TRPA1 channels transduce chemical, inflammatory, and neuropathic pain and are modulated by both electrophilic and nonelectrophilic compounds. Samanta et al. show that these modulators cause conformational rearrangements in the N-terminal ankyrin repeats, the pre-S1 helix, the TRP-like domain, and the linkers region of the channel.
Abstract
Pain, though serving the beneficial function of provoking a response to dangerous situations, is an unpleasant sensory and emotional experience. Transient receptor potential ankyrin 1 (TRPA1) is a member of the transient receptor potential (TRP) cation channel family and is localized in “nociceptors,” where it plays a key role in the transduction of chemical, inflammatory, and neuropathic pain. TRPA1 is a Ca2+-permeable, nonselective cation channel that is activated by a large variety of structurally unrelated electrophilic and nonelectrophilic chemical compounds. Electrophilic ligands are able to activate TRPA1 channels by interacting with critical cysteine residues on the N terminus of the channels via covalent modification and/or disulfide bonds. Activation by electrophilic compounds is dependent on their thiol-reactive moieties, accounting for the structural diversity of the group. On the other hand, nonelectrophilic ligands do not interact with critical cysteines on the channel, so the structural diversity of this group is unexplained. Although near-atomic-resolution structures of TRPA1 were resolved recently by cryo-electron microscopy, in the presence of both agonists and antagonists, detailed mechanisms of channel activation and inhibition by these modulators could not be determined. Here, we investigate the effect of both electrophilic and nonelectrophilic ligands on TRPA1 channel conformational rearrangements with limited proteolysis and mass spectrometry. Collectively, our results reveal that channel modulation results in conformational rearrangements in the N-terminal ankyrin repeats, the pre-S1 helix, the TRP-like domain, and the linker regions of the channel.
Introduction
Transient receptor potential ankyrin 1 (TRPA1), the lone member of the mammalian TRPA subfamily, is a significant transducer of chemical, neuropathic, and inflammatory pain signals (Story et al., 2003; Bandell et al., 2004; Bautista et al., 2005, 2006; Story and Gereau, 2006). It is expressed predominantly in small and medium-sized peptidergic primary afferent neurons of the sensory ganglia (Story et al., 2003; Kobayashi et al., 2005; Huang et al., 2012). TRPA1 is also expressed in various nonneuronal tissue types and organs, including epithelial cells, fibroblasts, and smooth muscle cells (Jaquemar et al., 1999; Streng et al., 2008). TRPA1 is a homotetrameric, nonselective cationic channel composed of a transmembrane domain (TMD) and a large cytosolic domain. Each monomer of the TRPA1 protein is made up of six transmembrane helices (S1–S6) and a reentrant pore loop in the TMD, which is preceded by a large N terminus and followed by a coiled coil–structured C terminus. Although TRPA1 has a structurally conserved TMD like other transient receptor potential (TRP) channels, it is unique among mammalian TRP channels in having a large number of ankyrin repeats (16 total) at its N terminus.
TRPA1 is best known as a chemonociceptor in the body (Macpherson et al., 2007b; Nilius et al., 2011, 2012). It can be activated by a multitude of structurally unrelated natural compounds like allyl isothiocyanate (in mustard oil), diallyl disulfide (in garlic), irritants like acrolein (in cigarette smoke), vehicle exhaust, metabolic byproducts of chemotherapeutic drugs, and endogenous inflammatory molecules (Bandell et al., 2004; Jordt et al., 2004; Bautista et al., 2005, 2006; Wang et al., 2008; Takahashi et al., 2011; Ogawa et al., 2012; Alpizar et al., 2013). This wide range of TRPA1 ligands can be broadly classified into two groups: electrophilic modulators and nonelectrophilic modulators. The electrophilic agonists activate TRPA1 via a cluster of cysteine residues present at the N terminus of the channel (Hinman et al., 2006; Macpherson et al., 2007a; Takahashi et al., 2008). Binding of these electrophilic agonists to the channel leads to disulfide bond formation between these critical cysteine residues, which triggers conformational changes at the N terminus and the opening of the TRPA1 channel (Hinman et al., 2006; Macpherson et al., 2007a; Takahashi et al., 2008; Wang et al., 2012). The activation mechanisms for nonelectrophilic ligands are still unknown. Recently, the structure of human TRPA1 was resolved at 4 to 4.5 Å resolution in the presence of either an electrophilic agonist (allyl isothiocyanate) or nonelectrophilic antagonists (HC-030031 and A-967079). Although the potential binding site for one antagonist, A-967079, has been visualized, no conformational changes could be resolved among these cryo-electron microscopy (cryo-EM) structures in activated and inhibited states (Paulsen et al., 2015). Therefore, the molecular mechanism of how TRPA1 modulators affect the conformation of the tertiary structure of the protein to either open or close its gates is still elusive.
The N terminus of TRPA1 contains 16 ankyrin repeats (AR1–AR16), which are 30- to 34-amino-acid-long helix-turn-helix motifs. The ankyrin repeats are arranged in tandem, forming an elongated ankyrin repeat domain (ARD), which is connected to the TMD via the pre-S1 region, and are usually involved in protein–protein and protein–ligand interactions (Li et al., 2006; Voronin and Kiseleva, 2007). Chimeric and mutagenesis studies have suggested that modulations in the ARD can be translated to the pore, leading to opening or closure of the channel (Cordero-Morales et al., 2011; Jabba et al., 2014). Chimeric studies have also shown that the ARD can be divided into two parts: a primary module composed of AR10–AR15 and an enhancer module composed of AR3–AR8 (Cordero-Morales et al., 2011). Mutagenesis studies have suggested that thiol-reactive activators of TRPA1 interact with the sulfhydryl groups of specific, conserved cysteine residues (C415, C422, C622, C642, C666, C174, C193, C634, and C859; numbering is for mouse TRPA1; Hinman et al., 2006; Macpherson et al., 2007a; Takahashi et al., 2011), and it has been shown that these critical cysteines undergo disulfide-bond formation or rearrangements, leading to N-terminal conformational changes, channel activation, and/or desensitization (Wang et al., 2012; Ibarra and Blair, 2013). Furthermore, the C terminus of TRPA1, which is linked to the transmembrane region by unstructured loops, β-sheets, and a TRP-like domain, has also been suggested to play a role in channel gating (Paulsen et al., 2015).
To determine the structural basis of TRPA1 channel modulation by ligands, we used a classic technique: limited proteolysis. Limited proteolysis is a well-established, simple biochemical technique used to probe information regarding protein structure and conformational changes (Hubbard and Klee, 1989; Fontana et al., 1997a,b; Hubbard, 1998). The theory underlying limited proteolysis is that a protein is incubated with a relatively low concentration of a protease of choice, which induces cuts at exposed recognition sites throughout the protein, mostly at loops and other flexible regions. Substrate binding, or activation and inhibition by other kinds of stimuli, induces conformational changes in the protein, changing the regions that are exposed and thus the pattern of protease cleavage. Subsequent analysis of the cleaved fragments by mass spectrometry (MS) results in an extremely robust technique for the evaluation of ligand-induced structural changes in various proteins (Schopper et al., 2017). These changes are sometimes so minor that they are difficult to determine by conventional biophysical methods, short of resolving a structure at atomic resolution. In these cases, biochemical assays like limited proteolysis can provide useful and accurate information about the change in the topology of the protein. Therefore, in this study, we used limited proteolysis coupled with in-solution MS to identify the regions of TRPA1 that undergo conformational changes during activation or inhibition by both electrophilic and nonelectrophilic ligands.
