Durai et al. demonstrate that the less severe DC deficiency in Flt3–/– mice compared to Flt3l–/– mice is due to the enhanced sensitivity of Flt3–/– progenitors to compensatory cytokines that support DC development.
Abstract
The receptor Flt3 and its ligand Flt3L are both critical for dendritic cell (DC) development, but DC deficiency is more severe in Flt3l−/− mice than in Flt3−/− mice. This has led to speculation that Flt3L binds to another receptor that also supports DC development. However, we found that Flt3L administration does not generate DCs in Flt3−/− mice, arguing against a second receptor. Instead, Flt3−/− DC progenitors matured in response to macrophage colony–stimulating factor (M-CSF) or stem cell factor, and deletion of Csf1r in Flt3−/− mice further reduced DC development, indicating that these cytokines could compensate for Flt3. Surprisingly, Flt3−/− DC progenitors displayed enhanced M-CSF signaling, suggesting that loss of Flt3 increased responsiveness to other cytokines. In agreement, deletion of Flt3 in Flt3l−/− mice paradoxically rescued their severe DC deficiency. Thus, multiple cytokines can support DC development, and the discrepancy between Flt3−/− and Flt3l−/− mice results from the increased sensitivity of Flt3−/− progenitors to these cytokines.
Introduction
Dendritic cells (DCs) are immune cells with critical functions in both the innate and adaptive immune responses that develop from hematopoietic progenitor cells (Liu et al., 2007; Mildner and Jung, 2014). The earliest committed progenitor with DC fate potential is the macrophage/DC progenitor (MDP; Fogg et al., 2006; Auffray et al., 2009), which develops into a common DC progenitor (CDP) that can give rise to plasmacytoid DCs (pDCs) as well as the classical DC (cDC) subsets, cDC1 and cDC2 (Naik et al., 2007; Onai et al., 2007). Committed cDC progenitors restricted to only the cDC1 or the cDC2 lineage have recently been identified in mice (Grajales-Reyes et al., 2015; Schlitzer et al., 2015) and in humans (Breton et al., 2015; Lee et al., 2015; See et al., 2017).
The development of DCs is dependent on the class III receptor tyrosine kinase (RTK) Fms-like tyrosine kinase 3 (Flt3) and its ligand Flt3L (McKenna et al., 2000; Waskow et al., 2008). Flt3 was first identified as a gene enriched in hematopoietic stem cells that encoded a protein homologous to the receptor c-Kit (Matthews et al., 1991). It was later recognized to be expressed on mature DCs and their progenitors as well (Miller et al., 2012). Flt3 shares structural properties and downstream signaling pathways with c-Kit and CSF1R, other members of the class III RTK family that are also expressed by committed DC progenitors (Onai et al., 2007; Verstraete and Savvides, 2012; Grajales-Reyes et al., 2015). The ligand for Flt3, Flt3L, was subsequently cloned and found to induce proliferation in early bone marrow (BM) progenitors (Lyman et al., 1993). Later, a role for Flt3L in DC homeostasis was uncovered from the expansion of DCs in mice and humans who were administered this cytokine (Maraskovsky et al., 1996, 2000). Furthermore, treatment of BM progenitors in vitro with Flt3L also supports the development of mature DCs (Brasel et al., 2000; Naik et al., 2005), and Flt3+ progenitors preferentially gave rise to DCs in vivo (D’Amico and Wu, 2003). Finally, genetic inactivation of the Flt3 (Mackarehtschian et al., 1995) or Flt3l (McKenna et al., 2000) genes in mice was observed to decrease the numbers of DCs (McKenna et al., 2000; Waskow et al., 2008), confirming their importance in DC homeostasis.
These original studies of Flt3−/− and Flt3l−/− mice surprisingly appeared to find DC deficiencies of varying severity in these two strains. Flt3l−/− mice analyzed between 5 and 14 wk of age had a 4- to 10-fold reduction in splenic CD8α– DCs and a 6- to 14-fold reduction in splenic CD8α+ DCs (McKenna et al., 2000). Meanwhile, an analysis of Flt3−/− mice found that although all DCs were reduced by 85% at 2 wk of age, they were reduced by only 43% (cDCs) or 65% (pDCs) at 9 wk of age (Waskow et al., 2008). Another study that examined both strains between 8 and 12 wk of age similarly found more severe reductions in CD8α+ DCs and CD11b+ DCs in Flt3l−/− mice compared with Flt3−/− mice (Ginhoux et al., 2009). This discrepancy has also been noted in the development of pre-pro-B cells, with Flt3l−/− mice demonstrating a severe reduction in this population, whereas Flt3−/− mice had only a twofold reduction (Mackarehtschian et al., 1995; Sitnicka et al., 2002, 2003; Nagasawa, 2006). However, no study has directly compared Flt3−/− mice with Flt3l−/− mice over time, so it is unknown whether the defects caused by Flt3L deficiency improve with age, as with Flt3 deficiency, or whether Flt3−/− mice and Flt3l−/− mice simply have distinct phenotypes. Nevertheless, these apparent discrepancies in phenotypes have led to suggestions that Flt3L could act on a second receptor other than Flt3 to support DC or B cell development, explaining the more severe defects in Flt3l−/− mice (Nagasawa, 2006; Helft et al., 2010; Satpathy et al., 2011; Moore and Anderson, 2013; Tsapogas et al., 2017).
Cytokines such as Flt3L can play supportive and instructive roles in the development of hematopoietic lineages (Enver et al., 1998; Metcalf, 1998), and the actions of a given cytokine may depend upon the progenitor stage and cell type it acts upon (Rieger et al., 2009; Mossadegh-Keller et al., 2013). For DCs, an instructive role for Flt3 was suggested by the redirection of megakaryocyte-erythrocyte progenitors (MEPs) toward DC fate upon forced Flt3 overexpression (Onai et al., 2006) and by transgenic mice in which Flt3L overexpression leads to a robust expansion of DCs, lymphocytes, and myeloid cells at the expense of erythrocytes and platelets (Tsapogas et al., 2014). Nonetheless, Flt3L administration does not increase in vivo DC output from early Flt3+ progenitors such as common lymphoid progenitors, common myeloid progenitors, or granulocyte-monocyte progenitors (Karsunky et al., 2003), and the continued presence of DCs in Flt3−/− and Flt3l−/− mice, albeit at reduced numbers, suggests that DC development can occur through some unidentified mechanism in the absence of this cytokine pathway. Thus, it remains unclear what specific role Flt3L plays in DC progenitor specification and development, as well as whether any other cytokines can contribute to this process.
To address the apparent discrepancy in phenotypes between Flt3−/− and Flt3l−/− mice, we directly compared DC development in these strains over time. We confirmed that Flt3l−/− mice show a severe and persistent DC defect, whereas Flt3−/− mice have a less severe defect at all ages analyzed. However, we were unable to demonstrate activity for Flt3L on a second receptor as has been proposed. Instead, we found that DC progenitors developed normally without instructional cues from Flt3 signaling and, surprisingly, that these progenitors could mature in Flt3−/− mice in response to macrophage colony–stimulating factor (M-CSF) and stem cell factor (SCF), ligands for other class III RTKs expressed by these cells. Deletion of Csf1r in Flt3−/− mice also led to a further reduction in DC development in a cell-intrinsic manner, indicating that this cytokine pathway was partially compensating for the loss of Flt3 in vivo. Although the expression of CSF1R and c-Kit was not altered in Flt3−/− progenitors, we found that these progenitors were more responsive to stimulation by M-CSF, suggesting that the absence of Flt3 potentiates signaling by other class III RTKs. To test whether this effect was responsible for the discrepancy between Flt3−/− and Flt3l−/− mice, we generated Flt3−/−Flt3l−/− mice and found that the additional deletion of Flt3 paradoxically restored DC numbers in Flt3l−/− mice. We conclude that multiple cytokine pathways can support the development of DCs, and that the discrepancy between Flt3−/− and Flt3l−/− mice is a result of the enhanced sensitivity of Flt3−/− BM progenitors to signaling by these alternative cytokines.
Results
Flt3l−/− mice have a more severe DC deficiency than Flt3−/− mice
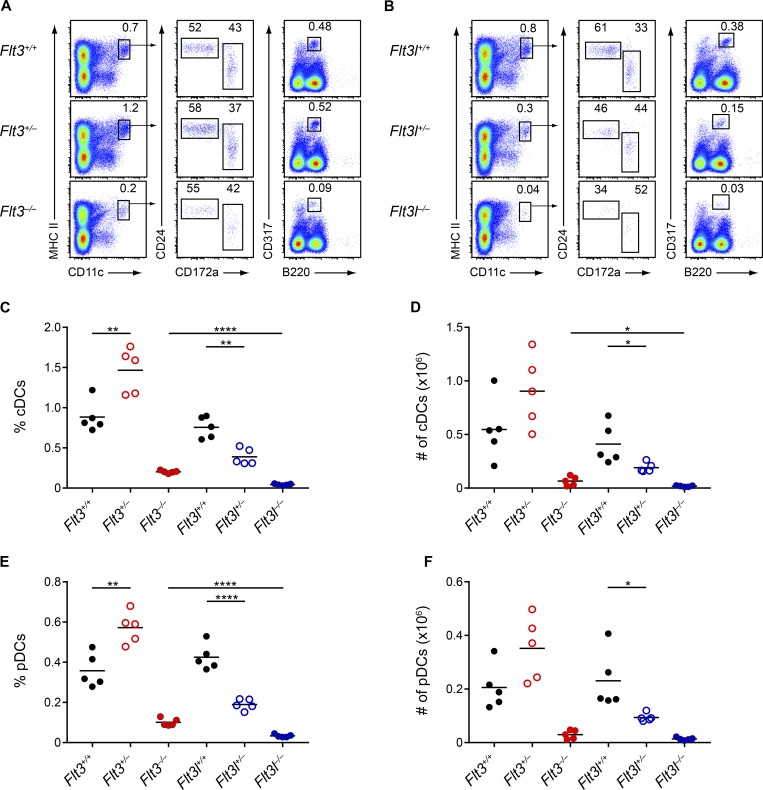
The original studies that analyzed Flt3−/− and Flt3l−/− mice did not directly compare these strains at similar ages, so the previously noted phenotypic discrepancy may simply have been a result of mismatched comparisons (McKenna et al., 2000; Waskow et al., 2008). We therefore examined matched sets of Flt3−/− and Flt3l−/− mice at both 2 and 8 wk of age (Figs. 1 and 2). We confirmed the previous report of severe reductions in the percentage (Fig. 1, C and E) and numbers (Fig. 1, D and F) of splenic cDCs and pDCs at 2 wk in both Flt3−/− and Flt3l−/− mice. We also found severe reductions in both populations in the skin-draining lymph nodes (SLNs; Fig. S1, B and C) and lungs (Fig. S1 G) of these strains. Even at this age, however, the defect in cDCs and pDCs was more severe in Flt3l−/− mice than in Flt3−/− mice in all organs analyzed. We also observed previously unrecognized heterozygous phenotypes in both Flt3+/− and Flt3l+/− mice compared with WT mice. Flt3l+/− mice showed a twofold reduction in DC numbers relative to WT mice, whereas, surprisingly, Flt3+/− mice had significantly more DCs than WT mice (Fig. 1, C–F).
Figure 1.
Flt3l−/− mice have a more severe DC deficiency than Flt3−/− mice at two weeks of age. (A and B) Splenocytes from mice of the indicated genotypes at 2-wk of age were analyzed by flow cytometry for DC populations. Shown are representative two-color histograms of live cells. Numbers specify the percentage of cells within the indicated gates. (C–F) Summary data for the percentage (C) and number (D) of splenic cDCs and the percentage (E) and number (F) of splenic pDCs in mice of the indicated genotypes. Dots represent biological replicates; small horizontal lines indicate the mean. Data are pooled from seven independent experiments (n = 5 mice per genotype). *, P < 0.05; **, P < 0.01; ****, P < 0.0001 using unpaired, two-tailed Student's t test.
Figure 2.
