Type I interferons have been implicated in the pathogenesis of tuberculosis. Herein, Moreira-Teixeira et al. discuss mechanistic and contextual factors that determine the role of type I interferons during Mycobacterium tuberculosis infection, from human disease to experimental models of tuberculosis.
Abstract
Tuberculosis remains one of the leading causes of mortality worldwide, and, despite its clinical significance, there are still significant gaps in our understanding of pathogenic and protective mechanisms triggered by Mycobacterium tuberculosis infection. Type I interferons (IFN) regulate a broad family of genes that either stimulate or inhibit immune function, having both host-protective and detrimental effects, and exhibit well-characterized antiviral activity. Transcriptional studies have uncovered a potential deleterious role for type I IFN in active tuberculosis. Since then, additional studies in human tuberculosis and experimental mouse models of M. tuberculosis infection support the concept that type I IFN promotes both bacterial expansion and disease pathogenesis. More recently, studies in a different setting have suggested a putative protective role for type I IFN. In this study, we discuss the mechanistic and contextual factors that determine the detrimental versus beneficial outcomes of type I IFN induction during M. tuberculosis infection, from human disease to experimental mouse models of tuberculosis.
Introduction
Although cytokines play critical roles in host defense and immune homeostasis, they can also be key mediators of inflammatory pathology. This paradoxical aspect of cytokine biology is perhaps best exemplified in the dichotomous functions of type I IFN in health and disease. The type I IFN family is a multigene cytokine family that encodes numerous partially homologous IFN-α subtypes, a single IFN-β, and several other poorly defined single genes products (Pestka et al., 2004). All type I IFN share a ubiquitously expressed heterodimeric receptor composed of the subunits IFNAR1 and IFNAR2, which signal through STAT1 and STAT2 to activate a broad family of IFN-stimulated genes (ISGs; Pestka et al., 2004; Ivashkiv and Donlin, 2014). Dominant IFNAR1 expression has been shown to favor IFN-β ligation and activation of the protein kinase B (also known as Akt) pathway over the classical JAK–STAT pathway (de Weerd et al., 2013). ISGs either stimulate or inhibit immune function and in so doing have host-protective or detrimental effects. Thus, although type I IFN is well known for its antiviral activity and stimulation of effector T cell responses (Yan and Chen, 2012; Crouse et al., 2015), it has also been implicated in autoimmune diseases (Hall and Rosen, 2010) and exacerbation of bacterial and even certain viral infections (Decker et al., 2005; Trinchieri, 2010; Davidson et al., 2015; McNab et al., 2015).
The host beneficial protective effects of type I IFN have been most thoroughly studied in viral infections where these cytokines stimulate the production of innate antiviral proteins and promote effector CD8+ T cell responses (Crouse et al., 2015). The deleterious effects of type I IFN seen in autoimmune diseases such as lupus or in the genetically based interferonopathies appear to stem largely from dysregulated cytokine synthesis (Banchereau and Pascual, 2006; Crow, 2015). Nevertheless, cytokine overproduction does not provide a unifying explanation for the deleterious activity of type I IFN in infections with bacteria such as Listeria monocytogenes, Brucella abortus, and Staphylococcus aureus (McNab et al., 2015; Stifter and Feng, 2015).
The present review focuses on the role of type I IFN in the immune response to Mycobacterium tuberculosis. This topic has received great attention (O’Garra et al., 2013; Mayer-Barber and Sher, 2015; Donovan et al., 2017; Sabir et al., 2017) both because of the global public health importance of this pathogen and the now numerous studies linking type I IFN expression with tuberculosis. However, recent data suggest that even in the prominent example of tuberculosis, the “foe”-like properties of type I IFN are not ironclad and the same cytokines can display “friendly” protective functions under different settings of host–pathogen encounter. Deciphering the mechanisms underlying these opposing activities of type I IFN in tuberculosis should contribute enormously to our understanding of the pathogenesis of this important disease while providing new insights into how this major cytokine pathway can be manipulated to ensure beneficial rather than deleterious outcomes for the host.
Role of cytokines in the immune response in tuberculosis
Despite being one of the oldest diseases, tuberculosis remains a devastating public health problem. Worldwide, 6.3 million new cases and 1.67 million deaths (of which 0.37 million were in HIV-positive coinfected individuals) were reported in 2016 alone (World Health Organization, 2017). Although most individuals exposed to M. tuberculosis generate an effective immune response and remain clinically asymptomatic (latent infection), ∼10% of the infected individuals will progress to active disease at some stage of their lifetime, presenting with clinical signs and symptoms of tuberculosis and, in a significant proportion of patients, cultivable bacilli in the sputum (Lawn and Zumla, 2011; O’Garra et al., 2013; Getahun et al., 2015). Several mechanisms have been described that result in the development of a protective immune response that controls infection (Flynn and Chan, 2001; North and Jung, 2004; Cooper, 2009; O’Garra et al., 2013); however, the host immune mechanisms underlying progression to active disease remain poorly understood.
The role of cytokines in host protection against M. tuberculosis infection is well established and was first demonstrated in experimental mouse models of infection that established critical roles for IFN-γ (Cooper et al., 1993; Flynn et al., 1993), TNF-α (Flynn et al., 1995a), and IL-12 (Cooper et al., 1995, 1997; Flynn et al., 1995b) in controlling infection. Importantly, these early discoveries were corroborated by studies of human tuberculosis establishing the requirement of the same cytokines for protection. Patients with rheumatoid arthritis or Crohn’s disease who were latently infected with M. tuberculosis, when treated with anti–TNF-α antibodies or soluble receptor for TNF, showed an increased rate of progression to active tuberculosis (Keane et al., 2001), corroborating a critical role for TNF-α in human tuberculosis. Moreover, human Mendelian susceptibility to mycobacterial disease resulting from deficient IL-12 or IFN-γ signaling confers high susceptibility to M. tuberculosis and other mycobacterial infections (Newport et al., 1996; Altare et al., 1998; de Jong et al., 1998; Jouanguy et al., 1999; Fortin et al., 2007). Finally, it is well known that HIV-positive individuals with reduced CD4+ T cells, the main source of IFN-γ during M. tuberculosis infection, also display increased susceptibility to tuberculosis (Post et al., 1995). The mechanisms underlying the role of these cytokines was later described as an IL-12/IFN-γ axis in which IL-12, produced early in infection by APCs, promotes the differentiation of CD4+ T helper 1 (Th1) cells and IFN-γ production. IFN-γ further activates macrophages to produce TNF-α and other protective cytokines, promoting intracellular killing of the pathogen through the production of reactive oxygen and nitrogen species (Flynn and Chan, 2001; North and Jung, 2004; Cooper, 2009; O’Garra et al., 2013).
Type I IFN–inducible transcriptional signature in human tuberculosis
In contrast to the now well-established protective function of IFN-γ, the pathogenic role of type I IFN in tuberculosis has only recently been appreciated. The first evidence revealing a role for type I IFN in the pathogenesis of human tuberculosis was provided a few years ago by a transcriptomic study of patients with active disease and latently infected or healthy individuals from the UK and South Africa (Berry et al., 2010). Blood transcriptional profiles of patients with active tuberculosis were dominated by a type I IFN-inducible gene signature that correlated with the extent of lung radiographic disease and diminished with successful treatment (Berry et al., 2010). Several other studies have since verified these findings in additional patient cohorts from different geographic regions with diverse host genetic and tuberculosis epidemiological backgrounds (Maertzdorf et al., 2011a,b, 2012; Bloom et al., 2012, 2013; Ottenhoff et al., 2012; Cliff et al., 2013; Roe et al., 2016; Zak et al., 2016; Sambarey et al., 2017b; Singhania et al., 2017; Esmail et al., 2018; Table 1). In addition, integration and meta-analysis of diverse human tuberculosis datasets confirmed the reproducibility of the type I IFN–inducible blood transcriptional signature in human tuberculosis (Joosten et al., 2013; Blankley et al., 2016; Sambarey et al., 2017a; Singhania et al., 2017; Table 1). Analysis of purified cells from the blood of patients with active disease showed overexpression of IFN-inducible genes in neutrophils and monocytes, but not in CD4+ or CD8+ T cells (Berry et al., 2010; Bloom et al., 2013), suggesting that overactivation of monocytes and neutrophils by type I IFN during infection may contribute to disease pathogenesis.
Table 1. Type I IFN–inducible blood transcriptional signature in human tuberculosis.
| Original study | Geographic location | Sample type | Cohort size | ● Type I IFN signature shown in original study | o Type I IFN signature reported in subsequent analysis | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Active disease | Correlated radiographical disease | Treatment response | Joosten et al., 2013 | Blankley et al., 2016 | Sambarey et al., 2017a | Singhania et al., 2017 | ||||
| Jacobsen et al., 2007 | Germany | PBMCs | Active TB (n = 9); LTBI (n = 9) | o | ||||||
| Mistry et al., 2007 | South Africa | Whole blood | Active, recurrent, or cured TB; LTBI (n = 10/group) | o | ||||||
| Berry et al., 2010a | UK | Whole blood (and sorted cells) | Active TB (n = 21); LTBI (n = 21); HCs (n = 12) | ● | ● | ● | o | o | o | o |
| South Africa | Whole blood | Active TB (n = 20); LTBI (n = 31) | ● | o | o | o | o | |||
| Maertzdorf et al., 2011a | South Africa | Whole blood | Active TB (n = 46); LTBI (n = 25); HC (n = 37) | ● | o | o | ||||
| Maertzdorf et al., 2011b | The Gambia | Whole blood | Active TB (n = 33); LTBI (n = 34); HC (n = 9) | ● | o | |||||
| Bloom et al., 2012 | South Africa | Whole blood | active TB (n = 33); LTBI (n = 34); HC (n = 9) | ● | ● | o | o | |||
| Maertzdorf et al., 2012 | Germany | Whole blood | Active TB (n = 8); LTBI (n = 4); HC (n = 14) | ● | o | o | ||||
| Ottenhoff et al., 2012 | Indonesia | PBMCs | Active TB over time during treatment (n = 23); HC (n = 23) | ● | ● | o | o | |||
| Cliff et al., 2013 | South Africa | Whole blood | Active TB over time during treatment (n = 27) | ● | ● | o | o | o | ||
| Bloom et al., 2013 | UK | Whole blood (and sorted cells) | Active TB (n = 35); HCs (n = 113) | ● | o | o | o | |||
| Kaforou et al., 2013 | South Africa, Malawi | Whole blood | Active TB (HIV−/+; n = 195); LTBI (HIV−/+; n = 167) | o | o | |||||
| Cai et al., 2014 | China | PBMCs | active TB (n = 9); LTBI (n = 6); HC (n = 6) | o | ||||||
| Anderson et al., 2014 | Kenya | Whole blood | Active TB (n = 79); LTBI (n = 14) | o | ||||||
| South Africa, Malawi | Active TB (n = 110); LTBI (n = 54) | o | ||||||||
| Roe et al., 2016 | UK | Whole blood | Active TB (n = 46); postrecovery (n = 31) | ● | ● | |||||
| Zak et al., 2016 | South Africa | Whole blood | Progressors (n = 40); nonprogressors (n = 104) | ● | o | |||||
| Sambarey et al., 2017b | India | Whole blood | Active TB (n = 19); LTBI (n = 13); HCs (n = 15) | ● | ||||||
| Singhania et al., 2017 | UK | Whole blood | Active TB (n = 53); LTBI (n = 49); HCs (n = 50) | ● | ||||||
| Esmail et al., 2018 | South Africa | Whole blood | Active TB (n = 15); subclinical TB (n = 10); latent TB (n = 25) | ● | ||||||
Type I IFN signature reported in original study (●) and/or in subsequent analysis by others (o). HC, healthy control; LTBI, latent tuberculosis infection; PBMC, peripheral blood mononuclear cell; TB, tuberculosis.
