Puljung reviews recent cryo-EM KATP channel structures and proposes a mechanism by which ligand binding results in channel opening.
Abstract
Adenosine triphosphate (ATP)–sensitive K+ (KATP) channels are molecular sensors of cell metabolism. These hetero-octameric channels, comprising four inward rectifier K+ channel subunits (Kir6.1 or Kir6.2) and four sulfonylurea receptor (SUR1 or SUR2A/B) subunits, detect metabolic changes via three classes of intracellular adenine nucleotide (ATP/ADP) binding site. One site, located on the Kir subunit, causes inhibition of the channel when ATP or ADP is bound. The other two sites, located on the SUR subunit, excite the channel when bound to Mg nucleotides. In pancreatic β cells, an increase in extracellular glucose causes a change in oxidative metabolism and thus turnover of adenine nucleotides in the cytoplasm. This leads to the closure of KATP channels, which depolarizes the plasma membrane and permits Ca2+ influx and insulin secretion. Many of the molecular details regarding the assembly of the KATP complex, and how changes in nucleotide concentrations affect gating, have recently been uncovered by several single-particle cryo-electron microscopy structures of the pancreatic KATP channel (Kir6.2/SUR1) at near-atomic resolution. Here, the author discusses the detailed picture of excitatory and inhibitory ligand binding to KATP that these structures present and suggests a possible mechanism by which channel activation may proceed from the ligand-binding domains of SUR to the channel pore.
Introduction
The recent revolution in cryo-electron microscopy (cryo-EM), enabled by direct electron detection, fast camera readouts, and sophisticated computational methods, has produced an embarrassment of new structures (at or near atomic resolution) of proteins resistant to characterization by x-ray crystallography (Cheng, 2015; Cheng et al., 2015). Ion channels and transporters with sufficient mass to provide good contrast in cryo-EM were the immediate beneficiaries of this improved technology. To that end, the ATP-sensitive K+ (KATP) channel, a massive (>800 kD) heteromultimeric complex, was an attractive target; the recent publication of four cryo-EM structures of KATP by three different groups demonstrates just how attractive (Lee et al., 2017; Li et al., 2017; Martin et al., 2017a,b).
K+ currents inhibited by millimolar ATP concentrations were first recorded from heart muscle (Noma, 1983). Despite being one of the most abundant channels in the cardiac sarcolemma, the role of KATP in the heart is relatively obscure. Its main function appears to be to attenuate the rise in intracellular Ca2+ during ischemia, thus limiting the degree of cell death and tissue damage (Nichols, 2016). Perhaps better understood is the role of KATP in triggering insulin secretion. The rise in blood sugar after a meal shuts KATP channels in the plasma membrane of pancreatic β cells (Ashcroft et al., 1984). The sudden reduction in K+ permeability, coupled with the high input resistance of β cells, depolarizes their plasma membranes, opening voltage-dependent Ca2+ channels and thus enabling the exocytosis of insulin granules (Rorsman and Trube, 1985; Arkhammar et al., 1987; Nelson et al., 1987). The central role of KATP in β cell excitability makes it an attractive target for pharmacological intervention for type 2 diabetes mellitus. Indeed, the commonly prescribed antidiabetic sulfonylurea (SU) drugs are KATP antagonists (Ashcroft et al., 2017).
The pore of KATP comprises four inward rectifier K+ channel subunits (Kir6.1 or Kir6.2; Fig. 1 A). Each Kir is associated with an SU receptor (SUR) subunit (SUR1, SUR2A, and SUR2B; Inagaki et al., 1997; Shyng and Nichols, 1997). The SUR is unusual in that it is part of a subgroup of the ATP-binding cassette (ABC) exporter family of proteins (ABCC) but lacks any intrinsic transport activity (Aguilar-Bryan et al., 1995; Tusnády et al., 1997). Rather, it has evolved as a modulatory accessory subunit for the KATP pore. The β cell isotype of KATP is formed by Kir6.2 and SUR1 (Inagaki et al., 1995; Sakura et al., 1995). The two subunits are closely associated even at the chromosomal level (both residing at position 11p15.1) and must properly coassemble to exit the ER/Golgi and traffic to the plasma membrane (Inagaki et al., 1995; Zerangue et al., 1999).
Figure 1.
Structure of KATP in the presence of ATP and glibenclamide. (A) Transmembrane topology of the pancreatic β cell KATP complex. For clarity, only two (of four) Kir6.2 subunits and one (of four) SUR1 subunits are shown. (B) Side view of the 3.63-Å structure of KATP (PDB accession no. 6BAA) in the presence of ATP and glibenclamide. For clarity, the pore domains of two of the Kir6.2 subunits have been removed, and only one SUR1 subunit is shown. Kir6.2 subunits are shown in yellow and brown. SUR1 is color-coded as follows: lavender, TMD0; orange, L0; blue, TMD1-NBD1; and green, TMD2-NBD2. The ochre blocks represent the approximate location of the lipid bilayer.
The β cell response to glucose is mediated by changes in the turnover of intracellular adenine nucleotides (ATP and ADP). Response of KATP to nucleotides is mediated by three classes of nucleotide-binding site (NBS, 12 sites in all) on the cytoplasmic side of the channel (Vedovato et al., 2015). The first class of site is located directly on Kir6.2 (Tucker et al., 1997). Binding of adenine nucleotides (ATP and ADP) to this site, in a reaction that does not require Mg2+ as a cofactor, inhibits KATP. The other two NBSs are located on SUR1 (Bernardi et al., 1992; Gribble et al., 1998). Like other ABC transporters, SUR1 has two cytoplasmic nucleotide-binding domains (NBDs; Aguilar-Bryan et al., 1995). As in bona fide ABC transporters, the NBDs of SUR1 form a head-to-tail dimer in the presence of Mg2+ and nucleotides, with two NBSs formed at the dimer interface (Smith et al., 2002; Masia and Nichols, 2008; ter Beek et al., 2014). Binding of Mg nucleotides (ATP or ADP) to these sites activates KATP (Gribble et al., 1998). The tension between the inhibitory and stimulatory inputs from the three classes of NBS determine KATP’s response to metabolic changes. Indeed, mutations in any NBS can result in diseases of insulin secretion, including neonatal diabetes or persistent hyperinsulinemic hyperglycemia of infancy (PHHI; Quan et al., 2011; Ashcroft et al., 2017).
In addition to its modulation by adenine nucleotides, KATP gating is also activated by anionic phospholipids. Like every other eukaryotic inward rectifier, binding of phosphoinositides to KATP, in particular phosphoinositol 4,5-bisphosphate (PIP2), is required to maintain channels in the open state (Fan and Makielski, 1997). This results in the unfortunate (from the electrophysiologist’s perspective) tendency for KATP currents to “run down” in excised membrane patches because of the activity of membrane-embedded lipid phosphatases and phospholipases (Hilgemann and Ball, 1996; Proks et al., 2016). This effect can be reduced to a great extent by the addition of EDTA in the bath solution (Lin et al., 2003). In addition to PIP2, KATP activity is increased by the binding of several other anionic lipids, including PI 3,4,5-P3 and PI-3,4-P2 (Rohács et al., 2003), PI-4-P (Fan and Makielski, 1997), phosphatidic acid (Fan et al., 2003), and long-chain acyl-coenzyme A esters (Bränström et al., 1998; Rohács et al., 2003; Schulze et al., 2003).
The influence of ligand binding on the open–closed equilibrium of KATP is schematized in Fig. 2. This model is an adaptation of the modular gating scheme proposed by Horrigan and Aldrich (2002) to describe the influence of different domains on the gating of BK channels and is presented here as a heuristic to describe the convergence of agonist and antagonist influences on the open probability (Po) of KATP. The model incorporates elements of those previously used to discuss the influence of PIP2 or nucleotides on KATP gating (Enkvetchakul and Nichols, 2003; Vedovato et al., 2015). The pore domain is simplified here to undergo a simple open–closed transition described by the equilibrium constant L. At the single-channel level, KATP exhibits bursting behavior with at least two closed states: a short intraburst closed state and a longer-lived interburst closed state. KATP agonists and antagonists primarily affect gating by prolonging or shortening the duration of the interburst closures (Rorsman and Trube, 1985; Ashcroft et al., 1988; Fan and Makielski, 1999; Li et al., 2002). The inhibitory NBSs exist in two states: a nucleotide-free “permissive” state and a nucleotide-bound “inhibited” state with an equilibrium affinity constant, KIB. Occupancy of each inhibitory site affects the open–closed transition of the pore by a coupling factor D (where D < 1). The PIP2 site exists in an unoccupied “resting” state and a PIP2-bound “activated” state, with an affinity constant, KPIP. Each PIP2-binding event favors the open–closed transition by a factor C. Finally, the NBDs are considered as a single site with an equilibrium affinity constant KNBD. Upon dimerization, the NBDs transition from resting to activated, favoring channel opening by a factor of E for each occupied dimer.
Figure 2.
Modular gating scheme for KATP. The open–closed transition of the pore domain of KATP (symbolized by the equilibrium constant L) is energetically coupled to three classes of ligand-binding site: four inhibitory nucleotide (either ADP or ATP, symbolized as ANP) binding sites on Kir6.2 (equilibrium association constant KIB); four stimulatory PIP2 binding sites on Kir6.2 (equilibrium association constant KPIP); and four stimulatory MgANP sites formed by the dimerization of the NBDs of SUR1 (equilibrium association constant KNBD). C, D, and E are coupling factors describing the interaction between the PIP2 site, the inhibitory ANP site, and the NBDs with the channel pore, respectively.
SUR1 affects the pore-forming subunit in several distinct ways: (a) SUR1 increases the unliganded Po of the pore domain (possibly a direct effect on L; Babenko and Bryan, 2003; Chan et al., 2003; Fang et al., 2006); (b) SUR1 increases the apparent affinity for nucleotide inhibition at Kir6.2 (affecting either KIB or D; Tucker et al., 1997); (c) SUR1 confers Mg-nucleotide activation on Kir6.2 (coupling factor E; Tucker et al., 1997; Gribble et al., 1998); (d) SUR1 confers sensitivity to pharmacological inhibitors (SUs) and activators (affecting L, E, or both; Tucker et al., 1997); and (e) SUR1 may increase the affinity for PIP2 or the ability of PIP2 to stabilize the open channel pore (affecting KPIP or C; Pratt et al., 2011). Through truncation, mutation, and pharmacological manipulation, many or perhaps all of these processes have been shown to be functionally separable. However, a complete understanding of the complicated pas de deux between Kir6.2 and SUR1 must begin with an understanding of the structure of each subunit and the nature of their assembly into a complex. Below, I will discuss the new cryo-EM structures of KATP, which represent the channel complex in a drug-inhibited state and two preopen conformations with Mg nucleotides bound at SUR1. I will describe in detail the protein–ligand interactions at Kir6.2 and SUR1, the resulting conformational changes in these subunits, and how a rearrangement of the interface between SUR1 and Kir6.2 may result in opening of the channel pore.
Structure and assembly of KATP
Until recently, only low-resolution structural information was available for the KATP complex, including a negative-stain EM structure of concatenated SUR1-Kir6.2 (18 Å), the EM structure of a tetrameric SUR2B (21 Å), and small-angle x-ray scattering and nuclear magnetic resonance studies of the NBDs (Mikhailov et al., 2005; Park and Terzic, 2010; de Araujo et al., 2011, 2015; López-Alonso et al., 2012; Fotinou et al., 2013). The ∼6-Å structure of the pancreatic β cell KATP (Kir6.2/SUR1) was initially solved in the presence of the SU glibenclamide by two different groups using cryo-EM (Table 1; Protein Data Bank [PDB] accession numbers 5WUA and 5TWV; Li et al., 2017; Martin et al., 2017b). The two structures were remarkably similar, despite the presence of an (unmodeled) GFP tag on the C terminus of Kir6.2 in one of the constructs used for structure determination (Li et al., 2017) and the presence of ATP bound to Kir6.2 in the other (Martin et al., 2017b). Subsequent optimization of sample freezing conditions improved the image contrast significantly and allowed for the refinement of the inhibited KATP structure down to 3.63-Å resolution (Table 1; PDB accession no. 6BAA; Figs. 1 B and 3; Martin et al., 2017a). The resulting structural model confirms many of the features expected from homologous Kir and ABC structures and years of structure–function studies on KATP.
