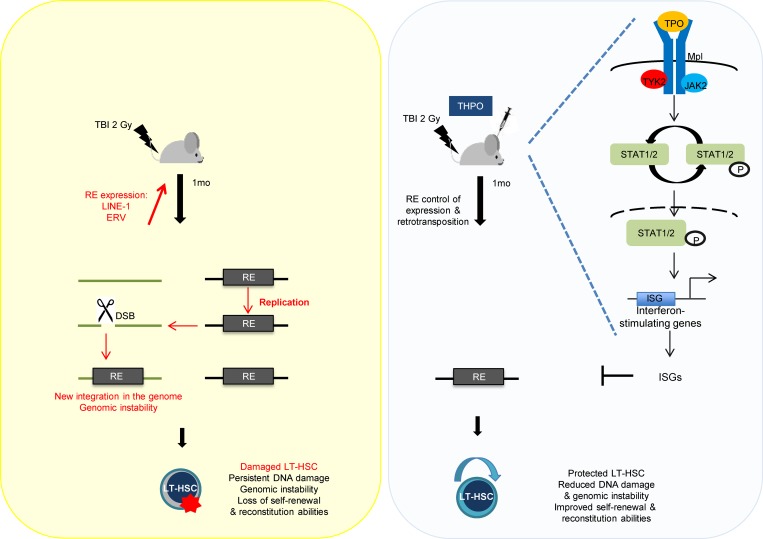
Propagation of retrotransposons induces genomic instability. Their roles in HSCs remain poorly studied. Barbieri et al. show that retrotransposon expression and mobilization are involved in long-lasting HSC impairment upon irradiation. These effects are counteracted by the self-renewal cytokine THPO through induction of interferon-like response.
Abstract
Maintenance of genomic integrity is crucial for the preservation of hematopoietic stem cell (HSC) potential. Retrotransposons, spreading in the genome through an RNA intermediate, have been associated with loss of self-renewal, aging, and DNA damage. However, their role in HSCs has not been addressed. Here, we show that mouse HSCs express various retroelements (REs), including long interspersed element-1 (L1) recent family members that further increase upon irradiation. Using mice expressing an engineered human L1 retrotransposition reporter cassette and reverse transcription inhibitors, we demonstrate that L1 retransposition occurs in vivo and is involved in irradiation-induced persistent γH2AX foci and HSC loss of function. Thus, RE represents an important intrinsic HSC threat. Furthermore, we show that RE activity is restrained by thrombopoietin, a critical HSC maintenance factor, through its ability to promote a potent interferon-like, antiviral gene response in HSCs. This uncovers a novel mechanism allowing HSCs to minimize irradiation-induced injury and reinforces the links between DNA damage, REs, and antiviral immunity.
Graphical Abstract
Introduction
Hematopoietic stem cells (HSCs) maintain homeostasis and replenish blood and the immune system throughout life. Maintenance of genomic integrity is crucial for the preservation of these functions. DNA damage in HSCs is associated with a reduced ability to reconstitute hematopoiesis and with an altered lymphoid/myeloid lineage potential (Nijnik et al., 2007; Rossi et al., 2007). The mechanisms underlying these effects are still poorly understood. This is, however, of major clinical concern. It is also crucial to understand why after radiotherapy or with age there is an accrued risk of developing bone marrow aplasia or secondary myelodysplastic syndromes. Double-strand breaks (DSBs), which are the most harmful form of DNA damage, can be generated by exogenous treatments such as ionizing radiations (IR) or internally by products of metabolism or as a result of genome replication or alteration of repair mechanisms (Rossi et al., 2007; Hoeijmakers, 2009).
Another highly dangerous, albeit poorly studied, source of endogenous DNA damage could come from the mobilization of retroelements (REs; Mita and Boeke, 2016). These sequences represent 30–50% of human and mouse genomes and can spread through an RNA intermediate using a “copy–paste” mechanism. REs can be classified into two major groups: long terminal repeat (LTR) elements, which comprise endogenous retrovirus (ERV), and non-LTR elements. This latter group includes long interspersed element-1 (LINE-1 or L1) and short interspersed elements (SINEs). ERVs exhibit relatively high activity in the mouse, whereas in humans, only the non-LTR elements are believed to be capable of retrotransposition. L1s continue to diversify genomes, on their own and through their ability to mobilize SINEs. A full-length L1 element consists of a 5′-untranslated region (5′-UTR) containing an internal promoter and two open reading frames encoding ORF1 and ORF2 proteins. ORF1 has chaperone and nucleic acid binding properties and ORF2 carries the L1 endonuclease and reverse transcription activities.
Propagation of REs in the genome requires DNA disruption. L1s are particularly strong inducers of DNA damage. Indeed, even the ORF2 protein alone, or abortive retrotransposition, can induce widespread DSBs, chronic DNA damage, and senescence (Gasior et al., 2006; Belancio et al., 2010). Derepression and mobilization of REs can lead to deletions and translocations and represent an increasingly recognized source of genomic instability (Gilbert et al., 2002, 2005; Symer et al., 2002; Iskow et al., 2010; Erwin et al., 2016). They also have a profound influence on the transcriptome and contribute to the wiring of regulatory networks in a cell-specific fashion (Han et al., 2004; Faulkner et al., 2009; Xie et al., 2013; Elbarbary et al., 2016). Given this harmful potential, RE expression is under tight control. ERVs and L1s are highly expressed in embryonic stem cells (ESCs) and germ cells, and L1 retrotransposition occurs during embryogenesis (Martin and Branciforte, 1993; Garcia-Perez et al., 2007; Kano et al., 2009; Mita and Boeke, 2016). Recent studies have also described somatic expression of L1 mRNA, as well as de novo insertions, particularly during neuronal progenitor differentiation and in the human brain (Muotri et al., 2005, 2010; Coufal et al., 2009; Belancio et al., 2010; Baillie et al., 2011; Evrony et al., 2012). Furthermore, increased L1 expression and new somatic insertions have been detected in various tumors (Iskow et al., 2010; Lee et al., 2012; Solyom et al., 2012). Previous studies have also shown that genotoxic stress can induce RE mobilization in different cell lines (Ishihara et al., 2000; Hagan et al., 2003; Farkash et al., 2006).
Similar to HSCs from irradiated animals, aged HSCs display persistent DNA damage. We and others have recently demonstrated that aged human and mouse HSCs display up-regulated expression of L1, SINE, and intracisternal A-particle (IAP) REs (Sun et al., 2014; Djeghloul et al., 2016). However, in spite of its possible relevance to HSC genomic instability, the mechanistic link between RE expression/mobilization and the accumulation of DNA damage has not been addressed. We show here that HSCs express various REs, including young L1 elements that represent an important intrinsic source of DNA damage. Indeed, L1s can successfully mobilize in vivo in HSCs upon total body irradiation (TBI) and are responsible for the long-lasting DNA damage induced by this treatment. We have previously shown that thrombopoietin (THPO), a critical HSC self-renewal factor (Qian et al., 2007; Yoshihara et al., 2007), limits TBI-induced HSC DNA damage and injury by improving DSB repair (de Laval et al., 2013). In this study, we uncover a novel mechanism by which THPO can control HSC genomic stability by restraining RE expression and mobilization. This activity is mediated by its ability to trigger a potent antiviral and IFN-like, STAT1- and STAT2-dependent signaling in HSCs. IFNs are critical for the cellular defense against viruses and are produced abundantly mainly during infections. Thus, the ability of HSCs to mount an antiviral innate immune state in response to a self-renewal cytokine may represent an important constitutive means to resist to RE-induced threat.
Results
Irradiation increases retrotransposon expression in HSCs
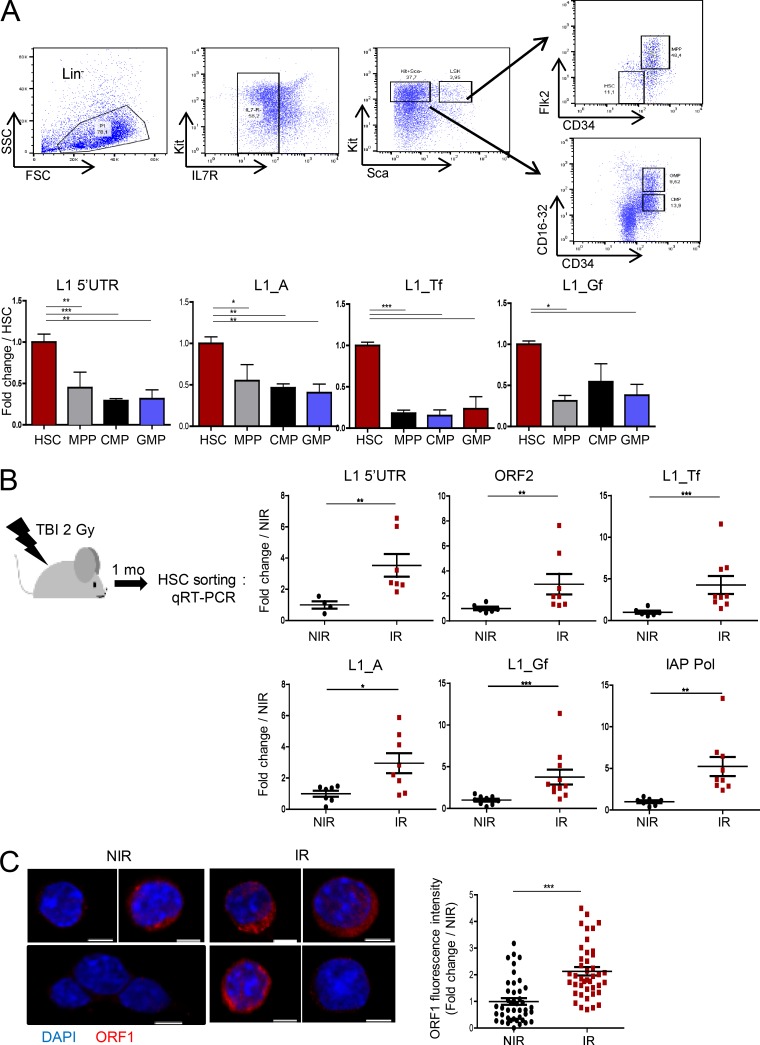
REs can be viewed as stress response genes and have been linked to DNA damage. To determine whether they could be involved in HSC genomic instability after genotoxic stress, we first assessed expression of the various REs in HSCs and various progenitors sorted by FACS using quantitative reverse transcriptase (qRT)-PCR. Analyses using primers recognizing various regions of L1 elements (5′-UTR and ORF2) or specific for active evolutionary recent mouse L1 family members (L1_A, Tf, and Gf) showed that HSCs (LSK-CD34−Flk2−) express L1 mRNA levels, including recent elements, at a significantly higher level than multipotent (LSK-CD34+Flk2+), common myeloid progenitors (CMP), and granulocyte-macrophage progenitors (GMP) myeloid progenitors or LSK cells, a mixed population of HSCs and progenitor cells (HSPCs; Fig. 1 A and Fig. S1 A). HSCs also express significant levels of two other movable REs, SINE B1 and IAP (Fig. S1 A). Quantitative PCR (qPCR) analysis performed on RNA samples before reverse transcription gave very low or undetermined cycle threshold (Ct) values, indicating that the higher RE expression found in HSCs was not a result of genomic contamination (data not depicted). In addition, expression of L1s and IAP was much lower in myeloid progenitor populations than in HSCs even though all the samples were prepared and tested together.
Figure 1.
Irradiation increases RE transcript levels in HSCs. (A) qRT-PCR analysis of L1 expression in LSK-CD34−Flk2− HSCs and progenitors. Ct values were normalized to β-actin. Results are expressed as fold change from the mean value of HSCs. Means ± SEM, n = 6 (5′-UTR and L1_A) or 3 (L1_Tf and Gf) pools of six to eight mice; two independent experiments. One-way ANOVA with Dunnett’s multiple comparison test. (B) Experimental design and RE mRNA expression in LSK-CD34−Flk2− HSCs cells isolated 1 mo after 2 Gy TBI (IR) or nontreated (NIR). Results are expressed as fold change from the NIR mean value after normalization as in A. Each dot represents an individual mouse. Means ± SEM from two (upper panels) and three (lower panels) independent experiments. Mann-Whitney test. (C) Representative images and quantification of L1 ORF1p staining in HSCs (LSK-CD34−Flk2−) isolated 1 mo after TBI or left untreated as in B. Bars, 3 µm. ImageJ was used for quantification. For each cell the fluorescence intensity was normalized to the total cell surface and then to the mean intensity of NIR cells. Each dot represents a cell. Means ± SEM from two independent experiments. Mann-Whitney test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
L1 fragments are frequently embedded into genes. The limited numbers of HSCs precluded us from performing Northern blot analysis. Thus, to confirm that HSCs express full-length L1 mRNA rather than truncated forms that would be transcribed from other genomic sites, purified RNA was reversed transcribed using a sense-strand L1–specific primer recognizing the 3′ end of ORF2 (Fig. S1 B), allowing the detection of only sense-strand L1 RNA transcripts, as described (Wissing et al., 2012). ESCs, known to express high levels of L1 elements were used as a positive control. As shown in Fig. S1 B, primers detecting L1 5′-UTR and ORF2 could amplify a 2.3 kB fragment from both ESC and HSC ORF2-directed cDNAs. No band was amplified when the reverse transcription step was omitted. This suggests the presence of full-length L1 RNAs species in HSCs.
