Abstract
In a genetic screen to identify genes that promote GLP-1/Notch signaling in Caenorhabditis elegans germline stem cells, we found a single mutation, om40, defining a gene called ego-3. ego-3(om40) causes several defects in the soma and the germline, including paralysis during larval development, sterility, delayed proliferation of germline stem cells, and ectopic germline stem cell proliferation. Whole genome sequencing identified om40 as an allele of hsp-90, previously known as daf-21, which encodes the C. elegans ortholog of the cytosolic form of HSP90. This protein is a molecular chaperone with a central position in the protein homeostasis network, which is responsible for proper folding, structural maintenance, and degradation of proteins. In addition to its essential role in cellular function, HSP90 plays an important role in stem cell maintenance and renewal. Complementation analysis using a deletion allele of hsp-90 confirmed that ego-3 is the same gene. hsp-90(om40) is an I→N conservative missense mutation of a highly conserved residue in the middle domain of HSP-90. RNA interference-mediated knockdown of hsp-90 expression partially phenocopied hsp-90(om40), confirming the loss-of-function nature of hsp-90(om40). Furthermore, reduced HSP-90 activity enhanced the effect of reduced function of both the GLP-1 receptor and the downstream LAG-1 transcription factor. Taken together, our results provide the first experimental evidence of an essential role for HSP90 in Notch signaling in development.
Keywords: C. elegans, HSP90, germline stem cells, Notch, GLP-1
The HSP90 molecular chaperone plays critical roles in protein homeostasis (proteostasis), participating in the folding, maturation, and degradation of hundreds of substrate proteins, known as clients (Eckl and Richter 2013; Li et al. 2013; Karagöz and Rüdiger 2015; Haase and Fitze 2016; Schopf et al. 2017). Metazoans encode several highly conserved HSP90 proteins with specific isoforms localized to mitochondria, chloroplasts, endoplasmic reticulum, and the cytosol (Eckl and Richter 2013; Röhl et al. 2013; Haase and Fitze 2016; Schopf et al. 2017). Operating in conjunction with more than 20 co-chaperones, the HSP90 homodimer is an ATP-dependent molecular machine that binds to partially folded proteins to assist in their maturation through a yet-to-be-elucidated mechanism (Eckl and Richter 2013; Li et al. 2013; Röhl et al. 2013; Mayer and Le Breton 2015; Haase and Fitze 2016; Pearl 2016; Bar-Lavan et al. 2016; Schopf et al. 2017; Avellaneda et al. 2017). In addition, HSP90 plays an important role in directing misfolded proteins for proteasomal degradation (Taipale et al. 2014; Labbadia and Morimoto 2015; Schopf et al. 2017). Although HSP90 is absent from Archea, it is found throughout Eubacteria and Eukarya and is an essential protein in numerous eukaryotes, including S. cerevisiae, C. elegans, D. melanogaster, and vertebrates (Borkovich et al. 1989; Rutherford and Lindquist 1998; Lele et al. 1999; Voss et al. 2000; Birnby et al. 2000).
HSP90 has been implicated in numerous human diseases, including neurodegenerative diseases and cancer. With respect to neurodegeneration, several disorders, including Alzheimer’s Disease, Parkinson’s Disease, and Huntington’s Disease, involve misfolding and aggregation of proteins, perhaps as a result of HSP90 dysfunction (Pratt et al. 2015; Lackie et al. 2017). In many types of cancer, HSP90 and other components of the molecular chaperone network are overexpressed, enabling the maturation of many mutant proliferative signaling kinases and transcription factors (i.e., oncoproteins), thereby contributing to the growth factor independent growth and unregulated proliferation that are two of the hallmarks of cancer (Jarosz 2016; Calderwood and Gong 2016; Wu et al. 2017). In their wild-type state, these regulatory proteins are also HSP90 clients, pointing to a key role for HSP90 in control of normal cellular proliferation in growth and development (Rutherford et al. 2007a, 2007b; Schopf et al. 2017).
The Notch pathway, an important signaling pathway, is also disrupted in a variety of genetic diseases and cancers in humans (Aster et al. 2017; Mašek and Andersson 2017; Siebel and Lendahl 2017). Notch has arguably been best-studied in the context of development in D. melanogaster and C. elegans (Greenwald and Kovall 2013; Kimble and Seidel 2014; Kovall et al. 2017; Siebel and Lendahl 2017). In C. elegans, germline proliferation requires inductive signaling from the somatic gonad to the germline mediated by GLP-1, one of two C. elegans Notch orthologs along with LIN-12 (Greenwald and Kovall 2013). Critical components of this signaling pathway include: DSL-type ligands, LAG-2 and APX-1; GLP-1/Notch receptor; and downstream transcriptional regulators, LAG-1 and SEL-8/LAG-3 (Hansen and Schedl 2013; Kimble and Seidel 2014). Upon ligand-receptor binding, a pair of proteolytic cleavages releases the Notch intracellular domain (NICD) for transport to the nucleus where it nucleates formation of a transcriptional activator complex (Greenwald and Kovall 2013). When GLP-1/Notch signaling is absent or reduced in the C. elegans gonad, germline stem cells prematurely exit mitosis, enter meiosis, and form gametes (Hansen and Schedl 2013; Greenwald and Kovall 2013; Kimble and Seidel 2014).
Genetic studies in C. elegans have identified many of the core components of the Notch signaling pathway and numerous regulators of Notch signaling (Greenwald and Kovall 2013). Interestingly, several of these regulators are predicted to be components of the proteostasis network, including proteasome subunit PAS-5 and ubiquitin E3 ligases UBR-5, SEL-10, and RFP-1 (Hubbard et al. 1997; MacDonald et al. 2008; Gupta et al. 2015; Safdar et al. 2016). One fruitful approach for identifying Notch pathway regulators has been to use a sensitized genetic background, temperature-sensitive glp-1(bn18) mutants raised at semi-permissive temperature, to recover genetic enhancers of the glp-1 reduced germline proliferation phenotype (ego mutants) (Qiao et al. 1995; Maine et al. 2004; Liu and Maine 2007; She et al. 2009). This approach has identified factors that promote germline proliferation, either as components or regulators of GLP-1/Notch signaling or in parallel with GLP-1/Notch signaling.
Here we describe the molecular identification of another positive regulator of germline proliferation identified by this approach, ego-3. The single ego-3 allele recovered in initial screens, ego-3(om40), has a pleiotropic phenotype suggesting the ego-3 gene product is active in numerous tissues throughout the course of development (Qiao et al. 1995). We report here that ego-3 encodes the C. elegans ortholog of the cytosolic form of the HSP90 molecular chaperone. Our molecular, genetic, and phenotypic analyses show that HSP-90 is required for GLP-1/Notch signaling in C. elegans germline stem cells and provide the first evidence that HSP90 plays an essential role in Notch signaling in development.
Materials and Methods
Nematode strains and culture
Worms were maintained on E. coli strain OP50 seeded on NGM-Lite agar plates (Sun and Lambie 1997) under standard conditions (Epstein and Shakes 1995). Wild-type strains used were C. elegans variant Bristol (N2) and CB4856 (Hawaiian strain). Mutant strains used in this study were: BE63 sqt-3(sc63), CB1489 him-8(e1489), DG2389 glp-1(bn18), DR96 unc-76(e911), EL44 unc-32(e189) glp-1(bn18), EL129 hsp-90(om40) unc-76(e911)/nT1[unc-?(n754) let-?], EL160 lag-1(om13ts)/nT1[unc-?(n754) let-?]; hsp-90(om40)/nT1[unc-?(n754) let-?], EL301 lag-1(om13ts), EL375 sqt-3(sc63) unc-76(e911), EL384 sqt-3(sc63) hsp-90(om118) unc-76(e911)/sdc-3(y52y180) unc-76, EL418 hsp-90(om118) unc-76(e911)/nT1[unc-?(n754) let-?], EL490 unc-39(e257) hsp-90(om40)/unc-39(e257) unc-76(e911), EL660 lag-1(om13ts)/mIs11him-8(e1489), JL48 unc-32(e189) glp-1(bn18); hsp-90(om40)/nT1[qIs51], JL49 hsp-90(om40)/nT1[qIs51], JL54 unc-32(e189) glp-1(bn18); hsp-90(p673)/nT1[qIs51], JT6130 hsp-90(p673), TY1470 yDf8/nT1[unc-?(n754) let-?], VC914 hsp-90(ok1333)/nT1[qIs51], YY216 eri-9(gg106). Note: The daf-21 gene name was recently changed to hsp-90. Accordingly, the strains listed here are referred to using hsp-90 rather than daf-21 and ego-3.