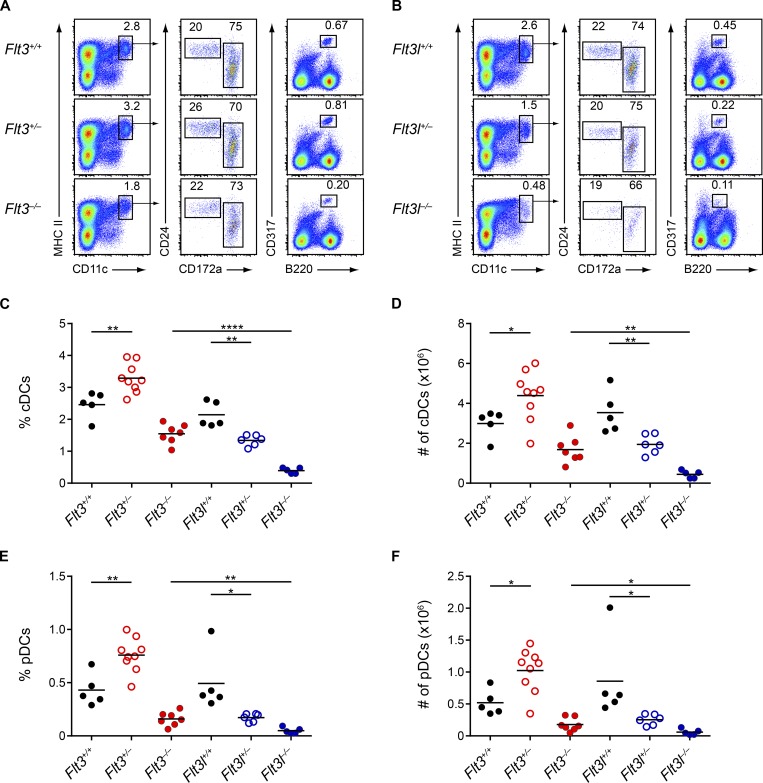
Flt3l−/− mice continue to have a more severe DC deficiency than Flt3−/− mice at eight weeks of age. (A and B) Splenocytes from mice of the indicated genotypes at 8-wk of age were analyzed by flow cytometry for DC populations. Shown are representative two-color histograms of live cells. Numbers specify the percentage of cells within the indicated gates. (C–F) Summary data for the percentage (C) and number (D) of splenic cDCs and the percentage (E) and number (F) of splenic pDCs in mice of the indicated genotypes. Dots represent biological replicates; small horizontal lines indicate the mean. Data are pooled from nine independent experiments (n = 5–9 mice per genotype). *, P < 0.05; **, P < 0.01; ****, P < 0.0001 using unpaired, two-tailed Student's t test.
In contrast, at 8 wk, Flt3−/− mice showed only a mild deficiency in the percentage and number of splenic DCs, whereas Flt3l−/− mice continued having severely reduced DC populations compared with both WT and Flt3−/− mice (Fig. 2, A–F). Findings were similar for cDCs and pDCs in the SLNs (Fig. S1, D and E) and for cDCs in the lungs (Fig. S1 H). Heterozygous phenotypes were again observed at 8 wk, with Flt3l+/− mice showing a twofold reduction in DCs compared with WT mice and Flt3+/− mice having consistently greater numbers of DCs compared with WT mice (Fig. 2, A–F). In summary, the more severe DC defect in Flt3l−/− mice compared with Flt3−/− mice appears to be an inherent difference between these strains.
Flt3L does not support DC development from Flt3−/− BM or in Flt3−/− mice
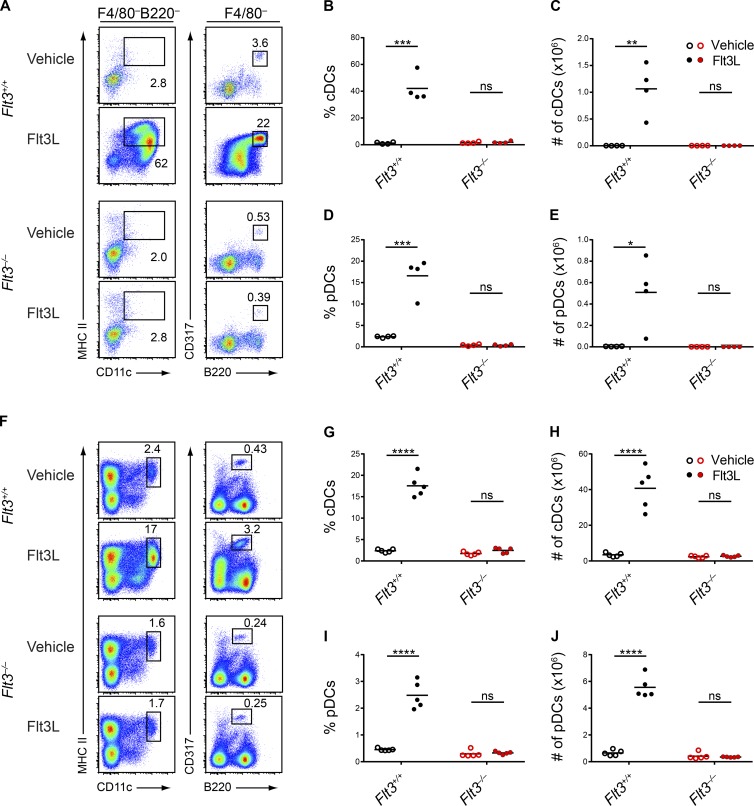
To explain the discrepancy between Flt3−/− and Flt3l−/− mice, some have suggested that Flt3L could support DC development in Flt3−/− mice by acting on a second receptor, thus explaining the less severe phenotype of Flt3−/− mice (Nagasawa, 2006; Helft et al., 2010; Satpathy et al., 2011; Moore and Anderson, 2013; Tsapogas et al., 2017). To test for evidence of a second Flt3L receptor, we cultured WT and Flt3−/− BM in vitro with Flt3L and assessed DC development, hypothesizing that Flt3L could bind to its second receptor in Flt3−/− BM and generate DCs. However, although Flt3L supported robust DC development from WT BM, it did not generate DCs from Flt3−/− BM (Fig. 3, A–E). Thus, in vitro, we were unable to provide evidence for a second receptor responding to Flt3L. We next administered Flt3L in vivo (Fig. 3, F–J). Flt3L administration to WT mice caused a significant increase in the percentage and numbers of splenic DCs, as expected (Maraskovsky et al., 1996), but failed to increase DCs in Flt3−/− mice (Fig. 3, F–J). In summary, we find no evidence for actions of Flt3L on a second receptor.
Figure 3.
Flt3L fails to generate DCs in Flt3−/− mice. (A–E) BM cells from WT (Flt3+/+) or Flt3−/− mice were treated with vehicle or Flt3L and cultured for 9 d. Live cells were subsequently analyzed by flow cytometry for DC populations. (A) Shown are representative two-color histograms of live cells pregated as indicated above the plots. Numbers specify the percentage of cells within the indicated gates. (B–E) Summary data for the percentage (B) and number (C) of cDCs in each culture and for the percentage (D) and number (E) of pDCs in each culture. Dots represent biological replicates; small horizontal lines indicate the mean. Data are pooled from three independent experiments (n = 4 mice per genotype). (F–J) WT (Flt3+/+) and Flt3−/− mice were administered vehicle or Flt3L subcutaneously for 8 d and splenocytes were subsequently analyzed by flow cytometry on day 9 for DC populations. (F) Shown are representative two-color histograms of live cells. Numbers specify the percentage of cells within the indicated gates. (G–J) Summary data for the percentage (G) and number (H) of splenic cDCs and the percentage (I) and number (J) of splenic pDCs. Dots represent biological replicates; small horizontal lines indicate the mean. Data are pooled from two independent experiments (n = 5 mice per group). ns, not significant (P > 0.05); *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 using unpaired, two-tailed Student's t test.
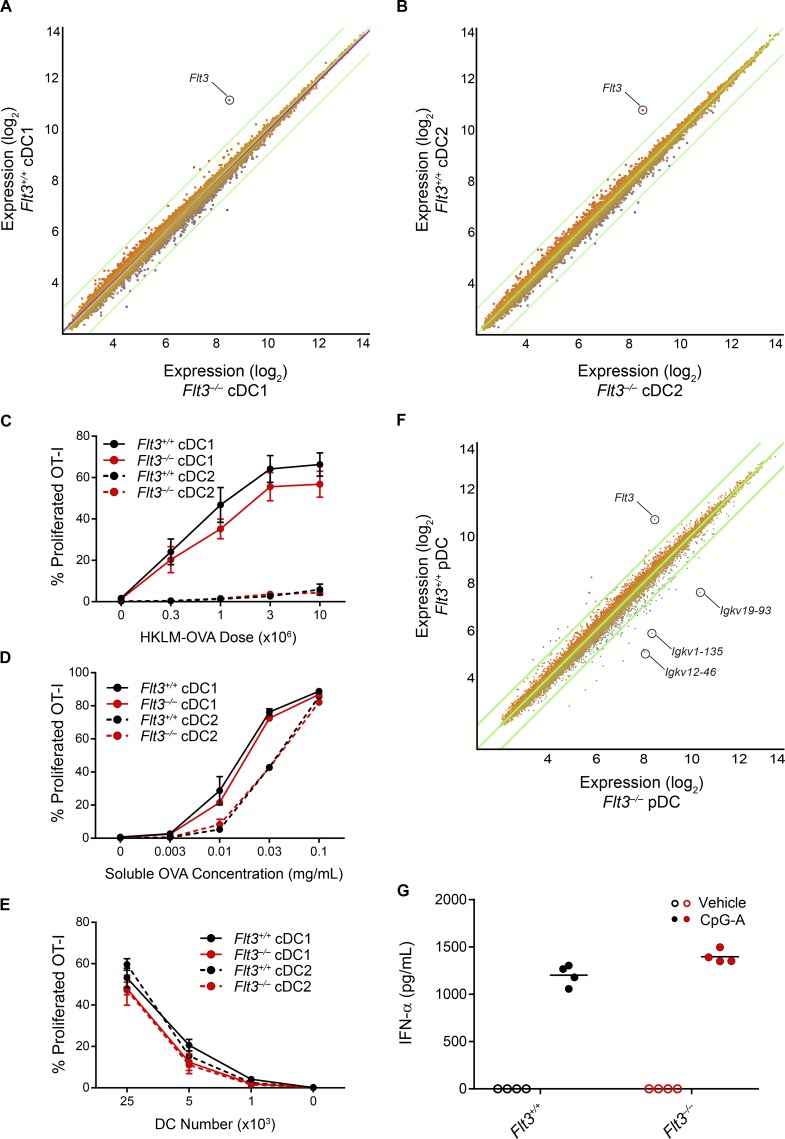
Because DCs in Flt3−/− mice appeared to arise independently of Flt3L, we wondered whether they were developmentally or functionally impaired. We therefore examined the transcriptional profile of splenic cDC1s and cDC2s from WT and Flt3−/− mice using gene expression microarrays. We found no substantial changes in gene expression in either cDC subset, with the only annotated transcript showing a greater-than fourfold change being Flt3 itself (Fig. 4, A and B). In addition, we found that Flt3−/− cDC1s were able to cross-present cell-associated and soluble antigen to CD8 T cells as efficiently as WT cDC1s (Fig. 4, C–E), suggesting they were functionally similar. We next examined the transcriptional profile of splenic pDCs from WT and Flt3−/− mice (Fig. 4 F). We again found only minor differences between these populations, with Flt3 being the only annotated transcript showing a greater-than fourfold increase in expression in WT pDCs and with several genes encoding Igκ segments being more highly expressed in Flt3−/− pDCs. pDCs have been found to develop from both myeloid and lymphoid progenitors (Sathe et al., 2013), with pDCs derived from lymphoid progenitors showcasing rearrangements of D-J segments in the immunoglobulin genes. The increased expression of these genes encoding Igκ segments in Flt3−/− pDCs could therefore indicate that the pDCs in Flt3−/− mice preferentially arise from lymphoid rather than myeloid progenitors. Other than these genes, we found that the expression of the vast majority of transcripts was similar between WT and Flt3−/− pDCs (Fig. 4 F). Finally, we stimulated WT and Flt3−/− pDCs with CpG oligodeoxynucleotides and found that both populations produced a comparable amount of IFN-α (Fig. 4 G). Together, these results suggest that the DCs in Flt3−/− mice are transcriptionally and functionally similar to WT DCs, implying that Flt3 signaling is not essential for generating functional DCs.
Figure 4.
