Abstract
Fusarium wilt, a soil-borne disease caused by the fungal pathogen Fusarium oxysporum f. sp. fragariae, threatens strawberry (Fragaria × ananassa) production worldwide. The spread of the pathogen, coupled with disruptive changes in soil fumigation practices, have greatly increased disease pressure and the importance of developing resistant cultivars. While resistant and susceptible cultivars have been reported, a limited number of germplasm accessions have been analyzed, and contradictory conclusions have been reached in earlier studies to elucidate the underlying genetic basis of resistance. Here, we report the discovery of Fw1, a dominant gene conferring resistance to Fusarium wilt in strawberry. The Fw1 locus was uncovered in a genome-wide association study of 565 historically and commercially important strawberry accessions genotyped with 14,408 SNP markers. Fourteen SNPs in linkage disequilibrium with Fw1 physically mapped to a 2.3 Mb segment on chromosome 2 in a diploid F. vesca reference genome. Fw1 and 11 tightly linked GWAS-significant SNPs mapped to linkage group 2C in octoploid segregating populations. The most significant SNP explained 85% of the phenotypic variability and predicted resistance in 97% of the accessions tested—broad-sense heritability was 0.96. Several disease resistance and defense-related gene homologs, including a small cluster of genes encoding nucleotide-binding leucine-rich-repeat proteins, were identified in the 0.7 Mb genomic segment predicted to harbor Fw1. DNA variants and candidate genes identified in the present study should facilitate the development of high-throughput genotyping assays for accurately predicting Fusarium wilt phenotypes and applying marker-assisted selection.
Keywords: Fragaria, Fusarium wilt, strawberry, innate immunity, polyploid
Cultivated strawberry (Fragaria × ananassa Duchesne ex Rozier) plant health and yield are adversely impacted by several soil-borne diseases (Maas 1998). One of the greatest threats to strawberry production worldwide has been Fusarium wilt, a soil-borne disease caused by the fungal pathogen Fusarium oxysporum f. sp. fragariae (Winks and Williams 1965; Okamoto et al. 1970; Mena et al. 1975; Castro-Franco and Davalos-Gonzalez 1990; Huang et al. 2005, Abdet-Sattar et al. 2008, Arroyo et al. 2009; Koike et al. 2009; Gordon 2017; Henry et al. 2017). Historically, strawberry fruit and nursery stock growers have relied on powerful soil fumigants to suppress F. oxysporum f. sp. fragariae and other soil-borne pathogens, allowing for monocultures or very tight crop rotation cycles (Goodhue et al. 2005; Lloyd and Gordon 2016; Tourte et al. 2016). Because fruit and nursery production are typically concentrated in unique coastal and high-elevation environments in California, the availability of land for crop rotations is limited, often necessitating continuous cropping (Guthman 2016).
Until 2005, the most widely used soil fumigant in strawberry production was methyl bromide (MeBr), an ozone-layer depleting chemical compound banned by a global treaty enacted to protect Earth’s atmosphere (Montzka et al. 2003; Velders et al. 2007). MeBr was commonly applied in combination with the fumigant chloropicrin. This combination was highly effective and provided growers with predictable control of fungi and weeds (Lloyd and Gordon 2016). The efficacy of fumigation with MeBr and chloropicrin, coupled with sophisticated production practices, and the introduction of increasingly higher yielding cultivars, doubled per-acre yields and increased production 10-fold in the US from 1970 to 2016 (United States Department of Agriculture (USDA) 2017a,b). The progressive phaseout of MeBr, pursuant to the Montreal Protocol (Montzka et al. 2003; Velders et al. 2007), concluded in 2016, the last year exemptions granted for strawberry growers (Downing 2016). The fumigant mixtures available to growers have failed to effectively suppress populations of soil-borne pathogens compared to previous MeBr mixtures. Thus far, chemical and non-chemical alternatives as effective as MeBr + chloropicrin fumigation have not emerged (Downing 2016; Goodhue et al. 2016; Guthman 2016; Guthman and Brown 2016a,b).
Fusarium wilt was first discovered in strawberry in California in 2006 (Koike et al. 2009), in fields where fully effective fumigation practices had not been used for several years in succession (Gordon et al. 2016). The pathogen has since become more widespread and currently appears to be found throughout the state (Henry et al. 2017). Consequently, Fusarium wilt has become an increasingly serious threat to strawberry production in California, as in many other parts of the world (Paynter et al. 2014; Koike and Gordon 2015; Paynter et al. 2016; Gordon et al. 2016; Henry et al. 2017). The development and deployment of resistant cultivars might be the only strategy that can provide reliable and adequate control of Fusarium wilt in strawberry.
The identification of genes conferring resistance to Fusarium wilt and the development of Fusarium wilt resistant cultivars has a long history in tomato (Solanum lycopersicum L.) and other economically important plants (Michielse and Rep 2009; Takken and Rep 2010). Several novel resistance (R) genes have been discovered, cloned, functionally characterized, and deployed in tomato (Ori et al. 1997; Houterman et al. 2008; Takken and Rep 2010; Andolfo et al. 2014; Gonzalez-Cendales et al. 2016; Catanzariti et al. 2017). The most extensively studied resistance (R) genes belong to super-families with highly conserved nucleotide binding (NB) and C-terminal leucine-rich repeat (LRR) domains (Martin et al. 2003; Wulff et al. 2011; Dangl et al. 2013). These have supplied several loci and alleles for developing tomato cultivars resistant to F. oxysporum f. sp. lycopersici (Andolfo et al. 2014; Gonzalez-Cendales et al. 2016; Catanzariti et al. 2017). While the R-genes described in tomato have been fairly durable, some have been overcome by the emergence of virulent races of the pathogen, the outcome of an ongoing evolutionary arms race (Anderson et al. 2010), which has necessitated the identification and deployment of novel R-genes (Houterman et al. 2009; Gonzalez-Cendales et al. 2016; Catanzariti et al. 2015, 2017).
Thus far, insights into the genetics of resistance to Fusarium wilt in strawberry have been limited. Several strawberry cultivars have been reported to be either highly susceptible or moderately to highly resistant to F. oxysporum f. sp. fragariae (Takahashi et al. 2003; Dávalos-González et al. 2004; Hutton and Gomez 2006; Takahashi et al. 2006; Fang et al. 2012a,b, 2013; Paynter et al. 2014; Koike and Gordon 2015; Husaini and Neri 2016; Borrero et al. 2017). While a full range of disease symptoms have been reported, including apparent immunity, genetic factors underlying resistance to Fusarium wilt have not been identified, and contradictory conclusions have been reached in previous studies (Mori et al. 2005; Paynter et al. 2014). Mori et al. (2005) observed the segregation of a dominant gene in a population developed from a cross between resistant (Asuka Wave) and susceptible (Sanchiigo) cultivars but observed significant phenotypic variability within resistant and susceptible classes and concluded that resistance was caused by qualitative and quantitative genetic factors. Paynter et al. (2014), in a study of segregating populations developed from crosses between resistant and susceptible cultivars, concluded that resistance to Fusarium wilt was quantitative, and reported an intermediate heritability (0.49). To explore these questions and develop insights into the genetics of resistance to Fusarium wilt in strawberry, we analyzed 565 germplasm accessions in the University of California, Davis (UCD) germplasm collection. Using genome-wide association and genetic mapping approaches, we identified and genetically and physically mapped a dominant gene (Fw1) that confers resistance to Fusarium wilt in strawberry. Our study was enabled by the availability of a high-density single-nucleotide polymorphism (SNP) genotyping array (Bassil et al. 2015; Verma et al. 2017) and a high-quality reference genome for woodland strawberry (F. vesca (A. Heller) Staudt), a diploid (2n = 2x = 14) ancestor of F. × ananassa (Edger et al. 2018).
Materials and Methods
Plant Material
The UC-Davis Strawberry Germplasm Collection was the source of the plant material investigated in our studies. We selected 565 F. × ananassa germplasm accessions for phenotypic screening and genome-wide association studies (GWAS). These included 50 UCD cultivars released since the inception of the breeding program in 1927, and numerous previously undocumented and uncharacterized germplasm accessions representing genetic diversity in the collection (File S2). To develop populations for quantitative trait locus (QTL) mapping, S1 progeny were developed by self-pollinating greenhouse-grown plants of the cultivars Fronteras and Portola. These cultivars were identified as highly resistant in the GWAS. Both were self-pollinated prior to knowing if they were heterozygous or homozygous for the Fusarium wilt resistance gene described herein. Genotypes of the parents and grandparents (Fronteras = 04C018P004/05C165P001 and Portola = 97C093P007/97C209P001) were subsequently inferred from the haplotypes of SNPs in linkage disequilibrium with the Fusarium wilt resistance gene. Fruit originating from self-pollination were harvested and macerated in a pectinase solution (0.6 g/L) to separate achenes (seeds) from receptacles. S1 seeds were scarified with a concentrated sulfuric acid solution (36 Normal) for 14 min, rinsed in water, dried on blotter paper, and germinated at room temperature (approximately 22-24°) in June 2016. S1 seedlings were grown in kiln-dried artificial media (2-parts vermiculite, 1-part sand) in a shade house in Winters, CA from June to October 2016.
Field Experiments
Germplasm accessions were phenotyped in experiments planted in the spring of 2016 and fall of 2016 in separate fields at the UC-Davis Plant Pathology Farm, Davis, CA. S1 populations were phenotyped in the fall-planted experiment. Neither field had ever been planted with strawberries. The soil type was a Yolo loam based on information provided by the Web Soil Survey (WSS), USDA Natural Resources Conservation Service (https://websoilsurvey.sc.egov.usda.gov/). The spring-planted field was not fumigated, whereas the fall-planted field was flat-fumigated in July 2016 with a chloropicrin-based fumigant (Pic-Clor 60, Cardinal Professional Products, Woodland, CA) at 500 lbs/acre. The fumigated field was sealed with a totally impermeable film (TIF) tarp for one week.