Original study providing the first data in human disease to support a role for type I IFN in the pathogenesis of tuberculosis.
Similar findings have since been observed in other mycobacterial infections. In human leprosy, caused by Myobacterium leprae, type I IFN and their downstream genes, were preferentially expressed in lesions of the disseminated and progressive lepromatous form at the site of disease, whereas IFN-γ and its downstream genes were preferentially expressed in the lesions from patients with the self-healing tuberculoid form (Teles et al., 2013). Overexpression of IFN-inducible genes in blood has also been reported in patients with other pulmonary diseases such as sarcoidosis and acute influenza infection but has not been detected in other bacterial infections (Berry et al., 2010; Maertzdorf et al., 2012; Bloom et al., 2013; Singhania et al., 2017). However, distinct gene patterns have been detected between tuberculosis and sarcoidosis (Bloom et al., 2013) and tuberculosis and influenza infection (Singhania et al., 2017).
In tuberculosis, overexpression of IFN response genes, including STAT1, IFITs, GBPs, MX1, OAS1, IRF1, and other genes, were also detected early in tuberculosis contacts who progressed to active disease (Zak et al., 2016; Scriba et al., 2017; Singhania et al., 2017; Esmail et al., 2018), suggesting that peripheral activation of the type I IFN response precedes the onset of active disease and clinical manifestations of tuberculosis. These changes apparently preceded the up-regulation of other innate immune responses and down-regulation of genes associated with specific lymphocyte cell populations (Scriba et al., 2017; Singhania et al., 2017), although this conclusion requires more detailed studies. The type I IFN–inducible signature was also present in 10–25% of latently infected patients who are asymptomatic (Berry et al., 2010; Singhania et al., 2017), suggesting that these patients may be at the highest risk to progress to active disease.
Several clinical studies have reported reactivation of tuberculosis in patients undergoing IFN-α based therapy for chronic viral hepatitis (Sabbatani et al., 2006; Farah and Awad, 2007; Telesca et al., 2007; Belkahla et al., 2010; Guardigni et al., 2012; Abutidze et al., 2016; de Oliveira Uehara et al., 2016; Matsuoka et al., 2016). In addition, a very recent study showed that impaired type I IFN signaling due to a rare East Asian functional mutation in the IFNAR1 gene was associated with increased resistance to tuberculosis (Zhang et al., 2018). As the control population consisted of healthy individuals, it is presently unclear whether this polymorphism influences susceptibility to infection as opposed to disease progression in latent infected individuals. In contrast, patients with an inherited deficiency in the gene encoding ISG15, who displayed immunological and clinical signs of enhanced type I IFN responses (Zhang et al., 2015), were shown to be more susceptible to mycobacterial infections (Bogunovic et al., 2012). Together, these findings provide strong evidence that type I IFN signaling correlates with impaired control of M. tuberculosis and other mycobacterial infections and underlies an increased risk of tuberculosis in humans.
Foe-like pathogenic role of high and sustained type I IFN in tuberculosis: Evidence from mouse models
Following on from the observations in human tuberculosis, whole-genome blood transcriptional profiling in animal models of tuberculosis have since shown up-regulation of type I IFN response related genes in response to M. tuberculosis infection in both nonhuman primates (Gideon et al., 2016) and mice (Domaszewska et al., 2017). However, different effects of type I IFN in either protection or pathogenesis in experimental models of tuberculosis have been reported as we discuss herein. Several studies have reported reduced bacterial loads (Ordway et al., 2007; Stanley et al., 2007; Mayer-Barber et al., 2011; Dorhoi et al., 2014) and/or improved host survival (Manca et al., 2005; Dorhoi et al., 2014; Kimmey et al., 2017) upon M. tuberculosis infection of IFNAR-deficient (Ifnar1−/−) mice compared with WT controls although this phenotype has not been universally observed (Cooper et al., 2000; Antonelli et al., 2010; Desvignes et al., 2012; McNab et al., 2013; Redford et al., 2014; Moreira-Teixeira et al., 2016, 2017). The reasons underlying the discrepancies in the results of these studies (summarized in Table 2) are presently unclear but may relate to differences in the M. tuberculosis strains used for challenge, host genetic background of the different colonies of Ifnar1−/− mice used, or environmental differences.
Table 2. Summary of outcomes reported following aerosol infection with M. tuberculosis in Ifnar−/− mice.
| Mtb strain | Dose (CFU) | Mouse background | Bacterial loadsa | Lung pathologya | Survivala | Reference | |
|---|---|---|---|---|---|---|---|
| Lung | Spleen | ||||||
| H37Rv | 100 | C57BL/6 (R) | Increased (day 28 p.i.) | — | — | — | Antonelli et al., 2010 |
| H37Rv | 100–150 | C57BL/6 (R) | Decreased (day 28 p.i.) | — | — | — | Mayer-Barber et al., 2011 |
| H37Rv | 100 | C57BL/6 (R) | Transiently decreased (day 18 p.i.) but similar at day 25 p.i. | — | — | Similar (day 70 p.i.) | Desvignes et al., 2012 |
| H37Rv | 50–150 | C57BL/6 (R) | Similar (day 27 p.i.) | — | — | — | Redford et al., 2014 |
| H37Rv | 100–150 | C57BL/6 (R) | — | — | — | Similar (day 80 p.i.) | Mayer-Barber et al., 2014 |
| H37Rv | 500 (200 for survival) | C57BL/6 (R) | Decreased (day 42 p.i.) | — | — | Similar (day 80 p.i.) | Dorhoi et al., 2014 |
| 200 | 129S2 (S) | Decreased (day 21 p.i.) | — | Decreased (day 21 p.i.) | Increased | ||
| H37Rv | 50–100 | KO 129 (S); WT C57BL/6 (R) | Decreased (chronic phase, > day 75 p.i.) | — | — | — | Ordway et al., 2007 |
| Erdman | 106 (i.v.) | C57BL/6 (R) | Similar (days 10, 21 p.i.) | Decreased (days 10, 21 p.i.) | — | — | Stanley et al., 2007 |
| Erdman | 100 | C57BL/6 (R) | Transiently decreased (day 7 p.i.) but similar at days 21, 77, 100, 245 p.i. | Transiently decreased at day 77 p.i. but similar at other time points | Similar (days 100, 275 p.i.) | Increased | Kimmey et al., 2017 |
| Erdman | 100 | B6/129 (S) | Transiently increased (days 10, 20, 40 p.i.) but similar by day 80 p.i. | — | — | — | Cooper et al., 2000 |
| Erdman | 50–100 | KO 129 (S); WT C57BL/6 (R) | Decreased (> day 25 p.i.) | — | — | Similar (day 200 p.i.) | Ordway et al., 2007 |
| CSU93 | 50–100 | KO 129 (S); WT C57BL/6 (R) | Decreased (> day 25 p.i.) | — | — | Similar (day 200 p.i.) | Ordway et al., 2007 |
| HN878 | 100–200 | C57BL/6 (R) | Similar (days 28, 60 p.i.) | — | — | — | Moreira-Teixeira et al., 2017 |
| HN878 | 30 | C57BL/6 (R) | Similar (day 48 p.i.) | — | — | — | McNab et al., 2013 |
| HN878 | 100–200 | 129 (S) | — | — | — | Increased | Manca et al., 2005 |
| HN878 | 50–100 | KO 129 (S); WT C57BL/6 (R) | Decreased (> day 50 p.i.) | — | — | Similar (day 200 p.i.) | Ordway et al., 2007 |
| BTB 02-171 | 30 | C57BL/6 (R) | Similar (day 56 p.i.) | — | — | — | McNab et al., 2013 |
| BTB 02-171 | 100–200 | C57BL/6 (R) | Similar (days 20, 24, 27 p.i.) | — | Similar (day 27 p.i.) | Similar (> day 200 p.i.) | Moreira-Teixeira et al., 2016 |
Dashes indicate no data reported. Mtb, M. tuberculosis; p.i., postinfection; (R), M. tuberculosis–resistant mouse strain; (S), M. tuberculosis–susceptible mouse strain.
Compared with WT mice.
Elevated production of type I IFN has been associated with the virulence of M. tuberculosis strains and increased host susceptibility. Studies of infection with hypervirulent M. tuberculosis clinical isolates (e.g., HN878 and BTB 02–171) compared with the less virulent laboratory strain (H37Rv) showed a positive correlation between increased levels of type I IFN and increased mycobacterial virulence (Manca et al., 2001, 2005; Ordway et al., 2007; Carmona et al., 2013). However, similar bacterial loads and survival were reported in Ifnar1−/− and WT controls in the M. tuberculosis–resistant C57BL/6 genetic background (McNab et al., 2013; Moreira-Teixeira et al., 2017). IFNAR deficiency in mice with a M. tuberculosis–susceptible genetic background (A129, 129S2) enhanced host survival following infection with not only the hypervirulent strain HN878 (Manca et al., 2005) but also the less virulent H37Rv strain (Dorhoi et al., 2014). A stronger up-regulation of type I IFN response related genes has recently been reported in the blood of the highly susceptible 129S2 mouse strain compared with the resistant C57BL/6 mouse strain early after infection with the M. tuberculosis strain H37Rv (Domaszewska et al., 2017). This increased expression of type I IFN–inducible genes detected in 129S2 compared with C57BL/6 mice is consistent with the results showing that the detrimental effect of type I IFN during M. tuberculosis (laboratory strain H37Rv) infection is more pronounced in the highly susceptible 129S2 strain than the resistant C57BL/6 strain (Dorhoi et al., 2014). However, a transient protective effect of type I IFN has also been observed during M. tuberculosis (Erdmann strain) infection using the 129 strain (Cooper et al., 2000). Differing findings may relate to the genetic variation among 129 substrains (Simpson et al., 1997; Festing et al., 1999) in addition to M. tuberculosis strain type and the host environment. Experiments in which Ifnar1−/− mice of either susceptible or resistant genetic backgrounds are studied side by side are needed to investigate the basis of the discrepancy across all the studies.