Flt3−/− DCs are transcriptionally and functionally similar to WT DCs. (A and B) Microarray analysis of gene expression in cDC1s (A) or cDC2s (B) from Flt3+/+ and Flt3−/− mice, presented as M-plots. Colors indicate higher (red) or lower (blue) expression in Flt3+/+ cells than in Flt3−/− cells. Annotated genes with a greater than fourfold change in expression are specified (n = 2 biological replicates per subset per genotype). (C) Sorted cDC subsets from Flt3+/+ or Flt3−/− mice were cultured for 3 d with CFSE-labeled OT-I cells and different doses of heat-killed LM-OVA (HKLM-OVA) and then assayed for OT-I proliferation (CFSE–CD44+). Summary data for the percentage of proliferated OT-I cells in cultures with cDC subsets from the indicated genotypes are shown. Data are pooled from two independent experiments (n = 4 mice per genotype). (D) Sorted cDC subsets from Flt3+/+ or Flt3−/− mice were cultured for 3 d with CFSE-labeled OT-I cells and different doses of soluble OVA as antigen and then assayed for OT-I proliferation (CFSE–CD44+). Summary data for the percentage of proliferated OT-I cells in cultures with cDC subsets from the indicated genotypes are shown. Data are pooled from two independent experiments (n = 4 mice per genotype). (E) Sorted cDC subsets from Flt3+/+ or Flt3−/− mice were incubated with soluble OVA, washed, and then different numbers of cDCs were cultured with CFSE-labeled OT-I cells for 3 d after which OT-I proliferation was assayed. Summary data for the percentage of proliferated OT-I cells in cultures with the cDC subsets from the indicated genotypes are shown. Dots represent the mean; error bars represent the SEM. Data are pooled from two independent experiments (n = 4 mice per genotype). (F) Microarray analysis of gene expression in pDCs from Flt3+/+ and Flt3−/− mice, presented as M-plots. Colors indicate higher (red) or lower (blue) expression in Flt3+/+ cells than in Flt3−/− cells. Annotated genes with a greater than fourfold change in expression are specified (n = 2 biological replicates per subset per genotype). (G) Sorted pDCs from Flt3+/+ or Flt3−/− mice were stimulated with vehicle or CpG-A oligodeoxynucleotides overnight and then the cell supernatant was analyzed for IFN-α production. Summary data for the quantity of IFN-α produced by pDCs of the indicated genotypes are shown. Dots represent biological replicates; small horizontal lines indicate the mean. Data are pooled from two independent experiments (n = 4 mice per genotype).
Committed DC progenitors develop in Flt3−/− BM and can mature in response to M-CSF and SCF
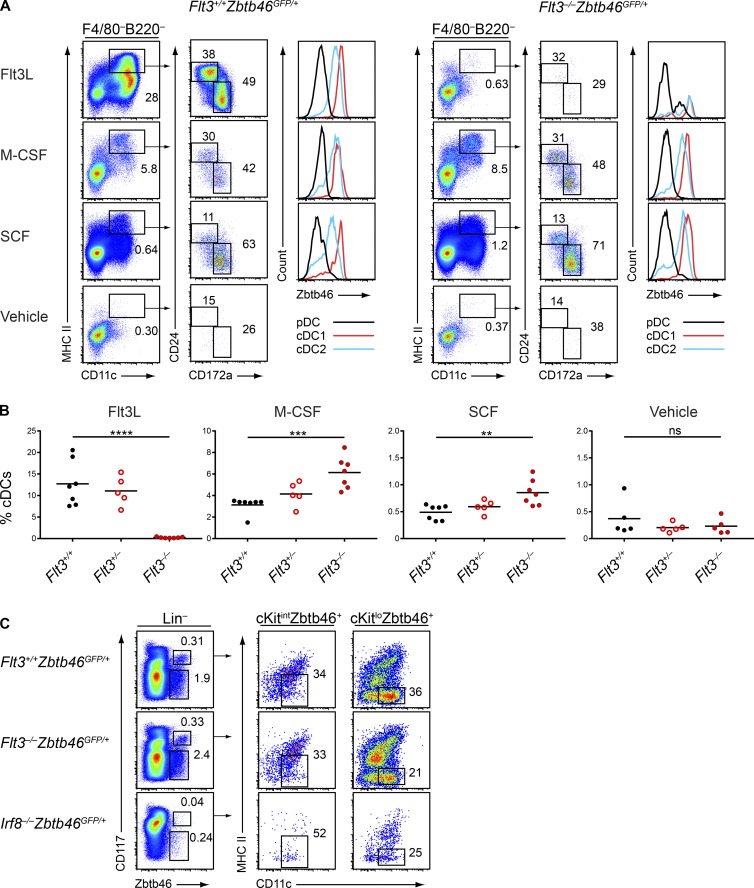
We next considered whether Flt3−/− DCs could be maturing in response to alternative cytokines. The receptors for M-CSF (CD115/CSF1R) and for SCF (CD117/c-Kit) are both expressed by the MDP and CDP (Liu et al., 2009), suggesting that these cytokines could act on these progenitors, and M-CSF has also been reported to support the development of some DC subsets in vitro (Fancke et al., 2008). To help identify cDCs in the Flt3−/− background, we crossed Flt3−/− mice to Zbtb46GFP/+ mice to generate Flt3−/−Zbtb46GFP/+ mice, in which we could use Zbtb46GFP expression to distinguish cDCs from other lineages (Meredith et al., 2012; Satpathy et al., 2012; Wu et al., 2016). We then cultured bulk BM from WT (Flt3+/+Zbtb46GFP/+), Flt3+/−Zbtb46GFP/+, and Flt3−/−Zbtb46GFP/+ mice with Flt3L, M-CSF, or SCF and assessed DC development. Surprisingly, we found that all three cytokines supported the development of cDC1s and cDC2s from WT and Flt3+/− BM, whereas M-CSF and SCF also supported cDC development from Flt3−/− BM (Fig. 5, A and B). Notably, M-CSF and SCF generated a greater percentage (Fig. 5 B) and number (Fig. S2 A) of cDCs from Flt3−/− BM than from WT BM, suggesting that Flt3−/− BM may be more responsive to these cytokines than WT BM. Importantly, the cDCs generated in all cultures were uniformly positive for Zbtb46GFP expression (Fig. 5 A), confirming their identity as bona fide cDCs (Meredith et al., 2012; Satpathy et al., 2012; Wu et al., 2016).
Figure 5.
Flt3−/− BM generates mature DCs in response to M-CSF or SCF and contains committed DC progenitors. (A and B) BM cells from mice of the indicated genotypes were treated with vehicle, Flt3L, M-CSF, or SCF and cultured for 7 d. Live cells were subsequently analyzed by flow cytometry for development of DCs. (A) Shown are representative two-color histograms of live cells pregated as indicated above the plots and one-color histograms of Zbtb46GFP expression in cells of the indicated subtype. Numbers represent the percentage of cells within the indicated gates. (B) Summary data for the percentage of cDCs in each culture. Dots represent biological replicates; small horizontal lines indicate the mean. Data are pooled from five independent experiments (n = 5–7 mice per genotype). (C) BM cells from mice of the indicated genotypes were analyzed by flow cytometry for DC progenitor populations; lineage markers include CD3, CD19, CD105, Ter119, and Ly-6G. Representative two-color histograms are shown of live cells pregated as indicated above the plots. Numbers specify the percentage of cells within the indicated gates. Data are representative of four independent experiments (n = 7 mice per genotype). ns, not significant (P > 0.05); **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 using unpaired, two-tailed Student's t test.
To further characterize these cells and to identify any heterogeneity in the populations, we examined the expression of a variety of functional markers in cDCs from each culture condition. We found that the cDCs generated in response to each cytokine generally expressed similar levels of costimulatory molecules such as CD86, PD-L2, CD40, and CD205; surface markers such as CD24, CD172a, and CD11b; and DC-specific markers such as Flt3/CD135 (the cDCs from Flt3L cultures expressed low levels of this receptor, potentially caused by down-regulation after exposure to Flt3L), CCR7, and CD117, and that they all expressed similarly low levels of macrophage markers such as CD115 and CD64 (Fig. S2 B). Thus, we concluded that the cDCs that mature in response to M-CSF and SCF are generally similar in phenotype to those that are generated by Flt3L. Finally, we found that Flt3L, M-CSF, and SCF could all support pDC development from WT BM and that M-CSF and SCF could also support pDC development from Flt3−/− BM (Fig. S3 A).
Because mature DCs could be generated from the BM of Flt3−/− mice, we wondered whether DC progenitor specification was occurring normally in these mice. Identifying the CDP relies on Flt3 expression (Onai et al., 2007) and is therefore not possible in Flt3−/− mice. However, two restricted cDC progenitors downstream of the CDP, pre-cDC1 and pre-cDC2, can be identified with Zbtb46GFP expression and additional markers without the need for Flt3 (Grajales-Reyes et al., 2015). We therefore analyzed BM from Flt3−/−Zbtb46GFP/+ mice to identify these progenitors and found normal frequencies of pre-cDC1s and pre-cDC2s compared with Flt3+/+Zbtb46GFP/+ mice (Fig. 5 C). As a control, BM from Irf8−/−Zbtb46GFP/+ mice lacked pre-cDC1s and had reduced pre-cDC2s, as expected (Grajales-Reyes et al., 2015). Thus, cDC progenitor specification appears to be independent of Flt3 signaling.
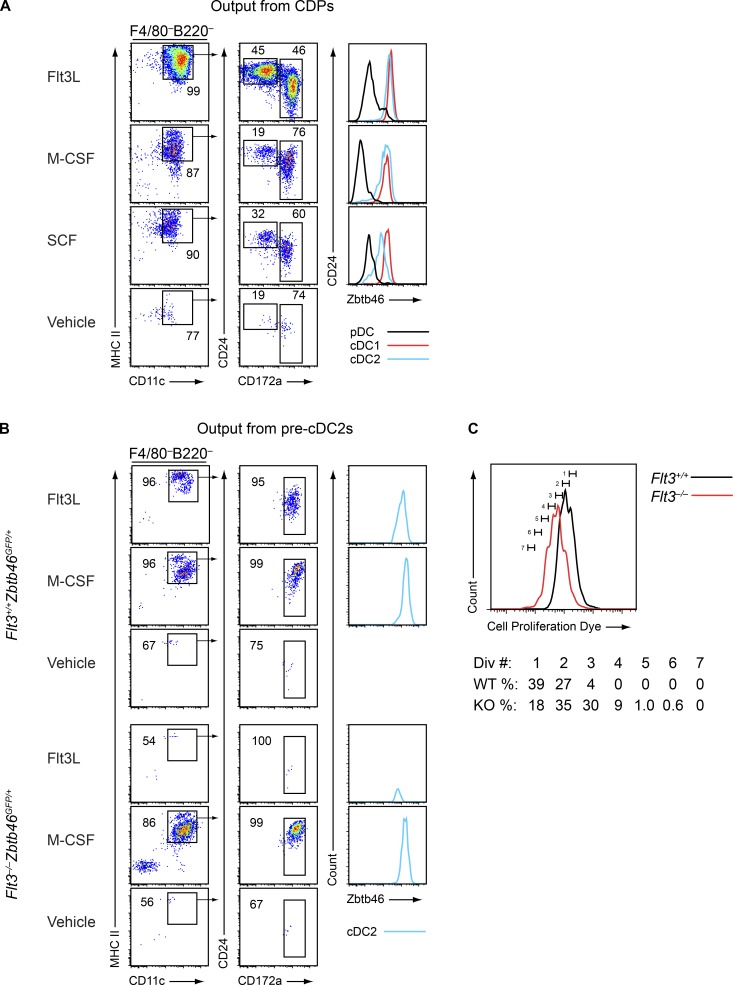
We next asked whether M-CSF and SCF could act directly on committed DC progenitors to support their maturation. To address this question, we sort purified WT CDPs, which express Flt3, CSF1R, and c-Kit, and cultured them with Flt3L, M-CSF, or SCF. These cytokines each supported development of the CDP into mature Zbtb46GFP-positive cDC1s and cDC2s (Fig. 6 A) as well as pDCs (Fig. S3 B). In addition, pre-cDC2s, which normally express CSF1R and Flt3, were isolated from Flt3+/+Zbtb46GFP/+ mice and Flt3−/−Zbtb46GFP/+ mice and cultured with Flt3L or M-CSF. As expected, Flt3L supported development of the pre-cDC2 into mature Zbtb46GFP-positive cDC2s from WT progenitors but not from Flt3−/− progenitors (Fig. 6 B). In contrast, M-CSF supported development of the pre-cDC2 into Zbtb46GFP-positive cDC2s from both WT and Flt3−/− progenitors (Fig. 6 B). When we analyzed cell divisions using cell proliferation dye tracing, we found that, when cultured with M-CSF, Flt3−/− pre-cDC2s proliferated more than WT pre-cDC2s (Fig. 6 C), again suggesting that Flt3−/− progenitors were more responsive to M-CSF than WT progenitors. Thus, our data suggest that specified DC progenitors that develop in the presence or absence of Flt3 signaling can mature in response to cytokines other than Flt3L.
