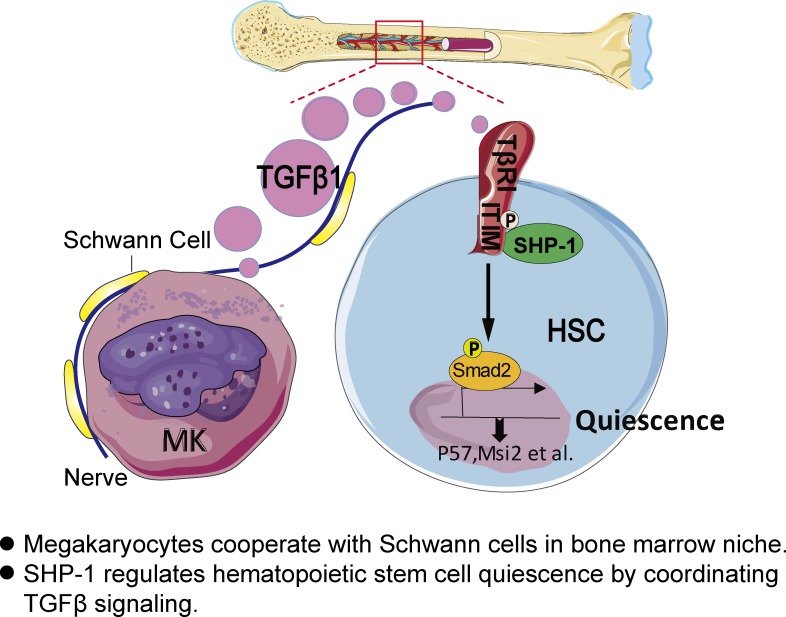
Megakaryocytes are known for hematopoietic stem cell (HSC) regulation. Here, Jiang et al. show that megakaryocytes cooperate with Schwann cells in bone marrow niche and that SHP-1 regulates HSC quiescence by coordinating TGF-β signaling in megakaryocyte niche.
Abstract
Cell cycle quiescence is critical for hematopoietic stem cell (HSC) maintenance. TGF-β signaling in bone marrow niche has been identified in regulating HSC quiescence; however, the intrinsic regulatory mechanisms remain unclear. This study reports that Shp-1 knockout HSCs have attenuated quiescence and impaired long-term self-renewal. SHP-1–activated HSCs are surrounded by megakaryocytes, which regulate HSC quiescence by producing TGF-β1. Mechanistically, SHP-1 interacts with the immunoreceptor tyrosine-based inhibition motif on TGF-β receptor 1 and is critical for TGF-β signaling activation in HSCs. Functionally, Shp-1 knockout HSCs do not respond to TGF-β–enforced HSC quiescence regulation, both in vitro and in vivo. Therefore, we identify TGF-β–SHP-1 as a novel intrinsic regulatory mechanism for HSC quiescence maintenance.
Graphical Abstract
Introduction
Hematopoietic stem cells (HSCs) produce differentiated cells to replenish the blood system through the lifetime. The quiescence of HSCs is a defining characteristic for their maintenance during homeostasis and regeneration post-injury (Wilson et al., 2008). Several bone marrow (BM) niche cells, such as mesenchymal stem cells (MSCs), nonmyelinating Schwann cells, and megakaryocytes (MKs) are known to maintain HSC quiescence (Yamazaki et al., 2011; Kunisaki et al., 2013; Bruns et al., 2014; Zhao et al., 2014; Itkin et al., 2016). Although the quiescent signals from MSCs need further characterization, it is clear that MKs and nonmyelinating Schwann cells regulate HSC quiescence by coordinating TGF-β signaling (Yamazaki et al., 2011; Zhao et al., 2014). TGF-β is a key signal for HSC quiescence regulation (Yamazaki et al., 2009; Blank and Karlsson, 2015); however, it is unclear how this niche signal regulates HSC quiescence through its intrinsic mechanisms.
SHP-1 is an SH2 domain–containing protein tyrosine phosphatase that controls the intracellular phosphotyrosine levels (Wu et al., 2003b; Lorenz, 2009). SHP-1 is expressed in all hematopoietic cells and attenuates receptor tyrosine kinase pathways initiated by growth factors and cytokines (Neel et al., 2003). SHP-1 inhibits cell growth and suppresses their oncogenic potentials in lymphocytes (Tibaldi et al., 2011; Viant et al., 2014; Chen et al., 2015). Loss of SHP-1 expression in B cells or dendritic cells results in elevated B-1a or Th1 cell differentiation and induces autoimmunity (Pao et al., 2007; Kaneko et al., 2012). Loss of SHP-1 expression in tumor-specific T cells, or natural killer cells, promotes their immune responsiveness and antitumor function (Stromnes et al., 2012; Viant et al., 2014). Our data suggest that SHP-1 might be involved in hematopoiesis and leukemogenesis, by interacting with immunoreceptor tyrosine-based inhibition motif (ITIM)–bearing receptors such as LAIR1 and LILRB2 (Zheng et al., 2012; Kang et al., 2015, 2016). However, whether SHP-1 directly contributes to HSC regulation is unknown. In this work, we found that SHP-1 is critical for TGF-β–mediated HSC quiescence control.
Results and discussion
Loss of SHP-1 results in HSC activation and subsequent exhaustion
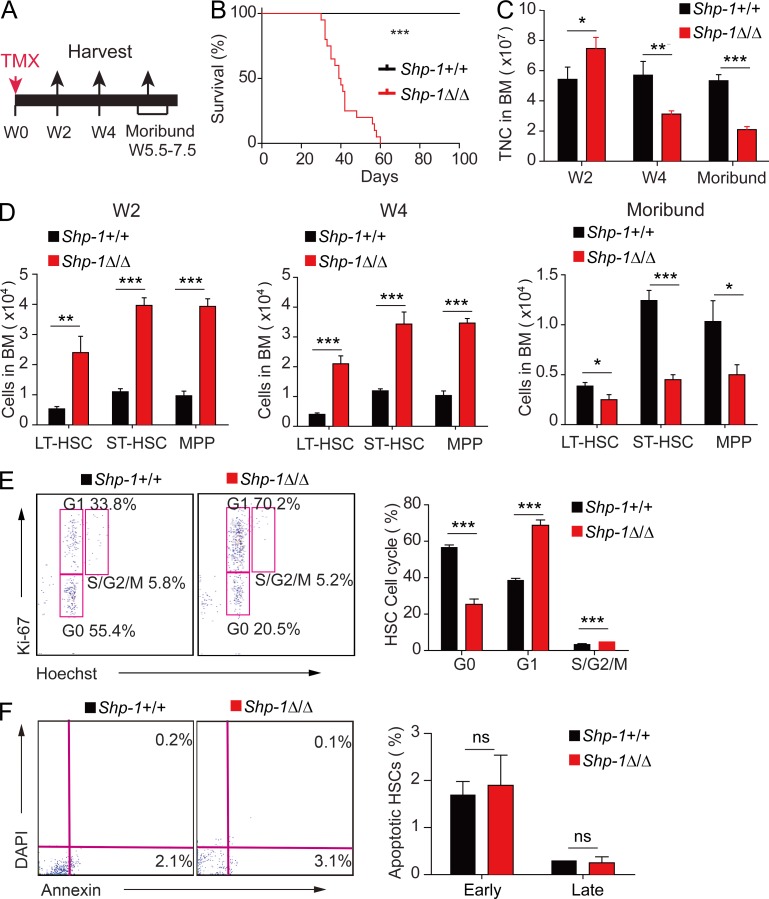
To obtain an inducible loss-of-function model for SHP-1 in HSCs, we crossed Shp-1fl/fl mice (Sacchetti et al., 2007) with transgenic mice expressing the tamoxifen-inducible Cre recombinase under the control of the stem cell leukemia (Scl) enhancer, which enabled knockout of floxed genes in HSCs and hematopoietic progenitors, upon tamoxifen treatment (Göthert et al., 2005). The resultant Scl-CreER+; Shp-1fl/fl (Shp-1Δ/Δ) mice and the control mice Scl-CreER–; Shp-1fl/fl (Shp-1+/+) were treated with tamoxifen to initiate Shp-1 knockout in HSCs (Fig. 1 A). The control mice had a normal lifespan. However, Shp-1Δ/Δ mice began to die ∼40 d after tamoxifen treatment (Fig. 1 B). Furthermore, we found that the total number of BM cells in Shp-1Δ/Δ mice was increased ∼37% at 2 wk, but reduced ∼45% at 4 wk and further reduced ∼60% at ∼7 wk (at moribund), after tamoxifen treatment (Fig. 1 C). The dynamic change of BM cell numbers indicated that there was a transient activation with subsequent exhaustion of hematopoiesis as a result of SHP-1 knockout in HSCs.