Materials and methods
Expression and purification of TRPA1
Mouse TRPA1 was expressed in the S. cerevisiae yeast strain BJ5457 and plasma membranes were isolated as described previously (Cvetkov et al., 2011). In brief, mouse TRPA1-expressing yeast cells were lysed using a microfluidizer (M-110Y; Microfluidics), and the lysate was cleared of cell debris by centrifuging at 3,000 g for 10 min; the supernatant was further centrifuged at 14,000 g for 35 min. Plasma membranes were obtained by using an ultracentrifuge to spin down the supernatant at 100,000 g for 45 min and storing the pelleted membranes at −80°C for future use. For protein purification, all the following steps were performed at 4°C. Membranes were solubilized in a buffer containing 20 mM HEPES, pH 8, 500 mM NaCl, 10% glycerol, 8 mM FC-12 (Anatrace), 0.4 mg/ml soybean polar lipids, 1 mM PMSF, and 10 mM Tris(2-carboxyethyl)phosphinehydrochloride supplemented with a protease inhibitor cocktail tablet mini (Roche) for 1 h. Detergent-insoluble material was removed by centrifuging at 100,000 g for 45 min. 1D4 antibody–coupled CnBr-activated Sepharose beads were added to the supernatant and incubated for 2 h. A column was packed with the above beads and washed with buffer containing 20 mM HEPES, pH 8.0, 150 mM NaCl, 10 mM Tris(2-carboxyethyl)phosphinehydrochloride, 10% glycerol, and 0.25% A8-35 (amphipol). TRPA1 was eluted with the same buffer supplemented with 10 mg/ml 1D4 peptide (GenScript USA). The protein was concentrated by spin concentrator (Millipore Amicon Ultra 50K Ultracel) and subjected to size-exclusion chromatography on a Superose 6 column (GE Healthcare) with a buffer containing 20 mM HEPES, pH 8.0, and 150 mM NaCl.
Limited proteolysis of TRPA1
Purified TRPA1 was concentrated to ∼1 mg/ml in a buffer suitable for limited proteolysis (20 mM HEPES, pH 8.0, and 150 mM NaCl). The protein was then pretreated with either 2 mM NMM (N-methylmaleimide), 333 µM A-967079 (Pfizer), 1 mM menthol, 333 µM PF-4840154 (Pfizer), or buffer for the apo state. Freshly prepared MS-grade trypsin was added at a protease/TRPA1 mol/mol ratio of 1:300 at room temperature for 15 min, and the reaction was quenched by adding 10 mM 4-(2-aminoethyl) benzenesulfonyl fluoride, and fivefold excess of soybean trypsin inhibitor II-S (Sigma) and placing on ice. All samples were precipitated with 10% trichloroacetic acid/acetone overnight at −20°C, washed three times with acetone, and air dried. The precipitation procedure was repeated twice. Air dried samples were reconstituted in 10 µl of 50 mM Tris, pH 8.0, and then reduced and alkylated with 10 mM dithiothreitol and 25 mM iodoacetamide, respectively. Subsequently, all samples were fully digested with Asp-N at an enzyme-to-protein ratio of 1:10 at 37°C overnight, followed by LC coupled with high-resolution MS.
LC-MS analysis
LC-MS analysis of digested samples was performed on the Orbitrap Elite mass spectrometer (Thermo Electron) interfaced with a Waters nanoAcquity UPLC system. Approximately 300 ng proteolytic peptide for each sample was loaded on a trap column (180 µm × 20 mm packed with C18 Symmetry, 5 µm, 100 Å; Waters) for desalting and then separated on a reverse-phase column (75 µm × 250 mm nano column, packed with C18 BEH130, 1.7 µm, 130 Å; Waters) using a gradient of 2–42% mobile phase B (0.1% formic acid in acetonitrile) versus mobile phase A (0.1% formic acid in water) over a period of 60 min with a flow rate of 300 nl/min. Peptides eluting from the column were introduced into the nanoelectrospray source using a capillary voltage of 2.5 kV. For MS analysis, a full scan was recorded for eluted peptides (m/z range of 380–1,700) in the Fourier transform mass analyzer with resolution of 120,000 followed by MS/MS of the 20 most intense peptide ions scanned in the ion trap mass analyzer. All MS data were acquired in the positive ion mode. The resulting MS and MS/MS data were searched against a TRPA1 protein sequence database using Mass Matrix software (Xu and Freitas, 2007) to identify peptide sequence and overall TRPA1 sequence coverage. In particular, MS and MS/MS spectra were searched for peptides derived from the dual digestion of TRPA1 with trypsin and Asp-N enzymes using mass accuracy values of 10 ppm and 0.8 D, respectively, with allowed variable modifications including carbamidomethylation for cysteines and oxidative modifications for methionines. The total number of MS/MS ion scans (spectral counting) for each identified peptide was used to compare peptide abundances (Liu et al., 2004).
Online supplemental information
Fig. S1 details the size exclusion chromatography of TRPA1 purification, showing symmetrical peak corresponding to the tetrameric TRPA1. This figure also reports Coomassie-stained SDS-PAGE gels showing the time course of limited proteolysis of purified TRPA1 with trypsin for different time periods. For different time periods, we see the generation of large stable peptide fragments. The other gel shows the resulting peptide fragments of limited proteolysis of TRPA1 without drug treatment and with drug application. Figs. S2 and S3 show representative MS/MS spectra of some of the cysteines that were modified by NMM treatment.
Results
Limited proteolysis and in-solution MS to determine ligand-induced conformational changes in TRPA1
To identify ligand-induced conformational changes of the full-length mouse TRPA1 ion channel, we used Saccharomyces cerevisiae as the heterologous expression system to express the full-length protein (Cvetkov et al., 2011; Moiseenkova-Bell and Wensel, 2011). Although inositol hexakisphosphate (IP6) was required for the purification of human TRPA1 in a previous cryo-EM study (Paulsen et al., 2015), the purification method used here has been proven to produce functional mouse TRPA1 and other TRP channels that can be used for structural, biochemical, and biophysical studies (Cvetkov et al., 2011; Moiseenkova-Bell and Wensel, 2011). The protein was purified using detergent and amphipol A8-35, and the peak corresponding to tetrameric TRPA1 (Fig. S1 A) was collected and used for all described studies.
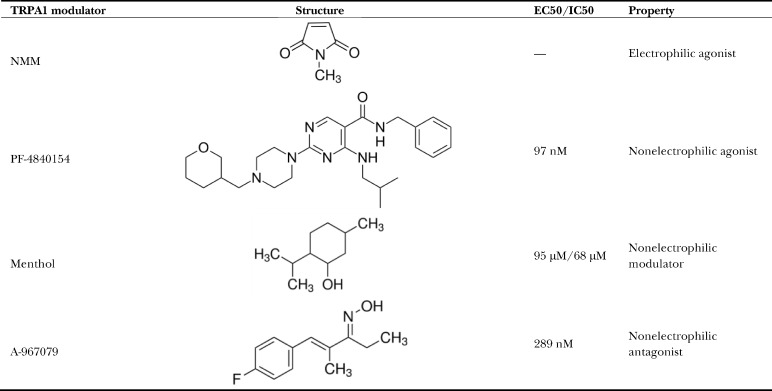
For limited proteolysis, the first digestion is usually performed using a protease with broad specificity. Complete digestion of a denatured protein with such an enzyme will thus yield relatively small peptides. However, when used for a short period of time at a low substrate-to-enzyme ratio, digestion of a protein in its native state will result in the formation of larger, stable peptides because of the inaccessibility of buried or protected sites by the protease. Here, we used the serine protease trypsin at a 1:300 trypsin/TRPA1 ratio and checked three time points over 30 min by SDS-PAGE (Fig. S1 C), which showed large, stable peptides appearing quickly and surviving for the entire time course. Based on this time course, we decided to use this ratio of trypsin and an intermediate digestion time of 15 min for all experiments. Using the above ratio of trypsin to protein and digesting for 15 min yielded a distinct cleavage pattern for apo-TRPA1 and drug-treated TRPA1 (Fig. S1 B). Therefore, these conditions were used for all experiments. Electrophilic and nonelectrophilic agonists and antagonists that have been used in this study are outlined in Table 1.
