H3K27me3 is known to silence gene expression. Zhang et al. reveal that, rather than functioning as a repressor, Ezh2 or H3K27me3 specifically mediates toll-like receptor (TLR)-induced proinflammatory responses in macrophages/microglia, in which Ezh2 directly targets Socs3, consequently mediates TLR-induced NF-κB activation, and facilitates autoimmune inflammation.
Abstract
Histone 3 Lys27 (H3K27) trimethyltransferase Ezh2 is implicated in the pathogenesis of autoimmune inflammation. Nevertheless, the role of Ezh2 in macrophage/microglial activation remains to be defined. In this study, we identified that macrophage/microglial H3K27me3 or Ezh2, rather than functioning as a repressor, mediates toll-like receptor (TLR)-induced proinflammatory gene expression, and therefore Ezh2 depletion diminishes macrophage/microglial activation and attenuates the autoimmune inflammation in dextran sulfate sodium–induced colitis and experimental autoimmune encephalomyelitis. Mechanistic characterizations indicated that Ezh2 deficiency directly stimulates suppressor of cytokine signaling 3 (Socs3) expression and therefore enhances the Lys48-linked ubiquitination and degradation of tumor necrosis factor receptor–associated factor 6. As a consequence, TLR-induced MyD88-dependent nuclear factor κB activation and the expression of proinflammatory genes in macrophages/microglia are compromised in the absence of Ezh2. The functional dependence of Ezh2 for Socs3 is further illustrated by the rescue experiments in which silencing of Socs3 restores macrophage activation and rescues autoimmune inflammation in macrophage/microglial Ezh2-deficient mice. Together, these findings establish Ezh2 as a macrophage lineage-specific mediator of autoimmune inflammation and highlight a previously unknown mechanism of Ezh2 function.
Introduction
Macrophages, the most plastic cells within the hematopoietic system, exert diversified functions such as maintaining homeostasis, tissue repair, and immune regulation (Gordon and Taylor, 2005; Schulz et al., 2012; Wynn et al., 2013; Ginhoux et al., 2016). When tissues are damaged after infection or injury, peripheral and tissue-resident macrophages are rapidly motivated and exhibit a proinflammatory phenotype (Geissmann et al., 2010; Nathan and Ding, 2010) and contribute to disease progression in many autoimmune diseases such as colitis and multiple sclerosis (Ponomarev et al., 2005; Friese and Fugger, 2007; Gelderman et al., 2007; Kamada et al., 2008; Platt et al., 2010). Microglia are a type of resident macrophage within the central nervous system (CNS; Ginhoux et al., 2010; Saijo and Glass, 2011) and play critical roles during the pathogenesis of multiple sclerosis and its animal model, experimental autoimmune encephalomyelitis (EAE; Heppner et al., 2005; Ponomarev et al., 2005, 2011; Friese and Fugger, 2007). We and others have demonstrated that inhibition of microglial activation suppressed the induction of autoimmune inflammation in the CNS (Heppner et al., 2005; Ponomarev et al., 2011; Xiao et al., 2013).
The pattern-recognition receptors–induced signal transduction pathways are the major initiator for the activation of macrophages/microglia. The TLR family is one of the best-characterized pattern-recognition receptor families that highly expresses on macrophages/microglia, recognizing various microbial components or endogenous damage-associated molecular patterns released from apoptotic cells or wounded tissues during the course of autoimmune inflammation (Akira et al., 2006; Prinz et al., 2006; Kawai and Akira, 2007; Gambuzza et al., 2011). After stimulation by their ligands, TLRs elicit cascades of signaling events in macrophages/microglia that lead to activation of NF-κB, activator protein 1, and IFN regulatory factors. These transcription factors act cooperatively to induce the expression of multiple genes involved in inflammatory responses (Akira et al., 2006). In particular, the TLR pathway that is dependent on the myeloid differentiation response factor MyD88, which transduces signal cascades from all the TLRs except TLR3, is critical for the induction of autoimmune inflammation such as inflammatory bowel disease (Asquith et al., 2010) and EAE (Miranda-Hernandez et al., 2011). TRAF6, a member of the TNF receptor–associated factor (TRAF) family, functions as a shared adaptor to mediate the signal transduction of MyD88-dependent pathway to activate NF-κB and MAPKs (Lomaga et al., 1999; Takeda and Akira, 2004; Kawai and Akira, 2007).
The epigenetic regulation has emerged as one of the key mechanisms of controlling proper gene expression (Medzhitov and Horng, 2009). The methylation of histone 3 Lys27 (H3K27) is mediated by the polycomb repressive complex-2 (PRC2), composed of Ezh2, Suz12, and Eed, and is linked to the silencing of gene expression (De Santa et al., 2007; Schuettengruber et al., 2007; Wei et al., 2009). In particular, Ezh2 is a key component of PRC2 mediating H3K27 trimethylation (H3K27me3) and has been suggested to play important roles in regulating immune cell functions (Su et al., 2003; Mandal et al., 2011; Tumes et al., 2013; Gunawan et al., 2015). In this study, we uncovered that inhibiting H3K27me3 or Ezh2 deficiency attenuates the inflammatory responses in macrophages, leading to the suppression of autoimmune inflammation. We also provide molecular and genetic evidence that Ezh2 serves as a negative regulator of suppression of cytokine signaling 3 (Socs3), which in turn promotes the Lys48-linked ubiquitination and degradation of TRAF6, therefore mediating the activation of TLR-induced MyD88-dependent signaling pathway.
Results
H3K27me3 inhibition specifically suppresses MyD88-dependent proinflammatory responses in macrophages/microglia
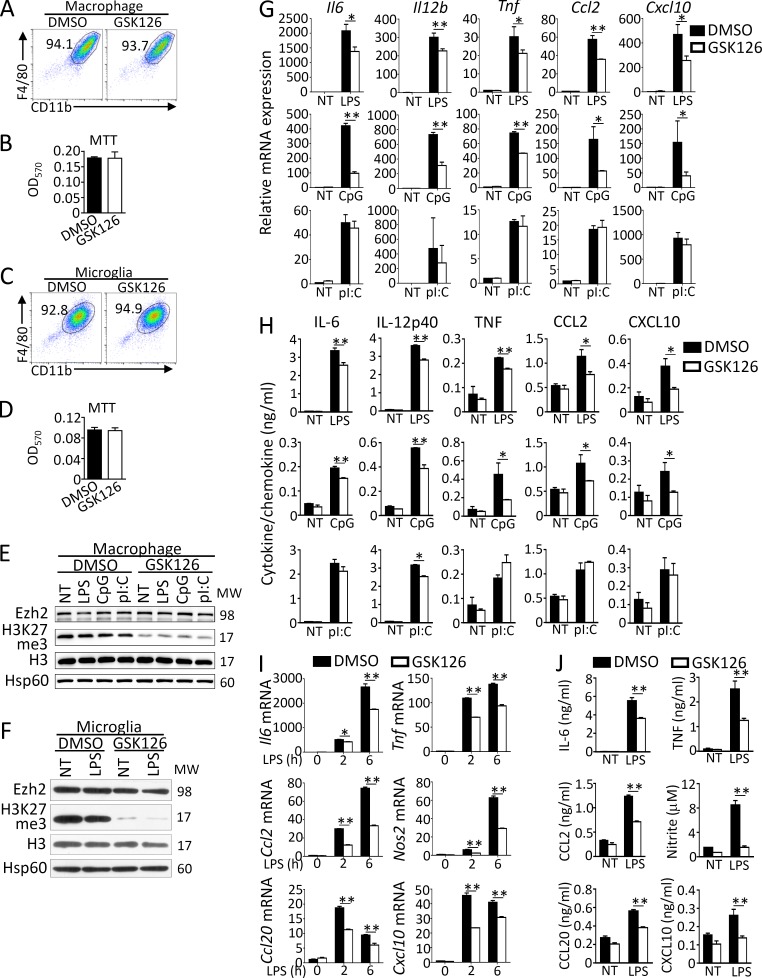
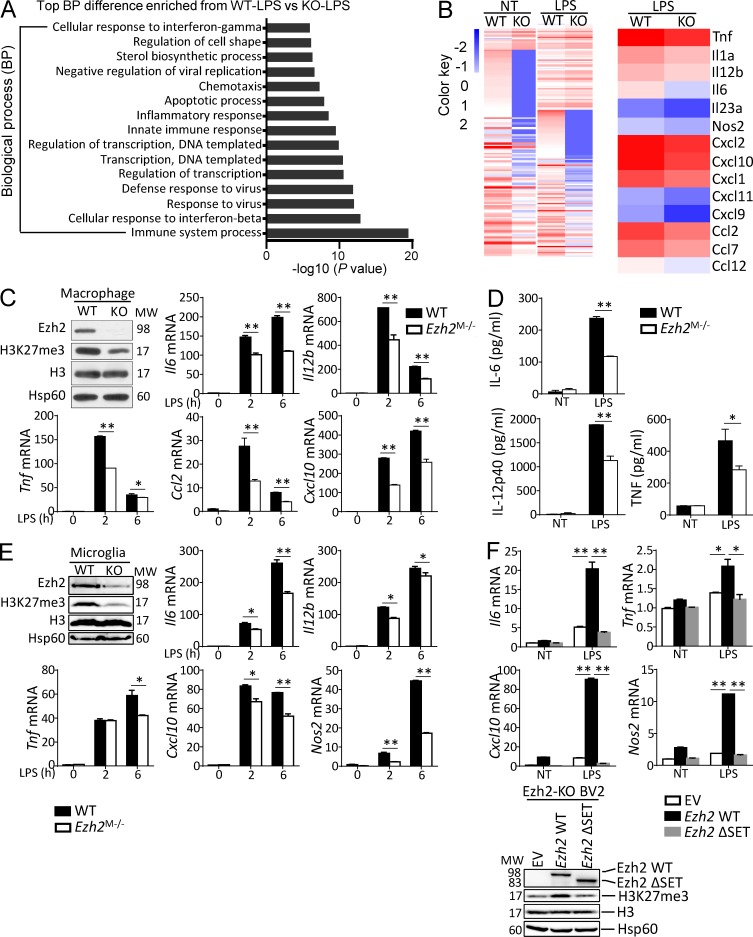
To study the role of H3K27me3 on macrophage/microglial activation, we examined the effect of GSK126, a selective Ezh2 inhibitor that inhibits H3K27me3, on TLR-mediated gene induction in primary cultured bone marrow–derived macrophages or microglia. The results revealed that GSK126 neither alters the expression of surface cell markers (CD11b, F4/80) nor affects the cell viability of macrophages and microglia (Fig. 1, A–D), whereas it indeed suppressed the trimethylation of H3K27 without affecting Ezh2 protein levels in macrophages and microglia with or without TLR ligand stimulation (Fig. 1, E and F). In addition, we detected significantly reduced mRNA expression of proinflammatory genes such as Il6, Il12b, Tnf, Ccl2, and Cxcl10 and suppressed production of these cytokine/chemokine proteins in GSK126-treated macrophages stimulated with TLR4 ligand LPS, TLR9 ligand CpG (Fig. 1, G and H), or TLR1/2 ligand Pam3Csk4 (data not depicted) compared with that of DMSO-treated cells. In contrast, TLR3 ligand polyinosinic:polycytidylic acid (poly I:C) induced comparable expression of proinflammatory genes at both mRNA and protein levels in DMSO- or GSK126-treated macrophages (Fig. 1, G and H). Consistently, GSK126 treatment also impaired LPS-induced proinflammatory gene expression at both mRNA and protein levels in primary cultured microglia (Fig. 1, I and J). These results suggest that H3K27me3 or Ezh2 specifically mediates TLR-induced MyD88-dependent proinflammatory gene expression in peripheral macrophages and microglia.
