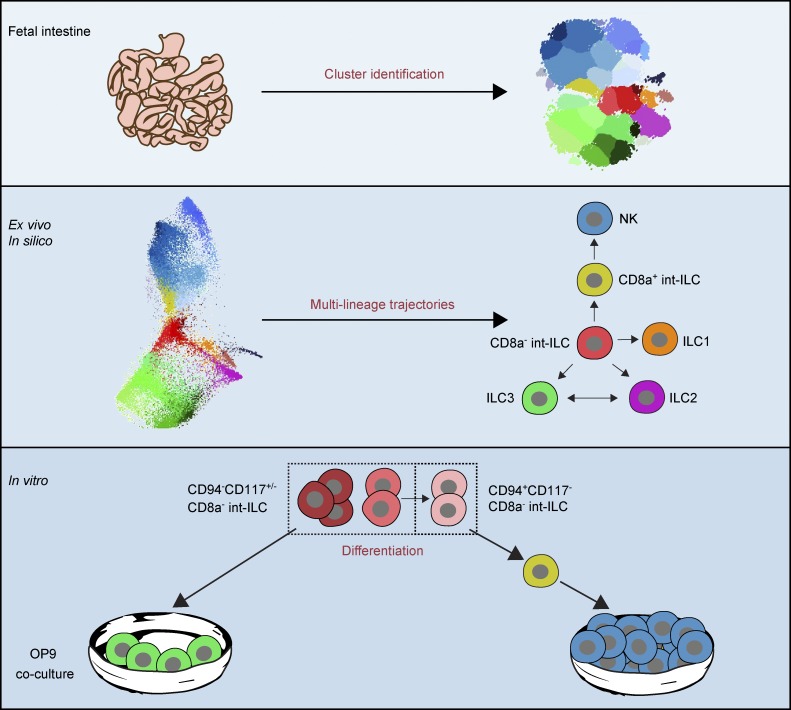
Li et al. apply mass cytometry to delineate the fetal gut innate lymphoid cell (ILC) population and utilize a t-SNE–based approach to predict potential differentiation trajectories. They identify an int-ILC subset that differentiates into NK cells or ILC3s in vitro.
Abstract
Innate lymphoid cells (ILCs) are abundant in mucosal tissues and involved in tissue homeostasis and barrier function. Although several ILC subsets have been identified, it is unknown if additional heterogeneity exists, and their differentiation pathways remain largely unclear. We applied mass cytometry to analyze ILCs in the human fetal intestine and distinguished 34 distinct clusters through a t-SNE–based analysis. A lineage (Lin)−CD7+CD127−CD45RO+CD56+ population clustered between the CD127+ ILC and natural killer (NK) cell subsets, and expressed diverse levels of Eomes, T-bet, GATA3, and RORγt. By visualizing the dynamics of the t-SNE computation, we identified smooth phenotypic transitions from cells within the Lin−CD7+CD127−CD45RO+CD56+ cluster to both the NK cells and CD127+ ILCs, revealing potential differentiation trajectories. In functional differentiation assays, the Lin−CD7+CD127−CD45RO+CD56+CD8a− cells could develop into CD45RA+ NK cells and CD127+RORγt+ ILC3-like cells. Thus, we identified a previously unknown intermediate innate subset that can differentiate into ILC3 and NK cells.
Graphical Abstract
Introduction
Innate lymphoid cells (ILCs) lack expression of T-cell receptors but otherwise are a functional counterpart of cytotoxic and T helper (Th) cell subsets. Helper ILCs are classified into three groups: ILC1, ILC2, and ILC3 (Spits et al., 2013). ILC1s are mainly characterized as lineage (Lin)−CD161+CD127+CRTH2−CD117−, express the transcription factor T-bet, and produce Th1 cell–associated cytokines. ILC2s are Lin−CD161+CD127+CRTH2+, express GATA3, and produce Th2 cell–associated cytokines. ILC3s, including fetal lymphoid tissue–inducer (LTi) cells, are Lin−CD161+CD127+CRTH2−CD117+ and RORγt+, and secrete Th17/Th22 cell–associated cytokines (Spits et al., 2013; Hazenberg and Spits, 2014). A fraction of human ILC3s expresses natural cytotoxicity receptors such as NKp44, NKp46, and NKp30, and neural cell adhesion molecule CD56, similar to natural killer (NK) cells (Cella et al., 2009; Cupedo et al., 2009). NK cells are a cytotoxic subset of ILCs that express the transcription factor T-bet and/or Eomes and produce IFN-γ, granzymes, and perforin (Spits et al., 2013). Also, ILCs are most abundant and reside in mucosal tissues such as the tonsil, lung, and intestine, where they can expand locally (Gasteiger et al., 2015).
Several studies have reported the differentiation pathways of ILCs in a variety of tissues in both mice and humans (Ishizuka et al., 2016b; Juelke and Romagnani, 2016). For example, in mouse fetal liver and adult intestine, a CXCR6+RORγt+α4β7+ subset has been identified that can differentiate into ILC3s and NK cells (Possot et al., 2011). As this subset was not found in adult bone marrow, it might migrate to the intestine during fetal development. In humans, RORγt+CD34+ progenitor cells were identified in the tonsil and intestine, but these were absent in peripheral blood, umbilical cord blood, bone marrow, and thymus (Montaldo et al., 2014; Scoville et al., 2016). Because these progenitors could differentiate into helper ILCs and NK cells, mucosal organs might be the preferential sites for ILC differentiation. In addition, a CD127+CD117+ ILC precursor (ILCP) has been identified in cord blood, peripheral blood, and tissues, including fetal liver, adult lung, and adult tonsil, that can generate all ILC subsets in situ and could represent an intermediate between precursor cells and mature ILCs (Lim et al., 2017). Also, previous studies have observed ILC plasticity mainly in mucosal tissues, such as the small intestine (Bernink et al., 2013, 2015; Bal et al., 2016; Lim et al., 2016), suggesting that environmental cues may play an important role in cell fate decision. So far, most of the studies on human ILC differentiation used CD34+ progenitors and mature types of ILCs (Juelke and Romagnani, 2016), whereas the intermediates or transitional stages connecting the CD34+ populations to mature types of ILCs have not been fully identified.
High-dimensional mass cytometry provides an opportunity to analyze the heterogeneity and potential differentiation pathways of human ILCs in an unbiased and data-driven fashion based on the simultaneous measurement of over 30 cellular markers at single-cell resolution (Bandura et al., 2009). Although the sensitivity of metal reporters in mass cytometry is not as sensitive as some of the brightest fluorochromes in flow cytometry, the advantage of including many more markers in a single antibody panel offers unique opportunities to evaluate the composition of the immune system with unprecedented resolution. Until recently, analysis of flow cytometry data were mainly performed with gating strategies based on primarily bimodal expression patterns. The incorporation of over 30 markers in mass cytometry antibody panels is not well compatible with such an analysis approach. Instead, t–distributed stochastic neighbor embedding (t-SNE)—based approaches are currently becoming the standard in the field as they allow the simultaneous analysis of all marker expression profiles in an unbiased fashion. Hierarchical SNE, for example, allows efficient analysis of mass cytometry datasets on tens of millions of cells at the single-cell level (van Unen et al., 2017). Here, we applied mass cytometry to analyze the ILC compartment in the human fetal intestine and provide evidence for previously unrecognized heterogeneity within this compartment. Moreover, we used a t-SNE–based computational approach to predict potential differentiation trajectories in silico, and provide evidence for the existence of a previously unrecognized innate cell subset that can differentiate into both NK cells and ILC3 in vitro.
Results
High-dimensional analysis reveals previously unrecognized heterogeneity in the ILC compartment
We developed a 35-metal isotope-tagged monoclonal antibody panel (Table S1) to identify the six major immune lineages (B cells, myeloid cells, CD4+, CD8+, γδ T cells, and Lin−CD7+ cells; the latter hereafter referred to as ILCs) and heterogeneity within those lineages. For this purpose, the panel included lineage markers and markers linked to cell differentiation, activation, trafficking, and responsiveness to humoral factors. With this panel, single-cell suspensions prepared from seven fetal intestines were analyzed individually. Single, live CD45+ cells were discriminated by event length, DNA stainings, and CD45 antibody staining (Fig. S1 A). All antibodies showed clear discrimination between antibody-positive and -negative cells (Fig. S1 B). Similar to our previous study (van Unen et al., 2016), we applied a combined t-SNE (van der Maaten and Hinton, 2008)–ACCENSE (Shekhar et al., 2014) data analysis approach to the six major cell lineages (Fig. S1 C), which revealed a large degree of heterogeneity within these lineages.
