Fassier et al. identify Fidgetin-like 1 (Fignl1) as a key growth cone (GC)-enriched microtubule (MT)-associated protein in motor circuit wiring. They show that Fignl1 modulates motor GC morphology and steering behavior by down-regulating EB binding at MT plus ends and promoting MT depolymerization beneath the cell cortex.
Abstract
During neural circuit assembly, extrinsic signals are integrated into changes in growth cone (GC) cytoskeleton underlying axon guidance decisions. Microtubules (MTs) were shown to play an instructive role in GC steering. However, the numerous actors required for MT remodeling during axon navigation and their precise mode of action are far from being deciphered. Using loss- and gain-of-function analyses during zebrafish development, we identify in this study the meiotic clade adenosine triphosphatase Fidgetin-like 1 (Fignl1) as a key GC-enriched MT-interacting protein in motor circuit wiring and larval locomotion. We show that Fignl1 controls GC morphology and behavior at intermediate targets by regulating MT plus end dynamics and growth directionality. We further reveal that alternative translation of Fignl1 transcript is a sophisticated mechanism modulating MT dynamics: a full-length isoform regulates MT plus end–tracking protein binding at plus ends, whereas shorter isoforms promote their depolymerization beneath the cell cortex. Our study thus pinpoints Fignl1 as a multifaceted key player in MT remodeling underlying motor circuit connectivity.
Introduction
The execution of coordinated movements requires the precise and temporal activation of distinct muscles by specific groups of motor neurons (Goulding, 2009). A crucial step in the assembly of motor circuits and thereby in the execution of complex movements relies on the accurate navigation of developing motor axons toward their correct muscle targets. Although several guidance signaling pathways are known to be critical for motor axon targeting, the myriad of molecules that integrate these extracellular signals into complex cytoskeletal changes underlying axon navigation decisions (Lowery and Van Vactor, 2009; Kolodkin and Tessier-Lavigne, 2011; Vitriol and Zheng, 2012) remain largely abstruse.
At the tip of each axon, the growth cone (GC) responds to multiple sources of spatial information as an acute sensor constantly probing its environment for the detection of these fine-tuned signals by extending or retracting actin-based protrusions. Microtubules (MTs) lying in the central domain of the GC dynamically explore the peripheral regions along actin filaments (F-actin) and drive the directional extension of the axon by invading and consolidating these actin protrusions (Schaefer et al., 2002; Suter et al., 2004). Historically, because of the peripheral location of F-actin and its role in cell motility (Pollard and Borisy, 2003), the actin cytoskeleton was attributed the leading role in GC directional steering, although MT remodeling was assumed to occur secondarily. However, subsequent studies demonstrated that modulation of MT dynamics on one side of the GC was sufficient to induce GC turning (Buck and Zheng, 2002; Rothenberg et al., 2003). Furthermore, like F-actin, MTs are direct targets of guidance cues in the regulation of axon extension and navigation (Kalil and Dent, 2004; Lee et al., 2004; Zhou et al., 2004; Schaefer et al., 2008; Shao et al., 2017).
The growing evidence for the MT instructive role in GC steering decisions has shed new light on the involvement of MT-interacting proteins in axon navigation (Dent et al., 2011). Several classes of MT-associated proteins including plus end–tracking proteins (+TIPs; Kalil and Dent, 2004; Lee et al., 2004; Erdogan et al., 2017), MT-stabilizing proteins (Bouquet et al., 2004; Del Río et al., 2004; Deuel et al., 2006; Deloulme et al., 2015), molecular motors (Phillis et al., 1996; Myers et al., 2006; Nadar et al., 2008), or other regulators of MT dynamics (Lewcock et al., 2007) were shown to play a pivotal role in axon guidance. However, for most of them, and notably for MT-severing enzymes, their clear implication in axon guidance decisions remains elusive and poorly investigated in vivo.
MT-severing enzymes katanin, spastin, and fidgetin (Fign) are critical regulators of MT length and dynamics in neurons and thereby of axon elongation and homeostasis (Karabay et al., 2004; Sherwood et al., 2004; Tarrade et al., 2006; Wood et al., 2006; Butler et al., 2010; Fassier et al., 2013; Leo et al., 2015). These MT-severing enzymes belong to the meiotic clade of the ATPases associated with diverse cellular activities (AAA+) superfamily together with their subsequently discovered paralogues, katanin-like 1, katanin-like 2, fidgetin-like 1 (Fignl1), and fidgetin-like 2 (Hanson and Whiteheart, 2005; Yang et al., 2005; Roll-Mecak and McNally, 2010; Sharp and Ross, 2012). Among these paralogues, Fignl1 was shown to bind MTs in vitro and to participate in MT-dependent cellular processes such as mitosis (Luke-Glaser et al., 2007) and ciliogenesis (Zhao et al., 2016). However, its exact mode of action and its role in vertebrate nervous system development remained poorly explored.
In this study, we unravel the key role of this AAA+ protein in vertebrate motor circuit wiring using loss- and gain-of-function analyses in zebrafish embryos. Overall, our data pinpoint Fignl1 as a multifaceted regulator of MT plus end dynamics in motor axon navigation and uncover the specific roles of its isoforms in regulating +TIP binding at MT plus ends or cortical MT depolymerization.
Results
Fignl1 is an MT-associated ATPase enriched in growing axons and navigating GCs
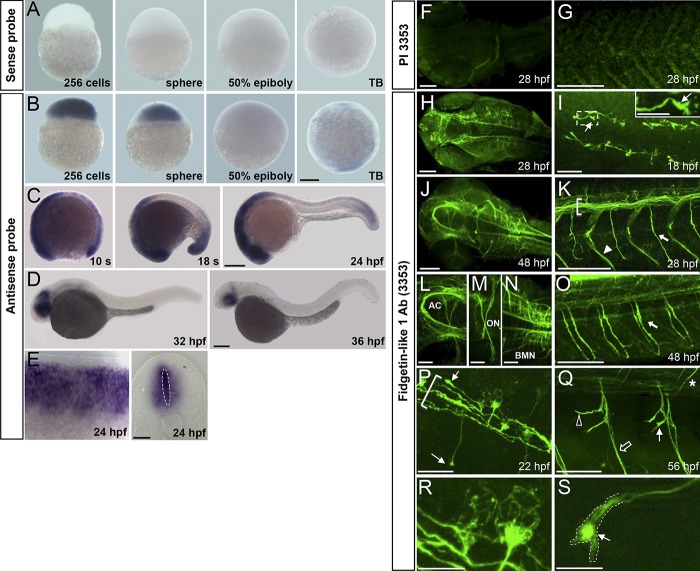
To investigate the role of Fignl1 during zebrafish development, we first assessed its expression pattern by wholemount in situ hybridization (Fig. 1, A–E). Fignl1 transcript was maternally deposited and strongly expressed during the cleavage period (256-cell and sphere stages). Its expression then decreased as gastrulation proceeds (50% epiboly, 5.3 h postfertilization [hpf]; Fig. 1 B) to be finally restricted to the prospective head and tail bud of the embryo at the end of gastrulation (Fig. 1 B, tailbud stage). During somitogenesis, Fignl1 transcript became enriched in the whole nervous system (Fig. 1 C). From 32 hpf, Fignl1 was progressively confined to the eyes, midbrain, and hindbrain (Fig. 1 D) and drastically reduced in 48-hpf larvae. In the developing spinal cord, Fignl1 expression reached its peak between 24 and 28 hpf and encompassed the central progenitor domain of spinal neurons (Fig. 1 E, dotted ellipse) and the postmitotic neurons located at the periphery of the neural tube (Fig. 1 E).
Figure 1.
Spatiotemporal expression of Fignl1 during zebrafish development. (A–E) Wholemount in situ hybridization with Fignl1 sense (A) and antisense (B–E) riboprobes at various developmental stages. Lateral views of the embryo with (A and B) animal pole to the top or (C and D) anterior to the left. (E) Lateral (left) and transversal (right) views of the spinal cord. (F–S) Immunolabeling of 18- to 56-hpf embryos with Fignl1 preimmune serum (PI 3353; F and G) or antibody (3353; H–S). Dorsal views of the head (F, H, J, and L–N), dorsal (I), and lateral (G, K, and O–S) views of the trunk; anterior is to the left, are shown. (I) Inset is a higher magnification of the boxed region. (I, P, Q, and S) Thin arrows show Fignl1 enrichment in navigating GCs of pMN (I, P, and S) and sMN (Q) axons. (K, O, and P) Brackets, thick arrows, and arrowhead indicate the ventral longitudinal fasciculus, SMN axons, and lateral myosepta, respectively. (Q) The open arrow, open arrowhead, and asterisk show ventral, rostral, and dorsal motor nerves, respectively. (R and S) MT-like pattern of Fignl1 in SMN soma and GCs. AC, anterior commissure; BMN, branchiomotor neuron; ON, optic nerve; s, somite; TB, tailbud. Bars: (A–D) 200 µm; (E and L–N) 50 µm; (F–H, J, K, O, and Q) 100 µm; (I and P) 25 µm; (R and S) 10 µm.
We next performed in toto immunostaining experiments from 18 to 56 hpf using a homemade Fignl1 antibody (3353). Staining specificity was confirmed with preimmune serum (Fig. 1, F and G) and antigen-blocking peptide (not depicted). Fignl1 was highly expressed in all commissures and axon tracts of the embryonic brain (Fig. 1, H, J, and L–N), whereas its expression was restricted to the ventral longitudinal fasciculus (Fig. 1, K and P, brackets) and the axons of primary and secondary motor neurons (pMNs and sMNs, respectively) in the spinal cord (Fig. 1, I, K, and O–S). Notably, high levels of Fignl1 were detected in the GC of navigating pioneer (Fig. 1, I, P, and S, arrows) and secondary motor axons (Fig. 1 Q, filled arrow). Higher magnifications of spinal motor neuron soma and GC revealed a filamentous staining pattern for this ATPase (Fig. 1, R and S).
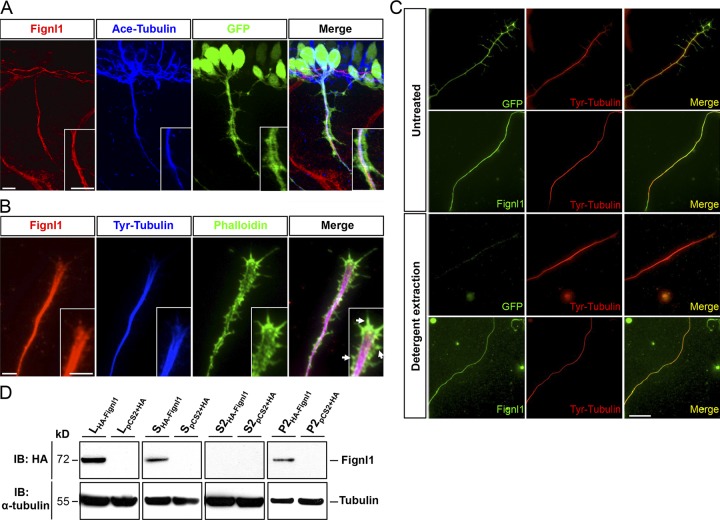
Double immunolabeling of Fignl1 and acetylated α-tubulin in 26-hpf Tg(Hb9:GFP) embryos showed that Fignl1 colocalized with MTs all along the axon shaft of spinal motor neurons (SMNs; Fig. 2 A). Moreover, Fignl1 decorated dynamic tyrosinated MTs that invade the peripheral domain of the GC in cultured Tg(Hb9:GFP) spinal neurons (Fig. 2 B). To confirm the link between Fignl1 and MTs, we used a detergent-extraction procedure on cultured Tg(Hb9:GFP) SMNs to remove all the proteins that were not associated with the cytoskeleton. After extraction, cytosolic GFP (expressed from the Hb9:GFP transgene) was eliminated, whereas Fignl1 was retained and appeared strongly colocalized with axonal MTs (Fig. 2 C). Finally, we performed MT cosedimentation assays from protein extracts of COS-7 cells overexpressing an HA-tagged version of zebrafish Fignl1 (HA-Fignl1), which further established that Fignl1 physically binds to MTs (Fig. 2 D).
Figure 2.
Fignl1 associates with MTs in spinal motor axons. (A) Immunodetection of Fignl1 (red), acetylated α-tubulin (blue), and GFP (green) in 26-hpf Tg(Hb9:GFP) transgenic embryos. Lateral views of the trunk. (B) GC from cultured Tg(Hb9:GFP) SMN labeled with phalloidin (green), tyrosinated tubulin (blue), and Fignl1 (red) antibodies. Arrows indicate actin protrusions. (C) Detergent extraction of soluble proteins from WT or Tg(Hb9:GFP) spinal neurons immunostained by tyrosinated tubulin (red) and Fignl1 or GFP (green). Bars: (A and B) 10 µm; (C) 20 µm. (D) MT copelleting assay from COS-7 cells transfected with pCS2+-HA or pCS2+-HA-Fignl1 (HA-Fignl1). Supernatants (S) were incubated with taxol and GTP to induce MT polymerization and centrifuged on a sucrose cushion to pellet MTs and their associated proteins (P2). Soluble tubulins and proteins that did not bind MTs remained in the S2 supernatant. WBs assessed Fignl1 and α-tubulin levels in different fractions. L, total lysate.
Fignl1 knockdown impairs larval locomotion
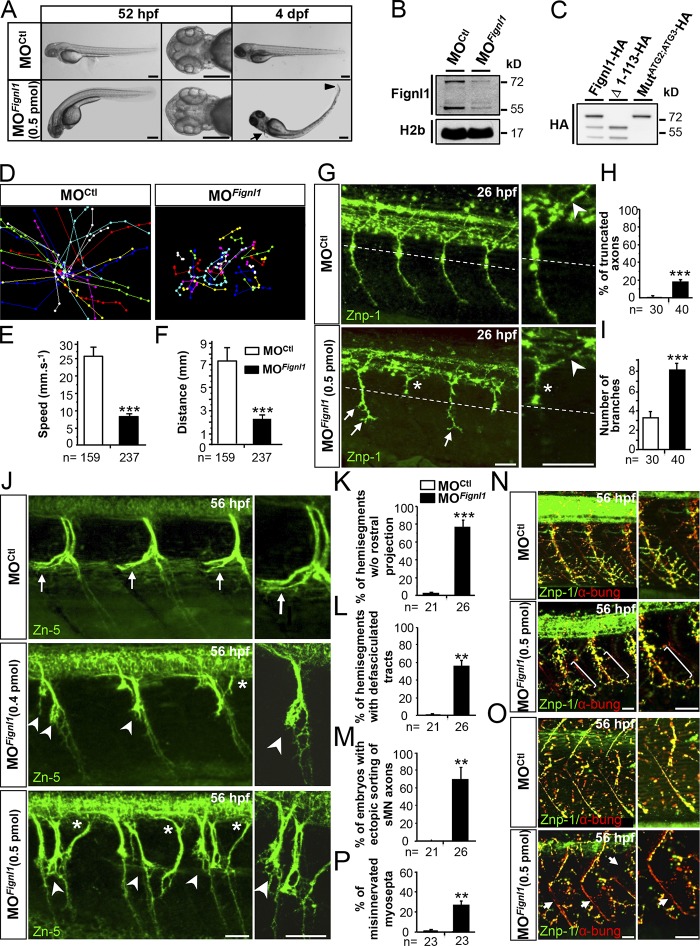
To explore Fignl1 function in developing axons, we undertook loss-of-function analyses during development of the zebrafish (Fig. 3) using two morpholino oligonucleotides (MOs) targeting Fignl1 translation initiation site (MOFignl1 and MOFignl1BIS; Figs. 3 and S1) as well as two control MOs, a 5-bp mismatch (MOCtl; Fig. 3) and a universal scramble (MOScramble; Fig. S1) MO. Fignl1 knockdown efficiency was assessed by Western blot (WB) analysis by using Fignl1 3353 antibody, which revealed two specific bands of 72 and 57 kD as shown by control experiments (preimmune serum and antigen-blocking peptides; Fig. S2 A). These two bands were significantly reduced in MOFignl1-injected embryos (Figs. 3 B and S2, A and B) and corresponded with the predicted molecular weight of full-length Fignl1 (FL-Fignl1) and a shorter isoform, which could result from the alternative use of additional in-frame translation start sites: methionine 114 (ATG2) or 174 (ATG3; Fig. S2 C). The exogenous expression of zebrafish Fignl1 cDNA fused to an HA tag in its C-terminal part and mutated or not on ATG2 and ATG3 codons confirmed that alternative translation of Fignl1 transcript generated N-terminally truncated isoforms (Fig. 3 C).
