Cells are thought to adapt to proteasome inhibition by using alternative pathways for degradation such as autophagy. Sha et al. now report that cells rapidly induce GABARAPL1 and p62 upon proteasome inhibition, but this promotes cell survival by sequestering ubiquitinated and sumoylated proteins long before the cells induce other Atg genes and activate autophagy.
Abstract
Proteasome inhibitors are used as research tools and to treat multiple myeloma, and proteasome activity is diminished in several neurodegenerative diseases. We therefore studied how cells compensate for proteasome inhibition. In 4 h, proteasome inhibitor treatment caused dramatic and selective induction of GABARAPL1 (but not other autophagy genes) and p62, which binds ubiquitinated proteins and GABARAPL1 on autophagosomes. Knockdown of p62 or GABARAPL1 reduced cell survival upon proteasome inhibition. p62 induction requires the transcription factor nuclear factor (erythroid-derived 2)-like 1 (Nrf1), which simultaneously induces proteasome genes. After 20-h exposure to proteasome inhibitors, cells activated autophagy and expression of most autophagy genes by an Nrf1-independent mechanism. Although p62 facilitates the association of ubiquitinated proteins with autophagosomes, its knockdown in neuroblastoma cells blocked the buildup of ubiquitin conjugates in perinuclear aggresomes and of sumoylated proteins in nuclear inclusions but did not reduce the degradation of ubiquitinated proteins. Thus, upon proteasome inhibition, cells rapidly induce p62 expression, which enhances survival primarily by sequestering ubiquitinated proteins in inclusions.
Introduction
Most protein breakdown in mammalian cells is catalyzed by the 26S proteasome, which selectively hydrolyzes proteins attached with ubiquitin (Ub) chains. Proteasomal degradation is essential for cell viability, and proteasome inhibitors can induce apoptosis (Manasanch and Orlowski, 2017). Multiple myeloma is a cancer of plasma cells that is particularly dependent on proteasome function because these cells produce and continually degrade large amounts of abnormal Igs (Goldberg, 2012). Consequently, these cells are particularly sensitive to proteasome inhibitors, and the introduction of bortezomib (BTZ) and carfilzomib (CFZ) dramatically improved myeloma treatment. However, a major limitation with these agents is the emergence of resistant cells by mechanisms still unexplained (Manasanch and Orlowski, 2017). Therefore, understanding cellular adaptations that enhance survival upon proteasome inhibition may lead to improved therapies, and may also increase our understanding of various neurodegenerative diseases, where the buildup of misfolded, aggregation-prone proteins can impair proteasome activities and cause a failure of protein homeostasis and loss of neuronal viability (Myeku et al., 2016). Because proteasome inhibitors are very widely used as research tools, knowledge of these cellular adaptations should also be of wide interest to biologists.
One important cellular adaptation to reduced proteasome activity is to increase the production of new proteasomes by stimulating the transcription of genes for proteasome subunits and the p97–VCP complex via the transcription factor nuclear factor (erythroid-derived 2)-like 1 (Nrf1; Radhakrishnan et al., 2010). Cells also degrade cytosolic proteins via autophagy. In this process, a portion of the cytoplasm or organelles are enclosed in a double-membrane structure, the autophagosome, which then fuses with lysosomes. More than 30 autophagy-related proteins (Atgs) function sequentially in the formation of the autophagosome (Wang and Klionsky, 2003). Although autophagy was initially viewed as a nonspecific process that provides nutrients, especially during starvation (Klionsky and Ohsumi, 1999), it also selectively degrades protein aggregates, viruses, bacteria, and organelles if they are tagged with a Ub chain. In mammalian cells, four proteins, p62, Nbr1, NDP52, and optineurin (OPTN), can bind ubiquitinated proteins and facilitate their degradation in autophagosomes (Rogov et al., 2014). These Ub receptors form homo- or heterooligomers and thus promote the formation of centrosome-localized inclusions, often termed aggresomes (Strnad et al., 2008; Richter-Landsberg and Leyk, 2013; Lu et al., 2017). Inclusion formation may limit the toxicity of these nondegraded proteins (Kopito, 2000; Nakaso et al., 2004; Richter-Landsberg and Leyk, 2013), but their degradation is also facilitated by Ub receptors that bind to the various Atg8 proteins (LC3A/B/C, GABARAP, and GABARAPL1/L2) on immature autophagosomes (Pankiv et al., 2007). Because the autophagy process consumes these Ub receptors and Atg8 proteins (Rogov et al., 2014), their continual production appears crucial for cells to sustain the capacity of autophagy.
Activation of autophagy can thus be a compensatory mechanism to help cells eliminate Ub conjugates that accumulate after proteasome inhibition. Many investigators have reported activation of autophagy in cells treated with proteasome inhibitors (Fels et al., 2008; Harada et al., 2008; Ding et al., 2009; Hoang et al., 2009; Milani et al., 2009; Belloni et al., 2010; Zhu et al., 2010). However, others reported no increase in lysosomal protein degradation upon BTZ treatment for many hours (Tsvetkov et al., 2015). It is also unclear whether this activated autophagy enhances Ub conjugate clearance and promotes survival, or whether it is a pathological response linked to autophagic cell death (Hoang et al., 2009; Belloni et al., 2010). Furthermore, it is unclear whether proteasome inhibition causes cells to induce the expression of certain Atg genes, especially Atg8 genes and Ub receptors. No studies have systematically measured the induction of all of them.
We therefore investigated whether, upon proteasome inhibition, cells enhance the expression of some or all Atg genes and Ub receptors (e.g., p62) to degrade ubiquitinated proteins. We performed this systematic transcriptional analysis primarily in myeloma cells, because proteasome inhibitors are used worldwide to treat myeloma patients, and neuroblastoma cells, because the buildup of misfolded proteins in neurons often impairs proteasome activity in neurodegenerative diseases. Therefore, the adaptations to decreased proteasome function are important to define in these lines that differ markedly in their susceptibility to proteasome inhibitors. Our goals were to determine when autophagy was activated after proteasome inhibition, whether specific Atg genes are induced, whether these responses enhance the degradation of the Ub conjugates, and whether they influence cell killing. We also investigated the transcription factors involved in these adaptations. Although several transcription factors have been reported to induce the expression of certain Atg genes or p62 under various stressful conditions, it is unclear whether they also function upon proteasome inhibition and whether genes for autophagy, p62, and proteasome subunits are coordinately induced under these conditions.
Results
Proteasome inhibition rapidly induced p62 and GABARAPL1
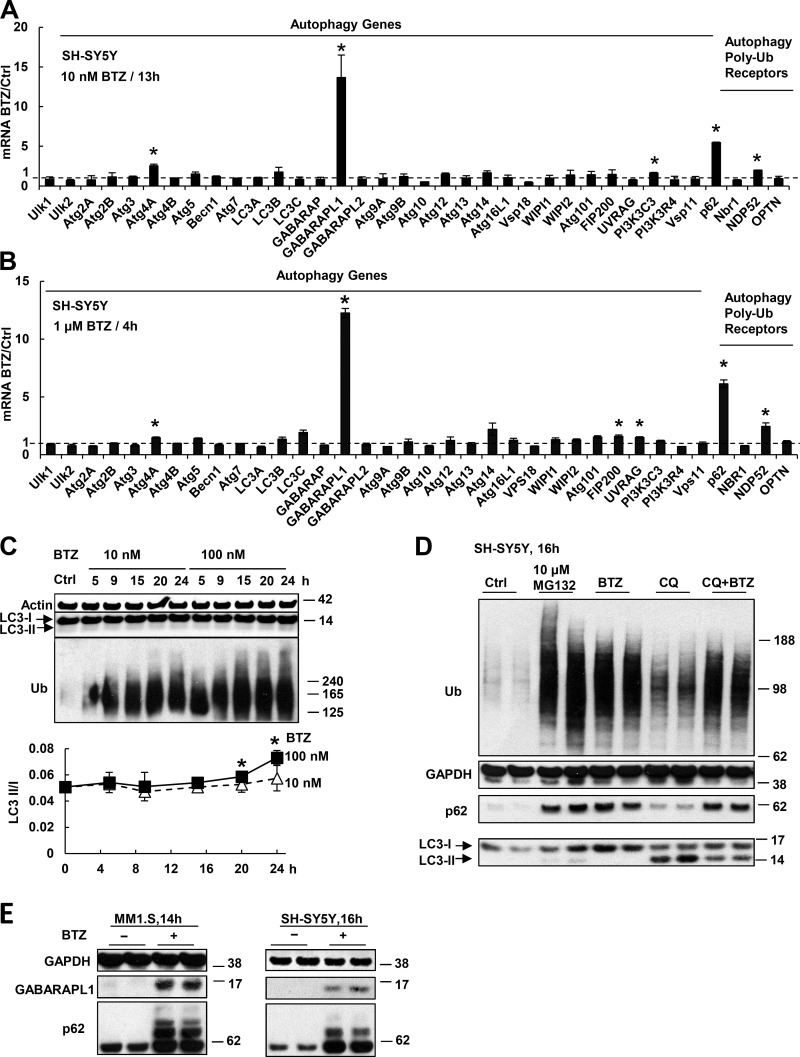
We tested whether proteasome inhibition, which reportedly activates autophagy (Iwata et al., 2005; Pandey et al., 2007), stimulated the expression of autophagy genes, p62, and related Ub receptors. When SH-SY5Y neuroblastoma cells were treated with a low concentration (10 nM) of BTZ for 13 h, they induced the mRNAs of p62 5–10-fold, and one of its binding proteins on the autophagosome, GABARAPL1, >10-fold (Fig. 1 A). When these cells were treated with a much higher concentration (1 µM) of BTZ, GABARAPL1 and p62 were induced to a similar extent, but within 4 h (Fig. 1 B). There was also a much weaker induction of Atg4A (1.5–2-fold) and NDP52 (three- to fourfold; Fig. 1, A and B). By 4 h, SH-SY5Y cells also induced the mRNAs of all proteasome subunits and the p97–VCP complex approximately two- to fourfold (Sha and Goldberg, 2014). In contrast, none of the 30 other Atg genes was induced significantly by either treatment (Fig. 1, A and B). We then measured autophagic activity by assaying the ratio of the lipidated autophagosome-bound LC3-II to nonlipidated LC3-I. SH-SY5Y cells have a low basal LC3-II/I ratio of 0.05, which was strikingly not elevated after treatment with 10 nM BTZ for 24 h, and with 100 nM BTZ, it increased by only 20% at 20 h and 40% at 24 h (Fig. 1 C). When substrate flux through autophagy was measured by treating SH-SY5Y cells with chloroquine, which raises lysosomal pH and inhibits the degradation of LC3-II, its level also did not increase after treatment with 10 nM BTZ for 16 h (Fig. 1 D). Therefore, the induction of p62 and GABARAPL1 mRNAs precedes by many hours the activation of autophagy.
Figure 1.
Proteasome inhibitor treatment causes rapid induction of p62 and GABARAPL1 but not other Atg genes or Ub receptors. (A and B) SH-SY5Y cells were treated with 10 nM BTZ for 13 h (A) or 1 µM for 4 h (B). The mRNAs of all Atg proteins or Ub receptors were measured in this and other figures by real time RT-PCR. (C) Autophagy was activated only after prolonged treatment with a high concentration of proteasome inhibitor. SH-SY5Y cells were treated with 10 or 100 nM BTZ for 5–24 h. To measure autophagy, the levels of LC3-I and lipidated (autophagosome-bound) LC3-II were measured (upper) by Western blotting, and the LC3-II/I ratio was quantified (lower). *, LC3-II/I ratio in cells treated with 100 nM BTZ is higher than that in untreated cells (P < 0.05; n = 2). (D) Treating SH-SY5Y cells for 16 h with 50 µM chloroquine (CQ) prevented the degradation of LC3-II, but cells treated with BTZ (10 nM) and CQ did not accumulate more LC3-II than cells treated only with CQ. (E) After SH-SY5Y or MM1.S cells were treated with 10 nM BTZ, levels of p62 and GABARAPL1 proteins were greatly increased. *, P < 0.05. Molecular masses are given in kilodaltons. Error bars indicate SD.
