular pH values were measured 15 min after the addition of test substances to 1-mL aliquots of the cell suspension. For these assays, cells were incubated in 24-well tissue-culture plates on a rotary shaker at 200 rpm.
Induction of ethylene biosynthesis was assayed as described previously (Felix et al., 1991). One-milliliter aliquots of the cell suspension (approximately 0.2 g fresh weight) were transferred to 6-mL glass tubes and the elicitors to be tested were added. The tubes were closed with rubber septa and ethylene accumulating in the free air space was measured by GC after 4 h of incubation on a rotary shaker.
For testing the induction of necrosis, preparations were injected through the lower leaf surfaces of tomato (5 weeks old) or tobacco (7 weeks old) plants with a 1-mL plastic syringe without a needle. For every site of injection, leaves were treated with 30 μL of the preparation to be tested and photographed after incubation for 6 d at 25°C (12 h of light/12 h of dark).
Reproducibility
The results shown in the figures represent single experiments representative of at least three independent repetitions.
RESULTS
Wild-Type and Mutant Forms of Endo-β- Xylanase from T. reesei
The XYN2 gene of T. reesei encodes a 223-amino acid proprotein (Saarelainen et al., 1993) (Fig. 1). The N-terminal sequence codes for a 33 amino acid secretion signal that is proteolytically cleaved off, resulting in a 190-amino acid mature xylanase II protein. Like other family 11 type endoxylanases, the enzyme of T. reesei has two conserved Glu residues at positions 119 and 210 (Saarelainen et al., 1993) (Fig. 1). For xylanases from Bacillus pumilus and Bacillus circulans, these Glu residues have been shown to be crucial for enzymatic activity (Ko et al., 1992; Wakarchuk et al., 1994). We used site-directed mutagenesis to change the codon GAG coding for the Glu residue at position 210 to either TCG, coding for Ser, or to GAC, coding for Asp (Fig. 1). The mutated XYN2 genes were then inserted into the expression vector pDLG1 (La Grange et al., 1996) to form plasmids pE210S and pE210D, respectively.
Figure 1.
Primary structure of xylanase II proprotein. A, Primary structure of the xylanase II proprotein from T. reesei as deduced from cDNA sequence (database accession no. U24191 [La Grange et al., 1996]). The first 33 amino acid residues (underlined) constitute the cleavable export signal. The two Glu residues essential for enzyme activity and conserved in all family 11 xylanases are highlighted (positions 119 and 210). B, Sequence of cDNA coding for xylanase II in the region coding for Glu 210. Primers used for inverse PCR to change the codon for Glu 210 to codons for Asp (E210D) or Ser (E210S) are indicated by arrows.
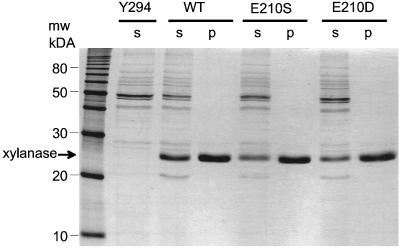
Wild-type and mutated xylanase II proteins (termed E210D and E210S) were expressed in S. cerevisiae grown in liquid culture. When analyzed by SDS-PAGE, the culture filtrates of these transformed yeast cells were found to contain proteins migrating with apparent molecular masses of 26 and 21 kD that were not present in the supernatants of untransformed yeast cultures (Fig. 2). Expression of XYN2 from T. reesei in yeast cells has previously been shown to cause the secretion of a glycosylated form of xylanase II migrating with an apparent molecular mass of 26 to 27 kD (La Grange et al., 1996). Deglycosylation of this protein resulted in a protein migrating at 21 kD, which corresponds to the molecular mass of the mature polypeptide with 190 amino acids. For further experiments the major, glycosylated forms of the xylanase II molecules were purified by cation-exchange chromatography on a MonoS FPLC column (Fig. 2).
Figure 2.
Wild-type and mutated xylanase II expressed in yeast cells. SDS-PAGE of proteins from culture medium of non-transformed yeast (Y294), and yeast expressing wild-type (WT) and mutated (E210S and E210D) forms of xylanase II. Lanes were loaded with 20 μg of total protein present in cell supernatants (S) or with 5 μg of protein after purification by cation-exchange chromatography. Standard molecular mass markers are indicated on the left.