Figure 1.
Loss of Shp-1 results in HSC activation and subsequent exhaustion. (A) Schema for tamoxifen treatment and sample analysis time points. TMX, tamoxifen; W, week. (B) Survival curves of Scl-CreER–;Shp-1fl/fl (Shp-1+/+) and Scl-CreER+;Shp-1fl/fl (Shp-1Δ/Δ) mice treated with tamoxifen (n = 23 mice; P < 0.0001, log-rank test). (C) Total BM cell numbers in Shp-1+/+ and Shp-1Δ/Δ mice at indicated time points after tamoxifen treatment (n = 3 mice). TNC, total nucleated cells. (D) Comparison of LT-HSC, ST-HSC, and MPP numbers in Shp-1+/+ and Shp-1Δ/Δ mice at three time points after tamoxifen treatment (n = 10 mice). (E) Flow cytometry analysis of cell cycle stage of BM cells from Shp-1+/+ and Shp-1Δ/Δ mice harvested 4 wk after tamoxifen treatment. Left panel shows the representative flow cytometry plots. Right panel plots percentages of Shp-1+/+ and Shp-1Δ/Δ cells in each stage of the cell cycle (n = 3 mice). (F) Flow cytometry analysis of early (detected as Annexin V+/7-AAD− staining) and late (detected as Annexin V+/7-AAD+ staining) apoptotic HSCs from BM of tamoxifen-treated Shp-1 KO mice (n = 3 mice). *, P < 0.05; **, P < 0.01; ***, P < 0.001. ns, not significant. Error bars show mean ± SEM.
To explore the effect of Shp-1 knockout in HSCs, we investigated the phenotypic HSCs and hematopoietic progenitors in the BM from the Shp-1Δ/Δ and Shp-1+/+ control mice. We found that long-term HSCs (LT-HSCs), identified as Lin–Sca-1+Kit+CD34–Flk2– cells, were increased ∼4.5-fold at 2 wk after tamoxifen treatment in Shp-1Δ/Δ mice compared with their control counterparts. Similarly, short-term HSCs (ST-HSCs; Lin–Sca-1+Kit+CD34+Flk2–) and hematopoietic multipotent progenitors (MPPs; Lin–Sca-1+Kit+CD34+Flk2+) were increased ∼3.6-fold and ∼4.1-fold, respectively, in Shp-1Δ/Δ mice compared with control Shp-1+/+ mice. Furthermore, LT-HSC, ST-HSC, and MPPs were also increased ∼5.2-fold, 2.9-fold, and 3.4-fold, respectively, in Shp-1Δ/Δ mice compared with controls at 4 wk after tamoxifen treatment. However, LT-HSCs were decreased ∼35%, and ST-HSCs and MPPs were decreased ∼63.8% and ∼51.6%, respectively, in Shp-1Δ/Δ mice compared with controls when Shp-1Δ/Δ mice were moribund (Fig. 1 D). The data suggest that HSCs were activated for proliferation at the early stage when Shp-1 was knocked out; however, this activation led to HSC exhaustion, which is responsible for the total BM cell reduction and moribund phenotype.
To identify whether Shp-1 deletion truly triggered HSC activation, we analyzed the cell cycle profile of HSCs at 4 wk after tamoxifen treatment. We found that the percentage of HSCs in G0 phase was reduced by 55.4%, whereas the G1 phase showed a ∼1.8-fold increase in Shp-1Δ/Δ mice compared with the control, which indicated the greatly decreased quiescence of HSCs after SHP-1 knockout (Fig. 1 E). We did not observe a significant difference in apoptosis in HSCs between Shp-1Δ/Δ and Shp-1+/+ mice (Fig. 1 F). The absence of Shp-1 causes activation of differentiated hematopoietic cells, which may compromise HSC functions. We further analyzed Shp-1Δ/Δ mice at 1 wk after tamoxifen treatment, when no activation of mature hematopoietic cells was observed, and found that the HSC quiescence was greatly decreased (40% reduction in G0) before HSC number increased (Fig. S1). Additionally, MKs have functional ITIM receptors that activate SHP-1, and Shp-1–null MKs show abnormal development and function (Mazharian et al., 2013). Then, we further used platelet factor 4-cre (Pf4-cre) to specifically delete Shp-1 in MKs by generating the Pf4-cre; Shp-1fl/fl line (Tiedt et al., 2007). We found that deletion of Shp-1 in MKs did not influence HSCs either in number or their cell cycle (Fig. S2). Overall, our data suggested that SHP-1 prevents exhaustion by maintaining HSC quiescence.
Loss of SHP-1 reduced HSC long-term self-renewal
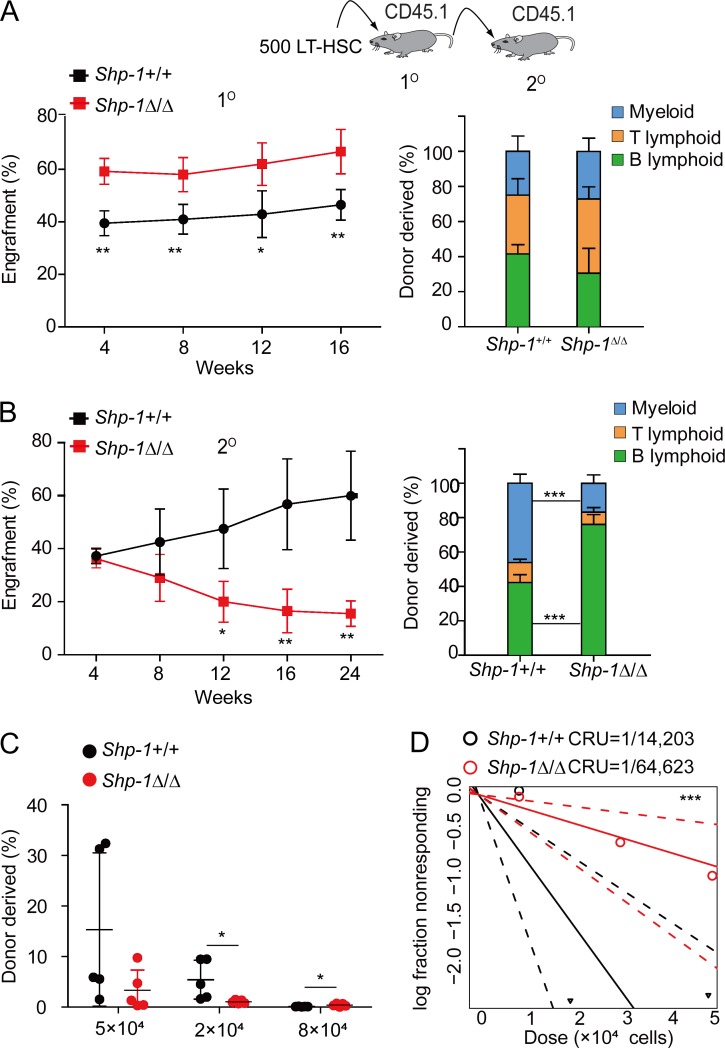
To evaluate the HSC functional change after Shp-1 knockout, we performed competitive transplantation experiments. As shown in Fig. 2 A, we transplanted 500 LT-HSCs from tamoxifen-treated Shp-1Δ/Δ or Shp-1+/+ control animals with competitor cells into lethally irradiated recipients. We found that HSCs from Shp-1Δ/Δ mice showed a significantly higher level of donor cell reconstitution (∼1.4-fold at 16 wk after transplantation, P = 0.0074) than their counterparts. We did not observe any biased lineage reconstitution (Fig. 2 A). To further explore whether loss of Shp-1 affects HSC long-term self-renewal, we performed a secondary transplantation. Interestingly, we found that Shp-1–null HSCs gradually lost their ability for hematopoietic reconstitution. The donor cell reconstitution of Shp-1–null HSCs was significantly reduced at 24 wk after secondary transplantation (74.2% reduction, P = 0.0022) compared with Shp-1+/+ HSCs. We also observed that Shp-1–null HSCs generated more B lymphocytes than control HSCs (46.2% vs. 16.8%), but less myeloid cells (42.2% vs. 76.1%; Fig. 2 B). Overall, this indicated that the loss of Shp-1 in HSCs caused increased activation and proliferation but impaired long-term self-renewal.
Figure 2.