Table 1. TRPA1 ligands used in this paper.
 |
Instead of using bands cut out from SDS-PAGE gels for mass spectrometric analysis, we used in-solution MS to get better overall coverage. Upon full digestion with endoproteinase Asp-N and subsequent analysis with in-solution MS, the sequence coverage for all the experiments was ≥90%, thus enabling us to identify the regions of conformational changes with confidence. To achieve this coverage and reliable peptide quantification, we used the spectral counting approach, which relies on comparing the number of identified tandem mass spectrometry (MS/MS) spectra from the same protein in each of the multiple liquid chromatography (LC) MS/MS datasets to quantify the number of identified nonredundant peptides (Zhu et al., 2010). The increase in protein abundance correlates with the increase in the number of its proteolytic peptides, which subsequently results in an increase in the number of identified unique peptides and the number of identified total MS/MS spectral count for a specific protein (Washburn et al., 2001; Liu et al., 2004; Zhu et al., 2010). Thus, spectral count can be used as a simple but reliable index for relative peptide/protein quantification.
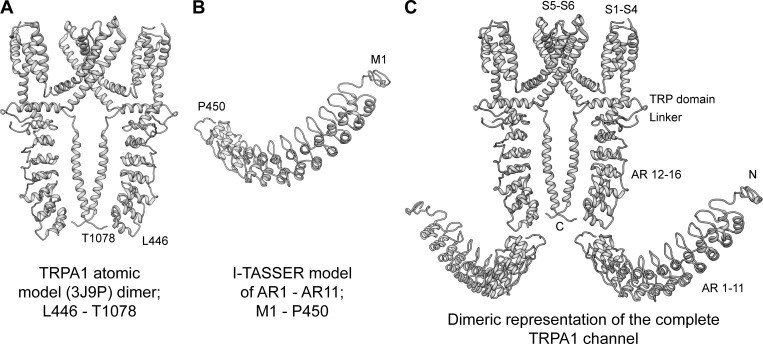
To illustrate TRPA1 conformational changes in the presence of various modulators, we created a TRPA1 channel dimer model representation (Fig. 1). For this, we downloaded the published atomic model (Protein Data Bank accession no. 3J9P; Fig. 1 A; Paulsen et al., 2015) and used ITASSER software (Roy et al., 2010; Yang et al., 2015) to build a homology model for AR1–AR11 (Fig. 1 B). Next, we overlaid these two parts of the structure in accordance to the proposed “propeller and independent wings” model by David Julius’s group (Paulsen et al., 2015) to get the complete structural representation (Fig. 1 C).
Figure 1.
Construction of a representation of the full-length TRPA1 channel. (A) Model 3J9P downloaded from the Protein Data Bank and depicted as a dimer. (B) Homology model of residues M1 to P450 generated by I-TASSER software. (C) The two models were aligned to generate a dimeric representation of the full channel.
Analysis of NMM (electrophilic agonist)–induced conformational change in TRPA1
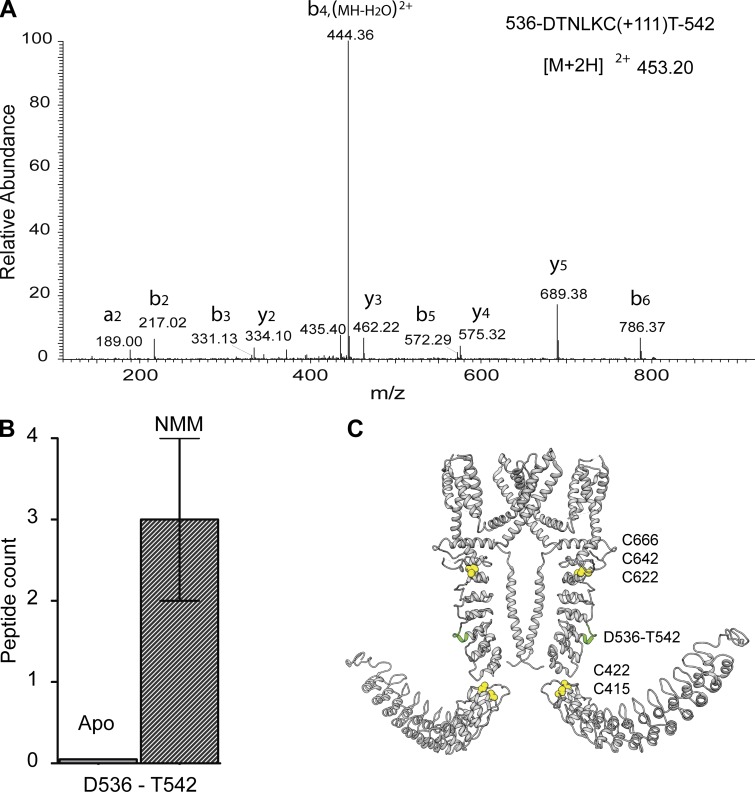
NMM is an electrophilic agonist that is commonly used to study TRPA1 ion channel activation (Hinman et al., 2006; Wang et al., 2012). It has been proposed that NMM and other electrophilic agonists covalently modify cysteine residues C415, C422, C622, C642, and C666 on the N terminus of the channel, which leads to channel opening (Hinman et al., 2006; Macpherson et al., 2007a). Purified full-length TRPA1 was incubated with 2 mM NMM for 10 min, and then the reaction was quenched with 10 mM dithiothreitol before performing limited proteolysis with trypsin. Partially digested apo-TRPA1 and NMM-activated TRPA1 were precipitated, reconstituted, alkylated, reduced, and fully digested with endoproteinase Asp-N before MS analysis. Apo- and NMM-treated TRPA1 showed different cleavage patterns in the MS analysis (Fig. 2); peptide DTNLKCT (536–542) was not present in apo-TRPA1 but appeared in the NMM-activated TRPA1 sample with a peptide count of two and four in two separate experiments (Fig. 2, A and B). Region D536 to T542 is located at the N terminus and is part of AR14 and the loop connecting AR14 and AR15 (Fig. 2 C). This region is positioned between two clusters of cysteines residues: (a) C415 and C422 and (b) C622, C642, and C666, which have been implicated in channel activation by NMM and other electrophilic agonists (Fig. 2 C). Also, the mass spectrometric analysis revealed that several these critical cysteines (e.g., C415, C622, C634, and C666) were modified by NMM, indicating that these cysteine residues are available for interaction with NMM in the full-length TRPA1 protein. A few of these modifications are shown in Figs. S2 and S3.
Figure 2.
Effect of NMM on TRPA1 conformation. (A) MS/MS spectra of doubly protonated ions for peptide 536-DTNLKCT-542 containing cysteine modified with NMM. (B) Graph of the occurrence of peptide 536-DTNLKCT-542 in apo- and NMM-treated TRPA1. Error bar represents mean ± SEM. (C) Peptide 536-DTNLKCT-542 (shown in green) mapped onto the dimeric representation of TRPA1; the cysteines necessary for channel activation by electrophilic ligands are shown as spheres in yellow.