Table 1. Published cryo-EM structures of KATP.
| Structure | Construct | Ligands | Resolution | PDB | EMDB | Reference |
|---|---|---|---|---|---|---|
| Å | ||||||
| Inhibited | Hamster SUR1 (Q608K), mouse Kir6.2-GFP | Glibenclamide | 5.6 | 5WUA | EMD-6689 | Li et al., 2017 |
| Inhibited | Hamster SUR1 (FLAG tag), rat Kir6.2 | Glibenclamide, ATP | 5.8 | 5TWV | EMD-8470 | Martin et al., 2017b |
| Inhibited | Hamster SUR1 (FLAG tag), rat Kir6.2 | Glibenclamide, ATP | 3.63 | 6BAA | EMD-7073 | Martin et al., 2017a |
| Closed, SUR activated (propeller) | Human SUR1–(Ser-Ala)3–human Kir6.2 (concatenated) | ATP (ADP), Mg2+, diC8 PIP2 | 5.6 | 6C3P | EMD-7339 | Lee et al., 2017 |
| Closed, SUR activated (quatrefoil) | Human SUR1–(Ser-Ala)3–human Kir6.2 (concatenated) | ATP (ADP), Mg2+, diC8 PIP2 | 3.9 | 6C3O | EMD-7338 | Lee et al., 2017 |
EMDB, Electron Microscopy Data Bank.
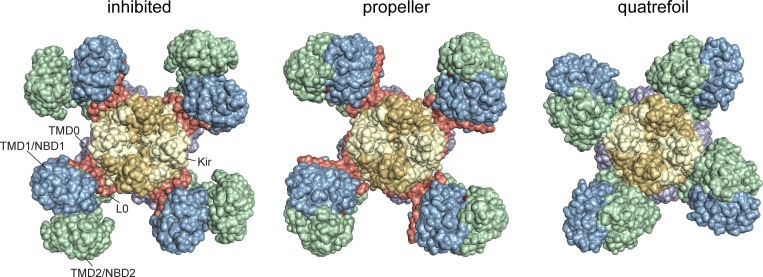
Figure 3.
Three structures of KATP. Surface representations of the entire KATP complex viewed from the cytoplasmic side (color-coded as in Fig. 1) in the SU-inhibited conformation (PDB accession no. 6BAA) and two structures with nucleotides bound to both the inhibitory Kir6.2 site and to the NBDs of SUR1: the propeller form (PDB accession no. 6C3P) and the quatrefoil form (PDB accession no. 6C3O).
Kir6.2 is a very typical inward rectifier (Fig. 1 B). It has two transmembrane helices (M1 and M2) separated by a reentrant pore helix and loop, the latter of which contains a K+ channel signature sequence (TIGFG), conferring ion selectivity (Heginbotham et al., 1994). The cytoplasmic half of the pore is primarily lined by residues from the M2 helix, and residues at the bottom of this helix (in particular F168) come together to form a tight seal, indicating that the pore domain is closed in the SU-inhibited structure. N- and C-terminal extensions from the pore form a large, basket-shaped, cytoplasmic domain crowned by the G-loop gate, which is also closed. There are extensive interactions between subunits in the cytoplasmic domain (Figs. 1 B and 3). Mutations expected to disrupt these interactions destabilize the open state of the channel, causing inactivation (Shyng et al., 2000; Lin et al., 2003; Borschel et al., 2017). This includes several mutations associated with PHHI (Lin et al., 2008).
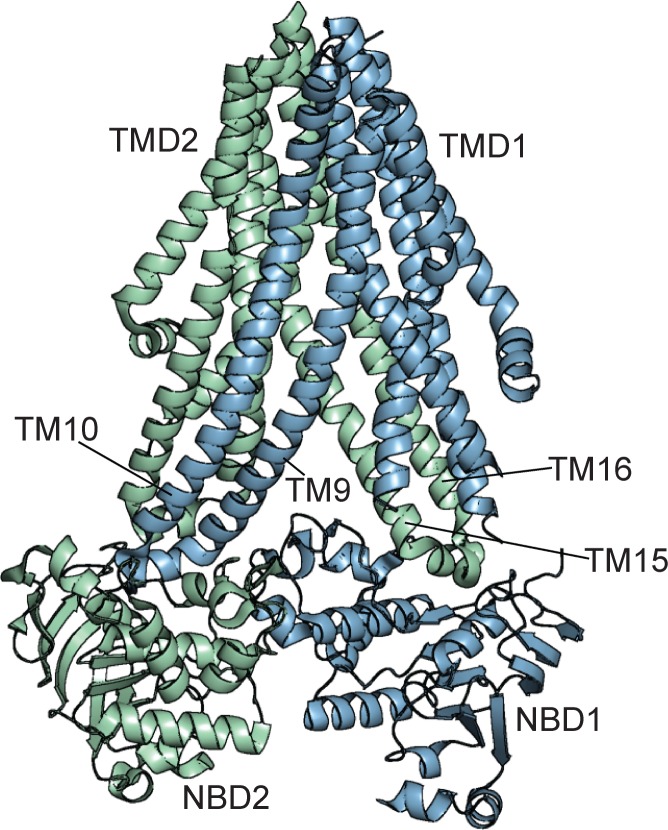
Typical of ABC exporters, SUR1 has two, six-helix transmembrane domains (TMDs; TMD1 and TMD2; Figs. 1 B and 4), each followed by an NBD. The ABC core of SUR1, as in other ABC proteins, is domain swapped, such that each half transporter is composed of helices from TMD1 and TMD2 (ter Beek et al., 2014). The first comprises TM6–8, TM11, and TM15–16; the second comprises TM9–10, TM12–14, and TM17. A short helix separates the domain swapped helices of each half-transporter module (TM15–16 in the first half transporter and TM9–10 in the second half transporter) and contacts a groove on the opposite NBD; the helix between TM15–16 contacts NBD1, and the helix between TM9–10 contacts NBD2 (Fig. 4).
Figure 4.
The ABC core structure of SUR1. Structure of TMD1-NBD1–TMD2-NBD2 of SUR1 in the inhibited state (PDB accession no. 6BAA). TMD1-NBD1 is colored blue. TMD2-NBD2 is colored green. The domain-swapped helices (TM9–10 and TM15–16) are labeled.
In a typical reaction cycle, ABC transporters transition between nucleotide-free, inward-facing states (access to the binding site of carried substrate on the cytoplasmic side) and nucleotide-bound, outward-facing states (release of substrate to the extracellular milieu; Higgins and Linton, 2004). In the SU-inhibited KATP structure, the NBDs are unoccupied (ATP, but not Mg2+, was included in the sample) and spaced far apart (Fig. 4), resembling the “inward-facing” apo structures of the ABC transporters P-glycoprotein (PDB accession no. 4M2T; Li et al., 2014) and MsbA (PDB accession no. 3B5W; Ward et al., 2007), as well as the fellow ABCC family members transporter associated with antigen processing (TAP; PDB accession no. 5U1D; Oldham et al., 2016), multidrug-resistance protein 1 (MRP1; PDB accession no. 5UJ9; Johnson and Chen, 2017), and the unphosphorylated state of the cystic fibrosis transmembrane conductance regulator (CFTR; PDB accession nos. 5UAR and 5UAK; Zhang and Chen, 2016; Liu et al., 2017). In addition to being physically separated, there is a misalignment of the NBDs, which is not present in the symmetric P-glycoprotein or MsbA structures but is visible to a lesser extent in the unphosphorylated structure of CFTR.
SUR1 has a bundle of five transmembrane helices (TMD0) N terminal to the ABC core (Fig. 1 B). A similar motif is present in the structure of MRP1 (Johnson and Chen, 2017). However, the resolution in this region of MRP1 was not sufficient to build a complete de novo atomic model. Interestingly, whereas there does appear to be some degree of conservation of the overall protein fold, there is little to no sequence conservation between MRP1 and SUR1 in this region. TMD0 is connected to TMD1 via an intracellular loop known as L0 (sometimes called the CL3 linker). The structure of the C-terminal two thirds of this loop is conserved in MRP1 and is also present at the N terminus of CFTR, where it is known as the lasso domain (Zhang and Chen, 2016; Liu et al., 2017; Zhang et al., 2017). In the SU-inhibited structure of KATP, the primary contact between Kir6.2 and SUR1 is mediated via a series of hydrophobic interactions between the M1 (outer) helix of Kir6.2 and the TM1 helix of the TMD0 domain (Fig. 1 B). Additional contacts between L0 and the cytoplasmic domain of Kir6.2 are also evident. In the presence of glibenclamide, the ABC core domains of KATP are tilted in the membrane plane and splayed outward from the pore, with NBD2 farthest away.
Our understanding of KATP structure and assembly was recently enhanced by the publication of two further structures of a concatenated SUR1–Kir6.2 construct (Ser-Ala-Ser-Ala-Ser-Ala linker) in the presence of Mg2+, ATP, and dioctanoyl PIP2 (diC8 PIP2; Table 1 and Fig. 3; Lee et al., 2017). These represent active (nucleotide-bound) conformations of SUR1 and suggest a mechanism by which occupancy of the NBSs of SUR1 may be related to the channel pore.
Modulation of Kir6.2 by PIP2 and ATP
Central to KATP’s function as a metabolic sensor is its inhibition by increased intracellular ATP concentrations after glucose uptake by pancreatic β cells. The inhibitory action of ATP is independent of Mg2+ and was shown to be intrinsic to Kir6.2 by expression of a C-terminally truncated subunit (Kir6.2-ΔC) that traffics to the plasma membrane in the absence of SUR (Tucker et al., 1997). Also intrinsic to the Kir6.2 subunit is PIP2 modulation (Fan and Makielski, 1997; Shyng et al., 2000). Binding of PIP2 antagonizes ATP inhibition (Baukrowitz et al., 1998; Shyng and Nichols, 1998; Fan and Makielski, 1999). Channel rundown, likely caused by degradation of PIP2 by endogenous lipid phosphatases and phospholipases, increases the apparent affinity for ATP (Baukrowitz et al., 1998; Ribalet et al., 2000). The effects of PIP2 and ATP are well modeled by a scheme that assumes that binding of PIP2 and ATP are mutually exclusive (Enkvetchakul and Nichols, 2003). However, it remains a possibility that the two ligands antagonize each other allosterically (i.e., through changes in channel Po; changing L in Fig. 2).
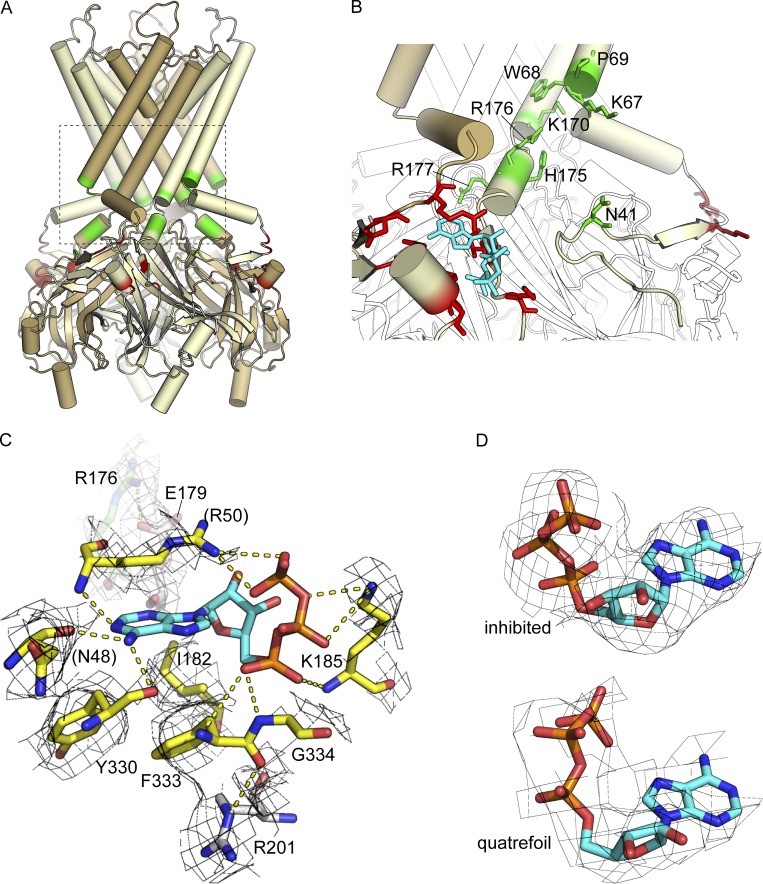
Putative PIP2-binding residues in Kir6.2 can be identified through structural and sequence alignment with Kir3.2 (GIRK2), the structure of which has been solved in the presence of PIP2 (PDB accession no. 3SYA; Whorton and MacKinnon, 2011). Phospholipid-binding residues are located on regions of Kir6.2 at the membrane–water interface, including the ends of the M1 and M2 helices, the loop connecting the slide helix (an N-terminal amphipathic helix) to M1, and the helix immediately after M2. Many of these residues (Fig. 5, A and B, green) are conserved between Kir3.2 and Kir6.2. The exceptions are N41 (lysine in Kir3.2) and H175/R176 (both lysines in Kir3.2). R177 of Kir6.2 has also been implicated in PIP2 binding (Fan and Makielski, 1997; Shyng et al., 2000), but it seems to be oriented away from the putative PIP2-binding surface in the structure (Fig. 5 B). Whereas diC8 PIP2 was included by Lee et al. (2017) in their samples, no density in the putative PIP2-binding site was detected in their structures. This is likely the result of antagonistic binding of ATP at the inhibitory site, which may be responsible for the relative constriction in the PIP2-binding pocket compared with that of Kir3.2. Alignment of the Lee et al. (PDB accession nos. 6C3O and 6C3P) structures with the SU-inhibited structure determined in the absence of PIP2 (PDB accession no. 6BAA) reveals no significant differences at the putative PIP2 site.