Neuroblastoma cells are resistant to killing by BTZ and do not lose viability by 36-h treatment with 500 nM BTZ. In contrast, half the myeloma (MM1.S) cells were killed by 20-h treatment with 10 nM BTZ (Fig. S1 A). Nevertheless, treatment of MM1.S cells with 10 nM BTZ for 13 h also induced 10-fold mRNAs for p62 and GABARAPL1 but not other Atg genes or Ub adapters (Fig. S1 B). This selective induction of p62 and GABARAPL1 preceded the massive cell death (Fig. S1 A) and the activation of autophagy, as shown by LC3-II level (Fig. S1 C). Treatment of these cells with other proteasome inhibitors CFZ or CEP18770, like BTZ, also caused a dramatic induction of p62 and GABARAPL1 but not other Atg genes (Fig. S1 D). Thus, the rapid induction of p62 and GABARAPL1 is part of a general response to proteasome inhibition that occurs long before the activation of autophagy. These large changes in gene expression dramatically elevated p62 and GABARAPL1 proteins in SH-SY5Y and MM1.S cells (Fig. 1 E).
In these BTZ-treated cells, some p62 migrated more slowly, apparently because of its ubiquitination, as was also reported recently (Lee et al., 2017; Peng et al., 2017). Accordingly, treatment of the cell lysate with the deubiquitinase USP2 eliminated the slowly migrating forms (Fig. S1 E). p62 ubiquitination was not simply a consequence of its dramatic buildup because blocking translation with cycloheximide failed to prevent the ubiquitination of preexistent p62 in BTZ-treated cells (Fig. S1 F). The ubiquitination of p62 was proposed to increase its ability to sequester Ub conjugates in aggregates (Peng et al., 2017). However, after BTZ treatment, both ubiquitinated and unmodified p62 are primarily in the aggregate fraction (10,000 g pellet in 1% Triton X-100), whereas the percentage of p62 ubiquitination was only slightly higher than in the soluble fraction (Fig. S1 G).
Prolonged proteasome inhibition causes induction of all autophagy genes and certain lysosomal components but not HDAC6
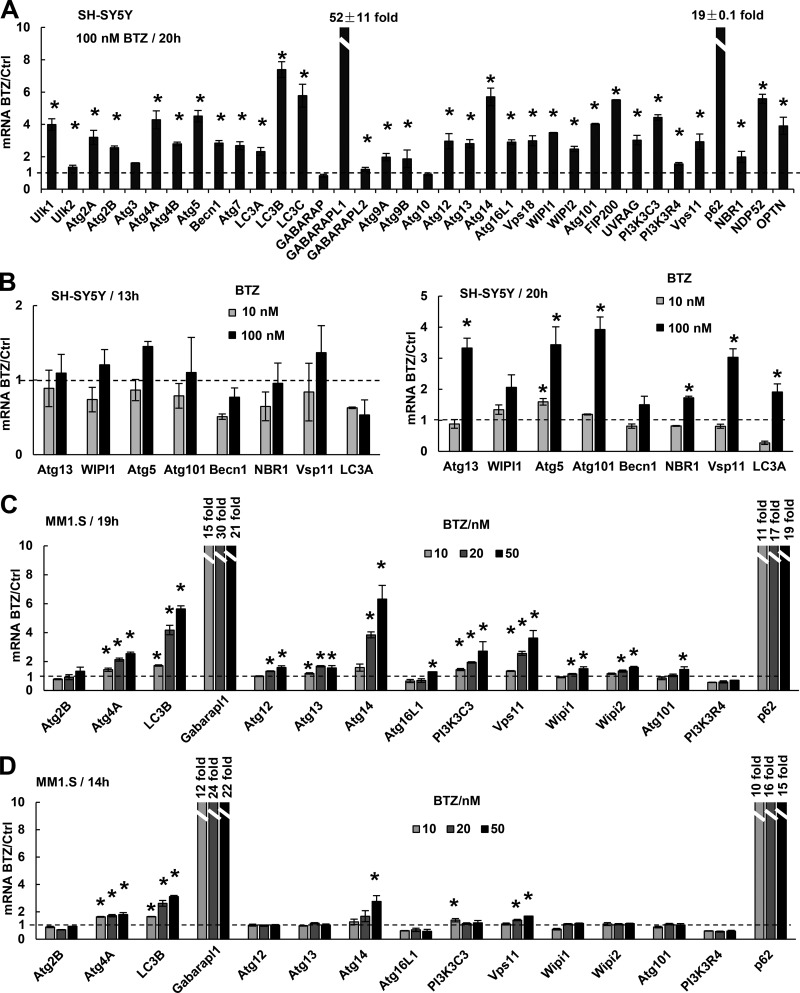
Proteasome inhibitors were reported to cause induction of the Atg genes Atg5, Atg7 (Zhu et al., 2010), and LC3B (Milani et al., 2009), none of which was rapidly induced in our research. Therefore, we tested whether these genes are induced with longer treatments or higher concentrations. When SH-SY5Y cells were exposed to 100 nM BTZ for 20 h, the mRNAs for all Atg genes except GABARAP and Atg10 were induced severalfold, although still much less than GABARAPL1 (52-fold) and p62 (19-fold; Fig. 2 A). These various Atg genes were induced by 30 nM BTZ (Fig. S2 A), which inhibited the chymotrypsin-like and caspase-like activities by >80% (Fig. S2 B) and should decrease the overall rate of protein degradation in cells by >50% (Kisselev et al., 2006). However, 10 nM BTZ, which inhibited these peptidase activities much less, did not induce Atg genes. Their induction became evident only at 20 h even when cells were treated with 100 nM BTZ (Fig. 2 B). Unlike neuroblastoma lines, MM1.S cells are very sensitive to proteasome inhibition, and >50% were killed when exposed to 20 nM BTZ for 19 h or 50 nM for 14 h (Fig. S2 C). These prolonged treatments with 20 or 50 nM but not 10 nM BTZ were necessary to induce the mRNAs for most Atg genes (Fig. 2, C and D). Thus, in both myeloma and neuroblastoma cells, to stimulate expression of all Atg genes, proteasome inhibition must be strong and prolonged.
Figure 2.
Prolonged and strong inhibition of the proteasome causes cells to induce the mRNAs for nearly all Atg genes and Ub receptors. (A) When treated with 100 nM BTZ for 20 h, SH-SY5Y cells induced the mRNAs for nearly all Atg genes and Ub receptors but less than did p62 and GABARAPL1. (B) SH-SY5Y cells induced the mRNAs of most Atg genes when treated with 100 nM BTZ for 20 h (right). Reducing the BTZ concentration to 10 nM (right) or the treatment time to 13 h (left) prevented induction. (C and D) MM1.S cells induced the mRNAs of most Atg genes when treated with 20 or 50 nM BTZ for 19 h, although much less than GABARAPL1 and p62 (C). Reducing the BTZ concentration to 10 nM (C) or the treatment time to 14 h (D) failed to induce most genes. In this figure and Fig. 3, the exact fold increase is shown for p62 and GABARAPL1 because their mRNAs increase >10-fold. *, P < 0.05. Error bars indicate SD.
Because proteolysis by autophagy requires lysosomal enzymes and lysosomal genes are expressed coordinately with autophagy genes under some conditions (Settembre et al., 2011), we assayed mRNAs for three lysosomal proteases, cathepsin A, D, and F, and the nonhydrolytic lysosomal components Lamp1, mColn1, and Clcn7 after treating SH-SY5Y cells with BTZ. Exposure to 1 µM BTZ for 4 h, which induced p62 and GABARAPL1 mRNAs, did not induce these lysosomal genes (Fig. S2 D). However, most of these lysosomal genes were induced upon treatment with 100 nM BTZ for 20 h (Fig. S2 E), together with all Atg genes. Another protein reported to stimulate the degradation of Ub conjugates by autophagy is HDAC6 (Kawaguchi et al., 2003; Lee et al., 2010). However, HDAC6 mRNA decreased in MM1.S and SH-SY5Y cells treated with 10 nM BTZ for 13 h (Fig. S2 F) and in SH-SY5Y cells treated with 100 nM BTZ for 20 h (Fig. S2 G).
Similar observations were obtained in the myeloma line RPMI 8226
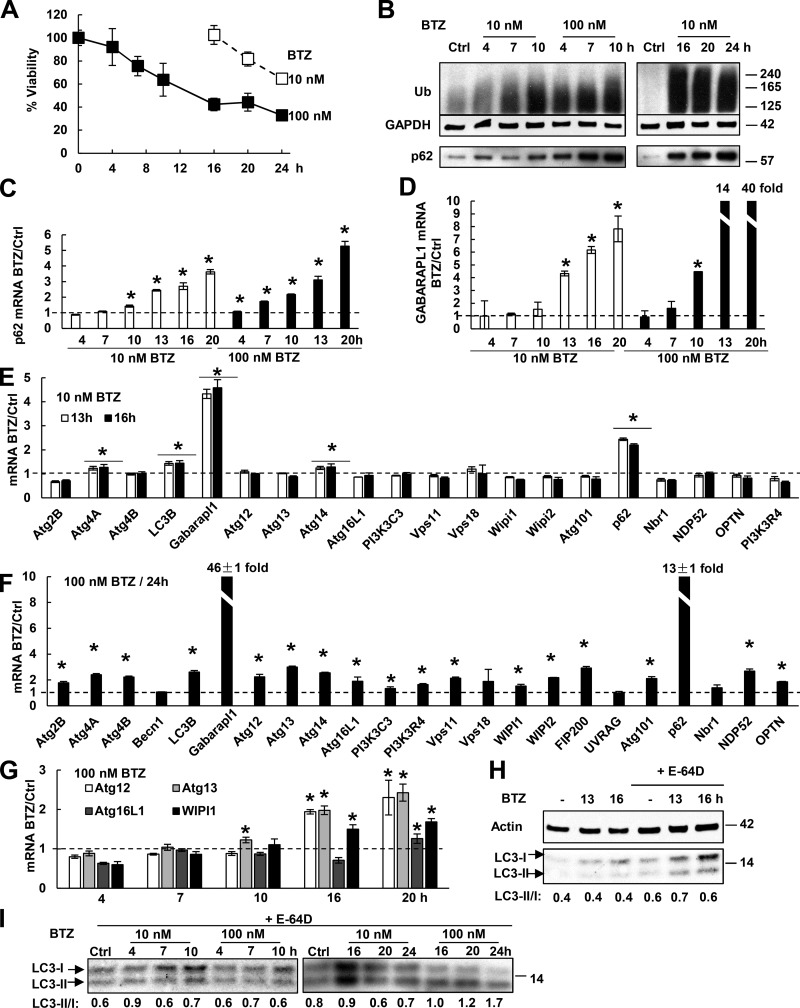
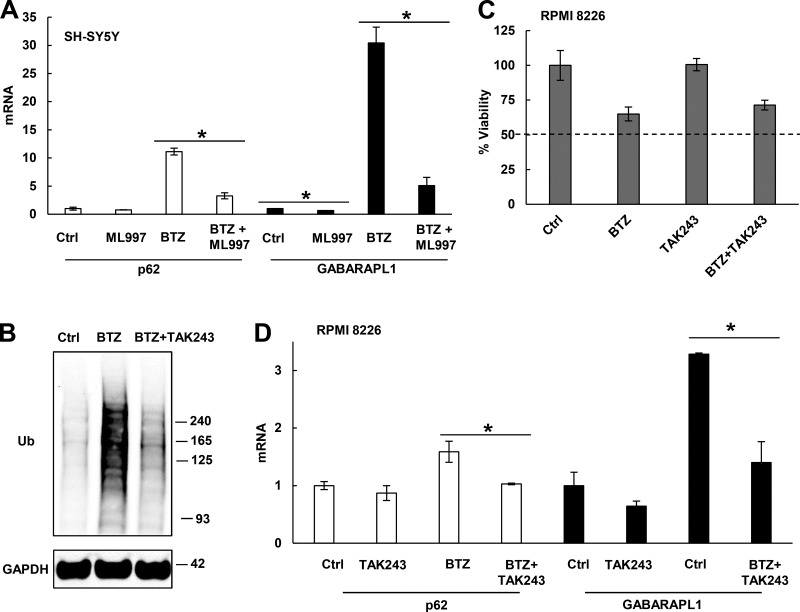
To test whether these findings also apply to other myeloma cells, we studied RPMI 8226 cells, which are less sensitive to killing by BTZ than MM1.S cells and retain >60% viability in 10 nM BTZ for 24 h and >40% viability in 100 nM BTZ for 20 h (Fig. 3 A). When these cells were treated with 10 nM BTZ for 10 h or with 100 nM BTZ for 4 h, p62 mRNA and protein levels were elevated (Fig. 3, B and C). GABARAPL1 mRNA increased more than fourfold when treated with 10 nM BTZ for 13 h or 100 nM BTZ for 10 h (Fig. 3 D). However, when treated with 10 nM BTZ, mRNAs of other Atg genes or Ub receptors did not increase (despite a small increase in Atg4A, LC3B, and Atg14; Fig. 3 E). In contrast, RPMI 8226 cells treated with 100 nM BTZ for 24 h induced mRNAs of nearly all Atg genes and Ub receptors (Fig. 3 F). The expression of most (three of four) Atg genes rose only by 16-h exposure to these high concentrations (Fig. 3 G). To measure the activation of autophagy in the BTZ-treated cells, lysosomal proteolysis was arrested for 1 h with E-64D (10 µM), which inhibited major lysosomal cathepsins and prevented LC3-II degradation (Fig. 3 H). The LC3-II/I ratio rose twofold upon treatment with 100 nM BTZ for 20 h and almost threefold by 24 h (Fig. 3 I), but did not change with a shorter treatment or with a low concentration (10 nM) for 24 h.