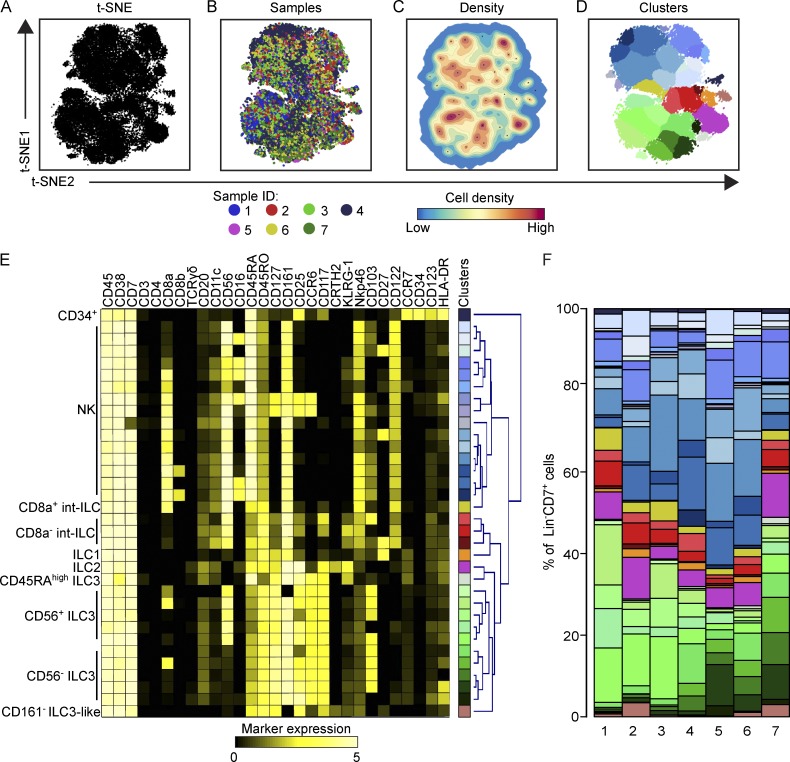
We next focused on the ILC compartment (Fig. S2 A), which comprised 20.4% ± 7.8% of the CD45+ cells. We pooled the data from the seven samples and performed a t-SNE analysis in Cytosplore (Höllt et al., 2016). This provided a two-dimensional map where cells are positioned based on the similarity in expression of all markers simultaneously (Fig. 1, A and B). Based on the density features of the t-SNE–embedded cells, we identified 34 phenotypically distinct clusters (Fig. 1, C and D) using the Gaussian mean shift clustering and generated a heat map showing the distinct marker expression profiles for each cluster (Fig. 1 E). Unbiased hierarchical clustering revealed distinct clusters including a group of CD34+ cells expressing CD45RA and CD117, a larger cluster of several types of NK cells, and a CD127+ ILC cluster with cells expressing markers corresponding to ILC1, ILC2, CD45RAhigh ILC3, subsets of several types of CD56+ and CD56− ILC3 (Hazenberg and Spits, 2014; Björklund et al., 2016), and a CD161− ILC3-like population (Simoni et al., 2017). In addition, several unrecognized cell clusters with a Lin−CD7+CD127−CD45RO+CD56+ phenotype (referred to as intermediate ILC [int-ILC] hereafter) were identified that clustered between the NK cells and CD127+ ILCs (Fig. 1, C–E). Although the majority of these int-ILCs (6.6% of ILCs ± 2.3%) were CD8a−, a smaller related population (3.0% ± 1.6%) was CD8a+ (Fig. 1 F). Importantly, analysis of the composition of the cluster frequencies in the individual fetal samples demonstrated that even though quantitative differences exist, most of the identified clusters, including the int-ILCs, were present in all seven samples (Fig. 1 F).
Figure 1.
High-dimensional mass cytometry analysis reveals previously unrecognized heterogeneity in the fetal intestinal ILC compartment. (A) A t-SNE embedding showing the collective ILCs (4.5 × 104 cells) derived from seven fetal intestines. Each black dot represents a single cell. (B) A t-SNE embedding as shown in A. Colors represent the different samples. (C) A density map describing the local probability density of t-SNE–embedded cells where black dots represent centroids of identified clusters using Gaussian mean shift clustering. (D) A t-SNE plot showing cluster partitions in different colors. (E) Heat map showing the median ArcSinh5-transformed marker expression values (black-to-yellow scale) of the clusters identified in A with description of 10 annotated categories based on preexisting literature, and hierarchical clustering thereof. (F) Composition of the ILC compartment in the individual fetal intestines (n = 7) represented in vertical bars where the size of the colored segments represents the proportion of cells as a percentage of total ILC in the sample. Colors as in A.
Together, these data indicate that all known NK and CD127+ ILC cell clusters could be identified simultaneously, and evidence for the existence of previously unrecognized clusters was obtained, as well.
Visualization of the t-SNE computation dynamics predicts potential differentiation trajectories in the ILC compartment
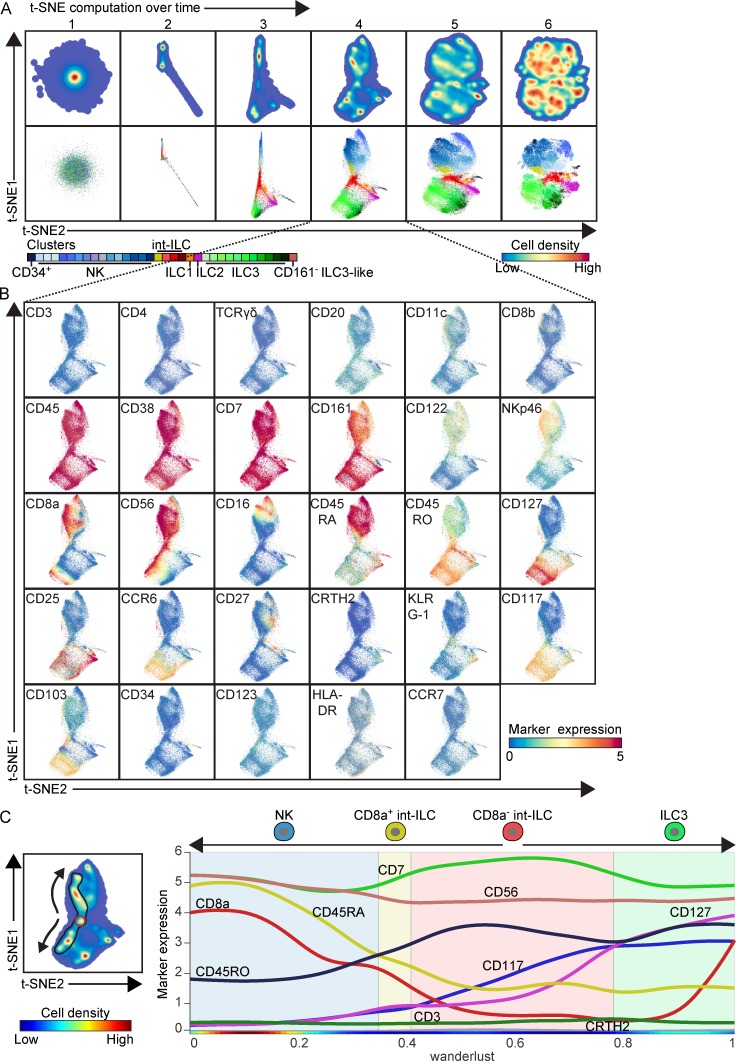
The cell surface phenotype of int-ILCs (i.e., CD127−CD45RO+) places them in between the CD127+ ILCs and the NK cells (Fig. 1, C–E), suggesting potential relationships with both. To investigate this in more detail, we sought to visualize potential relationships between cell populations without prior designation of a user-defined starting cell type in silico. To this end, we exploited the ability of Cytosplore to visualize the evolution of the t-SNE map (Höllt et al., 2016). Separating the computational modeling into six stages revealed how distinct cell clusters were formed, and their high-dimensional similarities were projected onto a two-dimensional map and linked to each other based on marker expression profiles (Fig. 2 A). Because the initial positions in the t-SNE map are assigned randomly, at the first stage of the t-SNE computation, all cells were unordered around a single density peak. Shortly thereafter, the CD34+ lymphoid precursor cells separated from the other cells (stage 2), and the first formation of the NK and CD127+ ILC clusters became apparent (stages 2 and 3). These early events were based on relatively large and highly discriminatory differences in the expression profiles between cell clusters, like the unique combination of CD34 and HLA-DR expression by CD34+ cells. At stage 4 of the t-SNE computation, the int-ILC cluster was positioned in the center with several distinct strands of cells forming trajectories toward the NK, CD27+ ILC1, KLRG-1+ ILC2, and CD103+ ILC3 clusters. In addition, a trajectory between the ILC2 and ILC3 clusters was visible (stage 4). Furthermore, cells from the CD8a− int-ILC population connected via the CD8a+ int-ILC population with NK cells, further supporting the notion that these two CD8a− and CD8a+ int-ILC populations are highly related (Fig. 2 A and Fig. S2 B). At the final stage of the t-SNE computation, the 34 clusters were defined, whereas the connections between the individual clusters were less clear as the t-SNE algorithm eventually assigns cells between two clusters to either one of the two. Individual marker expression patterns at stage 4 of the t-SNE computation gave insight into the separations of and the connections between clusters (Fig. 2 B). Here, the NK cluster was characterized by the coexpression of CD45RA, CD56, CD122, and NKp46, whereas the CD127+ ILC cluster expressed CD45RO, CD117, CD127, CD25 and, to a lesser extent, CCR6 and CD103. Similarly, connections between int-ILC to ILC1, ILC2, ILC3, and NK cells were marked by gradients of expression of CD27, KLRG-1, CD103, and CD56, respectively. The absence of CD45RA, CD127, and CCR6 in combination with the presence of CD45RO and CD56, and divergent expression of CD117, CD122, and CD25, positioned the int-ILC in between the NK cell and ILC clusters. Interestingly, CD56 expression linked the ILC3 to the CD8a− int-ILC, and the CD8a+ int-ILC to the NK cells.
Figure 2.
Monitoring t-SNE computation dynamics predicts potential differentiation trajectories of ILCs. (A) t-SNE embeddings of the collective ILC single-cell data derived from seven fetal intestines showing density features (top) and single cells (bottom) at six stages over the course of the t-SNE computation. Colors represent the local probability density of t-SNE–embedded cells (top) or cluster partitions (bottom), as described in Fig. 1 A. (B) t-SNE embeddings at stage 4 of the optimization phase as described in A. Colors of the cells represent ArcSinh5-transformed expression values of indicated markers. (C) Left: t-SNE embedding at stage 4 as in A. Colors represent density features, and black encirclement indicates the trajectory of cells along the CD56 expression continuum shown in B. Right: Wanderlust graph (trajectory 0–1.0) of the CD56-positive cells in the left panel showing median ArcSinh5-transformed expression of CD8a, CD45RO, CD45RA, CD7, CD3, CD56, CD117, CD127, and CRTH2 from the CD8a− int-ILCs (shaded red box) via the CD8a+ int-ILC (shaded yellow box) to NK cells (shaded blue box), and from the CD8a− int-ILCs to ILC3s (shaded green box). The rainbow color bar indicates relative cell density.
