Abstract
Set1 and Jhd2 regulate the methylation state of histone H3 lysine-4 (H3K4me) through their opposing methyltransferase and demethylase activities in the budding yeast Saccharomyces cerevisiae. H3K4me associates with actively transcribed genes and, like both SET1 and JHD2 themselves, is known to regulate gene expression diversely. It remains unclear, however, if Set1 and Jhd2 act solely through H3K4me. Relevantly, Set1 methylates lysine residues in the kinetochore protein Dam1 while genetic studies of the S. pombe SET1 ortholog suggest the existence of non-H3K4 Set1 targets relevant to gene regulation. We interrogated genetic interactions of JHD2 and SET1 with essential genes involved in varied aspects of the transcription cycle. Our findings implicate JHD2 in genetic inhibition of the histone chaperone complexes Spt16-Pob3 (FACT) and Spt6-Spn1. This targeted screen also revealed that JHD2 inhibits the Nrd1-Nab3-Sen1 (NNS) transcription termination complex. We find that while Jhd2’s impact on these transcription regulatory complexes likely acts via H3K4me, Set1 governs the roles of FACT and NNS through opposing H3K4-dependent and -independent functions. We also identify diametrically opposing consequences for mutation of H3K4 to alanine or arginine, illuminating that caution must be taken in interpreting histone mutation studies. Unlike FACT and NNS, detailed genetic studies suggest an H3K4me-centric mode of Spt6-Spn1 regulation by JHD2 and SET1. Chromatin immunoprecipitation and transcript quantification experiments show that Jhd2 opposes the positioning of a Spt6-deposited nucleosome near the transcription start site of SER3, a Spt6-Spn1 regulated gene, leading to hyper-induction of SER3. In addition to confirming and extending an emerging role for Jhd2 in the control of nucleosome occupancy near transcription start sites, our findings suggest some of the chromatin regulatory functions of Set1 are independent of H3K4 methylation.
Keywords: histone methylation, SET1, JHD2, transcription, histone mutants
Methylation of histone H3 lysine-4 (H3K4me) represents one of the most comprehensively studied chromatin modifications. Like most histone lysine methylations, H3K4 exhibits mono, di, and tri-methylated states (H3K4me1, H3K4me2, and H3K4me3) that have distinctive regulatory outputs reflecting the recruitment of effector proteins (santos-rosa et al. 2003; taverna et al. 2006; kim and buratowski 2009). In the budding yeast Saccharomyces cerevisiae, H3K4me levels are controlled by the opposing functions of two highly conserved enzymes belonging to the Trithorax/MLL and JARID1 families: the Set1 methyltransferase and the Jhd2 demethylase (krogan et al. 2002; ingvarsdottir et al. 2007; liang et al. 2007). Unless otherwise stated, whenever we refer to SET1 and JHD2, we are signifying the budding yeast genes specifically. While Jhd2 appears to act alone, Set1, like Trithorax/MLL, functions within COMPASS, a conserved complex of proteins that function in Set1 targeting and regulation (krogan et al. 2002). Surprisingly, despite the crucial role of Trithorax/MLL and JARID during development and disease in animals, neither SET1 nor JHD2 are essential for cell viability in budding yeast (benevolenskaya et al. 2005; dey et al. 2008; lopez-bigas et al. 2008). Moreover, JHD2 null mutants (jhd2∆) have surprisingly limited phenotypic impact in otherwise wild-type (WT) mitotic cells grown in the presence of glucose, confounding the study of this important chromatin regulatory protein using yeast (lenstra et al. 2011; xu et al. 2012).
Previously, we found that JHD2 exhibits specific functions during the gametogenesis phase of the budding yeast life cycle, also known as sporulation (xu et al. 2012). The formation of robust spores requires that JHD2 maintain a period of productive transcription in the face of encroaching and developmentally programmed transcriptional quiescence. During this period, JHD2 globally demethylates H3K4 at intergenic regions upstream of transcription start sites (TSSs) and overlapping with transcription termination sites (TTSs). Associated with these H3K4me defects, JHD2 also represses the accumulation of noncoding intergenic transcripts genome-wide (xu et al. 2012). These findings suggest roles for JHD2 in diverse aspects of transcription, including those related to termination and its associated mRNA processing pathways. Indeed, a recent study found that Jhd2 physically interacts with CPF (cleavage and polyadenylation factor) and causes defects in mRNA 3′ untranslated region length for some genes (blair et al. 2016). More recently, we determined that H3K4 demethylation by Jhd2 occurs not only during sporulation, but also in mitotic cells in response to non-fermentable carbon sources or to nitrogen manipulations that lead to increased levels of alpha-ketoglutarate, an intermediary metabolite essential for demethylase activity of Jhd2 and the entire Jumonji family of demethylases to which JARIDs belong (liang et al. 2007; soloveychik et al. 2016).
Our previous studies identified developmental/nutritional contexts during which JHD2 impacts gene expression, but the mechanisms remain opaque. To gain further insight into genetic pathways by which JHD2 and SET1 impact gene expression, we used a targeted screening approach. Because genome-wide screens have been unsuccessful in the identification of gene deletions that exhibit interactions with jhd2∆ ((costanzo et al. 2016), our unpublished findings), we selected essential components of the transcriptional machinery and tested whether temperature sensitive (TS) mutants of these components genetically interact with jhd2∆, and in most cases, set1∆ as well. Using this approach, we find that JHD2 genetically inhibits the essential RNA PolII transcription regulatory complexes Spt6-Spn1, FACT (facilitates chromatin transcription), and NNS (Nab3-Nrd1-Sen1) (formosa et al. 2001; vasiljeva and buratowski 2006; mcdonald et al. 2010). More detailed genetic experiments reveal that JHD2’s inhibitory role can be attributed solely to reversal of H3K4me. Accordingly, genetic interrogation of SET1 confirms a positive regulatory impact of H3K4me, but also illuminates a counterbalancing and inhibitory role of SET1 that acts independent of H3K4me. Using molecular biological experiments, we determine that Jhd2 opposes the positioning of a Spt6-Spn1-deposited nucleosome near the 3′ of the SRG1 non-coding transcript, and accordingly impacts the induction of the downstream SER3 gene known to be regulated by SRG1, perhaps explaining at least some of the basis for the genetic interactions. In summary, our findings identify three transcriptional regulatory complexes subject to regulation by Jhd2 and Set1, and illuminate that SET1 impinges upon them through H3K4me dependent and independent means.
Materials and Methods
Yeast Strains and Plasmids
Standard yeast genetic methods were used for construction of all strains. Yeast strains and plasmids used in this study are listed in Table S1. All strains were constructed through genetic crosses followed by dissections the BY4742 background. Yeast strains were inoculated into several mL of YPD (1% yeast extract, 2% peptone, and 2% glucose) and grown overnight at room temperature (23°). Each strain was diluted to an OD600 = 0.4, serially diluted five times and spotted on synthetic complete media (YNB media (Multicell Wisent) containing 5 g/L of ammonium sulfate and either 2% glucose or 2% galactose.
Western blots
Exponential cultures with OD600 between 1-2 were lysed by vortexing with acid washed glass beads in SUMEB buffer as described previously (Xu et al. 2012). Total protein concentration was quantified using an RC/DC assay (BioRad). Equal amounts of protein were electrophoresed on 12% SDS-PAGE gels, and transferred onto Amersham Hybond-P membranes (GE). Immunoblot analysis was performed using standard procedures. All blots were scanned with a ChemiDoc XRS+ Imaging System. Band intensities were quantified using ImageJ 1.51 v software.
Chromatin immunoprecipitation
Strains were grown at 30° in YPD to OD600 0.8. Crosslinking was performed in 1% formaldehyde for 15 min at room temperature (23°) and then immediately quenched with glycine at a final concentration of 125 mM. Cells were pelleted and washed 2 times with ice cold PBS. Cell pellets were snap frozen in 2 mL screwcap tubes using liquid nitrogen and stored at -80°. Frozen pellets were resuspended in lysis buffer (50 mM HEPES pH 7.5, 140 mM NaCl, 1 mM EDTA pH 8.3, 1% Triton X-100, 0.1% NaDOC) without thawing and topped off with glass beads. Cells were lysed through multiple rounds of beadbeating and then sonicated to shear the chromatin to fragment sizes of approximately 200 to 500 bp. Cross-linked chromatin fragments were immunoprecipitated with antibody overnight. Protein A-Sepharose beads were then added to the samples, and samples were incubated for 90 min. The immunoprecipitated complexes were washed with lysis buffer, lysis buffer containing 500 mM NaCl, wash buffer (10 mM Tris-HCl pH 8.0, 0.25M LiCl, 0.5% NP40, 0.5% NaDOC, 1mM EDTA pH 8.3), and TE (10 mM Tris-HCl pH 8.0, 1 mM EDTA). Next, the immunoprecipitated chromatin was eluted from beads with elution buffer (50 mM Tris-HCl pH 8.0, 10 mM EDTA, 1% SDS) and then eluted with TE/0.67% SDS (10 mM Tris-HCl pH 8.0, 1 mM EDTA pH 8.3, 0.67% SDS, 0.334 mg/mL proteinase K). The two elutions were combined. Whole cell extract samples were topped off with TE/1% SDS (10 mM Tris-HCl pH 8.0, 1mM EDTA, pH 8.3, 1% SDS, 0.25 mg/mL proteinase K). Formaldehyde cross-linking was reversed by incubating the eluates at 65° overnight. DNA from the eluates was treated with RNAse A and purified with a QIAquick PCR purification kit (Qiagen). Immunoprecipitated fractions (IP) and whole-cell extracts (Input) containing DNA were analyzed by real-time PCR as described previously (xu et al. 2012) using the primers listed in Table S2. SYBR green signal was measured using the BioRad iQ5 Multicolor Real Time PCR Detection System.