Robust evidence for the role of high and sustained levels of type I IFN in M. tuberculosis persistence and disease pathogenesis has been further provided by experimental models of type I IFN overexpression. Direct instillation of purified murine IFN-α/β into the lungs of M. tuberculosis–infected B6D2/F1 mice resulted in increased lung bacterial loads and reduced survival of infected mice (Manca et al., 2001). Prolonged induction of high levels of type I IFN by intranasal administration of the TLR3 ligand poly-ICLC during M. tuberculosis infection impaired control of bacterial growth and exacerbated pulmonary immunopathology in WT mice, but not in Ifnar−/− mice (Antonelli et al., 2010; Mayer-Barber et al., 2014). Host coinfection with influenza A virus, another well-known inducer of type I IFN, has also been shown to reduce host resistance to M. tuberculosis infection via the action of type I IFN, because this was observed in WT, but not in IFNAR-deficient, mice (Redford et al., 2014). Likewise, abrogation of negative regulators of type I IFN signaling leading to elevated levels of type I IFN resulted in impaired M. tuberculosis clearance (McNab et al., 2013; Dauphinee et al., 2014). Specifically, deletion of the MAPK kinase kinase 8 (MAP3K8; also known as TPL2), a negative regulator of type I IFN induction downstream of TLR, resulted in increased bacterial loads upon M. tuberculosis or L. monocytogenes infection, which was not observed in the absence of IFNAR (McNab et al., 2013). Impaired control of the bacterial loads in the absence of TPL2 was correlated with type I IFN–dependent induction of IL-10 and suppression of IL-12 production during infection (McNab et al., 2013). Mice carrying a loss-of function mutation within the ubiquitin-specific peptidase 18 (Usp18) gene, which results in increased levels of type I IFN production and hyperactivation of type I IFN signaling during bacterial infection, were also shown to be more susceptible to M. tuberculosis infection, showing increased bacterial burdens and decreased survival (Dauphinee et al., 2014). Collectively, these studies performed in experimental mouse models of M. tuberculosis infection point to a detrimental role of high and sustained levels of type I IFN in exacerbating tuberculosis.
Mechanisms of type I IFN induction in M. tuberculosis infection
Because type I IFN has been associated with disease pathogenesis, identifying mechanisms regulating type I IFN induction during M. tuberculosis infection has been an active area of research (Donovan et al., 2017; Sabir et al., 2017). In vitro studies in human and murine cells have shown that distinct mycobacterial molecules and signaling pathways may be involved in the induction of this family of cytokines during M. tuberculosis infection (Fig. 1). High levels of type I IFN are preferentially induced by virulent strains of M. tuberculosis (Manca et al., 2001, 2005; Novikov et al., 2011; Carmona et al., 2013), and this property seems to depend on the well-known mycobacterial virulence factor ESX-1 protein secretion system (Stanley et al., 2007; Manzanillo et al., 2012; Wassermann et al., 2015).
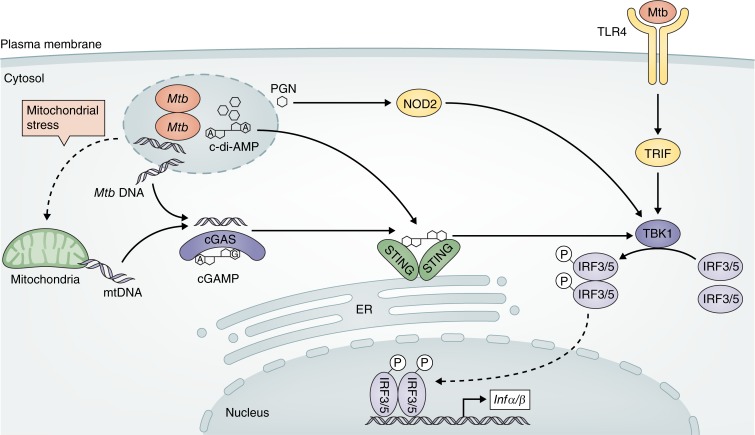
Figure 1.
Alternative pathways of type I IFN induction during M. tuberculosis infection. Recognition of mycobacterial products by a range of cell surface and cytosolic PRR, including TLR4, NOD2, and STING, activates the kinase TBK1 leading to phosphorylation (P) and dimerization of IRF3 or IRF5, which translocates into the nucleus and promotes transcription of type I IFN genes. Release of mycobacterial or mitochondrial DNA in the cytosol activates cGAS, which synthesizes cGAMP. Host-derived cGAMP and/or mycobacterial-derived c-di-AMP activates the STING pathway and the downstream TBK1–IRF3 signaling axis. Peptidoglycan fragments can be sensed by NOD2 in the cytosol, activating the TBK1–IRF5 signaling pathway. Detection of extracellular M. tuberculosis and/or its products by TLR4 triggers TRIF-TBK1-IRF3–dependent induction of type I IFN by certain strains. Mtb, M. tuberculosis; mtDNA, mitochondrial DNA; PGN, peptidoglycan.
Recognition of mycobacterial products in the cytosol, such as peptidoglycan fragments, by the cytosolic sensor nucleotide-binding oligomerization containing protein 2 (NOD2) has been shown to induce type I IFN expression by infected murine macrophages (Leber et al., 2008; Pandey et al., 2009). NOD2 activates the downstream serine/threonine-protein kinase (TBK1)–IFN regulatory factor 5 (IRF5) signaling pathway, leading to the production of type I IFN (Pandey et al., 2009). Three independent studies have identified a central role for the cytosolic DNA sensor nucleotidyltransferase cyclic GMP-AMP synthase (cGAS) in the detection of mycobacterial DNA in the host cytosol and induction of type I IFN transcription in macrophages (Collins et al., 2015; Wassermann et al., 2015; Watson et al., 2015). The sensing of DNA by cGAS leads to the synthesis of the second messenger cyclic di–GMP-AMP (cGAMP), which activates the stimulator of IFN genes (STING) and the downstream TBK1–IRF3 signaling pathway, culminating in the transcription of type I IFN (Manzanillo et al., 2012; Collins et al., 2015; Wassermann et al., 2015; Watson et al., 2015). Mycobacterial DNA in the host cytosol can also be sensed by absent in melanoma 2 (AIM-2) protein, which partially contributes to the activation of the NLRP3-inflammasome, promoting the maturation of the protective cytokine IL-1β (Shah et al., 2013; Wassermann et al., 2015). This AIM-2–IL-1β signaling pathway has been recently reported to negatively regulate the STING-dependent type I IFN production in macrophages and dendritic cells (DCs) by inhibiting the association between STING and TBK1 (Yan et al., 2018).
A more recent study suggested that cGAS/STING–dependent type I IFN induction in macrophages can also be triggered by mitochondrial DNA released into the cytosol due to mitochondrial stress caused by M. tuberculosis infection (Wiens and Ernst, 2016). Production of cyclic di-AMP (c-di-AMP) by M. tuberculosis has also been shown to promote type I IFN induction in a STING-dependent but cGAS-independent mechanism (Dey et al., 2015). STING-dependent induction of type I IFN can be regulated by mycobacterial and host phosphodiesterases, which inhibit STING activation and consequent induction of type I IFN by hydrolysis of both mycobacterial-derived c-di-AMP and host-derived cGAMP (Dey et al., 2017).
TLRs are another set of pattern recognition receptors (PRRs) that are important for sensing M. tuberculosis infection (Stamm et al., 2015). Whereas some M. tuberculosis strains activate mainly TLR2, others also activate TLR4 and the downstream MyD88-independent TIR domain–containing adapter-inducing IFN-β (TRIF), resulting in different macrophage responses characterized by varying production of type I IFN (Carmona et al., 2013; Moreira-Teixeira et al., 2016). Sensing of M. tuberculosis infection by C-type lectin receptors has also been reported to induce type I IFN production by B cells (Benard et al., 2017) and to amplify type I IFN responses in DCs (Troegeler et al., 2017). In B cells, induction of type I IFN during M. tuberculosis infection has also been shown to require the STING signaling pathway and to be negatively regulated by MyD88 (Benard et al., 2017).
These studies indicate that different M. tuberculosis strains can induce type I IFN by triggering multiple cell surface and cytosolic PRR and downstream signaling pathways (Fig. 1), which may contribute to the differential levels of type I IFN and virulence induced by distinct strains. Further studies are required to determine the relative importance of each of these pathways in vivo and determine how widely they function in different cell types, as they could provide novel targets for tuberculosis prevention and therapy.
Pathogenic effects of type I IFN signaling during M. tuberculosis infection
Studies performed in patients and mouse models of infection collectively point to a harmful role of high and sustained type I IFN in tuberculosis (discussed above). However, the mechanisms by which type I IFN signaling exacerbates M. tuberculosis infection are not yet fully understood (Mayer-Barber and Sher, 2015; McNab et al., 2015; Donovan et al., 2017; Sabir et al., 2017). Early studies with hypervirulent strains of M. tuberculosis associated the induction of higher levels of type I IFN with the suppression of proinflammatory cytokines and impaired antibacterial Th1 responses (Manca et al., 2001, 2005; Ordway et al., 2007). The inhibition of host-protective cytokines and innate cell responsiveness in addition to IFN-γ–driven antibacterial effects by type I IFN has since been verified by several other studies both in human cells (Mayer-Barber et al., 2011; Novikov et al., 2011; de Paus et al., 2013; Teles et al., 2013) and mouse models (Mayer-Barber et al., 2011; McNab et al., 2013, 2014; Dorhoi et al., 2014). In addition, type I IFN has also been shown to promote early cell death of alveolar macrophages (Dorhoi et al., 2014) and boost the local accumulation of permissive myeloid cells, which contribute to the spread of infection and pulmonary inflammation (Antonelli et al., 2010; Dorhoi et al., 2014), underlining the complex role of type I IFN during M. tuberculosis infection (Fig. 2).
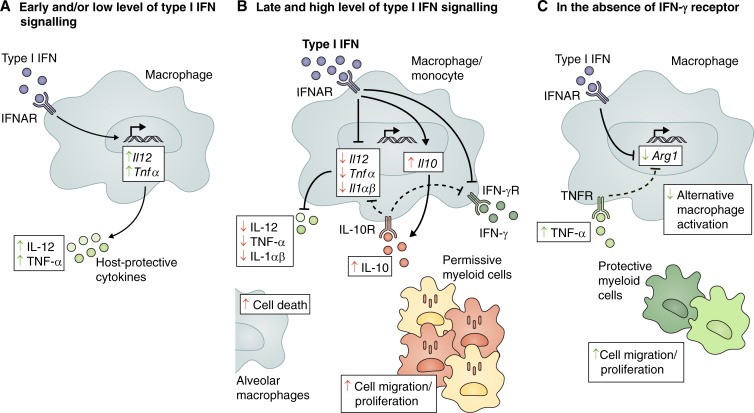
Figure 2.
Foe- and friendly-like effects of type I IFN during M. tuberculosis infection. Type I IFN has been reported to play both negative (red arrows) and positive (green arrows) functions during M. tuberculosis infection. (A) Tonic levels of autocrine type I IFN signaling prime the production of protective cytokines IL-12 and TNF-α. (B) However, high and sustained levels of type I IFN promote the production of IL-10 and inhibit the production of protective cytokines IL-12, TNF-α, IL-1α, and IL-1β. IL-10 mediates a suppressive feedback loop, contributing to the decreased production of IL-12 and TNF-α. Type I IFN also inhibits myeloid cell responsiveness to IFN-γ by both IL-10–dependent and independent mechanisms, suppressing IFN-γ–dependent host-protective immune responses. In addition, type I IFN can promote cell death in alveolar macrophages and accumulation of permissive myeloid cells at the site of infection. (C) In the absence of the IFN-γ receptor, type I IFN inhibits Arg1 expression directly or indirectly by increasing TNF-α levels, thus regulating macrophage activation toward a more protective phenotype. Type I IFN signaling can also promote the recruitment, differentiation, and/or survival of protective myeloid cells that control pathology at the site of infection. Arg1, arginase 1; IFNγR, IFN-γ receptor; IL-10R, IL-10 receptor; TNFR, TNF-α receptor.