Figure 5.
The PIP2- and ATP-binding sites of Kir6.2. (A) Tetrameric structure of Kir6.2 (PDB accession no. 6BAA). Alternate subunits are colored yellow and brown. The inhibitory nucleotide binding site is colored red. Putative PIP2-binding residues are colored green. (B) Close-up view of the boxed region from A with putative PIP2-binding residues (green sticks) labeled. ATP is shown in cyan. ATP-binding residues are shown as red sticks. (C) Close-up view of the ATP-binding site. Residues that contact ATP directly are shown as yellow sticks. Amino acids in white are those previously identified as affecting the apparent affinity for ATP but that do not contribute directly to ATP binding in the structure. R176 from the putative PIP2 site is colored green. Residues in parentheses are contributed by the adjacent subunit (brown in B). The EM density shown for these residues was contoured at 1.5 σ. (D) EM density (contoured at 3 σ) for ATP bound to Kir6.2 in the inhibited structure (PDB accession no. 6BAA, top) and quatrefoil structure (PDB accession no. 6C3O, bottom).
In Kir3.2, PIP2 binding is accompanied by a 15° clockwise rotation (when viewed from the intracellular side) of the cytoplasmic domain, which pulls the pore open (model derived from PDB accession no. 3SYQ; Whorton and MacKinnon, 2011). Both Martin et al. (2017b) and Li et al. (2017) report a subset of molecules in their micrographs for which two (of four) cytoplasmic domains of the Kir6.2 subunits are rotated 9–14° relative to their position in the majority of the molecules used for structural analysis. Li et al. identified a small density in the putative PIP2 site of this subset of particles that they speculate may have been endogenous PIP2 that copurified with the channel protein. However, the resolution of this structure was quite limited (8.5 Å), so any such assignments are tentative and should be interpreted with caution.
In all but one of the KATP structures determined (the Li et al. structure was determined in the absence of ATP; Li et al., 2017), there is strong density corresponding to ATP bound at the inhibitory site on Kir6.2, encompassing residues from the cytoplasmic N and C termini (Fig. 5 C; Lee et al., 2017; Martin et al., 2017a,b). As for the PIP2 site, alignment of the inhibitory ATP-binding sites in all three structures reveals no significant changes. ATP adopts an unusual conformation in these structures with the phosphates curled back toward the purine ring (Fig. 5, C and D). This configuration of ATP resembles that in the noncanonical ATP binding site on P2X4 (Hattori and Gouaux, 2012). The γ phosphate of ATP is exposed to solvent and adopts different rotamers in the different structures (PDB accession no. 6BAA vs. 6C3O; Fig. 5 D). Such flexibility may explain why ADP, as well as nucleotides with substitutions at this position, still bind to and inhibit KATP (Ämmälä et al., 1991; Wang et al., 2002; Proks et al., 2010).
ATP-binding residues are highlighted in red in Fig. 5 (A and B) and shown as yellow sticks in Fig. 5 C. Several of these have been identified by previous site-directed mutagenesis experiments (Drain et al., 1998; Tucker et al., 1998; Proks et al., 1999; Ribalet et al., 2003; Antcliff et al., 2005). In particular, substitutions at positions G334 and I182 have been shown to almost completely disrupt nucleotide binding without affecting the intrinsic open–closed transition of the Kir6.2 pore (Li et al., 2000, 2005). G334D, which is nearly completely insensitive to ATP inhibition at concentrations <10 mM, has been extensively exploited to study nucleotide activation of KATP in the absence of any inhibition (Drain et al., 1998; Li et al., 2002; Proks et al., 2010, 2014). I182 and G334 mutations have been interpreted as exerting their effects by preventing nucleotide binding (affecting KIB). Inspection of the structure suggests that this interpretation is correct.
The overall structure of the ATP-binding site and the majority of the ATP-binding residues identified in this structure are conserved in Kir3.2, with the exception of I182 (conservatively substituted to valine), K185 (threonine in Kir3.2), and G334 (H in Kir3.2). Why is Kir3.2 not inhibited by ATP? The increased bulk at the position equivalent to 334 in Kir3.2 (H357) would be sufficient to prevent binding of ATP. Furthermore, whereas an isoleucine to valine substitution at the position equivalent to 182 in Kir3.2 (V205) may be conservative, even conservative substitutions at position 182 (e.g., I182L) disrupt ATP binding to Kir6.2 (Li et al., 2000). Two other residues, E179 and R201, were suggested from previous modeling and mutagenesis to contribute directly to ATP binding (Shyng et al., 2000; Antcliff et al., 2005). The structure of the ATP binding site (Fig. 5 C) clearly demonstrates that neither residue binds nucleotide. R201 is a potential hydrogen bonding partner for the backbone carbonyl of F333 and thus may stabilize the short helix encompassing F333 and the critically important G334. E179 appears to form a salt bridge with R176 of the PIP2 site, so the effect of mutating E179 on ATP inhibition is likely to be allosteric.
Strikingly, the inhibitory ATP-binding site is positioned very close to the putative PIP2 site (Fig. 5, A and B). Residue N41 from the PIP2 site is located on the same N-terminal loop of Kir6.2 as residues N48 and R50 of the ATP-binding site. K185, which coordinates the phosphates of ATP, is just downstream of the helix that contains H175, R176, and R177 of the PIP2 site. The distinct nature of these binding sites eliminates the suggestion that ATP and PIP2 may directly compete for the same site (MacGregor et al., 2002). However, it is tempting to speculate that binding of one ligand (e.g., ATP) can distort the binding site for the other, indicating a direct interaction between the sites, rather than the two being allosterically coupled solely through the pore domain (i.e., via “?” in Fig. 2, rather than changes in L). Indeed, Lee et al. (2017) point out that the constriction of the PIP2 site in ATP-bound Kir6.2 relative to Kir3.2 is caused by ATP-binding residues N48 and R50. Further experiments that directly measure nucleotide and/or PIP2 binding to intact channels are required to test this model and determine whether the two ligand-binding sites function as a “PIP switch” to activate or shut the channel.
Contribution of SUR1 to ATP/PIP2 binding and the intrinsic gate of Kir6.2
In all of the available KATP structures, the major contact between Kir6.2 and SUR1 is between the M1 helix of Kir6.2 and the first transmembrane helix of TMD0. What is the functional consequence of this interaction? Coexpression of SUR1 with Kir6.2-ΔC increases the intrinsic Po (Fig. 2, L). Unexpectedly, the increase in Po is accompanied by an increase in the apparent affinity for inhibition by ATP rather than a decrease that would be expected for a ligand that stabilizes the closed state of a channel (change in KIB; Tucker et al., 1997; Babenko and Bryan, 2003; Chan et al., 2003; Fang et al., 2006). This suggests that residues from SUR1 may contribute directly to the ATP- or PIP2-binding sites. Fig. 6 shows that residues from TMD0 and L0 can be found in close apposition to the ATP-binding site. In particular, residues Q52 at the end of the slide helix of Kir6.2 and E203 from L0 are within 5 Å of one another. When both residues are mutated to cysteine, they can form a disulfide bond that locks the channels closed, indicating that interactions in this region directly impact channel gating (Pratt et al., 2012). R176 and R177 are also located close to this interface. Both of these residues have been implicated in transduction of nucleotide binding signals from SUR1 to Kir6.2 (John et al., 2001). A mutation (E128K) in TMD0 prevents the increase in Po that accompanies coexpression of SUR1 with Kir6.2. This residue faces away from the binding interface of SUR1 and Kir6.2 but may affect gating indirectly by upsetting the interface between TM3 and the L0 loop (Pratt et al., 2011). E128K channels do not respond to PIP2 stimulation, suggesting that this domain may increase Po by stabilizing interactions between Kir6.2 and PIP2 (Fig. 2, KPIP or C).
Figure 6.
Interactions between TMD0-L0 and the cytoplasmic domains of Kir6.2. Interface between the ATP-binding site on Kir6.2 and TMD0/L0 of SUR1. Kir6.2-Q52 and SUR1-E203 are shown as sticks and are separated by <5 Å.
One could speculate that the interactions described between TMD0–L0 and Kir6.2 in Fig. 6 stabilize ATP binding to the inhibitory site. However, this interpretation does not account fully for results from the expression of “mini KATP” channels formed by Kir6.2-ΔC and TMD0 or TMD0/L0. Kir6.2-ΔC has a low intrinsic Po (0.09–0.15; Babenko and Bryan, 2003; Chan et al., 2003). Coexpression with TMD0 (residues 1–195) enhances the Po by increasing the single-channel burst duration and lowers the apparent affinity for ATP inhibition (Babenko and Bryan, 2003). The decrease in apparent affinity is the expected result if ATP is considered to be an allosteric blocker that preferentially stabilizes the closed state of the channel (or destabilizes the open state; Li et al., 2002). Coexpression of Kir6.2-ΔC with TMD0 and the first portion of L0 (residues 1–232) results in channels that are nearly constitutively active (Po > 90%) and much less sensitive to ATP (Babenko and Bryan, 2003). As this construct encompasses the entire site depicted in Fig. 6, it is unlikely that such interactions directly stabilize ATP binding. However, it should be noted that coexpression of Kir6.2-ΔC with TMD0–L0 constructs longer than 232 amino acids progressively decreases Po, although not to the level of Kir6.2-ΔC alone (Babenko and Bryan, 2003). Residues in NBD2 and the distal C terminus of SUR1 and SUR2A have been shown to contribute to the increased apparent affinity for inhibition of KATP by ATP (Babenko et al., 1999b). Whether this represents a direct effect on ATP binding (KIB in Fig. 2) or an allosteric effect on the binding site is not known. The interactions between TMD0–L0 and Kir6.2 highlighted in Fig. 6 may explain the increased Po resulting from coexpression of Kir6.2 with SUR1, but the nature of the increased apparent affinity for ATP inhibition remains a mystery.
Nucleotide binding to the NBDs of SUR1
Lee et al. were able to resolve two different structures of KATP in the presence of Mg2+, ATP, and PIP2 (Fig. 3). These structures, termed “propeller” (PDB accession no. 6C3P, 5.6 Å) and “quatrefoil” (four leafed, PDB accession no. 6C3O, 3.9 Å), based on their appearance when viewed perpendicular to the membrane plane, differ mainly in a rigid-body movement of the ABC core domain. In both structures, the NBDs are dimerized with Mg nucleotides bound to each site. Fig. 7 shows the NBDs of the quatrefoil structure.
Figure 7.
MgATP and MgADP bound at the interface of NBD1 and NBD2 of SUR1. NBD dimer from the quatrefoil structure in Lee et al. (2017) (PDB accession no. 6C3O). The two NBSs (NBS1 and NBS2) are formed at the interface of the NBDs. MgATP is bound to NBS1, and MgADP is bound to NBS2. Important nucleotide-binding motifs are highlighted. A loops (orange) interact with the purine ring of ATP/ADP via π-stacking interactions (W688 in NBS1 and Y1353 in NBS2). WalkerA and WalkerB motifs are shown in red. WalkerA lysines (K719 and K1384) and acidic residues from the WalkerB motifs (D853, D854, D1505, and E1506) are shown as sticks. The ABC signature sequences are shown in magenta.