Figure 3.
Upon BTZ treatment, RPMI 8226 cells rapidly induce p62 and GABARAPL1, but only after prolonged treatment do they induce other Atg genes and activate autophagy. (A) The effect of BTZ treatment on the viability of RPMI 8226 cells was measured by MTS assay. n = 3. (B) BTZ treatment (especially with high concentrations) caused Ub conjugate buildup within 4–7 h and increased p62 protein levels. (C and D) Upon BTZ treatment, cells rapidly induced mRNAs for p62 (C) and GABARAPL1 (D). (E) Cells induced mRNAs for p62 and GABARAPL1, but not most other Atg genes when treated with 10 nM BTZ for 13 or 16 h. (F) Cells treated with 100 nM BTZ for 24 h induced almost all Atg genes. (G) Cells treated with 100 nM BTZ induced most Atg genes only at 16 or 20 h. (H) Treating control or BTZ-treated cells with 10 µM E-64D for 1 h to inhibit lysosomal proteases increased the LC3-II level and the LC3-II/I ratio. (I) The LC3-II/I ratio was measured after E-64D treatment as in H to evaluate autophagic activity. For this ratio to rise twofold, cells need to be treated with a high concentration (100 nM) of BTZ for ≥20 h. *, P < 0.05. Molecular masses are given in kilodaltons. Error bars indicate SD.
Thus, in all cells studied, proteasome inhibition causes two distinct transcriptional responses: (1) even a partial inhibition stimulated rapid expression of GABARAPL1 and p62 without increasing autophagosome formation; and (2) after prolonged and more complete inhibition, all Atg and some lysosomal genes are coordinately up-regulated, and there is increased autophagosome formation as indicated by elevated LC3-II content. By this time, myeloma cells show appreciable cell death, but none was evident in neuroblastoma lines.
GABARAPL1 and p62 but not Nbr1 promote cell survival upon proteasome inhibition
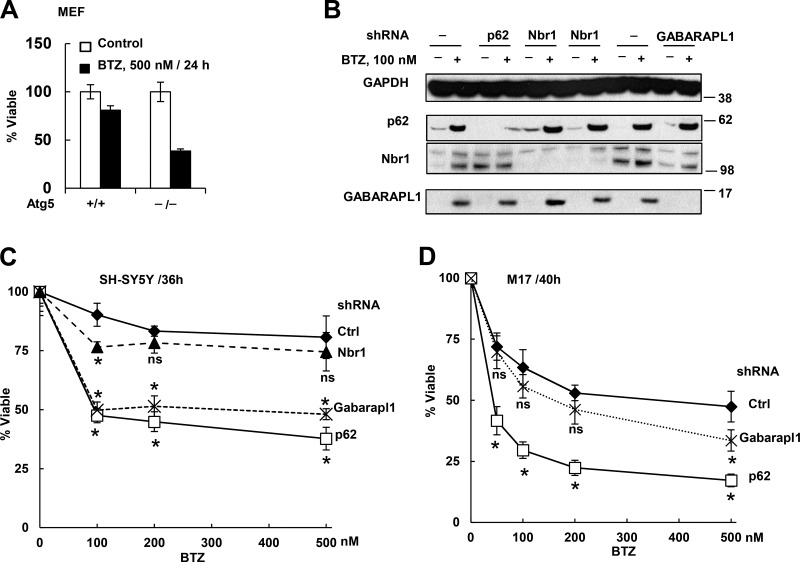
Despite its delayed activation after proteasome inhibition, autophagy may promote cell survival by helping eliminate the nondegraded Ub conjugates, or it may contribute to cell death occurring with prolonged proteasome inhibition. A protective role for the activation of autophagy seems more likely because mouse embryonic fibroblasts (MEFs) that cannot undergo autophagy as a result of the knockout of Atg5 are much more sensitive to killing by BTZ treatment than WT MEFs (Fig. 4 A). Also, no death was evident even when SH-SY5Y cells were treated with 100 nM BTZ for 20 h (Sha and Goldberg, 2014). Thus, the increased autophagy is not linked to apoptosis, even though in the MM1.S cells, the increase in autophagy after 20-h treatment with BTZ (10 nM) occurred simultaneously with widespread cell death (Fig. S1, A and C).
Figure 4.
Autophagy deficiency (Atg5−/− MEFs) or knockdown of p62 or GABARAPL1 but not Nbr1 increased cell sensitivity to killing by BTZ. (A) Atg5−/− MEFs lost viability much more than WT cells after BTZ treatment. (B) Confirmation of stable knockdown of p62, Nbr1, or GABARAPL1 by shRNA in untreated or BTZ-treated (16 h) SH-SY5Y cells. Molecular masses are given in kilodaltons. (C) SH-SY5Y cells expressing sh-p62 or sh-GABARAPL1 but not sh-Nbr1 lost viability more than WT cells did when treated with BTZ for 36 h. (D) Knockdown of p62 or GABRAPL1 also caused M17 cells to lose viability more than WT cells when treated with BTZ for 40 h. Viability was measured with the MTS assay. *, P < 0.05 compared with WT cells treated with the same BTZ concentration. n = 3. Error bars indicate SD.
The rapid induction of p62 and GABARAPL1 suggests that they are particularly important for cell survival upon proteasome inhibition. To test this possibility, we constructed in SH-SY5Y (Fig. 4 B) and M17 (not depicted) stable cells deficient in p62 and GABARAPL1, and as a control, Nbr1, the Ub receptor, which is not induced rapidly. The SH-SY5Y and M17 cells deficient in p62 or GABARAPL1 upon BTZ treatment lost viability (measured by mitochondria function and plasma membrane integrity) faster than WT cells (Fig. 4, C and D; and Fig. S3). In contrast, knockdown of Nbr1 did not enhance susceptibility to BTZ. p62 knockdown in myeloma cells was also reported to reduce viability and enhance susceptibility to BTZ (Milan et al., 2015). Therefore, induction of both p62 and GABARAPL1 is likely to enhance survival upon proteasome inhibition.
Ub conjugation is important for the induction of p62 and GABARAPL1
Proteasome inhibition caused rapid buildup of Ub conjugates. Therefore, we tested whether their buildup may help signal the induction of p62 and GABARAP1. To deplete SH-SY5Y cells of Ub conjugates, we used ML-997, an inhibitor of the Ub-activating enzyme E1 (Chen et al., 2011). ML-997 reduced expression of p62 and GABARAPL1 in cells treated with BTZ (Fig. 5 A). Similarly, when RPMI 8226 cells were treated for 12 h with 20 nM BTZ and a different E1 inhibitor, TAK243, to block Ub conjugation (Fig. 5 B) without causing >50% death (Fig. 5 C), the induction of p62 and GABARAPL1 mRNAs by BTZ was much reduced (Fig. 5 D). Thus, Ub conjugation is important for the activation of p62 and GABARAPL1 transcription.
Figure 5.
Upon proteasome inhibition, Ub conjugation is required for the induction of p62 and GABARAPL1. (A) Inhibiting Ub conjugation with 0.5 µM ML-997 greatly suppressed the induction of p62 and GABARAPL1 mRNAs in SH-SY5Y cells treated for 16 h with 100 nM BTZ. (B–D) In RPMI 8226 cells treated for 12 h with 20 nM BTZ, inhibiting Ub conjugation with 0.5 µM TAK-243 (B) depleted Ub conjugates (C) without causing widespread cell death (measured by MTS assay) and suppressed the induction of p62 and GABARAPL1 (D). *, P < 0.05. Molecular masses are given in kilodaltons. Error bars indicate SD.