Figure 1.
GSK126 suppresses MyD88-dependent proinflammatory responses in macrophages/microglia. (A–D) Flow cytometry of the surface CD11b and F4/80 expression and MTT analysis of primary cultured bone marrow–derived macrophages (A and B) or microglia (C and D) that were pretreated with DMSO or GSK126 (4 µM) for 3 d. (E–G and I) Immunoblot analysis (E and F) of Ezh2, H3K27me3, H3, and Hsp60 (loading control) in whole-cell lysates and real-time qRT-PCR analysis (G and I) of the indicated proinflammatory genes of macrophages (E and G) or microglia (F and I) that were pretreated with DMSO or GSK126 (4 µM) for 3 d and then left nontreated (NT) or stimulated for 6 h with the ligands of different TLRs: TLR4 (LPS, 100 ng/ml), TLR9 (CpG, 2.5 µM), and TLR3 (pI:C, 20 µg/ml). (H and J) ELISA showing the production of indicated proinflammatory cytokines/chemokines in the culture supernatants of macrophages (H) or microglia (J) that were pretreated with DMSO or GSK126 (4 µM) for 3 d and then left nontreated (NT) or treated for 24 h with the indicated TLR ligands. The qRT-PCR data were normalized to a reference gene Actb (β-actin), and other data were shown as mean ± SD based on three independent experiments. *, P < 0.05; **, P < 0.01 determined by Student’s t test or two-way ANOVA with post hoc test.
Ezh2 deficiency in peripheral macrophages suppresses dextran sulfate sodium–induced colitis
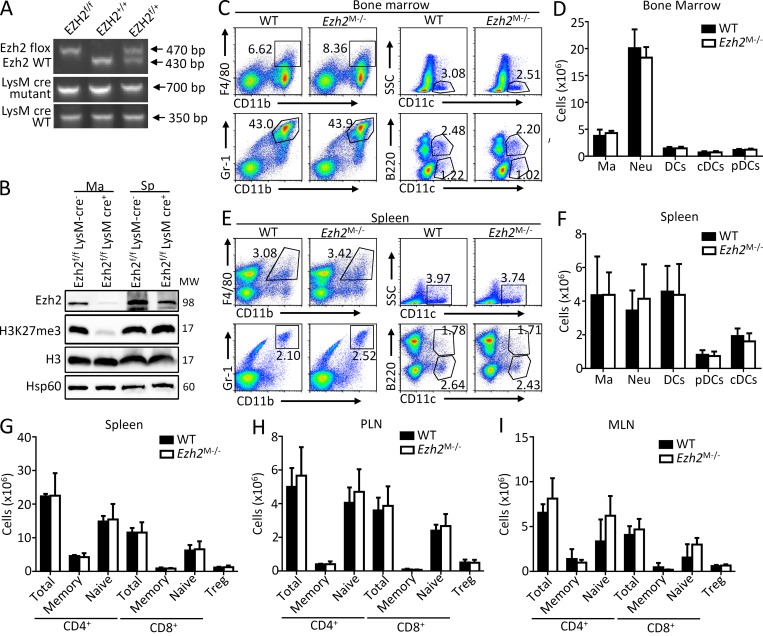
To further assess the role of Ezh2 in macrophages, we crossed the Ezh2f/f mice (Su et al., 2003) with mice expressing Cre recombinase from the myeloid cell lineage-specific lysozyme M promoter (LysM-Cre) to generate mice with conditional deletion of Ezh2 in myeloid cells such as macrophages and microglia (Ezh2f/fLysM-Cre, hereafter called Ezh2M−/−; Fig. 2 A). Immunoblot analysis revealed specifically the loss of Ezh2 protein expression and reduction of H3K27me3 in macrophages of the Ezh2M−/− mice (Fig. 2 B). The Ezh2M−/− mice were born at the expected Mendelian ratio and exhibited normal growth and survival. The development and maturation of myeloid cells appeared to be normal, because Ezh2M−/− mice and their WT littermates produced similar frequencies of macrophages, neutrophils, and dendritic cells in bone marrow and spleens (Fig. 2, C–F). In addition, Ezh2 deficiency in myeloid cells did not affect T cell development in thymus and the maturation and activation of peripheral lymphoid cells (data not depicted; Fig. 2, G–I).
Figure 2.
Ezh2 deficiency neither affects the development and maturation of myeloid cells nor influences the activation of peripheral lymphoid cells. (A) Genotyping PCR analysis of tail DNA from Ezh2f/f, Ezh2+/+, Ezh2f/+, and LysM-cre mice. (B) Immunoblot analysis of Ezh2, H3K27me3, H3, and Hsp60 in bone marrow macrophages and splenocytes from Ezh2f/f LysM-cre− (WT) and Ezh2f/f LysM-cre+ (Ezh2M−/−) mice. (C–F) Flow cytometry analysis of CD11b+F4/80+ macrophages (Ma), CD11b+Gr-1+ neutrophils (Neu), total CD11c+ DCs (DCs), CD11c+B220− conventional dendritic cells (cDCs), and CD11c+B220+ plasmacytoid dendritic cells (pDCs) in bone marrow (C and D) and in spleen (E and F) from WT and Ezh2M−/− mice. Data are presented as representative plots (C and E) and summary graphs (D and F). (G–I) Flow cytometry analysis of the cell numbers of total CD4+ and CD8+ T cells, CD4+ and CD8+ memory (CD62LlowCD44hi) and naive (CD62LhiCD44low) T cells, and CD4+CD25+Foxp3+ regulatory T (Treg) cells in spleen (G), PLN (peripheral lymph node; H), and MLN (mesenteric lymph node; I) from WT and Ezh2M−/− mice. The data are shown as mean ± SD based on three independent experiments. *, P < 0.05; **, P < 0.01 determined by Student’s t test.
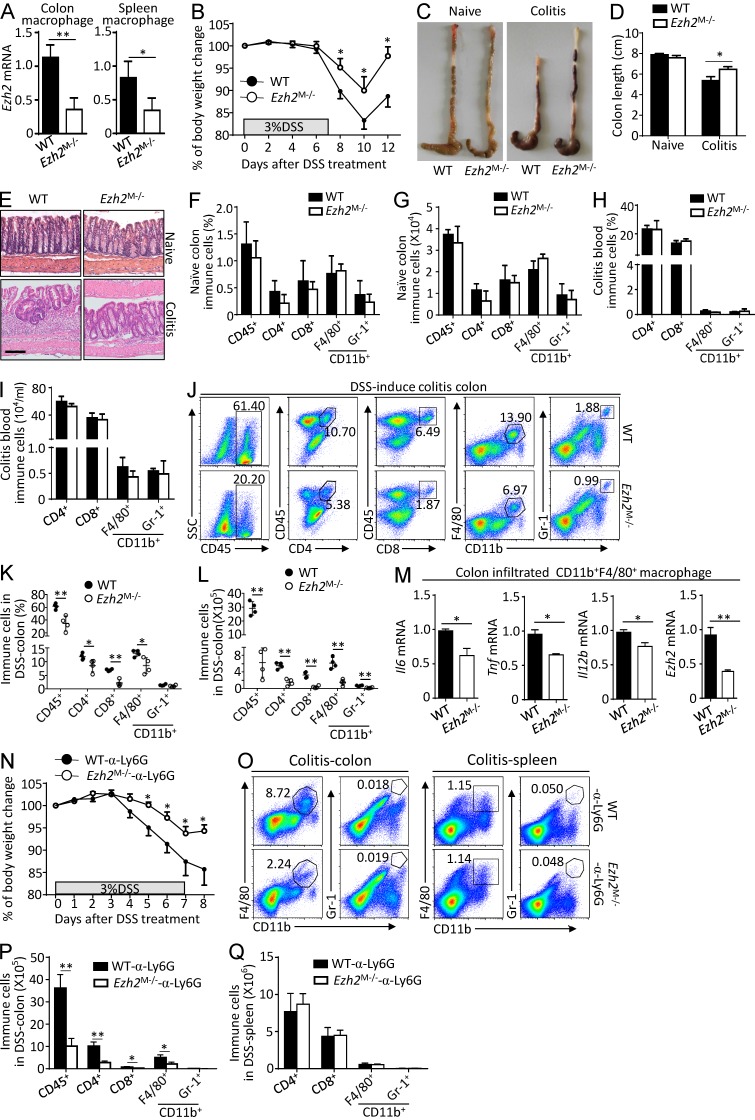
To investigate the in vivo function of Ezh2 in regulating peripheral macrophage-mediated autoimmune inflammation, WT and Ezh2M−/− mice were challenged with 3% dextran sulfate sodium (DSS) to induce a colitis model, which is commonly used to study the innate immune cell function (Chassaing et al., 2014) and mimic human autoimmune disease inflammatory bowel disease. In Ezh2M−/− mice, we confirmed the deletion deficiency of Ezh2 both in colonic and splenic macrophages (Fig. 3 A). In addition, macrophage-specific deficiency of Ezh2 did not affect the morphological and histological features of the colon, whereas it significantly inhibited the susceptibility of DSS-induced colitis, characterized by significantly less body-weight loss, longer colons, and reduced inflammatory infiltration with a less severe disruption of the mucosal epithelium in response to DSS treatment compared with WT control cohorts (Fig. 3, B–E). Consistently, we detected comparable frequencies and absolute numbers of colon-resident immune cells in naive WT and Ezh2M−/− mice (Fig. 3, F and G) and similar frequencies and numbers of blood-circulating immune cells in DSS-challenged WT and Ezh2M−/− mice (Fig. 3, H and I). In contrast, the frequencies and absolute numbers of colon-infiltrating total immune cells (CD45+), CD4+, and CD8+ T cells, macrophages (CD11b+F4/80+), and neutrophils (CD11b+Gr-1+) in Ezh2M−/− mice were dramatically reduced compared with those in WT mice after DSS challenge (Fig. 3, J–L). Accordingly, quantitative RT-PCR (qRT-PCR) assays revealed the reduced expression of proinflammatory cytokine genes in the colon-infiltrating macrophages of the Ezh2M−/− mice (Fig. 3 M). Moreover, we found that in the absence of neutrophils, Ezh2 deficiency in macrophages alone still suppressed the weight loss in colitis mice (Fig. 3, N–Q). Thus, these results establish Ezh2 as a pivotal mediator of colitis that functions in peripheral macrophages.
Figure 3.
