In this issue of JEM, Zhang et al. show that the suppressive epigenetic enzyme Ezh2 is an important regulator of macrophage activation. The absence of Ezh2 leads to reduced cytokine secretion and suppresses macrophage-dependent disease development. They identify the antiinflammatory factor Socs3 as an important target for Ezh2 and thus show that regulation of suppressive histone modifications controls macrophage activation in disease.
Abstract
In this issue of JEM, Zhang et al. (https://doi.org/10.1084/jem.20171417) show that the suppressive epigenetic enzyme Ezh2 is an important regulator of macrophage activation. The absence of Ezh2 leads to reduced cytokine secretion and suppresses macrophage-dependent disease development. They identify the antiinflammatory factor Socs3 as an important target for Ezh2 and thus show that regulation of suppressive histone modifications controls macrophage activation in disease.
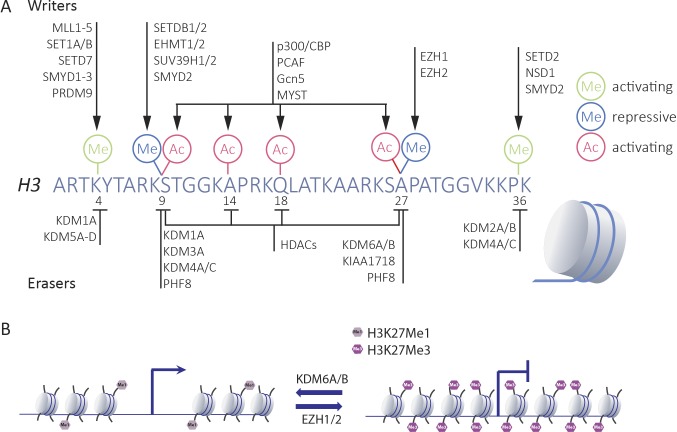
Histone tail modifications control the accessibility of chromatin and are regulated by a series of enzymes. These enzymes either deposit or remove activating and repressive marks, particularly at the tail of histone H3, thereby affecting a combinatorial repertoire of modifications that defines local chromatin behavior (Fig. 1). The Polycomb group protein (PcG) complexes play an important role in repression of genes. They are best known for their function as transcriptional repressors in development and cell fate determination. They were originally identified in Drosophila melanogaster as regulators of Hox gene expression and were shown to control developmental gene programs. Two complexes can be discriminated in the PcG family, Polycomb repressive complex 1 and 2 (PRC1 and PRC2), of which PRC1, through its E3 ligase activity, mediates ubiquitination of histone H2A at lysine 119, and PRC2 acts on methyl groups of histone H3 at lysine 27 (H3K27), leading to the formation of mono-, di-, or trimethylated positions (H3K27Me, H3K27Me2, and H3K27Me3). The latter modifications are catalyzed by the core proteins of the PRC2 complexes, the enhancers of zeste homologues Ezh1 and Ezh2 (Blackledge et al., 2015). The SET domain of the proteins confers methyltransferase activity and mediates methylation of histone tails. Although both Ezh1 and Ezh2 can be components of the PRC2 complex, Ezh2 seems to be most dominant in exerting H3K27 methyltransferase activity. Previous studies have identified an important role for Ezh2 in controlling immune cell function. It was shown that STAT5-mediated recruitment of Ezh2 to the Ig-κ locus mediates repression and maintenance of the proliferative capacity of B cells (Su et al., 2003; Mandal et al., 2011). Moreover, the absence of Ezh2 in T cells led to spontaneous differentiation of CD4+ T cells into both Th1 and Th2 commitment, showing that Ezh2 is required for maintenance of an unspecified state (Tumes et al., 2013). More recently, Ivashkiv and colleagues showed that IFN-γ activation of macrophages led to Ezh2-mediated suppression of a set of antiinflammatory genes in macrophages (Qiao et al., 2016).

Insights from Annette E. Neele and Menno P.J. de Winther.