We next applied Wanderlust (Bendall et al., 2014) to determine changes in marker expression along this CD56 continuum (Fig. 2 C), which demonstrated that the expression of CD127, CD117, and CD45RO gradually decreased whereas that of CD45RA and CD8a increased moving from CD8a− int-ILC to NK cells, via CD8a+ int-ILC; and the expression of CD127 and CD117 gradually increased from CD8a− int-ILC to ILC3. Altogether, these results suggest that these t-SNE–based trajectories may reflect potential differentiation pathways.
Expression of cytokines, transcription factors, and CD94 distinguish int-ILCs from mature CD127+ ILCs and NK cells
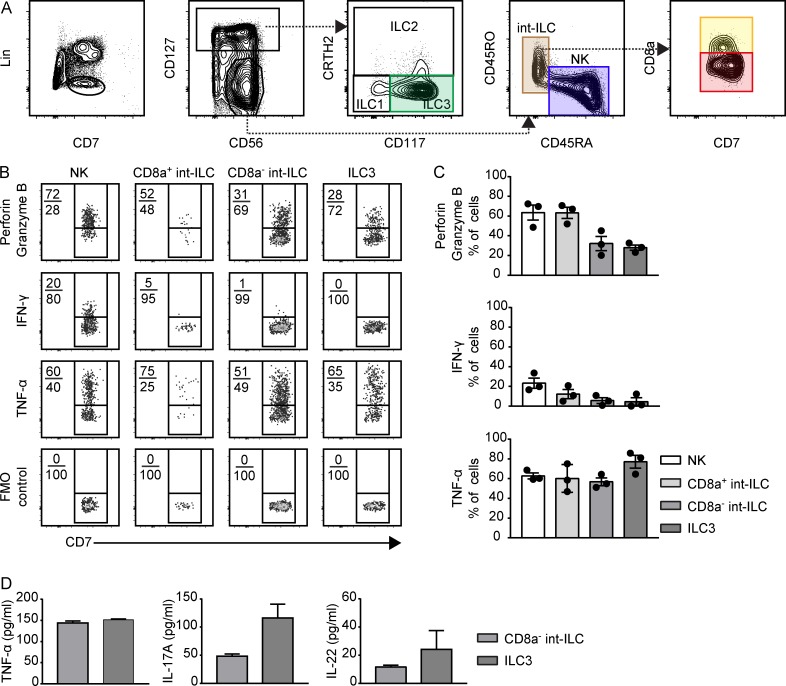
To further characterize the int-ILC population, we used the mass cytometry data (Fig. S3 A) to design a minimal antibody panel to distinguish the CD127−CD45RO+ int-ILCs from CD45RA+ NK cells, and to identify the mature CD127+ ILC types through differential expression of CD117 and CRTH2 (Fig. 3 A). Subsequently, we analyzed the proliferative state and examined the capacity of the subsets to produce cytokines and express markers linked to cytolytic potential by flow cytometry. For the former we stained the cells with the proliferation marker Ki-67 ex vivo. The highest percentage of Ki-67–positive cells was present in the CD8a+ int-ILC population (43.9%), whereas a mean of 20% of cells in the other subsets were Ki-67–positive (Fig. S3 B). Upon stimulation with PMA and ionomycin, perforin/granzyme B was detectable in all subsets, but more profoundly in the NK cells and CD8a+ int-ILCs, compared with the CD8a− int-ILCs and ILC3s (Fig. 3, B and C). Moreover, all subsets expressed high levels of TNF-α, whereas IFN-γ was detected mainly in the NK cells and CD8a+ int-ILCs but hardly in the CD8a− int-ILCs and ILC3s (Fig. 3, B and C). ILC2s expressed IL-4, IL-5, and IL-13, but ILC1s, very little IFN-γ (not depicted). In contrast, IL-4, IL-5, and IL-13 were undetectable in any of the other subsets (not depicted), whereas IL-17A and IL-22 expression was higher by ILC3s than CD8a− int-ILCs (Fig. 3 D).
Figure 3.
Cytokine production profiles of fetal intestinal ILCs ex vivo. (A) Representative biaxial plots depicting the gating strategy for ILC1, ILC2, ILC3, NK, and int-ILC subsets derived from a human fetal intestine analyzed by flow cytometry. The antibody cocktail contains Lin markers (CD3, CD19, CD11c, and CD14), and CD7, CD127, CD56, CRTH2, CD117, CD45RA, CD45RO, and CD8a to allow distinction of the ILC subsets. (B and C) Expression of cytotoxic molecules (perforin + granzyme B) and cytokines (IFN-γ and TNF-α) by the indicated subsets defined in A after stimulation with PMA and ionomycin for 6 h. The biaxial plots (B) depict one representative experiment, and the bar graphs (C) depict quantification of data obtained from three different human fetal intestines (three independent experiments). Error bar shows mean ± SEM. (D) Bar plots depict the secretion of TNF-α, IL-17A, and IL-22 by CD8a− int-ILC and ILC3, after stimulation with IL-2, IL-1β, and IL-23 for 4 d, using Luminex bead–based assay of an experiment with three intestines and duplicate wells (two independent experiments). Error bar shows mean ± SD. FMO, fluorescence-minus-one control.
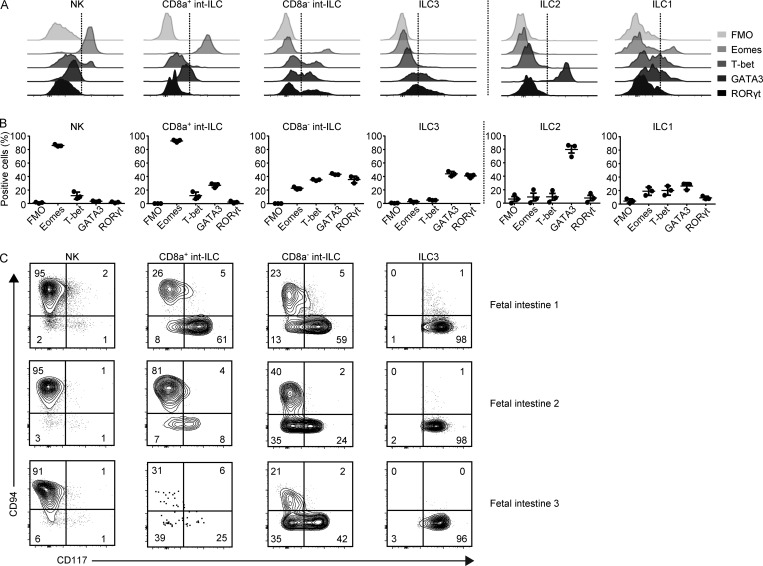
Next we determined the expression of key transcription factors associated with ILC development and phenotype. The expression of ID2, TCF7, AHR, NFIL3, ZBTB16, and TOX did not discriminate between the subsets (Fig. S3 C). In line with previous work (Artis and Spits, 2015), the ILC2 subset was strongly GATA3–positive and RORγt–negative, whereas ILC3s were GATA3– and RORγt–positive (Fig. 4, A and B; and Fig. S3 D). However, we found only low levels of T-bet expression by ILC1 (Fig. 4, A and B). Notably, both mature NK cells and CD8a+ int-ILCs expressed high levels of Eomes (Fig. 4, A and B; and Fig. S3, E and F). In contrast, the CD8a− int-ILCs were heterogeneous with respect to the expression of the four transcription factors, which were all expressed by a proportion of the cells (Fig. 4, A and B), an expression profile that does not correspond to those found in mature CD127+ ILCs. Furthermore, multiple lineage transcription factors could be simultaneously expressed by CD8a− int-ILCs, such as T-bet and GATA3 (26.1% of CD8a− int-ILCs; Fig. S3, G–I). Finally, the frequency of cells expressing Eomes decreased along the potential differentiation trajectory linking the NK cells to CD8a+ int-ILCs, CD8a− int-ILCs, and ILC3s, whereas that of RORγt increased (Fig. 4, A and B).
Figure 4.
Transcription factors and CD94 expression profiles of fetal intestinal ILCs ex vivo. (A and B) Flow cytometric determination of the expression of the transcription factors Eomes, T-bet, GATA3, and RORγt by the indicated ILC subsets as defined in Fig. 3 A. Histograms (A) depict the results with one representative human fetal intestine, and the graphs (B) depict quantification of data obtained from three different human fetal intestines (three independent experiments). Error bar shows mean ± SD. (C) Biaxial plots showing the expression of CD94 and CD117 by the indicated subsets. Results on three human fetal intestines are shown (three independent experiments). FMO, fluorescence-minus-one control.
To investigate the relationship between the int-ILCs, NK cells, and ILC3s further, we evaluated the expression of CD62L, CD57, CD5, and the NK cell–associated C-type lectin receptor CD94. Here, expression of CD62L, CD57, and CD5 was almost lacking and did not discriminate between the subsets (not depicted), whereas CD94 expression was high on NK cells but virtually absent from ILC3s (Fig. 4 C), in agreement with previous studies (Hazenberg and Spits, 2014; Scoville et al., 2016). In contrast, only part of the int-ILCs were CD94–positive with a higher expression level of CD94 on CD8a+ int-ILCs compared with CD8a− int-ILCs (Fig. 4 C), a result in line with the lower expression of Eomes in the latter. Furthermore, in contrast with CD94− cells, CD94+ cells lacked the expression of CD117, similar to mature NK cells (Fig. 4 C).