RNAi feeding assay
RNAi of hsp-90 was performed by the feeding method (Timmons et al. 2001). A 2258 bp genomic region containing nearly the entire hsp-90 protein-coding region was amplified by polymerase chain reaction from wild-type N2 genomic DNA using 5′-TGTCCGAGAACGCCGAAA-3′ and 5′-GTCGACCTCCTCCATGCG-3′ as primers. The resulting amplicon was cloned into RNAi feeding vector L4440 and then transformed into E. coli RNAi feeding strain HT115 (Timmons et al. 2001). RNAi was performed by placing animals of a particular strain and stage onto hsp-90 feeding plates at a specified temperature as described in the Results. Adults were examined by differential interference contrast microscopy or were stained with DAPI to visualize DNA and examined by fluorescence microscopy (see below). An E. coli feeding strain expressing GFP dsRNA was used as a negative control.
Isolation of om118 by non-complementation screen
om40 non-complementation screens were performed using either UV or EMS mutagenesis in an effort to isolate additional alleles of ego-3. UV mutagenesis was performed as described (Yandell et al. 1994) and yielded a single mutation, om118, that failed to complement the om40 germline and somatic phenotypes. In brief, sqt-3(sc63) unc-76(e911) hermaphrodites were treated with 30 ug/ml trimethylpsoralen (TMP) in M9 buffer for 15 min in the dark and irradiated with a long-wave UV source (Model UVGL-25, UVP, Inc.) at distance of 10 cm for 20 sec. The sqt-3(sc63) mutation confers recessive squat (Sqt) and dominant roller (Rol) phenotypes. Single candidate sqt-3(sc63) ego-3(omx) unc-76(e911)/sqt-3(sc63) ego-3(+) unc-76(e911) F1 hermaphrodites were mated with ego-3(om40) unc-76(e911)/++ males. Rol Unc cross-progeny (sqt-3(sc63) ego-3(omx) unc-76(e911)/sqt-3(+) ego-3(om40) unc-76(e911) or sqt-3(sc63) ego-3(+) unc-76(e911)/sqt-3(+) ego-3(om40) unc-76(e911)) were screened for the Ego-3 sterile phenotype, which would presumably be of the genotype sqt-3(sc63) ego-3(omx) unc-76(e911)/+ ego-3(om40) unc-76(e911). The om118 mutation was identified from a total of 30,000 haploid genomes and recovered from fertile sqt-3(sc63) ego-3(om118) unc-76(e911)/+++ Rol non-Unc siblings.
Single nucleotide polymorphism (SNP) mapping
We mapped ego-3(om40) relative to SNPs as described (Maine et al. 2004). We put unc-39(e257) ego-3(om40) and ego-3(om40) unc-76(e911) chromosomes, generated in the N2 strain background, over chromosomes from the polymorphic wild-type strain, CB4856, and picked unc-39(e257) non-ego-3(om40) and unc-76(e911) non-ego-3(om40) recombinants, respectively. Using data available in WormBase (www.wormbase.org), we identified candidate SNPs in the region and verified their identity by PCR amplification and Sanger sequencing. We isolated homozygous recombinants from each Unc non-Ego recombinant line, PCR amplified the SNP region, and assayed by differential restriction digest or Sanger sequencing, as appropriate. This approach mapped ego-3(om40) to the right of two SNPs in the region covered by cosmid C54G10 and to the left of three SNPs in the region covered by Y50E8A, placing it within a 70 kb interval.
Whole genome sequencing (WGS)
Growth and isolation of ego-3(om40) homozygotes:
Strain JL49, ego-3(om40)/nT1[qIs51], contains ego-3(om40) balanced by a variant of the reciprocal translocation nT1 marked with an integrated myo-2::GFP construct that confers pharyngeal GFP expression. JL49 exhibits pharyngeal GFP and segregates sterile non-GFP ego-3(om40) homozygotes and GFP heterozygotes as the live progeny. The homozygous balancer is recessive lethal, and the strain also segregates a large percentage of nonviable aneuploid progeny.
JL49 was harvested from a starved 60 mm plate using M9 buffer supplemented with 0.01% Triton X-100, pelleted by centrifugation, and replated on 100 mm rich agarose plates (standard NGM recipe with 8X peptone and substituting low-EEO agarose at 1:1 for agar) seeded with lawns of E. coli strain OP50 or χ1666. Populations were allowed to grow at 20° until dense with growing, well-fed worms of mixed stage, then harvested by M9/Triton X-100 wash and centrifugation. Excess bacterial food was removed by three to five further rounds of washing and pelleting.
The resulting population was flow-sorted using a Copas Biosort 250 with fluorescence detection and Biosorter software version 5.25.2. A sorter screening window was chosen to select worms at the L4 stage or older. This set was further restricted to animals not expressing pharyngeal GFP, using the fluorescence-detection capability of the Copas sorter, resulting in a very highly enriched sample of ego-3(om40) homozygotes for whole genome sequencing.
The sorted homozygotes were replated on standard 60 mm NGM plates, and any remaining GFP animals were removed by hand picking. The contamination rate for GFP animals was very low, approximately 40 GFP animals in 15,000 total sorted worms.
Library preparation and sequencing:
The sample of ego-3(om40) homozygotes was frozen at -80°. Genomic DNA was prepared using a standard protocol of Proteinase K digestion, treatment with RNAse A and 6 M NaCl, precipitation in isopropanol, washing, and resuspension in distilled water. A sequencing library was generated using the Nextera XT library preparation protocol, and the resulting library was sequenced on an Illumina HiSeq machine in the laboratory of Dr. Corey Nislow (Department of Pharmaceutical Sciences, University of British Columbia). Paired-end reads of 100 bp were generated, resulting in total genome coverage of 57X.
Sequence analysis:
Single-nucleotide variants (SNVs) were called with the analysis pipeline and filters developed for the Million Mutation Project (Thompson et al. 2013). Briefly, sequence read pairs were aligned to the C. elegans reference genome version WS230 (www.wormbase.org) using the short-read aligner Phaster (P. Green, personal communication). SNVs were then identified with SAMtools (Li et al. 2009) and annotated with a custom Perl script.
Sanger DNA sequencing
The om40 molecular lesion identified by WGS was confirmed by PCR amplification of the region containing the putative mutation from om40 homozygotes followed by Sanger sequencing. The om118 molecular lesion was identified by amplifying each predicted hsp-90 exon, including exon-intron junctions, by PCR from ego-3(om118) unc-76(e911)/nT1[unc-?(n754) let-?] heterozygotes followed by Sanger DNA sequencing. Sequence data suggested the presence of an insertion mutation in hsp-90. To determine unambiguously the location and sequence of the insertion, the region containing the putative insertion was again amplified from ego-3(om118) unc-76(e911)/nT1[unc-?(n754) let-?] heterozygotes, and PCR products were cloned into pUC19 using standard techniques. Plasmid DNA was isolated from 12 independent clones and sequenced. All clones contained either wild-type sequence or the om118 insertion mutation. PCR primer sequences and their locations can be found in Table S3 and Figure S1 in File S1, respectively.
Microscopy and DAPI methods
Microscopy was carried out with a Zeiss Axioskop equipped with differential interference contrast (DIC) optics and epifluorescence. For fluorescence visualization of nuclei, intact worms were fixed in -20° methanol, and dissected gonads were fixed with 3% paraformaldehyde. Fixed tissue was incubated for 10-15 min in 0.2 ug/ml DAPI at room temperature and mounted in Vectashield (Vector Laboratories).
Data availability
All C. elegans strains and plasmid constructs used in this study are available upon request. The raw sequence data from the WGS have been submitted to the NCBI BioProject (http://www.ncbi.nlm.nih.gov/bioproject) under accession number PRJNA428227 and can be accessed from the Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) with accession number SRP127800. File S1 contains Figure S1, Locations of primers used to amplify and sequence regions of the hsp-90 gene to identify mutations; Figures S2–S4 in File S1, Sanger DNA sequencing chromatograms of wild-type and mutant sequences; Table S1 in File S1, Gonad transcriptome data for protein-coding genes in the ego-3 region; Table S2 in File S1, Codon and splicing changes detected in ego-3(om40) by whole genome sequence analysis; and Table S3 in File S1, Primers used to amplify and sequence regions of the hsp-90 gene to identify mutations.
Results
One goal of the glp-1 enhancer screen was to isolate partial loss-of-function alleles of genes that promote GLP-1/Notch signaling (Qiao et al. 1995). The rationale was to recover mutations that cause premature loss of all germline stem cells in a sensitized genetic background where GLP-1 activity is suboptimal, but do not cause the loss of germline stem cells when GLP-1 activity is normal. For example, glp-1(bn18ts); ego-3(om40) adults do not maintain a population of proliferative germ cells, but instead all germ cells enter meiosis prematurely and undergo gametogenesis (Qiao et al. 1995). In a glp-1(+) background, in contrast, ego-3(om40) adults maintain a proliferative germ line. During development, ego-3(om40) alone also causes a number of defects in somatic and germline tissues including delayed development, paralyzed movement (an Unc phenotype), and “early” and “late” germline defects (Qiao et al. 1995). Notably, the Unc phenotype is much less severe in adults than in larvae. The early germline phenotype is a transient pause in larval germline proliferation from early L3 to early/mid-L4 stages; the late germline phenotype includes delayed onset of meiosis and gametogenesis, sterility, and development of a proximal germ cell tumor. Most aspects of the ego-3(om40) phenotype appear to be loss-of-function based on analysis of ego-3(om40)/yDf8 individuals (Qiao et al. 1995). However, the proximal tumor phenotype is milder in ego-3(om40)/yDf8 animals, suggesting this aspect of the phenotype may not be due to a strict loss-of-function.