In the current issue of JEM, Zhang et al. identify Ezh2 as an important regulator of macrophage activation and autoimmune inflammation. Using approaches with specific small molecule inhibitors and cell-specific knockouts for Ezh2, they show that blockade of Ezh2 leads to suppressed activation of bone marrow–derived macrophages and isolated microglia when triggered by a series of TLR ligands. Interestingly, although TLR1/2, TLR4, and TLR9 responses decreased in these experiments, blockade of Ezh2 did not affect responses to the TLR3 stimulus poly I:C, indicating specific effects on MyD88-dependent signaling pathways. Their subsequent work focused on studying Ezh2 in disease models and establishment of mechanisms. When myeloid-specific Ezh2 knockouts were applied to a mouse model for colitis, Zhang et al. (2018) found that the absence of Ezh2 strongly suppressed disease development characterized by a major reduction of recruited immune cells in the gut. These effects seemed independent of neutrophil-Ezh2 because depletion of neutrophils did not have major effects on the difference in disease development between wild-type and myeloid Ezh2-deficient mice.
Epigenetic enzymes regulate activating and repressive histone marks. (A) Key enzymes that control chromatin accessibility by regulating methyl and acetyl marks at the tail of histone H3 by either depositing (i.e., the writers) or removing (i.e., the erasers) histone modifications. (B) The repressive methyl mark at H3K27 is regulated by the writers EZH1/EZH2 and the erasers KDM6A/KDM6B.
Likewise, Ezh2 affected central nervous system (CNS) autoimmune inflammation. In an MOG (myelin oligodendrocyte glycoprotein) antigen–induced EAE model for multiple sclerosis, they found that the myeloid absence of Ezh2 led to a reduced incidence of EAE and reduced symptom development. Although isolated T cells from myeloid Ezh2-deficient mice showed normal responses to antigen, diseased CNS was characterized by strongly suppressed recruitment of T cells, myeloid cells, and activated microglia. To identify whether the reduced disease development was attributable to peripheral myeloid cells or tissue-resident cells, Zhang et al. (2018) next performed studies using bone marrow chimeras, and this identified that particularly Ezh2 in tissue-resident microglia contributes to disease, whereas Ezh2 in circulating myeloid cells is less relevant.
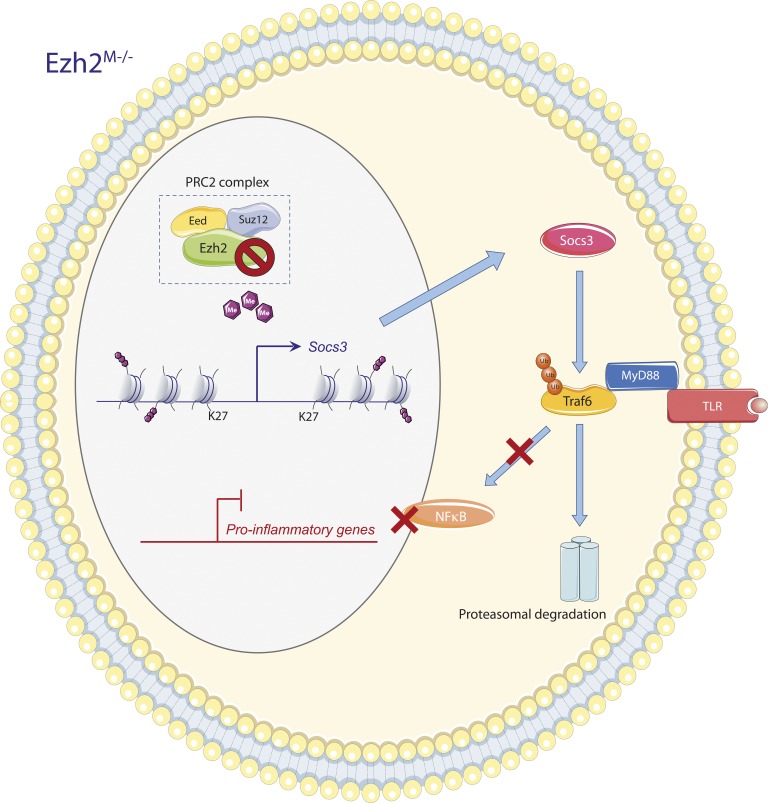
Because of the clear function of Ezh2 in mediating transcriptional silencing, the authors next reasoned that the absence of Ezh2 might lead to the expression of a suppressive factor resulting in the observed reduced macrophage activation and subsequent disease development. By performing chromatin immunoprecipitation (ChIP) and RNA sequencing analysis, Zhang et al. (2018) defined Ezh2-binding sites and dynamics in response to macrophage activation. They identified several genes that were strongly up-regulated in the absence of Ezh2 and also contained Ezh2-binding sites. A key suppressive factor that was derived from this work was Socs3, an antiinflammatory gene and inhibitor of JAK/STAT signaling (Qin et al., 2012). They showed that Socs3 targets the