The cGAS–cGAMP–STING pathway mediates immune and inflammatory responses to cytosolic DNA. This review summarizes recent findings on how genomic instability leads to cGAS activation and how this pathway critically connects DNA damage to autoinflammatory diseases, cellular senescence, and cancer.
Abstract
Detection of microbial DNA is an evolutionarily conserved mechanism that alerts the host immune system to mount a defense response to microbial infections. However, this detection mechanism also poses a challenge to the host as to how to distinguish foreign DNA from abundant self-DNA. Cyclic guanosine monophosphate (GMP)–adenosine monophosphate (AMP) synthase (cGAS) is a DNA sensor that triggers innate immune responses through production of the second messenger cyclic GMP-AMP (cGAMP), which binds and activates the adaptor protein STING. However, cGAS can be activated by double-stranded DNA irrespective of the sequence, including self-DNA. Although how cGAS is normally kept inactive in cells is still not well understood, recent research has provided strong evidence that genomic DNA damage leads to cGAS activation to stimulate inflammatory responses. This review summarizes recent findings on how genomic instability and DNA damage trigger cGAS activation and how cGAS serves as a link from DNA damage to inflammation, cellular senescence, and cancer.
Introduction
Genomic instability, a major driving force of cancer and age-related diseases, poses an ominous threat to human health and longevity. Nevertheless, several stringent and intricate cellular programs maintain genome integrity and prevent cells from becoming malignant. Cells may return to normal function if the genetic lesions are successfully repaired, enter a state of permanent cell-cycle arrest known as senescence if the damage is persistent but tolerable, or undergo programmed cell death to destroy an intolerably damaged genome.
Although DNA damage response (DDR) was long thought to mainly regulate genome integrity and cell fates, accumulating evidence indicates that genomic instability also triggers inflammatory response (Fig. 1). In tissue culture systems, DNA damaging agents such as topoisomerase inhibitors and ionizing irradiation induce the expression of type I IFNs and other cytokines (Fenech and Morley, 1986; Schlegel et al., 1986; Coppé et al., 2008; Rodier et al., 2009; Brzostek-Racine et al., 2011; Fenech et al., 2011; Kondo et al., 2013; Ahn et al., 2014b; Lan et al., 2014; Härtlova et al., 2015; Xia et al., 2016a; Harding et al., 2017; Luthra et al., 2017). The degree of inflammatory gene induction by genomic DNA damage is usually lower than that induced by DNA transfection or viral infection. Nonetheless, cells that sustain nuclear DNA damage also become more resistant to viral infections (Mboko et al., 2012; Härtlova et al., 2015; Luthra et al., 2017). Consistent with these in vitro findings, in vivo studies revealed that chemotherapy (Sistigu et al., 2014) and radiation treatment (Burnette et al., 2011; Lim et al., 2012; Deng et al., 2014) induce type I IFN signaling in tumors to promote antitumor immunity.
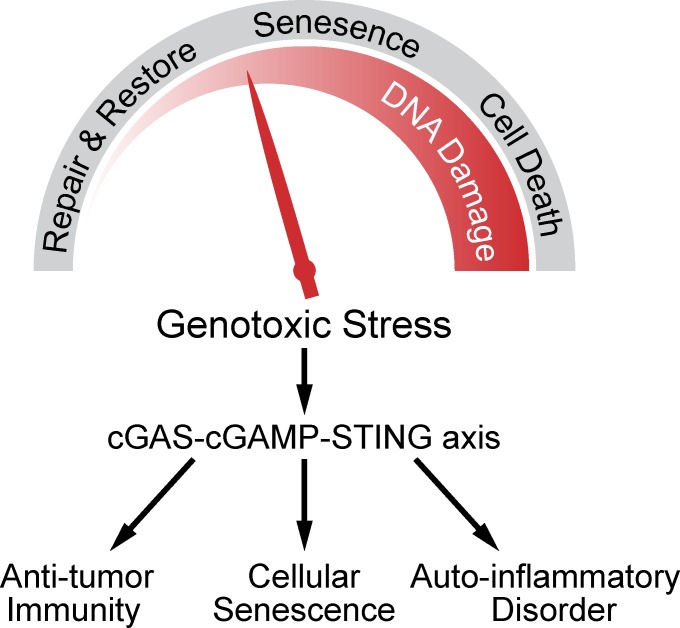
Figure 1.
Inflammatory response is another biological outcome of genomic instability. Genotoxic stress leads to DNA damage repair, cellular senescence, and cell death in a manner that depends on the severity of the DNA damage. The cGAS–cGAMP–STING pathway is activated by DNA damage to mediate antitumor immunity, senescence, and inflammatory responses.
In addition to inducing cytokines, DNA damage also enhances the expression of ligands of natural killer (NK) cells such as NKG2D ligands (Gasser et al., 2005; Lam et al., 2014). These surface proteins attract NKG2D-positive NK cells and activated CD8 T lymphocytes to target damaged cells for elimination by the immune system (Bauer et al., 1999). The expression of NKG2D ligands is likely a result of type I IFN induction by DNA damage (Zhang et al., 2008; Lam et al., 2014).
Recent studies have provided mechanistic insights into how DNA damage induces type I IFNs and other immune-regulatory cytokines (Erdal et al., 2017; Glück et al., 2017; Harding et al., 2017; Mackenzie et al., 2017; Yang et al., 2017). A cytosolic DNA sensing pathway has emerged as the major link between DNA damage and innate immunity (Fig. 2). DNA normally resides in the nucleus and mitochondria; hence, its presence in the cytoplasm serves as a danger-associated molecular pattern (DAMP) to trigger immune responses. Cyclic guanosine monophosphate (GMP)–adenosine monophosphate (AMP) synthase (cGAS) is the sensor that detects DNA as a DAMP and induces type I IFNs and other cytokines (Sun et al., 2013). DNA binds to cGAS in a sequence-independent manner; this binding induces a conformational change of the catalytic center of cGAS such that this enzyme can convert guanosine triphosphate (GTP) and ATP into the second messenger cyclic GMP-AMP (cGAMP; Wu et al., 2013). The cGAMP produced by cGAS contains two phosphodiester bonds: one between the 2’-hydroxyl group of GMP and 5′-phosphate of AMP and the other between the 3′-hydroxyl of AMP and 5′-phosphate of GMP (Ablasser et al., 2013; Diner et al., 2013; Gao et al., 2013a; Zhang et al., 2013). This cGAMP molecule, termed 2’3′-cGAMP, is an endogenous high-affinity ligand for the adaptor protein Stimulator of IFN Gene (STING, also known as MITA, MPYS, and ERIS; Ishikawa and Barber, 2008; Jin et al., 2008; Zhong et al., 2008; Sun et al., 2009).
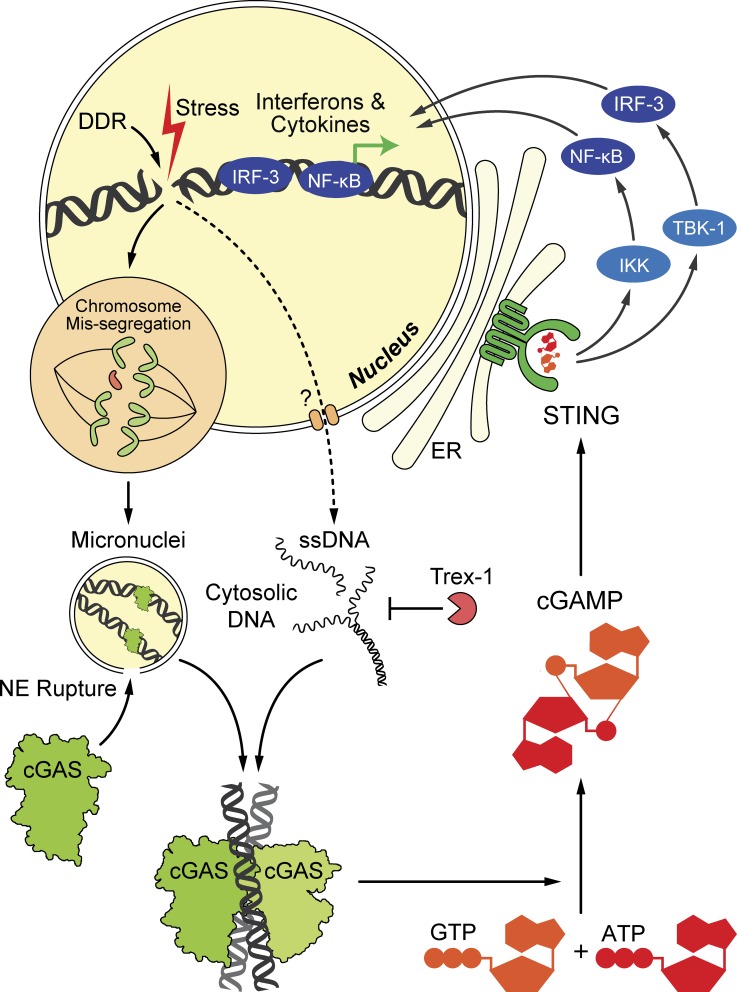
Figure 2.
The cGAS–cGAMP–STING pathway detects cytoplasmic DNA after DNA damage and activate type I IFNs and other cytokines. Like DDR, the immune response is induced by various forms of genotoxic stress, ranging from ionizing radiation, DNA-damaging drugs, oxidative stress, oncogenic signaling, telomere shortening, and chromosome missegregation to viral infections and activation of endogenous retroelements. Nuclear DNA damage can generate cytoplasmic DNA in two possible ways. First, certain genomic damage causes chromosome to missegregate in subsequent cell division; the chromosome failing to partition into the new nuclei will form micronuclei. When the nuclear evelope (NE) of micronuclei ruptures, the DNA content is exposed to cGAS surveillance. Second, nuclear DNA damage can also generate ssDNA in the cytoplasm in a less understood process (dashed line and question mark). Such cytoplasmic DNA is degraded by Trex1, the loss of which leads to cGAS activation. Active cGAS dimerizes to synthesize cGAMP from GTP and ATP. cGAMP acts as a second messenger to activate STING on the ER surface. STING then activates transcription factors IRF3 and NF-κB via kinases TBK1 and IKK, respectively. IRF3 and NF-κB translocate into the nucleus to elicit the expression of IFNs and other cytokines. Damage to mitochondria or mitochondrial DNA can also lead to accumulation of mitochondrial DNA in the cytosol, resulting in cGAS activation and sterile inflammation (not depicted).
STING forms a transmembrane homodimer that localizes to the ER and binds cyclic dinucleotides (CDNs), such as cGAMP and the bacterial second messengers c-di-GMP and c-di-AMP (Burdette et al., 2011; Wu et al., 2013). cGAMP binding induces a conformational change in STING (Gao et al., 2013b; Zhang et al., 2013), which subsequently translocates from the ER to the Golgi apparatus (Ishikawa and Barber, 2008; Saitoh et al., 2009). This process is thought to liberate the STING carboxyl terminus to subsequently recruit and activate TANK-binding kinase 1 (TBK1) and IFN regulatory factor 3 (IRF3) via a phosphorylation-dependent mechanism (Tanaka and Chen, 2012; Liu et al., 2015). STING also activates NF-κB, which functions together with IRF3 to turn on the transcription of type I IFNs and other cytokines (Fig. 2).
