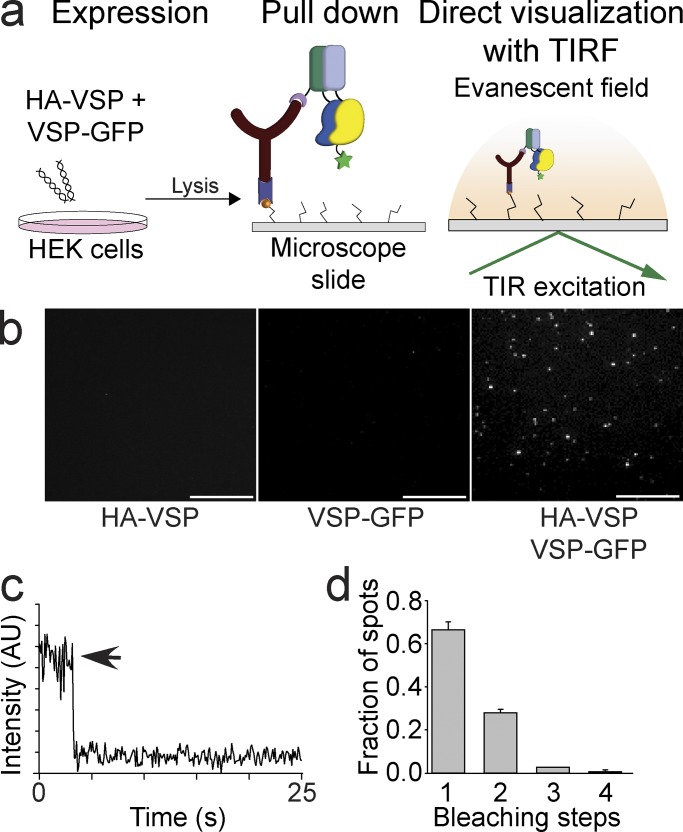
The Ciona intestinalis voltage-sensing phosphatase (Ci-VSP) was not thought to multimerize. Rayaprolu et al. show that Ci-VSP exists as a dimer and that this interaction lowers the voltage dependence of activation and alters substrate specificity.
Abstract
Multimerization is a key characteristic of most voltage-sensing proteins. The main exception was thought to be the Ciona intestinalis voltage-sensing phosphatase (Ci-VSP). In this study, we show that multimerization is also critical for Ci-VSP function. Using coimmunoprecipitation and single-molecule pull-down, we find that Ci-VSP stoichiometry is flexible. It exists as both monomers and dimers, with dimers favored at higher concentrations. We show strong dimerization via the voltage-sensing domain (VSD) and weak dimerization via the phosphatase domain. Using voltage-clamp fluorometry, we also find that VSDs cooperate to lower the voltage dependence of activation, thus favoring the activation of Ci-VSP. Finally, using activity assays, we find that dimerization alters Ci-VSP substrate specificity such that only dimeric Ci-VSP is able to dephosphorylate the 3-phosphate from PI(3,4,5)P3 or PI(3,4)P2. Our results indicate that dimerization plays a significant role in Ci-VSP function.
Introduction
The discovery of voltage-sensing phosphatases (VSPs) changed how we think about the regulation of phosphatidylinositol phosphates (PIPs). PIPs are crucial lipid second messengers, responsible for regulating many basic cellular functions including cell proliferation, migration, development, synaptic regulation, and ion channel modulation (Di Paolo and De Camilli, 2006; Balla et al., 2009; Logothetis et al., 2010; Koch and Claesson-Welsh, 2012). VSPs link voltage sensing and enzyme catalysis in a unique way. They combine an N-terminal voltage-sensing domain (VSD) resembling that of ion channels and a C-terminal phosphatase domain (PD) homologous to PTEN (phosphatase and tensin homologue deleted on chromosome 10). The VSD responds to changes in membrane potential and then activates the PD to dephosphorylate both the 3- and 5-position phosphates from PIPs (Murata et al., 2005; Iwasaki et al., 2008; Halaszovich et al., 2009; Kohout et al., 2010; Kurokawa et al., 2012; Castle et al., 2015; Grimm and Isacoff, 2016). The VSP from Ciona intestinalis was the first to be well characterized (Murata et al., 2005), but VSPs are conserved across many species, including zebrafish (Hossain et al., 2008), mice (Wu et al., 2001; Rosasco et al., 2015), and humans (Walker et al., 2001). Until the discovery of these VSPs, the connection between voltage sensing and PIP signaling was thought to be indirect. We now know that VSPs are a direct link between the electrical and chemical signaling cascades in the cell. Interestingly, the cellular processes requiring voltage regulation of PIP concentrations are still unknown. Thus, understanding how VSPs function will shed light on two critical signaling cascades and their cellular processes.
An important characteristic for any protein function is whether it multimerizes. Proteins form multimers for many reasons, including to enable, enhance, and regulate function. Some proteins, like the serotonin transporter SERT, form multimers in a dynamic and concentration-dependent manner (Anderluh et al., 2017). GABAB receptors heterodimerize to function (Jones et al., 1998; Kaupmann et al., 1998; White et al., 1998) while also forming dynamic tetramers (Calebiro et al., 2013) and heteromultimers with auxiliary proteins (Schwenk et al., 2010). Metabotropic glutamate receptors function as either homo- or heterodimers (Kunishima et al., 2000; Doumazane et al., 2011). Other proteins are less variable. Receptor tyrosine kinases, like the epidermal growth factor receptor, dimerize upon ligand binding (Yarden, 2001; Schlessinger, 2014). Voltage-gated potassium channels are obligate or strict tetramers where the pore domains from four subunits assemble to form a central pore (Liman et al., 1992; Perozo et al., 1993; Doyle et al., 1998). Interestingly, the voltage-gated proton channel, Hv, functions as a dimer, but because the pore is located within each subunit, it also conducts protons as a monomer when the dimer interface is disrupted (Tombola et al., 2010). In addition, the Hv subunits are allosterically coupled in their dimer state, creating a positive cooperativity between the subunits (Tombola et al., 2010). These differences in multimerization are important for understanding how any protein functions on a molecular level and in its physiological context.
The same is true for C. intestinalis VSP (Ci-VSP), where knowing whether the VSDs and/or the PDs cooperate with each other is important for understanding how the protein will respond to cellular signals. Although one study suggested Ci-VSP may cluster into domains (Villalba-Galea et al., 2013), most researchers have not considered multimerization when analyzing Ci-VSP function, because our previous study concluded that Ci-VSP functions as a monomer (Kohout et al., 2008). The N and C termini of Ci-VSP were even used in an Hv/VSP chimera to monomerize the Hv channel (Tombola et al., 2010). However, new research sparked a reevaluation of our model. Specifically, recent studies show that the well-known tumor suppressor and homologue of VSP, PTEN, dimerizes, and an inactivating mutation in one subunit of the dimer has a dominant-negative effect on function (Papa et al., 2014; Heinrich et al., 2015). Other studies have shown that the Hv proton channel VSDs directly interact with each other, forming a functional dimer, independent of a separate pore domain (Koch et al., 2008; Tombola et al., 2010). In addition, the crystal structure of the WT VSP VSD found four subunits in the asymmetric unit organized as two dimers (Li et al., 2014). Collectively, these results suggest both the VSP VSD and PD could interact and function as more than just a monomer.
In this study, we find that VSP subunits interact with each under high-concentration conditions, forming stable dimers. We also find that these dimers are functionally relevant. One subunit of the dimer is able to influence the VSD motions of the other subunit and the enzymatic activity changes when the dimer is favored at high-protein concentrations. We propose a model for VSP dimerization where the orientation of each subunit is important for catalysis.
Materials and methods
Molecular biology
The Ci-VSP in pSD64TF vector was provided by Y. Okamura (Osaka University, Osaka, Japan) and subcloned into pNice for the HEK293T experiments. PLC-PH–based Förster resonance energy transfer (FRET) sensor (fPLC) and TAPP-PH–based FRET sensor (fTAPP) were both provided by E.Y. Isacoff (University of California, Berkeley, Berkeley, CA). ASIC1A was provided by E. Lingueglia (Université Côte d’Azur, Valbonne, France). All mutations and N-terminal epitope tags were added using Pfu Turbo polymerase (Agilent). All DNA was confirmed by sequencing. Complementary RNA (cRNA) was transcribed using SP6 mMessage mMachine (Ambion) kits. The HEK293T cell line was verified by DNA fingerprinting by the Leibniz-Institut, Germany, and was confirmed to be a pure human cell culture with no detectable contamination of mitochondrial sequences from mouse, rat, or Chinese and Syrian hamster cells.
Voltage-clamp fluorometry (VCF)
VCF was performed as described previously (Castle et al., 2015). In brief, surgically removed Xenopus laevis oocytes were injected with 50 nl of 0.8–2.0 µg/µl cRNA. The concentrations varied based on the type of experiment and the construct being injected. Cells were incubated in ND-96 (96 mM NaCl, 2 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 50 mg/ml gentamicin, 2.5 mM sodium pyruvate, and 10 mM HEPES, pH 7.6) at 18°C for 24–36 h.
A Leica DM IRBE inverted microscope with a Leica HC Pl APO 20×/0.7 fluorescence objective was used with a Dagan CA-1B amplifier and illuminated with a Lumen Dynamics X-Cite XLED1 light source. Intensity was measured with a ThorLabs photomultiplier tube (PMT). The amplifier and light-emitting diode were controlled by a Digidata-1440A board and pClamp10.7 software package (Axon Instruments). For tetramethylrhodamine maleimide (TMRM) experiments, light was filtered through an HQ531/40 excitation filter, an HQ593/40 emission filter, and a Q562LP dichroic (Semrock). Fluorescence signals were low-pass filtered at 2 kHz through an eight-pole Bessel filter (Frequency Devices) for VCF and at 500 Hz for FRET.
On the day of the experiment, oocytes were transferred to a high-potassium solution (92 mM KCl, 0.75 mM CaCl2, 1 mM MgCl2, and 10 mM HEPES, pH 7.5). TMRM (Invitrogen) was added at a final concentration of 25 µM, and the cells were left in the dark on ice for 30 min. Labeled oocytes were then extensively washed and stored in ND-96′ (ND-96 without gentamicin or pyruvate) at 12–18°C until the end of the experiment. The VCF voltage protocol consisted of 10-mV steps starting at −150 mV and ending at 200 mV with a holding potential of −80 mV. The measured fluorescence was then plotted against the applied voltage to generate the fluorescence–voltage relationship.