Isolation of a strong ego-3 loss-of-function allele
To identify additional alleles of ego-3, we carried out a non-complementation screen using UV/trimethylpsoralen (TMP) mutagenesis. One mutation, ego-3(om118), was identified that fails to complement ego-3(om40) for the germline and somatic developmental defects described above and for glp-1 enhancement. In contrast, ego-3(om118) homozygotes and ego-3(om118)/yDf8 transheterozygotes die as embryos, thus ego-3(om118) appears to be a more severe loss-of-function allele than ego-3(om40). Consistent with this hypothesis, the ego-3(om118/om40) Unc phenotype is more severe than for ego-3(om40) animals, persisting into adulthood rather than being limited to larval stages.
ego-3 is allelic to daf-21 and encodes HSP90
ego-3(om40) maps just to the left of unc-61 on LGV (Qiao et al. 1995). To determine the molecular identity of ego-3, we performed additional three-factor and SNP mapping to refine the location of ego-3. Three-factor mapping placed ego-3(om40) to the right of or very close to the left of daf-21, which was recently renamed hsp-90 (www.wormbase.org). Specifically, 23/23 Unc non-Ego recombinants from an hsp-90(p673)/ego-3(om40) unc-76(e911) strain were hsp-90(p673) unc-76(e911)/ego-3(om40) unc-76(e911). Note that hsp-90(p673) is a recessive gain-of-function mutation causing a dauer-constitutive (Daf-c) phenotype (Birnby et al. 2000), and therefore would be expected to complement a loss-of-function mutation in the same gene.
We next mapped ego-3(om40) relative to SNPs in the unc-39 to unc-76 interval, further localizing it to an ∼70 kb region containing 14 predicted protein-coding genes and several non-coding RNA genes (Figure 1). Gonad transcriptome data (Guo et al. 2015) indicated that nine of these genes are expressed in the gonad (Table S1 in File S1). As none was an obvious candidate for involvement in GLP-1/Notch signaling, we undertook to identify the ego-3(om40) mutation by whole genome sequencing (WGS).
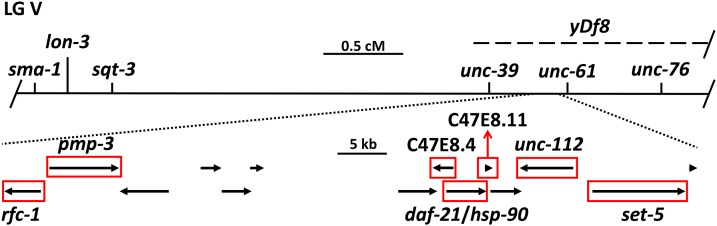
Figure 1.
Genetic and physical maps of ego-3 region. The genetic map of the ego-3 region is shown at the top along with the location of the yDf8 deletion used in this study. SNP mapping localized ego-3 to a region to the left of unc-61. The physical map of this region, obtained from WormBase WS259 and showing only protein-coding genes, is shown below the genetic map. Arrows indicate location, length, and orientation of genes. Red boxes indicate genes with substantial expression in the gonad (Table S1 in File S1).
WGS of ego-3(om40) homozygotes identified one sequence variant within the protein-coding region of one gene in the mapped region, hsp-90, which encodes the C. elegans ortholog of the cytoplasmic form of the molecular chaperone HSP90 (Table S2 in File S1) (Birnby et al. 2000). The om40 sequence variant is a T→A transition causing an I→N non-conservative missense mutation (Figure 2A), which we confirmed by Sanger sequencing (Figure S2 in File S1). Subsequently, we amplified and Sanger sequenced all hsp-90 exons from ego-3(om118) to locate the om118 lesion. We discovered a 20 bp non-tandem duplication within the same exon containing the om40 mutation, which is predicted to cause a frameshift and to introduce a premature termination codon (Figure 2B and Figure S4 in File S1). The inserted sequence occurs only once in the C. elegans wild-type genome and is therefore highly likely to have originated from within the hsp-90 gene. These data are strong evidence that hsp-90 and ego-3 are the same gene.
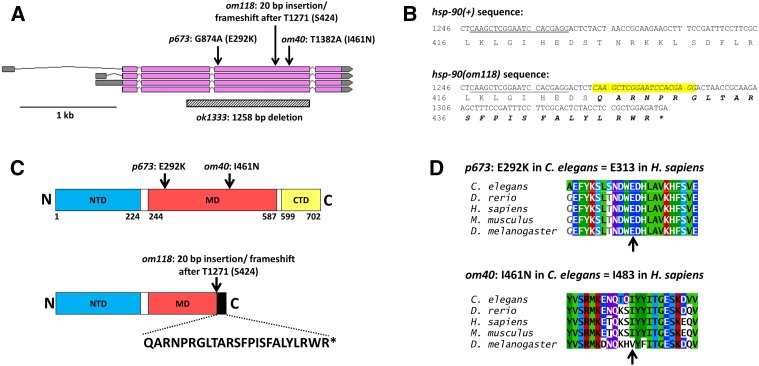
Figure 2.
Molecular analysis of hsp-90 alleles. A. hsp-90 is predicted to encode four transcript isoforms, all of which have the same predicted amino acid sequence. Arrows indicate locations of new and previously identified hsp-90 alleles. The locations of p673, om118, and om40 were determined by Sanger sequencing, and the location of ok1333 was obtained from WormBase. The nucleotide positions of the mutations are numbered from the first nucleotide of the protein-coding sequence; numbering does not include nucleotides in introns. Amino acid positions in parentheses are numbered from the initiator methionine. Numbering is from WormBase WS259. B. Partial DNA and amino acid sequences of hsp-90(+) and hsp-90(om118) insertion/frameshift mutation. Numbers indicate nucleotide positions in the protein-coding sequence and amino acid positions in the predicted translation product. The underlined nucleotide sequence is duplicated and inserted 5 bp downstream in hsp-90(om118); the inserted sequence is highlighted in yellow. Amino acids changed as a result of the insertion are shown in bold italics. *Premature termination codon. C. Schematic diagram of C. elegans HSP-90 domain organization and location of mutations. Top, schematic diagram showing locations of p673 and om40 missense mutations. Bottom, schematic diagram showing location of om118 insertion mutation. The black box indicates extent of the amino acid sequence changes caused by the insertion and resulting frameshift. The sequence of amino acids added to HSP-90 after S424 are shown. The predicted length of the mutant protein is 448 amino acids. Amino acid sequences of C. elegans HSP-90 (NP_506626.1) and H. sapiens HSP90AA1 (NP_005339.3) were aligned using NCBI Protein BLAST (blastp) to identify corresponding amino acid residues and HSP90AA1 domain boundaries from Haase and Fitze 2016 were used to identify the corresponding boundaries in HSP-90. Numbers indicate amino acid residues in C. elegans HSP-90. NTD, N-terminal domain; MD, middle domain; CTD, C-terminal domain. D. Partial amino acid sequence alignment of HSP90 from C. elegans and selected metazoans. Top, alignment of region containing the p673 mutation. Bottom, alignment of region containing the om40 mutation. The corresponding amino acid number in H. sapiens for each mutation is shown. Arrow, location of amino acid affected by the indicated mutations.
To confirm that ego-3 and hsp-90 are indeed the same gene, we tested for complementation between ego-3(om40) and hsp-90(ok1333), a deletion and presumed null allele (Figure 2A) (C. elegans Deletion Mutant Consortium 2012). hsp-90(ok1333) worms have a paralyzed Unc phenotype and arrest development during mid to late larval stages (www.wormbase.org). We crossed ego-3(om40)/nT1[qIs51] males with hsp-90(ok1333)/nT1[qIs51] hermaphrodites at 20°. The nT1[qIs51] balancer chromosome has pharyngeal GFP expression, so non-GFP progeny will be hsp-90(ok1333) homozygous self-progeny and hsp-90(ok1333)/ego-3(om40) cross-progeny. Large numbers of GFP males, presumably ego-3(om40)/nT1[qIs51] cross-progeny, were produced, indicating that the ego-3(om40)/nT1[qIs51] males mated efficiently.
The vast majority of non-GFP offspring from the cross were paralyzed Uncs and arrested as larvae, as expected if hsp-90(ok1333) and ego-3(om40) do not complement. A few non-GFP worms survived to adulthood, with most developing into sterile, severe Unc hermaphrodites and severe Unc males. Therefore, ego-3(om40) fails to complement hsp-90(ok1333), and ego-3 is allelic to hsp-90, consistent with our molecular analysis of ego-3 alleles.