Rapid induction of p62 and GABARAPL1 does not rely on transcription factors reported to induce p62 or autophagy genes
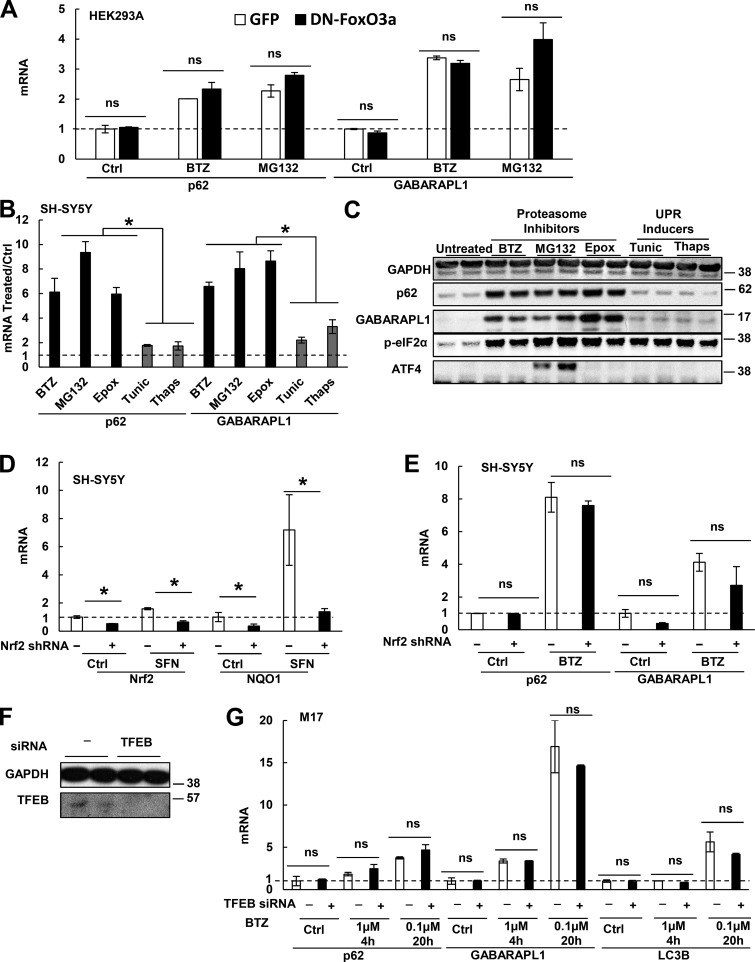
Several transcription factors have been reported to mediate the expression of autophagy genes or p62 under various conditions. Therefore, we tested whether any of them may mediate the rapid induction of p62 and GABARAPL1. To overexpress or knock down a transcription factor, we used HEK293A cells, which can be efficiently transfected. Our laboratory showed that during muscle atrophy, FoxO3a transcribes most Atg genes, including GABARAPL1, and thus promotes lysosomal proteolysis (Zhao et al., 2007). However, expression of a dominant-negative (DN) FoxO3a-ΔCT (DN-FoxO3a) in HEK293A cells (Fig. S4 A) did not inhibit the induction of p62 or GABARAPL1 by exposure to 10 nM BTZ or 10 µM MG132 for 16 h (Fig. 6 A). Furthermore, treatment of MM1.S and HEK293A cells with BTZ actually increased phosphorylation of FoxO3a at Thr32 (Fig. S4 B), which inactivates FoxO3a and excludes it from the nucleus. Thus, upon proteasome inhibition, FoxO3a is neither activated nor required for p62 and GABARAPL1 induction.
Figure 6.
FoxO3a, p-eIF2α, ATF4, Nrf2, or TFEB are not responsible for the induction of p62 and GABARAPL1 upon proteasome inhibition. (A) Expression of DN-FoxO3a did not affect the ability of HEK293A cells to induce p62 and GABARAPL1 mRNAs upon treatment with BTZ (10 nM) or MG132 (10 µM) for 16 h. (B and C) Unlike proteasome inhibitors, UPR inducers (tunicamycin and thapsigargin) did not cause a large (more than fivefold) induction of p62 or GABARAPL1. SH-SY5Y cells were treated for 16 h with BTZ (10 nM), MG132 (10 µM), epoxomicin (Epox; 50 nM), tunicamycin (Tunic; 10 µg/ml), or thapsigargin (Thaps; 300 nM), and mRNAs (B) and proteins (C) were measured. (D) To confirm successful knockdown of Nrf2, WT or stable Nrf2 knockdown (by shRNA) SH-SY5Y cells were treated for 16 h with sulforaphane (SFN; 10 µM) to activate Nrf2, and mRNAs of Nrf2 and NQO1, whose expression requires Nrf2, were measured. (E) sh-Nrf2 cells were not defective in inducing p62 or GABARAPL1 mRNAs upon BTZ treatment (10 nM for 16 h). (F and G) Knockdown of TFEB by siRNA in M17 cells was validated by Western blotting (F), but did not reduce the cells’ ability to rapidly induce p62 and GABARAPL1 mRNAs upon BTZ treatment or to induce LC3B mRNA when treated with 0.1 µM BTZ for 20 h (G). *, P < 0.05; ns, P > 0.05. Molecular masses are given in kilodaltons. Error bars indicate SD.
As part of the unfolded protein response (UPR), the translation initiation factor eIF2α is phosphorylated, which inhibits translation generally but promotes the translation of the transcription factor ATF4 (Fels and Koumenis, 2006). It was reported that proteasome inhibition in a prostate cancer line led to eIF2α phosphorylation and induction of Atg5 and Atg7 (Zhu et al., 2010) and that ATF4 can induce LC3B in a breast cancer line (Milani et al., 2009) and p62 in HeLa cells (Demishtein et al., 2017). However, our data do not support a role for either factor in the rapid induction of p62 and GABARAPL1. All cells when treated with nanomolar concentrations of BTZ similarly induced p62 and GABARAPL1, but the p-eIF2α content was raised only in SH-SY5Y cells (Fig. 6 C), but was decreased in MM1.S, HEK293A (Fig. S4 C), U266 (myeloma), and HAP1 cells (Fig. S4 D); and hardly accumulated in RPMI 8226 cells (Fig. S4 D). Even for SH-SY5Y cells, p-eIF2α is unlikely to be important for the induction of p62 and GABARAPL1 because tunicamycin and thapsigargin, which induce the UPR and raise p-eIF2α content, caused a much smaller induction of p62 or GABARAPL1 than proteasome inhibitors (Fig. 6, B and C). In addition, p62 and GABARAPL1 induction could occur without any accumulation of ATF4 (Fig. 6, B and C). For example, in SH-SY5Y cells treated with low concentrations of BTZ (10 nM) or epoxomicin (50 nM), ATF4 does not build up (Fig. 6 C), and blocking p-eIF2α–mediated translation of ATF4 with integrated stress response inhibitor (ISRIB; Sidrauski et al., 2015) did not suppress p62 or GABARAPL1 induction by 10 nM BTZ (Fig. S4 E). BTZ treatment of cholangiocarcinoma cells was also reported to induce the UPR without raising p-eIF2α and ATF4 contents (Vaeteewoottacharn et al., 2013). However, high concentrations of MG132 (e.g., 10 µM), which also induce p62 and GABARAPL1 mRNA and protein (Fig. 6, B and C), did cause ATF4 buildup in SH-SY5Y cells (Fig. 6 C), and ISRIB treatment suppressed the induction of p62 and GABARAPL1 by 10 µM MG132 (Fig. S4 E). Thus, although ATF4 can induce p62 expression, p-eIF2α-ATF4 signaling is not activated by low concentrations of proteasome inhibitors in most cells tested, and thus it does not play a major role in the rapid induction of p62 and GABARAPL1 under these conditions.
Upon oxidative stress, Nrf2 simulates p62 expression (Jain et al., 2010). However, knockdown of Nrf2 in SH-SY5Y cells did not affect p62 or GABARAPL1 induction after BTZ treatment (Fig. 6, D and E), and Nrf2 overexpression in HEK293A cells increased the expression of its canonical target gene, NQO1, but not p62 or GABARAPL1 (Fig. S4, F and G). KLF4 was reported to induce p62 in CFZ-treated myeloma cells (Riz et al., 2015). However, KLF4 knockdown did not reduce p62 mRNA in untreated or BTZ-treated HEK293A cells (Fig. S4 H). Although nuclear factor (NF)-κB has also been reported to promote p62 expression (Ling et al., 2012), NF-κB activation is inhibited by proteasome inhibition (Palombella et al., 1994). Furthermore, p62 induction in HEK293A and M17 cells was not inhibited when NF-κB activity was prevented by overexpression of an IκBα superrepressor (Fig. S4 I; Boehm et al., 2007). During starvation, transcription factor EB (TFEB) promotes the coordinate expression of many lysosomal and Atg genes as well as p62 (Settembre et al., 2011). However, knockdown of TFEB in M17 cells did not affect the rapid induction of p62 and GABARAPL1 by BTZ treatment or the induction of LC3B by prolonged BTZ treatment (Fig. 6, F and G).
p62 expression is mediated by transcription factors Nrf1 and NF-E2
Because the dramatic induction of p62 and GABARAPL1 upon proteasome inhibition appeared independent of all transcription factors previously reported to promote their expression, we tried to identify additional ones by searching for binding elements in the promoter region of p62. TRANSFAC database predicted that the nucleotides between −280 and −290 (TGCTGAGTCAT) can bind NF-E2 (and we therefore termed this element NFE2-E; Fig. 7 A), but they may also be recognized by the related factors Nrf1 and Nrf2. NF-E2 is essential for erythrocyte and platelet maturation (Andrews and Orkin, 1994; Fujita et al., 2013) and is believed to express primarily in blood cells, although we detected some expression in HEK293A and M17 cells (Fig. 7 B and not depicted).
Figure 7.
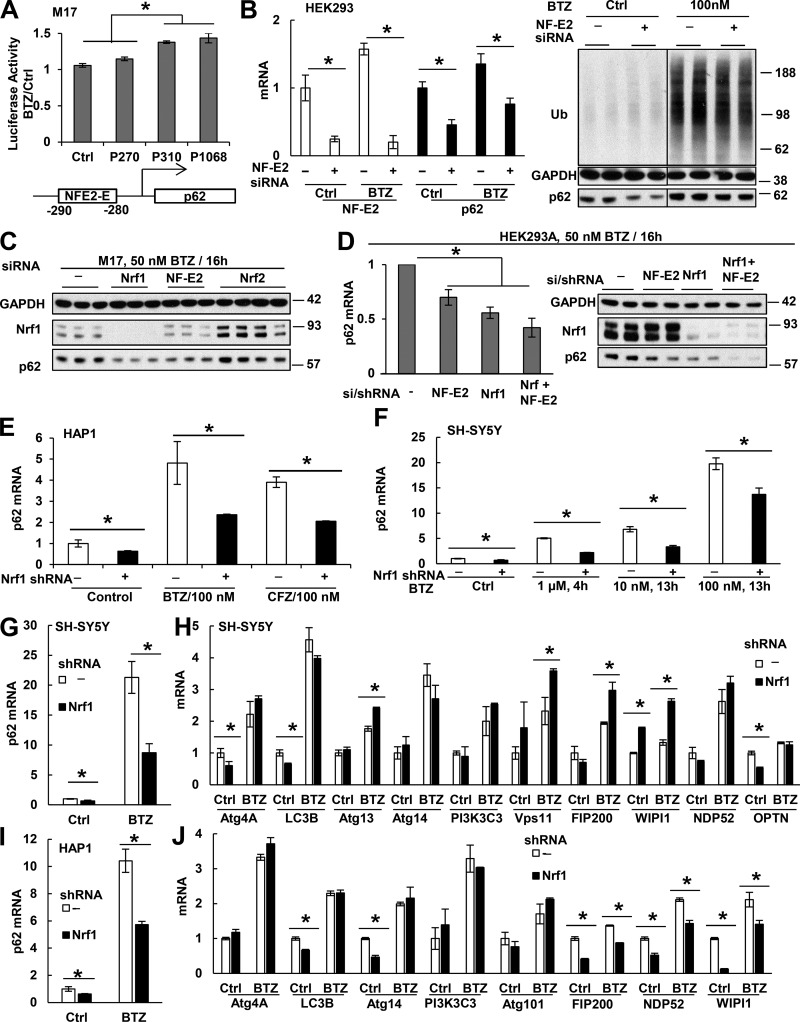
Nrf1 and NF-E2 activate p62 transcription in control cells and upon proteasome inhibition. (A) When different p62 promoter segments were fused to luciferase and expressed in M17 cells, the segments (P310 [−310 to 37] and P1068) that contain NFE2-E increased luciferase expression by ∼40% upon BTZ (0.1 µM) treatment for 16 h. However, the segment (P270) lacking NFE2-E showed no effect. n = 4. (B) NF-E2 knockdown by siRNA in HEK293A cells (verified by NF-E2 mRNA measurement in the left panel) reduced p62 mRNA (left) and protein (right) in cells treated or not for 16 h with 0.1 µM BTZ. (C) In M17 cells, knockdown of Nrf1 or NF-E2, but not Nrf2, reduced p62 level after BTZ treatment. (D) In HEK293A cells, knockdown of both NF-E2 (by siRNA) and Nrf1 (by stably expressing shRNA) caused additive reduction of p62 mRNA (left) and protein (right) after BTZ treatment. Molecular masses are given in kilodaltons. (E and F) Stable knockdown of Nrf1 with shRNA (E) suppressed p62 mRNA in untreated HAP1 cells or cells treated for 16 h with 100 nM BTZ or CFZ, and in SH-SY5Y cells treated with BTZ at indicated conditions (F). (G–J) Knockdown of Nrf1 in SH-SY5Y (G and H) or HAP1 (I and J) cells suppressed p62 mRNA when treated with 100 nM BTZ for 20 h (G and I) but did not suppress the mRNAs for most other Atg genes and Ub receptors (H and J). *, P < 0.05. Error bars indicate SD.