We next examined whether ionizing radiation could affect RE expression. Attempts to assess short-time effects of irradiation led to unreproducible results, probably as a result of stress-induced variations imposed by the culture conditions in vitro. It was also impossible to assess RE expression in vivo short term after IR because TBI, even at low doses, has been shown to induce a rapid decrease in c-Kit and an increase in Sca1 (Simonnet et al., 2009), impeding HSC sorting. Therefore, we chose to analyze RE expression 1 mo after mice were subjected to a single low dose of irradiation (2 Gy; Fig. 1 B), a time and dose at which expression of these markers has recovered. In addition, under these conditions, irradiated HSCs still present DNA damage, as shown by increased γH2AX foci (Simonnet et al., 2009; de Laval et al., 2013). HSCs harvested 1 mo after TBI displayed a general increase in L1 elements, as observed with 5′-UTR and ORF2 primers and of recent L1_A, Tf, and Gf family members. TBI also increases IAP elements, as compared with nontreated cells. To confirm increased expression of active L1 elements in HSCs, we performed immunofluorescence (IF) analysis using specific antibodies directed against the mouse L1 ORF1 protein (ORF1p; Martin and Branciforte, 1993; Malki et al., 2014). This antibody was able to detect ORF1p in ESCs, but not in mouse embryonic fibroblasts (Fig. S1 C). Although few cells expressing low amounts of ORF1p with the characteristic punctate cytoplasmic staining (Malki et al., 2014) could be detected in LSK-CD34−Flk2− HSCs, ORF1p expression was significantly increased 1 mo after TBI (Fig. 1 C). This indicates that TBI induces long-lasting change in expression of L1 elements in HSCs.
Irradiation induces L1 retrotransposition in vivo in HSCs
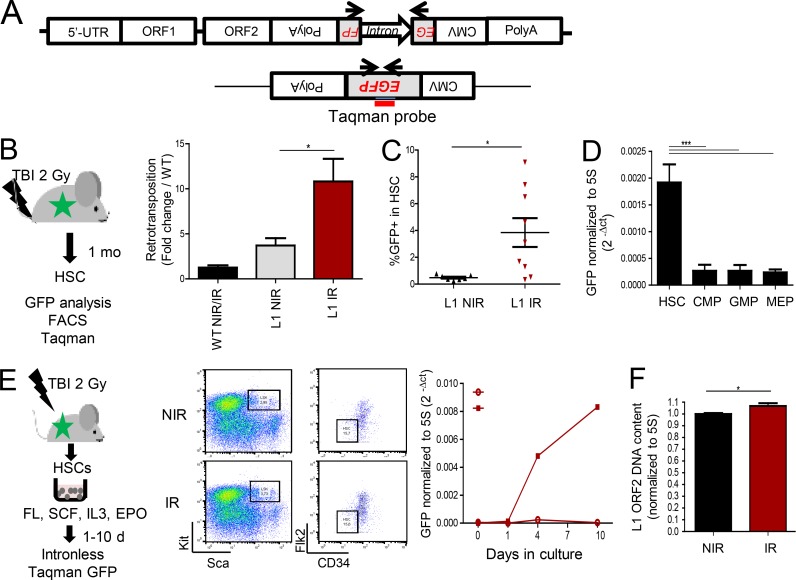
The increase of both L1 mRNA and ORF1p, which is required for retrotransposition, suggests that L1 mobilization may take place in HSCs. To test this hypothesis in vivo, we made use of a transgenic mouse model that carries an engineered human L1 harboring a GFP-based retrotransposition reporter cassette (referred to here as L1-GFP), where the human L1 transgene is under the control of its native 5′-UTR promoter (Fig. 2 A). In this model, GFP is expressed only if L1 is transcribed, spliced, reverse transcribed, and reintegrated in the genome (Okudaira et al., 2011). Retrotransposition can be monitored using a Taqman qPCR assay with GFP exon–exon junction primers and probe (Fig. 2 A). Retrotransposition was detected in LSK-CD34−Flk2− HSCs isolated from nonirradiated L1-GFP mice when compared with WT mice, albeit at low levels (Fig. 2 B). However, retrotransposition was greatly increased 1 mo after TBI (Fig. 2 B). Confirming these results, 1 mo after TBI, GFP could also be detected by FACS analysis in HSCs defined by the LSK-CD48−CD150+ phenotype (Fig. 2 C and Fig. S1 D) and in LSK cells (Fig. S1, D and E). As for L1 mRNA, the engineered human L1 retrotransposed at a higher level in HSCs than in myeloid progenitors at the basal state (Fig. 2 D). To analyze the kinetics of retrotransposition despite the loss of HSC markers 1–10 d after irradiation, HSCs were sorted immediately after TBI and cultured in vitro (Fig. 2 E, left panel). Under these conditions, no change in Sca and Kit marker expression or LSK and HSC numbers and repartition were observed (Fig. 2 E, middle panel; and Fig. S1 F). Monitoring GFP by the sensitive Taqman assay at different times of the culture showed that retrotransposition events could be detected as early as 4 d after irradiation and increased by day 10 (Fig. 2 E, right panel). Likewise, the presence of GFP could be detected in the Lin− cell population isolated 4 and 7 d after different times (Fig. S1 G). These results indicate that the increased retrotransposition is a result of a direct effect of irradiation on HSCs.
Figure 2.
Irradiation promotes active retrotransposition in HSCs. (A) The huL1-GFP transgene and position of the primers used to detect retrotransposition. (B–D) Experimental design and analysis of L1 retrotransposition by Taqman (B and D) or FACS (C) in HSCs and progenitors of L1-GFP mice before (NIR; B–D) or after TBI (IR; B and C). (B) Means ± SEM, n = 3 (WT), 7 (L1-GFP NIR), and 13 (L1-GFP IR) mice from three independent experiments. Mann-Whitney test. (C) Each triangle represents an individual mouse. Means ± SEM from two independent experiments. Mann-Whitney test. (D) n = 7 (HSC, CMP, and GMP) and 6 (MEP) mice from two independent experiments; ANOVA with Dunnett’s multiple comparison test. (E) Kinetics of L1 retrotransposition after TBI by Taqman-based qPCR assay. Left, experimental design; middle, representative FACS images of HSC sorting just after TBI; right, GFP expression normalized on 5S rDNA. Means from two independent cultures. Open circles, NIR; closed squares; IR. (F) qPCR analysis of the number of genomic copies of L1 ORF2 in HSCs 1 mo after TBI (IR) or not (NIR), normalized to 5S rDNA. n = 6 (NIR) and 8 (IR) mice. Means ± SEM from two independent experiments. Mann-Whitney test. *, P < 0.05; ***, P < 0.001.
Finally, we determined whether endogenous L1 can also mobilize after TBI, using an established qPCR assay for detecting de novo RE insertion (Muotri et al., 2010). We found that HSCs isolated from WT mice 1 mo after TBI had increased ORF2 genomic copy number (Fig. 2 F), suggesting that endogenous L1 retrotransposition could be promoted in HSCs in vivo upon irradiation.
L1 retrotransposition is involved in persistent DNA damage and loss of function of HSCs upon irradiation
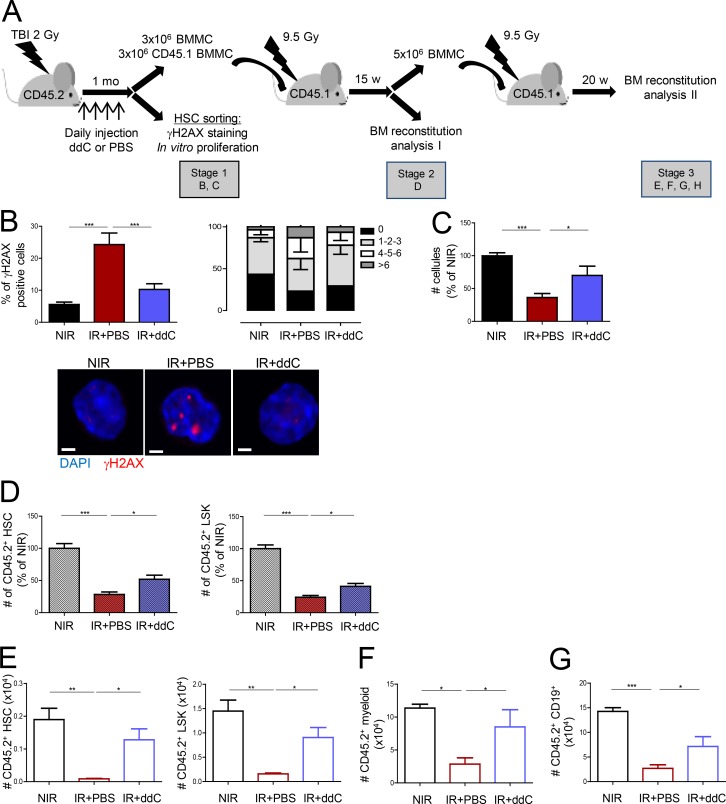
L1 mobilization in cell lines has been shown to induce widespread DSBs, as measured by the presence of γH2AX foci and senescence (Gasior et al., 2006; Belancio et al., 2010). We and others have shown that TBI induces γH2AX foci in HSCs that persist for several weeks. This is accompanied by a loss of HSC function (Simonnet et al., 2009; de Laval et al., 2013). Thus, we next analyzed whether L1-increased expression and retrotransposition could be involved in these phenomena. L1 mobilization requires its reverse transcription activity, which is carried by ORF2p (Mita and Boeke, 2016). This activity is sensitive to reverse transcription inhibitors (RTIs), including the nucleoside analogue 3′dideoxycytidine (ddC; Dai et al., 2011). Mice were subjected to TBI and treated with ddC or PBS daily for 1 mo (Fig. 3 A). As shown in Fig. 3 B, IR-induced persistent γH2AX foci in HSCs were significantly decreased upon ddC treatment. ddC also partially restored the loss of proliferation in vitro of irradiated HSCs (Fig. 3 C). Similar results were observed upon treatment with a nonnucleoside RTI, Efavirenz (EFV; Fig. S2, A and B). To determine whether inhibition of active retrotransposition could also rescue HSC proliferation in vivo, competitive transplantation experiments were performed with ddC-treated bone marrow (Fig. 3 A). At the end of treatment (stage 1), the numbers of HSCs and progenitors and their repartition in the LSK compartment was greatly altered in the irradiated mice, as compared with their nontreated counterparts (Fig. S2, C–E). This is in agreement with previous data (Shao et al., 2014; Fleenor et al., 2015). However, no difference was observed between the PBS- and ddC-treated groups at that stage. In contrast, 15 wk after reconstitution, the absolute numbers (Fig. 3 D) and the relative frequencies (Fig. S2 F) of donor LSK and HSCs found in the BM were greatly improved in the ddC-treated group, when compared with the PBS group. Secondary transplants confirmed that the capacity of HSCs to reconstitute hematopoiesis after TBI could be restored by ddC treatment (Fig. 3, E–G; and Fig. S2 G). This shows that TBI-induced retrotransposition leads to persistent γH2AX foci and negatively impacts HSC self-renewal.
Figure 3.