Loss of Shp-1 reduced HSC long-term self-renewal. (A and B) Competitive reconstitution analysis using 500 HSCs from tamoxifen-treated Shp-1+/+ and Shp-1Δ/Δ mice. Peripheral blood analysis for total engrafted donor cells at the indicated number of weeks after primary transplantation (1°; A) and secondary transplantation (2°; B) and the percentage of donor-derived B, T, and myeloid lineage cells at 16 wk of primary transplantation (right panel of A) and at 24 wk of secondary transplantation (right panel of B; n = 5 mice per group). (C and D) Limiting dilution assays comparing the frequencies of HSC in Shp-1+/+ and Shp-1Δ/Δ mice. BM cells were collected 4 wk after tamoxifen treatment. The repopulation percentages were detected 20 wk after transplant of Shp-1+/+ and Shp-1Δ/Δ mice (C) and plot of competitive repopulating units (CRUs) calculated using ELDA (P = 0.009; D). n = 5 mice per group. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Error bars show mean ± SEM.
To quantify the number of functional HSCs in Shp-1Δ/Δ mice, we performed a competitive repopulation unit (CRU) assay. 50,000, 20,000, and 8,000 BM cells from Shp-1Δ/Δ mice or Shp-1+/+ controls were transplanted into irradiated recipients with 200,000 competitor cells. The percentage of donor cell reconstitution was measured at 40 wk after transplantation (Fig. 2 C) to quantify the number of functional HSCs per mice. We found that the frequency of HSCs in Shp-1Δ/Δ mice was 1 per 64,623, which was ∼78% less than the frequency (1 per 14,203) in Shp-1+/+ control mice (P = 0.009; Fig. 2 D). Overall, our data demonstrated that the loss of SHP-1 induced transient HSC activation and proliferation, but reduced functional long-term self-renewal of HSCs.
SHP-1 interacts with TGF-β receptor 1 (TβRI) and coordinates TGF-β signaling.
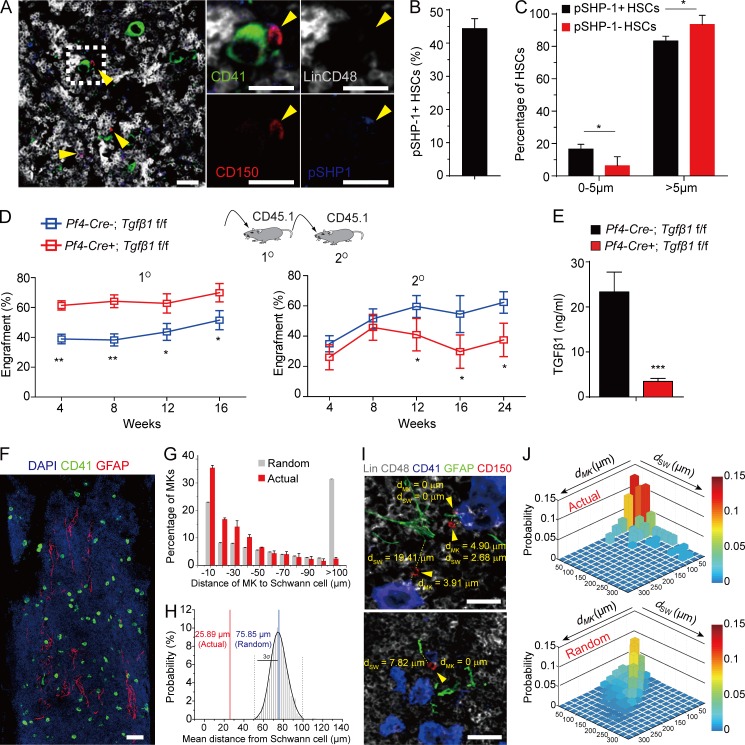
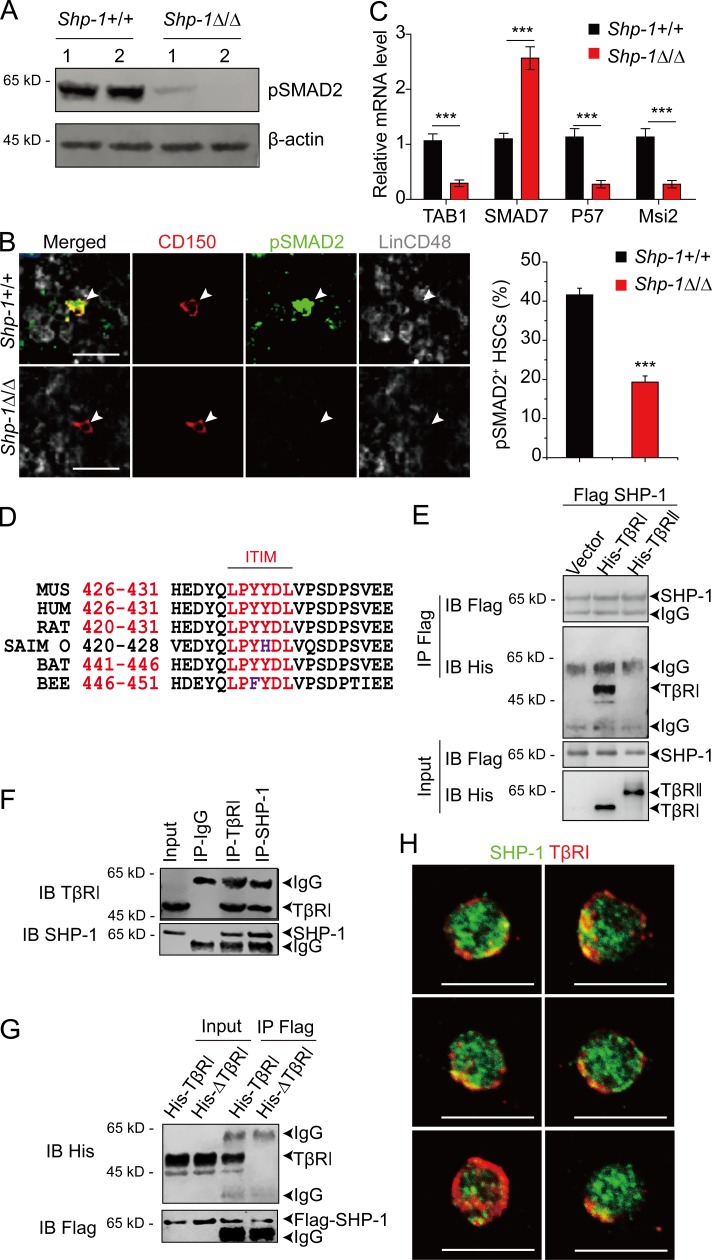
To understand the mechanism of SHP-1 in regulating HSC quiescence, we performed immunostaining to investigate how SHP-1–activated HSCs are maintained in the BM niche. We found that ∼44% of HSCs are pSHP-1+ and interestingly, pSHP-1+ HSCs were significantly associated with MKs compared with pSHP-1– HSCs (P = 0.023; Fig. 3, A–C and Fig. S3). This finding is in line with our study that MKs maintain HSC quiescence by TGF-β signaling (Zhao et al., 2014). Furthermore, we hypothesized that SHP-1 might regulate HSC quiescence by coordinating TGF-β signaling. Consistent with Shp1 knockout the loss of TGF-β signaling resulted in transient HSC activation, but repressed long-term self-renewal potential. Our data showed that the knockout of TGF-β1 from MKs resulted in increased reconstitution of BM cells from Pf4-cre+; TGF-β1f/f compared with Pf4-cre–; TGF-β1f/f control mice (1.35-fold increase at 16 wk after transplantation, P = 0.028; Fig. 3 D, left). However, the reconstitution was decreased by 40% at 24 wk after secondary transplantation (P = 0.038; Fig. 3 D, right). We further found that TGF-β1 protein expression in BM was remarkably reduced (85.2% reduction) by ELISA in Pf4-cre+; TGF-β1f/f mice compared with control Pf4-cre–; TGF-β1f/f mice (Fig. 3 E). This indicates that MKs are the major source of TGF-β1 protein in BM. Because MKs produce TGF-β1 in latent form and Schwann cells are the major activator for latent TGF-β1 in BM (Yamazaki et al., 2011), we further analyzed the association of MKs with Schwann cells in BM and found that a considerable fraction of MKs (35.4%, <10 µm) were associated with Schwann cells (Fig. 3, F and G). To test the significance of this association, we compared the actual MKs and simulations of randomly placed MKs in relation to Schwann cells (Fig. 3 G). The observed mean distance of MKs to Schwann cells (25.89 µm) was statistically different from the mean distance of randomly placed MKs to Schwann cells (75.85 µm) (Fig. 3 H). Our data show that the observed association of MKs with Schwann cells is statistically different from a random distribution (P = 2.7 × 10−14 by two-sample Kolmogorov-Smirnov [KS] test; Fig. 3 G). This suggests a niche surrounding MKs and Schwann cells in BM dedicated to HSC maintenance. To explore the spatial arrangement of HSCs in MK–Schwann cell niche, we measured the distances of actual and randomly placed HSCs to MKs (dMK) and to Schwann cells (dSW), respectively (Fig. 3 I). The actual HSCs were significantly closer to MK–Schwann cell clusters compared with randomly placed HSCs (P = 1.6 × 10−20 by two-dimensional KS test with two samples; Fig. 3, I and J). Overall, our data suggest that MKs and Schwann cells may synergistically form a unique niche to coordinate TGF-β signaling for HSC regulation in BM.