SER3 RT-qPCR analysis
5-10 OD equivalents were harvested for RNA extraction. For RT-qPCR, RNA was extracted with acidic phenol at 65° for 30 min. RNA was then purified, precipitated and resuspended in RNAse free water. cDNA was prepared as described previously using random priming (xu et al. 2012). Quantitative PCR quantification of cDNA using SYBR green was measured using the BioRad iQ5 Multicolor Real Time PCR Detection System as described previously (xu et al. 2012). Primer sequences for SER3 and SCR1 are shown in Table S2. Transcript levels for each primer pair tested were normalized to the reference transcript SCR1.
SER3 induction assays
Yeast strains were inoculated into several mL of YPD and grown overnight at room temperature (23°). Each strain was diluted to an OD600 = 0.4 in synthetic complete media and grown to OD600 = 2 at 30°. At this time, cells were harvested by centrifugation, washed with water, and resuspended in synthetic media lacking serine. Growth of cells was resumed with constant shaking at 30°. Cell samples were collected at specific intervals and frozen using liquid nitrogen for analysis by RT-qPCR as described previously (xu et al. 2012). Primer sequences for SER3 and SCR1 are shown in Table S2. RNA isolation, cDNA synthesis, and qPCR is described above.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Strains are available upon request.
Results
Genetic interactions of JHD2 with essential transcription regulatory factors
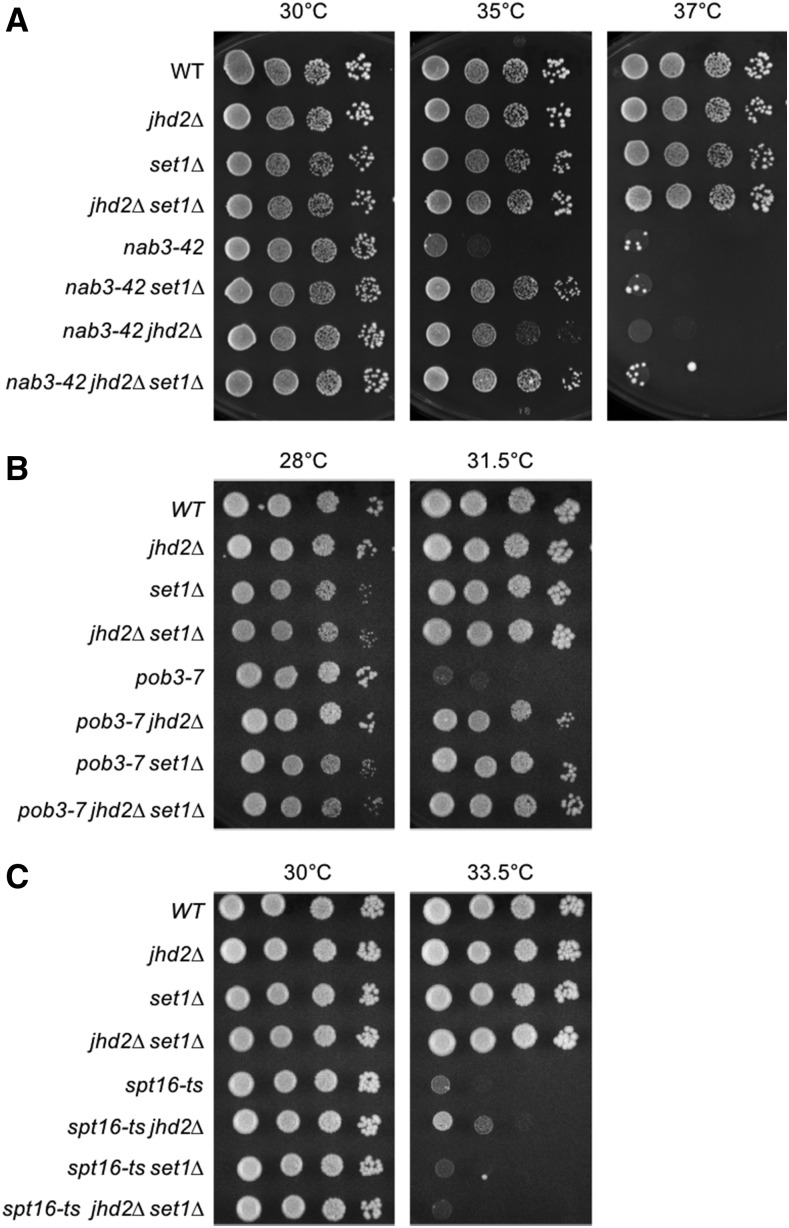
We used a targeted screening approach to identify mutants that exhibit genetic interactions with jhd2∆. Because Jhd2 reverses a histone modification associated with active transcription, we evaluated temperature sensitive (TS) alleles of essential genes related to transcriptional control as interacting candidates of jhd2∆. We used strains sourced from a library created and curated previously (li et al. 2011), with the only exceptions being nab3-42 and spt16-319, which we obtained separately (o’donnell et al. 2004; darby et al. 2012). All strains described here were engineered using genetic crosses and tetrad dissection. Strain fitness was assessed using spot assays to compare growth rates at varied temperatures. Deletion of JHD2 or SET1 did not cause growth defects under these conditions (Figure 1A, B, and C). For all results shown, we isolated at least 2 independently constructed strain replicates through tetrad dissection. Though replicates and WT control strains are not always shown in the interest of space, all results we report here are upheld in these replicates.
Figure 1.
TS alleles of NNS and FACT subunits exhibit genetic interactions with H3K4 modifying enzymes. Yeast strains with the indicated genotypes were serially diluted ten-fold, spotted onto agar plates containing synthetic complete media, and grown at indicated temperatures. Genetic interactions of jhd2∆ and set1∆ with different temperature sensitive alleles of NNS and FACT subunits are shown: (A) nab3-42 (B) pob3-7 (C) spt16-ts.
Given the physical association of Jhd2 with CPF/CF, we first evaluated jhd2∆ genetic interactions with TS mutations in 10 different subunits of this large complex (blair et al. 2016). Somewhat surprisingly, we failed to identify any reproducible interactions with CPF/CF (Table 1). NNS functions alongside CPF as one of two distinct mRNA processing pathways in budding yeast. Nab3 and Nrd1 encode RRM domain containing proteins that recognize specific sequences in nascent transcripts (steinmetz and brow 1996; carroll et al. 2004; creamer et al. 2011). The Sen1 subunit is homologous to the human helicase protein Senataxin and provides the activity that physically dislodges PolII from the DNA (porrua and libri 2013; chen et al. 2014). While the majority of mRNA 3′ end processing of protein coding genes occurs through CPF, NNS controls the transcriptional termination and processing of noncoding RNAs. The most widely studied targets of NNS are snRNAs, snoRNAs, and cryptic unstable transcripts (CUTS) (ursic et al. 1997; steinmetz et al. 2001; arigo et al. 2006; thiebaut et al. 2006).
Table 1. Genetic interactions of JHD2 with essential transcription regulatory factors.
| Allele | Protein Group | Genetic Interaction |
|---|---|---|
| ctf2-1 | CPF/CF | N.I. |
| pti1-ts7 | CPF/CF | N.I. |
| fip1-433 | CPF/CF | N.I. |
| fip1-ph | CPF/CF | N.I. |
| pap1-1 | CPF/CF | N.I. |
| ssu72-2 | CPF/CF | N.I. |
| clp1-ph | CPF/CF | N.I. |
| hrp1-7 | CPF/CF | N.I. |
| hrp1-4 | CPF/CF | N.I. |
| rna15-58 | CPF/CF | N.I. |
| rpb2-6 | Pol2/CTD | N.I. |
| rpo21-1 | Pol2/CTD | N.I. |
| rpb3-2 | Pol2/CTD | N.I. |
| rpb5-H147R | Pol2/CTD | N.I. |
| fcp1-1 | Pol2/CTD | N.I. |
| kin28-ts | Pol2/CTD | N.I. |
| CTD truncation | Pol2/CTD | N.I. |
| ess1-H146R | Pol2 CTD isomerase | ++ |
| spt15-P65F | TBP | N.I. |
| mot1-1033 | TBP | N.I. |
| taf5-15 | TBP | N.I. |
| med7-163 | Mediator | N.I. |
| med7-141 | Mediator | N.I. |
| med6-ts | Mediator | N.I. |
| med4-54 | Mediator | N.I. |
| med4-6 | Mediator | N.I. |
| med8-39 | Mediator | N.I. |
| rox3-182 | Mediator | N.I. |
| nab3-11 | NNS | ++ |
| nab3-42 | NNS | +++ |
| sen1-1 | NNS | + |
| spt6-14 | Spt6-Spn1 | ++ |
| spn1-K192N | Spt6-Spn1 | +++ |
| spt16-ts | FACT | + |
| spt16-319 | FACT | +++ |
| pob3-7 | FACT | +++ |
| pob3-L78R | FACT | ++ |
| mtr4-1 | TRAMP | + |
| mtr3-ts | Exosome | — |
Specific TS allele is listed in the left column and the associated protein complex of each factor is shown in the corresponding middle column. In the right column, any observed genetic interactions with jhd2∆ are documented. N.I. no interaction, + positive interaction (+ weakest, ++ medium, +++ strong suppression), - weak negative interaction.