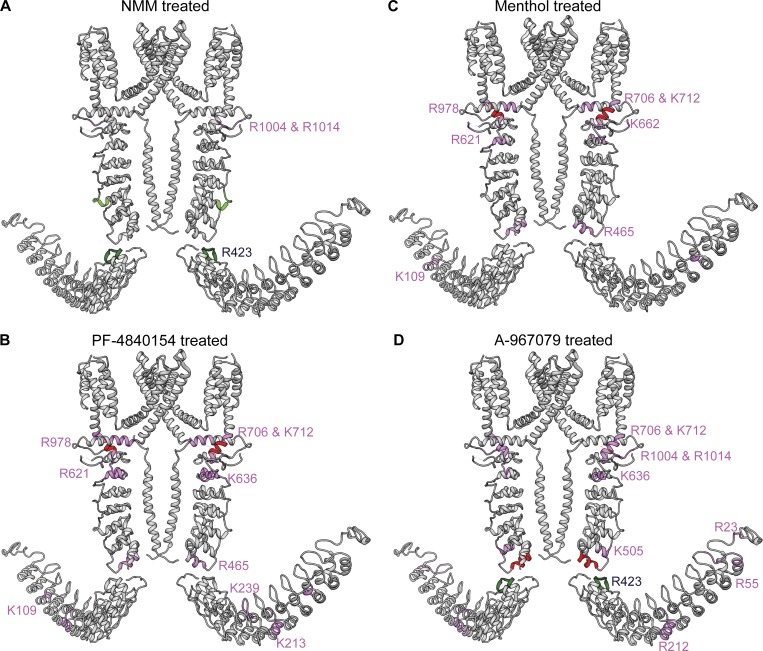
Additionally, there were also a couple of regions of missed trypsin cleavages in the NMM-TRPA1 sample at residues R1004 and R1014, located in the linker domain (Fig. 5 A). Interestingly, residue R423 was protected from being cleaved by trypsin in apo-TRPA1 but was exposed for cleavage in the presence of NMM (Fig. 5 A).
Figure 5.
Dimer representations of TRPA1 showing additional regions of miscleavage upon ligand interaction. Miscleaved regions are mapped onto the dimeric representation of TRPA1. (A) Residue R423 (highlighted in dark green) was protected, and residues R1004 and R1014 (highlighted in pink) were exposed for miscleavage upon treatment with NMM. (B and C) Residues that were exposed for miscleavage upon treatment with PF-4840154 (B) or menthol (C) are highlighted in pink. (D) Residue R423 (highlighted in dark green) was protected from miscleavage, and several other residues were exposed for miscleavage (highlighted in pink) upon treatment with A-967079.
Collectively, our data show that TRPA1 modification by NMM leads to N-terminal conformational changes in the ARD11 to ARD15 region of the protein. As NMM is an agonist and mediates channel opening, it seems likely that the conformational changes we observe in ARD11 to ARD15 are related to the opening of the channel.
Analysis of PF-4840154 (nonelectrophilic agonist)–induced conformational change in TRPA1
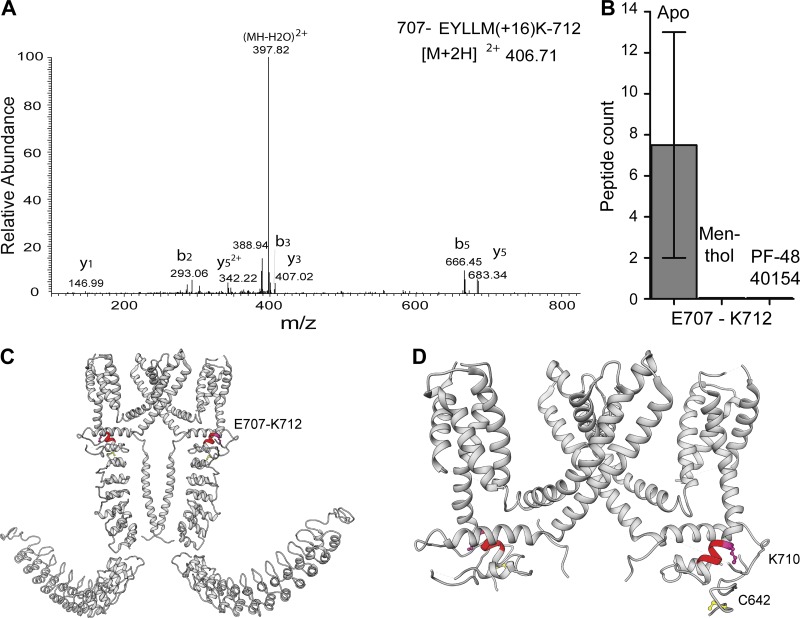
TRPA1 is a genuine polymodal channel, as it can be activated not only by structurally unrelated electrophilic ligands but also by a large number of nonelectrophilic ligands (Bandell et al., 2004; Bautista et al., 2006; Story and Gereau, 2006; Talavera et al., 2009; Zygmunt and Högestätt, 2014). Recently, a potent nonelectrophilic agonist (PF-4840154) was developed by Pfizer that has been shown to selectively activate TRPA1 and elicit a pain sensation (Ryckmans et al., 2011). Purified TRPA1 was incubated with 333 µM PF-4840154 for 10 min before performing limited proteolysis with trypsin. Partially digested apo-TRPA1 and PF-4840154–activated TRPA1 were precipitated, reconstituted, alkylated, reduced, and then fully digested with endoproteinase Asp-N and subjected to LC-MS analysis. Analysis of the peptide products by MS revealed that peptide EYLLMK (707–712), located in the pre-S1 helix, was detected in apo-TRPA1 with a peptide count of 2 and 13 in two separate experiments, but it was not present in the PF-4840154–treated sample (Fig. 3, A and B). Intriguingly, the E707–K712 peptide was also present in the NMM-activated TRPA1 samples (Fig. 3 B), suggesting that electrophilic agonist activation of the channel differs profoundly from activation by nonelectrophilic agonists such as PF-4840154.
Figure 3.
Effect of nonelectrophilic modulators on TRPA1 conformation. (A) MS/MS spectra of doubly protonated ions for peptide 707-EYLLMK-712 with oxidized methionine for PF-4840154–treated TRPA1. (B) Graph of the occurrence of peptide 707-EYLLMK-712 in apo-, menthol-, and PF-4840154–treated TRPA1. Error bar represents mean ± SEM. (C) Peptide 707-EYLLMK-712 (shown in red) mapped onto the dimeric representation of TRPA1. (D) Magnified view of the linker region with cysteine and lysine residue highlighted.
The cryo-EM structure revealed that the pre-S1 region of the channel resides in close proximity to the TRP-like domain at the C terminus of the channel and harbors critical cysteine and lysine residues that are involved in channel activation (Fig. 3 D). The E707–K712 peptide belongs to the pre-S1 helix (Fig. 3, C and D), which has been implicated in allosteric modulation of the channel by electrophilic ligands through its interaction with the TRP-like domain and transmission of the signal to the pore of the channel (Paulsen et al., 2015). Additionally, it has been suggested that the TRP-like domain interacts with the pre-S1 helix and the S4–S5 linker via hydrophobic interactions that would be affected upon ligand binding (Paulsen et al., 2015). Recently, the S4–S5 linker has been shown to play a critical role in activation or inhibition of another “pain” sensor, transient receptor potential vanilloid 1 (TRPV1; Gao et al., 2016). It has been suggested that TRPV1 modulators (activator capsaicin and inhibitor capsazepine) interact with the S4–S5 linker and induce conformational changes in this region that open or close the channel.