Ezh2 deficiency in myeloid cells suppresses DSS-induced colitis. (A) qRT-PCR analysis of Ezh2 mRNA in FACS-sorted CD11b+F4/80+macrophages from the colon and spleen of naive WT and Ezh2M−/− mice. (B) The body-weight loss of WT and Ezh2M−/− mice that were challenged with DSS in drinking water for 7 d. (C–E) The colon images (C), colon lengths (D), and representative H&E-stained images of proximal colon cross sections (E) of naive and DSS-challenged WT and Ezh2M−/− mice at day 12. Bars, 100 µm. (F and G) The frequencies (F) and absolute numbers (G) of the indicated immune cell populations in the colon of naive WT and Ezh2M−/− mice based on flow cytometry analysis. (H–L) Flow cytometry analysis of blood-circulating (H and I) and colon-infiltrated immune cell (J–L) of DSS-challenged WT and Ezh2M−/− mice (n = 4 mice per group) at day 6. Data are presented as representative plots (J) and summary graphs (H, I, K, and L). (M) qRT-PCR analysis of the indicated proinflammatory genes and Ezh2 mRNA in FACS-sorted colon infiltrated CD11b+F4/80+macrophages from DSS-challenged WT and Ezh2M−/− mice (n = 4 mice per group) at day 6. (N) The body-weight loss of WT and Ezh2M−/− mice that were deleted of CD11b+Gr-1+ neutrophils with anti-Ly6G antibody (α-Ly6G) and then challenged with DSS in drinking water for 7 d. (O–Q) Flow cytometry analysis of colon-infiltrated and spleen immune cells of DSS-challenged WT and Ezh2M−/− mice (n = 4 mice per group) that were deleted of neutrophils as described in N at day 6. The qRT-PCR data were normalized to a reference gene Actb (β-actin), and other data are shown as mean ± SD based on three independent experiments. *, P < 0.05; **, P < 0.01 determined by Student’s t test.
Microglia Ezh2 facilitates CNS autoimmune inflammation
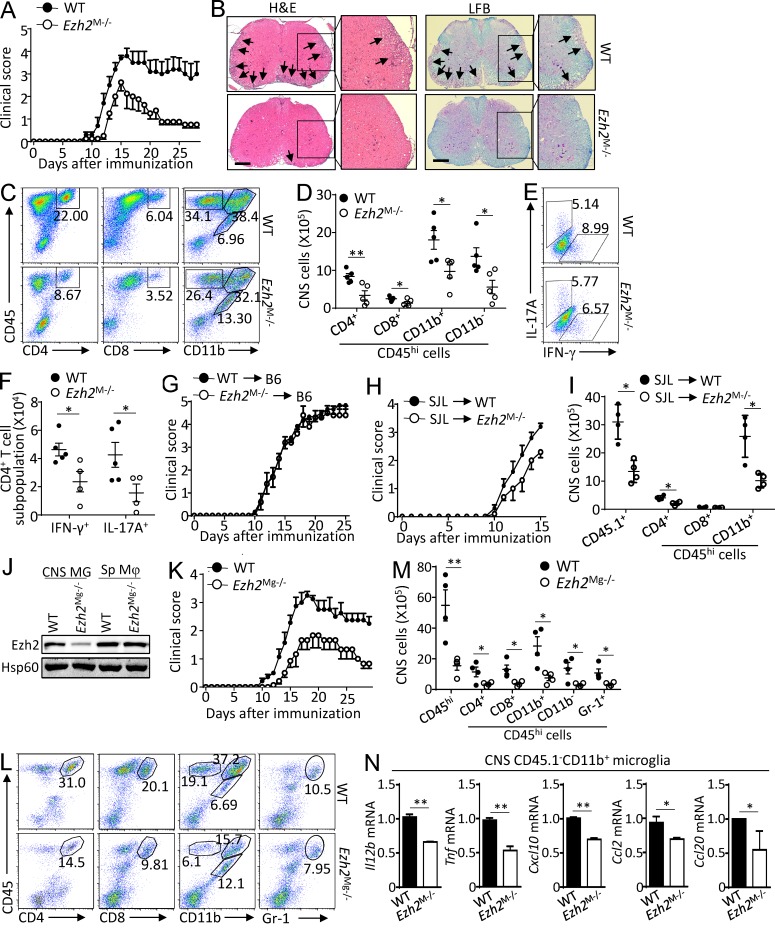
To examine the role of Ezh2 in tissue-resident macrophages such as microglia, we immunized WT and Ezh2M−/− mice with a myelin oligodendrocyte glycoprotein (MOG) peptide (MOG35–55) along with pertussis toxin to induce CNS autoimmunity model EAE, in which microglia are critical for the disease initiation and progression (Heppner et al., 2005; Chastain et al., 2011; Gao and Tsirka, 2011; Ponomarev et al., 2011; Xiao et al., 2013). As expected, the WT mice developed severe clinical symptoms that were characterized by a gradual increase in the severity of paralysis (Fig. 4 A), inflammatory cell infiltration into the CNS, and demyelination (Fig. 4 B). Despite the competent responses of T cells in the Ezh2M−/− mice to antigen stimulation (Fig. S1, A–F), Ezh2 mutant mice displayed decreased incidence of EAE, and if EAE developed, the symptom was significantly milder as indicated by lower clinical scores (Fig. 4, A and B). Likewise, Ezh2M−/− mice displayed reduced frequencies and numbers of CNS-infiltrating T cells (CD4+ and CD8+), myeloid cells, and activated resident microglial cells (CD11b+CD45hi) and total lymphocytes (CD11b−CD45hi) with a concomitant increase in the frequency and number of resting microglial cells (CD11b+CD45lo; Fig. 4, C and D). In addition, the absolute numbers of IL-17+ Th17 cells and IFN-γ+ Th1 cells among the CD4+ T cells infiltrating the CNS were significantly lower in Ezh2M−/− mice than that in Ezh2-sufficient mice (Fig. 4, E and F).
Figure 4.
Microglia Ezh2 facilitates CNS autoimmune inflammation. (A) Mean clinical scores of age- and sex-matched WT and myeloid cell Ezh2-deficient (Ezh2M−/−) mice after the induction of EAE with MOG35–55 (n = 5 mice per group). (B) H&E and LFB staining of spinal cord sections from MOG35–55-immunized WT and Ezh2M−/− mice visualizing immune-cell infiltration and demyelination (arrows), respectively. Bars, 100 µm. Magnification of enlarged images, ×100. (C and D) Flow cytometry analysis of immune-cell infiltration into the CNS (brain and spinal cord) of MOG35–55-immunized WT and Ezh2M−/− mice (n = 5 mice per group) at day 14 after immunization. Data are presented as representative plots (C) and summary graphs (D). The numbers in the plots are the percentages of each gated cell population among the total CNS-infiltrated cells. (E and F) Frequency and absolute number of IFN-γ– and IL-17–producing effector T cells in the CNS (brain and spinal cord) of day 14 MOG35–55-immunized WT and Ezh2M−/− mice, shown as representative plots (E) and summary graphs (F). (G and H) Clinical scores after the induction of EAE in B6 mice that were adoptively transferred with WT or Ezh2M−/− bone marrow cells (n = 7 mice per group; G) or clinical scores after the induction of EAE in WT and Ezh2M−/− mice adoptively transferred with B6-SJL bone marrow cells (n = 5 mice per group; H). (I) Flow cytometry analysis of CNS-infiltrating immune cells of the MOG35–55-immunized WT and Ezh2M−/− SJL-chimeric mice at day 15 after immunization as described in H, showing a summary graph of the absolute cell numbers. (J) Immunoblot analysis of Ezh2 and Hsp60 expression in FACS-sorted CNS microglia (MG) and spleen CD11b+F4/80+ macrophages (Sp Mφ) from WT (Ezh2+/+Cx3cr1-creER-EYFP+) and microglia Ezh2-deficient (Ezh2Mg−/−, Ezh2f/fCx3cr1-creER-EYFP+) mice. (K) Mean clinical scores of age- and sex-matched WT and microglia Ezh2-deficient (Ezh2Mg−/−) mice that exposed with tamoxifen 30 d before immunization and then applied for EAE induction (n = 5 mice per group). (L and M) Flow cytometry analysis of immune-cell infiltration into the CNS (brain and spinal cord) of MOG35–55-immunized WT and Ezh2Mg−/− mice (n = 5 mice per group) at day 14 after immunization. Data are presented as representative plots (L) and summary graphs (M). (N) qRT-PCR analysis of the indicated genes in FACS-sorted CNS CD45.1−CD11b+ microglia of MOG35–55-immunized WT and Ezh2M−/− SJL-chimeric mice (n = 5 mice per group) on day 14 after immunization. The qRT-PCR data were normalized to a reference gene Actb (β-actin), and other data are shown as the mean ± SD based on three independent experiments. *, P < 0.05; **, P < 0.01 determined by Student’s t test.
To determine whether Ezh2 deficiency in peripheral myeloid cells contributes to the suppression of EAE, the radiation bone marrow chimeric mice were generated by reconstituting lethally irradiated WT mice with bone marrow cells isolated from WT or Ezh2M−/− mice. We observed comparable EAE disease scores between these two chimeric mice (Fig. 4 G). In contrast, a reverse bone marrow transfer experiment by reconstituting lethally irradiated WT or Ezh2M−/− mice with WT bone marrow cells isolated from B6/SJL-transgenic mice (cells expressing CD45.1 congenic marker) revealed that the WT SJL-chimeric mice were highly susceptible to EAE induction, whereas the Ezh2-deficienct SJL-chimeric mice were refractory (Fig. 4 H). Additionally, the recruitment of peripheral CD45.1+ leukocytes into the CNS of Ezh2-deficienct SJL-chimeric mice was inhibited compared with that of WT control mice (Fig. 3 I). Thus, Ezh2 in radioresistant microglia but not in peripheral myeloid cells participated in the pathogenesis of EAE.
To further confirm the pathological role of Ezh2 in microglia to mediate EAE pathogenesis, we crossed the Ezh2f/f mice with Cx3cr1-CreER-EYFP mice to generate microglia-conditional Ezh2-knockout (hereafter called Ezh2Mg−/−) mice (Fig. S1 G). By using the tamoxifen-inducible system, Ezh2 is specifically deleted in microglia of Ezh2Mg−/− mice because microglia persist throughout the entire life of the organism, whereas short-lived blood monocytes are replaced by their WT monocyte progeny within 1 mo (Fig. S1 H; Goldmann et al., 2013). The Ezh2Mg−/− mice and their WT littermates were treated with tamoxifen, and 1 mo later we observed specific deletion of Ezh2 in CNS microglia but not in splenic macrophages in Ezh2Mg−/− mice (Fig. 4 J and Fig. S1 I), which were then immunized with MOG35-55 to induce EAE. We found that mice with specific deletion of Ezh2 in microglia displayed significantly delayed onset and reduced severity of EAE disease, as well as substantially less infiltration of immune cells into the CNS, relative to that of their Ezh2-sufficient counterparts (Fig. 4, K and L). Moreover, The Ezh2-deficient microglia that isolated from EAE-induced Ezh2-deficient SJL-chimera mice, in which monocyte-derived macrophages are CD45.1+CD11b+ cells and radioresistant CNS microglia are CD45.1−CD11b+ cells, also exhibited impaired expression of proinflammatory cytokine and chemokine genes (Fig. 4 N). These data collectively suggest a microglia-specific role for Ezh2 in mediating CNS autoimmune inflammation and EAE induction.
Ezh2 mediates MyD88-dependent inflammatory responses in macrophages/microglia
To dissect the mechanistic role of Ezh2 in macrophage/microglia-mediated autoimmune inflammation, we conducted gene expression profile analysis using WT and Ezh2-deficient bone marrow–derived macrophages. Gene ontology (GO) analysis revealed the most prominent biological processes altered in macrophages upon LPS stimulation were associated with immune system process (Fig. 5 A), and the heat map results summarized that Ezh2 deficiency impaired LPS-induced proinflammatory cytokines and chemokines gene expression (Fig. 5 B). Consistent with H3K27me3 inhibition data, quantitative RT–PCR assays confirmed that Ezh2 deficiency severely attenuated proinflammatory gene induction by MyD88-dependent TLR ligands but not TLR3-induced gene expression (Fig. 5 C and Fig. S2 A). Similar results were obtained by ELISA assessing the production of secreted cytokines (Fig. 5 D) and by qRT-PCR analysis of primary cultured microglia that stimulated with the TLR4 ligand LPS and TLR1/2 ligand Pam3Csk4 (Fig. 5 E and Fig. S2 B). In addition, the role of Ezh2 in mediating TLR-induced gene expression was dependent on its C-terminal methyltransferase SET domain, because stable expression of WT Ezh2, but not the empty lentiviral vector (EV) control protein or a SET domain deleted Ezh2 mutant, Ezh2ΔSET, restored H3K27me3 and the LPS-induced gene expression in the Ezh2-KO BV2 microglia cells (Fig. 5 F). These results establish Ezh2 as a pivotal methyltransferase that mediates MyD88-dependent proinflammatory responses specifically in peripheral macrophages and microglia.