Entries in both experiments were arranged in balanced square lattice experiment designs with four single-plant replications per entry. The configuration was 24 × 24 (= 576 entries) in the spring-planted experiment and 31 × 31 (= 960 entries) in the fall-planted experiment. The number of entries and lattice configuration were greater in the fall-planted experiment because additional germplasm accessions were tested for a study to be reported elsewhere. The incomplete block assignments and randomizations were generated in the R-package ‘agricolae’ (De Mendiburu 2015). Bare-root plants were planted in 15.25 cm high raised beds in a single row with 30.5 cm spacing between plants and 76.2 cm spacing between beds center-to-center. We installed drip irrigation and covered the beds with black plastic mulch prior to planting. These experiments were sub-surface drip-irrigated as needed to maintain adequate soil moisture. Fertilization was done via injection through the drip system with approximately 198 kg/ha of nitrogen applied over the growing season.
The spring-planted GWAS experiment was phenotyped in the spring and summer of 2016, whereas the fall-planted GWAS experiment was phenotyped in the spring and summer of 2017. For the spring-planted 2016 field experiment, clonal propagules were produced from stolons of mother plants grown in a low-elevation (41m) nursery in Winters, CA, harvested in January 2016, and stored at -3°. The plants were removed from the freezer and stored at 3.5° for one week prior to planting in the field in April 2016. For the fall-planted 2016-2017 field experiment, clonal propagules of germplasm accessions were produced from stolons of mother plants grown in a high-elevation (1,294 m) nursery in Dorris, CA. The mother plants were harvested from a low-elevation field nursery in January 2016, stored at -3°, and planted in April 2016 at the high-elevation nursery. Clonal propagules were harvested in October 2016 and stored at 3.5° for two weeks before pathogen-inoculation and planting. For S1 populations phenotyped in the fall-planted 2016-2017 experiment, seeds were germinated in June 2016, transplanted to peat pellets, and grown in a shade house in Winters, CA before being pathogen-inoculated and transplanted to the field in October 2016.
Disease Resistance Phenotyping
Seventeen-week-old S1 seedlings and clonal propagules of the germplasm accessions were inoculated with a virulent isolate of F. oxysporum f. sp. fragariae (AMP132) immediately before planting. To produce spores (inoculum), the pathogen was grown on potato dextrose agar (PDA) under continuous fluorescent lighting at room temperature for 30 days, as described by Gordon et al. (2016). Spores were freed from the surface by adding sterile deionized (DI) water with 0.5% potassium chloride (KCl) to the growth plates and scraping the edge of the media with a sterile glass slide. The resulting suspension was filtered through two layers of sterilized cheesecloth. Spore densities were estimated using a hemocytometer. Spore suspensions were diluted with 0.1% water agar to a final density of 5 × 106 spores/ml. Spore inoculum was prepared one day prior to planting and stored at 4° overnight. The suspension was shaken to re-suspend spores before inoculation. Seedlings and bare-root plants were dipped in the spore suspension and immediately planted.
For each study, phenotyping was initiated as soon as symptoms appeared and continued on a weekly basis in the spring of 2016 (4-9 weeks post-inoculation) and bi-weekly basis in the spring of 2017 (26-36 weeks post-inoculation). We utilized the progression of disease symptoms as a guide for both initiating and terminating phentoyping. Symptoms had fully progressed and were most extreme among resistant and susceptible checks in the final time point in each year (9-weeks post-inoculation in 2016 and 36-weeks post-inoculation in 2017). We used a previously described 1-5 rating scale to phenotype disease symptoms, where symptomless plants were scored as 1, stunted plants were scored as 2, chlorotic plants were scored as 3, wilting plants were scored as 4, and plants killed by the pathogen were scored as 5 (Gordon et al. 2016; Henry et al. 2017).
DNA Marker Genotyping
For DNA isolation, newly emerging leaves were harvested from field grown plants of the germplasm accessions and shade house-grown seedlings of S1 populations. Leaf tissue was placed into 1.1 ml tubes, freeze-dried in a Benchtop Pro (VirTis SP Scientific, Stone Bridge, NY), and ground using stainless steel beads in a Mini 1600 (SPEX Sample Prep, Metuchen, NJ). Genomic DNA (gDNA) was extracted from powdered leaf samples using the E-Z 96 Plant DNA Kit (Omega Bio-Tek, Norcross, GA, USA) according to the manufacturer’s instructions. To enhance the quality of the DNA and reduce polysaccharide carry-through, the protocol was modified with a Proteinase K treatment, a separate RNase treatment, an additional spin, and heated incubation steps during elution. DNA quantification was performed using Quantiflor dye (Promega, Madison, WI) on a Synergy HTX (Biotek, Winooski, VT).
SNP genotyping with the Affymetrix IStraw35 Axiom Array (Bassil et al. 2015; Verma et al. 2017) was performed by Affymetrix (Santa Clara, CA) on a GeneTitan HT Microarray System using gDNA samples that passed quality and quantity control standards. SNP genotypes were automatically called with the Affymetrix Axiom Analysis Suite software (v1.1.1.66, Affymetrix, Santa Clara, CA). Samples with a call-rate greater than 94% were retained. The quality metrics output by the Affymetrix Axiom Analysis Suite, custom R scripts, and the R-package ‘SNPRelate’ (Zheng et al. 2012) were utilized to filter SNPs; 14,408 SNPs with high-quality bi-allelic clusters and < 5% missing data were selected for subsequent analyses. The R-packages ‘SNPRelate’ (Zheng et al. 2012) and ‘GWASTools’ (Gogarten et al. 2012) were used to generate genotypic input files for GWAS from raw genotyping reads.
Genome-Wide Association Study
Type III analysis of variance (ANOVA) was performed using a mixed linear model for the lattice experiment design with incomplete and complete blocks as random effects and entries as fixed effects. Statistical analyses were performed using the R-packages ‘lme4’ and ‘car’ (Fox and Weisberg 2011; Bates et al. 2015). The recovery of intra-block error information from the lattice experiment designs was negligible and failed to increase efficiency relative to randomized complete block (RCB) experiment designs; hence, we utilized linear models for RCB experiment designs for subsequent analyses. Least square means for entries were estimated using the R-package ‘lsmeans’ with complete blocks as a random effect and entries as a fixed effect (Lenth 2016) and were subsequently used as phenotypic input for GWAS. REML variance component and broad-sense heritabilities (Lynch and Walsh 1998) were estimated using the R-package ‘lme4’ (Bates et al. 2015), with entries, complete blocks, and years as random effects.
Because the germplasm collection we studied included numerous closely related individuals, we investigated and accounted for population structure using principal components analysis in related samples (PCAiR) with ‘GENESIS’ (http://bioconductor.org/packages/release/bioc/vignettes/GENESIS/inst/doc/pcair.html; Conomos et al. 2015, 2016; Conomos and Thornton 2016). The p-value inflation factors (λ), ignoring population structure, were 3.09 for the 2016 and 3.69 for the 2017 GWAS experiments (Clayton 2015). We subsequently used the first three principal components (PCs) from PC-AiR as input for calculating the kinship matrix, which was done using PC-relate in ‘GENESIS’ (Conomos and Thornton 2016). The resultant kinship matrix was used in a mixed linear model GWAS analysis, assuming a Gaussian distribution of the dependent variable. Wald tests were performed as implemented in ‘GENESIS’ using default parameters (Conomos and Thornton 2016). Because an octoploid reference genome was unavailable, we utilized a diploid reference genome for F. vesca (Edger et al. 2018) for GWAS, plotting p-values against physical positions (Mb). SNP probe sequences from the Affymetrix IStraw35 Axiom Array (Bassil et al. 2015; Verma et al. 2017) were physically mapped to the diploid reference genome using the Burrows-Wheeler Aligner (BWA v.0.7.15; Li and Durbin 2009). The ancestry-adjusted p-value inflation factors were 0.75 for the 2016 and 0.85 for the 2017 GWAS experiments, which suggested that the population structure corrections in the mixed linear model GWAS were effective (Hinrichs et al. 2009).
Genetic and Quantitative Trait Locus Mapping
Because the parents and grandparents of the S1 populations were outbred, genetic mapping was performed using the full-sib mapping algorithm of JoinMap 4.1 (van Ooijen 2006), which utilizes the general maximum-likelihood (ML) algorithm of Wu et al. (2002) for simultaneously estimating linkage and linkage phases in full-sib families. Because we selfed a single individual descended from two outbred parents (grandparents of the S1 offspring), heterozygous loci were expected to segregate 1 AA: 2 AB: 1 BB in the Fronteras and Portola S1 populations, where A is the allele inherited from one grandparent and B is the allele inherited from the other grandparent. S1 individuals were genotyped with the Affymetrix IStraw35 Axiom Array (Bassil et al. 2015; Verma et al. 2017). SNPs that produced co-dominant (bi-allelic) segregation patterns identified using the Affymetrix Axiom Analysis Suite were selected for subsequent analyses. For genetic mapping, we identified and selected 5,673 SNPs in the Fronteras S1 population and 7,345 SNPs in the Portola S1 population. Numerous SNPs were in complete LD across the genome. To reduce the dimensions of the data and more robustly order loci, co-segregating SNPs were assigned to bins and one SNP from each bin was selected for inclusion in the analysis. Once linkage phases were estimated, SNPs were recoded according to the inferred linkage phase, analogous to an F2 population developed from a cross between inbred parents. Loci were grouped using a minimum likelihood odds (LOD) threshold of 8.0 and ordered using the multi-point ML algorithm in JoinMap 4.1 with default parameters and three rounds of locus ordering (van Ooijen 2006). Genetic distances were calculated using the Kosambi mapping function. By cross-referencing previously mapped iStraw35 and iStraw 90 SNPs (Verma et al. 2017), linkage groups identified in the present study were aligned with 28 linkage groups previously described by Bassil et al. (2015) and Mangandi et al. (2017). The linkage group numbers and orientations in Bassil et al. (2015) and Mangandi et al. (2017) trace their origin to van Dijk et al. (2014).
We assigned S1 offspring to resistant and susceptible phenotypic classes and tested the hypothesis of the segregation of a single gene using standard goodness-of-fit statistics. Offspring with Fusarium wilt scores < 2.5 were classified as resistant, whereas offspring with Fusarium wilt score ≥ 2.5 were classified as susceptible. The observed segregation ratios were tested for goodness-of-fit to the expected segregation ratio of three resistant to one susceptible using Chi-square statistics with the R-function ‘chisq.test’.