Figure 3.
Fignl1 knockdown impairs larval locomotion and spinal motor axon targeting. (A) Lateral and dorsal views of 52-hpf (left) and 4-dpf (right) larvae injected with Fignl1 morpholino (MOFignl1) or a control morpholino (MOCtl). Arrow and arrowhead indicate the atrophy of the pectoral fins and the tail tip. Bars, 250 µm. (B) WB analysis of Fignl1 knockdown efficiency. H2b was a loading control. Full-length blots with the use of preimmune serum or blocking peptides are shown in Fig. S2. (C) WB analysis of Fignl1 isoforms from protein extracts of COS-7 cells expressing HA-tagged WT (Fignl1-HA), mutated (MutATG2;ATG3-HA), or truncated (Δ1–113-HA) versions of Fignl1 with an HA antibody. Fignl1 isoforms are schematized in Fig. S2. (D–F) Tracking analysis of 56-hpf control and Fignl1 morphant larvae in a touch-escape response test. (D) Plot line representation of larval escape behavior. Each line represents the trajectory of one larva. (E) Mean swimming speed. (F) Mean covered distance. (G) Immunolabeling of pMN axons in 26-hpf MOCtl and MOFignl1 embryos with Znp-1 antibody. CaP axons of MOFignl1 embryos were truncated and stalled (asterisks) at the horizontal myoseptum (dotted line) or showed aberrant distal branches (arrows), whereas middle pMN axons were unaffected (arrowheads). (H and I) Quantification of pMN axon defects. (J) Immunolabeling of sMNs in 56-hpf MOCtl and MOFignl1 larvae with Zn-5 antibody. Arrows show control rostral nerves. Arrowheads indicate the absence of rostral nerves and the aberrant defasciculation of MOFignl1 motor tracts. Asterisks point out the ectopic sorting of MOFignl1 sMN axons. (K–M) Quantifications of sMN axon defects. (N and O) Immunolabeling of neuromuscular junctions (yellow, merge staining) in 56-hpf MOCtl and MOFignl1 larvae using pre- (Znp-1, green) and postsynaptic (AchR by α-bungarotoxin, red) markers. MOFignl1 larvae showed defective innervation of lateral myotome (N, brackets) and myosepta (O, arrows). (P) Percentage of misinnervated lateral myosepta. Quantifications were performed on 12 hemisegments around the yolk tube. **, P ≤ 0.01; ***, P ≤ 0.001; unpaired two-tailed t test. Error bars are SEM. The number (n) of embryos analyzed in three independent experiments is indicated under the corresponding histogram bar. (G, J, N, and O) Lateral views of the trunk; anterior is to the left. Right panels are higher magnifications of left panels. Bars, 25 µm.
Fignl1 morphants (i.e., embryos injected with MOFignl1 or MOFignl1BIS) developed normally compared with their control siblings (i.e., injected with MOCtl or MOScramble). However, ∼98% of them failed to hatch from their chorion at larval stages and displayed a curved-tail phenotype associated with a slight atrophy of the pectoral fins (Fig. 3 A, arrow) and the tail tip (Fig. 3 A, arrowhead). Moreover, although Fignl1 morphants responded to touch in a touch-escape response test (Fig. 3, D–F; and Videos 1 and 2), they showed striking locomotor deficits, which were characterized by reduced swimming velocity (Fig. 3 E) and distances (Fig. 3 F) compared with control larvae. These deficits were partially rescued by human FIGNL1 overexpression (Fig. S1, N and O; and Video 3). Our results thus unveiled a specific and evolutionarily conserved role for Fignl1 in vertebrate locomotion.
Fignl1 knockdown dramatically affects spinal motor axon outgrowth and pathfinding
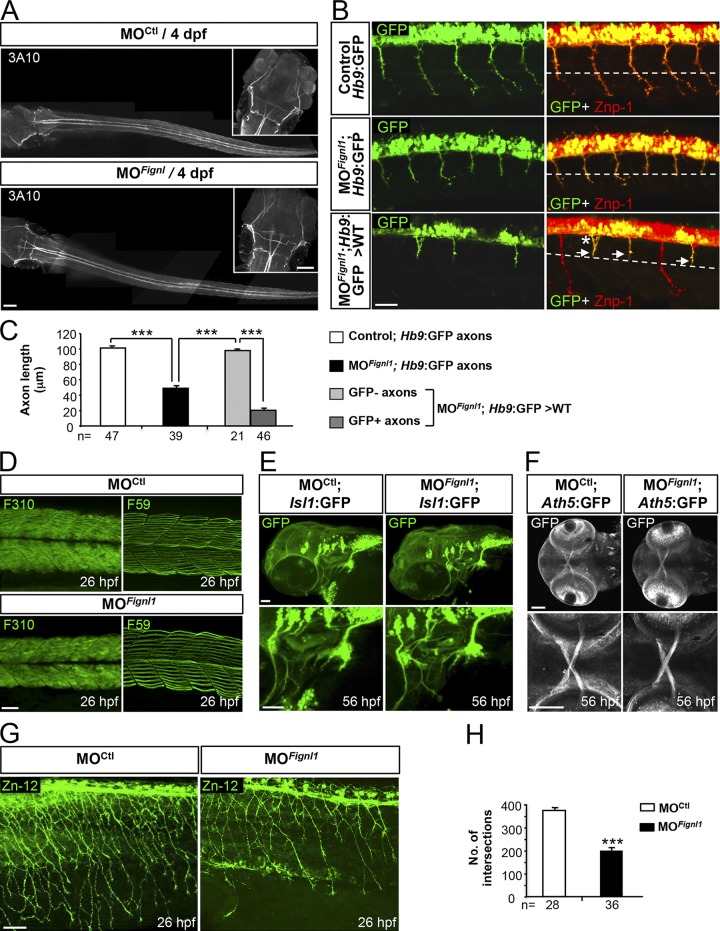
To explore whether Fignl1 morphant locomotor deficits could be attributed to motor circuit wiring defects, we analyzed the development of two key players in touch-evoked escape behavior: the SMNs and Mauthner reticulospinal neurons. Fignl1 depletion led to substantial defects of both pMN and sMN axon targeting without affecting the Mauthner fibers (Fig. 3, G–P; and Fig. 4 A). About 20% of ventrally projecting morphant caudal pMN (CaP) axons were abnormally truncated (Figs. 3 G and S1 I, asterisks) and stalled at the horizontal myoseptum (i.e., a guidance choice point indicated by a dashed line in Fig. 3 G), whereas those that reached the ventral myotome showed an increased number of distal branches (Figs. 3 G and S1 I, arrows) compared with control CaP axons (Fig. 3, H and I; and Fig. S1, H and J). Conversely, dorsally projecting pMN axons were unaffected in morphant embryos (Fig. 3 G, arrowhead). Furthermore, 56-hpf morphant sMN axons showed prominent pathfinding defects that occurred more frequently at the first intermediate target (Figs. 3 J and S1 L). Morphant sMN axons failed to grow rostrally along the horizontal myoseptum in most spinal hemisegments and appeared abnormally defasciculated at this choice point (Figs. 3 J, arrowheads, and S1 L, arrows) compared with control sMN axons (Fig. 3, J–L, arrows; and Fig. S1, K and M). Moreover, 53% of Fignl1 morphant larvae showed multiple aberrant exit points of sMN axons from the spinal cord (Fig. 3 J, asterisks), which were never observed in control larvae (0%; Fig. 3 M). In vivo time-lapse recordings of sMN axons in control and morphant Tg(Olig2:GFP) transgenic larvae showed that Fignl1 knockdown dramatically affected the navigational behavior of these axons at the horizontal myoseptum. At this intermediate target, morphant sMN axons endlessly explored their environment in random directions, which strikingly contrasted with the highly stereotyped turning behavior of control sMN axons (Videos 4 and 5). This inability of morphant sMN axons in finding their way led to massive defasciculation of spinal motor tracts (Fig. 3 J and Video 5). Subsequently, morphant larvae showed abnormal innervation of the myotome (Fig. 3 N, brackets in the bottom panels) and lateral myosepta (Fig. 3 O, arrows in the bottom panels) compared with control larvae (Fig. 3, N and O, top, and P) as illustrated by the reduced colocalization between pre (znp1; green) and postsynaptic markers (α-bungarotoxin; red). Furthermore, early embryonic transplant experiments revealed that grafted morphant Tg(Hb9:GFP) motor neurons failed to develop properly in a WT environment and displayed axon pathfinding defects similar to those of morphant embryos (Fig. 4, B and C), indicating that Fignl1 acts cell-autonomously to control SMN axon targeting. Moreover, these pathfinding errors could not be assigned to SMN specification, localization, or survival defects (Fig. S3), nor to muscle fiber alterations (Fig. 4 D) and were significantly rescued by human FIGNL1 overexpression (Fig. S1, P–R). Because Fignl1 is expressed in most developing axons, we extended our analysis to other neuronal populations (including mechanosensory Rohon-Beard [RB] neurons, branchiomotor neurons, and retinal ganglion cells; Fig. 4, E–H), which revealed a reduced innervation of the skin by RB peripheral axons compared with controls (Fig. 4, G and H). This defective cutaneous innervation of Fignl1 morphants was correlated with a reduction in the RB neuron population (Fig. S3, A–C).
Figure 4.
Fignl1 is mainly required for spinal motor axon targeting. (A) Immunolabeling of the Mauthner fibers in 4-dpf MOCtl (n = 17) and MOFignl1 (n = 19) larvae using the 3A10 antibody. Dorsal views of the larvae; anterior is to the left. Insets are higher magnifications of the head. (B) Immunostaining of SMN with Znp-1 and GFP antibodies in 30-hpf nontransplanted control (Hb9:GFP), morphant (MOFignl1; Hb9:GFP), and WT embryos transplanted with SMN precursors from MOFignl1 Tg(Hb9:GFP) embryos. The dashed line indicates the horizontal myoseptum. Arrows and asterisk show stalled or misrouted SMN axons, respectively. (C) Mean length of SMN axons. The number of axons analyzed is indicated under each histogram bar. (D) Immunostaining of slow and fast muscle fibers in 26-hpf MOCtl (n = 15) and MOFignl1 (n = 15) embryos using F59 and F310 antibodies. (E) Analysis of branchiomotor axons in 56-hpf MOCtl (n = 20) and MOFignl1 (n = 25) Tg(Islet1:GFP) larvae. Lateral views of the head; anterior is to the left. (F) Analysis of the optic nerve in 56-hpf MOCtl (n = 20) and MOFignl1(n = 20) Tg(Ath5:GFP) larvae. Ventral views of the head; anterior is to the left. Bottom panels are higher magnifications of top panels. (G) Immunostaining of mechanosensory RB neurons in 26-hpf MOCtl and MOFignl1 embryos. Skin innervation by RB axons is reduced in morphant embryos. (B, D, and G) Lateral views of the trunk; anterior is to the left. (H) Quantification of skin innervation using a stereological method (Rønn et al., 2000). The number of intersections between RB axons and a grid of 11 parallel lines spaced every 22 µm was estimated in 26-hpf MOCtl (n = 28) and MOFignl1 embryos (n = 36) from three independent experiments. ***, P ≤ 0.0001; unpaired two-tailed t test. Error bars are SEM. Bars: (A) 100 µm; (B) 25 µm; (D) 40 µm; (E and G) 50 µm; (F) 60 µm.
Fignl1 overexpression impairs spinal motor axon targeting
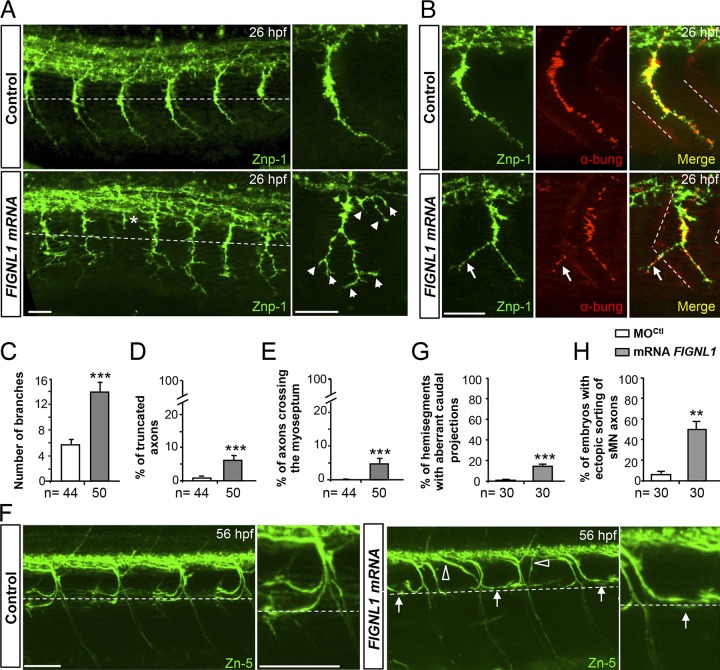
We next performed gain-of-function experiments by injecting zebrafish Fignl1 or human FIGNL1 transcripts into WT embryos. FIGNL1 overexpression led to various defects in SMN axon extension, branching, and pathfinding that were conspicuously different from those of morphants (Fig. 5). FIGNL1-overexpressing CaP axons exhibited aberrant collateral branches all along their length (Fig. 5, A, arrowheads, and C), which occasionally crossed the lateral myoseptum to ectopically innervate the muscle fibers of neighboring somites (Fig. 5 B, arrows). This was never observed in control and morphant CaP axons (Fig. 5, A and E; and Fig. 3 G). Moreover, a few FIGNL1-overexpressing pMN axons stopped soon after exiting the spinal cord and never reached the horizontal myoseptum, unlike control (Fig. 5 A, asterisk, and D) and morphant axons (Fig. 3 G). FIGNL1-overexpressing larvae also showed misguided sMN axons that project caudally instead of rostrally along the horizontal myoseptum (Fig. 5 F, arrows) or exit ectopically from the spinal cord (Fig. 5 F, open arrowheads). These defects were rarely observed in control larvae (Fig. 5, G and H), and the caudal orientation of sMN axons was never observed in Fignl1 morphants (Fig. 5 F vs. Fig. 3 J). Overall, both loss- and gain-of-function analyses revealed the importance of fine-tuning Fignl1 expression for zebrafish motor circuit wiring.
Figure 5.
FIGNL1 overexpression leads to spinal motor axon pathfinding defects. (A) Immunolabeling of pMN axons in 26-hpf control and human FIGNL1 mRNA–injected embryos with the use of Znp-1 antibody. Arrowheads and asterisk indicate hyperbranched or truncated pMN axons. (B) Analysis of neuromuscular junctions in 26-hpf control and FIGNL1 mRNA–injected embryos with the use of pre- (Znp-1, green) and postsynaptic markers. α-bung, α-bungarotoxin (red). Arrows indicate aberrant synapses between CaP axons and muscle fibers from the neighboring somite. (C) Mean number of branches. (D) Percentage of truncated axons. (E) Percentage of axons crossing the lateral myoseptum. (F) Immunolabeling of sMN axons with Zn-5 antibody in 56-hpf control and human FIGNL1 mRNA–injected larvae. Arrows show misguided rostral pMN–like sMNs. Open arrowheads indicate aberrant sMN axon exit points. (A, B, and F) Lateral views of the trunk; anterior is to the left. Right panels are higher magnifications of one SMN axon from the left panels. Dashed lines indicate the horizontal myotome (A and F) or lateral myosepta (B). Bars: (A and B) 25 µm; (F) 50 µm. (G) Percentage of hemisegments with aberrant caudal projections. (H) Percentage of embryos with ectopic sorting of sMN axons. The total number (n) of embryos from three independent experiments is indicated under the corresponding histogram bar. 24 hemisegments were analyzed per animal. **, P ≤ 0.01; ***, P ≤ 0.001; unpaired two-tailed t test. Error bars are SEM.
