PDGFRβ translocates to the nucleus in a ligand-dependent manner tethered by TATA element–modifying factor 1 (TMF-1). Papadopoulos et al. show that PDGFRβ interacts with TMF-1 and Fer kinase in the nucleus, regulating chromatin remodeling by the SWI–SNF complex and controlling proliferation via a p21-dependent mechanism.
Abstract
Translocation of full-length or fragments of receptors to the nucleus has been reported for several tyrosine kinase receptors. In this paper, we show that a fraction of full-length cell surface platelet-derived growth factor (PDGF) receptor β (PDGFRβ) accumulates in the nucleus at the chromatin and the nuclear matrix after ligand stimulation. Nuclear translocation of PDGFRβ was dependent on PDGF-BB–induced receptor dimerization, clathrin-mediated endocytosis, β-importin, and intact Golgi, occurring in both normal and cancer cells. In the nucleus, PDGFRβ formed ligand-inducible complexes with the tyrosine kinase Fer and its substrate, TATA element–modifying factor 1 (TMF-1). PDGF-BB stimulation decreased TMF-1 binding to the transcriptional regulator Brahma-related gene 1 (Brg-1) and released Brg-1 from the SWI–SNF chromatin remodeling complex. Moreover, knockdown of TMF-1 by small interfering RNA decreased nuclear translocation of PDGFRβ and caused significant up-regulation of the Brg-1/p53-regulated cell cycle inhibitor CDKN1A (encoding p21) without affecting PDGFRβ-inducible immediate-early genes. In conclusion, nuclear interactions of PDGFRβ control proliferation by chromatin remodeling and regulation of p21 levels.
Introduction
PDGF family members stimulate mitogenesis and chemotaxis of fibroblasts, pericytes, and smooth muscle cells (Heldin and Westermark, 1999) and exert their effects via binding to α- and β-tyrosine kinase receptors (PDGFRα and PDGFRβ, respectively). Binding of ligands to the extracellular domains of PDGF receptors (PDGFRs) triggers dimerization of the receptors and autophosphorylation within their intracellular domains, leading to activation of multiple signaling pathways; their signaling is disrupted in various pathological conditions, including cancer (Papadopoulos and Lennartsson, 2017; Heldin et al., 2018). PDGFRs are internalized from the plasma membrane via receptor-mediated endocytosis (Lemmon and Schlessinger, 2010) and continue to assemble signaling complexes and transmit signals while internalized in endosomes (Miaczynska et al., 2004; Miaczynska, 2013). Notably, internalized growth factor receptors may activate different signaling molecules depending on their various intracellular localizations (Schlessinger and Lemmon, 2006; Kermorgant and Parker, 2008; Sigismund et al., 2008; Choudhary et al., 2009). Moreover, there is increasing evidence suggesting that membrane receptors not only signal from the plasma membrane and intracellular vesicles, but are able to traffic to the nucleus in a ligand-dependent manner and transmit signals by direct binding to DNA and/or by participating in other nuclear events (Carpenter and Liao, 2013). Among prominent examples are EGF receptor (EGFR) family members (Lo et al., 2006; Wang et al., 2010a, 2012; De Angelis Campos et al., 2011) and insulin growth factor receptor 1 (IGF-1R; Aleksic et al., 2010; Packham et al., 2015).
Nuclear receptor tyrosine kinases (RTKs) have been found to transactivate promoters of target genes (Lin et al., 2001), interact with transcription factors (Wang et al., 2010b), affect DNA replication and damage repair (Wang et al., 2006), bind to putative enhancer elements on genomic DNA (Sehat et al., 2010), and regulate transcription of ribosomal RNA genes independently of canonical activation of downstream phosphatidylinositol-3-kinase (PI3-kinase) and Erk MAP-kinase pathways (Li et al., 2011). Recently, IGF-1R was shown to phosphorylate histone H3 on tyrosine 41, leading to stabilization of the Brahma-related gene (Brg-1) chromatin binding (Warsito et al., 2016).
In the nucleus, genomic DNA is packaged into nucleosomes that are organized in higher order chromatin structures forming functional compartments and chromosomal territories of active and repressed chromatin (Strouboulis and Wolffe, 1996). It has been shown that transcriptionally active DNA is tightly associated with the nuclear skeleton (or nuclear matrix), whereas inactive loci are not (Jackson et al., 1993). The SWI–SNF chromatin remodeling complex is enriched at the active chromatin and associated with the nuclear matrix (Reyes et al., 1997). It is a large protein complex that provides coordinate regulation of gene expression programs. The SWI–SNF complex consists of multiple subunits, including mutually exclusive DNA helicase ATPases Brahma homologue (BRM) and Brg-1, “core” elements Brg-1–associated factors 155 and 170 (BAF155 and BAF170), and variable modulatory subunits (Wilson and Roberts, 2011). SWI–SNF chromatin remodeling complexes were found to act as tumor suppressors; their subunit proteins are deleted or mutated in ∼20% of human cancers, exhibiting a broad mutation pattern similar to that of TP53 (Kadoch et al., 2013). Interestingly, activation of T lymphocytes with phosphatidylinositol 4,5-bisphosphate led to rapid changes in chromatin binding of SWI–SNF complexes, thus demonstrating a direct interface between signaling at the membrane and chromatin regulation (Zhao et al., 1998; Rando et al., 2002).
TATA element–modifying factor 1 (TMF-1), also named androgen receptor activator 160 kD (ARA160), is a Golgi protein that mediates intracellular transport by tethering vesicles (Fridmann-Sirkis et al., 2004; Yamane et al., 2007). In the nucleus, TMF-1 competes with TATA-binding protein for binding to some RNA polymerase II TATA box–containing promoters (Garcia et al., 1992), serves as a coactivator of the androgen receptor in human prostate cells (Hsiao and Chang, 1999), and has been copurified with the SWI–SNF chromatin remodeling complex (Euskirchen et al., 2011). TMF-1 can be tyrosine phosphorylated by the nuclear nonreceptor tyrosine kinase Fer (Schwartz et al., 1998), which we previously reported to interact with PDGFRβ and to play a critical role in PDGF-BB–induced STAT3 activation and cell transformation (Lennartsson et al., 2013).
Here, we show that PDGFRβ rapidly translocates to the nucleus and localizes to the chromatin and nuclear matrix in response to PDGF-BB stimulation in human BJhTERT fibroblasts and other cell lines. Nuclear interaction of PDGFRβ with nonreceptor tyrosine kinase Fer and TMF-1 leads to reassembly of Brg-1–containing SWI–SNF complexes, subsequent chromatin remodeling, and changes in the expression of CDKN1A mRNA and its encoded protein p21, affecting regulation of cell proliferation.
Results
PDGF stimulation causes nuclear accumulation of full-length PDGFRβ
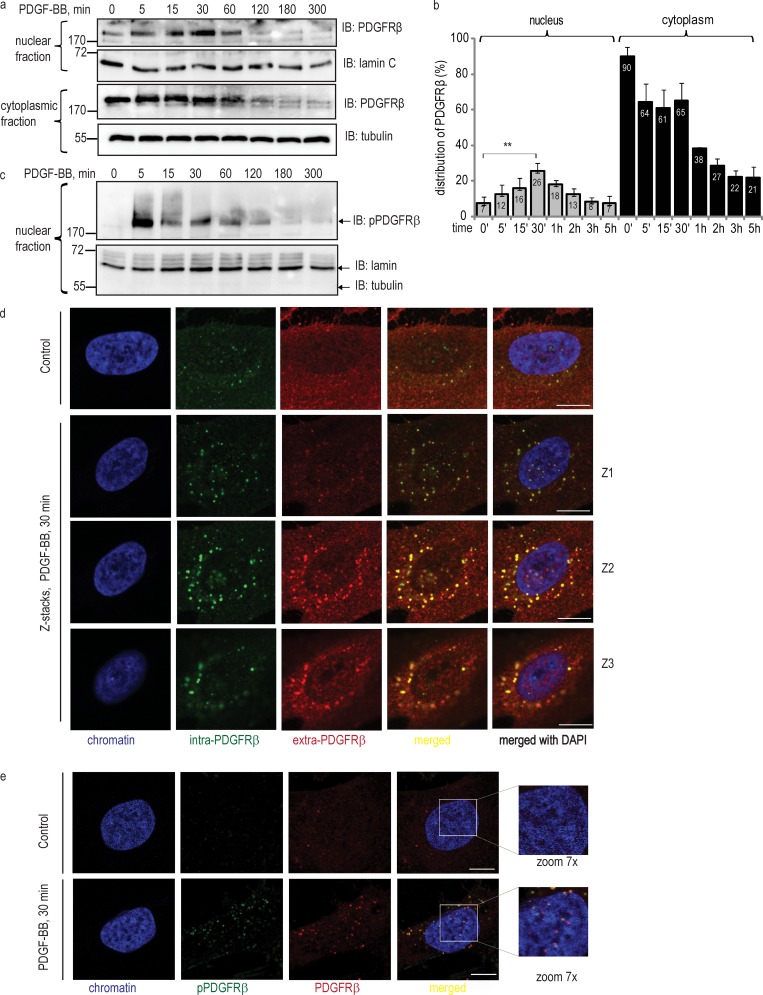
To explore whether PDGFRβ is translocated to the nucleus, we isolated nuclei and found that in response to PDGF-BB stimulation, there was an accumulation of PDGFRβ in the nucleus in a time-dependent manner (Fig. 1, a and b). Nuclei of unstimulated cells contained a basic level of PDGFRβ, calculated as 7% relative to the total amount of the receptor in the cells. The observed nuclear accumulation was increased already after 5 min of PDGF-BB stimulation, reached a peak of 26% of the total receptor amount at 30 min, and then gradually declined, returning to baseline after ∼2 h of PDGF-BB stimulation. The decrease was concomitant with a decrease in cytoplasmic levels of PDGFRβ during the course of stimulation (Fig. 1 b), reflecting subsequent degradation of PDGFRβ. Based on the migration properties in an SDS-PAGE gel, as well as using different antibodies recognizing either the extracellular or intracellular regions of the PDGFRβ, we concluded that the full-length receptor enters the nucleus. Immunoblotting with a phosphotyrosine antibody (Fig. 1 c) revealed that the nuclear PDGFRβ was phosphorylated. To exclude the possibility that nuclei were contaminated with cytoplasmic organelles, we optimized our protocol and analyzed nuclear extracts (using lamin A/C as the nuclear marker) for purity from plasma membrane (using CD49b as a marker), lipid rafts (marker caveolin), Golgi (marker GM130), ER (marker Bip/GRP78), soluble cytoplasm (marker tubulin), lysosomes (marker Lamp-1), and early endosomes (marker EEA1; Fig. S1).
Figure 1.
Nuclear translocation of PDGFRβ in response to ligand stimulation. (a) Immunoblotting analysis of nuclear and cytoplasmic distribution of PDGFRβ. Nuclear and cytoplasmic fractions were immunoblotted for total PDGFRβ, showing nuclear accumulation of PDGFRβ (top) versus cytoplasmic localization (bottom). Lamin A/C and tubulin were used as markers for nuclear and cytoplasmic fractions, respectively. (b) The distribution of PDGFRβ in the nucleus and in the cytoplasm was quantified as emission values of the immunoblot signal, normalized to the signal of the markers. Total amount of receptor in the cell was calculated as the sum of signal intensity values detected in nuclear and cytoplasmic fractions before stimulation with PDGF-BB (0 min), which was taken as 100%. The signal values obtained during the course of stimulation were calculated relative to this value. Error bars indicate SD. Quantification was based on three independent experiments, and t test statistical analysis was performed for the difference between PDGFRβ signal in unstimulated nuclei and at 30 min of stimulation with PDGF-BB; **, P = 0.0092. (c) Immunoblotting of nuclear fractions using a phosphotyrosine antibody, detecting phosphorylated PDGFRβ (top). The purity of the fractionation was verified (bottom). (d) Nuclear localization of PDGFRβ as detected by costaining with two types of PDGFRβ antibodies. PDGFRβ was stained with intra-PDGFRβ antibody (green) and with extra-PDGFRβ antibody (red); chromatin was stained with DAPI (blue). Merged images are shown with (red, green, and blue) and without the nucleus (red and green). Z1, Z2, and Z3 rows of images show stacks through the nucleus of cells simulated with PDGF-BB for 30 min. Bars, 10 µm. (e) Colocalization of immunostaining for total and phospho-specific PDGFRβ antibodies. Cells were immunostained with total PDGFRβ antibody (red) and pTyr (PY99) antibody (green); chromatin was stained with DAPI (blue). Images were taken with the focus on the nucleus; yellow indicates colocalization. Molecular mass was measured in kilodaltons. Bars, 10 µm. IB, immunoblotting.