Enzyme Activity of Wild-Type and Mutant Forms of Xylanase II
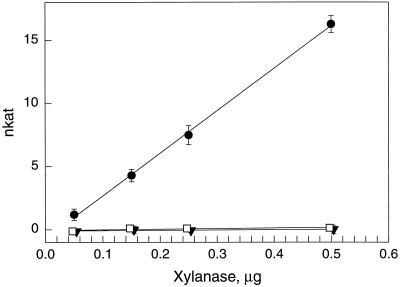
The purified wild-type and mutant forms of the T. reesei xylanase II were assayed for their endo-β-1,4-xylanase activity by detecting the production of reducing sugars from 4-O-methyl glucuronoxylan. Mutant forms of xylanase had drastically reduced enzymatic activities compared with the wild type (Fig. 3). In further experiments with highly increased concentrations of purified proteins (10 μg), low but significant residual activities could be detected in the mutated forms of xylanase (Table I). The specific enzyme activity of mutant E210D was 0.8% and that of E210S was 0.1% of the wild-type activity.
Figure 3.
Enzyme activity of wild-type and mutated xylanase II. Endo-β-1,4-xylanase activity of wild-type (●), mutant E210S (▾), and mutant E210D (□) xylanase II purified from transgenic yeast cultures. Symbols represent the average of three measurements and bars represent sd.
Table I.
Specific β-1,4-xylanase activity of wild-type and mutant forms of xylanase II
| Xylanase II | β-1,4-Xylanase Activity | |
|---|---|---|
| pkat μg−1 proteina | % of wild- type activity | |
| Wild type | 7,776 ± 628 | 100 |
| E210S | 8 ± 1 | 0.1 |
| E210D | 61 ± 3 | 0.8 |
Values represent averages from three independent measurements each with three replicates ± sd.
Early Elicitor Responses Induced by Wild-Type and Mutant Forms of Xylanase II
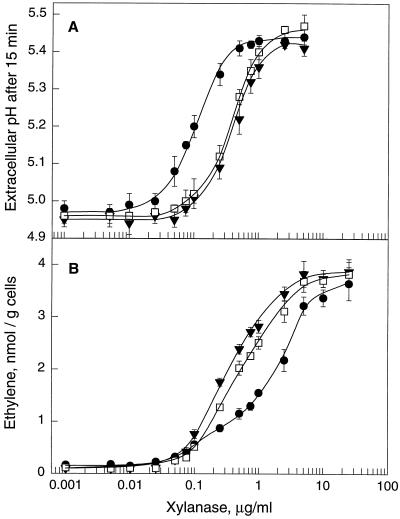
Medium alkalinization has been reported as a rapid response of suspension-cultured cells to treatment with xylanase preparations from Trichoderma viride (Bailey et al., 1992; Felix et al., 1993). Medium alkalinization was also observed after treatment of suspension-cultured tomato cells with preparations from yeast transgenic for the wild-type and mutant forms of xylanase II from T. reesei but not after treatment with corresponding protein preparations from non-transformed yeast cultures (data not shown). Alkalinization of the culture medium, measured as the extracellular pH reached after 15 min of treatment, was a steady, sigmoid function of the dose of elicitor applied (Fig. 4A). The wild-type form of xylanase II stimulated the alkalinization response half-maximally at 0.12 μg/mL (Fig. 4A; Table II) or, based on an estimated molecular mass of 26 kD, at a concentration of approximately 5 nm.
Figure 4.
Alkalinization response and induction of ethylene biosynthesis in suspension-cultured tomato cells. A, Extracellular pH in tomato cells treated for 15 min with different doses of wild-type (●), mutant E210S (▾), and mutant E210D (□) xylanase II. Symbols represent the average of three measurements and bars represent sd. B, Induction of ethylene biosynthesis in tomato cells by different doses of wild-type (●), mutant E210S (▾), and mutant E210D (□) xylanase II. Ethylene accumulating in the free-air phase of the cultures was measured 4 h after of treatment. Symbols represent the average of three measurements and bars represent sd.
Table II.
Xylanase concentration for half-maximal induction of alkalinization response and ethylene biosynthesis
| Xylanase II | Alkalinizationa | Ethylene Biosynthesisa |
|---|---|---|
| μg/mL | ||
| Wild type | 0.12 ± 0.07 | 1.40 ± 0.57 |
| E210S | 0.48 ± 0.22 | 0.28 ± 0.07 |
| E210D | 0.46 ± 0.23 | 0.45 ± 0.07 |
Values represent averages from three independent dose response curves ± sd. Every dilution step of each curve was measured with three replicates.