MyD88 signaling pathways in macrophages by mediating the ubiquitination of TRAF6, an adapter molecule critical for MyD88-dependent signaling. The absence of Ezh2 significantly enhanced TRAF6 ubiquitination and degradation, thereby suppressing MyD88 signaling and subsequent activation of the transcription factor pathway NF-κB. These studies thus define Ezh2 as being important in controlling macrophage activation by specifically targeting the antiinflammatory gene Socs3 (Fig. 2). Because Socs3 is also an important regulator of a series of (cytokine) receptors (e.g., IL-6 receptor; Carow and Rottenberg, 2014), it will be very interesting to determine how additional inflammatory pathways are affected by the Ezh2 deletion–mediated induction of Socs3.
The authors show a novel role for Ezh2 in macrophage activation and inflammation, but the H3K27 demethylase Kdm6b (also known as Jmjd3) has been already extensively studied. Both LPS and IL-4 induce Kdm6b expression (De Santa et al., 2007; Ishii et al., 2009), and Kdm6b targets a large proportion of LPS-induced genes. Interestingly, Kdm6b deficiency suppresses only a subset of these, including the inflammatory genes Il6, Il12b, and Ccl5 (De Santa et al., 2009). Most of the genes affected by Kdm6b deficiency were not associated with changes in H3K27Me3 levels, which indicates that Kdm6b controls the expression of LPS-activated macrophages in a H3K27 demethylation–independent manner (De Santa et al., 2009). Accordingly, dual KDM6A and KDM6B inhibition with the small molecule inhibitor GSK-J4 in human macrophages also suppressed the LPS-induced inflammatory response of macrophages (Kruidenier et al., 2012). Besides regulating proinflammatory responses, Kdm6b also affects alternative activation during helminth infection and responses to chitin (Satoh et al., 2010). Again, many of the infection-induced genes down-regulated by Kdm6b deficiency (e.g., Arg1, Chi3l3, and Mrc1) did not show changes in H3K27Me3 levels. However, Irf4 was identified as one of the direct targets of Kdm6b-mediated demethylation, and this transcription factor is crucial for alternative activation of macrophages. Kdm6b is thus regulated in response to various triggers, where it affects several states of macrophage activation. Although Zhang et al. (2018) focused on the regulation of inflammatory responses by Ezh2 in macrophages, they also show that Ezh2 deficiency down-regulates the expression of IL-4 responsive genes. It is likely that other, non–Socs3-dependent mechanisms are at play here, and it will be highly interesting to investigate the consequences of Ezh2 deficiency on disease processes that are more dependent on alternative macrophage activation, such as parasite infection or allergic asthma.
The fact that genetic ablation and inhibition of Ezh2 display similar effects on inflammation as suppression of Kdm6b indicates that distinct mechanisms regulate the inflammatory response, because both act on H3K27 methylation. The authors speculate that Ezh2-mediated trimethylation or Kdm6b-mediated demethylation of H3K27 may have their preference to specifically target different genes, resulting in functional diversity of macrophage activation.