Fluorescence measurement of activity
FRET-based PIP sensors (Grimm and Isacoff, 2016) were used to measure either PI(4,5)P2 (phosphatidylinositol-4,5-bisphosphate) or PI(3,4)2 (phosphatidylinositol-3,4-bisphosphate). Specifically, we used the pleckstrin homology (PH) domain from PLC for PI(4,5)P2 and the PH domain from the tandem PH domain containing protein 1 (TAPP) for PI(3,4)P2. The FRET sensors were designed with an N-terminal CFP and a C-terminal YFP and were called fPLC and fTAPP, respectively. fPLC cRNA was injected at 0.8 µg/µl, and fTAPP was injected at 0.4 µg/µl. For testing the N-terminal tagged Ci-VSPs, 0.8 µg/µl of tagged VSP cRNA was mixed with either the fTAPP or fPLC cRNA and injected. For the concentration-dependence activity assay, cRNA concentrations ranging from 0.02 to 1.6 µg/µl encoding His-tagged Ci-VSP (His-VSP) were mixed with fTAPP cRNA and injected. For the mixture of active and inactive VSP, His-VSP and the catalytically dead C363S mutant with a FLAG tag (FLAG-CS) were mixed at a ratio of 1:10 (0.1:1.0 µg/µl or 0.2:2.0 µg/µl) along with the fTAPP cRNA. In all experiments, 50 nl of the mixtures was injected into oocytes and then incubated in ND-96 for no less than 36 h.
On the day of the experiments, cells were labeled with TMRM as listed above. After incubation, oocytes were washed with ND-96′ and stored in ND-96′ containing 8 µM insulin to promote PI3 kinase activity and up-regulate PI(3,4,5)P3 levels. Ci-VSP expression was confirmed in each oocyte by VCF as described above. For PH domain experiments, light was filtered through a HQ436/20 excitation filter and directed to the objective with a 455LP dichroic (Chroma). The microscope cube did not contain an emission filter, because the ThorLabs PMT module contains its own cube. Thus, the emitted light was filtered before the PMTs with a 510-nm dichroic, an HQ480/40 emission filter for CFP, and an HQ535/30 emission filter for YFP (Chroma). The voltage protocol consisted of steps from −100 to 160 mV in irregular increments. Rest periods of 1 min between voltage steps were used to allow the cell to recover depleted PIP concentrations before the next voltage step. The resulting fluorescence was then plotted versus the voltage to generate the fluorescence versus voltage relationship.
Abs and Ab beads
6x-His epitope tag Ab (His.H8) was purchased from Invitrogen. Talon Cobalt affinity resin (635501) was purchased from Takara Bio Inc. c-Myc (9E10) Ab was purchased from Developmental Studies Hybridoma Bank. Monoclonal FLAG M2 Ab (F1804-50UG), anti-FLAG M2 affinity gel (A2220-1ML), and anti-c-Myc agarose conjugate (A7470) were purchased from Sigma-Aldrich. Biotinylated anti–HA (human influenza hemagglutinin) Ab (ab26228) was purchased from Abcam. Secondary anti-mouse conjugated to horseradish peroxidase was purchased from GE Healthcare (NA931-100UL).
Coimmunoprecipitation
Oocytes were injected with 50 nl of 0.4–0.8 µg/µl cRNA for each construct. For a mixture of constructs, injection concentrations for each construct were kept the same as the unmixed construct. Oocytes were incubated for 24–36 h in ND-96 at 18–21°C. Protein expression was confirmed using VCF as described above. After expression, four oocytes per construct were lysed in 200 µl lysis buffer (150 mM NaCl, 0.1% IGEPAL, and 20 mM Tris-base, pH 7.6, with protease inhibitors added [25 µl/ml, Pierce Protease inhibitor tablets without EDTA]). The lysate was centrifuged at 21,100 g for 15 min at 4°C. 180 µl of the supernatant was transferred into a fresh tube and centrifuged again for 5 min. 160 µl of the supernatant was split into three parts (two aliquots of 65 µl and one aliquot of 30 µl). Cobalt beads were added to one of the 65-µl aliquots, and anti-FLAG or anti-Myc beads were added to the other aliquot depending on which epitope tag was used. These samples were left to mix overnight at 4°C. The 30-µl aliquot was stored at −20°C and used as an input control. The samples with the beads were then centrifuged at 2,400 g for 5 min at 4°C to precipitate the beads. The supernatant was discarded, and the beads were washed with lysis buffer twice. After the last wash, 50 µl of 4× sample buffer was added directly to the beads and boiled for 3 min at 95°C. The samples were then centrifuged at 2,400 g for 5 min at room temperature. The supernatants were collected, and 6 µl of 1 M dithiothreitol was added to each. To the input control, 10 µl of 4× sample buffer and 5 µl of 1 M DTT were added. All samples were again boiled for 7 min at 95°C. The samples were then electrophoresed at 200 mV for 55 min on a 10% SDS-polyacrylamide gel.
Western blotting
The samples were transferred from the polyacrylamide gel onto a nitrocellulose membrane at 350 mA for 70 min using a sodium borate buffer system. After transfer, the membranes were incubated in blocking buffer (5% milk in 1× TBS-T) for 15–30 min. Blocking buffer was then replaced with the corresponding primary Ab solutions (1:2,000 dilution prepared in blocking buffer) and incubated overnight at 4°C with shaking. Membranes were then washed three times with 1× TBS-T (5–10 min). Secondary Ab solutions prepared in blocking buffer (1:4,000 dilution) were added to the membranes and incubated for 1 h at room temperature. Washes with 1× TBS-T were repeated three times (5–10 min each). Membranes were developed using WesternBright ECL reagents (Advansta).
SiMPull
For SiMPull experiments, 24 h after transfection using Lipofectamine 2000 (Invitrogen), HEK239T cells were harvested from 35-mm Petri dishes by incubating with Ca2+-free PBS buffer for 20–30 min, followed by gentle pipetting. Cells were pelleted and lysed in buffer containing 50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, protease inhibitor mixture (Thermo Scientific), and 1.5% (vol/vol) IGEPAL (Sigma). After a 30–60-min incubation at 4°C, lysate was centrifuged for 20 min at 15,000 g, and the supernatant was collected. Coverslips passivated with polyethylene glycol (PEG) (∼99%)/biotin-PEG (∼1%) and treated with Neutravidin (Thermo Scientific) were prepared as described previously (Jain et al., 2011). Biotinylated anti-HA (15 nM) was applied for 20 min and then washed out. Anti-HA dilutions and washes were done in T50 buffer containing 50 mM NaCl and 10 mM Tris, pH 7.5. Lysate, diluted at 1:10 in T50 with BSA, was then applied to the chamber and washed away after a brief incubation (∼2 min). Single molecules were imaged using a 488-nm argon laser on a total internal reflection fluorescence (TIRF) microscope with a 100× objective (Nikon). We recorded the emission light after an additional 3× magnification and passage through a double-dichroic mirror and an emission filter (525/50 for GFP) with a front-illuminated CMOS camera (Zyla 4.2 PLUS sCMOS; Andor). Movies of 250 frames were acquired at frame rates of 10–20 Hz. The imaged area was 13 × 13 µm2. Representative datasets are presented to compare conditions tested on the same day quantitatively.
Data analysis
Fluorescence traces were analyzed using Clampfit 10.7 (Molecular Devices), IGOR Pro (WaveMetrics), and Excel (Microsoft). Steady-state voltage-dependent traces were fit with either single or double Boltzmann equations. TMRM data were normalized to the maximum amplitude of the Boltzmann fits, whereas the PH domain data are shown as ΔF/F. Error bars indicate SEM. No explicit power analysis was used to predetermine the sample size for any experiments in the article, but the sample sizes used were comparable to those in previous publications. Fluorescence experiments were repeated on three different days using at least three different batches of oocytes (biological replicates) until a minimum n of 10 was reached (technical replicates). A batch of oocytes is defined as coming from a single frog. Coimmunoprecipitation experiments (from expression to Westerns) were repeated on three different days using at least three different batches of oocytes until a minimum n of 4 was reached.
Photobleaching data were analyzed using ImageJ (National Institutes of Health). Only single and diffraction-limited spots were included for analysis, and the number of spots per movie (768 × 768 pixels ∼100 µm2) was determined manually using ImageJ software. For each condition, we used at least four independent movies derived from at least four different HEK293T transfections (biological replicate). Then, the number of bleaching steps was manually determined using ImageJ software from at least three independent movies (∼50 spots per movie were quantified, technical replicate). Regions of clustered or not-fully diffraction limited spots were excluded, as well as spots that moved, showed extreme intensity fluctuations, or unequal bleaching steps. The bleaching step histograms present pooled data, taken from at least three different movies. For both, the error bars give the statistical uncertainty for a counting experiment, which is √n, for n being the number of counts (Reiner et al., 2012). For statistical analysis, when the distribution was normal and the variances of the two populations are assumed to be equal, we used the Student’s t test to determine significance.
Online supplemental material
Table S1 summarizes the voltage dependence of VSD motions. Fig. S1 shows that the catalytically inactive VSP (C363S) mutant does not alter PIP concentrations in oocytes. Fig. S2 shows the uncropped Western blots shown in Fig. 2. Fig. S3 shows Western blots from oocytes injected with increasing cRNA concentrations. The higher concentrations of cRNA led to higher protein expression, validating the protein concentrations used in Fig. 7 b. Fig. S4 shows Western blots from oocytes injected with a 1:10 ratio of WT and C363S, validating the relative protein concentration used in Fig. 7 d.