The mutants of xylanase II were potent elicitors of the alkalinization response as well, and the concentrations required for half-maximal induction were only approximately four times higher than for the wild-type form (Fig. 4A; Table II). Medium alkalinization induced by the wild-type and mutant forms of xylanase II showed the characteristics observed for the induction of this response by xylanase purified from T. viride (Felix et al., 1993). In particular, activity was completely inactivated by heat treatment (5 min at 95°C) and, when compared at saturating doses in the same batch of cells, alkalinization started after the same lag phase of approximately 3.5 min and resulted in the same maximal pH increase (data not shown).
Xylanase from T. viride has been observed to stimulate ethylene biosynthesis in tobacco plants (Fuchs et al., 1989) and in suspension-cultured cells of tobacco and tomato (Bailey et al., 1992; Felix et al., 1994). The purified xylanase II proteins from transgenic yeast were assayed for the induction of ethylene biosynthesis in tomato cell cultures (Fig. 4B). The wild-type and the two mutant forms of xylanase II were potent elicitors of ethylene biosynthesis (Fig. 4B), whereas no comparable induction of ethylene biosynthesis was observed with the corresponding protein preparations from non-transformed yeast cultures (data not shown). The dose-response relationships for the two mutant forms of xylanase II were very similar to those observed for induction of the alkalinization response described above, and half-maximal induction occurred at concentrations of 0.28 μg/mL for mutant E210S and 0.45 μg/mL for mutant E210D (Fig. 4B; Table II). In contrast, the wild-type form of xylanase II was less efficient as an inducer of ethylene biosynthesis, and half-maximal induction was only observed at a concentration of 1.40 μg/mL (Table II). Nevertheless, all three xylanase II proteins induced the same maximal level of ethylene biosynthesis when applied at saturating doses of >10 μg/mL (Fig. 4B).
Apparently, the enzymatic activity of xylanase negatively affected its activity as an inducer of ethylene biosynthesis. This effect was also apparent by comparing the ethylene-inducing activity of mutant E210D with that of mutant E210S. Mutant E210D retained a somewhat higher residual activity than mutant E210S, and was also slightly less efficient in inducing ethylene biosynthesis (Table II).
In summary, the wild-type form of xylanase II as well as the two mutant forms, E210S and E210D, were potent elicitors of rapid responses in tomato cells, and the minor differences in efficiency as elicitors did not reflect the major changes observed in enzyme activity.
Elicitation of Necrosis by Wild-Type and Mutant Forms of Xylanase II
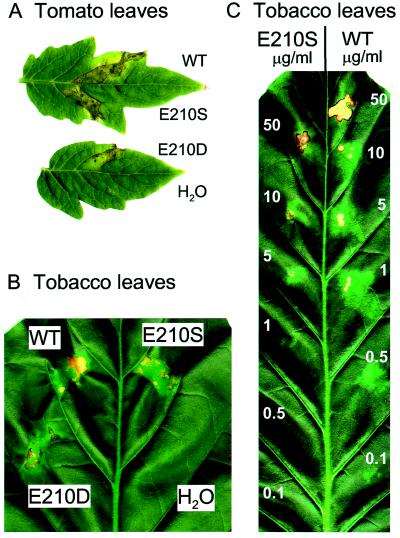
Xylanase from T. viride has been reported to induce necrosis when injected into tobacco leaves (Bailey et al., 1990). Purified wild-type and mutant forms of xylanase II were injected into tomato and tobacco leaves to test their capacity to induce necrosis. All three forms of xylanase II induced necrosis (Fig. 5, A and B). In tomato leaves necrotic areas became visible 2 d after injection, whereas in tobacco leaves the areas injected with xylanase II forms turned light green within 2 d and necrotic spots started to develop 3 d after injection. Injection of water did not result in any visible symptoms in tomato or tobacco leaves. Also, no symptoms were observed in leaves of either plant species injected with heat-denatured xylanase II forms (5 min at 95°C) or with corresponding protein preparations from non-transformed yeast cultures (data not shown). Injecting wild-type and mutant E210S xylanase II at different concentrations into tobacco leaves revealed that the lowest concentration that induced necrosis was 1 μg/mL for the wild-type form and 5 μg/mL for the mutant E210S (Fig. 5C). Thus, as observed for the alkalinization response, the mutant form appears to be slightly (approximately 5-fold) less effective than the wild-type form.