Socs3-mediated inhibition of inflammation by Ezh2 deficiency. Macrophage Ezh2 deficiency reduces suppressive H3K27Me3 marks at the Socs3 transcriptional start site and distal enhancer, resulting in increased Socs3 expression. Cytosolic Socs3 inhibits proinflammatory gene expression by targeting the TLR-induced MyD88–TRAF6–NF-κB signaling pathway. Socs3 enhances TRAF6 ubiquitination, resulting in its proteasomal degradation, and thereby suppresses activation of NF-κB–dependent inflammatory genes. The figure was made with use of Smart Servier Medical Art, licensed under a Creative Common Attribution 3.0 Unported License.
Besides their direct epigenetic function, histone-independent functions of these enzymes also exist. In various cancers, Ezh2 acts as a transcriptional activator, rather than as a repressor, an effect that is independent of the PRC2 complex (Kim and Roberts, 2016). Ezh2 can methylate nonhistone proteins and, depending on the target, thereby control nuclear localization of such proteins or target them for degradation (Hamamoto et al., 2015). Previous experiments have shown that deficiency of Ezh2 in dendritic cells (CD11c-cre) also improved the outcome of EAE, which was the result of impaired integrin-dependent trans-endothelial migration (Gunawan et al., 2015). This effect was also independent of H3K27 methylation but mediated through direct methylation of the extracellular protein Talin. It is likely that in the present study, H3K27 methylation–independent effects also contribute to the improved outcome in colitis and EAE. Future studies should shed light on the direct and indirect effects of these enzymes in immune cells by overlaying H3K27Me3, Ezh2, and Kdm6b ChIP data with RNA sequencing data in both wild-type and knockout mice. Possibly, investigation into the methylome of cells may also be of relevance to discriminate the epigenetic from nonepigenetic functions of these enzymes.
In various cancers, mutations in EZH2 have been identified, which include gain-of-function mutations and mutations leading to overexpression of EZH2 (Kim and Roberts, 2016). Therefore, several EZH2 inhibitors have been developed in recent years, as cancer therapeutics and phase 1/2 clinical trials are currently being performed with an orally bioavailable Ezh2 inhibitor (EPZ-6438) in patients with advanced solid tumors or B cell lymphomas. Besides induction of Ezh2, loss-of-function mutations in EZH2 can drive oncogenesis in specific types of cancer (e.g., MDS, MPN, and T-ALL), and therefore some caution is necessary with the use of these inhibitors in clinical trials. Zhang et al. (2018) show that Ezh2 inhibition by GSK126, a SAM-competitive inhibitor, blocks the inflammatory response of macrophages, highlighting the potential of pharmacological modulation of Ezh2 to alter macrophage activation and thereby control inflammation in autoimmune and inflammatory disease. The next step of using these inhibitors in mouse inflammatory disease models will thus be of great interest in order to test the therapeutic potential of epigenetic inhibition to control inflammation in disease.