Fignl1 influences GC morphology through MT cytoskeleton remodeling
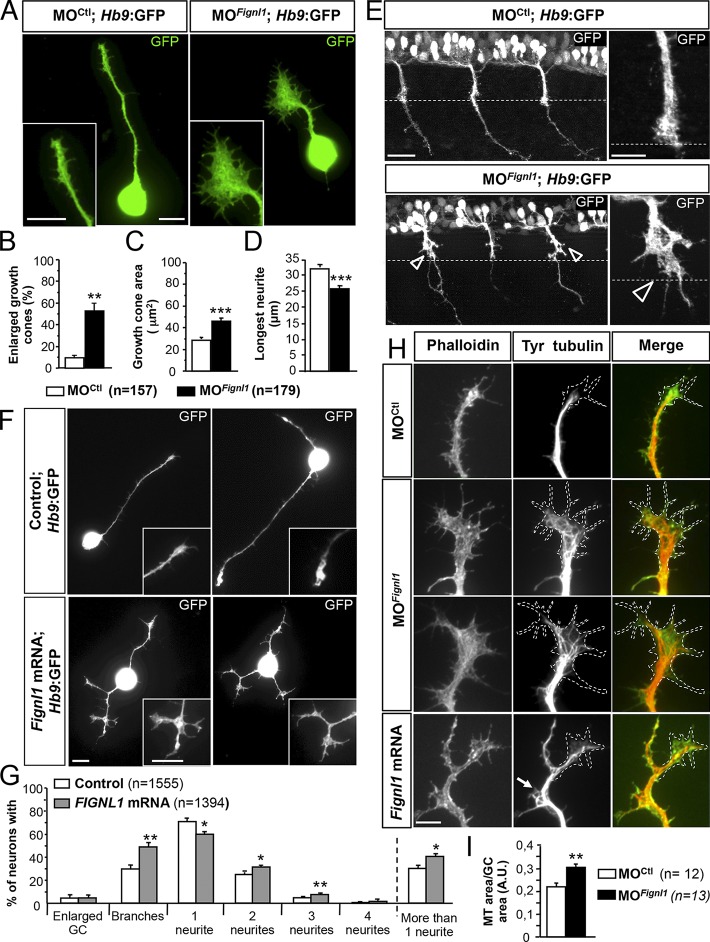
To identify the mechanism by which Fignl1 controls SMN axon pathfinding, we analyzed GC morphology and cytoskeleton organization in cultured SMNs from 24-hpf control, Fignl1-depleted, or -overexpressing Tg(Hb9:GFP) embryos. A majority (51.6%) of morphant SMN GCs appeared abnormally enlarged (vs. 10% in controls; Fig. 6, A and B; and Fig. S1 E) and showed a 40% increase of their surface compared with control GCs (Fig. 6 C). This aberrant expansion of morphant GC area was tightly correlated with a significant reduction in the longest neurite length compared with controls (Fig. 6 D). The enlarged GC phenotype also occurred in vivo at the horizontal myoseptum of Tg(Hb9:GFP) morphant embryos (Fig. 6 E, open arrowheads) as well as in cultured mammalian neurons transfected with Fignl1-shRNA (Fig. S4). In contrast, Fignl1-overexpressing SMN GCs were abnormally split and branched (Fig. 6 F, insets), which was correlated with their increased number of branches in vitro (Fig. 6, F and G) and in vivo (Fig. 5, A–C). Furthermore, Fignl1-overexpressing SMNs showed a slight but significant increase in the number of neurites compared with controls (Fig. 6, F and G).
Figure 6.
Fignl1 modulates SMN morphology via MT remodeling. (A) SMN primary cultures from 24-hpf MOCtl or MOFignl1 Tg(Hb9:GFP) embryos labeled at 9 hpp with a GFP antibody. (B) Percentage of enlarged GCs. (C) Mean GC area. (D) Mean length of the longest neurite. (B–D) 179 MOCtl and 157 MOFignl1 SMNs were analyzed in three independent experiments. (E) Immunostaining of 26-hpf MOCtl and MOFignl1 Tg(Hb9:GFP) embryos using a GFP antibody. Lateral views of the trunk; anterior is to the left. Right images are higher magnifications of spinal motor CGs on facing left images. Open arrowheads indicate enlarged GCs. Dashed lines outline the horizontal myoseptum. (F) SMN primary culture from 24-hpf control and Fignl1 mRNA–injected Tg(Hb9:GFP) embryos labeled 9 hpp with GFP antibody. (A and F) Insets show higher magnifications of GCs. (G) Percentage of SMNs with enlarged GCs, collateral branches, and one, two, three, or more neurites. Quantifications were performed on 1,300 control and Fignl1-injected SMN in four independent experiments. (H) Immunodetection of MTs (red) and F-actin (green) in cultured control, Fignl1-depleted, or Fignl1-overexpressing Tg(Hb9:GFP) SMNs at 9 hpp. Dashed lines outline the GC surface. Bars, 10 µm. (I) GC area occupied by MTs. Quantifications on at least 13 MOCtl and MOFignl1 SMNs selected from three independent experiments. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; unpaired two-tailed t test. Error bars are SEM.
Interestingly, MTs in enlarged morphant GCs were splayed, looped, or bent. They occupied a larger GC surface and markedly invaded the peripheral domain (Fig. 6, H, middle, and I; and Fig. S4) compared with MTs of control GCs, which appeared tightly bundled, restricted to the central domain and oriented in axon growth direction (Fig. 6 H, top, and Fig. S4). In contrast, MTs of Fignl1-overexpressing neurons were debundled at nascent or mature ectopic branch points but not in GCs (Fig. 6 H, bottom). Conversely, the F-actin network seemed unaltered by variations in Fignl1 expression levels (Fig. 6 H). Our data thus suggest that Fignl1 controls motor GC morphology and behavior through MT remodeling.
Fignl1 knockdown alters MT dynamics in spinal motor axons and GCs in vivo
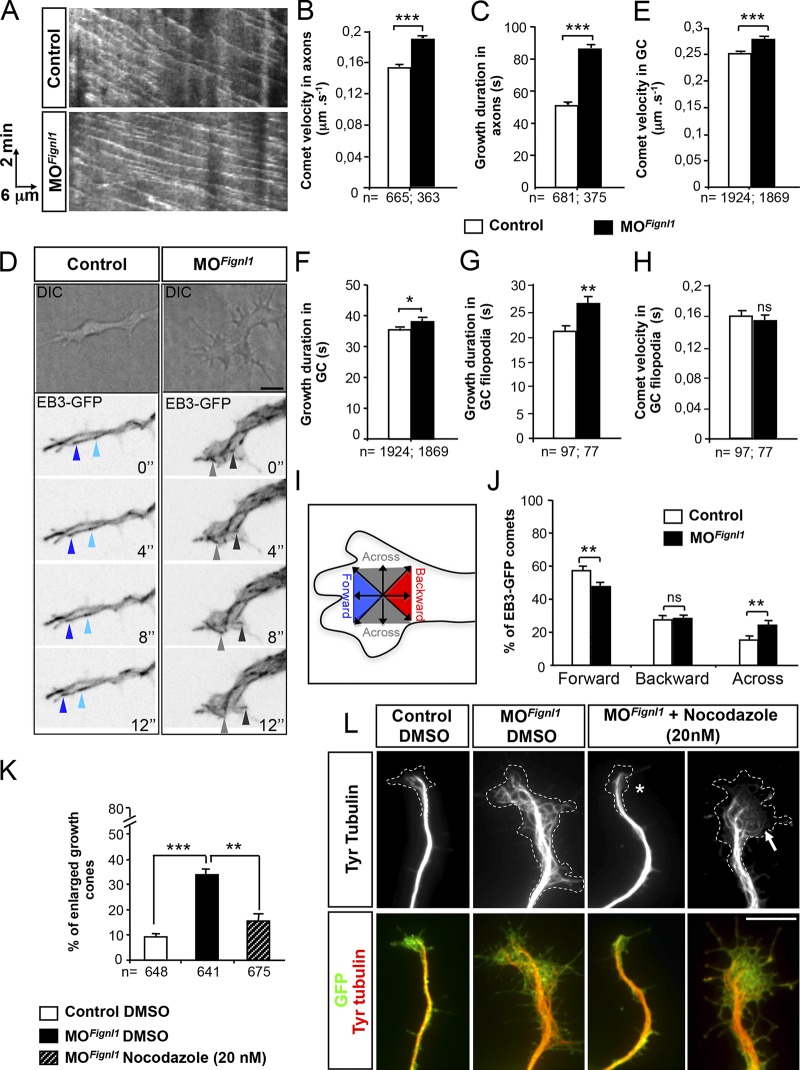
To assess whether Fignl1 influences MT dynamics and/or growth directionality, we used time-lapse videomicroscopy to monitor MT plus end behavior in vivo in growing axons. We imaged 52-hpf control and MOFignl1-injected Tg(Mnx1:Gal4;UAS:EB3-GFP) transgenic larvae that selectively expressed the GFP-tagged +TIP EB3 in SMNs. Kymograph analyses of time-lapse recordings showed that EB3-GFP-comet velocity was significantly increased in morphant SMN axons compared with controls (Fig. 7, A and B; Videos 6 and 7). Moreover, EB3-GFP comets could be tracked over longer time periods in morphant SMN axons compared with controls (Fig. 7, A and C; Videos 6 and 7), which suggested that Fignl1 knockdown reduced MT dynamic instability and/or increased MT sliding in vivo. The monitoring of EB3-GFP comets in navigating GCs was more complex in vivo because of the difficulty in acquiring images with the high speed required for EB3 movement recording and the z stack thickness imposed by EB3-comet tracking in the whole GC area. To overcome these technical limitations, EB3-GFP comets were monitored within GCs of cultured SMNs from 24-hpf control and morphant Tg(Hb9:Gal4;UAS:EB3-GFP) embryos (Fig. 7, D–J). By automatically tracking EB3-GFP displacement, we showed that EB3-GFP comet velocity and duration were also significantly increased in morphant GCs compared with controls (Fig. 7, E and F; Videos 8 and 9). Furthermore, the kymograph analysis of EB3-GFP comets within GC filopodia revealed that morphant MTs spent more time growing along F-actin than control MTs (Fig. 7 G), although their growth rate was not significantly increased in these protrusions (Fig. 7 H). Finally, Fignl1 knockdown also altered MT growth directionality as they entered the GC. Indeed, the percentage of comets moving forward toward the leading edge of the GC was decreased, whereas the fraction of comets moving across the GC was significantly increased (Fig. 7 D, gray arrows, and Video 9) in morphant GCs compared with controls (Fig. 7, D, blue arrows, and I and J; Video 8). Contrastingly, the percentage of comets moving backward was unaffected by Fignl1 depletion (Fig. 7 J).
Figure 7.
Fignl1 depletion alters MT plus end dynamics in navigating motor axons and GCs. (A–C) Time-lapse recordings of EB3-GFP comets in SMN axons of 52-hpf control and MOFignl1 Tg(Hb9:Gal4;UAS:EB3-GFP) larvae (related to Videos 6 and 7). (A) Representative kymograms of a 5-min EB3-GFP recording from control and morphant SMN axons. Horizontal bar, 6 µm; vertical bar, 2 min. (B and C) Mean comet velocity (B) and growth duration (C) in SMN axons. (D–J) Time-lapse recordings of EB3-GFP comets in cultured control and MOFignl1 Tg(Hb9:Gal4;UAS:EB3-GFP) SMNs. (D) Top panels are differential interference contrast (DIC) images illustrating GC morphology. Bottom panels are still z projection images extracted from EB3-GFP time-lapse videos (related to Videos 8 and 9). Blue and gray arrowheads point at EB3 comets moving forward toward the leading edge of the GC or across the GC, respectively. (E–H) Mean comet velocity (E and H) and growth duration (F and G) in GCs (E and F) and GC filopodia (G and H). (B, C, and E–H) EB3 comets were monitored in SMN axons of at least 10 control and MOFignl1 larvae (B and C) or in 15 control and MOFignl1 GCs (E–J) from three independent experiments. (I) Diagram illustrating the assessment of comet directionality in GCs. (J) Quantification of comet directionality in GCs. (K and L) Primary cultures of SMNs from 24-hpf control and MOFignl1 Tg(Hb9:GFP) embryos treated 3 hpp with 20 nM nocodazole or DMSO and labeled at 9 hpp with GFP and tyrosinated tubulin (Tyr tubulin) antibodies (L). Dotted line indicates GC surface. Nocodazole treatment rescues the GC area (asterisk) and reduces MT invasion in GC (arrow) of morphant SMNs. (K) Percentage of enlarged motor GCs. The total number (n) of analyzed comets or neurons from three independent experiments is mentioned under the histogram bar. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; unpaired two-tailed t test (B, C, and E–H) and Kruskal–Wallis ANOVA test with Dunn’s post hoc test (K). Error bars are SEM. Bars: (D) 5 µm; (L) 10 µm.
To explore whether a sustained MT polymerization associated with Fignl1 knockdown could contribute to the abnormal GC morphology, cultured morphant Tg(Hb9:GFP) SMNs were treated with low doses (20 nM) of the MT-depolymerizing drug nocodazole or the DMSO carrier. Nocodazole treatment markedly reduced the proportion of enlarged GCs in morphant SMN cultures compared with DMSO treatment and evened up the ratio of enlarged versus normal GCs with that of controls (Fig. 7, K and L). Furthermore, the overfilling of morphant enlarged GCs by splayed MTs was rescued in a majority of nocodazole-treated GCs (Fig. 7 L, asterisk), although it appeared more limited in the remaining enlarged GCs in which MTs were partially uncoiled (Fig. 7 L, arrow). These data strongly suggest that the altered morphology and navigational behavior of Fignl1-depleted GCs are mainly linked to abnormal MT dynamics.