Figure 3.
The spatial relationship of pSHP-1+ HSCs in BM niche. (A) Representative image of a BM section from a C57BL/6J mouse showing pSHP-1+ HSCs were surrounded by MKs. The yellow arrowheads indicate HSCs. Gray, lineage cells; green, MKs; red, HSCs; blue, pSHP-1. Dashed box indicates area of focus. Bars, 25 µm. (B) Percentage of pSHP-1+ HSCs in BM. (C) Distance between pSHP-1+ HSCs and pSHP-1– HSCs to MKs as indicated (n = 311 HSCs from four mice in four independent experiments). (D) Competitive reconstitution analysis of functional HSCs from Pf4-cre–; TGF-β1f/f or from Pf4-cre+; TGF-β1f/f mice. Plots of peripheral blood analysis for total engrafted donor cells at the indicated number of weeks after primary transplantation (1°) and secondary transplantation (2°; n = 10). (E) TGF-β1 protein levels as determined by ELISA in BM from Pf4-cre; TGF-β1f/f mice (cre−, n = 4 mice; cre+, n = 5 mice). The spatial relationship of MKs and Schwann cells. (F) Representative image of a BM section showing CD41+ MKs (green) and GFAP+ Schwann cells (red). Bar, 100 µm. (G) Distance between MKs and Schwann cells. The numbers on the x axis indicate intervals of 10 µm (−10 indicates the interval 0–10; −20 indicates 10–20, and so on). P = 2.7 × 10−14 by two-sample KS test. (H) Probability distributions of the mean distances between MKs and Schwann cells derived from simulations of randomly positioned MKs and actual Schwann on maps of BM. Mean distances observed in situ (red line) are shown in relation to the grand mean (mean of the means) ± 3 SD (3σ, blue and dotted black lines, respectively). Probability P (μ < 25.89 µm) = 2.2 × 10−16. n = 365 MKs from three mice in three independent experiments. (I) The spatial relationship of HSCs (red), MKs (blue), and Schwann cells (green). dMK and dSW indicate the distance between the HSC to the closest MK and Schwann cell, respectively. The arrowheads indicate HSCs. Bars, 25 µm. (J) 2D probability distribution of the distances between HSCs to MKs (dMK) and Schwann cells (dSW) in the BM. Upper panel, HSC distribution in the BM (n = 95 HSCs, from three mice); lower panel, randomized HSC distribution models generated by computational simulations of random HSC localization (P = 1.57 × 10−20 by 2D KS test with two samples). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The consistent phenotype between TGF-β1 knockout from niche cells and SHP-1 knockout from HSCs suggested that SHP-1 might be involved in TGF-β signaling regulation. To investigate this hypothesis, we first measured the level of pSMAD2 (downstream effectors of TGF-β signaling). We found that the level of pSMAD2 was remarkably reduced in BM cells from Shp-1Δ/Δ mice than from control Shp-1+/+ mice (Fig. 4 A). Consistently, the frequency of pSMAD2+ HSCs from Shp-1Δ/Δ mice was reduced by 53.6% compared with HSCs from Shp-1+/+ controls (Fig. 4 B). Loss of SHP-1 resulted in decreased expression of TGF-β activated kinase binding protein 1 (TAB1; 72.5% reduction) and increased expression of TGF-β inhibitor SMAD7 (2.57-fold increase) in HSCs, which indicated the dampened TGF-β signal activity in HSCs. Furthermore, SHP-1–null HSCs have low expression of P57 (75.6% reduction), which is a SMAD2/3 downstream factor and is critical for TGF-β–mediated cell cycle arrest in HSCs (Scandura et al., 2004; Yamazaki et al., 2006, 2009). SHP-1–null HSCs also have a low expression of Musashi-2 (Msi2; 75.6% reduction), which controls TGF-β signaling in HSCs (Fig. 4 C; Park et al., 2014). Overall, these data indicated that SHP-1–null HSCs had reduced TGF-β signaling, which may account for their loss of quiescence.
Figure 4.
SHP-1 interacts with TβRI and coordinates TGF-β signaling. (A) Immunoblotting for pSMAD2 protein in BM Lin− cells. Actin was used as a loading control. 1 and 2 indicate two individual mice. (B) Representative image and frequency of pSMAD2 in HSCs from Shp-1+/+ or Shp-1Δ/Δ mice (n = 112 HSCs from Shp-1+/+ mice; n = 166 HSCs from Shp-1Δ/Δ mice from three mice). The arrowheads indicate HSCs. Bars, 25 µm. (C) RT-qPCR analysis of genes in HSCs from Shp-1+/+ or Shp-1Δ/Δ mice. (D) ITIM motif in TβRI is highly conserved among different species. (E) Coexpression of Flag SHP-1 WT and vector, or HA-TβRI, or HA-TβRII in 293T cells. Cell lysate were precipitated with Flag beads; both input and precipitation were immune-blotted with Flag or His antibodies. (F) SHP-1 binds with TβRI endogenously in THP1 cells in bidirectional pull-down assay. (G) SHP-1 binds with TβRI by the ITIM motif. ITIM motif deleted TβRI or WT TβRI was used in the coimmunoprecipitation assay in 293 T cells. (H) Representative images of sorted LT-HSCs immunostained with SHP-1 (green) and TβRI (red). Bars, 10 µm. ***, P < 0.001. Error bars show mean ± SEM.
Motif analysis showed that TβRI had a very conserved ITIM motif (Fig. 4 D; Staub et al., 2004). SHP-1 is known to interact with and activate the ITIM-containing receptors (Long, 1999; Billadeau and Leibson, 2002; Schlichter et al., 2014). To test whether SHP-1 physically interacts with TβRI, we performed the immunoprecipitation assay. Interestingly, our results showed that SHP-1 specifically interacted with TβRI, but not with TβRII, which does not have an ITIM motif (Fig. 4 E). This interaction can also be confirmed at endogenous level (Fig. 4 F), which rules out the possibility of artificial interaction as a result of protein overexpression. To further investigate whether the ITIM domain is essential for the interaction between SHP-1 and TβRI, we generated a ΔTβRI mutant with the ITIM domain deleted. As hypothesized, ΔTβRI mutant failed to interact with SHP-1 (Fig. 4 G). We also found that a portion of SHP-1 was localized in close proximity to TβRI in HSCs (Fig. 4 H). Overall, our data demonstrated that SHP-1 interacted with TβRI via the ITIM motif of TβRI and indicated that SHP-1 might regulate HSC quiescence by coordinating TGF-β signal.
SHP-1 regulates HSC quiescence by cooperating TGF-β signal
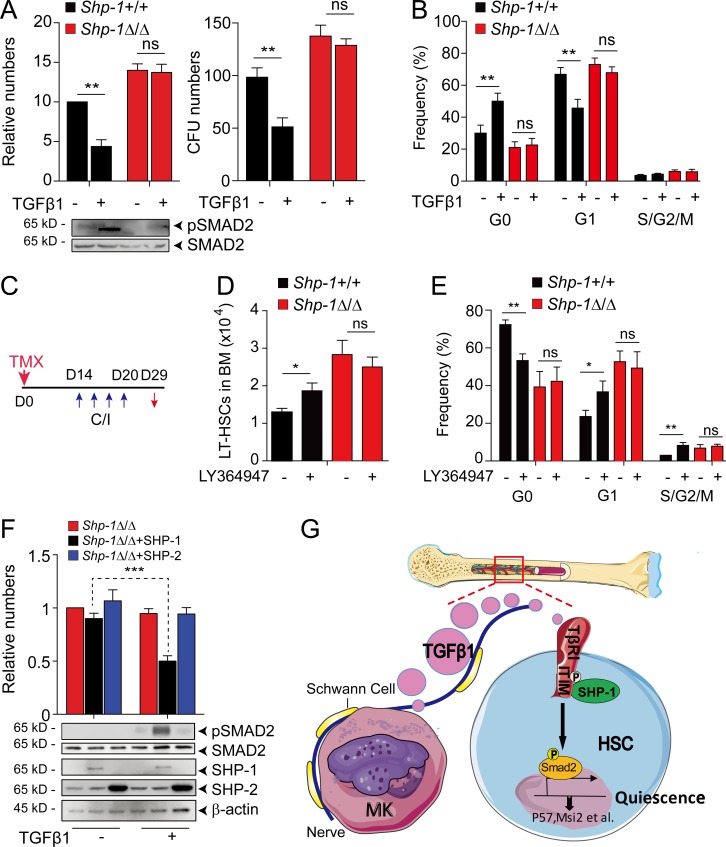
Because TGF-β1 is known to suppress HSC proliferation in vitro (Larsson et al., 2003; Yamazaki et al., 2009), we further investigated whether SHP-1 can regulate this process. As expected, TGF-β1 treatment significantly suppressed HSC proliferation (56% reduction), colony forming capability (48% reduction in CFU activity), and the pSMAD2 level in HSCs from Shp-1+/+ control mice. However, no significant difference was observed in HSCs from Shp-1Δ/Δ mice after TGF-β1 treatment (Fig. 5 A). This demonstrated that TGF-β cannot suppress HSC proliferation in Shp-1 null HSCs and indicated that SHP-1 might be critical for TGF-β signaling in regulating HSC quiescence. Next, we analyzed the cell cycle profile of HSCs after TGF-β1 treatment. Consistently, we found that TGF-β1 treatment significantly increased G0 phase (1.7-fold, P = 0.008) and reduced G1 phase (31.7%, P = 0.0002) HSCs in control SHP-1+/+ mice. However, no significant cell cycle difference was observed in HSCs from Shp-1Δ/Δ mice after TGF-β1 treatment (Fig. 5 B).