We found that jhd2∆ robustly suppressed 2 different alleles of NAB3, nab3-42 and nab3-11 (Table 1, Figure 1A and Figure S1A). Deletion of JHD2 also modestly suppressed sen1-1 (Table 1, Figure S1B). As shown, incubation at high temperature was sufficient to eliminate growth of nab3-42, confirming that jhd2∆ did not provide bypass suppression and that Jhd2 therefore acted to inhibit nab3-42 function at semi-permissive temperatures (Figure 1A). Though we only show the lack of bypass suppression for nab3-42 in the interest of space, this was the case with all interactions we report here. NNS termination is coupled with polyadenylation by the TRAMP complex and RNA processing by Exosome (vasiljeva and buratowski 2006; vanacova and stefl 2007). We observed a similar suppressive interaction of jhd2∆ with the sole essential TRAMP component, MTR4 (Table 1 and data not shown). Interestingly, the only exosome subunit encoding gene we investigated, MTR3, provided the solitary example of a negative interaction with JHD2; jhd2∆ caused a modest but reproducible enhancement of the mtr3-ts growth defect (Table 1 and data not shown).
Among the numerous other essential genes we interrogated, including those contributing to the functions of mediator, RNAPII and its associated C-terminal domain modifications, and TATA binding protein (TBP), only temperature sensitive alleles of genes encoding Spt6-Spn1 and FACT subunits exhibited genetic interactions with jhd2∆ (Table 1, Figure 1B and 1C). Spt6-Spn1 and FACT are conserved heterodimeric histone chaperone complexes that enable RNA PolII transcription through chromatin, deposit nucleosomes in the wake of elongating polymerase, and suppress spurious intragenic transcription (kaplan et al. 2003; duina 2011; Hainer et al. 2011). While Spt6-Spn1 and FACT have many common and overlapping functions, they exhibit differences in histone binding specificity and in their abilities to disrupt nucleosomes during transcription (bortvin and winston 1996; orphanides et al. 1999; belotserkovskaya et al. 2003; mccullough et al. 2015). FACT can disrupt and reassemble nucleosomes, while Spt6-Spn1 only has the ability to reassemble them (orphanides et al. 1998; orphanides et al. 1999; belotserkovskaya et al. 2003). We also confirmed a previous finding that the temperature sensitive growth defect of ess1-H146R, which encodes a CTD-associated prolyl-isomerase, was suppressed by jhd2∆ (ma et al. 2012).
H3K4me dependent and independent modulation of NNS and FACT by JHD2 and SET1
As JHD2 reverses H3K4me, we hypothesized that increased H3K4me caused by jhd2∆ accounted for the genetic interactions described above. Indeed, H3K4me3 has been suggested to promote NNS function, with set1∆ enhancing the growth and termination phenotypes caused by alleles of NRD1 that delete portions of its N-terminus (terzi et al. 2011). Terzi et al. also showed that set1∆ enhanced termination defects caused by nab3-11, though they did not evaluate SET1’s impact on nab3-11 growth properties (terzi et al. 2011). We therefore tested if jhd2∆ suppression of NNS mutations could occur in the absence of SET1. Unexpectedly, set1∆ suppressed the growth defect of nab3-42 to a degree that exceeded jhd2∆ suppression, with the triple mutants resembling nab3-42 set1∆ (Figure 1A). Terzi et al. (2011) found that H3K4me3 promotes NNS function using truncation alleles of NRD1 (terzi et al. 2011), but the suppression of nab3-42 by set1∆ presented here suggested that H3K4me antagonizes NNS function. To rule out some unusual allele specific interaction of nab3-42 with set1∆, we replicated the set1∆ and jhd2∆ interactions with NAB3 using the nab3-11 allele (Figure S1A). Furthermore, and consistent with the NAB3 results, sen1-1 was also suppressed by both jhd2∆ and set1∆, again with the triple mutants resembling sen1-1 set1∆ (Figure S1B). While none of our experiments overlap with those of Terzi et al., their findings would not have predicted ours nor vice versa. In particular, our finding that set1∆ suppressed the nab3-11 growth defect is somewhat discordant with Terzi et al.’s finding of an aggravating consequence of set1∆ on nab3-11 for transcriptional termination. Among the many possible explanations for this discrepancy, a simple view is that the essential functions of NAB3 may not be directly reflected by termination defects described by Terzi et al.
Our targeted screen also identified the FACT subunit encoding alleles pob3-7 and spt16-ts as exhibiting alleviating genetic interactions with jhd2∆ (Table 1, Figure 1B and 1C). Using the same experimental approach as with NNS subunits, we tested if jhd2∆ suppression of pob3-7 and spt16-ts required SET1. Curiously, and unlike the case with NNS subunits, pob3-7 and spt16-ts exhibited distinct genetic interactions in combination with set1∆, even though both factors function in the FACT complex. We found that set1∆ suppressed the growth defect of pob3-7 mutants to a level comparable to the suppression conferred by jhd2∆, with the triple mutants showing no evidence of synergy (Figure 1B). In contrast, the spt16-ts temperature sensitive growth defect was suppressed by jhd2∆ in a SET1 dependent manner, suggesting that H3K4me promotes Spt16 function (Figure 1C). We confirmed these genetic interactions with additional alleles of POB3 and SPT16, pob3-L78R and spt16-319 (Figure S1C and D). The differences in SET1 genetic interaction phenotypes observed for SPT16 and POB3 mutants suggest complexities in nucleosome interactions, or that their encoded proteins may exhibit separable functions apart from the FACT complex.
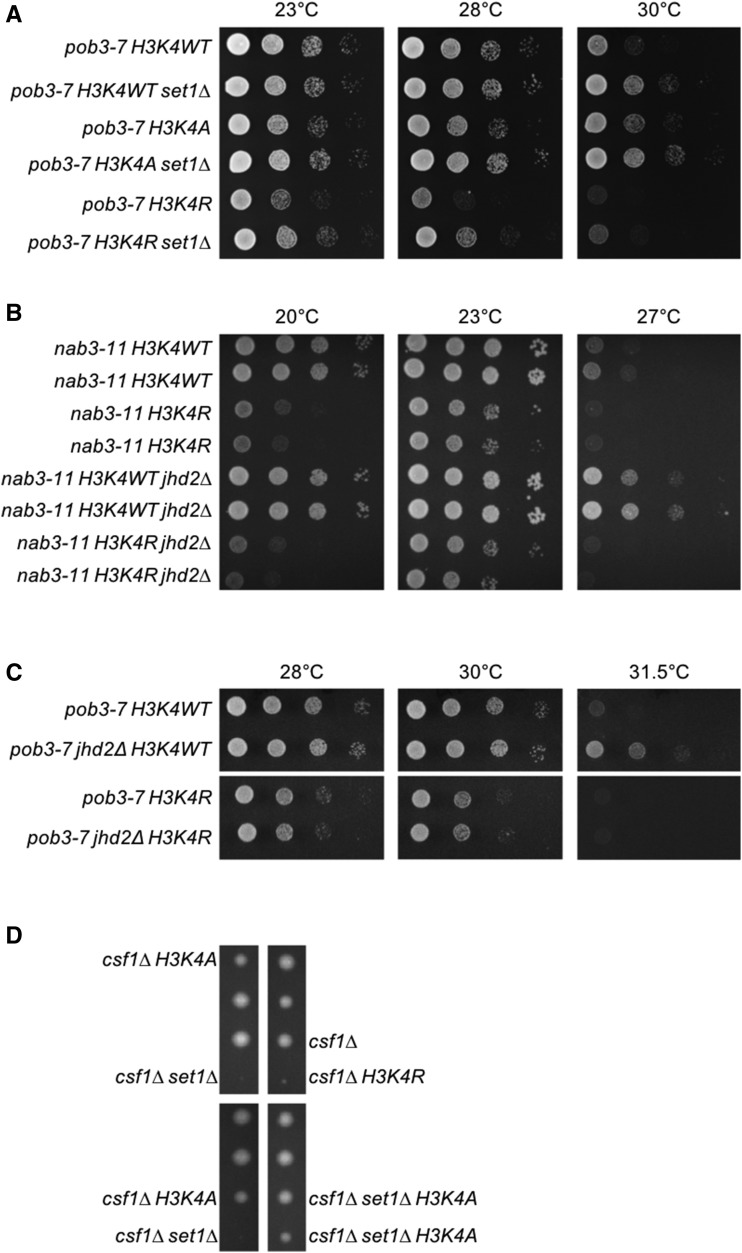
To investigate how both set1∆ and jhd2∆ can rescue certain temperature sensitive growth defects, we employed strains that express H3K4 amino acid substitution mutants from a chromosomal locus in place of wild type H3 (dai et al. 2008). The most widely utilized amino acid substitutions for this approach are alanine (H3K4A) and arginine (H3K4R). We first confirmed the histone mutations by sequencing and then determined that H3K4A and H3K4R alone or in combination with set1∆ did not cause any growth phenotypes under the conditions of our spotting assays (Figure S2A). Unexpectedly, we found that H3K4A and H3K4R modified pob3-7 and nab3-11 mutants in opposing fashion. H3K4A suppressed pob3-7 and nab3-11 temperature sensitivity (Figure 2A and Figure S2B). Suppression by H3K4A and set1∆ did not display any synergy. H3K4R caused the opposite effect of H3K4A, exacerbating the temperature sensitive growth defect of nab3-11 and pob3-7 (Figure 2A, 2B, 2C, Figure S2B, and Figure S3A). We note that this latter result is consistent with previous findings suggesting a positive impact for H3K4me3 on NNS function (terzi et al. 2011).