We confirmed the presence of PDGFRβ in the nucleus by confocal microscopy of immunofluorescent staining with antibodies raised against the intracellular (Fig. S2 a) and extracellular (Fig. S2 c), parts of PDGFRβ, as well as by costaining with these antibodies (Fig. 1 d). After PDGF-BB stimulation, PDGFRβ (Fig. S2, in red) stained as punctuate, dot-like structures both in the cytoplasm and in the nucleus (also see magnification of the nuclear area), presumably as a consequence of dimerization and clustering of the receptor molecules. We performed Z stacks through the nucleus (Fig. S2 b), which confirmed the presence of PDGFRβ in the middle of the nucleus. Additionally, we performed stacks through the nucleus of the cells costained with two PDGFRβ antibodies, thus demonstrating localization in the middle of the nucleus for the fraction of PDGFRβ that was recognized by both antibodies (Fig. 1 d). To further confirm our findings, we performed double staining of PDGFRβ with an antibody against the total receptor and a phosphotyrosine antibody, detecting autophosphorylated PDGFRβ. Analysis of images showed colocalization of the signals and the presence of phosphorylated PDGFRβ in the nucleus (Fig. 1 e). Altogether, these findings indicate that the full-length PDGFRβ translocates to the nucleus in response to PDGF-BB stimulation.
PDGFRβ kinase activity promotes, but is not necessary for, PDGFRβ nuclear accumulation
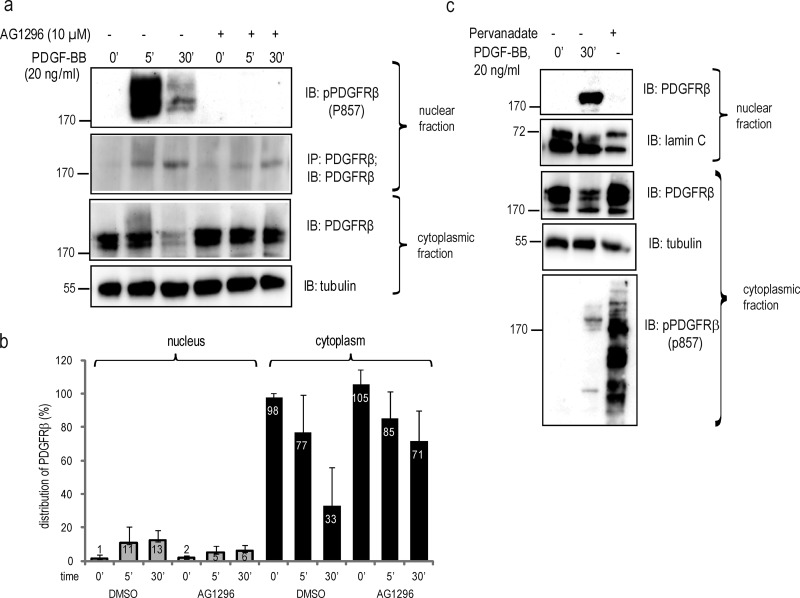
To determine whether the kinase activity of PDGFRβ is required for the nuclear translocation, we used the PDGFR kinase inhibitor AG1296. Although AG1296 treatment resulted in reduced nuclear translocation by ∼50%, PDGFRβ still appeared in the nucleus after PDGF-BB stimulation (Fig. 2, a and b). Similar results were obtained by the unrelated PDGFR kinase inhibitor imatinib (not depicted). Most probably, the reduction in the nuclear translocation of PDGFRβ reflects the decrease in internalization of the cell surface receptor after inhibition of its kinase activity (Sorkin et al., 1991). To further understand the translocation mechanism, we treated cells with the general phosphatase inhibitor sodium pervanadate, which induces strong receptor phosphorylation in the absence of ligand-induced receptor dimerization. Despite achieving PDGFRβ phosphorylation, as determined by a phospho-specific PDGFRβ antibody, P857 (Fig. 2 c, bottom), pervanadate treatment was not sufficient to provoke nuclear translocation (Fig. 2 c, top), suggesting that ligand-induced dimerization of PDGFRβ, but not receptor phosphorylation, is necessary for nuclear translocation.
Figure 2.
PDGF-BB–induced nuclear transport of PDGFRβ is not quenched by receptor kinase inhibition. (a) PDGFRβ translocation to the nucleus upon inhibition of PDGFRβ kinase activity. Nuclear extracts were immunoblotted for pTyr857-PDGFRβ (top) or immunoprecipitated and immunoblotted with the PDGFRβ antibody ctβ (second panel). Cytoplasmic extracts were immunoblotted for PDGFRβ with the ctβ antibody (third panel) or α-tubulin (bottom). (b) Quantification of PDGFRβ distribution upon inhibition of receptor tyrosine kinase activity with AG1296. Immunoblots were quantified as described in Fig. 1. Error bars indicate SD. Quantification was based on four independent experiments. (c) Cells were stimulated with PDGF-BB or phosphatase activity was inhibited with sodium pervanadate, and lysates were immunoblotted with total PDGFR antibody (Y92) or lamin A/C for nuclear fractions (top two panels) or total PDGFRβ, α-tubulin, and pPDGFRβ antibody for cytoplasmic fractions (three bottom panels). Molecular mass was measured in kilodaltons. IB, immunoblotting; IP, immunoprecipitation.
Characterization of the nuclear translocation pathway
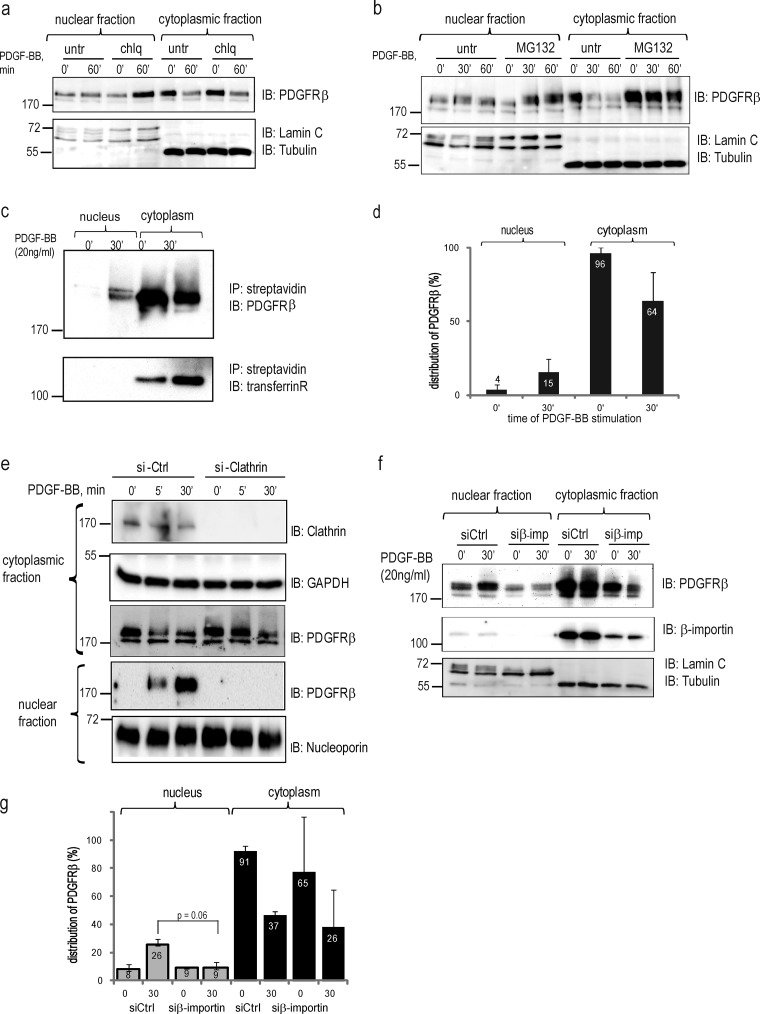
Internalization of PDGFRβ leads to its subsequent degradation via proteasomal (Mori et al., 1992) and lysosomal (Haglund et al., 2003) pathways. We investigated whether inhibition of degradation affected the nuclear accumulation of PDGFRβ. Interestingly, lysosomal inhibition by chloroquine (Fig. 3 a) resulted in selective nuclear accumulation of PDGFRβ, whereas proteasomal inhibition by MG132 led to a concomitant increase in PDGFRβ levels in the cytoplasm and the nucleus (Fig. 3 b). This suggests that the sorting pathways of PDGFR toward the nucleus prevent lysosomal degradation of PDGFRβ, whereas proteasomal degradation occurs ubiquitously in both the cytoplasm and the nucleus.
Figure 3.
Characterization of nuclear translocation pathways. (a) PDGFRβ accumulates in the nucleus after inhibition of the endosomal–lysosomal pathway. Nuclear and cytoplasmic fractions treated or not with chloroquine (chlq) were immunoblotted with total PDGFR antibody (top), lamin A/C, and α-tubulin (bottom). A representative experiment of three biological repeats is shown. (b) Accumulation of PDGFRβ in the nucleus upon inhibition of proteasomal degradation. Nuclear and cytoplasmic fractions treated or not with MG132 were immunoblotted with total PDGFRβ antibody (top) or nuclear and cytoplasmic markers (bottom). (c) Biotinylated cell surface receptor is translocated to the nucleus. Cytoplasmic and nuclear fractions were prepared from biotinylated cells stimulated with PDGF-BB. Biotinylated proteins were precipitated with streptavidin agarose and immunoblotted for total PDGFRβ (top) or transferrin receptor (bottom). (d) Quantification of the distribution of biotinylated PDGFRβ was performed as described in Fig. 1 b. Error bars indicate SD. (e) Nuclear translocation of PDGFRβ is inhibited by siRNA knockdown of clathrin. The efficiency of clathrin siRNA knockdown was determined with an anticlathrin antibody (top). The levels of PDGFRβ in the cytoplasmic and nuclear fractions were determined by immunoblotting (middle). The purity of the fractions was confirmed by immunoblotting for GAPDH (a cytoplasmic marker) or lamin A/C. (f) Nuclear translocation of PDGFRβ is decreased upon siRNA knockdown of β-importin. The level of PDGFRβ in the cytoplasmic and nuclear fractions was determined by immunoblotting with total PDGFR antibody (top); the purity of fractionations was confirmed by blotting for lamin A/C and α–tubulin (bottom). The efficiency of the knockdown was determined by immunoblotting with anti–β-importin antibody (middle). (g) Quantification of PDGFRβ distribution upon β-importin siRNA knockdown was performed as described above and was based on three independent experiments. Molecular mass was measured in kilodaltons. Error bars indicate SD. IB, immunoblotting; IP, immunoprecipitation.
To determine whether the receptors seen in the nucleus represented either receptors internalized from the cell surface or newly synthesized receptors, we biotinylated cell surface proteins before PDGF-BB treatment and analyzed the presence of biotinylated PDGFRβ in the nuclear fraction. Indeed, we detected an accumulation of biotinylated, plasma membrane–derived PDGFRβ in the nucleus (Fig. 3 c, top) with comparable kinetics as observed for the unlabeled PDGFRβ (Fig. 3 d). As a negative control, we used the transferrin receptor, which is a transmembrane protein that does not respond to PDGF-BB stimulation; this receptor was not translocated to the nucleus (Fig. 3 c, bottom).
We proceeded to block common subcellular trafficking pathways that are known to be involved in sorting RTKs. PDGFRβ internalizes from the cell surface to a large extent through clathrin-coated pits (Kapeller et al., 1993) and to some extent through lipid rafts via caveolae-containing vesicles (Liu et al., 1996). Indeed, depletion of cells of clathrin with siRNA efficiently blocked nuclear PDGFRβ accumulation after PDGF-BB stimulation (Fig. 3 e), supporting the notion that the cell surface PDGFRβ is sorted toward the nucleus via clathrin-coated vesicles.
Interaction with β-importin is required by many proteins for their entry into the nucleus. We found that when β-importin levels were down-regulated by 85% by siRNA, there was a significant inhibition of nuclear PDGFRβ accumulation (Fig. 3, f and g), suggesting that translocation of PDGFRβ to the nucleus occurs in a β-importin–dependent manner.