Figure 5.
SHP-1 regulates HSC quiescence by cooperating TGF-β signaling. (A and B) Shp-1–deficient HSPCs are resistant to the inhibitory effect of TGF-β1 in vitro. LSK cells isolated from Shp-1+/+ and Shp-1Δ/Δ mice were treated with or without 5 ng/ml TGF-β1 in ex vivo expansion medium. 5 d later, the cells were used in CFU assay (A, right) or calculated for total numbers (A, left). pSMAD2 and SMAD protein were measured (A, bottom). These cells were also used for HSC cell cycle analysis (B). (C–E) Scheme for LY364947 injection to Shp-1+/+ and Shp-1Δ/Δ mice after tamoxifen administration (C). Comparison of LT-HSC numbers (n = 5 mice; D) and cell cycle stage of HSCs (n = 5 mice; E) from Shp-1+/+ and Shp-1Δ/Δ mice as indicated. (F) LSK cells from Shp-1Δ/Δ mice were treated with or without TGF-β1 after overexpression of SHP-1 or SHP-1. Cells were quantified for cell number (upper panel) and protein expression as indicated (lower panel). (G) Schematic showing the role of SHP-1 in TGF-β–mediated HSC quiescence regulation. *, P < 0.05; **, P < 0.01; ***, P < 0.001. ns, not significant. Error bars show mean ± SEM.
To further investigate whether SHP-1 is critical for TGF-β regulated HSC quiescence in vivo, we treated mice with a small molecule inhibitor of TGF-β signaling (LY364947; Fig. 5 C). We found that in vivo inhibiting TGF-β signaling in control Shp-1+/+ mice significantly promoted HSC proliferation (P = 0.013), but not in Shp-1Δ/Δ mice (Fig. 5 D). Coordinately, LY364947-treated Shp-1+/+ animals have prolonged cycling of LT-HSCs (30% reduction of G0, P = 0.0016; a 1.5-fold increase of G1, P = 0.027; and a 2.8-fold increase of S/G2/M, P = 0.0038). However, Shp-1Δ/Δ mice did not respond to LY364947 treatment (Fig. 5 E). Furthermore, we found that SHP-1, but not SHP-2, can rescue the proliferation inhibition response to TGF-β1 treatment (47% reduction) in Shp-1 null HSCs (Fig. 5 F). Thus, our in vivo data proved that SHP-1 is critical for TGF-β signaling enforced regulation of HSC quiescence.
SHP-1 is suggested as a negative signal regulator and plays a suppressive role in myeloid immune cells (Lorenz et al., 1996; Paulson et al., 1996; Jiao et al., 1997; Tapley et al., 1997; Wu et al., 2003a; Xiao et al., 2010; Stadtmann et al., 2015); however, its role in HSCs is less clear. Using conditional knockout and BM reconstitution assay, we demonstrated that SHP-1 is essential for HSC quiescence and long-term self-renewal maintenance. Our previous work identified that ∼20% HSCs are surrounded by MKs, which produce TGF-β and other signals such as CXCL4 for HSC quiescence maintenance (Bruns et al., 2014; Zhao et al., 2014; Nakamura-Ishizu et al., 2015). However, the identity of this MK-adjacent HSC subpopulation and their intrinsic regulatory mechanisms are largely unknown. In this work, we found that HSCs with active SHP-1 are surrounded by MKs. Our functional data further demonstrated that SHP-1 is a critical downstream effector of TGF-β signaling in regulating HSCs. First, either Shp-1 knockout from HSCs or TGF-β1 knockout from MKs led to increased activation, but attenuated long-term self-renewal of HSCs. Second, SHP-1 physically interacts with ITIM receptor TβRI and is critical for activation of SMAD2, a TGF-β downstream effector. Third, SHP-1 knockout HSCs do not respond to TGF-β signaling in cell cycle regulation both in vitro and in vivo. The data clearly indicated that SHP-1 is indispensable for TGF-β mediated HSC quiescence maintenance as illustrated in Fig. 5 G. SHP-1 deficient HSCs have increased expression of Smad7, which promotes HSPC proliferation in a retroviral overexpression system (Blank et al., 2006). However, the in vivo function of Smad7 in TGF-β–SHP-1 pathway for HSC regulation need further characterization.
The data also suggested that SHP-1 activates TβRI by overcoming the ITIM-mediated negative effect. Paired immunoglobulin-like receptor B, an ITIM receptor, is important for both normal HSC self-renewal and leukemia development potentially through interacting with SHP-2 and SHP-1 (Zheng et al., 2012). We previously showed that the LAIR1–SHP-1 pathway is essential for acute myeloid leukemia development and acute myeloid leukemia stem cell regulation, but LAIR1-deficient mice maintain normal hematopoiesis and HSC function (Kang et al., 2015). Here, we identified that TβRI–SHP-1 regulates the maintenance of HSC quiescence. This indicated that ITIM receptors have overlapping yet distinct functions in regulating hematopoiesis and leukemogenesis. Because SHP-1 is suggested to be involved in leukemogenesis (Xiao et al., 2010; Chen et al., 2015), further characterization is needed to understand the function of TGF-β–SHP-1 pathway in leukemogenesis. Overall, our observation provided a novel intrinsic molecular mechanism to understand how HSC quiescence is regulated by niche signals.
Materials and methods
Animals
B6.129P2-Ptpn6tm1Rsky/J (Shp-1fl/fl), Tgfb1tm2.1Doe/J (Tgfb1fl/fl), and C57BL/6-Tg(Pf4-cre)Q3Rsko/J (Pf4-cre) mice were obtained from the Jackson Laboratory. Scl-CreER mice were provided by J.R. Goethert. Scl-CreER mice were mated with the Shp-1fl/fl line to generate Scl-CreER; Shp-1fl/fl mice. For induction of Cre-ER recombinase, mice were administered tamoxifen by oral gavage (10 mg/0.5 ml sunflower oil; Sigma-Aldrich). Tgfb1fl/fl mice were mated with the Pf4-cre line to generate Pf4-cre; Tgfb1fl/fl mice (Tiedt et al., 2007). All experimental mice were a mix of male and female 6–8-wk-old mice. TGF-β1 inhibitor LY364947 (S2805; Selleckchem) was intraperitoneally injected at 1 mg/kg. All mouse strains used in this study had a C57BL/6J genetic background. Animals were randomly included in the experiments according to genotyping results. Animals were blindly selected. The figure legends detail the number of animals used per study. All animal experiments were performed according to protocols approved by the institutional animal care and use committee.
PCR genotyping
Tissues were lysed in lysis buffer (Tissue PCR kit; Sigma-Aldrich) according to the manufacturer’s instruction. Mice were genotyped by PCR for 3 min at 95°C (20 s at 95°C, 30 s at 60°C, and 30 s at 72°C) 30 times and 3 min at 72°C. Shp-1 genotyping PCR primers were 5′-ACCCTCCAGCTCCTCTTC-3′, 5′-TGAGGTCCCGGTGAAACC-3′, and 5′-TGTTATGCATGTGTGTATCG-3′. Scl-CreER genotyping PCR primers were 5′-GAACCTGAAGATGTTCGCGAT-3′ and 5′-ACCGTCAGTACGTGAGATATC-3′. Tgfb1fl/fl genotyping PCR primers were 5′-AAGACCTGGGTTGGAAGTG-3′ and 5′-CTTCTCCGTTTCTCTGTCACCCTAT-3′. Pf4-cre genotyping PCR primers were 5′-CCAAGTCCTACTGTTTCTCACTC-3′ and 5′-TGCACAGTCAGCAGGTT-3′.