Fignl1 full-length isoform negatively regulates +TIP binding at MT plus ends through specific interactions with EBs
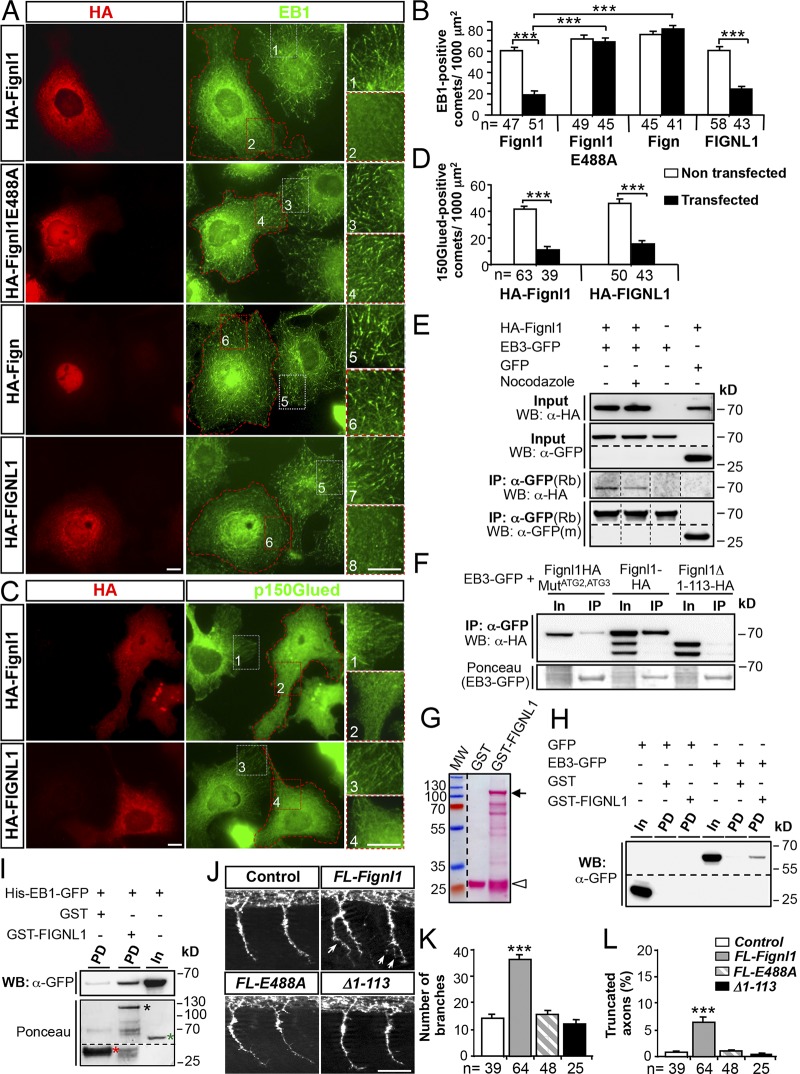
To decipher the role of Fignl1 isoforms in MT dynamicity in a simple model system, HA- or GFP-tagged zebrafish Fignl1 or human FIGNL1 full-coding cDNAs were transfected in COS-7 cells. Overexpression of zebrafish or human Fignl1 failed to dismantle the MT network unlike spastin overexpression (Fig. S5, A and B). Contrastingly, it led to a striking reduction in both EB1- and p150Glued-positive comets and concomitantly to an enhanced diffuse staining of these proteins in the cytoplasm compared with NT cells or cells expressing an ATPase-deficient Fignl1 mutant (HA-Fignl1E488A; substitution of a highly conserved glutamate residue in the Walker B motif required for ATP hydrolysis; Hanson and Whiteheart, 2005; Yakushiji et al., 2006), Fign, or GFP alone (Fig. 8, A–D; and Fig. S5, C–E). Notably, EB mislocalization in cells overexpressing Fignl1 was not associated with an overall stabilization of the MT network as shown by the quantification of the mean acetylated-tubulin fluorescence intensity (Fig. S5, F and G), or by nocodazole susceptibility assay (Fig. S5, H and I). This suggested that Fignl1 negatively regulated EB binding at MT plus ends in an ATP-dependent manner. Coimmunoprecipitation (co-IP) and GST pulldown assays further showed that Fignl1 directly bound to EB1 and EB3 (Fig. 8, E–I; and Fig. S5 J), unlike spastin and Fign (Fig. S5 K). Furthermore, EB–Fignl1 interactions were favored by the MT lattice because nocodazole-induced MT depolymerization reduced the amount of HA-Fignl1 that copelleted with EB3-GFP (Fig. 8 E). However, because both WT and E488A Fignl1 similarly bound EB1, this interaction may not be regulated by ATP hydrolysis (Fig. S5 J). Interestingly, N-terminally truncated Fignl1 isoforms failed to bind EBs in co-IP assays (Fig. 8 F), which suggested a specific role for Fignl1 full-length isoform in the regulation of MT plus end complexes. Finally, overexpression analyses in zebrafish embryos revealed that Fignl1E488A or EB-binding-defective Fignl1Δ1–113 did not affect pMN axon targeting unlike their WT counterpart (Fig. 8, J–L). This demonstrates that the cooperative interaction between FL-Fignl1 and the core +TIP EBs is critical for motor circuit connectivity.
Figure 8.
FL-Fignl1 isoform specifically binds EB1/3 and negatively regulates +TIP binding at MT plus ends. (A–D) COS-7 cells overexpressing HA-tagged WT (FL-Fignl1–HA; A–D) and mutated (HA-Fignl1E488A; A and B) zebrafish Fignl1, zebrafish Fign (HA-Fign; A and B), and human Fignl1 (HA-FIGNL1; A–D) were immunolabeled with HA (A and C: red) and EB1 (A: green) or p150Glued (C: green) antibodies. (A and C) Red dashed lines indicate transfected cells. Right panels represent higher magnifications of boxed regions in corresponding panels. (B and D) Mean number of EB1- (B) or p150Glued-positive (D) comets per 1,000 µm2. (E) Co-IP of HA-Fignl1 with EB3-GFP. COS-7 cells were cotransfected with HA-Fignl1 and EB3-GFP or GFP and treated with 20 µM nocodazole or DMSO 24 h posttransfection (hpt). (F) Co-IP of Fignl1 isoforms with EB3-GFP. COS-7 cells were cotransfected with Fignl1-HA, Fignl1MutATG2-ATG3-HA, or Fignl1Δ1–113-HA and EB3-GFP. Ponceau staining shows immunoprecipitated EB3-GFP. (E and F) Co-IP assays were performed 24 hpt with a GFP antibody. (G) Ponceau staining shows the expression of recombinant GST (open arrowhead) and GST-FIGNL1 (arrow) in BL21 E. coli purified on glutathione Sepharose beads. Lower bands result from protein degradation. MW, molecular weight. (H and I) GST pulldown assays. EB3-GFP from COS-7 cells or recombinant His-EB1-GFP (I) were incubated with either GST or GST-FIGNL1–bound beads. (I) Ponceau staining (bottom) shows GST (red asterisk), GST-FIGNL1 (black asterisk), and His-EB1-GFP (green asterisk) recombinant proteins. (E, F, H, and I) Immunoprecipitated or pulled-down proteins were revealed by using HA or GFP antibody. In, input; IP, immunoprecipitation; m, mouse; PD, pulldown; Rb, rabbit. (J) Immunolabeling of pMNs in 26-hpf control and WT Fignl1 (FL-Fignl1), E488A Fignl1 (FL-E488A), or Fignl1Δ1–113 (Δ1–113) mRNA–injected embryos with the use of Znp-1 antibody. Arrows point at aberrant pMN collateral branches. (K) Mean number of branches. (L) Percentage of truncated axons. Quantifications on 12 spinal hemisegments per embryo. The total number (n) of analyzed cells or embryos per condition in three independent experiments is indicated under the histogram bars. ***, P ≤ 0.001; unpaired two-tailed t test (B and D) or Kruskal–Wallis ANOVA test with Dunn’s post hoc test (K and L). Error bars are SEM. Bars: (A and C) 20 µm; (J) 50 µm.
N-terminally truncated Fignl1 isoforms have a cortical MT-depolymerizing activity
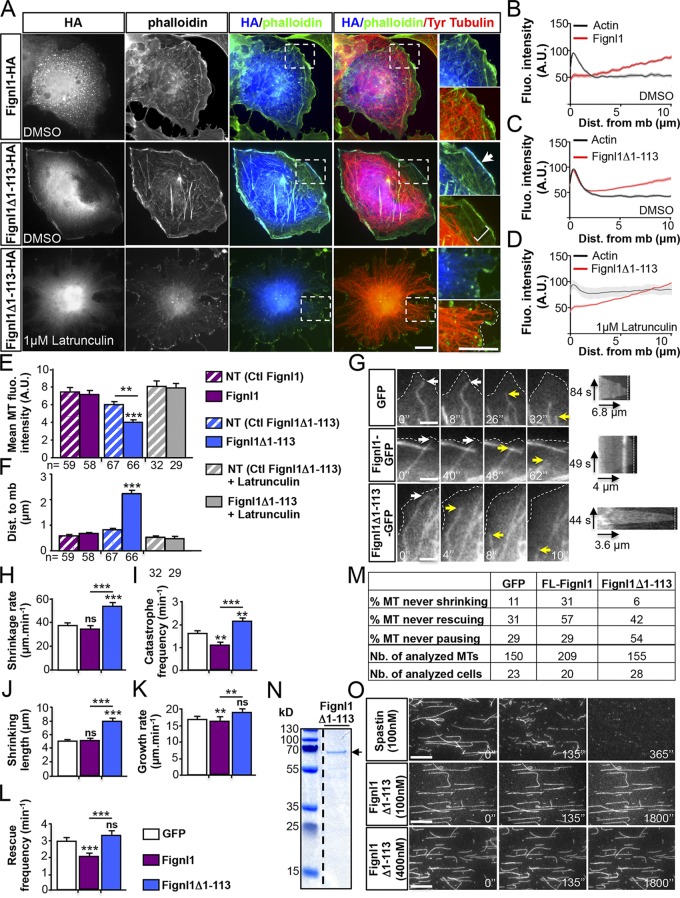
To characterize the role of N-terminally truncated Fignl1 isoforms, Fignl1Δ1–113-HA (or -GFP) cDNAs were transfected in COS-7 cells. Although Fignl1-HA (or -GFP) showed a cytoplasmic punctate distribution (Fig. 9, A and B), Fignl1Δ1–113-HA (or -GFP) was diffused in the cytosol and strikingly enriched at the cell membrane where it strongly colocalized with the actin cortex (Fig. 9, A, arrow, and C). This cortical localization was dependent on F-actin because it disappeared after a treatment with the F-actin–depolymerizing drug, latrunculin B (Fig. 9, A and D). Furthermore, the cortical enrichment of Fignl1Δ1–113-HA (or -GFP) was associated with a significant reduction in MT density beneath the cell cortex, which contrasted with the dense subcortical MT array of Fignl1-HA–transfected or mock cells (Fig. 9, A and E). Moreover, the distance between MT ends and the cell cortex was markedly increased in Fignl1Δ1–113-HA–transfected cells compared with Fignl1-HA–expressing or mock cells (Fig. 9, A and F). Latrunculin B–mediated delocalization of cortical Fignl1Δ1–113 rescued both MT density and contacts at the cell cortex of Fignl1Δ1–113-HA–expressing cells (Fig. 9, A, E, and F). We next used total internal reflection fluorescence (TIRF) microscopy to monitor MT behavior beneath the cell cortex of mouse embryonic fibroblasts (MEFs) cotransfected with mCherry α-tubulin and Fignl1-GFP, Fignl1Δ1–113-GFP, or GFP constructs (Fig. 9 G). MEFs are large, flat cells ideally suited for live imaging of individual MTs. Fignl1 overexpression in MEFs led to similar MT phenotypes to those of COS-7 cells. Kymograph analyses of MT ends located within 12 µm of the cell cortex revealed that Fignl1Δ1–113-GFP increased MT catastrophe frequency as well as shrinkage rate and length compared with GFP alone or Fignl1-GFP (Fig. 9, H–J). Contrastingly, MTs of Fignl1-GFP–transfected cells were less dynamic and showed reduced catastrophe and rescue frequencies as well as growth rate compared with mock cells or Fignl1Δ1–113-GFP–expressing cells (Fig. 9, I and K–M). Furthermore, the time length of MT persistence at the cell cortex was abnormally increased for some MTs of Fignl1-GFP–expressing cells, although slightly reduced for MTs of Fignl1Δ1–113-GFP–expressing MEFs compared with MTs of control cells (GFP) as shown on kymograms (Fig. 9 G). Because MT growth and shrinkage rates in interphasic cells were shown to be 5 to 10 times higher than the MT sliding rate (Oladipo et al., 2007; Jolly et al., 2010), the contribution of MT sliding to the growth/shrinkage rates associated with Fignl1 overexpression was considered to be minor in these interphasic MEFs.
Figure 9.
The N-terminally truncated Fignl1 isoforms promote MT depolymerization at the cell cortex. (A) COS-7 cells transfected with Fignl1-HA or Fignl1Δ1–113-HA were treated with DMSO or latrunculin B 24 h posttransfection and labeled with phalloidin, HA, and tyrosinated tubulin (Tyr tubulin) antibodies. Right: Higher magnifications of boxed regions in corresponding panels. Dotted line outlines the plasma membrane. Arrow points out the colocalization between Fignl1Δ1–113 and cortical F-actin. Bracket indicates the reduced density of MTs beneath the cell cortex of Fignl1Δ1–113-HA–expressing cells. (B–D) Mean fluorescence intensity of F-actin and Fignl1-HA (B) or Fignl1Δ1–113-HA (C and D) in COS-7 cells treated with DMSO (B and C) or latrunculin B (D). Dist. from mb, distance from membrane. (E) MT fluorescence intensity beneath the cell cortex. NT, nontransfected. (F) Mean distance between MT plus ends and the plasma membrane (Dist. to mb). (G–M) Live TIRF recordings of MT behavior beneath the cell membrane of MEFs expressing α-tubulin–mCherry and Fignl1-GFP, Fignl1Δ1–113-GFP, or GFP. (G) Representative time series (left) and kymograms (right) of individual MT behavior at the cell membrane (dashed lines). White and yellow arrows, respectively, point at MT ends contacting the membrane or undergoing catastrophe. Black arrows are kymogram scale bars. (H–L and M) MT dynamic instability analyses. Catastrophe and rescue frequencies (I and L), shrinkage and growth rates (H and K), shrinking length (J), and the percentage of MT that never shrunk, grew, or paused (M) were calculated from kymograph analysis of at least 150 MTs per condition. **, P ≤ 0.01; ***, P ≤ 0.001; Kruskal–Wallis ANOVA test with Dunn’s post hoc test. Error bars are SEM. The total number of cells (E, F, and M) or MTs (M) analyzed per condition in three independent experiments is indicated under the histogram bar (E and F) or in the table (M). Nb., number. (N) Coomassie blue–stained SDS-PAGE gel showing purified His-Fignl1Δ1–113 (arrow). (O) In vitro MT-severing assay. Atto 565–labeled and biotinylated taxol-stabilized MTs were immobilized in perfusion chambers. GFP-spastin (top) or His-Fignl1Δ1–113 (middle and bottom) were diluted to 100 or 400 nM and perfused into the chambers. Panels are representative time frames of TIRF recordings acquired every 5 min over 30 min. Bars: (A) 20 µm; (G) 3 µm; (O) 5 µm.
To test whether Fignl1 promoted MT catastrophe through MT severing, we performed in vitro MT-severing assays using recombinant GFP-spastin as a positive control. Atto 565–labeled and taxol-stabilized MTs were rapidly severed and completely disassembled within 360 s after 100 nM GFP-spastin perfusion, whereas no severing events were observed after 100 or 400 nM His-Fignl1Δ1–113 perfusions even after 1,800 s (Fig. 9, N and O). Overall, these results demonstrated that Fignl1 isoforms differentially affected MT dynamics and suggested that N-terminally truncated Fignl1 isoforms promoted MT disassembly beneath the cell cortex through a depolymerizing rather than a severing process.
Discussion
Altogether, our work in zebrafish and mammalian cells unveils the evolutionarily conserved role of Fignl1 in neuronal connectivity. Fignl1 controls GC morphology and steering behaviors at intermediate guidance targets through its dual ability to displace the core +TIP EBs from MT plus ends and promote MT depolymerization at the interface with the actin cortex. Our study thus unveils a novel function for the “meotic clade” ATPase subfamily in the regulation of MT plus end complexes and provides new insight into the molecular mechanisms underlying motor axon navigation.
Fignl1, a multifaceted regulator of MT plus end dynamics
Fignl1 belongs to the meiotic clade of AAA+ ATPases together with the well-known MT-severing enzymes spastin, katanin, and fidgetin that make internal breaks in the MT lattice (Roll-Mecak and McNally, 2010; Mukherjee et al., 2012; Sharp and Ross, 2012; Monroe and Hill, 2016). However, unlike its extensively studied homologues, the cellular and developmental functions of Fignl1 have been rarely explored and never in the nervous system (Yang et al., 2005; Luke-Glaser et al., 2007; Park et al., 2007; L’Hôte et al., 2011; Yuan and Chen, 2013; Girard et al., 2015; Zhao et al., 2016). Our analysis of zebrafish Fignl1 during vertebrate development revealed its key role in SMN axon targeting as reported for spastin and katanin (Wood et al., 2006; Butler et al., 2010). MT-severing enzymes are required for common cellular processes through distinct mechanisms, for example, in chromosome poleward motility during Drosophila melanogaster mitosis (Zhang et al., 2007) or axon branching processes in cultured mammalian neurons (Yu et al., 2008). However, their functional diversity so far lies in their different subcellular distribution and the regulation of their severing activity by MT posttranslational modifications (Lacroix et al., 2010; Sudo and Baas, 2010; Leo et al., 2015; Hu et al., 2017) or MT-associated proteins (Yu et al., 2008). Our study thus provides new molecular insight into their functional specificity by unraveling a crucial link between FL-Fignl1 and the core +TIPs EB1 and EB3. It further reveals that Fignl1 short isoforms promote MT disassembly through a depolymerization rather than a severing process.