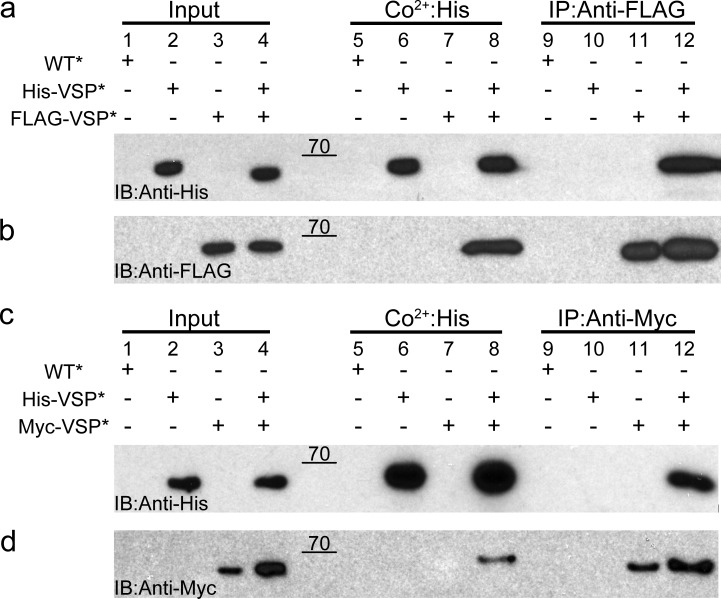
Figure 2.
Ci-VSP subunits interact with each other. (a and b) Oocytes expressing WT*, His-VSP*, FLAG-VSP*, or a His-VSP*/FLAG-VSP* mixture were processed and pulled down with Co2+ beads or coimmunoprecipitated with anti-FLAG beads. Subsequent Western blots were stained with anti-His (a) and anti-FLAG (b). The His blot shows His-VSP* coimmunoprecipitated from the His/FLAG mixture using anti-FLAG beads (lane 12), whereas the FLAG blot shows FLAG-VSP* pulled down from the His/FLAG mixture using Co2+ beads (lane 8). (c and d) Oocytes expressing WT*, His-VSP*, Myc-VSP*, or a His-VSP*/Myc-VSP* mixture were processed and pulled down with Co2+ beads or coimmunoprecipitated with anti-Myc beads. Western blots were stained with anti-His (c) and anti-Myc (d). The His blot shows His-VSP* coimmunoprecipitated from the His/Myc mixture using anti-Myc beads (lane 12), whereas the Myc blot shows Myc-VSP* pulled down from the His/Myc mixture using Co2+ beads (lane 8). These pull-downs indicate that the individual Ci-VSP subunits interact with each and form multimers on the membrane. Blots are representative of n = 4 experiments. Raw data for the Western blots are shown in Fig. S2. IB, immunoblotting; IP, immunoprecipitation.
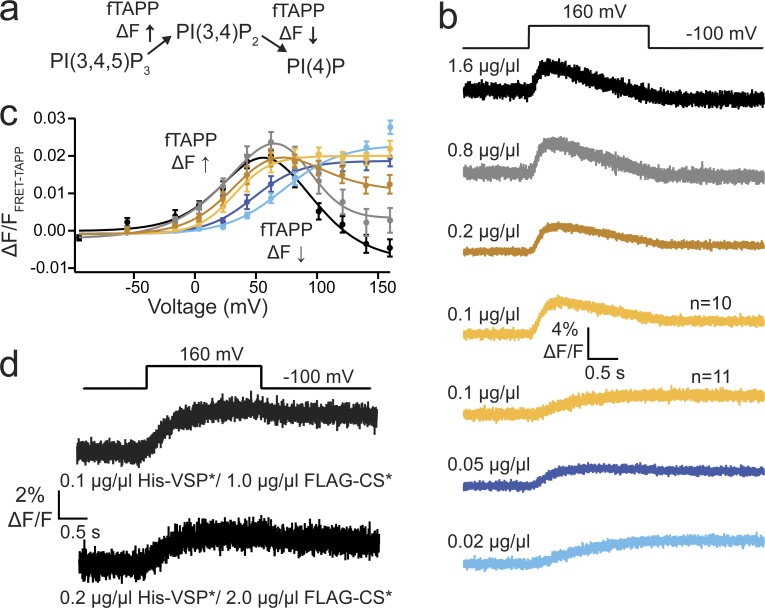
Figure 7.
Ci-VSP dimerization responsible for 3-phosphatase activity. (a) Schematic of fTAPP assay where the fTAPP FRET sensor increases FRET when PI(3,4)P2 is produced and decreases FRET when PI(3,4)P2 is depleted. (b) Averaged fTAPP FRET traces over time during a voltage step from a holding potential of −100 to 160 mV for fTAPP coexpressed with varying amounts of His-VSP cRNA: 1.6, 0.8, 0.2, 0.1, 0.05, and 0.02 µg/µl. Higher His-VSP* concentrations display both 5- and 3-phosphatase activity, whereas the lower concentrations only show 5-phosphatase activity. The 0.1 µg/µl samples were evenly split with half the cells showing both activities and half the cells showing only one. n ≥ 11. (c) ΔF/F fTAPP FRET ratio versus voltage relationships for all six cRNA concentrations. As the concentration of His-VSP* decreases, the voltage dependence of the FRET increase shifts to higher voltages whereas the FRET signal decrease disappears. Error bars represent ±SEM; n ≥ 11. Data fit with a double or single Boltzmann equation. (d) Averaged fTAPP FRET traces over time for fTAPP coexpressed with 1:10 mixtures of active and inactive VSP: 0.1:1.0 µg/µl, His-VSP*:FLAG-CS* and 0.2:2.0 µg/µl, His-VSP*:FLAG-CS*. In both cases, only a FRET increase is observed, even though the 0.2 µg/µl concentration alone in b always show both an increase and a decrease, and the 0.1 µg/µl concentration alone in b was evenly split. n ≥ 10.
Results
N-terminal tags do not alter Ci-VSP function
To differentiate the potential subunits within a Ci-VSP complex, we attached N-terminal epitope tags: a 6xHis tag, a FLAG tag, a Myc tag, and an HA tag. This allowed us to use validated Abs in our pull-downs and Westerns. We tested both the VSD motions and the catalytic activity of each tagged VSP to ensure the tags did not interfere with normal VSP function. First, the VSD motions were tested using VCF, a technique that uses environmentally sensitive fluorophores to track protein conformations in a voltage-dependent manner (Mannuzzu et al., 1996). In brief, each protein was engineered with an external cysteine in the S3-S4 loop, G214C, for labeling with TMRM (labeled proteins denoted with an asterisks). The kinetics of the VSD motions (Fig. 1 a) and the voltage dependence of those motions (Fig. 1 b and Table S1) were similar between WT Ci-VSP (WT*), His-VSP*, FLAG-VSP*, Myc-VSP*, and HA-VSP*. The N-terminal tags do not change how the VSD from Ci-VSP responds to voltage.
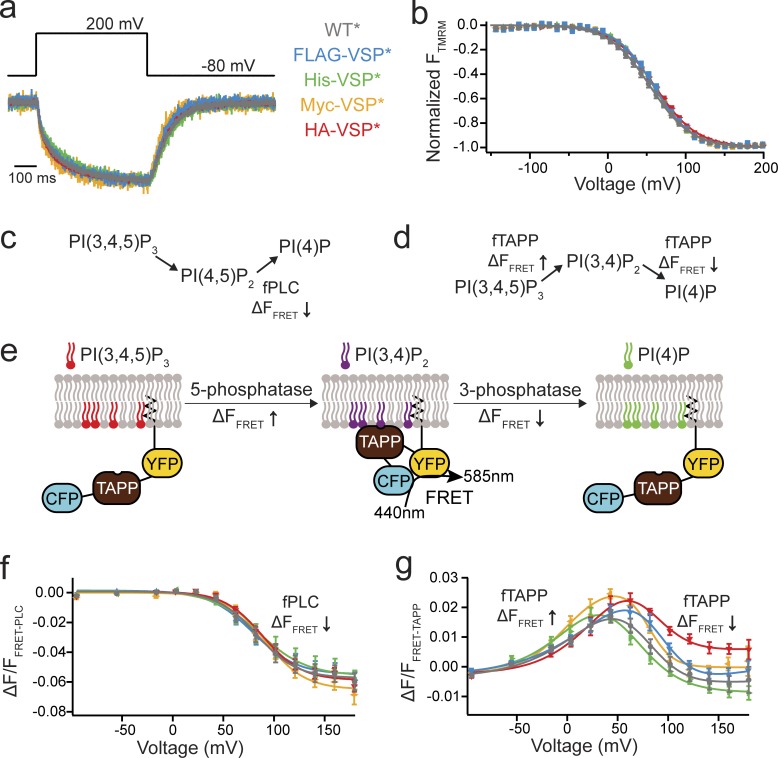
Figure 1.
N-terminal tags do not alter Ci-VSP function. (a) Representative TMRM fluorescence traces during a step from a holding potential of −80 to 200 mV for WT*, FLAG-VSP*, His-VSP*, Myc-VSP*, and HA-VSP* (where the * indicates TMRM labeling). Traces are normalized to the maximal fluorescence change. The voltage trace shows the actual voltage recorded during acquisition. (b) Normalized TMRM fluorescence versus voltage relationship. Data fit to single Boltzmann equations. None of the N-terminal tags alter the kinetics or the voltage dependence of the VSD motions, indicating they are not influencing VSP function. n ≥ 11. Some errors bars are smaller than the size of the symbols. (c and d) Schematics of the VSP reactions where the fPLC monitors the PI(4,5)P2 reactions and the fTAPP monitors the PI(3,4)P2 reactions. (e) Cartoon representation of fTAPP binding and release with increasing and decreasing PI(3,4)P2 concentrations. The binding of fTAPP to PI(3,4)P2 results in a conformational change that increases the FRET signal. Similarly, a reduction of PI(3,4)P2 results in a decrease in the FRET signal. (f) ΔF/F fPLC FRET ratio versus voltage relationship shows a fluorescence decrease (net 5-phosphatase reaction). n ≥ 9. Data fit with a single Boltzmann equation. (g) ΔF/F fTAPP FRET ratio versus voltage relationship with the fluorescence increase (net 5-phosphatase reaction) predominating at lower voltages and the fluorescence decrease (net 3-phosphatase reaction) predominating at higher voltages. n ≥ 11. Data fit with a double Boltzmann equation. Error bars are ±SEM.