Flow cytometry
For phenotype analysis, hematopoietic cells were harvested from BM (femur and tibia). In brief, two million BM cells were resuspended in 100 µl cold PBS with 2% FBS and were blocked with PE-Cy7 anti–mouse CD16/CD32 (93; eBioscience; 1:200). After 15 min, 2 µl biotin anti–mouse Lineage depletion cocktail (BD Biosciences) was added, and the samples were kept on ice for 30 min. Cells were spun down, resuspended in 100 µl cold PBS with 2% FBS, and stained with streptavidin PerCP-Cyc5.5 (eBioscience, 1:200), APC-eFluor 780 anti-mouse c-Kit (eBioscience, 2B8, 1:200), eFluor 450 anti-mouse CD34 (eBioscience, RAM34, 1:50), eFluor 660 anti-mouse CD127 (eBioscience, A7R34, 1:50), FITC anti–mouse Sca-1 (E13-161.7; BD Biosciences; 1:200), or PE-CF594 anti–mouse CD135 (A2F10.1; BD Biosciences; 1:100). Samples were kept on ice for 1 h after addition of antibody and then washed with 100 µl of cold PBS with 2% FBS. Cells were resuspended with 300–500 µl cold PBS with 2% FBS for flow cytometry analysis. For cell cycle analysis, the cells were fixed and permeabilized with Foxp3/Transcription Factor Staining Buffer Set (eBioscience) according to the manufacturer’s protocol. In brief, 150 µl Foxp3 Fixation/Permeabilization working solution was added to each well, and samples were incubated for 30 to 60 min. Cells were spun down and washed twice with Permeabilization Buffer. Cells were then resuspended in 100 µl Permeabilization Buffer with 10 µg/ml Hoechst-33342 and 0.3 µl PE anti–mouse/rat Ki-67 (SolA15; eBioscience) was added. Samples were incubated in dark at room temperature for at least 30 min. Cells were washed with 200 µl Permeabilization Buffer and resuspended in 300 to 500 µl of cold PBS with 2% FBS for flow cytometry analysis as previously described (Huynh et al., 2011; Zheng et al., 2011a). For apoptosis assay, the cells were stained with PE Annexin V and 7-AAD (BD Biosciences). Cell sorting and analysis were performed using a cell sorter (FACSARIA III; BD Biosciences) or flow cytometer (Attune NxT; Thermo Fisher).
Mouse competitive reconstitution analyses
The indicated numbers of CD45.2 total BM cells or Lin−Sca-1+Kit+ Flk2−CD34− (LSKFC) cells from Scl-CreER+; Shp-1fl/fl mice and Scl-CreER–; Shp-1fl/fl control mice treated with tamoxifen; 2 × 105 BM cells from Pf4-cre–; TGF-β1f/f or from Pf4-cre+; TGF-β1f/f mice (CD45.2) were mixed with 2 × 105 CD45.1 competitor BM cells, and the mixture was injected intravenously into 6–8-wk-old CD45.1 mice irradiated with a total dose of 10 Gy. To measure reconstitution, peripheral blood was collected at the indicated time points after transplantation, and CD45.1+ and CD45.2+ cells in lymphoid and myeloid compartments were measured as described before (Huynh et al., 2011; Zheng et al., 2011b). CRU assay was measured by ELDA (Hu and Smyth, 2009).
Colony assays
BM cells were diluted to the indicated concentration in Iscove’s modified Dulbecco’s medium with 2% FBS and were seeded into methylcellulose medium M3434 (StemCell Technologies) for CFU-GM, CFU-GEMM, and BFU-E colony formation assays and into M3630 (StemCell Technologies) for the CFU–Pre-B colony formation assay according to the manufacturer’s protocols. (Zheng et al., 2012; Kang et al., 2015).
Immunostaining
Femurs or tibias were perfused before removal and fixed by 4% paraformaldehyde. Frozen sections were retrieved using 1 µg/ml proteinase K in TE buffer (100 mM Tris-HCl, pH 8.0, and 50 mM EDTA) at 37°C for 30 min. Blocking was done with Universal Blocking Reagent (HK112-5K; BioGenex). Anti–mouse pSHP-1 (ab51171; rabbit; Abcam; 1:100) and anti–mouse pSMAD2 (04-953; rabbit; Millipore; 1:100) and anti–mouse glial fibrillary acidic protein (GFAP; IR524; rabbit; Dako) were used. Secondary staining was done with donkey anti–rabbit AF680 (Invitrogen; 1:1,000) and donkey anti–rabbit AF488 (Invitrogen; 1:1,000 dilution). For HSC staining in bone sections, we used anti–mouse CD150 (TC15-12F12.2; Biolegend; 1:100), anti–mouse CD48-biotin (HM48-1; Biolegend; 1:100), FITC anti–mouse CD41 (MWReg30; Thermo Fisher; 1:100), APC anti–mouse CD41, and biotin anti–mouse Lineage (biotin anti–mouse B220, CD3, CD4, CD8, Gr1, IgM, Mac-1, and Ter119; rat; Biolegend; 1:50 dilution). Secondary staining was done with donkey anti–rat Dylight 550 (SA5-10027; Invitrogen; 1:1,000) and streptavidin-Brilliant violet 421 (Biolegend; 1:1,000). For sorted HSCs staining, cells were fixed with precooled methanol. Anti–mouse SHP-1 (ab131537; rabbit; Abcam; 1:100) and anti–mouse TβRI (sc-101574; rat; Santa Cruz; 1:100) were used. For high-resolution three-dimensional images, z-stack collected images from Nikon C2plus, or Zeiss LSM-710 confocal microscopy were analyzed with Imaris software (Bitplane) and ImageJ (National Institutes of Health).
Computational modeling of random MK and HSC localization
Images of femur frozen sections were used to produce spatial maps of Schwann’s cells onto which 85 MKs or 20 HSCs were randomly placed in the marrow. After delimiting the boundaries of the marrow according to the bone-to-marrow interface, background staining was removed to accurately represent Schwann’s cells, MKs, or HSCs. 85 random MKs or 20 HSCs were selected from the defined region, and the shortest Euclidean distance was calculated for each MKs to Schwann’s cell, or HSC to MKs and Schwann’s cell. 1,000 times simulations were performed for each model. Random and actual distance distributions were compared using the modified nonparametric two-dimensional (2D) KS test as described (Bruns et al., 2014).
TGF-β1 in vitro treatment assay
LSK cells isolated from Shp-1+/+ and Shp-1Δ/Δ mice were cultured in StemSpan medium (StemCell Technologies) supplemented with 50 ng/ml rmTHPO and 50 ng/ml rmSCF and treated with or without 5 ng/ml TGF-β1. 5 d later, the cells were used in CFU assay (M3434), calculated for total cell numbers, or harvested for Western blots.
Quantitative RT-PCR (RT-qPCR)
RT-qPCR was performed as described previously (Kang et al., 2015); RT-PCR was performed on 5 ng total RNA with specific primers and a QIAGEN One Step RT-PCR kit (210210; Qiagen) according to the manufacturer’s protocol. β-actin was used as the reference gene to normalize the relative expression for quantitative RT-PCR analysis. RT-qPCR primers used in this study were as follows: Tab1: forward (5′-ACAGCAAGCTCTACGTTGCC-3′) and reverse (5′-GTAACCTGTAACCCATCCACTG-3′); Smad7: forward (5′-GCATTCCTCGGAAGTCAAGAG-3′) and reverse (5′-CCAGGGGCCAGATAATTCGT-3′); Msi2: forward (5′-ACCTCACCAGATAGCCTTAGAG-3′) and reverse (5′-AGCGTTTCGTTGTGGGATCTC-3′); P57KIP2: forward (5′-GCAGGACGAGAATCAAGAGCA-3′) and reverse (5′-GCTTGGCGAAGAAGTCGTT-3′)’; β-actin: forward (5′-GCTCTTTTCCAGCCTTCCTT-3′) and reverse (5′-CTTCTGCATCCTGTCAGCAA-3′); and β-actin: forward (5′-GCTCTTTTCCAGCCTTCCTT-3′) and reverse (5′-CTTCTGCATCCTGTCAGCAA-3′).