Interestingly, the ego-3(om40) larval phenotype resembles the daf-21(ok1333) phenotype in that animals are paralyzed, develop slowly, and some aspects of development arrest during L2-L3 stage (www.wormbase.org; Qiao et al. 1995). However, both somatic and germline development arrest in hsp-90(ok1333) mutants whereas only germline development arrests in ego-3(om40). We interpret the viability and complementation of om40/p673 transheterozygotes (constructed during genetic mapping, see Materials and Methods) as resulting from the previously reported gain-of-function nature of the p673 allele (Birnby et al. 2000).
Additionally, we determined the nucleotide sequence change in hsp-90(p673), which had not been previously reported although the amino acid sequence change for this allele had been (Birnby et al. 2000). We found a G→A transition mutation at the expected location, confirming the presence of the previously identified non-conservative E→K missense mutation (Figure 2A and Figure S3 in File S1).
The om40, om118, and p673 alleles are all predicted to cause amino acid sequence changes affecting the middle domain (MD) of HSP90 (Figure 2C). Both the p673 and om40 alleles are non-conservative missense mutations of highly conserved residues in metazoans (Figure 2D) (Birnby et al. 2000). Furthermore, om118 is predicted to lead to loss of much of the middle domain and all of the carboxy-terminal domain (CTD) of HSP90. The MD is essential for interactions with both client proteins and co-chaperones, and it contains a catalytic loop that contributes to ATP hydrolysis (Röhl et al. 2013; Haase and Fitze 2016; Schopf et al. 2017). The CTD is the dimerization domain of HSP90, and it contains a highly conserved tetratricopeptide repeat (TPR) domain binding site required for interactions with many co-chaperones (Röhl et al. 2013; Haase and Fitze 2016; Schopf et al. 2017).
Based on our molecular and genetic data, we conclude that ego-3 and hsp-90 are the same gene. In keeping with chaperone nomenclature in other organisms, the ego-3 and daf-21 gene names have been changed to hsp-90 (www.wormbase.org).
hsp-90(p673) does not enhance glp-1(bn18)
We hypothesized that hsp-90(p673) should not enhance glp-1(bn18), because hsp-90(p673) is a gain-of-function allele (Birnby et al. 2000), and it also complements hsp-90(om40) (see above). To test this, we constructed an unc-32(e189) glp-1(bn18); hsp-90(p673)/nT1[qIs51] strain. unc-32(e189) glp-1(bn18); hsp-90(p673) offspring are viable and fertile at 20° and produce a large proportion of dauer offspring, as expected for hsp-90(p673) animals. We also used DIC microscopy to examine the germlines of unc-32(e189) glp-1(bn18); hsp-90(p673) worms raised at 20° to see if hsp-90(p673) enhanced the mild germline proliferation defect in glp-1(bn18). hsp-90(p673) germlines resemble N2 wild-type germlines with extensive germline proliferation (Figure 3, A and C). In contrast, unc-32(e189) glp-1(bn18) worms raised at 20° have substantially smaller germlines because of reduced GLP-1/Notch signaling (Figure 3B) (Qiao et al. 1995). unc-32(e189) glp-1(bn18); hsp-90(p673) germlines closely resemble those of unc-32(e189) glp-1(bn18) animals (Figure 3D), therefore hsp-90(p673) does not enhance glp-1(bn18). Taken together, our results show that the hsp-90(om40) and hsp-90(p673) alleles both retain substantial HSP-90 activity such that the heteroallelic combination has approximately normal HSP-90 function. We further conclude that only a subset of hsp-90 mutations have the ability to enhance glp-1.
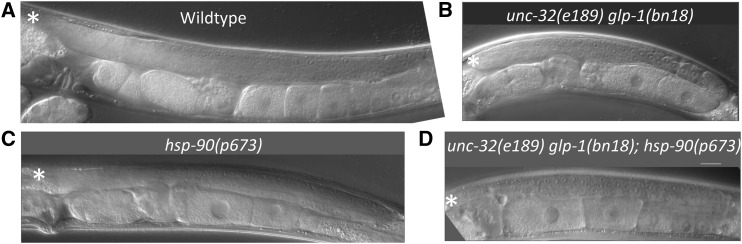
Figure 3.
hsp-90(p673) does not enhance the germline proliferation defect in glp-1(bn18). Germline morphology of animals raised at 20°. Adult hermaphrodites were photographed at the same magnification with DIC microscopy; one gonad arm from representative animals is shown in each panel. Asterisk indicates the distal end of the germline. A. N2 (wild-type) adult hermaphrodite gonad has a large germline with extensive proliferation. B. unc-32(e189) glp-1(bn18ts) hermaphrodites are fertile but have approximately half as many germ cells as wild type (N2) (Qiao et al. 1995). unc-32(e189) is a visible genetic marker that does not affect germline development. C. hsp-90(p673) hermaphrodites are fertile with a wild-type germline morphology. D. unc-32(e189) glp-1(bn18ts); hsp-90(p673) hermaphrodites are fertile and have a germline similar in size to the unc-32(e189) glp-1(bn18ts) germline.
The glp-1(bn18) phenotype is enhanced by the loss of HSP90 activity
Our previous study determined that the glp-1(bn18) germline proliferation defect is enhanced in hsp-90(om40)/yDf8; glp-1(bn18) animals (Qiao et al. 1995), strongly suggesting a loss of HSP-90 activity is responsible for the enhancement. In addition, the early germline (arrested proliferation) and uncoordinated phenotypes were similar in hsp-90(om40)/yDf8; glp-1(+) and hsp-90(om40/om40) mutants, consistent with those defects arising from reduced HSP-90 activity. However, the late germline phenotype, including proximal germline proliferation (Pro phenotype) and delayed gametogenesis, was less severe in hsp-90(om40)/yDf8 than in hsp-90(om40/om40) adults. At the time, we hypothesized this result might indicate that hsp-90(om40) has some gain-of-function character.
Molecular identification of the ego-3 gene has now allowed us to investigate the nature of the om40 allele in more detail. The lethal hsp-90(ok1333) allele appears to be null (as described above) and the yDf8 deletion completely uncovers the hsp-90 gene; therefore, the viability of hsp-90(om40/ok1333) and hsp-90(om40)/yDf8 confirm that hsp-90(om40) is not a null allele. At the same time, similarities among hsp-90(om40), hsp-90(om40/ok1333), and hsp-90(om40)/yDf8 phenotypes suggest that hsp-90(om40) predominantly causes a loss of gene function. To confirm this interpretation and to investigate the nature of the proximal proliferation defect, we knocked down HSP-90 via RNAi and examined the impact on development. Our first strategy was to place L4 wild-type (N2) hermaphrodites onto feeding plates containing bacteria expressing hsp-90 dsRNA at 20° and evaluate their F1 progeny. This strategy partially phenocopied hsp-90(om40), yielding offspring that were sterile, many with very reduced germline proliferation. Knockdown also caused some embryonic lethality in the F1, a phenotype observed in hsp-90(ok1333) and hsp-90(om118), and a protruding vulva phenotype. We observed similar results in unc-32(e189) glp-1(bn18); hsp-90(RNAi) animals with the additional observation of some embryonic lethality. We also performed hsp-90(RNAi) in an enhanced RNAi mutant, eri-9(gg106), and saw the same phenotypes, with the addition that some larvae arrested development. We next placed L1 larvae onto hsp-90 RNAi plates at 20° and examined the adults three days later. In a wild-type background, hsp-90(RNAi) adults were sterile with protruding vulvas and had very reduced germlines; the same results were obtained for unc-32(e189) glp-1(bn18); hsp-90(RNAi) although a smaller proportion of animals had a protruding vulva. eri-9(gg106) hsp-90(RNAi) adults were also sterile, but most animals were Unc and very sick with several having died in early adulthood. In no case did we observe the proximal germline proliferation (Pro) characteristic of hsp-90(om40).
As a follow up experiment, we assayed the effect of beginning hsp-90 RNAi at L3 stage, reasoning that allowing some germline development to occur prior to knocking down HSP-90 might increase the chance of observing the Pro phenotype observed in hsp-90(om40) adults (Figure 4A). We performed this study with a him-8(e1489) strain in order to evaluate both males and hermaphrodites. Untreated him-8(e1489) worms have normal germlines (Figure 4C). The majority of hsp-90(RNAi) adults produced in this study had germline developmental defects, often resembling either the hsp-90(om40) early or late germline defects (Figure 4, B, D, and E). Approximately 73% of germlines were small and germ cell nuclei had an enlarged morphology similar to the hsp-90(om40) early germline phenotype (n = 52; Figure 4D); this hsp-90(RNAi) phenotype is consistent with previous observations (Waters et al. 2010). The remaining ∼27% of germlines had more extensive proliferation and produced gametes. A subset of these, ∼6% of germlines assayed, had proximal proliferation (Figure 4, B and E) resembling the hsp-90(om40) Pro phenotype (Figure 4A). Therefore, we conclude that the hsp-90(om40) Pro phenotype results from reduced HSP90 activity.