Retrotransposition induces HSC damage. (A) Experimental design for RTI treatment. BM, bone marrow; BMMC, BM mononuclear cell. (B and C) γH2AX foci positive cells, foci repartition, and representative images (B) and ex vivo proliferation of LSK-CD34−Flk2− HSCs isolated at stage 1 (C). Means ± SEM, n = 7 (NIR and PBS) and 8 (ddC) mice from two independent experiments. ANOVA with Dunnett’s multiple comparison test. Bars, 30 µm. (D) Total CD45.2+ donor contribution in HSCs and LSK cells at stage 2 in mice transplanted with cells from mice irradiated and treated with ddC or not. Results were normalized to the total numbers of CD45.2+ HSCs or LSK cells found in the NIR controls and represent the means ± SEM from 7 (NIR), 9 (IR+PBS), and 10 (IR+ddC) mice from two independent experiments. One-way ANOVA with Dunnett’s multiple comparison test. (E, F, and G) Total CD45.2+ donor contribution at stage 3 in HSCs and LSK cells (E), myeloid CD11b+ and Gr1+ cells (F), and CD19+ B cells (G). Means ± SEM, n = 4 (NIR and IR+PBS) and 5 (ddC) mice from one representative experiment out of two performed. One-way ANOVA with Dunnett’s multiple comparison test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Thrombopoietin restrains L1 expression and retrotransposition in HSCs
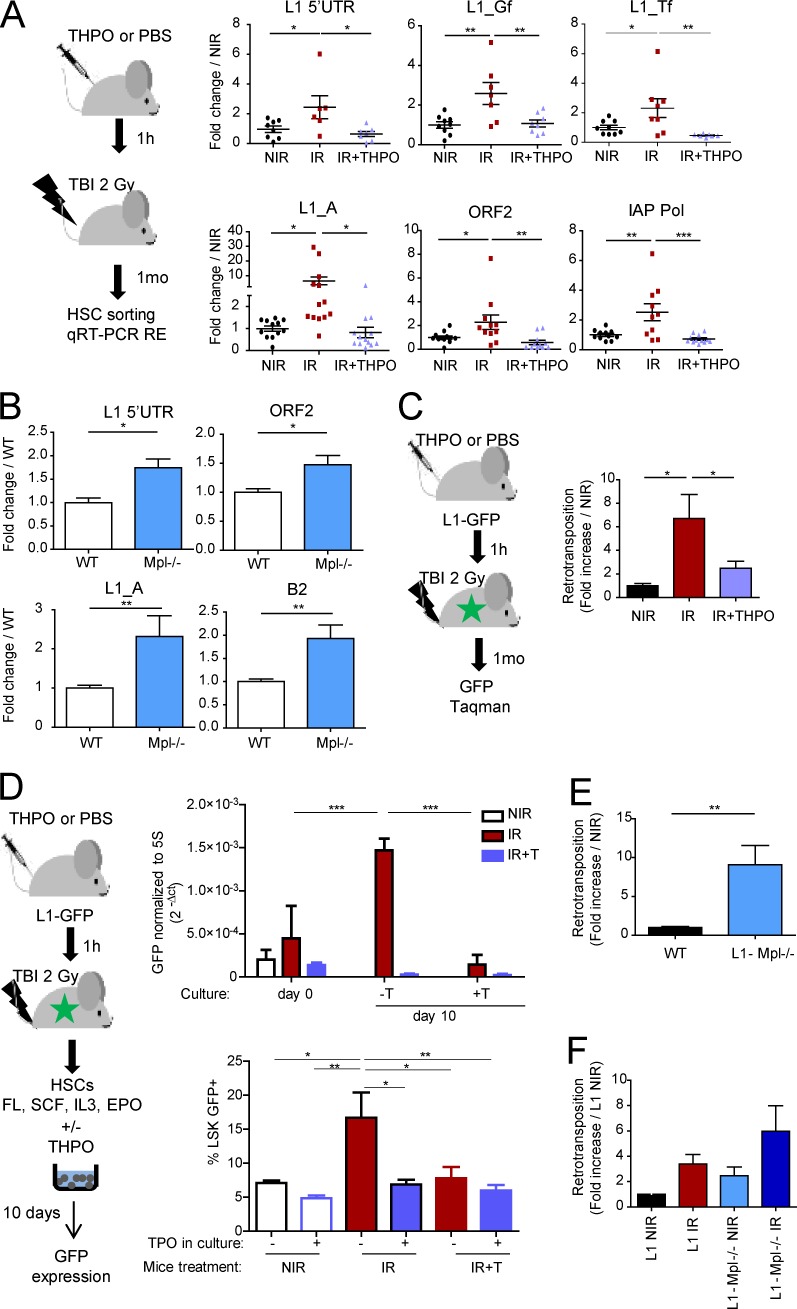
The above data suggest that HSCs may need means to protect themselves against the possible harmful potential of transcribed active transposable elements. We previously showed that one injection of THPO before irradiation could limit TBI-induced accumulation of γH2AX foci and loss of HSC function (de Laval et al., 2013). To examine whether this could be linked to an effect on RE expression and/or mobilization, mice were injected with a single dose of THPO 1 h before TBI, and RE expression was assessed 1 mo later (Fig. 4 A). We found that this treatment prevented TBI-induced increased expression of recent L1 members and IAP in HSCs. Similar results were observed in LSK cells (Fig. S3 A). Conversely, the basal RE mRNA levels were significantly increased in Mpl−/− HSCs deficient for the THPO receptor (Fig. 4 B) and in Thpo−/− LSK cells (Fig. S3 B).
Figure 4.
THPO signaling controls L1 expression and retrotransposition in vivo in HSCs. (A) Experimental design and RE mRNA expression in HSCs isolated 1 mo after 2 Gy TBI with (IR+THPO) or without (IR) THPO injection or nontreated (NIR). Each point represents an individual mouse. Results represent means ± SEM and are expressed as fold change from the NIR mean value after normalization. Data are pooled from three (upper panels) or four (lower panels) independent experiments. One-way ANOVA with Dunnett’s multiple comparison test. (B) RE expression in WT and Mpl−/− HSCs. Means ± SEM normalized to the mean values of WT mice. n = 10 (5′-UTR, ORF2, and L1_A) and 7 (B2) pools of three WT or five Mpl−/− mice from three to four independent experiments. Mann-Whitney test. (C) Experimental design and GFP Taqman assays in HSCs isolated from L1-GFP mice 1 mo after TBI with or without THPO injection. Means ± SEM, n = 7 (NIR), 10 (IR), and 11 (IR+THPO) mice from two independent experiments. One-way ANOVA with Dunnett’s multiple comparison test. (D) Experimental design and GFP expression in total cell progeny and in the LSK compartment (FACS) of L1-GFP HSCs isolated immediately after TBI with or without THPO injection and cultured for 10 d in vitro in the presence (+T) or absence of THPO (−T); two (IR) and three (NIR and IR+THPO) independent cultures. Means ± SEM. One-way ANOVA with multiple comparison tests. (E and F) L1 retrotransposition in HSCs and progenitors from L1-Mpl−/−, either nontreated or 1 mo after TBI, as indicated. Means ± SEM. (E) n = 12 (WT) and 13 (L1-Mpl−/−) mice from four independent experiments. Mann-Whitney test. (F) n = 7 (L1 and L1-Mpl−/− NIR), 9 (L1 IR), and 8 (L1-Mpl−/− IR) mice from two independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
THPO could also inhibit in vivo L1 mobilization in L1-GFP mice, as shown by the significantly reduced level of retrotransposition detected by Taqman assay 1 mo after TBI in HSCs from mice that had received THPO instead of PBS 1 h before TBI (Fig. 4 C). To determine whether this was a result of a direct effect of THPO on HSCs, HSCs were sorted immediately after TBI, with or without THPO treatment, and cultured in vitro in the presence or absence of THPO (Fig. 4 D, left panel). After 10 d, GFP Taqman assays showed that the highest levels of retrotransposition were detected in the progeny of HSCs isolated from irradiated mice cultured in the absence of THPO (Fig. 4 D, upper panel). THPO completely blocked TBI-induced retrotransposition, whether it was injected 1 h before TBI or added after TBI in the culture, demonstrating its direct effect on HSCs in vivo and in vitro. Similar results were observed by measuring GFP by FACS in the LSK compartment (Fig. 4 D, bottom panel). In contrast, THPO had no effect on the total cell number and the percentage of LSK cells and HSCs recovered at the end of the culture in vitro (Fig. S3, C and D).
L1 retrotransposition was also significantly increased in both LSK-CD34−Flk2− and LSK-CD48−CD150+ HSCs from L1-Mpl−/− (Fig. 4 E and Fig. S3 E) and L1-Thpo−/− mice (Fig. S3 F), even under steady-state conditions. It was further enhanced in L1-Mpl−/− HSCs 1 mo after TBI (Fig. 4 F). Thus, THPO signaling in vivo is required to limit RE expression and L1 mobilization in HSCs, under both steady-state conditions and irradiation stress.
THPO induces an early antiviral, IFN-like gene expression response in HSCs
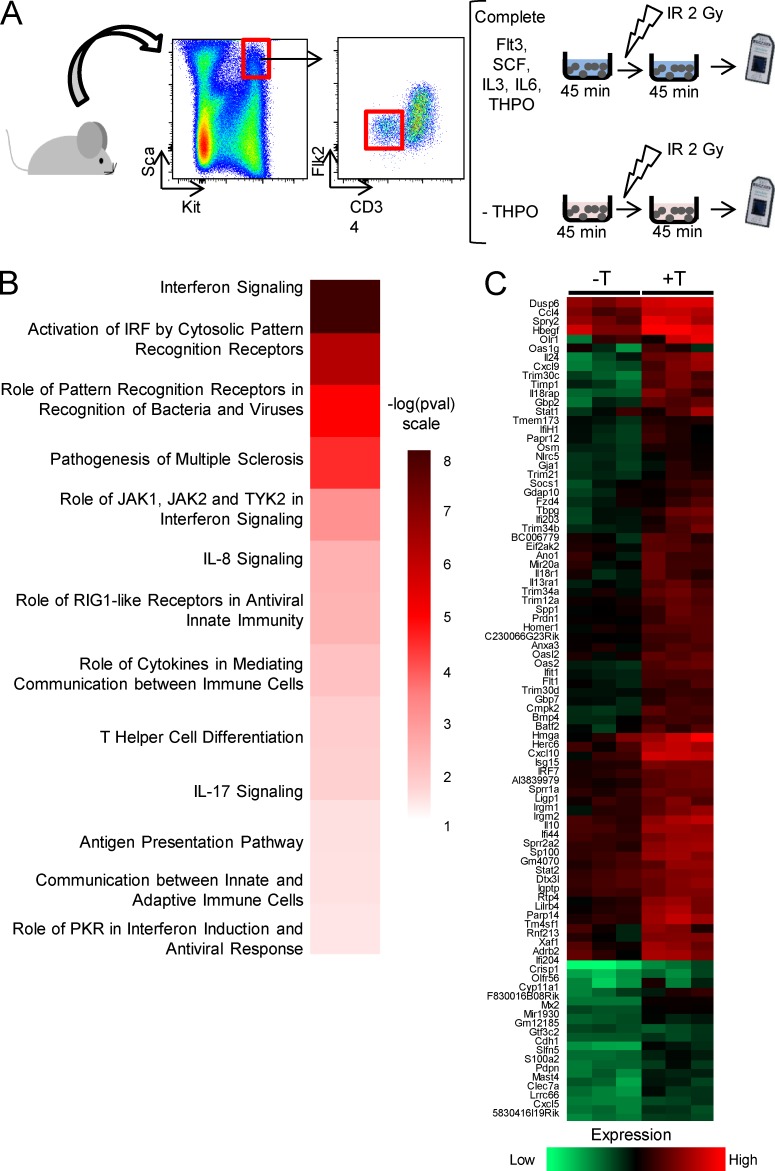
We next thought to determine by which mechanism THPO could limit RE expression and activity in HSCs. Because only one injection of THPO 1 h before TBI is sufficient to protect HSCs from increased RE expression and retrotransposition, we hypothesized that THPO-mediated control of REs takes place at early time after THPO stimulation. We previously showed that a short preincubation of purified HSCs in medium containing THPO before irradiation in vitro could fully recapitulate the effect of THPO injection in vivo on HSC genomic stability and function (de Laval et al., 2013). To analyze whether the ability of THPO to prevent TBI-induced RE expression was a result of THPO-induced specific transcriptional changes during this preincubation time, purified LSK-CD34−Flk2− HSCs were cultured in vitro for 45 min with or without THPO before irradiation, and microarray analyses were performed 45 min later (Fig. 5 A). 338 differentially expressed genes (Table S1; fold change ≥ 1.5, P values ≤ 0.05) specifically regulated by THPO were identified. Ingenuity pathway analysis revealed pathways related to IFN signaling and antiviral innate immunity as top significantly THPO-activated canonical pathways (Fig. 5 B). More than 60% of the up-with-THPO gene list (Fig. 5 C and Table S1) is composed of IFN-stimulated genes (ISGs) with characteristics of IFN type I response genes that are found in the Interferome database (Rusinova et al., 2013). Indeed, qRT-PCR assays showed that these genes are induced by IFN-α in LSK cells (Fig. S4 A).
Figure 5.
THPO induces an IFN signature in HSCs. (A) Experimental design for microarray analysis of THPO differentially regulated genes in HSCs. Gray objects represent Affimetrix Gene Chips (microarrays) used to measure gene expression in cells after treatments. (B) Heat map of ingenuity pathway analysis p-values for the top canonical pathways of differentially regulated genes, obtained using the Perseus software (MaxQuant). (C) Means of triplicate values for up-regulated genes calculated and clusterized using the Perseus software.