Second, we tested the catalytic activity of the tagged VSPs. Ci-VSP functions as both a 3- and 5-phosphatase, catalyzing four different reactions (Fig. 1, c and d; Murata et al., 2005; Iwasaki et al., 2008; Halaszovich et al., 2009; Kohout et al., 2010; Kurokawa et al., 2012; Castle et al., 2015; Grimm and Isacoff, 2016). To monitor all Ci-VSP reactions, PH domains that bind specific PIPs were used (Stauffer et al., 1998; Halaszovich et al., 2009; Castle et al., 2015; Grimm and Isacoff, 2016). In particular, we used a FRET-based PH domain assay where CFP and YFP are fused to a PH domain anchored to the membrane through a prenylation site at the C terminus (Fig. 1 e; Sato et al., 2003; Grimm and Isacoff, 2016). As the PH domains from these sensors bind to their respective PIPs, a conformational change is induced, which results in an increased FRET signal between the CFP and YFP proteins (Fig. 1 e). As the appropriate PIP is depleted, the PH domain unbinds and the FRET signal decreases. We chose the PH domains from PLC and TAPP because they bind specifically to PI(4,5)P2 and PI(3,4)P2, respectively. Both fPLC and fTAPP change their FRET signals with changing concentrations of their respective PIPs (Grimm and Isacoff, 2016). For fPLC and WT*, the expected increase in fPLC FRET, after the 3-phosphatase activity, was too small to accurately analyze (<0.1% ΔF/F); however, we observed a robust decrease in fPLC FRET after the 5-phosphatase activity (Fig. 1 f). When testing the N-terminal tagged proteins (His-VSP*, FLAG-VSP*, Myc-VSP*, and HA-VSP*) using fPLC, we found the catalytic activity indistinguishable from WT* (Fig. 1 e). For fTAPP and WT*, the expected increase and decrease in fTAPP FRET, after the known 5- and 3-phosphatase activities, was observed (Fig. 1 f). When using fTAPP with the tagged VSPs, minor shifts in voltage dependence were seen between the tagged VSP*s and WT* (Fig. 1 g). To control for the endogenous VSP present in X. laevis oocytes, we tested both sensors with catalytically dead Ci-VSP, C363S (CS*), and found minimal changes in FRET, even at the highest voltages tested (Fig. S1). Overall, the N-terminal tags do not adversely influence either the VSD motions or catalytic activity of Ci-VSP, allowing us to use them in various assays to probe for Ci-VSP multimerization.
Coimmunoprecipitation indicates higher-order multimers in Ci-VSP
We next performed coimmunoprecipitations with the N-terminal tagged Ci-VSPs to determine whether VSP multimers are present. The cRNA for both His-VSP and FLAG-VSP or His-VSP and Myc-VSP were coinjected into X. laevis oocytes, and protein expression was confirmed by VCF 24–36 h later. Cells injected with His-VSP alone, FLAG-VSP alone, or Myc-VSP alone were tested as positive controls and to ensure no cross-reactivity existed between the N-terminal tags. Untagged Ci-VSP (WT) was also tested as a negative control. Expressing cells were lysed and tagged proteins pulled down with Co2+ beads or immunoprecipitated with anti-FLAG–coated beads or anti-Myc–coated beads to be tested by Western blot. The Co2+, anti-FLAG–, and anti-Myc–coated beads all pulled down a protein band corresponding to the expected molecular weight for monomeric Ci-VSP, ∼66 kD (Fig. 2, a and c, lanes 6 and 8; Fig. 2 b, lanes 11 and 12; Fig. 2 d, lanes 11 and 12). No nonspecific interactions were observed with any of the pull-down controls; for example, no His-VSP* was observed in the His-VSP*–only sample when pulled down with anti-FLAG beads (Fig. 2 a, lane 10). When the mixtures were pulled down using Co2+ beads, the FLAG-VSP* from the His/FLAG mixture (Fig. 2 b, lane 8) and the Myc-VSP* from the His/Myc mixture (Fig. 2 d, lane 8) showed consistent bands at ∼66 kD, indicating they interact with the His-VSP*. The reverse was also observed where the anti-FLAG–coated beads coimmunoprecipitated the His-VSP* from the His/FLAG mixture (Fig. 2 a, lane 12), and the anti-Myc–coated beads coimmunoprecipitated the His-VSP* from the His/Myc mixture (Fig. 2 c, lane 12). These results show that at least two Ci-VSP subunits interact with each other, suggesting Ci-VSP exists as multimers.
SiMPull indicates two subunits in VSP complex
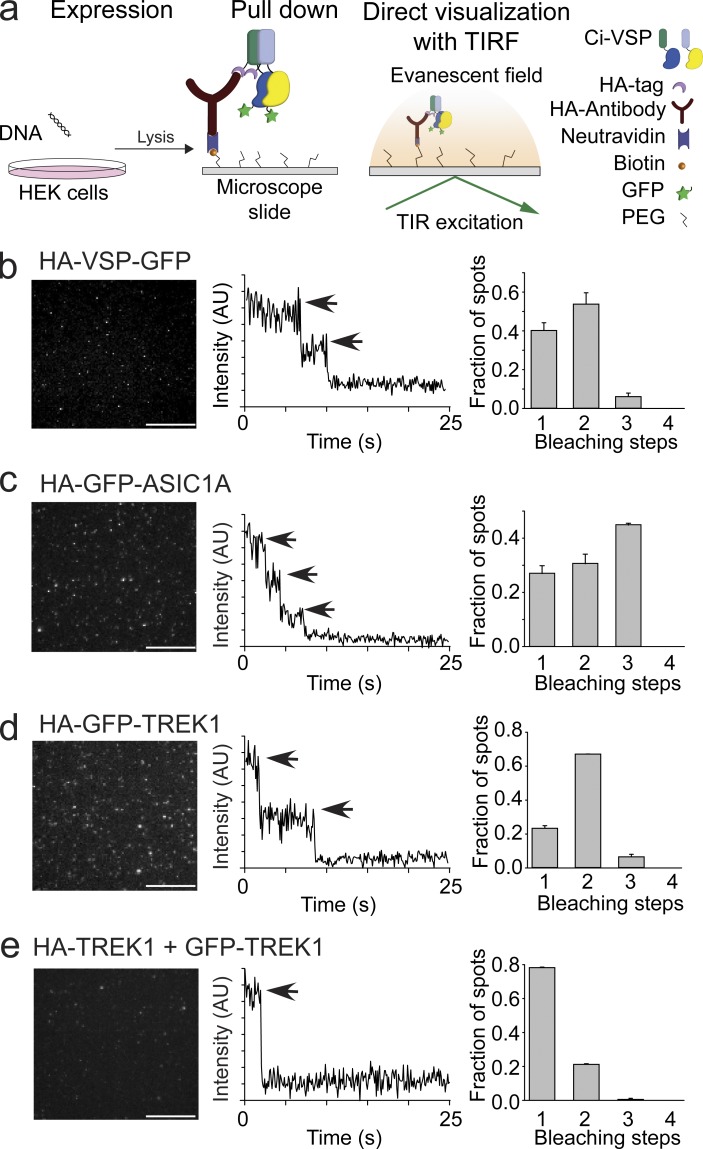
If Ci-VSP subunits are capable of interacting with each other, then a single molecule pull-down (SiMPull) assay will determine the exact number of subunits within a Ci-VSP complex. SiMPull is a recently developed technique that maintains high plasma membrane expression levels of Ci-VSP and then dilutes the lysates to count single molecules via TIRF (Fig. 3 a; Jain et al., 2011; Levitz et al., 2016). We first counted the number of subunits in a Ci-VSP complex using an N-terminal HA and C-terminal GFP tag on the same protein (HA-VSP-GFP). Neither the HA tag (Fig. 1) nor the GFP tag (Kohout et al., 2008; Ratzan et al., 2011) alters Ci-VSP function. HEK293T cells were transfected with HA-VSP-GFP, and after 24 h of expression, dilute cell lysates were flowed over coverslips previously passivated and coated with anti-HA at very low concentrations (Fig. 3 a). Because of the lysate dilution, the final concentration of bound HA-VSP-GFP at the surface of the coverslip is low enough for single-molecule subunit counting, whereas the formation of the complex was under typical, high-expression conditions. To count the subunits in the complex, the GFP fluorescence was bleached, and the resulting fluorescence intensity drops were recorded over time (see Materials and methods). The majority of HA-VSP-GFP spots bleached in two steps, indicating dimers (52%), whereas the remaining spots bleached in single steps (40%) or in three steps (8%; Fig. 3 b). In parallel with the HA-VSP-GFP, we also tagged control proteins with HA and GFP to quantitate known multimerization states: ASIC1A channels with the GFP on all subunits (trimers), TREK1 channels with the GFP on all subunits (dimers), and TREK1 channels with the GFP on only one subunit (monomer). For ASIC1A channel spots, 44% bleached in three steps, 30% bleached in two steps, and 26% bleached in a single step (Fig. 3 c). For TREK1 channel spots with the GFP on all the subunits, 69% bleached in two steps, 24% bleached in single steps, and 7% bleached in three steps (Fig. 3 d). For TREK1 channel spots with the GFP on only one subunit, 80% bleached in single steps, and 20% bleached in two steps (Fig. 3 e). This distribution of bleaching in our controls indicates a GFP maturation probability of ∼80%, which follows a binomial distribution. Ci-VSP more closely resembles the distribution seen for TREK1 channels, indicating that Ci-VSP does form dimeric complexes on the membrane. However, Ci-VSP does not follow the distribution for an obligate dimer. The larger number of VSP monomers suggests that Ci-VSP is in a dynamic equilibrium between the dimer and monomer states.
Figure 3.
Ci-VSP forms dimers. (a) Schematic of the SiMPull subunit counting assay depicting the HA-VSP-GFP expression in HEK293 cells followed by pull down of the HA tag with anti-HA on coverslips and recording of the GFP fluorescence using TIRF. TIR, total internal reflection. (b) Left: TIRF images of HA-VSP-GFP single molecules. Middle: Representative trace showing two-step photobleaching of HA-VSP-GFP. AU, arbitrary units. Right: Summary of photobleaching step distribution for HA-VSP-GFP. Two-step bleaching events indicate VSP forms dimers on the membrane. (c) SiMPull data for trimer control, HA-GFP-ASIC1A. Left: TIRF images. Middle: Representative trace showing three-step photobleaching. Right: Summary of photobleaching step distribution. (d) SiMPull data for dimer control, HA-GFP-TREK1. Left: TIRF images. Middle: Representative trace showing two-step photobleaching. Right: Summary of photobleaching step distribution. (e) SiMPull data for monomer control, HA-TREK1 pull down of TREK1-GFP. Left: TIRF images. Middle: Representative trace showing single-step photobleaching. Right: Summary of photobleaching step distribution. Error bars are ±SEM. Bars, 2 μm.