The NBSs of SUR1 are asymmetric. Like other ABCC family members, SUR1 has one catalytically (ATP hydrolysis) competent NBS (NBS2, the consensus site) and one site that is unable to catalyze hydrolysis of ATP (NBS1, the degenerate site; ter Beek et al., 2014; Vedovato et al., 2015). Interestingly, whereas the samples were prepared in the presence of ATP, NBS2 is occupied by MgADP rather than MgATP in both the propeller and quatrefoil structures. This suggests that in the time between sample preparation and flash freezing, the ATPase activity of NBS2 was sufficient to hydrolyze bound ATP. Alternatively, the ADP may have been present as an impurity in the original ATP stock, and occupancy of NBS2 by ADP may simply reflect a higher binding affinity for ADP over ATP (Lee et al., 2017). Absent any direct measurement of real-time binding affinity of the two NBSs, it is difficult to differentiate between these two possibilities.
The overall structure of the NBD dimer is similar to that of other ABC transporters and to a previous homology model of the NBDs based on the structure of MJ0796 (Smith et al., 2002; Masia and Nichols, 2008). The two NBSs are formed at the dimer interface with portions of each site contributed by each NBD. The A loop (orange), WalkerA, and WalkerB motifs (red) of NBS1 are contributed by NBD1, whereas the ABC signature sequence (magenta) of NBS1 is contributed by NBD2. Likewise, the A loop and Walker motifs of NBS2 are contributed by NBD2, and the signature sequence of NBS2 is located on NBD1. The purine rings of ATP/ADP are coordinated via π-stacking interactions with aromatic residues of the A loop of each binding site (W688 in NBS1 and Y1353 of NBS2). The WalkerA lysine residues (K719 in NBS1 and K1384 in NBS2) coordinate the β and γ phosphates of ATP in NBS1 and the β phosphate of ADP in NBS2 as expected (Smith et al., 2002; ter Beek et al., 2014). The WalkerB domains contain pairs of acidic residues (D853/D854 in NBS1 and D1505/E1506 in NBS2). The first residue of each pair coordinates Mg2+. The second binds and polarizes the attacking water molecule in the hydrolysis reaction in typical ABC transporters (ter Beek et al., 2014). The ABC signature sequence of NBS2 (LSGGQ, residues on NBD1) is intact, but it is replaced by FSQGQ in NBS1, perhaps explaining why that site is unable to hydrolyze ATP (Fig. 7; Matsuo et al., 1999).
Each NBD is composed of a RecA subdomain (678–781 in NBD1 and 1,343–1,434 in NBD2) and a helical subdomain (782–877 in NBD1 and 1,435–1,498 in NBD2). In typical ABC transporters, the RecA and helical subdomains within each NBD rotate toward one another upon nucleotide binding, assuming a more compact conformation (ter Beek et al., 2014). When NBD1 and NBD2 of the quatrefoil form of SUR1 are aligned with the NBDs of the SU-inhibited structure at their respective helical subdomains, it becomes apparent that NBD2 adopts a more compact conformation in the presence of agonist, but NBD1 does not. This results in a large gap between MgADP and the signature sequence in NBS2. Lee et al. (2017) speculate that this gap may be sufficient to allow nucleotide dissociation from NBS2, allowing MgADP to equilibrate at this binding site from solution, even when the NBDs are dimerized.
ABC exporters use the hydrolysis of ATP as a “power stroke” to change conformation and move solutes across the plasma membrane (Higgins and Linton, 2004). Previous work, including electrophysiological studies and photoaffinity labeling experiments, suggests that KATP is activated by ATP binding (with or without Mg2+) to NBS1 and MgADP binding to NBS2 (Matsuo et al., 1999; Vedovato et al., 2015). It is widely believed that KATP activation by MgATP requires hydrolysis of ATP to ADP (Zingman et al., 2001). However, whereas NBS2 of SUR1 is competent to hydrolyze ATP (Matsuo et al., 1999; de Wet et al., 2007), it remains unclear whether ATP hydrolysis occurs on the timescale of channel gating. The measured specific hydrolysis rate of the concatenated SUR1-Kir6.2 construct used by Lee et al. for structural characterization was 0.02 s−1 (50 s for one reaction cycle to occur), a value close to that previously reported for SUR1 (0.03 s−1; de Wet et al., 2007). The specific hydrolysis rate for the bona fide ABC transporter P-glycoprotein (with two active ATPase sites) was much faster (0.62 s−1) when measured using the same technique used by Lee et al. (Kim and Chen, 2018). Interestingly, the hydrolysis rate for purified CFTR, an ABCC family member that is also an ion channel, was 0.06 s−1, only threefold faster than that for KATP (Liu et al., 2017). It is important to note, however, that ATP hydrolysis by CFTR results in the termination of opening bursts (Csanády et al., 2010), not channel activation, as has been proposed for KATP. The hydrolysis rate for the SUR1-Kir6.2 concatemer was much slower than the rate at which macroscopic KATP current appears after MgATP application to Kir6.2-G334D/SUR1 channels (which are not inhibited by nucleotides; Proks et al., 2010). However, it should be noted that the time course of the increase in KATP current after MgATP application depends on multiple rate constants (binding, dissociation, conformational changes, and hydrolysis, if present), so direct comparison of the macroscopic activation rate to the ATP hydrolysis rate is not possible absent further information.
KATP channel current is directly activated by MgADP binding in the absence of ATP, indicating that the act of hydrolysis in and of itself is not required to drive an activating conformational change in SUR1 (Proks et al., 2010). Saturating concentrations of ATP activate Kir6.2-G334D/SUR1 channels to the same extent as ADP, and differences in the relative ability of the two ligands to gate the channel can be explained by differences in their binding affinities (Proks et al., 2010; Vedovato et al., 2015). The CFTR gating cycle involves hydrolysis of ATP. As a consequence, single-channel analysis of CFTR currents indicates a step in the gating process that is not at equilibrium (i.e., irreversible; Csanády et al., 2010). In contrast to this, single-channel analysis of KATP gating shows no evidence of nonequilibrium gating (Choi et al., 2008). Thus, it remains an open question whether ATP must be hydrolyzed to ADP to activate Kir6.2 via the NBDs of SUR1, and further investigation is required.
Glibenclamide binding and inhibition of KATP
The higher-resolution inhibited structure of KATP shows clear density for glibenclamide bound in the transmembrane region of SUR1 (Fig. 8 A; Martin et al., 2017a). The drug is sandwiched between residues on helices TM7, TM8, and TM11 on one side and TM15 and TM17 on the other (Fig. 8 C). Most of the amino acid residues at this binding interface are conserved between SUR1 and SUR2A with the exception of S1238 (Y in SUR2A) and T1242 (S in SUR2A). These substitutions may explain the relative insensitivity of Kir6.2/SUR2A channels to glibenclamide and tolbutamide compared with Kir6.2/SUR1 (Venkatesh et al., 1991). Previous studies using SUR1/SUR2A chimeras, taking advantage of this difference in SU apparent affinity, implicated TM16–TM17 as part of the binding site (Ashfield et al., 1999; Babenko et al., 1999a). In the structure, residue S1238 of TM16 is positioned close to the cyclohexyl moiety at one end of glibenclamide (Fig. 8 C). Substitution of S1238 with tyrosine (the equivalent SUR2A residue) in SUR1 reduces the apparent SU binding affinity, whereas replacing the tyrosine in SUR2A with serine confers higher sensitivity to the SU gliclazide (Ashfield et al., 1999; Proks et al., 2014). The structure suggests that the introduction of a bulkier residue at that position may create a steric clash that destabilizes SUs at this site. The SU tolbutamide has a butyl chain at the same position as the cyclohexyl moiety of glibenclamide. This aliphatic chain would also be expected to clash with a tyrosine residue at a position analogous to S1238 in the SU binding site (i.e., as in SUR2A). Many of the residues in close apposition to glibenclamide in Fig. 8 C had not previously been implicated in drug binding. As an additional test of their structure, Martin et al. (2017a) used site-directed mutagenesis coupled with patch-clamp and flux assays to verify the contribution of these residues to SU inhibition.
Figure 8.
The glibenclamide binding site. (A) SU-inhibited SUR1 structure (PDB accession no. 6BAA) showing only TMD1-NBD1 and TMD2-NBD2. Glibenclamide is depicted as magenta spheres. (B) Surface representation of the glibenclamide binding pocket in the SU-inhibited structure (PDB accession no. 6BAA) and the same region from the partially activated propeller structure (PDB accession no. 6C3P). The structure of glibenclamide from PDB accession no. 6BAA is shown in both. The two structures were aligned at TMD1. (C) Glibenclamide-interacting residues. The panel on the left shows only the portion of the glibenclamide site contributed by TMD1, and the panel on the right (rotated 180°) shows only the interactions between TMD2 and glibenclamide.
SUs affect KATP channels formed by Kir6.2/SUR1 in two distinct ways: they prevent nucleotide-dependent stimulation, and they reduce the intrinsic Po of KATP (Proks et al., 2014). These two effects may be physically separable. In Kir6.2/SUR2A, SUs only affect the Po in the absence of nucleotides (L), not nucleotide stimulation (Fig. 2, E or KNBD; Proks et al., 2014). The structure of KATP bound to glibenclamide suggests that SUs may prevent nucleotide stimulation of KATP by wedging themselves in between the two half-transporter domains of the ABC core, thus preventing association of the NBDs. However, this observation does not explain the effect of SUs on the intrinsic Po of KATP. Fig. 8 B shows a surface representation of the glibenclamide binding site. Glibenclamide is nestled in a pocket that follows its contours very closely. By comparison, when the same region is shown in the propeller form of KATP, the site collapses, such that it can no longer accommodate drug binding. This is an expected consequence of a model in which glibenclamide prevents NBD association by disrupting the movement of TMD1 and TMD2 and explains the observation that nucleotide binding to the NBDs destabilizes SU binding (Bernardi et al., 1992; Ueda et al., 1999). Nucleotide binding has the opposite effect on a class of KATP-selective K+ channel openers (KCOs), slowing their dissociation from SUR2A/B (Gribble et al., 2000; Reimann et al., 2000). Binding studies have implicated TM16–TM17 and the loop between TM13 and TM14 in KCO binding (Uhde et al., 1999; Hambrock et al., 2004).
Mutagenesis and binding studies describe a near continuous surface in L0 that was proposed to form an SU-binding site (Mikhailov et al., 2001; Vila-Carriles et al., 2007; Ashcroft et al., 2017). In light of this information, Li et al. (2017) initially suggested that there was density in their 5.6-Å structure that might correspond to bound glibenclamide at this position. The 3.63-Å structure of KATP bound to glibenclamide revealed this density to be previously unmodeled residues. The effects of mutating L0 on glibenclamide binding are likely to be either allosteric through an effect on channel opening (L0 has been shown to affect intrinsic Po, i.e., L, of Kir6.2; Babenko and Bryan, 2003) or via an indirect destabilization of the glibenclamide site by L0.
SUR1 is not a bona fide ABC exporter
Whereas SUR1 has no known transport function, it remains a possibility that it transports some as-yet-unidentified ligand. However, comparison of the nucleotide-bound (quatrefoil) structure of the ABC core of SUR1 with that of the ABC transporter Sav1866 in the presence of ADP (PDB accession no. 2HYD) suggests that SUR1 is not capable of moving solutes across the plasma membrane (Fig. 9; Dawson and Locher, 2006). In the presence of ADP, Sav1866 adopts an obviously outward-facing conformation, which is necessary to release transported solute molecules into the extracellular space. In contrast to this, the quatrefoil form and propeller form of SUR1 are closed on the extracellular side, despite association of the NBDs. Therefore, any transport function for SUR1 remains unlikely.
Figure 9.
SUR1 is not a transporter. Top: Structure of the ABC transporter Sav1866 bound to ADP (PDB accession no. 2HYD) adopting an outward-facing conformation. Bottom: Structure of the ABC core of SUR1 bound to MgATP and MgADP (quatrefoil form, PDB accession no. 6C3O).