Figure 2.
Genetic suppression of NNS and FACT by set1∆ and jhd2∆ dependent and independent of H3K4me. Plate spot assays (as described in Figure 1) were used to compare the growth of the indicated strains. Isogenic strains were constructed through genetic crosses with H3K4A and H3K4R substitution mutants from the Dharmacon Non Essential Histone H3 & H4 Mutant Collection. (A) Genetic interactions of set1∆, H3K4A, and H3K4R with pob3-7. (B) Genetic interactions of jhd2∆ and H3K4R with nab3-11. Growth of two independent isolates of each genotype is shown. (C) Genetic interactions of jhd2∆ and H3K4R with pob3-7. (D) Diploid strains containing csf1∆, set1∆, and either H3K4A or H3K4R were sporulated and dissected onto YPD medium. A representative field of the haploid ascospores recovered and their respective genotypes are shown.
To shed light on the confounding differences in H3K4A and H3K4R phenotypes, we performed additional studies exploiting an annotated synthetic lethal interaction between set1∆ and csf1∆ (costanzo et al. 2016). CSF1 (cold sensitive for fermentation) encodes a conserved yet poorly characterized protein required for growth on glucose at low temperatures and is involved in secretory protein maturation and amino acid metabolism. (tokai et al. 2000; copic et al. 2009; costanzo et al. 2016). In support of the interpretation that csf1∆ set1∆ lethality was caused by loss of H3K4me, we found that jhd2∆ suppressed csf1∆ and that JHD2 overexpression using an integrated galactose-inducible allele enhanced csf1∆ (Figure S2C and D). The csf1∆ enhancement by GAL-JHD2 was abrogated by mutation of histidine-427 to alanine (H427A), which renders Jhd2 catalytically inactive (ingvarsdottir et al. 2007; liang et al. 2007) (Figure S2C and D).
We constructed diploid strains heterozygous for csf1∆, set1∆, and H3K4A or H3K4R mutants and interrogated them using tetrad dissections. In addition to confirming csf1∆ set1∆ synthetic lethality, we found that H3K4R exhibited synthetic lethality with csf1∆ (Figure 2D). In stark contrast, H3K4A had no consequence for csf1∆ viability, and remarkably, suppressed the synthetic lethality of csf1∆ set1∆ (Figure 2D). We dissected dozens of tetrads and confirmed that this pattern of inheritance was highly stereotypical and not due to random spore inviability (Table 2). It is noteworthy that while both H3K4A and H3K4R abolish H3K4 methylation, arginine retains the positively charged characteristic of lysine in contrast to the aliphatic alanine residue. Accordingly, arginine much more closely resembles the structure of a lysine residue compared with alanine. These results suggest that H3K4R more reliably recapitulates the consequences of set1∆ for chromatin structure and point toward neomorphic characteristics of H3K4A, which seem likely to manifest differently depending on genetic context. We therefore proceeded with our studies using the H3K4R mutant.
Table 2. Observed and expected genetic frequencies of csf1∆ with histone H3K4A and H3K4R amino acid substitution mutants.
| H3K4 | ||||
|---|---|---|---|---|
| Genotype | WT | A | R | Expected frequency |
| csf1Δ | 12 (0.16) | 7 (0.09) | 0 (0.00) | 0.125 |
| set1Δ csf1Δ | 0 (0.00) | 9 (0.11) | 0 (0.00) | 0.125 |
| Total number of spores | 73 | 79 | 71 | |
Diploid strains mmy7658 (H3K4R), mmy7659 (H3K4WT), and mmy7660 (H3K4A) were sporulated and dissected onto YPD medium. These strains are heterozygous for set1∆, csf1∆, and the corresponding histone amino acid substitution. The observed frequency of each genotype was calculated by dividing to the total number of spores counted and is shown in the parentheses.
To reconcile the H3K4R mutant phenotype with suppression of NNS and FACT subunits by set1∆, we constructed strains combining H3K4WT or H3K4R with set1∆ and NNS/FACT mutants. We found that set1∆ conferred suppression of the growth phenotypes of pob3-7, nab3-11, and sen1-1 even in the context of H3K4R albeit not to the extent seen in H3K4WT (Figure 2A, Figure S2B, Figure S3A, and B). Thus, the suppression of NNS and FACT subunits by set1∆ in H3K4R mutants suggests that some activity of Set1 acted in opposition to NNS and FACT independent of H3K4me (Figure 3).
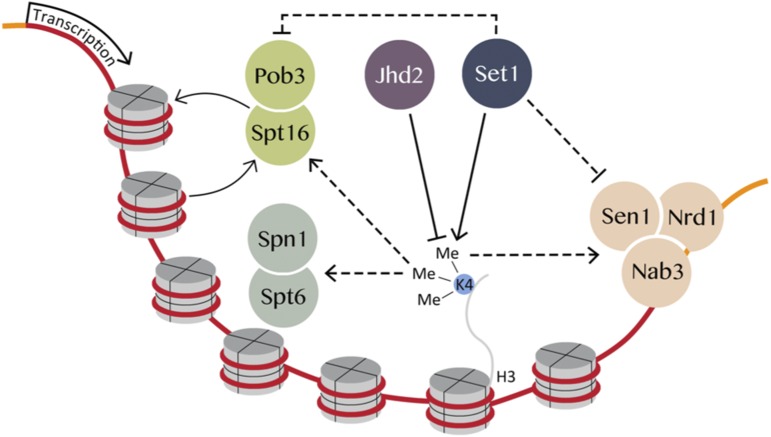
Figure 3.
Genetic model of FACT and NNS regulation by Set1 and Jhd2. Methylation of H3K4 promoted FACT and NNS function while Set1 exhibited an inhibitory function independent of H3K4. Spt6-Spn1 is positively regulated by H3K4me3.
To further evaluate the positive impact of H3K4me on NNS and FACT and the role of JHD2 in this, we performed analogous experiments with jhd2∆. Suppression of nab3-11, pob3-7, and sen1-1 by jhd2∆ could no longer occur in the context of H3K4R, arguing that Jhd2 opposed NNS and FACT function solely through reversal of H3K4me (Figure 2B, 2C, and Figure S3C). Based on these results as well as previous findings from Terzi et al., we propose counterbalancing regulation of NNS and FACT by Set1 and Jhd2, with H3K4me promoting their functions and an unknown inhibitory impact conferred by Set1 in a H3K4 independent manner (Figure 3).
To gain insight into how Set1 opposed FACT independent of H3K4, we constructed strains that had the endogenous SET1 locus replaced with SET1-G951S, an allele that abolishes Set1 enzymatic activity, but does not disrupt the integrity of COMPASS (nagy et al. 2002; schibler et al. 2016). We found that SET1-G951S suppressed pob3-7 to an extent that was equivalent to set1∆ in strains expressing a WT allele of H3K4 (Figure S4). As enzymatically dead alleles of SET1 lead to a depletion of Set1 protein levels, this was to be expected (soares et al. 2014). Curiously, in strains expressing H3K4R, which do not deplete Set1 levels to nearly the extent seen with enzymatically dead alleles of Set1 (soares et al. 2014), SET1-G951S failed to suppress pob3-7 at 30°, but did suppress equivalently to set1∆ at the more stringent temperature of 31.5° (Figure S4). Similar results were obtained with nab3-11 (data not shown). These results, while complex, suggest that the H3K4-independent inhibition of pob3-7 by Set1 may involve both enzymatic and non-enzymatic roles of this protein (Figure 3).