On one hand, we showed that FL-Fignl1 negatively regulates EB binding at MT plus ends as described for MAP1B (Tortosa et al., 2013) and Tau (Sayas et al., 2015). However, in contrast with these MT stabilizers, FL-Fignl1 may not compete off or sequester EB proteins from MT plus ends as the MT lattice favors Fignl1 interactions with EBs but would rather dissociate them from growing plus tips in an ATPase-dependent manner. Indeed, Fignl1E488A, harboring a mutation in the Walker B motif required for ATP hydrolysis (Hanson and Whiteheart, 2005; Yakushiji et al., 2006), fails to delocalize EB proteins from MT plus ends, although it binds these +TIPs. AAA+ enzymes conventionally use the energy released by ATP hydrolysis to induce conformational changes of their substrates, therefore promoting the assembly or disassembly of macromolecular complexes (White and Lauring, 2007). Based on this paradigm, FL-Fignl1 may trigger the dissociation of MT plus end complexes via EB conformational remodeling. As a consequence, Fignl1 would modify +TIP-related MT behaviors such as dynamics, cortical interaction, and cross talk with the actin cytoskeleton, which are all critical for polarized migration of cells and GC navigation (Akhmanova and Steinmetz, 2008; Bearce et al., 2015; Cammarata et al., 2016). Furthermore, the acidic tail of EB1 (Mishima et al., 2007) may provide a substrate for Fignl1 long isoform as shown for the acidic tubulin C-terminal tail in tubulin conformational remodeling by spastin or katanin (Roll-Mecak and Vale, 2008; Johjima et al., 2015).
On the other hand, our analysis revealed that N-terminally truncated Fignl1 isoforms reduce MT plus end contacts with the cortex by promoting their catastrophe, as shown for katanin (Zhang et al., 2011). Moreover, Fignl1 enrichment at the cell cortex is dependent on F-actin and required for its depolymerase activity. These results identify Fignl1 as a novel coordinator of MT remodeling at the interface with the actin cytoskeleton. However, the molecular mechanism underlying its targeting and activation at the cell cortex remains to be clarified. The inability of FL-Fignl1 to promote cortical MT depolymerization suggests that its specific N-terminal domain (amino-acids 1–113) might inhibit its cortical recruitment. Although the hypothesis that N-terminally truncated isoforms may sever MTs at their tips cannot be ruled out, our in vitro and ex vivo experiments suggest that they may induce MT disassembly through a depolymerizing rather than a severing process. Interestingly, katanin and Fign have both severing and end-depolymerizing activities in vitro (Zhang et al., 2011; Mukherjee et al., 2012), although the mechanism underlying this depolymerizing activity remained undetermined. The lack of Fignl1Δ1–113–mediated end depolymerization in vitro (Fig. 9) strengthens the possibility that Fignl1-depolymerizing activity relies on its cortical recruitment by specific binding partners and/or in vivo signaling pathways. Finally, the strong oligomerization capacity of Fignl1 (Peng et al., 2013) implies that both isoforms may also act in concert. Indeed, transient oligomerization between cortical Fignl1Δ1–113 and EB-bound FL-Fignl1 may aid Fignl1Δ1–113 in capturing EB-bound growing plus ends and/or promoting its depolymerizing activity by removing +TIP complexes from MT plus ends.
Fignl1-driven MT plus end dynamics are required for motor axon navigation at intermediate targets
Numerous studies have shown that changes in GC behavior initially elicited by extracellular guidance signals ultimately rely on the coordinated and asymmetric remodeling of both actin and MT cytoskeleton (Schaefer et al., 2008; Lowery and Van Vactor, 2009; Dent et al., 2011; Kalil and Dent, 2014). However, the numerous players that coordinate MT remodeling during axon navigation remain largely unknown. In this study, we identify Fignl1 as a key regulator of MT dynamics in motor GC morphology and navigational behavior at intermediate targets as shown for the ubiquitin ligase PHR1 (Lewcock et al., 2007). As they reach intermediate targets, GCs pause and undergo changes in morphology, signaling properties, and cytoskeleton organization (Rothenberg et al., 2003; Lee et al., 2004; Hendricks and Jesuthasan, 2009) that are precisely controlled by local axon guidance cues and required for navigation decisions. The large number of morphant motor axons showing the characteristic features of paused GCs (i.e., enlarged GCs filled with looped MTs) or defective steering behavior at guidance choice points suggest that MT remodeling by Fignl1 is crucial for exiting the paused state and driving directional turning. Based on impaired MT dynamicity, abnormal organization, and invasion of the peripheral domain in morphant GCs as well as their increased persistence in filopodia, Fignl1 isoforms may act in concert to drive GC turning by asymmetrically impeding MT invasion and stabilization in the GC peripheral domain. If Fignl1 short isoforms promote cortical MT depolymerization, FL-Fignl1 could play several concomitant roles by dissociating the scaffolding +TIP EBs and their associated proteins from MT plus ends. Indeed, FL-Fignl1 might (A) reduce MT invasion in filopodia, which critically relies on EB interactions with the F-actin–bound drebrin (Geraldo et al., 2008), (B) alter APC-mediated MT growth directionality (Purro et al., 2008) and stabilization (Zhou et al., 2004), (C) reduce CLASP-mediated MT–F-actin cross-linking (Hur et al., 2011; Marx et al., 2013), or (D) favor MT coupling with actin retrograde flow by reducing LIS-1 recruitment at MT plus ends (Grabham et al., 2007). Notably, uncoupling F-actin–MT interactions leads MTs to progress further into the periphery and increases MT lateral movements (Burnette et al., 2007) as shown for morphant GCs. Furthermore, Fignl1 multifunctionality in MT dynamics could also include a role in MT sliding, which is essential for MT-polarized invasion into the GC peripheral domain (Kahn and Baas, 2016; Lu and Gelfand, 2017) and may be associated with its depolymerizing activity as demonstrated for some molecular motors during mitotic spindle elongation (Brust-Mascher et al., 2004; Cheerambathur et al., 2007; Wang et al., 2010).
Because axon terminal branching is linked to GC pausing (Kalil et al., 2000), the abnormal pausing behavior of morphant GCs could underlie the distal hyperbranching of morphant axons. However, this phenotype may also arise from defective pruning of supernumerary branches, which relies on MT depolymerization (Schuldiner and Yaron, 2015; Brill et al., 2016). Furthermore, the increased neurite number of Fignl1-overexpressing SMNs also suggests a role for Fignl1 in neuronal polarization. Indeed, several +TIPs and associated proteins play key roles in neuronal polarity (Neukirchen and Bradke, 2011; Beffert et al., 2012; Fréal et al., 2016). Characterizing the fine-tuned control of vertebrate motor axon specification, outgrowth and navigation by +TIP regulatory partners would provide important insight into the instructive role of MTs in neuronal connectivity and ultimately into the pathogenic mechanisms underlying human disorders associated with axonal MT defects (Falnikar and Baas, 2009; Sayas and Ávila, 2014).
Materials and methods
Zebrafish maintenance
Zebrafish embryos were obtained from natural spawning of WT or transgenic Tg(Isl1:GFP) (Higashijima et al., 2000), Tg(Hb9:GFP) (Flanagan-Steet et al., 2005), Tg(Olig2:GFP) (Shin et al., 2003), Tg(Mnx1:Gal4) (Zelenchuk and Brusés, 2011), Tg(UAS:EB3-GFP) (Revenu et al., 2014), and Tg(Ath5:GFP) (Masai et al., 2003) lines. All embryos were maintained in E3 medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4, and 0.00001% [wt/vol] Methylene blue) at 28°C and staged by hpf and gross morphology according to Kimmel et al. (1995). To prevent pigment formation, 0.2 mM 1-phenyl-2-thiourea (Sigma-Aldrich) was added to the E3 media 24 hpf.
Cloning, site-targeted mutagenesis, and plasmid constructs
Zebrafish full-coding Fignl1 cDNA was isolated from a home-made prim-5 total embryo cDNA library by using the Advantage 2 amplification kit (Invitrogen) with the following primers (including or not including an HA tag): forward, 5′-ATATAGAATTCATGTACCCATACGATGTTCCAGATTACGCTAGCAGAGCACACCTGGACGAGTG-3′ and 5′-ATATAGAATTCATGAGCAGAGCACACCTGGACGAGTG-3′; and reverse, 5′-ATATATCTAGATTAACGGCCACAACCAAAAGTCTTGTTCCAC-3′. Human full-coding FIGNL1 cDNA was similarly isolated from a human skeletal muscle Marathon-ready cDNA collection (Invitrogen) with the following primers (with or without an HA tag): forward, 5′-ATATAGAATTCATGTACCCATACGATGTTCCAGATTACGCTCAGACCTCCAGCTCTAGATCTG-3′ and 5′-ATATAGAATTCATGCAGACCTCCAGCTCTAGATCTG-3′; and reverse, 5′-ATATACTCGAGTTACTTTCCACAACCAAAAGTTTTGTTCCAG-3′. PCR products were then cloned into the TOPO TA cloning pcr4 vector (Invitrogen), sequenced, and subcloned into pCS2+ by using EcoRI–XbaI and EcoRI–XhoI restriction sites for zebrafish Fignl1 (HA-Fignl1) and human FIGNL1 (HA-FIGNL1), respectively. E488A point mutation was introduced in HA-Fignl1 construct by using the QuikChange site-directed mutagenesis kit (Agilent Technologies) and a set of complementary primers (forward, 5′-CCTGCAGTTATCTTCATTGATGCGATTGATTCTCTTCTTTCACAACG-3′; and reverse, 5′-CGTTGTGAAAGAAGAGAATCAATCGCATCAATGAAGATAACTGCAGG-3′).
FL-Fignl1–HA (available from GenBank under accession no. NM_001128751) and Fignl1Δ1–113-HA cDNAs were synthesized and cloned in pCS2+ vector (EcoRI–XbaI restriction sites) by GeneCust. The targeted mutagenesis of methionines 114 and 174 in FL-Fignl1–HA construct was performed by GeneCust (FL-Fignl1–HAMutATG2, ATG3).
FL-Fignl1 and Fignl1Δ1–113 cDNAs were PCR-amplified from FL-Fignl1–HA construct and cloned into pEGFP-N1 and pET28a+ vectors by using the In-Fusion HD cloning kit (Thermo Fisher Scientific) and the following primers: forward zFignl1-EGFP (EcoRI), 5′-CTCAAGCTTCGAATTCGCCACCATGAGCAGAGCACACCTGG-3′; reverse zFignl1-EGFP (BamHI), 5′-GGCGACCGGTGGATCCCGGCCACAACCAAAAGTCTTG-3′; forward zFignl1-D113-EGFP (EcoRI), 5′-CTCAAGCTTCGAATTCGCCACCATGATACAGGCCAGGTCAGATGG-3′; reverse zFignl1-D113-EGFP (EcoNI), 5′-GTTCTTCCTCTTGGAGGGGTTTG-3′; forward His-Fignl1-His (NcoI), 5′-AGGAGATATACCATGGGCAGCAGCCATCATCATCATCATCACATGAGCAGAGCACACCTGGAC-3′; forward His–Fignl1-D1–113–His (NcoI), 5′-AGGAGATATACCATGGGCAGCAGCCATCATCATCATCATCACATGATACAGGCCAGGTCAGATG-3′; and reverse His–Fignl1/Fignl1-D1–113–His (XhoI), 5′-GGTGGTGGTGCTCGAGTGGACGGCCACAACCAAAAGTCTTG-3′.
Zebrafish spastin cDNA was amplified and HA-tagged by PCR (forward, 5′-ATACTCGAGCAAGCTTGATTTAGGTGA-3′; and reverse, 5′-GGCTCTAGATCAAGCGTAGTCAGGCACGTCGTAAGGGTAACTAGCGCCTACGCCAGTCGTGTCTCCGT-3′) from a clone provided by F. Giudicelli (Institut de Biologie Paris-Seine, Paris, France) and subcloned into pCS2+ by using the XhoI–XbaI restriction sites. Zebrafish Fign and mouse fignl1 full-length cDNAs were reverse transcribed, PCR-amplified, and HA-tagged by using the SuperScript III One-Step RT-PCR system with Platinum Taq High Fidelity (Thermo Fisher Scientific) from total RNA extract of 24-hpf zebrafish embryos or E16.5 mouse embryonic brain by following the manufacturer’s instructions. The following primers were used: forward EcoRI-zFign, 5′-ATCCGGAATTCATGCAGTGGACCCCAGAGCATG-3′; reverse XbaI-zFign-HA, 5′-CTAGTCTAGATTAAGCGTAATCTGGAACATCGTATGGGTATTACTGGCTACAGCCAAACATCTTATTCC-3′; forward EcoRI-mouseFignl1, 5′-ATCCGGAATTCATGGAGACGTCCAGCTCCATGTCTGTGG-3′; And reverse XhoI-mFignl1_3′UTR-HA, 5′-TCCGCTCGAGAGTAAAATCATATACATATATGTTGTATACAGGACTG-3′.
Control-shRNA (5′-GTTCGCAACCCGTCTAATA-3′) and Fignl1-shRNA (5′-TTTGACGGAGTTGCTTCAT-3′) were synthesized by Eurogentec and cloned in pSUPER.neo + GFP (OligoEngine) by using the HindIII and BglII restriction sites.
All clones were sequenced before use, and the plasmids were purified by using a QIAfilter EndoFree Plasmid Maxi kit (QIAGEN). EB3-GFP and α-tubulin–mCherry constructs were provided by A. Andrieux (Grenoble Institut Neurosciences, Grenoble, France; Peris et al., 2009).
Antibodies
A rabbit polyclonal Fignl1 antibody (3353) was generated against a mix of two synthetic peptides located in the N-terminal region of zebrafish Fignl1 (ARNQKNHSDKWESSL and QSNVACTSRSNVHNQP) by using the antipeptide 28-d Speedy protocol from Eurogentec. Final bleed was affinity purified.
The following primary antibodies were used at the respective dilutions: mouse Znp-1, F59, and F310 (1:100; ZIRC; University of Oregon), mouse Zn-5 and Zn-12 (1:250; ZIRC; University of Oregon), mouse 3A10 (1:100; AB_531874; Developmental Studies Hybridoma Bank), rabbit GABA (1:1,000; A2052; Sigma-Aldrich), rabbit α-Pax2 (1:100; PRB-276P; Covance), mouse Islet1/2 (1:200; 39.4D5; Developmental Studies Hybridoma Bank), mouse HuC/D (1:200, A21271; Invitrogen), mouse acetylated α-tubulin (1:1,000; T7451; Sigma-Aldrich), mouse α-tubulin (1:20,000; T6074; Sigma-Aldrich), rabbit GFP (1:1,000; A11122; Invitrogen), mouse GFP (1:1,000; Invitrogen), phospho–histone H3 (1:200; Upstate), rabbit caspase 3 active (1:400, AF835; R&D Systems), Fignl1 (1:200), mouse tyrosinated tubulin (1:4,000; T9028; Sigma-Aldrich), Alexa Fluor 488–conjugated phalloidin (1:200; A12379; Invitrogen), rat HA (1:100; 11867423001; Roche), mouse EB1 (1:300; 610535; Transduction Laboratories), rat EB3 (1:200; ab53360; Abcam), p150Glued (1:100; 610473; Transduction Laboratories), H2b (1:16,000; ab1790; Abcam), and GAPDH (1:20,000; sc-32233; Santa Cruz Biotechnology, Inc.).