Figure 6.
Committed DC progenitors can mature in response to M-CSF and SCF. (A) CDPs (Lin–CD117intCD135+CD115+CD11c–MHCII–) were sorted from Zbtb46GFP/+ mice and cultured with vehicle, Flt3L, M-CSF, or SCF for 5 d. Cells were subsequently analyzed by flow cytometry for development of DCs. Shown are representative two-color histograms of live cells pregated as indicated above the plots and one-color histograms of Zbtb46GFP expression. Numbers specify the percentage of cells within the indicated gates. Data are representative of three independent experiments (sorted cells from three to five mice were pooled in each individual experiment). (B) Pre-cDC2s (Lin–CD115+CD11c+MHCII–Zbtb46+) were sorted from Flt3+/+Zbtb46GFP/+ or Flt3−/−Zbtb46GFP/+ mice as indicated and cultured with vehicle, Flt3L, or M-CSF for 5 d. Cells were subsequently analyzed by flow cytometry for development of DCs. Shown are representative two-color histograms of live cells pregated as indicated above the plots and one-color histograms of Zbtb46GFP expression in cells of the indicated subtype. Numbers specify the percentage of cells within the indicated gates. Data are representative of three independent experiments (sorted cells from three to five mice per genotype were pooled in each individual experiment). (C) Pre-cDC2s were sorted from Flt3+/+Zbtb46GFP/+ or Flt3−/−Zbtb46GFP/+ mice, labeled with Cell Proliferation Dye (CPD), and cultured with M-CSF for 5 d. Cells were subsequently analyzed by flow cytometry for dilution of CPD to determine the number of divisions they underwent. Shown is a representative one-color histogram of CPD levels and the percentage of cells at each division. Data are representative of two independent experiments (sorted cells from three mice per genotype were pooled in each individual experiment).
DC development in Flt3−/− mice is substantially dependent on CSF1R
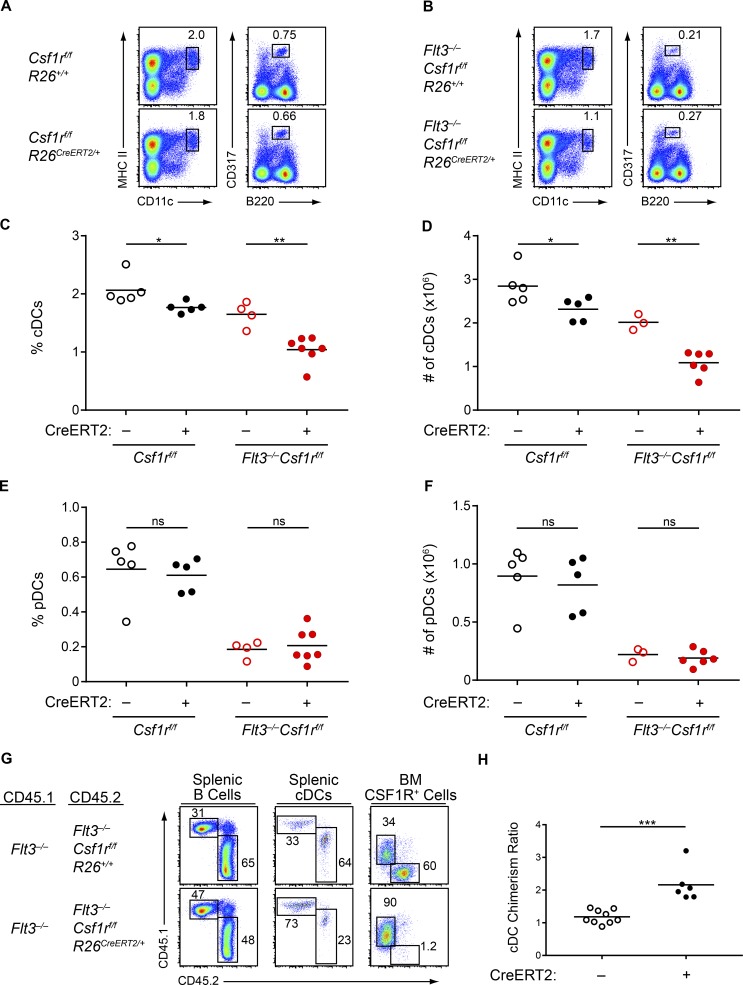
To determine whether M-CSF supported the in vivo DC development observed in Flt3−/− mice, we deleted Csf1r (Li et al., 2006) in WT and Flt3−/− mice using a tamoxifen-inducible Cre recombinase (Ventura et al., 2007). Deletion of Csf1r in WT mice caused a small decrease in splenic cDC percentage and numbers, consistent with a previous study describing a minor role for CSF1R in cDC homeostasis (Fig. 7, A, C, and D; Ginhoux et al., 2009). In contrast, deletion of Csf1r in Flt3−/− mice led to a substantial reduction in cDCs (Fig. 7, B–D), confirming that cDCs developing in Flt3−/− mice arise in part through the actions of CSF1R. pDC development, however, was unaffected by the additional deletion of Csf1r (Fig. 7, E and F), consistent with prior studies showing that pDC progenitors are enriched in the CSF1R– fraction of BM progenitors (Onai et al., 2013) and that almost all pDCs are derived from an IL-7R+ progenitor (Schlenner et al., 2010). Our gene expression data had also indicated that the pDCs in Flt3−/− mice may be preferentially developing from lymphoid progenitors (Fig. 4 F), which, combined with the lack of a role for CSF1R, suggests that the pDCs in Flt3−/− mice may be developing in response to compensatory signaling from a receptor expressed on lymphoid progenitors such as IL-7R. In summary, a substantial portion of cDC but not pDC development in Flt3−/− mice is dependent on CSF1R.
Figure 7.
cDC development in Flt3−/− mice is dependent on CSF1R. (A–F) Mice of the indicated genotypes were treated with tamoxifen to delete Csf1r. After treatment, splenocytes were analyzed by flow cytometry for DC populations. (A and B) Shown are representative two-color histograms of live cells. Numbers specify the percentage of cells within the indicated gates. (C–F) Summary data of the percentage (C) and number (D) of splenic cDCs and for the percentage (E) and number (F) of splenic pDCs in mice of the indicated genotypes. Dots represent biological replicates; small horizontal lines indicate the mean. Data are pooled from seven independent experiments. (n = 3–7 mice per genotype). (G and H) WT CD45.1+ mice were lethally irradiated and reconstituted with Flt3−/− CD45.1+:Flt3−/−Csf1rf/fR26+/+CD45.2+ BM at a 1:1 ratio (CreERT2–) or Flt3−/−CD45.1+:Flt3−/−Csf1rf/fR26CreERT2/+CD45.2+ BM at a 1:1 ratio (CreERT2+). After reconstitution and tamoxifen treatment to delete Csf1r, spleens and BM were analyzed by flow cytometry. (G) Shown are representative two-color histograms (pregating, above plots; splenic B cells, B220+CD24+; splenic cDCs, CD11c+MHCII+; BM CSF1R+ cells, CD115+). Numbers specify the percentage of cells within the indicated gates. (H) Summary data of the chimerism ratio of splenic cDCs presented as the ratio of CD45.1+ to CD45.2+ cDCs normalized to the ratio of CD45.1+ to CD45.2+ B cells within the same mouse. Dots represent individual mice; small horizontal lines indicate the mean. Data are pooled from four independent experiments (n = 6–9 chimeras per genotype). ns, not significant (P > 0.05); *, P < 0.05; **, P < 0.01; ***, P < 0.001 using unpaired, two-tailed Student's t test.
To test whether this action of CSF1R was cell-intrinsic to DC progenitors, we generated mixed BM chimeras with a 50:50 mix of Flt3−/− BM and either Flt3−/−Csf1rf/fR26+/+ BM (CreERT2–) or Flt3−/−Csf1rf/fR26CreERT2/+ BM (CreERT2+). Although all BM progenitors in these chimeras were deficient in Flt3, tamoxifen treatment led to the additional deletion of CSF1R in half of the BM cells in CreERT2+ chimeras but not in CreERT2– chimeras (Fig. 7 G). Thus, we could directly compare the DC potential of progenitors lacking both Flt3 and CSF1R with progenitors lacking only Flt3. In CreERT2+ chimeras, cDCs preferentially developed from Flt3−/− BM cells, rather than from Flt3−/−Csf1r−/− BM, whereas in CreERT2– chimeras there was no skew in DC development between Flt3−/− and Flt3−/−Csf1rf/fR26+/+ BM (Fig. 7, G and H). This result suggests that the contribution of CSF1R to DC development in Flt3−/− progenitors is cell-intrinsic.
Flt3−/− progenitors express normal levels of CSF1R and c-Kit but display enhanced signaling from these receptors
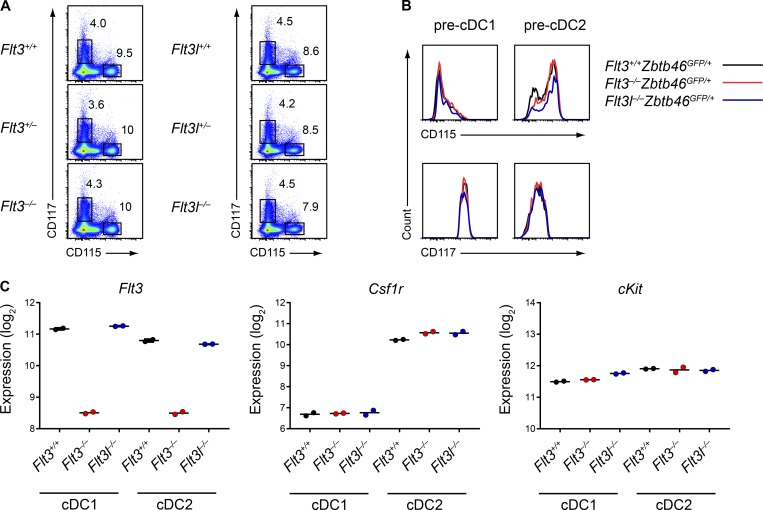
Because M-CSF and SCF could support DC development independently of Flt3, we wondered whether the discrepancy between Flt3−/− and Flt3l−/− mice might be a result of differences in the expression or signaling of CSF1R or c-Kit in these different strains. First, we found no differences in surface expression of CSF1R or c-Kit in total BM isolated from WT, Flt3−/−, and Flt3l−/− mice (Fig. 8 A). We next sought to quantify the expression of these receptors in early DC progenitors, but because Flt3 is required to identify the MDP and CDP (Onai et al., 2007), we could not analyze these progenitors in Flt3−/− mice. We therefore generated Flt3l−/−Zbtb46GFP/+ mice to identify the pre-cDC1 and pre-cDC2 in Flt3l−/− mice, which would allow us to compare these progenitors in WT, Flt3−/−, and Flt3l−/− mice. We found that both progenitors still developed in Flt3l−/− mice, but were substantially fewer in number relative to WT and Flt3−/− mice (Fig. S4 A). This was similar to the reported deficiency in MDPs and CDPs in Flt3l−/− mice (Kingston et al., 2009), suggesting that all DC progenitors are affected by the loss of Flt3L. We found, however, that the expression of CSF1R and c-Kit was not different in pre-cDC1s or pre-cDC2s from Flt3−/− or Flt3l−/− mice relative to WT mice (Fig. 8 B), again suggesting that changes in receptor expression did not explain the differences between these strains. Finally, we sought to determine the expression of these receptors in mature splenic cDCs from these strains, but as collagenase digestion of the spleen impairs detection of these receptors by FACS (Merad et al., 2013), we instead used gene expression microarrays. We found no differences in Csf1r or Kit gene expression in splenic cDC1s or cDC2s from WT, Flt3−/−, and Flt3l−/− mice (Fig. 8 C). Thus, we excluded differences in receptor expression as a cause of the discrepancy between Flt3−/− and Flt3l−/− mice.