The molecular details of the cGAS–STING pathway and its pivotal roles in eliciting effective immunity against various microbial pathogens have been extensively reviewed elsewhere (Xiao and Fitzgerald, 2013; Cai et al., 2014; Wu and Chen, 2014; Chen et al., 2016b; Ma and Damania, 2016; Tao et al., 2016; Kato et al., 2017). In this review, we primarily focus on its emerging function in mediating DNA damage–induced inflammatory responses. We further discuss recent findings on how the cGAS–STING axis contributes to the outcome of autoinflammatory diseases, senescence, and cancer.
cGAS senses cytoplasmic DNA as a consequence of nuclear DNA damage
DNA damage in the nucleus results in the accumulation of cytoplasmic DNA, notably in the form of micronuclei (Fig. 2). Micronuclei are small, DNA-containing organelles that resemble satellites to the primary nuclei (Fenech et al., 2011). They are products of chromosome damage as a result of genotoxic stress and chromosome missegregation in subsequent cell division. For instance, centromere-deficient chromosome fragments may result from the error-prone nonhomologous end-joining (NHEJ) repair and are unable to segregate normally, thereby forming micronuclei outside the newly formed nuclei. Similarly, whole chromosomes left behind by the spindle because of centromere hypomethylation or kinetochore dysfunction also end up as micronuclei after mitosis (Fenech et al., 2011). Since their early identification as “Howell-Jolly bodies” more than a century ago, micronuclei have been strongly associated with DNA damage and were adopted as a sensitive biomarker of genotoxicity (Fenech and Morley, 1985, 1986; Schlegel et al., 1986). The cytokinesis-block micronucleus (CBMN) assay, which scores the frequency of micronuclei, has been a widely used method to assess the genotoxicity of different chemical or radioactive mutagens (Fenech, 2007).
Micronuclei are originally formed with a nuclear envelope (NE), but more than half the micronuclei lose compartmentalization after mitosis as their NE ruptures (Hatch et al., 2013). NE breaks were found to negatively correlate with lamina integrity because lamin networks critically contribute to NE structural resilience (Vargas et al., 2012; Hatch et al., 2013). It was recently reported that NE of the nucleus can be rapidly repaired by the endosomal sorting complexes required for transport III (ESCRT-III) complex (Denais et al., 2016; Raab et al., 2016), and this mechanism may provide similar protection to NEs of micronuclei. These observations imply that micronuclei NEs may have compromised lamina function or membrane repair capacity so that it ruptures easily, although the detailed mechanisms underlying NE rupture await further study. A consequence of micronuclei rupture is that chromosomal DNA become accessible to cGAS (Bartsch et al., 2017; Dou et al., 2017; Glück et al., 2017; Harding et al., 2017; Mackenzie et al., 2017; Yang et al., 2017).
DNA damage also induces the accumulation of cytoplasmic DNA into “speckles,” which are less aggregated than micronuclei (Ahn et al., 2014b; Lan et al., 2014; Härtlova et al., 2015; Shen et al., 2015; Erdal et al., 2017). Cytoplasmic DNA speckles contain mostly single-stranded DNA (ssDNA; Härtlova et al., 2015; Shen et al., 2015; Erdal et al., 2017), but may also contain small amounts of double-stranded DNA (dsDNA; Shen et al., 2015). ssDNA in general induces very little type I IFNs compared with dsDNA (Ishii et al., 2006) because cGAS doesn’t bind ssDNA. However, stem-loop–forming ssDNA sequences in HIV reverse transcripts were shown to have a high potency of cGAS activation (Herzner et al., 2015). Accumulation of ssDNA in cells deficient in the Trex1 exonuclease is also associated with chronic IFN induction and autoinflammatory phenotypes (Yang et al., 2007; Stetson et al., 2008; Gao et al., 2015; Wolf et al., 2016). Among the ssDNA species generated after DNA damage, those that form double-stranded secondary structures may activate cGAS. Trex1 has also been shown to degrade nicked dsDNA (Chowdhury et al., 2006), and such DNA may accumulate in Trex1-deficient cells to activate cGAS.
Little is known about how DNA damage leads to DNA accumulation in the cytosol. MUS81 is an endonuclease that may play a role in this process. The MUS81-EME1 complex belongs to the family of structure-specific endonucleases that resolve interstrand DNA structures such as stalled replication forks and Holliday junctions (Dehé and Gaillard, 2017). MUS81 is required for genomic integrity and tumor suppression, as mus81−/− mice display increased chromosomal aberrations in T cells and are predisposed to lymphomas and other cancers (McPherson et al., 2004). It was found that MUS81 is required for the generation of cytoplasmic DNA in prostate cancer cells (Ho et al., 2016). The amount of cytoplasmic DNA positively correlates with MUS81 protein level and the number of MUS81 nuclear foci, suggesting that MUS81 is engaged in the process that converts nuclear DNA into cytoplasmic forms. Consistently, MUS81 promotes type I IFNs and thus immune rejection of prostate tumor (Ho et al., 2016). In contrast, however, it was also reported that MUS81 is a negative regulator of spontaneous type I IFN production (Laguette et al., 2014). Thus, the exact roles of MUS81 in regulating cytoplasmic DNA sensing remain to be elucidated. A recent study reported that extracellular telomere repeat DNA (ECTR), generated by the alternative lengthening of telomeres (ALT) mechanism, accumulates in the cytoplasm of ALT cell lines and activates IFN-β via the cGAS–STING pathway (Chen et al., 2017). As MUS81 is necessary for ALT (Zeng et al., 2009), the cytoplasmic DNA derived from MUS81 activity may include ECTR DNA.
Multiple studies have recently demonstrated that induction of cytokines by DNA damage is mediated through the cGAS–STING pathway (Lan et al., 2014; Härtlova et al., 2015; Bartsch et al., 2017; Erdal et al., 2017; Glück et al., 2017; Harding et al., 2017; Mackenzie et al., 2017; Yang et al., 2017). After DNA damage, cGAS is recruited to micronuclei (Glück et al., 2017; Harding et al., 2017; Mackenzie et al., 2017; Yang et al., 2017). NE rupture of micronuclei appears to precede cGAS recruitment (Harding et al., 2017; Mackenzie et al., 2017). cGAS in the micronuclei is presumably activated by the chromosomal DNA fragments to synthesize cGAMP, but direct evidence is still lacking. Interestingly, although cGAS usually localizes in the cytoplasm of nondividing cells, a majority of cGAS associates with chromatin after nuclear membrane breakdown during mitosis (Harding et al., 2017; Mackenzie et al., 2017; Yang et al., 2017). Because there is no evidence of cGAS activation during cell division, it will be interesting to determine how cGAS activity is modulated during chromatin association. Similarly, it remains unclear what form of chromatin DNA in the micronuclei may activate cGAS. Because cGAS-associated micronuclei stain positive for phosphorylated γ-H2A, a DNA damage marker (Mackenzie et al., 2017; Yang et al., 2017), DNA damage itself may contribute to cGAS sensing. In this regard, a recent proteomic study showed that cGAS is associated with a ribonuclear complex dubbed HEXIM1-DNA–protein kinase (PK)–paraspeckle components–ribonucleoprotein complex (HDP–RNP) that contains DNA-PK (Morchikh et al., 2017). It will be interesting to determine whether this complex plays a role in the regulation of cGAS activity by DNA damage in the primary nucleus and in micronuclei.
Together, these new findings have demonstrated that the cGAS–cGAMP–STING pathway connects genotoxic stress to cytokine expression by sensing cytoplasmic DNA as a common consequence of nuclear DNA damage.
The cGAS–cGAMP–STING axis connects DNA damage to autoinflammatory diseases
Genetic deficiencies that compromise DDR functions also induce cytokines and lead to autoinflammatory diseases. An underlying mechanism is the aberrant activation of the cGAS–STING pathway. For instance, Ataxia-Telangiectasia Mutated (ATM) is a critical kinase for DDR initiation. ATM mutations result in dysfunctional V(D)J recombination and ultimately the immunodeficiency syndrome Ataxia-Telangiectasia (A-T), because V(D)J recombination is essential for antibody production and T cell development. A-T patients are also prone to autoinflammation. Elevated type I IFN signaling was associated with ATM deficiency in cultured cells (Siddoo-Atwal et al., 1996; Sugihara et al., 2011), as well as in sera of human A-T patients (Härtlova et al., 2015). The autoinflammatory phenotype of A-T is abrogated in atm−/− sting−/− double knockout mice and significantly reduced after cGAS knockdown, indicating that the cGAS–STING pathway plays an essential role in this disease (Härtlova et al., 2015).
Aicardi-Goutières syndrome (AGS) is an inheritable neurological disease that leads to microcephaly, intellectual retardation, and childhood death (Crow and Rehwinkel, 2009). This debilitating disease is driven by chronic IFN signaling caused by recessive mutation in one of a few genes involved in nucleic acid metabolism, such as trex1, rnaseH2a, rnaseH2b, and samhd1 (Crow and Rehwinkel, 2009). For instance, the trex1 gene encodes a 3′–5′ exonuclease that salvages DNA fragments generated from nuclear DNA damage (Yang et al., 2007; Erdal et al., 2017). Similar to human AGS patients with trex1 deficiency, trex1−/− mice also develop life-shortening inflammatory phenotypes (Crow et al., 2006a), which can be rescued by genetic ablation of either cGas or sting (Gall et al., 2012; Ahn et al., 2014a; Gao et al., 2015; Gray et al., 2015). In tissues from trex1−/− mice, elevated cGAMP was detected by mass spectrometry; cGAMP detection in these mice was also dependent on wild-type cGAS, directly demonstrating that cGAS is activated in trex1−/− mice (Gao et al., 2015).
RNaseH2 is a nuclear enzyme responsible for removing ribonucleotides that are misincorporated into genomic DNA (Reijns et al., 2012). Genetic defects in any of the three subunits of RNaseH2 leads to chronic DNA damage, elevated type I IFNs and perinatal lethality in mice (Hiller et al., 2012; Reijns et al., 2012; Mackenzie et al., 2016; Pokatayev et al., 2016), and the AGS phenotypes in humans (Crow et al., 2006b). In RnaseH2b−/− cells, persistent genome instability promotes the formation of micronuclei, which recruit cGAS and activate type I IFN signaling (Bartsch et al., 2017; Mackenzie et al., 2017). Genetic removal of cGAS or STING can experimentally reverse the ensuing inflammation and autoimmune phenotypes in RNaseH2-deficient mice (Mackenzie et al., 2016; Pokatayev et al., 2016). AGS is also caused by mutations in human SAMHD1 (Rice et al., 2009), a deoxyribonucleotide triphosphate triphosphatase that balances nuclear levels of deoxyribonucleotide triphosphates (Franzolin et al., 2013) and genome stability (Kretschmer et al., 2015). Although SAMHD1-deficient mice do not show any overt autoinflammatory phenotype, their myeloid cells express elevated levels of IFN-stimulated genes that are dependent on cGAS and STING (Maelfait et al., 2016). Together, these observations demonstrate the central role of the DNA-sensing pathway in mediating DNA damage–induced inflammatory diseases.
ERCC1-XPF is a structure-specific endonuclease critical for multiple genome maintenance mechanisms such as nucleotide excision repair and dsDNA break repair. Mice lacking ERCC1 display increased genome instability and chronic expression of inflammatory cytokines, leading to premature aging and early death (Melton et al., 1998; Niedernhofer et al., 2006; Karakasilioti et al., 2013). Notably, ERCC1 deficiency in humans causes the severe developmental disorder Cerebro-Oculo-Facio-Skeletal syndrome (COFS). Key features of the COFS syndrome, such as microcephaly with calcification, are similar to those of AGS and the Cockayne syndrome, the latter being caused by mutations in ercc8 or ercc6 (Troelstra et al., 1992; Henning et al., 1995). While the aging phenotype of ERCC1 deficiency requires NF-κB in mice (Tilstra et al., 2012), it remains to be determined whether aberrant cGAS–STING activity may also underpin these inheritable disorders.