Wholemount in situ hybridization
A digoxigenin-labeled Fignl1 antisense RNA probe was synthesized from the linearized Fignl1 cDNA construct in TOPO TA cloning pcr4 vector by using T3 RNA polymerase (Promega) according to the supplier’s instructions. Wholemount in situ hybridization experiments were performed at different developmental stages from the one-cell stage to 52 hpf by using standard procedures (Macdonald et al., 1994). Pictures were acquired with a binocular stereomicroscope (M165C; Leica Microsystems) combined with a high-definition camera (IC80 HD; Leica Microsystems) or an Axiovert 200M microscope (Axiovision; ZEISS). Pictures were adjusted for brightness and contrast with ImageJ software (National Institutes of Health).
Morpholino and RNA injections
MO blocking Fignl1 translation initiation site (i.e., MOFignl1, 5′-TCGTCCAGGTGTGCTCTGCTCATGC-3′; and MOFignl1BIS, 5′-CTGGCCTGCTCCAGCCTGCATAAGA-3′) as well as Fignl1-mismatch (i.e., MOCtl, 5′-TCATCCAAGTGTACTCTGCTAATAC-3′) or control scramble morpholinos (i.e., MOScramble, 5′-CCTCTTACCTCAGTTACAATTTATA-3′) were designed and produced by Gene Tools and injected at 0.4 or 0.5 pmol/embryo at the two-cell stage, whereas RNA injections were performed at the one-cell stage. Human FIGNL1 and zebrafish FL-Fignl1, Fignl1E488A, and Fignl1Δ1–113 mRNAs were in vitro transcribed from linearized pCS2+ constructs by using the SP6 mMessage mMachine kit (Ambion) and injected at 100–200 pg for rescue and overexpression experiments. Rescue analyses were performed by coinjecting human FIGNL1 transcript with any of the two MOs.
Touch-response test and manual tracking
To assess the motor behavior of control (including control, MOCtl-, and MOScramble-injected) and morphant (MOFignl1- or MOFignl1BIS-injected) larvae, we performed a touch-escape response test by applying a tactile stimulus with a pair of forceps and analyzing their escape behavior under a binocular stereomicroscope (M165FC; Leica Microsystems) equipped with a Power Shot A520 camera (Canon). The swimming speed and distance were quantified by using the manual tracking plugin of ImageJ software on control (n = 159) and MOFignl1-injected (n = 237) larvae from five independent experiments.
Wholemount immunohistochemistry
Embryos were fixed in 4% paraformaldehyde for 2 h at room temperature, washed four times with PBST1% (1% Triton X-100 in PBS), permeabilized in a 0.25% trypsin solution (at 25°C) after 24 hpf, blocked for 2 h in PBST1% supplemented with 10% of normal goat serum, and subsequently incubated overnight at 4°C with primary antibodies diluted in blocking solution. After several washes in PBST1%, embryos were incubated overnight at 4°C with the appropriate secondary antibody (Invitrogen) and/or Alexa Fluor 555 α-bungarotoxin (1:1,000; B35451; Invitrogen). Another experimental procedure was used for Fignl1, Zn-12, GABA, α-Pax2, Islet1/2, F310, F59, and HuC/D antibodies. In this protocol, embryos were fixed in 4% paraformaldehyde for 4 h at 4°C, blocked in PBS supplemented with 5% normal goat serum, BSA (4 mg/ml), and 0.5% triton X-100, and incubated overnight at 4°C with the appropriate antibodies diluted in the blocking buffer. Images were acquired by using a fluorescence microscope equipped with an Apotome module (Axiovert 200M), the Axiocam MRm camera, and Axiovision software (ZEISS). Images were processed with ImageJ software. Each figure corresponds with a projection image from a z stack of 2-µm sections. Each experiment was reproduced three times independently, and >15 control and morphant embryos were analyzed in each of them.
Transplant experiments
Mosaic embryos were generated by homotopic transplantation as previously described (Fassier et al., 2010). In brief, ∼40–50 dorsoposterior epiblast cells were collected by capillary suction around the posterior midline from a 70–80% epiboly MOFignl1-injected Tg(Hb9:GFP) donor embryo and placed at the equivalent location in the same stage WT host embryo. Around half of these transplants led to incorporation of most of the donor cells in the ventral spinal cord. Mosaic embryos were selected at 30 hpf for GFP-positive motor axons in at least 14 somites on each side, fixed, and immunolabeled with Znp-1 and GFP antibodies.
In vivo time-lapse videomicroscopy
Control and MOFignl1-injected Tg(Olig2:GFP) or Tg(Mnx1:Gal4; UAS:EB3-GFP) larvae were anesthetized at 40 hpf with tricaine and embedded in 0.8% low–melting point agarose in a 35-mm glass-based dish (Iwaki). Time-lapse video recording of sMN axon outgrowth was performed at 28°C in E3 medium supplemented with tricaine using a DMI 6000B inverted spinning-disk microscope with a 40× 1.25 NA immersion objective (Leica Microsystems). Z stacks were taken every 8 min over a 30-h period with an electron-multiplying charge-coupled device (EMCCD) camera (QuantEM 512SC; Photometrics) and MetaMorph software (Molecular Devices) and were compiled into time-lapse videos. Each video panel represents a single-plane projection of a z stack from sixty 1-µm-thick planes taken at the indicated time interval. Time-lapse recordings of EB3-GFP comets were performed in the same imaging medium with the same microscope on 12 control and morphant Tg(Mnx1:Gal4; UAS:EB3-GFP) by using a 63× 1.4 NA immersion objective. A z stack of twenty three 0.8-µm-thick planes was acquired every 4 s over a 5-min period. Comet velocity and growth duration were estimated on single-plane z projection videos by using kymograph analysis (FIJI; ImageJ) with lines drawn along the axon (Fassier et al., 2013).
Primary cultures of zebrafish spinal neurons, immunocytochemistry, time-lapse videomicroscopy, and data analysis
Primary cultures of zebrafish spinal neurons were prepared from 24-hpf embryos as detailed by Fassier et al. (2010). In brief, trunks of dechorionated embryos were isolated by removing the yolk and the head, incubated for 20 min in a 0.25% trypsin solution, and mechanically dissociated with a Pasteur pipette in l-15 medium (Leibovitz; Invitrogen) supplemented with 1% FCS. Cells were plated onto poly d-ornithine/laminin-coated dishes at a density of seven spinal cords per 35-mm dish and maintained at room temperature for 9 h postplating (hpp). Zebrafish spinal neurons were fixed in 4% PFA/4% sucrose for 13 min at room temperature, permeabilized for 5 min in PBS/0.1% Triton X-100, blocked in PBS/3% BSA/5% normal goat serum, and incubated with the appropriate antibodies. For detergent extraction assay, cultured spinal neurons were washed with PHEM buffer (60 mM Pipes, 25 mM Hepes, 10 mM EGTA, and 2 mM MgCl2, pH 6.9) and permeabilized with PHEM buffer supplemented with 0.05% Triton X-100 and 10 µM taxol for 3 min. Cultures were fixed in 2% PFA and 0.05% glutaraldehyde in PHEM, blocked in PBS/5% normal goat serum, and incubated overnight with primary antibodies. Control and morphant motor neuron GC area and neuritic length were measured by using ImageJ software in three independent experiments. More than 60 SMNs were analyzed in each experiment. Morphological analysis of control and Fignl1-overexpressing motor neurons was performed on >1,300 neurons in four independent experiments. For rescue experiments, 20 nM nocodazole (in DMSO; Sigma-Aldrich) or DMSO alone was added 3 hpp to control and MOFignl1-cultured spinal neurons. The percentage of enlarged GCs was estimated at 9 hpp on at least 150 neurons per condition in four independent experiments.
Time-lapse recordings of EB3-GFP comets were performed on 15 control and morphant Tg(Mnx1:Gal4; UAS:EB3-GFP)-cultured SMNs by using a DMI 6000B inverted spinning-disk microscope with a 63× 1.4 NA immersion objective. Z stacks were acquired every 2 s over 5 min with a step size of 0.3 µm with an EMCCD camera (QuantEM 512SC) and MetaMorph software and were combined into time-lapse videos. EB3-GFP comet velocity and growth duration were automatically analyzed on maximum z projection videos focused on GCs by using the TrackMate plugin in FIJI (http://fiji.sc/TrackMate). The growth directionality of each comet was estimated from TrackMate data by using a home-made plugin, which calculated the angle of comet displacement according to the main GC axis as previously described (Purro et al., 2008).
Mammalian cell cultures, transfection, immunocytochemistry, live imaging, and data analysis
COS-7 cell lines were cultured in DMEM supplemented with 10% FCS at 37°C under 5% CO2 and transfected with the different cDNA constructs by using Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s instructions with a DNA/Lipofectamine ratio of 1:1.5. Cells were fixed 24 h posttransfection in ice-cold methanol at −20°C for 10 min and postfixed in 4% PFA/4% sucrose solution for 20 min at room temperature or directly fixed in 4% PFA/4% sucrose for 10 min. Cells were next blocked in PBS/5% normal goat serum and incubated overnight with primary antibodies.
For nocodazole susceptibility assays or F-actin depolymerization experiments, cells were treated with 20 µM nocodazole (Sigma-Aldrich) for 15 min or 1 µM latrunculin B (Sigma-Aldrich) for 45 min before fixation. The carrier alone (DMSO) was used as a control.
MT density was calculated either in the whole cell or beneath the cell cortex by estimating the mean tyrosinated-tubulin fluorescence intensity with respect to the cell area or in a 25-µm-wide × 5-µm-high region area under the plasma membrane. The mean number of EB1-, EB3-, or p150Glued-positive comets was estimated with respect to the cell area. The mean fluorescence intensity profile of F-actin and FL-Fignl1–HA or Fignl1Δ1–113-HA was estimated along four different 10-µm-long lanes (spaced by at least 8 µm) per cell. Each lane was drawn from and oriented perpendicular to the cell membrane. At least 20 cells from three independent experiments were analyzed per condition.
MEFs were prepared from E14.5 embryos by following standard procedures and cultured in DMEM supplemented with 10% FBS. MEFs were electroporated with α-tubulin–mCherry and FL-Fignl1–GFP, Fignl1Δ1–113-GFP, or GFP constructs by using 4D-Nucleofector and the P4 Primary Cell 4D-Nucleofector X kit (Lonza). Live imaging of fluorescent MTs was performed by using a TIRF microscope (Eclipse Ti2; Nikon) with a 100× 1.49 NA oil-immersion objective. Images were captured every 2 s over a 5-min period with an iXon Ultra 897 camera (Andor) and NIS software (Nikon). Drift in z was minimized with the Nikon Perfect Focus system z drift correction. Kymograph analyses of MT dynamics were performed by using the kymograph tool plugin (FIJI). Only MTs with growing plus ends located within 12 µm beneath the cell cortex at the beginning of the video were included in these analyses. At least 20 cells and 150 individual MTs were analyzed in three independent experiments. MT dynamic instability parameters were calculated from kymograms by using the Slope and Speed macro designed by E. Denarier (Grenoble Institut Neurosciences, Grenoble, France). Catastrophe frequency was calculated by dividing the number of catastrophe events by the time that MT spent growing or pausing. Reciprocally, rescue frequency was obtained by dividing the number of rescue events by the time MTs spent shrinking or pausing.
Primary hippocampal neurons were isolated from E18 rat brains by following standard procedures. Neurons were seeded on coverslips coated with poly-l-lysine (37.5 µg/ml) and laminin (5 µg/ml) in neurobasal medium supplemented with 2% B27, 0.5 mM glutamine, 15.6 µM glutamate, and 1% penicillin/streptomycin. Hippocampal neurons were transfected at day in vitro 1 (DIV1) by using Lipofectamine 2000 (DNA/Lipofectamine ratio of 1.8:3.3; Thermo Fisher Scientific) and fixed at DIV3 for 10 min in 4% PFA/4% sucrose. Cells were washed in PBS, sequentially incubated with primary and secondary antibodies diluted in GDB buffer (0.1% BSA, 0.45 M NaCl, 0.3% Triton X-100, and 16.7 mM phosphate buffer, pH 7.4) and mounted in Vectashield medium (Vector Labs). The percentage of neurons with enlarged GCs was estimated on at least 150 transfected neurons per condition from three independent experiments. GC area and axonal length were measured on at least 30 neurons from three independent experiments by using ImageJ software.
MT cosedimentation experiments
COS-7 cells transfected with pCS2+-HA or pCS2+-HA-Fignl1 were collected in 120 µl MEM buffer (0.1 M 2-morpholinoethane sulfonic acid, 1 mM EGTA, and 0.5 mM MgCl2, pH 6.9) supplemented with a cocktail of protease inhibitors (Roche) and sonicated (total lysate). Homogenates were spun at 150,000 g for 10 min at 4°C in an Airfuge ultracentrifuge (Beckman Coulter). The supernatant was collected and supplemented with 20 µM taxol (Sigma-Aldrich) and 1 mM GTP. MTs were left to polymerize for 30 min at 37°C. The resulting extract was layered onto a 10% (wt/vol) sucrose cushion (one third of the final volume) in MEM and spun at 180,000 g for 15 min at 37°C. After centrifugation, the supernatant was carefully removed, and the pellet was resuspended in MEM buffer supplemented with protease inhibitors. The different fractions were assessed for Fignl1 and α-tubulin detection by WB by using HA and α-tubulin antibodies, respectively.
MT severing assays
Perfusion chambers (5 µl) were prepared as previously described (Portran et al., 2013) with the exception that they were built between two coverslips, one being functionalized with silane–polyethylene glycol (PEG; 30 kD; Creative PEGWorks) and the other with silane-PEG-biotin (3.5 kD; Laysan Bio). Chambers were subsequently coated with 5 µl NeutrAvidin (25 mg/ml in 1% BSA in BRB80; i.e., 80 mM Pipes, 1 mM EDTA, and 1 mM MgCl2, pH 6.8), passivated for 30 s with 50 µl poly-L-lysine graft–PEG (2 kD; 0.1 mg/ml in 10 mM Hepes, pH 7.4; Jenkem) and washed with 300 µl 1% pluronic F-127 (Sigma-Aldrich) and 1% BSA in BRB80. Atto 565–labeled and biotinylated taxol-stabilized MTs were prepared by incubating a mix of 90 µM bovine brain tubulin containing 15% Atto 565–labeled tubulin and 20% biotinylated tubulin with 1 mM GTP in BRB80 buffer for 35 min at 35°C. 100 µM taxol was then added, and the incubation was resumed after 30 min. MTs were diluted in BRB80 supplemented with 1% BSA and 10 µM taxol and were perfused in the chambers. After a 5-min incubation, chambers were washed with 300 µl BRB80/1% BSA/10 µM taxol and perfused with 20 µl imaging buffer (50 mM KCl, 1% BSA, 5 mM ATP, 3 mM MgCl2, 10 µM taxol, 10 mM DTT, 150 µg/ml catalase, 2 mg/ml glucose, and 1 mg/ml glucose oxidase in BRB80). Chambers were perfused with imaging buffer containing 100 or 400 nM His-Fignl1Δ1–113 or GFP-spastin (provided by M. Vantard and J. Gaillard, Grenoble Institut Neurosciences, Grenoble, France). Samples were visualized on an inverted microscope (Eclipse Ti; Nikon) equipped with an iLas2 TIRF system (Roper Scientific) by using an Apochromat 60× 1.49 NA oil-immersion objective. Images were captured every 5 s over 30 min with an EMCCD camera (Evolve 512; Photometrics) controlled by MetaMorph software (version 7.7.5). These assays were reproduced three times independently.
WB analysis
Zebrafish protein extracts were prepared from 28-hpf control and morphant embryos in SDS sample buffer complemented with protease inhibitors (0.5 µl per embryo; 1 M Tris-HCl pH 6.8, 10% glycerol, 5% β-mercaptoethanol, 3.5% SDS, 5 mM aprotinin, 5 mM leupeptin, and 1 mM PMSF). COS-7 cell lysates were prepared by using an SDS lysis buffer (25 mM sodium sulfate, pH 7.2, 5 mM EDTA, and 1% SDS) supplemented with 1 mM PMSF and a cocktail of protease inhibitors (Roche). Zebrafish (20 µg) and COS-7 (5 µg) protein extracts were electrophoresed into 10% SDS-PAGE gel and transferred to nitrocellulose membranes. Immunoblotting was performed after overnight incubation at 4°C with Fignl1 antibody, its corresponding preimmune serum (1:1,000), HA, or GFP antibodies. H2b or GAPDH was used as the loading control. Fignl1 levels were estimated by quantifying band density normalized to H2b values (ImageJ software) in three independent experiments.