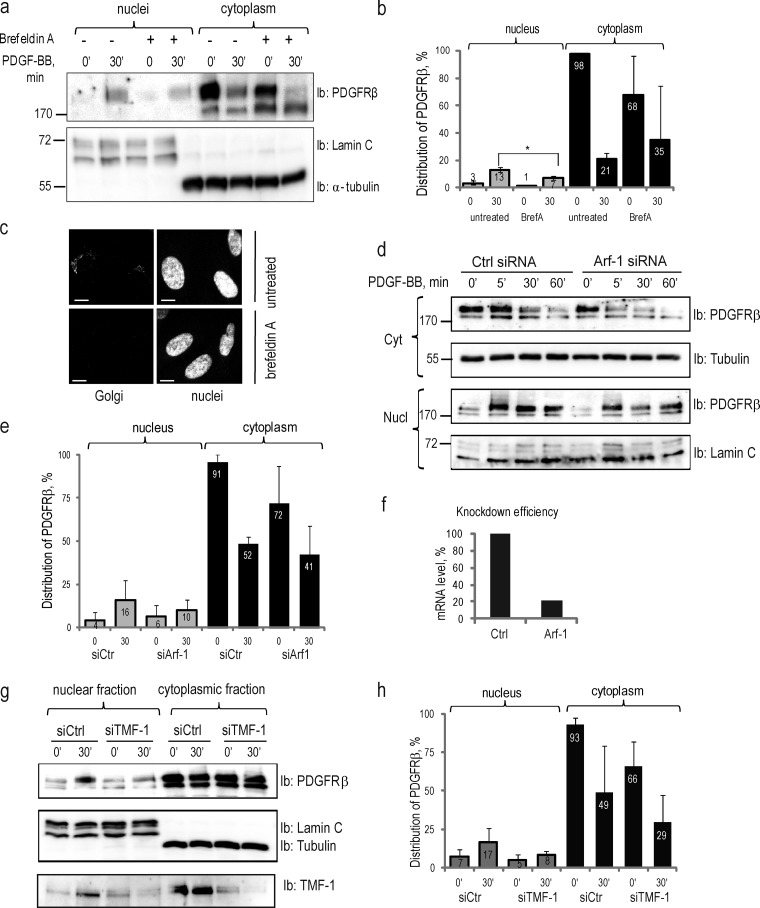
Recently, it has been proposed that EGFR accumulates in the nucleus by using the retrograde transport machinery from Golgi to the ER. We investigated the importance of Golgi for the nuclear transport of PDGFRβ by inhibiting all trans-Golgi trafficking with brefeldin A (Fig. 4, a–c) and redistributing the Golgi complex to the ER with ADP ribosylation factor 1 (Arf-1) siRNA (Fig. 4, d–f). In both cases, we observed some reduction in the nuclear accumulation of PDGFRβ, and cytoplasmic levels of mature PDGFRβ were also decreased. Because we found an interaction between PDGFRβ and TMF-1 in the nucleus (see below), and because TMF-1 was reported to play an important role in intra-Golgi trafficking (Wong and Munro, 2014) and in the retrograde transport from Golgi to ER (Yamane et al., 2007), we investigated whether TMF-1 is necessary for the nuclear translocation of PDGFRβ. Indeed, when BJhTERT fibroblasts were depleted of TMF-1 with siRNA, a certain decrease of nuclear accumulation of PDGFRβ at 30 min of PDGF-BB stimulation was observed (Fig. 4, g and h). The cytoplasmic levels of PDGFRβ were also affected, which suggests that TMF-1 is involved in maintaining cytoplasmic levels of PDGFRβ as well as participating in its nuclear function.
Figure 4.
Nuclear translocation of PDGFRβ is dependent on intact Golgi and retrograde transport system. (a) Inhibition of nuclear translocation of PDGFRβ upon brefeldin A treatment. Nuclear and cytoplasmic fractions of cells treated or not with brefeldin A were immunoblotted for PDGFRβ (top) and markers (bottom). (b) Quantification of the distribution of PDGFR in the nucleus and the cytoplasm upon treatment with brefeldin A was performed, as described above, for three independent experiments. *, P < 0.05, two-tailed t test. (c) The integrity of the Golgi apparatus was lost upon treatment with brefeldin A. Golgi (top left) was absent after the treatment with brefeldin A (bottom left); nuclei are presented on images to the right. Bars, 10 µm. (d) PDGFRβ translocation to the nucleus is decreased upon siRNA knockdown of Arf-1. Cytoplasmic (two top panels) and nuclear (two bottom panels) fractions were immunoblotted for PDGFRβ; the purity of the fractions was determined by immunoblotting for α-tubulin and lamin C. (e) Nuclear distribution of PDGFRβ upon Arf-1 knockdown was quantified based on three independent experiments. (f) The efficiency of Arf-1 depletion was confirmed by quantitative PCR. The mean mRNA level in knockdown samples was calculated as a percentage relative to the mean mRNA levels in control samples. (g) Nuclear translocation of PDGFRβ is decreased upon siRNA knockdown of TMF-1. Nuclear and cytoplasmic fractions were immunoblotted with PDGFRβ antibody (top) or lamin A/C and α-tubulin (middle). The efficiency of TMF-1 knockdown was demonstrated by immunoblotting with TMF-1 antibody (bottom). (h) The distribution of PDGFRβ in the nucleus and cytoplasm upon depletion of TMF-1 was quantified as described above based on three independent experiments. Molecular mass was measured in kilodaltons. Error bars indicate SD. Ib, immunoblotting.
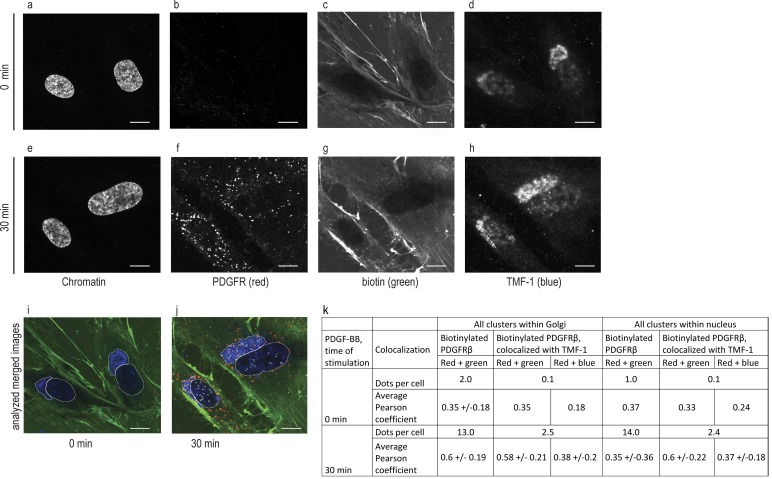
To further clarify the involvement of TMF-1–positive Golgi vesicles in the nuclear transport of PDGFRβ, we biotinylated cell surface PDGFRβ and investigated triple colocalization between biotin, PDGFRβ, and TMF-1, thus detecting biotinylated cell surface–derived PDGFRβ interacting with TMF-1 in the area of Golgi and in the nucleus (Fig. 5). After quantification of colocalization (Fig. 5 k), we detected on average 13 biotinylated PDGFRβ clusters per cell (n = 29) within Golgi and 14 within the nucleus after 30 min of stimulation with mean Pearson coefficients of 0.6 ± 0.19 and 0.35 ± 0.36, respectively. A Pearson coefficient >0 indicates probability of colocalization. Although most biotinylated PDGFRβ clusters positively correlated with TMF-1, on average 2.5 dots per cell in the Golgi and 2.4 dots per cell in the nucleus displayed triple positive correlation with a Pearson coefficient >0.2. These findings suggest that biotinylated PDGFRβ traffics to the nucleus via TMF-1–positive Golgi vesicles.
Figure 5.
Biotinylated PDGFR traffics to the nucleus via TMF-1–positive Golgi vesicles. Biotinylated cells were stimulated (e–h) or not (a–d) with PDGF-BB and immunostained for biotin, PDGFRβ, TMF-1, and DAPI. Original black and white images (a–h) were uploaded into an automatic pipeline at Cell Profiler for quantification of colocalization. The merged image that was created by the software is presented, where Golgi area (as detected by TMF-1) and nucleus (as detected by DAPI) were isolated and PDGFRβ clusters were analyzed within these areas for unstimulated (i) and stimulated (j) cells. The mean number of dots and mean Pearson correlation coefficients for these dots (pairwise for red, green, and blue channels) were calculated and presented in the table (k). Bars, 10 µm.
Nuclear translocation of PDGFRβ occurs in both normal and cancer cells
To assess the generality of translocation of PDGFRβ, we analyzed several different cell lines expressing endogenous PDGFRβ, such as primary fibroblasts AG1523 (Fig. S3 a), glioblastoma U105MG (Fig. S3 b), and osteosarcoma U2OS (Fig. S3 c) cells. We found that nuclear translocation of PDGFRβ occurred in both normal and cancer cells. The effect of the receptor kinase inhibitor AG1296 on primary fibroblasts and on glioblastoma and osteosarcoma cancer cell lines was similar to that observed in BJhTERT fibroblasts, i.e., nuclear translocation of PDGFRβ was decreased, but not completely blocked. In the same way, the effects of brefeldin and MG132 were reproduced in the osteosarcoma cell line U2OS (Fig. S3 c).
PDGFRβ interacts with the Fer nonreceptor tyrosine kinase and its substrate TMF-1 in a ligand-inducible manner, affecting the composition of the SWI–SNF remodeling complex
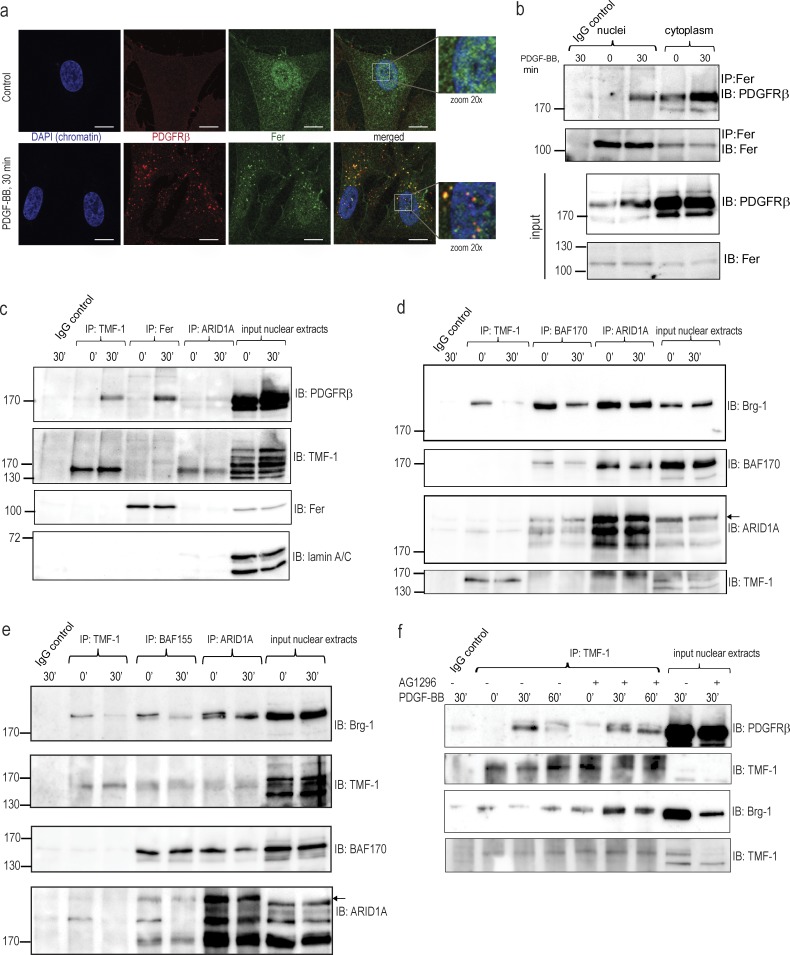
We could not detect any robust and reproducible binding of PDGFRβ to the promoters of known PDGF-BB–inducible genes. Instead, we found that activated PDGFRβ colocalized with the Fer kinase in the characteristic punctuate structures formed by PDGFRβ after 30 min of PDGF-BB stimulation (Fig. 6 a). We identified an interaction of PDGFRβ with Fer in both the cytoplasm and in the nucleus (Fig. 6 b), which coincided with the peak of nuclear accumulation of PDGFRβ. This suggests that nuclear PDGFRβ may transmit nuclear signals via interaction with the Fer kinase, which could further interact with chromatin (Hao et al., 1991) or chromatin-modifying proteins. Because TMF-1 was reported to be one of the substrates for Fer, we investigated interactions between PDGFRβ, Fer, and TMF-1. By coimmunoprecipitation, we demonstrated a complex between PDGFRβ and TMF-1 in the nucleus at the peak of PDGF-BB stimulation, independently of its interaction with Fer (Fig. 6 c). Immunoprecipitation (IP) of the auxiliary component of SWI–SNF complex ARID1A showed no interaction with Fer or PDGFRβ and weak interaction with TMF-1. We also detected an interaction of PDGFRβ with TMF-1 in the cytoplasm (not depicted), which suggests that TMF-1 may be important for the cytoplasmic function of PDGFRβ as well.
Figure 6.