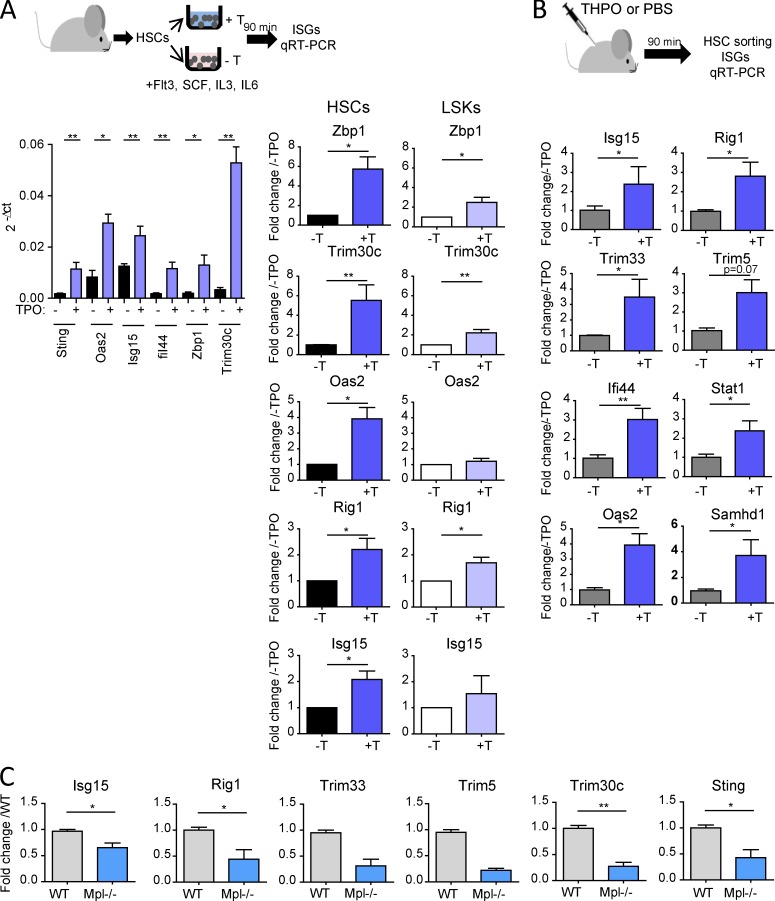
qRT-PCR analyses on LSK-CD34−Flk2− HSCs stimulated in vitro with THPO confirmed the microarray results (Fig. 6 A). Importantly, increased ISG expression was also detected in HSCs isolated 90 min after THPO injection in mice, showing that THPO could also induce an early IFN-like gene response in vivo (Fig. 6 B). The response is specific for THPO, as shown by the absence of significant ISG induction upon incubation of HSCs in medium without THPO, despite the presence of the other cytokines (Fig. S4 B). Furthermore, the induction of ISG expression by THPO was similar in irradiated and nonirradiated samples (Fig. S4 C). In agreement with the highest Mpl expression in HSCs (Qian et al., 2007), THPO-mediated ISG induction was more potent in HSCs than in LSKs (Fig. 6 A, right panel). THPO was able to trigger ISG expression in vitro in HSCs from mice deficient for IFN-I receptor Ifnar1 (Fig. S4 D), indicating that THPO-induced, IFN-like gene expression response does not result from increased IFN secretion or cross talk with IFN-type I receptor. Confirming these results, injection of MAR1-5A3, an anti-IFNAR1 blocking antibody, could reverse IFN-α– but not THPO-induced ISG expression in HSCs in vivo (Fig. S4 E). Finally, confirming THPO’s ability to induce an antiviral transcriptional response in vivo, Mpl−/− HSCs displayed decreased levels of THPO-induced ISGs (Fig. 6 C). Altogether, these results show that THPO is a potent inducer of antiviral response genes in HSCs/HSPCs in vitro and in vivo and can behave as an IFN-like factor.
Figure 6.
THPO induces IFN-stimulating gene expression in HSCs. (A) qRT-PCR analysis for THPO–up-regulated genes in HSCs and LSK incubated for 90 min in vitro in a medium containing THPO (+T) or not (−T). Data normalized to β-actin and/or Gapdh levels. Left, means ± SEM of 2-ΔCt values from n = 4–5 pools of three to six mice in two independent experiments. Mann-Whitney test. Right, comparison of ISG expression in HSCs and LSK cells stimulated with (+T) or without (−T) THPO. Data were normalized as above and are presented on the same scale. Means ± SEM. n = 4–6 pools of mice for HSCs and 8–11 for LSK cells; three to five independent experiments. Paired t test. (B) qRT-PCR analysis in HSCs isolated from mice 90 min after THPO injection in vivo (+T) or not (−T). Data are expressed as fold change from the nontreated mice mean value after normalization. Means ± SEM, n = 5 (−T) and 7 (+T) mice from two independent experiments. Mann-Whitney test. (C) Basal ISG mRNA expression in WT and Mpl−/− HSCs. Means ± SEM normalized to the mean of WT HSCs, n = 3 (Isg15, Rig1, Trim30c, and Sting) or 2 (Trim33 and Trim5) pools of three (WT) to six (Mpl−/−) mice from three independent experiments. Mann-Whitney test. *, P < 0.05; **, P < 0.01.
THPO-induced, IFN-like signaling in HSCs is required to limit retrotransposon expression upon irradiation
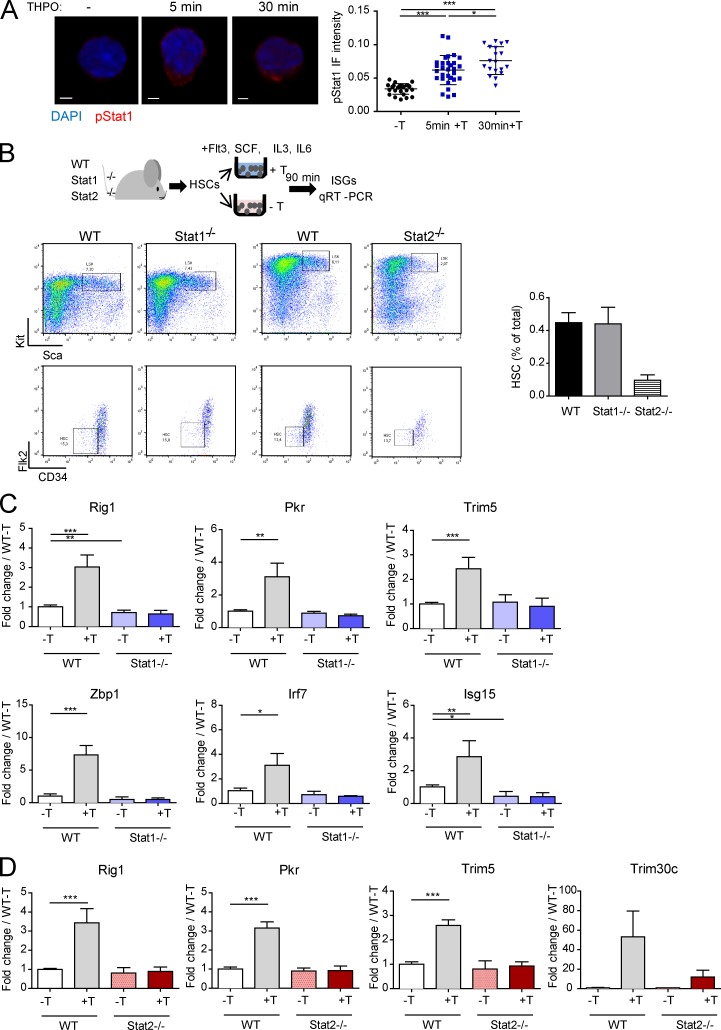
Numerous ISGs are viral restriction factors (Schneider et al., 2014). Thus, we next examined whether this signaling could be linked to its capacity to restrain REs in HSCs. Like THPO, all IFNs signal via the JAK/STAT pathway. The main IFN type I signaling involves STAT1 and STAT2, which form a transcriptional complex with IRF9, called ISGF3, that binds to ISG promoters (Schneider et al., 2014). Fig. 7 A shows that THPO induces a rapid and sustained STAT1 phosphorylation in HSCs. We could not assess Stat2 activation in HSCs as a result of the lack of antibody recognizing mouse phospho-STAT2 in IF or cytometry. However, THPO could induce phosphorylation of both STAT1 and STAT2, together with ISG expression, in a human cell line expressing Mpl (Fig. S5, A and B). THPO-induced ISG expression was completely abolished in both Stat1−/− and Stat2−/− HSCs (Fig. 7, B, C, and D), supporting the possibility that THPO can mimic IFN type I signaling. No significant change in Mpl mRNA levels were observed in HSCs from these mice (Fig. S5 C).
Figure 7.
STAT1 and STAT2 are required for THPO-mediated regulation of RE expression and DNA damage upon irradiation. (A) Representative images and quantification of pSTAT1 staining in HSCs, with or without stimulation with THPO in vitro for the indicated times. Each point represents a cell. Means ± SEM. One-way ANOVA with Dunnett’s multiple comparison test. Bars, 30 µm. (B) Experimental design and representative FACS images of WT, Stat1−/−, and Stat2−/− HSC sorting of the experiments shown in C and D. The graph on the right shows the total HSC frequency in WT, Stat1−/−, and Stat2−/− mice. Representative experiment with cells from four (WT) and two pools of six mice (Stat1−/− and Stat2−/−). Means ± SEM. (C and D) ISG mRNA expression in WT, Stat1−/− (C), and Stat2−/− HSCs (D) after stimulation for 90 min in vitro with (+T) or without THPO (−T). Data are normalized to the mean value of WT HSCs cultured in the absence of THPO. Means ± SEM, n = 4 pools of five to six mice from two independent experiments. Paired t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
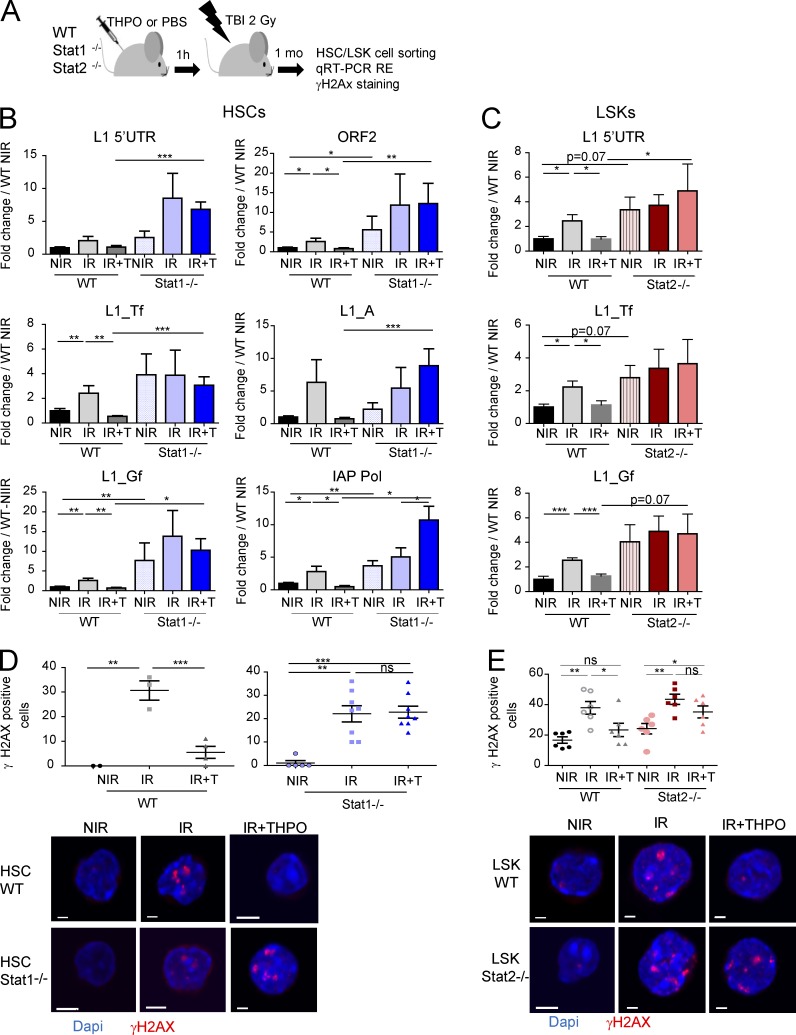
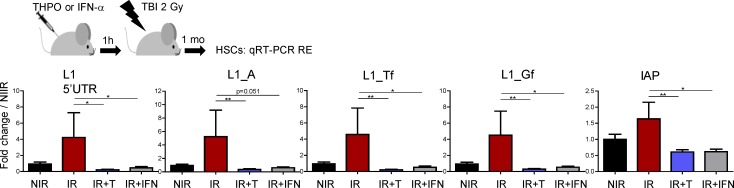
Strikingly, THPO injection in Stat1−/− mice could not prevent TBI-induced increase in L1 and IAP expression in HSCs (Fig. 8, A and B). Similar results were observed using Stat2−/− LSK cells (Fig. 8, A and C). In agreement with a role of STAT1 and STAT2 in controlling THPO-mediated RE expression under basal conditions in vivo, HSCs from nontreated Stat1−/− and Stat2−/− mice, as Mpl−/− HSCs, express slightly higher levels of IAP and L1 REs than WT HSCs. Furthermore, both Stat1−/− and Stat2−/− cells displayed decreased THPO-mediated resorption of γH2AX foci at 1 mo after TBI (Fig. 8, D and E). These results show that THPO-induced IFN type I signaling in HSCs is required to prevent long-lasting accumulation of REs and RE-induced persistent γH2AX foci upon irradiation. Interestingly, injection of IFN-α before TBI could also block irradiation-induced increased RE expression in HSCs (Fig. 9). This supports the possibility that IFN-type I signaling may have an unexpected protective role on HSCs upon genotoxic stress.