To test whether NFE2-E is important for p62 induction, we tested whether NFE2-E could drive luciferase expression in M17 cells upon BTZ exposure. BTZ (100 nM; 16 h) caused <10% increase in luciferase expression whether or not it was fused to the p62 promoter fragment (−270 to 37), which does not contain NFE2-E (Fig. 7 A). However, when luciferase was fused to the p62 promoter fragments (−310 to 37 or −1,068 to 37), which include NFE2-E, then BTZ caused a 40% increase in its expression (Fig. 7 A). To further test whether NF-E2 may be involved in this induction of p62, we knocked down NF-E2 with siRNA in HEK293A (Fig. 7 B) or M17 (Fig. 7 C) cells. Although the level of NF-E2 is very low in these cells, it was still reduced by >80% with siRNA (Fig. 7 B). NF-E2 knockdown decreased the basal and BTZ-induced level of p62 mRNA by >50% (Fig. 7 B) and also reduced p62 protein content (Fig. 7, B and C). NF-E2 belongs to a family of oxidative stress–activated transcription factors that also includes Nrf1 and Nrf2. Unlike knockdown of Nrf2 (Fig. 6, D and E; and Fig. 7 C), Nrf1 knockdown in M17 (Fig. 7 C), HEK293A (Fig. 7 D), HAP1 (Fig. 7 E), or SH-SY5Y (Fig. 7 F) cells reduced the level of p62 mRNA and protein in control cells and ones treated with BTZ or CFZ. Furthermore, expressing NF-E2 siRNA in a HEK293A cell line stably expressing Nrf1 shRNA further suppressed p62 expression upon BTZ treatment (50 nM; 16 h; Fig. 7 D). Thus, NF-E2 and Nrf1 both activate p62 expression.
Thus, upon proteasome inhibition, Nrf1 is critical for the induction of not only proteasome genes and p97, but also p62. NF-E2 appears to be less important than Nrf1 in these compensatory responses because its knockdown reduced p62 expression less than Nrf1 knockdown (Fig. 7, C and D) and did not affect the expression of proteasome genes (not depicted). Although the AP-1 transcription factor has been suggested to also recognize this NFE2-E (Vadlamudi and Shin, 1998; Ling et al., 2012), the induction of p62 and GABARAPL1 mRNAs in SH-SY5Y cells by BTZ (100 nM; 16 h) was not affected when AP1 was inhibited by the JNK inhibitor SP600125, which blocks the phosphorylation that activates the AP1 subunit c-Jun (Fig. S4 J; Park et al., 2004).
The induction of GABARAPL1 upon proteasome inhibition, like that of p62, is greatly suppressed by the inhibition of ubiquitination (Fig. 5). However, knockdown of Nrf1 or NF-E2 did not significantly reduce GABARAPL1 expression, and no NF-E2 or Nrf1-binding elements were found in its promoter region. Therefore, some unknown transcription factor must trigger GABARAPL1 expression upon proteasome inhibition. In addition, we examined whether Nrf1 also causes the coordinated expression of all Atg genes and Ub receptors after prolonged proteasome inhibition (Fig. 7, G–J). SH-SY5Y and HAP1 cells stably expressing Nrf1 shRNA were treated with 100 nM BTZ for 20 h to induce these genes. As expected, knockdown of Nrf1 suppressed the induction of p62 in both lines (Fig. 7, G and I). It also reduced the basal expression of some Atg genes in both lines (Fig. 7, H and J) but did not reduce the BTZ-induced expression of any of these genes in SH-SY5Y cells (Fig. 7 H) and most (five of eight) genes in HAP1 cells (Fig. 7 J). Thus, Nrf1 does not play an important role in the induction of all Atg genes and Ub receptors after prolonged proteasome inhibition.
p62 promotes the sequestration of ubiquitinated proteins but not their degradation in neuroblastoma cells
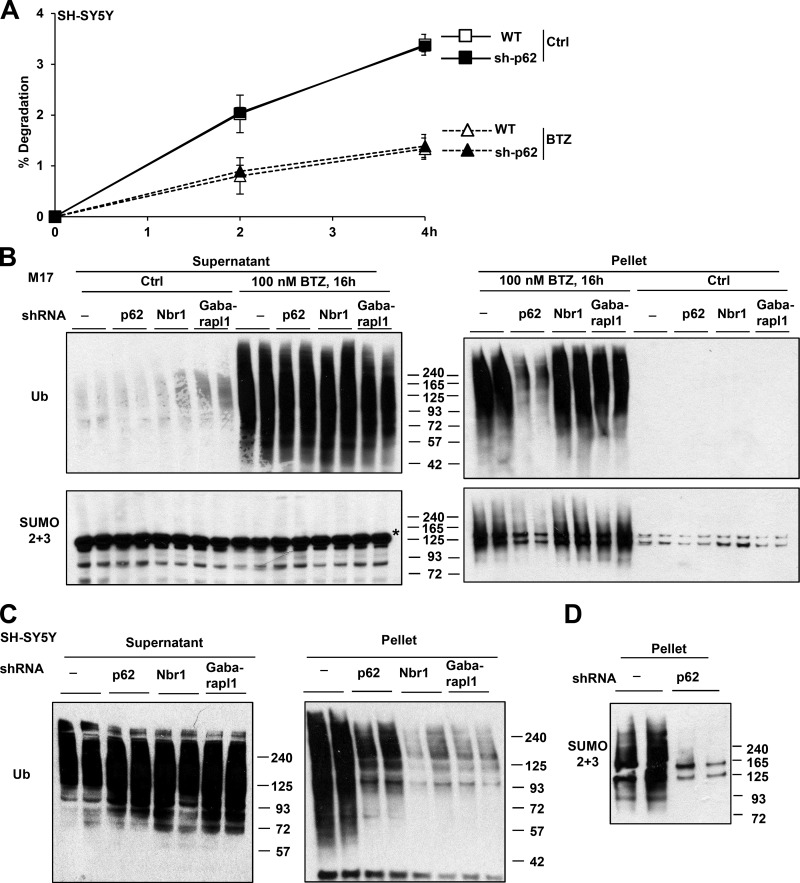
Because p62 is a Ub adapter for autophagy and GABARAPL1 is one of its binding sites on the autophagic vacuole, it seemed likely that these proteins promote survival by enhancing the degradation of ubiquitinated proteins in autophagosomes. To test this hypothesis, we examined whether p62 knockdown in SH-SY5Y cells reduced their ability to degrade cell proteins labeled with [3H]phenylalanine (Zhao et al., 2015). Surprisingly, cells deficient in p62 degraded the bulk of long-lived cell proteins at rates similar to WT cells (Fig. 8 A). p62 knockdown also did not decrease the remaining lysosomal degradation of long-lived proteins when cells were treated with 1 µM BTZ for 4 h, which completely blocked proteasomal degradation (Fig. 8 A). Thus, the induction of p62 after exposure of these cells to proteasome inhibitors does not enhance the degradation of Ub conjugates by the UPS or autophagy. Therefore, p62 must promote survival by a distinct mechanism.
Figure 8.
p62 knockdown in neuroblastoma cells did not reduce protein degradation or increase levels of Ub conjugates but impaired the buildup of polyubiquitinated and polysumoylated proteins in inclusions. (A) p62 knockdown (sh-p62) did not affect the ability of SH-SY5Y cells to degrade long-lived proteins when treated or not with 1 µM BTZ to completely inhibit the proteasome. n = 6. (B) In M17 cells treated with BTZ, stable knockdown of p62 but not Nbr1 or GABARAPL1 reduced the buildup of Ub or SUMO2/3 conjugates in the pellet that contains inclusions. In the supernatant, there are no SUMO2/3-conjugated proteins, and the bands recognized by the SUMO2/3 antibody (asterisk) are not specific. (C and D) In SH-SY5Y cells treated for 16 h with 100 nM BTZ, knockdown of p62 reduced Ub (C) and SUMO2/3 (D) conjugates in the pellet. (C) Knockdown of Nbr1 and GABARAPL1 also reduced Ub conjugates in the pellet. Molecular masses are given in kilodaltons. Error bars indicate SD.
Before its association with Atg8 proteins, p62 sequesters Ub conjugates in insoluble cytosolic inclusions often termed aggresomes, which may limit toxicity and promote survival. To examine both the degradation and aggregation of Ub conjugates, we extracted cell proteins with buffer containing 1% Triton X-100 and centrifuged at 10,000 g for 10 min. Treatment of SH-SY5Y and M17 cells with BTZ for 4 h caused a buildup of Ub conjugates in supernatant, but in the next 4–8 h, conjugates accumulated in the pellet (Fig. S5 A and not depicted) that correspond with protein aggregates. In both cell lines, knockdown of p62 did not affect the amount of Ub conjugates in the supernatant but significantly reduced the buildup of insoluble conjugates (Fig. 8, B and C; and Fig. S5 A). Even after treatment with 100 nM BTZ for 20–24 h, when almost all Atg genes were induced (Fig. 2 A), and autophagy increased (Fig. 1 C), p62 deficient SH-SY5Y cells still did not accumulate more Ub conjugates in the supernatant than WT cells (Fig. S5 B).
Thus, in neuroblastoma cells, the major role of the p62 induction after proteasome inhibition seems to be sequestering the Ub conjugates in insoluble inclusions rather than to promote their degradation by autophagy. In contrast, in BTZ-treated myeloma cells, p62 knockdown was reported to cause a buildup of Ub conjugates (Milan et al., 2015). One possible explanation for this difference is that in myeloma cells, where the production of misfolded proteins is especially high, autophagy is more active, as indicated by a much higher level of LC3-II in MM1.S (Fig. S1 C) and RPMI 8226 (Fig. 3 H) cells than in SH-SY5Y cells (Fig. 1 C). Thus, p62 may be more actively facilitating aggregate degradation by autophagy. In contrast, in BTZ-treated SH-SY5Y cells, there appears to be relatively little autophagy-mediated destruction of Ub conjugates or p62 because inhibition of autophagy by concanamycin A caused almost no further buildup of Ub conjugates (Fig. S5 C) or p62 (Fig. S5 D).