Together, these data indicate that the int-ILC subset is distinct from mature ILCs, where the expression pattern of the cytokines, CD94, and transcription factors link the CD8a+ int-ILCs to NK cells and the CD8a− int-ILCs to ILC3s.
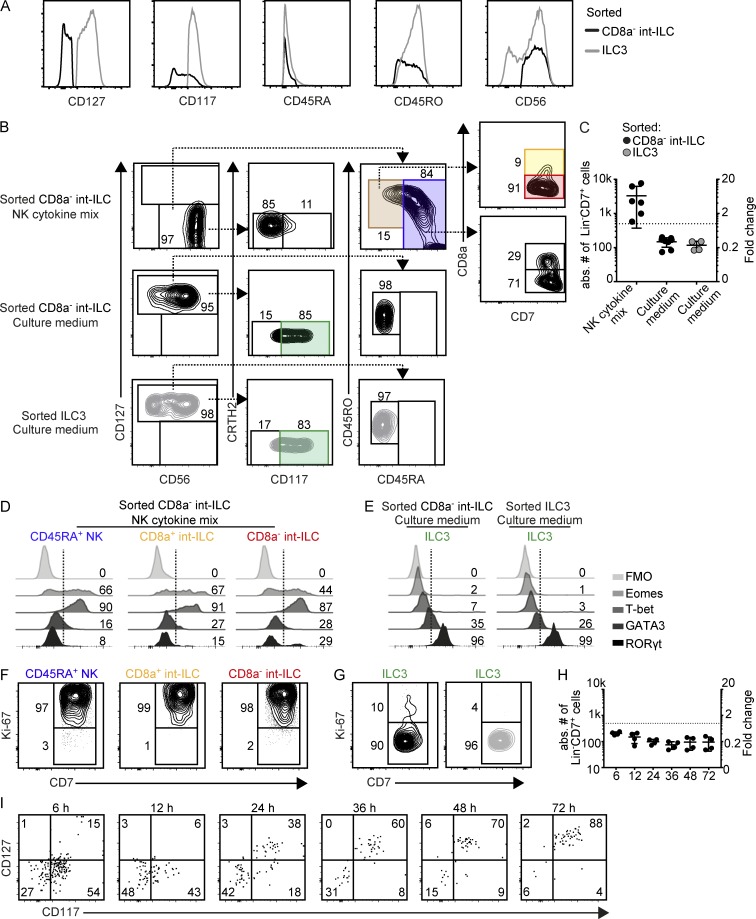
int-ILC can differentiate into CD45RA+ NK cells
To test the hypothesis that the int-ILC subset may differentiate into CD127+ ILCs and/or NK cells, we first purified the CD8a− int-ILCs by flow cytometry (Fig. 5 A) and performed functional differentiation assays by coculturing these with OP9 stromal cells expressing the Notch ligand Delta-like 1 (OP9-DL1). After 7 d of culture in medium containing stem cell factor (SCF), IL-7, IL-2, and IL-15 (hereafter referred to as NK cytokine mix), the majority of the int-ILCs acquired a CD45RA+ phenotype (Fig. 5 B) and expanded substantially (Fig. 5 C). Also, these cells up-regulated CD94 (42% positive; Fig. S4 A) and displayed expression of Eomes and/or T-bet, but no RORγt or GATA3 (Fig. 5 D), all similar to mature NK cells. Furthermore, part of these cells expressed CD8a (Fig. 5 B), a marker expressed by most fetal NK cells ex vivo (Fig. 2 B), where most of the CD8a+ cells displayed the highest expression of Eomes (Fig. S4 B). Moreover, a small fraction of generated cells maintained the CD45RA−CD45RO+ int-ILC phenotype, and a fraction of them also acquired CD8a (Fig. 5 B). In line with the suggested differentiation trajectory (Fig. 2 C), the expression of ILC3-associated RORγt decreased from the CD8a− int-ILCs to CD8a+ int-ILCs to CD45RA+ NK cells, whereas all populations expressed high levels of NK cell–associated Eomes and/or T-bet (Fig. 5 D). Also, the cells became uniformly Ki-67–positive (Fig. 5 F), consistent with the observed increase in cell numbers (Fig. 5 C). As similar results were obtained when purified CD8a− int-ILCs were cocultured with OP9 stromal cells without Delta-like 1, Notch signaling appears not to be involved (Fig. S4 C). In addition, upon culture on OP9-DL1, purified CD8a+ int-ILCs also acquired the CD45RA+ NK cell phenotype, maintained CD8a expression, and expressed high levels of CD94 (84% positive; Fig. S4 D). Finally, purified CD8a− int-ILCs cocultured with OP9-DL1 and IL-15 cytokine only similarly expanded (not depicted), acquired CD45RA, up-regulated CD94 (41% positive), and became in part CD8a–positive (Fig. S4 E). However, under these conditions, ∼60% of these generated cells remained CD117+ (Fig. S4 E), suggesting an incomplete conversion to the mature NK–cell phenotype (Freud et al., 2006). Together these data indicate that in the presence of NK cytokines, proliferative CD45RA+ NK cells are generated from int-ILC.
Figure 5.
int-ILC can differentiate into CD45RA+ NK cells and ILC3. (A) Representative histograms depicting the expression of CD127, CD117, CD45RA, CD45RO, and CD56 by flow cytometry–purified CD8a− int-ILCs (black line) and ILC3s (gray line) from human fetal intestines. Data are representative of six independent experiments. (B–G) Purified CD8a− int-ILCs and ILC3s were cocultured in 96-well plates at 500 cells/well with irradiated OP9-DL1 stromal cells for 7 d with culture medium alone or supplemented with NK cytokine mix. Generated cells were analyzed by flow cytometry. Duplicated wells were included for each condition. Representative plots show a single duplicate. (B) Representative biaxial plots depict the phenotypes of generated Lin−CD7+ cells based on the gating strategy for ILC1, ILC2, ILC3, NK, and int-ILC subsets as shown in Fig. 3 A, for three different combinations of sorted cell populations (int-ILC in black contours, ILC3 in gray contours) and culture conditions as indicated (three to five independent experiments). (C) Quantification of the generated Lin−CD7+ cells in B in absolute cell number (left axis) and fold change (right axis) compared with the number of initially sorted cells (dashed line; two to four independent experiments). Error bar shows mean ± SD. (D and E) Histograms depict the expression of transcription factors Eomes, T-bet, GATA3, and RORγt by the indicated subsets generated from sorted CD8a− int-ILCs with NK cytokine mix and (D) sorted CD8a− int-ILCs or ILC3s with culture medium (E). Numbers indicate the percentage of positive cells. Combined data on five human fetal intestines. (F and G) Biaxial plots depict the expression of Ki-67 by indicated subsets generated from the combinations of sorted cell populations (int-ILC in black contours, ILC3 in gray contours) and culture conditions as in D and E. Combined data on five human fetal intestines. (H and I) Purified CD8a− int-ILCs were cocultured at 500 cells/well with irradiated OP9-DL1 stromal cells in culture medium and harvested at the time points indicated in hours. Duplicated wells were included in each experiment (two independent experiments). (H) Quantification of the generated Lin−CD7+ cells in absolute cell number (left axis) and fold change (right axis) compared with the number of initially sorted cells (dashed line). (I) Representative biaxial plots show the expression of CD127 and CD117 by the generated Lin−CD7+ cells. FMO, fluorescence-minus-one control.
CD8a− int-ILC can differentiate into ILC3
In marked contrast, when purified CD8a− int-ILCs and ILC3s (Fig. 5 A) were individually cocultured with OP9-DL1 in cytokine-free culture medium, the ILC3s retained their phenotype, whereas the CD8a− int-ILCs acquired an ILC3 phenotype as they became CD127+CD117+ (Fig. 5 B), remained CD45RA−CD45RO+ (Fig. 5 B), and remained CD8a− (not depicted), in the absence of cell expansion and proliferation (Fig. 5, C and G). This phenotype was also stable during prolonged culture (Fig. S4 F). In addition, these cells homogeneously expressed RORγt, but no Eomes or T-bet (Fig. 5 E), suggesting an established ILC3 population (Hazenberg and Spits, 2014). As similar results were observed when we cocultured the CD8a− int-ILCs with OP9 stromal cells that lacked Notch ligand Delta-like 1, Notch signaling appears not to be involved (Fig. S4, G and H). Unlike CD8a− int-ILCs and ILC3s, both purified CD45RA+ NK cells and CD8a+ int-ILCs did not survive under these conditions. To exclude that the generation of ILC3 by int-ILCs was a result of outgrowth of contaminating ILC3s, we determined cell numbers and the acquisition of CD127 and CD117 at various time points during culture. After 24 and 72 h of culture, 38% and 88% of purified CD8a− int-ILCs had acquired both CD127 and CD117, respectively, whereas no increase in cell numbers was observed (Fig. 5, H and I). Together with the observation that only a very small proportion of both the purified mature ILC3s and differentiated ILC3s from int-ILCs were Ki-67+ (Fig. 5 G), this indicates that it is highly unlikely that selective outgrowth of contaminating ILC3s could explain the appearance of cells with an ILC3 phenotype in the CD8a− int-ILC/OP9 cocultures. Thus, these results indicate that the CD8a− int-ILC population can differentiate into ILC3 in vitro.