Conformational changes in KATP: The pore domain
The acquisition of KATP structures in three distinct conformations (inhibited, propeller, and quatrefoil) allows for some speculation about the conformational changes associated with channel gating. All of these structures were solved in the presence of ATP, which was resolved in the inhibitory site of Kir6.2 of each form (Lee et al., 2017; Martin et al., 2017a,b). Therefore, one might reasonably expect the pore domain to be closed in all three structures. Fig. 10 A clearly demonstrates that whereas there may be global differences in the conformation of the three different structures of KATP (Fig. 3), there are no apparent structural changes in the pore domain. The channel gate is clearly closed in all three structures, with F168 occluding the pore at the M2 bundle crossing. The cytoplasmic domain of Kir6.2, on the other hand, rotates 11.5° clockwise (when viewed from the intracellular side) between the inhibited and quatrefoil structures (Fig. 10 B). A similar rotation (15° clockwise) underlies the putative opening transition of Kir3.2 (Whorton and MacKinnon, 2011). A comparison of various crystal forms of KirBac3.1 highlights a 23° rotation of the cytoplasmic domains (Clarke et al., 2010). In two structures of KirBac3.1 in the nontwist form (clockwise rotation of the cytoplasmic domains relative to the twist form, when viewed from the intracellular side), the selectivity filter is in a conductive conformation (i.e., there is density for four K+ ions). Structures in which the cytoplasmic domain is in the twist form all have changes in ion occupancy, which the authors believe to reflect blocked or subconductance states. Interestingly, these changes in ion occupancy occur with no concomitant change in the opening of the bundle-crossing gate. The model of the quatrefoil state of KATP (PDB accession no. 6C3O) features three K+ ions in the selectivity filter. The other structural models in the PDB (accession nos. 6BAA and 6C3P) do not include any ions in the selectivity filter, but inspection of the EM density in the pore region (contoured at 5 σ) shows that there may, in fact, be ions in the selectivity filters of all three structures (Fig. 10 C). It is tempting to speculate that these putative changes in ion occupancy may result from rotations in the cytoplasmic domains and that the pore structure in the propeller and quatrefoil forms may be described as adopting a preopen closed state. However, it should be noted that cytoplasmic domain of a mutant KirBac3.1 (S129R) with an open bundle crossing gate is in the twist form, arguing against the idea that a clockwise rotation of the cytoplasmic domain results in channel opening, at least in prokaryotic Kirs (Bavro et al., 2012).
Figure 10.
Conformational differences in Kir6.2 between inhibited and partially activated states. (A) Two (of four) pore domains and slide helices of Kir6.2 (residues 51–179) from the SU-inhibited structure (yellow, PDB accession no. 6BAA), the propeller form (light cyan, PDB accession no. 6C3P), and the quatrefoil form (magenta, PDB accession no. 6C3O) of KATP. F168 is shown as spheres to mark the bundle-crossing gate. (B) Rotation of the cytoplasmic domain of Kir6.2 between the SU-inhibited structure (yellow) and the quatrefoil form (magenta). Spheres mark the location of the α-carbons of residues D323 (outer ring) and D350 (inner ring). (C) Selectivity filter structures (only showing two diagonally opposed subunits) of the inhibited state (PDB accession no. 6BAA), the propeller form (PDB accession no. 6C3P), and the quatrefoil form (PDB accession no. 6C3O). The density within the pore of each has been contoured at 5 σ and shows the potential location of K+ ions.
Conformational changes in KATP: SUR1
The ABC core domain of SUR1 in the presence of glibenclamide adopts an inward-facing conformation with the NBDs spaced very far apart and offset relative to one another (Figs. 4 and 11). Subsequent to binding Mg nucleotides, the NBDs of SUR1 align, forming a dimer, and stabilizing a conformational change that brings TMD1 and TMD2 closer together (Fig. 11). This “activated” conformation of the SUR1 ABC core is very similar in the propeller and quatrefoil forms. Currently, there is no structure for a nucleotide- and SU-free apo state of SUR1. It remains a possibility that the inhibited structure may represent distortions introduced by drug binding between the two half transporters of the core domain. Thus, Fig. 11 also includes a speculative apo state of SUR1, based on the apo structure of the bacterial transporter TM287/288 (PDB accession no. 4Q4H; Hohl et al., 2014). This structure was chosen because the NBDs of TM287/288, like SUR1, contain one consensus NBS and one degenerate site. In the apo structure of TM287/288, the NBDs remain partially associated in an open-dimer conformation, even in the absence of nucleotides. A similar structure was observed for the transporter MsbA. The crystal structure of MsbA was solved in two different apo states, one of which (closed apo, PDB accession no. 3B5X) is inward facing, but with the NBDs in close apposition (Ward et al., 2007). In this closed apo state, the NBDs are offset as in the inhibited structure of KATP, introducing a twist at the TMDs. It has also been suggested that the NBDs of CFTR remain partially associated throughout many gating cycles, with ATP bound to NBS1 (Basso et al., 2003). Whether SUR1 adopts a similar structure in the apo state is an open question.
Figure 11.
Nucleotide-dependent conformational change in SUR1. Top, from left: Structure of SUR1 (residues 195–1,581, L0-TMD1-NBD1-TMD2-NBD2) in the SU-inhibited form (PDB accession no. 6BAA), putative apo state of SUR1 based on the structure of TM287/288 (PDB accession no. 4Q4H), activated state of SUR1 in the propeller form (PDB accession no. 6C3P), and activated state of SUR1 in the quatrefoil form (PDB accession no. 6C3O). Bottom: Conformational differences in the NBDs of the above structures. MgATP/ADP are shown as spheres.
Conformational changes in KATP: Subunit rearrangements
All of the structures to date of the KATP complex represent closed channels with ATP bound to the inhibitory site on Kir6.2. As such, the open state of KATP remains elusive. The interface formed between TMD0 and M1 of Kir6.2 is essentially unchanged in the three different structures (inhibited, propeller, and quatrefoil). Therefore, the mechanism by which TMD0/L0 modulates the intrinsic Po of Kir6.2 (represented by L in Fig. 2) or increases the apparent ATP affinity at the inhibitory site (affecting KIB or D) is still unknown. However, a comparison of the three available structures suggests a possible mechanism by which nucleotide occupancy of the NBDs of SUR may be communicated to the pore domain (Fig. 2, coupling factor E).
The overall topology of KATP is very similar in the inhibited state and propeller structure (Figs. 3 and 12). The NBDs of SUR align and dimerize upon nucleotide binding, with no substantial rearrangement of the channel complex. The transition from the propeller to quatrefoil structures, however, involves a large rotation and translation of ABC core domain of SUR1 (Figs. 3 and 12). When viewed from the cytoplasm, the NBD dimer rotates nearly 90°, bringing NBD2 in close apposition with the cytoplasmic domain of Kir6.2. This creates a new putative polar interface that may stabilize the 11.5° rotation of the cytoplasmic domain of Kir6.2 (Fig. 13 A). Whereas much of L0 is unresolved in the quatrefoil structure, it is likely that such a large rotation of the ABC core domain would also disrupt the interface between L0 and Kir6.2, which could potentially promote channel opening (Pratt et al., 2012).
Figure 12.
Putative conformational changes in the KATP complex upon SUR1 activation. Cartoon representation showing the transition from the inhibited state of KATP through the propeller form to the quatrefoil form. In the inhibited form, the NBDs are out of alignment and spaced far apart. In the presence of Mg2+ and nucleotides, the NBDs first dimerize (propeller form), but the overall conformation of the complex remains unaffected. In the quatrefoil form, the dimerized NBDs rotate (along with TMD1 and TMD2) such that a new interface is formed between NBD2 and the cytoplasmic domain of Kir6.2. The interface between TMD0 and Kir6.2 remains largely unaffected.
Figure 13.
New interfaces formed in the quatrefoil structure (PDB accession no. 6C3O). (A) Potential polar contacts between NBD2 (green) and Kir6.2 (yellow) shown as sticks. MgADP is shown as cyan spheres. The EM density for the residues at the interface has been contoured at 3 σ. (B) Putative interface formed between TMD0 and TMD2 in the quatrefoil structure. On the left, helices TMD3 and TM15 are in the foreground. The image on the right is rotated 160° to show interactions between TM16 and TM2.
Is there any evidence for such a large rotation of the ABC core domain? Is the observed polar interface formed between Kir6.2 and NBD2 physiologically relevant? It is certainly possible that the quatrefoil structure is distorted by the rather short linker (only six amino acids) between the C terminus of SUR1 and the N terminus of Kir6.2. Lee et al. (2017) provide some evidence that their concatenated construct forms functional channels that are inhibited by ATP and tolbutamide and activated by diazoxide. Introduction of the Kir6.2-G334D ATP-binding mutant into their concatenated construct allowed them to demonstrate activation by 1 mM MgATP, so the functional connection between Kir6.2 and SUR1 was at least partially intact. However, it should be noted that a similar concatenated construct (6-glycine linker) showed reduced ATP sensitivity compared with wild-type channels (Shyng and Nichols, 1997; Cartier et al., 2001).
What evidence is there for a new interface between NBD2 and the cytoplasmic domains of Kir6.2? Whereas the resolution in the NBDs of the quatrefoil form is lower than that of the TMDs, Fig. 13 A shows that there is reasonably good density for many of the amino acid residues at this putative interaction site (in particular, H276 and H278 of Kir6.2 and R1352 of SUR1). Earlier structure–function studies demonstrated that mutations at this interface affect nucleotide activation of KATP. A nearby mutation on the external surface of NBD2, G1400R (based on the numbering in the quatrefoil structure; Fig. 13 A), completely abolishes nucleotide activation of KATP (de Wet et al., 2012). Mutations of R1352 are associated with PHHI (R1352P; Verkarre et al., 1998; Saint-Martin et al., 2015) and leucine-sensitive hypoglycemia (R1352H; Magge et al., 2004). R1352P channels do not traffic properly to the plasma membrane (Saint-Martin et al., 2015), whereas R1352H affects channel function by reducing the extent of MgADP and diazoxide activation compared with wild-type (Magge et al., 2004).
Other studies have more broadly supported the existence of interactions between the cytoplasmic regions of SUR1 and Kir6.2. Babenko et al. (1999b) showed that the distal C terminus of SUR1 contributes to ATP inhibition, suggesting proximity to the cytoplasmic domain of Kir6.2. Lodwick et al. (2014) used thermodynamic mutant cycle analysis to identify a salt bridge between K338 in Kir6.2 and E1318 in NBD2 of SUR2A. Breaking this salt bridge potentiates the effects of the KCO pinacidil and antagonizes the effects of glibenclamide. The equivalent position on SUR1 (D1354 in the quatrefoil structure) is ∼20 Å from K338, which is too far away for a salt bridge. Still, the ability to form a salt bridge, even transiently, between NBD2 and the cytoplasmic domains of Kir6.2 in functional channels suggests that a rotation of the ABC core domain of SUR1 relative to Kir6.2 of the scale suggested by the quatrefoil structure is possible during the KATP gating cycle.
The quatrefoil form also predicts the formation of a novel interface between TMD0 and TMD2. Fig. 13 B shows the interactions (mostly hydrophobic) between helices TM2–3 of TMD0 and TM15–16 of TMD2. TM15–16 are the two helices that are domain swapped in the ABC core structure and the helix between TM15 and TM16 directly contacts NBD1 (Fig. 4), suggesting a potential pathway by which occupancy of the NBSs may be transmitted through TMD0 to the pore. Further investigations are necessary to test whether these new interfaces form during KATP gating and whether such interactions are associated with channel opening.
Conclusions and open questions
The structures of KATP discussed in this review are both corroborative and revelatory. As expected, the structure of Kir6.2 is very similar to other published structures of inward rectifiers and SUR1 is a fairly typical ABC exporter. However, several features of these models are novel and/or unexpected. The structure of TMD0 was unknown before the publication of the first inhibited-state structures. It was well accepted that TMD0 could directly associate with Kir6.2. However, the precise nature of this interaction was unclear (Chan et al., 2003). Localization of interacting sites between TMD0/L0 and Kir6.2 help explain how these residues may contribute to the increase in Po when coexpressed with the pore subunit (changes in L; Fig. 2). Site-directed mutagenesis and careful electrophysiology had determined the approximate location of the inhibitory ATP binding site on Kir6.2 (reflected in KIB; Fig. 2), but the details of binding were not well modeled, probably because of the unexpected and unusual conformation adopted by ATP (Antcliff et al., 2005; Lee et al., 2017; Martin et al., 2017a). The new structures reveal the precise location of the inhibitory site relative to the putative PIP2-binding site (KPIP) and suggest a mechanism by which the two ligands may interact (either directly or through changes in pore gating; Fig. 2, parameters C and D). Perhaps most exciting is that comparison of the structures provides a new speculative model to describe activation of KATP by Mg nucleotides (Fig. 2, E) and how the increase in Po brought about by nucleotide binding can be distinct from the direct increase in Po (L) from interactions between TMD0 and Kir6.2.