Figure 8.
Flt3−/− and Flt3l−/− BM progenitors and cDCs express normal levels of CSF1R and c-Kit. (A) BM cells from mice of the indicated genotypes were analyzed by flow cytometry for cytokine receptor expression. Representative two-color histograms of live cells are shown. Numbers specify the percentage of cells within the indicated gates. Data are representative of at least three independent experiments (n = 4–7 mice per genotype). (B) Pre-cDC1s and pre-cDC2s from mice of the indicated genotypes were analyzed by flow cytometry for cytokine receptor expression levels. Representative one-color histograms of each receptor are shown. Data are representative of three independent experiments (n = 3–7 mice per genotype). (C) Expression of the indicated genes in splenic cDC1s and cDC2s from mice of the indicated genotypes was determined by gene expression microarray analysis. Dots represent biological replicates (n = 2 mice per subset per genotype).
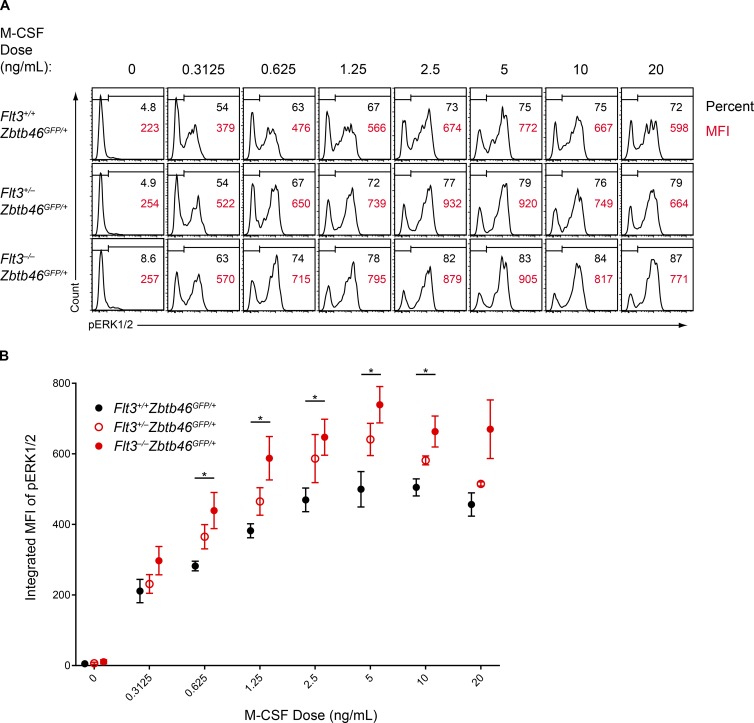
The in vitro DC development supported by M-CSF and SCF was greater in Flt3−/− BM relative to WT BM (Fig. 5 B), and Flt3−/− pre-cDC2s proliferated more than WT pre-cDC2s in response to M-CSF (Fig. 6 C), suggesting there was some alteration in receptor sensitivity between WT and Flt3−/− mice. Flt3, CSF1R, and c-Kit are all class III RTKs that share signaling adaptor molecules such as Shc and Grb2 (Dosil et al., 1993; Rottapel et al., 1994; Verstraete and Savvides, 2012; Tsapogas et al., 2017). We wondered whether the loss of Flt3 might increase the availability of these adaptors and thereby enhance signaling from CSF1R or c-Kit. To test this, we quantified the sensitivity of CSF1R signaling in BM progenitors. We treated serum-starved BM cells from Flt3+/+, Flt3+/−, or Flt3−/− mice with varying concentrations of M-CSF and quantified phosphorylation of the MAPK pathway protein Erk1/2 in pre-cDC2s (Fig. 9 A). We examined this progenitor because it expresses CSF1R and can be identified independently of Flt3 by using Zbtb46-GFP expression (Grajales-Reyes et al., 2015). Over a range of concentrations, both the percentage of pre-cDC2s responding to M-CSF and the intensity of signaling in each progenitor was greater in Flt3−/− pre-cDC2s compared with Flt3+/+ pre-cDC2 cells, with Flt3+/− pre-cDC2s displaying an intermediate phenotype (Fig. 9, A and B). Thus, the loss of Flt3 directly enhances the sensitivity and strength of CSF1R signaling in response to M-CSF. Importantly, no differences were observed in M-CSF signaling sensitivity in Flt3l−/− pre-cDC2s relative to WT for most concentrations of M-CSF (Fig. S4 B), indicating that the increased sensitivity to M-CSF was specific to Flt3−/− mice and could possibly explain the difference between these mice and Flt3l−/− mice.
Figure 9.
Flt3−/− DC progenitors display increased sensitivity to M-CSF. (A and B) Serum-starved BM from mice of the indicated genotypes was treated with M-CSF and assayed for phosphorylated Erk1/2 (pErk1/2) by intracellular flow cytometry. (A) Shown are representative one-color histograms of pErk1/2 in pre-cDC2s stimulated with the indicated concentrations of M-CSF. Numbers in black indicate the percentage of cells within the gate, and numbers in red indicate the geometric MFI of pErk1/2 in the gated population. Data are representative of three independent experiments. (B) Summary data presented as the integrated MFI of pErk1/2 in pre-cDC2s from mice of the indicated genotypes stimulated with the indicated concentration of M-CSF. Dots indicate the mean from three independent experiments; error bars indicate the SEM (n = 3 mice per genotype). *, P < 0.05 using Student's t test.
Deletion of Flt3 in Flt3l−/− mice paradoxically restores DC development
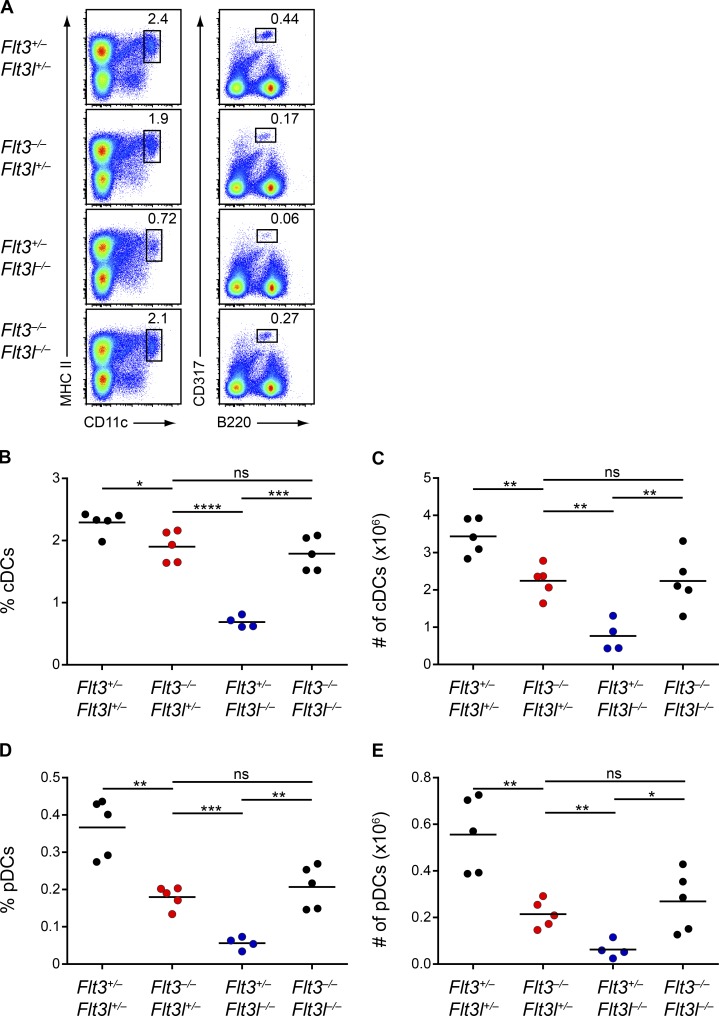
The increased sensitivity of CSF1R signaling in Flt3−/− DC progenitors could explain the discrepancy in DC numbers between Flt3−/− and Flt3l−/− mice. Flt3−/− DC progenitors could respond more robustly to alternative cytokines such as M-CSF or SCF and thus develop greater numbers of DCs. This hypothesis predicts that the increased CSF1R sensitivity in Flt3−/− mice should occur regardless of the presence of Flt3L and thus that loss of Flt3 in the Flt3l−/− background should enhance CSF1R signaling and potentially increase DC numbers. To test this hypothesis, we crossed Flt3−/− and Flt3l−/− mice to generate Flt3−/−Flt3l−/− mice and characterized DC development in the resulting littermates. We first found that, unlike Flt3+/− mice, Flt3+/−Flt3l+/− mice had mildly reduced DC numbers (Fig. 10, A–E). This suggests that the decrease in DCs we found in Flt3l+/− mice (Figs. 1 and 2) was more impactful than the increase in DCs observed in Flt3+/− mice (Figs. 1 and 2) and that together these alleles resulted in a net decrease in DC numbers in Flt3+/−Flt3l+/− mice. But most surprisingly, we found that Flt3−/−Flt3l−/− mice had an increase in splenic cDC and pDC percentage and numbers relative to Flt3l−/− mice and were indistinguishable from Flt3−/− mice (Fig. 10, A–E). This increase was also observed in cDCs and pDCs in the SLNs (Fig. S5, A and B), as well as in cDCs in the lung (Fig. S5 C). Thus, paradoxically, the severe DC deficiency observed in Flt3l−/− mice was rescued by the additional loss of Flt3. These results suggest that the discrepancy observed between Flt3−/− and Flt3l−/− mice is, at least in part, a result of an inherent alteration in the response of DC progenitors to alternative cytokines secondary to the loss of Flt3.
Figure 10.
Deletion of Flt3 rescues the severe DC defect in Flt3l−/− mice. (A–E) Splenocytes from mice of the indicated genotypes at 8-wk of age were analyzed by flow cytometry for DC populations. (A) Shown are representative two-color histograms of live cells. Numbers specify the percentage of cells within the indicated gates. (B–E) Summary data for the percentage (B) and number (C) of splenic cDCs and the percentage (D) and number (E) of splenic pDCs in mice of the indicated genotypes. Dots represent biological replicates; small horizontal lines indicate the mean. Data are pooled from six independent experiments (n = 4–5 mice per genotype). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 using unpaired, two-tailed Student's t test.
Discussion
In this study, we have identified the previously unknown cytokine pathways that support the residual DC development seen in Flt3−/− and Flt3l−/− mice, and we provide an explanation for the puzzling discrepancy in phenotypes between these two strains. Although both Flt3 and Flt3L have long been recognized to play major roles in DC homeostasis (McKenna et al., 2000; Waskow et al., 2008), it had remained unclear through what mechanism DCs continue to arise in mice deficient in either of these factors. We have demonstrated here that M-CSF and SCF, two cytokines whose receptors are expressed in DC progenitors, are capable of generating DCs from both WT and Flt3−/− BM progenitors in vitro. We also found that a substantial fraction of the DCs that arise in vivo in Flt3−/− mice are dependent on CSF1R for their development, with c-Kit possibly responsible for generating the remainder of the DCs.
These findings, along with the continued presence of committed DC progenitors in Flt3−/− mice, suggest that DC lineage commitment may not require the Flt3 pathway and that multiple cytokines can provide the supportive signals necessary for the maturation of these progenitors. Furthermore, the DCs that develop in Flt3−/− mice were functional and had transcriptional profiles indistinguishable from WT DCs, suggesting that the actions of Flt3L in DC development can be compensated entirely by other cytokines. Though we had originally hypothesized that WT DCs would possess a gene signature indicative of their maturation in response to Flt3 signaling, that we did not see such a signature or a corresponding gene signature suggestive of M-CSF signaling in the Flt3−/− DCs suggests that these cytokines may not impose any long-lasting genetic instruction upon mature DCs, but instead simply support their development. DCs are therefore another example of a lineage that continues to develop and function in the absence of its principal growth factor, similar to granulocytes (Lieschke et al., 1994), erythrocytes (Wu et al., 1995), and lymphocytes (Peschon et al., 1994; von Freeden-Jeffry et al., 1995). Findings such as these help elucidate the precise role of cytokines in hematopoietic lineage commitment and suggest at least some level of promiscuity between cytokine pathways and cell types.