Type I IFN has been reported to inhibit the production of IL-1α and IL-1β, which are critical for host defense against M. tuberculosis infection (Mayer-Barber et al., 2010, 2011), both in vitro in infected human (Mayer-Barber et al., 2011; Novikov et al., 2011; de Paus et al., 2013) and mouse myeloid cells (Mayer-Barber et al., 2011; McNab et al., 2014) and in vivo in mouse models (Mayer-Barber et al., 2011; Dorhoi et al., 2014). Prostaglandin E2 (PGE2) has been shown to be a critical downstream mediator of IL-1–dependent host resistance, and accordingly, type I IFN has been shown to limit PGE2 in vitro in human and mouse cells and in vivo (Mayer-Barber et al., 2014). PGE2 is known to prevent necrosis of M. tuberculosis–infected macrophages by promoting apoptosis, a cell death modality that contains the pathogen, limiting its dissemination (Chen et al., 2008; Divangahi et al., 2009). Thus, an additional mechanism by which type I IFN could enhance M. tuberculosis infection and disease is through promotion of necrosis as a consequence of PGE2 inhibition.
In turn, it has recently been demonstrated that there is a reciprocal control of type I IFN regulation by IL-1β through a PGE2-mediated mechanism (Mayer-Barber et al., 2014). Increasing the levels of PGE2 during M. tuberculosis infection, by direct administration of this prostanoid or by increasing its level through 5-lipoxygenase blockade with zileuton, limited type I IFN induction and conferred host resistance to infection in mice (Mayer-Barber et al., 2014). These findings suggest zileuton as a drug that could be used for host-directed therapy of tuberculosis by limiting detrimental type I IFN overproduction.
The production of other proinflammatory cytokines such as TNF-α and IL-12 has also been shown to be negatively affected by the addition of exogenous IFN-α or IFN-β to human monocytes (de Paus et al., 2013) and murine macrophages (McNab et al., 2014) in in vitro cultures. Type I IFN has also been reported to induce the production of the immunosuppressive cytokine IL-10 in vitro in macrophages (Mayer-Barber et al., 2011; McNab et al., 2014) and in vivo in CD4+ T cells (Moreira-Teixeira et al., 2017), which has been shown to increase susceptibility to M. tuberculosis infection (Redford et al., 2011). In addition, the increased susceptibility of M. tuberculosis–infected mice given the double-stranded RNA homologue poly-ICLC, which led to enhanced type I IFN production, was shown to be dependent on IL-10 production (Mayer-Barber et al., 2014). The inhibitory effect of type I IFN on the production of IL-12 and TNF-α by M. tuberculosis–infected macrophages is abrogated in the absence of IL-10 (McNab et al., 2014). However, the inhibitory effect of type I IFN on IL-1β production is only slightly affected by IL-10 deficiency (Mayer-Barber et al., 2011; McNab et al., 2014), indicating that type I IFN suppresses the macrophage response to M. tuberculosis infection by both IL-10–dependent and –independent mechanisms.
In addition, type I IFN has been shown to repress macrophage/monocyte responsiveness to the antibacterial effects of IFN-γ during mycobacterial infections (de Paus et al., 2013; Teles et al., 2013; McNab et al., 2014). In both mouse and human cells, type I IFN has been shown to suppress the ability of monocytes and macrophages to up-regulate antimycobacterial effector molecules and to restrict bacterial growth in response to both M. tuberculosis and M. leprae (Teles et al., 2013; McNab et al., 2014). This inhibitory effect of type I IFN on the antimycobacterial activity of IFN-γ in human macrophages has shown to be mediated by IL-10 (Teles et al., 2013). However, the inhibition of IFN-γ–induced cytokine production by type I IFN in murine macrophages seems to be mediated by both IL-10–dependent and –independent mechanisms (McNab et al., 2014). Down-regulation of IFN-γ–driven inducible nitric oxide synthase and IL-12/23 p40 by type I IFN has also been reported in lung myeloid cells during M. tuberculosis infection in vivo (Mayer-Barber et al., 2011; Mayer-Barber and Sher, 2015). Although induction of IL-10 by type I IFN has been reported during M. tuberculosis infection in vivo (Moreira-Teixeira et al., 2017), it is not yet clear whether IL-10 is responsible for the suppressive effects of type I IFN on IFN-γ function in vivo.
Friendly protective functions of type I IFN in tuberculosis
Although a role for type I IFN in the pathogenesis of tuberculosis is strongly supported by studies in both human tuberculosis (Zhang et al., 2018) and studies in experimental mouse models of M. tuberculosis infection (Antonelli et al., 2010; McNab et al., 2013; Dorhoi et al., 2014; Mayer-Barber et al., 2014; Redford et al., 2014), there is also evidence that type I IFN can display protective functions under specific conditions. Several clinical case reports have described improved clinical symptoms and decreased bacterial burden after coadministration of IFN-α together with antimycobacterial chemotherapy to patients with active tuberculosis who were not responsive to conventional treatment and/or had recurrent disease (Giosué et al., 1998; Palmero et al., 1999; Giosuè et al., 2000; Mansoori et al., 2002; Zarogoulidis et al., 2012). These clinical studies of IFN-α adjunct therapy were designed before the now numerous studies linking type I IFN expression with active tuberculosis, and the mechanisms underlying the beneficial effects of IFN-α administration in these patients remain unclear. Therapeutic effects of IFN-α have also been reported in young patients suffering from mycobacterial infections with complete or partial IFN-γ receptor (IFNGR) signaling deficiencies when administered together with antimycobacterial chemotherapy (Ward et al., 2007; Bax et al., 2013).
A protective role for type I IFN in the absence of IFN-γ signaling has also been proposed in mouse models of M. tuberculosis infection (Desvignes et al., 2012; Moreira-Teixeira et al., 2016), suggesting that the dominant suppressive effect of type I IFN on IFN-γ antimycobacterial activity may mask potentially protective functions of this family of cytokines (Fig. 2). These studies reported increased pulmonary pathology and early mortality following M. tuberculosis infection in mice deficient in both type I IFN and IFN-γ receptors (Ifngr1−/−Ifnar1−/−) compared with single IFNGR-deficient mice (Ifngr1−/−; Desvignes et al., 2012; Moreira-Teixeira et al., 2016). In contrast to what has been reported in the immunocompetent host, where type I IFN signaling may promote local accumulation of permissive myeloid cells that contribute to the spread of infection and pulmonary inflammation (Antonelli et al., 2010; Dorhoi et al., 2014), in the absence of IFN-γ signaling, type I IFN may facilitate the recruitment, differentiation, and/or survival of myeloid cells that control pathology (Desvignes et al., 2012). Although no difference in the lung bacterial burden was observed during infection with the less virulent laboratory strain H37Rv (Desvignes et al., 2012), infection with the hypervirulent strain BTB 02–171, shown to induce high levels of type I IFN (Carmona et al., 2013), revealed the ability of type I IFN to also control bacterial replication (Moreira-Teixeira et al., 2016). Following infection with BTB 02–171, increased lung bacterial loads were observed in double Ifngr1−/−Ifnar1−/− mice, which are deficient in both type I IFN and IFN-γ receptors, compared with single Ifngr1−/−mice. Increased control of bacterial replication in the absence of IFN-γ signaling correlated with suppression of alternative macrophage activation by type I IFN, likely by direct regulation of macrophage activation as well as by regulation of cytokine expression in the infected lungs (Moreira-Teixeira et al., 2016). These findings may help to explain some of the mechanisms underlying the beneficial effects of IFN-α treatment, at least in patients with compromised IFN-γ responses, and provide new avenues for host-directed therapies in tuberculosis.
Type I IFN has also been reported to increase host resistance in mouse models of infection against avirulent mycobacterial strains, such as Mycobacterium smegmatis (Ruangkiattikul et al., 2017), Mycobacterium avium (Denis, 1991), and Mycobacterium bovis bacillus Calmette-Guérin (BCG; Kuchtey et al., 2006). In addition, it has been suggested that type I IFN may improve the immunogenicity of BCG vaccination against M. tuberculosis infection in mouse and guinea pig models (Bottai et al., 2015; Gröschel et al., 2017; Rivas-Santiago and Guerrero, 2017). Recombinant expression of the ESX-1 protein secretion system in the attenuated BCG vaccine has been shown to increase the protective effect of vaccination against M. tuberculosis infection (Bottai et al., 2015; Gröschel et al., 2017). This increased protection correlated with enhanced IFN-β production by macrophages infected with recombinant ESX-1–expressing BCG (Gröschel et al., 2017). However, the contribution of type I IFN in the protection conferred by ESX-1–expressing BCG vaccine still requires investigation. In support for a role of type I IFN in increasing BCG vaccine efficacy, an independent study has shown that administration of IFN-α during BCG vaccination promotes the production of host-protective cytokines (e.g., IFN-γ, TNF-α, and IL-12) and confers increased protection against M. tuberculosis infection over that observed with BCG alone (Rivas-Santiago and Guerrero, 2017). These findings suggest that type I IFN may play a friendly protective role in the context of BCG-induced immunity and could be targeted to improve preventive vaccination against tuberculosis.
In vitro experiments in human peripheral blood cells have shown that treatment with type I IFN can increase DC maturation and IL-12 production following BCG infection, which then promotes T cell priming and production of IFN-γ (Giacomini et al., 2009), and may help to explain the positive effects of type I IFN during vaccination. Whereas addition of IFN-β at the time of infection has been shown to negatively affect proinflammatory cytokine production via IL-10–dependent and –independent mechanisms (Mayer-Barber et al., 2011; McNab et al., 2014), basal type I IFN signaling has been shown to be required for maximal production of IL-12 and TNF-α by macrophages in response to M. tuberculosis infection (McNab et al., 2014). Indeed, Ifnar1−/− macrophages have been shown to produce lower levels of IL-12 and TNF-α in response to M. tuberculosis infection compared with WT macrophages (McNab et al., 2014). Tonic type I IFN signaling has also been reported to be important for optimal IL-12 production by macrophages (Howes et al., 2016) and DCs (Gautier et al., 2005) in other contexts. Moreover, pretreatment of WT macrophages with IFN-β for >8 h before M. tuberculosis infection enhanced the production of IL-12 and TNF-α (McNab et al., 2014), indicating that the timing of type I IFN signaling may be important in determining the effects of type I IFN during M. tuberculosis infection. Thus, these findings suggest that early tonic type I IFN signaling can elicit friendly functions during M. tuberculosis infection by priming innate immune cells for the production of proinflammatory host-protective cytokines (Fig. 2).