Spt6-Spn1 was opposingly governed by JHD2 and SET1 through H3K4me3
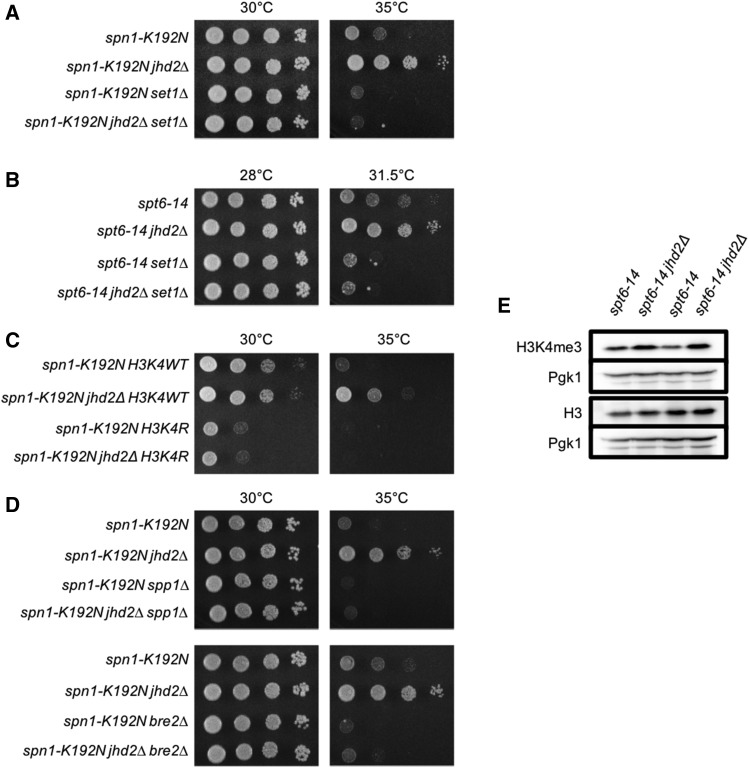
The complexity of interactions exhibited by NNS and FACT with SET1 rendered them difficult to further study without a more detailed understanding of what regulatory functions SET1 has independent of H3K4. We therefore extended our genetic analysis to Spt6-Spn1. As mentioned above, we determined that TS alleles of both Spt6-Spn1 subunits, spt6-14 and spn1-K192N, were suppressed by jhd2∆ (Table 1, Figure 4A and 4B). In contrast to NNS and FACT, the temperature sensitive growth defects of both Spt6-Spn1 subunits were suppressed by jhd2∆ in a SET1 and H3K4 methylation dependent manner (Figure 4A, B, C, and data not shown). Moreover, and consistent with the interpretation that SET1-mediated H3K4me promoted Spt6-Spn1 function, both set1∆ and H3K4R enhanced the growth defects of spt6-14 and spn1-K192N (Figure 4A, B, C, and data not shown).
Figure 4.
Spt6-Spn1 complex temperature sensitive mutants are rescued by an increase in H3K4me3 through jhd2∆. Plate spot assays (as described in Figure 1) were used to compare the growth of the indicated strains. Genetic interactions of jhd2∆ and set1∆ with Spt6-Spn1 are shown: (A) spt6-14 (B) spn1-K192N. (C) Isogenic strains were constructed through genetic crosses with the H3K4R substitution mutant from the Dharmacon Non Essential Histone H3 & H4 Mutant Collection. Genetic interactions of jhd2∆ and H3K4R with spn1-K192N. (D) Genetic interactions of jhd2∆, nonessential COMPASS subunits spp1∆ and bre2∆, and spn1-K192N. (E) Western blot analysis of H3K4me3, Histone H3, and Pgk1 were performed on biological duplicates of spt6-14 and spt6-14 jhd2∆ grown at 30°C. Pgk1 serves as a loading control.
To validate that Jhd2 demethylation of H3K4 opposed Spt6-Spn1, we complemented the jhd2∆-suppressed strains with previously described JHD2 expressing plasmids (mersman et al. 2009). Cells harboring these plasmids express JHD2 under the control of a constitutive PYK1 promoter, which leads to modestly increased levels of Jhd2 and associated decreased H3K4me3 levels. These effects on H3K4me could be abrogated by the H427A mutation or by deletion of its conserved PHD domain, which may enable Jhd2 recruitment to chromatin (mersman et al. 2009). As expected, we found that jhd2∆ suppression of spt6-14 and spn1-K192N could be reverted by JHD2 complementation and that the growth defects of spt6-14 and spn1-K192N were in fact enhanced by the JHD2 overexpressing plasmids (Figure S5A and B). Importantly, this suppression was lost in the H427A and PHD domain mutants (which both encode stable proteins (mersman et al. 2009)) (Figure S5A and B). Our findings support the conclusion that suppression of Spt6-Spn1 alleles by jhd2∆ was due to the loss of H3K4 demethylation.
To gain insights into which H3K4 methylation species accounted for suppression of Spt6-Spn1, we genetically interrogated COMPASS complex subunits known to specifically perturb H3K4me3 or H3K4me3 and -me2. The effects of bre2∆ and spp1∆ on H3K4me have been extensively described. Deletion of BRE2 results in a complete loss of H3K4me3 and a significant decrease of H3K4me2 but no change in H3K4me1, while spp1∆ results in a substantial decrease of H3K4me3 with no detectable consequence on bulk H3K4me2 or -me1 (schneider et al. 2005; Dehé et al. 2006; takahashi et al. 2009; margaritis et al. 2012). As with set1∆, we found that both bre2∆ and spp1∆ enhanced the growth defect of spn1-K192N (Figure 4D). Moreover, jhd2∆ suppression of spn1-K192N was completely dependent on BRE2 and SPP1 (Figure 4D). We compared bulk H3K4me3 levels by western blot in spt6-14 and spt6-14 jhd2∆ at a semi-permissive temperature. Consistent with the genetic data, we observed a modest increase in bulk H3K4me3 levels in the spt6-14 jhd2∆ double mutant (Figure 4E). We confirmed the reproducibility of this by quantifying the relative abundance of H3K4me3/H3 as well as of H3/Pgk1 from three biological replicates of spt6-14 and spt6-14 jhd2∆ (Figure S6). Collectively, our data support the conclusion that JHD2 and SET1 opposingly governed the function of Spt6-Spn1 through H3K4me3 (Figure 3).
JHD2 repressed TSS-associated nucleosome occupancy and transcriptional induction of a Spt6-Spn1 regulated gene
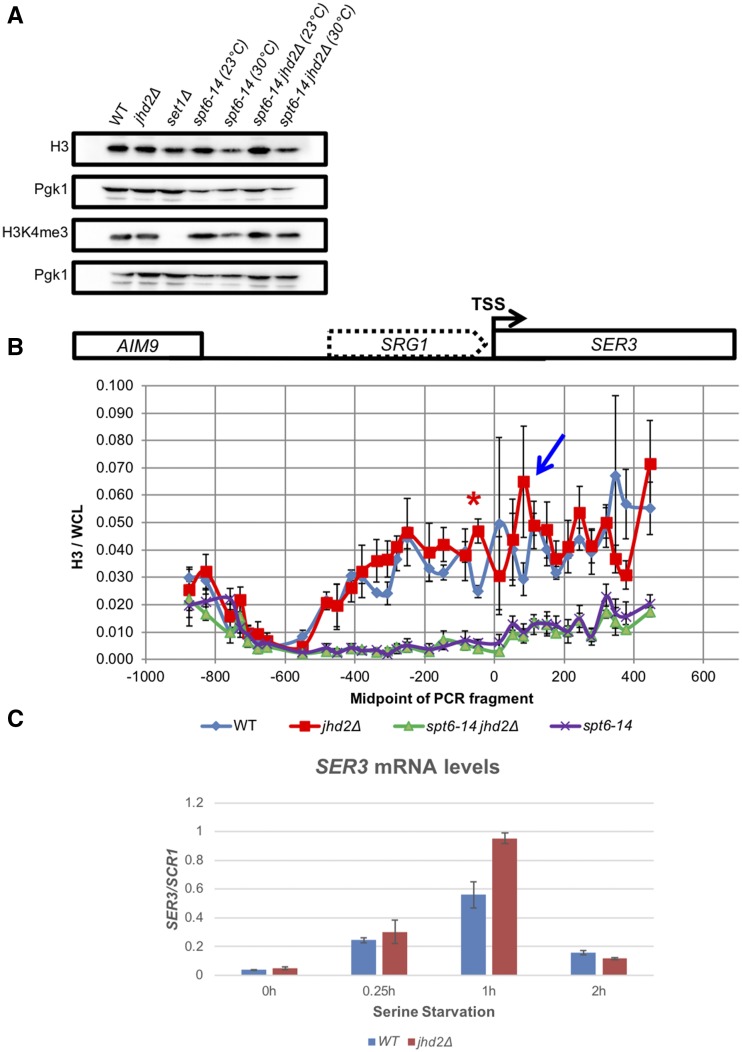
Reflecting its role in the recycling of nucleosomes in the wake of elongating PolII, Spt6-Spn1 mutation causes reduced histone occupancy genome-wide (ivanovska et al. 2011). We asked if jhd2∆ suppression of Spt6-Spn1 involved a reversal of these histone occupancy defects. First, we used western blots to measure bulk histone H3 levels and found that jhd2∆, set1∆, and spt6-14 mutants showed no apparent defects in histone H3 levels at the permissive temperature of 23° (Figure 5A). At the semi-permissive temperature of 30° spt6-14 exhibited a dramatic reduction in H3 levels, consistent with its role as a histone chaperone (Figure 5A). This reduction was not reversed by jhd2∆ suggesting that suppression did not occur by generically improving Spt6-Spn1 histone recycling or chaperone function (Figure 4E, 5A, and Figure S6).