Figure 8.
THPO-induced IFN type I signaling is required for THPO-mediated regulation of RE expression and DNA damage upon irradiation. (A) Experimental design for the experiments shown in B–E. (B and C) RE mRNA in WT, Stat1−/− HSCs (B), and Stat2−/− LSK cells (C) from nonirradiated mice (NIR) or 1 mo after TBI with (+T) or without THPO (−T) injection. Data are normalized to the mean values of nontreated WT mice. Means ± SEM. (B) n = 8–12 mice. (C) n = 5–7 mice pooled two independent experiments. One-way ANOVA with Dunnett’s multiple comparison test and Mann-Whitney tests. (D and E) Representative images and quantification of WT, Stat1−/− HSCs (D), WT, and Stat2−/− LSK cells (E) expressing more than four γH2AX foci 1 mo after TBI in the presence or absence of THPO. Each point represents an individual mouse. Means ± SEM. One-way ANOVA with Dunnett’s multiple comparison test. (D) WT, representative experiments from three similar performed; Stat1−/−, data pooled from two independent experiments. (E) Data pooled from two independent experiments. One-way ANOVA with Dunnett’s multiple comparison test. Bars, 30 µm. *, P < 0.05; **, P < 0.01; ***, P < 0.001. ns, not significant.
Figure 9.
IFN-α limits RE expression in HSCs upon irradiation. Experimental design and RE mRNA expression in HSCs isolated 1 mo after 2 Gy TBI with prior injection of one dose of THPO or IFN-α as indicated. Results represent means ± SEM and are expressed as fold change from the NIR mean value after normalization. n = 6 (NIR, IR, and IR+THPO) or 7 (IR+IFN) mice per condition, pooled from two independent experiments. Mann-Whitney test. *, P < 0.05; **, P < 0.01.
Discussion
Because of their lifelong potential, HSCs need to be protected from endogenous and exogenous insults that may trigger genomic instability to ensure their long-term functional activity and prevent their transformation. Intrinsic HSC-protective mechanisms, including low metabolism, high xenobiotic efflux activity, quiescence, and activation of a strong DNA damage response, as well as environmental factors such as the hypoxic nature of the niche or the action of cytokine-controlling repair pathway activity have been shown to contribute to minimize accumulation of DNA damage and allow preservation of HSC potential (Bakker and Passegué, 2013). We show here that RE expression and mobilization can also constitute an endogenous source of HSC genomic instability that increases upon genotoxic stress. We described the ability of HSCs to mount a constitutive IFN-like antiviral response in response to THPO as a novel HSC intrinsic protective mechanism against this form of damage.
Recent data have highlighted the causal relationships between the L1- or LTR-containing RE activity and IFN-induced antiviral response. Indeed, many viral restriction factors are IFN-regulated genes (Schneider et al., 2014). Mutations or deficiencies in several of these genes that we found induced by THPO in HSCs, such as Samhd1, lead to abnormal RE accumulation and retrotransposition. Mutant SAMHD1 of Aicardi-Goutières syndrome patients are defective in L1 inhibition, leading to abnormal L1 and Alu/SVA accumulation and mobilization (Zhao et al., 2013). REs in turn can serve as a source of endogenous signal that triggers type I IFN immune response, eventually leading to autoimmune diseases (Stetson et al., 2008; Zhao et al., 2013). Conversely, it has been recently reported that IFN-α stimulation or overexpression of several ISGs reduce RE expression and L1 propagation in cell lines (Goodier et al., 2012, 2015; Koito and Ishizaka, 2013; Zhang et al., 2014; Hu et al., 2015; Yu et al., 2015). This shows that IFN contributes to the immune control of both REs and exogenous pathogens. Using HSCs deficient for Stat1 or Stat2, which are both required for IFN-I signaling and antiviral immunity (Durbin et al., 1996), we demonstrate here that this pathway is also crucial to confer on THPO the ability to regulate RE expression in HSCs and therefore limit HSC threat upon irradiation.
The mechanism underlying this inhibitory effect remains to be determined. Recent studies have shown that various ISGs can restrict L1 and IAP expression by different means (Pizarro and Cristofari, 2016). This includes interaction with ORF1p or L1 RNA in cytoplasmic stress granules, disruption of ribonucleoprotein particle integrity, RNA degradation, or processing by the RNA-induced silencing complex (Goodier et al., 2012, 2015; Zhang et al., 2014; Hu et al., 2015; Pizarro and Cristofari, 2016). This suggests that THPO-induced ISGs may be required to prevent L1 accumulation by acting mainly at the posttranscriptional level. At the transcriptional level, RE repression is controlled mainly by epigenetic mechanisms (Mita and Boeke, 2016). We have shown recently that H3K9-trimethylation catalyzed by Suv39h1 is required to repress both L1 and IAP expression in HSCs during the course of aging (Djeghloul et al., 2016). Differences in H3K9 methylation and chromatin condensation between HSCs and progenitors (Ugarte et al., 2015) could explain the high basal expression levels of L1 and IAP REs in the former. Interestingly, IFN-α has been shown to inhibit hepatitis B virus transcription by inducing a STAT1/2-dependent epigenetic regulation (Belloni et al., 2012). ISGs involved in transcriptional silencing, such as Trim33, were found among THPO–up-regulated genes (Rajsbaum et al., 2008). Interestingly, Trim33 has been shown to control expression of different LTR-containing REs (Herquel et al., 2013; Isbel et al., 2015). Thus, the THPO-induced STAT1/2 signaling pathway might restrain RE accumulation in HSCs through transcriptional regulatory mechanisms. This could explain the long-lasting effect of a single THPO injection on RE expression. Whether one specific or several ISGs are required to control RE expression in HSCs requires further investigation.
In agreement with previous reports showing increased RE mobilization upon stress (Ishihara et al., 2000; Hagan et al., 2003; Farkash et al., 2006), we found that TBI triggers not only increased L1 expression, but also successful de novo L1 retrotransposition in vivo in primary HSCs. Very recently, Macia et al. (2017) reported that human CD34+ cells do not express L1 and are unable to support L1 retrotransposition, suggesting that L1 mobilization in somatic healthy tissues is restricted to neuronal precursor cells. The discrepancy between this study and ours could be a result of differences in the cell population tested and/or to the treatment applied. Indeed, the CD34+ cells used by Macia et al. is a population composed mainly of progenitors, whereas we found higher L1 expression and retrotransposition levels in HSCs than in LSKs and progenitors. In addition, in the basal state, HSCs express low ORF1p and display low retrotransposition levels, both phenomena being greatly increased upon irradiation.
THPO injection before TBI reversed irradiation-induced L1 expression and L1 mobilization in HSCs. Addition of THPO in the culture after irradiation could also restrain L1 mobilization, demonstrating a direct effect of the cytokine on HSCs. In addition, L1 mobilization was enhanced in HSCs from L1-Mpl−/− and L1-Thpo−/− mice, showing that THPO signaling is required to control L1 retrotransposition in HSCs, in vivo, after genotoxic stress, as well as under steady-state conditions. Although ddC had no effect on TBI-induced changes in LSK and HSC numbers (Shao et al., 2014; Fleenor et al., 2015), its ability to rescue both γH2AX foci accumulation and HSC reconstitution ability after TBI further demonstrates that endogenous retrotransposition plays a role in the long-lasting HSC injury induced by irradiation. This suggests that THPO-mediated L1 mobilization repression plays an important role in its ability to maintain HSC genomic stability. We have previously shown that THPO restrains HSC DNA damage by up-regulating the efficiency of the classical DNA-PK–dependent nonhomologous end joining (NHEJ) DNA repair (de Laval et al., 2013). These two mechanisms are not mutually exclusive. Indeed, RE insertion requires the creation but also the repair of broken DNA and is modulated by DNA repair pathways (Coufal et al., 2011; White et al., 2015). L1, but also SINE and IAP REs, can integrate at preformed DSBs and repair them (Lin and Waldman, 2001; Morrish et al., 2002; Onozawa et al., 2014). In the case of L1, insertions at preexisting DSBs are independent of ORF2p endonuclease activity and increase in cells deficient or mutated for NHEJ factors (Morrish et al., 2002). Interestingly, such repair is highly mutagenic and often shows features of alternative-NHEJ with the presence of short stretches of microhomologies (Onozawa et al., 2014), similarly to what is observed in irradiated cells in the absence of THPO signaling (de Laval et al., 2013). Thus, by modulating DNA repair, THPO may also regulate the extent and the type of RE insertions in HSCs and their ability to induce genomic instability. Because integration of viruses into host DNA induces DNA damage, some ISG products with antiviral activity, such as SAMHD1 and TREX, have developed the ability to regulate DNA damage and repair responses (Yang et al., 2007; Clifford et al., 2014). This suggests that the THPO-induced IFN-like response may constitute a positive retrocontrol pathway improving HSC DNA damage responses.
THPO has been shown to activate STAT1, STAT5, and STAT3 in cell lines and megakaryocytes (Drachman et al., 1997; Rouyez et al., 2005). We show that THPO also induces a rapid activation of STAT1 in HSCs. THPO also triggers STAT1 and STAT2 phosphorylation in a human cell line expressing Mpl. The requirement of both STAT1 and STAT2 for THPO-mediated ISG induction and RE repression suggests that THPO activates an IFN-I–like signaling in HSCs. This is to our knowledge the first demonstration of the involvement of STAT2 in THPO signaling. Although intriguing, this recalls previous studies showing that IFNs and THPO can induce similar transcriptional complexes and that IFN-α can trigger megakaryopoiesis (Rouyez et al., 2005; Haas et al., 2015). Chronic IFN-I exposure in vivo has been shown to induce transient HSC proliferation and/or apoptosis (Essers et al., 2009; Pietras et al., 2014), whereas THPO is required for HSC maintenance through regulation of survival, quiescence, or self-renewal divisions and favors transplantation (Qian et al., 2007; Yoshihara et al., 2007; Kovtonyuk et al., 2016). However, our results suggest that under some instances, such as genotoxic stress, IFN-α and THPO may similarly protect HSCs against REs. This function might be important to prevent DNA damage in HSCs during emergency myelopoiesis. In addition to STAT1/STAT2 activation, each cytokine also activates its own specific signaling pathways leading to unique gene and functional programs that may explain their different final effects on HSCs. For example, THPO stimulates integrin inside-out signaling and adhesion as well as pathways blocking oxidative stress that have been shown to be required for HSC function (Kirito et al., 2005; Umemoto et al., 2012). It also induces the expression of quiescence genes such as p57 (Yoshihara et al., 2007), whereas IFN-α exposure induces cell cycle genes (Essers et al., 2009) or a transient decrease of quiescence regulators, including p57 (Pietras et al., 2014). Although constitutive IFN-I secretion does occur in healthy mice, IFNs are secreted in abundance, primarily in response to viral infection. The ability of THPO to behave as a stronger inducer of ISGs in HSCs than in LSKs, together with its restricted expression in the hematopoietic system, suggests that it has evolved as a constitutive IFN, more specifically dedicated to HSCs and allowing their protection against RE-induced threat while maintaining their self-renewal ability. By inducing Stat1 and Stat2 expression, THPO may also prime HSCs to respond to IFNs upon infection.
Transcriptional derepression and increased mobilization of REs occur in the genome of aging somatic cells, including HSCs (De Cecco et al., 2013; Sun et al., 2014; Van Meter et al., 2014; Djeghloul et al., 2016). Interestingly, in HSCs, the same retrotransposon mRNA species were found to be increased with TBI and age, e.g. evolutionary recent active L1 family members (A, Tf, and Gf), as well as IAP retroviruses. This suggests that the deregulation of RE expression with age, as after irradiation, could be involved in HSC loss of function and persistent DNA damage. Flach et al. (2014) have recently shown that the γH2AX foci present in old HSCs are resulting from residual replication stress on ribosomal DNA and colocalize with the nucleolus. We were not able to demonstrate such a nucleolar localization of γH2AX foci persisting 1 mo after sublethal TBI (Fig. S5 D). Thus, whether the deregulation of REs plays a direct role in HSC aging remains to be investigated thoroughly.