Closing remarks
Members of the type I IFN family were first discovered for their ability to interfere with viral replication and are well known for their antiviral responses (Yan and Chen, 2012; Crouse et al., 2015). Unlike their largely protective role in viral infections, this family of cytokines plays a less predictable role in bacterial infection (Trinchieri, 2010; McNab et al., 2015; Stifter and Feng, 2015; Mayer-Barber and Yan, 2017). Over the past decade, numerous studies have uncovered a foe-like pathogenic role of type I IFN in tuberculosis. Patients with active tuberculosis have a blood transcriptional gene signature dominated by type I IFN–related genes that is correlated with disease severity and is down-regulated following successful treatment (Table 1). In experimental models, elevated levels of type I IFN are associated with the virulence of M. tuberculosis strains and increased host susceptibility. Several mechanisms underlying the pathogenic role of type I IFN in tuberculosis have been described, including induction of IL-10 and negative regulation of the IL-12/IFN-γ and IL-1β/PGE2 host-protective responses. However, there is also evidence that type I IFN may play a friendly role in certain contexts, highlighting the complex role of type I IFN in tuberculosis (Fig. 2). Although high levels of type I IFN have negative effects during the course of M. tuberculosis infection, tonic I IFN signaling or low levels of type I IFN in the context of low mycobacterial loads may in turn have positive effects by priming host-protective responses. This may explain the protective effects of IFN-α therapy together with anti-mycobacterial drugs in patients with active disease. Thus, it is likely that a balanced induction of this family of cytokines is required for optimal protection. Various signaling pathways have been described to induce type I IFN production in response to M. tuberculosis infection (Fig. 1). Differential activation of these pathways and/or high or low engagement of these signaling pathways during infection may contribute to the induction of distinct levels of type I IFN and the differential virulence of M. tuberculosis strains. In addition, potential differences in temporal and spatial induction of individual IFN-α subtypes and IFN-β during infection could contribute to variations in disease outcome and determine foe or friend features of the type I IFN response. A further unexplored area is the possible differential role of IFNAR subunits (IFNAR1 and IFNAR2) in determining the outcome of type I IFN signaling. Dominant IFNAR1 expression has been shown to favor IFN-β ligation and activation of the protein kinase B (also known as Akt) pathway over the classical JAK–STAT pathway (de Weerd et al., 2013). Therefore, differential expression of IFNAR subunits on the relevant responding cells during M. tuberculosis infection could be another factor regulating detrimental versus protective activities of type I IFN signaling during infection. Deciphering the mechanisms underlying the differential induction of type I IFN and what determines the outcome of type I IFN signaling during M. tuberculosis infection, from induction of IL-10 to the regulation of the IL-12/IFN-γ and IL-1β/PGE2 host-protective responses, may offer new avenues for host-directed therapies for tuberculosis. In addition, such research may yield important basic information about type I IFN induction and function that will enhance our broader understanding of how this major cytokine family impacts on disease outcomes.
Acknowledgments
We thank Akul Singhania for critically reading the manuscript.
L. Moreira-Teixeira and A. O’Garra were supported by The Francis Crick Institute (FC001126), which receives its core funding from Cancer Research UK, the UK Medical Research Council, and the Wellcome Trust. K. Mayer-Barber and A. Sher were supported by the Intramural Research Program of the National Institutes of Allergy and Infectious Diseases.
The authors declare no competing financial interests.
Author contributions: L. Moreira-Teixeira and A. O'Garra designed the manuscript. L. Moreira-Teixeira developed the figures and tables and wrote and edited the manuscript. K. Mayer-Barber, A. Sher, and A. O'Garra contributed to the critical review of the literature and reviewed the article before final submission. All authors read and approved the manuscript prior to submission.
References
- Abutidze A., Bolokadze N., Chkhartishvili N., Sharvadze L., and Tsertsvadze T.. 2016. Incidence of Tuberculosis among Hiv/Hcv Co-Infected Patients Receiving Hepatitis C Treatment with Pegylated Interferon and Ribavirin in Georgia. Georgian Med. News. 252:10–15. [PMC free article] [PubMed] [Google Scholar]
- Altare F., Durandy A., Lammas D., Emile J.F., Lamhamedi S., Le Deist F., Drysdale P., Jouanguy E., Döffinger R., Bernaudin F., et al. 1998. Impairment of mycobacterial immunity in human interleukin-12 receptor deficiency. Science. 280:1432–1435. 10.1126/science.280.5368.1432 [DOI] [PubMed] [Google Scholar]
- Anderson S.T., Kaforou M., Brent A.J., Wright V.J., Banwell C.M., Chagaluka G., Crampin A.C., Dockrell H.M., French N., Hamilton M.S., et al. 2014. Diagnosis of childhood tuberculosis and host RNA expression in Africa. N. Engl. J. Med. 370:1712–1723. 10.1056/NEJMoa1303657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonelli L.R., Gigliotti Rothfuchs A., Gonçalves R., Roffê E., Cheever A.W., Bafica A., Salazar A.M., Feng C.G., and Sher A.. 2010. Intranasal Poly-IC treatment exacerbates tuberculosis in mice through the pulmonary recruitment of a pathogen-permissive monocyte/macrophage population. J. Clin. Invest. 120:1674–1682. 10.1172/JCI40817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banchereau J., and Pascual V.. 2006. Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity. 25:383–392. 10.1016/j.immuni.2006.08.010 [DOI] [PubMed] [Google Scholar]
- Bax H.I., Freeman A.F., Ding L., Hsu A.P., Marciano B., Kristosturyan E., Jancel T., Spalding C., Pechacek J., Olivier K.N., et al. 2013. Interferon alpha treatment of patients with impaired interferon gamma signaling. J. Clin. Immunol. 33:991–1001. 10.1007/s10875-013-9882-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkahla N., Kchir H., Maamouri N., Ouerghi H., Hariz F.B., Chouaib S., Chaabouni H., and Mami N.B.. 2010. [Reactivation of tuberculosis during dual therapy with pegylated interferon and ribavirin for chronic hepatitis C]. Rev. Med. Interne. 31:e1–e3. 10.1016/j.revmed.2009.11.017 [DOI] [PubMed] [Google Scholar]
- Benard A., Sakwa I., Schierloh P., Colom A., Mercier I., Tailleux L., Jouneau L., Boudinot P., Al-Saati T., Lang R., et al. 2017. B Cells Producing Type I Interferon Modulate Macrophage Polarization in Tuberculosis. Am. J. Respir. Crit. Care Med. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry M.P., Graham C.M., McNab F.W., Xu Z., Bloch S.A., Oni T., Wilkinson K.A., Banchereau R., Skinner J., Wilkinson R.J., et al. 2010. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 466:973–977. 10.1038/nature09247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankley S., Graham C.M., Levin J., Turner J., Berry M.P., Bloom C.I., Xu Z., Pascual V., Banchereau J., Chaussabel D., et al. 2016. A 380-gene meta-signature of active tuberculosis compared with healthy controls. Eur. Respir. J. 47:1873–1876. 10.1183/13993003.02121-2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom C.I., Graham C.M., Berry M.P., Wilkinson K.A., Oni T., Rozakeas F., Xu Z., Rossello-Urgell J., Chaussabel D., Banchereau J., et al. 2012. Detectable changes in the blood transcriptome are present after two weeks of antituberculosis therapy. PLoS One. 7:e46191 10.1371/journal.pone.0046191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom C.I., Graham C.M., Berry M.P., Rozakeas F., Redford P.S., Wang Y., Xu Z., Wilkinson K.A., Wilkinson R.J., Kendrick Y., et al. 2013. Transcriptional blood signatures distinguish pulmonary tuberculosis, pulmonary sarcoidosis, pneumonias and lung cancers. PLoS One. 8:e70630 10.1371/journal.pone.0070630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogunovic D., Byun M., Durfee L.A., Abhyankar A., Sanal O., Mansouri D., Salem S., Radovanovic I., Grant A.V., Adimi P., et al. 2012. Mycobacterial disease and impaired IFN-γ immunity in humans with inherited ISG15 deficiency. Science. 337:1684–1688. 10.1126/science.1224026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottai D., Frigui W., Clark S., Rayner E., Zelmer A., Andreu N., de Jonge M.I., Bancroft G.J., Williams A., Brodin P., and Brosch R.. 2015. Increased protective efficacy of recombinant BCG strains expressing virulence-neutral proteins of the ESX-1 secretion system. Vaccine. 33:2710–2718. 10.1016/j.vaccine.2015.03.083 [DOI] [PubMed] [Google Scholar]
- Cai Y., Yang Q., Tang Y., Zhang M., Liu H., Zhang G., Deng Q., Huang J., Gao Z., Zhou B., et al. 2014. Increased complement C1q level marks active disease in human tuberculosis. PLoS One. 9:e92340 10.1371/journal.pone.0092340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona J., Cruz A., Moreira-Teixeira L., Sousa C., Sousa J., Osorio N.S., Saraiva A.L., Svenson S., Kallenius G., Pedrosa J., et al. 2013. Mycobacterium tuberculosis Strains Are Differentially Recognized by TLRs with an Impact on the Immune Response. PLoS One. 8:e67277 10.1371/journal.pone.0067277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Divangahi M., Gan H., Shin D.S.J., Hong S., Lee D.M., Serhan C.N., Behar S.M., and Remold H.G.. 2008. Lipid mediators in innate immunity against tuberculosis: Opposing roles of PGE2 and LXA4 in the induction of macrophage death. J. Exp. Med. 205:2791–2801. 10.1084/jem.20080767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliff J.M., Lee J.S., Constantinou N., Cho J.E., Clark T.G., Ronacher K., King E.C., Lukey P.T., Duncan K., Van Helden P.D., et al. 2013. Distinct phases of blood gene expression pattern through tuberculosis treatment reflect modulation of the humoral immune response. J. Infect. Dis. 207:18–29. 10.1093/infdis/jis499 [DOI] [PubMed] [Google Scholar]