Figure 5.
JHD2 deletion does not rescue Spt6-Spn1 mutants by compensating for defects in chromatin structure. (A) Western blot detection of H3K4me3, pan-H3, and Pgk1 are shown from extracts of wildtype and mutant cells grown at 23°C (permissive) and 30°C (semi-permissive temperature). Pgk1 serves as a loading control. (B) Histone H3 ChIP was performed on strains grown at 30°C (semi-permissive temperature) across the SRG1-SER3 locus (schematic of locus shown above graph). H3 ChIP signal was normalized to signal from whole cell lysate (WCL) DNA. The transcription start site of SER3 is positioned at 0 and for SRG1 at -480. Error bars represent standard error of the mean. Significance as calculated by a two-tailed student’s t-test is shown where *P < 0.05. A blue arrow denotes an additional H3 ChIP peak in jhd2∆ that was not statistically significant. (C) SER3 transcript level was measured by RT-qPCR at specific intervals after serine withdrawal in wildtype and jhd2∆ mutants. SER3 signal was normalized to SCR1.
To gain more detailed insight into the relationship between Jhd2 and Spt6-Spn1, we measured chromatin-associated histone H3 in WT, jhd2∆, spt6-14, and spt6-14 jhd2∆ cells at the SRG1-SER3 locus using chromatin immunoprecipitation with an anti-H3 antibody followed by real time PCR quantification of the immunoprecipitated DNA (ChIP-qPCR). In order to evaluate nucleosome positions at high resolution across SRG1-SER3, we used 33 closely spaced PCR amplicons to measure immunoprecipitated DNA (Hainer et al. 2011). The averaged results of 3 independent biological experiments are shown (Figure 5B). As previously described, an abundance of H3 was detected throughout the SRG1 transcript unit extending into the SER3 gene in WT cells. These H3 levels were dramatically depleted in spt6-14 mutants grown at the semi-permissive temperature (30°C), confirming that their deposition was Spt6 dependent (Hainer et al. 2011) (Figure 5B). Consistent with our western blotting results, spt6-14 jhd2∆ mutants did not exhibit improved H3 deposition at SRG1-SER3 at the semi-permissive temperature we used (Figure 5B). In WT and jhd2∆ cells with normal SPT6 function, the H3 ChIP profiles were nearly superimposable throughout the entire SRG1-SER3 region with one conspicuous exception: jhd2∆ caused a highly significant increase in H3 abundance at a single distinctive peak near the 3′ end of the SRG1 transcript unit, presumably reflecting a well-positioned nucleosome (Figure 5B, peak denoted with a red asterisk, P < 0.05). This JHD2-repressed nucleosome was positioned ∼75 base pairs upstream of the SER3 TSS, and positioned ∼75 base pairs downstream of this TSS we observed an additional H3 ChIP peak in jhd2∆, though this second peak was not statistically significant (Figure 5B, denoted with a blue arrow). Spanning the SER3 TSS, H3 ChIP signal was noisy in both WT and jhd2∆, suggesting a dynamic state of nucleosomal occupancy at this region (Figure 5B).
We performed our ChIP experiments from cells grown in rich media, a condition in which SER3 transcription is repressed in a manner dependent on Spt6-Spn1-deposited nucleosomes across the SER3 TSS in response to SRG1 transcription (pruneski and martens 2011) (Figure 5B). A simple encapsulation of our findings posits that JHD2 prevented Spt6-Spn1-deposited nucleosomes from accumulating at positions on the flanks of the SER3 TSS, resulting in dynamic and noisy nucleosome occupancy across the SER3 TSS (Figure 5B). We detected no differences in SER3 abundance from WT and jhd2∆ cells grown in rich media, suggesting that this altered chromatin architecture did not impact SER3 repression (data not shown). To determine if it instead impacted SER3 induction, we subjected WT and jhd2∆ cells to serine starvation and compared their relative SER3 mRNA abundance using quantitative PCR amplification of reverse transcribed RNA (RT-qPCR). We found that SER3 was maximally expressed 1 hr after serine withdrawal, and that SER3 mRNA accumulated to ∼1.7 increased levels in jhd2∆ at this timepoint (Figure 5C). Following 2 hr of serine starvation, yeast cells adapt and repress SER3 induction, and jhd2∆ had no consequence for this repression (martens et al. 2005) (Figure 5C). Thus, the phased nucleosomes flanking the SER3 TSS we observed in jhd2∆ were associated with a hyper-induction of SER3.
Discussion
In this work, we identify novel genetic interactions between the H3K4 demethylase JHD2 and genes encoding subunits of the essential transcription regulatory complexes NNS, FACT, and Spt6-Spn1. Our genetic findings support the conclusion that reversal of H3K4me by Jhd2 opposes the functions of these complexes (Figure 3). In the case of NNS, our findings are in good agreement with a previous study suggesting that H3K4me3 promotes NNS function (terzi et al. 2011). A simple prediction from our findings is that set1∆, which abolishes all H3K4 methylation, should cause the opposite phenotype as, and exhibit epistasis to, jhd2∆. While this was indeed the case for Spt6-Spn1 and the SPT16 subunit of FACT, TS alleles of SEN1, NAB3, and POB3 were each suppressed by set1∆ to an extent equivalent to, or exceeding that by jhd2∆. More detailed genetic studies revealed that SET1 opposed the functions of SEN1, NAB3, and POB3 in a manner that was independent of H3K4me. We point out that the Swd2 subunit of COMPASS also functions within the NNS-related termination complex APT, which could potentially explain the different NNS interactions with SET1 deletion vs. K4 mutation shown in this study (Nedea et al. 2003; Soares and Buratowski 2012). Our genetic model posits that H3K4me positively influences NNS and FACT functions, and that Set1 exhibits an additional counterbalancing H3K4me-independent activity that negatively impacts these complexes (Figure 3). For Spt6-Spn1, our genetic data suggested a positive impact by H3K4me3 with no repressive H3K4-indpendent role for Set1 (Figure 3). Our molecular studies showed that Jhd2, and by inference H3K4me reversal, negatively regulated the accumulation of Spt6-Spn1 deposited nucleosomes flanking the SER3 TSS, resulting in hyper-induction of SER3 upon serine starvation.
In contrast to our findings and what has been reported previously (Hainer et al. 2011), Ramakrishnan et al. found that both jhd2∆ and set1∆ caused dramatic down-regulation of SER3 in rich serine-replete conditions (ramakrishnan et al. 2016). Under such conditions SER3 is normally strongly repressed, and we found that levels were so low as to be nearly undetectable with no difference between WT and jhd2∆ (Figure 5C) (martens et al. 2004; martens et al. 2005; Hainer et al. 2011). To reconcile these discrepancies, we point out that all of the experiments in the Ramakrishnan et al. study were performed in a strain background that utilized plasmid expressed histone H2A-H2B, with the endogenous H2A-H2B genes deleted, a condition known to reduce histone dosage and alter transcription (clark-adams et al. 1988). This is not to say we reject the findings of Ramakrishnan et al.; rather, the findings of Ramakrishnan et al. seem to illuminate a facile way to investigate chromatin biology in yeast, by ‘sensitizing’ the chromatin through reduced histone dosage. Whatever the case may be, our findings uphold an emerging role for Jhd2 in the control of nucleosome occupancy at TSS’s and TTS’s. JHD2’s known developmental role in opposing nucleosome accumulation at these regions may be attributable to it antagonistic impact on Spt6-Spn1 (xu et al. 2012). As Spt6-Spn1, Set1, Jhd2, and H3K4me are conserved throughout eukaryotes, these mechanisms may have broad significance.