Our results also suggest that maintenance of a certain level of THPO signaling may be required to prevent radiotherapy-induced myeloid malignancies. This is in agreement with the fact that patients with MPL loss-of-function mutations have an increased propensity of developing myelodysplastic syndromes (Maserati et al., 2008). These observations could also be highly relevant in the context of myeloproliferative neoplasms in which IFN-I administration is used as a treatment. IFN-α causes preferential depletion of Jak2V617F-mutated HSCs compared with normal HSCs (Hasan et al., 2013; Mullally et al., 2013). On the other hand, with progression to myelofibrosis, patients become resistant to this treatment. Our results showing that increased THPO signaling and ISG expression can protect HSCs from RE-induced DNA damage may explain this resistance. In that context, chronic IFN-I treatment and STAT1/2 induction were shown to induce chemotherapy resistance in certain cancers (Khodarev et al., 2004; Cheon et al., 2013). Because massive unleashing of REs has been shown to induce cancer stem cell apoptosis (Chiappinelli et al., 2015), strategies combining TBI and STAT1/2 blockade could also be considered.
Materials and methods
Mice and mice treatments
All mice are on a C57BL/6 background. Mpl−/− and Thpo−/− mice were described previously (de Laval et al., 2013). Stat1−/− (Durbin et al., 1996) and Stat2−/− (Park et al., 2000) mice were obtained from M. Muller (University of Veterinary Medicine, Vienna, Austria) and T. Kolbe (University Center for Biomodels, Vienna, Austria), respectively. WT C57BL/6J CD45.2 and CD45.1 mice were from Envigo and Charles River Laboratories, respectively. The transgenic mice expressing an engineered human L1 harboring a GFP-based retrotransposition reporter cassette (L1-GFP, strain 67; Okudaira et al., 2011) was from by T. Okamura and Y. Ishizaka (National Center for Global Health and Medicine, Department of Infection Diseases, Tokyo, Japan). L1-GFP mice were crossed with Mpl−/− and Thpo−/− (referred to as L1-Mpl−/− and L1-Thpo−/− mice, respectively). All the mice were housed in a specific pathogen-free environment. All procedures were reviewed and approved by the Animal Care Committee no. 26 approved by the French Ministry for Research (agreement number no. 01773.03). Unless otherwise specified, mice of 6- to 10-wk of age were used. Mice were treated with one dose of THPO (16 µg/kg body weight, i.v.) or IFN-α (50,000 U, i.v.), anti-IFNAR blocking monoclonal antibody (200 µg; MAR1-5A3) before sublethal TBI (2 Gy; RX irradiator X-RAD 320). ddC, EFV (10 mg/kg weight, s.c.; Sigma) or their respective diluent alone (PBS for ddC and DMSO for EFV) were injected s.c. 1 h before TBI (2 Gy) and then daily for 1 mo.
Cells and cell culture in vitro
Lin−Sca+kit+ cells (referred to as LSK or HSPC), HSCs (LSK-CD34−Flk2− or LSK-CD150+CD48−), multipotent progenitors (MPP, LSK-CD34+Flk2+), CMPs (Lin−Sca1−c-Kit+CD34hiCD16/32−) and GMPs (Lin−Sca1−c-Kit+CD34lowCD16/32+) were sorted using a cell sorter (Influx; BD). HSCs were cultured in serum-free expansion medium (StemSpan; StemCell Technologies) supplemented with recombinant Flt3-Ligand (FLT3-L; 100 ng/ml), IL-3 (10 ng/ml), IL-6 (10 ng/ml), and stem cell factor (SCF, 100 ng/ml) in the presence (+THPO) or absence (−THPO) of 100 ng/ml THPO or of 100 ng/ml IFN-α. All cytokines were from Peprotech. When the cells were irradiated in vitro, THPO was added to the medium 1 h before, as described (de Laval et al., 2013). For in vitro growth, duplicate samples of 100 HSCs were sorted in 96-well plates in proliferation medium: IMDM medium supplemented with 10% FBS, 1% penicillin-streptomycin, and 1 mM glutamine and containing 25 ng/ml FLT3-L, 10 ng/ml IL-3, 25 ng/ml SCF, 25 ng/ml THPO, 4 U/ml erythropoietin (EPO), and 50 µM β-mercaptoethanol. Cell numbers were evaluated at different times. UT7-Mpl cells were grown in α-MEM medium supplemented with 10% FCS and 2 u/ml EPO. The cells were stimulated by adding directly the THPO mimetic peptide GW395058, as described previously (de Laval et al., 2013).
Bone marrow reconstitution
For bone marrow reconstitution experiments, 3 × 106 CD45.2+ bone marrow cells isolated from mice subjected to TBI (2 Gy) and treated with or without ddC for 1 mo were injected in lethally irradiated (9.5 Gy) C57BL/6 CD45.1 congenic mice, together with 3 × 106 competitor CD45.1+ cells. Bone marrows were collected 4 mo later. After red blood cell lysis, the cells were stained with antibodies against CD45.2, CD45.1, and HSC markers and analyzed by FACS. Secondary reconstitutions were performed by injecting 5 × 106 BM cells from the first recipients in lethally irradiated CD45.1+ recipients. BM was harvested 5 mo later.
qRT-PCR
Total RNA was extracted using the RNeasy plus micro kit (Qiagen) or the Direct-Zol RNA micro prep kit (Zymo Research; Proteigene) and reverse transcribed with superscript Vilo (Thermo Fisher). Real time PCR was performed using SYBR qPCR premix Ex Taq (Takara) on a real time PCR machine (7500; Applied Biosystems). Quantification was done using the ΔΔCt-method with normalization on β-actin, Gapdh, and/or Hprt expression. For analysis of RE expression, the extracted RNAs were subjected to an additional treatment with RNase-free DNase (Ambion) according to manufacturer instructions, and the samples were tested for qPCR before reverse transcription to rule out detection of contaminating DNA. Only the samples giving Ct values close to the no template control were further analyzed. Primer sequences are shown in Table S1.
Retrotransposition assays
L1-GFP mice were treated with THPO or PBS and subjected to TBI (2 Gy) 1 h later. 1 mo after TBI, total bone marrow was stained with lineage (CD11b, Gr1, CD19, CD3, and Ter119)-APC, Kit-PercpCy5.5, Sca-PECy7, CD48-PB, and CD150-PE antibodies, and GFP was analyzed in the LSK- and HSC-gated populations. Alternatively, the presence of GFP was assessed by Taqman-based qPCR assay using primers and probe spanning the GFP exon–exon junction (Iijima et al., 2013). In brief, 700–1,000 cells were FACS-sorted directly in 10 µl of prepGEM tissue extraction buffer (ZyGEM) and lysed according to the manufacturer’s instructions. A nested PCR using GFP-specific primers was used to preamplify the L1 transgene, and the presence of the exon–exon junction was then assessed by qPCR using primers and an exon–exon junction probe specific for GFP (Table S1). Data were normalized to RNA 5S DNA. To test retrotransposition in vitro, 700 HSCs were isolated from L1-GFP mice immediately after TBI (2 Gy) or THPO injection and TBI treatment and cultured at 37°C in IMDM medium supplemented with 10% FBS, 1% penicillin-streptomycin, and 1 mM glutamine, and containing 25 ng/ml FLT3-L, 10 ng/ml IL-3, 25 ng/ml SCF, 25ng/ml THPO, 4U/ml EPO, and 50 µM β-mercaptoethanol supplemented, or not, with 25 ng/ml THPO. Cells were harvested at different times and DNA was extracted as above. The presence of GFP was analyzed by exon–exon Taqman assay. At day 10 of culture, the cells were stained with Kit-PrcpCy5.5 and Sca-PECy7 antibodies and GFP expression was analyzed by FACS, gating on the LSK cell compartment.
Microarray analysis
Triplicate samples of 10,000–15,000 LSK-CD34−Flk2− HSCs were sorted and incubated in vitro for 45 min in complete medium, with or without THPO before irradiation at 2 Gy. RNA was purified 45 min later using the RNeasy Plus micro kit (Qiagen) and were hybridized onto whole genome arrays (Affymetrix MouseGene2.0ST). Raw data were normalized using the robust Multichip Algorithm (RMA) in Bioconductor R. Quality controls and statistics were performed using Partek GS. A classical ANOVA for each gene was performed and pairwise Tukey's ANOVA was applied to identify differentially expressed genes in THPO-treated and untreated samples. Functional enrichment analysis of differentially expressed genes was performed using ingenuity pathway analysis (Ingenuity Systems). All data have been submitted on GEO Omnibus site under the accession no. GSE84195.
Immunofluorescence
3,000–5,000 HSCs were cytospun on polylysine-coated glass slides, and IF was performed as previously described (de Laval et al., 2013). Monoclonal anti-γH2AX antibody (clone JBW301) was purchased from Millipore. The presence of the L1 ORF1 protein was assessed in HSCs using the rabbit anti–mouse L1 ORF1p antibody (Martin and Branciforte, 1993; Malki et al., 2014), a gift of A. Bortvin (Carnegie Institution for Science, Baltimore, MA). The antibody was used at a concentration of 3 µg/ml. To test STAT1 activation, freshly sorted HSCs were stimulated for various times in vitro in the presence of THPO (100 ng/ml), cytospun as above and stained with antiphospho-Tyr(701)STAT1 antibody (clone 58D6 rabbit mAb; Cell Signaling Technologies). Detection was performed using Alexa Fluor 555–coupled anti–rabbit secondary antibody. All slides were visualized using SPE confocal microscope (Leica). Pictures were analyzed using ImageJ software or CellProfiler.
qPCR of genomic LINE-1
Quantification of genomic insertions of L1 was performed as described (Coufal et al., 2009; Muotri et al., 2010). In brief, duplicate samples of 200 HSCs or LSK cells from WT or Mpl−/− mice were sorted directly in 10 µl of prepGEM Tissue DNA extraction buffer (ZyGEM) and lysed according to the manufacturer’s instructions. L1 ORF2 was amplified in triplicate using either Taqman- or SYBR-Green–based qRT-PCR. Relative genomic ORF2 content was normalized to the nonmobile 5S ribosomal RNA genomic content as described (Coufal et al., 2009; Muotri et al., 2010).
Analysis of expressed L1 mRNAs
RNA from HSCs and ESCs were extracted and subjected to RNase treatment as above. RNAs were reverse transcribed using SuperScript Reverse Transcriptase IV (Thermo Fisher) for 15 min at 55°C and 10 min at 80°C with 2 µM mORF2 reverse primer. PCR amplifying a 2,296 bp sequence was then performed using L1-5′-UTR forward and mORF2 reverse primers (Table S1). Actin expression was used as amplification control because unspecific annealing of RT primer produces cDNA from highly abundant RNAs (Parent et al., 2015).
Statistical analysis
Results were statistically evaluated using either one-way ANOVA or t test by Prism version 5.0 software (GraphPad Software Inc.).
Online supplemental material
Fig. S1 is related to Figs. 1 and 2. It shows increased L1 expression and retrotransposition in HSCs upon irradiation. Fig. S2, related to Fig. 3, shows that reverse transcription inhibitors rescue irradiation-induced persistent γH2AX foci HSC loss of function. Fig. S3, related to Fig. 4, shows increased RE expression and retrotransposition in Thpo−/− and Mpl−/− LSK cells and HSCs. Figs. S4 and S5, related to Figs. 5, 6, and 7 provides supplementary data on the induction of ISGs and Stat1/2 activation by THPO and IFN-α in HSCs, LSK, and human UT7-Mpl cells. Table S1 shows up- and down-regulated genes differentially expressed in THPO stimulated HSCs (fold change > 1.5; P > 0.05). Table S2 describes the primers used in this study.
Supplementary Material
Acknowledgments
We thank R. Dyunga for technical assistance; SEVIL Institut National de la Santé et de la Recherche Médicale (INSERM) UMS-33 and Gustave Roussy animal facilities; the Imaging and Cytometry Platform of Gustave Roussy for cell sorting and confocal analysis; Dr. T. Okamura, Dr. Y. Ishizaka, Dr. M. Muller, and Dr. T. Kolbe for providing mice; Dr. A. Bortvin (Carnegie Institution for Science, Baltimore, MA) for the gift of anti-mORF1 antibody; Dr. M. Goodhardt (INSERM UMRS-1126, Paris), Dr. M. Gaudry (INSERM U1170), and Dr. G. Cristofari (IRCAN, Nice) for helpful discussions and critical reading of the manuscript.
This work was supported by INSERM and grants from Fondation pour la Recherche Médicale (Equipe labellisée FRM DEQ20150331743) and Ligue Contre le Cancer (LNCC DM/CB/003-14 and LNCC DM/CB/103-14) to F. Porteu. Support for S.N. Constantinescu was from FRS-FNRS (grant WELBIO-CR-2017A-02), Salus Sanguinis, the Project ARC 16/21–073 of the Université Catholique de Louvain, Fondation Contre le Cancer, and Ludwig Institute for Cancer Research. D. Barbieri and E. Elvira-Matelot are recipients of fellowships from LNCC and FRM, respectively.
The authors declare no competing financial interests.