Linkage groups were scanned for quantitative trait loci (QTL) using the interval mapping function in MapQTL 6 (van Ooijen 2009). Several tightly linked SNPs on linkage group 2C, previously identified by GWAS, co-segregated with a QTL that was targeted in subsequent analyses. Significant SNP loci in the haploblock were individually used as fixed effects (independent variables) in linear model analyses to estimate additive (a) and dominance (d) effects, degree of dominance (d/a), and the proportion of the phenotypic variance associated with the additive and dominance effects of the SNP locus (Falconer and Mackay 1996; Lynch and Walsh 1998). SNPs were physically mapped in the diploid reference genome (Edger et al. 2018). We used linkage phases of SNP markers to infer the haplotypes of the parents (Fronteras and Portola). The inferred haplotypes were supported by the three-generation pedigree of Fronteras. The 05C165P001 parent of Fronteras was susceptible to Fusarium wilt and homozygous for the eight most significant SNPs, whereas the 04C018P004 was resistant to Fusarium wilt and heterozygous for the eight most significant SNPs (File S2).
Data Availability
All data required to replicate the analyses are available as supplements cited in-text or in Supplemental Data Files 1-4. Supplemental Data Files 1 and 2 contain the raw genotypic data for the germplasm accessions and mapping populations, respectively. Supplemental Data File 3 provides additional information regarding SNP nomenclature, alleles, and genomic locations. Supplemental Data File 4 provides the raw phenotypic (disease symptom) scores for every time point in 2016 and 2017 studies. Supplemental material available at Figshare: https://doi.org/10.25387/g3.6007715.
Results
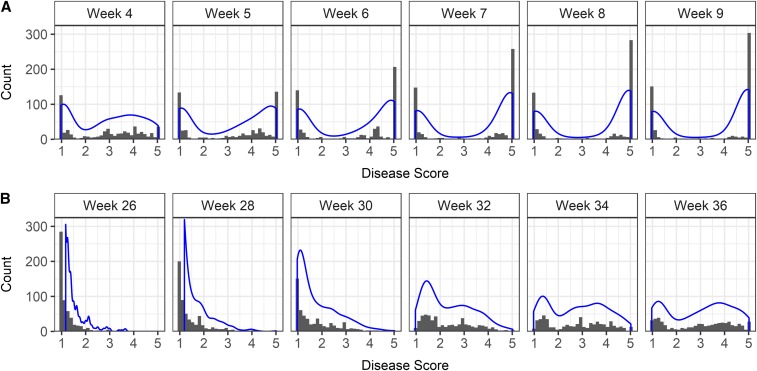
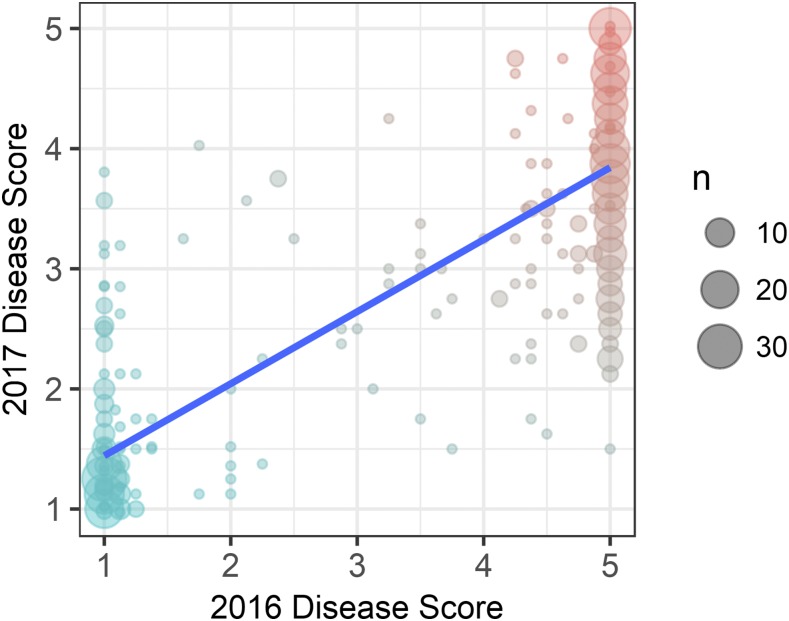
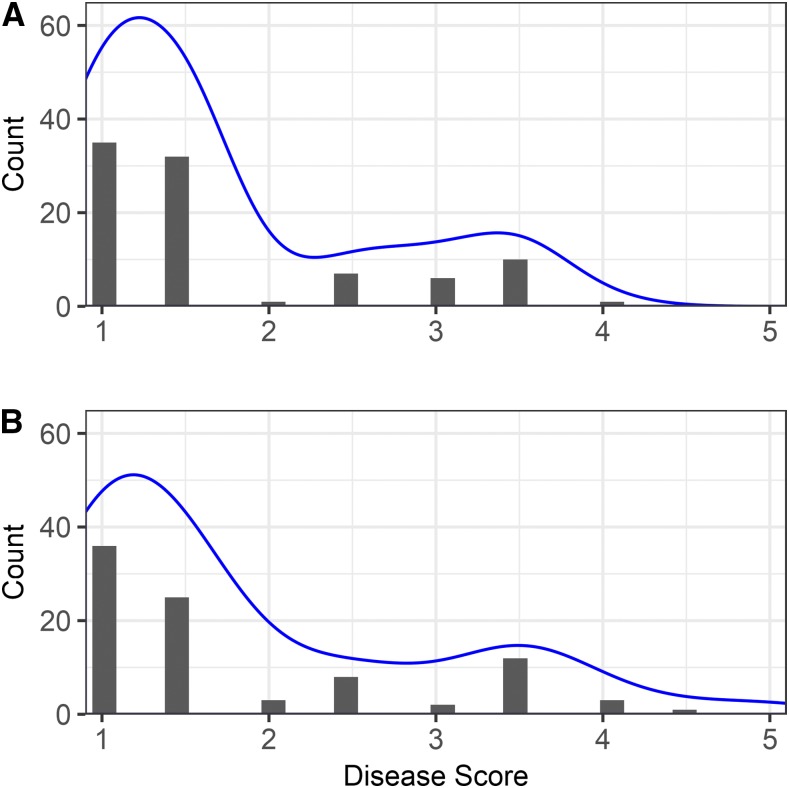
One-third of the germplasm accessions screened for resistance to the AMP132 isolate of F. oxysporum f. sp. fragariae were symptomless and classified as resistant (Figure 1). Apart from a small number of germplasm accessions with intermediate symptoms (<2% of those studied), nearly two-thirds developed severe vascular wilt symptoms, including stunting, chlorosis, wilting, browning, and dieback (File S2). To study the progression of disease symptoms and quantify changes in phenotypic distributions over time, plants were phenotyped over six weeks in 2016 and 10 weeks in 2017 (27,168 phenotypic observations were collected over the course of the study). As predicted a priori, the shapes of the phenotypic distributions changed as symptoms progressed and disease severity increased (Figure 1). Symptoms developed more rapidly and phenotypic differences were more extreme in 2016 than 2017. The final 2016 phenotypic distribution (9 weeks post-inoculation) was bi-modal with strong separation between resistant and susceptible accessions, whereas the final 2017 phenotypic distribution (36 weeks post-inoculation) was flatter with weaker separation between resistant and susceptible accessions (Figure 1; File S2). Symptom development was less severe for several susceptible accessions in 2017 than 2016. While the accession by year interaction was highly significant as a result (P < 0.001; File S1), the phenotypic correlation between years was positive and highly significant (r = 0.84; P < 0.001; Figure 2), broad-sense heritabilities exceeded 90% (H2 = 0.98 in 2016, 0.90 in 2017, and 0.96 across years), and the classification of accessions as resistant or susceptible was highly consistent over years (Figure 2; File S2).
Figure 1.
Phenotypic distributions for resistance to Fusarium wilt in a genome-wide association study (GWAS) in strawberry. Histograms are shown for phenotypes observed in (A) 2016 and (B) 2017 field experiments in Davis, California among 565 strawberry germplasm accessions artificially inoculated with isolate AMP132 of Fusarium oxysporum f. sp. fragariae. Phenotypes were observed four to nine weeks post-inoculation in 2016 and 26 to 36 weeks post-inoculation in 2017. Least square means were estimated from four clonal replicates per entry with entries arranged in a square lattice experiment design. Disease scores ranged from 1 to 5, where 1 = healthy and 5 = dead.
Figure 2.
Phenotypic correlation (Pearson’s r = 0.84, P < 0.001) between years for Fusarium wilt resistance phenotypes in strawberry. A fitted linear regression is shown in blue. Fusarium wilt resistance was phenotyped in 2016 and 2017 field experiments in Davis, California among 565 strawberry germplasm accessions artificially inoculated with isolate AMP132 of Fusarium oxysporum f. sp. fragariae. The phenotypes shown were observed nine weeks post-inoculation in 2016 (x-axis) and 36 weeks post-inoculation in 2017 (y-axis).