Western blotting
Cell lysates (100-µg samples) were prepared using Transmembrane Protein Extraction Reagent (TmPER-200; FIVEphoton Biochemicals) or in RIPA buffer. Proteins were separated by electrophoresis on 4–15% SDS-polyacrylamide gels, and the proteins were electroblotted onto a nitrocellulose membrane. The membrane was probed with the indicated primary antibodies for 1 h at room temperature or overnight at 4°C and then incubated with secondary antibody, which was detected by exposure to x-ray film, or by digital imaging with a charge-coupled device camera system (Odyssey Fc). The antibodies used included anti–mouse SHP-1 (ab2020; Abcam; 1:2,000 dilution), anti–mouse pSMAD2 (04-953; Millipore; 1:1,000 dilution), anti–mouse TβRI (sc-9048; Santa Cruz; 1:500 dilution), anti-FLAG (SIGI-25; Sigma-Aldrich; 1: 5,000), anti-His (Santa Cruz; 1:5,000), and anti–mouse actin (1A4; Sigma-Aldrich; 1:10,000 dilution), anti–mouse TGF-β1(ab92486; Abcam; 1:500 dilution). The images shown are representative of images from at least three experiments.
ELISAs
TGF-β1 was measured by Quantikine ELISA kit (R&D Systems) following the manufacturer’s instruction.
Statistical analyses
Data are expressed as means ± SEM. For all experiments except determination of survival, data were analyzed by Student’s t test, and differences were considered statistically significant if P < 0.05. The survival of the two groups were analyzed using a log-rank test, and differences were considered statistically significant if P < 0.05. Comparisons of probability distributions between two samples were done using two-sample KS test, and 2D distance distributions that were derived from the actual measurements and random simulations were compared using a nonparametric 2D KS test with two samples. Differences were considered statistically significant if P < 0.05. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Online supplemental material
Fig. S1 shows loss of Shp-1 results in HSC activation in early time point. Fig. S2 shows the loss of Shp-1 in MK niche does not compromise HSC quiescence. Fig. S3 shows the spatial relationship of pSHP-1+ HSCs with MKs in BM.
Supplementary Material
Acknowledgments
We would like to thank the National Key Research and Development Program of China (grants 2017YFA0103403), National Natural Science Foundation of China (grants 91749106 and 81670101), Guangdong Innovative and Entrepreneurial Research Team Program (grant 2016ZT06S029), National Institutes of Health (grant 1R01CA172268), the March of Dimes Foundation (grant 1-FY14-201), the Welch Foundation (grant I-1834), and Guangdong Province Key Laboratory of Malignant Tumor Epigenetics and Gene Regulation (grant 2017B030314026) for generous support. M. Zhao is supported by 1000 Youth Talents Plan.
The authors declare no competing financial interests.
Author contributions: L. Jiang, X. Han, and J. Wang designed and performed the experiments, analyzed the data, and generated figures. C. Wang, J. Xie, G. Wu, H. Phan, and Z. Liu provided technical support and reagents necessary for this study. X. Sun performed computational simulations. C. Zhang, M. Zhao, and X. Kang supervised the experiments and cowrote the manuscript.
References
- Billadeau D.D., and Leibson P.J.. 2002. ITAMs versus ITIMs: striking a balance during cell regulation. J. Clin. Invest. 109:161–168. 10.1172/JCI0214843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank U., and Karlsson S.. 2015. TGF-β signaling in the control of hematopoietic stem cells. Blood. 125:3542–3550. 10.1182/blood-2014-12-618090 [DOI] [PubMed] [Google Scholar]
- Blank U., Karlsson G., Moody J.L., Utsugisawa T., Magnusson M., Singbrant S., Larsson J., and Karlsson S.. 2006. Smad7 promotes self-renewal of hematopoietic stem cells. Blood. 108:4246–4254. 10.1182/blood-2006-02-005611 [DOI] [PubMed] [Google Scholar]
- Bruns I., Lucas D., Pinho S., Ahmed J., Lambert M.P., Kunisaki Y., Scheiermann C., Schiff L., Poncz M., Bergman A., and Frenette P.S.. 2014. Megakaryocytes regulate hematopoietic stem cell quiescence through CXCL4 secretion. Nat. Med. 20:1315–1320. 10.1038/nm.3707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Shojaee S., Buchner M., Geng H., Lee J.W., Klemm L., Titz B., Graeber T.G., Park E., Tan Y.X., et al. 2015. Signalling thresholds and negative B-cell selection in acute lymphoblastic leukaemia. Nature. 521:357–361. 10.1038/nature14231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göthert J.R., Gustin S.E., Hall M.A., Green A.R., Göttgens B., Izon D.J., and Begley C.G.. 2005. In vivo fate-tracing studies using the Scl stem cell enhancer: embryonic hematopoietic stem cells significantly contribute to adult hematopoiesis. Blood. 105:2724–2732. 10.1182/blood-2004-08-3037 [DOI] [PubMed] [Google Scholar]
- Hu Y., and Smyth G.K.. 2009. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods. 347:70–78. 10.1016/j.jim.2009.06.008 [DOI] [PubMed] [Google Scholar]
- Huynh H., Zheng J., Umikawa M., Zhang C., Silvany R., Iizuka S., Holzenberger M., Zhang W., and Zhang C.C.. 2011. IGF binding protein 2 supports the survival and cycling of hematopoietic stem cells. Blood. 118:3236–3243. 10.1182/blood-2011-01-331876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itkin T., Gur-Cohen S., Spencer J.A., Schajnovitz A., Ramasamy S.K., Kusumbe A.P., Ledergor G., Jung Y., Milo I., Poulos M.G., et al. 2016. Distinct bone marrow blood vessels differentially regulate haematopoiesis. Nature. 532:323–328. 10.1038/nature17624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao H., Yang W., Berrada K., Tabrizi M., Shultz L., and Yi T.. 1997. Macrophages from motheaten and viable motheaten mutant mice show increased proliferative responses to GM-CSF: detection of potential HCP substrates in GM-CSF signal transduction. Exp. Hematol. 25:592–600. [PubMed] [Google Scholar]
- Kaneko T., Saito Y., Kotani T., Okazawa H., Iwamura H., Sato-Hashimoto M., Kanazawa Y., Takahashi S., Hiromura K., Kusakari S., et al. 2012. Dendritic cell-specific ablation of the protein tyrosine phosphatase Shp1 promotes Th1 cell differentiation and induces autoimmunity. J. Immunol. 188:5397–5407. 10.4049/jimmunol.1103210 [DOI] [PubMed] [Google Scholar]
- Kang X., Lu Z., Cui C., Deng M., Fan Y., Dong B., Han X., Xie F., Tyner J.W., Coligan J.E., et al. 2015. The ITIM-containing receptor LAIR1 is essential for acute myeloid leukaemia development. Nat. Cell Biol. 17:665–677. 10.1038/ncb3158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang X., Kim J., Deng M., John S., Chen H., Wu G., Phan H., and Zhang C.C.. 2016. Inhibitory leukocyte immunoglobulin-like receptors: Immune checkpoint proteins and tumor sustaining factors. Cell Cycle. 15:25–40. 10.1080/15384101.2015.1121324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunisaki Y., Bruns I., Scheiermann C., Ahmed J., Pinho S., Zhang D., Mizoguchi T., Wei Q., Lucas D., Ito K., et al. 2013. Arteriolar niches maintain haematopoietic stem cell quiescence. Nature. 502:637–643. 10.1038/nature12612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson J., Blank U., Helgadottir H., Björnsson J.M., Ehinger M., Goumans M.J., Fan X., Levéen P., and Karlsson S.. 2003. TGF-beta signaling-deficient hematopoietic stem cells have normal self-renewal and regenerative ability in vivo despite increased proliferative capacity in vitro. Blood. 102:3129–3135. 10.1182/blood-2003-04-1300 [DOI] [PubMed] [Google Scholar]
- Long E.O. 1999. Regulation of immune responses through inhibitory receptors. Annu. Rev. Immunol. 17:875–904. 10.1146/annurev.immunol.17.1.875 [DOI] [PubMed] [Google Scholar]