Differentiation properties of CD8a− int-ILC subpopulations
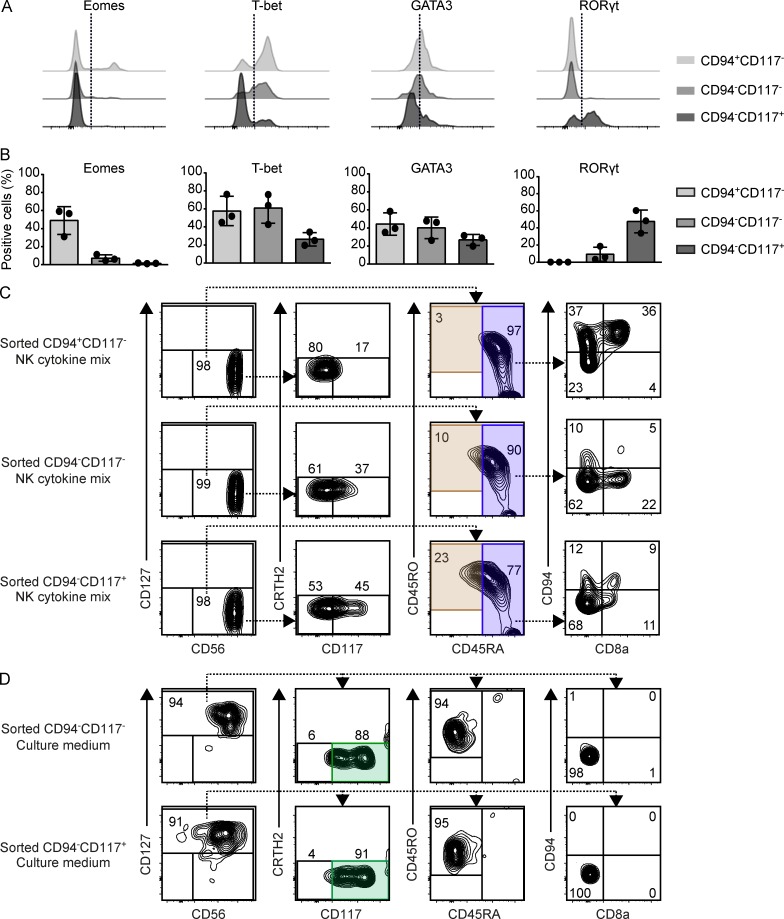
By the differential expression of CD94 and CD117, three distinct CD8a− int-ILC subpopulations could be distinguished: CD94+CD117−, CD94−CD117−, and CD94−CD117+ (Fig. 4 C). We therefore investigated whether these subsets could differentiate into either NK cells or ILC3s in vitro. For this purpose, we first examined the expression of transcription factors (Fig. 6, A and B). This revealed that Eomes was primarily present in the CD94+CD117− subset, whereas RORγt expression was most pronounced in the CD94−CD117+ subset. In addition, all subsets expressed GATA3 and T-bet, where T-bet expression was most pronounced by the CD117− subsets. Together, these results explain the heterogeneity in the expression of transcription factors by CD8a− int-ILCs and position the CD94−CD117− subset in between the CD94+CD117− and CD94−CD117+ subsets.
Figure 6.
Distinct differentiation properties of CD8a− int-ILC subpopulations. (A and B) Expression of the transcription factors Eomes, T-bet, GATA3, and RORγt by the indicated subpopulations of CD8a− int-ILC ex vivo. Histograms depict the results with one fetal intestine (A), and the graphs depict quantification of data obtained from three intestines (B; two independent experiments). Error bar shows mean ± SD. (C and D) Purified CD94+CD117−CD8a− int-ILC, CD94−CD117−CD8a− int-ILC, and CD94−CD117+CD8a− int-ILC populations were cocultured in 96-well plates at 500 cells/well with irradiated OP9-DL1 stromal cells for 7 d with (C) culture medium supplemented with NK cytokine mix or (D) culture medium alone. Generated cells were analyzed by flow cytometry. Representative biaxial plots depict the phenotypes of the generated Lin−CD7+ cells based on the gating strategy for ILC1, ILC2, ILC3, NK, and int-ILC subsets as shown in Fig. 3 A, for the five combinations of cell populations and culture conditions indicated. Duplicated wells were included in each condition. Representative plots show a single duplicate (three to four independent experiments).
Next, we purified CD94+CD117−, CD94−CD117−, and CD94−CD117+ subsets individually and cocultured them with OP9-DL1 stromal cells with either NK cytokine mix or cytokine-free medium. In the presence of NK cytokine mix (Fig. 6 C), virtually all of the CD94+CD117− cells acquired the CD45RA+ NK cell phenotype, maintained expression of high levels of CD94 (76%), acquired CD8a expression (43%), and expanded substantially (12-fold; not depicted). Similarly, most of the CD94−CD117− cells and the majority of the CD94−CD117+ cells also acquired the CD45RA+ NK cell phenotype, acquired expression of CD94 (17% and 25%, respectively) and CD8a (24% and 16%, respectively), and expanded (ninefold and sevenfold, respectively; not depicted). In contrast, in cytokine-free medium (Fig. 6 D), both the CD94−CD117− and CD94−CD117+ int-ILCs acquired an ILC3 phenotype as they became CD127+, remained CD45RO+CD45RA−CD94−CD8a−, and acquired or increased the levels of CD117 expression, respectively (Fig. 6 D). Similar to CD8a+ int-ILCs and CD45RA+ NK cells, the CD94+CD117− int-ILCs did not survive under these conditions.
Collectively, in these in vitro experiments, all three subpopulations of the CD8a− int-ILCs can differentiate into NK cells, whereas the CD94−CD117− and CD94−CD117+ cells, but not the CD94+CD117− cells, can differentiate into ILC3s.
Discussion
Numerous studies have reported substantial heterogeneity in the ILC compartment (Hazenberg and Spits, 2014; Björklund et al., 2016). In our mass cytometry–based approach, the NK, ILC1, ILC2, and ILC3 subsets could be readily identified in the human fetal intestine as well as substantial variability within those subsets, results that were highly consistent in several samples analyzed. Based on the marker expression profiles, we identified a large number of distinct NK cell clusters, whose biological significance needs to be investigated in the future studies. In addition, we found a small CD27+ ILC cluster that matches ILC1 criteria (Spits et al., 2013; Klose et al., 2014). However, in contrast to findings from Bernink et al. (2013), but in line with other studies (Björklund et al., 2016; Simoni et al., 2017), only a few of these ILC1s expressed T-bet. Furthermore, two additional CD127+ ILC1-like clusters were identified that clustered with the NK cells because of the expression of several NK cell–associated markers, including CD45RA, CD56, CD8a, and NKp46. Consistent with previous studies (Hazenberg and Spits, 2014; Bernink et al., 2015; Björklund et al., 2016), the ILC3 compartment was most frequent and heterogeneous, including CD45RO+ ILC3, CD45RA+ ILC3, HLA-DR+ ILC3, and CD56+/− ILC3 clusters. Further studies will be required to clarify the potential functional significance of observed heterogeneity in the ILC3 lineage. We could not distinguish LTi cells from ILC3s as no specific cell surface marker for human LTi cells was available at the time we performed our analysis. Moreover, although most of the human ILCs described express CD161 (Spits et al., 2013; Hazenberg and Spits, 2014), we also detected a recently described CD161−CD117+ ILC3-like cluster that clustered with the CD127+ ILCs (Simoni et al., 2017). In addition, we identified two previously unknown CD8a+ counterparts of ILC3s that warrant further investigation. We also observed a rare CD34+CD45RA+CD117+ population that resembles the CD34+ precursors recently described in human tonsils and intestines after birth (Montaldo et al., 2014; Scoville et al., 2016).
Finally, we identified a Lin−CD7+CD127−CD45RO+CD56+ group of cells that by unbiased clustering was positioned between the CD45RA+ NK cells and CD127+ ILCs, and was termed int-ILC. Although in previous studies (Spits et al., 2013; Hazenberg and Spits, 2014) such CD56+CD127− cells were classified as NK cells, the simultaneous use of CD45RA and CD45RO allowed us to distinguish these CD45RO+ cells from the CD45RA+ NK cells. It is important to note that these int-ILCs display variable expression of several surface markers, including CD8a, CD94, CD117, CD122, CD25, CD27, KLRG-1, and CD103, indicating that they are not a homogenous group of cells. Their unique position between the NK cells and CD127+ ILCs, however, prompted us to investigate potential relationships between these cell clusters.
In recent years, several studies have explored developmental pathways by applying computational approaches on mass cytometry datasets. Wanderlust accurately predicted B–cell lymphopoiesis (Bendall et al., 2014), and Wishbone was found to recover the T–cell developmental pathway (Setty et al., 2016) at single-cell resolution. However, Wanderlust is not suitable to predict multilineage differentiation trajectories. Although Wishbone can be used to identify bifurcating developmental trajectories, it needs the designation of a user-defined precursor cell type. As we wished to investigate potential developmental relationships without preassumptions, we developed a novel computational approach to visualize the evolution of the t-SNE map over the course of the optimization. t-SNE is a nonlinear dimensionality reduction algorithm that projects the high-dimensional similarities onto a two-dimensional map based on the concurrent marker expression profiles (van der Maaten and Hinton, 2008). Here t-SNE not only employs bimodal distribution patterns but also incorporates gradients of the expression of cellular markers and therefore offers superior resolution. Importantly, the Cytosplore framework (Höllt et al., 2016) can visualize every iteration by making use of the A-tSNE (Pezzotti et al., 2017). This allowed us to analyze potential relationships between the int-ILC, ILC1, ILC2, ILC3, and NK clusters through visualization of gradients along putative differentiation trajectories. Such gradients are clearly visible in our Cytosplore analysis (Fig. 2) and contribute to the generation of cell clusters. Our current results indicate that at least some of those gradual changes in marker expression profiles correlate with differentiation pathways of immune subsets. Consistent with the observed plasticity among ILCs (Bernink et al., 2015; Bal et al., 2016), the analysis revealed a clear trajectory between the ILC2 and ILC3 clusters. This may imply that the CD103− ILC2 can differentiate into CD103+ ILC3 locally depending on physiological or pathological conditions. Alternatively, the fetal ILC2s may leave the intestine, in line with previous studies concluding that ILC2s can be found in the peripheral blood (Mjösberg et al., 2011) but are virtually absent in the human intestine after birth (Bernink et al., 2015). In addition, a trajectory within the CD56+ cell compartment was revealed, where the CD8a- int-ILC was connected with the CD56+ ILC3 on one side and with the CD8a+ int-ILC and the NK cells on the other. Importantly, although the above analysis was performed on the innate cell population present in the seven fetal intestines collectively, similar relationships between cell populations were revealed when the innate cell compartment of each fetal sample was analyzed individually, attesting to the robustness of the approach (Fig. S2 B).