Co-IP and GST pulldown assays
For co-IP assays, transfected cells were treated or not with 20 µM nocodazole (Sigma-Aldrich) or DMSO for 12 min and lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 20 mM EGTA, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS) supplemented with a cocktail of protease inhibitors (Roche). Co-IPs were performed with 600 µg precleared protein extracts, 2 µg GFP antibody (ab6556-25; Abcam), and 20 µl protein G Sepharose (GE Healthcare). GST-FIGNL1 and His-Fignl1Δ1–113-His were induced in BL21 Escherichia coli and purified on glutathione Sepharose 4 Fast Flow beads (GE Healthcare) or Protino Ni-TED columns (MACHEREY-NAGEL) according to the manufacturer’s instructions. For GST pulldown assays, COS-7 cells overexpressing EB3-GFP or GFP alone were lysed in RIPA buffer. Protein lysates or purified His-EB1-GFP were incubated with glutathione Sepharose 4 Fast Flow beads previously bound to purified GST-FIGNL1 or GST alone for 4 h at 4°C. Beads were washed several times in RIPA buffer and resuspended in 2× Laemmli buffer. Proteins retained on the beads were analyzed by WB by using GFP antibody.
Statistical analysis
All data were obtained from at least three independent experiments and are presented as means ± SEM. Statistical significance of the data were evaluated by using either the parametric unpaired two-tailed t test, nonparametric Mann–Whitney test, or Kruskal–Wallis ANOVA test with Dunn’s post hoc test when analyzing more than two groups assuming non-Gaussian distribution. Data distribution was tested for normality by using the D’Agostino and Pearson omnibus normality test. All statistical analyses were performed using Prism (GraphPad Software).
Online supplemental material
Fig. S1 shows the specificity of the SMN and larval mobility defects of Fignl1 morphants described in Fig. 3 with the use of different control morpholinos and rescue experiments. Fig. S2 demonstrates the specificity of the Fignl1 antibody and provides quantifications of Fignl1 knockdown efficiency shown in Fig. 3 as well as a schematic representation of the Fignl1 constructs used in Figs. 3, 8, and 9. Fig. S3 shows that Fignl1 knockdown does not perturb SMN specification, localization, or survival. Fig. S4 establishes that Fignl1 knockdown in cultured mammalian neurons affects GC morphology and MT organization as in zebrafish motor neurons. Fig. S5 shows that overexpression of GFP-tagged human FIGNL1 in COS-7 cells alters +TIP localization at MT plus ends and that both WT and ATPase-deficient Fignl1 specifically binds to EB1 or EB3 in co-IP assays compared with its closest homologues spastin and Fign. Videos 1, 2, and 3 show the locomotor behavior of control, morphant, and rescued larvae, respectively, after touch stimuli. Videos 4 and 5 reveal the navigational behavior of control and Fignl1-depleted SMN axons at a guidance choice point. Videos 6 and 7 show EB3-GFP comet movements in vivo, which reflect MT plus end dynamics in navigating SMN axons of control and Fignl1 morphant larvae. Videos 8 and 9 reveal EB3-GFP comet movements in the developing GCs of control and morphant cultured SMNs.
Supplementary Material
Acknowledgments
We thank Richard Schwartzmann, Jean-François Gilles, and France Lam from the Institute of Biology Paris-Seine (Université Pierre et Marie Curie, Paris, France) imaging facility for their assistance in microscopy (TIRF and spinning disk) and image analysis, and we thank Alex Bois and Stéphane Tronche from the Institute of Biology Paris-Seine aquatic facility for fish care. We also thank Nicolas Jardin for his help in experimentation, Eric Denarier for providing us with the kymogram analysis plugin, and F. Nothias’s, S. Schneider-Maunoury’s, and A. Andrieux’s laboratories for fruitful discussions.
This work was supported by research grants from the Association Française contre les Myopathies, the Université Pierre et Marie Curie Emergence program, and the Association Strümpell-Lorrain. C. Fassier was a recipient of an Association Française contre les Myopathies postdoctoral fellowship before becoming an Institut National de la Santé et de la Recherche Medicale research fellow.
The authors declare no competing financial interests.
Author contributions: C. Fassier and J. Hazan conceived and designed the study. C. Fassier, A. Fréal, L. Gasmi, C. Delphin, D. Ten Martin, and M. Tambalo performed all experiments with the contributions from S. De Gois and C. Bosc for cloning and mutagenesis. S. Bolte carried out image processing. P. Mailly designed the plugin for comet tracking. C. Revenu provided the Tg(UAS:EB3-GFP) line for in vivo comet dynamic analysis. C. Houart carried out transplant experiments. J.-C. Larcher and L. Peris provided their expertise in MT cosedimentation and live imaging. S. Schneider-Maunoury, J.C. Larcher, F. Nothias, and C. Houart had insightful advice throughout the study. A. Andrieux provided intellectual input and expertise in neuronal cytoskeleton. C. Fassier and J. Hazan wrote the manuscript.
References
- Akhmanova A., and Steinmetz M.O.. 2008. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 9:309–322. 10.1038/nrm2369 [DOI] [PubMed] [Google Scholar]
- Bearce E.A., Erdogan B., and Lowery L.A.. 2015. TIPsy tour guides: How microtubule plus-end tracking proteins (+TIPs) facilitate axon guidance. Front. Cell. Neurosci. 9:241 10.3389/fncel.2015.00241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beffert U., Dillon G.M., Sullivan J.M., Stuart C.E., Gilbert J.P., Kambouris J.A., and Ho A.. 2012. Microtubule plus-end tracking protein CLASP2 regulates neuronal polarity and synaptic function. J. Neurosci. 32:13906–13916. 10.1523/JNEUROSCI.2108-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouquet C., Soares S., von Boxberg Y., Ravaille-Veron M., Propst F., and Nothias F.. 2004. Microtubule-associated protein 1B controls directionality of growth cone migration and axonal branching in regeneration of adult dorsal root ganglia neurons. J. Neurosci. 24:7204–7213. 10.1523/JNEUROSCI.2254-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill M.S., Kleele T., Ruschkies L., Wang M., Marahori N.A., Reuter M.S., Hausrat T.J., Weigand E., Fisher M., Ahles A., et al. 2016. Branch-specific microtubule destabilization mediates axon branch loss during neuromuscular synapse elimination. Neuron. 92:845–856. 10.1016/j.neuron.2016.09.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brust-Mascher I., Civelekoglu-Scholey G., Kwon M., Mogilner A., and Scholey J.M.. 2004. Model for anaphase B: Role of three mitotic motors in a switch from poleward flux to spindle elongation. Proc. Natl. Acad. Sci. USA. 101:15938–15943. 10.1073/pnas.0407044101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck K.B., and Zheng J.Q.. 2002. Growth cone turning induced by direct local modification of microtubule dynamics. J. Neurosci. 22:9358–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette D.T., Schaefer A.W., Ji L., Danuser G., and Forscher P.. 2007. Filopodial actin bundles are not necessary for microtubule advance into the peripheral domain of Aplysia neuronal growth cones. Nat. Cell Biol. 9:1360–1369. 10.1038/ncb1655 [DOI] [PubMed] [Google Scholar]
- Butler R., Wood J.D., Landers J.A., and Cunliffe V.T.. 2010. Genetic and chemical modulation of spastin-dependent axon outgrowth in zebrafish embryos indicates a role for impaired microtubule dynamics in hereditary spastic paraplegia. Dis. Model. Mech. 3:743–751. 10.1242/dmm.004002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammarata G.M., Bearce E.A., and Lowery L.A.. 2016. Cytoskeletal social networking in the growth cone: How +TIPs mediate microtubule-actin cross-linking to drive axon outgrowth and guidance. Cytoskeleton (Hoboken). 73:461–476. 10.1002/cm.21272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheerambathur D.K., Civelekoglu-Scholey G., Brust-Mascher I., Sommi P., Mogilner A., and Scholey J.M.. 2007. Quantitative analysis of an anaphase B switch: Predicted role for a microtubule catastrophe gradient. J. Cell Biol. 177:995–1004. 10.1083/jcb.200611113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deloulme J.C., Gory-Fauré S., Mauconduit F., Chauvet S., Jonckheere J., Boulan B., Mire E., Xue J., Jany M., Maucler C., et al. 2015. Microtubule-associated protein 6 mediates neuronal connectivity through Semaphorin 3E-dependent signalling for axonal growth. Nat. Commun. 6:7246 10.1038/ncomms8246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Río J.A., González-Billault C., Ureña J.M., Jiménez E.M., Barallobre M.J., Pascual M., Pujadas L., Simó S., La Torre A., Wandosell F., et al. 2004. MAP1B is required for Netrin 1 signaling in neuronal migration and axonal guidance. Curr. Biol. 14:840–850. 10.1016/j.cub.2004.04.046 [DOI] [PubMed] [Google Scholar]
- Dent E.W., Gupton S.L., and Gertler F.B.. 2011. The growth cone cytoskeleton in axon outgrowth and guidance. Cold Spring Harb. Perspect. Biol. 3:a001800 10.1101/cshperspect.a001800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuel T.A.S., Liu J.S., Corbo J.C., Yoo S.-Y., Rorke-Adams L.B., and Walsh C.A.. 2006. Genetic interactions between doublecortin and doublecortin-like kinase in neuronal migration and axon outgrowth. Neuron. 49:41–53. 10.1016/j.neuron.2005.10.038 [DOI] [PubMed] [Google Scholar]
- Erdogan B., Cammarata G.-M., Lee E.-J., Pratt B.-C., Francl A.-F., Rutherford E.-L., and Lowery L.-A.. 2017. The microtubule plus-end-tracking protein TACC3 promotes persistent axon outgrowth and mediates responses to axon guidance signals during development. Neural Dev. 12:3 10.1186/s13064-017-0080-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falnikar A., and Baas P.W.. 2009. Critical roles for microtubules in axonal development and disease. Results Probl. Cell Differ. 48:47–64. 10.1007/400_2009_2 [DOI] [PubMed] [Google Scholar]
- Fassier C., Hutt J.A., Scholpp S., Lumsden A., Giros B., Nothias F., Schneider-Maunoury S., Houart C., and Hazan J.. 2010. Zebrafish atlastin controls motility and spinal motor axon architecture via inhibition of the BMP pathway. Nat. Neurosci. 13:1380–1387. 10.1038/nn.2662 [DOI] [PubMed] [Google Scholar]
- Fassier C., Tarrade A., Peris L., Courageot S., Mailly P., Dalard C., Delga S., Roblot N., Lefèvre J., Job D., et al. 2013. Microtubule-targeting drugs rescue axonal swellings in cortical neurons from spastin knockout mice. Dis. Model. Mech. 6:72–83. 10.1242/dmm.008946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan-Steet H., Fox M.A., Meyer D., and Sanes J.R.. 2005. Neuromuscular synapses can form in vivo by incorporation of initially aneural postsynaptic specializations. Development. 132:4471–4481. 10.1242/dev.02044 [DOI] [PubMed] [Google Scholar]
- Fréal A., Fassier C., Le Bras B., Bullier E., De Gois S., Hazan J., Hoogenraad C.C., and Couraud F.. 2016. Cooperative interactions between 480 kDa Aankyrin-G and EB proteins assemble the axon initial segment. J. Neurosci. 36:4421–4433. 10.1523/JNEUROSCI.3219-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldo S., Khanzada U.K., Parsons M., Chilton J.K., and Gordon-Weeks P.R.. 2008. Targeting of the F-actin-binding protein drebrin by the microtubule plus-tip protein EB3 is required for neuritogenesis. Nat. Cell Biol. 10:1181–1189. 10.1038/ncb1778 [DOI] [PubMed] [Google Scholar]
- Girard C., Chelysheva L., Choinard S., Froger N., Macaisne N., Lemhemdi A., Mazel J., Crismani W., and Mercier R.. 2015. AAA-ATPase FIDGETIN-LIKE 1 and helicase FANCM antagonize meiotic crossovers by distinct mechanisms. PLoS Genet. 11:e1005369 10.1371/journal.pgen.1005369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulding M. 2009. Circuits controlling vertebrate locomotion: Moving in a new direction. Nat. Rev. Neurosci. 10:507–518. 10.1038/nrn2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabham P.W., Seale G.E., Bennecib M., Goldberg D.J., and Vallee R.B.. 2007. Cytoplasmic dynein and LIS1 are required for microtubule advance during growth cone remodeling and fast axonal outgrowth. J. Neurosci. 27:5823–5834. 10.1523/JNEUROSCI.1135-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson P.I., and Whiteheart S.W.. 2005. AAA+ proteins: Have engine, will work. Nat. Rev. Mol. Cell Biol. 6:519–529. 10.1038/nrm1684 [DOI] [PubMed] [Google Scholar]
- Hendricks M., and Jesuthasan S.. 2009. PHR regulates growth cone pausing at intermediate targets through microtubule disassembly. J. Neurosci. 29:6593–6598. 10.1523/JNEUROSCI.1115-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashijima S., Hotta Y., and Okamoto H.. 2000. Visualization of cranial motor neurons in live transgenic zebrafish expressing green fluorescent protein under the control of the islet-1 promoter/enhancer. J. Neurosci. 20:206–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Feng J., Bo W., Wu R., Dong Z., Liu Y., Qiang L., and Liu M.. 2017. Fidgetin regulates cultured astrocyte migration by severing tyrosinated microtubules at the leading edge. Mol. Biol. Cell. 28:545–553. 10.1091/mbc.E16-09-0628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur E.M., Saijilafu B.D. Lee S.J. Kim W.L. Xu, and Zhou F.Q.. 2011. GSK3 controls axon growth via CLASP-mediated regulation of growth cone microtubules. Genes Dev. 25:1968–1981. 10.1101/gad.17015911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johjima A., Noi K., Nishikori S., Ogi H., Esaki M., and Ogura T.. 2015. Microtubule severing by katanin p60 AAA+ ATPase requires the C-terminal acidic tails of both α- and β-tubulins and basic amino acid residues in the AAA+ ring pore. J. Biol. Chem. 290:11762–11770. 10.1074/jbc.M114.614768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly A.L., Kim H., Srinivasan D., Lakonishok M., Larson A.G., and Gelfand V.I.. 2010. Kinesin-1 heavy chain mediates microtubule sliding to drive changes in cell shape. Proc. Natl. Acad. Sci. USA. 107:12151–12156. 10.1073/pnas.1004736107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn O.I., and Baas P.W.. 2016. Microtubules and growth Ccones: Motors drive the turn. Trends Neurosci. 39:433–440. 10.1016/j.tins.2016.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalil K., and Dent E.W.. 2004. Hot +TIPS: Guidance cues signal directly to microtubules. Neuron. 42:877–879. 10.1016/j.neuron.2004.06.009 [DOI] [PubMed] [Google Scholar]
- Kalil K., and Dent E.W.. 2014. Branch management: Mechanisms of axon branching in the developing vertebrate CNS. Nat. Rev. Neurosci. 15:7–18. 10.1038/nrn3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalil K., Szebenyi G., and Dent E.W.. 2000. Common mechanisms underlying growth cone guidance and axon branching. J. Neurobiol. 44:145–158. [DOI] [PubMed] [Google Scholar]