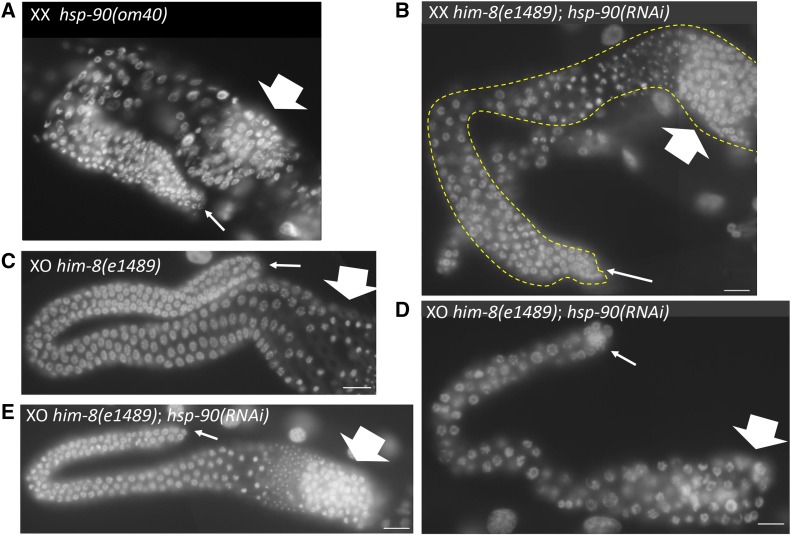
Figure 4.
hsp-90(RNAi) phenocopies hsp-90(om40) germline phenotype. Germline nuclei from adult worms were stained with DAPI to reveal chromosome morphology. Thick arrows and thin arrows indicated proximal and distal regions of the germline, respectively. Scale bar, 16 um. A. Gonad from intact hsp-90(om40) adult hermaphrodite (XX) showing the Pro phenotype characteristic of hsp-90(om40). Mitotic nuclei are visible in both distal and proximal regions. B. Gonad dissected from a him-8(e1489); hsp-90(RNAi) adult hermaphrodites (XX). Gonad outlined with yellow dashed line shows the Pro phenotype. C. Gonad dissected from a him-8(e1489) adult male (XO) displays wild-type germline nuclear morphology. Mitotic nuclei are visible in the distal region and sperm (smallest bright spots) are present in the proximal region. D. Gonad dissected from a him-8(e1489); hsp-90(RNAi) adult male (XO) showing large nuclei in the distal region. E. Gonad dissected from a him-8(e1489); hsp-90(RNAi) adult male (XO) showing the Pro phenotype.
Loss of HSP-90 activity also enhances lag-1(om13ts)
To further explore the role of HSP-90 in promoting Notch signaling, we asked whether hsp-90(om40) enhances the loss of lag-1 function in the germline. LAG-1 is a CSL-type transcription factor active downstream of GLP-1 in the C. elegans germline (Hansen and Schedl 2013; Kimble and Seidel 2014). Here, we used a temperature sensitive allele, lag-1(om13ts), that is fertile at 20° (Qiao et al. 1995; Safdar et al. 2016). We reasoned that the combination of partial loss-of-function mutations in two genes involved in GLP-1/Notch signaling should substantially reduce germline proliferation. We generated a lag-1(om13ts)/nT1[unc-?(n754) let-?]; hsp-90(om40)/nT1[unc-?(n754) let-?] strain and evaluated germline proliferation in the hsp-90(om40); lag-1(om13ts) progeny raised at 20°. hsp-90(om40) and lag-1(om13ts) single mutants, each obtained from balanced strains, were assayed in parallel; all animals were stained with DAPI as ∼1 day old adults. As expected based on our previously published data, germline proliferation was retained in 100% of hsp-90(om40) and lag-1(om13ts) single mutants (Table 1) (Qiao et al. 1995; Safdar et al. 2016). In contrast, 100% of lag-1(om13ts); hsp-90(om40) adult hermaphrodite germlines contained sperm, but lacked proliferating and meiotic germ cells (Table 1). Hence, in these double mutants, all proliferating germ cells prematurely exit mitosis, enter meiosis, and undergo gametogenesis. The germ cell precursors in these lag-1(om13ts); hsp-90(om40) animals apparently divided only a few times, as an average of only 26 sperm were observed per gonad arm (range 10-46; n = 12), corresponding to an average of 6-7 germ cells. This result is consistent with a severe reduction in GLP-1 signaling in the lag-1(om13); hsp-90(om40) double mutant and further supports our conclusion that HSP-90 activity promotes GLP-1 pathway signaling activity.
Table 1. hsp-90(om40) enhances lag-1(om13ts) in the germline.
| Genotype | Na | % with distal mitotic GCb | % with meiotic GCb | % with gametes (type) |
|---|---|---|---|---|
| lag-1(om13); hsp-90(om40) | 62 | 0 | 0 | 100 (sperm) |
| lag-1(om13ts) | 30 | 100 | 100 | 100 (sperm + oocytes) |
| hsp-90(om40) | 32 | 100 | 94 | 78 (sperm +/or oocytes)c |
N, number of gonad arms assayed.
GC, germ cells.
The phenotype of ∼1 day old adult hsp-90(om40) hermaphrodites was variable with respect to gamete production and presence of a proximal tumor, as previously described (Qiao et al. 1995).
Assays were performed at 20°. Mutations were maintained as balanced heterozygotes. Homozygous mutant offspring were identified and assayed within the first 24 hr of adulthood.
discussion
By various genetic and molecular criteria, we have determined the identity of ego-3 as hsp-90, which encodes the C. elegans ortholog of the molecular chaperone HSP90. Our findings that hsp-90(om40) enhances mutations in both the GLP-1/Notch receptor and the LAG-1 downstream effector demonstrate that HSP-90 activity directly or indirectly promotes GLP-1/Notch signaling and is therefore required for proper germline stem cell function. While HSP90 has long been known to participate in a variety of developmental as well as proliferative/oncogenic signaling pathways in metazoans (Whitesell and Lindquist 2005; Rutherford et al. 2007b; Green et al. 2011; Haslbeck et al. 2012; Garg et al. 2016; Calderwood and Gong 2016; Wu et al. 2017; Chatterjee and Burns 2017), our results provide the first evidence that HSP90 plays a role in Notch signaling in development. Recently, direct physical interaction between HSP90 and Notch intracellular domain (NICD) has been demonstrated in cultured human cells (Deskin et al. 2016; Wang et al. 2017), consistent with our results and with a direct role for HSP90 in Notch signaling.
Our discovery that HSP-90 modulates GLP-1/Notch signaling in C. elegans adds to the number of proteostasis network components known to regulate Notch function in various organisms. These components are also important in stem cell function in general (Lee et al. 2017; Noormohammadi et al. 2017; Werner et al. 2017) and in C. elegans germline stem cell function in particular. In addition to direct interaction between NICD and HSP90, Notch signaling is altered by loss of or reduced function of ubiquitin E3 ligases and the proteasome (MacDonald et al. 2008; Gupta et al. 2015; Safdar et al. 2016; Carrieri and Dale 2017; Wang et al. 2017; Perez-Mockus and Schweisguth 2017). Our data do not allow us to determine which aspect or aspects of HSP90’s function in proteostasis of GLP-1/Notch, i.e., folding, maturation, interactions with other proteins, or degradation, are necessary for GLP-1/Notch signaling. Systematic investigation of the hundreds of proteins that make up the proteostasis network regulating synthesis, maturation, function, and degradation of the proteome is likely to yield additional factors regulating Notch signaling.
In C. elegans, hsp-90 has previously been shown to be an essential gene that produces a variety of phenotypes when mutated or knocked down by RNAi including sterility, embryonic and larval lethality, constitutive dauer formation, defective motility, and various germline defects (Vowels and Thomas 1994; Piano et al. 2000; Rual et al. 2004; Gaiser et al. 2009, 2011; Waters et al. 2010; C. elegans Deletion Mutant Consortium 2012; Frumkin et al. 2014). These pleiotropic effects are consistent with ubiquitous expression of hsp-90 in most tissues (except for mature sperm) and throughout the nematode lifecycle from embryonic through adult stages (Yamaguchi et al. 1983; Inoue et al. 2003; Waters et al. 2010; Klosin et al. 2017).
The germline phenotypes seen in hsp-90(om40) and hsp-90(RNAi) animals are consistent with previously observed strong expression of HSP-90 in the developing and mature C. elegans germline (Yamaguchi et al. 1983; Inoue et al. 2003), where the GLP-1/Notch receptor is expressed (Crittenden et al. 1994). A germline role for HSP-90 in GLP-1/Notch signaling is also indicated by the observation that the hsp-90(om40) larval germline phenotype is epistatic to glp-1(oz112gf/q224), a heteroallelic combination that results in germline overproliferation (Qiao et al. 1995; Berry et al. 1997). The GLP-1/Notch protein encoded by glp-1(oz112) is constitutively active, yet reduction of HSP-90 activity in glp-1(oz112gf/q224); hsp-90(om40) larvae is sufficient to reduce GLP-1/Notch signaling and produce the hsp-90(om40) phenotype instead. Hence, Qiao et al. (1995) concluded that HSP-90 acts downstream of GLP-1/Notch. Furthermore, the recently reported physical interaction between NICD and HSP90 also supports the germline as a location of HSP-90 action in GLP-1/Notch signaling in C. elegans (Deskin et al. 2016; Wang et al. 2017). Of course, none of these observations rules out a separate role for HSP-90 in the signaling cell (e.g., distal tip cell).