Based on these recent discoveries, we speculate that the absence of E707–K712 peptide in the PF-4840154–treated sample could be caused by disruption of the hydrophobic interactions between the TRP-like domain and the pre-S1 helix upon PF-4840154 binding to the S4–S5 linker in TRPA1 (Fig. 3 C), thus triggering channel activation.
Analysis of menthol (nonelectrophilic modulator)–induced conformational change in TRPA1
Menthol is a widely used naturally occurring nonelectrophilic modulator of TRPA1 that has been shown to have a bimodal effect on rodent TRPA1 channels (Karashima et al., 2007). Specifically, it has been shown that 1 mM menthol reversibly blocks the channel, whereas a submicromolar to low-micromolar range of menthol concentrations activates the channel. Purified TRPA1 was incubated with 2 mM menthol for 10 min before performing limited proteolysis with trypsin. Partially digested apo-TRPA1 and menthol-blocked TRPA1 were precipitated, reconstituted, alkylated, reduced, and then fully digested with endoproteinase Asp-N and subjected to MS analysis. With 2 mM menthol, we expected to capture the channel in an inhibited state, but surprisingly, we found a cleavage pattern very similar to PF-4840154–activated TRPA1, with peptide E707–K712 absent in menthol-treated TRPA1 (Fig. 3, A and B). Analysis of missed cleavages revealed that although a majority of the same residues (K109, R465, R621, R706, K712, and R978) were shielded from cleavage by trypsin in both menthol-treated and PF-4840154–activated TRPA1, there were a few exceptions. Residue K662, which missed cleavage in the menthol-treated sample, was not affected by treatment with PF-4840154, whereas residues K213, K239, and K636 were not cleaved in PF-4840154–bound TRPA1 but were trypsinized in both apo- and menthol-bound TRPA1 (Fig. 5 C). The majority of these differences were observed in the N-terminal linker domain and in the flexible N-terminal ARs.
Given these similarities to the PF-4840154–activated state and that menthol is an agonist at low concentrations, we suggest that with 2 mM menthol, the channel was first activated and then desensitized and that we captured the desensitized state of TRPA1. However, it is entirely possible that we captured TRPA1 in an inhibited state and that there is a very similar global mechanism of TRPA1 activation, desensitization, and inhibition by nonelectrophilic ligands that could possibly be conserved among other TRP channels.
Analysis of A-967079 (nonelectrophilic antagonist)–induced conformational change in TRPA1
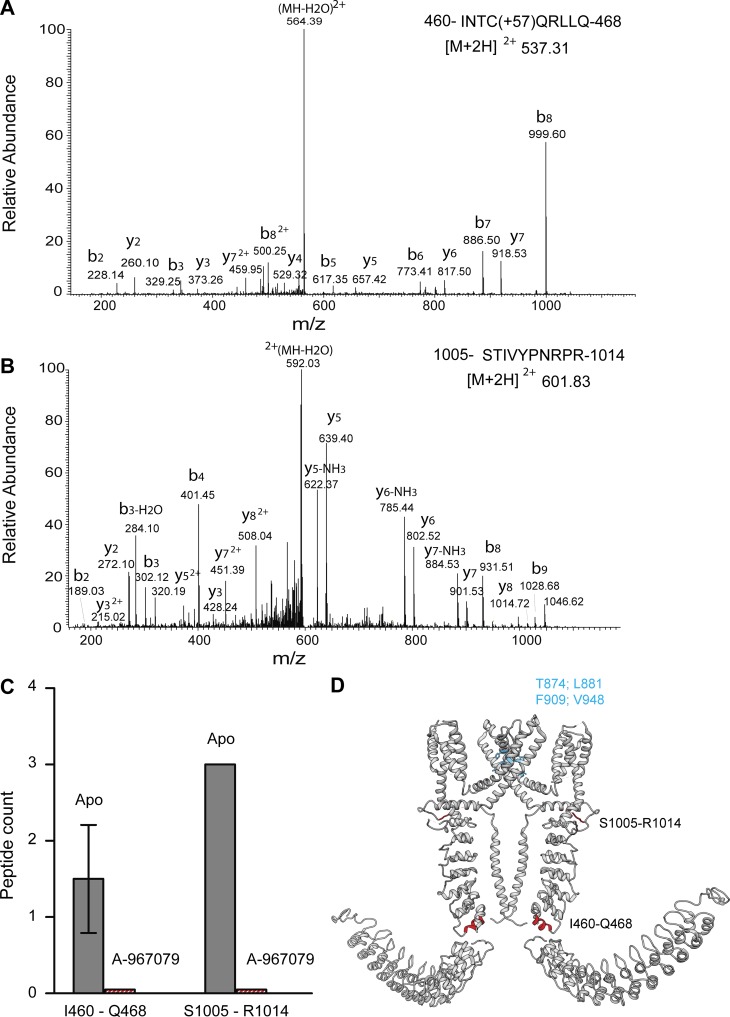
To determine whether a nonelectrophilic antagonist causes conformational changes in similar regions of full-length TRPA1, we used A-967079, a commonly used inhibitor of TRPA1 activity (McGaraughty et al., 2010; Chen et al., 2011; Banzawa et al., 2014). Purified TRPA1 was pretreated with 333 µM A-967079 for 10 min before performing limited proteolysis with trypsin. Partially digested apo-TRPA1 and A-967079–inactivated TRPA1 were precipitated, reconstituted, alkylated, reduced, and then fully digested with endoproteinase Asp-N and subjected to MS analysis. In this case, we detected two different peptides that were present in the apo-TRPA1 state but absent in the A-967079–treated sample: INTCQRLLQ (460–468), with a peptide count of one and two, and STIVYPNRPR (1,005–1,014), with a peptide count of three and three in two separate experiments (Fig. 4, A–C). Structurally, I460–Q468 forms one of the helices of the helix-turn-helix of AR12, and S1005–R1014 is a part of the flexible linker that connects the C terminus to the TMD (Fig. 4 D). The published cryo-EM structure of human TRPA1 revealed the binding pocket of A-967079 to be surrounded by transmembrane helices S5 and S6 and pore helix 1. Because this is quite far from the regions of missed peptide cleavages, this suggests that channel inhibition by A-967079 occurs through a global conformational change. Additionally, as AR1 to AR11 were too flexible to be resolved in the cryo-EM TRPA1 structure, AR12 is the first of them visible in that structure and so is at the transition point between the stable and dynamic portions of TRPA1. As one of the missed cleavage sites is found in AR11 (Fig. 5 D), this suggests the intriguing possibility that A-967079 inactivation of TRPA1 may involve modulating the interaction between the relatively stationary main channel and the flexible N-terminal AR region.
Figure 4.
Effect of A-967079 on TRPA1 conformation. (A and B) MS/MS spectra of doubly protonated ions for A, peptide 460-INTCQRLLQ-468 with carbamidomethylated cysteine (A) and peptide 1005-STIVYPNRPR-1014 (B). (C) Graph of the occurrence of peptide 460-INTCQRLLQ-468 and peptide 1005-STIVYPNRPR-1014 in apo- and A-967079–treated TRPA1. Error bar represents mean ± SEM. (D) The two peptides (shown in red) are mapped onto the dimeric representation of TRPA1; the binding pocket for A-967079 is shown in blue.