Figure 5.
Induction of necrosis in tomato and tobacco leaves. Tomato (A) and tobacco (B) leaves injected with wild-type (WT) and mutant forms (E210S and E210D) of xylanase II and water as a control. Leaves were injected with xylanase preparations at a concentration of 1 mg/mL for tomato leaves and 10 μg/mL for tobacco leaves. C, Tobacco leaf injected with different concentrations of wild-type and mutant E210S xylanase as indicated.
DISCUSSION
Plants have evolved systems to monitor the intactness of their cells or tissues, and they respond to wounding or pathogen attack with the induction of protective responses. Detection of injury can occur via endogenous, plant-derived products such as pectic fragments (Collmer and Keen, 1986), cutin monomers (Schweizer et al., 1996), or, in the case of solanaceous plants, specific wound hormones such as systemin (Ryan, 1992). Detection of cell injury could also occur via detection of cell wall loosening, e.g. by sensing changes in turgor pressure or via the activation of stretch-activated channels. Yet another possibility to detect potential pathogenic invaders lies in the perception of chemical cues, also termed elicitors, that are characteristic for the invading microorganisms. Highly specific and sensitive perception systems for a variety of elicitors have been described (Boller, 1995).
The goal of this study was to determine whether enzymatic activity of fungal endo-β-1,4-xylanase is necessary for its elicitor activity. We mutated amino acid 210 of xylanase II from T. reesei from Glu to either Ser or Asp. While these mutations drastically reduced the enzyme activity, they did not affect elicitor activity in a comparable manner. The mutated forms of xylanase II were still active as elicitors of extracellular alkalinization and ethylene biosynthesis of tomato cell cultures, and of necrosis in tomato and tobacco leaves. Although the mutations of Glu 210 resulted in slightly altered elicitor activities (e.g. approximately 4-fold lower for induction of the alkalinization response) these changes did not reflect the much bigger reductions in enzyme activities (>100-fold). These results clearly demonstrate that enzymatic activity of fungal endo-β-1,4-xylanase is not necessary for its elicitor activity and, together with the recent observation of a specific binding site for endo-β-1,4-xylanase on the surface of tobacco cells (Hanania and Avni, 1997), they show that the elicitor activity is solely based on the specific recognition of the xylanase protein.
The mutant forms of xylanase II were slightly less active in eliciting extracellular alkalinization of tomato cell cultures and necrosis in tobacco leaves. At present we have no explanation for this effect, but mutation of the Glu residue might cause an alteration in the overall structure of the protein, or the Glu residue might be part of the recognition site used by the putative plant receptor for xylanase. Structural analyses of xylanases from B. circulans and B. pumilus mutated at the corresponding Glu residues have demonstrated no major alterations in the overall structure of the proteins (Ko et al., 1992; Wakarchuk et al., 1994).
Interestingly, the mutant forms of xylanase II were more effective than the wild-type form as elicitors of ethylene biosynthesis. The dose-response curves for the two mutant forms were very similar for induction of both extracellular alkalinization and stimulation of ethylene biosynthesis. In contrast, induction by the wild-type form had a different dose dependence, and the concentration necessary for half-maximal induction of ethylene biosynthesis was 10-fold higher than for induction of the alkalinization response. A closer look at the dose-response curve shows that wild-type xylanase did not follow a simple sigmoid function for ethylene induction. Rather, the wild-type form was as active as the mutant forms at lower concentrations and less active at higher concentrations. Apparently, induction of ethylene production appears to represent a combination of a positive elicitor effect of the xylanase II protein and a negative effect of xylanase II enzymatic activity.
There are two mechanisms that could explain a negative effect of enzymatic activity. First, xylanase activity might release harmful or inhibitory fragments from the cell wall. Indeed, the release of toxic cell wall fragments has been reported in maize cells treated with a xylanase isolated from Magnaporthe grisea (Bucheli et al., 1990). Second, the enzyme might harm or inhibit the cells by changing the integrity of certain components in the cell wall or at the plasma membrane interface without actually releasing active fragments. However, the enzymatically active wild-type form of xylanase did induce full induction of ethylene biosynthesis when added at higher doses, a finding that argues against a generally inhibitory or toxic effect of the xylanase activity. Alternatively, one could speculate that putative xylanase substrates in the plant cell walls could act as binding sites for the enzymatically active xylanase and adsorb part of the xylanase molecules. However, since the wild-type form of xylanase is actually more efficient in the induction of the alkalinization response, this does not seem to provide a straightforward explanation of the experimental data. Differences in the kinetics of induction or in the stability of the different forms of xylanase could provide explanations for differences in the rapid alkalinization assay measured after 15 min and the slower induction of ethylene biosynthesis measured after 4 h. Further investigations are necessary to determine whether there is really a negative effect of xylanase activity on plant cells in culture and whether such an effect also occurs in planta.