To further investigate the nature of this Ci-VSP dimer, we split the tags between two different subunits: an N-terminal HA-tagged Ci-VSP (HA-VSP) and a C-terminal GFP-tagged Ci-VSP (VSP-GFP). HA-VSP and VSP-GFP were transfected individually or mixed together in HEK293T cells (Fig. 4 a). Under these conditions, the HA-VSP will bind to the passivated coverslip and VSP-GFP will not. As expected, when each construct is expressed alone, no visible fluorescence is observed for either protein (Fig. 4 b, left and middle). When coexpressing HA-VSP with VSP-GFP, we observed fluorescent spots on the coverslips, indicating that the VSP-GFP interacts with the HA-VSP (Fig. 4 b, right). These spots were also bleached and counted (Fig. 4 c). The majority of spots (65%) were monomers (Fig. 4 d) resembling the distribution of spots for the single tagged TREK1 (Fig. 3 e, right). These results further support our conclusion that Ci-VSP subunits form dimers when present under normal cell conditions.
Figure 4.
Ci-VSP subunits pull each other down consistent with a dimer complex. (a) Schematic of the SiMPull assay where HA-VSP is coexpressed with VSP-GFP in HEK293 cells followed by pull down of the HA-VSP with anti-HA on coverslips. Any HA-VSP dimers will not be visible, and VSP-GFP dimers do not interact and are washed away, leaving the HA-VSP with VSP-GFP dimer as the only visible fluorescent spot using TIRF. TIR, total internal reflection. (b) SiMPull TIRF images of the HA-VSP alone (left), VSP-GFP alone (middle), and HA-VSP/VSP-GFP mixture (right). Fluorescent spots are only visible in the HA-VSP/VSP-GFP sample. Bars, 2 μm. (c) Representative trace showing single-step photobleaching of the HA-VSP/VSP-GFP spots in b. AU, arbitrary units. (d) Summary of the photobleaching step distribution of the HA-VSP/VSP-GFP spots in b. As expected, monomers predominate the mixture, indicating that a HA-VSP subunit pulls down a VSP-GFP subunit, forming a dimer complex. Error bars are ±SEM.
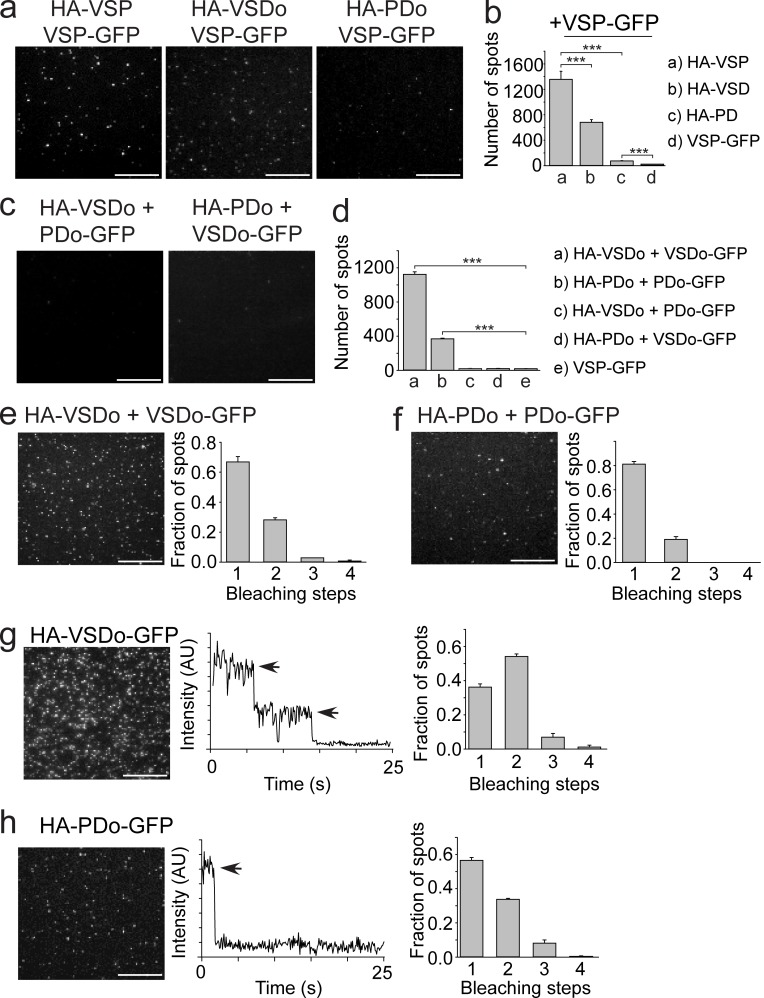
Multimerization mediated mainly by the VSD
Both the VSD and the PD are known to fold and function as independent domains (Liu et al., 2012; Li et al., 2014). To determine whether one or both domains were responsible for mediating the observed Ci-VSP subunit interactions, Ci-VSP was split into the VSD and the PD. Specifically, we tested the VSD-only protein (VSDo) and the phosphatase-only protein (PDo) using SiMPull to pull down full-length VSP-GFP. The HA-VSDo pulled down about half of the VSP-GFP (Fig. 5, a [middle] and b) compared with full-length HA-VSP (Fig. 5, a [left] and b), whereas only a small but significant fraction (P < 0.001) of VSP-GFP was pulled down by the HA-PDo (Fig. 5, a [right] and b). These results indicate that the VSD may be the main driving force behind dimer formation. Next, the ability of each domain to pull each other or a copy of itself down was tested (VSD-PD, VSD–VSD, and PD-PD). When HA-PDo is coexpressed with VSDo-GFP (Fig. 5, c [left] and d) or when HA-VSDo is coexpressed with GFP-PDo (Fig. 5 c [right] and d), no spots were detected, showing that VSD and PD are not interacting. When the HA-VSDo was coexpressed with VSDo-GFP, the GFP is clearly pulled down, giving 1,121 ± 29 spots with 65% bleaching in a single step (Fig. 5, d and e). When HA-PDo is coexpressed with PDo-GFP, we also observed spots but fewer in number (368 ± 8), and 80% bleach in a single step (Fig. 5, d and f). In both cases, the VSDo pull down and the PDo pull down, reinforcing the dimer organization on a domain level. To further characterize the domain interactions, we determined the number of subunits in HA-VSDo-GFP and HA-PDo-GFP. The HA-VSDo-GFP bleached in a combination of two steps (55%) and one step (37%), mimicking the distribution observed for full-length VSP (Fig. 5 g). The HA-PDo-GFP also bleached in one and two steps, but the distribution favored single steps (57% for single, 34% for double; Fig. 5 h). From our results, we suggest that the VSD is the main driving force behind Ci-VSP dimerization. The PDs may contribute, but that contribution is likely much weaker than the interaction between VSDs.
Figure 5.
Ci-VSP dimerization mediated mainly by VSD. (a) TIRF images of a VSP-GFP pull-down using either HA-VSP (full length, left, reused from Fig. 4 b, right), HA-VSDo (middle), or HA-PDo (right). (b) Summary of the number of single-molecule spots for each pull-down condition. The HA-VSDo was able to pull down almost half of the number of VSP-GFPs compared with full-length HA-VSP. VSP-GFP pull down with HA-PDo was significantly higher than pull down with VSP-GFP alone. Student’s t tests, ***, P < 0.001. (c) TIRF images of HA-VSDo pull-down of PDo-GFP (left) and HA-PDo pull-down of VSDo-GFP (right). No spots were detected in either experiment as expected. (d) Summary of the number of single-molecule spots for each pull-down condition shown in c, e, and f. Student’s t tests, ***, P < 0.001. (e) HA-VSDo pull-down of VSDo-GFP. TIRF single-molecule image is shown on the left, and a summary of the photobleaching step distribution is shown on the right. Spots bleached in mostly single steps, indicating that a VSD is able to pull down another. (f) HA-PDo pull-down of PDo-GFP. TIRF single-molecule image is shown on the left, and a summary of the photobleaching step distribution is shown on the right. Spots bleached in mostly single steps, indicating that a PD is able to pull down another. (g) SiMPull data for HA-VSDo-GFP. Left: TIRF image of single molecules. Middle: Representative trace showing two-step photobleaching. Right: Summary of photobleaching step distribution. Two-step bleaching events indicate VSD–VSDs form dimers on the membrane. (h) SiMPull data for HA-PDo-GFP. Left: TIRF image of single molecules. Middle: Representative trace showing single-step photobleaching. Right: Summary of photobleaching step distribution. PD–PD dimers are weaker than the VSD–VSD dimers. Error bars are ±SEM. Bars, 2 μm.
Is the dimerization of VSP important for function?
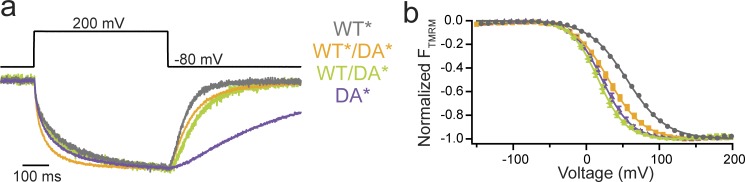
Voltage sensing
We tested this question by investigating the VSD motions. Because the VSDs help mediate the dimer interaction, each VSD within the dimer is expected to cooperate with its neighboring VSD as seen in the Hv channel (Tombola et al., 2010). VSD cooperativity was investigated with a previously characterized mutation, D331A, that significantly altered VSD kinetics and voltage dependence when compared with WT (Fig. 6 and Table S1; Kohout et al., 2010). In particular, we mixed WT* (where the * indicates TMRM labeling) with D331A (DA*) or WT (unlabeled) with DA* and tested whether the VSD motions were influenced by the mixtures. In both cases, the deactivation kinetics of the mixtures were closer to that of WT than of DA, indicating that the VSD from the WT subunit (either WT* or WT) accelerates the VSD repolarization of the DA* subunit (Fig. 6 a). Interestingly, the voltage dependences of the mixtures were shifted to lower voltages, closer to DA* than to WT* alone, suggesting that the DA subunit lowers the energy needed to move the WT subunit VSD (Figs. 6 b and S1).
Figure 6.