We also examined the effect of knocking down GABARAPL1 or Nbr1 in M17 and SH-SY5Y cells. Although their knockdown also reduced the amount of pelleted Ub conjugates in SH-SY5Y cells (Fig. 8 C), this effect was not observed in M17 cells (Fig. 8 B). The basis for these different responses in these two lines is unclear.
p62 is required for the formation of perinuclear aggregates that contain Ub chains and nuclear aggregates that contain SUMO 2/3 chains
Upon BTZ treatment, there is a buildup not only of Ub conjugates but also of SUMO2/3 conjugates (Schimmel et al., 2008) presumably because protein sumoylation can trigger ubiquitination and degradation (Guo et al., 2014). After 8-h exposure of M17 and SH-SY5Y cells to BTZ, sumoylated proteins accumulated in the 10,000-g pellets simultaneously with Ub conjugates but did not build up in the supernatant (Fig. 8, B and D; and Fig. S5 A). Surprisingly, the buildup of SUMO2/3 conjugates in the pellet, like Ub conjugates, was reduced by the knockdown of p62 in both cell types (Fig. 8, B and D). SUMO2/3 modifies mostly nuclear proteins (Martin et al., 2007). Misfolded proteins in the nucleus tend to associate in promyelocytic leukemia (PML) bodies and are modified with poly-SUMO2/3 chains, but they subsequently undergo ubiquitination and proteasomal degradation (Guo et al., 2014). These SUMO2/3-positive nuclear bodies thus appear to be quite different from the Ub-positive p62-containing aggregates found in the cytoplasm.
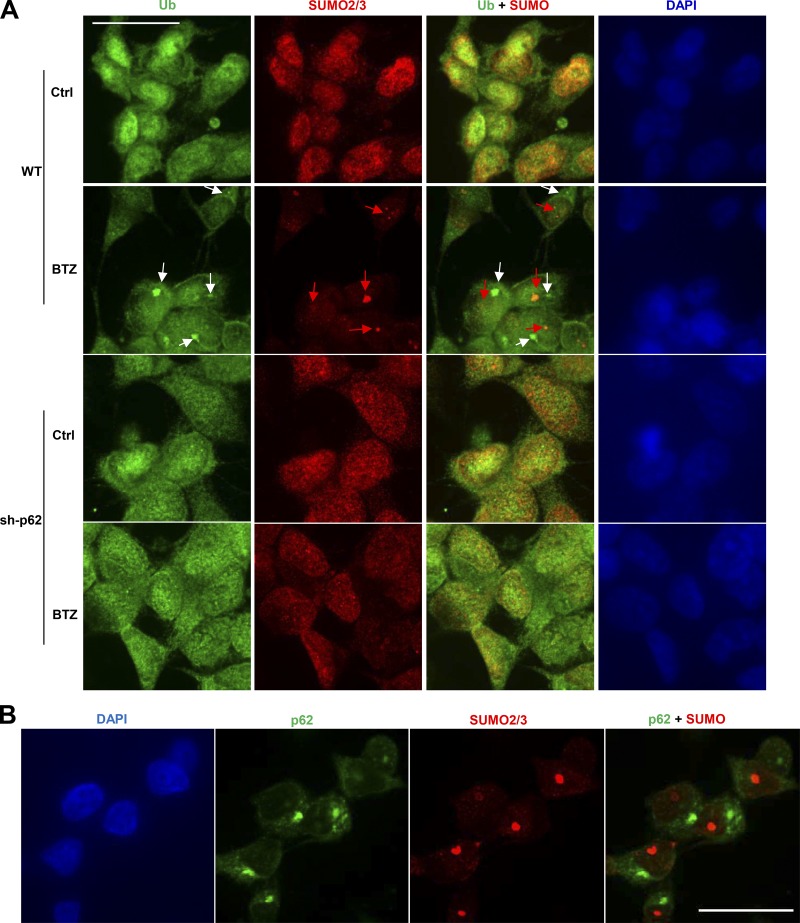
To determine whether p62 also promotes the sumoylated inclusions in the nucleus, we immunostained WT or sh-p62 M17 cells after BTZ treatment (100 nM) for 8 h to detect Ub or SUMO2/3 (Fig. 9 A). Under these conditions, two spatially distinct types of aggregates were evident: SUMO-positive aggregates in the nucleus (red arrows) and Ub-positive aggregates in the perinuclear cytoplasm (white arrows), and knockdown of p62 reduced the formation of both (Fig. 9 A). To determine whether p62 also associates with the SUMO-positive nuclear inclusions, we immunostained WT M17 cells with antibodies against p62 or SUMO2/3. Upon BTZ treatment (100 nM for 8 h), p62 became concentrated in the perinuclear region as expected. However, there was no detectable localization of p62 in the nucleus and no colocalization with sumoylated nuclear aggregates (Fig. 9 B). Therefore, p62 somehow is important for the formation of nuclear sumoylated aggregates without itself accumulating in them.
Figure 9.
p62 promotes the formation of cytosolic inclusions containing Ub conjugates and nuclear inclusions containing sumoylated proteins, but p62 accumulates only in cytosolic inclusions. (A) M17 cells were treated with 100 nM BTZ for 8 h and immunostained with Ub and SUMO2/3 antibodies. SUMO-positive aggregates (red arrows) and Ub-positive aggregates (white arrows) both build up in BTZ-treated WT cells but do not colocalize. The buildup of both aggregates was greatly reduced in sh-p62 cells. (B) WT M17 cells were treated for 8 h with 100 nM BTZ and immunostained with p62 and SUMO2/3 antibodies. There was no colocalization of p62 and SUMO2/3. Bars, 25 µm.
Discussion
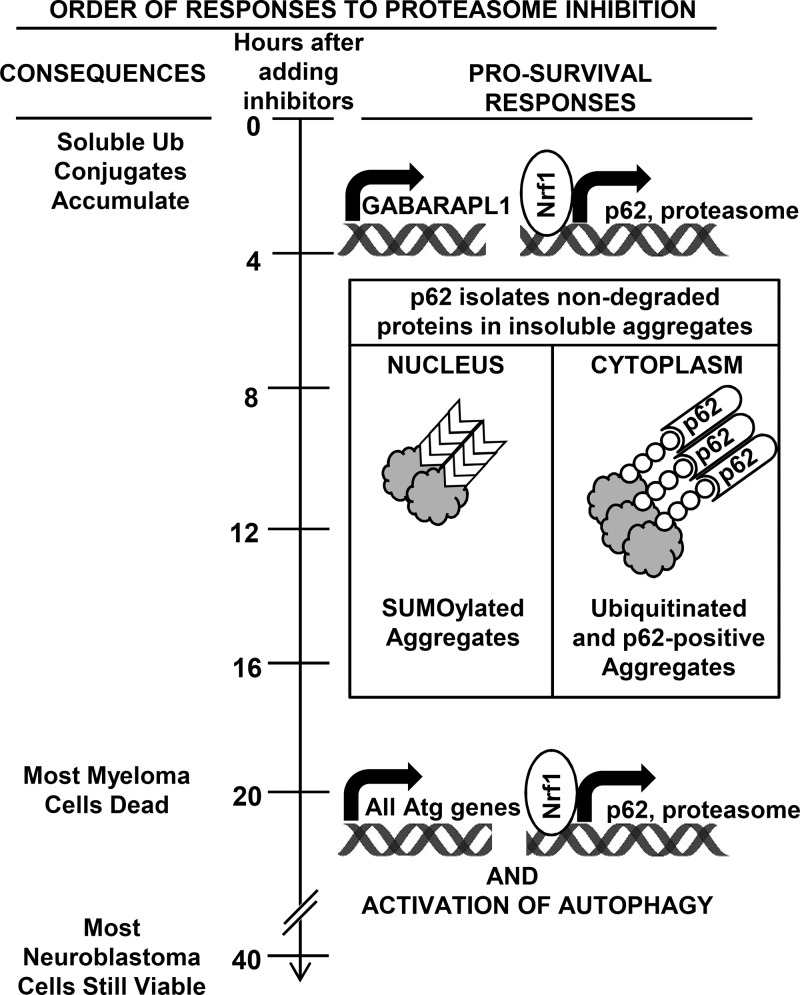
Our study uncovered two distinct transcriptional responses to proteasome inhibition that appear similar in very different cell types (Fig. 10). First, there is a rapid (within 4 h), large, and selective induction of p62 and GABARAPL1 without any rise in autophagosome formation or increase in the expression of its many components. This rapid response is evident in both myeloma cells, which generate large amounts of misfolded proteins and are exceptionally sensitive to proteasome inhibition, and neuroblastoma cells, which are much more resistant. Therefore, these responses seem likely to occur in all mammalian cells, although this conclusion will require studies of additional cell types. The rapid induction of p62 and GABARAPL1 enhances survival. p62 appears essential for subsequent sequestration of Ub conjugates in large cytosolic inclusions.
Figure 10.
Summary of responses to proteasome inhibitors described in this study. When treated with proteasome inhibitors, which rapidly cause the buildup of soluble Ub conjugates, >50% of myeloma cells are killed by 20 h, but most neuroblastoma cells are viable at 40 h. Within 4 h, however, both cell types induce p62 and GABARAPL1 but not most Atg genes. The induction of p62 occurs simultaneously with that of all proteasome genes and also requires Nrf1. p62 promotes survival by sequestering nondegraded proteins in large cytoplasmic ubiquitinated and nuclear sumoylated aggregates, which are evident by 8 h. p62 accumulates only in cytoplasmic aggregates. Many hours later, prolonged treatment with a high concentration of inhibitors causes induction of all Atg genes and some lysosomal genes and activation of autophagy.
Second, after a longer and stronger proteasome inhibition, there is coordinate induction of almost all Atg genes and Ub receptors, and autophagosome formation is clearly elevated. These slower responses also appear to be adaptive rather than causing autophagic cell death because autophagy activation and Atg gene induction were evident in BTZ-treated neuroblastoma cells at 20 h, long before widespread cell death became evident. Thus, the activation of autophagy by enhancing the clearance of potentially toxic Ub conjugates after they are bound by p62 in aggresomes appears to help cells compensate for the reduced proteasomal degradation. Unlike neuroblastoma lines, when BTZ-treated myeloma cells activated autophagy (at 20 h), there was already widespread cell death. Myeloma cells, which produce a very large amount of abnormal Igs, are particularly dependent on ER-associated degradation and proteasomes, and presumably, the enhancement of their autophagic capacity is insufficient and too slow to compensate for the loss of proteasomal function and prevent cell death.
During autophagy, p62 and GABARAPL1 are consumed with their cargo of Ub conjugates. Thus, when autophagy is most active, cells must increase the production of these key proteins to prevent their depletion and to sustain a high capacity for autophagy. In fact, although p62 and GABARAPL1 were markedly induced by 4 h, their induction continued to increase and reached very high levels (19–50-fold mRNA) after 20-h exposure to BTZ when autophagy was activated and other Atg genes were also induced (to a much lower extent).
Our data also indicate that, in addition to enhancing Ub conjugate destruction by lysosomes, p62 has another important prosurvival role probably related to its ability to sequester Ub conjugates in inclusions. Knockdown of p62 did not reduce protein breakdown either in control SH-SY5Y cells or after complete proteasome inhibition. Actually, in a mammary epithelial cell line even after maximal proteasome inhibition, there is no clear rise in lysosomal proteolysis for 20 h (Tsvetkov et al., 2015), suggesting that increasing lysosomal degradation is not an essential prosurvival adaptation. Furthermore, p62 knockdown did not cause a greater buildup of Ub conjugates in BTZ-treated neuroblastoma cells as would be anticipated if p62 promotes their degradation, and similar findings were reported in HeLa cells (Demishtein et al., 2017). Thus, p62 promotes survival of neuroblastoma cells upon proteasome inhibition but does not play an important role in promoting destruction of Ub conjugates by either proteasome or autophagy as originally proposed (Pankiv et al., 2007), presumably because of the low activity of autophagy in these cells. In contrast, myeloma cells, in addition to their high proteasome dependence, have a high basal autophagy rate (Hoang et al., 2009), which presumably also promotes clearance of the aggregated Ub conjugates. Knocking down p62 in myeloma cells was reported to cause further buildup of Ub conjugates upon proteasome inhibition (Milan et al., 2015). Nevertheless, in myeloma cells, autophagy rate also did not rise until after 20 h of proteasome inhibition (Figs. 3 I and S1 C). In myeloma patients treated with BTZ, which is cleared from the circulation in a few hours, this delayed activation of autophagy may not even occur because cells can quickly produce new proteasomes to restore their degradative capacity.