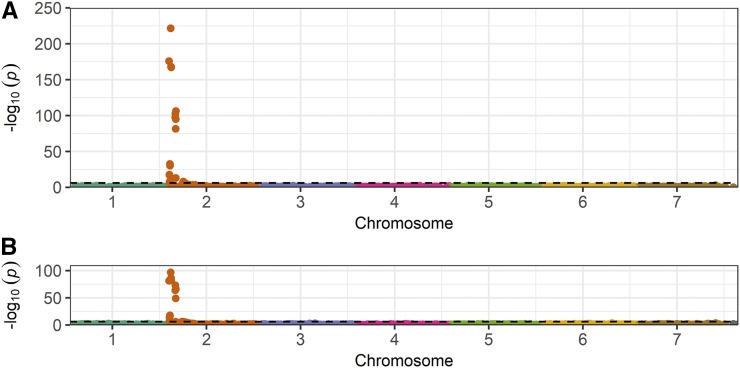
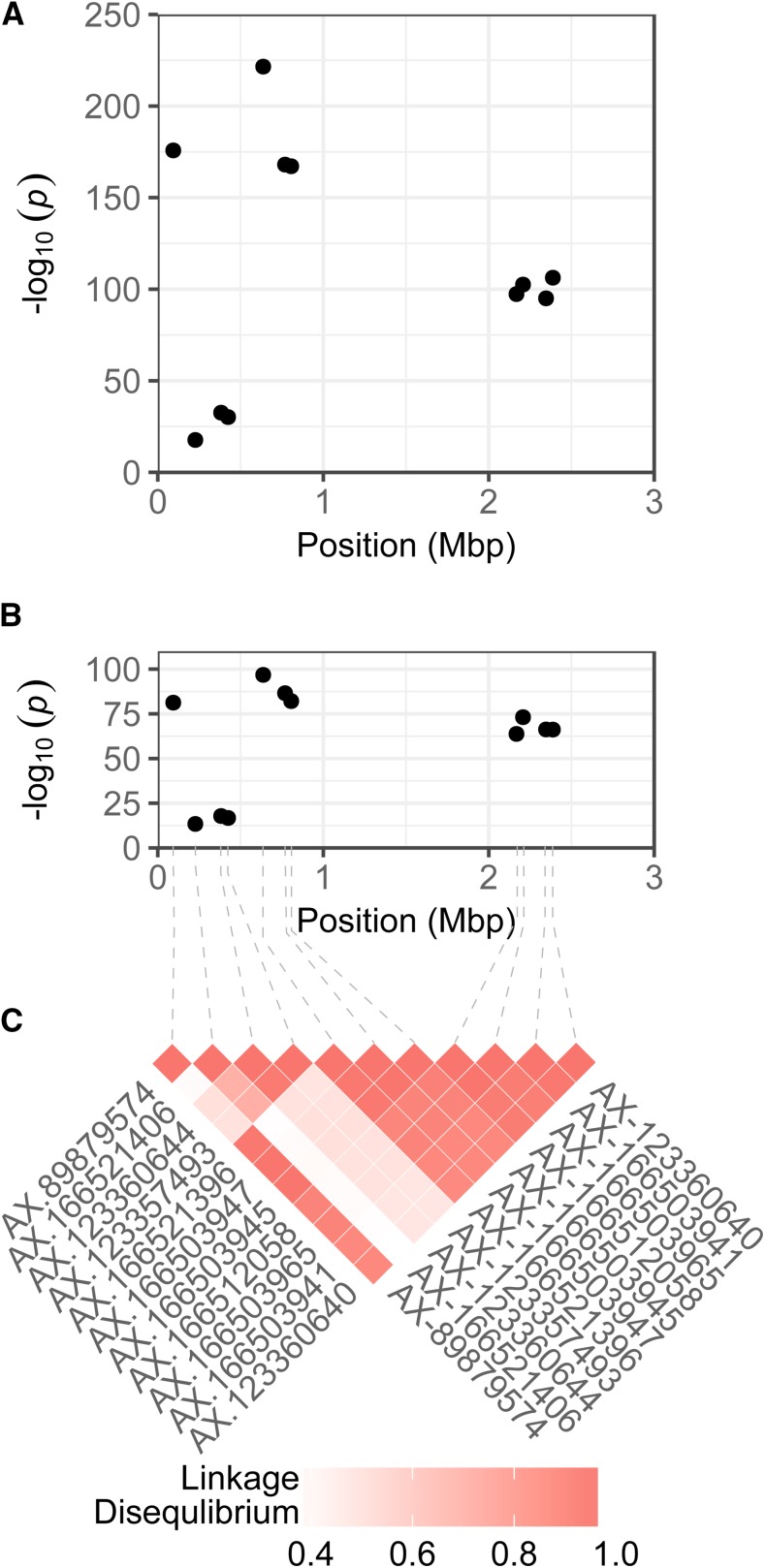
Genome-wide association studies were conducted using 14,408 SNPs mapped against chromosome positions in a diploid (2n = 2x = 14) F. vesca reference genome (Edger et al. 2018). GWAS identified 14 SNPs in a 2.3 Mb genomic segment on the upper arm of chromosome 2 that were in linkage disequilibrium with Fusarium wilt resistance phentoypes (Figure 3; File S2). The -log10 p-values for significant SNPs greatly exceeded conservative genome-wide statistical significance thresholds, ranging from 9.18 × 10−9 for AX-123360644 in the 2017 experiment to 2.95 × 10−222 for AX-166521396 in the 2016 experiment (Figure 3; File S2). Significant SNPs were not identified elsewhere in the genome. When genomic locations were examined in greater depth, the 14 SNPs were discovered to have mapped to 0.71 Mb and 0.22 Mb genomic segments separated by a 1.36 Mb segment that was devoid of significant SNPs (Figure 4; File S2). The most significant SNP in the study (AX-166521396) was located in the upper 0.7 Mb genomic segment (Figure 4; File S2).
Figure 3.
Genome-wide association study for resistance to Fusarium wilt in octoploid strawberry using chromosome positions from the diploid (x = 7) F. vesca reference genome (Edger et al. 2018). Manhattan plots are for phenotypes observed in 2016 (A) and 2017 (B) experiments. The horizontal dashed line identifies a 0.01 Bonferroni-corrected significance threshold.
Figure 4.
SNPs in linkage disequilibrium with a Fusarium wilt resistance gene (Fw1) that were genetically mapped to chromosome 2C in octoploid strawberry. Manhattan plots are shown for phenotypes observed in 2016 (A) and 2017 (B) GWAS experiments with -log10 p-values for nine SNPs plotted against chromosome positions in a diploid (x = 7) F. vesca reference genome (Edger et al. 2018). Pairwise marker linkage disequilibrium statistics are shown for the GWAS panel (C).
From the strength of the GWAS signal and short physical distance spanned by significant SNPs on chromosome 2 in the diploid reference genome (Figures 3-4), we hypothesized that the SNP haploblock was in linkage disequilibrium with a gene conferring resistance to Fusarium wilt on a chromosome 2 homeolog in the octoploid genome. To investigate this, genetic and QTL mapping studies were conducted in S1 populations developed by self-pollinating cultivars (Fronteras and Portola) inferred to be heterozygous for the hypothesized resistance gene (File S2). The phenotypic distributions for both S1 populations were bi-modal with fairly distinct separation between resistance and susceptible classes (Figure 5). For hypothesis testing, progeny with phenotypic scores < 2.5 were classified as homozygous or heterozygous resistant (R_), whereas progeny with phenotypic scores ≥ 2.5 were classified as homozygous susceptible (rr). Using these classifications, the observed phenotypic ratios were not significantly different from 3 R_: 1 rr, the phenotypic ratio expected for the segregation of a dominant gene. We observed 68 R_: 24 rr among Fronteras S1 progeny (χ2 = 0.06; P = 0.81) and 64 R_: 28 rr among Portola S1 progeny (χ2 = 1.45; P = 0.23). Shifting the threshold upward to 3 or downward to 2 did not change the statistical inference.
Figure 5.
Distributions for Fusarium wilt resistance phenotypes observed in octoploid segregating populations. Histograms are shown for Fusarium wilt resistance phenotypes in segregating populations developed by self-pollinating the F. × ananassa cultivars (A) Fronteras and (B) Portola. Parents (Fronteras and Portola) and grandparents (04C018P004/05C165P001 and 97C093P007/97C209P001) of the S1 populations and 93 S1 individuals from each population were artificially inoculated with Fusarium oxysporum f. sp. fragariae isolate AMP132. Phenotypes were observed 36 weeks post-inoculation in a 2017 field experiment in Davis, California.
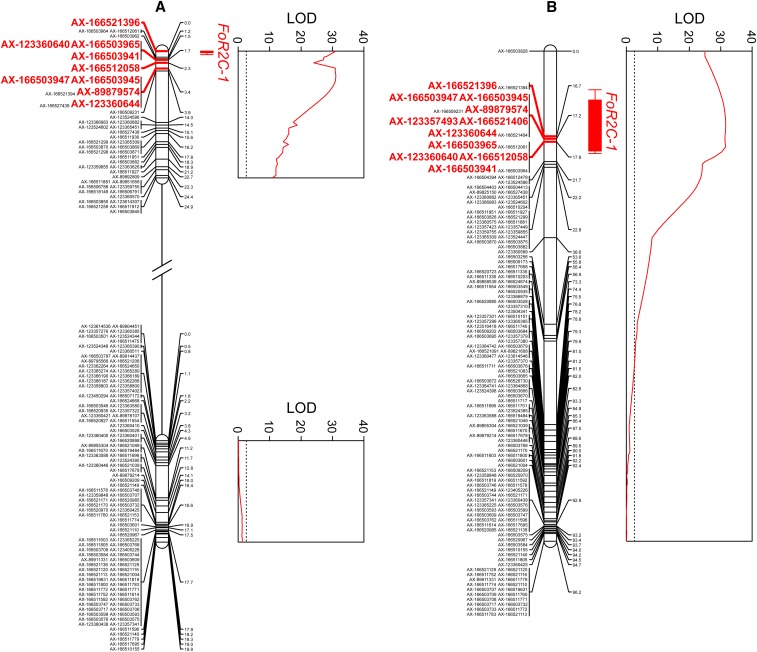
Using a high-density SNP array (Bassil et al. 2015; Verma et al. 2017), 5,673 co-dominant SNPs were genotyped and mapped in the Fronteras S1 population, whereas 7,345 co-dominant SNPs were genotyped and mapped in the Portola S1 population (Supplemental Files 3-4). Because SNP marker densities were low in several genomic regions, the number of linkage groups (40 in the Fronteras S1 and 50 in the Portola S1 population) exceeded the haploid chromosome number (28). Nevertheless, by cross-referencing SNPs previously mapped by Mangandi et al. (2017), sub-linkage groups were aligned and oriented with 28 previously identified F. × ananassa linkage groups numbered using the nomenclature of van Dijk et al. (2014). Using interval mapping, a single large-effect QTL was identified on linkage group 2C in both S1 populations (Figure 6). Eleven of the 14 GWAS-identified SNPs segregated, mapped to a short interval on the upper arm of linkage group 2C, and co-segregated with the QTL (Figure 6; Supplemental Files 3-4). Two of the 11 SNPs (AX-166521406 and AX-123357493) only segregated in the Fronteras S1 population. Additive and dominance effects for individual SNPs were highly significant (P < 0.001) and nearly identical across the linkage group 2C haploblock within each population (File S3). The additive and dominance effects for the AX-166521396 SNP locus were -0.85 and -0.86 in the Fronteras S1 population and -1.12 and -0.94 in the Portola S1 population (File S3). The AX-166521396 SNP explained 85% of the phenotypic variation for resistance to Fusarium wilt in both S1 populations (Figure 6; File S3). Significant QTL were not identified elsewhere in the genome (File S4). The QTL was completely dominant in one population and nearly completely dominant in the other—the degree of dominance (|d/a|) for the AX-166521396 SNP was 1.01 in the Fronteras and 0.84 in the Portola S1 population (File S3). We concluded that the QTL was caused by the segregation of a dominant gene conferring resistance to Fusarium wilt, hereafter identified as Fw1. One recombinant individual was observed between the upper and lower SNP haploblocks among 186 S1 individuals. The recombinant individual (16S408P105) was susceptible to Fusarium wilt, homozygous for the susceptible haplotype in the upper haploblock, and heterozygous for the resistant haplotype in the lower haploblock; hence, Fw1 appears to be located upstream of the lower haploblock within or near the upper haploblock harboring the AX-166521396 SNP locus (Figures 4 and 6).