An important feature of our findings relates to functions of Set1 that are independent H3K4 methylation. The only confirmed non-H3K4 enzymatic target of Set1, or indeed of any member of this highly conserved family of enzymes, is the essential kinetochore protein Dam1. Dam1 lysine residue methylation by Set1 regulates the phosphorylation status of neighboring serine residues by mediating the opposing activities of Ipl1 and Glc7 kinase and phosphatase (zhang et al. 2005). Schizosaccharomyces pombe Set1 (spSet1) controls transcription at silent mating loci, telomeric regions, and retrotransposon (Tf) elements, as well as the clustering of diverse genomic regions encoding Tf elements. Interestingly, spSet1 accomplishes these sundry functions in a manner dependent on differing domains of Set1 that do, or do not, have consequences for H3K4me. The two key regulatory roles of spSet1 appear dependent on spSet1’s enzymatic activity and on its RNA binding RRM2 domain, a domain found in both fission and budding yeast Set1 (Trésaugues et al. 2006; lorenz et al. 2014; mikheyeva et al. 2014). Consistent with the RRM2 having important regulatory significance independent of H3K4me, a recent study identified many mRNA targets bound by the budding yeast Set1 RRM2 domain (sayou et al. 2017). Thus, the RRM2 domain may underlie at least some of the H3K4me-independent regulatory functions of Set1 we identify here. Our genetic results, however, also suggest that enzymatic activity of Set1 directed toward a non-H3K4 substrate has regulatory significance, as may be the case for S. pombe Set1 (lorenz et al. 2014; mikheyeva et al. 2014). It seems highly plausible that chromatin-associated factors may be targets of regulatory lysine methylation by Set1 and other histone methyltransferases.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.118.200151/-/DC1.
Acknowledgments
This work was supported by CIHR grant MOP-89996 to M.D.M. We are particularly grateful to Dr. Charlie Boone and Dr. Brenda Andrews for access to their temperature-sensitive mutant library and other reagents. We thank Dr. Jeffry L. Corden for the nab3-42 mutant. We thank Dr. Scott D. Briggs for PYK1-JHD2, PYK1-JHD2(H427A), and PYK1-JHD2(PHDΔ) plasmids. The SET1-G951S and spt16-319 mutant strains were kindly provided by Dr. Sharon Dent and Dr. Richard A. Singer.
Footnotes
Communicating editor: N. Rhind
Literature Cited
- Arigo J. T., Eyler D. E., Carroll K. L., Corden J. L., 2006. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol. Cell 23(6): 841–851. 10.1016/j.molcel.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Belotserkovskaya R., Oh S., Bondarenko V. A., Orphanides G., Studitsky V. M., et al. , 2003. FACT facilitates transcription-dependent nucleosome alteration. Science 301(5636): 1090–1093. 10.1126/science.1085703 [DOI] [PubMed] [Google Scholar]
- Benevolenskaya E. V., Murray H. L., Branton P., Young R. A., Kaelin W. G., Jr, 2005. Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Mol. Cell 18(6): 623–635. 10.1016/j.molcel.2005.05.012 [DOI] [PubMed] [Google Scholar]
- Blair L. P., Liu Z., Labitigan R. L., Wu L., Zheng D., et al. , 2016. KDM5 lysine demethylases are involved in maintenance of 3′UTR length. Sci. Adv. 2(11): e1501662 10.1126/sciadv.1501662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortvin A., Winston F., 1996. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science 272(5267): 1473–1476. 10.1126/science.272.5267.1473 [DOI] [PubMed] [Google Scholar]
- Carroll K. L., Pradhan D. A., Granek J. A., Clarke N. D., Corden J. L., 2004. Identification of cis elements directing termination of yeast nonpolyadenylated snoRNA transcripts. Mol. Cell. Biol. 24(14): 6241–6252. 10.1128/MCB.24.14.6241-6252.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Muller U., Sundling K. E., Brow D. A., 2014. Saccharomyces cerevisiae Sen1 as a model for the study of mutations in human Senataxin that elicit cerebellar ataxia. Genetics 198(2): 577–590. 10.1534/genetics.114.167585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Adams C. D., Norris D., Osley M. A., Fassler J. S., Winston F., 1988. Changes in histone gene dosage alter transcription in yeast. Genes Dev. 2(2): 150–159. 10.1101/gad.2.2.150 [DOI] [PubMed] [Google Scholar]
- Copic A., Dorrington M., Pagant S., Barry J., Lee M. C., et al. , 2009. Genomewide analysis reveals novel pathways affecting endoplasmic reticulum homeostasis, protein modification and quality control. Genetics 182(3): 757–769. 10.1534/genetics.109.101105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M., VanderSluis B., Koch E. N., Baryshnikova A., Pons C., et al. , 2016. A global genetic interaction network maps a wiring diagram of cellular function. Science 353: aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creamer T. J., Darby M. M., Jamonnak N., Schaughency P., Hao H., et al. , 2011. Transcriptome-wide binding sites for components of the Saccharomyces cerevisiae non-poly(A) termination pathway: Nrd1, Nab3, and Sen1. PLoS Genet. 7(10): e1002329 10.1371/journal.pgen.1002329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J., Hyland E. M., Yuan D. S., Huang H., Bader J. S., et al. , 2008. Probing nucleosome function: a highly versatile library of synthetic histone H3 and H4 mutants. Cell 134(6): 1066–1078. 10.1016/j.cell.2008.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby M. M., Serebreni L., Pan X., Boeke J. D., Corden J. L., 2012. The Saccharomyces cerevisiae Nrd1-Nab3 transcription termination pathway acts in opposition to Ras signaling and mediates response to nutrient depletion. Mol. Cell. Biol. 32(10): 1762–1775. 10.1128/MCB.00050-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehé P. M., Dichtl B., Schaft D., Roguev A., Pamblanco M., et al. , 2006. Protein interactions within the Set1 complex and their roles in the regulation of histone 3 lysine 4 methylation. J. Biol. Chem. 281(46): 35404–35412. 10.1074/jbc.M603099200 [DOI] [PubMed] [Google Scholar]
- Dey B. K., Stalker L., Schnerch A., Bhatia M., Taylor-Papidimitriou J., et al. , 2008. The histone demethylase KDM5b/JARID1b plays a role in cell fate decisions by blocking terminal differentiation. Mol. Cell. Biol. 28(17): 5312–5327. 10.1128/MCB.00128-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duina A. A., 2011. Histone Chaperones Spt6 and FACT: Similarities and Differences in Modes of Action at Transcribed Genes. Genet. Res. Int. 2011: 625210 10.4061/2011/625210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa T., Eriksson P., Wittmeyer J., Ginn J., Yu Y., et al. , 2001. Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J. 20(13): 3506–3517. 10.1093/emboj/20.13.3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainer S. J., Pruneski J. A., Mitchell R. D., Monteverde R. M., Martens J. A., 2011. Intergenic transcription causes repression by directing nucleosome assembly. Genes Dev. 25(1): 29–40. 10.1101/gad.1975011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingvarsdottir K., Edwards C., Lee M. G., Lee J. S., Schultz D. C., et al. , 2007. Histone H3 K4 demethylation during activation and attenuation of GAL1 transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 27(22): 7856–7864. 10.1128/MCB.00801-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovska I., Jacques P. E., Rando O. J., Robert F., Winston F., 2011. Control of chromatin structure by spt6: different consequences in coding and regulatory regions. Mol. Cell. Biol. 31(3): 531–541. 10.1128/MCB.01068-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan C. D., Laprade L., Winston F., 2003. Transcription elongation factors repress transcription initiation from cryptic sites. Science 301(5636): 1096–1099. 10.1126/science.1087374 [DOI] [PubMed] [Google Scholar]
- Kim T., Buratowski S., 2009. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell 137(2): 259–272. 10.1016/j.cell.2009.02.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N. J., Dover J., Khorrami S., Greenblatt J. F., Schneider J., et al. , 2002. COMPASS, a histone H3 (Lysine 4) methyltransferase required for telomeric silencing of gene expression. J. Biol. Chem. 277(13): 10753–10755. 10.1074/jbc.C200023200 [DOI] [PubMed] [Google Scholar]
- Lenstra T. L., Benschop J. J., Kim T., Schulze J. M., Brabers N. A., et al. , 2011. The specificity and topology of chromatin interaction pathways in yeast. Mol. Cell 42(4): 536–549. 10.1016/j.molcel.2011.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Vizeacoumar F. J., Bahr S., Li J., Warringer J., et al. , 2011. Systematic exploration of essential yeast gene function with temperature-sensitive mutants. Nat. Biotechnol. 29(4): 361–367. 10.1038/nbt.1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., Klose R. J., Gardner K. E., Zhang Y., 2007. Yeast Jhd2p is a histone H3 Lys4 trimethyl demethylase. Nat. Struct. Mol. Biol. 14(3): 243–245 (erratum: Nat. Struct. Mol. Biol. 14: 351) 10.1038/nsmb1204 [DOI] [PubMed] [Google Scholar]