Recent studies have shown that cGAS activation by self DNA is also linked to more common and complex diseases such as myocardial infarction (MI) and age-related macular degeneration (AMD). MI is known to drive inflammation and exacerbate lethality. The massive death of cardiomyocytes activates in heart macrophages strong type I IFN signaling that is mediated by the cGAS–STING pathway (King et al., 2017; Cao et al., 2018). Mice lacking components of the pathway, including cGAS, IRF3, and IFNAR1, had significantly improved early survival in experimental MI model, compared with wild-type animals. Similarly, cGAS also facilitates inflammation in the eyes (Kerur et al., 2017). In an Alu-RNA–induced model of macular degeneration in mice, retinal pigment epithelium (RPE) degeneration was shown to rely on caspase 4/11 and Gasdermin D of the inflammasome pathway, as well as cGAS, STING, IRF3, and IFNAR1. Although it is unclear how the cGAS–STING pathway leads to inflammasome activation in RPE cells, it has been recently reported that STING-mediated cell death can activate the NLRP3 inflammasome in some human myeloid cells (Gaidt et al., 2017).
While accumulation of cytosolic DNA through nuclear DNA damage, loss-of-function mutations of DNA degrading enzymes, or tissue damage leads to autoinflammatory diseases, gain-of-function mutations in STING are sufficient to cause an autoinflammatory disorder termed STING-associated vasculopathy with onset in infancy (SAVI; Liu et al., 2014; Melki et al., 2017). These mutations lead to constitutive STING activation and type I IFN signaling. Interestingly, the resulting vascular and pulmonary syndrome of SAVI resemble those of some DDR deficiency–associated autoinflammation, such as A-T and Artemis deficiency (Gul et al., 2017), consistent with the notion that these genetic disorders affect the same signaling pathway.
Besides damaged nuclear DNA, cGAS also responds to other self-DNA that mislocalizes to the cytoplasm and drives autoinflammation. Haplodeficiency of TFAM (transcription factor A, mitochondria), a protein that packages mitochondria DNA (mtDNA) into nucleoid, was reported to promote releases of mtDNA into the cytosol, which activate cGAS to confer IFN-mediated viral resistance (West et al., 2015). In response to apoptotic stimuli, mitochondria not only releases cytochrome c to activate caspases to execute apoptosis, but also mtDNA to the cytosol, which could have otherwise activated cGAS to trigger inflammation if there were no concurrent activation of caspases (Rongvaux et al., 2014; White et al., 2014). Through an unknown mechanism caspase activation in apoptotic cells inactivate the cGAS–STING pathway, thereby suppressing sterile inflammation. DNaseII is a lysosomal endonuclease that is responsible for degrading phagocytosed exogenous DNA from dead cells or expelled nuclei of developing red blood cells. Mice lacking DNaseII are embryonically lethal as a result of type I IFN–driven cell death (Kawane et al., 2003), which can be partially rescued by genetically ablating type I IFN receptor (Yoshida et al., 2005) and nearly completely rescued by deleting STING (Ahn et al., 2012) or cGAS (Gao et al., 2015). In DNaseII-deficient but not normal fetal livers, cGAMP can be detected by mass spectrometry (Gao et al., 2015). Therefore, cGAS also senses phagocytosed DNA that are inadequately digested in the lysosome. Human patients with loss of function mutations of DNaseII have recently been identified, and these patients exhibit symptoms of type I interferonopathy, including tissue inflammation and elevated anti-DNA antibodies (Rodero et al., 2017). Together, these findings strongly support the central role of the cGAS–cGAMP–STING pathway in the pathogenesis of a variety of type I IFN–mediated monogenic diseases. Therefore, components of this pathway represent promising targets for developing pharmacological intervention for AGS and other autoinflammatory disorders.
cGAS is essential for DNA damage–induced cellular senescence
Cellular senescence is a state of irreversible cell cycle arrest, induced by a variety of external or internal stress such as telomere shortening, oxidative damage, and oncogenic signaling. Although the causes of and phenotypes generated by cellular senescence are manifold, persistent DDR is thought to be the common mechanism that is critical for the establishment and maintenance of senescence phenotypes (d’Adda di Fagagna, 2008). Specifically, DDR activates the p53–p21 and p16 INK4a–Rb pathways to block the cell cycle and implement senescence programs.
Recent research has provided strong evidence that cGAS also has an essential role in promoting cellular senescence (Dou et al., 2017; Glück et al., 2017; Yang et al., 2017). The initial clue came from spontaneous immortalization of murine embryonic fibroblasts (MEFs). When primary MEFs are serially passaged, the majority of cells gradually senesce, and only a small fraction overcome the growth crisis and become immortalized. Compared with wild-type MEFs, the cGas−/− MEFs are immortalized more rapidly, suggesting that cGAS inhibits cell proliferation (Glück et al., 2017; Yang et al., 2017). Oxidative DNA damage was previously shown to limit spontaneous immortalization (Parrinello et al., 2003). The antiproliferative effect of cGAS is ameliorated at lower oxygen levels, suggesting cGAS functions downstream of oxidative DNA damage (Glück et al., 2017). Consistent with its role in limiting cell replication, cGAS is also required for the expression of senescence markers, such as p16INK4a and senescence-associated β-galactosidase (SA-β-Gal) during MEF immortalization. Therefore, cGAS plays an indispensable role in cellular senescence in response to oxidative stress. Similarly, irradiation, DNA-damaging drugs, or oncogene activation all induce cellular senescence in a cGAS-dependent manner. The activation of oncogenes, such as NRasV12, leads to DNA hyperreplication, increased replication errors, and initiation of the DNA damage response (Di Micco et al., 2006). In cells under these genotoxic stresses, cGAS is recruited to micronuclei. In oncogene-induced senescent cells, cGAMP is detectable by LC-MS (Dou et al., 2017). Therefore, cGAS senses micronuclei as the result of genomic DNA damage to promote senescent phenotypes.
How does the cGAS–STING pathway regulate cellular senescence, given that it is not an integral component of either the p53–p21 or the p16 INK4a–Rb pathway? A mechanism is likely through the senescence-associated secretory phenotype (SASP), which refers to the phenomenon that senescent cells produce and release a range of cytokines, chemokines, and proteases to the extracellular milieu to modulate senescent cells themselves and their microenvironment (Kuilman and Peeper, 2009). After various forms of senescing stimuli, such as oxidation, radiation, chemotherapeutic drugs, or mitogenic stress, cells produce in a cGAS-dependent manner a range of cytokines and chemokines, such as IFN-β, IL-1β, IL-6, and IL-8 (Dou et al., 2017; Glück et al., 2017; Yang et al., 2017). Some of these cytokines, namely IL-8 and IL-6, are known to feedback to the secreting cells to reinforce senescence signaling (Acosta et al., 2008; Kuilman et al., 2008). Type I IFNs are also prosenescence and antiproliferative, as they induce DNA damage and elevate the p53 level (Takaoka et al., 2003; Moiseeva et al., 2006). Therefore, the cGAS–STING pathway provides a critical paracrine signal that is necessary for sustaining cellular senescence.
Besides promoting senescence cell-autonomously, SASP can signal to the immune system and modulate the tissue microenvironment. Previously, it was demonstrated in a liver cancer model that senescent liver cells attract via SASP a variety of immune cells, specifically NK lymphocytes and neutrophils, to clear tumor cells (Xue et al., 2007). In a liver fibrosis model, SASP was shown to recruit and activate NK cells to eliminate senescent cells and resolve fibrosis (Krizhanovsky et al., 2008). The cGAS–STING pathway is also required for SASP-mediated antitumor effect. Dou et al. used an NRasV12-expressing vector to induce liver tumors and found in the STING null mice attenuated infiltration of immune cells, reduced clearance of senescent cells at the early stage, and increased tumor growth at later stages (Dou et al., 2017). Therefore, senescence can promote immune surveillance to suppress malignancy via SASP.
Although evidence cited above illustrates SASP as an antitumor component of senescence, it should be noted that SASP can also promote tumorigenesis (Coppé et al., 2010). SASP from senescent fibroblasts can strongly induce epithelium cells to proliferate, transform, and metastasize (Krtolica et al., 2001; Coppé et al., 2008). Particularly, IL-6 and IL-8 can induce invasiveness of epithelium cells (Coppé et al., 2008); matrix metalloprotease can remodel tissue microenvironment to increase the availability of growth factors (Liu and Hornsby, 2007); and vasculature endothelium growth factor (VEGF) can promote endothelium cell migration and tumor angiogenesis (Coppé et al., 2006). Therefore, SASP can mediate both positive and negative effects of senescence on cancer. Such dichotomous functions of senescence and the role of the cGAS–STING pathway in these functions should be further clarified in future studies.
Senescence also regulates stem cell functions and aging. Previously, type I IFN signaling was shown to mediate a variety of stem cell defects in telomerase-deficient mice as the result of persistent DNA damage (Yu et al., 2015). Deletion of the IFN α/β receptor 1 (IFNAR1) rescues the senescence phenotypes in Terc−/− mice. As the role of IFN-β in stress-induced senescence is reminiscent of the roles of cGAS and STING (Katlinskaya et al., 2016), it would be interesting to investigate the cGAS–STING pathway in the process of stem cell senescence under stress conditions. Recent evidence shows that STING null mice display remarkably less hair graying months after irradiation (Dou et al., 2017). Radiation-induced genomic damage was previously shown to cause a loss of renewal of melanocyte stem cells (Inomata et al., 2009); therefore, STING-mediated SASP may contribute to regulating these stem cells. Remarkably, experimental elimination of p16INK4a-positive senescent cells in mice can attenuate aging phenotypes in organs such as the kidney and the heart, significantly prolonging mouse life span and health span (Baker et al., 2016). Given that the cGAS–STING pathway plays critical roles in cellular senescence, inhibitors of cGAS or STING may offer similar benefits to treat senescence and age-related diseases.
DNA damage activates the cGAS–cGAMP–STING pathway in cancer
Crucial links between DNA damage and cancer have long been recognized. On one hand, DNA damage can promote antitumor immunity in both natural rejection and therapies. On the other hand, genome instability is a hallmark as well as an important driving force of cancer. As the cGAS–cGAMP–STING pathway critically determines the immunological outcomes of DNA damage, it also plays prominent roles in both aspects of cancer.
Effective antitumor immunity relies on cross-presentation of tumor antigens by APCs to CD8 T lymphocytes. The activation of APCs requires type I IFN signaling initiated by innate immune sensors (Diamond et al., 2011; Fuertes et al., 2011). Recent evidence has shown that the cGAS–STING pathway, activated in APCs by cytosolic DNA, provides a critical source of such a priming signal. It was shown that the STING-deficient mice fail to reject the growth of inoculated tumor cells spontaneously (Woo et al., 2014) after local radiation therapy (Deng et al., 2014) or after immune checkpoint blockades using antibodies against PD-L1 (Wang et al., 2017) or CD47 (Xu et al., 2017). Similarly, cGas−/− mice also show defective tumor rejection (Wang et al., 2017; Xu et al., 2017). In line with these findings, the cGAS–STING pathway in dendritic cells (DCs) is required to prime DCs in coculture with tumor cells for cross-presentation to activate CD8 T cells (Deng et al., 2014; Wang et al., 2017; Xu et al., 2017). Consistent with tumor suppression by the cGAS–STING pathway, treatment with exogenous cGAMP or other STING agonists can also enhance immunity and promote tumor regression (Li et al., 2013; Chandra et al., 2014; Corrales et al., 2015; Demaria et al., 2015). Importantly, cGAMP therapy can further enhance the antitumor effect of ionizing irradiation (Deng et al., 2014) or chemotherapy (Li et al., 2016) and synergize with immune checkpoint blockade (Demaria et al., 2015; Wang et al., 2017).