Several functionally important regions of KATP remain disordered or otherwise unresolved in all of the structures. The N terminus of Kir6.2 is unresolved up to position 32 in all of the structures. The first 14 amino acids of Kir6.2 may be important for coupling to SUR1. Deleting or disrupting this region abrogates the increase in apparent affinity for nucleotide inhibition and high-affinity SU block usually conferred by SUR1 (Babenko et al., 1999a; Giblin et al., 1999; Reimann et al., 1999). Both the C terminus of Kir6.2 and the loop between TMD1 and NBD1 of SUR1 contain RKR ER retention motifs (Zerangue et al., 1999). Neither region is resolved in any of the available structures. However, based on the available structural evidence, the two regions are very far apart, disfavoring a mechanism by which the two regions mutually obscure one another to allow exit from the ER. This is consistent with data suggesting that the RKR of Kir6.2 is masked by SUR1, whereas the RKR of SUR1 binds 14–3-3 proteins to enable forward trafficking (Heusser et al., 2006).
Very few protein structures answer more questions than they raise. In keeping with this, the KATP structures provide ample fodder for further experiments as many key questions remain unanswered. What does the apo state of KATP look like? How is the structure of SUR2A different from that of SUR1 and why is its regulation by metabolism and its interaction with Kir6.2 different? How does SUR contribute to the increased apparent affinity of Kir6.2 for inhibitory nucleotide binding? How do SUs affect the intrinsic Po of Kir6.2? Most importantly, all of the structures to date represent closed channels. How do the various domains of KATP rearrange to allow for channel opening?
Isolating the KATP complex in a stable open state may prove to be difficult. Most open-state structures of Kirs were solved using channel mutations that stabilize opening (Cuello et al., 2010a,b; Whorton and MacKinnon, 2011; Bavro et al., 2012; Zubcevic et al., 2014). It is possible that the presence of a positively modulating, MgADP-bound SUR1 and specific KCOs may allow for a wild-type open KATP structure to be solved, as the presence of a Ca2+ bound RCK domain has allowed for the solution of an open-state structure of MthK (Jiang et al., 2002). However, the introduction of mutations that either reduce inhibitory binding of nucleotides to Kir6.2 (e.g., G334D; Drain et al., 1998) or increase the intrinsic Po (e.g., Kir6.2-C166S; Trapp et al., 1998) may prove necessary if an open-state structure is to be obtained.
The locations of all three classes of NBS has been unequivocally established along with the inhibitory SU site and a putative site for PIP2 binding. However, understanding the details of ligand binding alone does not illuminate the mechanism by which these ligands affect Po, how different binding events converge energetically on the channel pore to influence its activity, or how binding of one ligand may influence that of another. Future efforts may produce cryo-EM structures of still more gating intermediates of KATP. However, to fully understand the behavior of such a motley protein complex with several different modules acting in concert to change Po, structural snapshots must be supplemented with lower-resolution approaches that can follow the dynamics of functional channels. Now that multiple interacting sites between Kir6.2 and SUR1 have been described structurally, directed attempts may be made to stabilize or disrupt these interfaces to determine which properties these sites confer from SUR1 to Kir6.2 (e.g., increase in intrinsic Po, enhanced nucleotide inhibition, nucleotide activation, and PIP2 stabilization). It is time to emerge from the liquid ethane, dust off those old voltage clamps, cross-linkers, and FRET pairs, and get to work.
Acknowledgments
I would like to thank Prof. Frances Ashcroft, Dr. Phillip Stansfeld, Mr. Samuel Usher, and Dr. Natascia Vedovato for frequent discussions regarding the structure and function of KATP. All figures were prepared using PyMol (Schrodinger, LLC) and Illustrator (Adobe).
This work was supported by the European Research Council (grant 332620) and the Biotechnology and Biological Sciences Research Council (BB/R002517/1).
The author declares no competing financial interests.
Lesley C. Anson served as editor.
Note added in proof
While this manuscript was in revision, a preprint was uploaded to bioRxiv (Wu et al., 2018. bioRxiv. https://doi.org/10.1101/283440) describing three additional cryo-EM structures of a concatenated SUR1-Kir6.2 with a 39–amino acid linker. The structures were solved in the presence of (a) ATP-γ-S, (b) ATP-γ-S and glibenclamide, and (c) MgADP, vanadate, diC8 PIP2, and a KCO. The first two states are similar to the glibenclamide inhibited form (PDB accession no. 6BAA), and the structure in the presence of MgADP is most similar to the propeller structure (PDB accession no. 6C3P). At the time of this writing, the EM densities and PDB models have been submitted, but not released.
References
- Aguilar-Bryan L., Nichols C.G., Wechsler S.W., Clement J.P. IV, Boyd A.E. III, González G., Herrera-Sosa H., Nguy K., Bryan J., and Nelson D.A.. 1995. Cloning of the beta cell high-affinity sulfonylurea receptor: a regulator of insulin secretion. Science. 268:423–426. 10.1126/science.7716547 [DOI] [PubMed] [Google Scholar]
- Ämmälä C., Bokvist K., Galt S., and Rorsman P.. 1991. Inhibition of ATP-regulated K(+)-channels by a photoactivatable ATP-analogue in mouse pancreatic beta-cells. Biochim. Biophys. Acta. 1092:347–349. 10.1016/S0167-4889(97)90011-2 [DOI] [PubMed] [Google Scholar]
- Antcliff J.F., Haider S., Proks P., Sansom M.S., and Ashcroft F.M.. 2005. Functional analysis of a structural model of the ATP-binding site of the KATP channel Kir6.2 subunit. EMBO J. 24:229–239. 10.1038/sj.emboj.7600487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkhammar P., Nilsson T., Rorsman P., and Berggren P.O.. 1987. Inhibition of ATP-regulated K+ channels precedes depolarization-induced increase in cytoplasmic free Ca2+ concentration in pancreatic beta-cells. J. Biol. Chem. 262:5448–5454. [PubMed] [Google Scholar]
- Ashcroft F.M., Harrison D.E., and Ashcroft S.J.. 1984. Glucose induces closure of single potassium channels in isolated rat pancreatic beta-cells. Nature. 312:446–448. 10.1038/312446a0 [DOI] [PubMed] [Google Scholar]
- Ashcroft F.M., Ashcroft S.J., and Harrison D.E.. 1988. Properties of single potassium channels modulated by glucose in rat pancreatic beta-cells. J. Physiol. 400:501–527. 10.1113/jphysiol.1988.sp017134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashcroft F.M., Puljung M.C., and Vedovato N.. 2017. Neonatal Diabetes and the KATP Channel: From Mutation to Therapy. Trends Endocrinol. Metab. 28:377–387. 10.1016/j.tem.2017.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashfield R., Gribble F.M., Ashcroft S.J., and Ashcroft F.M.. 1999. Identification of the high-affinity tolbutamide site on the SUR1 subunit of the K(ATP) channel. Diabetes. 48:1341–1347. 10.2337/diabetes.48.6.1341 [DOI] [PubMed] [Google Scholar]
- Babenko A.P., and Bryan J.. 2003. Sur domains that associate with and gate KATP pores define a novel gatekeeper. J. Biol. Chem. 278:41577–41580. 10.1074/jbc.C300363200 [DOI] [PubMed] [Google Scholar]
- Babenko A.P., Gonzalez G., and Bryan J.. 1999a The tolbutamide site of SUR1 and a mechanism for its functional coupling to K(ATP) channel closure. FEBS Lett. 459:367–376. 10.1016/S0014-5793(99)01215-6 [DOI] [PubMed] [Google Scholar]
- Babenko A.P., Gonzalez G., and Bryan J.. 1999b Two regions of sulfonylurea receptor specify the spontaneous bursting and ATP inhibition of KATP channel isoforms. J. Biol. Chem. 274:11587–11592. 10.1074/jbc.274.17.11587 [DOI] [PubMed] [Google Scholar]
- Basso C., Vergani P., Nairn A.C., and Gadsby D.C.. 2003. Prolonged nonhydrolytic interaction of nucleotide with CFTR’s NH2-terminal nucleotide binding domain and its role in channel gating. J. Gen. Physiol. 122:333–348. 10.1085/jgp.200308798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baukrowitz T., Schulte U., Oliver D., Herlitze S., Krauter T., Tucker S.J., Ruppersberg J.P., and Fakler B.. 1998. PIP2 and PIP as determinants for ATP inhibition of KATP channels. Science. 282:1141–1144. 10.1126/science.282.5391.1141 [DOI] [PubMed] [Google Scholar]
- Bavro V.N., De Zorzi R., Schmidt M.R., Muniz J.R., Zubcevic L., Sansom M.S., Vénien-Bryan C., and Tucker S.J.. 2012. Structure of a KirBac potassium channel with an open bundle crossing indicates a mechanism of channel gating. Nat. Struct. Mol. Biol. 19:158–163. 10.1038/nsmb.2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi H., Fosset M., and Lazdunski M.. 1992. ATP/ADP binding sites are present in the sulfonylurea binding protein associated with brain ATP-sensitive K+ channels. Biochemistry. 31:6328–6332. 10.1021/bi00142a023 [DOI] [PubMed] [Google Scholar]
- Borschel W.F., Wang S., Lee S., and Nichols C.G.. 2017. Control of Kir channel gating by cytoplasmic domain interface interactions. J. Gen. Physiol. 149:561–576. 10.1085/jgp.201611719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bränström R., Leibiger I.B., Leibiger B., Corkey B.E., Berggren P.O., and Larsson O.. 1998. Long chain coenzyme A esters activate the pore-forming subunit (Kir6. 2) of the ATP-regulated potassium channel. J. Biol. Chem. 273:31395–31400. 10.1074/jbc.273.47.31395 [DOI] [PubMed] [Google Scholar]
- Cartier E.A., Conti L.R., Vandenberg C.A., and Shyng S.L.. 2001. Defective trafficking and function of KATP channels caused by a sulfonylurea receptor 1 mutation associated with persistent hyperinsulinemic hypoglycemia of infancy. Proc. Natl. Acad. Sci. USA. 98:2882–2887. 10.1073/pnas.051499698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.W., Zhang H., and Logothetis D.E.. 2003. N-terminal transmembrane domain of the SUR controls trafficking and gating of Kir6 channel subunits. EMBO J. 22:3833–3843. 10.1093/emboj/cdg376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y. 2015. Single-Particle Cryo-EM at Crystallographic Resolution. Cell. 161:450–457. 10.1016/j.cell.2015.03.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Grigorieff N., Penczek P.A., and Walz T.. 2015. A primer to single-particle cryo-electron microscopy. Cell. 161:438–449. 10.1016/j.cell.2015.03.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K.H., Tantama M., and Licht S.. 2008. Testing for violations of microscopic reversibility in ATP-sensitive potassium channel gating. J. Phys. Chem. B. 112:10314–10321. 10.1021/jp712088v [DOI] [PubMed] [Google Scholar]
- Clarke O.B., Caputo A.T., Hill A.P., Vandenberg J.I., Smith B.J., and Gulbis J.M.. 2010. Domain reorientation and rotation of an intracellular assembly regulate conduction in Kir potassium channels. Cell. 141:1018–1029. 10.1016/j.cell.2010.05.003 [DOI] [PubMed] [Google Scholar]
- Csanády L., Vergani P., and Gadsby D.C.. 2010. Strict coupling between CFTR’s catalytic cycle and gating of its Cl- ion pore revealed by distributions of open channel burst durations. Proc. Natl. Acad. Sci. USA. 107:1241–1246. 10.1073/pnas.0911061107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuello L.G., Jogini V., Cortes D.M., Pan A.C., Gagnon D.G., Dalmas O., Cordero-Morales J.F., Chakrapani S., Roux B., and Perozo E.. 2010a Structural basis for the coupling between activation and inactivation gates in K(+) channels. Nature. 466:272–275. 10.1038/nature09136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuello L.G., Jogini V., Cortes D.M., and Perozo E.. 2010b Structural mechanism of C-type inactivation in K(+) channels. Nature. 466:203–208. 10.1038/nature09153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson R.J., and Locher K.P.. 2006. Structure of a bacterial multidrug ABC transporter. Nature. 443:180–185. 10.1038/nature05155 [DOI] [PubMed] [Google Scholar]