PDGFRβ forms ligand-dependent complex with Fer and TMF-1, affecting composition of the SWI–SNF remodeling complex. (a) Immunofluorescence staining of colocalization of PDGFRβ and Fer kinase. PDGFRβ was immunostained with extra PDGFRβ (red), chromatin with DAPI (blue), and the Fer kinase was stained in green. Yellow indicates colocalization between PDGFRβ and Fer. Bars, 10 µm. (b) Coimmunoprecipitation of PDGFRβ and Fer kinase in the nucleus and in the cytoplasm. Immunoprecipitated complexes were immunoblotted for PDGFRβ (top) or Fer (second panel). 10% of the input material was analyzed by immunoblotting for PDGFRβ and Fer (two bottom panels). (c) PDGFRβ forms PDGF-BB–inducible complexes with TMF-1 and Fer in the nucleus. TMF-1, Fer, and ARID1A immunocomplexes were immunoblotted for PDGFRβ, TMF-1, and Fer. 10% of the input lysate was loaded on the last two lanes on each blot; nuclear TMF-1 was poorly detected by direct immunoblotting as opposite to immunoblotting after IP. The membrane was reblotted for lamin A/C as a loading control. (d) Dissociation of TMF-1–Brg-1 and Brg-1–BAF170 nuclear complexes upon PDGF-BB stimulation. TMF-1, BAF170 and ARID1A immunocomplexes were immunoblotted for Brg-1 and reblotted for BAF170 or blotted for ARID1A and reblotted for TMF-1. 10% of the input lysate was loaded on each gel. Arrows indicate ARID1A protein. (e) Dissociation of TMF-1–Brg-1 and Brg-1–BAF155 nuclear complexes upon PDGF-BB stimulation. This experiment reproduces the result from d, but an antibody against the BAF155 core subunit of SWI–SNF chromatin remodeling complex was used instead of BAF170 for IP. Arrows indicate ARID1A protein. (f) Kinase-inactive PDGFRβ binds TMF-1 and prevents PDGF-BB–inducible dissociation of the SWI–SNF complex. Cells were pretreated with AG1296 and stimulated with PDGF-BB. TMF-1 immunocomplexes from nuclear extracts were blotted for PDGFRβ (top) and reblotted for TMF-1 (second) or blotted for Brg-1 (third) and reblotted for TMF-1 (bottom). Molecular mass was measured in kilodaltons. IB, immunoblotting; IP, immunoprecipitation.
TMF-1 has been reported to interact with two ATPases of the SWI–SNF remodeling complex, i.e., BRM and Brg-1 in a yeast two-hybrid system (Mori and Kato, 2002). Here, we demonstrate an interaction between TMF-1 and Brg-1 in serum-starved unstimulated fibroblasts, which was lost upon PDGF-BB stimulation (Fig. 6, d and e, IP: TMF-1), concomitant with decreased binding of Brg-1 to the core subunits of the SWI–SNF remodeling complex, BAF170 (Fig. 6 d, IP: BAF170) and BAF155 (Fig. 6 e, IP: BAF155). The interaction between Brg-1 and another subunit of the SWI–SNF remodeling complex, ARID1A (Fig. 6, d and e, IP: ARID1A), was decreased, but to a lesser extent. Thus, we suggest that by interacting with Brg-1, TMF-1 may support the transcriptionally regulatory function of the SWI–SNF complex in serum-starved cells, which becomes disassembled upon PDGF-BB stimulation, leading to activation or repression of target genes.
To investigate the importance of the kinase activity of PDGFRβ for its nuclear interactions, we performed coimmunoprecipitations between PDGFRβ, TMF-1, and Bgr-1 in the absence or presence of AG1296. We found that the binding of PDGFRβ to TMF-1 was not inhibited by AG1296; rather, it was stabilized at 60 min of PDGF-BB stimulation (Fig. 6 f, top two blots) when PDGFRβ levels and the interaction with TMF-1 declined under normal conditions. In the same way, the complex between TMF-1 and Brg-1 was stabilized in the presence of AG1296 (Fig. 6 f). These findings suggest that the kinase activity of PDGFRβ is needed for a dynamic turnover of the repressive SWI–SNF complex over its target promoters.
Nuclear PDGFRβ is localized in the nuclear matrix
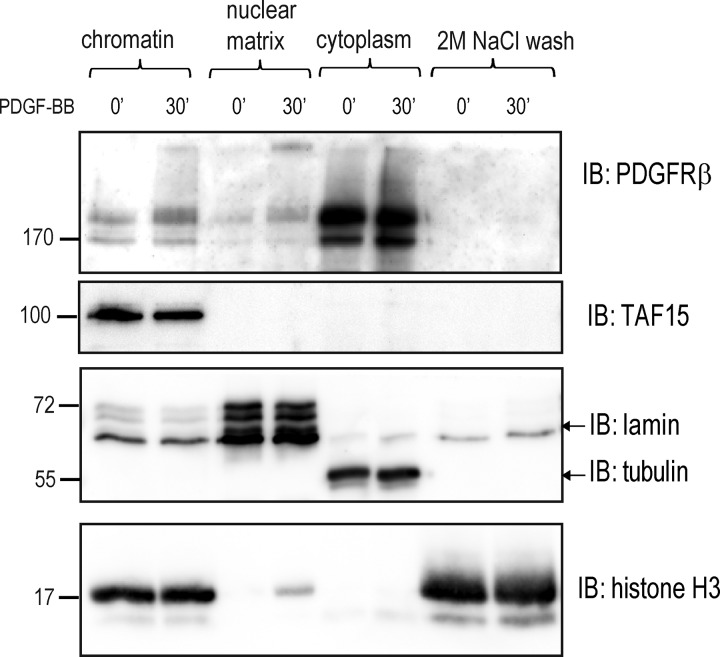
Having found a connection between nuclear PDGFRβ and the matrix-associated SWI–SNF chromatin remodeling complex, we investigated whether PDGFRβ can itself localize to the nuclear matrix. Nuclear matrix is defined as the nuclear structure that remains after salt extraction of nuclease-treated nuclei (Reyes et al., 1997), which consists of a peripheral lamina pore complex and an internal network of filamentous ribonucleoproteins, forming a scaffold for DNA attachment (Berezney et al., 1995). When a fractionation protocol was used that allowed for separation of soluble chromatin proteins from insoluble hydrophobic nuclear matrix, we discovered that a fraction of PDGFRβ remained bound to the nuclear matrix even after the nuclear pellet was washed with 2 M NaCl (Fig. 7, top), a treatment that was used to remove all soluble proteins and unbound histones from the matrix after DNase I digestion (Fig. 7, bottom, shows a large amount of free histones in the salt wash fraction). These findings suggest that after PDGF-BB stimulation, PDGFRβ traffics from the cell surface to the nuclear matrix, where it may form a point of contact with the chromatin and interact with the transcriptional machinery and chromatin-modifying proteins.
Figure 7.
PDGFRβ in the nucleus is localized to the chromatin and nuclear matrix. Chromatin, nuclear matrix, cytoplasmic fractions, and 2 M NaCl wash eluate were immunoblotted for PDGFRβ. RNase II cofactor TATA-associated factor 15 (TAF15), lamin A/C, α-tubulin, and histone H3 were used as markers for transcriptionally active chromatin, nuclear matrix, cytoplasm, and soluble chromatin, respectively. Molecular mass was measured in kilodaltons. IB, immunoblotting.
TMF-1–mediated chromatin remodeling leads to changes in gene transcription specific for nuclear interactions of PDGFRβ
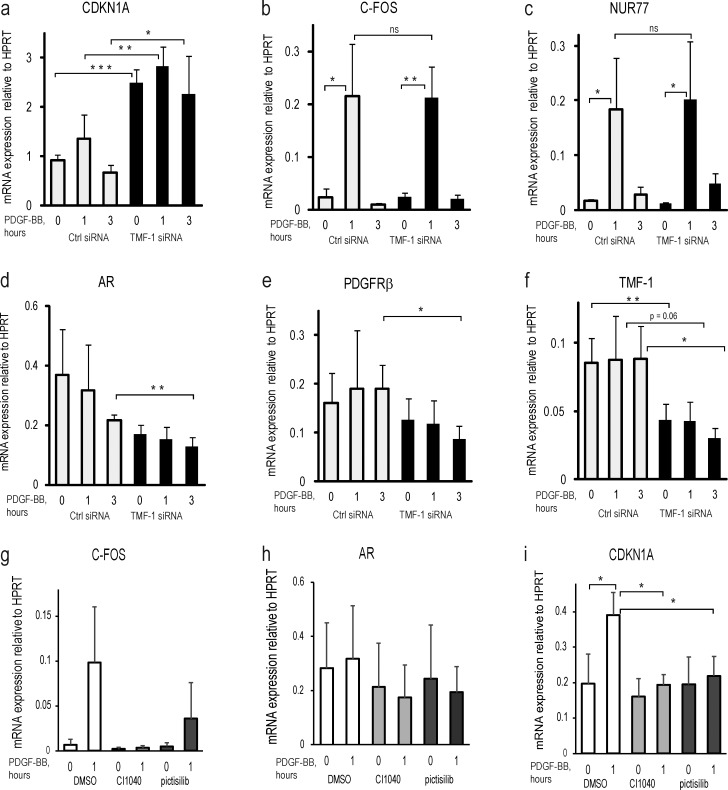
To determine the functional consequences of TMF-1–dependent rearrangement of the SWI–SNF chromatin remodeling complex, we depleted BJhTERT fibroblasts of TMF-1 with siRNA and performed RNA expression analysis of potential Brg-1 target CDKN1A and PDGF-BB–inducible genes based on the finding that the Brg-1–ARID1A–p53 complex serves as a key regulator of transcription of CDKN1A (Guan et al., 2011). We observed a transient increase in CDKN1A mRNA expression in response to PDGF-BB stimulation and a significant up-regulation of its transcription in cells depleted of TMF-1 (Fig. 8 a). Interestingly, up-regulation of immediate early response genes in response to PDGF-BB was not affected by the TMF-1 knockdown, as exemplified by the known PDGF-BB–responsive genes C-FOS (Fig. 8 b) and NUR77, an orphan nuclear receptor gene with tumor-suppressing activity (Fig. 8 c; Bian et al., 2017) that we previously reported to be regulated by PDGF-BB (Eger et al., 2014). We found that although PDGF-BB stimulation suppressed the mRNA expression of AR in control cells, the levels of TMF-1–activated AR were down-regulated even more after the knockdown of TMF-1 (Fig. 8 d), as were the levels of PDGFRβ itself (Fig. 8 e). The efficiency of siTMF-1 mRNA knockdown is presented in Fig. 8 f. These results suggest an important novel suppressive function of TMF-1 in the regulation of expression of the cell cycle regulator CDKN1A, which is modulated after activation of PDGFRβ.
Figure 8.
Transcriptional analysis of target genes during PDGF-BB stimulation at TMF-1 knockdown or inhibition of PDGFRβ kinase activity and cytoplasmic signaling via Erk1/2 and PI3-kinase. Cells were transfected with control (white bars) or TMF-1 (black bars) siRNA (a–f) or pretreated with CI1040 (light gray bars) and pictilisib inhibitor (dark gray bars; g–i) before stimulation with PDGF-BB. mRNA expression of the following genes was analyzed by quantitative PCR to test for the knockdown efficiency: CDKN1A, C-FOS, nuclear receptor NUR77, AR, PDGFRβ, and TMF-1. ΔΔCt was calculated as a log2 of the difference between cycle threshold values of a reference gene HPRT and a gene of interest. The levels of expression represent an average signal obtained from four biological replicates with SD indicated on the graph. Two-tailed t test statistical analysis was performed for the difference between mRNA expression levels of CDKN1A in control cells versus siTMF knockdown cells. *, P < 0.05; **, P < 0.01; ***, P < 0.001. ns, not significant.
We further investigated the effect of canonical cytoplasmic signaling of PDGFRβ on transcription of PDGF-BB–inducible genes. After blocking Erk MAP-kinases and PI3-kinase with CI1040 and pictilisib, respectively, the transcriptional response of C-FOS (Fig. 8 g) was inhibited, whereas the AR level was unaffected (Fig. 8 h). Interestingly, the steady-state levels of CDKN1A were not affected, whereas its induction after 1 h of PDGF-BB stimulation was inhibited by CI1040 and pictilisib (Fig. 8 i), demonstrating that inhibition of PDGFRβ cytoplasmic signaling does not up-regulate p21, as does the knockdown of TMF-1. Thus, regulation of p21 levels during PDGFRβ activation occurs independently of cytoplasmic signaling via Erk MAP-kinase or PI3-kinase, although it is controlled by the nuclear interactors of PDGFRβ.