Figure 6.
Genetic mapping of a Fusarium wilt resistance gene (Fw1) in octoploid segregating populations. Likelihood-odds (LOD) statistics and linkage group positions (cM) are shown for a quantitative trait locus (QTL) for Fusarium wilt resistance identified by interval mapping in (A) Portola and (B) Fronteras S1 mapping populations. The parents (Fronteras and Portola) and grandparents (04C018P004/05C165P001 and 97C093P007/97C209P001) of the S1 populations and 93 S1 individuals from each population were genotyped with the iStraw35 SNP array and artificially inoculated with isolate AMP132 of Fusarium oxysporum f. sp. fragariae at planting. Phenotypes were observed 36 weeks post-inoculation in a 2017 field experiment in Davis, California. The Fw1 QTL mapped to identical locations on the upper arm of chromosome 2C in both populations. One- and two-LOD confidence intervals are shown. Highlighted SNPs (bold red) were significant in genome-wide association studies.
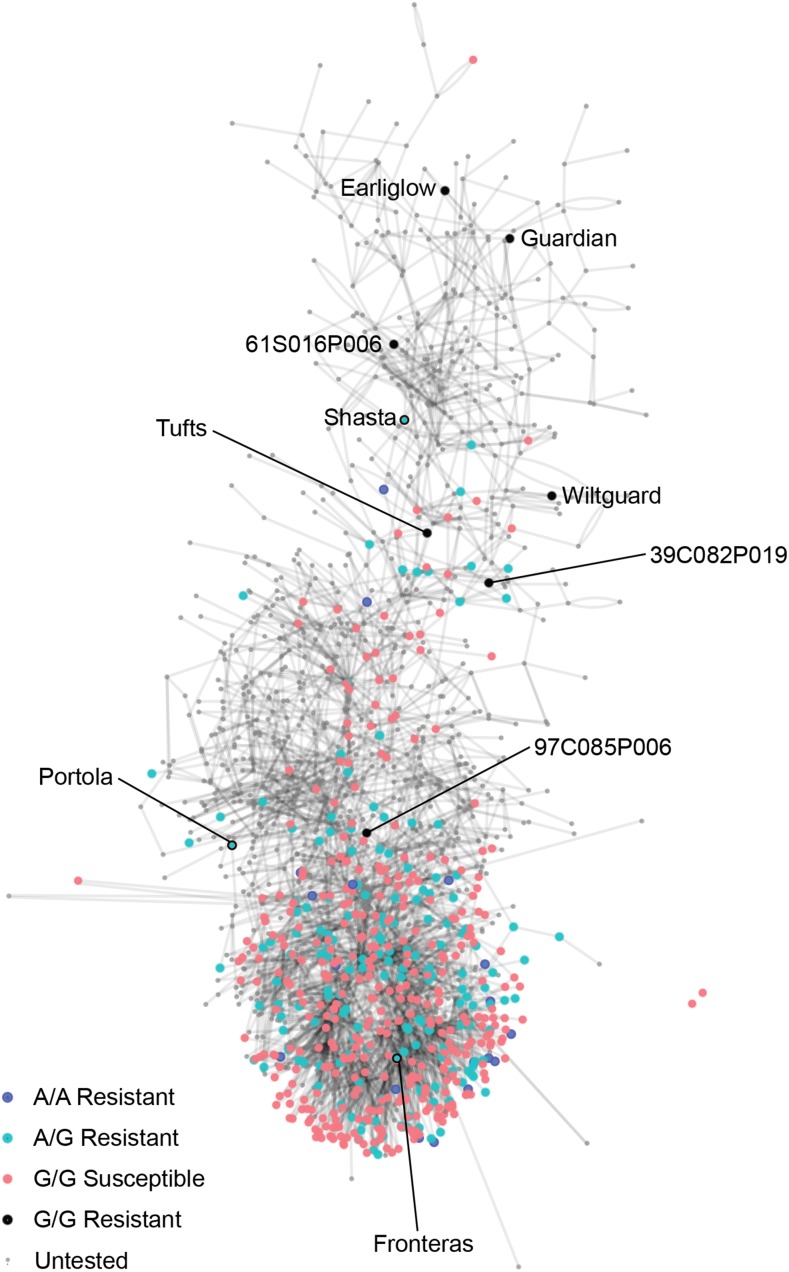
Haplotypes for SNPs in linkage disequilibrium with Fw1 were inferred from pedigree records and linkage phases in segregating populations (File S2). The haplotype associated with the resistant Fw1 allele was observed in 161 accessions (28.4%), whereas the haplotype associated with the susceptible Fw1 allele was observed in 368 accessions (65.0%). Within the upper haploblock, which harbors AX-166521396, the resistant haplotype was observed in 189 accessions (33.4%), whereas the susceptible haplotype was observed in 370 accessions (65.4%). To retrace the breeding history of Fw1, we assembled and analyzed pedigree records for F. × ananassa germplasm accessions originating between 1814 and 2014 (Figure 7; File S5). The earliest sources of the resistant Fw1 allele among the UCD germplasm accessions tested were cultivars developed in 1935—Solana (PI 551665) and Shasta (PI 551663)—both of which were heterozygous (Figure 7). Resistant Fw1 homozygotes were rare in the germplasm studied (<3% of the accessions). Soquel (PI 666602) was the only homozygous resistant cultivar out of 50 tested (Figure 7; File S2).
Figure 7.
Pedigree network for 1,663 F. × ananassa germplasm accessions with birth years ranging from 1814 to 2012. The north to south orientation of the network is approximately chronological. The pedigree records are supplied in Supplemental File 5. Nodes represent germplasm accessions, whereas connecting lines represent first-degree relatives (parent-offspring). The color of the node signifies a combination of the Fusarium wilt resistance phenotype and the AX-166521396 SNP marker genotype for 565 germplasm accessions. The other 1,098 germplasm accessions in the pedigree network were untested (small light gray nodes). The AX-166521396 SNP marker was in linkage disequilibrium with the Fw1 gene conferring resistance to Fusarium wilt. The A allele was associated with the resistant allele (Fw1), whereas the G allele was associated with the susceptible allele (fw1). AX-166521396 SNP marker genotypes predicted Fusarium wilt resistance phenotypes in 97% of the germplasm accessions tested: most A/A and A/G genotypes were resistant (blue and cyan filled circles, respectively), whereas most G/G genotypes were susceptible (salmon filled circles). Seven G/G genotypes were resistant and predicted to carry novel Fusarium wilt resistance genes (black filled circles).
Of 11 genetically mapped SNPs in linkage disequilibrium with Fw1 on linkage group 2C (Figure 6), eight accurately predicted phenotypes in 93.8–97.3% of the germplasm accessions tested (Figure 4; Supplemental Files 2 and 6). The A allele for the most predictive SNP (AX-166521396) was associated with the resistant Fw1 allele, had a frequency of 0.18 in the UCD germplasm collection, and was homozygous (A/A) or heterozygous (A/G) in 190 accessions (Figure 7; File S6). The resistant Fw1 allele appears to have survived by chance and to be randomly distributed over generations and pedigrees (Figure 7).
AX-166521396 accurately predicted Fusarium wilt resistance phenotypes in 97.3% of the germplasm accessions tested in the 2016 experiment: one out of 16 accessions with the A/A genotype and three out of 174 accessions with the A/G genotype were susceptible, whereas seven out of 371 accessions with the G/G genotype were resistant. The seven outliers (resistant accessions with G/G genotypes) included the cultivars Guardian (PI 551407), Wiltguard (52C016P007), Earliglow (PI 551394), and Tufts (63C120P001) and UCD germplasm accessions 39C082P019, 61S016P006, and 97C085P006 (Figure 7; File S2). Because these accessions were both highly resistant and homozygous for the susceptible Fw1 allele (G/G), we suspect that they harbor novel Fusarium wilt resistance genes. These germplasm accessions, with one exception (97C085P006), originated between 1939 and 1975, upstream of apparent bottlenecks in the UCD breeding program (Figure 7).