The presence of p62 in inclusions was the original basis for it being named the sequestosome (Shin, 1998). Presumably, its capacity to bind and sequester Ub conjugates minimizes their toxic potential. Accordingly, the formation of inclusions by p62 correlates with longer survival of cells treated with MG132 (Nakaso et al., 2004). Importantly, p62-dependent inclusions were evident after 8–12 h of proteasome inhibition when there was no apparent elevation of autophagy. p62 is also a major component of cytosolic inclusions found in neurodegenerative diseases, which presumably form by mechanisms similar to the aggresomes resulting from proteasome inhibition. In a cellular model of Huntington’s disease, caused by a polyglutamine expansion in huntingtin, there is clear evidence that this formation of cytosolic inclusions reduces neuronal death (Arrasate et al., 2004), as suggested by our findings. Thus, blocking p62-mediated aggregate formation rather than blocking autophagy might enhance the toxicity of proteasome inhibitors and may be achieved by preventing posttranslational modifications of p62. Upon proteasome inhibition, cells enhance aggregate formation not only by producing more p62 but also by stimulating p62 phosphorylation by TBK1 or casein kinase II (Matsumoto et al., 2011; Pilli et al., 2012). Also, upon proteasome inhibition, some p62 becomes ubiquitinated (Fig. S1, E and F), which increases its ability to bind Ub and drive inclusion formation (Peng et al., 2017).
The selective induction of p62 and GABARAPL1 but not other Ub receptors or Atg proteins along with their abilities to reduce cell death indicate clearly that Ub receptors and Atg8 proteins can serve distinct roles. Surprisingly, Nbr1, which forms mixed complexes with p62 (Lamark et al., 2009) and is found in most p62-positive inclusions (Kirkin et al., 2009), was not induced rapidly after proteasome inhibition, and Nbr1 knockdown had little effect on viability. In addition to its protective role in segregating Ub conjugates, p62 induction may also be beneficial in other ways such as the regulation of NF-κB (Sanz et al., 2000; Duran et al., 2008), mTOR (Duran et al., 2011; Linares et al., 2013), Nrf2, and c-myc (Umemura et al., 2016).
More surprising was the selective and dramatic induction of GABARAPL1 because (a) GABARAPL1 does not bind p62 as tightly as LC3B does (Rozenknop et al., 2011) and thus does not appear to be the preferred p62-binding protein on the autophagosome; (b) all three LC3 genes are induced when neuroblastoma cells activate autophagosome formation (Fig. 2 A), thus arguing against GABARAPL1 playing a distinct role in Ub conjugate degradation; and (c) knockdown of GABARAPL1 reduced inclusion formation in SH-SY5Y cells, even though Atg8 proteins are not known to bind Ub conjugates or promote their aggregation (Fig. 8 C). Because GABARAPL1 is the only member of the large Atg family that was induced with p62 without an increase in autophagy, it may enhance viability in some unknown manner.
Although various transcription factors were reported to induce p62 or Atg genes upon proteasome inhibition (ATF4 and KLF4) or in other conditions (FoxO3a, Nrf2, AP1, NF-κB, and TFEB), none of them appears to catalyze the transcriptional adaptations to partial proteasome inhibition (e.g., with nanomolar BTZ) described in this study. In contrast, Nrf1 appears to be particularly important in the rapid induction of p62. This new role in p62 regulation complements Nrf1’s well-established role in the “bounce-back” expression of new proteasomes and p97, which occurred simultaneously with p62 induction after proteasome inhibition. These coordinated responses all enhance cell survival and compensate for the reduced proteolytic capacity. After loss of proteasome function, Nrf1 is activated through proteolytic cleavage of its ER-bound precursor by the aspartyl protease Ddi2 (Koizumi et al., 2016; Lehrbach and Ruvkun, 2016) after Nrf1 ubiquitination (Sha and Goldberg, 2014). This novel mechanism may explain the suppression of p62 induction when ubiquitination is inhibited (Fig. 5). Nrf1 belongs to a family of transcription factors that also includes NF-E2 and Nrf2 (Hayes and McMahon, 2001). Upon oxidative stress, Nrf2 was reported to induce proteasome genes (Kwak et al., 2003; Sebens et al., 2011) and p62 (Jain et al., 2010). However, upon proteasome inhibition, Nrf2 is not important for the induction of proteasomes (Radhakrishnan et al., 2010; Steffen et al., 2010) or p62 (Fig. 6, D and E). NF-E2 also contributes to this increase in p62 expression in neuroblastoma cells (Fig. 7), but its role is less important than that of Nrf1. Despite its major role in the rapid response, Nrf1 does not mediate the coordinate induction of all Atg genes and Ub receptors upon prolonged proteasome inhibition.
Surprisingly, upon proteasome inhibition, p62 is also required for the formation of nuclear inclusions containing proteins bearing SUMO2/3 chains, although these nuclear aggregates do not contain detectable p62. p62 is mainly localized in the cytoplasm, but it can shuttle in and out of the nucleus, where it promotes the buildup of Ub conjugates in the PML bodies (Pankiv et al., 2010). There was no detectable colocalization of p62 or Ub in these nuclear bodies, presumably because of p62’s rapid transit in and out of the nucleus (Pankiv et al., 2010) and perhaps the rapid removal of Ub chains by deubiquitinases. PML bodies are the major sites of sumoylation in cells, and their main component, the PML protein, catalyzes sumoylation of aggregated proteins. These sumoylated proteins eventually are ubiquitinated by RNF4, a SUMO-targeted Ub ligase, before degradation by the proteasome (Guo et al., 2014). Thus, PML bodies seem to function as nuclear sequestration sites for misfolded proteins (Rockel et al., 2005), and the formation of these nuclear SUMO-positive bodies appears to be another mechanism by which p62 isolates the nondegraded proteins in inclusions before their eventual degradation.
Materials and methods
Cell lines and growth conditions
Neuroblastoma cells M17 (CRL-2267; ATCC) and SH-SY5Y (CRL-2266) were cultured in DMEM-F12 (1:1; 10-092-CV; Mediatech) and HEK293A cells in DMEM (10-013-CV; Mediatech). Myeloma cells MM1.S (provided by T. Hideshima, Dana Farber Cancer Institute, Boston, MA), RPMI 8226, U266, and KMS-12-BM (provided to us by G. Bianchi, Dana Farber Cancer Institute, Boston, MA) were cultured in RPMI 1640 (10-040-CV; Mediatech). Atg5KO (Atg5−/−) MEFs and their parental cell lines were described by Mizushima et al. (2001). WT HAP1 cells (Horizon) were cultured in Iscove’s modified Dulbecco’s medium (12440061; Thermo Fisher Scientific). All media contained 10% FBS (F6178, 100 ml; Sigma-Aldrich) and 1% penicillin-streptomycin solution (15070-063; Thermo Fisher Scientific). All cells were maintained in a humidified incubator at 37°C and 5% CO2.
Overexpression of DN-FoxO3a, HA-Nrf2, and IκBSR in HEK293A or M17 cells
Plasmid (1796) expressing the DN-FoxO3a (pECE-FoxO3aΔCT) and plasmid (15264) expressing the IκBα super repressor (IκBSR) were purchased from Addgene. Plasmid expressing HA-Nrf2 (pCI-HA-Nrf2) was provided by M. Hannink (University of Missouri, Columbia, MO; Zhang and Hannink, 2003). HEK293A cells were seeded in six-well plates until 80% confluence. Transfection mixture containing 10 µg DNA, 5 µl Lipofectamine 2000 (11668-019; Thermo Fisher Scientific) was prepared in 500 µl Opti-MEM I Reduced-Serum Medium (51985-034; Thermo Fisher Scientific), allowed to mix at room temperature for 20 min, and added to each well containing 2.5 ml complete medium. After 8 h, the transfection mixture was replaced with fresh medium. M17 cells were seeded in 12-well plates until 50% confluence. Transfection mixture containing 2 µg DNA and 6 µl Fugene HD (E2311; Promega) was prepared in 100 µl Opti-MEM, allowed to mix at room temperature for 15 min, and added to each well containing 1 ml complete medium. Cells were normally incubated for 48–72 h before assays. Mock transfection was normally performed with plasmid expressing GFP (pCMV-GFP; Kobayashi et al., 2002).
Transient knockdown of NF-E2, Nrf1, Nrf2, TFEB, and KLF4 in HEK293A cells or M17 cells by siRNA
NF-E2 siRNA was purchased from Santa Cruz Biotechnology, Inc. (sc-36046), Nrf1 siRNA and TFEB siRNA from Thermo Fisher Scientific (L-019733-00-0005 and L-009798-00-0005), Nrf2 siRNA from Thermo Fisher Scientific (S9492), and KLF4 siRNA from GE Healthcare (L-005089-00-0005). To knock down genes in HEK293A cells, transfection mixture containing 20 pmol siRNA and 1 µl Lipofectamine 2000 was prepared in 100 µl Opti-MEM and allowed to mix at room temperature for 20 min before addition to the cells (cultured with 500 µl penicillin/streptomycin-free DMEM in a 24-well plate until 30–40% confluence). To knock down genes in M17 cells, transfection mixture containing 40 pmol siRNA and 1 µl DharmaFECT 4 reagent (T-2004-01; GE Healthcare) was prepared in 100 µl Opti-MEM and allowed to mix at room temperature for 20 min before adding to the cells (cultured with 500 µl penicillin/streptomycin-free DMEM/F12 in a 24-well plate until 30–40% confluence). As a control, we prepared a mixture with only the transfection reagent and no siRNA.
Construction of stable knockdown cell lines for Nrf1, Nrf2, p62, Nbr1, and GABARAPL1
Stable knockdown cell lines for Nrf1 (HEK293A and HAP1), Nrf2 (SH-SY5Y), p62 (M17 and SH-SY5Y), Nbr1 (M17 or SH-SY5Y), and GABARAPL1 (M17 or SH-SY5Y) were constructed using lentiviral particles expressing shRNAs for Nrf1 (sc-43575-V; Santa Cruz Biotechnology, Inc.), Nrf2 (sc-156128-V; Santa Cruz Biotechnology, Inc.), p62 (TRCN0000007235; Sigma-Aldrich), Nbr1 (TRCN0000123161; Sigma-Aldrich), or GABARAPL1 (TRCN0000060673; Sigma-Aldrich). As controls, cells were infected with lentiviral particles expressing cop-GFP (sc-108084; Santa Cruz Biotechnology, Inc.). Lentivirus infection was set up in 24-well format with 2.5 × 104 cells seeded in each well. 5 × 104 lentiviral particles were mixed with the complete medium to a total volume of 250 µl. Polybrene (sc-134220; Santa Cruz Biotechnology, Inc.) was added at a final concentration of 8 µg/ml. The mixture was applied to each well for 6 h, and 350 µl fresh complete medium were added to each well. 24 h after infection, stable clones were selected in the presence of 1 µg/ml puromycin.