Nuclear signaling of PDGFRβ promotes proliferation via a p21-dependent mechanism
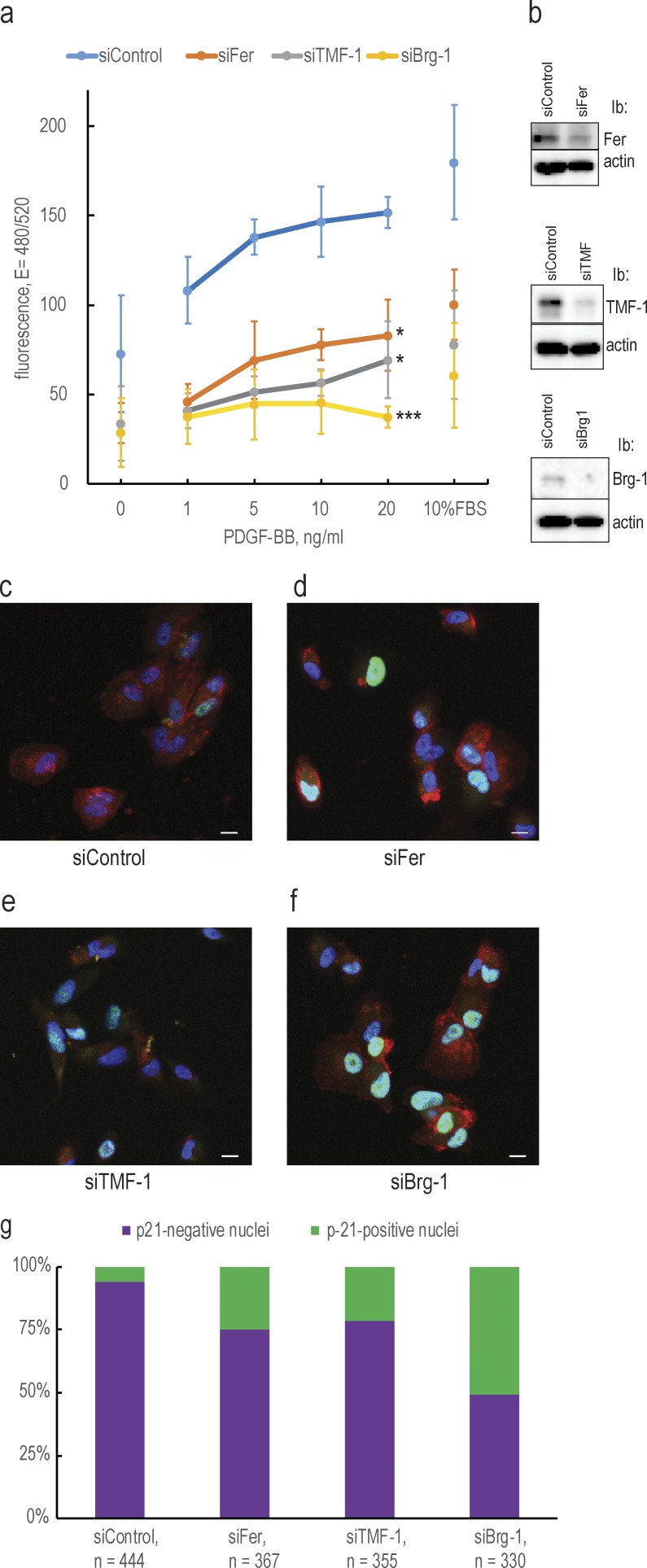
To uncover a functional significance of nuclear traffic and nuclear interactions of PDGFRβ, we depleted BJhTERT fibroblasts of Fer, TMF, or Brg-1 proteins and analyzed proliferation in response to PDGF-BB (Fig. 9 a); knockdown efficiencies are presented in Fig. 9 b. We found that the proliferative response of cells was severely inhibited by the knockdown of each of the interacting proteins. We also investigated a possible functional mechanism of inhibition of proliferation and found an increased number of cells expressing p21 protein after knockdown of Fer (21%; Fig. 9 d), TMF-1 (25%; Fig. 9 e), or Brg-1 (50%; Fig. 9 f) as compared with the control cells (6.5%; Fig. 9 c); quantification is presented in Fig. 9 g. These findings demonstrate the importance of the Fer–TMF-1–Brg-1 axis for the regulation of p21 levels and proper progression of the cell cycle and establishes a function of PDGFRβ in the regulation of the cell cycle by its nuclear interactions with Fer and TMF-1.
Figure 9.
The Fer–TMF-1–Brg-1 axis regulates proliferation by controlling p21 protein levels. (a) Knockdown of Fer, TMF-1, and Brg-1 inhibit proliferation in response to PDGF-BB and serum. Proliferative response was evaluated by measuring incorporation of a fluorescent DNA-binding dye; fluorescence values were plotted against increasing concentrations of PDGF-BB. The statistical significance of the difference between control and knockdown cells at 20 ng/ml PDGF-BB was analyzed with two-tailed t test. Error bars indicate SD. *, P < 0.05; ***, P < 0.001. (b) The efficiencies of the knockdowns were determined by immunoblotting. Ib, immunoblotting. (c–f) Immunostaining of p21 protein expression upon Fer, TMF-1, and Brg-1 knockdown. Cells treated with control siRNA or siRNA for Fer, TMF-1, or Brg-1 were stained for p21 protein (green) and PDGFRβ (red); chromatin was stained with DAPI (blue). Bars, 10 µm. (g) The percentage of p21-expressing cells was counted using the automatic pipeline in Cell Profiler.
Discussion
In addition to the well-characterized signaling from the plasma membrane and endosomes by RTKs, ligand-induced nuclear localization has been described for several of the members (Carpenter and Liao, 2013). Full-length receptors found in the nucleus include EGFR, ErbB2, ErbB3, FGF receptors (FGFR1, FGFR2), IGF-1R, Met, and VEGF receptors (VEGFR1 and VEGFR2; Carpenter and Liao, 2013). In this study, we showed that in response to PDGF-BB stimulation, the full-length PDGFRβ translocates to the nucleus. Nuclear translocation was not absolutely dependent on the receptor kinase activity of PDGFRβ, and PDGFRβ phosphorylation, promoted by phosphatase inhibition in the absence of ligand binding, did not lead to nuclear translocation. Together, these observations suggest that dimerization of receptors is essential for nuclear translocation, whereas receptor kinase activity is not; however, the kinase activity has a functional role in the nucleus. Our data suggest that the nuclear receptor is derived from the cell surface and is dependent on clathrin coating of vesicles and TMF-1 tethering. Similarly, EGFR (De Angelis Campos et al., 2011) and IGF-1R (Aleksic et al., 2010) translocate to the nucleus in clathrin-dependent manners. Clathrin plays an essential role in budding off vesicles within the intra-Golgi network, whereas TMF-1 acts by capturing and tethering cargo vesicles, connecting them to their destination organelles. In relocation studies, TMF-1 demonstrated specificity for Golgi resident proteins and for some of the endosomes-to-Golgi cargo (Wong and Munro, 2014). TMF-1 also plays a critical role in the Rab6-dependent retrograde transport of Shiga toxin from early recycling endosomes to the trans-Golgi network and from Golgi to the ER (Yamane et al., 2007; Miller et al., 2013). Thus, it seems feasible that internalized PDGFRβ is directed to the nucleus via TMF-1–positive Golgi vesicles, representing the retrograde transport machinery.
Additionally, we observed a decrease in nuclear translocation of PDGFRβ after efficient knockdown of β-importin, which is one of the major mediators of protein nuclear transport via the nuclear pore complex (Kimura and Imamoto, 2014). A role of β-importin in the nuclear translocation was also shown for EGFR (Lo et al., 2006), ErbB-2 (Giri et al., 2005), c-Met (Gomes et al., 2008), IGF-R1 (Packham et al., 2015), and FGFR-1 (Stachowiak et al., 2007), suggesting that β-importin plays a general role in the translocation of cell surface RTKs to the nucleus. As was suggested for EGFR, β-importin may interact with retrograde-transported vesicles and facilitate their traffic toward the nucleus (Wang et al., 2010c); thus, there may be no direct interaction of β-importin with PDGFRβ of the kind that occurs for soluble proteins.
Despite the accumulating data about nuclear traffic of transmembrane receptors, it has been difficult to explain the presence and the function of hydrophobic receptors in the soluble nucleoplasm. Our findings offer a possible explanation for the nuclear presence and function of transmembrane receptors. The nuclear matrix serves as the internal scaffold of the nucleus, anchoring the origin of replication sites, core enhancers, transcription factors, and matrix- and scaffold-associated DNA regions that function as control elements for transcriptional domains (Laemmli et al., 1992; Forrester et al., 1994; Boulikas, 1995). In addition, full-length FGFR1 and several unidentified proteins with tyrosine kinase activity have been localized to the nuclear matrix (Stachowiak et al., 1996a,b), including nonreceptor tyrosine kinases of the Src family (Radha et al., 1996; Nakayama et al., 2006). It was also shown that ATP-dependent release of the SWI–SNF complex from its nuclear targets is a result of a cascade of kinase activities, and hyperphosphorylation of Brg-1 and BRM leads to their release from the tight association with the nuclear matrix that coincides with the start of mitosis (Reyes et al., 1997). The nuclear matrix may thus serve as a docking site for PDGFRβ in the nucleus, whereas the intracellular part of the receptor may freely interact with either its binding partners or phosphorylation targets, or directly with the chromatin. Interestingly, in our fractionation experiments, a certain amount of PDGFRβ was detected not only at the nuclear matrix but also in the chromatin fraction, suggesting that PDGFRβ could also maintain direct interactions with the chromatin.
In this work, we have uncovered an inducible nuclear protein complex of PDGFRβ with both nonreceptor tyrosine kinase Fer and its substrate TMF-1, suggesting that the effect of PDGFRβ on chromatin may be partially mediated through these interactions. Fer caused tyrosine phosphorylation of TMF-1, with no direct interaction between endogenous Fer and TMF-1 being detected (Schwartz et al., 1998), consistent with our observations. Thus, the interaction between Fer and TMF-1 could be transient, or phosphorylation of TMF-1 by Fer could be mediated by PDGFRβ.
We have demonstrated that the formation of a PDGFRβ–TMF-1 complex correlates with changes in the composition of the SWI–SNF remodeling complex, which loses its binding to TMF-1 concomitant with the release of Brg-1 after PDGF-BB stimulation. The transient character of this interaction is dependent on the kinase activity of PDGFRβ because its inhibition led to stabilization of PDGFRβ binding to TMF-1 and TMF-1 to the SWI–SNF chromatin remodeling complex. The SWI–SNF complex is extensively regulated by different signaling pathways (Simone, 2006) and can repress or activate multiple genes depending on which transcription factors and modulators it recruits. A possible mechanism behind the changes in SWI–SNF composition could be that phosphorylation of TMF-1 by Fer and/or PDGFRβ promotes disruption of the TMF-1–containing repressive SWI–SNF complex; this may lead to release of Brg-1 followed by its binding to p53, which activates the transcription of CDKN1A, although interaction with the kinase inactive receptor may maintain a repressive SWI–SNF complex and inhibit p21 expression. Transient up-regulation of p21 in response to PDGF-BB may represent a feedback mechanism regulating proper cell cycle progression, which also has been observed in NIH 3T3 (Yu et al., 2003). We speculate that when Fer, TMF-1, or Brg-1 was depleted, the inhibitory SWI–SNF protein complex on the p21 promoter could not be maintained, which led to up-regulation of mRNA transcription even in unstimulated cells. Of note, we observed that the knockdown of Fer, TMF-1, or Brg-1 significantly decreased proliferation of fibroblasts in response to PDGF-BB, which was explained by up-regulation of the number of cells expressing p21 that may cause cell cycle block. TMF-1 has been reported to act as a tumor suppressor in cancer models (Abrham et al., 2009) and to suppress proliferation (Perry et al., 2004; Volpe et al., 2006), although we found a growth-suppressing effect in the absence of TMF-1, which demonstrates the complexity of pro- and antiproliferation signaling involving TMF-1–mediated growth control. Our results suggest a novel function of the Fer–TMF-1–Brg-1 axis in the regulation of tumor suppressor p21, which controls the progression of the cell cycle and is transiently regulated by the nuclear presence of PDGFRβ.
Interestingly, PDGF-BB–induced up-regulation of immediate early gene expression was not affected by the knockdown of TMF-1, as it was by inhibition of cytoplasmic PDGFRβ signaling via Erk MAP-kinase and PI3-kinase. It was postulated that the large group of immediate early genes is constitutively posed for transcription and does not require SWI–SNF-dependent chromatin remodeling to be transcribed (Ramirez-Carrozzi et al., 2009; Fowler et al., 2011). Thus, nuclear translocation of PDGFRβ and its interaction with TMF-1 implies that transcription of cell cycle regulators and subsequent progression of the cell cycle is not solely dependent on activation of cytoplasmic signaling pathways. In conclusion, we show that PDGFRβ traffics to the nucleus after PDGF-BB treatment. The presence of PDGFRβ in the nucleus may catalyze specific regulatory events at the nuclear matrix and chromatin that are necessary for modulation of gene transcription in response to growth stimuli. These findings have implications for our understanding of the mechanisms whereby PDGFRs regulate cell growth and proliferation.