To identify candidate genes for Fw1, we examined gene annotations in the 2.3 Mb segment spanned by GWAS-significant SNPs: 93,425 to 2,387,499 bp on chromosome 2 in the diploid reference genome (Edger et al. 2018; Supplemental Files 2 and 7). Several recurrent defense-related genes reside in the target segment (File S7), including several with conserved domains common to disease resistance genes in plants, e.g., “extracellular membrane-anchored leucine-rich repeat (LRR) receptor-like proteins” and “nucleotide binding LRR proteins” (Martin et al. 2003; Chisholm et al. 2006; Wulff et al. 2011; Dangl et al. 2013). The strongest candidates for Fw1 are Toll/interleukin-1 receptor (TIR) NB-LRR encoding genes (FvH4_2g00540, FvH4_2g00550, and FvH4_2g00570) found in a small cluster located between 577,691 and 606,648 bp on chromosome 2 in the F. vesca genome (Edger et al. 2018; File S7), immediately upstream of the most significant SNP marker (AX-166521396; 636,941 bp). The TAIR annotation for these genes returned RPP13, a TIR-NB-LRR encoding gene that confers resistance to downy mildew in Arabidopsis, a disease caused by the oomycete Peronospora parasitica (Bittner-Eddy et al. 1999, 2000; Rose et al. 2004; The Arabidopsis Information Resource (TAIR) 2015). The other defense-related genes in the target segment included glycosyl hydrolase (GH) and transferase genes (FvH4_2g00020, FvH4_2g00340, FvH4_2g00600, FvH4_2g02970), in addition to homologs of a vacuolar sorting protein (VPS52; FvH4_2g00140), Whirly (FvH4_2g00250), powdery mildew resistance 5 (PMR5; FvH4_2g02780), histidyl-tRNA synthetase (FvH4_2g00780), aspartyl-tRNA synthetase (FvH4_2g03040), and TARGET OF RAPAMYCIN (TOR; FvH4_2g03080). While none of these can be ruled out, Fw1 has the hallmark of a gene encoding one of the well-known classes of R-genes that trigger innate immunity in plants (Martin et al. 2003; Chisholm et al. 2006; Wulff et al. 2011; Dangl et al. 2013).
Discussion
We identified a dominant gene (Fw1) in octoploid strawberry that confers resistance to a virulent isolate of F. oxysporum f. sp. fragariae found in California (Gordon et al. 2016; Henry et al. 2017). Our findings were consistent with the hypothesis that gene-for-gene resistance to Fusarium wilt might be operating in strawberry, as previously suggested by Mori et al. (2005) in a study where a segregating population was screened for resistance to a Japanese isolate of F. oxysporum f. sp. fragariae (91060510). While Mori et al. (2005) concluded that resistance was caused by the segregation of both “qualitative and quantitative genes”, the evidence for the segregation of a dominant gene was compelling. Similar to Mori et al. (2005), we observed phenotypic variability among progeny classified as resistant or susceptible; however, the Fw1 locus was sufficient to explain phenotypic variability for resistance to Fusarium wilt in our study (Figures 5-6; Supplemental Files 2-3). We found no evidence for the segregation of additional loci (Figure 3; File S4)—non-genetic variability was negligible and broad-sense heritabilities were in the 0.90 to 0.98 range, double the estimate reported by Paynter et al. (2014). Paynter et al. (2014) screened segregating progeny for resistance to a mixture of two virulent Australian isolates of F. oxysporum f. sp. fragariae (N13581 and N15309). The effectiveness of Fw1 against these isolates and other isolates of the pathogen is unknown. Moreover, several factors, including the absence of characterized resistance genes, has precluded the assignment of isolates to races through the study of differential host-pathogens interactions. The present study opens the way to exploring the race structure of F. oxysporum f. sp. fragariae isolates by testing host differentials (Gordon and Martyn 1997; Takken and Rep 2010).
Several strawberry cultivars have been identified as resistant or susceptible to Fusarium wilt in previous studies conducted using a variety of screening protocols, pathogen isolates, and environments (Takahashi et al. 2003; Dávalos-González et al. 2004; Takahashi et al. 2006; Fang et al. 2012a,b, 2013; Paynter et al. 2014; Gordon et al. 2016; Husaini and Neri 2016; Borrero et al. 2017). The resistance classifications of the cultivars tested in our study were consistent with those previously reported. Of the 50 cultivars we screened, eight were previously screened for resistance to a mixture of four isolates of F. oxysporum f. sp. fragariae excluding AMP132, and yielded phenotypic classifications identical to those reported here for the AMP132 isolate (Gordon et al. 2016).
The potential for new races of F. oxysporum f. sp. fragariae to emerge in California and the durability of the Fw1 R-gene are uncertain. Genes that confer vertical resistance to pathogens are commonly defeated by the loss or mutation of effector alleles in the pathogen (Chisholm et al. 2006; Jones and Dangl 2006; Ronald and Beutler 2010; Takken and Rep 2010; Monaghan and Zipfel 2012), which may or may not play a role in Fw1-mediated resistance. As reported by Henry et al. (2017), a single lineage currently dominates populations of F. oxysporum f. sp. fragariae in California. Although Fusarium oxysporum formae speciales seem to evolve new races more slowly than many other pathogens, high concentrations of inoculum, human-aided dispersal, and selection pressure increase the probability that new races will emerge (Gordon and Martyn 1997; McDonald and Linde 2002a,b). As our study shows, most of the Fusarium wilt resistant cultivars developed by UCD over the last 90 years (30% of those tested) carry a single R-gene (Fw1)—the other 70% were susceptible to Fusarium wilt. With the emergence of the pathogen in California over the last decade (Koike et al. 2009; Koike and Gordon 2015), we expect the frequency of Fusarium wilt resistant cultivars to greatly increase in California, which, when coupled with shifting fumigation practices, could increase selection pressure on the pathogen. Tomato provides a model for predicting what might eventually transpire in strawberry (Takken and Rep 2010). Several structurally and functionally diverse Fusarium wilt resistance genes (e.g., I, I-2, I-3, and I-7) have been described in tomato (Gonzalez-Cendales et al. 2016; Catanzariti et al. 2017). The tomato I gene (discovered in 1939) was less durable than the I-2 gene (discovered in 1954); however, both were eventually defeated by the emergence of new F. oxysporum f. sp. lycopersici races (Bohn and Tucker 1939; Alexander and Tucker 1945; Takken and Rep 2010; Catanzariti et al. 2017). These R-genes operate by different mechanisms that appear to affect durability and collectively provide greater safety against pathogen evolution than the individual R-genes (Catanzariti et al. 2015, 2017). Consequently, tomato breeders have identified and pyramided multiple Fusarium wilt R-genes in the arms race with the pathogen (Takken and Rep 2010).
The development and deployment of Fusarium wilt resistant cultivars in strawberry will be critically important as the pathogen spreads and increases in importance in California and other parts of the world (Koike et al. 2009; Paynter et al. 2014; Gordon et al. 2016). The array-based SNP markers and candidate genes described here provide the foundation for developing high-throughput genotyping assays for the application of marker-assisted selection, which can accurately predict Fusarium wilt phenotypes and accelerate breeding efforts. Because the Fw1 R-allele was present in 97% of the resistant germplasm accessions tested, including several cultivars spanning the breeding history of strawberry in California (Figure 7; File S2), this allele is probably found in breeding programs around the world. Similar to tomato (Houterman et al. 2008; Gonzalez-Cendales et al. 2016; Catanzariti et al. 2015, 2017), our study suggests that multiple Fusarium wilt resistance genes exist in strawberry (Figure 7; Supplement File 4). These will undoubtedly be needed to slow the emergence of new races of the pathogen and should be identified and preemptively deployed to minimize genetic vulnerability in strawberry. Several important questions remain to be answered. The number of unique loci and alleles involved in resistance to Fusarium wilt is unknown, and the genes encoding Fw1 and the other R-genes predicted by our study remain to be identified, cloned, and characterized. The identification and characterization of effector genes in F. oxysporum f. sp. fragariae will be critical for understanding the interaction between the strawberry and F. oxysporum f. sp. fragariae and the co-evolution of resistance and avirulence genes (Takken and Rep 2010).
ACKNOWLEDGMENTS
This research was supported by grants to S.J.K. from the United Stated Department of Agriculture (http://dx.doi.org/10.13039/100000199) National Institute of Food and Agriculture (NIFA) Specialty Crops Research Initiative (#2017-51181-26833) and California Strawberry Commission (http://dx.doi.org/10.13039/100006760), in addition to funding from the University of California, Davis (http://dx.doi.org/10.13039/100007707).
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.6007715.
Communicating editor: E. Huang
Literature Cited
- Abdet-Sattar M., El-Marzoky H., Mohamed A., 2008. Occurrence of soilborne diseases and root knot nematodes in strawberry plants grown on compacted rice straw bales compared with naturally infested soils. J. Plant Prot. Res. 48(2): 223–235. 10.2478/v10045-008-0026-5 [DOI] [Google Scholar]
- Alexander L. J., Tucker C. M., 1945. Physiologic specialization in the tomato wilt fungus Fusarium oxysporum f. sp. lycopersici. J. Agric. Res. 70: 303–313. [Google Scholar]
- Anderson J. P., Gleason C. A., Foley R. C., Thrall P. H., Burdon J. B., et al. , 2010. Plants vs. pathogens: an evolutionary arms race. Funct. Plant Biol. 37(6): 499–512. 10.1071/FP09304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfo G., Jupe F., Witek K., Etherington G. J., Ercolano M. R., et al. , 2014. Defining the full tomato NB-LRR resistance gene repertoire using genomic and cDNA RenSeq. BMC Plant Biol. 14(1): 120 10.1186/1471-2229-14-120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Information Resource (TAIR), 2015 Locus: AT3G46530 Available at: http://www.arabidopsis.org/servlets/TairObject?id=35741&type=locus. Accessed: February 2, 2018.