The putative link between the int-ILC and the mature ILC and NK cells is further strengthened by several other observations. First, both the cytokine production profiles and the CD94 expression profile ex vivo link the CD8a+ int-ILCs to NK cells and the CD8a− int-ILCs to the ILC3s. Second, the expression pattern of the transcription factors by int-ILCs is heterogeneous with features of both NK cells (Eomes and T-bet) and ILC3s (GATA3 and RORγt), where the CD8a+ int-ILCs resemble the NK cells whereas the CD8a− int-ILCs are closer to CD127+ ILCs. Finally, in the OP9-based coculture system, in the presence of NK cytokines, purified int-ILC expanded and displayed a CD45RA+Eomes+/T-bet+ NK–cell phenotype, whereas in the absence of cytokines, the cells did not expand but acquired a stable CD127+CD117+RORγt+ ILC3 phenotype.
Further dissection of the CD8a− int-ILC compartment demonstrated the existence of three distinct subpopulations based on differential expression of CD94 and CD117: CD94+CD117−, CD94−CD117−, and CD94−CD117+. In our in vitro experiments, both the Eomes-expressing CD94+CD117− and non–Eomes-expressing CD94−CD117− and CD94−CD117+ subpopulations could differentiate into CD45RA+ NK cells when cultured with NK cytokine mix. Furthermore, the differentiation into CD45RA+ NK cells was most efficient in the case of the CD94+CD117− cells and least for the CD94−CD117+ cells, compatible with a model where the CD94+CD117− cells are positioned close to NK cells, the CD94−CD117+ cells most distant, and the CD94−CD117− cells in between. The acquisition of CD8a by the CD94−CD8a− int-ILCs in these cultures indicates that the CD8a+ int-ILC may also be an intermediate stage toward NK–cell differentiation. In the absence of cytokines, CD117+CD127+ ILC3-like cells could be generated from both CD94−CD117+ and CD94−CD117− cells but not from CD94+CD117− cells. Together this indicates that CD94+CD117− cells can exclusively differentiate into NK cells whereas the other two populations can differentiate into both NK cells and ILC3s, at least under the in vitro conditions used.
In the absence of cytokines, the int-ILC changed into ILC3-like cells without signs of cell expansion, cell division, or cell death, arguing that the generation of ILC3 from the int-ILC is not a result of selective outgrowth of contaminating ILC3s. In agreement, we did not observe any proliferative response of flow cytometry–purified ILC3 under the same experimental conditions.
In mice, it has been shown that developmental hierarchy of ILCs goes from the common lymphoid progenitor to mature ILCs via α4β7-expressing lymphoid progenitor, early innate lymphoid progenitor, common helper ILC progenitor, and the ILCP (Ishizuka et al., 2016b). Here early innate lymphoid progenitor has been distinguished from other progenitors by the lack of CD127 and ILC lineage-specific transcription factors (Yang et al., 2015), whereas the ILCP exhibits coexpression of transcription factors associated with ILC1, ILC2, and ILC3 subsets (Ishizuka et al., 2016a). Finally, a mouse fetal transitional CD127+ ILCP has been identified in the intestine which expresses varying amount of T-bet, GATA3, and RORγt (Bando et al., 2015). In humans, two studies have shown that ILC3s can be generated from RORγt+CD34+ progenitors from tonsils and intestines (Montaldo et al., 2014; Scoville et al., 2016), but little is known about the intermediate stages. Interestingly, Scoville et al. (2016) generated both NK cells and the three types of helper ILCs from these CD34+ progenitors in vitro; however, the generated cells did not express CD127 but rather CD161 and intracellular ILC-related cytokine profiles. Also, an NK cell lineage–restricted CD34+ progenitor was identified in the human fetal liver and bone marrow (Renoux et al., 2015). In contrast, CD127− int-ILCs in our study express CD45RO and variable levels of Eomes, T-bet, GATA3, and RORγt, but no CD34 and CD45RA, markers expressed by most human progenitors. Also, with our mass cytometry approach, we have been unable to identify cells with an int-ILC phenotype in the human fetal liver and fetal spleen (not depicted), indicating that the int-ILC may specifically reside in mucosal tissues. Together, the cell surface phenotype, tissue distribution, and transcription factor profiles of CD127− int-ILC suggest that these cells are distinct from previously identified ILC progenitors and may be in an intermediate differentiation stage among ILC lineages. Finally, we observed that the ILC3s and NK cells derived from the CD8a− int-ILC could partly revert their phenotype upon prolonged culture, indicating that the int-ILC may represent an intermediate between two plastic lineages (not depicted).
It has been shown that environmental cues including OP9/OP9-DL1 stromal cells and cytokines such as IL-7 and SCF play an important role in driving ILC3 differentiation (Satoh-Takayama et al., 2010; Montaldo et al., 2014; Scoville et al., 2016). Although the addition of SCF and IL-7 did promote significant expansion of the CD8a− int-ILCs, they did not differentiate into other types of cells (not depicted). Instead, the differentiation of CD8a− int-ILCs toward ILC3s occurred in cytokine-free medium. In mice, Notch signaling for ILC3 development is necessary in adults but not in fetuses (Possot et al., 2011), whereas in humans, the differentiation of CD34+ progenitors to ILC3s can occur without Notch signaling (Montaldo et al., 2014; Scoville et al., 2016). Consistent with these observations, the generation of ILC3s from the CD8a− int-ILCs was Notch-independent.
In conclusion, we delineated the heterogeneity of ILCs in the human fetal intestine and developed a computational model to predict potential differentiation trajectories based on mass cytometry data. This allowed the identification of a previously unidentified innate cell cluster that harbors cells that can differentiate into NK cells and ILC3-like cells in vitro. This may provide plasticity in the human fetal intestine in response to external stimuli.
Materials and methods
Human fetal intestine and cell isolation
Human fetal intestines from elective abortions were collected after informed consent. Approval by the medical ethical commission of the Leiden University Medical Center (protocol P08.087) was obtained in accordance with the local ethical guidelines and the Declaration of Helsinki. The gestational age ranged from 16 to 22 wk. Single-cell suspensions from fetal intestines were prepared as previously described. In brief, the mesentery, colon part, and meconium were removed from the fetal intestine. The intestines were then cut into small fragments and treated with 1 mM 1,4-dithiothreitol (Fluka) in 15 ml of HBSS (Sigma-Aldrich) for 2 × 10 min (replacing buffer) at room temperature (rT) to dissolve the mucus and subsequently with 1 mM EDTA (Merck) in 15 ml of HBSS under rotation for 2 × 1 h (replacing buffer) at 37°C to separate the epithelium from the lamina propria fraction. To obtain single-cell suspensions from the lamina propria, the intestines were rinsed with HBSS and incubated with 15 ml IMDM (Lonza) supplemented with 10% FCS, 10 U/ml collagenase IV (Worthington), and 200 µg/ml DNase I grade II (Roche Diagnostics), at 37°C overnight, after which cell suspensions were filtered through a 70-µm nylon cell strainer. Finally, the immune cells were isolated with a Percoll (GE Healthcare) gradient and stored in liquid nitrogen.
Mass cytometry antibody staining and data acquisition
Details on antibodies used are listed in Table S1. Conjugation of the purified antibodies with metal reporters was performed with the MaxPar X8 antibody labeling kit (Fluidigm Sciences) according to the manufacturer's instructions. Procedures for mass cytometry antibody staining and data acquisition were performed as previously described (van Unen et al., 2016). In brief, cells from fetal intestinal lamina propria were thawed and incubated with 1 ml 500× diluted 500 µM Cell-ID intercalator-103Rh (Fluidigm Sciences) for 15 min at rT to identify dead cells. Cells were then stained with metal-conjugated antibodies for 45 min at rT. After staining, cells were labeled with 1 ml 1,000× diluted 125 µM Cell-ID intercalator-Ir (Fluidigm Sciences) to stain all cells in MaxPar Fix and Perm Buffer (Fluidigm Sciences) overnight at 4°C. Finally, cells were acquired by CyTOF 2 mass cytometer (Fluidigm Sciences). Data were normalized by using EQ Four Element Calibration Beads (Fluidigm Sciences) with the reference EQ passport P13H2302 during the course of each experiment.
Mass cytometry data analysis
The biaxial plots showing antibody staining patterns in Fig. S1 were generated in Cytobank (Kotecha et al., 2010). Data for single, live CD45+ cells gated from each fetal intestine individually using Cytobank (Kotecha et al., 2010) as shown in Fig. S1 A were sample-tagged and hyperbolic arcsinh–transformed with a cofactor of 5 before t-SNE analysis. The 907 clusters shown in Fig. S1 C were identified by analyzing the entire immune system (CD45+ cells) using the t-SNE–ACCENSE analysis pipeline as described before (van Unen et al., 2016). Next, t-SNE was performed for the ILC dataset using A-tSNE (Pezzotti et al., 2017) in Cytosplore (Höllt et al., 2016). t-SNE was performed with default parameters (perplexity, 30; iterations, 1,000). All t-SNE plots were generated in Cytosplore. Hierarchical clustering of the heat map was created with Pearson Correlation and mean linkage clustering in MultiExperiment Viewer (http://www.tm4.org). Wanderlust analysis was performed on cells that were selected along the linear CD56 expression continuum at stage 4 of the t-SNE computation with the CD56+CD8a+ ILC3 cluster as starting point (as this cluster is located at the outer end of the CD56 trajectory), using Cyt in Matlab (Bendall et al., 2014).