Figure 5.
Ezh2 mediates MyD88-dependent inflammatory responses in macrophages/microglia. (A) GO term analysis of Ezh2 function in macrophages treated with LPS through DAVID informatics shows that the most significantly enriched biological process is related to immune system process. (B) RNA-Seq analysis by using WT and Ezh2-deficient macrophages left nontreated (NT) or stimulated for 2 h with LPS, showing the heat maps of genes with adjusted P value <0.05, false discovery rate <0.05, and log2 fold-change >1.2 (left) and the indicated proinflammatory cytokine and chemokine genes down-regulated in Ezh2-deficient macrophages relative to WT cells that treated with LPS for 2 h (right). The source RNA-Seq data were deposited into the Gene Expression Omnibus with the accession no. GSE101316. (C and E) Immunoblot analysis of Ezh2, H3K27me3, H3, and Hsp60, showing the efficiency of Ezh2 deletion and H3K27me3 inhibition in Ezh2-deficient macrophages (C) or microglia (E) compared with WT cells, and qRT-PCR determining the relative expression of indicated proinflammatory genes in macrophages (C) or microglia (E) from WT and Ezh2M−/− mice treated with LPS at the indicated time points. (D) ELISA showing the production of IL-6, IL-12p40, and TNF in the culture supernatants of WT and Ezh2-deficient macrophages that were left nontreated (NT) or treated for 24 h with LPS. (F) qRT-PCR analysis of the indicated genes (left) and IB analysis of Ezh2, H3K27me3, H3, and Hsp60 (right) in CRISPR/Cas9-mediated Ezh2 knockout BV2 microglial cells that were infected with EV or lentiviral vector encoding WT Ezh2 or its SET domain deletion mutant (ΔSET), left nontreated (NT), or treated for 6 h with LPS. Data were normalized to a reference gene, Actb (β-actin), and shown as mean ± SD based on three independent experiments. *, P < 0.05; **, P < 0.01 determined by Student’s t test.
Ezh2 epigenetically controls Socs3 expression in macrophages/microglia
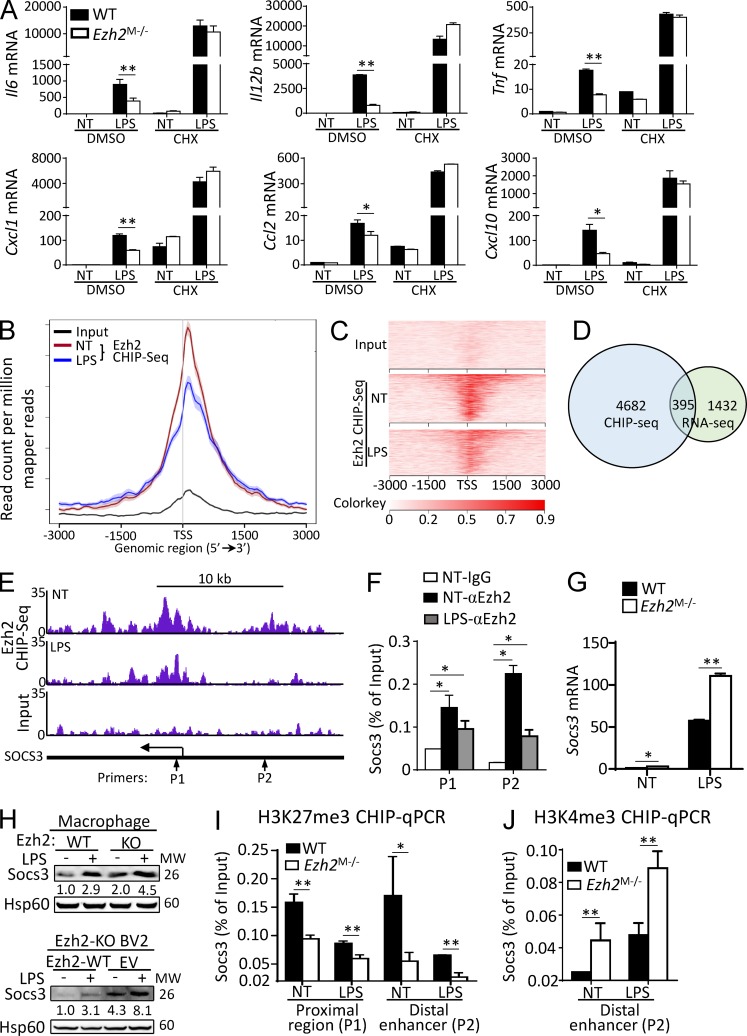
Previous studies have suggested that Ezh2-mediated H3K27me3 is associated with gene transcriptional silencing (De Santa et al., 2007; Schuettengruber et al., 2007; Wei et al., 2009). However, Ezh2 deficiency or H3K27me3 inhibition impaired proinflammatory gene expression in macrophages/microglia, and we reasoned that Ezh2 may control the production of antiinflammatory mediators to regulate the macrophage/microglial activation and autoimmune inflammation. Indeed, when the cells were pretreated with protein synthesis inhibitor cycloheximide, the expression of TLR-induced proinflammatory genes was comparable between WT and Ezh2-deficient macrophages (Fig. 6 A). To identify the potential antiinflammatory genes that are regulated by Ezh2-mediated H3K27me3, we immunoprecipitated chromatin from WT macrophages stimulated with or without LPS and analyzed the precipitated DNA with deep chromatin immunoprecipitation (ChIP) sequencing (ChIP-Seq). In line with the roles of EZH2 in the regulation of gene transcriptions, annotation of the peak in genomic positions to the closest genes demonstrated that many peaks were positioned immediately after the transcription start site (TSS). Intriguingly, we also noticed LPS treatments greatly attenuated Ezh2 binding in the mouse genome (Fig. 6, B and C), suggesting that reductions of Ezh2 occupancies in the subset of genes might be required for LPS-induced inflammatory gene transcriptions. Focusing on LPS-treated macrophages, we identified 5,077 genes harboring Ezh2 occupancies within the core transcription regions (−3 to 3 kb from TSS; Fig. 6, B–D). To correlate chromatin binding with direct gene regulations, we integrated the Ezh2-dependent transcriptome with a primary focus on the genes displaying up-regulation in the absence of Ezh2. Among this group of 1,827 genes, vein diagrams indicated there were ∼395 genes harboring Ezh2 occupancies as well (Fig. 6 D), suggesting they might be directly regulated by Ezh2.
Figure 6.
Ezh2 epigenetically controls Socs3 expression in macrophages/microglia. (A) qRT-PCR analysis of the indicated genes in WT and Ezh2-deficient macrophages that were pretreated with DMSO or cycloheximide (CHX) for 1 h and then left nontreated (NT) or stimulated with LPS for 6 h. (B) Mean Ezh2 ChIP signal in WT macrophages that were left nontreated (NT) or stimulated for 2 h with LPS (100 ng/ml). (C) Heat maps of Input and Ezh2 ChIP-Seq signals at TSSs (±3 kb) in WT bone marrow–derived macrophage left nontreated (NT) or stimulated for 2 h with LPS. The source ChIP-Seq data were deposited into the Gene Expression Omnibus with the accession no. GSE101320. (D) Venn diagram showing the numbers of genes harboring Ezh2 peaks and displaying up-regulation in LPS-stimulated macrophages. (E) Snapshot of the Ezh2 ChIP-Seq signal at the Socs3 loci in WT macrophages left nontreated (NT) or stimulated for 2 h with LPS; the arrows indicate the location of the ChIP primer pairs (P1 and P2). (F) ChIP-qPCR analysis of Ezh2 binding to the Socs3 loci in WT macrophages left nontreated (NT) or stimulated for 2 h with LPS. (G and H) qRT-PCR and IB analysis of Socs3 mRNA and protein levels in WT and Ezh2-deficient macrophages (G, upper panel of H) and Ezh2-KO BV2 cells that infected with EV or lentiviral vector encoding WT Ezh2 (lower panel of H), left nontreated (NT) or stimulated with LPS for 6 h (qPCR) or 12 h (IB). The relative expression of Socs3 compared with Hsp60 was quantified and presented below the Socs3 IB panels. (I and J) ChIP-qPCR analysis of H3K27me3 (I) and H3K4me3 (J) modifications at the Socs3 loci in WT and Ezh2-deficient macrophages left nontreated (NT) or stimulated for 2 h with LPS. The qRT-PCR data were normalized to a reference gene, Actb (β-actin), and other data are shown as mean ± SD based on three independent experiments. *, P < 0.05; **, P < 0.01 determined by Student’s t test.
Among these genes, we selected the most up-regulated genes that may also modulate macrophage function based on reported data, through which several candidate genes, including PPAR-γ, ATF3, Sphk1, and Socs3, were chosen for further analysis. Interestingly, we identified Socs3, a previously known antiinflammatory gene, harbored several Ezh2 intervals near the TSS and distal enhancer regions (Fig. 6 E). Such occupancies of Ezh2 in the Socs3 gene locus were validated by ChIP coupled with quantitative PCR (ChIP-qPCR) assays (Fig. 6 F). Indeed, we detected both mRNA and protein levels of Socs3 were significantly increased in Ezh2-deficient macrophages and BV2 microglial cells compared with those of WT cells (Fig. 6, G and H). Accordingly, Ezh2 depletion caused a substantial reduction of H3K27me3 levels both at the TSS proximal region and distal enhancer, whereas the histone markers of H3K4me3 were further enhanced at the distal enhancer in comparison with control cells as predicted (Fig. 6, I and J). Collectively, these results establish Socs3, an antiinflammatory gene, as a direct target that is fine-tuned by Ezh2-mediated H3K27me3 in macrophages/microglia.