The molecular identities of the two known hsp-90 missense mutations, hsp-90(p673) and hsp-90(om40), which cause non-conservative amino acid substitutions of highly conserved residues in the middle domain HSP-90, do not suggest obvious mechanisms by which they might produce their phenotypic effects. hsp-90(p673) is a recessive gain-of-function mutation producing a dauer-constitutive phenotype (Vowels and Thomas 1994; Birnby et al. 2000). The gain-of-function nature of this allele is consistent with the results of numerous hsp-90(RNAi) experiments, including our own, that fail to show an increase in dauer formation (Piano et al. 2000; Rual et al. 2004; Gaiser et al. 2009, 2011; Waters et al. 2010; Melo and Ruvkun 2012; Frumkin et al. 2014). It is also consistent with our genetic analysis showing that hsp-90(p673) retains substantial HSP-90 activity and with the biochemical and biophysical analysis of bacterially-expressed wild-type and E292K (corresponding to hsp-90(p673)) C. elegans HSP-90 showing that this mutation only slightly reduces function of the protein (Gaiser et al. 2011). All of these data support the gain-of-function character of hsp-90(p673). Specifically, Gaiser et al. (2011) demonstrated that the ATPase activity of the HSP-90(E292K) mutant protein is slightly reduced at 25°, but not at lower temperature. Moreover, the E292K substitution does not seem to alter the structural stability of the protein nor its binding of two HSP-90 co-chaperones, p23 (encoded by ZC395.10) and AHA-1, although binding to the STI-1 co-chaperone was slightly reduced (Gaiser et al. 2011).
With respect to hsp-90(om40), Qiao et al. (1995) described a complex suite of phenotypes produced by hsp-90(om40), leading the authors to conclude that hsp-90(om40) might have both loss- and gain-of-function features. For example, the Pro phenotype is less severe in hsp-90(om40/null) adults, a result usually interpreted to mean that a phenotype is a consequence of a gain-of-function. However, nearly all hsp-90(om40) phenotypes (Unc, arrested germline mitosis, delayed gametogenesis, oocyte defects, large germline nuclei, and sterility), including Pro, are recapitulated by creating hsp-90(om40) transheterozygotes with two severe loss-of-function alleles, hsp-90(om118) and hsp-90(ok1333) and by hsp-90(RNAi) knockdown (Piano et al. 2000; Rual et al. 2004; Gaiser et al. 2009, 2011; Waters et al. 2010; Frumkin et al. 2014). Thus, we conclude that hsp-90(om40), which causes an I461N substitution, is a loss-of-function mutation and that the I461N substitution reduces HSP-90 activity.
HSP90 is assisted in its protein folding function of a plethora of clients by more than 20 different co-chaperones (Li et al. 2013; Schopf et al. 2017). Thus, the highly varied and complex phenotypes associated with different hsp-90 alleles and with hsp-90(RNAi) are not surprising and are likely in part a consequence of disrupting both HSP-90’s interaction with any of a number of co-chaperones and the folding of at least several of the hundreds of client proteins with which HSP90 assists (https://www.picard.ch/downloads/Hsp90interactors.pdf). Furthermore, co-chaperones display a range of tissue expression patterns, including in C. elegans, suggesting that even wild-type HSP-90 activities will vary in different tissues (Haslbeck et al. 2012). This dizzying complexity of the HSP90 chaperone system may explain some of the unusual features of hsp-90(om40). For example, a curious aspect of the hsp-90(om40) phenotype is a reversal of larval germline mitotic arrest and severe Unc defects when mutant animals reach late larval stages and adulthood (Qiao et al. 1995). Perhaps HSP-90(I461N) has impaired interactions with co-chaperones and/or client proteins expressed in larval germline and muscle, but has increased activity in older animals because of differences in co-chaperones and client proteins produced during late larval and adult stages.
In wild-type C. elegans, mitotic proliferation of germline stem cells is restricted to the distal region of the germline. However, one of the unusual loss-of-function phenotypes seen in hsp-90(om40) and hsp-90(RNAi) worms is proliferation in the proximal region of the germline (Pro phenotype) leading to formation of a proximal germline tumor. How does reduction of HSP-90 function, which appears to reduce GLP-1/Notch signaling and therefore reduce distal germline mitotic proliferation, paradoxically promote proximal germline mitotic proliferation? Whatever the mechanism, it seems to involve GLP-1/Notch signaling because proximal tumor formation is suppressed by the loss of glp-1 function (Qiao et al. 1995). In hsp-90(om40) and hsp-90(RNAi) hermaphrodites, mitotic arrest during larval germline development delays the entry of germ cells into meiosis and exposes them to a proposed latent sheath cell niche, thereby stimulating their proliferation via GLP-1/Notch signaling, as described for mutations in several other genes (McCarter et al. 1997; Killian and Hubbard 2005; McGovern et al. 2009; Korta and Hubbard 2010). It is not clear, though, what the mechanism is for proximal tumor formation in males because males do not have gonad sheath cells (Wolf et al. 1978), but the numerous membrane-bound and secreted Notch ligands expressed in C. elegans are possible candidates for activating proximal proliferation in males as well as in hermaphrodites (Chen and Greenwald 2004; Komatsu et al. 2008; Greenwald and Kovall 2013). Further research into the HSP-90 chaperone system, including the roles and expression patterns of specific co-chaperones, should help to illuminate the complex functions of HSP-90 reported here.
Supplementary Material
Supplemental Material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.118.300551/-/DC1.
ACKNOWLEDGMENTS
Some strains were provided by the Caenorhabditis Genetics Center (CGC), which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440), and we made extensive use of WormBase. VC914 (hsp-90(ok1333) V/nT1[qIs51](IV;V)) was provided through the CGC by the C. elegans Deletion Mutant Consortium 2012. We thank the following for technical assistance: Xia Xu, construction of GFP RNAi feeding construct; Jill Spoerke, assistance with the om40 non-complementation screen; Alyson Wolk and Brianna Lajeunesse, assistance with PCR; and Rebecca Rohwer, assistance with complementation analysis. J.L.L. thanks Dr. Peter Harte, Dept. of Genetics and Genome Sciences, Case Western Reserve University School of Medicine, in whose lab some of the experiments were conducted during J.L.L.’s sabbatical. The authors acknowledge funding from John Carroll University to J.L.L., from the National Science Foundation (Grants #IBN-9318709 and #IBN-0077172) and Syracuse University to E.M.M., and from the Canadian Institute for Health Research (Grant #PJT-148549) and the National Institutes of Health (Grant #5P40OD010440) to D.G.M.