Antibodies and flow cytometry
FITC-conjugated anti-CD11c (3.9), PerCP-Cy5.5–conjugated anti-CD45RO (UCHL1), PE/Dazzle 594–conjugated anti-CD45RA (HI100), PE-Cy7–conjugated anti-CD127 (A019D5), Brilliant Violet 605–conjugated anti-CRTH2 (BM16), anti–T-bet (4B10), PE-conjugated anti–T-bet (4B10), anti–IFN-γ (4S.B3), anti–IL-5 (TRFK5), anti–IL-13 (JES10-5A2), anti–IL-17A (BL168), and anti–TNF-α (Mab11) were purchased from BioLegend. The following monoclonal antibodies were purchased from BD: FITC-conjugated anti-CD3 (SK7), anti-CD19 (4G7), anti-CD14 (MϕP9), APC-conjugated anti-CD117 (YB5.B8), APC-R700–conjugated anti-CD56 (NCAM16.2), V450-conjugated anti-CD7 (M-T701), Brilliant Violet 605–conjugated anti-CD94 (HP-3D9), PE-conjugated anti-CD94 (HP-3D9), anti-RORγt (Q21-559), anti–Ki-67 (B56), anti-perforin (δG9), and anti–IL-4 (3010.211). PE-conjugated anti-Eomes (WD1928), anti-GATA3 (TWAJ), anti–granzyme B (GB11), and eFluor 660–conjugated anti-GATA3 (TWAJ) were purchased from eBioscience. PE-conjugated anti–IL-22 (IC7821P) was purchased from R&D Systems. Pacific Orange–conjugated anti-CD8a (3B5) was purchased from Life Technologies.
For the cell surface staining, cells were incubated with fluorochrome-conjugated antibodies and human FC block (BioLegend) for 30–45 min at 4°C. The transcription factor staining was performed by using Foxp3 Staining Buffer Set (eBioscience) according to the manufacturer’s instructions. For the intracellular cytokine staining/cytotoxic molecule, cells were stimulated with 0.1 mg/ml PMA (Sigma-Aldrich) and 1 µg/ml ionomycin (Sigma-Aldrich) for 6 h at 37°C, and GolgiPlug (BD Biosciences) was added for the final 4 h, after which cells were stained by using Fixation Buffer and Intracellular Staining Perm Wash Buffer (BioLegend). Cells were acquired on an LSR II cytometer (BD Biosciences) or sorted on a FACSAria III sorter (BD Biosciences) based on the gating strategy as shown in Fig. 3 A. Data were analyzed with FlowJo V10 software.
Quantitative real-time PCR
RNA extraction was performed with the NucleoSpin RNA XS kit (Macherey-Nagel). cDNA was synthesized with the High Capacity cDNA Reverse Transcription kit (Applied Biosystems). Real-time PCR was performed in a StepOnePlus Real-Time PCR Systems (Applied Biosystems) with FastStart Universal SYBR Green Master Mix (Roche). ΔCt values were calculated using GAPDH as reference gene. The sequences of real-time PCR primers are as follows: GAPDH, forward primer: 5′-GTCTCCTCTGACTTCAACAGCG-3′; reverse primer: 5′-ACCACCCTGTTGCTGTAGCCAA-3′; AHR, forward primer: 5′-CTTAGGCTCAGCGTCAGTTA-3′; reverse primer: 5′-GTAAGTTCAGGCCTTCTCTG-3′; ID2, forward primer: 5′-TTGTCAGCCTGCATCACCAGAG-3′; reverse primer: 5′-AGCCACACAGTGCTTTGCTGTC-3′; NFIL3, forward primer: 5′-TGGAGAAGACGAGCAACAGGTC-3′; reverse primer: 5′-CTTGTGTGGCAAGGCAGAGGAA-3′; ZBTB16, forward primer: 5′-GAGCTTCCTGATAACGAGGCTG-3′; reverse primer: 5′-AGCCGCAAACTATCCAGGAACC-3′; TOX, forward primer: 5′-AGCATACAGAGCCAGCCTTG-3′; reverse primer: 5′-TGCATGGCAGTTAGGTGAGG-3′; and TCF1, forward primer: 5′-TGCAGCTATACCCAGGCTGG-3′; reverse primer: 5′-CCTCGACCGCCTCTTCTTC-3′.
Cell culture and differentiation assays
OP9-DL1 or OP9 stromal cells were maintained in Minimum Essential Medium α (Lonza) supplemented with 10% FCS. Flow cytometry–purified CD8a− int-ILCs or CD94+CD117−, CD94−CD117−, and CD94−CD117+ subpopulations (500 cells/well) or CD8a+ int-ILCs (100 cells/well) were cocultured with irradiated OP9 or OP9-DL1 stromal cells (1,500 rad, 5,000 cells/well) in a 96-well plate (Corning) and maintained in culture medium (IMDM supplemented with 10% human serum) or in culture medium containing 25 ng/ml SCF (Miltenyi Biotec), 25 ng/ml IL-7 (Peprotech), 10 U/ml IL-2 (Novartis), and 10 ng/ml IL-15 (R&D Systems) or only IL-15. The phenotype of generated progeny was determined by flow cytometry.
Cytokine secretion
CD8a− int-ILCs and ILC3 (2,000 cells/well) were stimulated with 10 U/ml IL-2 (Novartis), 50 ng/ml IL-1β (Peprotech), and 50 ng/ml IL-23 (Peprotech) for 4 d. TNF-α, IL-17A, and IL-22 were measured by using Bio-Plex Pro human cytokine 17-plex panel kit and Bio-Plex Pro human Treg cytokine panel 12-plex kit (Bio-Rad).
Online supplemental material
Fig. S1 shows the mass cytometry–based analysis of the entire immune system in the human fetal intestine. Fig. S2 shows t-SNE–based analysis of the Lin−CD7+ innate immune compartment in the human fetal intestine. Fig. S3 shows Ki-67 and transcription factor expression profiles of fetal intestinal ILCs ex vivo. Fig. S4 is related to Fig. 5, and shows that CD8a− int-ILC can differentiate into CD45RA+ NK cells and ILC3. Table S1 lists the information of antibodies used in the mass cytometry data.
Supplementary Material
Acknowledgments
We thank the Center for Contraception, Abortion and Sexuality in Leiden and The Hague for their efforts in collecting and providing the fetal material; K. Lodder, T. van Herwaarden, Dr. M. Bialecka, Dr. F. Wang, and M. Gomes Fernandes for the dissection of the fetal intestines; E. de Haas for flow cytometric cell sorting; and E. van Beelen for the Luminex assay. We thank Drs. F.J.T. Staal, K. Schepers, and T. Cupedo for critical review of the manuscript.
This research was supported by the Leiden University Medical Center and the Netherlands Organization for Scientific Research (ZonMW grant 91112008 and Applied Technical Sciences Visual Analysis in Population Imaging Research [VAnPIRe] grant 12720). N. Li was supported by the China Scholarship Council.
The authors declare no competing financial interests.
Author contributions: N. Li, V. van Unen, and F. Koning conceived the study and wrote the manuscript. N. Li performed most experiments with the help of V. van Unen. T. Höllt, N. Pezzotti, V. van Unen, E. Eisemann, A. Vilanova, and B.P.F. Lelieveldt developed the Cytosplore application. V. van Unen and N. Li performed mass cytometry analyses. A. Thompson performed the real-time PCR. S.M.C. de Sousa Lopes provided human fetal intestines. J. van Bergen provided conceptual input. All authors discussed the results and commented on the manuscript.