Ezh2 regulates TRAF6 degradation and its downstream signaling
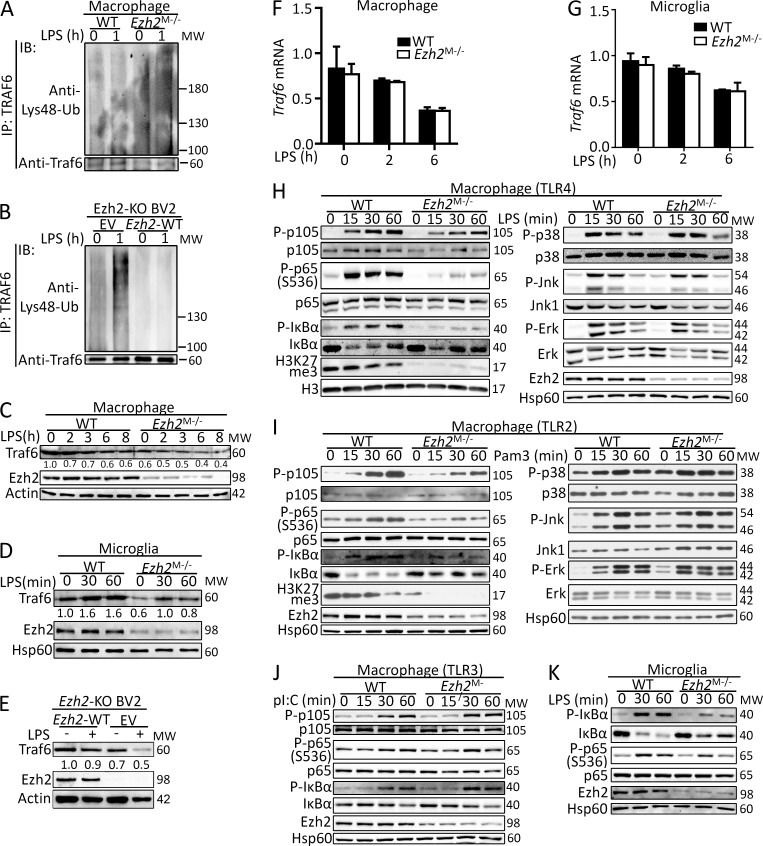
It is reported that Socs3 directly mediates the Lys48-linked ubiquitination and degradation of TRAF6 (Williams et al., 2014; Zhou et al., 2015), which is a critical adaptor essential for MyD88-dependent TLR-induced downstream signaling activation (Lomaga et al., 1999; Takeda and Akira, 2004). As expected, we found that Ezh2 deficiency markedly promoted Lys48-ubiquitination of TRAF6 in Ezh2-deficient macrophages and BV2 microglial cells (Fig. 7, A and B). Accordingly, TRAF6 protein levels were significantly lower in Ezh2-deficient macrophages, microglia, and BV2 cells compared with WT cells (Fig. 7, C–E), whereas TRAF6 mRNA transcription was comparable between the two groups of cells (Fig. 7, F and G). We next examined the effect of Ezh2 deficiency on TLR-induced activation of NF-κB and MAPK, which are the downstream signaling controlled by TRAF6 in MyD88-dependent pathway. The results revealed that loss of Ezh2 in macrophages impaired LPS- and Pam3CSK4-induced activation of NF-κB through MyD88-dependent TLR (Fig. 7, H and I). In contrast, Ezh2 was dispensable for NF-κB activation by the Trif-dependent TLR ligand poly I:C (Fig. 7 J), and Ezh2-deficient macrophages did not show an appreciable defect in the MyD88-dependent TLR-induced activation of the MAPKs p38, ERK, and JNK (Fig. 7, H and I). Similarly, in microglia, Ezh2 deficiency also impaired LPS-induced activation of NF-κB (Fig. 7 K). These data collectively establish Ezh2 as a pivotal regulator of TRAF6 stability, mediating MyD88-dependent NF-κB activation.
Figure 7.
Ezh2 regulates TRAF6 degradation and its downstream signaling. (A and B) Analysis of TRAF6 Lys48-linked ubiquitination in WT and Ezh2-deficient macrophages (A) or in Ezh2-KO BV2 cell that infected with EV or lentiviral vector encoding WT Ezh2 (B) that was stimulated with LPS at the indicated time points. (C–E) IB analysis of TRAF6 protein levels in WT and Ezh2-deficient macrophages (C), in microglia (D), or in Ezh2-KO BV2 cells that were infected with EV or lentiviral vector encoding WT Ezh2 (E) that was stimulated without or with LPS at the indicated time points. The relative expression of Traf6 compared with loading controls (Actin or Hsp60) was quantified and presented below the Traf6 IB panels. (F and G) qRT-PCR analysis of Traf6 mRNA expression in WT and Ezh2-deficient macrophages (F) or microglia (G) that was stimulated with LPS at the indicated time points. (H–K) Immunoblot analysis of phosphorylated (P-) and total NF-κB and MAPKs signaling proteins, H3K27me3, H3, Ezh2, or Hsp60 (loading control) in whole-cell lysates of WT and Ezh2-deficient macrophages that were left unstimulated or stimulated with LPS (100 ng/ml; H) or Pam3CSK4 (100 ng/ml; I) or poly I:C (pI:C, 20 µg/ml; J), and IB of the indicated signal protein in the whole-cell lysates of WT and Ezh2-deficient primary microglia that were left unstimulated or stimulated with LPS (100 ng/ml; K) at the indicated time points. The qRT-PCR data were normalized to a reference gene, Actb (β-actin), and other data are shown as mean ± SD based on three independent experiments.
Inhibition of Socs3 restores autoimmune inflammation in Ezh2-deficient mice
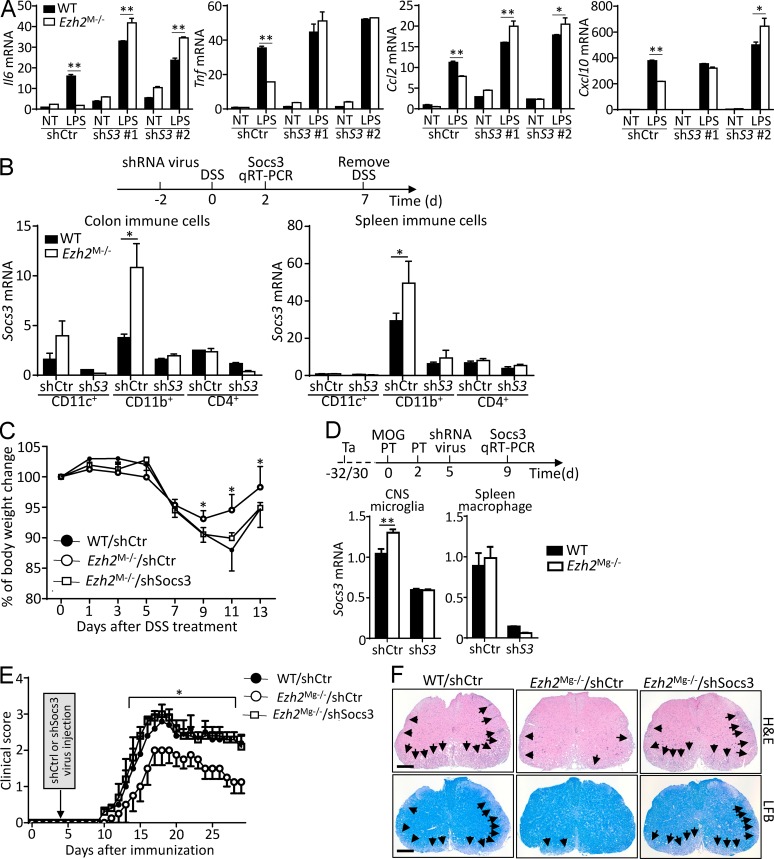
Socs3 is an antiinflammatory gene that negatively regulates macrophage activation, and genetic ablation of Socs3 in myeloid cells aggravated the disease severity of LPS-induced septic shock and EAE (Qin et al., 2012a,b). To assess the functional importance of Ezh2-mediated Socs3 suppression in mediating macrophage activation and autoimmune inflammation, we knocked down Socs3 in WT and Ezh2-deficient macrophages. Interestingly, Socs3 silencing promoted LPS-induced proinflammatory gene expression and erased the differences between WT and Ezh2-deficient cells (Fig. 8 A). In addition, we found that the expression of Socs3 was significantly higher in colonic and splenic macrophages that isolated from Ezh2M−/− mice than that from WT mice (Fig. 8 B), which confirmed the in vivo function of Ezh2 deficiency in promoting Socs3 expression in macrophages. Expectedly, injection of the Ezh2M−/− mice with retrovirus carrying a Socs3-specific shRNA significantly suppressed Socs3 expression in both colonic and splenic CD11b+ macrophages (Fig. 8 B) and largely restored DSS-induced colitis accordingly, characterized by the increased body-weight loss similar to that in WT mice receiving retrovirus carrying control shRNA (Fig. 8 C). Meanwhile, compared with that in WT mice, Socs3 expression was only higher in CNS microglia but not in splenic macrophages of Ezh2Mg−/− mice (Fig. 8 D); Ezh2 is specifically deleted in CNS microglia but not in peripheral splenic macrophages. Accordingly, knockdown of Socs3 in CNS microglia also significantly rescued the defects of Ezh2Mg−/− mice in EAE induction and in the inflammatory infiltration and demyelination in the CNS (Fig. 8, D–F). Collectively, these in vitro and in vivo studies highlight the functional importance of Ezh2-mediated Socs3 suppression in mediating macrophage/microglial activation and autoimmune inflammation.
Figure 8.
Inhibition of Socs3 restores autoimmune inflammation in Ezh2-deficient mice. (A) qRT-PCR determining the relative expression of the indicated genes in WT and Ezh2-deficient macrophages that infected with retrovirus carrying control vector (shCtr) or Socs3 shRNA (shS3 #1 and shS3 #2), left nontreated (NT) or stimulated with LPS for 6 h. (B) Upper panel: scheme of how Socs3 was knocked down in vivo in WT and Ezh2M−/− mice that applied for DSS challenge. Lower panel: qRT-PCR determining Socs3 mRNA expression in FACS-sorted colon and spleen immune-cell populations in WT and Ezh2M−/− mice that were i.v. injected with retrovirus carrying control vector (shCtrl) or Socs3 shRNA (shSocs3). (C) The body-weight loss of WT and Ezh2M−/− mice that were knocked down Socs3 as described in B and then challenged with DSS in drinking water for 7 d. (D) Upper panel: scheme of how Socs3 was knocked down in vivo in WT and Ezh2Mg−/− mice that applied for EAE induction. Ta, tamoxifen. Lower panel: qRT-PCR determining Socs3 mRNA expression in FACS-sorted CNS YFP+ microglia and spleen CD11b+F4/80+ macrophages in WT (Ezh2+/+Cx3cr1-creER-EYFP+) and Ezh2Mg−/− (Ezh2f/fCx3cr1-creER-EYFP+) mice that were exposed by tamoxifen and then were immunized with MOG peptide to induce EAE and i.v. injected with retrovirus carrying control vector (shCtrl) or Socs3 shRNA (shSocs3) at the indicated time points. (E) Mean clinical scores of WT and Ezh2Mg−/− mice that were knocked down Socs3 and applied for EAE induction as described in D. (F) H&E and LFB staining of spinal cord sections from MOG35–55-immunized mice as described in D, visualizing immune-cell infiltration and demyelinization (arrows), respectively. Bars, 100 µm. The qRT-PCR data were normalized to a reference gene, Actb (β-actin), and the other data are shown as mean ± SD based on three independent experiments. *, P < 0.05; **, P < 0.01 determined by Student’s t test.
Discussion
Macrophages form a major type of innate immune cells to sense pathogen invasion and tissue injury, and excess production of proinflammatory cytokines by activated macrophages contributes to the pathogenesis of autoimmune inflammation. We and others have shown that depletion or functional inhibition of macrophages exhibits promising effects in attenuating the severity of several autoimmune diseases (Heppner et al., 2005; Marshall et al., 2007; Ponomarev et al., 2011; Xiao et al., 2013). Thus, a large number of clinical trials were tested in autoimmune diseases such as colitis, multiple sclerosis, and arthritis by targeting macrophage activation (Hamilton et al., 2016). Here we identified Ezh2 as a pivotal mediator in both peripheral macrophages and CNS-resident microglia, through which it regulates the pathogenesis of autoimmune inflammation. It is reported that peripheral monocyte-derived and gut-resident macrophages may be functionally different in modulating colitis pathogenesis (Zigmond et al., 2012). Nevertheless, considering the universal effect of Ezh2 in regulating the activation of macrophage lineage cells, we speculated that Ezh2 deficiency may impair the inflammatory responses both in peripheral monocyte-derived and gut-resident macrophages, which collectively contributed to the inhibition of colitis pathology in Ezh2M−/− mice.