Footnotes
Communicating editor: M. Walhout
Literature Cited
- Aster J. C., Pear W. S., Blacklow S. C., 2017. The varied roles of Notch in cancer. Annu. Rev. Pathol. Mech. Dis. 12(1): 245–275. 10.1146/annurev-pathol-052016-100127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avellaneda M. J., Koers E. J., Naqvi M. M., Tans S. J., 2017. The chaperone toolbox at the single-molecule level: From clamping to confining. Protein Sci. 26(7): 1291–1302. 10.1002/pro.3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Lavan Y., Shemesh N., Ben-Zvi A., 2016. Chaperone families and interactions in metazoa. Essays Biochem. 60(2): 237–253. 10.1042/EBC20160004 [DOI] [PubMed] [Google Scholar]
- Berry L. W., Westlund B., Schedl T., 1997. Germ-line tumor formation caused by activation of glp-1, a Caenorhabditis elegans member of the Notch family of receptors. Development 124: 925–936. [DOI] [PubMed] [Google Scholar]
- Birnby D. A., Link E. M., Vowels J. J., Tian H., Colacurcio P. L., et al. , 2000. A transmembrane guanylyl cyclase (DAF-11) and Hsp90 (DAF-21) regulate a common set of chemosensory behaviors in Caenorhabditis elegans. Genetics 155: 85–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkovich K. A., Farrelly F. W., Finkelstein D. B., Taulien J., Lindquist S., 1989. hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol. Cell. Biol. 9(9): 3919–3930. 10.1128/MCB.9.9.3919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood S. K., Gong J., 2016. Heat shock proteins promote cancer: It’s a protection racket. Trends Biochem. Sci. 41(4): 311–323. 10.1016/j.tibs.2016.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrieri F. A., Dale J. K., 2017. Turn it down a Notch. Front. Cell Dev. Biol. 4: 151 10.3389/fcell.2016.00151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- C. elegans Deletion Mutant Consortium, 2012. Large-scale screening for targeted knockouts in the Caenorhabditis elegans genome. G3 2: 1415–1425. 10.1534/g3.112.003830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S., Burns T., 2017. Targeting heat shock proteins in cancer: A promising therapeutic approach. Int. J. Mol. Sci. 18(12): 1978 10.3390/ijms18091978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Greenwald I., 2004. The lateral signal for LIN-12/Notch in C. elegans vulval development comprises redundant secreted and transmembrane DSL proteins. Dev. Cell 6(2): 183–192. 10.1016/S1534-5807(04)00021-8 [DOI] [PubMed] [Google Scholar]
- Crittenden S. L., Troemel E. R., Evans T. C., Kimble J., 1994. GLP-1 is localized to the mitotic region of the C. elegans germ line. Development 120: 2901–2911. [DOI] [PubMed] [Google Scholar]
- Deskin B., Lasky J., Zhuang Y., Shan B., 2016. Requirement of HDAC6 for activation of Notch1 by TGF-β1. Sci. Rep. 6(1): 31086 10.1038/srep31086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckl J. M., Richter K., 2013. Functions of the Hsp90 chaperone system: Lifting client proteins to new heights. Int. J. Biochem. Mol. Biol. 4: 157. [PMC free article] [PubMed] [Google Scholar]
- Epstein H. F., Shakes D. C., 1995. Caenorhabditis elegans: Modern Biological Analysis of an Organism. Academic Press, San Diego. [Google Scholar]
- Frumkin A., Dror S., Pokrzywa W., Bar-Lavan Y., Karady I., et al. , 2014. Challenging muscle homeostasis uncovers novel chaperone interactions in Caenorhabditis elegans. Front. Mol. Biosci. 1: 21 10.3389/fmolb.2014.00021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiser A. M., Brandt F., Richter K., 2009. The non-canonical Hop protein from Caenorhabditis elegans exerts essential functions and forms binary complexes with either Hsc70 or Hsp90. J. Mol. Biol. 391(3): 621–634. 10.1016/j.jmb.2009.06.051 [DOI] [PubMed] [Google Scholar]
- Gaiser A. M., Kaiser C. J. O., Haslbeck V., Richter K., 2011. Downregulation of the Hsp90 system causes defects in muscle cells of Caenorhabditis elegans. PLoS One 6(9): e25485 10.1371/journal.pone.0025485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg G., Khandelwal A., Blagg B. S. J., 2016. Anticancer inhibitors of Hsp90 function, pp. 51–88 in Advances in Cancer Research. Elsevier, Amsterdam. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R. A., Kao H.-L., Audhya A., Arur S., Mayers J. R., et al. , 2011. A high-resolution C. elegans essential gene network based on phenotypic profiling of a complex tissue. Cell 145(3): 470–482. 10.1016/j.cell.2011.03.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald, I., and R. Kovall, 2013 Notch signaling: genetics and structure (January 17, 2013). WormBook, ed. The C. elegans Research Community WormBook, doi/10.1895/wormbook.1.10.2, http://wormbook.org 10.1895/wormbook.1.10.2 [DOI] [PMC free article] [PubMed]
- Guo Y., Yang B., Li Y., Xu X., Maine E. M., 2015. Enrichment of H3K9me2 on unsynapsed chromatin in Caenorhabditis elegans does not target de novo sites. G3 (Bethesda) 5(9): 1865–1878. 10.1534/g3.115.019828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P., Leahul L., Wang X., Wang C., Bakos B., et al. , 2015. Proteasome regulation of the chromodomain protein MRG-1 controls the balance between proliferative fate and differentiation in the C. elegans germ line. Development 142(2): 291–302. 10.1242/dev.115147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase M., Fitze G., 2016. HSP90AB1: Helping the good and the bad. Gene 575(2): 171–186. 10.1016/j.gene.2015.08.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen D., Schedl T., 2013. Stem cell proliferation versus meiotic fate decision in Caenorhabditis elegans, pp. 71–99 in Germ Cell Development in C. elegans, edited by Schedl T. Springer, New York. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbeck V., Kaiser C. J. O., Richter K., 2012. Hsp90 in non-mammalian metazoan model systems. Biochim. Biophys. Acta BBAMol. Cell Res. 1823(3): 712–721. 10.1016/j.bbamcr.2011.09.004 [DOI] [PubMed] [Google Scholar]
- Hubbard E. J. A., Wu G., Kitajewski J., Greenwald I., 1997. sel-10, a negative regulator of lin-12 activity in Caenorhabditis elegans, encodes a member of the CDC4 family of proteins. Genes Dev. 11(23): 3182–3193. 10.1101/gad.11.23.3182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., Takamura K., Yamae H., Ise N., Kawakami M., et al. , 2003. Caenorhabditis elegans DAF-21 (HSP90) is characteristically and predominantly expressed in germline cells: Spatial and temporal analysis. Dev. Growth Differ. 45(4): 369–376. 10.1046/j.1440-169X.2003.00706.x [DOI] [PubMed] [Google Scholar]
- Jarosz D., 2016. Hsp90: a global regulator of the genotype-to-phenotype map in cancers, pp. 225–247 in Advances in Cancer Research. Elsevier, Amsterdam. [DOI] [PubMed] [Google Scholar]
- Karagöz G. E., Rüdiger S. G. D., 2015. Hsp90 interaction with clients. Trends Biochem. Sci. 40(2): 117–125. 10.1016/j.tibs.2014.12.002 [DOI] [PubMed] [Google Scholar]
- Killian D. J., Hubbard E. J. A., 2005. Caenorhabditis elegans germline patterning requires coordinated development of the somatic gonadal sheath and the germ line. Dev. Biol. 279(2): 322–335. 10.1016/j.ydbio.2004.12.021 [DOI] [PubMed] [Google Scholar]
- Kimble, J., and H. Seidel, 2014 C. elegans germline stem cells and their niche, StemBook. Harvard Stem Cell Institute, Cambridge, MA. [PubMed] [Google Scholar]
- Klosin A., Casas E., Hidalgo-Carcedo C., Vavouri T., Lehner B., 2017. Transgenerational transmission of environmental information in C. elegans. Science 356(6335): 320–323. 10.1126/science.aah6412 [DOI] [PubMed] [Google Scholar]
- Komatsu H., Chao M. Y., Larkins-Ford J., Corkins M. E., Somers G. A., et al. , 2008. OSM-11 facilitates LIN-12 Notch signaling during Caenorhabditis elegans vulval development. PLoS Biol. 6(8): e196 10.1371/journal.pbio.0060196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korta D. Z., Hubbard E. J. A., 2010. Soma-germline interactions that influence germline proliferation in Caenorhabditis elegans. Dev. Dyn. 239: 1449–1459. 10.1002/dvdy.22268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovall R. A., Gebelein B., Sprinzak D., Kopan R., 2017. The canonical Notch signaling pathway: Structural and biochemical insights into shape, sugar, and force. Dev. Cell 41(3): 228–241. 10.1016/j.devcel.2017.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbadia J., Morimoto R. I., 2015. The biology of proteostasis in aging and disease. Annu. Rev. Biochem. 84(1): 435–464. 10.1146/annurev-biochem-060614-033955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackie R. E., Maciejewski A., Ostapchenko V. G., Marques-Lopes J., Choy W.-Y., et al. , 2017. The Hsp70/Hsp90 chaperone machinery in neurodegenerative diseases. Front. Neurosci. 11: 254 10.3389/fnins.2017.00254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. J., Gutierrez-Garcia R., Vilchez D., 2017. Embryonic stem cells: a novel paradigm to study proteostasis? FEBS J. 284(3): 391–398. 10.1111/febs.13810 [DOI] [PubMed] [Google Scholar]
- Lele Z., Hartson S. D., Martin C. C., Whitesell L., Matts R. L., et al. , 1999. Disruption of zebrafish somite development by pharmacologic inhibition of Hsp90. Dev. Biol. 210(1): 56–70. 10.1006/dbio.1999.9262 [DOI] [PubMed] [Google Scholar]