- Collins A.C., Cai H., Li T., Franco L.H., Li X.D., Nair V.R., Scharn C.R., Stamm C.E., Levine B., Chen Z.J., and Shiloh M.U.. 2015. Cyclic GMP-AMP Synthase Is an Innate Immune DNA Sensor for Mycobacterium tuberculosis. Cell Host Microbe. 17:820–828. 10.1016/j.chom.2015.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A.M. 2009. Cell-mediated immune responses in tuberculosis. Annu. Rev. Immunol. 27:393–422. 10.1146/annurev.immunol.021908.132703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A.M., Dalton D.K., Stewart T.A., Griffin J.P., Russell D.G., and Orme I.M.. 1993. Disseminated tuberculosis in interferon γ gene-disrupted mice. J. Exp. Med. 178:2243–2247. 10.1084/jem.178.6.2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A.M., Roberts A.D., Rhoades E.R., Callahan J.E., Getzy D.M., and Orme I.M.. 1995. The role of interleukin-12 in acquired immunity to Mycobacterium tuberculosis infection. Immunology. 84:423–432. [PMC free article] [PubMed] [Google Scholar]
- Cooper A.M., Magram J., Ferrante J., and Orme I.M.. 1997. Interleukin 12 (IL-12) is crucial to the development of protective immunity in mice intravenously infected with Mycobacterium tuberculosis. J. Exp. Med. 186:39–45. 10.1084/jem.186.1.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A.M., Pearl J.E., Brooks J.V., Ehlers S., and Orme I.M.. 2000. Expression of the nitric oxide synthase 2 gene is not essential for early control of Mycobacterium tuberculosis in the murine lung. Infect. Immun. 68:6879–6882. 10.1128/IAI.68.12.6879-6882.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouse J., Kalinke U., and Oxenius A.. 2015. Regulation of antiviral T cell responses by type I interferons. Nat. Rev. Immunol. 15:231–242. 10.1038/nri3806 [DOI] [PubMed] [Google Scholar]
- Crow Y.J. 2015. Type I interferonopathies: mendelian type I interferon up-regulation. Curr. Opin. Immunol. 32:7–12. 10.1016/j.coi.2014.10.005 [DOI] [PubMed] [Google Scholar]
- Dauphinee S.M., Richer E., Eva M.M., McIntosh F., Paquet M., Dangoor D., Burkart C., Zhang D.E., Gruenheid S., Gros P., et al. 2014. Contribution of increased ISG15, ISGylation and deregulated type I IFN signaling in Usp18 mutant mice during the course of bacterial infections. Genes Immun. 15:282–292. 10.1038/gene.2014.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson S., Maini M.K., and Wack A.. 2015. Disease-promoting effects of type I interferons in viral, bacterial, and coinfections. J. Interferon Cytokine Res. 35:252–264. 10.1089/jir.2014.0227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker T., Müller M., and Stockinger S.. 2005. The yin and yang of type I interferon activity in bacterial infection. Nat. Rev. Immunol. 5:675–687. 10.1038/nri1684 [DOI] [PubMed] [Google Scholar]
- de Jong R., Altare F., Haagen I.A., Elferink D.G., Boer T., van Breda Vriesman P.J., Kabel P.J., Draaisma J.M., van Dissel J.T., Kroon F.P., et al. 1998. Severe mycobacterial and Salmonella infections in interleukin-12 receptor-deficient patients. Science. 280:1435–1438. 10.1126/science.280.5368.1435 [DOI] [PubMed] [Google Scholar]
- Denis M. 1991. Recombinant murine beta interferon enhances resistance of mice to systemic Mycobacterium avium infection. Infect. Immun. 59:1857–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Uehara S.N., Emori C.T., Perez R.M., Mendes-Correa M.C., de Souza Paiva Ferreira A., de Castro Amaral Feldner A.C., Silva A.E., Filho R.J., de Souza E Silva I.S., and Ferraz M.L.. 2016. High incidence of tuberculosis in patients treated for hepatitis C chronic infection. Braz. J. Infect. Dis. 20:205–209. 10.1016/j.bjid.2015.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Paus R.A., van Wengen A., Schmidt I., Visser M., Verdegaal E.M., van Dissel J.T., and van de Vosse E.. 2013. Inhibition of the type I immune responses of human monocytes by IFN-α and IFN-β. Cytokine. 61:645–655. 10.1016/j.cyto.2012.12.005 [DOI] [PubMed] [Google Scholar]
- Desvignes L., Wolf A.J., and Ernst J.D.. 2012. Dynamic roles of type I and type II IFNs in early infection with Mycobacterium tuberculosis. J. Immunol. 188:6205–6215. 10.4049/jimmunol.1200255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Weerd N.A., Vivian J.P., Nguyen T.K., Mangan N.E., Gould J.A., Braniff S.J., Zaker-Tabrizi L., Fung K.Y., Forster S.C., Beddoe T., et al. 2013. Structural basis of a unique interferon-β signaling axis mediated via the receptor IFNAR1. Nat. Immunol. 14:901–907. 10.1038/ni.2667 [DOI] [PubMed] [Google Scholar]
- Dey B., Dey R.J., Cheung L.S., Pokkali S., Guo H., Lee J.H., and Bishai W.R.. 2015. A bacterial cyclic dinucleotide activates the cytosolic surveillance pathway and mediates innate resistance to tuberculosis. Nat. Med. 21:401–406. 10.1038/nm.3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey R.J., Dey B., Zheng Y., Cheung L.S., Zhou J., Sayre D., Kumar P., Guo H., Lamichhane G., Sintim H.O., and Bishai W.R.. 2017. Inhibition of innate immune cytosolic surveillance by an M. tuberculosis phosphodiesterase. Nat. Chem. Biol. 13:210–217. 10.1038/nchembio.2254 [DOI] [PubMed] [Google Scholar]
- Divangahi M., Chen M., Gan H., Desjardins D., Hickman T.T., Lee D.M., Fortune S., Behar S.M., and Remold H.G.. 2009. Mycobacterium tuberculosis evades macrophage defenses by inhibiting plasma membrane repair. Nat. Immunol. 10:899–906. 10.1038/ni.1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domaszewska T., Scheuermann L., Hahnke K., Mollenkopf H., Dorhoi A., Kaufmann S.H.E., and Weiner J.. 2017. Concordant and discordant gene expression patterns in mouse strains identify best-fit animal model for human tuberculosis. Sci. Rep. 7:12094 10.1038/s41598-017-11812-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan M.L., Schultz T.E., Duke T.J., and Blumenthal A.. 2017. Type I Interferons in the Pathogenesis of Tuberculosis: Molecular Drivers and Immunological Consequences. Front. Immunol. 8:1633 10.3389/fimmu.2017.01633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorhoi A., Yeremeev V., Nouailles G., Weiner J. III, Jörg S., Heinemann E., Oberbeck-Müller D., Knaul J.K., Vogelzang A., Reece S.T., et al. 2014. Type I IFN signaling triggers immunopathology in tuberculosis-susceptible mice by modulating lung phagocyte dynamics. Eur. J. Immunol. 44:2380–2393. 10.1002/eji.201344219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmail H., Lai R.P., Lesosky M., Wilkinson K.A., Graham C.M., Horswell S., Coussens A.K., Barry C.E. III, O’Garra A., and Wilkinson R.J.. 2018. Complement pathway gene activation and rising circulating immune complexes characterize early disease in HIV-associated tuberculosis. Proc. Natl. Acad. Sci. USA. 115:E964–E973. 10.1073/pnas.1711853115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farah R., and Awad J.. 2007. The association of interferon with the development of pulmonary tuberculosis. Int. J. Clin. Pharmacol. Ther. 45:598–600. 10.5414/CPP45598 [DOI] [PubMed] [Google Scholar]
- Festing M.F., Simpson E.M., Davisson M.T., and Mobraaten L.E.. 1999. Revised nomenclature for strain 129 mice. Mamm. Genome. 10:836 10.1007/s003359901099 [DOI] [PubMed] [Google Scholar]
- Flynn J.L., and Chan J.. 2001. Immunology of tuberculosis. Annu. Rev. Immunol. 19:93–129. 10.1146/annurev.immunol.19.1.93 [DOI] [PubMed] [Google Scholar]
- Flynn J.L., Chan J., Triebold K.J., Dalton D.K., Stewart T.A., and Bloom B.R.. 1993. An essential role for interferon gamma in resistance to Mycobacterium tuberculosis infection. J. Exp. Med. 178:2249–2254. 10.1084/jem.178.6.2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn J.L., Goldstein M.M., Chan J., Triebold K.J., Pfeffer K., Lowenstein C.J., Schreiber R., Mak T.W., and Bloom B.R.. 1995a Tumor necrosis factor-alpha is required in the protective immune response against Mycobacterium tuberculosis in mice. Immunity. 2:561–572. 10.1016/1074-7613(95)90001-2 [DOI] [PubMed] [Google Scholar]
- Flynn J.L., Goldstein M.M., Triebold K.J., Sypek J., Wolf S., and Bloom B.R.. 1995b IL-12 increases resistance of BALB/c mice to Mycobacterium tuberculosis infection. J. Immunol. 155:2515–2524. [PubMed] [Google Scholar]
- Fortin A., Abel L., Casanova J.L., and Gros P.. 2007. Host genetics of mycobacterial diseases in mice and men: forward genetic studies of BCG-osis and tuberculosis. Annu. Rev. Genomics Hum. Genet. 8:163–192. 10.1146/annurev.genom.8.080706.092315 [DOI] [PubMed] [Google Scholar]
- Gautier G., Humbert M., Deauvieau F., Scuiller M., Hiscott J., Bates E.E., Trinchieri G., Caux C., and Garrone P.. 2005. A type I interferon autocrine-paracrine loop is involved in Toll-like receptor-induced interleukin-12p70 secretion by dendritic cells. J. Exp. Med. 201:1435–1446. 10.1084/jem.20041964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getahun H., Matteelli A., Chaisson R.E., and Raviglione M.. 2015. Latent Mycobacterium tuberculosis infection. N. Engl. J. Med. 372:2127–2135. 10.1056/NEJMra1405427 [DOI] [PubMed] [Google Scholar]
- Giacomini E., Remoli M.E., Gafa V., Pardini M., Fattorini L., and Coccia E.M.. 2009. IFN-beta improves BCG immunogenicity by acting on DC maturation. J. Leukoc. Biol. 85:462–468. 10.1189/jlb.0908583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gideon H.P., Skinner J.A., Baldwin N., Flynn J.L., and Lin P.L.. 2016. Early Whole Blood Transcriptional Signatures Are Associated with Severity of Lung Inflammation in Cynomolgus Macaques with Mycobacterium tuberculosis Infection. J. Immunol. 197:4817–4828. 10.4049/jimmunol.1601138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giosué S., Casarini M., Alemanno L., Galluccio G., Mattia P., Pedicelli G., Rebek L., Bisetti A., and Ameglio F.. 1998. Effects of aerosolized interferon-alpha in patients with pulmonary tuberculosis. Am. J. Respir. Crit. Care Med. 158:1156–1162. 10.1164/ajrccm.158.4.9803065 [DOI] [PubMed] [Google Scholar]
- Giosuè S., Casarini M., Ameglio F., Zangrilli P., Palla M., Altieri A.M., and Bisetti A.. 2000. Aerosolized interferon-alpha treatment in patients with multi-drug-resistant pulmonary tuberculosis. Eur. Cytokine Netw. 11:99–104. [PubMed] [Google Scholar]
- Gröschel M.I., Sayes F., Shin S.J., Frigui W., Pawlik A., Orgeur M., Canetti R., Honoré N., Simeone R., van der Werf T.S., et al. 2017. Recombinant BCG Expressing ESX-1 of Mycobacterium marinum Combines Low Virulence with Cytosolic Immune Signaling and Improved TB Protection. Cell Reports. 18:2752–2765. 10.1016/j.celrep.2017.02.057 [DOI] [PubMed] [Google Scholar]
- Guardigni V., Fabbri G., Grilli A., and Contini C.. 2012. Successful antiviral treatment of chronic hepatitis C in patients with rare comorbidities. Two case-reports. Ann. Hepatol. 11:404–408. [PubMed] [Google Scholar]