Ezh2 is critical in mediating the formation of H3K27me3, which is associated with gene transcriptional suppression (De Santa et al., 2007; Schuettengruber et al., 2007; Wei et al., 2009; Liu et al., 2017). However, we found that Ezh2 in macrophages/microglia exhibits opposite effects, reflected by impaired TLR-induced proinflammatory gene expression in the absence of Ezh2 or H3K27me3. This effect of Ezh2 in macrophages/microglia is due to the suppressed expression of Socs3, an antiinflammatory gene directly targeted by Ezh2-mediated H3K27me3. Additionally, Ezh2 does not affect TLR-induced inflammatory responses in dendritic cells (unpublished data), although Ezh2 deficiency in dendritic cells suppressed CNS autoimmunity (Gunawan et al., 2015). Thus, it is more likely that Ezh2 and/or H3K27me3 has a unique regulatory pattern specifically in macrophage lineage cells.
It is reported that the inhibitor of H3K27 demethylase Jmjd3 repressed LPS-induced expression of proinflammatory genes in activated macrophages (Kruidenier et al., 2012). However, genetic ablation of Jmjd3 in macrophages did not affect the gene expression induced by various TLR ligands but impaired macrophage M2 polarization (Satoh et al., 2010). Our study demonstrates that the selective inhibitor or genetic ablation of Ezh2 exhibits similar effects in macrophages/microglia in which Ezh2 or H3K27me3 mediate TLR-induced proinflammatory gene expression. In addition, Ezh2 deficiency also slightly impaired M2 polarization (Fig. S3). These data combined with published studies suggested that Ezh2- or Jmjd3-mediated modulation of H3K27me3 diversely regulates inflammatory gene expression through different molecular mechanisms in macrophages and that the functional diversity of H3K27me3 may be due to the delicate regulatory difference by its methyltransferase and demethylase. Because either Ezh2-mediated trimethylation or Jmjd3-mediated demethylation of H3K27 is a universal effect on the whole genome, it is reasonable to speculate these two enzymes may have their preference to target some specific genes, resulting in functional diversity of macrophage activation. In T cells, Ezh2 deficiency promoted Th1/Th2 cell differentiation in nonpolarized T cells through enhanced expression of Tbx21 and Gata3 (Tumes et al., 2013), implying a cell-specific function of Ezh2 and H3K27me3 in a different immune cell subset.
Previous studies have demonstrated the critical roles of monocyte-derived macrophages in regulating EAE pathogenesis in which CNS-infiltrated macrophages are highly phagocytic and inflammatory (Ajami et al., 2011; Yamasaki et al., 2014). Nevertheless, we and others have suggested that the pathogenesis of EAE and multiple sclerosis also involves microglia (Heppner et al., 2005; Chastain et al., 2011; Gao and Tsirka, 2011; Ponomarev et al., 2011; Xiao et al., 2013). Here we found that Ezh2 in monocyte-derived macrophages is dispensable for EAE induction by using the bone marrow chimeric EAE mouse models in which the bone marrow from WT and Ezh2M−/− mice exhibits comparable capability to induce EAE, suggesting Ezh2 does not directly function in peripheral macrophages to affect EAE disease progression, although it impairs monocyte infiltration in actively induced Ezh2M−/− EAE mice. However, Ezh2-specific deletion in microglia but not in peripheral macrophages by using the Ezh2Mg−/− mice significantly suppressed CNS autoimmune inflammation after EAE induction, which suggested a unique role of Ezh2 in microglia to regulate EAE pathogenesis. Therefore, the suppressed activation of microglia but not peripheral macrophages was the reason for impaired monocyte infiltration and ameliorated disease severity in actively induced Ezh2M−/− EAE mice. In addition, a recently published microarray dataset suggests an elevated tendency of Ezh2 expression in the brain tissues of multiple sclerosis patients compared with that of control donors (Gene Expression Omnibus accession no. GSE32915; Fig. S4), implying a pathological role of Ezh2 in microglia to mediate the pathogenesis of human multiple sclerosis.
Socs3, a well-known antiinflammatory mediator, has been shown to negatively regulate TLR-induced macrophage activation and autoimmune inflammation (Marshall et al., 2007; Qin et al., 2012a,b). In this study, we identified that Ezh2-mediated H3K27me3 directly targets Socs3 for its transcriptional inhibition. Thus, Ezh2 deficiency elevated Socs3 expression in vitro and in vivo, which explains suppressed TLR-induced inflammatory responses in macrophages/microglia and inhibited autoimmune inflammation in the absence of Ezh2 or H3K27me3. In addition, Ezh2 deficiency impaired TLR-induced MyD88-dependent NF-κB activation because of promoted Lys48-linked ubiquitination and degradation of TRAF6, which is mediated by the elevated expression of Socs3. We noticed that Ezh2 deficiency inhibited TLR-induced NF-κB activation without affecting MAPK activation because of decreased TRAF6 levels, implying that the activation of NF-κB is more sensitive to the alteration of Traf6 levels than MAPK or there may exist other cross-talked signaling pathways to compensate the activation of MAPK. In addition, silencing of Socs3 restores TLR-induced inflammatory responses in Ezh2-deficient macrophages, and Socs3 shRNA retrovirus rescues the induction of autoimmune inflammation in both colitis and EAE models. Given the facts that Socs3 knockdown was also observed in other cell types, we cannot fully exclude the possibilities that Socs3 shRNA retrovirus injection may function in other immune cells to regulate the autoimmune inflammation in macrophage/microglial Ezh2-deficient mice.
In summary, our work establishes Ezh2 as a mediator of macrophage/microglial activation and autoimmune inflammation. On the basis of our data, we propose a model in which Ezh2-mediated H3K27me3 directly targets and suppresses the expression of Socs3. Ezh2 deficiency–induced up-regulation of Socs3 mediates the Lys48-linked ubiquitination and degradation of TRAF6, leading to the suppression of TLR-induced MyD88-dependent NF-κB activation and the inhibition of genes involved in macrophage/microglial activation and autoimmune inflammation (Fig. 9). Thus, the development of therapeutic strategies to specifically target Ezh2 or H3K27me3 in macrophage lineage cells might be useful for manipulating autoimmune diseases.
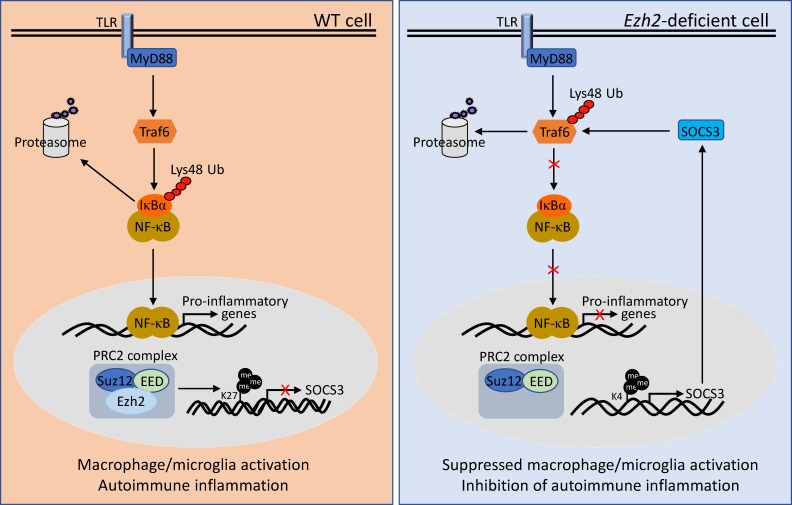
Figure 9.
Model of Ezh2 in the regulation of macrophages/microglia activation and autoimmune inflammation. In WT macrophages/microglia, Ezh2, together with EED and Suz12 form PRC2 complex, which directly target Socs3 to promote the H3K27me3 and suppress the expression of Socs3, lead to uncontrolled TLR-induced NF-κB activation and increased proinflammatory gene expression, therefore finally promoting the autoimmune inflammation. Whereas in Ezh2-deficient cells, Ezh2 depletion caused a substantial reduction of H3K27me3 levels, incensement of H3K4me3 at the Socs3 gene enhancer region induced the up-regulation of Socs3. Socs3 in turn mediated the Lys48-linked ubiquitination and degradation of TRAF6, leading to the suppression of TLR-induced MyD88-dependent NF-κB activation and the inhibition of genes involved in macrophage/microglial activation and autoimmune inflammation.
Materials and methods
Mice
Ezh2 floxed mice were crossed with lysozyme M-Cre mice (LysM-Cre; The Jackson Laboratory) to produce myeloid cell–conditional Ezh2 knockout mice (Ezh2flox/flox LysM-Cre; termed Ezh2M−/−) or with Cx3cr1-CreER-EYFP mice (The Jackson Laboratory) to produce microglia–conditional Ezh2 knockout mice (Ezh2flox/flox Cx3cr1-CreER-EYFP, termed Ezh2Mg−/−). All mice were maintained in a specific pathogen–free facility, and all animal experiments were in accordance with protocols approved by the institutional Biomedical Research Ethics Committee, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences.
Plasmids, antibodies, and reagents
Flag-Ezh2 and Flag-Ezh2ΔSET were generated by the insertion of cDNA encoding full-length Ezh2 and its SET domain–deficient mutant (ΔSET) into pLVX-GFP vector. The control and Socs3 shRNA–expressing pLKO.1 plasmids were purchased from OriGene. Antibodies for Ezh2 (5246s), H3K27me3 (9733s), H3 (9715s), P-p105 (4806s), P-p65 S536 (3033s), P-p38 (9215s), P-IκBα (2859s), P-Jnk (4668s), and Socs3 (2932s) were purchased from Cell Signaling Technology. Antibodies for IκBα (sc-371), p38 (sc-7149), Jnk1 (sc-474), Erk (sc-94), P-Erk (sc-7383), p105 (sc-1190), p65 (sc-109), and Traf6 (sc-7221) were purchased from Santa Cruz Biotechnology. Lys48-Ub antibody was purchased from Merck Millipore. Antibodies used for flow cytometry were from eBioscience. The Ezh2 inhibitor GSK126 (HY-13470) was from MedChem Express. LPS was from Sigma Aldrich. CpG, poly I:C, and Pam3CSK4 were from InvivoGen.
DSS-induced colitis
To induce colitis, mice (6–8 wk old) were fed with 3% DSS (36–50 kD; MP Biomedicals) for 7 d followed by regular drinking water. For the neutrophil-deletion experiment, mice were injected i.v. with anti–mouse Ly6G antibody (BE0075-1; Bio X Cell) at the dose of 100 µg/mouse starting 1 d before DSS challenge and then injected with the antibody another three times at 2-d intervals. During the experiment, body weights, stool consistency, and gastrointestinal bleeding were monitored daily. Mice were sacrificed at the indicated time points, and colons were collected immediately for measuring colon length, histology analysis, and immune cell isolation from the mucosa.