- Lorenz U. 2009. SHP-1 and SHP-2 in T cells: two phosphatases functioning at many levels. Immunol. Rev. 228:342–359. 10.1111/j.1600-065X.2008.00760.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz U., Bergemann A.D., Steinberg H.N., Flanagan J.G., Li X., Galli S.J., and Neel B.G.. 1996. Genetic analysis reveals cell type-specific regulation of receptor tyrosine kinase c-Kit by the protein tyrosine phosphatase SHP1. J. Exp. Med. 184:1111–1126. 10.1084/jem.184.3.1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazharian A., Mori J., Wang Y.J., Heising S., Neel B.G., Watson S.P., and Senis Y.A.. 2013. Megakaryocyte-specific deletion of the protein-tyrosine phosphatases Shp1 and Shp2 causes abnormal megakaryocyte development, platelet production, and function. Blood. 121:4205–4220. 10.1182/blood-2012-08-449272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura-Ishizu A., Takubo K., Kobayashi H., Suzuki-Inoue K., and Suda T.. 2015. CLEC-2 in megakaryocytes is critical for maintenance of hematopoietic stem cells in the bone marrow. J. Exp. Med. 212:2133–2146. 10.1084/jem.20150057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neel B.G., Gu H., and Pao L.. 2003. The 'Shp’ing news: SH2 domain-containing tyrosine phosphatases in cell signaling. Trends Biochem. Sci. 28:284–293. 10.1016/S0968-0004(03)00091-4 [DOI] [PubMed] [Google Scholar]
- Pao L.I., Lam K.P., Henderson J.M., Kutok J.L., Alimzhanov M., Nitschke L., Thomas M.L., Neel B.G., and Rajewsky K.. 2007. B cell-specific deletion of protein-tyrosine phosphatase Shp1 promotes B-1a cell development and causes systemic autoimmunity. Immunity. 27:35–48. 10.1016/j.immuni.2007.04.016 [DOI] [PubMed] [Google Scholar]
- Park S.M., Deering R.P., Lu Y., Tivnan P., Lianoglou S., Al-Shahrour F., Ebert B.L., Hacohen N., Leslie C., Daley G.Q., et al. 2014. Musashi-2 controls cell fate, lineage bias, and TGF-β signaling in HSCs. J. Exp. Med. 211:71–87. 10.1084/jem.20130736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson R.F., Vesely S., Siminovitch K.A., and Bernstein A.. 1996. Signalling by the W/Kit receptor tyrosine kinase is negatively regulated in vivo by the protein tyrosine phosphatase Shp1. Nat. Genet. 13:309–315. 10.1038/ng0796-309 [DOI] [PubMed] [Google Scholar]
- Sacchetti B., Funari A., Michienzi S., Di Cesare S., Piersanti S., Saggio I., Tagliafico E., Ferrari S., Robey P.G., Riminucci M., and Bianco P.. 2007. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell. 131:324–336. 10.1016/j.cell.2007.08.025 [DOI] [PubMed] [Google Scholar]
- Scandura J.M., Boccuni P., Massagué J., and Nimer S.D.. 2004. Transforming growth factor beta-induced cell cycle arrest of human hematopoietic cells requires p57KIP2 up-regulation. Proc. Natl. Acad. Sci. USA. 101:15231–15236. 10.1073/pnas.0406771101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlichter L.C., Jiang J., Wang J., Newell E.W., Tsui F.W., and Lam D.. 2014. Regulation of hERG and hEAG channels by Src and by SHP-1 tyrosine phosphatase via an ITIM region in the cyclic nucleotide binding domain. PLoS One. 9:e90024 10.1371/journal.pone.0090024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtmann A., Block H., Volmering S., Abram C., Sohlbach C., Boras M., Lowell C.A., and Zarbock A.. 2015. Cross-Talk between Shp1 and PIPKIγ Controls Leukocyte Recruitment. J. Immunol. 195:1152–1161. 10.4049/jimmunol.1500606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub E., Rosenthal A., and Hinzmann B.. 2004. Systematic identification of immunoreceptor tyrosine-based inhibitory motifs in the human proteome. Cell. Signal. 16:435–456. 10.1016/j.cellsig.2003.08.013 [DOI] [PubMed] [Google Scholar]
- Stromnes I.M., Fowler C., Casamina C.C., Georgopolos C.M., McAfee M.S., Schmitt T.M., Tan X., Kim T.D., Choi I., Blattman J.N., and Greenberg P.D.. 2012. Abrogation of SRC homology region 2 domain-containing phosphatase 1 in tumor-specific T cells improves efficacy of adoptive immunotherapy by enhancing the effector function and accumulation of short-lived effector T cells in vivo. J. Immunol. 189:1812–1825. 10.4049/jimmunol.1200552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapley P., Shevde N.K., Schweitzer P.A., Gallina M., Christianson S.W., Lin I.L., Stein R.B., Shultz L.D., Rosen J., and Lamb P.. 1997. Increased G-CSF responsiveness of bone marrow cells from hematopoietic cell phosphatase deficient viable motheaten mice. Exp. Hematol. 25:122–131. [PubMed] [Google Scholar]
- Tibaldi E., Brunati A.M., Zonta F., Frezzato F., Gattazzo C., Zambello R., Gringeri E., Semenzato G., Pagano M.A., and Trentin L.. 2011. Lyn-mediated SHP-1 recruitment to CD5 contributes to resistance to apoptosis of B-cell chronic lymphocytic leukemia cells. Leukemia. 25:1768–1781. 10.1038/leu.2011.152 [DOI] [PubMed] [Google Scholar]
- Tiedt R., Schomber T., Hao-Shen H., and Skoda R.C.. 2007. Pf4-Cre transgenic mice allow the generation of lineage-restricted gene knockouts for studying megakaryocyte and platelet function in vivo. Blood. 109:1503–1506. 10.1182/blood-2006-04-020362 [DOI] [PubMed] [Google Scholar]
- Viant C., Fenis A., Chicanne G., Payrastre B., Ugolini S., and Vivier E.. 2014. SHP-1-mediated inhibitory signals promote responsiveness and anti-tumour functions of natural killer cells. Nat. Commun. 5:5108 10.1038/ncomms6108 [DOI] [PubMed] [Google Scholar]
- Wilson A., Laurenti E., Oser G., van der Wath R.C., Blanco-Bose W., Jaworski M., Offner S., Dunant C.F., Eshkind L., Bockamp E., et al. 2008. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 135:1118–1129. 10.1016/j.cell.2008.10.048 [DOI] [PubMed] [Google Scholar]
- Wu C., Guan Q., Wang Y., Zhao Z.J., and Zhou G.W.. 2003a SHP-1 suppresses cancer cell growth by promoting degradation of JAK kinases. J. Cell. Biochem. 90:1026–1037. 10.1002/jcb.10727 [DOI] [PubMed] [Google Scholar]
- Wu C., Sun M., Liu L., and Zhou G.W.. 2003b The function of the protein tyrosine phosphatase SHP-1 in cancer. Gene. 306:1–12. 10.1016/S0378-1119(03)00400-1 [DOI] [PubMed] [Google Scholar]
- Xiao W., Ando T., Wang H.Y., Kawakami Y., and Kawakami T.. 2010. Lyn- and PLC-beta3-dependent regulation of SHP-1 phosphorylation controls Stat5 activity and myelomonocytic leukemia-like disease. Blood. 116:6003–6013. 10.1182/blood-2010-05-283937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki S., Iwama A., Takayanagi S., Morita Y., Eto K., Ema H., and Nakauchi H.. 2006. Cytokine signals modulated via lipid rafts mimic niche signals and induce hibernation in hematopoietic stem cells. EMBO J. 25:3515–3523. 10.1038/sj.emboj.7601236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki S., Iwama A., Takayanagi S., Eto K., Ema H., and Nakauchi H.. 2009. TGF-beta as a candidate bone marrow niche signal to induce hematopoietic stem cell hibernation. Blood. 113:1250–1256. 10.1182/blood-2008-04-146480 [DOI] [PubMed] [Google Scholar]
- Yamazaki S., Ema H., Karlsson G., Yamaguchi T., Miyoshi H., Shioda S., Taketo M.M., Karlsson S., Iwama A., and Nakauchi H.. 2011. Nonmyelinating Schwann cells maintain hematopoietic stem cell hibernation in the bone marrow niche. Cell. 147:1146–1158. 10.1016/j.cell.2011.09.053 [DOI] [PubMed] [Google Scholar]
- Zhao M., Perry J.M., Marshall H., Venkatraman A., Qian P., He X.C., Ahamed J., and Li L.. 2014. Megakaryocytes maintain homeostatic quiescence and promote post-injury regeneration of hematopoietic stem cells. Nat. Med. 20:1321–1326. 10.1038/nm.3706 [DOI] [PubMed] [Google Scholar]
- Zheng J., Huynh H., Umikawa M., Silvany R., and Zhang C.C.. 2011a Angiopoietin-like protein 3 supports the activity of hematopoietic stem cells in the bone marrow niche. Blood. 117:470–479. 10.1182/blood-2010-06-291716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Umikawa M., Zhang S., Huynh H., Silvany R., Chen B.P., Chen L., and Zhang C.C.. 2011b Ex vivo expanded hematopoietic stem cells overcome the MHC barrier in allogeneic transplantation. Cell Stem Cell. 9:119–130. 10.1016/j.stem.2011.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Umikawa M., Cui C., Li J., Chen X., Zhang C., Huynh H., Kang X., Silvany R., Wan X., et al. 2012. Inhibitory receptors bind ANGPTLs and support blood stem cells and leukaemia development. Nature. 485:656–660. 10.1038/nature11095 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.