Materials and methods
Cell culture, drug treatments, and PDGF stimulation
Normal human foreskin fibroblasts BJhTERT (Clontech) were used in all experiments except Fig. S3. Cells were cultured in DMEM (GIBCO BRL) supplemented with 10% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin and were routinely tested for mycoplasma contamination and found negative. Cells were seeded in 100-mm Petri dishes, starved in DMEM, supplemented with 0.1% FBS overnight, and stimulated at 50% confluence with 20 ng/ml of recombinant human PDGF-BB (gift from Amgen) for the indicated periods of time. To interfere with activation of PDGFRβ, cells were pretreated with the specific PDGFRβ kinase inhibitor AG1296 (10 µM) for 1 h before PDGF-BB stimulation. To enhance receptor phosphorylation in the absence of PDGF-BB stimulation, cells were treated with sodium pervanadate (1 mM hydrogen peroxide and 0.1 mM vanadate, freshly mixed) for 30 min. To block Golgi to ER retrograde traffic, cells were pretreated with 500 ng/ml brefeldin A (Sigma) for 1.5 h before PDGF-BB stimulation. To block lysosomal or proteasomal degradation, cells were pretreated for 3 h with 100 µM chloroquine or 25 µM MG132, respectively. To inhibit cytoplasmic signaling of PDGFRβ, cells were pretreated with 3 µM Erk1/2 MAP-kinase inhibitor CI1040 (Selleckchem) or 2 µM PI3-kinase inhibitor pictilisib (Selleckchem) before stimulation with PDGF-BB.
Antibodies
The following antibodies were used for the detection of total PDGFRβ: homemade rabbit polyclonal antibody, raised against a GST fusion of the C-terminal part of PDGFRβ (denoted ctβ; 78) and purified on a protein A agarose column (Pierce; used for IP and immunoblotting of streptavidin pull-downs); rabbit monoclonal anti-PDGFRβ clone Y92 (ab32570, Abcam; raised against the intracellular part of the PDGFRβ-abbreviated intra-PDGFRβ antibody, used for all immunoblots and immunofluorescence in Figs. 1 d and S1); polyclonal rabbit anti-PDGFRβ (sc-339; Santa Cruz, discontinued; used for immunofluorescence in Fig. 1 e); goat anti-PDGFRβ (AF358; R&D; raised against the extracellular part of PDGFRβ-abbreviated extra-PDGFRβ antibody, used for immunofluorescence in Figs. 1 d, 5 a, 9, and S2 c); mouse anti-PDGFRβ antibody (MAB1262; R&D; used for immunofluorescence in Fig. 5); and antiphospho-PDGFRβ site pTyr857 (Cell Signaling; used for immunoblotting). PDGFRβ detection in the nucleus was consistently reproduced by all antibodies used. Phosphotyrosine-specific mouse monoclonal antibody PY99, mouse anticlathrin heavy chain, mouse antikaryopherin (Kpn or β-importin), p21, and rabbit Fer antibody were from Santa Cruz. Antibodies against lamin A/C, Histone H3, Brg-1, BAF170, BAF155, and ARID1A were purchased from Cell Signaling, antibodies against TMF-1 were from ProteinTech, α-tubulin was from Sigma, and the anti-mouse organelle detection kit was from BD Transduction. Fluorescent antibodies were streptavidin–Alexa Fluor 488, anti–mouse IgG–Alexa Fluor 488 (green), anti–rabbit IgG–Alexa Fluor 546 (red), anti–rabbit Alexa Fluor 594 (red), and anti–rabbit Alexa Fluor 633 (far red) from ThermoScientific.
siRNA knockdown
For the knockdown of clathrin, cells were seeded in 100-mm Petri dishes and transfected the next day at ∼50% confluence with 20 nM clathrin heavy chain Silencer Select siRNA (Invitrogen) using 5 µl of SilentFect reagent (BioRad) in 10% FBS in DMEM. The next day, the media were replaced with 0.1% FBS in DMEM, and on day 2, cells were replated and retransfected with 20 nM siRNA, serum starved for 3 d, stimulated with PDGF-BB, and collected for nuclear fractionation. As control knockdown, stealth RNAi negative control #12935112 (Invitrogen) was used. For the knockdown of β-importin, cells were seeded as above and transfected with a mix of three siRNAs (4 nM each) from the Tri-silencer siRNA kit (Origene) using 5 µl of SilentFect reagent; the negative control was used as provided in the kit. The medium was replaced with 0.1% FBS in DMEM, and cells were stimulated with PDGF at 6 d after knockdown and collected for nuclear fractionation. For the knockdown of Arf-1, Fer, TMF-1, and Brg-1, cells were transfected with Tri-silencer siRNA kits in the same ways as for Kpn, but analyzed at 3 d after knockdown. Levels of the knockdown were analyzed by immunoblotting (clathrin, Fer, TMF-1, Brg-1, and β-importin) or quantitative PCR (Arf-1 and TMF-1).
Subcellular fractionation
Cells were seeded, starved, and stimulated as described above, washed two times with cold PBS, lysed in 1 ml of cytoplasmic lysis buffer (10 mM MES, pH 6.2, 10 mM NaCl, 1.5 mM MgCl2, 1 mM EDTA, 5 mM DTT, 1% Triton X-100, 1 mM pefablock [Sigma], and 1 mM NaF) for 10 min on ice, scraped into Eppendorf tubes, and centrifuged for 10 min at 3,000 rpm in a bench-top centrifuge. The supernatant was recentrifuged at 13,000 rpm for 20 min and collected as the cytoplasmic fraction. The pellet from the first centrifugation (nuclei) was washed three times with 1 ml of cytoplasmic lysis buffer supplemented with 1% NP-40 and once with cytoplasmic lysis buffer without addition of detergents. The purified nuclear pellet was resuspended in 0.5 ml of nuclear extraction buffer (25 mM Tris-HCl, pH 10.5, 1 mM EDTA, 0.5 M NaCl, 5 mM β-mercaptoethanol, and 0.5% Triton X-100), vortexed for 10 min at 4°C, and centrifuged for 20 min at 13,000 rpm, and the supernatant was collected as a nuclear fraction. This protocol was specifically optimized and regularly tested for the nuclear fraction not to contain any contamination of Golgi, ER, lipid rafts, and other organelles.
Immunoblotting
Equal volumes of cytoplasmic and nuclear extracts (adjusted by blotting with cytoplasmic and nuclear markers) were boiled in SDS sample buffer containing 10 mM DTT and separated by SDS-PAGE. Proteins were electrotransferred to polyvinylidene difluoride membrane (Immobilon P) and blocked for 1 h in 5% BSA in PBS containing 0.04% Tween 20. Membranes were incubated with primary antibody, diluted in 1% BSA in PBS at 4°C overnight, washed three times in PBS, and incubated with horseradish peroxidase–conjugated anti–rabbit (1:40,000 dilution) or anti–mouse (1:25,000 dilution) antibody (Amersham Pharmacia). Proteins were visualized using Super Signal West Dura ECL substrate (ThermoScientific), and membranes were scanned with a charge-coupled device camera Intelligent Dark Box II (Fujifilm). Raw data files were exported as TIFF files from Aida software, and level autocontrast was applied to the blots using Photoshop (Adobe).
IP and coimmunoprecipitation
Nuclear extracts were prepared as described in the subcellular fractionation protocol with the following modifications. Cytoplasmic buffer was 10 mM MES, pH 6.2, 10 mM NaCl, 1.5 mM MgCl2, 1 mM EDTA, 1% Triton X-100, and 1× Halt protease inhibitor cocktail (ThermoScientific); washing buffer was the same as cytoplasmic but without detergents; and nuclear extraction buffer was 25 mM Tris-HCl, pH 7.5, 1 mM EDTA, 0.5 M NaCl, 0.5% Triton X-100, and 1× Halt protease inhibitor cocktail. Nuclear extracts were diluted two times with neutralization buffer (100 mM Tris-HCl, pH 7.4, and 0.5% Triton X-100), and equal volumes of cytoplasmic (where applicable) or nuclear fractions were incubated overnight at 4°C with the antibodies at 1:100 dilution. 50 µl of 50% protein G–Sepharose slurry was added, and incubation was prolonged for 1 h at 4°C; beads were washed three times in IP washing buffer (10 mM Tris-HCl, pH 7.4, 250 mM NaCl, and 0.5% Triton X-100) and eluted in 40 µl of sample buffer containing 10 mM DTT. Eluates were resolved by SDS-PAGE and subjected to immunoblotting as described above.
Biotinylation of cell surface PDGFRβ
Cells were seeded in Petri dishes as above and starved overnight in DMEM supplemented with 0.1% FBS and washed two times in cold PBS. Cell-surface proteins were then biotinylated by incubation with 0.2 µg/ml EZ-link Sulfo-SS-Biotin (ThermoScientific). After 1 h at 4°C, biotinylation was blocked with 50 mM Tris-HCl, pH 7.5, warm media was replaced, and cells were stimulated with PDGF-BB for the indicated time periods. Cells were lysed and processed for nuclear fractionation. For analysis by immunofluorescence (Fig. 5), remaining cell surface biotin was stripped with 100 mM sodium 2-mercaptoethanesulfonate (MESNA) in 10 mM Tris, pH 7.0, 100 mM NaCl, and 1 mM EDTA at RT for 5 min; slides were fixed and processed as described below.
Immunofluorescence
Cells were grown on 10 × 10–mm coverslips, starved, and stimulated with 20 ng/ml PDGF-BB. Cells were then fixed in 3.5% paraformaldehyde for 10 min at RT, washed two times in PBS, and immunostained. For that, coverslips were permeabilized in 1 ml PBS, supplemented with 1% BSA and 0.1% SDS for 15 min at RT, and then blocked in 1 ml PBS supplemented with 1% BSA at RT for 45 min. Primary antibody at a 1:100 dilution was applied in PBS supplemented with 1% BSA overnight at 4°C. Coverslips were washed five times in PBS and incubated with a 1:200 dilution of fluorescent secondary antibody in 1% BSA in PBS for 50 min. Coverslips were washed five times with PBS, mounted with one drop of Vectashield mounting media with DAPI (Vector Labs), and analyzed by confocal microscopy.
Microscope image acquisition and processing
Images were acquired using ZEISS LSM510 (Figs. 1 g and S1) and LSM700 (Figs. 1 d, 4 c, 5 a–j, 6 a, 9 c–f, and S2 c) inverted confocal microscopes with numerical aperture 1.4 oil objectives at the Biological Visualization Facility at 512 × 512 and 1,128 × 1,128 pixels, respectively, at RT using Zen black software and a high-resolution AxioCam microscope camera (ZEISS). Images were exported as merged TIFF files with 8-bit resolution. Images were separated according to their colors in Photoshop software, and adjustment of brightness of individual color channels was performed equally on all images within each experiment.
For quantification of colocalization (Fig. 5), original black and white individual channel images were uploaded into Cell Profiler imaging software (Carpenter et al., 2006), and an automatic pipeline was created at the SciLife BioImage Informatics Facility, whereby TMF-1 staining was used to isolate regions of Golgi and DAPI staining was used to isolate the nucleus. Dot-like clusters of PDGFRβ were circled by the pipeline, and pairwise correlation was analyzed for PDGFR-biotin and PDGFR–TMF-1 channels within the circled areas. Correlation was evaluated by calculating the Pearson correlation coefficient, which is >0 for the possibility of colocalization and equal to 1 when the correlation is perfect. To increase the credibility of identified triple colocalized dots, the cutoff for triple colocalizations was placed at 0.2, meaning that the Pearson coefficient for each pair of channels for a triple colocalized dot was >0.2. The images presented in the figure were merged into an RGB image by the software.
For counting p21-expressing cells (Fig. 9), an automatic pipeline for Cell Profiler was created with the help of the SciLife BioImage Informatics Facility, whereby all DAPI-positive nuclei were counted versus p21-expressing nuclei (Fig. 9, c–g, colored in green) on RGB confocal fluorescent images, taken as described above.