Induction and assessment of EAE
The induction of active EAE was as previously described (Xiao et al., 2013). In brief, age- and sex-matched mice (8–10 wk old) were injected s.c. with 200 µg MOG35–55 peptide in CFA containing 5 mg/ml heat-killed Mycobacterium tuberculosis H37Ra (Difco). On the day of immunization and 48 h later, the mice were also injected i.p. with pertussis toxin (200 ng/mouse; List Biological Laboratories) in PBS. For the Ezh2Mg−/− mice and their littermates, 5- to 6-wk-old mice were treated orally with 4 mg tamoxifen (T5648; Sigma Aldrich) solved in 200 µl corn oil (C8267; Sigma Aldrich) via gavage at two time points 48 h apart for the induction of Cre recombinase in these Cx3cr1-CreER-EYFP mice. 1 mo later, tamoxifen-exposed mice were applied for the EAE induction. The immunized mice were examined daily and scored for disease severity by using the standard method: 0, no clinical signs; 0.5, partially limp tail; 1, paralyzed tail; 2, loss in coordinated movement, hind limb paresis; 2.5, one hind limb paralyzed; 3, both hind limbs paralyzed; 3.5, hind limbs paralyzed, weakness in forelimbs; 4, forelimbs paralyzed; 5, moribund or death. After the onset of EAE, food and water were provided on the cage floor.
Bone marrow chimeras
Bone marrow cells (10 × 106) derived from donor mice were i.v. injected into lethally irradiated (950 rad) recipient mice. The lethal-dose irradiation would eliminate the bone marrow and peripheral immune cells without affecting the radioresistant CNS-resident cells, thus the chimeric mice would have their peripheral immune system reconstituted with bone marrow cells derived from donor mice. After 8 wk, the chimeric mice were immunized for EAE induction.
Histological analysis
Spinal cords dissected from mice transcardially perfused with 4% paraformaldehyde were fixed in 4% paraformaldehyde overnight, embedded in paraffin, and sectioned for staining with H&E and Luxol fast blue (LFB).
Isolation and analysis of immune cells from CNS or colon
For preparation of immune cells in the CNS or colon, CNS (brains and spinal cords) from naive and MOG35–55-immunized mice or colons from naive or DSS-induced colitis mice were excised and digested at 37°C with collagenase IV (0.5 mg/ml; Gibco) and DNase I (10 µg/ml; Roche) in RPMI 1640. Dispersed cells were layered onto a Percoll density gradient and isolated by collection of the interface fraction between 37% and 70% Percoll. After intensive washing, the frequencies of immune cell subpopulations in the CNS or colon were analyzed by flow cytometry. These frequencies were then multiplied by the total cell numbers of the Percoll-isolated cells from each mouse to get the absolute numbers of immune cell subpopulations. The colonic macrophages and CNS microglia were prepared by using the abovementioned protocol to collect the Percoll-isolated cells, which were then applied for FACS sorting by using specific antibodies against related cell markers. Inflammatory gene expression in CNS microglia or colonic macrophages was analyzed by qRT-PCR.
Real-time qRT-PCR
Total RNA was isolated by using TRIzol reagent (Invitrogen) and subjected to cDNA synthesis. qRT-PCR was performed in triplicate by using SYBR Green Supermix (Roche). The expression of individual genes was calculated by a standard curve method and normalized to the expression of Actb. The gene-specific PCR primers (all for mouse genes) are shown in Table S1.
Flow cytometry
Cell suspensions were subjected to flow cytometry analyses as previously described (Xiao et al., 2013) by using a Beckman Gallios or BD Aria2 flow cytometer. For the gating strategy, FSC/SSC was initially applied to the gate for live cells and then used for the antibodies with specific fluorochrome to make the subsequent gates. All the samples in the same experiments and comparisons were gated under the same parameters.
Cell culture
For culturing the primary microglia, the meninges of the brains from newborn mice (1–2 d old) were removed and then dissociated with 0.25% trypsin at 37°C, filtered with a 70-µm mesh, and the cells were cultured in complete DMEM medium. After 2 wk of cultivation (with medium changes at 24 h and then every 3 d), microglia were collected by intensive washing and shaking the culture (250 rpm) for 1 h at 37°C. For culturing the primary macrophages, bone marrow cells were isolated from femurs and tibias of adult mice and cultured for 5 d in a DMEM medium supplemented with macrophage CSF conditional medium.
Immunoblot, immunoprecipitation, and ubiquitination assays
Whole-cell lysates or whole-cell extracts were prepared and subjected to immunoblot (IB) and immunoprecipitation (IP) assays as described (Xiao et al., 2013). The samples were resolved by 8.25% SDS–PAGE. After electrophoresis, separated proteins were transferred onto nitrocellulose membrane (Millipore). For IB assays, the nitrocellulose membrane was blocked with 5% nonfat milk, incubated with a specific primary antibody, horseradish peroxidase–conjugated secondary antibody, and detected based on ECL reactive signal. For Lys48-linked ubiquitination assays, the cells were pretreated with MG132 (C2211; Sigma Aldrich) for 2 h, stimulated with LPS for the indicated time, and then lysed for IP followed by detecting ubiquitination by IB by using anti-Lys48 ubiquitin.
RNA-Seq analysis
Total RNA isolated from WT and Ezh2M−/− macrophages left nontreated or stimulated with LPS were subjected to HiSeq RNA-Seq performed by BGI Tech Solutions. Transcriptomic reads from the RNA-Seq experiments were mapped to a reference genome (mm10) by using Bowtie. Gene expression levels were quantified by using the RSEM software package. Significant genes were defined by the p-value and false discovery rate of cutoff of 0.05 and fold changes ≥1.5. Differentially expressed genes were subsequently analyzed by using the DAVID bioinformatics platform and Ingenuity Pathway Analysis. The genes used for the GO term analyses are the up- and down-regulated genes in LPS-stimulated WT macrophages versus LPS-stimulated Ezh2-deficient macrophages (P < 0.05, fold change ≥ 1.2).
ChIP-Seq assay
The chromatin of WT macrophages left nontreated or treated with LPS was prepared and subjected to ChIP-Seq analysis performed by Active Motif. 75 nucleotide reads generated by Illumina sequencing were mapped to the genome by using the BWA algorithm with default settings. Only reads that passed the Illumina purity filter, aligned with no more than two mismatches, and mapped uniquely to the genome were used in the subsequent analysis. The heat maps and average profile for RefSeq gene bodies were generated by using ngsplot v2.61.
CHIP-qPCR
Macrophages (10 × 106) were stimulated for 2 h with LPS, fixed in 1% formaldehyde, and then sonicated. Lysates were subjected to IP with the indicated antibodies, and the precipitated DNA was then purified by columns (Millipore) and quantified by qPCR by using primers listed in Table S1. The precipitated DNA is presented as percentage of the total input DNA (y axis).
Statistical analysis
One-way or two-way ANOVA, where applicable, was performed to determine whether an overall statistically significant change existed before the Student’s t test was used to analyze the difference between any two groups. Data are presented as means ± SD. P < 0.05 was considered statistically significant.
Data availability
Sequencing data are deposited into the Gene Expression Omnibus (accession no. GSE101383), which includes RNA-Seq data no. GSE101316 and ChIP-Seq data no. GSE101320. Source data files for Figs. 4 and 5 are available online, and there is no restriction on data availability.
Online supplemental material
Fig. S1 shows that Ezh2 deficiency in macrophages does not affect peripheral T cell responses during EAE process and the identification of Ezh2 deletion efficiency in microglia of Ezh2Mg−/− mice. Fig. S2 shows that Ezh2 deficiency impairs TLR2-mediated proinflammatory gene expression in primary cultured macrophages and microglia. Fig. S3 shows that Ezh2 deficiency slightly impairs M2 macrophage polarization. Fig. S4 shows Ezh2 expression in brain samples from non-multiple sclerosis (MS) donors and MS patients. Table S1 shows primers used for real-time quantitative PCR or CHIP-qPCR.
Supplementary Material
Acknowledgments
This research was supported by grants from the National Natural Science Foundation of China (81571545 and 81770567), the Jiangsu Provincial Key Laboratory and Development Program (BE2016722), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB19000000), the Thousand Talents Plan of China, the CAS Key Laboratory of Stem Cell Biology, the Collaborative Innovation Center of Systems Biomedicine, the European Research Council (695714 IMMUNOALZHEIMER and 693606 IMPEDE), and the National Multiple Sclerosis Society (RG-1501-02926).
The authors declare no competing financial interests.
Author contributions: X. Zhang designed and performed the experiments, prepared the figures, and wrote part of the manuscript. Y. Wang and J. Yuan contributed to the experiments of colitis model and signaling analysis. N. Li contributed to the data analysis of RNA-Seq and ChIP-Seq. S. Pei, J. Xu, X. Luo, C. Mao, J. Liu, T. Yu, S. Gan, G. Constantin, and J. Jin contributed to part of the experiments. Q. Zheng and Y. Liang contributed to the construction of Ezh2 knockout BV2 cells. W. Guo provided the Cx3cr1-creER transgenic mice. J. Qiu provided the LysM-cre transgenic mice. J. Qin provided the Ezh2 flox mice and contributed to data analysis and manuscript editing. Y. Xiao designed and supervised the work, prepared the figures, and wrote the manuscript.