Ci-VSP VSD motions influence each other across the multimer complex. (a) Representative TMRM fluorescence traces during a step from a holding potential of −80 to 200 mV for WT* (G214C), DA* (G214C D331A), WT*/DA*, and WT/DA*. Traces are normalized to the maximal fluorescence change. The deactivation kinetics of the mixtures more closely resemble the WT* alone than the DA* alone, indicating that the WT* VSD influences the VSD kinetics from the DA* subunit. (b) Normalized TMRM fluorescence versus voltage relationship. Data fit to single Boltzmann equations. The voltage dependence of the mixtures more closely resembles the DA* alone than the WT* alone, suggesting that the DA* VSD more strongly influences the VSD voltage dependence. Error bars represent ±SEM; n ≥ 12. Some errors bars are smaller than the size of the symbols.
Enzymatic activity
If Ci-VSP subunit interactions change the VSD cooperativity, then catalytic activity is also expected to change as a result of dimer formation. Because the Ci-VSP dimers may not be obligate as suggested by the SiMPull results, we hypothesized that the dimers are concentration dependent. To test this hypothesis, we used the fTAPP activity assay (Fig. 7 a) under two different conditions: decreasing concentrations of active Ci-VSP alone, and mixtures between active and inactive Ci-VSPs. If the dimers are concentration dependent, the monomer should be favored at lower concentrations. For the first set of conditions, oocytes were injected with several concentrations of active His-VSP cRNA (0.02, 0.05, 0.1, 0.2, 0.8, and 1.6 µg/µl) mixed with fTAPP cRNA. As expected, after 36 h expression, all cells expressed His-VSP though in lower amounts when injected with lower cRNA concentrations (Fig. S3). For the higher His-VSP cRNA concentrations (0.2, 0.8, and 1.6 µg/µl), the fTAPP FRET assay showed the characteristic 5- and 3-phosphatase activities that have been previously reported (Murata et al., 2005; Iwasaki et al., 2008; Halaszovich et al., 2009; Kohout et al., 2010; Kurokawa et al., 2012; Castle et al., 2015; Grimm and Isacoff, 2016; Fig. 7 b, top three). At the next concentration (0.1 µg/µl), the 3-phosphatase activity was more variable, with about half the tested cells showing 3-phosphatase activity (n = 10) and half showing no 3-phosphatase activity (n = 11; Fig. 7 b, two yellow traces). The 5-phosphatase activity stayed robust regardless of concentration. At the lowest concentrations tested (0.02 and 0.05 µg/µl), only 5-phosphatase activity was observed (Fig. 7 b, bottom two). Based on these strong results, we propose that the 3-phosphatase activity is dependent on dimerization, whereas the 5-phosphatase activity is inherent in the monomer. The voltage dependence of the 5-phosphatase activity also shifted to higher voltages when Ci-VSP was at lower concentrations, suggesting a cooperative interaction between the PDs (Fig. 7 c). Under our second set of conditions, we included the inactivating mutant, Ci-VSP-C363S (CS), where we mixed active His-VSP* and inactive FLAG-CS* at a 1:10 ratio. By using a 1:10 ratio, the majority of the dimers should contain, at maximum, one His-VSP* active subunit. The N-terminal tags allowed us to confirm the mixtures were strongly biased toward the inactive FLAG-CS protein (Fig. S4). Repeating the fTAPP FRET assay with His-VSP:FLAG-CS (0.1:1.0 or 0.2:2.0 µg/µl), we detected only 5-phosphatase activity (Fig. 7 d), even though the His-VSP alone at 0.2 µg/µl showed both 3- and 5-phosphatase activity (Fig. 7 b, brown trace) and at 0.1 µg/µl showed a mix between both 3- and 5-phosphatase activity and 5-phosphatase only (Fig. 7 b, yellow traces). This result suggests the mixed active/inactive dimer either is not functional (dominant negative) and only the monomeric His-VSP is functional or two active subunits are required for the 3-phosphatase activity and the mixed active/inactive dimers are only capable of 5-phosphatase activity.
Discussion
Ci-VSP allows the cell to convert an electrical signal into a chemical signal because of its unique architecture, combining a VSD with a PD. We set out to determine whether Ci-VSP exists only as a monomer or whether Ci-VSP multimerizes into a higher-order complex. We demonstrated that Ci-VSP does form dimers. The next question we addressed was whether the dimerization is functionally important. We found that the dimers influence the VSD motions and appear to be responsible for Ci-VSP’s 3-phosphatase activity. Specifically, at low concentrations, Ci-VSP only displayed 5-phosphatase activity, whereas at higher concentrations, favoring dimerization, it showed both 3- and 5-phosphatase activity. From our activity data, not only are dimers responsible for part of Ci-VSP’s activity, but dimer formation is also driven by concentration, making Ci-VSP dimerization important for its enzymatic activity.
Multiple complementary approaches were used to address the question of Ci-VSP multimerization. Initially, we tested for interactions between Ci-VSP subunits using coimmunoprecipitation. Regardless of which epitope tag we used to differentiate Ci-VSP subunits, we found one Ci-VSP subunit could pull down another Ci-VSP subunit (Fig. 2), indicating that at a minimum, Ci-VSP could form dimers if not higher-order complexes. Our SiMPull results further indicate interactions between Ci-VSP subunits (Fig. 4) and show that the complex is a dimer (Fig. 3). On the surface, our current results appear to contradict our previous study, which suggested that Ci-VSP functions as a monomer (Kohout et al., 2008). In that work, the monomer interpretation was based on TIRF experiments where the Ci-VSP was kept at single-molecule concentrations in live oocytes. Subunit counting at high concentrations was not tested at that time because TIRF subunit counting in living cells is limited to low concentrations. This limitation is not important when testing proteins that are obligate multimers, like channels, because they will multimerize during protein processing in the ER before trafficking to the plasma membrane. However, not all proteins are obligate multimers in this way. Some proteins form multimers in a concentration- or ligand-dependent way (Yarden, 2001; Schwenk et al., 2010; Calebiro et al., 2013; Anderluh et al., 2017). As a result, the TIRF concentration restrictions limit the interpretation of the original subunit counting experiment. Here, we turned to SiMPull, giving us the advantage of exogenous concentrations of Ci-VSP on the plasma membrane followed by a diluted lysate to lower the concentration enough to count individual complexes. Under these conditions, Ci-VSP forms a mixture of monomers and dimers. In fact, the number of bleaching steps observed does not follow the same ratio of one and two bleaching steps as a protein known to only exist on the plasma membrane as a strict dimer, TREK1 (Levitz et al., 2016). Because TREK1 is an obligate dimer, we suggest that Ci-VSP is not an obligate dimer; instead, it exists on the plasma membrane as a mixture of monomers and dimers. Given that the main difference between our current results and our previous results is concentration, we propose that Ci-VSP forms dimers in a concentration-dependent manner.
To test our concentration hypothesis as well as to determine whether the dimerization was functionally relevant, we used an activity assay and manipulated the relative Ci-VSP concentrations from low to high by injecting six different concentrations of cRNA into oocytes (Fig. S3). At low concentrations, only 5-phosphatase activity was observed, whereas at high concentrations, both 3- and 5-phosphatase activities were present, as has been reported previously (Murata et al., 2005; Iwasaki et al., 2008; Halaszovich et al., 2009; Kohout et al., 2010; Kurokawa et al., 2012; Castle et al., 2015; Grimm and Isacoff, 2016; Fig. 7). This functional difference between low and high concentrations of Ci-VSP supports our hypothesis that Ci-VSP dimer formation is driven by its concentration in the membrane. It also indicates that Ci-VSP dimerization is functionally relevant. By manipulating the concentration of Ci-VSP, the enzymatic activity and any cellular signaling driven by Ci-VSP will be altered. This suggests that Ci-VSP function in a cell is controlled by expression. The physiological function of Ci-VSP remains elusive and concentration dependence could lead to a better understanding of how it contributes to cellular signaling.
Other factors may also contribute to dimerization. Previous studies have suggested Ci-VSP is regulated by PI(4,5)P2 binding to the linker between the VSD and the PD (Villalba-Galea et al., 2009; Kohout et al., 2010). How PI(4,5)P2 concentrations may impact dimerization or selectivity remain to be determined. Another factor stems from X. laevis oocyte expression of small amounts of an endogenous VSP (Xl-VSP1 and 2). Although we cannot rule out heterodimerization between Ci-VSP and Xl-VSP1 or Xl-VSP2, the overexpressed Ci-VSP is expected to overwhelm the Xl-VSP1 and 2 proteins, minimizing any contribution from theoretical heterodimers. HEK293 cells do not express an endogenous VSP (Mavrantoni et al., 2015). Further experiments are needed to explore lipid and heterodimerization influences.
We next separated Ci-VSP into its component domains, the VSD and the PD, to determine whether one or both are responsible for the dimerization. Our SiMPull results show that the VSDo can efficiently pull down full-length Ci-VSP when compared with the PDo (Fig. 5, a and b). We also found that HA-VSDo can pull down VSDo-GFP (Fig. 5 c) and that HA-VSDo-GFP forms dimers (Fig. 5 e), though not strict dimers like TREK. These data strongly support the VSD–VSD interactions being the main driving force behind Ci-VSP dimer organization. Functionally, this interaction seems to facilitate the activation of Ci-VSP because the voltage dependence of the 5-phosphatase activity is shifted to lower voltages when Ci-VSP is dimeric (Fig. 7 c). In addition, it suggests the motions of both VSDs are coordinated within the dimer in response to depolarization. This result is confirmed by the fact that the D331A mutant VSD shifts the voltage dependence of the WT VSD toward hyperpolarized values similar to those observed for the D331A alone (Fig. 6 b).