References
- Artis D., and Spits H.. 2015. The biology of innate lymphoid cells. Nature. 517:293–301. 10.1038/nature14189 [DOI] [PubMed] [Google Scholar]
- Bal S.M., Bernink J.H., Nagasawa M., Groot J., Shikhagaie M.M., Golebski K., van Drunen C.M., Lutter R., Jonkers R.E., Hombrink P., et al. 2016. IL-1β, IL-4 and IL-12 control the fate of group 2 innate lymphoid cells in human airway inflammation in the lungs. Nat. Immunol. 17:636–645. 10.1038/ni.3444 [DOI] [PubMed] [Google Scholar]
- Bando J.K., Liang H.-E., and Locksley R.M.. 2015. Identification and distribution of developing innate lymphoid cells in the fetal mouse intestine. Nat. Immunol. 16:153–160. 10.1038/ni.3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandura D.R., Baranov V.I., Ornatsky O.I., Antonov A., Kinach R., Lou X., Pavlov S., Vorobiev S., Dick J.E., and Tanner S.D.. 2009. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal. Chem. 81:6813–6822. 10.1021/ac901049w [DOI] [PubMed] [Google Scholar]
- Bendall S.C., Davis K.L., Amir A.D., Tadmor M.D., Simonds E.F., Chen T.J., Shenfeld D.K., Nolan G.P., and Pe’er D.. 2014. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell. 157:714–725. 10.1016/j.cell.2014.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernink J.H., Peters C.P., Munneke M., te Velde A.A., Meijer S.L., Weijer K., Hreggvidsdottir H.S., Heinsbroek S.E., Legrand N., Buskens C.J., et al. 2013. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat. Immunol. 14:221–229. 10.1038/ni.2534 [DOI] [PubMed] [Google Scholar]
- Bernink J.H., Krabbendam L., Germar K., de Jong E., Gronke K., Kofoed-Nielsen M., Munneke J.M., Hazenberg M.D., Villaudy J., Buskens C.J., et al. 2015. Interleukin-12 and -23 Control Plasticity of CD127(+) Group 1 and Group 3 Innate Lymphoid Cells in the Intestinal Lamina Propria. Immunity. 43:146–160. 10.1016/j.immuni.2015.06.019 [DOI] [PubMed] [Google Scholar]
- Björklund Å.K., Forkel M., Picelli S., Konya V., Theorell J., Friberg D., Sandberg R., and Mjösberg J.. 2016. The heterogeneity of human CD127(+) innate lymphoid cells revealed by single-cell RNA sequencing. Nat. Immunol. 17:451–460. 10.1038/ni.3368 [DOI] [PubMed] [Google Scholar]
- Cella M., Fuchs A., Vermi W., Facchetti F., Otero K., Lennerz J.K.M., Doherty J.M., Mills J.C., and Colonna M.. 2009. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 457:722–725. 10.1038/nature07537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cupedo T., Crellin N.K., Papazian N., Rombouts E.J., Weijer K., Grogan J.L., Fibbe W.E., Cornelissen J.J., and Spits H.. 2009. Human fetal lymphoid tissue-inducer cells are interleukin 17-producing precursors to RORC+ CD127+ natural killer-like cells. Nat. Immunol. 10:66–74. 10.1038/ni.1668 [DOI] [PubMed] [Google Scholar]
- Freud A.G., Yokohama A., Becknell B., Lee M.T., Mao H.C., Ferketich A.K., and Caligiuri M.A.. 2006. Evidence for discrete stages of human natural killer cell differentiation in vivo. J. Exp. Med. 203:1033–1043. 10.1084/jem.20052507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger G., Fan X., Dikiy S., Lee S.Y., and Rudensky A.Y.. 2015. Tissue residency of innate lymphoid cells in lymphoid and nonlymphoid organs. Science. 350:981–985. 10.1126/science.aac9593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazenberg M.D., and Spits H.. 2014. Human innate lymphoid cells. Blood. 124:700–709. 10.1182/blood-2013-11-427781 [DOI] [PubMed] [Google Scholar]
- Höllt T., Pezzotti N., van Unen V., Koning F., Eisemann E., Lelieveldt B., and Vilanova A.. 2016. Cytosplore: Interactive Immune Cell Phenotyping for Large Single-Cell Datasets. Comput. Graph. Forum. 35:171–180. 10.1111/cgf.12893 [DOI] [Google Scholar]
- Ishizuka I.E., Chea S., Gudjonson H., Constantinides M.G., Dinner A.R., Bendelac A., and Golub R.. 2016a Single-cell analysis defines the divergence between the innate lymphoid cell lineage and lymphoid tissue-inducer cell lineage. Nat. Immunol. 17:269–276. 10.1038/ni.3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka I.E., Constantinides M.G., Gudjonson H., and Bendelac A.. 2016b The Innate Lymphoid Cell Precursor. Annu. Rev. Immunol. 34:299–316. 10.1146/annurev-immunol-041015-055549 [DOI] [PubMed] [Google Scholar]
- Juelke K., and Romagnani C.. 2016. Differentiation of human innate lymphoid cells (ILCs). Curr. Opin. Immunol. 38:75–85. 10.1016/j.coi.2015.11.005 [DOI] [PubMed] [Google Scholar]
- Klose C.S.N., Flach M., Möhle L., Rogell L., Hoyler T., Ebert K., Fabiunke C., Pfeifer D., Sexl V., Fonseca-Pereira D., et al. 2014. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell. 157:340–356. 10.1016/j.cell.2014.03.030 [DOI] [PubMed] [Google Scholar]
- Kotecha N., Krutzik P.O., and Irish J.M.. 2010. Web-based analysis and publication of flow cytometry experiments. Curr. Protoc. Cytom. Chapter 10:Unit 10.17 10.1002/0471142956.cy1017s53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim A.I., Menegatti S., Bustamante J., Le Bourhis L., Allez M., Rogge L., Casanova J.-L., Yssel H., and Di Santo J.P.. 2016. IL-12 drives functional plasticity of human group 2 innate lymphoid cells. J. Exp. Med. 213:569–583. 10.1084/jem.20151750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim A.I., Li Y., Lopez-Lastra S., Stadhouders R., Paul F., Casrouge A., Serafini N., Puel A., Bustamante J., Surace L., et al. 2017. Systemic Human ILC Precursors Provide a Substrate for Tissue ILC Differentiation. Cell. 168:1086–1100.e10. 10.1016/j.cell.2017.02.021 [DOI] [PubMed] [Google Scholar]
- Mjösberg J.M., Trifari S., Crellin N.K., Peters C.P., van Drunen C.M., Piet B., Fokkens W.J., Cupedo T., and Spits H.. 2011. Human IL-25- and IL-33-responsive type 2 innate lymphoid cells are defined by expression of CRTH2 and CD161. Nat. Immunol. 12:1055–1062. 10.1038/ni.2104 [DOI] [PubMed] [Google Scholar]
- Montaldo E., Teixeira-Alves L.G., Glatzer T., Durek P., Stervbo U., Hamann W., Babic M., Paclik D., Stölzel K., Gröne J., et al. 2014. Human RORγt(+)CD34(+) cells are lineage-specified progenitors of group 3 RORγt(+) innate lymphoid cells. Immunity. 41:988–1000. 10.1016/j.immuni.2014.11.010 [DOI] [PubMed] [Google Scholar]
- Pezzotti N., Lelieveldt B.P.F., Van Der Maaten L., Hollt T., Eisemann E., and Vilanova A.. 2017. Approximated and User Steerable tSNE for Progressive Visual Analytics. IEEE Trans. Vis. Comput. Graph. 23:1739–1752. 10.1109/TVCG.2016.2570755 [DOI] [PubMed] [Google Scholar]
- Possot C., Schmutz S., Chea S., Boucontet L., Louise A., Cumano A., and Golub R.. 2011. Notch signaling is necessary for adult, but not fetal, development of RORγt(+) innate lymphoid cells. Nat. Immunol. 12:949–958. 10.1038/ni.2105 [DOI] [PubMed] [Google Scholar]
- Renoux V.M., Zriwil A., Peitzsch C., Michaëlsson J., Friberg D., Soneji S., and Sitnicka E.. 2015. Identification of a Human Natural Killer Cell Lineage-Restricted Progenitor in Fetal and Adult Tissues. Immunity. 43:394–407. 10.1016/j.immuni.2015.07.011 [DOI] [PubMed] [Google Scholar]
- Satoh-Takayama N., Lesjean-Pottier S., Vieira P., Sawa S., Eberl G., Vosshenrich C.A.J., and Di Santo J.P.. 2010. IL-7 and IL-15 independently program the differentiation of intestinal CD3-NKp46+ cell subsets from Id2-dependent precursors. J. Exp. Med. 207:273–280. 10.1084/jem.20092029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoville S.D., Mundy-Bosse B.L., Zhang M.H., Chen L., Zhang X., Keller K.A., Hughes T., Chen L., Cheng S., Bergin S.M., et al. 2016. A Progenitor Cell Expressing Transcription Factor RORγt Generates All Human Innate Lymphoid Cell Subsets. Immunity. 44:1140–1150. 10.1016/j.immuni.2016.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty M., Tadmor M.D., Reich-Zeliger S., Angel O., Salame T.M., Kathail P., Choi K., Bendall S., Friedman N., and Pe’er D.. 2016. Wishbone identifies bifurcating developmental trajectories from single-cell data. Nat. Biotechnol. 34:637–645. 10.1038/nbt.3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhar K., Brodin P., Davis M.M., and Chakraborty A.K.. 2014. Automatic Classification of Cellular Expression by Nonlinear Stochastic Embedding (ACCENSE). Proc. Natl. Acad. Sci. USA. 111:202–207. 10.1073/pnas.1321405111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoni Y., Fehlings M., Kløverpris H.N., McGovern N., Koo S.-L., Loh C.Y., Lim S., Kurioka A., Fergusson J.R., Tang C.-L., et al. 2017. Human Innate Lymphoid Cell Subsets Possess Tissue-Type Based Heterogeneity in Phenotype and Frequency. Immunity. 46:148–161. 10.1016/j.immuni.2016.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spits H., Artis D., Colonna M., Diefenbach A., Di Santo J.P., Eberl G., Koyasu S., Locksley R.M., McKenzie A.N.J., Mebius R.E., et al. 2013. Innate lymphoid cells--a proposal for uniform nomenclature. Nat. Rev. Immunol. 13:145–149. 10.1038/nri3365 [DOI] [PubMed] [Google Scholar]
- van der Maaten L., and Hinton G.. 2008. Visualizing Data using t-SNE. J. Mach. Learn. Res. 9:2579–2605. [Google Scholar]
- van Unen V., Li N., Molendijk I., Temurhan M., Höllt T., van der Meulen-de Jong A.E., Verspaget H.W., Mearin M.L., Mulder C.J., van Bergen J., et al. 2016. Mass Cytometry of the Human Mucosal Immune System Identifies Tissue- and Disease-Associated Immune Subsets. Immunity. 44:1227–1239. 10.1016/j.immuni.2016.04.014 [DOI] [PubMed] [Google Scholar]
- van Unen V., Höllt T., Pezzotti N., Li N., Reinders M.J.T., Eisemann E., Koning F., Vilanova A., and Lelieveldt B.P.F.. 2017. Visual analysis of mass cytometry data by hierarchical stochastic neighbour embedding reveals rare cell types. Nat. Commun. 8:1740 10.1038/s41467-017-01689-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Li F., Harly C., Xing S., Ye L., Xia X., Wang H., Wang X., Yu S., Zhou X., et al. 2015. TCF-1 upregulation identifies early innate lymphoid progenitors in the bone marrow. Nat. Immunol. 16:1044–1050. 10.1038/ni.3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.