References
- Ajami B., Bennett J.L., Krieger C., McNagny K.M., and Rossi F.M.. 2011. Infiltrating monocytes trigger EAE progression, but do not contribute to the resident microglia pool. Nat. Neurosci. 14:1142–1149. 10.1038/nn.2887 [DOI] [PubMed] [Google Scholar]
- Akira S., Uematsu S., and Takeuchi O.. 2006. Pathogen recognition and innate immunity. Cell. 124:783–801. 10.1016/j.cell.2006.02.015 [DOI] [PubMed] [Google Scholar]
- Asquith M.J., Boulard O., Powrie F., and Maloy K.J.. 2010. Pathogenic and protective roles of MyD88 in leukocytes and epithelial cells in mouse models of inflammatory bowel disease. Gastroenterology. 139:519–529, 529.e1–529.e2. 10.1053/j.gastro.2010.04.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassaing B., Aitken J.D., Malleshappa M., and Vijay-Kumar M.. 2014. Dextran sulfate sodium (DSS)-induced colitis in mice. Curr. Protoc. Immunol. 104:15.25.1–15.25.14 [DOI] [PMC free article] [PubMed]
- Chastain E.M., Duncan D.S., Rodgers J.M., and Miller S.D.. 2011. The role of antigen presenting cells in multiple sclerosis. Biochim. Biophys. Acta. 1812:265–274. 10.1016/j.bbadis.2010.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F., Totaro M.G., Prosperini E., Notarbartolo S., Testa G., and Natoli G.. 2007. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 130:1083–1094. 10.1016/j.cell.2007.08.019 [DOI] [PubMed] [Google Scholar]
- Friese M.A., and Fugger L.. 2007. T cells and microglia as drivers of multiple sclerosis pathology. Brain. 130:2755–2757. 10.1093/brain/awm246 [DOI] [PubMed] [Google Scholar]
- Gambuzza M., Licata N., Palella E., Celi D., Foti Cuzzola V., Italiano D., Marino S., and Bramanti P.. 2011. Targeting Toll-like receptors: emerging therapeutics for multiple sclerosis management. J. Neuroimmunol. 239:1–12. 10.1016/j.jneuroim.2011.08.010 [DOI] [PubMed] [Google Scholar]
- Gao Z., and Tsirka S.E.. 2011. Animal models of MS reveal multiple roles of microglia in disease pathogenesis. Neurol. Res. Int. 2011:383087 10.1155/2011/383087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissmann F., Manz M.G., Jung S., Sieweke M.H., Merad M., and Ley K.. 2010. Development of monocytes, macrophages, and dendritic cells. Science. 327:656–661. 10.1126/science.1178331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelderman K.A., Hultqvist M., Pizzolla A., Zhao M., Nandakumar K.S., Mattsson R., and Holmdahl R.. 2007. Macrophages suppress T cell responses and arthritis development in mice by producing reactive oxygen species. J. Clin. Invest. 117:3020–3028. 10.1172/JCI31935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F., Greter M., Leboeuf M., Nandi S., See P., Gokhan S., Mehler M.F., Conway S.J., Ng L.G., Stanley E.R., et al. 2010. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 330:841–845. 10.1126/science.1194637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F., Schultze J.L., Murray P.J., Ochando J., and Biswas S.K.. 2016. New insights into the multidimensional concept of macrophage ontogeny, activation and function. Nat. Immunol. 17:34–40. 10.1038/ni.3324 [DOI] [PubMed] [Google Scholar]
- Goldmann T., Wieghofer P., Müller P.F., Wolf Y., Varol D., Yona S., Brendecke S.M., Kierdorf K., Staszewski O., Datta M., et al. 2013. A new type of microglia gene targeting shows TAK1 to be pivotal in CNS autoimmune inflammation. Nat. Neurosci. 16:1618–1626. 10.1038/nn.3531 [DOI] [PubMed] [Google Scholar]
- Gordon S., and Taylor P.R.. 2005. Monocyte and macrophage heterogeneity. Nat. Rev. Immunol. 5:953–964. 10.1038/nri1733 [DOI] [PubMed] [Google Scholar]
- Gunawan M., Venkatesan N., Loh J.T., Wong J.F., Berger H., Neo W.H., Li L.Y., La Win M.K., Yau Y.H., Guo T., et al. 2015. The methyltransferase Ezh2 controls cell adhesion and migration through direct methylation of the extranuclear regulatory protein talin. Nat. Immunol. 16:505–516. 10.1038/ni.3125 [DOI] [PubMed] [Google Scholar]
- Hamilton J.A., Cook A.D., and Tak P.P.. 2016. Anti-colony-stimulating factor therapies for inflammatory and autoimmune diseases. Nat. Rev. Drug Discov. 16:53–70. 10.1038/nrd.2016.231 [DOI] [PubMed] [Google Scholar]
- Heppner F.L., Greter M., Marino D., Falsig J., Raivich G., Hövelmeyer N., Waisman A., Rülicke T., Prinz M., Priller J., et al. 2005. Experimental autoimmune encephalomyelitis repressed by microglial paralysis. Nat. Med. 11:146–152. 10.1038/nm1177 [DOI] [PubMed] [Google Scholar]
- Kamada N., Hisamatsu T., Okamoto S., Chinen H., Kobayashi T., Sato T., Sakuraba A., Kitazume M.T., Sugita A., Koganei K., et al. 2008. Unique CD14 intestinal macrophages contribute to the pathogenesis of Crohn disease via IL-23/IFN-gamma axis. J. Clin. Invest. 118:2269–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T., and Akira S.. 2007. TLR signaling. Semin. Immunol. 19:24–32. 10.1016/j.smim.2006.12.004 [DOI] [PubMed] [Google Scholar]
- Kruidenier L., Chung C.W., Cheng Z., Liddle J., Che K., Joberty G., Bantscheff M., Bountra C., Bridges A., Diallo H., et al. 2012. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature. 488:404–408. 10.1038/nature11262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Peng J., Sun T., Li N., Zhang L., Ren J., Yuan H., Kan S., Pan Q., Li X., et al. 2017. Epithelial EZH2 serves as an epigenetic determinant in experimental colitis by inhibiting TNFα-mediated inflammation and apoptosis. Proc. Natl. Acad. Sci. USA. 114:E3796–E3805. 10.1073/pnas.1700909114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomaga M.A., Yeh W.C., Sarosi I., Duncan G.S., Furlonger C., Ho A., Morony S., Capparelli C., Van G., Kaufman S., et al. 1999. TRAF6 deficiency results in osteopetrosis and defective interleukin-1, CD40, and LPS signaling. Genes Dev. 13:1015–1024. 10.1101/gad.13.8.1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M., Powers S.E., Maienschein-Cline M., Bartom E.T., Hamel K.M., Kee B.L., Dinner A.R., and Clark M.R.. 2011. Epigenetic repression of the Igk locus by STAT5-mediated recruitment of the histone methyltransferase Ezh2. Nat. Immunol. 12:1212–1220. 10.1038/ni.2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall D., Cameron J., Lightwood D., and Lawson A.D.. 2007. Blockade of colony stimulating factor-1 (CSF-I) leads to inhibition of DSS-induced colitis. Inflamm. Bowel Dis. 13:219–224. 10.1002/ibd.20055 [DOI] [PubMed] [Google Scholar]
- Medzhitov R., and Horng T.. 2009. Transcriptional control of the inflammatory response. Nat. Rev. Immunol. 9:692–703. 10.1038/nri2634 [DOI] [PubMed] [Google Scholar]
- Miranda-Hernandez S., Gerlach N., Fletcher J.M., Biros E., Mack M., Körner H., and Baxter A.G.. 2011. Role for MyD88, TLR2 and TLR9 but not TLR1, TLR4 or TLR6 in experimental autoimmune encephalomyelitis. J. Immunol. 187:791–804. 10.4049/jimmunol.1001992 [DOI] [PubMed] [Google Scholar]
- Nathan C., and Ding A.. 2010. Nonresolving inflammation. Cell. 140:871–882. 10.1016/j.cell.2010.02.029 [DOI] [PubMed] [Google Scholar]
- Platt A.M., Bain C.C., Bordon Y., Sester D.P., and Mowat A.M.. 2010. An independent subset of TLR expressing CCR2-dependent macrophages promotes colonic inflammation. J. Immunol. 184:6843–6854. 10.4049/jimmunol.0903987 [DOI] [PubMed] [Google Scholar]
- Ponomarev E.D., Shriver L.P., Maresz K., and Dittel B.N.. 2005. Microglial cell activation and proliferation precedes the onset of CNS autoimmunity. J. Neurosci. Res. 81:374–389. 10.1002/jnr.20488 [DOI] [PubMed] [Google Scholar]
- Ponomarev E.D., Veremeyko T., Barteneva N., Krichevsky A.M., and Weiner H.L.. 2011. MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-α-PU.1 pathway. Nat. Med. 17:64–70. 10.1038/nm.2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz M., Garbe F., Schmidt H., Mildner A., Gutcher I., Wolter K., Piesche M., Schroers R., Weiss E., Kirschning C.J., et al. 2006. Innate immunity mediated by TLR9 modulates pathogenicity in an animal model of multiple sclerosis. J. Clin. Invest. 116:456–464. 10.1172/JCI26078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H., Holdbrooks A.T., Liu Y., Reynolds S.L., Yanagisawa L.L., and Benveniste E.N.. 2012a SOCS3 deficiency promotes M1 macrophage polarization and inflammation. J. Immunol. 189:3439–3448. 10.4049/jimmunol.1201168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H., Yeh W.I., De Sarno P., Holdbrooks A.T., Liu Y., Muldowney M.T., Reynolds S.L., Yanagisawa L.L., Fox T.H. III, Park K., et al. 2012b Signal transducer and activator of transcription-3/suppressor of cytokine signaling-3 (STAT3/SOCS3) axis in myeloid cells regulates neuroinflammation. Proc. Natl. Acad. Sci. USA. 109:5004–5009. 10.1073/pnas.1117218109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo K., and Glass C.K.. 2011. Microglial cell origin and phenotypes in health and disease. Nat. Rev. Immunol. 11:775–787. 10.1038/nri3086 [DOI] [PubMed] [Google Scholar]
- Satoh T., Takeuchi O., Vandenbon A., Yasuda K., Tanaka Y., Kumagai Y., Miyake T., Matsushita K., Okazaki T., Saitoh T., et al. 2010. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat. Immunol. 11:936–944. 10.1038/ni.1920 [DOI] [PubMed] [Google Scholar]
- Schuettengruber B., Chourrout D., Vervoort M., Leblanc B., and Cavalli G.. 2007. Genome regulation by polycomb and trithorax proteins. Cell. 128:735–745. 10.1016/j.cell.2007.02.009 [DOI] [PubMed] [Google Scholar]
- Schulz C., Gomez Perdiguero E., Chorro L., Szabo-Rogers H., Cagnard N., Kierdorf K., Prinz M., Wu B., Jacobsen S.E., Pollard J.W., et al. 2012. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 336:86–90. 10.1126/science.1219179 [DOI] [PubMed] [Google Scholar]
- Su I.H., Basavaraj A., Krutchinsky A.N., Hobert O., Ullrich A., Chait B.T., and Tarakhovsky A.. 2003. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat. Immunol. 4:124–131. 10.1038/ni876 [DOI] [PubMed] [Google Scholar]
- Takeda K., and Akira S.. 2004. TLR signaling pathways. Semin. Immunol. 16:3–9. 10.1016/j.smim.2003.10.003 [DOI] [PubMed] [Google Scholar]
- Tumes D.J., Onodera A., Suzuki A., Shinoda K., Endo Y., Iwamura C., Hosokawa H., Koseki H., Tokoyoda K., Suzuki Y., et al. 2013. The polycomb protein Ezh2 regulates differentiation and plasticity of CD4(+) T helper type 1 and type 2 cells. Immunity. 39:819–832. 10.1016/j.immuni.2013.09.012 [DOI] [PubMed] [Google Scholar]
- Wei G., Wei L., Zhu J., Zang C., Hu-Li J., Yao Z., Cui K., Kanno Y., Roh T.Y., Watford W.T., et al. 2009. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 30:155–167. 10.1016/j.immuni.2008.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J.J., Munro K.M., and Palmer T.M.. 2014. Role of ubiquitylation in controlling suppressor of cytokine signalling 3 (Socs3) function and expression. Cells. 3:546–562. 10.3390/cells3020546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn T.A., Chawla A., and Pollard J.W.. 2013. Macrophage biology in development, homeostasis and disease. Nature. 496:445–455. 10.1038/nature12034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Jin J., Chang M., Chang J.H., Hu H., Zhou X., Brittain G.C., Stansberg C., Torkildsen Ø., Wang X., et al. 2013. Peli1 promotes microglia-mediated CNS inflammation by regulating Traf3 degradation. Nat. Med. 19:595–602. 10.1038/nm.3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki R., Lu H., Butovsky O., Ohno N., Rietsch A.M., Cialic R., Wu P.M., Doykan C.E., Lin J., Cotleur A.C., et al. 2014. Differential roles of microglia and monocytes in the inflamed central nervous system. J. Exp. Med. 211:1533–1549. 10.1084/jem.20132477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Liu Z., Cheng X., Zheng Y., Zeng F., and He Y.. 2015. Socs1 and Socs3 degrades Traf6 via polyubiquitination in LPS-induced acute necrotizing pancreatitis. Cell Death Dis. 6:e2012 10.1038/cddis.2015.342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond E., Varol C., Farache J., Elmaliah E., Satpathy A.T., Friedlander G., Mack M., Shpigel N., Boneca I.G., Murphy K.M., et al. 2012. Ly6C hi monocytes in the inflamed colon give rise to proinflammatory effector cells and migratory antigen-presenting cells. Immunity. 37:1076–1090. 10.1016/j.immuni.2012.08.026 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data are deposited into the Gene Expression Omnibus (accession no. GSE101383), which includes RNA-Seq data no. GSE101316 and ChIP-Seq data no. GSE101320. Source data files for Figs. 4 and 5 are available online, and there is no restriction on data availability.