- Li J., Buchner J., 2013. Structure, function and regulation of the Hsp90 machinery. Biomed. J. 36(3): 106–117. 10.4103/2319-4170.113230 [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25(16): 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Maine E. M., 2007. The Bro1-domain protein, EGO-2, promotes Notch Signaling in Caenorhabditis elegans. Genetics 176(4): 2265–2277. 10.1534/genetics.107.071225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald L. D., Knox A., Hansen D., 2008. Proteasomal regulation of the proliferation vs. meiotic entry decision in the Caenorhabditis elegans germ line. Genetics 180(2): 905–920. 10.1534/genetics.108.091553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maine E. M., Hansen D., Springer D., Vought V. E., 2004. Caenorhabditis elegans atx-2 promotes germline proliferation and the oocyte fate. Genetics 168(2): 817–830. 10.1534/genetics.104.029355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mašek J., Andersson E. R., 2017. The developmental biology of genetic Notch disorders. Development 144(10): 1743–1763. 10.1242/dev.148007 [DOI] [PubMed] [Google Scholar]
- Mayer M. P., Le Breton L., 2015. Hsp90: Breaking the Symmetry. Mol. Cell 58(1): 8–20. 10.1016/j.molcel.2015.02.022 [DOI] [PubMed] [Google Scholar]
- McCarter J., Bartlett B., Dang T., Schedl T., 1997. Soma–germ cell interactions in Caenorhabditis elegans: multiple events of hermaphrodite germline development require the somatic sheath and spermathecal lineages. Dev. Biol. 181(2): 121–143. 10.1006/dbio.1996.8429 [DOI] [PubMed] [Google Scholar]
- McGovern M., Voutev R., Maciejowski J., Corsi A. K., Hubbard E. J. A., 2009. A “latent niche” mechanism for tumor initiation. Proc. Natl. Acad. Sci. USA 106(28): 11617–11622. 10.1073/pnas.0903768106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo J. A., Ruvkun G., 2012. Inactivation of conserved C. elegans genes engages pathogen- and xenobiotic-associated defenses. Cell 149(2): 452–466. 10.1016/j.cell.2012.02.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noormohammadi A., Calculli G., Gutierrez-Garcia R., Khodakarami A., Koyuncu S., et al. , 2017. Mechanisms of protein homeostasis (proteostasis) maintain stem cell identity in mammalian pluripotent stem cells. Cell. Mol. Life Sci.. 10.1007/s00018-017-2602-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl L. H., 2016. Review: The HSP90 molecular chaperone—an enigmatic ATPase. Biopolymers 105: 594–607. 10.1002/bip.22835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Mockus G., Schweisguth F., 2017. Cell polarity and Notch signaling: Linked by the E3 ubiquitin ligase Neuralized? BioEssays 39(11): 1700128 10.1002/bies.201700128 [DOI] [PubMed] [Google Scholar]
- Piano F., Schetter A. J., Mangone M., Stein L., Kemphues K. J., 2000. RNAi analysis of genes expressed in the ovary of Caenorhabditis elegans. Curr. Biol. 10(24): 1619–1622. 10.1016/S0960-9822(00)00869-1 [DOI] [PubMed] [Google Scholar]
- Pratt W. B., Gestwicki J. E., Osawa Y., Lieberman A. P., 2015. Targeting Hsp90/Hsp70-based protein quality control for treatment of adult onset neurodegenerative diseases. Annu. Rev. Pharmacol. Toxicol. 55(1): 353–371. 10.1146/annurev-pharmtox-010814-124332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao L., Lissemore J. L., Shu P., Smardon A., Gelber M. B., et al. , 1995. Enhancers of glp-1, a gene required for cell-signaling in Caenorhabditis elegans, define a set of genes required for germline development. Genetics 141: 551–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röhl A., Rohrberg J., Buchner J., 2013. The chaperone Hsp90: changing partners for demanding clients. Trends Biochem. Sci. 38(5): 253–262. 10.1016/j.tibs.2013.02.003 [DOI] [PubMed] [Google Scholar]
- Rual J.-F., Ceron J., Koreth J., Hao T., Nicot A.-S., et al. , 2004. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 14(10b): 2162–2168. 10.1101/gr.2505604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford S., Hirate Y., Swalla B. J., 2007a The Hsp90 capacitor, developmental remodeling, and evolution: The robustness of gene networks and the curious evolvability of metamorphosis. Crit. Rev. Biochem. Mol. Biol. 42(5): 355–372. 10.1080/10409230701597782 [DOI] [PubMed] [Google Scholar]
- Rutherford S., Knapp J. R., Csermely P., 2007b Hsp90 and developmental networks, pp. 190–197 in Molecular Aspects of the Stress Response: Chaperones, Membranes and Networks, edited by Csermely P., Vígh L. Springer, New York. [Google Scholar]
- Rutherford S. L., Lindquist S., 1998. Hsp90 as a capacitor for morphological evolution. Nature 396(6709): 336–342. 10.1038/24550 [DOI] [PubMed] [Google Scholar]
- Safdar K., Gu A., Xu X., Au V., Taylor J., et al. , 2016. UBR-5, a conserved HECT-type E3 ubiquitin ligase, negatively regulates Notch-type signaling in Caenorhabditis elegans. G3 (Bethesda) 6(7): 2125–2134. 10.1534/g3.116.027805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopf F. H., Biebl M. M., Buchner J., 2017. The HSP90 chaperone machinery. Nat. Rev. Mol. Cell Biol. 18(6): 345–360. 10.1038/nrm.2017.20 [DOI] [PubMed] [Google Scholar]
- She X., Xu X., Fedotov A., Kelly W. G., Maine E. M., 2009. Regulation of heterochromatin assembly on unpaired chromosomes during Caenorhabditis elegans meiosis by components of a small RNA-mediated pathway. PLoS Genet. 5(8): e1000624 10.1371/journal.pgen.1000624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebel C., Lendahl U., 2017. Notch signaling in development, tissue homeostasis, and disease. Physiol. Rev. 97(4): 1235–1294. 10.1152/physrev.00005.2017 [DOI] [PubMed] [Google Scholar]
- Sun A. Y., Lambie E. J., 1997. gon-2, a gene required for gonadogenesis in Caenorhabditis elegans. Genetics 147: 1077–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale M., Tucker G., Peng J., Krykbaeva I., Lin Z.-Y., et al. , 2014. A quantitative chaperone interaction network reveals the architecture of cellular protein homeostasis pathways. Cell 158(2): 434–448. 10.1016/j.cell.2014.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson O., Edgley M., Strasbourger P., Flibotte S., Ewing B., et al. , 2013. The million mutation project: A new approach to genetics in Caenorhabditis elegans. Genome Res. 23(10): 1749–1762. 10.1101/gr.157651.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L., Court D. L., Fire A., 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263(1-2): 103–112. 10.1016/S0378-1119(00)00579-5 [DOI] [PubMed] [Google Scholar]
- Voss A. K., Thomas T., Gruss P., 2000. Mice lacking HSP90beta fail to develop a placental labyrinth. Development 127: 1–11. [DOI] [PubMed] [Google Scholar]
- Vowels J. J., Thomas J. H., 1994. Multiple chemosensory defects in daf-11 and daf-21 mutants of Caenorhabditis elegans. Genetics 138: 303–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Hu Y., Xiao D., Wang J., Liu C., et al. , 2017. Stabilization of Notch1 by the Hsp90 chaperone is crucial for T-cell leukemogenesis. Clin. Cancer Res. 23(14): 3834–3846. 10.1158/1078-0432.CCR-16-2880 [DOI] [PubMed] [Google Scholar]
- Waters K., Yang A. Z., Reinke V., 2010. Genome-wide analysis of germ cell proliferation in C. elegans identifies VRK-1 as a key regulator of CEP-1/p53. Dev. Biol. 344(2): 1011–1025. 10.1016/j.ydbio.2010.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner A., Manford A. G., Rape M., 2017. Ubiquitin-dependent regulation of stem cell biology. Trends Cell Biol. 27(8): 568–579. 10.1016/j.tcb.2017.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitesell L., Lindquist S. L., 2005. HSP90 and the chaperoning of cancer. Nat. Rev. Cancer 5(10): 761–772. 10.1038/nrc1716 [DOI] [PubMed] [Google Scholar]
- Wolf N., Hirsh D., McIntosh J. R., 1978. Spermatogenesis in males of the free-living nematode, Caenorhabditis elegans. J. Ultrastruct. Res. 63(2): 155–169. 10.1016/S0022-5320(78)80071-9 [DOI] [PubMed] [Google Scholar]
- Wu J., Liu T., Rios Z., Mei Q., Lin X., et al. , 2017. Heat shock proteins and cancer. Trends Pharmacol. Sci. 38(3): 226–256. 10.1016/j.tips.2016.11.009 [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y., Murakami K., Furusawa M., Miwa J., 1983. Germline-specific antigens identified by monoclonal antibodies in the nematode Caenorhabditis elegans. Dev. Growth Differ. 25(2): 121–131. 10.1111/j.1440-169X.1983.00121.x [DOI] [PubMed] [Google Scholar]
- Yandell M. D., Edgar L. G., Wood W. B., 1994. Trimethylpsoralen induces small deletion mutations in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 91(4): 1381–1385. 10.1073/pnas.91.4.1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All C. elegans strains and plasmid constructs used in this study are available upon request. The raw sequence data from the WGS have been submitted to the NCBI BioProject (http://www.ncbi.nlm.nih.gov/bioproject) under accession number PRJNA428227 and can be accessed from the Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) with accession number SRP127800. File S1 contains Figure S1, Locations of primers used to amplify and sequence regions of the hsp-90 gene to identify mutations; Figures S2–S4 in File S1, Sanger DNA sequencing chromatograms of wild-type and mutant sequences; Table S1 in File S1, Gonad transcriptome data for protein-coding genes in the ego-3 region; Table S2 in File S1, Codon and splicing changes detected in ego-3(om40) by whole genome sequence analysis; and Table S3 in File S1, Primers used to amplify and sequence regions of the hsp-90 gene to identify mutations.