Nuclear matrix isolation
Isolation of chromatin and nuclear matrix fractions was performed according to Reyes et al. (1997) with some modifications. Cells were grown in 100-mm Petri dishes, starved overnight at 50% of confluence, and stimulated with 20 ng/ml PDGF-BB for 30 min. After two washes in PBS, cells were extracted in 1 ml cytoplasmic extraction buffer (as described in the subcellular fractionation protocol) and washed once with the same buffer without detergents. The pellet was resuspended in 200 µl cytoplasmic lysis buffer (10 mM MES, pH 6.8, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl, 1 mM EGTA, 1 mM EDTA, 0.5% Triton X-100, 1 mM Pefablock, and 1 mM NaF), and chromatin was solubilized by DNA digestion with 20 U DNase I (New England Biolabs) for 15 min at 37°C. Proteins were extracted by addition of 50 µl of 1 M ammonium sulfate to a final concentration of 200 mM, and samples were pelleted again after 5 min on ice. Soluble chromatin fraction was collected, and the pellet was further extracted with 250 µl of 2 M NaCl in cytoplasmic lysis buffer for 5 min on ice and then centrifuged. This treatment removed DNA and histones from the nucleus and the supernatant was collected as a 2 M salt wash fraction. The remaining pellet was solubilized in 250 µl of 8 M urea, 0.1 M NaH2PO4, and 10 mM Tris, pH 8, and considered as a nuclear matrix–containing fraction. The chromatin, 2 M salt wash, and nuclear matrix fractions were diluted two times and analyzed by immunoblot along with the collected cytoplasmic fraction.
mRNA expression analysis
BJhTERT fibroblasts were transfected with 12 nM TMF-1 siRNA mix or 12 nM nontargeting negative control siRNA (Origene). After 48 h of transfection, cells were serum starved overnight and stimulated or not with 20 ng/ml PDGF-BB for 1 h and 3 h. Cells were lysed, and RNA was prepared with the NucleoSpin RNA Plus kit (Macherey Nagel) according to the manufacturer’s instructions. cDNA was prepared from 1 µg RNA using the PCR Biosystems cDNA kit, diluted five times, and used for quantitative PCR with the following primers: HPRT forward 5′-CCTGGCGTCGTGATTAGTGAT-3′, HPRT reverse 5′-AGACGTTCAGTCCTGTCCATAA-3′, CDKN1A forward 5′-TGTGAGCAGCTGCCGAAGTCA-3′, CDKN1A reverse 5′-TGACATGGCGCCTCCTCTGAGT-3′, C-FOS forward 5′-CGGGGATAGCCTCTCTTACT-3′, C-FOS reverse 5′-CCAGGTCCGTGCAGAAGTC-3′, NUR77 forward 5′-CTCTGGAGGTCATCCGCAAG-3′, NUR77 reverse 5′-CTGGCTTAGACCTGTACGCC-3′, AR forward 5′-GACGACCAGATGGCTGTCATT-3′, AR reverse 5′-GGGCGAAGTAGAGCATCCTG-3′, PDGFRβ forward 5′-AGCACACTGCGTCTGCAGCA-3′, and PDGFRβ reverse 5′-TGAGCACCACCAGGGCCAG-3′. The expression levels were calculated as the difference between the cycle threshold value for the control gene and the cycle threshold value of the test gene, taken to the power of 2. Expression levels of tested genes and the level of the TMF-1 knockdown were plotted as the mean of four biological replicates; standard deviation is indicated. T test statistical analysis was performed for the difference between mRNA expression levels of CDKN1A in control cells versus siTMF knockdown cells.
Proliferation assay
BJhTERT fibroblasts were seeded in triplicates in a 48-well plate (5,000 cells per well) 3 d after the transfection with Fer, TMF-1, or Brg-1 siRNAs and allowed to attach overnight. The media were replaced with 0.1% FBS in DMEM with increasing concentrations of PDGF-BB up to 20 ng/ml. Full-growth media (10% FBS in DMEM) was used as a control. On day 3 of incubation with PDGF-BB, cells were washed, frozen, and assayed with the CyQuant Cell Proliferation kit (ThermoScientific) as described by the manufacturer. The amount of DNA-incorporated fluorescent dye reflecting the number of cells was analyzed by measuring fluorescence at 480/520 nm with the EnSpire Multimode Reader (Perkin Elmer).
Online supplemental material
Fig. S1 contains controls for the purity of the nuclear extracts. Validation of nuclear traffic of PDGFR is presented in Fig. S2, as detected separately by two types of PDGFR antibodies used for costaining in Fig. 1 d. The generality of PDGFR nuclear traffic was confirmed for primary fibroblasts AG1523 and cancer cell lines glioblastoma U105MG and osteosarcoma U2OS, as presented in Fig. S3.
Supplementary Material
Acknowledgments
We thank Petter Ranefall from the SciLife BioImage Informatics Facility for creating automatic pipelines for quantification of immunofluorescence images and Maria Tsioumpekou for helpful discussions.
This work was supported by the Ludwig Institute for Cancer Research, the Swedish Cancer Society (grants 2016/445 and 140332), and the Swedish Research Council (grant 2015-02757).
The authors declare no competing financial interests.
Author contributions: N. Papadopoulos designed and performed the experiments and wrote the manuscript. J. Lennartsson and C.-H. Heldin supervised the work and corrected the manuscript.
References
- Abrham G., Volpe M., Shpungin S., and Nir U.. 2009. TMF/ARA160 downregulates proangiogenic genes and attenuates the progression of PC3 xenografts. Int. J. Cancer. 125:43–53. 10.1002/ijc.24277 [DOI] [PubMed] [Google Scholar]
- Aleksic T., Chitnis M.M., Perestenko O.V., Gao S., Thomas P.H., Turner G.D., Protheroe A.S., Howarth M., and Macaulay V.M.. 2010. Type 1 insulin-like growth factor receptor translocates to the nucleus of human tumor cells. Cancer Res. 70:6412–6419. 10.1158/0008-5472.CAN-10-0052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezney R., Mortillaro M.J., Ma H., Wei X., and Samarabandu J.. 1995. The nuclear matrix: a structural milieu for genomic function. Int. Rev. Cytol. 162A:1–65. [DOI] [PubMed] [Google Scholar]
- Bian X.-L., Chen H.-Z., Yang P.-B., Li Y.-P., Zhang F.-N., Zhang J.-Y., Wang W.-J., Zhao W.-X., Zhang S., Chen Q.-T., et al. 2017. Nur77 suppresses hepatocellular carcinoma via switching glucose metabolism toward gluconeogenesis through attenuating phosphoenolpyruvate carboxykinase sumoylation. Nat. Commun. 8:14420 10.1038/ncomms14420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulikas T. 1995. Chromatin domains and prediction of MAR sequences. Int. Rev. Cytol. 162A:279–388. [DOI] [PubMed] [Google Scholar]
- Carpenter G., and Liao H.-J.. 2013. Receptor tyrosine kinases in the nucleus. Cold Spring Harb. Perspect. Biol. 5:a008979 10.1101/cshperspect.a008979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter A.E., Jones T.R., Lamprecht M.R., Clarke C., Kang I.H., Friman O., Guertin D.A., Chang J.H., Lindquist R.A., Moffat J., et al. 2006. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7:R100 10.1186/gb-2006-7-10-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C., Olsen J.V., Brandts C., Cox J., Reddy P.N.G., Böhmer F.D., Gerke V., Schmidt-Arras D.-E., Berdel W.E., Müller-Tidow C., et al. 2009. Mislocalized activation of oncogenic RTKs switches downstream signaling outcomes. Mol. Cell. 36:326–339. 10.1016/j.molcel.2009.09.019 [DOI] [PubMed] [Google Scholar]
- De Angelis Campos A.C., Rodrigues M.A., de Andrade C., de Goes A.M., Nathanson M.H., and Gomes D.A.. 2011. Epidermal growth factor receptors destined for the nucleus are internalized via a clathrin-dependent pathway. Biochem. Biophys. Res. Commun. 412:341–346. 10.1016/j.bbrc.2011.07.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eger G., Papadopoulos N., Lennartsson J., and Heldin C.-H.. 2014. NR4A1 promotes PDGF-BB-induced cell colony formation in soft agar. PLoS One. 9:e109047 10.1371/journal.pone.0109047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euskirchen G.M., Auerbach R.K., Davidov E., Gianoulis T.A., Zhong G., Rozowsky J., Bhardwaj N., Gerstein M.B., and Snyder M.. 2011. Diverse roles and interactions of the SWI/SNF chromatin remodeling complex revealed using global approaches. PLoS Genet. 7:e1002008 10.1371/journal.pgen.1002008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester W.C., van Genderen C., Jenuwein T., and Grosschedl R.. 1994. Dependence of enhancer-mediated transcription of the immunoglobulin mu gene on nuclear matrix attachment regions. Science. 265:1221–1225. 10.1126/science.8066460 [DOI] [PubMed] [Google Scholar]
- Fowler T., Sen R., and Roy A.L.. 2011. Regulation of primary response genes. Mol. Cell. 44:348–360. 10.1016/j.molcel.2011.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridmann-Sirkis Y., Siniossoglou S., and Pelham H.R.B.. 2004. TMF is a golgin that binds Rab6 and influences Golgi morphology. BMC Cell Biol. 5:18 10.1186/1471-2121-5-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia J.A., Ou S.H., Wu F., Lusis A.J., Sparkes R.S., and Gaynor R.B.. 1992. Cloning and chromosomal mapping of a human immunodeficiency virus 1 “TATA” element modulatory factor. Proc. Natl. Acad. Sci. USA. 89:9372–9376. 10.1073/pnas.89.20.9372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri D.K., Ali-Seyed M., Li L.-Y., Lee D.-F., Ling P., Bartholomeusz G., Wang S.-C., and Hung M.-C.. 2005. Endosomal transport of ErbB-2: mechanism for nuclear entry of the cell surface receptor. Mol. Cell. Biol. 25:11005–11018. 10.1128/MCB.25.24.11005-11018.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes D.A., Rodrigues M.A., Leite M.F., Gomez M.V., Varnai P., Balla T., Bennett A.M., and Nathanson M.H.. 2008. c-Met must translocate to the nucleus to initiate calcium signals. J. Biol. Chem. 283:4344–4351. 10.1074/jbc.M706550200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan B., Wang T.-L., and Shih IeM.. 2011. ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodeling, is a tumor suppressor in gynecologic cancers. Cancer Res. 71:6718–6727. 10.1158/0008-5472.CAN-11-1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund K., Sigismund S., Polo S., Szymkiewicz I., Di Fiore P.P., and Dikic I.. 2003. Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat. Cell Biol. 5:461–466. 10.1038/ncb983 [DOI] [PubMed] [Google Scholar]
- Hao Q.L., Ferris D.K., White G., Heisterkamp N., and Groffen J.. 1991. Nuclear and cytoplasmic location of the FER tyrosine kinase. Mol. Cell. Biol. 11:1180–1183. 10.1128/MCB.11.2.1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldin C.H., and Westermark B.. 1999. Mechanism of action and in vivo role of platelet-derived growth factor. Physiol. Rev. 79:1283–1316. 10.1152/physrev.1999.79.4.1283 [DOI] [PubMed] [Google Scholar]
- Heldin C.-H., Lennartsson J., and Westermark B.. 2018. Involvement of platelet-derived growth factor ligands and receptors in tumorigenesis. J. Intern. Med. 283:16–44. 10.1111/joim.12690 [DOI] [PubMed] [Google Scholar]
- Hsiao P.W., and Chang C.. 1999. Isolation and characterization of ARA160 as the first androgen receptor N-terminal-associated coactivator in human prostate cells. J. Biol. Chem. 274:22373–22379. 10.1074/jbc.274.32.22373 [DOI] [PubMed] [Google Scholar]
- Jackson D.A., Hassan A.B., Errington R.J., and Cook P.R.. 1993. Visualization of focal sites of transcription within human nuclei. EMBO J. 12:1059–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoch C., Hargreaves D.C., Hodges C., Elias L., Ho L., Ranish J., and Crabtree G.R.. 2013. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 45:592–601. 10.1038/ng.2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapeller R., Chakrabarti R., Cantley L., Fay F., and Corvera S.. 1993. Internalization of activated platelet-derived growth factor receptor-phosphatidylinositol-3′ kinase complexes: potential interactions with the microtubule cytoskeleton. Mol. Cell. Biol. 13:6052–6063. 10.1128/MCB.13.10.6052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermorgant S., and Parker P.J.. 2008. Receptor trafficking controls weak signal delivery: a strategy used by c-Met for STAT3 nuclear accumulation. J. Cell Biol. 182:855–863. 10.1083/jcb.200806076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., and Imamoto N.. 2014. Biological significance of the importin-β family-dependent nucleocytoplasmic transport pathways. Traffic. 15:727–748. 10.1111/tra.12174 [DOI] [PubMed] [Google Scholar]