Loss of a receptor and its unique ligand would generally be expected to produce the same phenotype. However, our direct comparison of Flt3−/− and Flt3l−/− mice clearly documents that loss of the ligand causes a more severe and durable DC deficiency than loss of the receptor. We were unable to find evidence to support previous suggestions that Flt3L might be acting at a second receptor beside Flt3 (Nagasawa, 2006; Helft et al., 2010; Satpathy et al., 2011; Moore and Anderson, 2013; Tsapogas et al., 2017). Instead, we found that Flt3−/− DC progenitors exhibit an intrinsic alteration in the sensitivity of other cytokine receptors compared with WT progenitors. This may allow compensatory cytokines such as M-CSF and SCF to generate a greater number of DCs in Flt3−/− mice relative to WT or Flt3l−/− mice. We therefore predicted, and then validated, the paradoxical result that the severe DC defect in Flt3l−/− mice is actually restored upon deletion of Flt3. Thus, it appears that the discrepancy in DC development between Flt3−/− and Flt3l−/− mice arises secondary to a phenotype of increased sensitivity of Flt3−/− DC progenitors to alternative cytokines. It is possible that a similar increase in sensitivity to IL-7 in early B cell progenitors could explain the previously identified discrepancy in B cell development between these strains (Nagasawa, 2006).
The increased sensitivity of Flt3+/− progenitors to M-CSF (Fig. 9) might also explain the increased DC numbers we observed in Flt3+/− mice (Figs. 1 and 2). Flt3+/− BM produced DCs in response to Flt3L as well as to M-CSF (Fig. 5 B), so the increased DC numbers seen in Flt3+/− mice conceivably results from the combination of both normal Flt3L signaling and enhanced M-CSF signaling contributing to DC development in these mice. In contrast, in Flt3−/− mice only enhanced M-CSF signaling, but not Flt3L signaling, contributes to DC development, and these mice therefore have fewer DCs despite their even greater sensitivity to M-CSF.
We have not explored the biochemical basis for the altered sensitivity observed in Flt3−/− DC progenitors. Conceivably, loss of Flt3 increases the availability of shared signaling adaptor proteins such as Shc and Grb2 used by other class III RTKs (Dosil et al., 1993; Rottapel et al., 1994; Verstraete and Savvides, 2012; Tsapogas et al., 2017). The increased availability of these adaptors could then allow other receptors acting through them to activate downstream pathways more quickly and potently. Although the exact mechanism remains to be uncovered, our results highlight the potential for unintended consequences that may result when manipulating the levels of signaling molecules in complex biological systems. The deletion of Flt3 appears to have ramifications beyond simply the loss of signaling by this receptor, and the study of other receptor knockout mice may have similar caveats that should be considered.
Materials and methods
Mice
All mice were bred and maintained on the C57BL/6 background in a specific pathogen-free animal facility following institutional guidelines and with protocols approved by the Animal Studies Committee at Washington University in St. Louis. Unless otherwise specified, experiments were performed with mice 6- to 10-wk of age. No differences were observed between male and female mice in any assays and so mice of both genders were used throughout this study. Within individual experiments, mice used were age- and sex-matched littermates whenever possible.
Flt3−/− mice (Mackarehtschian et al., 1995) were a gift from E. Camilla Forsberg (University of California, Santa Cruz, Santa Cruz, CA). Zbtb46GFP/+ mice have been previously described (Satpathy et al., 2012; Grajales-Reyes et al., 2015). The following mice were purchased from The Jackson Laboratory: Flt3l−/− (C57BL/6-Flt3ltm1Imx/TacMmjax), CD45.1+ (B6.SJL-Ptprca Pepcb/BoyJ), OT-I (C57BL/6-Tg(TcraTcrb)1100Mjb/J), Csf1rf/f (B6.Cg-Csf1rtm1.2Jwp/J), and R26CreERT2/+ (B6.129-Gt(ROSA)26Sortm1(cre/ERT2)Tyj/J).
DC preparation
Lymphoid and nonlymphoid organ DCs were harvested and prepared as described previously (Satpathy et al., 2012). In brief, spleens and SLNs (inguinal, axillary, and brachial) were minced and digested in 5 ml IMDM + 10% FCS (cIMDM) with 250 µg/ml collagenase B (Roche) and 30 U/ml DNase I (Sigma-Aldrich) for 45 min at 37°C with stirring. Lungs were minced and digested in 5 ml of cIMDM with 4 mg/ml collagenase D (Roche) and 30 U/ml DNase I (Sigma-Aldrich) for 1.5 h at 37°C with stirring. After digestion, cell suspensions from all organs were passed through 70-µm strainers and red blood cells were then lysed with ammonium chloride-potassium bicarbonate lysis buffer. Cells were counted with a Vi-CELL analyzer (Beckman Coulter) and 3–5 × 106 cells were used per antibody-staining reaction.
For experiments requiring sorting of splenic cDC subsets, cDCs were first enriched from total spleen cells with CD11c microbeads (Miltenyi Biotec). cDCs were then sorted on a flow cytometer (FACSAria Fusion; BD Biosciences). cDC1s were sorted as CD11c+MHC-II+CD24+CD172a–F4/80–, and cDC2s were sorted as CD11c+MHCII+CD24–CD172a+F4/80–. Sort purity of >95% was confirmed by post-sort analysis before cells were used for further experiments.
For experiments requiring sorting of splenic pDC subsets, spleen cells were first depleted of CD3-, B220-, and Ly6G-expressing cells by staining with the corresponding biotinylated antibodies followed by depletion with MagniSort Streptavidin Negative Selection beads (Thermo Fisher). pDCs were then sorted on a FACSAria Fusion flow cytometer as B220+CD317+. Sort purity of >95% was confirmed by post-sort analysis before cells were used for further experiments.
Antibodies and flow cytometry
Samples were stained at 4°C in MACS buffer (DPBS + 0.5% BSA + 2 mM EDTA) in the presence of Fc block (2.4G2; BD Biosciences). The following antibodies were purchased from BD Biosciences: CD8α V450 (53-6.7); CD11b PE (M1/70); CD19 Biotin (1D3); CD44 APC-Cy7 (IM7); CD45R Alexa Fluor 488; V450 (RA3-6B2); CD45.2 PerCP-Cy5.5 (104); CD64 Alexa Fluor 647 (X54-5/7.1); CD103 BV421 (M290); CD117 BUV395 (2B8); CD127 BV421 (SB/199); CD135 PE-CF594 (A2F10.1); Ly-6C Alexa Fluor 700 (AL-21); and I-A/I-E V500 (M5/114.15.2). From eBioscience: CD11c PE (N418); CD44 APC (IM7); CD105 Biotin (MJ7/18); CD117 PE/Cy7 (2B8); CD172a PerCP-eFluor 710 (P84); CD317 APC (eBio927); and F4/80 APC-eFluor 780 (BM8). From Invitrogen: TCR Vα2 PE (B20.1) and CD172a PerCP-eFluor 710 (P84). From Tonbo Biosciences: CD3e Biotin (145-2C11); CD11c APC-Cy7 (N418); CD45.1 PE-Cy7 (A20); and I-A/I-E V450 (M5/114.15.2). From BioLegend: CD4 APC (RM4-5); CD24 PE/Cy7 (M1/69); CD45.1 BV605 (A20); CD45.2 Alexa Fluor 700 (104); CD115 BV711 (ASF98); Ly-6G Biotin (1A8); GFP Alexa Fluor 488 (FM264G); Siglec H PE (551); Ter-119 Biotin (TER-119); TCR Vα2 PE; and PerCP-Cy5.5 (B20.1). From Cell Signaling: P-p44/42 MAPK (Erk1/2) (T202/Y204) PE (197G2). In general, all antibodies were used at a dilution of 1:200 unless specified otherwise by the manufacturer. Cells were analyzed on a FACSCanto II analyzer or FACSAria Fusion. Data were analyzed with FlowJo software (TreeStar).
BM isolation and cell culture
BM was harvested from the femur, tibia, and pelvis. Bones were fragmented by mortar and pestle in MACS buffer, and debris was removed by passing cells through a 70-µm strainer. Red blood cells were subsequently lysed with ammonium chloride-potassium bicarbonate lysis buffer. Cells were counted on a Vi-CELL analyzer (Beckman Coulter), and 3–5 × 106 cells were stained for analysis. For Flt3L culture experiments, whole BM (8 × 106 cells in 4 ml cIMDM) was cultured at 37°C with 100 ng/ml Flt3L (Peprotech) or equivalent volume PBS (vehicle) for 9 d. For comparative cytokine culture experiments, whole BM (6 × 106 cells in 4 ml of cIMDM) was cultured at 37°C with 100 ng/ml Flt3L (Peprotech), 20 ng/ml M-CSF (Peprotech), 50 ng/ml SCF (Peprotech), or equivalent volume PBS (vehicle) for 7 d. For all culture experiments, loosely adherent and suspension cells were harvested by gentle pipetting at the indicated time point and stained with fluorescent antibodies for analysis by flow cytometry.
In vivo Flt3L administration
Flt3+/+ or Flt3−/− mice were injected subcutaneously with sterile saline (vehicle) or 10 µg of purified Flt3L-Fc fusion protein (generated in-house) daily for 8 d and analyzed on the following day.
Expression microarray analysis
Total RNA was extracted from sorted splenic DCs with an RNAqueous-Micro kit (Ambion), amplified with an Ovation Pico WTA System (NuGEN), and hybridized to Mouse Gene 1.0 ST arrays (Affymetrix). Expression values were analyzed after robust multiarray average summarization and quantile normalization using ArrayStar 4 software (DNASTAR). All gene expression microarray data have been deposited in the NCBI Gene Expression Omnibus database with accession nos. GSE110789 and GSE110790.
Antigen presentation assays
In vitro cell-associated cross-presentation assays have been described previously (Kretzer et al., 2016). In brief, Listeria monocytogenes (LM)-OVA (a gift from H. Shen, University of Pennsylvania, Philadelphia, PA) was grown in brain–heart infusion broth at 37°C for 6 h and then frozen overnight after dilution plating for titer enumeration. Bacteria were subsequently thawed and washed three times with PBS before heat killing at 80°C for 1 h and then freezing at −80°C. OT-I T cells were meanwhile sorted from spleens of OT-I mice as B220–CD4–CD11c–CD45.1+CD8+Vα2+ and labeled with CFSE (eBioscience). Sort purity of >95% was confirmed by post-sort analysis before cells were used for assays. 10,000 sorted splenic cDCs of the indicated subset from Flt3+/+ or Flt3−/− mice and 25,000 sorted CFSE-labeled OT-I T cells were incubated with various doses of heat-killed LM-OVA for 3 d at 37°C. After incubation was complete, OT-I cells were assayed for CFSE dilution and CD44 expression.
For soluble OVA presentation assays, 10,000 sorted splenic cDCs of the indicated subset from Flt3+/+ or Flt3−/− mice and 25,000 sorted CFSE-labeled OT-I T cells were incubated with the indicated concentration of OVA (Worthington Biochemical Corporation) for 3 d at 37°C. After incubation was complete, OT-I T cells were assayed for CFSE dilution and CD44 expression.
Alternatively, sorted splenic cDCs of the indicated subset from Flt3+/+ or Flt3−/− mice were incubated with 0.1 mg/ml of soluble OVA (Worthington Biochemical Corporation) for 45 min at 37°C, washed three times with PBS, and then different numbers of cDCs were incubated with 25,000 sorted CFSE-labeled OT-I T cells for 3 d at 37°C. After incubation was complete, OT-I T cells were assayed for CFSE dilution and CD44 expression.
Type I interferon production from pDCs
25,000 sorted pDCs from Flt3+/+ or Flt3−/− mice were incubated overnight at 37°C in cIMDM with 100 ng/ml Flt3L plus either 6 µg/ml of CpG-A oligodeoxynucleotides or the equivalent volume of PBS (vehicle). The next day cells were spun down and the supernatant was collected. IFN-α in the supernatant was measured using the PBL Verikine Mouse IFN Alpha All Subtype ELISA kit, High Sensitivity (PBL Assay Science).