Xylanase II proteins described in this report and xylanase purified from T. viride induce elicitor responses such as medium alkalinization and ethylene biosynthesis with similar kinetics and maximal responses. However, on a per-weight basis, xylanase from T. viride appeared to be approximately five to 10 times more efficient than the xylanase II of T. reesei expressed in yeast. Xylanase from T. viride and xylanase II from T. reesei are structurally very similar, and the amino acid sequences differ at eight positions only (database accession nos. A44595 and S67387). At present, we cannot distinguish whether the differences in elicitor activity are due to differences in the amino acid sequences or whether they originate from differences in protein modification, such as glycosylation in T. viride and yeast. Although preliminary results with xylanase II from T. reesei expressed in E. coli indicate that glycosylation is not essential for elicitor activity (J. Enkerli, unpublished results), glycosylation might have a modifying effect on the elicitor activity of xylanase. Detailed studies (e.g. replacing single amino acids in xylanase II from T. reesei) will be necessary to investigate the basis for the differences in elicitor activity between the various xylanase proteins.
Xylan is part of the complex hemicellulose fraction of plant cell walls. The actual composition of hemicellulose varies from species to species, and so far it is unclear whether tomato or tobacco cells grown in suspension cultures contain xylan in their cell walls that could serve as a substrate for endo-β-1,4-xylanase. Efforts to release water-soluble reducing sugars from tobacco plants (Dean and Anderson, 1991) or suspension-cultured tomato cells (M. Bürgin, personal communication) by the action of endo-β-1,4-xylanase have not been successful. The inhibitory activity of enzymatically active xylanase on ethylene biosynthesis observed in this report could provide the first evidence for a possible substrate present in the cell walls of tomato cells.
Several microbial cell wall-degrading enzymes have been recognized as elicitors of plant defense reactions (Walton, 1994). Among these, fungal xylanase is the only example known so far that exhibits its elicitor activity not through its enzymatic activity but rather by being recognized directly. Identification of the epitope of xylanase II protein of T. reesei responsible for its elicitor activity will provide the basis for an understanding of the interaction of proteinaceous elicitors with their respective binding proteins in plant cells.
ACKNOWLEDGMENTS
We thank Dr. W.H. van Zyl for providing plasmid pDLG5 and S. cerevisiae strain Y294. We also thank Dr. Margaret A. Collinge for critical reading of the manuscript.
LITERATURE CITED
- Apel-Birkhold PC, Walton JD. Cloning, disruption, and expression of two endo-β 1,4-xylanase genes, XYL2 and XYL3, from Cochliobolus carbonum. Appl Environ Microbiol. 1996;62:4129–4135. doi: 10.1128/aem.62.11.4129-4135.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avni A, Avidan N, Eshed Y, Zamir D, Bailey BA, Stommel JR, Anderson JD. The response of Lycopersicon esculentum to fungal xylanase is controlled by a single dominant gene (abstract no. 872) Plant Physiol. 1994;105:S-158. [Google Scholar]
- Bailey BA, Dean JFD, Anderson JD. An ethylene biosynthesis-inducing endoxylanase elicits electrolyte leakage and necrosis in Nicotiana tabacum cv Xanthi leaves. Plant Physiol. 1990;94:1849–1854. doi: 10.1104/pp.94.4.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey BA, Korcak RF, Anderson JD. Alterations in Nicotiana tabacum L. cv Xanthi cell membrane function following treatment with an ethylene biosynthesis-inducing endoxylanase. Plant Physiol. 1992;100:749–755. doi: 10.1104/pp.100.2.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey MJ, Biely P, Poutanen K. Interlaboratory testing of methods for assay of xylanase activity. J Biotechnol. 1992;23:257–270. [Google Scholar]