Discussion
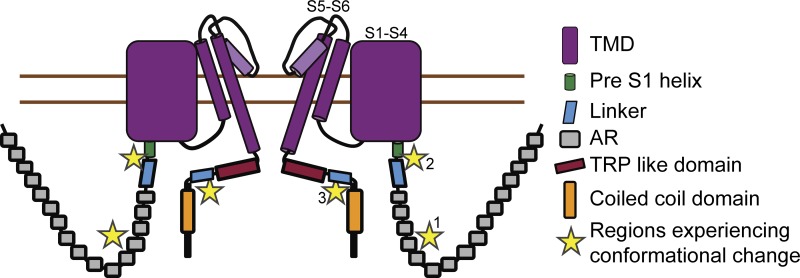
The mechanism for how a multitude of structurally diverse and unrelated compounds can modulate the polymodal TRPA1 has remained elusive. In this study, we reveal the critical regions of TRPA1 that undergo conformational rearrangement upon ligand interaction to accomplish the opening and closing of the ion conducting pore. We examined the topological changes of TRPA1 upon the addition of a diverse group of modulators encompassing both electrophilic and nonelectrophilic ligands by limited proteolysis and MS. It has been demonstrated before that certain cysteine and lysine residues in the cytosolic domain of the protein are critical for activation of the channel by electrophilic compounds (Hinman et al., 2006; Macpherson et al., 2007a); however, how modification of these residues gets translated to the pore is still unclear. The molecular mechanism by which the nonelectrophilic ligands control the gating mechanism is also unknown. In this study, we illustrated that across all of the modulators tested, TRPA1 saw topological rearrangements stretching from the primary module of the ARD (AR10–AR15; Cordero-Morales et al., 2011) through the linker region composing the loops, β-sheets, pre-S1 helix, and TRP-like domain, suggesting that these are the critical regions involved in the opening and closing of the pore.
The primary module of the ARD (Fig. 6, star 1) undergoes changes for all tested modulators. Of particular interest are the changes seen on treatment with A-967079 and NMM. On A-967079 treatment, peptide I460–Q468, which forms the second helix of AR12, becomes protected from cleavage. This is far away from the binding site of A-967079 and the pore, indicating that binding of this ligand induces allosteric conformational changes in the protein. From recent cryo-EM data of the TRP channel no mechanoreceptor potential C (Jin et al., 2017) and the different subpopulations resolved, it was proposed that the force experienced by the channel in the N terminus could be transduced to the linker region via the spring-like ARs. Our limited proteolysis data corroborate these structural observations. In the NMM-treated TRPA1 sample, peptide D536–T542, found in AR14 and the AR14-AR15 linker, becomes accessible for cleavage, indicating that NMM induces conformational rearrangement in this region. It is interesting to note that this region lies in between the two clusters of cysteine residues that were implicated by two separate groups (Hinman et al., 2006; Macpherson et al., 2007a) to be necessary for channel activation by electrophilic ligands. Moreover, mass spectrometric analysis of the NMM-treated TRPA1 sample showed that a lot of these critical cysteines (Cys-193, Cys-415, Cys-463, Cys-622, Cys-634, and Cys-666), along with some other cysteines (Cys-31, Cys-45, Cys-66, Cys-89, Cys-105, Cys-214, Cys-259, Cys-274, Cys-541, Cys-609, and Cys-1087), were modified by NMM, indicating that NMM binding to TRPA1 could lead to disruption of some existing disulfide bonds and formation of new disulfide bonds (Wang et al., 2012), making the 536–542 region accessible to trypsin cleavage.
Figure 6.
Cartoon representation of TRPA1 showing the regions involved in channel gating. The regions critical for channel gating are highlighted with numbered stars.
For all of the modulators, the data show that the linker regions between the TMD and the two cytosolic domains play a major role in channel gating and experience conformational changes. On the N-terminal side (Fig. 6, star 2), the pre-S1 helix and a linker domain made of two helix-turn-helix motifs connected by two antiparallel β sheets harbor some of the cysteine and lysine residues (C622, C642, C666, and K710; Fig. 3 D) that are required for activation by electrophilic ligands (Hinman et al., 2006; Macpherson et al., 2007a). The nonelectrophilic modulators also modify this region, as menthol, PF-4840154, and A-967079 treatment all cause a conformational change in the pre-S1 helix, as peptide E707–K712, comprising the majority of the pre-S1 helix, is protected from the proteolytic effect of trypsin in all three cases. On the C-terminal side, the linker regions and TRP-like domain form the connection to the TMD. Peptide S1005–R1014, which is found in the linker connecting the C-terminal coiled coil to the TRP-like domain, is protected from cleavage in the presence of both NMM and A-967079 despite being highly flexible. This is intriguing, because the binding site of A-967079 is very close to the pore in the TMD, but it somehow causes structural rearrangement in the linker region (Fig. 6, star 3). These behaviors of TRPA1 can potentially be explained by the structural proximity of the linkers connecting the N terminus and the C terminus to the TMD, as seen in the cryo-EM structure. The TRP-like domain, which lies parallel and close to the inner leaflet of the membrane, can interact with the pre-S1 helix and the S4–S5 linker. Furthermore, in the case of TRPV1, it has been proposed that the area encompassing S4, the S4–S5 linker, S6, and the TRP domain is critical for opening or closing the channel. Both agonist (RTX/DkTx) and antagonist (capsazepine) bind to the same pocket but trigger different structural rearrangements, one leading to channel opening and the other to its closing (Gao et al., 2016). It is possible that a very similar mechanism also exists for TRPA1 modulation by different ligands, and our data validate this by revealing that across different classes of ligand, the entire linker region is vital in governing the ion-conducting path. It is likely that this mechanism is conserved throughout the TRP channel family.
Although our study highlights the conformational changes specific to different gating states of the mouse TRPA1 ion channel, it has some limitations because of sample preparation. The mouse TRPA1 channel has been shown to require Ca2+ ions and polyphosphates for its activation (Cavanaugh et al., 2008), but in our study, we used purified full-length mouse TRPA1 that had been reconstituted in amphipol A8-35 without the addition of IP6, and only trace amounts of Ca2+ were present in our buffers. The human TRPA1 channel purification conditions in the cryo-EM study were very similar to ours, with the exception of the presence of IP6 in the buffers. Based on our previous work (Cvetkov et al., 2011; Wang et al., 2012), the presence of IP6 is not necessary for channel purification, but it would be important in our future experiments to test how IP6, together with Ca2+, influences the conformational changes in the channel in the presence of different modulators. Like other TRP channel structural biology groups, we also used amphipol A8-35 to stabilize purified protein for our analysis; however, we could not resolve conformational differences in the transmembrane region of the channel using in-solution MS. This is likely because of the presence of A8-35 in the mouse TRPA1 transmembrane region, shielding it from proteolysis.
It is also very important to note that the methods used in this current study do not fully allow us to interpret how local conformational changes in the mouse TRPA1 result in the overall conformational changes of the channel. Although further cryo-EM and MS studies are required to visualize the molecular mechanism of TRPA1 gating, our data suggest that, for all ligand classes, the primary module of the ARD, the pre-S1 helix, the TRP-like domain, and the linker regions of the channel are vital for conformation changes in the channel that lead to channel opening.
Supplementary Material
Acknowledgments
We thank Denice Major for assistance with hybridoma and cell culture at Department of Ophthalmology and Visual Sciences.
The Department of Ophthalmology and Visual Sciences was supported by the National Institutes of Health (Core Grant P30EY11373). This work was supported by grants from the National Institutes of Health (R01GM103899 to V.Y. Moiseenkova-Bell).
The authors declare no competing financial interests.
Author contributions: A. Samanta conducted all biochemical studies, including protein purification, sample preparation, data analysis, and interpretation. J. Kiselar performed MS data collection and analysis. S. Han assisted with data analysis. V.Y. Moiseenkova-Bell designed and supervised the execution of all experiments in this article. A. Samanta and V.Y. Moiseenkova-Bell wrote the manuscript. A. Samanta, R.A. Pumroy, and V.Y. Moiseenkova-Bell edited the manuscript. All authors reviewed the final manuscript.