- Hall J.C., and Rosen A.. 2010. Type I interferons: crucial participants in disease amplification in autoimmunity. Nat. Rev. Rheumatol. 6:40–49. 10.1038/nrrheum.2009.237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howes A., Taubert C., Blankley S., Spink N., Wu X., Graham C.M., Zhao J., Saraiva M., Ricciardi-Castagnoli P., Bancroft G.J., and O’Garra A.. 2016. Differential Production of Type I IFN Determines the Reciprocal Levels of IL-10 and Proinflammatory Cytokines Produced by C57BL/6 and BALB/c Macrophages. J. Immunol. 197:2838–2853. 10.4049/jimmunol.1501923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashkiv L.B., and Donlin L.T.. 2014. Regulation of type I interferon responses. Nat. Rev. Immunol. 14:36–49. 10.1038/nri3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen M., Repsilber D., Gutschmidt A., Neher A., Feldmann K., Mollenkopf H.J., Ziegler A., and Kaufmann S.H.. 2007. Candidate biomarkers for discrimination between infection and disease caused by Mycobacterium tuberculosis. J. Mol. Med. (Berl.). 85:613–621. 10.1007/s00109-007-0157-6 [DOI] [PubMed] [Google Scholar]
- Joosten S.A., Fletcher H.A., and Ottenhoff T.H.. 2013. A helicopter perspective on TB biomarkers: pathway and process based analysis of gene expression data provides new insight into TB pathogenesis. PLoS One. 8:e73230 10.1371/journal.pone.0073230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouanguy E., Lamhamedi-Cherradi S., Lammas D., Dorman S.E., Fondanèche M.C., Dupuis S., Döffinger R., Altare F., Girdlestone J., Emile J.F., et al. 1999. A human IFNGR1 small deletion hotspot associated with dominant susceptibility to mycobacterial infection. Nat. Genet. 21:370–378. 10.1038/7701 [DOI] [PubMed] [Google Scholar]
- Kaforou M., Wright V.J., Oni T., French N., Anderson S.T., Bangani N., Banwell C.M., Brent A.J., Crampin A.C., Dockrell H.M., et al. 2013. Detection of tuberculosis in HIV-infected and -uninfected African adults using whole blood RNA expression signatures: a case-control study. PLoS Med. 10:e1001538 10.1371/journal.pmed.1001538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane J., Gershon S., Wise R.P., Mirabile-Levens E., Kasznica J., Schwieterman W.D., Siegel J.N., and Braun M.M.. 2001. Tuberculosis associated with infliximab, a tumor necrosis factor alpha-neutralizing agent. N. Engl. J. Med. 345:1098–1104. 10.1056/NEJMoa011110 [DOI] [PubMed] [Google Scholar]
- Kimmey J.M., Campbell J.A., Weiss L.A., Monte K.J., Lenschow D.J., and Stallings C.L.. 2017. The impact of ISGylation during Mycobacterium tuberculosis infection in mice. Microbes Infect. 19:249–258. 10.1016/j.micinf.2016.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchtey J., Fulton S.A., Reba S.M., Harding C.V., and Boom W.H.. 2006. Interferon-alphabeta mediates partial control of early pulmonary Mycobacterium bovis bacillus Calmette-Guérin infection. Immunology. 118:39–49. 10.1111/j.1365-2567.2006.02337.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawn S.D., and Zumla A.I.. 2011. Tuberculosis. Lancet. 378:57–72. 10.1016/S0140-6736(10)62173-3 [DOI] [PubMed] [Google Scholar]
- Leber J.H., Crimmins G.T., Raghavan S., Meyer-Morse N.P., Cox J.S., and Portnoy D.A.. 2008. Distinct TLR- and NLR-mediated transcriptional responses to an intracellular pathogen. PLoS Pathog. 4:e6 10.1371/journal.ppat.0040006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertzdorf J., Ota M., Repsilber D., Mollenkopf H.J., Weiner J., Hill P.C., and Kaufmann S.H.. 2011a Functional correlations of pathogenesis-driven gene expression signatures in tuberculosis. PLoS One. 6:e26938 10.1371/journal.pone.0026938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertzdorf J., Repsilber D., Parida S.K., Stanley K., Roberts T., Black G., Walzl G., and Kaufmann S.H.. 2011b Human gene expression profiles of susceptibility and resistance in tuberculosis. Genes Immun. 12:15–22. 10.1038/gene.2010.51 [DOI] [PubMed] [Google Scholar]
- Maertzdorf J., Weiner J. III, Mollenkopf H.J., Bauer T., Prasse A., Müller-Quernheim J., Kaufmann S.H., and Kaufmann S.H.. TBornotTB Network . 2012. Common patterns and disease-related signatures in tuberculosis and sarcoidosis. Proc. Natl. Acad. Sci. USA. 109:7853–7858. 10.1073/pnas.1121072109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manca C., Tsenova L., Bergtold A., Freeman S., Tovey M., Musser J.M., Barry C.E. III, Freedman V.H., and Kaplan G.. 2001. Virulence of a Mycobacterium tuberculosis clinical isolate in mice is determined by failure to induce Th1 type immunity and is associated with induction of IFN-α/β. Proc. Natl. Acad. Sci. USA. 98:5752–5757. 10.1073/pnas.091096998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manca C., Tsenova L., Freeman S., Barczak A.K., Tovey M., Murray P.J., Barry C., and Kaplan G.. 2005. Hypervirulent M. tuberculosis W/Beijing strains upregulate type I IFNs and increase expression of negative regulators of the Jak-Stat pathway. J. Interferon Cytokine Res. 25:694–701. 10.1089/jir.2005.25.694 [DOI] [PubMed] [Google Scholar]
- Mansoori D., Tavana S., Mirsaeidi M., Yazdanpanah M., and Sohrabpour H.. 2002. The efficacy of interferon-α in the treatment of multidrug resistant tuberculosis. Tanaffos. 1:29–34. [Google Scholar]
- Manzanillo P.S., Shiloh M.U., Portnoy D.A., and Cox J.S.. 2012. Mycobacterium tuberculosis activates the DNA-dependent cytosolic surveillance pathway within macrophages. Cell Host Microbe. 11:469–480. 10.1016/j.chom.2012.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S., Fujikawa H., Hasegawa H., Ochiai T., Watanabe Y., and Moriyama M.. 2016. Onset of Tuberculosis from a Pulmonary Latent Tuberculosis Infection during Antiviral Triple Therapy for Chronic Hepatitis C. Intern. Med. 55:2011–2017. 10.2169/internalmedicine.55.6448 [DOI] [PubMed] [Google Scholar]
- Mayer-Barber K.D., and Sher A.. 2015. Cytokine and lipid mediator networks in tuberculosis. Immunol. Rev. 264:264–275. 10.1111/imr.12249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer-Barber K.D., and Yan B.. 2017. Clash of the Cytokine Titans: counter-regulation of interleukin-1 and type I interferon-mediated inflammatory responses. Cell. Mol. Immunol. 14:22–35. 10.1038/cmi.2016.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer-Barber K.D., Barber D.L., Shenderov K., White S.D., Wilson M.S., Cheever A., Kugler D., Hieny S., Caspar P., Núñez G., et al. 2010. Caspase-1 independent IL-1beta production is critical for host resistance to mycobacterium tuberculosis and does not require TLR signaling in vivo. J. Immunol. 184:3326–3330. 10.4049/jimmunol.0904189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer-Barber K.D., Andrade B.B., Barber D.L., Hieny S., Feng C.G., Caspar P., Oland S., Gordon S., and Sher A.. 2011. Innate and adaptive interferons suppress IL-1α and IL-1β production by distinct pulmonary myeloid subsets during Mycobacterium tuberculosis infection. Immunity. 35:1023–1034. 10.1016/j.immuni.2011.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer-Barber K.D., Andrade B.B., Oland S.D., Amaral E.P., Barber D.L., Gonzales J., Derrick S.C., Shi R., Kumar N.P., Wei W., et al. 2014. Host-directed therapy of tuberculosis based on interleukin-1 and type I interferon crosstalk. Nature. 511:99–103. 10.1038/nature13489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab F.W., Ewbank J., Rajsbaum R., Stavropoulos E., Martirosyan A., Redford P.S., Wu X., Graham C.M., Saraiva M., Tsichlis P., et al. 2013. TPL-2-ERK1/2 signaling promotes host resistance against intracellular bacterial infection by negative regulation of type I IFN production. J. Immunol. 191:1732–1743. 10.4049/jimmunol.1300146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab F.W., Ewbank J., Howes A., Moreira-Teixeira L., Martirosyan A., Ghilardi N., Saraiva M., and O’Garra A.. 2014. Type I IFN induces IL-10 production in an IL-27-independent manner and blocks responsiveness to IFN-γ for production of IL-12 and bacterial killing in Mycobacterium tuberculosis-infected macrophages. J. Immunol. 193:3600–3612. 10.4049/jimmunol.1401088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab F., Mayer-Barber K., Sher A., Wack A., and O’Garra A.. 2015. Type I interferons in infectious disease. Nat. Rev. Immunol. 15:87–103. 10.1038/nri3787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry R., Cliff J.M., Clayton C.L., Beyers N., Mohamed Y.S., Wilson P.A., Dockrell H.M., Wallace D.M., van Helden P.D., Duncan K., and Lukey P.T.. 2007. Gene-expression patterns in whole blood identify subjects at risk for recurrent tuberculosis. J. Infect. Dis. 195:357–365. 10.1086/510397 [DOI] [PubMed] [Google Scholar]
- Moreira-Teixeira L., Sousa J., McNab F.W., Torrado E., Cardoso F., Machado H., Castro F., Cardoso V., Gaifem J., Wu X., et al. 2016. Type I IFN Inhibits Alternative Macrophage Activation during Mycobacterium tuberculosis Infection and Leads to Enhanced Protection in the Absence of IFN-γ Signaling. J. Immunol. 197:4714–4726. 10.4049/jimmunol.1600584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira-Teixeira L., Redford P.S., Stavropoulos E., Ghilardi N., Maynard C.L., Weaver C.T., Freitas do Rosário A.P., Wu X., Langhorne J., and O’Garra A.. 2017. T Cell-Derived IL-10 Impairs Host Resistance toMycobacterium tuberculosisInfection. J. Immunol. 199:613–623. 10.4049/jimmunol.1601340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport M.J., Huxley C.M., Huston S., Hawrylowicz C.M., Oostra B.A., Williamson R., and Levin M.. 1996. A mutation in the interferon-gamma-receptor gene and susceptibility to mycobacterial infection. N. Engl. J. Med. 335:1941–1949. 10.1056/NEJM199612263352602 [DOI] [PubMed] [Google Scholar]
- North R.J., and Jung Y.J.. 2004. Immunity to tuberculosis. Annu. Rev. Immunol. 22:599–623. 10.1146/annurev.immunol.22.012703.104635 [DOI] [PubMed] [Google Scholar]
- Novikov A., Cardone M., Thompson R., Shenderov K., Kirschman K.D., Mayer-Barber K.D., Myers T.G., Rabin R.L., Trinchieri G., Sher A., and Feng C.G.. 2011. Mycobacterium tuberculosis triggers host type I IFN signaling to regulate IL-1β production in human macrophages. J. Immunol. 187:2540–2547. 10.4049/jimmunol.1100926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Garra A., Redford P.S., McNab F.W., Bloom C.I., Wilkinson R.J., and Berry M.P.. 2013. The immune response in tuberculosis. Annu. Rev. Immunol. 31:475–527. 10.1146/annurev-immunol-032712-095939 [DOI] [PubMed] [Google Scholar]
- Ordway D., Henao-Tamayo M., Harton M., Palanisamy G., Troudt J., Shanley C., Basaraba R.J., and Orme I.M.. 2007. The hypervirulent Mycobacterium tuberculosis strain HN878 induces a potent TH1 response followed by rapid down-regulation. J. Immunol. 179:522–531. 10.4049/jimmunol.179.1.522 [DOI] [PubMed] [Google Scholar]