References
- Blackledge N.P., Rose N.R., and Klose R.J.. 2015. Targeting Polycomb systems to regulate gene expression: modifications to a complex story. Nat. Rev. Mol. Cell Biol. 16:643–649. 10.1038/nrm4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carow B., and Rottenberg M.E.. 2014. SOCS3, a Major Regulator of Infection and Inflammation. Front. Immunol. 5:58 10.3389/fimmu.2014.00058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F., Totaro M.G., Prosperini E., Notarbartolo S., Testa G., and Natoli G.. 2007. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 130:1083–1094. 10.1016/j.cell.2007.08.019 [DOI] [PubMed] [Google Scholar]
- De Santa F., Narang V., Yap Z.H., Tusi B.K., Burgold T., Austenaa L., Bucci G., Caganova M., Notarbartolo S., Casola S., et al. 2009. Jmjd3 contributes to the control of gene expression in LPS-activated macrophages. EMBO J. 28:3341–3352. 10.1038/emboj.2009.271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawan M., Venkatesan N., Loh J.T., Wong J.F., Berger H., Neo W.H., Li L.Y.J., La Win M.K., Yau Y.H., Guo T., et al. 2015. The methyltransferase Ezh2 controls cell adhesion and migration through direct methylation of the extranuclear regulatory protein talin. Nat. Immunol. 16:505–516. 10.1038/ni.3125 [DOI] [PubMed] [Google Scholar]
- Hamamoto R., Saloura V., and Nakamura Y.. 2015. Critical roles of non-histone protein lysine methylation in human tumorigenesis. Nat. Rev. Cancer. 15:110–124. 10.1038/nrc3884 [DOI] [PubMed] [Google Scholar]
- Ishii M., Wen H., Corsa C.A.S., Liu T., Coelho A.L., Allen R.M., Carson W.F. IV, Cavassani K.A., Li X., Lukacs N.W., et al. 2009. Epigenetic regulation of the alternatively activated macrophage phenotype. Blood. 114:3244–3254. 10.1182/blood-2009-04-217620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.H., and Roberts C.W.M.. 2016. Targeting EZH2 in cancer. Nat. Med. 22:128–134. 10.1038/nm.4036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruidenier L., Chung C.-W., Cheng Z., Liddle J., Che K., Joberty G., Bantscheff M., Bountra C., Bridges A., Diallo H., et al. 2012. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature. 488:404–408. 10.1038/nature11262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M., Powers S.E., Maienschein-Cline M., Bartom E.T., Hamel K.M., Kee B.L., Dinner A.R., and Clark M.R.. 2011. Epigenetic repression of the Igk locus by STAT5-mediated recruitment of the histone methyltransferase Ezh2. Nat. Immunol. 12:1212–1220. 10.1038/ni.2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y., Kang K., Giannopoulou E., Fang C., and Ivashkiv L.B.. 2016. IFN-γ Induces Histone 3 Lysine 27 Trimethylation in a Small Subset of Promoters to Stably Silence Gene Expression in Human Macrophages. Cell Reports. 16:3121–3129. 10.1016/j.celrep.2016.08.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H., Holdbrooks A.T., Liu Y., Reynolds S.L., Yanagisawa L.L., and Benveniste E.N.. 2012. SOCS3 deficiency promotes M1 macrophage polarization and inflammation. J. Immunol. 189:3439–3448. 10.4049/jimmunol.1201168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T., Takeuchi O., Vandenbon A., Yasuda K., Tanaka Y., Kumagai Y., Miyake T., Matsushita K., Okazaki T., Saitoh T., et al. 2010. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat. Immunol. 11:936–944. 10.1038/ni.1920 [DOI] [PubMed] [Google Scholar]
- Su I.-H., Basavaraj A., Krutchinsky A.N., Hobert O., Ullrich A., Chait B.T., and Tarakhovsky A.. 2003. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat. Immunol. 4:124–131. 10.1038/ni876 [DOI] [PubMed] [Google Scholar]
- Tumes D.J., Onodera A., Suzuki A., Shinoda K., Endo Y., Iwamura C., Hosokawa H., Koseki H., Tokoyoda K., Suzuki Y., et al. 2013. The polycomb protein Ezh2 regulates differentiation and plasticity of CD4(+) T helper type 1 and type 2 cells. Immunity. 39:819–832. 10.1016/j.immuni.2013.09.012 [DOI] [PubMed] [Google Scholar]
- Zhang X., Wang Y., Yuan J., Li N., Pei S., Xu J., Luo X., Mao C., Liu J., Yu T., et al. 2018. Macrophage/microglial Ezh2 facilitates autoimmune inflammation through inhibition of Socs3. J. Exp. Med. 10.1084/jem.20171417 [DOI] [PMC free article] [PubMed] [Google Scholar]