Sharona E. Gordon served as editor.
References
- Alpizar Y.A., Gees M., Sanchez A., Apetrei A., Voets T., Nilius B., and Talavera K.. 2013. Bimodal effects of cinnamaldehyde and camphor on mouse TRPA1. Pflugers Arch. 465:853–864. 10.1007/s00424-012-1204-x [DOI] [PubMed] [Google Scholar]
- Bandell M., Story G.M., Hwang S.W., Viswanath V., Eid S.R., Petrus M.J., Earley T.J., and Patapoutian A.. 2004. Noxious cold ion channel TRPA1 is activated by pungent compounds and bradykinin. Neuron. 41:849–857. 10.1016/S0896-6273(04)00150-3 [DOI] [PubMed] [Google Scholar]
- Banzawa N., Saito S., Imagawa T., Kashio M., Takahashi K., Tominaga M., and Ohta T.. 2014. Molecular basis determining inhibition/activation of nociceptive receptor TRPA1 protein: a single amino acid dictates species-specific actions of the most potent mammalian TRPA1 antagonist. J. Biol. Chem. 289:31927–31939. 10.1074/jbc.M114.586891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bautista D.M., Movahed P., Hinman A., Axelsson H.E., Sterner O., Högestätt E.D., Julius D., Jordt S.E., and Zygmunt P.M.. 2005. Pungent products from garlic activate the sensory ion channel TRPA1. Proc. Natl. Acad. Sci. USA. 102:12248–12252. 10.1073/pnas.0505356102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bautista D.M., Jordt S.E., Nikai T., Tsuruda P.R., Read A.J., Poblete J., Yamoah E.N., Basbaum A.I., and Julius D.. 2006. TRPA1 mediates the inflammatory actions of environmental irritants and proalgesic agents. Cell. 124:1269–1282. 10.1016/j.cell.2006.02.023 [DOI] [PubMed] [Google Scholar]
- Cavanaugh E.J., Simkin D., and Kim D.. 2008. Activation of transient receptor potential A1 channels by mustard oil, tetrahydrocannabinol and Ca2+ reveals different functional channel states. Neuroscience. 154:1467–1476. 10.1016/j.neuroscience.2008.04.048 [DOI] [PubMed] [Google Scholar]
- Chen J., Joshi S.K., DiDomenico S., Perner R.J., Mikusa J.P., Gauvin D.M., Segreti J.A., Han P., Zhang X.F., Niforatos W., et al. 2011. Selective blockade of TRPA1 channel attenuates pathological pain without altering noxious cold sensation or body temperature regulation. Pain. 152:1165–1172. 10.1016/j.pain.2011.01.049 [DOI] [PubMed] [Google Scholar]
- Cordero-Morales J.F., Gracheva E.O., and Julius D.. 2011. Cytoplasmic ankyrin repeats of transient receptor potential A1 (TRPA1) dictate sensitivity to thermal and chemical stimuli. Proc. Natl. Acad. Sci. USA. 108:E1184–E1191. 10.1073/pnas.1114124108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvetkov T.L., Huynh K.W., Cohen M.R., and Moiseenkova-Bell V.Y.. 2011. Molecular architecture and subunit organization of TRPA1 ion channel revealed by electron microscopy. J. Biol. Chem. 286:38168–38176. 10.1074/jbc.M111.288993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana A., Polverino de Laureto P., De Filippis V., Scaramella E., and Zambonin M.. 1997a Probing the partly folded states of proteins by limited proteolysis. Fold. Des. 2:R17–R26. 10.1016/S1359-0278(97)00010-2 [DOI] [PubMed] [Google Scholar]
- Fontana A., Zambonin M., Polverino de Laureto P., De Filippis V., Clementi A., and Scaramella E.. 1997b Probing the conformational state of apomyoglobin by limited proteolysis. J. Mol. Biol. 266:223–230. 10.1006/jmbi.1996.0787 [DOI] [PubMed] [Google Scholar]
- Gao Y., Cao E., Julius D., and Cheng Y.. 2016. TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature. 534:347–351. 10.1038/nature17964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinman A., Chuang H.H., Bautista D.M., and Julius D.. 2006. TRP channel activation by reversible covalent modification. Proc. Natl. Acad. Sci. USA. 103:19564–19568. 10.1073/pnas.0609598103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D., Li S., Dhaka A., Story G.M., and Cao Y.Q.. 2012. Expression of the transient receptor potential channels TRPV1, TRPA1 and TRPM8 in mouse trigeminal primary afferent neurons innervating the dura. Mol. Pain. 8:66 10.1186/1744-8069-8-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard S.J. 1998. The structural aspects of limited proteolysis of native proteins. Biochim. Biophys. Acta. 1382:191–206. 10.1016/S0167-4838(97)00175-1 [DOI] [PubMed] [Google Scholar]
- Hubbard M.J., and Klee C.B.. 1989. Functional domain structure of calcineurin A: mapping by limited proteolysis. Biochemistry. 28:1868–1874. 10.1021/bi00430a066 [DOI] [PubMed] [Google Scholar]
- Ibarra Y., and Blair N.T.. 2013. Benzoquinone reveals a cysteine-dependent desensitization mechanism of TRPA1. Mol. Pharmacol. 83:1120–1132. 10.1124/mol.112.084194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabba S., Goyal R., Sosa-Pagán J.O., Moldenhauer H., Wu J., Kalmeta B., Bandell M., Latorre R., Patapoutian A., and Grandl J.. 2014. Directionality of temperature activation in mouse TRPA1 ion channel can be inverted by single-point mutations in ankyrin repeat six. Neuron. 82:1017–1031. 10.1016/j.neuron.2014.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaquemar D., Schenker T., and Trueb B.. 1999. An ankyrin-like protein with transmembrane domains is specifically lost after oncogenic transformation of human fibroblasts. J. Biol. Chem. 274:7325–7333. 10.1074/jbc.274.11.7325 [DOI] [PubMed] [Google Scholar]
- Jin P., Bulkley D., Guo Y., Zhang W., Guo Z., Huynh W., Wu S., Meltzer S., Cheng T., Jan L.Y., et al. 2017. Electron cryo-microscopy structure of the mechanotransduction channel NOMPC. Nature. 547:118–122. 10.1038/nature22981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordt S.E., Bautista D.M., Chuang H.H., McKemy D.D., Zygmunt P.M., Högestätt E.D., Meng I.D., and Julius D.. 2004. Mustard oils and cannabinoids excite sensory nerve fibres through the TRP channel ANKTM1. Nature. 427:260–265. 10.1038/nature02282 [DOI] [PubMed] [Google Scholar]
- Karashima Y., Damann N., Prenen J., Talavera K., Segal A., Voets T., and Nilius B.. 2007. Bimodal action of menthol on the transient receptor potential channel TRPA1. J. Neurosci. 27:9874–9884. 10.1523/JNEUROSCI.2221-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K., Fukuoka T., Obata K., Yamanaka H., Dai Y., Tokunaga A., and Noguchi K.. 2005. Distinct expression of TRPM8, TRPA1, and TRPV1 mRNAs in rat primary afferent neurons with adelta/c-fibers and colocalization with trk receptors. J. Comp. Neurol. 493:596–606. 10.1002/cne.20794 [DOI] [PubMed] [Google Scholar]