How is cGAS activated in APCs? Cell death is known to occur commonly in tumors as a consequence of genomic instability, hypoxia, or other tumor-associated stresses (Lowe and Lin, 2000), providing ample DNA that may activate cGAS in APCs (Xu et al., 2017). As infiltrating APCs sample tumor tissue by phagocytosis, they also pick up tumor DNA. Through an undefined mechanism, tumor DNA may escape phagosomes and enter the cytoplasm to activate cGAS and STING (Woo et al., 2014). Together, the data suggest that cytoplasmic DNA sensing is an early and critical step in APC activation that determines the outcome of antitumor immunity.
Besides APCs, the cGAS pathway is also active in some tumor cells to promote antitumor immunity. The activity is likely triggered by increased genome instability in tumor cells. For instance, genome-derived DNA was shown to accumulate in the cytoplasm of prostate cancer cells as the result of nuclear MUS81 activity, inducing STING-dependent type I IFNs and tumor rejection (Ho et al., 2016). Radiation further promotes DNA damage and DNA sensing in tumor cells. It was shown that irradiated B16 cells can be used as a cancer vaccine to promote systematic tumor rejection in combination with immune checkpoint blockade. However, STING deficiency in the irradiated tumor cells was found to abolish the efficacy of this combination therapy (Harding et al., 2017). Therefore, DNA damage intrinsically engages the cGAS–STING pathway in tumor cells to prevent malignancy. In addition to elevating antitumor immunity, this pathway is also implicated in shaping the tumor microenvironment. Sting−/− mice are more susceptible to colitis-associated cancer induced by chronic DNA damage and inflammation (Zhu et al., 2014; Ahn et al., 2015). Persistent activation of the cGAS–STING pathway also lead to cell death, adding another layer of resistance to tumorigenesis (Li et al., 2016; Tang et al., 2016; Gaidt et al., 2017; Gulen et al., 2017; Larkin et al., 2017). For example, 5,6-dimethylxanthenon-4-acetic acid (DMXAA), a potent agonist of mouse STING (Conlon et al., 2013), can effectively induce tumor hemorrhagic necrosis and promote tumor regression in mice (Zwi et al., 1994). Moreover, cGAMP treatment can also trigger the activation of NLRP3 inflammasome (Gaidt et al., 2017; Swanson et al., 2017). In summary, the cGAS–STING pathway acts as an intrinsic barrier to tumorigenesis by linking DNA damage to several antitumor mechanisms: immune surveillance, cellular senescence, and cell death. Therefore, this pathway represents an attractive pharmaceutical target to harness these intrinsic antitumor mechanisms for cancer therapy.
In response to the tumor suppressive functions of cGAS and STING, cancer cells frequently adapt to down-regulate these proteins to promote malignancy (Xia et al., 2016a,b). Likewise, oncogenic DNA viruses, such as human papilloma virus and adenovirus, evolved specific viral proteins to directly antagonize STING (Lau et al., 2015). As a counter measure to these immune evasive mechanisms, engineered oncolytic viruses, which preferentially replicate in tumor cells defective in type I IFN signaling, can be exploited to treat late-stage malignant cancer (Xia et al., 2016b; de Queiroz et al., 2017).
On the other side of the dichotomous relationship between DNA damage and cancer are protumor functions of the cGAS–STING pathway induced by genotoxic stress. 7,12-dimethylbenz(a)anthracene (DMBA) is a strong DNA-damaging agent that induces inflammation-driven skin carcinogenesis. As DMBA activates the cGAS–STING pathway, DMBA-treated sting−/− mice, unlike other cancer models, are actually more resistant to the growth of DMBA-induced skin cancer (Ahn et al., 2014b). Adoptive transfer assays implicated STING in both hematopoietic and nonhematopoietic cells for tumorigenesis in the DMBA model. Another study shows that cancer cells metastasizing to the brain can also use the cGAS–STING pathway to promote tumor progression (Chen et al., 2016a). These cancer cells transfer cGAMP via gap junctions into astrocytes; subsequent paracrine signaling from astrocytes feedback to cancer cells to promote malignancy (Chen et al., 2016a). It is possible that the protumor effect of the cGAS–STING pathway may be linked in part to inflammatory cytokines, such as TNF-α. Although TNF-α can induce potent necrosis in tumors, it is also an important driver of inflammatory carcinogenesis (Balkwill, 2009). TNF-α can promote epithelium cells to survive under stress, induce blood vessel growth (Leibovich et al., 1987), and enhance metastasis (Malik et al., 1990; Orosz et al., 1993). tnf-α−/− mice were shown to be resistant to carcinogen-induced skin cancer (Moore et al., 1999). Therefore, an active cGAS–STING axis can exacerbate certain cancers. Such findings add a note of caution to the development of anticancer strategies that aim to activate the cGAS–STING pathway.
In summary, genomic instability engages the cGAS–cGAMP–STING pathway on both sides of cancer. On the bright side, this pathway is instrumental for antitumor immunity and is an intrinsic barrier in tumor cells that blocks malignancy. On the dark side, this pathway can promote inflammation-driven carcinogenesis and metastasis. As mounting evidence suggests that the cGAS–cGAMP–STING pathway makes fundamental contributions to at least three major cancer therapies, radiation therapy, chemotherapy, and immunotherapy, it represents a promising pharmaceutical target to enhance our current treatment regimens. A major task in cancer therapies is to develop new approaches to maximize the bright side while minimizing the dark side of the contributions of the cGAS pathway in cancer.
Closing remarks and future directions
It is now well established that cGAS is a major sensor of microbial pathogens that contain DNA or generate DNA in their life cycles. New studies have illuminated an equally critical role of cGAS in the surveillance of self-DNA that mislocalizes to the cytoplasm under pathological conditions. Although decades of research have led to a good understanding of how genotoxic stress and genomic instability regulate DNA damage repair, cell cycle checkpoints, and programmed cell death, the mechanism by which DNA damage leads to inflammation and autoimmunity was not well understood until recently. In retrospect, it is logical that cGAS is well positioned to connect nuclear DNA damage to immune responses because cGAS can be activated by any dsDNA that enters the cytoplasm. It is interesting to note that in addition to chemical and radiation damage to DNA, many other biological processes such as oncogene activation, telomere shortening, and cell division errors all impinge on DNA. Although NF-κB is well known to play an important role in mediating inflammatory responses to DNA damage (Janssens and Tschopp, 2006), the link between DNA and NF-κB was not clear. Now cGAS has emerged as an important link between DNA damage and activation of NF-κB, IRF3, and likely other transcription factors.
Many important questions remain. For example, it is not clear how cytoplasmic DNA is generated from damaged nuclear genome. Although MUS81 has been shown to regulate generation of cytoplasmic DNA from genomic DNA (Ho et al., 2016), additional factors may be required for this process. The exact cellular mechanism responsible for transporting genomic DNA fragments from the nucleus to the cytoplasm remains to be elucidated. Also unclear is how GAS is activated in micronuclei. Although cGAS was found to associate with the chromatin during mitosis (Yang et al., 2017), there is no evidence that cGAS is activated during cell cycle transition. Thus, it is important to determine how cGAS is kept inactive when it is associated with the chromosome during mitosis and how cGAS is activated by chromatin fragments within the micronuclei. How cGAS is activated in APCs by tumor DNA also requires further investigation. After APCs phagocytose tumor cells, tumor antigens are delivered from the phagosome to the cytosol for proteasomal processing and cross-presentation of antigenic peptides. It is possible that tumor cell DNA enters the cytosol along with tumor protein antigens, but details regarding the cytosolic delivery of tumor proteins and DNA remain to be delineated. As tumor DNA is also chromatinized, it is unknown whether it requires any further processing to become an active cGAS ligand.
The knowledge gained from understanding the cGAS–STING pathway in DDRs may be translated into innovative therapies for a variety of human diseases. Antagonists of cGAS or STING are potentially useful in blocking uncontrolled cytokine expression that leads to inflammation and autoimmune diseases. Because these antagonists may inhibit cellular senescence, they may also be used for treating senescence-associated diseases. Several attempts have been reported to find compounds that inhibit cGAS. So far the reported molecules are active against cGAS in vitro, but haven’t demonstrated desirable potency in vivo (An et al., 2015; Hall et al., 2017; Vincent et al., 2017). A concern about using antagonists of cGAS or STING in treatments of inflammation or senescence-associated diseases is a potential increase of susceptibility to infectious diseases as well as cancer. Hopefully, redundancy in the human immune system and selection of appropriate doses of drugs that partially inhibit the cGAS–STING pathway would allow these drugs to provide therapeutic benefits to the patients without compromising their immunity to infections and tumorigenesis.
Agonists of the cGAS–STING pathway such as cGAMP and its analogues, on the other hand, can be applied to enhance antitumor immunity in conjunction with other cancer therapies. These small molecules can be further improved by preventing hydrolysis of the phosphodiester bonds (Li et al., 2014). Several STING agonists have already entered clinical trials in human, and more potent and selective agonists of STING are expected in the next few years. Although these STING agonists will strongly enhance antitumor responses, including in those “cold” tumors that are so far refractory to immune checkpoint blockade, a major concern of using these compounds systemically is the cytokine “storm,” the acute production of multiple inflammatory cytokines that could cause severe toxicity and even death. Thus, it remains to be seen whether there is a sufficient therapeutic window that allows STING agonists to be used at certain doses to provide efficacious antitumor activity while minimizing immunotoxicity to patients. Innovative approaches, such as nanoparticles (Hanson et al., 2015; Luo et al., 2017; Wilson et al., 2018), viral particles (Bridgeman et al., 2015; Gentili et al., 2015), or targeted deliveries, could allow precise delivery of STING agonists to tumors to generate localized and specific antitumor immune responses and significantly improve the therapeutic window of using these compounds for cancer immunotherapies.
Acknowledgments
We thank Minghao Li and Aline McKenzie for their help in manuscript editing and Jose Cabrera for graphic assistance.
Work in the Chen laboratory is supported by grants from the Welch Foundation (grant I-1389), Cancer Prevention and Research Institute of Texas (grants RP120718 and RP150498), and Lupus Research Alliance (grant LRI-2014). Tuo Li is supported by the Postdoctoral Fellowship from Cancer Research Institute. Zhijian J. Chen is an Investigator of the Howard Hughes Medical Institute.
The authors declare no competing financial interests.