Progenitor sorting and culture
For sorting experiments, BM was isolated as described above and depleted of CD3-, CD19-, CD105-, Ter119-, and Ly6G-expressing cells by staining with the corresponding biotinylated antibodies followed by depletion with MagniSort Streptavidin Negative Selection beads (Thermo Fisher). The entirety of the remaining BM was then stained with fluorescent antibodies before sorting. Gates used to define CDPs, pre-cDC1s, and pre-cDC2s were based on previous studies (Onai et al., 2007; Grajales-Reyes et al., 2015). CDPs were identified as Lin–CD117intCD135+CD115+CD11c–MHCII–; pre-cDC1s as Lin–CD117intCD11c+MHCIIintZbtb46-GFP+; and pre-cDC2s as Lin–CD117loCD115+CD11c+MHCII–Zbtb46-GFP+. Lineage markers included CD3, CD19, CD105, Ter119, and Ly6G. A FACSAria Fusion was used for sorting, and cells were sorted into cIMDM. Sort purity of >95% was confirmed by post-sort analysis before cells were used for further experiments. Sorted cells were cultured at 37°C in 200 µl total volume of cIMDM with 100 ng/ml Flt3L (Peprotech), 20 ng/ml M-CSF (Peprotech), 50 ng/ml SCF (Peprotech), or equivalent volume of PBS (vehicle) for 5 d.
For cell proliferation studies, sorted pre-cDC2s were washed three times with PBS to remove serum and then incubated with 10 µM of eBioscience Cell Proliferation Dye eFluor 450 (Thermo Fisher) at 37°C for 10 min. Reaction was quenched by the direct addition of FCS, and cells were washed three times with cIMDM. Cells were subsequently cultured at 37°C in 200 µl total volume of cIMDM with 20 ng/ml M-CSF (Peprotech) for 5 d.
Tamoxifen administration
Mice were orally gavaged with 4 mg tamoxifen (Sigma-Aldrich) dissolved in corn oil (Sigma-Aldrich) for an initial loading dose and then placed on tamoxifen citrate chow (Envigo) for 4–5 wk. Mice were given up to 2 d of regular chow per week if significant weight loss was observed. After treatment, mice were rested on regular chow for one week before analysis. Deletion of CSF1R in the BM was confirmed in all mice.
BM chimeras
BM cells from donor mice were collected as described above. To generate mixed BM chimeras, BM cells were isolated from mice of the appropriate genotypes, counted with a Vi-CELL analyzer (Beckman Coulter), and mixed at a ratio of 1:1 before transplantation. Recipient mice received a single dose of 1050 rads of whole-body irradiation and then received a transplant of 10 × 106 total BM cells the following day. Mice were allowed to reconstitute for 8–10 wk before further experiments or analyses were conducted.
BM progenitor cytokine stimulation
BM was isolated and lineage-depleted as described above. Lin– BM cells were then aliquoted at 106 cells per condition and serum-starved in plain IMDM for 4 h at 37°C. After starvation, M-CSF was added at the indicated concentrations, and cells were incubated at 37°C for 5 min for stimulation. Cells were then fixed for 15 min at room temperature by adding paraformaldehyde (Electron Microscopy Sciences) directly to cells to a final concentration of 2%. Cells were washed twice with MACS buffer and then permeabilized by adding ice-cold methanol directly to cells with gentle vortexing, followed by incubation at 4°C for 30 min. Cells were again washed twice with MACS buffer and then stained with fluorescent antibodies against cell surface and intracellular antigens for 30 min at room temperature. Antibodies were previously tested individually to ensure their functionality in fixed and permeabilized cells. Flow cytometric analyses were then performed on stained cells. For analysis, we used integrated mean fluorescence intensity (iMFI), which combines frequency and MFI into a single measure of total functional response (Darrah et al., 2007; Shooshtari et al., 2010).
Statistical analysis
All statistical analyses were performed using Prism 7 (GraphPad Software). Statistical significance was evaluated using unpaired two-tailed Student's t test unless otherwise noted. Differences with P ≤ 0.05 were considered significant. n represents the number of biological replicates.
Online supplemental material
Fig. S1 shows the percentage of cDCs and pDCs in SLNs and lungs from the mice strains analyzed in Figs. 1 and 2. Fig. S2 shows the number of cDCs developing from the in vitro cytokine cultures from Fig. 5 and the expression of various costimulatory molecules on these cDCs. Fig. S3 shows the development of pDCs from the in vitro cytokine cultures from Fig. 5 and the development of pDCs from sorted CDPs treated with various cytokines from Fig. 6. Fig. S4 shows the continued development of pre-cDC1s and pre-cDC2s in Flt3l−/− mice and that the pre-cDC2s from Flt3l−/− mice are no more sensitive to M-CSF than the pre-cDC2s from Flt3l+/+ mice. Fig. S5 depicts that the deletion of Flt3 in Flt3l−/− mice reverses the severe DC defect in SLNs and lungs.
Supplementary Material
Acknowledgments
We thank Dr. E. Camilla Forsberg for Flt3–/– mice and the Alvin J. Siteman Cancer Center at Washington University School of Medicine for use of the Center for Biomedical Informatics and Multiplex Gene Analysis Genechip Core Facility. We thank Ansuman T. Satpathy and Xiaodi Wu for their helpful suggestions on this manuscript. We thank Hannah L. Miller for technical help with pDC assays.
This work was supported by the Howard Hughes Medical Institute (K.M. Murphy), the US National Institutes of Health (grant F30DK108498 to V. Durai, grant T32 AI007163-40 to D.J. Theisen, and grant T32 CA 009621 to J.T. Davidson IV), and the US National Science Foundation (grant DGE-1143954 to P. Bagadia).
The authors declare no competing financial interests.
Author contributions: V. Durai, T.L. Murphy, and K.M. Murphy designed the study with advice from D.H. Fremont; V. Durai and P. Bagadia performed experiments related to cell sorting, cell culture, and flow cytometry with advice from C.G. Briseño and J.T. Davidson IV; V. Durai, C.G. Briseño, and M. Gargaro performed in vivo manipulations of mice with advice from A. Iwata; V. Durai and D.J. Theisen performed cross-presentation assays; V. Durai, T.L. Murphy, and K.M. Murphy wrote the manuscript with contributions from all authors.
References
- Auffray C., Fogg D.K., Narni-Mancinelli E., Senechal B., Trouillet C., Saederup N., Leemput J., Bigot K., Campisi L., Abitbol M., et al. 2009. CX3CR1+ CD115+ CD135+ common macrophage/DC precursors and the role of CX3CR1 in their response to inflammation. J. Exp. Med. 206:595–606. 10.1084/jem.20081385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasel K., De Smedt T., Smith J.L., and Maliszewski C.R.. 2000. Generation of murine dendritic cells from flt3-ligand-supplemented bone marrow cultures. Blood. 96:3029–3039. [PubMed] [Google Scholar]
- Breton G., Lee J., Zhou Y.J., Schreiber J.J., Keler T., Puhr S., Anandasabapathy N., Schlesinger S., Caskey M., Liu K., and Nussenzweig M.C.. 2015. Circulating precursors of human CD1c+ and CD141+ dendritic cells. J. Exp. Med. 212:401–413. 10.1084/jem.20141441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amico A., and Wu L.. 2003. The early progenitors of mouse dendritic cells and plasmacytoid predendritic cells are within the bone marrow hemopoietic precursors expressing Flt3. J. Exp. Med. 198:293–303. 10.1084/jem.20030107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrah P.A., Patel D.T., De Luca P.M., Lindsay R.W., Davey D.F., Flynn B.J., Hoff S.T., Andersen P., Reed S.G., Morris S.L., et al. 2007. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat. Med. 13:843–850. 10.1038/nm1592 [DOI] [PubMed] [Google Scholar]
- Dosil M., Wang S., and Lemischka I.R.. 1993. Mitogenic signalling and substrate specificity of the Flk2/Flt3 receptor tyrosine kinase in fibroblasts and interleukin 3-dependent hematopoietic cells. Mol. Cell. Biol. 13:6572–6585. 10.1128/MCB.13.10.6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enver T., Heyworth C.M., and Dexter T.M.. 1998. Do stem cells play dice? Blood. 92:348–351, discussion :352. [PubMed] [Google Scholar]
- Fancke B., Suter M., Hochrein H., and O’Keeffe M.. 2008. M-CSF: a novel plasmacytoid and conventional dendritic cell poietin. Blood. 111:150–159. 10.1182/blood-2007-05-089292 [DOI] [PubMed] [Google Scholar]
- Fogg D.K., Sibon C., Miled C., Jung S., Aucouturier P., Littman D.R., Cumano A., and Geissmann F.. 2006. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science. 311:83–87. 10.1126/science.1117729 [DOI] [PubMed] [Google Scholar]
- Ginhoux F., Liu K., Helft J., Bogunovic M., Greter M., Hashimoto D., Price J., Yin N., Bromberg J., Lira S.A., et al. 2009. The origin and development of nonlymphoid tissue CD103+ DCs. J. Exp. Med. 206:3115–3130. 10.1084/jem.20091756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grajales-Reyes G.E., Iwata A., Albring J., Wu X., Tussiwand R., Kc W., Kretzer N.M., Briseño C.G., Durai V., Bagadia P., et al. 2015. Batf3 maintains autoactivation of Irf8 for commitment of a CD8α(+) conventional DC clonogenic progenitor. Nat. Immunol. 16:708–717. 10.1038/ni.3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helft J., Ginhoux F., Bogunovic M., and Merad M.. 2010. Origin and functional heterogeneity of non-lymphoid tissue dendritic cells in mice. Immunol. Rev. 234:55–75. 10.1111/j.0105-2896.2009.00885.x [DOI] [PubMed] [Google Scholar]
- Karsunky H., Merad M., Cozzio A., Weissman I.L., and Manz M.G.. 2003. Flt3 ligand regulates dendritic cell development from Flt3+ lymphoid and myeloid-committed progenitors to Flt3+ dendritic cells in vivo. J. Exp. Med. 198:305–313. 10.1084/jem.20030323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston D., Schmid M.A., Onai N., Obata-Onai A., Baumjohann D., and Manz M.G.. 2009. The concerted action of GM-CSF and Flt3-ligand on in vivo dendritic cell homeostasis. Blood. 114:835–843. 10.1182/blood-2009-02-206318 [DOI] [PubMed] [Google Scholar]
- Kretzer N.M., Theisen D.J., Tussiwand R., Briseño C.G., Grajales-Reyes G.E., Wu X., Durai V., Albring J., Bagadia P., Murphy T.L., and Murphy K.M.. 2016. RAB43 facilitates cross-presentation of cell-associated antigens by CD8α+ dendritic cells. J. Exp. Med. 213:2871–2883. 10.1084/jem.20160597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Breton G., Oliveira T.Y., Zhou Y.J., Aljoufi A., Puhr S., Cameron M.J., Sékaly R.P., Nussenzweig M.C., and Liu K.. 2015. Restricted dendritic cell and monocyte progenitors in human cord blood and bone marrow. J. Exp. Med. 212:385–399. 10.1084/jem.20141442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Chen K., Zhu L., and Pollard J.W.. 2006. Conditional deletion of the colony stimulating factor-1 receptor (c-fms proto-oncogene) in mice. Genesis. 44:328–335. 10.1002/dvg.20219 [DOI] [PubMed] [Google Scholar]
- Lieschke G.J., Grail D., Hodgson G., Metcalf D., Stanley E., Cheers C., Fowler K.J., Basu S., Zhan Y.F., and Dunn A.R.. 1994. Mice lacking granulocyte colony-stimulating factor have chronic neutropenia, granulocyte and macrophage progenitor cell deficiency, and impaired neutrophil mobilization. Blood. 84:1737–1746. [PubMed] [Google Scholar]
- Liu K., Waskow C., Liu X., Yao K., Hoh J., and Nussenzweig M.. 2007. Origin of dendritic cells in peripheral lymphoid organs of mice. Nat. Immunol. 8:578–583. 10.1038/ni1462 [DOI] [PubMed] [Google Scholar]