Interestingly, we found that HA-PDo pulled down PDo-GFP (Fig. 5 d) and HA-PDo-GFP formed dimers (Fig. 5 f) even though HA-PDo was not able to efficiently pull down full-length Ci-VSP (Fig. 5 b). This apparent discrepancy between the ability of the PD to pull down another PD (Fig. 5 f) and its inability to pull down the full-length Ci-VSP may stem from the PD–PD interaction being inherently weaker than the VSD–VSD interaction. However, if the PD–PD interaction is weaker, then a similar number of spots should be visible (compare Fig. 5 a, right, to Fig. 5 f). Instead, more spots are observed when just the PD pulls down another PD. Although the VSD and PD do fold independently, there is a tight coupling between the VSD and PD in full-length Ci-VSP (Villalba-Galea et al., 2009; Kohout et al., 2010). Thus, it is possible that in full-length Ci-VSP, the PD is in a resting conformation not adopted by the PDo. This is consistent with activity data because full-length Ci-VSP is off at resting potentials, whereas the Ci-VSP PDo is constitutively active. This conformational difference could reduce the affinity of the soluble PD for the PD in full-length Ci-VSP. However, this does not mean that the PDs are not interacting within the Ci-VSP dimer. Our functional data support the PDs interacting with each other within the full-length Ci-VSP dimer because only 5-phosphatase activity is observed at low concentrations, where monomers are favored, compared with 3- and 5-phosphatase activity at high concentrations, where dimers are favored. This change in activity indicates that the PDs are likely interacting and influencing the substrate specificity of their immediate neighbor.
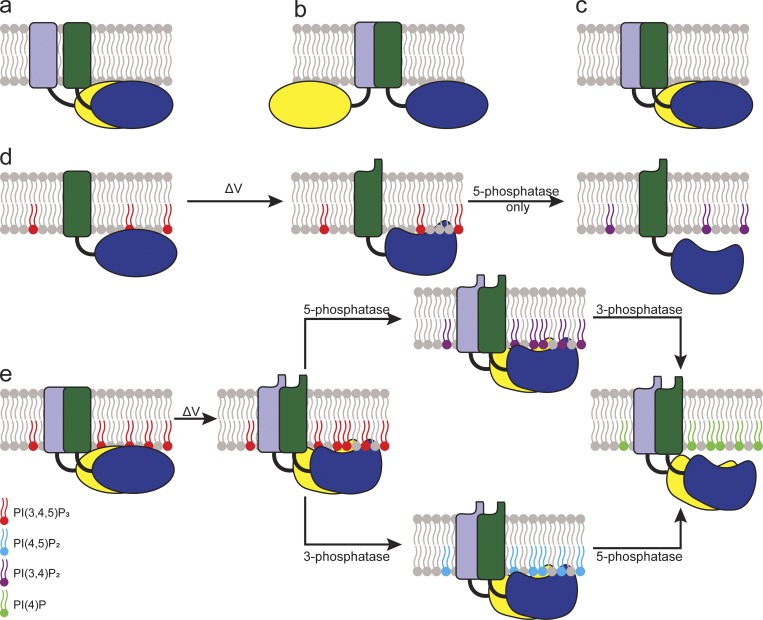
Based on our results, we suggest a model for how the individual subunits within a Ci-VSP dimer may be organized. Because we see a strong contribution of the VSD, a PD-only organization can be ruled out (Fig. 8 a). A VSD-only organization is also unlikely (Fig. 8 b) because we see PD–PD pull-down and a dimerization-dependent substrate selectivity different than that of the monomer. We cannot rule out extensive allosteric conformational changes caused by the VSD dimers leading to changes in the individual PDs. However, it is more likely that Ci-VSP subunits organize in a side-by-side fashion (Fig. 8 c). In this model, the VSD induces the dimerization of Ci-VSP in a concentration-dependent manner. This dimerization alters the PD conformations, allowing their interaction. The VSDs in the dimer cooperate, causing a coordinated motion after a membrane depolarization. The PD dimer interaction then changes the active site in each PD, allowing the 3-phosphatase activity in addition to the 5-phosphatase activity (Fig. 8, d and e).
Figure 8.
Cartoon schematics presenting possible Ci-VSP dimer organizations. (a) PD-based dimer. (b) VSD-based dimer. (c) Both VSD and PD contribute to dimer interactions. The side-by-side organization most closely agrees with our data. (d) Model for the 5-phosphatase reaction catalyzed by VSP monomer. (e) Model for the 5- and 3-phosphatase reactions catalyzed by VSP dimer.
Although the organization of Ci-VSP is important for understanding its function, it is also important for its use as a tool. Specifically, VSPs are used to manipulate PIP concentrations when studying PIP-dependent processes. By changing VSP concentrations, PIP concentrations can now be more finely tuned, targeting only 5-phosphates instead of both 3- and 5-phosphate. In addition, VSP domains have been used to generate biosensors to noninvasively track biologically relevant signaling processes within live cells (Dimitrov et al., 2007; Lundby et al., 2008; Jung et al., 2017; Lee et al., 2017). For many years, making a sensor to track membrane voltage was stymied because the sensors were not properly trafficked to the plasma membrane. When Ci-VSP was discovered, the field exploded because the Ci-VSP VSD overcame those expression issues. When the monomer result was published, this also simplified the interpretation of the voltage sensor results, because any fluorescence signal was the result of a single sensor. However, with our dimer results, we now know that the voltage biosensors using the Ci-VSP VSD form dimers in a concentration-dependent manner. This possibility will have significant implications on biosensor design and needs to be considered when interpreting the biosensor fluorescence signals.
In conclusion, we have shown that Ci-VSP dimerizes in a concentration-dependent manner. The Ci-VSP monomer is a 5-phosphatase, whereas the dimer is both a 5- and 3-phosphatase. This functional impact of dimerization on enzymatic activity could indicate that Ci-VSP signaling is regulated by expression and may ultimately play a role in its physiological function.
Supplementary Material
Acknowledgments
We thank Y. Okamura for providing the Ci-VSP cDNA, E.Y. Isacoff for the fPLC and fTAPP, and E. Lingueglia for ASIC1A.
This work was supported by start-up funds from the Vice President for Research, Creativity and Technology Transfer at Montana State University (S.C. Kohout); National Institute of General Medical Science of the National Institutes of Health grants R01GM111685 (S.C. Kohout) and P20GM103474 (K. Stengel); Fondation pour la Recherche Médicale Equipe FRM grant DEQ20170336753 (G. Sandoz); ATIP-AVENIR grants (G. Sandoz); and Fondation NRJ-Institut de France and Agence Nationale de la Recherche (Laboratory of Excellence “Ion Channel Science and Therapeutics” grant ANR-11LABX-0015-01).
The authors declare no competing financial interests.
Author contributions: G. Sandoz and S.C. Kohout conceived the experiments. V. Rayaprolu, K. Stengel, G. Sandoz, and S.C. Kohout designed experiments. V. Rayaprolu, K. Stengel, P. Royal, and S.C. Kohout collected and analyzed data. S.C. Kohout, G. Sandoz, and V. Rayaprolu interpreted data and drafted and revised the manuscript. All authors approved the final version of the manuscript.
Sharona E. Gordon served as editor.
References
- Anderluh A., Hofmaier T., Klotzsch E., Kudlacek O., Stockner T., Sitte H.H., and Schütz G.J.. 2017. Direct PIP2 binding mediates stable oligomer formation of the serotonin transporter. Nat. Commun. 8:14089 10.1038/ncomms14089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T., Szentpetery Z., and Kim Y.J.. 2009. Phosphoinositide signaling: new tools and insights. Physiology (Bethesda). 24:231–244. 10.1152/physiol.00014.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calebiro D., Rieken F., Wagner J., Sungkaworn T., Zabel U., Borzi A., Cocucci E., Zürn A., and Lohse M.J.. 2013. Single-molecule analysis of fluorescently labeled G-protein-coupled receptors reveals complexes with distinct dynamics and organization. Proc. Natl. Acad. Sci. USA. 110:743–748. 10.1073/pnas.1205798110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle P.M., Zolman K.D., and Kohout S.C.. 2015. Voltage-sensing phosphatase modulation by a C2 domain. Front. Pharmacol. 6:63 10.3389/fphar.2015.00063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov D., He Y., Mutoh H., Baker B.J., Cohen L., Akemann W., and Knöpfel T.. 2007. Engineering and characterization of an enhanced fluorescent protein voltage sensor. PLoS One. 2:e440 10.1371/journal.pone.0000440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo G., and De Camilli P.. 2006. Phosphoinositides in cell regulation and membrane dynamics. Nature. 443:651–657. 10.1038/nature05185 [DOI] [PubMed] [Google Scholar]
- Doumazane E., Scholler P., Zwier J.M., Trinquet E., Rondard P., and Pin J.-P.. 2011. A new approach to analyze cell surface protein complexes reveals specific heterodimeric metabotropic glutamate receptors. FASEB J. 25:66–77. 10.1096/fj.10-163147 [DOI] [PubMed] [Google Scholar]
- Doyle D.A., Morais Cabral J., Pfuetzner R.A., Kuo A., Gulbis J.M., Cohen S.L., Chait B.T., and MacKinnon R.. 1998. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 280:69–77. 10.1126/science.280.5360.69 [DOI] [PubMed] [Google Scholar]
- Grimm S.S., and Isacoff E.Y.. 2016. Allosteric substrate switching in a voltage-sensing lipid phosphatase. Nat. Chem. Biol. 12:261–267. 10.1038/nchembio.2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halaszovich C.R., Schreiber D.N., and Oliver D.. 2009. Ci-VSP is a depolarization-activated phosphatidylinositol-4,5-bisphosphate and phosphatidylinositol-3,4,5-trisphosphate 5′-phosphatase. J. Biol. Chem. 284:2106–2113. 10.1074/jbc.M803543200 [DOI] [PubMed] [Google Scholar]
- Heinrich F., Chakravarthy S., Nanda H., Papa A., Pandolfi P.P., Ross A.H., Harishchandra R.K., Gericke A., Lösche M., and Lö Sche M.. 2015. The PTEN Tumor Suppressor Forms Homodimers in Solution. Structure. 23:1952–1957. 10.1016/j.str.2015.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain M.I., Iwasaki H., Okochi Y., Chahine M., Higashijima S., Nagayama K., and Okamura Y.. 2008. Enzyme domain affects the movement of the voltage sensor in ascidian and zebrafish voltage-sensing phosphatases. J. Biol. Chem. 283:18248–18259. 10.1074/jbc.M706184200 [DOI] [PubMed] [Google Scholar]