- Karabay A., Yu W., Solowska J.M., Baird D.H., and Baas P.W.. 2004. Axonal growth is sensitive to the levels of katanin, a protein that severs microtubules. J. Neurosci. 24:5778–5788. 10.1523/JNEUROSCI.1382-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., and Schilling T.F.. 1995. Stages of embryonic development of the zebrafish. Dev. Dyn. 203:253–310. 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- Kolodkin A.L., and Tessier-Lavigne M.. 2011. Mechanisms and molecules of neuronal wiring: a primer. Cold Spring Harb. Perspect. Biol. 3:a001727 10.1101/cshperspect.a001727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- L’Hôte D., Vatin M., Auer J., Castille J., Passet B., Montagutelli X., Serres C., and Vaiman D.. 2011. Fidgetin-like1 is a strong candidate for a dynamic impairment of male meiosis leading to reduced testis weight in mice. PLoS One. 6:e27582 10.1371/journal.pone.0027582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix B., van Dijk J., Gold N.D., Guizetti J., Aldrian-Herrada G., Rogowski K., Gerlich D.W., and Janke C.. 2010. Tubulin polyglutamylation stimulates spastin-mediated microtubule severing. J. Cell Biol. 189:945–954. 10.1083/jcb.201001024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Engel U., Rusch J., Scherrer S., Sheard K., and Van Vactor D.. 2004. The microtubule plus end tracking protein Orbit/MAST/CLASP acts downstream of the tyrosine kinase Abl in mediating axon guidance. Neuron. 42:913–926. 10.1016/j.neuron.2004.05.020 [DOI] [PubMed] [Google Scholar]
- Leo L., Yu W., D’Rozario M., Waddell E.-A., Marenda D.-R., Baird M.-A., Davidson M.-W., Zhou B., Wu B., Baker L., et al. 2015. Vertebrate Fidgetin restrains axonal growth by severing labile domains of Mmicrotubules. Cell Reports. 12:1723–1730. 10.1016/j.celrep.2015.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewcock J.W., Genoud N., Lettieri K., and Pfaff S.L.. 2007. The ubiquitin ligase Phr1 regulates axon outgrowth through modulation of microtubule dynamics. Neuron. 56:604–620. 10.1016/j.neuron.2007.09.009 [DOI] [PubMed] [Google Scholar]
- Lowery L.A., and Van Vactor D.. 2009. The trip of the tip: understanding the growth cone machinery. Nat. Rev. Mol. Cell Biol. 10:332–343. 10.1038/nrm2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W., and Gelfand V.I.. 2017. Moonlighting motors: Kinesin, dynein, and cell polarity. Trends Cell Biol. 27:505–514. 10.1016/j.tcb.2017.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke-Glaser S., Pintard L., Tyers M., and Peter M.. 2007. The AAA-ATPase FIGL-1 controls mitotic progression, and its levels are regulated by the CUL-3MEL-26 E3 ligase in the C. elegans germ line. J. Cell Sci. 120:3179–3187. 10.1242/jcs.015883 [DOI] [PubMed] [Google Scholar]
- Macdonald R., Xu Q., Barth K.A., Mikkola I., Holder N., Fjose A., Krauss S., and Wilson S.W.. 1994. Regulatory gene expression boundaries demarcate sites of neuronal differentiation in the embryonic zebrafish forebrain. Neuron. 13:1039–1053. 10.1016/0896-6273(94)90044-2 [DOI] [PubMed] [Google Scholar]
- Marx A., Godinez W.J., Tsimashchuk V., Bankhead P., Rohr K., and Engel U.. 2013. Xenopus cytoplasmic linker-associated protein 1 (XCLASP1) promotes axon elongation and advance of pioneer microtubules. Mol. Biol. Cell. 24:1544–1558. 10.1091/mbc.E12-08-0573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai I., Lele Z., Yamaguchi M., Komori A., Nakata A., Nishiwaki Y., Wada H., Tanaka H., Nojima Y., Hammerschmidt M., et al. 2003. N-cadherin mediates retinal lamination, maintenance of forebrain compartments and patterning of retinal neurites. Development. 130:2479–2494. 10.1242/dev.00465 [DOI] [PubMed] [Google Scholar]
- Mishima M., Maesaki R., Kasa M., Watanabe T., Fukata M., Kaibuchi K., and Hakoshima T.. 2007. Structural basis for tubulin recognition by cytoplasmic linker protein 170 and its autoinhibition. Proc. Natl. Acad. Sci. USA. 104:10346–10351. 10.1073/pnas.0703876104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe N., and Hill C.P.. 2016. Meiotic clade AAA ATPases: Protein polymer disassembly machines. J. Mol. Biol. 428:1897–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Diaz Valencia J.D., Stewman S., Metz J., Monnier S., Rath U., Asenjo A.B., Charafeddine R.A., Sosa H.J., Ross J.L., et al. 2012. Human Fidgetin is a microtubule severing the enzyme and minus-end depolymerase that regulates mitosis. Cell Cycle. 11:2359–2366. 10.4161/cc.20849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers K.A., Tint I., Nadar C.-V., He Y., Black M.-M., and Baas P.-W.. 2006. Antagonistic forces generated by cytoplasmic dynein and myosin-II during growth cone turning and axonal retraction. Traffic. 7:1333–1351. 10.1111/j.1600-0854.2006.00476.x [DOI] [PubMed] [Google Scholar]
- Nadar V.C., Ketschek A., Myers K.A., Gallo G., and Baas P.W.. 2008. Kinesin-5 is essential for growth-cone turning. Curr. Biol. 18:1972–1977. 10.1016/j.cub.2008.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neukirchen D., and Bradke F.. 2011. Neuronal polarization and the cytoskeleton. Semin. Cell Dev. Biol. 22:825–833. 10.1016/j.semcdb.2011.08.007 [DOI] [PubMed] [Google Scholar]
- Oladipo A., Cowan A., and Rodionov V.. 2007. Microtubule motor Ncd induces sliding of microtubules in vivo. Mol. Biol. Cell. 18:3601–3606. 10.1091/mbc.E06-12-1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.J., Kim S.J., Rhee Y., Byun J.H., Kim S.H., Kim M.H., Lee E.J., and Lim S.-K.. 2007. Fidgetin-like 1 gene inhibited by basic fibroblast growth factor regulates the proliferation and differentiation of osteoblasts. J. Bone Miner. Res. 22:889–896. 10.1359/jbmr.070311 [DOI] [PubMed] [Google Scholar]
- Peng W., Lin Z., Li W., Lu J., Shen Y., and Wang C.. 2013. Structural insights into the unusually strong ATPase activity of the AAA domain of the Caenorhabditis elegans fidgetin-like 1 (FIGL-1) protein. J. Biol. Chem. 288:29305–29312. 10.1074/jbc.M113.502559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peris L., Wagenbach M., Lafanechère L., Brocard J., Moore A.T., Kozielski F., Job D., Wordeman L., and Andrieux A.. 2009. Motor-dependent microtubule disassembly driven by tubulin tyrosination. J. Cell Biol. 185:1159–1166. 10.1083/jcb.200902142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillis R., Statton D., Caruccio P., and Murphey R.K.. 1996. Mutations in the 8 kDa dynein light chain gene disrupt sensory axon projections in the Drosophila imaginal CNS. Development. 122:2955–2963. [DOI] [PubMed] [Google Scholar]
- Pollard T.D., and Borisy G.G.. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 112:453–465. 10.1016/S0092-8674(03)00120-X [DOI] [PubMed] [Google Scholar]
- Portran D., Zoccoler M., Gaillard J., Stoppin-Mellet V., Neumann E., Arnal I., Martiel J.L., and Vantard M.. 2013. MAP65/Ase1 promote microtubule flexibility. Mol. Biol. Cell. 24:1964–1973. 10.1091/mbc.E13-03-0141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purro S.A., Ciani L., Hoyos-Flight M., Stamatakou E., Siomou E., and Salinas P.C.. 2008. Wnt regulates axon behavior through changes in microtubule growth directionality: A new role for adenomatous polyposis coli. J. Neurosci. 28:8644–8654. 10.1523/JNEUROSCI.2320-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenu C., Streichan S., Donà E., Lecaudey V., Hufnagel L., and Gilmour D.. 2014. Quantitative cell polarity imaging defines leader-to-follower transitions during collective migration and the key role of microtubule-dependent adherens junction formation. Development. 141:1282–1291. 10.1242/dev.101675 [DOI] [PubMed] [Google Scholar]
- Roll-Mecak A., and McNally F.J.. 2010. Microtubule-severing enzymes. Curr. Opin. Cell Biol. 22:96–103. 10.1016/j.ceb.2009.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll-Mecak A., and Vale R.D.. 2008. Structural basis of microtubule severing by the hereditary spastic paraplegia protein spastin. Nature. 451:363–367. 10.1038/nature06482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rønn L.C., Ralets I., Hartz B.P., Bech M., Berezin A., Berezin V., Møller A., and Bock E.. 2000. A simple procedure for quantification of neurite outgrowth based on stereological principles. J. Neurosci. Methods. 100:25–32. 10.1016/S0165-0270(00)00228-4 [DOI] [PubMed] [Google Scholar]
- Rothenberg M.E., Rogers S.L., Vale R.D., Jan L.Y., and Jan Y.-N.. 2003. Drosophila pod-1 crosslinks both actin and microtubules and controls the targeting of axons. Neuron. 39:779–791. 10.1016/S0896-6273(03)00508-7 [DOI] [PubMed] [Google Scholar]
- Sayas C.L., and Ávila J.. 2014. Crosstalk between axonal classical microtubule-associated proteins and end binding proteins during axon extension: possible implications in neurodegeneration. J. Alzheimers Dis. 40(Suppl 1):S17–S22. [DOI] [PubMed] [Google Scholar]
- Sayas C.L., Tortosa E., Bollati F., Ramírez-Ríos S., Arnal I., and Avila J.. 2015. Tau regulates the localization and function of End-binding proteins 1 and 3 in developing neuronal cells. J. Neurochem. 133:653–667. 10.1111/jnc.13091 [DOI] [PubMed] [Google Scholar]
- Schaefer A.W., Kabir N., and Forscher P.. 2002. Filopodia and actin arcs guide the assembly and transport of two populations of microtubules with unique dynamic parameters in neuronal growth cones. J. Cell Biol. 158:139–152. 10.1083/jcb.200203038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A.W., Schoonderwoert V.T., Ji L., Mederios N., Danuser G., and Forscher P.. 2008. Coordination of actin filament and microtubule dynamics during neurite outgrowth. Dev. Cell. 15:146–162. 10.1016/j.devcel.2008.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner O., and Yaron A.. 2015. Mechanisms of developmental neurite pruning. Cell. Mol. Life Sci. 72:101–119. 10.1007/s00018-014-1729-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Q., Yang T., Huang H., Alarmanazi F., and Liu G.. 2017. Uncoupling of UNC5C with polymerized TUBB3 in microtubules mediates Nnetrin-1 repulsion. J. Neurosci. 37:5620–5633. 10.1523/JNEUROSCI.2617-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp D.J., and Ross J.L.. 2012. Microtubule-severing enzymes at the cutting edge. J. Cell Sci. 125:2561–2569. 10.1242/jcs.101139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood N.T., Sun Q., Xue M., Zhang B., and Zinn K.. 2004. Drosophila spastin regulates synaptic microtubule networks and is required for normal motor function. PLoS Biol. 2:e429 10.1371/journal.pbio.0020429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Park H.-C., Topczewska J.M., Mawdsley D.J., and Appel B.. 2003. Neural cell fate analysis in zebrafish using olig2 BAC transgenics. Methods Cell Sci. 25:7–14. 10.1023/B:MICS.0000006847.09037.3a [DOI] [PubMed] [Google Scholar]
- Sudo H., and Baas P.W.. 2010. Acetylation of microtubules influences their sensitivity to severing by katanin in neurons and fibroblasts. J. Neurosci. 30:7215–7226. 10.1523/JNEUROSCI.0048-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter D.M., Schaefer A.W., and Forscher P.. 2004. Microtubule dynamics are necessary for SRC family kinase-dependent growth cone steering. Curr. Biol. 14:1194–1199. 10.1016/j.cub.2004.06.049 [DOI] [PubMed] [Google Scholar]
- Tarrade A., Fassier C., Courageot S., Charvin D., Vitte J., Peris L., Thorel A., Mouisel E., Fonknechten N., Roblot N., et al. 2006. A mutation of spastin is responsible for swellings and impairment of transport in a region of axon characterized by changes in microtubule composition. Hum. Mol. Genet. 15:3544–3558. 10.1093/hmg/ddl431 [DOI] [PubMed] [Google Scholar]
- Tortosa E., Galjart N., Avila J., and Sayas C.L.. 2013. MAP1B regulates microtubule dynamics by sequestering EB1/3 in the cytosol of developing neuronal cells. EMBO J. 32:1293–1306. 10.1038/emboj.2013.76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitriol E.A., and Zheng J.Q.. 2012. Growth cone travel in space and time: The cellular ensemble of cytoskeleton, adhesion, and membrane. Neuron. 73:1068–1081. 10.1016/j.neuron.2012.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Brust-Mascher I., Cheerambathur D., and Scholey J.M.. 2010. Coupling between microtubule sliding, plus-end growth and spindle length revealed by kinesin-8 depletion. Cytoskeleton (Hoboken). 67:715–728. 10.1002/cm.20482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White S.R., and Lauring B.. 2007. AAA+ ATPases: Achieving diversity of function with conserved machinery. Traffic. 8:1657–1667. 10.1111/j.1600-0854.2007.00642.x [DOI] [PubMed] [Google Scholar]
- Wood J.D., Landers J.A., Bingley M., McDermott C.J., Thomas-McArthur V., Gleadall L.J., Shaw P.J., and Cunliffe V.T.. 2006. The microtubule-severing protein Spastin is essential for axon outgrowth in the zebrafish embryo. Hum. Mol. Genet. 15:2763–2771. 10.1093/hmg/ddl212 [DOI] [PubMed] [Google Scholar]
- Yakushiji Y., Nishikori S., Yamanaka K., and Ogura T.. 2006. Mutational analysis of the functional motifs in the ATPase domain of Caenorhabditis elegans fidgetin homologue FIGL-1: fFirm evidence for an intersubunit catalysis mechanism of ATP hydrolysis by AAA ATPases. J. Struct. Biol. 156:93–100. 10.1016/j.jsb.2006.03.001 [DOI] [PubMed] [Google Scholar]
- Yang Y., Mahaffey C.L., Bérubé N., Nystuen A., and Frankel W.N.. 2005. Functional characterization of fidgetin, an AAA-family protein mutated in fidget mice. Exp. Cell Res. 304:50–58. 10.1016/j.yexcr.2004.11.014 [DOI] [PubMed] [Google Scholar]
- Yu W., Qiang L., Solowska J.M., Karabay A., Korulu S., and Baas P.W.. 2008. The microtubule-severing proteins spastin and katanin participate differently in the formation of axonal branches. Mol. Biol. Cell. 19:1485–1498. 10.1091/mbc.E07-09-0878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., and Chen J.. 2013. FIGNL1-containing protein complex is required for efficient homologous recombination repair. Proc. Natl. Acad. Sci. USA. 110:10640–10645. 10.1073/pnas.1220662110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelenchuk T.A., and Brusés J.L.. 2011. In vivo labeling of zebrafish motor neurons using an mnx1 enhancer and Gal4/UAS. Genesis. 49:546–554. 10.1002/dvg.20766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Rogers G.C., Buster D.W., and Sharp D.J.. 2007. Three microtubule severing enzymes contribute to the “Pacman-flux” machinery that moves chromosomes. J. Cell Biol. 177:231–242. 10.1083/jcb.200612011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Grode K.D., Stewman S.F., Diaz-Valencia J.D., Liebling E., Rath U., Riera T., Currie J.D., Buster D.W., Asenjo A.B., et al. 2011. Drosophila katanin is a microtubule depolymerase that regulates cortical-microtubule plus-end interactions and cell migration. Nat. Cell Biol. 13:361–370. 10.1038/ncb2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Jin M., Wang M., Sun L., Hong X., Cao Y., and Wang C.. 2016. Fidgetin-like 1 is a ciliogenesis-inhibitory centrosome protein. Cell Cycle. 15:2367–2375. 10.1080/15384101.2016.1204059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F.Q., Zhou J., Dedhar S., Wu Y.-H., and Snider W.D.. 2004. NGF-induced axon growth is mediated by localized inactivation of GSK-3beta and functions of the microtubule plus end binding protein APC. Neuron. 42:897–912. 10.1016/j.neuron.2004.05.011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.