- de Araujo E.D., Ikeda L.K., Tzvetkova S., and Kanelis V.. 2011. The first nucleotide binding domain of the sulfonylurea receptor 2A contains regulatory elements and is folded and functions as an independent module. Biochemistry. 50:6655–6666. 10.1021/bi200434d [DOI] [PubMed] [Google Scholar]
- de Araujo E.D., Alvarez C.P., López-Alonso J.P., Sooklal C.R., Stagljar M., and Kanelis V.. 2015. Phosphorylation-dependent changes in nucleotide binding, conformation, and dynamics of the first nucleotide binding domain (NBD1) of the sulfonylurea receptor 2B (SUR2B). J. Biol. Chem. 290:22699–22714. 10.1074/jbc.M114.636233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wet H., Mikhailov M.V., Fotinou C., Dreger M., Craig T.J., Vénien-Bryan C., and Ashcroft F.M.. 2007. Studies of the ATPase activity of the ABC protein SUR1. FEBS J. 274:3532–3544. 10.1111/j.1742-4658.2007.05879.x [DOI] [PubMed] [Google Scholar]
- de Wet H., Shimomura K., Aittoniemi J., Ahmad N., Lafond M., Sansom M.S., and Ashcroft F.M.. 2012. A universally conserved residue in the SUR1 subunit of the KATP channel is essential for translating nucleotide binding at SUR1 into channel opening. J. Physiol. 590:5025–5036. 10.1113/jphysiol.2012.236075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drain P., Li L., and Wang J.. 1998. KATP channel inhibition by ATP requires distinct functional domains of the cytoplasmic C terminus of the pore-forming subunit. Proc. Natl. Acad. Sci. USA. 95:13953–13958. 10.1073/pnas.95.23.13953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enkvetchakul D., and Nichols C.G.. 2003. Gating mechanism of KATP channels: function fits form. J. Gen. Physiol. 122:471–480. 10.1085/jgp.200308878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z., and Makielski J.C.. 1997. Anionic phospholipids activate ATP-sensitive potassium channels. J. Biol. Chem. 272:5388–5395. 10.1074/jbc.272.9.5388 [DOI] [PubMed] [Google Scholar]
- Fan Z., and Makielski J.C.. 1999. Phosphoinositides decrease ATP sensitivity of the cardiac ATP-sensitive K(+) channel. A molecular probe for the mechanism of ATP-sensitive inhibition. J. Gen. Physiol. 114:251–269. 10.1085/jgp.114.2.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z., Gao L., and Wang W.. 2003. Phosphatidic acid stimulates cardiac KATP channels like phosphatidylinositols, but with novel gating kinetics. Am. J. Physiol. Cell Physiol. 284:C94–C102. 10.1152/ajpcell.00255.2002 [DOI] [PubMed] [Google Scholar]
- Fang K., Csanády L., and Chan K.W.. 2006. The N-terminal transmembrane domain (TMD0) and a cytosolic linker (L0) of sulphonylurea receptor define the unique intrinsic gating of KATP channels. J. Physiol. 576:379–389. 10.1113/jphysiol.2006.112748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotinou C., Aittoniemi J., de Wet H., Polidori A., Pucci B., Sansom M.S., Vénien-Bryan C., and Ashcroft F.M.. 2013. Tetrameric structure of SUR2B revealed by electron microscopy of oriented single particles. FEBS J. 280:1051–1063. 10.1111/febs.12097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giblin J.P., Leaney J.L., and Tinker A.. 1999. The molecular assembly of ATP-sensitive potassium channels. Determinants on the pore forming subunit. J. Biol. Chem. 274:22652–22659. 10.1074/jbc.274.32.22652 [DOI] [PubMed] [Google Scholar]
- Gribble F.M., Tucker S.J., Haug T., and Ashcroft F.M.. 1998. MgATP activates the beta cell KATP channel by interaction with its SUR1 subunit. Proc. Natl. Acad. Sci. USA. 95:7185–7190. 10.1073/pnas.95.12.7185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribble F.M., Reimann F., Ashfield R., and Ashcroft F.M.. 2000. Nucleotide modulation of pinacidil stimulation of the cloned K(ATP) channel Kir6.2/SUR2A. Mol. Pharmacol. 57:1256–1261. [PubMed] [Google Scholar]
- Hambrock A., Kayar T., Stumpp D., and Osswald H.. 2004. Effect of two amino acids in TM17 of Sulfonylurea receptor SUR1 on the binding of ATP-sensitive K+ channel modulators. Diabetes. 53(Suppl 3):S128–S134. 10.2337/diabetes.53.suppl_3.S128 [DOI] [PubMed] [Google Scholar]
- Hattori M., and Gouaux E.. 2012. Molecular mechanism of ATP binding and ion channel activation in P2X receptors. Nature. 485:207–212. 10.1038/nature11010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heginbotham L., Lu Z., Abramson T., and MacKinnon R.. 1994. Mutations in the K+ channel signature sequence. Biophys. J. 66:1061–1067. 10.1016/S0006-3495(94)80887-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heusser K., Yuan H., Neagoe I., Tarasov A.I., Ashcroft F.M., and Schwappach B.. 2006. Scavenging of 14-3-3 proteins reveals their involvement in the cell-surface transport of ATP-sensitive K+ channels. J. Cell Sci. 119:4353–4363. 10.1242/jcs.03196 [DOI] [PubMed] [Google Scholar]
- Higgins C.F., and Linton K.J.. 2004. The ATP switch model for ABC transporters. Nat. Struct. Mol. Biol. 11:918–926. 10.1038/nsmb836 [DOI] [PubMed] [Google Scholar]
- Hilgemann D.W., and Ball R.. 1996. Regulation of cardiac Na+,Ca2+ exchange and KATP potassium channels by PIP2. Science. 273:956–959. 10.1126/science.273.5277.956 [DOI] [PubMed] [Google Scholar]
- Hohl M., Hürlimann L.M., Böhm S., Schöppe J., Grütter M.G., Bordignon E., and Seeger M.A.. 2014. Structural basis for allosteric cross-talk between the asymmetric nucleotide binding sites of a heterodimeric ABC exporter. Proc. Natl. Acad. Sci. USA. 111:11025–11030. 10.1073/pnas.1400485111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horrigan F.T., and Aldrich R.W.. 2002. Coupling between voltage sensor activation, Ca2+ binding and channel opening in large conductance (BK) potassium channels. J. Gen. Physiol. 120:267–305. 10.1085/jgp.20028605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki N., Gonoi T., Clement J.P. IV, Namba N., Inazawa J., Gonzalez G., Aguilar-Bryan L., Seino S., and Bryan J.. 1995. Reconstitution of IKATP: an inward rectifier subunit plus the sulfonylurea receptor. Science. 270:1166–1170. 10.1126/science.270.5239.1166 [DOI] [PubMed] [Google Scholar]
- Inagaki N., Gonoi T., and Seino S.. 1997. Subunit stoichiometry of the pancreatic beta-cell ATP-sensitive K+ channel. FEBS Lett. 409:232–236. 10.1016/S0014-5793(97)00488-2 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Lee A., Chen J., Cadene M., Chait B.T., and MacKinnon R.. 2002. Crystal structure and mechanism of a calcium-gated potassium channel. Nature. 417:515–522. 10.1038/417515a [DOI] [PubMed] [Google Scholar]
- John S.A., Weiss J.N., and Ribalet B.. 2001. Regulation of cloned ATP-sensitive K channels by adenine nucleotides and sulfonylureas: interactions between SUR1 and positively charged domains on Kir6.2. J. Gen. Physiol. 118:391–405. 10.1085/jgp.118.4.391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson Z.L., and Chen J.. 2017. Structural Basis of Substrate Recognition by the Multidrug Resistance Protein MRP1. Cell. 168:1075–1085.e9. 10.1016/j.cell.2017.01.041 [DOI] [PubMed] [Google Scholar]
- Kim Y., and Chen J.. 2018. Molecular structure of human P-glycoprotein in the ATP-bound, outward-facing conformation. Science. 359:915–919. 10.1126/science.aar7389 [DOI] [PubMed] [Google Scholar]
- Lee K.P.K., Chen J., and MacKinnon R.. 2017. Molecular structure of human KATP in complex with ATP and ADP. eLife. 6:e32481 10.7554/eLife.32481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Jaimes K.F., and Aller S.G.. 2014. Refined structures of mouse P-glycoprotein. Protein Sci. 23:34–46. 10.1002/pro.2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Wang J., and Drain P.. 2000. The I182 region of k(ir)6.2 is closely associated with ligand binding in K(ATP) channel inhibition by ATP. Biophys. J. 79:841–852. 10.1016/S0006-3495(00)76340-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Geng X., and Drain P.. 2002. Open state destabilization by ATP occupancy is mechanism speeding burst exit underlying KATP channel inhibition by ATP. J. Gen. Physiol. 119:105–116. 10.1085/jgp.119.1.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Geng X., Yonkunas M., Su A., Densmore E., Tang P., and Drain P.. 2005. Ligand-dependent linkage of the ATP site to inhibition gate closure in the KATP channel. J. Gen. Physiol. 126:285–299. 10.1085/jgp.200509289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Wu J.X., Ding D., Cheng J., Gao N., and Chen L.. 2017. Structure of a Pancreatic ATP-Sensitive Potassium Channel. Cell. 168:101–110.e10. 10.1016/j.cell.2016.12.028 [DOI] [PubMed] [Google Scholar]
- Lin Y.W., Jia T., Weinsoft A.M., and Shyng S.L.. 2003. Stabilization of the activity of ATP-sensitive potassium channels by ion pairs formed between adjacent Kir6.2 subunits. J. Gen. Physiol. 122:225–237. 10.1085/jgp.200308822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.W., Bushman J.D., Yan F.F., Haidar S., MacMullen C., Ganguly A., Stanley C.A., and Shyng S.L.. 2008. Destabilization of ATP-sensitive potassium channel activity by novel KCNJ11 mutations identified in congenital hyperinsulinism. J. Biol. Chem. 283:9146–9156. 10.1074/jbc.M708798200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Zhang Z., Csanády L., Gadsby D.C., and Chen J.. 2017. Molecular Structure of the Human CFTR Ion Channel. Cell. 169:85–95.e8. 10.1016/j.cell.2017.02.024 [DOI] [PubMed] [Google Scholar]
- Lodwick D., Rainbow R.D., Rubaiy H.N., Al Johi M., Vuister G.W., and Norman R.I.. 2014. Sulfonylurea receptors regulate the channel pore in ATP-sensitive potassium channels via an intersubunit salt bridge. Biochem. J. 464:343–354. 10.1042/BJ20140273 [DOI] [PubMed] [Google Scholar]
- López-Alonso J.P., de Araujo E.D., and Kanelis V.. 2012. NMR and fluorescence studies of drug binding to the first nucleotide binding domain of SUR2A. Biochemistry. 51:9211–9222. 10.1021/bi301019e [DOI] [PubMed] [Google Scholar]
- MacGregor G.G., Dong K., Vanoye C.G., Tang L., Giebisch G., and Hebert S.C.. 2002. Nucleotides and phospholipids compete for binding to the C terminus of KATP channels. Proc. Natl. Acad. Sci. USA. 99:2726–2731. 10.1073/pnas.042688899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magge S.N., Shyng S.L., MacMullen C., Steinkrauss L., Ganguly A., Katz L.E., and Stanley C.A.. 2004. Familial leucine-sensitive hypoglycemia of infancy due to a dominant mutation of the beta-cell sulfonylurea receptor. J. Clin. Endocrinol. Metab. 89:4450–4456. 10.1210/jc.2004-0441 [DOI] [PubMed] [Google Scholar]
- Martin G.M., Kandasamy B., DiMaio F., Yoshioka C., and Shyng S.L.. 2017a Anti-diabetic drug binding site in a mammalian KATP channel revealed by Cryo-EM. eLife. 6:e31054 10.7554/eLife.31054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G.M., Yoshioka C., Rex E.A., Fay J.F., Xie Q., Whorton M.R., Chen J.Z., and Shyng S.L.. 2017b Cryo-EM structure of the ATP-sensitive potassium channel illuminates mechanisms of assembly and gating. eLife. 6:e24149 10.7554/eLife.24149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masia R., and Nichols C.G.. 2008. Functional clustering of mutations in the dimer interface of the nucleotide binding folds of the sulfonylurea receptor. J. Biol. Chem. 283:30322–30329. 10.1074/jbc.M804318200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo M., Kioka N., Amachi T., and Ueda K.. 1999. ATP binding properties of the nucleotide-binding folds of SUR1. J. Biol. Chem. 274:37479–37482. 10.1074/jbc.274.52.37479 [DOI] [PubMed] [Google Scholar]