- Iwasaki H., Murata Y., Kim Y., Hossain M.I., Worby C.A., Dixon J.E., McCormack T., Sasaki T., and Okamura Y.. 2008. A voltage-sensing phosphatase, Ci-VSP, which shares sequence identity with PTEN, dephosphorylates phosphatidylinositol 4,5-bisphosphate. Proc. Natl. Acad. Sci. USA. 105:7970–7975. 10.1073/pnas.0803936105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A., Liu R., Ramani B., Arauz E., Ishitsuka Y., Ragunathan K., Park J., Chen J., Xiang Y.K., and Ha T.. 2011. Probing cellular protein complexes using single-molecule pull-down. Nature. 473:484–488. 10.1038/nature10016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K.A., Borowsky B., Tamm J.A., Craig D.A., Durkin M.M., Dai M., Yao W.J., Johnson M., Gunwaldsen C., Huang L.Y., et al. . 1998. GABA(B) receptors function as a heteromeric assembly of the subunits GABA(B)R1 and GABA(B)R2. Nature. 396:674–679. 10.1038/25348 [DOI] [PubMed] [Google Scholar]
- Jung A., Rajakumar D., Yoon B.-J., and Baker B.J.. 2017. Modulating the Voltage-sensitivity of a Genetically Encoded Voltage Indicator. Exp. Neurobiol. 26:241–251. 10.5607/en.2017.26.5.241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaupmann K., Malitschek B., Schuler V., Heid J., Froestl W., Beck P., Mosbacher J., Bischoff S., Kulik A., Shigemoto R., et al. . 1998. GABA(B)-receptor subtypes assemble into functional heteromeric complexes. Nature. 396:683–687. 10.1038/25360 [DOI] [PubMed] [Google Scholar]
- Koch S., and Claesson-Welsh L.. 2012. Signal transduction by vascular endothelial growth factor receptors. Cold Spring Harb. Perspect. Med. 2:a006502 10.1101/cshperspect.a006502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch H.P., Kurokawa T., Okochi Y., Sasaki M., Okamura Y., and Larsson H.P.. 2008. Multimeric nature of voltage-gated proton channels. Proc. Natl. Acad. Sci. USA. 105:9111–9116. 10.1073/pnas.0801553105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohout S.C., Ulbrich M.H., Bell S.C., and Isacoff E.Y.. 2008. Subunit organization and functional transitions in Ci-VSP. Nat. Struct. Mol. Biol. 15:106–108. 10.1038/nsmb1320 [DOI] [PubMed] [Google Scholar]
- Kohout S.C., Bell S.C., Liu L., Xu Q., Minor D.L. Jr., and Isacoff E.Y.. 2010. Electrochemical coupling in the voltage-dependent phosphatase Ci-VSP. Nat. Chem. Biol. 6:369–375. 10.1038/nchembio.349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunishima N., Shimada Y., Tsuji Y., Sato T., Yamamoto M., Kumasaka T., Nakanishi S., Jingami H., and Morikawa K.. 2000. Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor. Nature. 407:971–977. 10.1038/35039564 [DOI] [PubMed] [Google Scholar]
- Kurokawa T., Takasuga S., Sakata S., Yamaguchi S., Horie S., Homma K.J., Sasaki T., and Okamura Y.. 2012. 3′ Phosphatase activity toward phosphatidylinositol 3,4-bisphosphate [PI(3,4)P2] by voltage-sensing phosphatase (VSP). Proc. Natl. Acad. Sci. USA. 109:10089–10094. 10.1073/pnas.1203799109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Geiller T., Jung A., Nakajima R., Song Y.-K., and Baker B.J.. 2017. Improving a genetically encoded voltage indicator by modifying the cytoplasmic charge composition. Sci. Rep. 7:8286 10.1038/s41598-017-08731-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitz J., Royal P., Comoglio Y., Wdziekonski B., Schaub S., Clemens D.M., Isacoff E.Y., and Sandoz G.. 2016. Heterodimerization within the TREK channel subfamily produces a diverse family of highly regulated potassium channels. Proc. Natl. Acad. Sci. USA. 113:4194–4199. 10.1073/pnas.1522459113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Wanderling S., Paduch M., Medovoy D., Singharoy A., McGreevy R., Villalba-Galea C.A., Hulse R.E., Roux B., Schulten K., et al. . 2014. Structural mechanism of voltage-dependent gating in an isolated voltage-sensing domain. Nat. Struct. Mol. Biol. 21:244–252. 10.1038/nsmb.2768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liman E.R., Tytgat J., and Hess P.. 1992. Subunit stoichiometry of a mammalian K+ channel determined by construction of multimeric cDNAs. Neuron. 9:861–871. 10.1016/0896-6273(92)90239-A [DOI] [PubMed] [Google Scholar]
- Liu L., Kohout S.C., Xu Q., Müller S., Kimberlin C.R., Isacoff E.Y., Minor D.L. Jr., Müller S., Kimberlin C.R., Isacoff E.Y., and Minor D.L.. 2012. A glutamate switch controls voltage-sensitive phosphatase function. Nat. Struct. Mol. Biol. 19:633–641. 10.1038/nsmb.2289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logothetis D.E., Petrou V.I., Adney S.K., and Mahajan R.. 2010. Channelopathies linked to plasma membrane phosphoinositides. Pflugers Arch. 460:321–341. 10.1007/s00424-010-0828-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundby A., Mutoh H., Dimitrov D., Akemann W., and Knöpfel T.. 2008. Engineering of a genetically encodable fluorescent voltage sensor exploiting fast Ci-VSP voltage-sensing movements. PLoS One. 3:e2514 10.1371/journal.pone.0002514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannuzzu L.M., Moronne M.M., and Isacoff E.Y.. 1996. Direct physical measure of conformational rearrangement underlying potassium channel gating. Science. 271:213–216. 10.1126/science.271.5246.213 [DOI] [PubMed] [Google Scholar]
- Mavrantoni A., Thallmair V., Leitner M.G., Schreiber D.N., Oliver D., and Halaszovich C.R.. 2015. A method to control phosphoinositides and to analyze PTEN function in living cells using voltage sensitive phosphatases. Front. Pharmacol. 6:68 10.3389/fphar.2015.00068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata Y., Iwasaki H., Sasaki M., Inaba K., and Okamura Y.. 2005. Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature. 435:1239–1243. 10.1038/nature03650 [DOI] [PubMed] [Google Scholar]
- Papa A., Wan L., Bonora M., Salmena L., Song M.S., Hobbs R.M., Lunardi A., Webster K., Ng C., Newton R.H., et al. . 2014. Cancer-associated PTEN mutants act in a dominant-negative manner to suppress PTEN protein function. Cell. 157:595–610. 10.1016/j.cell.2014.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perozo E., MacKinnon R., Bezanilla F., and Stefani E.. 1993. Gating currents from a nonconducting mutant reveal open-closed conformations in Shaker K+ channels. Neuron. 11:353–358. 10.1016/0896-6273(93)90190-3 [DOI] [PubMed] [Google Scholar]
- Ratzan W.J., Evsikov A.V., Okamura Y., and Jaffe L.A.. 2011. Voltage sensitive phosphoinositide phosphatases of Xenopus: their tissue distribution and voltage dependence. J. Cell. Physiol. 226:2740–2746. 10.1002/jcp.22854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner A., Arant R.J., and Isacoff E.Y.. 2012. Assembly stoichiometry of the GluK2/GluK5 kainate receptor complex. Cell Reports. 1:234–240. 10.1016/j.celrep.2012.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosasco M.G., Gordon S.E., and Bajjalieh S.M.. 2015. Characterization of the Functional Domains of a Mammalian Voltage-Sensitive Phosphatase. Biophys. J. 109:2480–2491. 10.1016/j.bpj.2015.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M., Ueda Y., Takagi T., and Umezawa Y.. 2003. Production of PtdInsP3 at endomembranes is triggered by receptor endocytosis. Nat. Cell Biol. 5:1016–1022. 10.1038/ncb1054 [DOI] [PubMed] [Google Scholar]
- Schlessinger J. 2014. Receptor tyrosine kinases: legacy of the first two decades. Cold Spring Harb. Perspect. Biol. 6:1–13. 10.1101/cshperspect.a008912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk J., Metz M., Zolles G., Turecek R., Fritzius T., Bildl W., Tarusawa E., Kulik A., Unger A., Ivankova K., et al. . 2010. Native GABA(B) receptors are heteromultimers with a family of auxiliary subunits. Nature. 465:231–235. 10.1038/nature08964 [DOI] [PubMed] [Google Scholar]
- Stauffer T.P., Ahn S., and Meyer T.. 1998. Receptor-induced transient reduction in plasma membrane PtdIns(4,5)P2 concentration monitored in living cells. Curr. Biol. 8:343–346. 10.1016/S0960-9822(98)70135-6 [DOI] [PubMed] [Google Scholar]
- Tombola F., Ulbrich M.H., Kohout S.C., and Isacoff E.Y.. 2010. The opening of the two pores of the Hv1 voltage-gated proton channel is tuned by cooperativity. Nat. Struct. Mol. Biol. 17:44–50. 10.1038/nsmb.1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalba-Galea C.A., Miceli F., Taglialatela M., and Bezanilla F.. 2009. Coupling between the voltage-sensing and phosphatase domains of Ci-VSP. J. Gen. Physiol. 134:5–14. 10.1085/jgp.200910215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalba-Galea C.A., Frezza L., Sandtner W., and Bezanilla F.. 2013. Sensing charges of the Ciona intestinalis voltage-sensing phosphatase. J. Gen. Physiol. 142:543–555. 10.1085/jgp.201310993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker S.M., Downes C.P., and Leslie N.R.. 2001. TPIP: a novel phosphoinositide 3-phosphatase. Biochem. J. 360:277–283. 10.1042/bj3600277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.H., Wise A., Main M.J., Green A., Fraser N.J., Disney G.H., Barnes A.A., Emson P., Foord S.M., and Marshall F.H.. 1998. Heterodimerization is required for the formation of a functional GABA(B) receptor. Nature. 396:679–682. 10.1038/25354 [DOI] [PubMed] [Google Scholar]
- Wu Y., Dowbenko D., Pisabarro M.T., Dillard-Telm L., Koeppen H., and Lasky L.A.. 2001. PTEN 2, a Golgi-associated testis-specific homologue of the PTEN tumor suppressor lipid phosphatase. J. Biol. Chem. 276:21745–21753. 10.1074/jbc.M101480200 [DOI] [PubMed] [Google Scholar]
- Yarden Y. 2001. The EGFR family and its ligands in human cancer. signalling mechanisms and therapeutic opportunities. Eur. J. Cancer. 37(Suppl 4):S3–S8. 10.1016/S0959-8049(01)00230-1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.