- Li J., Mahajan A., and Tsai M.D.. 2006. Ankyrin repeat: a unique motif mediating protein-protein interactions. Biochemistry. 45:15168–15178. 10.1021/bi062188q [DOI] [PubMed] [Google Scholar]
- Liu H., Sadygov R.G., and Yates J.R. III. 2004. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76:4193–4201. 10.1021/ac0498563 [DOI] [PubMed] [Google Scholar]
- Macpherson L.J., Dubin A.E., Evans M.J., Marr F., Schultz P.G., Cravatt B.F., and Patapoutian A.. 2007a Noxious compounds activate TRPA1 ion channels through covalent modification of cysteines. Nature. 445:541–545. 10.1038/nature05544 [DOI] [PubMed] [Google Scholar]
- Macpherson L.J., Xiao B., Kwan K.Y., Petrus M.J., Dubin A.E., Hwang S., Cravatt B., Corey D.P., and Patapoutian A.. 2007b An ion channel essential for sensing chemical damage. J. Neurosci. 27:11412–11415. 10.1523/JNEUROSCI.3600-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaraughty S., Chu K.L., Perner R.J., Didomenico S., Kort M.E., and Kym P.R.. 2010. TRPA1 modulation of spontaneous and mechanically evoked firing of spinal neurons in uninjured, osteoarthritic, and inflamed rats. Mol. Pain. 6:14 10.1186/1744-8069-6-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moiseenkova-Bell V., and Wensel T.G.. 2011. Functional and structural studies of TRP channels heterologously expressed in budding yeast. Adv. Exp. Med. Biol. 704:25–40. 10.1007/978-94-007-0265-3_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilius B., Prenen J., and Owsianik G.. 2011. Irritating channels: the case of TRPA1. J. Physiol. 589:1543–1549. 10.1113/jphysiol.2010.200717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilius B., Appendino G., and Owsianik G.. 2012. The transient receptor potential channel TRPA1: from gene to pathophysiology. Pflugers Arch. 464:425–458. 10.1007/s00424-012-1158-z [DOI] [PubMed] [Google Scholar]
- Ogawa H., Takahashi K., Miura S., Imagawa T., Saito S., Tominaga M., and Ohta T.. 2012. H(2)S functions as a nociceptive messenger through transient receptor potential ankyrin 1 (TRPA1) activation. Neuroscience. 218:335–343. 10.1016/j.neuroscience.2012.05.044 [DOI] [PubMed] [Google Scholar]
- Paulsen C.E., Armache J.P., Gao Y., Cheng Y., and Julius D.. 2015. Structure of the TRPA1 ion channel suggests regulatory mechanisms. Nature. 525:552 10.1038/nature14871 [DOI] [PubMed] [Google Scholar]
- Roy A., Kucukural A., and Zhang Y.. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5:725–738. 10.1038/nprot.2010.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryckmans T., Aubdool A.A., Bodkin J.V., Cox P., Brain S.D., Dupont T., Fairman E., Hashizume Y., Ishii N., Kato T., et al. 2011. Design and pharmacological evaluation of PF-4840154, a non-electrophilic reference agonist of the TrpA1 channel. Bioorg. Med. Chem. Lett. 21:4857–4859. 10.1016/j.bmcl.2011.06.035 [DOI] [PubMed] [Google Scholar]
- Schopper S., Kahraman A., Leuenberger P., Feng Y., Piazza I., Müller O., Boersema P.J., and Picotti P.. 2017. Measuring protein structural changes on a proteome-wide scale using limited proteolysis-coupled mass spectrometry. Nat. Protoc. 12:2391–2410. 10.1038/nprot.2017.100 [DOI] [PubMed] [Google Scholar]
- Story G.M., and Gereau R.W. IV. 2006. Numbing the senses: role of TRPA1 in mechanical and cold sensation. Neuron. 50:177–180. 10.1016/j.neuron.2006.04.009 [DOI] [PubMed] [Google Scholar]
- Story G.M., Peier A.M., Reeve A.J., Eid S.R., Mosbacher J., Hricik T.R., Earley T.J., Hergarden A.C., Andersson D.A., Hwang S.W., et al. 2003. ANKTM1, a TRP-like channel expressed in nociceptive neurons, is activated by cold temperatures. Cell. 112:819–829. 10.1016/S0092-8674(03)00158-2 [DOI] [PubMed] [Google Scholar]
- Streng T., Axelsson H.E., Hedlund P., Andersson D.A., Jordt S.E., Bevan S., Andersson K.E., Högestätt E.D., and Zygmunt P.M.. 2008. Distribution and function of the hydrogen sulfide-sensitive TRPA1 ion channel in rat urinary bladder. Eur. Urol. 53:391–399. 10.1016/j.eururo.2007.10.024 [DOI] [PubMed] [Google Scholar]
- Takahashi N., Mizuno Y., Kozai D., Yamamoto S., Kiyonaka S., Shibata T., Uchida K., and Mori Y.. 2008. Molecular characterization of TRPA1 channel activation by cysteine-reactive inflammatory mediators. Channels (Austin). 2:287–298. 10.4161/chan.2.4.6745 [DOI] [PubMed] [Google Scholar]
- Takahashi N., Kuwaki T., Kiyonaka S., Numata T., Kozai D., Mizuno Y., Yamamoto S., Naito S., Knevels E., Carmeliet P., et al. 2011. TRPA1 underlies a sensing mechanism for O2. Nat. Chem. Biol. 7:701–711. 10.1038/nchembio.640 [DOI] [PubMed] [Google Scholar]
- Talavera K., Gees M., Karashima Y., Meseguer V.M., Vanoirbeek J.A., Damann N., Everaerts W., Benoit M., Janssens A., Vennekens R., et al. 2009. Nicotine activates the chemosensory cation channel TRPA1. Nat. Neurosci. 12:1293–1299. 10.1038/nn.2379 [DOI] [PubMed] [Google Scholar]
- Voronin D.A., and Kiseleva E.V.. 2007. Functional role of proteins containing ankyrin repeats. Tsitologiia. 49:989–999. [PubMed] [Google Scholar]
- Wang L., Cvetkov T.L., Chance M.R., and Moiseenkova-Bell V.Y.. 2012. Identification of in vivo disulfide conformation of TRPA1 ion channel. J. Biol. Chem. 287:6169–6176. 10.1074/jbc.M111.329748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Dai Y., Fukuoka T., Yamanaka H., Kobayashi K., Obata K., Cui X., Tominaga M., and Noguchi K.. 2008. Phospholipase C and protein kinase A mediate bradykinin sensitization of TRPA1: a molecular mechanism of inflammatory pain. Brain. 131:1241–1251. 10.1093/brain/awn060 [DOI] [PubMed] [Google Scholar]
- Washburn M.P., Wolters D., and Yates J.R. III. 2001. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19:242–247. 10.1038/85686 [DOI] [PubMed] [Google Scholar]
- Xu H., and Freitas M.A.. 2007. A mass accuracy sensitive probability based scoring algorithm for database searching of tandem mass spectrometry data. BMC Bioinformatics. 8:133 10.1186/1471-2105-8-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Yan R., Roy A., Xu D., Poisson J., and Zhang Y.. 2015. The I-TASSER Suite: protein structure and function prediction. Nat. Methods. 12:7–8. 10.1038/nmeth.3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Smith J.W., and Huang C.M.. 2010. Mass spectrometry-based label-free quantitative proteomics. J. Biomed. Biotechnol. 2010:840518 10.1155/2010/840518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zygmunt P.M., and Högestätt E.D.. 2014. TRPA1. Handb. Exp. Pharmacol. 222:583–630. 10.1007/978-3-642-54215-2_23 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.