- Biely P, Vrsanska M, Tenkanen M, Kluepfel D. Endo-β-1,4-xylanase families: differences in catalytic properties. J Biotechnol. 1997;57:151–166. doi: 10.1016/s0168-1656(97)00096-5. [DOI] [PubMed] [Google Scholar]
- Boller T. Chemoperception of microbial signals in plant cells. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:189–214. [Google Scholar]
- Bucheli P, Doares SH, Albersheim P, Darvill A. Host-pathogen interactions. XXXVI. Partial purification and characterization of heat-labile molecules secreted by the rice blast pathogen that solubilize plant cell wall fragments that kill plant cells. Physiol Mol Plant Pathol. 1990;36:159–173. [Google Scholar]
- Campbell RL, Rose DR, Wakarchuk WW, To R, Sung W, Yagouchi M. A comparison of the structures of the 20 kd xylanases from Trichoderma harzianum and Bacillus circulans. In: Suominen P, Reinikainen T, editors. Proceedings of the Second TRICEL Symposium on Trichoderma reesei Cellulases and Other Hydrolases, Espoo, Finland, 1993. Helsinki: Foundation for Biotechnical and Industrial Fermentation Research; 1993. pp. 63–72. [Google Scholar]
- Clackson T, Güssow D, Jones PT. General applications of PCR to gene cloning and manipulation. In: Mcpherson MJ, Quirke P, Taylor GR, editors. PCR, a Practical Approach. Oxford: Oxford University Press; 1991. pp. 187–214. [Google Scholar]
- Collmer A, Keen NT. The role of pectic enzymes in plant pathogenesis. Annu Rev Phytopathol. 1986;24:383–409. [Google Scholar]
- Dean JFD, Anderson JD. Ethylene biosynthesis-inducing xylanase. II. Purification and physical characterization of the enzyme produced by Trichoderma viride. Plant Physiol. 1991;95:316–323. doi: 10.1104/pp.95.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean JFD, Gamble HR, Anderson JD. The ethylene biosynthesis-inducing xylanase: its induction in Trichoderma viride and certain plant pathogens. Phytopathology. 1989;79:1071–1078. [Google Scholar]
- Dean JFD, Gross KC, Anderson JD. Ethylene biosynthesis-inducing xylanase. III. Product characterization. Plant Physiol. 1991;96:571–576. doi: 10.1104/pp.96.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix G, Grosskopf DG, Regenass M, Basse CW, Boller T. Elicitor-induced ethylene biosynthesis in tomato cells: characterization and use as a bioassay for elicitor action. Plant Physiol. 1991;97:19–25. doi: 10.1104/pp.97.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix G, Regenass M, Boller T. Specific perception of subnanomolar concentrations of chitin fragments by tomato cells: induction of extracellular alkalinization, changes in protein phosphorylation, and establishment of a refractory state. Plant J. 1993;4:307–316. [Google Scholar]
- Felix G, Regenass M, Spanu P, Boller T. The protein phosphatase inhibitor calyculin A mimics elicitor action in plant cells and induces rapid hyperphosphorylation of specific proteins as revealed by pulse-labeling with [33P]phosphate. Proc Natl Acad Sci USA. 1994;91:952–956. doi: 10.1073/pnas.91.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluhr R, Sessa G, Sharon A, Ori N, Lotan T. Pathogenesis-related proteins exhibit both pathogen-induced and developmental regulation. In: Henneke H, Verma DSP, editors. Advances in Molecular Genetics of Plant-Microbe Interactions. Dordrecht, The Netherlands: Kluwer; 1991. pp. 387–394. [Google Scholar]
- Fuchs Y, Saxena A, Gamble HR, Anderson JD. Ethylene biosynthesis-inducing protein from cellulysin is an endoxylanase. Plant Physiol. 1989;89:138–143. doi: 10.1104/pp.89.1.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giesbert S, Lepping HB, Tenberge KB, Tudzynski P. The xylanolytic system of Claviceps purpurea: cytological evidence for secretion of xylanases in infected rey tissue and molecular characterization of two xylanase genes. Phytopathology. 1998;88:1020–1030. doi: 10.1094/PHYTO.1998.88.10.1020. [DOI] [PubMed] [Google Scholar]
- Gietz D, Jean AS, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanania U, Avni A. High-affinity binding site for ethylene-inducing xylanase elicitor on Nicotiana tabacum membranes. Plant J. 1997;12:113–120. [Google Scholar]