Author contributions: D. Barbieri, E. Elvira-Matelot, Y. Pelinksi, and L. Genève designed and performed the experiments and analyzed the results. B. de Laval performed microarray experiments. G. Yogarajah and C. Pecquet performed experiments. C. Pecquet and S.N. Constantinescu provided Stat2−/− mice. S.N. Constantinescu commented on the manuscript. F. Porteu designed and supervised the study and wrote the manuscript.
References
- Baillie J.K., Barnett M.W., Upton K.R., Gerhardt D.J., Richmond T.A., De Sapio F., Brennan P.M., Rizzu P., Smith S., Fell M., et al. 2011. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 479:534–537. 10.1038/nature10531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker S.T., and Passegué E.. 2013. Resilient and resourceful: genome maintenance strategies in hematopoietic stem cells. Exp. Hematol. 41:915–923. 10.1016/j.exphem.2013.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belancio V.P., Roy-Engel A.M., Pochampally R.R., and Deininger P.. 2010. Somatic expression of LINE-1 elements in human tissues. Nucleic Acids Res. 38:3909–3922. 10.1093/nar/gkq132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belloni L., Allweiss L., Guerrieri F., Pediconi N., Volz T., Pollicino T., Petersen J., Raimondo G., Dandri M., and Levrero M.. 2012. IFN-α inhibits HBV transcription and replication in cell culture and in humanized mice by targeting the epigenetic regulation of the nuclear cccDNA minichromosome. J. Clin. Invest. 122:529–537. 10.1172/JCI58847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheon H., Holvey-Bates E.G., Schoggins J.W., Forster S., Hertzog P., Imanaka N., Rice C.M., Jackson M.W., Junk D.J., and Stark G.R.. 2013. IFNβ-dependent increases in STAT1, STAT2, and IRF9 mediate resistance to viruses and DNA damage. EMBO J. 32:2751–2763. 10.1038/emboj.2013.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappinelli K.B., Strissel P.L., Desrichard A., Li H., Henke C., Akman B., Hein A., Rote N.S., Cope L.M., Snyder A., et al. 2015. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell. 162:974–986. 10.1016/j.cell.2015.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford R., Louis T., Robbe P., Ackroyd S., Burns A., Timbs A.T., Wright Colopy G., Dreau H., Sigaux F., Judde J.G., et al. 2014. SAMHD1 is mutated recurrently in chronic lymphocytic leukemia and is involved in response to DNA damage. Blood. 123:1021–1031. 10.1182/blood-2013-04-490847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coufal N.G., Garcia-Perez J.L., Peng G.E., Yeo G.W., Mu Y., Lovci M.T., Morell M., O’Shea K.S., Moran J.V., and Gage F.H.. 2009. L1 retrotransposition in human neural progenitor cells. Nature. 460:1127–1131. 10.1038/nature08248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coufal N.G., Garcia-Perez J.L., Peng G.E., Marchetto M.C., Muotri A.R., Mu Y., Carson C.T., Macia A., Moran J.V., and Gage F.H.. 2011. Ataxia telangiectasia mutated (ATM) modulates long interspersed element-1 (L1) retrotransposition in human neural stem cells. Proc. Natl. Acad. Sci. USA. 108:20382–20387. 10.1073/pnas.1100273108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L., Huang Q., and Boeke J.D.. 2011. Effect of reverse transcriptase inhibitors on LINE-1 and Ty1 reverse transcriptase activities and on LINE-1 retrotransposition. BMC Biochem. 12:18 10.1186/1471-2091-12-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cecco M., Criscione S.W., Peterson A.L., Neretti N., Sedivy J.M., and Kreiling J.A.. 2013. Transposable elements become active and mobile in the genomes of aging mammalian somatic tissues. Aging (Albany N.Y.). 5:867–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laval B., Pawlikowska P., Petit-Cocault L., Bilhou-Nabera C., Aubin-Houzelstein G., Souyri M., Pouzoulet F., Gaudry M., and Porteu F.. 2013. Thrombopoietin-increased DNA-PK-dependent DNA repair limits hematopoietic stem and progenitor cell mutagenesis in response to DNA damage. Cell Stem Cell. 12:37–48. 10.1016/j.stem.2012.10.012 [DOI] [PubMed] [Google Scholar]
- Djeghloul D., Kuranda K., Kuzniak I., Barbieri D., Naguibneva I., Choisy C., Bories J.C., Dosquet C., Pla M., Vanneaux V., et al. 2016. Age-Associated Decrease of the Histone Methyltransferase SUV39H1 in HSC Perturbs Heterochromatin and B Lymphoid Differentiation. Stem Cell Reports. 6:970–984. 10.1016/j.stemcr.2016.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drachman J.G., Sabath D.F., Fox N.E., and Kaushansky K.. 1997. Thrombopoietin signal transduction in purified murine megakaryocytes. Blood. 89:483–492. [PubMed] [Google Scholar]
- Durbin J.E., Hackenmiller R., Simon M.C., and Levy D.E.. 1996. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell. 84:443–450. 10.1016/S0092-8674(00)81289-1 [DOI] [PubMed] [Google Scholar]
- Elbarbary R.A., Lucas B.A., and Maquat L.E.. 2016. Retrotransposons as regulators of gene expression. Science. 351:aac7247 10.1126/science.aac7247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin J.A., Paquola A.C., Singer T., Gallina I., Novotny M., Quayle C., Bedrosian T.A., Alves F.I., Butcher C.R., Herdy J.R., et al. 2016. L1-associated genomic regions are deleted in somatic cells of the healthy human brain. Nat. Neurosci. 19:1583–1591. 10.1038/nn.4388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers M.A., Offner S., Blanco-Bose W.E., Waibler Z., Kalinke U., Duchosal M.A., and Trumpp A.. 2009. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. 458:904–908. 10.1038/nature07815 [DOI] [PubMed] [Google Scholar]
- Evrony G.D., Cai X., Lee E., Hills L.B., Elhosary P.C., Lehmann H.S., Parker J.J., Atabay K.D., Gilmore E.C., Poduri A., et al. 2012. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell. 151:483–496. 10.1016/j.cell.2012.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkash E.A., Kao G.D., Horman S.R., and Prak E.T.. 2006. Gamma radiation increases endonuclease-dependent L1 retrotransposition in a cultured cell assay. Nucleic Acids Res. 34:1196–1204. 10.1093/nar/gkj522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner G.J., Kimura Y., Daub C.O., Wani S., Plessy C., Irvine K.M., Schroder K., Cloonan N., Steptoe A.L., Lassmann T., et al. 2009. The regulated retrotransposon transcriptome of mammalian cells. Nat. Genet. 41:563–571. 10.1038/ng.368 [DOI] [PubMed] [Google Scholar]
- Flach J., Bakker S.T., Mohrin M., Conroy P.C., Pietras E.M., Reynaud D., Alvarez S., Diolaiti M.E., Ugarte F., Forsberg E.C., et al. 2014. Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature. 512:198–202. 10.1038/nature13619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleenor C.J., Rozhok A.I., Zaberezhnyy V., Mathew D., Kim J., Tan A.C., Bernstein I.D., and DeGregori J.. 2015. Contrasting roles for C/EBPα and Notch in irradiation-induced multipotent hematopoietic progenitor cell defects. Stem Cells. 33:1345–1358. 10.1002/stem.1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Perez J.L., Marchetto M.C., Muotri A.R., Coufal N.G., Gage F.H., O’Shea K.S., and Moran J.V.. 2007. LINE-1 retrotransposition in human embryonic stem cells. Hum. Mol. Genet. 16:1569–1577. 10.1093/hmg/ddm105 [DOI] [PubMed] [Google Scholar]
- Gasior S.L., Wakeman T.P., Xu B., and Deininger P.L.. 2006. The human LINE-1 retrotransposon creates DNA double-strand breaks. J. Mol. Biol. 357:1383–1393. 10.1016/j.jmb.2006.01.089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert N., Lutz-Prigge S., and Moran J.V.. 2002. Genomic deletions created upon LINE-1 retrotransposition. Cell. 110:315–325. 10.1016/S0092-8674(02)00828-0 [DOI] [PubMed] [Google Scholar]
- Gilbert N., Lutz S., Morrish T.A., and Moran J.V.. 2005. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol. Cell. Biol. 25:7780–7795. 10.1128/MCB.25.17.7780-7795.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier J.L., Cheung L.E., and Kazazian H.H. Jr. 2012. MOV10 RNA helicase is a potent inhibitor of retrotransposition in cells. PLoS Genet. 8:e1002941 10.1371/journal.pgen.1002941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier J.L., Pereira G.C., Cheung L.E., Rose R.J., and Kazazian H.H. Jr. 2015. The Broad-Spectrum Antiviral Protein ZAP Restricts Human Retrotransposition. PLoS Genet. 11:e1005252 10.1371/journal.pgen.1005252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas S., Hansson J., Klimmeck D., Loeffler D., Velten L., Uckelmann H., Wurzer S., Prendergast A.M., Schnell A., Hexel K., et al. 2015. Inflammation-Induced Emergency Megakaryopoiesis Driven by Hematopoietic Stem Cell-like Megakaryocyte Progenitors. Cell Stem Cell. 17:422–434. 10.1016/j.stem.2015.07.007 [DOI] [PubMed] [Google Scholar]
- Hagan C.R., Sheffield R.F., and Rudin C.M.. 2003. Human Alu element retrotransposition induced by genotoxic stress. Nat. Genet. 35:219–220. 10.1038/ng1259 [DOI] [PubMed] [Google Scholar]
- Han J.S., Szak S.T., and Boeke J.D.. 2004. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 429:268–274. 10.1038/nature02536 [DOI] [PubMed] [Google Scholar]
- Hasan S., Lacout C., Marty C., Cuingnet M., Solary E., Vainchenker W., and Villeval J.L.. 2013. JAK2V617F expression in mice amplifies early hematopoietic cells and gives them a competitive advantage that is hampered by IFNα. Blood. 122:1464–1477. 10.1182/blood-2013-04-498956 [DOI] [PubMed] [Google Scholar]
- Herquel B., Ouararhni K., Martianov I., Le Gras S., Ye T., Keime C., Lerouge T., Jost B., Cammas F., Losson R., and Davidson I.. 2013. Trim24-repressed VL30 retrotransposons regulate gene expression by producing noncoding RNA. Nat. Struct. Mol. Biol. 20:339–346. 10.1038/nsmb.2496 [DOI] [PubMed] [Google Scholar]
- Hoeijmakers J.H. 2009. DNA damage, aging, and cancer. N. Engl. J. Med. 361:1475–1485. 10.1056/NEJMra0804615 [DOI] [PubMed] [Google Scholar]
- Hu S., Li J., Xu F., Mei S., Le Duff Y., Yin L., Pang X., Cen S., Jin Q., Liang C., and Guo F.. 2015. SAMHD1 Inhibits LINE-1 Retrotransposition by Promoting Stress Granule Formation. PLoS Genet. 11:e1005367 10.1371/journal.pgen.1005367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima K., Okudaira N., Tamura M., Doi A., Saito Y., Shimura M., Goto M., Matsunaga A., Kawamura Y.I., Otsubo T., et al. 2013. Viral protein R of human immunodeficiency virus type-1 induces retrotransposition of long interspersed element-1. Retrovirology. 10:83 10.1186/1742-4690-10-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isbel L., Srivastava R., Oey H., Spurling A., Daxinger L., Puthalakath H., and Whitelaw E.. 2015. Trim33 Binds and Silences a Class of Young Endogenous Retroviruses in the Mouse Testis; a Novel Component of the Arms Race between Retrotransposons and the Host Genome. PLoS Genet. 11:e1005693 10.1371/journal.pgen.1005693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara H., Tanaka I., Furuse M., and Tsuneoka K.. 2000. Increased expression of intracisternal A-particle RNA in regenerated myeloid cells after X irradiation in C3H/He inbred mice. Radiat. Res. 153:392–397. 10.1667/0033-7587(2000)153[0392:IEOIAP]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Iskow R.C., McCabe M.T., Mills R.E., Torene S., Pittard W.S., Neuwald A.F., Van Meir E.G., Vertino P.M., and Devine S.E.. 2010. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 141:1253–1261. 10.1016/j.cell.2010.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano H., Godoy I., Courtney C., Vetter M.R., Gerton G.L., Ostertag E.M., and Kazazian H.H. Jr. 2009. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 23:1303–1312. 10.1101/gad.1803909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodarev N.N., Beckett M., Labay E., Darga T., Roizman B., and Weichselbaum R.R.. 2004. STAT1 is overexpressed in tumors selected for radioresistance and confers protection from radiation in transduced sensitive cells. Proc. Natl. Acad. Sci. USA. 101:1714–1719. 10.1073/pnas.0308102100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirito K., Fox N., Komatsu N., and Kaushansky K.. 2005. Thrombopoietin enhances expression of vascular endothelial growth factor (VEGF) in primitive hematopoietic cells through induction of HIF-1alpha. Blood. 105:4258–4263. 10.1182/blood-2004-07-2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koito A., and Ishizaka Y.. 2013. Retroviruses, retroelements and their restrictions. Front. Microbiol. 4:197 10.3389/fmicb.2013.00197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtonyuk L.V., Manz M.G., and Takizawa H.. 2016. Enhanced thrombopoietin but not G-CSF receptor stimulation induces self-renewing hematopoietic stem cell divisions in vivo. Blood. 127:3175–3179. 10.1182/blood-2015-09-669929 [DOI] [PubMed] [Google Scholar]