- Lopez-Bigas N., Kisiel T. A., Dewaal D. C., Holmes K. B., Volkert T. L., et al. , 2008. Genome-wide analysis of the H3K4 histone demethylase RBP2 reveals a transcriptional program controlling differentiation. Mol. Cell 31(4): 520–530. 10.1016/j.molcel.2008.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz D. R., Meyer L. F., Grady P. J., Meyer M. M., Cam H. P., 2014. Heterochromatin assembly and transcriptome repression by Set1 in coordination with a class II histone deacetylase. eLife 3: e04506 10.7554/eLife.04506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z., Atencio D., Barnes C., DeFiglio H., Hanes S. D., 2012. Multiple roles for the Ess1 prolyl isomerase in the RNA polymerase II transcription cycle. Mol. Cell. Biol. 32(17): 3594–3607. 10.1128/MCB.00672-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margaritis T., Oreal V., Brabers N., Maestroni L., Vitaliano-Prunier A., et al. , 2012. Two distinct repressive mechanisms for histone 3 lysine 4 methylation through promoting 3′-end antisense transcription. PLoS Genet. 8(9): e1002952 10.1371/journal.pgen.1002952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens J. A., Laprade L., Winston F., 2004. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429(6991): 571–574. 10.1038/nature02538 [DOI] [PubMed] [Google Scholar]
- Martens J. A., Wu P. Y., Winston F., 2005. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev. 19(22): 2695–2704. 10.1101/gad.1367605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough L., Connell Z., Petersen C., Formosa T., 2015. The Abundant Histone Chaperones Spt6 and FACT Collaborate to Assemble, Inspect, and Maintain Chromatin Structure in Saccharomyces cerevisiae. Genetics 201(3): 1031–1045. 10.1534/genetics.115.180794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald S. M., Close D., Xin H., Formosa T., Hill C. P., 2010. Structure and biological importance of the Spn1-Spt6 interaction, and its regulatory role in nucleosome binding. Mol. Cell 40(5): 725–735. 10.1016/j.molcel.2010.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mersman D. P., Du H. N., Fingerman I. M., South P. F., Briggs S. D., 2009. Polyubiquitination of the demethylase Jhd2 controls histone methylation and gene expression. Genes Dev. 23(8): 951–962. 10.1101/gad.1769209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikheyeva I. V., Grady P. J., Tamburini F. B., Lorenz D. R., Cam H. P., 2014. Multifaceted genome control by Set1 Dependent and Independent of H3K4 methylation and the Set1C/COMPASS complex. PLoS Genet. 10(10): e1004740 10.1371/journal.pgen.1004740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P. L., Griesenbeck J., Kornberg R. D., Cleary M. L., 2002. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc. Natl. Acad. Sci. USA 99(1): 90–94. 10.1073/pnas.221596698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedea E., He X., Kim M., Pootoolal J., Zhong G., et al. , 2003. Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′-ends. J. Biol. Chem. 278: 33000–33010. [DOI] [PubMed] [Google Scholar]
- O’Donnell A. F., Brewster N. K., Kurniawan J., Minard L. V., Johnston G. C., et al. , 2004. Domain organization of the yeast histone chaperone FACT: the conserved N-terminal domain of FACT subunit Spt16 mediates recovery from replication stress. Nucleic Acids Res. 32(19): 5894–5906. 10.1093/nar/gkh922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G., LeRoy G., Chang C. H., Luse D. S., Reinberg D., 1998. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell 92(1): 105–116. 10.1016/S0092-8674(00)80903-4 [DOI] [PubMed] [Google Scholar]
- Orphanides G., Wu W. H., Lane W. S., Hampsey M., Reinberg D., 1999. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature 400(6741): 284–288. 10.1038/22350 [DOI] [PubMed] [Google Scholar]
- Porrua O., Libri D., 2013. A bacterial-like mechanism for transcription termination by the Sen1p helicase in budding yeast. Nat. Struct. Mol. Biol. 20(7): 884–891. 10.1038/nsmb.2592 [DOI] [PubMed] [Google Scholar]
- Pruneski J. A., Martens J. A., 2011. Transcription of intergenic DNA deposits nucleosomes on promoter to silence gene expression. Cell Cycle 10(7): 1021–1022. 10.4161/cc.10.7.15167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan S., Pokhrel S., Palani S., Pflueger C., Parnell T. J., et al. , 2016. Counteracting H3K4 methylation modulators Set1 and Jhd2 co-regulate chromatin dynamics and gene transcription. Nat. Commun. 7: 11949 10.1038/ncomms11949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H., Schneider R., Bernstein B. E., Karabetsou N., Morillon A., et al. , 2003. Methylation of histone H3 K4 mediates association of the Isw1p ATPase with chromatin. Mol. Cell 12(5): 1325–1332. 10.1016/S1097-2765(03)00438-6 [DOI] [PubMed] [Google Scholar]
- Sayou C., Millan-Zambrano G., Santos-Rosa H., Petfalski E., Robson S., et al. , 2017. RNA binding by histone methyltransferases Set1 and Set2. Mol. Cell. Biol. 37: e00165–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler A., Koutelou E., Tomida J., Wilson-Pham M., Wang L., et al. , 2016. Histone H3K4 methylation regulates deactivation of the spindle assembly checkpoint through direct binding of Mad2. Genes Dev. 30: 1187–1197. 10.1101/gad.278887.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J., Wood A., Lee J. S., Schuster R., Dueker J., et al. , 2005. Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Mol. Cell 19(6): 849–856. 10.1016/j.molcel.2005.07.024 [DOI] [PubMed] [Google Scholar]
- Soares L. M., Buratowski S., 2012. Yeast Swd2 is essential because of antagonism between Set1 histone methyltransferase complex and APT (associated with Pta1) termination factor. J Biol Chem 287: 15219–15231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares L. M., Radman-Livaja M., Lin S. G., Rando O. J., Buratowski S., 2014. Feedback control of Set1 protein levels is important for proper H3K4 methylation patterns. Cell Reports 6(6): 961–972. 10.1016/j.celrep.2014.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soloveychik M., Xu M., Zaslaver O., Lee K., Narula A., et al. , 2016. Mitochondrial control through nutritionally regulated global histone H3 lysine-4 demethylation. Sci. Rep. 6(1): 37942 10.1038/srep37942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz E. J., Brow D. A., 1996. Repression of gene expression by an exogenous sequence element acting in concert with a heterogeneous nuclear ribonucleoprotein-like protein, Nrd1, and the putative helicase Sen1. Mol. Cell. Biol. 16(12): 6993–7003. 10.1128/MCB.16.12.6993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz E. J., Conrad N. K., Brow D. A., Corden J. L., 2001. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature 413(6853): 327–331. 10.1038/35095090 [DOI] [PubMed] [Google Scholar]
- Takahashi Y. H., Lee J. S., Swanson S. K., Saraf A., Florens L., et al. , 2009. Regulation of H3K4 trimethylation via Cps40 (Spp1) of COMPASS is monoubiquitination independent: implication for a Phe/Tyr switch by the catalytic domain of Set1. Mol. Cell. Biol. 29(13): 3478–3486. 10.1128/MCB.00013-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna S. D., Ilin S., Rogers R. S., Tanny J. C., Lavender H., et al. , 2006. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol. Cell 24(5): 785–796. 10.1016/j.molcel.2006.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzi N., Churchman L. S., Vasiljeva L., Weissman J., Buratowski S., 2011. H3K4 trimethylation by Set1 promotes efficient termination by the Nrd1-Nab3-Sen1 pathway. Mol. Cell. Biol. 31(17): 3569–3583. 10.1128/MCB.05590-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiebaut M., Kisseleva-Romanova E., Rougemaille M., Boulay J., Libri D., 2006. Transcription termination and nuclear degradation of cryptic unstable transcripts: a role for the nrd1-nab3 pathway in genome surveillance. Mol. Cell 23(6): 853–864. 10.1016/j.molcel.2006.07.029 [DOI] [PubMed] [Google Scholar]
- Tokai M., Kawasaki H., Kikuchi Y., Ouchi K., 2000. Cloning and characterization of the CSF1 gene of Saccharomyces cerevisiae, which is required for nutrient uptake at low temperature. J. Bacteriol. 182(10): 2865–2868. 10.1128/JB.182.10.2865-2868.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trésaugues L., Dehé P. M., Guérois R., Rodriguez-Gil A., Varlet I., et al. , 2006. Structural characterization of Set1 RNA recognition motifs and their role in histone H3 lysine 4 methylation. J. Mol. Biol. 359(5): 1170–1181. 10.1016/j.jmb.2006.04.050 [DOI] [PubMed] [Google Scholar]
- Ursic D., Himmel K. L., Gurley K. A., Webb F., Culbertson M. R., 1997. The yeast SEN1 gene is required for the processing of diverse RNA classes. Nucleic Acids Res. 25(23): 4778–4785. 10.1093/nar/25.23.4778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacova S., Stefl R., 2007. The exosome and RNA quality control in the nucleus. EMBO Rep. 8(7): 651–657. 10.1038/sj.embor.7401005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiljeva L., Buratowski S., 2006. Nrd1 interacts with the nuclear exosome for 3′ processing of RNA polymerase II transcripts. Mol. Cell 21(2): 239–248. 10.1016/j.molcel.2005.11.028 [DOI] [PubMed] [Google Scholar]
- Xu M., Soloveychik M., Ranger M., Schertzberg M., Shah Z., et al. , 2012. Timing of transcriptional quiescence during gametogenesis is controlled by global histone H3K4 demethylation. Dev. Cell 23(5): 1059–1071. 10.1016/j.devcel.2012.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Lin W., Latham J. A., Riefler G. M., Schumacher J. M., et al. , 2005. The Set1 methyltransferase opposes Ipl1 aurora kinase functions in chromosome segregation. Cell 122(5): 723–734. 10.1016/j.cell.2005.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Strains are available upon request.