- Laemmli U.K., Käs E., Poljak L., and Adachi Y.. 1992. Scaffold-associated regions: cis-acting determinants of chromatin structural loops and functional domains. Curr. Opin. Genet. Dev. 2:275–285. 10.1016/S0959-437X(05)80285-0 [DOI] [PubMed] [Google Scholar]
- Lemmon M.A., and Schlessinger J.. 2010. Cell signaling by receptor tyrosine kinases. Cell. 141:1117–1134. 10.1016/j.cell.2010.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennartsson J., Ma H., Wardega P., Pelka K., Engström U., Hellberg C., and Heldin C.-H.. 2013. The Fer tyrosine kinase is important for platelet-derived growth factor-BB-induced signal transducer and activator of transcription 3 (STAT3) protein phosphorylation, colony formation in soft agar, and tumor growth in vivo. J. Biol. Chem. 288:15736–15744. 10.1074/jbc.M113.476424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L.-Y., Chen H., Hsieh Y.-H., Wang Y.-N., Chu H.-J., Chen Y.-H., Chen H.-Y., Chien P.-J., Ma H.-T., Tsai H.-C., et al. 2011. Nuclear ErbB2 enhances translation and cell growth by activating transcription of ribosomal RNA genes. Cancer Res. 71:4269–4279. 10.1158/0008-5472.CAN-10-3504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.Y., Makino K., Xia W., Matin A., Wen Y., Kwong K.Y., Bourguignon L., and Hung M.C.. 2001. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat. Cell Biol. 3:802–808. 10.1038/ncb0901-802 [DOI] [PubMed] [Google Scholar]
- Liu P., Ying Y., Ko Y.G., and Anderson R.G.. 1996. Localization of platelet-derived growth factor-stimulated phosphorylation cascade to caveolae. J. Biol. Chem. 271:10299–10303. 10.1074/jbc.271.17.10299 [DOI] [PubMed] [Google Scholar]
- Lo H.-W., Ali-Seyed M., Wu Y., Bartholomeusz G., Hsu S.-C., and Hung M.-C.. 2006. Nuclear-cytoplasmic transport of EGFR involves receptor endocytosis, importin beta1 and CRM1. J. Cell. Biochem. 98:1570–1583. 10.1002/jcb.20876 [DOI] [PubMed] [Google Scholar]
- Miaczynska M. 2013. Effects of membrane trafficking on signaling by receptor tyrosine kinases. Cold Spring Harb. Perspect. Biol. 5:a009035 10.1101/cshperspect.a009035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miaczynska M., Pelkmans L., and Zerial M.. 2004. Not just a sink: endosomes in control of signal transduction. Curr. Opin. Cell Biol. 16:400–406. 10.1016/j.ceb.2004.06.005 [DOI] [PubMed] [Google Scholar]
- Miller V.J., Sharma P., Kudlyk T.A., Frost L., Rofe A.P., Watson I.J., Duden R., Lowe M., Lupashin V.V., and Ungar D.. 2013. Molecular insights into vesicle tethering at the Golgi by the conserved oligomeric Golgi (COG) complex and the golgin TATA element modulatory factor (TMF). J. Biol. Chem. 288:4229–4240. 10.1074/jbc.M112.426767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K., and Kato H.. 2002. A putative nuclear receptor coactivator (TMF/ARA160) associates with hbrm/hSNF2 alpha and BRG-1/hSNF2 beta and localizes in the Golgi apparatus. FEBS Lett. 520:127–132. 10.1016/S0014-5793(02)02803-X [DOI] [PubMed] [Google Scholar]
- Mori S., Heldin C.H., and Claesson-Welsh L.. 1992. Ligand-induced polyubiquitination of the platelet-derived growth factor beta-receptor. J. Biol. Chem. 267:6429–6434. [PubMed] [Google Scholar]
- Nakayama Y., Kawana A., Igarashi A., and Yamaguchi N.. 2006. Involvement of the N-terminal unique domain of Chk tyrosine kinase in Chk-induced tyrosine phosphorylation in the nucleus. Exp. Cell Res. 312:2252–2263. 10.1016/j.yexcr.2006.03.021 [DOI] [PubMed] [Google Scholar]
- Packham S., Warsito D., Lin Y., Sadi S., Karlsson R., Sehat B., and Larsson O.. 2015. Nuclear translocation of IGF-1R via p150(Glued) and an importin-β/RanBP2-dependent pathway in cancer cells. Oncogene. 34:2227–2238. 10.1038/onc.2014.165 [DOI] [PubMed] [Google Scholar]
- Papadopoulos N., and Lennartsson J.. 2017. The PDGF/PDGFR pathway as a drug target. Mol. Aspects Med.:S0098-2997(17)30140-1 10.1016/j.mam.2017.11.007 [DOI] [PubMed] [Google Scholar]
- Perry E., Tsruya R., Levitsky P., Pomp O., Taller M., Weisberg S., Parris W., Kulkarni S., Malovani H., Pawson T., et al. 2004. TMF/ARA160 is a BC-box-containing protein that mediates the degradation of Stat3. Oncogene. 23:8908–8919. 10.1038/sj.onc.1208149 [DOI] [PubMed] [Google Scholar]
- Radha V., Nambirajan S., and Swarup G.. 1996. Association of Lyn tyrosine kinase with the nuclear matrix and cell-cycle-dependent changes in matrix-associated tyrosine kinase activity. Eur. J. Biochem. 236:352–359. 10.1111/j.1432-1033.1996.00352.x [DOI] [PubMed] [Google Scholar]
- Ramirez-Carrozzi V.R., Braas D., Bhatt D.M., Cheng C.S., Hong C., Doty K.R., Black J.C., Hoffmann A., Carey M., and Smale S.T.. 2009. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 138:114–128. 10.1016/j.cell.2009.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando O.J., Zhao K., Janmey P., and Crabtree G.R.. 2002. Phosphatidylinositol-dependent actin filament binding by the SWI/SNF-like BAF chromatin remodeling complex. Proc. Natl. Acad. Sci. USA. 99:2824–2829. 10.1073/pnas.032662899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes J.C., Muchardt C., and Yaniv M.. 1997. Components of the human SWI/SNF complex are enriched in active chromatin and are associated with the nuclear matrix. J. Cell Biol. 137:263–274. 10.1083/jcb.137.2.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger J., and Lemmon M.A.. 2006. Nuclear signaling by receptor tyrosine kinases: the first robin of spring. Cell. 127:45–48. 10.1016/j.cell.2006.09.013 [DOI] [PubMed] [Google Scholar]
- Schwartz Y., Ben-Dor I., Navon A., Motro B., and Nir U.. 1998. Tyrosine phosphorylation of the TATA element modulatory factor by the FER nuclear tyrosine kinases. FEBS Lett. 434:339–345. 10.1016/S0014-5793(98)01003-5 [DOI] [PubMed] [Google Scholar]
- Sehat B., Tofigh A., Lin Y., Trocmé E., Liljedahl U., Lagergren J., and Larsson O.. 2010. SUMOylation mediates the nuclear translocation and signaling of the IGF-1 receptor. Sci. Signal. 3:ra10 10.1126/scisignal.2000628 [DOI] [PubMed] [Google Scholar]
- Sigismund S., Argenzio E., Tosoni D., Cavallaro E., Polo S., and Di Fiore P.P.. 2008. Clathrin-mediated internalization is essential for sustained EGFR signaling but dispensable for degradation. Dev. Cell. 15:209–219. 10.1016/j.devcel.2008.06.012 [DOI] [PubMed] [Google Scholar]
- Simone C. 2006. SWI/SNF: the crossroads where extracellular signaling pathways meet chromatin. J. Cell. Physiol. 207:309–314. 10.1002/jcp.20514 [DOI] [PubMed] [Google Scholar]
- Sorkin A., Westermark B., Heldin C.H., and Claesson-Welsh L.. 1991. Effect of receptor kinase inactivation on the rate of internalization and degradation of PDGF and the PDGF beta-receptor. J. Cell Biol. 112:469–478. 10.1083/jcb.112.3.469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachowiak M.K., Maher P.A., Joy A., Mordechai E., and Stachowiak E.K.. 1996a Nuclear localization of functional FGF receptor 1 in human astrocytes suggests a novel mechanism for growth factor action. Brain Res. Mol. Brain Res. 38:161–165. 10.1016/0169-328X(96)00010-1 [DOI] [PubMed] [Google Scholar]
- Stachowiak M.K., Maher P.A., Joy A., Mordechai E., and Stachowiak E.K.. 1996b Nuclear accumulation of fibroblast growth factor receptors is regulated by multiple signals in adrenal medullary cells. Mol. Biol. Cell. 7:1299–1317. 10.1091/mbc.7.8.1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachowiak M.K., Maher P.A., and Stachowiak E.K.. 2007. Integrative nuclear signaling in cell development--a role for FGF receptor-1. DNA Cell Biol. 26:811–826. 10.1089/dna.2007.0664 [DOI] [PubMed] [Google Scholar]
- Strouboulis J., and Wolffe A.P.. 1996. Functional compartmentalization of the nucleus. J. Cell Sci. 109:1991–2000. [DOI] [PubMed] [Google Scholar]
- Volpe M., Shpungin S., Barbi C., Abrham G., Malovani H., Wides R., and Nir U.. 2006. trnp: A conserved mammalian gene encoding a nuclear protein that accelerates cell-cycle progression. DNA Cell Biol. 25:331–339. 10.1089/dna.2006.25.331 [DOI] [PubMed] [Google Scholar]
- Wang S.-C., Nakajima Y., Yu Y.-L., Xia W., Chen C.-T., Yang C.-C., McIntush E.W., Li L.-Y., Hawke D.H., Kobayashi R., and Hung M.-C.. 2006. Tyrosine phosphorylation controls PCNA function through protein stability. Nat. Cell Biol. 8:1359–1368. 10.1038/ncb1501 [DOI] [PubMed] [Google Scholar]
- Wang Y.-N., Wang H., Yamaguchi H., Lee H.-J., Lee H.-H., and Hung M.-C.. 2010a COPI-mediated retrograde trafficking from the Golgi to the ER regulates EGFR nuclear transport. Biochem. Biophys. Res. Commun. 399:498–504. 10.1016/j.bbrc.2010.07.096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.-N., Yamaguchi H., Hsu J.-M., and Hung M.-C.. 2010b Nuclear trafficking of the epidermal growth factor receptor family membrane proteins. Oncogene. 29:3997–4006. 10.1038/onc.2010.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.-N., Yamaguchi H., Huo L., Du Y., Lee H.-J., Lee H.-H., Wang H., Hsu J.-M., and Hung M.-C.. 2010c The translocon Sec61beta localized in the inner nuclear membrane transports membrane-embedded EGF receptor to the nucleus. J. Biol. Chem. 285:38720–38729. 10.1074/jbc.M110.158659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.-N., Lee H.-H., Lee H.-J., Du Y., Yamaguchi H., and Hung M.-C.. 2012. Membrane-bound trafficking regulates nuclear transport of integral epidermal growth factor receptor (EGFR) and ErbB-2. J. Biol. Chem. 287:16869–16879. 10.1074/jbc.M111.314799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warsito D., Lin Y., Gnirck A.-C., Sehat B., and Larsson O.. 2016. Nuclearly translocated insulin-like growth factor 1 receptor phosphorylates histone H3 at tyrosine 41 and induces SNAI2 expression via Brg1 chromatin remodeling protein. Oncotarget. 7:42288–42302. 10.18632/oncotarget.9785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson B.G., and Roberts C.W.M.. 2011. SWI/SNF nucleosome remodellers and cancer. Nat. Rev. Cancer. 11:481–492. 10.1038/nrc3068 [DOI] [PubMed] [Google Scholar]
- Wong M., and Munro S.. 2014. Membrane trafficking. The specificity of vesicle traffic to the Golgi is encoded in the golgin coiled-coil proteins. Science. 346:1256898 10.1126/science.1256898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane J., Kubo A., Nakayama K., Yuba-Kubo A., Katsuno T., Tsukita S., and Tsukita S.. 2007. Functional involvement of TMF/ARA160 in Rab6-dependent retrograde membrane traffic. Exp. Cell Res. 313:3472–3485. 10.1016/j.yexcr.2007.07.010 [DOI] [PubMed] [Google Scholar]
- Yu J., Liu X.-W., and Kim H.-R.C.. 2003. Platelet-derived growth factor (PDGF) receptor-alpha-activated c-Jun NH2-terminal kinase-1 is critical for PDGF-induced p21WAF1/CIP1 promoter activity independent of p53. J. Biol. Chem. 278:49582–49588. 10.1074/jbc.M309986200 [DOI] [PubMed] [Google Scholar]
- Zhao K., Wang W., Rando O.J., Xue Y., Swiderek K., Kuo A., and Crabtree G.R.. 1998. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 95:625–636. 10.1016/S0092-8674(00)81633-5 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.