- Liu K., Victora G.D., Schwickert T.A., Guermonprez P., Meredith M.M., Yao K., Chu F.F., Randolph G.J., Rudensky A.Y., and Nussenzweig M.. 2009. In vivo analysis of dendritic cell development and homeostasis. Science. 324:392–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman S.D., James L., Vanden Bos T., de Vries P., Brasel K., Gliniak B., Hollingsworth L.T., Picha K.S., McKenna H.J., Splett R.R., et al. 1993. Molecular cloning of a ligand for the flt3/flk-2 tyrosine kinase receptor: a proliferative factor for primitive hematopoietic cells. Cell. 75:1157–1167. 10.1016/0092-8674(93)90325-K [DOI] [PubMed] [Google Scholar]
- Mackarehtschian K., Hardin J.D., Moore K.A., Boast S., Goff S.P., and Lemischka I.R.. 1995. Targeted disruption of the flk2/flt3 gene leads to deficiencies in primitive hematopoietic progenitors. Immunity. 3:147–161. 10.1016/1074-7613(95)90167-1 [DOI] [PubMed] [Google Scholar]
- Maraskovsky E., Brasel K., Teepe M., Roux E.R., Lyman S.D., Shortman K., and McKenna H.J.. 1996. Dramatic increase in the numbers of functionally mature dendritic cells in Flt3 ligand-treated mice: multiple dendritic cell subpopulations identified. J. Exp. Med. 184:1953–1962. 10.1084/jem.184.5.1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraskovsky E., Daro E., Roux E., Teepe M., Maliszewski C.R., Hoek J., Caron D., Lebsack M.E., and McKenna H.J.. 2000. In vivo generation of human dendritic cell subsets by Flt3 ligand. Blood. 96:878–884. [PubMed] [Google Scholar]
- Matthews W., Jordan C.T., Gavin M., Jenkins N.A., Copeland N.G., and Lemischka I.R.. 1991. A receptor tyrosine kinase cDNA isolated from a population of enriched primitive hematopoietic cells and exhibiting close genetic linkage to c-kit. Proc. Natl. Acad. Sci. USA. 88:9026–9030. 10.1073/pnas.88.20.9026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna H.J., Stocking K.L., Miller R.E., Brasel K., De Smedt T., Maraskovsky E., Maliszewski C.R., Lynch D.H., Smith J., Pulendran B., et al. 2000. Mice lacking flt3 ligand have deficient hematopoiesis affecting hematopoietic progenitor cells, dendritic cells, and natural killer cells. Blood. 95:3489–3497. [PubMed] [Google Scholar]
- Merad M., Sathe P., Helft J., Miller J., and Mortha A.. 2013. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu. Rev. Immunol. 31:563–604. 10.1146/annurev-immunol-020711-074950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith M.M., Liu K., Darrasse-Jeze G., Kamphorst A.O., Schreiber H.A., Guermonprez P., Idoyaga J., Cheong C., Yao K.H., Niec R.E., and Nussenzweig M.C.. 2012. Expression of the zinc finger transcription factor zDC (Zbtb46, Btbd4) defines the classical dendritic cell lineage. J. Exp. Med. 209:1153–1165. 10.1084/jem.20112675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf D. 1998. Lineage commitment and maturation in hematopoietic cells: the case for extrinsic regulation. Blood. 92:345–347, discussion :352. [PubMed] [Google Scholar]
- Mildner A., and Jung S.. 2014. Development and function of dendritic cell subsets. Immunity. 40:642–656. 10.1016/j.immuni.2014.04.016 [DOI] [PubMed] [Google Scholar]
- Miller J.C., Brown B.D., Shay T., Gautier E.L., Jojic V., Cohain A., Pandey G., Leboeuf M., Elpek K.G., Helft J., et al. Immunological Genome Consortium . 2012. Deciphering the transcriptional network of the dendritic cell lineage. Nat. Immunol. 13:888–899. 10.1038/ni.2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore A.J., and Anderson M.K.. 2013. Dendritic cell development: a choose-your-own-adventure story. Adv. Hematol. 2013:949513 10.1155/2013/949513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossadegh-Keller N., Sarrazin S., Kandalla P.K., Espinosa L., Stanley E.R., Nutt S.L., Moore J., and Sieweke M.H.. 2013. M-CSF instructs myeloid lineage fate in single haematopoietic stem cells. Nature. 497:239–243. 10.1038/nature12026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa T. 2006. Microenvironmental niches in the bone marrow required for B-cell development. Nat. Rev. Immunol. 6:107–116. 10.1038/nri1780 [DOI] [PubMed] [Google Scholar]
- Naik S.H., Proietto A.I., Wilson N.S., Dakic A., Schnorrer P., Fuchsberger M., Lahoud M.H., O’Keeffe M., Shao Q.X., Chen W.F., et al. 2005. Cutting edge: generation of splenic CD8+ and CD8- dendritic cell equivalents in Fms-like tyrosine kinase 3 ligand bone marrow cultures. J. Immunol. 174:6592–6597. 10.4049/jimmunol.174.11.6592 [DOI] [PubMed] [Google Scholar]
- Naik S.H., Sathe P., Park H.Y., Metcalf D., Proietto A.I., Dakic A., Carotta S., O’Keeffe M., Bahlo M., Papenfuss A., et al. 2007. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat. Immunol. 8:1217–1226. 10.1038/ni1522 [DOI] [PubMed] [Google Scholar]
- Onai N., Obata-Onai A., Tussiwand R., Lanzavecchia A., and Manz M.G.. 2006. Activation of the Flt3 signal transduction cascade rescues and enhances type I interferon-producing and dendritic cell development. J. Exp. Med. 203:227–238. 10.1084/jem.20051645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onai N., Obata-Onai A., Schmid M.A., Ohteki T., Jarrossay D., and Manz M.G.. 2007. Identification of clonogenic common Flt3+M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nat. Immunol. 8:1207–1216. 10.1038/ni1518 [DOI] [PubMed] [Google Scholar]
- Onai N., Kurabayashi K., Hosoi-Amaike M., Toyama-Sorimachi N., Matsushima K., Inaba K., and Ohteki T.. 2013. A clonogenic progenitor with prominent plasmacytoid dendritic cell developmental potential. Immunity. 38:943–957. 10.1016/j.immuni.2013.04.006 [DOI] [PubMed] [Google Scholar]
- Peschon J.J., Morrissey P.J., Grabstein K.H., Ramsdell F.J., Maraskovsky E., Gliniak B.C., Park L.S., Ziegler S.F., Williams D.E., Ware C.B., et al. 1994. Early lymphocyte expansion is severely impaired in interleukin 7 receptor–deficient mice. J. Exp. Med. 180:1955–1960. 10.1084/jem.180.5.1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieger M.A., Hoppe P.S., Smejkal B.M., Eitelhuber A.C., and Schroeder T.. 2009. Hematopoietic cytokines can instruct lineage choice. Science. 325:217–218. 10.1126/science.1171461 [DOI] [PubMed] [Google Scholar]
- Rottapel R., Turck C.W., Casteran N., Liu X., Birnbaum D., Pawson T., and Dubreuil P.. 1994. Substrate specificities and identification of a putative binding site for PI3K in the carboxy tail of the murine Flt3 receptor tyrosine kinase. Oncogene. 9:1755–1765. [PubMed] [Google Scholar]
- Sathe P., Vremec D., Wu L., Corcoran L., and Shortman K.. 2013. Convergent differentiation: myeloid and lymphoid pathways to murine plasmacytoid dendritic cells. Blood. 121:11–19. 10.1182/blood-2012-02-413336 [DOI] [PubMed] [Google Scholar]
- Satpathy A.T., Murphy K.M., and Kc W.. 2011. Transcription factor networks in dendritic cell development. Semin. Immunol. 23:388–397. 10.1016/j.smim.2011.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpathy A.T., Kc W., Albring J.C., Edelson B.T., Kretzer N.M., Bhattacharya D., Murphy T.L., and Murphy K.M.. 2012. Zbtb46 expression distinguishes classical dendritic cells and their committed progenitors from other immune lineages. J. Exp. Med. 209:1135–1152. 10.1084/jem.20120030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenner S.M., Madan V., Busch K., Tietz A., Läufle C., Costa C., Blum C., Fehling H.J., and Rodewald H.R.. 2010. Fate mapping reveals separate origins of T cells and myeloid lineages in the thymus. Immunity. 32:426–436. 10.1016/j.immuni.2010.03.005 [DOI] [PubMed] [Google Scholar]
- Schlitzer A., Sivakamasundari V., Chen J., Sumatoh H.R., Schreuder J., Lum J., Malleret B., Zhang S., Larbi A., Zolezzi F., et al. 2015. Identification of cDC1- and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow. Nat. Immunol. 16:718–728. 10.1038/ni.3200 [DOI] [PubMed] [Google Scholar]
- See P., Dutertre C.A., Chen J., Günther P., McGovern N., Irac S.E., Gunawan M., Beyer M., Händler K., Duan K., et al. 2017. Mapping the human DC lineage through the integration of high-dimensional techniques. Science. 356:eaag3009 10.1126/science.aag3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shooshtari P., Fortuno E.S. III, Blimkie D., Yu M., Gupta A., Kollmann T.R., and Brinkman R.R.. 2010. Correlation analysis of intracellular and secreted cytokines via the generalized integrated mean fluorescence intensity. Cytometry A. 77A:873–880. 10.1002/cyto.a.20943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitnicka E., Bryder D., Theilgaard-Mönch K., Buza-Vidas N., Adolfsson J., and Jacobsen S.E.. 2002. Key role of flt3 ligand in regulation of the common lymphoid progenitor but not in maintenance of the hematopoietic stem cell pool. Immunity. 17:463–472. 10.1016/S1074-7613(02)00419-3 [DOI] [PubMed] [Google Scholar]
- Sitnicka E., Brakebusch C., Martensson I.L., Svensson M., Agace W.W., Sigvardsson M., Buza-Vidas N., Bryder D., Cilio C.M., Ahlenius H., et al. 2003. Complementary signaling through flt3 and interleukin-7 receptor α is indispensable for fetal and adult B cell genesis. J. Exp. Med. 198:1495–1506. 10.1084/jem.20031152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsapogas P., Swee L.K., Nusser A., Nuber N., Kreuzaler M., Capoferri G., Rolink H., Ceredig R., and Rolink A.. 2014. In vivo evidence for an instructive role of fms-like tyrosine kinase-3 (FLT3) ligand in hematopoietic development. Haematologica. 99:638–646. 10.3324/haematol.2013.089482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsapogas P., Mooney C.J., Brown G., and Rolink A.. 2017. The Cytokine Flt3-Ligand in Normal and Malignant Hematopoiesis. Int. J. Mol. Sci. 18:1115 10.3390/ijms18061115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A., Kirsch D.G., McLaughlin M.E., Tuveson D.A., Grimm J., Lintault L., Newman J., Reczek E.E., Weissleder R., and Jacks T.. 2007. Restoration of p53 function leads to tumour regression in vivo. Nature. 445:661–665. 10.1038/nature05541 [DOI] [PubMed] [Google Scholar]
- Verstraete K., and Savvides S.N.. 2012. Extracellular assembly and activation principles of oncogenic class III receptor tyrosine kinases. Nat. Rev. Cancer. 12:753–766. 10.1038/nrc3371 [DOI] [PubMed] [Google Scholar]
- von Freeden-Jeffry U., Vieira P., Lucian L.A., McNeil T., Burdach S.E., and Murray R.. 1995. Lymphopenia in interleukin (IL)-7 gene-deleted mice identifies IL-7 as a nonredundant cytokine. J. Exp. Med. 181:1519–1526. 10.1084/jem.181.4.1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskow C., Liu K., Darrasse-Jèze G., Guermonprez P., Ginhoux F., Merad M., Shengelia T., Yao K., and Nussenzweig M.. 2008. The receptor tyrosine kinase Flt3 is required for dendritic cell development in peripheral lymphoid tissues. Nat. Immunol. 9:676–683. 10.1038/ni.1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Liu X., Jaenisch R., and Lodish H.F.. 1995. Generation of committed erythroid BFU-E and CFU-E progenitors does not require erythropoietin or the erythropoietin receptor. Cell. 83:59–67. 10.1016/0092-8674(95)90234-1 [DOI] [PubMed] [Google Scholar]
- Wu X., Briseño C.G., Durai V., Albring J.C., Haldar M., Bagadia P., Kim K.W., Randolph G.J., Murphy T.L., and Murphy K.M.. 2016. Mafb lineage tracing to distinguish macrophages from other immune lineages reveals dual identity of Langerhans cells. J. Exp. Med. 213:2553–2565. 10.1084/jem.20160600 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.