- Lee E., Iskow R., Yang L., Gokcumen O., Haseley P., Luquette L.J. III, Lohr J.G., Harris C.C., Ding L., Wilson R.K., et al. Cancer Genome Atlas Research Network . 2012. Landscape of somatic retrotransposition in human cancers. Science. 337:967–971. 10.1126/science.1222077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., and Waldman A.S.. 2001. Capture of DNA sequences at double-strand breaks in mammalian chromosomes. Genetics. 158:1665–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macia A., Widmann T.J., Heras S.R., Ayllon V., Sanchez L., Benkaddour-Boumzaouad M., Muñoz-Lopez M., Rubio A., Amador-Cubero S., Blanco-Jimenez E., et al. 2017. Engineered LINE-1 retrotransposition in nondividing human neurons. Genome Res. 27:335–348. 10.1101/gr.206805.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malki S., van der Heijden G.W., O’Donnell K.A., Martin S.L., and Bortvin A.. 2014. A role for retrotransposon LINE-1 in fetal oocyte attrition in mice. Dev. Cell. 29:521–533. 10.1016/j.devcel.2014.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S.L., and Branciforte D.. 1993. Synchronous expression of LINE-1 RNA and protein in mouse embryonal carcinoma cells. Mol. Cell. Biol. 13:5383–5392. 10.1128/MCB.13.9.5383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maserati E., Panarello C., Morerio C., Valli R., Pressato B., Patitucci F., Tassano E., Di Cesare-Merlone A., Cugno C., Balduini C.L., et al. 2008. Clonal chromosome anomalies and propensity to myeloid malignancies in congenital amegakaryocytic thrombocytopenia (OMIM 604498). Haematologica. 93:1271–1273. 10.3324/haematol.12748 [DOI] [PubMed] [Google Scholar]
- Mita P., and Boeke J.D.. 2016. How retrotransposons shape genome regulation. Curr. Opin. Genet. Dev. 37:90–100. 10.1016/j.gde.2016.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrish T.A., Gilbert N., Myers J.S., Vincent B.J., Stamato T.D., Taccioli G.E., Batzer M.A., and Moran J.V.. 2002. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat. Genet. 31:159–165. 10.1038/ng898 [DOI] [PubMed] [Google Scholar]
- Mullally A., Bruedigam C., Poveromo L., Heidel F.H., Purdon A., Vu T., Austin R., Heckl D., Breyfogle L.J., Kuhn C.P., et al. 2013. Depletion of Jak2V617F myeloproliferative neoplasm-propagating stem cells by interferon-α in a murine model of polycythemia vera. Blood. 121:3692–3702. 10.1182/blood-2012-05-432989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muotri A.R., Chu V.T., Marchetto M.C., Deng W., Moran J.V., and Gage F.H.. 2005. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 435:903–910. 10.1038/nature03663 [DOI] [PubMed] [Google Scholar]
- Muotri A.R., Marchetto M.C., Coufal N.G., Oefner R., Yeo G., Nakashima K., and Gage F.H.. 2010. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 468:443–446. 10.1038/nature09544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijnik A., Woodbine L., Marchetti C., Dawson S., Lambe T., Liu C., Rodrigues N.P., Crockford T.L., Cabuy E., Vindigni A., et al. 2007. DNA repair is limiting for haematopoietic stem cells during ageing. Nature. 447:686–690. 10.1038/nature05875 [DOI] [PubMed] [Google Scholar]
- Okudaira N., Goto M., Yanobu-Takanashi R., Tamura M., An A., Abe Y., Kano S., Hagiwara S., Ishizaka Y., and Okamura T.. 2011. Involvement of retrotransposition of long interspersed nucleotide element-1 in skin tumorigenesis induced by 7,12-dimethylbenz[a]anthracene and 12-O-tetradecanoylphorbol-13-acetate. Cancer Sci. 102:2000–2006. 10.1111/j.1349-7006.2011.02060.x [DOI] [PubMed] [Google Scholar]
- Onozawa M., Zhang Z., Kim Y.J., Goldberg L., Varga T., Bergsagel P.L., Kuehl W.M., and Aplan P.D.. 2014. Repair of DNA double-strand breaks by templated nucleotide sequence insertions derived from distant regions of the genome. Proc. Natl. Acad. Sci. USA. 111:7729–7734. 10.1073/pnas.1321889111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent J.S., Jauvion V., Bouché N., Béclin C., Hachet M., Zytnicki M., and Vaucheret H.. 2015. Post-transcriptional gene silencing triggered by sense transgenes involves uncapped antisense RNA and differs from silencing intentionally triggered by antisense transgenes. Nucleic Acids Res. 43:8464–8475. 10.1093/nar/gkv753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C., Li S., Cha E., and Schindler C.. 2000. Immune response in Stat2 knockout mice. Immunity. 13:795–804. 10.1016/S1074-7613(00)00077-7 [DOI] [PubMed] [Google Scholar]
- Pietras E.M., Lakshminarasimhan R., Techner J.M., Fong S., Flach J., Binnewies M., and Passegué E.. 2014. Re-entry into quiescence protects hematopoietic stem cells from the killing effect of chronic exposure to type I interferons. J. Exp. Med. 211:245–262. 10.1084/jem.20131043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizarro J.G., and Cristofari G.. 2016. Post-Transcriptional Control of LINE-1 Retrotransposition by Cellular Host Factors in Somatic Cells. Front. Cell Dev. Biol. 4:14 10.3389/fcell.2016.00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian H., Buza-Vidas N., Hyland C.D., Jensen C.T., Antonchuk J., Månsson R., Thoren L.A., Ekblom M., Alexander W.S., and Jacobsen S.E.. 2007. Critical role of thrombopoietin in maintaining adult quiescent hematopoietic stem cells. Cell Stem Cell. 1:671–684. 10.1016/j.stem.2007.10.008 [DOI] [PubMed] [Google Scholar]
- Rajsbaum R., Stoye J.P., and O’Garra A.. 2008. Type I interferon-dependent and -independent expression of tripartite motif proteins in immune cells. Eur. J. Immunol. 38:619–630. 10.1002/eji.200737916 [DOI] [PubMed] [Google Scholar]
- Rossi D.J., Bryder D., Seita J., Nussenzweig A., Hoeijmakers J., and Weissman I.L.. 2007. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature. 447:725–729. 10.1038/nature05862 [DOI] [PubMed] [Google Scholar]
- Rouyez M.C., Lestingi M., Charon M., Fichelson S., Buzyn A., and Dusanter-Fourt I.. 2005. IFN regulatory factor-2 cooperates with STAT1 to regulate transporter associated with antigen processing-1 promoter activity. J. Immunol. 174:3948–3958. 10.4049/jimmunol.174.7.3948 [DOI] [PubMed] [Google Scholar]
- Rusinova I., Forster S., Yu S., Kannan A., Masse M., Cumming H., Chapman R., and Hertzog P.J.. 2013. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 41(D1):D1040–D1046. 10.1093/nar/gks1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider W.M., Chevillotte M.D., and Rice C.M.. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 32:513–545. 10.1146/annurev-immunol-032713-120231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L., Feng W., Li H., Gardner D., Luo Y., Wang Y., Liu L., Meng A., Sharpless N.E., and Zhou D.. 2014. Total body irradiation causes long-term mouse BM injury via induction of HSC premature senescence in an Ink4a- and Arf-independent manner. Blood. 123:3105–3115. 10.1182/blood-2013-07-515619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonnet A.J., Nehmé J., Vaigot P., Barroca V., Leboulch P., and Tronik-Le Roux D.. 2009. Phenotypic and functional changes induced in hematopoietic stem/progenitor cells after gamma-ray radiation exposure. Stem Cells. 27:1400–1409. 10.1002/stem.66 [DOI] [PubMed] [Google Scholar]
- Solyom S., Ewing A.D., Rahrmann E.P., Doucet T., Nelson H.H., Burns M.B., Harris R.S., Sigmon D.F., Casella A., Erlanger B., et al. 2012. Extensive somatic L1 retrotransposition in colorectal tumors. Genome Res. 22:2328–2338. 10.1101/gr.145235.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson D.B., Ko J.S., Heidmann T., and Medzhitov R.. 2008. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 134:587–598. 10.1016/j.cell.2008.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D., Luo M., Jeong M., Rodriguez B., Xia Z., Hannah R., Wang H., Le T., Faull K.F., Chen R., et al. 2014. Epigenomic profiling of young and aged HSCs reveals concerted changes during aging that reinforce self-renewal. Cell Stem Cell. 14:673–688. 10.1016/j.stem.2014.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symer D.E., Connelly C., Szak S.T., Caputo E.M., Cost G.J., Parmigiani G., and Boeke J.D.. 2002. Human l1 retrotransposition is associated with genetic instability in vivo. Cell. 110:327–338. 10.1016/S0092-8674(02)00839-5 [DOI] [PubMed] [Google Scholar]
- Ugarte F., Sousae R., Cinquin B., Martin E.W., Krietsch J., Sanchez G., Inman M., Tsang H., Warr M., Passegué E., et al. 2015. Progressive Chromatin Condensation and H3K9 Methylation Regulate the Differentiation of Embryonic and Hematopoietic Stem Cells. Stem Cell Reports. 5:728–740. 10.1016/j.stemcr.2015.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemoto T., Yamato M., Ishihara J., Shiratsuchi Y., Utsumi M., Morita Y., Tsukui H., Terasawa M., Shibata T., Nishida K., et al. 2012. Integrin-αvβ3 regulates thrombopoietin-mediated maintenance of hematopoietic stem cells. Blood. 119:83–94. 10.1182/blood-2011-02-335430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Meter M., Kashyap M., Rezazadeh S., Geneva A.J., Morello T.D., Seluanov A., and Gorbunova V.. 2014. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat. Commun. 5:5011 10.1038/ncomms6011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T.B., Morales M.E., and Deininger P.L.. 2015. Alu elements and DNA double-strand break repair. Mob. Genet. Elements. 5:81–85. 10.1080/2159256X.2015.1093067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissing S., Muñoz-Lopez M., Macia A., Yang Z., Montano M., Collins W., Garcia-Perez J.L., Moran J.V., and Greene W.C.. 2012. Reprogramming somatic cells into iPS cells activates LINE-1 retroelement mobility. Hum. Mol. Genet. 21:208–218. 10.1093/hmg/ddr455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M., Hong C., Zhang B., Lowdon R.F., Xing X., Li D., Zhou X., Lee H.J., Maire C.L., Ligon K.L., et al. 2013. DNA hypomethylation within specific transposable element families associates with tissue-specific enhancer landscape. Nat. Genet. 45:836–841. 10.1038/ng.2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.G., Lindahl T., and Barnes D.E.. 2007. Trex1 exonuclease degrades ssDNA to prevent chronic checkpoint activation and autoimmune disease. Cell. 131:873–886. 10.1016/j.cell.2007.10.017 [DOI] [PubMed] [Google Scholar]
- Yoshihara H., Arai F., Hosokawa K., Hagiwara T., Takubo K., Nakamura Y., Gomei Y., Iwasaki H., Matsuoka S., Miyamoto K., et al. 2007. Thrombopoietin/MPL signaling regulates hematopoietic stem cell quiescence and interaction with the osteoblastic niche. Cell Stem Cell. 1:685–697. 10.1016/j.stem.2007.10.020 [DOI] [PubMed] [Google Scholar]
- Yu Q., Carbone C.J., Katlinskaya Y.V., Zheng H., Zheng K., Luo M., Wang P.J., Greenberg R.A., and Fuchs S.Y.. 2015. Type I interferon controls propagation of long interspersed element-1. J. Biol. Chem. 290:10191–10199. 10.1074/jbc.M114.612374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A., Dong B., Doucet A.J., Moldovan J.B., Moran J.V., and Silverman R.H.. 2014. RNase L restricts the mobility of engineered retrotransposons in cultured human cells. Nucleic Acids Res. 42:3803–3820. 10.1093/nar/gkt1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K., Du J., Han X., Goodier J.L., Li P., Zhou X., Wei W., Evans S.L., Li L., Zhang W., et al. 2013. Modulation of LINE-1 and Alu/SVA retrotransposition by Aicardi-Goutières syndrome-related SAMHD1. Cell Reports. 4:1108–1115. 10.1016/j.celrep.2013.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.