- Arroyo F. T., Llergo Y., Aguado A., Romero F., 2009. First report of Fusarium wilt caused by Fusarium oxysporum on strawberry in Spain. Plant Dis. 93(3): 323 10.1094/PDIS-93-3-0323B [DOI] [PubMed] [Google Scholar]
- Bassil N. V., Davis T. M., Zhang H., Ficklin S., Mittmann M., et al. , 2015. Development and preliminary evaluation of a 90 K Axiom SNP array for the allo-octoploid cultivated strawberry Fragaria × ananassa. BMC Genomics 16(1): 155 10.1186/s12864-015-1310-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D., Maechler M., Bolker B., Walker S., 2015. Fitting Linear Mixed-Effects Models using lme4. J. Stat. Software 67(1): 1–48. R package version 1.1–13. [Google Scholar]
- Bittner-Eddy P., Can C., Gunn N., Pinel M., Tör M., et al. , 1999. Genetic and physical mapping of the RPP13 locus, in Arabidopsis, responsible for specific recognition of several Peronospora parasitica (downy mildew) isolates. Mol. Plant Microbe Interact. 12(9): 792–802. 10.1094/MPMI.1999.12.9.792 [DOI] [PubMed] [Google Scholar]
- Bittner‐Eddy P. D., Crute I. R., Holub E. B., Beynon J. L., 2000. RPP13 is a simple locus in Arabidopsis thaliana for alleles that specify downy mildew resistance to different avirulence determinants in Peronospora parasitica. Plant J. 21(2): 177–188. 10.1046/j.1365-313x.2000.00664.x [DOI] [PubMed] [Google Scholar]
- Bohn G. W., Tucker C. M., 1939. Immunity to Fusarium wilt in the tomato. Science 89(2322): 603–604. 10.1126/science.89.2322.603 [DOI] [PubMed] [Google Scholar]
- Borrero C., Bascón J., Gallardo M. Á., Orta M. S., Avilés M., 2017. New foci of strawberry Fusarium wilt in Huelva (Spain) and susceptibility of the most commonly used cultivars. Sci. Hortic. (Amsterdam) 226: 85–90. 10.1016/j.scienta.2017.08.034 [DOI] [Google Scholar]
- Castro-Franco J., Davalos-Gonzalez P. A., 1990. Aetiology of “secadera” or root and top rot of strawberries in Irapuato, Gto. Rev. Mex. Fitopatol. 81: 81–86. [Google Scholar]
- Catanzariti A. M., Lim G. T., Jones D. A., 2015. The tomato I‐3 gene: a novel gene for resistance to Fusarium wilt disease. New Phytol. 207(1): 106–118. 10.1111/nph.13348 [DOI] [PubMed] [Google Scholar]
- Catanzariti A. M., Do H. T. T., Bru P., de Sain M., Thatcher L. F., et al. , 2017. The tomato I gene for Fusarium wilt resistance encodes an atypical leucine-rich repeat receptor-like protein whose function is nevertheless dependent on SOBIR1 and SERK3/BAK1. Plant J. 89(6): 1195–1209. 10.1111/tpj.13458 [DOI] [PubMed] [Google Scholar]
- Chisholm S. T., Coaker G., Day B., Staskawicz B. J., 2006. Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124(4): 803–814. 10.1016/j.cell.2006.02.008 [DOI] [PubMed] [Google Scholar]
- Clayton, D., 2015 snpStats: SnpMatrix and XSnpMatrix classes and methods. Available at: https://www.bioconductor.org/packages/release/bioc/html/snpStats.html. R package version 1.24.0.
- Conomos M. P., Miller M. B., Thornton T., 2015. Robust inference of population structure for ancestry prediction and correction of stratification in the presence of relatedness. Genet. Epidemiol. 39(4): 276–293. 10.1002/gepi.21896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conomos, M. P., and T. Thornton, 2016 GENetic EStimation and inference in structured samples (GENESIS): statistical methods for analyzing genetic data from samples with population structure and/or relatedness. Available at: https://bioconductor.org/packages/release/bioc/html/GENESIS.html. R package version 2.4.0.
- Conomos M. P., Reiner A. P., Weir B. S., Thornton T. A., 2016. Model-free estimation of recent genetic relatedness. Am. J. Hum. Genet. 98(1): 127–148. 10.1016/j.ajhg.2015.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl J. L., Horvath D. M., Staskawicz B. J., 2013. Pivoting the plant immune system from dissection to deployment. Science 341(6147): 746–751. 10.1126/science.1236011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dávalos-González P. A., Jofre-Garfias A. E., Hernandez-Razo A. R., Narro-Sánchez J., Castro-Franco J., et al. , 2004. Strawberry breeding for the Central Plateau of Mexico. Acta Hortic. (708): 547–552. [Google Scholar]
- De Mendiburu, F., 2015 Agricolae: statistical procedures for agricultural research. Available at: http://tarwi.lamolina.edu.pe/∼fmendiburu/. R package version 1.2–3.
- Downing J., 2016. A crossroads for strawberries. Calif. Agric. 70(3): 100 10.3733/ca.2016a0008 [DOI] [Google Scholar]
- Edger P. P., VanBuren R., Colle M., Poorten T. J., Wai C. M., et al. , 2018. Single-molecule sequencing and optical mapping yields an improved genome of woodland strawberry (Fragaria vesca) with chromosome-scale contiguity. Gigascience 7(2): 1–7. 10.1093/gigascience/gix124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X., Kuo J., You M. P., Finnegan P. M., Barbetti M. J., 2012a Comparative root colonisation of strawberry cultivars Camarosa and Festival by Fusarium oxysporum f. sp. fragariae. Plant Soil 358(1-2): 75–89. 10.1007/s11104-012-1205-8 [DOI] [Google Scholar]
- Fang X., Phillips D., Verheyen G., Li H., Sivasithamparam K., et al. , 2012b Yields and resistance of strawberry cultivars to crown and root diseases in the field, and cultivar responses to pathogens under controlled environment conditions. Phytopathol. Mediterr. 51(1): 69–84. [Google Scholar]
- Fang X., Jost R., Finnegan P. M., Barbetti M. J., 2013. Comparative proteome analysis of the strawberry-Fusarium oxysporum f. sp. fragariae pathosystem reveals early activation of defense responses as a crucial determinant of host resistance. J. Proteome Res. 12(4): 1772–1788. 10.1021/pr301117a [DOI] [PubMed] [Google Scholar]
- Falconer D. S., Mackay T. F., 1996. Introduction to Quantitative Genetics, Ed. 4th Pearson Education Limited, Essex. [Google Scholar]
- Fox, J., and S. Weisberg, 2011 An {R} Companion to Applied Regression (2nd edn.). SAGE Publications, Inc., Thousand Oaks. R package version 2.1–5. [Google Scholar]
- Gogarten S. M., Bhangale T., Conomos M. P., Laurie C. A., McHugh C. P., et al. , 2012. GWASTools: an R/Bioconductor package for quality control and analysis of genome-wide association studies. Bioinfo. 28(24): 3329–3331. R package version 1.20.0. 10.1093/bioinformatics/bts610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Cendales Y., Catanzariti A., Baker B., Mcgrath D. J., Jones D. A., 2016. Identification of I-7 expands the repertoire of genes for resistance to Fusarium wilt in tomato to three resistance gene classes. Mol. Plant Pathol. 17(3): 448–463. 10.1111/mpp.12294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodhue R. E., Fennimore S. A., Ajwa H. A., 2005. The economic importance of methyl bromide: does the California strawberry industry qualify for a critical use exemption from the methyl bromide ban? Rev. Agric. Econ. 27(2): 198–211. 10.1111/j.1467-9353.2005.00221.x [DOI] [Google Scholar]
- Goodhue R. E., Schweisguth M., Klonsky K. M., 2016. Revised chloropicrin use requirements impact strawberry growers unequally. Calif. Agric. 70(3): 116–123. 10.3733/ca.2016a0002 [DOI] [Google Scholar]
- Gordon T. R., 2017. Fusarium oxysporum and the Fusarium wilt Syndrome. Annu. Rev. Phytopathol. 55(1): 23–39. 10.1146/annurev-phyto-080615-095919 [DOI] [PubMed] [Google Scholar]
- Gordon T. R., Martyn R. D., 1997. The evolutionary biology of Fusarium oxysporum. Annu. Rev. Phytopathol. 35(1): 111–128. 10.1146/annurev.phyto.35.1.111 [DOI] [PubMed] [Google Scholar]
- Gordon T. R., Daugovish O., Koike S. T., Islas C. M., Kirkpatrick S. C., et al. , 2016. Options for management of Fusarium wilt of strawberry in California. Int. J. Fruit Sci. 16(sup1, S1) 160–168. 10.1080/15538362.2016.1219294 [DOI] [Google Scholar]
- Guthman J., 2016. Going both ways: more chemicals, more organics, and the significance of land in post-methyl bromide fumigation decisions for California’s strawberry industry. J. Rural Stud. 47: 76–84. 10.1016/j.jrurstud.2016.07.020 [DOI] [Google Scholar]
- Guthman J., Brown S., 2016a Midas’ not-so-golden touch: on the demise of methyl iodide as a soil fumigant in California. J. Environ. Policy Plann. 18(3): 324–341. 10.1080/1523908X.2015.1077441 [DOI] [Google Scholar]
- Guthman J., Brown S., 2016b Whose life counts: biopolitics and the “bright line” of chloropicrin mitigation in California’s strawberry industry. Sci. Tech. Hum. Val. 41(3): 461–482. 10.1177/0162243915606804 [DOI] [Google Scholar]
- Henry P. M., Kirkpatrick S. C., Islas C. M., Pastrana A. M., Yoshisato J. A., et al. , 2017. The population of Fusarium oxysporum f. sp. fragariae, cause of Fusarium wilt of strawberry, in California. Plant Dis. 101(4): 550–556. 10.1094/PDIS-07-16-1058-RE [DOI] [PubMed] [Google Scholar]
- Hinrichs A. L., Larkin E. K., Suarez B. K., 2009. Population stratification and patterns of linkage disequilibrium. Genet. Epidemiol. 33(S1): S88–S92. 10.1002/gepi.20478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houterman P. M., Cornelissen B. J. C., Rep M., 2008. Suppression of plant resistance gene-based immunity by a fungal effector. PLoS Pathog. 4(5): e1000061 10.1371/journal.ppat.1000061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houterman P. M., Ma L., van Ooijen G., de Vroomen M. J., Cornelissen B. J. C., et al. , 2009. The effector protein Avr2 of the xylem-colonizing fungus Fusarium oxysporum activates the tomato resistance protein I-2 intracellularly. Plant J. 58(6): 970–978. 10.1111/j.1365-313X.2009.03838.x [DOI] [PubMed] [Google Scholar]
- Huang Y., Zhen W., Zhang L., Zhang G., Tian L., et al. , 2005. Separation of causal organisms and biocontrol of strawberry (Fragaria ananassa Dach) replant disease. Biotech. 15(6): 74–76. [Google Scholar]
- Husaini A. M., Neri D., 2016. Strawberry: Growth, Development and Diseases, CABI, Oxfordshire: 10.1079/9781780646633.0000 [DOI] [Google Scholar]
- Hutton, D. M., and A. Gomez, 2006 The incidence of fusarium wilt in Queensland cultivars. HAL Project BS05003. Dept. Primary Ind. Fish., Queensland, Australia.