References
- Ablasser A., Goldeck M., Cavlar T., Deimling T., Witte G., Röhl I., Hopfner K.P., Ludwig J., and Hornung V.. 2013. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 498:380–384. 10.1038/nature12306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acosta J.C., O’Loghlen A., Banito A., Guijarro M.V., Augert A., Raguz S., Fumagalli M., Da Costa M., Brown C., Popov N., et al. 2008. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell. 133:1006–1018. 10.1016/j.cell.2008.03.038 [DOI] [PubMed] [Google Scholar]
- Ahn J., Gutman D., Saijo S., and Barber G.N.. 2012. STING manifests self DNA-dependent inflammatory disease. Proc. Natl. Acad. Sci. USA. 109:19386–19391. 10.1073/pnas.1215006109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J., Ruiz P., and Barber G.N.. 2014a Intrinsic self-DNA triggers inflammatory disease dependent on STING. J. Immunol. 193:4634–4642. 10.4049/jimmunol.1401337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J., Xia T., Konno H., Konno K., Ruiz P., and Barber G.N.. 2014b Inflammation-driven carcinogenesis is mediated through STING. Nat. Commun. 5:5166 10.1038/ncomms6166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J., Konno H., and Barber G.N.. 2015. Diverse roles of STING-dependent signaling on the development of cancer. Oncogene. 34:5302–5308. 10.1038/onc.2014.457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An J., Woodward J.J., Sasaki T., Minie M., and Elkon K.B.. 2015. Cutting edge: Antimalarial drugs inhibit IFN-β production through blockade of cyclic GMP-AMP synthase-DNA interaction. J. Immunol. 194:4089–4093. 10.4049/jimmunol.1402793 [DOI] [PubMed] [Google Scholar]
- Baker D.J., Childs B.G., Durik M., Wijers M.E., Sieben C.J., Zhong J., Saltness R.A., Jeganathan K.B., Verzosa G.C., Pezeshki A., et al. 2016. Naturally occurring p16(Ink4a)-positive cells shorten healthy lifespan. Nature. 530:184–189. 10.1038/nature16932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkwill F. 2009. Tumour necrosis factor and cancer. Nat. Rev. Cancer. 9:361–371. 10.1038/nrc2628 [DOI] [PubMed] [Google Scholar]
- Bartsch K., Knittler K., Borowski C., Rudnik S., Damme M., Aden K., Spehlmann M.E., Frey N., Saftig P., Chalaris A., and Rabe B.. 2017. Absence of RNase H2 triggers generation of immunogenic micronuclei removed by autophagy. Hum. Mol. Genet. 26:3960–3972. 10.1093/hmg/ddx283 [DOI] [PubMed] [Google Scholar]
- Bauer S., Groh V., Wu J., Steinle A., Phillips J.H., Lanier L.L., and Spies T.. 1999. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. 285:727–729. 10.1126/science.285.5428.727 [DOI] [PubMed] [Google Scholar]
- Bridgeman A., Maelfait J., Davenne T., Partridge T., Peng Y., Mayer A., Dong T., Kaever V., Borrow P., and Rehwinkel J.. 2015. Viruses transfer the antiviral second messenger cGAMP between cells. Science. 349:1228–1232. 10.1126/science.aab3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzostek-Racine S., Gordon C., Van Scoy S., and Reich N.C.. 2011. The DNA damage response induces IFN. J. Immunol. 187:5336–5345. 10.4049/jimmunol.1100040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdette D.L., Monroe K.M., Sotelo-Troha K., Iwig J.S., Eckert B., Hyodo M., Hayakawa Y., and Vance R.E.. 2011. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 478:515–518. 10.1038/nature10429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette B.C., Liang H., Lee Y., Chlewicki L., Khodarev N.N., Weichselbaum R.R., Fu Y.X., and Auh S.L.. 2011. The efficacy of radiotherapy relies upon induction of type i interferon-dependent innate and adaptive immunity. Cancer Res. 71:2488–2496. 10.1158/0008-5472.CAN-10-2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X., Chiu Y.H., and Chen Z.J.. 2014. The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Mol. Cell. 54:289–296. 10.1016/j.molcel.2014.03.040 [DOI] [PubMed] [Google Scholar]
- Cao D., Schiattarella G.G., Villalobos E., Jiang N., May H.I., Li T., Chen Z.J., Gillette T.G., and Hill J.A.. 2018. Cytosolic DNA Sensing Promotes Macrophage Transformation and Governs Myocardial Ischemic Injury. Circulation.:CIRCULATIONAHA.117.031046 10.1161/CIRCULATIONAHA.117.031046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra D., Quispe-Tintaya W., Jahangir A., Asafu-Adjei D., Ramos I., Sintim H.O., Zhou J., Hayakawa Y., Karaolis D.K.R., and Gravekamp C.. 2014. STING ligand c-di-GMP improves cancer vaccination against metastatic breast cancer. Cancer Immunol. Res. 2:901–910. 10.1158/2326-6066.CIR-13-0123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Boire A., Jin X., Valiente M., Er E.E., Lopez-Soto A., Jacob L., Patwa R., Shah H., Xu K., et al. 2016a Carcinoma-astrocyte gap junctions promote brain metastasis by cGAMP transfer. Nature. 533:493–498. 10.1038/nature18268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Sun L., and Chen Z.J.. 2016b Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat. Immunol. 17:1142–1149. 10.1038/ni.3558 [DOI] [PubMed] [Google Scholar]
- Chen Y.-A., Shen Y.-L., Hsia H.-Y., Tiang Y.-P., Sung T.-L., and Chen L.-Y.. 2017. Extrachromosomal telomere repeat DNA is linked to ALT development via cGAS-STING DNA sensing pathway. Nat. Struct. Mol. Biol. 24:1124–1131. 10.1038/nsmb.3498 [DOI] [PubMed] [Google Scholar]
- Chowdhury D., Beresford P.J., Zhu P., Zhang D., Sung J.S., Demple B., Perrino F.W., and Lieberman J.. 2006. The exonuclease TREX1 is in the SET complex and acts in concert with NM23-H1 to degrade DNA during granzyme A-mediated cell death. Mol. Cell. 23:133–142. 10.1016/j.molcel.2006.06.005 [DOI] [PubMed] [Google Scholar]
- Conlon J., Burdette D.L., Sharma S., Bhat N., Thompson M., Jiang Z., Rathinam V.A., Monks B., Jin T., Xiao T.S., et al. 2013. Mouse, but not human STING, binds and signals in response to the vascular disrupting agent 5,6-dimethylxanthenone-4-acetic acid. J. Immunol. 190:5216–5225. 10.4049/jimmunol.1300097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppé J.P., Kauser K., Campisi J., and Beauséjour C.M.. 2006. Secretion of vascular endothelial growth factor by primary human fibroblasts at senescence. J. Biol. Chem. 281:29568–29574. 10.1074/jbc.M603307200 [DOI] [PubMed] [Google Scholar]
- Coppé J.P., Patil C.K., Rodier F., Sun Y., Muñoz D.P., Goldstein J., Nelson P.S., Desprez P.Y., and Campisi J.. 2008. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 6:2853–2868. 10.1371/journal.pbio.0060301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppé J.P., Desprez P.Y., Krtolica A., and Campisi J.. 2010. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu. Rev. Pathol. 5:99–118. 10.1146/annurev-pathol-121808-102144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrales L., Glickman L.H., McWhirter S.M., Kanne D.B., Sivick K.E., Katibah G.E., Woo S.-R., Lemmens E., Banda T., Leong J.J., et al. 2015. Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell Reports. 11:1018–1030. 10.1016/j.celrep.2015.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow Y.J., and Rehwinkel J.. 2009. Aicardi-Goutieres syndrome and related phenotypes: linking nucleic acid metabolism with autoimmunity. Hum. Mol. Genet. 18(R2):R130–R136. 10.1093/hmg/ddp293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow Y.J., Hayward B.E., Parmar R., Robins P., Leitch A., Ali M., Black D.N., van Bokhoven H., Brunner H.G., Hamel B.C., et al. 2006a Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi-Goutières syndrome at the AGS1 locus. Nat. Genet. 38:917–920. 10.1038/ng1845 [DOI] [PubMed] [Google Scholar]
- Crow Y.J., Leitch A., Hayward B.E., Garner A., Parmar R., Griffith E., Ali M., Semple C., Aicardi J., Babul-Hirji R., et al. 2006b Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutières syndrome and mimic congenital viral brain infection. Nat. Genet. 38:910–916. 10.1038/ng1842 [DOI] [PubMed] [Google Scholar]
- d’Adda di Fagagna F. 2008. Living on a break: cellular senescence as a DNA-damage response. Nat. Rev. Cancer. 8:512–522. 10.1038/nrc2440 [DOI] [PubMed] [Google Scholar]
- Dehé P.-M., and Gaillard P.-H.L.. 2017. Control of structure-specific endonucleases to maintain genome stability. Nat. Rev. Mol. Cell Biol. 18:315–330. 10.1038/nrm.2016.177 [DOI] [PubMed] [Google Scholar]
- Demaria O., De Gassart A., Coso S., Gestermann N., Di Domizio J., Flatz L., Gaide O., Michielin O., Hwu P., Petrova T.V., et al. 2015. STING activation of tumor endothelial cells initiates spontaneous and therapeutic antitumor immunity. Proc. Natl. Acad. Sci. USA. 112:15408–15413. 10.1073/pnas.1512832112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denais C.M., Gilbert R.M., Isermann P., McGregor A.L., te Lindert M., Weigelin B., Davidson P.M., Friedl P., Wolf K., and Lammerding J.. 2016. Nuclear envelope rupture and repair during cancer cell migration. Science. 352:353–358. 10.1126/science.aad7297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L., Liang H., Xu M., Yang X., Burnette B., Arina A., Li X.D., Mauceri H., Beckett M., Darga T., et al. 2014. STING-Dependent Cytosolic DNA Sensing Promotes Radiation-Induced Type I Interferon-Dependent Antitumor Immunity in Immunogenic Tumors. Immunity. 41:843–852. 10.1016/j.immuni.2014.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Queiroz N.M.G.P., Xia T., and Barber G.N.. 2017. Defective STING signaling in ovarian cancer cells favor oncolytic virus action. J. Immunol. 198:130.28. [Google Scholar]
- Diamond M.S., Kinder M., Matsushita H., Mashayekhi M., Dunn G.P., Archambault J.M., Lee H., Arthur C.D., White J.M., Kalinke U., et al. 2011. Type I interferon is selectively required by dendritic cells for immune rejection of tumors. J. Exp. Med. 208:1989–2003. 10.1084/jem.20101158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Micco R., Fumagalli M., Cicalese A., Piccinin S., Gasparini P., Luise C., Schurra C., Garre’ M., Nuciforo P.G., Bensimon A., et al. 2006. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 444:638–642. 10.1038/nature05327 [DOI] [PubMed] [Google Scholar]
- Diner E.J., Burdette D.L., Wilson S.C., Monroe K.M., Kellenberger C.A., Hyodo M., Hayakawa Y., Hammond M.C., and Vance R.E.. 2013. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Reports. 3:1355–1361. 10.1016/j.celrep.2013.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Z., Ghosh K., Vizioli M.G., Zhu J., Sen P., Wangensteen K.J., Simithy J., Lan Y., Lin Y., Zhou Z., et al. 2017. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature. 550:402–406. 10.1038/nature24050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdal E., Haider S., Rehwinkel J., Harris A.L., and McHugh P.J.. 2017. A prosurvival DNA damage-induced cytoplasmic interferon response is mediated by end resection factors and is limited by Trex1. Genes Dev. 31:353–369. 10.1101/gad.289769.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenech M. 2007. Cytokinesis-block micronucleus cytome assay. Nat. Protoc. 2:1084–1104. 10.1038/nprot.2007.77 [DOI] [PubMed] [Google Scholar]