- Ottenhoff T.H., Dass R.H., Yang N., Zhang M.M., Wong H.E., Sahiratmadja E., Khor C.C., Alisjahbana B., van Crevel R., Marzuki S., et al. 2012. Genome-wide expression profiling identifies type 1 interferon response pathways in active tuberculosis. PLoS One. 7:e45839 10.1371/journal.pone.0045839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmero D., Eiguchi K., Rendo P., Castro Zorrilla L., Abbate E., and González Montaner L.J.. 1999. Phase II trial of recombinant interferon-alpha2b in patients with advanced intractable multidrug-resistant pulmonary tuberculosis: long-term follow-up. Int. J. Tuberc. Lung Dis. 3:214–218. [PubMed] [Google Scholar]
- Pandey A.K., Yang Y., Jiang Z., Fortune S.M., Coulombe F., Behr M.A., Fitzgerald K.A., Sassetti C.M., and Kelliher M.A.. 2009. NOD2, RIP2 and IRF5 play a critical role in the type I interferon response to Mycobacterium tuberculosis. PLoS Pathog. 5:e1000500 10.1371/journal.ppat.1000500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestka S., Krause C.D., and Walter M.R.. 2004. Interferons, interferon-like cytokines, and their receptors. Immunol. Rev. 202:8–32. 10.1111/j.0105-2896.2004.00204.x [DOI] [PubMed] [Google Scholar]
- Post F.A., Wood R., and Pillay G.P.. 1995. Pulmonary tuberculosis in HIV infection: radiographic appearance is related to CD4+ T-lymphocyte count. Tuber. Lung Dis. 76:518–521. 10.1016/0962-8479(95)90527-8 [DOI] [PubMed] [Google Scholar]
- Redford P.S., Murray P.J., and O’Garra A.. 2011. The role of IL-10 in immune regulation during M. tuberculosis infection. Mucosal Immunol. 4:261–270. 10.1038/mi.2011.7 [DOI] [PubMed] [Google Scholar]
- Redford P.S., Mayer-Barber K.D., McNab F.W., Stavropoulos E., Wack A., Sher A., and O’Garra A.. 2014. Influenza A virus impairs control of Mycobacterium tuberculosis coinfection through a type I interferon receptor-dependent pathway. J. Infect. Dis. 209:270–274. 10.1093/infdis/jit424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas-Santiago C.E., and Guerrero G.G.. 2017. IFN-αBoosting of Mycobacterium bovis Bacillus Calmette Güerin-Vaccine Promoted Th1 Type Cellular Response and Protection against M. tuberculosis Infection. BioMed Res. Int. 2017:8796760 10.1155/2017/8796760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe J.K., Thomas N., Gil E., Best K., Tsaliki E., Morris-Jones S., Stafford S., Simpson N., Witt K.D., Chain B., et al. 2016. Blood transcriptomic diagnosis of pulmonary and extrapulmonary tuberculosis. JCI Insight. 1:e87238 10.1172/jci.insight.87238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruangkiattikul N., Nerlich A., Abdissa K., Lienenklaus S., Suwandi A., Janze N., Laarmann K., Spanier J., Kalinke U., Weiss S., and Goethe R.. 2017. cGAS-STING-TBK1-IRF3/7 induced interferon-β contributes to the clearing of non tuberculous mycobacterial infection in mice. Virulence. 8:1303–1315. 10.1080/21505594.2017.1321191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbatani S., Manfredi R., Marinacci G., Pavoni M., Cristoni L., and Chiodo F.. 2006. Reactivation of severe, acute pulmonary tuberculosis during treatment with pegylated interferon-alpha and ribavirin for chronic HCV hepatitis. Scand. J. Infect. Dis. 38:205–208. 10.1080/00365540500263268 [DOI] [PubMed] [Google Scholar]
- Sabir N., Hussain T., Shah S.Z.A., Zhao D., and Zhou X.. 2017. IFN-β: A Contentious Player in Host-Pathogen Interaction in Tuberculosis. Int. J. Mol. Sci. 18:2725 10.3390/ijms18122725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambarey A., Devaprasad A., Baloni P., Mishra M., Mohan A., Tyagi P., Singh A., Akshata J.S., Sultana R., Buggi S., and Chandra N.. 2017a Meta-analysis of host response networks identifies a common core in tuberculosis. NPJ Syst. Biol. Appl. 3:4 10.1038/s41540-017-0005-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambarey A., Devaprasad A., Mohan A., Ahmed A., Nayak S., Swaminathan S., D’Souza G., Jesuraj A., Dhar C., Babu S., et al. 2017b Unbiased Identification of Blood-based Biomarkers for Pulmonary Tuberculosis by Modeling and Mining Molecular Interaction Networks. EBioMedicine. 15:112–126. 10.1016/j.ebiom.2016.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scriba T.J., Penn-Nicholson A., Shankar S., Hraha T., Thompson E.G., Sterling D., Nemes E., Darboe F., Suliman S., Amon L.M., et al. other members of the ACS cohort study team . 2017. Sequential inflammatory processes define human progression from M. tuberculosis infection to tuberculosis disease. PLoS Pathog. 13:e1006687 10.1371/journal.ppat.1006687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S., Bohsali A., Ahlbrand S.E., Srinivasan L., Rathinam V.A.K., Vogel S.N., Fitzgerald K.A., Sutterwala F.S., and Briken V.. 2013. Cutting edge: Mycobacterium tuberculosis but not nonvirulent mycobacteria inhibits IFN-β and AIM2 inflammasome-dependent IL-1β production via its ESX-1 secretion system. J. Immunol. 191:3514–3518. 10.4049/jimmunol.1301331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson E.M., Linder C.C., Sargent E.E., Davisson M.T., Mobraaten L.E., and Sharp J.J.. 1997. Genetic variation among 129 substrains and its importance for targeted mutagenesis in mice. Nat. Genet. 16:19–27. 10.1038/ng0597-19 [DOI] [PubMed] [Google Scholar]
- Singhania A., Verma R., Graham C.M., Lee J., Trang T., Richarsdon M., Lecine P., Leissner P., Berry M.P.R., Wilkinson R.J., et al. 2017. A modular transcriptional signature identifies phenotypic heterogeneity of human tuberculosis infection. bioRxiv. 10.1101/216879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm C.E., Collins A.C., and Shiloh M.U.. 2015. Sensing of Mycobacterium tuberculosis and consequences to both host and bacillus. Immunol. Rev. 264:204–219. 10.1111/imr.12263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley S.A., Johndrow J.E., Manzanillo P., and Cox J.S.. 2007. The Type I IFN response to infection with Mycobacterium tuberculosis requires ESX-1-mediated secretion and contributes to pathogenesis. J. Immunol. 178:3143–3152. 10.4049/jimmunol.178.5.3143 [DOI] [PubMed] [Google Scholar]
- Stifter S.A., and Feng C.G.. 2015. Interfering with immunity: detrimental role of type I IFNs during infection. J. Immunol. 194:2455–2465. 10.4049/jimmunol.1402794 [DOI] [PubMed] [Google Scholar]
- Teles R.M., Graeber T.G., Krutzik S.R., Montoya D., Schenk M., Lee D.J., Komisopoulou E., Kelly-Scumpia K., Chun R., Iyer S.S., et al. 2013. Type I interferon suppresses type II interferon-triggered human anti-mycobacterial responses. Science. 339:1448–1453. 10.1126/science.1233665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telesca C., Angelico M., Piccolo P., Nosotti L., Morrone A., Longhi C., Carbone M., and Baiocchi L.. 2007. Interferon-alpha treatment of hepatitis D induces tuberculosis exacerbation in an immigrant. J. Infect. 54:e223–e226. 10.1016/j.jinf.2006.12.009 [DOI] [PubMed] [Google Scholar]
- Trinchieri G. 2010. Type I interferon: Friend or foe? J. Exp. Med. 207:2053–2063. 10.1084/jem.20101664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troegeler A., Mercier I., Cougoule C., Pietretti D., Colom A., Duval C., Vu Manh T.P., Capilla F., Poincloux R., Pingris K., et al. 2017. C-type lectin receptor DCIR modulates immunity to tuberculosis by sustaining type I interferon signaling in dendritic cells. Proc. Natl. Acad. Sci. USA. 114:E540–E549. 10.1073/pnas.1613254114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward C.M., Jyonouchi H., Kotenko S.V., Smirnov S.V., Patel R., Aguila H., McSherry G., Dashefsky B., and Holland S.M.. 2007. Adjunctive treatment of disseminated Mycobacterium avium complex infection with interferon alpha-2b in a patient with complete interferon-gamma receptor R1 deficiency. Eur. J. Pediatr. 166:981–985. 10.1007/s00431-006-0339-1 [DOI] [PubMed] [Google Scholar]
- Wassermann R., Gulen M.F., Sala C., Perin S.G., Lou Y., Rybniker J., Schmid-Burgk J.L., Schmidt T., Hornung V., Cole S.T., and Ablasser A.. 2015. Mycobacterium tuberculosis Differentially Activates cGAS- and Inflammasome-Dependent Intracellular Immune Responses through ESX-1. Cell Host Microbe. 17:799–810. 10.1016/j.chom.2015.05.003 [DOI] [PubMed] [Google Scholar]
- Watson R.O., Bell S.L., MacDuff D.A., Kimmey J.M., Diner E.J., Olivas J., Vance R.E., Stallings C.L., Virgin H.W., and Cox J.S.. 2015. The Cytosolic Sensor cGAS Detects Mycobacterium tuberculosis DNA to Induce Type I Interferons and Activate Autophagy. Cell Host Microbe. 17:811–819. 10.1016/j.chom.2015.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiens K.E., and Ernst J.D.. 2016. The Mechanism for Type I Interferon Induction by Mycobacterium tuberculosis is Bacterial Strain-Dependent. PLoS Pathog. 12:e1005809 10.1371/journal.ppat.1005809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization 2017. Global Tuberculosis Report 2017. World Health Organization, Geneva: 262 pp. [Google Scholar]
- Yan N., and Chen Z.J.. 2012. Intrinsic antiviral immunity. Nat. Immunol. 13:214–222. 10.1038/ni.2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S., Shen H., Lian Q., Jin W., Zhang R., Lin X., Gu W., Sun X., Meng G., Tian Z., et al. 2018. Deficiency of the AIM2-ASC Signal Uncovers the STING-Driven Overreactive Response of Type I IFN and Reciprocal Depression of Protective IFN-γ Immunity in Mycobacterial Infection. J. Immunol. 200:1016–1026. 10.4049/jimmunol.1701177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zak D.E., Penn-Nicholson A., Scriba T.J., Thompson E., Suliman S., Amon L.M., Mahomed H., Erasmus M., Whatney W., Hussey G.D., et al. ACS and GC6-74 cohort study groups . 2016. A blood RNA signature for tuberculosis disease risk: a prospective cohort study. Lancet. 387:2312–2322. 10.1016/S0140-6736(15)01316-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarogoulidis P., Kioumis I., Papanas N., Manika K., Kontakiotis T., Papagianis A., and Zarogoulidis K.. 2012. The effect of combination IFN-alpha-2a with usual antituberculosis chemotherapy in non-responding tuberculosis and diabetes mellitus: a case report and review of the literature. J. Chemother. 24:173–177. 10.1179/1973947812Y.0000000005 [DOI] [PubMed] [Google Scholar]
- Zhang G., deWeerd N.A., Stifter S.A., Liu L., Zhou B., Wang W., Zhou Y., Ying B., Hu X., Matthews A.Y., et al. 2018. A proline deletion in IFNAR1 impairs IFN-signaling and underlies increased resistance to tuberculosis in humans. Nat. Commun. 9:85 10.1038/s41467-017-02611-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Bogunovic D., Payelle-Brogard B., Francois-Newton V., Speer S.D., Yuan C., Volpi S., Li Z., Sanal O., Mansouri D., et al. 2015. Human intracellular ISG15 prevents interferon-α/β over-amplification and auto-inflammation. Nature. 517:89–93. 10.1038/nature13801 [DOI] [PMC free article] [PubMed] [Google Scholar]