- Jones J. D., Dangl J. L., 2006. The plant immune system. Nature 444(7117): 323–329. 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- Koike S. T., Kirkpatrick S. C., Gordon T. R., 2009. Fusarium wilt of strawberry caused by Fusarium oxysporum in California. Plant Dis. 93(10): 1077 10.1094/PDIS-93-10-1077A [DOI] [PubMed] [Google Scholar]
- Koike S. T., Gordon T. R., 2015. Management of Fusarium wilt of strawberry. Crop Prot. 73: 67–72. 10.1016/j.cropro.2015.02.003 [DOI] [Google Scholar]
- Lenth R. V., 2016. Least-squares means: the R package lsmeans. J. Stat. Software 69(1): 1–33. R package version 2.27–2. [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinfo. 25(14): 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd M., Gordon T. R., 2016. Growing for the future: collective action, land stewardship and soilborne pathogens in California strawberry production. Calif. Agric. 70(3): 101–103. 10.3733/ca.2016a0009 [DOI] [Google Scholar]
- Lynch M., Walsh B., 1998. Genetics and Analysis of Quantitative Traits, Sinauer Associates, Inc., Sunderland. [Google Scholar]
- Maas J. L., 1998. Compendium of Strawberry Diseases, APS Press, St. Paul. [Google Scholar]
- Mangandi J., Verma S., Osorio L., Peres N. A., van de Weg E., et al. , 2017. Pedigree-based analysis in a multiparental population of octoploid strawberry reveals QTL alleles conferring resistance to Phytophthora cactorum. G3 (Bethesda) 7(6): 1707–1719. 10.1534/g3.117.042119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G. B., Bogdanove A. J., Sessa G., 2003. Understanding the functions of plant disease resistance proteins. Annu. Rev. Plant Biol. 54(1): 23–61. 10.1146/annurev.arplant.54.031902.135035 [DOI] [PubMed] [Google Scholar]
- McDonald B. A., Linde C., 2002a Pathogen population genetics, evolutionary potential, and durable resistance. Annu. Rev. Phytopathol. 40(1): 349–379. 10.1146/annurev.phyto.40.120501.101443 [DOI] [PubMed] [Google Scholar]
- McDonald B. A., Linde C., 2002b The population genetics of plant pathogens and breeding strategies for durable resistance. Euphytica 124(2): 163–180. 10.1023/A:1015678432355 [DOI] [Google Scholar]
- Mena A. J., Palacios de Garcia M. E., Gonzàles M. A., 1975. Root diseases of strawberry caused by Fusarium oxysporum Sch. fs fragariae Winks et Will and Rhizoctonia fragariae Husain et McKeen. Rev. Agro. Noroeste Arg. 12: 299–307. [Google Scholar]
- Michielse C. B., Rep M., 2009. Pathogen profile update: Fusarium oxysporum. Mol. Plant Pathol. 10(3): 311–324. 10.1111/j.1364-3703.2009.00538.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan J., Zipfel C., 2012. Plant pattern recognition receptor complexes at the plasma membrane. Curr. Opin. Plant Biol. 15(4): 349–357. 10.1016/j.pbi.2012.05.006 [DOI] [PubMed] [Google Scholar]
- Montzka S. A., Fraser P. J., Butler J. H., Connell P. S., Cunnold D. M., et al. , 2003. Controlled substances and other source gases, pp. 1–83 in Scientific Assessment of Ozone Depletion: 2002, edited by Nohende Ajvaon A., Albritton D. L., Megie G., Watson R. World Meteorological Organization, Geneva. [Google Scholar]
- Mori T., Kitamura H., Kuroda K., 2005. Varietal differences in Fusarium wilt resistance in strawberry cultivars and the segregation of this trait in F1 hybrids. J. Jpn. Soc. Hortic. Sci. 74(1): 57–59. 10.2503/jjshs.74.57 [DOI] [Google Scholar]
- Okamoto H., Fujii S., Kato K., Yoshioka A., 1970. A new strawberry disease ‘Fusarium wilt’. Plant Prot. 24: 231–235. [Google Scholar]
- Ori N., Eshed Y., Paran I., Presting G., Aviv D., et al. , 1997. The I2C family from the wilt disease resistance locus I2 belongs to the nucleotide binding, leucine-rich repeat superfamily of plant resistance genes. Plant Cell 9(4): 521–532. 10.1105/tpc.9.4.521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paynter M. L., Czislowski E., Herrington M. E., Aitken E. A., 2016. Differences in pathogenicity, genetic variability and cultivar responses among isolates of Fusarium oxysporum from strawberry in Australia. J. Am. Soc. Hortic. Sci. 141(6): 645–652. 10.21273/JASHS03888-16 [DOI] [Google Scholar]
- Paynter M. L., de Faveri J., Herrington M. E., 2014. Resistance to Fusarium oxysporum f. sp. fragariae and predicted breeding values in strawberry. J. Am. Soc. Hortic. Sci. 139(2): 178–184. [Google Scholar]
- Ronald P. C., Beutler B., 2010. Plant and animal sensors of conserved microbial signatures. Science 330(6007): 1061–1064. 10.1126/science.1189468 [DOI] [PubMed] [Google Scholar]
- Rose L. E., Bittner-Eddy P. D., Langley C. H., Holub E. B., Michelmore R. W., et al. , 2004. The maintenance of extreme amino acid diversity at the disease resistance gene, RPP13, in Arabidopsis thaliana. Genetics 166(3): 1517–1527. 10.1534/genetics.166.3.1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H., Yoshida Y., Kanda H., Furuya H., Matsmoto T., 2003. Breeding of Fusarium wilt-resistant strawberry cultivar suitable for field culture in Northern Japan. Acta Hortic. (626): 113–118. 10.17660/ActaHortic.2003.626.15 [DOI] [Google Scholar]
- Takahashi H., Yoshida Y., Kanda H., 2006. WB-B33, a new Fusarium wilt-resistant strawberry line. Acta Hortic. (760): 419–424. [Google Scholar]
- Takken F., Rep M., 2010. The arms race between tomato and Fusarium oxysporum. Mol. Plant Pathol. 11(2): 309–314. 10.1111/j.1364-3703.2009.00605.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourte L., Bolda M., Klonsky K. M., 2016. The evolving fresh market berry industry in Santa Cruz and Monterey counties. Calif. Agric. 70(3): 107–115. 10.3733/ca.2016a0001 [DOI] [Google Scholar]
- United States Department of Agriculture (USDA), 2017a Economic Research Service (ERS) Fruit and Tree Nut Yearbook: Dataset (89022). Available at: http://usda.mannlib.cornell.edu/MannUsda/viewDocumentInfo.do?documentID=1377. Accessed: February 2, 2018.
- United States Department of Agriculture (USDA), 2017b National Agricultural Statistics Service Agricultural Statistics Board (NASS SAB) Noncitrus Fruits and Nuts. Available at: http://usda.mannlib.cornell.edu/MannUsda/viewDocumentInfo.do?documentID=1113. Accessed: February 2, 2018.
- van Dijk T., Pagliarani G., Pikunova A., Noordijk Y., Yilmaz-Temel H., et al. , 2014. Genomic rearrangements and signatures of breeding in the allo-octoploid strawberry as revealed through an allele dose based SSR linkage map. BMC Plant Biol. 14(1): 55 10.1186/1471-2229-14-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooijen J. W., 2006. Joinmap 4, Software for the Calculation of Genetic Linkage Maps in Experimental Populations, Kyazma B.V., Wageningen, Netherlands. [Google Scholar]
- van Ooijen J. W., 2009. MapQTL 6, Software for the Mapping of Quantitative Trait Loci in Experimental Populations of Diploid Species, Kyazma B.V., Wageningen, Netherlands. [Google Scholar]
- Velders G. J., Andersen S. O., Daniel J. S., Fahey D. W., McFarland M., 2007. The importance of the Montreal Protocol in protecting climate. Proc. Natl. Acad. Sci. USA 104(12): 4814–4819. 10.1073/pnas.0610328104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S., Bassil N. V., Van De Weg E., Harrison R. J., Monfort A., et al. , 2017. Development and evaluation of the Axiom IStraw35 384HT array for the allo-octoploid cultivated strawberry Fragaria × ananassa. Acta Hortic. (1156): 75–82. 10.17660/ActaHortic.2017.1156.10 [DOI] [Google Scholar]
- Winks B. L., Williams Y. N., 1965. A wilt of strawberry caused by a new form of Fusarium oxysporum. Queensl. J. Agric. Anim. Sci 22: 475–479. [Google Scholar]
- Wu R., Ma C. X., Painter I., Zeng Z. B., 2002. Simultaneous maximum likelihood estimation of linkage and linkage phases in outcrossing species. Theor. Popul. Biol. 61(3): 349–363. 10.1006/tpbi.2002.1577 [DOI] [PubMed] [Google Scholar]
- Wulff B. B., Horvath D. M., Ward E. R., 2011. Improving immunity in crops: new tactics in an old game. Curr. Opin. Plant Biol. 14(4): 468–476. 10.1016/j.pbi.2011.04.002 [DOI] [PubMed] [Google Scholar]
- Zheng, X., D. Levine, J. Shen, S. M. Gogarten, C. Laurie et al., 2012 A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinforma. 28(24): 3326–3328. R package version 1.8.0. 10.1093/bioinformatics/bts606 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data required to replicate the analyses are available as supplements cited in-text or in Supplemental Data Files 1-4. Supplemental Data Files 1 and 2 contain the raw genotypic data for the germplasm accessions and mapping populations, respectively. Supplemental Data File 3 provides additional information regarding SNP nomenclature, alleles, and genomic locations. Supplemental Data File 4 provides the raw phenotypic (disease symptom) scores for every time point in 2016 and 2017 studies. Supplemental material available at Figshare: https://doi.org/10.25387/g3.6007715.