- Mikhailov M.V., Mikhailova E.A., and Ashcroft S.J.. 2001. Molecular structure of the glibenclamide binding site of the beta-cell K(ATP) channel. FEBS Lett. 499:154–160. 10.1016/S0014-5793(01)02538-8 [DOI] [PubMed] [Google Scholar]
- Mikhailov M.V., Campbell J.D., de Wet H., Shimomura K., Zadek B., Collins R.F., Sansom M.S., Ford R.C., and Ashcroft F.M.. 2005. 3-D structural and functional characterization of the purified KATP channel complex Kir6.2-SUR1. EMBO J. 24:4166–4175. 10.1038/sj.emboj.7600877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson T.Y., Gaines K.L., Rajan A.S., Berg M., and Boyd A.E. III. 1987. Increased cytosolic calcium. A signal for sulfonylurea-stimulated insulin release from beta cells. J. Biol. Chem. 262:2608–2612. [PubMed] [Google Scholar]
- Nichols C.G. 2016. Adenosine Triphosphate-Sensitive Potassium Currents in Heart Disease and Cardioprotection. Card. Electrophysiol. Clin. 8:323–335. 10.1016/j.ccep.2016.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma A. 1983. ATP-regulated K+ channels in cardiac muscle. Nature. 305:147–148. 10.1038/305147a0 [DOI] [PubMed] [Google Scholar]
- Oldham M.L., Grigorieff N., and Chen J.. 2016. Structure of the transporter associated with antigen processing trapped by herpes simplex virus. eLife. 5:e21829 10.7554/eLife.21829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., and Terzic A.. 2010. Quaternary structure of KATP channel SUR2A nucleotide binding domains resolved by synchrotron radiation X-ray scattering. J. Struct. Biol. 169:243–251. 10.1016/j.jsb.2009.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt E.B., Tewson P., Bruederle C.E., Skach W.R., and Shyng S.L.. 2011. N-terminal transmembrane domain of SUR1 controls gating of Kir6.2 by modulating channel sensitivity to PIP2. J. Gen. Physiol. 137:299–314. 10.1085/jgp.201010557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt E.B., Zhou Q., Gay J.W., and Shyng S.L.. 2012. Engineered interaction between SUR1 and Kir6.2 that enhances ATP sensitivity in KATP channels. J. Gen. Physiol. 140:175–187. 10.1085/jgp.201210803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proks P., Gribble F.M., Adhikari R., Tucker S.J., and Ashcroft F.M.. 1999. Involvement of the N-terminus of Kir6.2 in the inhibition of the KATP channel by ATP. J. Physiol. 514:19–25. 10.1111/j.1469-7793.1999.019af.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proks P., de Wet H., and Ashcroft F.M.. 2010. Activation of the K(ATP) channel by Mg-nucleotide interaction with SUR1. J. Gen. Physiol. 136:389–405. 10.1085/jgp.201010475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proks P., de Wet H., and Ashcroft F.M.. 2014. Sulfonylureas suppress the stimulatory action of Mg-nucleotides on Kir6.2/SUR1 but not Kir6.2/SUR2A KATP channels: a mechanistic study. J. Gen. Physiol. 144:469–486. 10.1085/jgp.201411222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proks P., Puljung M.C., Vedovato N., Sachse G., Mulvaney R., and Ashcroft F.M.. 2016. Running out of time: the decline of channel activity and nucleotide activation in adenosine triphosphate-sensitive K-channels. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371:20150426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan Y., Barszczyk A., Feng Z.P., and Sun H.S.. 2011. Current understanding of K ATP channels in neonatal diseases: focus on insulin secretion disorders. Acta Pharmacol. Sin. 32:765–780. 10.1038/aps.2011.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimann F., Tucker S.J., Proks P., and Ashcroft F.M.. 1999. Involvement of the n-terminus of Kir6.2 in coupling to the sulphonylurea receptor. J. Physiol. 518:325–336. 10.1111/j.1469-7793.1999.0325p.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimann F., Gribble F.M., and Ashcroft F.M.. 2000. Differential response of K(ATP) channels containing SUR2A or SUR2B subunits to nucleotides and pinacidil. Mol. Pharmacol. 58:1318–1325. 10.1124/mol.58.6.1318 [DOI] [PubMed] [Google Scholar]
- Ribalet B., John S.A., and Weiss J.N.. 2000. Regulation of cloned ATP-sensitive K channels by phosphorylation, MgADP, and phosphatidylinositol bisphosphate (PIP(2)): a study of channel rundown and reactivation. J. Gen. Physiol. 116:391–410. 10.1085/jgp.116.3.391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribalet B., John S.A., and Weiss J.N.. 2003. Molecular basis for Kir6.2 channel inhibition by adenine nucleotides. Biophys. J. 84:266–276. 10.1016/S0006-3495(03)74847-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohács T., Lopes C.M., Jin T., Ramdya P.P., Molnár Z., and Logothetis D.E.. 2003. Specificity of activation by phosphoinositides determines lipid regulation of Kir channels. Proc. Natl. Acad. Sci. USA. 100:745–750. 10.1073/pnas.0236364100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorsman P., and Trube G.. 1985. Glucose dependent K+-channels in pancreatic beta-cells are regulated by intracellular ATP. Pflugers Arch. 405:305–309. 10.1007/BF00595682 [DOI] [PubMed] [Google Scholar]
- Saint-Martin C., Zhou Q., Martin G.M., Vaury C., Leroy G., Arnoux J.B., de Lonlay P., Shyng S.L., and Bellanné-Chantelot C.. 2015. Monoallelic ABCC8 mutations are a common cause of diazoxide-unresponsive diffuse form of congenital hyperinsulinism. Clin. Genet. 87:448–454. 10.1111/cge.12428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakura H., Ammälä C., Smith P.A., Gribble F.M., and Ashcroft F.M.. 1995. Cloning and functional expression of the cDNA encoding a novel ATP-sensitive potassium channel subunit expressed in pancreatic beta-cells, brain, heart and skeletal muscle. FEBS Lett. 377:338–344. 10.1016/0014-5793(95)01369-5 [DOI] [PubMed] [Google Scholar]
- Schulze D., Rapedius M., Krauter T., and Baukrowitz T.. 2003. Long-chain acyl-CoA esters and phosphatidylinositol phosphates modulate ATP inhibition of KATP channels by the same mechanism. J. Physiol. 552:357–367. 10.1113/jphysiol.2003.047035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyng S., and Nichols C.G.. 1997. Octameric stoichiometry of the KATP channel complex. J. Gen. Physiol. 110:655–664. 10.1085/jgp.110.6.655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyng S.L., and Nichols C.G.. 1998. Membrane phospholipid control of nucleotide sensitivity of KATP channels. Science. 282:1138–1141. 10.1126/science.282.5391.1138 [DOI] [PubMed] [Google Scholar]
- Shyng S.L., Cukras C.A., Harwood J., and Nichols C.G.. 2000. Structural determinants of PIP(2) regulation of inward rectifier K(ATP) channels. J. Gen. Physiol. 116:599–608. 10.1085/jgp.116.5.599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P.C., Karpowich N., Millen L., Moody J.E., Rosen J., Thomas P.J., and Hunt J.F.. 2002. ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol. Cell. 10:139–149. 10.1016/S1097-2765(02)00576-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Beek J., Guskov A., and Slotboom D.J.. 2014. Structural diversity of ABC transporters. J. Gen. Physiol. 143:419–435. 10.1085/jgp.201411164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapp S., Proks P., Tucker S.J., and Ashcroft F.M.. 1998. Molecular analysis of ATP-sensitive K channel gating and implications for channel inhibition by ATP. J. Gen. Physiol. 112:333–349. 10.1085/jgp.112.3.333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker S.J., Gribble F.M., Zhao C., Trapp S., and Ashcroft F.M.. 1997. Truncation of Kir6.2 produces ATP-sensitive K+ channels in the absence of the sulphonylurea receptor. Nature. 387:179–183. 10.1038/387179a0 [DOI] [PubMed] [Google Scholar]
- Tucker S.J., Gribble F.M., Proks P., Trapp S., Ryder T.J., Haug T., Reimann F., and Ashcroft F.M.. 1998. Molecular determinants of KATP channel inhibition by ATP. EMBO J. 17:3290–3296. 10.1093/emboj/17.12.3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusnády G.E., Bakos E., Váradi A., and Sarkadi B.. 1997. Membrane topology distinguishes a subfamily of the ATP-binding cassette (ABC) transporters. FEBS Lett. 402:1–3. 10.1016/S0014-5793(96)01478-0 [DOI] [PubMed] [Google Scholar]
- Ueda K., Komine J., Matsuo M., Seino S., and Amachi T.. 1999. Cooperative binding of ATP and MgADP in the sulfonylurea receptor is modulated by glibenclamide. Proc. Natl. Acad. Sci. USA. 96:1268–1272. 10.1073/pnas.96.4.1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhde I., Toman A., Gross I., Schwanstecher C., and Schwanstecher M.. 1999. Identification of the potassium channel opener site on sulfonylurea receptors. J. Biol. Chem. 274:28079–28082. 10.1074/jbc.274.40.28079 [DOI] [PubMed] [Google Scholar]
- Vedovato N., Ashcroft F.M., and Puljung M.C.. 2015. The Nucleotide-Binding Sites of SUR1: A Mechanistic Model. Biophys. J. 109:2452–2460. 10.1016/j.bpj.2015.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh N., Lamp S.T., and Weiss J.N.. 1991. Sulfonylureas, ATP-sensitive K+ channels, and cellular K+ loss during hypoxia, ischemia, and metabolic inhibition in mammalian ventricle. Circ. Res. 69:623–637. 10.1161/01.RES.69.3.623 [DOI] [PubMed] [Google Scholar]
- Verkarre V., Fournet J.C., de Lonlay P., Gross-Morand M.S., Devillers M., Rahier J., Brunelle F., Robert J.J., Nihoul-Fékété C., Saudubray J.M., and Junien C.. 1998. Paternal mutation of the sulfonylurea receptor (SUR1) gene and maternal loss of 11p15 imprinted genes lead to persistent hyperinsulinism in focal adenomatous hyperplasia. J. Clin. Invest. 102:1286–1291. 10.1172/JCI4495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila-Carriles W.H., Zhao G., and Bryan J.. 2007. Defining a binding pocket for sulfonylureas in ATP-sensitive potassium channels. FASEB J. 21:18–25. 10.1096/fj.06-6730hyp [DOI] [PubMed] [Google Scholar]
- Wang C., Wang K., Wang W., Cui Y., and Fan Z.. 2002. Compromised ATP binding as a mechanism of phosphoinositide modulation of ATP-sensitive K+ channels. FEBS Lett. 532:177–182. 10.1016/S0014-5793(02)03671-2 [DOI] [PubMed] [Google Scholar]
- Ward A., Reyes C.L., Yu J., Roth C.B., and Chang G.. 2007. Flexibility in the ABC transporter MsbA: Alternating access with a twist. Proc. Natl. Acad. Sci. USA. 104:19005–19010. 10.1073/pnas.0709388104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whorton M.R., and MacKinnon R.. 2011. Crystal structure of the mammalian GIRK2 K+ channel and gating regulation by G proteins, PIP2, and sodium. Cell. 147:199–208. 10.1016/j.cell.2011.07.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerangue N., Schwappach B., Jan Y.N., and Jan L.Y.. 1999. A new ER trafficking signal regulates the subunit stoichiometry of plasma membrane K(ATP) channels. Neuron. 22:537–548. 10.1016/S0896-6273(00)80708-4 [DOI] [PubMed] [Google Scholar]
- Zhang Z., and Chen J.. 2016. Atomic Structure of the Cystic Fibrosis Transmembrane Conductance Regulator. Cell. 167:1586–1597.e9. 10.1016/j.cell.2016.11.014 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Liu F., and Chen J.. 2017. Conformational Changes of CFTR upon Phosphorylation and ATP Binding. Cell. 170:483–491.e8. 10.1016/j.cell.2017.06.041 [DOI] [PubMed] [Google Scholar]
- Zingman L.V., Alekseev A.E., Bienengraeber M., Hodgson D., Karger A.B., Dzeja P.P., and Terzic A.. 2001. Signaling in channel/enzyme multimers: ATPase transitions in SUR module gate ATP-sensitive K+ conductance. Neuron. 31:233–245. 10.1016/S0896-6273(01)00356-7 [DOI] [PubMed] [Google Scholar]
- Zubcevic L., Bavro V.N., Muniz J.R., Schmidt M.R., Wang S., De Zorzi R., Venien-Bryan C., Sansom M.S., Nichols C.G., and Tucker S.J.. 2014. Control of KirBac3.1 potassium channel gating at the interface between cytoplasmic domains. J. Biol. Chem. 289:143–151. 10.1074/jbc.M113.501833 [DOI] [PMC free article] [PubMed] [Google Scholar]