- Henrissat B, Bairoch A. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1993;293:781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko EP, Akatsuka H, Moriyama H, Shinmyo A, Hata Y, Katsube Y, Urabe I, Okada H. Site-directed mutagenesis at aspartate and glutamate residues of xylanase from Bacillus pumilus. Biochem J. 1992;288:117–121. doi: 10.1042/bj2880117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krengel U, Dijkstra BW. Three-dimensional structure of endo-1,4-β-xylanase I from Aspergillus niger: molecular basis for its low pH optimum. J Mol Biol. 1996;263:70–78. doi: 10.1006/jmbi.1996.0556. [DOI] [PubMed] [Google Scholar]
- La Grange DC, Pretorius IS, Van Zyl WH. Expression of a Trichoderma reesei β-xylanase gene (XYN2) in Saccharomyces cerevisiae. Appl Environ Microbiol. 1996;62:1036–1044. doi: 10.1128/aem.62.3.1036-1044.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan T, Fluhr R. Xylanase, a novel elicitor of pathogenesis-related proteins in tobacco, uses a non-ethylene pathway for induction. Plant Physiol. 1990;93:811–817. doi: 10.1104/pp.93.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttig M, Pretorius IS, Van Zyl WH. Cloning of two β-xylanase-encoding genes from Aspergillus niger and their expression in Saccharomyces cerevisiae. Biotechnol Lett. 1997;19:411–415. [Google Scholar]
- Ryan CA. The search for the proteinase-inhibitor inducing factor, PIIF. Plant Mol Biol. 1992;19:123–133. doi: 10.1007/BF00015610. [DOI] [PubMed] [Google Scholar]
- Saarelainen R, Paloheimo M, Fagerström R, Suominen PL, Navalainen KMH. Cloning, sequencing and enhanced expression of the Trichoderma reesei endoxylanase II (pI 9) gene xln2. Mol Gen Genet. 1993;241:497–503. doi: 10.1007/BF00279891. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: a Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schweizer P, Felix G, Buchala A, Muller C, Métraux JP. Perception of free cutin monomers by plant cells. Plant J. 1996;10:331–341. [Google Scholar]
- Sharon A, Fuchs Y, Anderson JD. The elicitation of ethylene biosynthesis by a Trichoderma xylanase is not related to the cell wall degradation activity of the enzyme. Plant Physiol. 1993;102:1325–1329. doi: 10.1104/pp.102.4.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Törrönen A, Mach RL, Messner R, Gonzalez R, Kalkkinen N, Harkki A, Kubicek CP. The two major xylanases from Trichoderma reesei: characterization of both enzymes and genes. Bio-Technology. 1992;10:1461–1465. doi: 10.1038/nbt1192-1461. [DOI] [PubMed] [Google Scholar]
- Törrönen A, Rouvinen J. Structural comparison of two major endo-1,4-xylanases from Trichoderma reesei. Biochemistry. 1995;34:847–856. doi: 10.1021/bi00003a019. [DOI] [PubMed] [Google Scholar]
- Wakarchuk WW, Campbell RL, Sung WL, Da Voodi J, Yaguchi M. Mutational and crystallographic analyses of the active site residues of the Bacillus circulans xylanase. Protein Sci. 1994;3:467–475. doi: 10.1002/pro.5560030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton JD. Deconstructing the cell wall. Plant Physiol. 1994;104:1113–1118. doi: 10.1104/pp.104.4.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KKY, Saddler YN. Trichoderma xylanases, their properties and application. Crit Rev Biotechnol. 1992;12:413–435. [Google Scholar]
- Wu SC, Kauffmann S, Darvill AG, Albersheim P. Purification, cloning and characterization of two xylanases from Magnaporthe grisea, the rice blast fungus. Mol Plant-Microbe Interact. 1995;8:506–514. doi: 10.1094/mpmi-8-0506. [DOI] [PubMed] [Google Scholar]
- Yagouchi M, Roy C, Ujiie M, Watson DC, Wakarchuk W. Amino acid sequence of the low-molecular-weight xylanase from Trichoderma viride. In: Visser J, Beldman G, Kusters-van Someren MA, Voragen AGJ, editors. Xylans and Xylanases. Amsterdam: Elsevier Science Publishers; 1992. pp. 149–154. [Google Scholar]
- Yano A, Suzuki K, Uchimiya H, Shinshi H. Induction of hypersensitive cell death by a fungal protein in cultures of tobacco cells. Mol Plant-Microbe Interact. 1998;11:115–123. [Google Scholar]