- Fenech M., and Morley A.A.. 1985. Measurement of micronuclei in lymphocytes. Mutat. Res. 147:29–36. 10.1016/0165-1161(85)90015-9 [DOI] [PubMed] [Google Scholar]
- Fenech M., and Morley A.A.. 1986. Cytokinesis-block micronucleus method in human lymphocytes: effect of in vivo ageing and low dose X-irradiation. Mutat. Res. 161:193–198. 10.1016/0027-5107(86)90010-2 [DOI] [PubMed] [Google Scholar]
- Fenech M., Kirsch-Volders M., Natarajan A.T., Surralles J., Crott J.W., Parry J., Norppa H., Eastmond D.A., Tucker J.D., and Thomas P.. 2011. Molecular mechanisms of micronucleus, nucleoplasmic bridge and nuclear bud formation in mammalian and human cells. Mutagenesis. 26:125–132. 10.1093/mutage/geq052 [DOI] [PubMed] [Google Scholar]
- Franzolin E., Pontarin G., Rampazzo C., Miazzi C., Ferraro P., Palumbo E., Reichard P., and Bianchi V.. 2013. The deoxynucleotide triphosphohydrolase SAMHD1 is a major regulator of DNA precursor pools in mammalian cells. Proc. Natl. Acad. Sci. USA. 110:14272–14277. 10.1073/pnas.1312033110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuertes M.B., Kacha A.K., Kline J., Woo S.R., Kranz D.M., Murphy K.M., and Gajewski T.F.. 2011. Host type I IFN signals are required for antitumor CD8+ T cell responses through CD8alpha+ dendritic cells. J. Exp. Med. 208:2005–2016. 10.1084/jem.20101159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidt M.M., Ebert T.S., Chauhan D., Ramshorn K., Pinci F., Zuber S., O’Duill F., Schmid-Burgk J.L., Hoss F., Buhmann R., et al. 2017. The DNA Inflammasome in Human Myeloid Cells Is Initiated by a STING-Cell Death Program Upstream of NLRP3. Cell. 171:1110–1124.e18. 10.1016/j.cell.2017.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall A., Treuting P., Elkon K.B., Loo Y.M., Gale M. Jr., Barber G.N., and Stetson D.B.. 2012. Autoimmunity initiates in nonhematopoietic cells and progresses via lymphocytes in an interferon-dependent autoimmune disease. Immunity. 36:120–131. 10.1016/j.immuni.2011.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D., Li T., Li X.D., Chen X., Li Q.Z., Wight-Carter M., and Chen Z.J.. 2015. Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proc. Natl. Acad. Sci. USA. 112:E5699–E5705. 10.1073/pnas.1516465112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P., Ascano M., Wu Y., Barchet W., Gaffney B.L., Zillinger T., Serganov A.A., Liu Y., Jones R.A., Hartmann G., et al. 2013a Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 153:1094–1107. 10.1016/j.cell.2013.04.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P., Ascano M., Zillinger T., Wang W., Dai P., Serganov A.A., Gaffney B.L., Shuman S., Jones R.A., Deng L., et al. 2013b Structure-function analysis of STING activation by c[G(2′,5′)pA(3′,5′)p] and targeting by antiviral DMXAA. Cell. 154:748–762. 10.1016/j.cell.2013.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser S., Orsulic S., Brown E.J., and Raulet D.H.. 2005. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature. 436:1186–1190. 10.1038/nature03884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentili M., Kowal J., Tkach M., Satoh T., Lahaye X., Conrad C., Boyron M., Lombard B., Durand S., Kroemer G., et al. 2015. Transmission of innate immune signaling by packaging of cGAMP in viral particles. Science. 349:1232–1236. 10.1126/science.aab3628 [DOI] [PubMed] [Google Scholar]
- Glück S., Guey B., Gulen M.F., Wolter K., Kang T.-W., Schmacke N.A., Bridgeman A., Rehwinkel J., Zender L., and Ablasser A.. 2017. Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat. Cell Biol. 19:1061–1070. 10.1038/ncb3586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray E.E., Treuting P.M., Woodward J.J., and Stetson D.B.. 2015. Cutting Edge: cGAS Is Required for Lethal Autoimmune Disease in the Trex1-Deficient Mouse Model of Aicardi-Goutières Syndrome. J. Immunol. 195:1939–1943. 10.4049/jimmunol.1500969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gul E., Sayar E.H., Gungor B., Eroglu F.K., Surucu N., Keles S., Guner S.N., Findik S., Alpdundar E., Ayanoglu I.C., et al. 2017. Type I IFN-related NETosis in ataxia telangiectasia and Artemis deficiency. J. Allergy Clin. Immunol.:S0091-6749(17)31762-1. [DOI] [PubMed] [Google Scholar]
- Gulen M.F., Koch U., Haag S.M., Schuler F., Apetoh L., Villunger A., Radtke F., and Ablasser A.. 2017. Signalling strength determines proapoptotic functions of STING. Nat. Commun. 8:427 10.1038/s41467-017-00573-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J., Brault A., Vincent F., Weng S., Wang H., Dumlao D., Aulabaugh A., Aivazian D., Castro D., Chen M., et al. 2017. Discovery of PF-06928215 as a high affinity inhibitor of cGAS enabled by a novel fluorescence polarization assay. PLoS One. 12:e0184843 10.1371/journal.pone.0184843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson M.C., Crespo M.P., Abraham W., Moynihan K.D., Szeto G.L., Chen S.H., Melo M.B., Mueller S., and Irvine D.J.. 2015. Nanoparticulate STING agonists are potent lymph node-targeted vaccine adjuvants. J. Clin. Invest. 125:2532–2546. 10.1172/JCI79915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding S.M., Benci J.L., Irianto J., Discher D.E., Minn A.J., and Greenberg R.A.. 2017. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature. 548:466–470. 10.1038/nature23470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Härtlova A., Erttmann S.F., Raffi F.A., Schmalz A.M., Resch U., Anugula S., Lienenklaus S., Nilsson L.M., Kröger A., Nilsson J.A., et al. 2015. DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity. 42:332–343. 10.1016/j.immuni.2015.01.012 [DOI] [PubMed] [Google Scholar]
- Hatch E.M., Fischer A.H., Deerinck T.J., and Hetzer M.W.. 2013. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell. 154:47–60. 10.1016/j.cell.2013.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henning K.A., Li L., Iyer N., McDaniel L.D., Reagan M.S., Legerski R., Schultz R.A., Stefanini M., Lehmann A.R., Mayne L.V., and Friedberg E.C.. 1995. The Cockayne syndrome group A gene encodes a WD repeat protein that interacts with CSB protein and a subunit of RNA polymerase II TFIIH. Cell. 82:555–564. 10.1016/0092-8674(95)90028-4 [DOI] [PubMed] [Google Scholar]
- Herzner A.M., Hagmann C.A., Goldeck M., Wolter S., Kübler K., Wittmann S., Gramberg T., Andreeva L., Hopfner K.P., Mertens C., et al. 2015. Sequence-specific activation of the DNA sensor cGAS by Y-form DNA structures as found in primary HIV-1 cDNA. Nat. Immunol. 16:1025–1033. 10.1038/ni.3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller B., Achleitner M., Glage S., Naumann R., Behrendt R., and Roers A.. 2012. Mammalian RNase H2 removes ribonucleotides from DNA to maintain genome integrity. J. Exp. Med. 209:1419–1426. 10.1084/jem.20120876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S.S., Zhang W.Y., Tan N.Y., Khatoo M., Suter M.A., Tripathi S., Cheung F.S., Lim W.K., Tan P.H., Ngeow J., and Gasser S.. 2016. The DNA Structure-Specific Endonuclease MUS81 Mediates DNA Sensor STING-Dependent Host Rejection of Prostate Cancer Cells. Immunity. 44:1177–1189. 10.1016/j.immuni.2016.04.010 [DOI] [PubMed] [Google Scholar]
- Inomata K., Aoto T., Binh N.T., Okamoto N., Tanimura S., Wakayama T., Iseki S., Hara E., Masunaga T., Shimizu H., and Nishimura E.K.. 2009. Genotoxic stress abrogates renewal of melanocyte stem cells by triggering their differentiation. Cell. 137:1088–1099. 10.1016/j.cell.2009.03.037 [DOI] [PubMed] [Google Scholar]
- Ishii K.J., Coban C., Kato H., Takahashi K., Torii Y., Takeshita F., Ludwig H., Sutter G., Suzuki K., Hemmi H., et al. 2006. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat. Immunol. 7:40–48. 10.1038/ni1282 [DOI] [PubMed] [Google Scholar]
- Ishikawa H., and Barber G.N.. 2008. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 455:674–678. 10.1038/nature07317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens S., and Tschopp J.. 2006. Signals from within: the DNA-damage-induced NF-kappaB response. Cell Death Differ. 13:773–784. 10.1038/sj.cdd.4401843 [DOI] [PubMed] [Google Scholar]
- Jin L., Waterman P.M., Jonscher K.R., Short C.M., Reisdorph N.A., and Cambier J.C.. 2008. MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol. Cell. Biol. 28:5014–5026. 10.1128/MCB.00640-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakasilioti I., Kamileri I., Chatzinikolaou G., Kosteas T., Vergadi E., Robinson A.R., Tsamardinos I., Rozgaja T.A., Siakouli S., Tsatsanis C., et al. 2013. DNA damage triggers a chronic autoinflammatory response, leading to fat depletion in NER progeria. Cell Metab. 18:403–415. 10.1016/j.cmet.2013.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katlinskaya Y.V., Katlinski K.V., Yu Q., Ortiz A., Beiting D.P., Brice A., Davar D., Sanders C., Kirkwood J.M., Rui H., et al. 2016. Suppression of Type I Interferon Signaling Overcomes Oncogene-Induced Senescence and Mediates Melanoma Development and Progression. Cell Reports. 15:171–180. 10.1016/j.celrep.2016.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K., Omura H., Ishitani R., and Nureki O.. 2017. Cyclic GMP-AMP as an Endogenous Second Messenger in Innate Immune Signaling by Cytosolic DNA. Annu. Rev. Biochem. 86:541–566. 10.1146/annurev-biochem-061516-044813 [DOI] [PubMed] [Google Scholar]
- Kawane K., Fukuyama H., Yoshida H., Nagase H., Ohsawa Y., Uchiyama Y., Okada K., Iida T., and Nagata S.. 2003. Impaired thymic development in mouse embryos deficient in apoptotic DNA degradation. Nat. Immunol. 4:138–144. 10.1038/ni881 [DOI] [PubMed] [Google Scholar]
- Kerur N., Fukuda S., Banerjee D., Kim Y., Fu D., Apicella I., Varshney A., Yasuma R., Fowler B.J., Baghdasaryan E., et al. 2017. cGAS drives noncanonical-inflammasome activation in age-related macular degeneration. Nat. Med. 24:50–61. 10.1038/nm.4450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King K.R., Aguirre A.D., Ye Y.-X., Sun Y., Roh J.D., Ng R.P. Jr., Kohler R.H., Arlauckas S.P., Iwamoto Y., Savol A., et al. 2017. IRF3 and type I interferons fuel a fatal response to myocardial infarction. Nat. Med. 23:1481–1487. 10.1038/nm.4428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T., Kobayashi J., Saitoh T., Maruyama K., Ishii K.J., Barber G.N., Komatsu K., Akira S., and Kawai T.. 2013. DNA damage sensor MRE11 recognizes cytosolic double-stranded DNA and induces type I interferon by regulating STING trafficking. Proc. Natl. Acad. Sci. USA. 110:2969–2974. 10.1073/pnas.1222694110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretschmer S., Wolf C., König N., Staroske W., Guck J., Häusler M., Luksch H., Nguyen L.A., Kim B., Alexopoulou D., et al. 2015. SAMHD1 prevents autoimmunity by maintaining genome stability. Ann. Rheum. Dis. 74:e17 10.1136/annrheumdis-2013-204845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizhanovsky V., Yon M., Dickins R.A., Hearn S., Simon J., Miething C., Yee H., Zender L., and Lowe S.W.. 2008. Senescence of activated stellate cells limits liver fibrosis. Cell. 134:657–667. 10.1016/j.cell.2008.06.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krtolica A., Parrinello S., Lockett S., Desprez P.-Y., and Campisi J.. 2001. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc. Natl. Acad. Sci. USA. 98:12072–12077. 10.1073/pnas.211053698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuilman T., and Peeper D.S.. 2009. Senescence-messaging secretome: SMS-ing cellular stress. Nat. Rev. Cancer. 9:81–94. 10.1038/nrc2560 [DOI] [PubMed] [Google Scholar]
- Kuilman T., Michaloglou C., Vredeveld L.C., Douma S., van Doorn R., Desmet C.J., Aarden L.A., Mooi W.J., and Peeper D.S.. 2008. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 133:1019–1031. 10.1016/j.cell.2008.03.039 [DOI] [PubMed] [Google Scholar]
- Laguette N., Brégnard C., Hue P., Basbous J., Yatim A., Larroque M., Kirchhoff F., Constantinou A., Sobhian B., and Benkirane M.. 2014. Premature activation of the SLX4 complex by Vpr promotes G2/M arrest and escape from innate immune sensing. Cell. 156:134–145. 10.1016/j.cell.2013.12.011 [DOI] [PubMed] [Google Scholar]
- Lam A.R., Bert N.L., Ho S.S., Shen Y.J., Tang L.F., Xiong G.M., Croxford J.L., Koo C.X., Ishii K.J., Akira S., et al. 2014. RAE1 ligands for the NKG2D receptor are regulated by STING-dependent DNA sensor pathways in lymphoma. Cancer Res. 74:2193–2203. 10.1158/0008-5472.CAN-13-1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y.Y., Londoño D., Bouley R., Rooney M.S., and Hacohen N.. 2014. Dnase2a deficiency uncovers lysosomal clearance of damaged nuclear DNA via autophagy. Cell Reports. 9:180–192. 10.1016/j.celrep.2014.08.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin B., Ilyukha V., Sorokin M., Buzdin A., Vannier E., and Poltorak A.. 2017. Cutting Edge: Activation of STING in T Cells Induces Type I IFN Responses and Cell Death. J. Immunol. 199:397–402. 10.4049/jimmunol.1601999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau L., Gray E.E., Brunette R.L., and Stetson D.B.. 2015. DNA tumor virus oncogenes antagonize the cGAS-STING DNA-sensing pathway. Science. 350:568–571. 10.1126/science.aab3291 [DOI] [PubMed] [Google Scholar]
- Leibovich S.J., Polverini P.J., Shepard H.M., Wiseman D.M., Shively V., and Nuseir N.. 1987. Macrophage-induced angiogenesis is mediated by tumour necrosis factor-α. Nature. 329:630–632. 10.1038/329630a0 [DOI] [PubMed] [Google Scholar]
- Li L., Yin Q., Kuss P., Maliga Z., Millán J.L., Wu H., and Mitchison T.J.. 2014. Hydrolysis of 2‘3’-cGAMP by ENPP1 and design of nonhydrolyzable analogs. Nat. Chem. Biol. 10:1043–1048. 10.1038/nchembio.1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Cheng H., Yuan H., Xu Q., Shu C., Zhang Y., Xu P., Tan J., Rui Y., Li P., and Tan X.. 2016. Antitumor Activity of cGAMP via Stimulation of cGAS-cGAMP-STING-IRF3 Mediated Innate Immune Response. Sci. Rep. 6:19049 10.1038/srep19049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.-D., Wu J., Gao D., Wang H., Sun L., and Chen Z.J.. 2013. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 341:1390–1394. 10.1126/science.1244040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J., Gerber S., and Lord E.. 2012. The importance of type I interferons in radiation-mediated antitumor responses. J. Immunol. 188:162.33. [Google Scholar]
- Liu D., and Hornsby P.J.. 2007. Senescent human fibroblasts increase the early growth of xenograft tumors via matrix metalloproteinase secretion. Cancer Res. 67:3117–3126. 10.1158/0008-5472.CAN-06-3452 [DOI] [PubMed] [Google Scholar]
- Liu S., Cai X., Wu J., Cong Q., Chen X., Li T., Du F., Ren J., Wu Y.-T., Grishin N.V., and Chen Z.J.. 2015. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 347:aaa2630 10.1126/science.aaa2630 [DOI] [PubMed] [Google Scholar]
- Liu Y., Jesus A.A., Marrero B., Yang D., Ramsey S.E., Sanchez G.A.M., Tenbrock K., Wittkowski H., Jones O.Y., Kuehn H.S., et al. 2014. Activated STING in a vascular and pulmonary syndrome. N. Engl. J. Med. 371:507–518. 10.1056/NEJMoa1312625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe S.W., and Lin A.W.. 2000. Apoptosis in cancer. Carcinogenesis. 21:485–495. 10.1093/carcin/21.3.485 [DOI] [PubMed] [Google Scholar]
- Luo M., Wang H., Wang Z., Cai H., Lu Z., Li Y., Du M., Huang G., Wang C., Chen X., et al. 2017. A STING-activating nanovaccine for cancer immunotherapy. Nat. Nanotechnol. 12:648–654. 10.1038/nnano.2017.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthra P., Aguirre S., Yen B.C., Pietzsch C.A., Sanchez-Aparicio M.T., Tigabu B., Morlock L.K., García-Sastre A., Leung D.W., Williams N.S., et al. 2017. Topoisomerase II Inhibitors Induce DNA Damage-Dependent Interferon Responses Circumventing Ebola Virus Immune Evasion. MBio. 8:e01611–17. 10.1128/mBio.00368-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z., and Damania B.. 2016. The cGAS-STING Defense Pathway and Its Counteraction by Viruses. Cell Host Microbe. 19:150–158. 10.1016/j.chom.2016.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie K.J., Carroll P., Lettice L., Tarnauskaitė Ž., Reddy K., Dix F., Revuelta A., Abbondati E., Rigby R.E., Rabe B., et al. 2016. Ribonuclease H2 mutations induce a cGAS/STING-dependent innate immune response. EMBO J. 35:831–844. 10.15252/embj.201593339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie K.J., Carroll P., Martin C.A., Murina O., Fluteau A., Simpson D.J., Olova N., Sutcliffe H., Rainger J.K., Leitch A., et al. 2017. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature. 548:461–465. 10.1038/nature23449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maelfait J., Bridgeman A., Benlahrech A., Cursi C., and Rehwinkel J.. 2016. Restriction by SAMHD1 Limits cGAS/STING-Dependent Innate and Adaptive Immune Responses to HIV-1. Cell Reports. 16:1492–1501. 10.1016/j.celrep.2016.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S.T., Naylor M.S., East N., Oliff A., and Balkwill F.R.. 1990. Cells secreting tumour necrosis factor show enhanced metastasis in nude mice. Eur. J. Cancer. 26:1031–1034. 10.1016/0277-5379(90)90044-T [DOI] [PubMed] [Google Scholar]
- Mboko W.P., Mounce B.C., Wood B.M., Kulinski J.M., Corbett J.A., and Tarakanova V.L.. 2012. Coordinate regulation of DNA damage and type I interferon responses imposes an antiviral state that attenuates mouse gammaherpesvirus type 68 replication in primary macrophages. J. Virol. 86:6899–6912. 10.1128/JVI.07119-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson J.P., Lemmers B., Chahwan R., Pamidi A., Migon E., Matysiak-Zablocki E., Moynahan M.E., Essers J., Hanada K., Poonepalli A., et al. 2004. Involvement of mammalian Mus81 in genome integrity and tumor suppression. Science. 304:1822–1826. 10.1126/science.1094557 [DOI] [PubMed] [Google Scholar]
- Melki I., Rose Y., Uggenti C., Van Eyck L., Frémond M.-L., Kitabayashi N., Rice G.I., Jenkinson E.M., Boulai A., Jeremiah N., et al. 2017. Disease-associated mutations identify a novel region in human STING necessary for the control of type I interferon signaling. J. Allergy Clin. Immunol. 140:543–552.e5. 10.1016/j.jaci.2016.10.031 [DOI] [PubMed] [Google Scholar]
- Melton D.W., Ketchen A.M., Núñez F., Bonatti-Abbondandolo S., Abbondandolo A., Squires S., and Johnson R.T.. 1998. Cells from ERCC1-deficient mice show increased genome instability and a reduced frequency of S-phase-dependent illegitimate chromosome exchange but a normal frequency of homologous recombination. J. Cell Sci. 111:395–404. [DOI] [PubMed] [Google Scholar]
- Moiseeva O., Mallette F.A., Mukhopadhyay U.K., Moores A., and Ferbeyre G.. 2006. DNA damage signaling and p53-dependent senescence after prolonged beta-interferon stimulation. Mol. Biol. Cell. 17:1583–1592. 10.1091/mbc.E05-09-0858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R.J., Owens D.M., Stamp G., Arnott C., Burke F., East N., Holdsworth H., Turner L., Rollins B., Pasparakis M., et al. 1999. Mice deficient in tumor necrosis factor-α are resistant to skin carcinogenesis. Nat. Med. 5:828–831. 10.1038/10552 [DOI] [PubMed] [Google Scholar]
- Morchikh M., Cribier A., Raffel R., Amraoui S., Cau J., Severac D., Dubois E., Schwartz O., Bennasser Y., and Benkirane M.. 2017. HEXIM1 and NEAT1 Long Non-coding RNA Form a Multi-subunit Complex that Regulates DNA-Mediated Innate Immune Response. Mol. Cell. 67:387–399.e5. 10.1016/j.molcel.2017.06.020 [DOI] [PubMed] [Google Scholar]
- Niedernhofer L.J., Garinis G.A., Raams A., Lalai A.S., Robinson A.R., Appeldoorn E., Odijk H., Oostendorp R., Ahmad A., van Leeuwen W., et al. 2006. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 444:1038–1043. 10.1038/nature05456 [DOI] [PubMed] [Google Scholar]
- Orosz P., Echtenacher B., Falk W., Rüschoff J., Weber D., and Männel D.N.. 1993. Enhancement of experimental metastasis by tumor necrosis factor. J. Exp. Med. 177:1391–1398. 10.1084/jem.177.5.1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrinello S., Samper E., Krtolica A., Goldstein J., Melov S., and Campisi J.. 2003. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat. Cell Biol. 5:741–747. 10.1038/ncb1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokatayev V., Hasin N., Chon H., Cerritelli S.M., Sakhuja K., Ward J.M., Morris H.D., Yan N., and Crouch R.J.. 2016. RNase H2 catalytic core Aicardi-Goutières syndrome-related mutant invokes cGAS-STING innate immune-sensing pathway in mice. J. Exp. Med. 213:329–336. 10.1084/jem.20151464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab M., Gentili M., de Belly H., Thiam H.R., Vargas P., Jimenez A.J., Lautenschlaeger F., Voituriez R., Lennon-Duménil A.M., Manel N., and Piel M.. 2016. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science. 352:359–362. 10.1126/science.aad7611 [DOI] [PubMed] [Google Scholar]
- Reijns M.A., Rabe B., Rigby R.E., Mill P., Astell K.R., Lettice L.A., Boyle S., Leitch A., Keighren M., Kilanowski F., et al. 2012. Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell. 149:1008–1022. 10.1016/j.cell.2012.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice G.I., Bond J., Asipu A., Brunette R.L., Manfield I.W., Carr I.M., Fuller J.C., Jackson R.M., Lamb T., Briggs T.A., et al. 2009. Mutations involved in Aicardi-Goutières syndrome implicate SAMHD1 as regulator of the innate immune response. Nat. Genet. 41:829–832. 10.1038/ng.373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodero M.P., Tesser A., Bartok E., Rice G.I., Della Mina E., Depp M., Beitz B., Bondet V., Cagnard N., Duffy D., et al. 2017. Type I interferon-mediated autoinflammation due to DNase II deficiency. Nat. Commun. 8:2176 10.1038/s41467-017-01932-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodier F., Coppé J.P., Patil C.K., Hoeijmakers W.A., Muñoz D.P., Raza S.R., Freund A., Campeau E., Davalos A.R., and Campisi J.. 2009. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat. Cell Biol. 11:973–979. 10.1038/ncb1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rongvaux A., Jackson R., Harman C.C., Li T., West A.P., de Zoete M.R., Wu Y., Yordy B., Lakhani S.A., Kuan C.-Y., et al. 2014. Apoptotic caspases prevent the induction of type I interferons by mitochondrial DNA. Cell. 159:1563–1577. 10.1016/j.cell.2014.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T., Fujita N., Hayashi T., Takahara K., Satoh T., Lee H., Matsunaga K., Kageyama S., Omori H., Noda T., et al. 2009. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc. Natl. Acad. Sci. USA. 106:20842–20846. 10.1073/pnas.0911267106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel R., MacGregor J.T., and Everson R.B.. 1986. Assessment of cytogenetic damage by quantitation of micronuclei in human peripheral blood erythrocytes. Cancer Res. 46:3717–3721. [PubMed] [Google Scholar]
- Shen Y.J., Le Bert N., Chitre A.A., Koo C.X., Nga X.H., Ho S.S., Khatoo M., Tan N.Y., Ishii K.J., and Gasser S.. 2015. Genome-derived cytosolic DNA mediates type I interferon-dependent rejection of B cell lymphoma cells. Cell Reports. 11:460–473. 10.1016/j.celrep.2015.03.041 [DOI] [PubMed] [Google Scholar]
- Siddoo-Atwal C., Haas A.L., and Rosin M.P.. 1996. Elevation of interferon beta-inducible proteins in ataxia telangiectasia cells. Cancer Res. 56:443–447. [PubMed] [Google Scholar]
- Sistigu A., Yamazaki T., Vacchelli E., Chaba K., Enot D.P., Adam J., Vitale I., Goubar A., Baracco E.E., Remédios C., et al. 2014. Cancer cell-autonomous contribution of type I interferon signaling to the efficacy of chemotherapy. Nat. Med. 20:1301–1309. 10.1038/nm.3708 [DOI] [PubMed] [Google Scholar]
- Stetson D.B., Ko J.S., Heidmann T., and Medzhitov R.. 2008. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 134:587–598. 10.1016/j.cell.2008.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugihara T., Murano H., Nakamura M., Ichinohe K., and Tanaka K.. 2011. Activation of interferon-stimulated genes by gamma-ray irradiation independently of the ataxia telangiectasia mutated-p53 pathway. Mol. Cancer Res. 9:476–484. 10.1158/1541-7786.MCR-10-0358 [DOI] [PubMed] [Google Scholar]
- Sun L., Wu J., Du F., Chen X., and Chen Z.J.. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 339:786–791. 10.1126/science.1232458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., Li Y., Chen L., Chen H., You F., Zhou X., Zhou Y., Zhai Z., Chen D., and Jiang Z.. 2009. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl. Acad. Sci. USA. 106:8653–8658. 10.1073/pnas.0900850106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson K.V., Junkins R.D., Kurkjian C.J., Holley-Guthrie E., Pendse A.A., El Morabiti R., Petrucelli A., Barber G.N., Benedict C.A., and Ting J.P.. 2017. A noncanonical function of cGAMP in inflammasome priming and activation. J. Exp. Med. 214:3611–3626. 10.1084/jem.20171749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaoka A., Hayakawa S., Yanai H., Stoiber D., Negishi H., Kikuchi H., Sasaki S., Imai K., Shibue T., Honda K., and Taniguchi T.. 2003. Integration of interferon-alpha/beta signalling to p53 responses in tumour suppression and antiviral defence. Nature. 424:516–523. 10.1038/nature01850 [DOI] [PubMed] [Google Scholar]
- Tanaka Y., and Chen Z.J.. 2012. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci. Signal. 5:ra20 10.1126/scisignal.2002521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C.-H.A., Zundell J.A., Ranatunga S., Lin C., Nefedova Y., Del Valle J.R., and Hu C.-C.A.. 2016. Agonist-Mediated Activation of STING Induces Apoptosis in Malignant B Cells. Cancer Res. 76:2137–2152. 10.1158/0008-5472.CAN-15-1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao J., Zhou X., and Jiang Z.. 2016. cGAS-cGAMP-STING: The three musketeers of cytosolic DNA sensing and signaling. IUBMB Life. 68:858–870. 10.1002/iub.1566 [DOI] [PubMed] [Google Scholar]
- Tilstra J.S., Robinson A.R., Wang J., Gregg S.Q., Clauson C.L., Reay D.P., Nasto L.A., St Croix C.M., Usas A., Vo N., et al. 2012. NF-κB inhibition delays DNA damage-induced senescence and aging in mice. J. Clin. Invest. 122:2601–2612. 10.1172/JCI45785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troelstra C., van Gool A., de Wit J., Vermeulen W., Bootsma D., and Hoeijmakers J.H.J.. 1992. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne’s syndrome and preferential repair of active genes. Cell. 71:939–953. 10.1016/0092-8674(92)90390-X [DOI] [PubMed] [Google Scholar]
- Vargas J.D., Hatch E.M., Anderson D.J., and Hetzer M.W.. 2012. Transient nuclear envelope rupturing during interphase in human cancer cells. Nucleus. 3:88–100. 10.4161/nucl.18954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent J., Adura C., Gao P., Luz A., Lama L., Asano Y., Okamoto R., Imaeda T., Aida J., Rothamel K., et al. 2017. Small molecule inhibition of cGAS reduces interferon expression in primary macrophages from autoimmune mice. Nat. Commun. 8:750 10.1038/s41467-017-00833-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Hu S., Chen X., Shi H., Chen C., Sun L., and Chen Z.J.. 2017. cGAS is essential for the antitumor effect of immune checkpoint blockade. Proc. Natl. Acad. Sci. USA. 114:1637–1642. 10.1073/pnas.1621363114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West A.P., Khoury-Hanold W., Staron M., Tal M.C., Pineda C.M., Lang S.M., Bestwick M., Duguay B.A., Raimundo N., MacDuff D.A., et al. 2015. Mitochondrial DNA stress primes the antiviral innate immune response. Nature. 520:553–557. 10.1038/nature14156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M.J., McArthur K., Metcalf D., Lane R.M., Cambier J.C., Herold M.J., van Delft M.F., Bedoui S., Lessene G., Ritchie M.E., et al. 2014. Apoptotic caspases suppress mtDNA-induced STING-mediated type I IFN production. Cell. 159:1549–1562. 10.1016/j.cell.2014.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D.R., Sen R., Sunshine J.C., Pardoll D.M., Green J.J., and Kim Y.J.. 2018. Biodegradable STING agonist nanoparticles for enhanced cancer immunotherapy. Nanomedicine (Lond.). 14:237–246. 10.1016/j.nano.2017.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf C., Rapp A., Berndt N., Staroske W., Schuster M., Dobrick-Mattheuer M., Kretschmer S., König N., Kurth T., Wieczorek D., et al. 2016. RPA and Rad51 constitute a cell intrinsic mechanism to protect the cytosol from self DNA. Nat. Commun. 7:11752 10.1038/ncomms11752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo S.R., Fuertes M.B., Corrales L., Spranger S., Furdyna M.J., Leung M.Y., Duggan R., Wang Y., Barber G.N., Fitzgerald K.A., et al. 2014. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity. 41:830–842. 10.1016/j.immuni.2014.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., and Chen Z.J.. 2014. Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 32:461–488. 10.1146/annurev-immunol-032713-120156 [DOI] [PubMed] [Google Scholar]
- Wu J., Sun L., Chen X., Du F., Shi H., Chen C., and Chen Z.J.. 2013. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 339:826–830. 10.1126/science.1229963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia T., Konno H., Ahn J., and Barber G.N.. 2016a Deregulation of STING Signaling in Colorectal Carcinoma Constrains DNA Damage Responses and Correlates With Tumorigenesis. Cell Reports. 14:282–297. 10.1016/j.celrep.2015.12.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia T., Konno H., and Barber G.N.. 2016b Recurrent Loss of STING Signaling in Melanoma Correlates with Susceptibility to Viral Oncolysis. Cancer Res. 76:6747–6759. 10.1158/0008-5472.CAN-16-1404 [DOI] [PubMed] [Google Scholar]
- Xiao T.S., and Fitzgerald K.A.. 2013. The cGAS-STING pathway for DNA sensing. Mol. Cell. 51:135–139. 10.1016/j.molcel.2013.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M.M., Pu Y., Han D., Shi Y., Cao X., Liang H., Chen X., Li X.-D., Deng L., Chen Z.J., et al. 2017. Dendritic Cells but Not Macrophages Sense Tumor Mitochondrial DNA for Cross-priming through Signal Regulatory Protein α Signaling. Immunity. 47:363–373.e5. 10.1016/j.immuni.2017.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W., Zender L., Miething C., Dickins R.A., Hernando E., Krizhanovsky V., Cordon-Cardo C., and Lowe S.W.. 2007. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 445:656–660. 10.1038/nature05529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Wang H., Ren J., Chen Q., and Chen Z.J.. 2017. cGAS is essential for cellular senescence. Proc. Natl. Acad. Sci. USA. 114:E4612–E4620. 10.1073/pnas.1705499114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.G., Lindahl T., and Barnes D.E.. 2007. Trex1 exonuclease degrades ssDNA to prevent chronic checkpoint activation and autoimmune disease. Cell. 131:873–886. 10.1016/j.cell.2007.10.017 [DOI] [PubMed] [Google Scholar]
- Yoshida H., Okabe Y., Kawane K., Fukuyama H., and Nagata S.. 2005. Lethal anemia caused by interferon-beta produced in mouse embryos carrying undigested DNA. Nat. Immunol. 6:49–56. 10.1038/ni1146 [DOI] [PubMed] [Google Scholar]
- Yu Q., Katlinskaya Y.V., Carbone C.J., Zhao B., Katlinski K.V., Zheng H., Guha M., Li N., Chen Q., Yang T., et al. 2015. DNA-damage-induced type I interferon promotes senescence and inhibits stem cell function. Cell Reports. 11:785–797. 10.1016/j.celrep.2015.03.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng S., Xiang T., Pandita T.K., Gonzalez-Suarez I., Gonzalo S., Harris C.C., and Yang Q.. 2009. Telomere recombination requires the MUS81 endonuclease. Nat. Cell Biol. 11:616–623. 10.1038/ncb1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Niu J., Zhang J., Wang Y., Zhou Z., Zhang J., and Tian Z.. 2008. Opposing effects of interferon-alpha and interferon-gamma on the expression of major histocompatibility complex class I chain-related A in tumors. Cancer Sci. 99:1279–1286. 10.1111/j.1349-7006.2008.00791.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Shi H., Wu J., Zhang X., Sun L., Chen C., and Chen Z.J.. 2013. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol. Cell. 51:226–235. 10.1016/j.molcel.2013.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B., Yang Y., Li S., Wang Y.Y., Li Y., Diao F., Lei C., He X., Zhang L., Tien P., and Shu H.B.. 2008. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 29:538–550. 10.1016/j.immuni.2008.09.003 [DOI] [PubMed] [Google Scholar]
- Zhu Q., Man S.M., Gurung P., Liu Z., Vogel P., Lamkanfi M., and Kanneganti T.-D.. 2014. Cutting edge: STING mediates protection against colorectal tumorigenesis by governing the magnitude of intestinal inflammation. J. Immunol. 193:4779–4782. 10.4049/jimmunol.1402051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwi L.J., Baguley B.C., Gavin J.B., and Wilson W.R.. 1994. Correlation between immune and vascular activities of xanthenone acetic acid antitumor agents. Oncol. Res. 6:79–85. [PubMed] [Google Scholar]