Treatment with proteasome inhibitors or other compounds
Stock solutions were prepared for the following proteasome inhibitors: MG132 (10 mM, DMSO; I-130; Boston Biochem), BTZ (100 mM, DMSO; B-1408; LC Laboratories), epoxomicin (200 µM, DMSO; 324800-100UG; EMD Millipore), CEP18770/delanzomib (10 mM, DMSO; A4009; Apexbio), and CFZ (20 mM, DMSO; F1300; UBPBio); inducers of the UPR: tunicamycin (10 mg/ml, DMSO; T7765-10MG; Sigma-Aldrich) and thapsigargin (100 µM, DMSO; T9033-0.5MG; Sigma-Aldrich); cycloheximide (10 mg/ml, DMSO; C7698; Sigma-Aldrich); NaAsO2 (10 mM, H2O; S-225; Thermo Fisher Scientific); NMS-859 (10 mM, DMSO; M60148; Xcess Biosciences); sulforaphane (10 mM, DMSO; 574215-25MG; EMD Bioscience); bAP15 (10 mM, DMSO; F2100; UBPBio); chloroquine (100 mM, H2O; C6628-25G; Sigma-Aldrich); E-64D (10 mM, DMSO; E8640-250UG; Sigma-Aldrich); concanamycin A (200 µM, DMSO; sc-202111A; Santa Cruz Biotechnology, Inc.); TAK243/MLN7243 (10 mM, DMSO; A1384; Active Biochem), and SP600125 (25 mM, DMSO; BML-EI305-0010; Enzo Life Sciences). The E1 inhibitor ML-997 (20 mM, DMSO) was provided by L. Dick (Takeda Pharmaceuticals, Cambridge, MA). The p-eIF2α inhibitor ISRIB (20 mM, DMSO) was provided by M. Soustek (Dana Farber Cancer Institute, Boston, MA).
Immunostaining
M17 cells were seeded in Lab-TekII eight-well chamber slides (154534; Thermo Fisher Scientific). After treatment with 0.1 µM BTZ, the cells were fixed with 100% methanol prestored at −20°C for 10 min and then rehydrated by washing 3× for 5 min in PBS. Immunostaining was performed with anti-p62 antibody (1:100; 610832; BD), anti-Ub antibody (P4D1, 1:100; sc-8017; Santa Cruz Biotechnology, Inc.), or anti-SUMO2/3 antibody (1:100; ab-3742; Abcam). The secondary antibodies were Alexa Fluor 488–conjugated anti–mouse secondary antibody (1:500; A-11001; Thermo Fisher Scientific) and Alexa Fluor 555–conjugated anti–rabbit secondary antibody (1:500; A-21428; Thermo Fisher Scientific). Nuclei were counterstained with DAPI contained in the mounting medium (P36966; Thermo Fisher Scientific). Images were taken with a Lucille spinning-disk confocal microscope (Nikon) equipped with an ORCA-ER cooled charge-coupled device camera (confocal for 488- and 555-nm channels and wide-field, for the DAPI channel; Hamamatsu Photonics) and a 63×/1.4 NA oil objective at room temperature, remotely controlled with MetaMorph image acquisition software (Molecular Devices).
Lysate preparation, fractionation, and Western blotting
Cells were lysed for 30 min in ice-cold 1% Triton X-100 lysis buffer (50 mM Tris-Cl, pH 7.4, 150 mM NaCl, 1 mM NaF, 1 mM EDTA, 1 mM Na3VO4, 1 mM DTT, 1% Triton X-100, and protease inhibitor cocktail tablet; Roche). After centrifugation (10,000 g for 10 min), except where specified, only the supernatant was used as cleared lysate for Western blotting. Pellets were solubilized in 2% SDS (in 50 mM Tris-Cl, pH 7.4) and sonicated. To detect proteins, we used antibodies against GAPDH (200 µl, 1:10,000; G8795; Sigma-Aldrich), p62 (1:1,000; 610832; BD), GABARAPL1 (1:1,000; 11010-1-AP; ProteinTech Group), Nbr1 (1:1,000; H00004077-B01P; Abnova), phospho-eIF2α (1:1,000; 3597S; Cell Signaling Technology), ATF4 (1:1,000; WH0000468M1; Sigma-Aldrich), TFEB (1:1,000; MBS120432; MyBiosource), Ub (1:2,000; sc-8017; Santa Cruz Biotechnology, Inc.), Nrf1 (1:1,000; 8052S; Cell Signaling Technology), SUMO2/3 (1:2,000; ab-3742; Abcam), LC3 (1:1,000; NB100-2220; Novus Biologicals), FoxO3 (1:1,000; 9467; Cell Signaling Technology), phospho-FoxO1/3 (1:1,000; 9494S; Cell Signaling Technology), eIF2α (1:1,000; 9722; Cell Signaling Technology), Nrf2 (1:1,000; sc-722; Santa Cruz Biotechnology, Inc.), and NQO1 (1:1,000; ab2346; Abcam). To detect via enhanced chemiluminescence, we used HRP-conjugated goat anti–mouse (1:10,000; W4021; Promega) or goat anti–rabbit (1:10,000; W4011; Promega) secondary antibodies and SuperSignal West Pico chemiluminescent substrate (34080; Thermo Fisher Scientific). To detect via the Odyssey CLx infrared imaging system, we used IRDye 680LT goat anti–mouse IgG (H+L; 1:10,000; 926-68020; LI-COR Biosciences) or IRDye 800CW donkey anti–rabbit IgG (H+L; 1:10,000; 926-32213; LI-COR Biosciences) secondary antibodies. Quantification of signals by densitometry was performed using the Odyssey CLx infrared imaging system (LI-COR Biosciences). Actin or GAPDH was used as the loading control for cell lysate soluble in 1% Triton X-100, and lamin was used as the loading control for the pellet fraction after centrifugation.
Real-time RT-PCR
Primers were designed for individual genes to allow quantification by real time RT-PCR. mRNA was extracted from cultured cells via TRIzol reagent (5596-018; Thermo Fisher Scientific) and precipitated with isopropanol, and cDNA was synthesized using MultiScribe reverse transcription (4311235; Applied Biosystems) according to product instructions. Real-time RT-PCR was performed using Perfecta SYBR green FastMix (101414-270; VWR International) on a C-1000 thermocycler (Bio-Rad Laboratories) using the following parameters: preheat 95°C for 5 min; 40 × (95°C for 28 s, 60°C for 28 s, and 72°C for 28 s); and elongation 72°C for 10 min. Two biological duplicates (separate wells treated independently) were measured to determine the mean ± SD of each measurement, and the values of biological duplicates were determined from three replicates in RT-PCR. Relative mRNA levels of each gene were normalized to PGK1 mRNA.
Primer sequences
Table 1 lists sequences of RT-PCR primers, and Table 2 lists sequences of primers for cloning p62 promoter segments.
Table 1. Sequences of RT-PCR primers.
| Gene | Forward primer (5′–3′) | Reverse primer (5′–3′) |
|---|---|---|
| PGK1 | AAAGTCAGCCATGTGAGCACT | CCACCCCAGGAAGGACTTTA |
| ULK1 | CTGTGCAGATGGTGCAGTC | CACAGATGCCAGTCAGCAG |
| ULK2 | CACAAATACTGCTTGGAAAGGA | CACCAAAAAGACAGAGTTGGG |
| Atg2A | CCGTGTATGACATCCTGTCC | ACGTCACAGATGGTCTGAGC |
| Atg2B | GCTGCTTCCTTTGGTACCTC | GTCTGAGCCGTGTCTGTGAT |
| Atg3 | AGAAGGAGGGGGAGAACTTG | GGTCAATGGTCACATCTATGG |
| Atg4A | AACCTGGATCCTTCAGTTGC | CAAACTCCTCCAGTTGCTCA |
| Atg4B | ACTGGGAAGATGGACGCAG | AGTATCCAAACGGGCTCTGA |
| Atg5 | TCAGAAGCTGTTTCGTCCTG | TGCAGAGGTGTTTCCAACAT |
| BECN1 | TGGACACGAGTTTCAAGATCC | CTCCTGGGTCTCTCCTGGTT |
| Atg7 | ATAATGTCCTTCCCGTCAGC | TCTCATCATCGCTCATGTCC |
| LC3A | CGACCGCTGTAAGGAGGTA | TTGACCAACTCGCTCATGTT |
| LC3B | GAGAAGACCTTCAAGCAGCG | TATCACCGGGATTTTGGTTG |
| LC3C | CTGACCATGACCCAGTTCCT | TGTACACGAAGCCATCCTCA |
| GABARAP | ATGTCATTCCACCCACCAGT | TTGAGCTTGAAGGAGGAGGA |
| GABARAPL1 | TCGGAAAAAGGAAGGAGAAA | TGGCCAACAGTAAGGTCAGA |
| GABARAPL2 | CTGTGGCTCAGTTCATGTGG | GTGTTCTCTCCGCTGTAGGC |
| Atg9A | CTGGAGCTCCTGAGACCACT | CCCTGCTCAGCCTTGTCTAT |
| Atg9B | CTGCAAGGAAAAGCTCTGG | ACAACCCCAAGGTGTGAGAC |
| Atg10 | CATTGTAGGGCCAGTTGTTG | CATTTGTGTCCAAGGGAAAA |
| Atg12 | TTGTGGCCTCAGAACAGTTG | CCATCACTGCCAAAACACTC |
| Atg13 | GGTGTCATCCTGTTCCTCCT | GGGACTCCTTCCTTCTTCCT |
| Atg14 | ACAACCACTGCATACCCTCA | CGCTCTCATCTGATTCTCCA |
| Atg16L1 | GGTTCTTTCAAAGCAGCACA | CTCTGGGAGGTCCATGCTAT |
| Vps18 | GCGCTTGTGTCCATGTCTAC | GCCATGGCTGTCTGTACATC |
| WIPI1 | GGGACCAGTGTGTCTTGATG | GTGGTTTAAGCAGTGGAGCA |
| WIPI2 | CTCGCTAGCCACAATTCAGA | CACCCAGGTCGTCTGTGTAG |
| Atg101 | CCCAGGATGTTGACTGTGAC | CTACCACATGCACCTTGACC |
| FIP200 | GACCCTGGGTACTTGGAAAA | AGCACTGCAGGACAAATCAG |
| UVRAG | CAAAAGGAGGGGAGAAGTTG | CCCCAAATATGGAGCTTTGT |
| PIK3C3 | CCTTGTCGATGAGCTTTGGT | CAGATGGATCAGAACCCACA |
| PI3KR4 | GGTCCTATGTTGTTGCAGGA | CACACCTTCACAATCCCATC |
| Vps11 | TAACAGTGCCTTGGAGTTGC | GAAAAGCTGTCATTGGAGCA |
| p62 | CCCGTCTACAGGTGAACTCC | CTGGGAGAGGGACTCAATCA |
| NBR1 | GTTGCTGCCTCTGCATACAA | TTTCTTCAGCAGCCGTAGGT |
| NDP52 | AAGGAGGCGCAAGACAAAAT | ATCTGCTGTTGCTCCAAGGT |
| OPTN | GTCCTTGATGGAGATGCAGA | AGGCAGAACCTCTCCACACT |
| CtsA | GGATTCCCTCAACCAGAAGA | TGGTCATCAGTATGGCTGCT |
| Lamp1 | TGCCTTTAAAGCTGCCAAC | TTCTCGTCCAGCAGACACTC |
| CtsD | CCAGAGGACTACACGCTCAA | CTGCTCTGGGACTCTCCTCT |
| CtsF | CTTTGGCATGCAGTTTTACC | GGACCCACGATGCAAGTAGT |
| MColn1 | TACATGGTGCTCAGCCTCTT | GTCGAATCAATTCACCAGCA |
| Clcn7 | AGTGCACCATGGACCTCTC | AGCTCCTCCAAGCCTCTCTT |
| Nrf2 | CGGTATGCAACAGGACATTG | AGAGGATGCTGCTGAAGGAA |
| NF-E2 | CTAGAGCCATCTGGGCTTTC | GCAGTAAGTTGTGGGTGGTG |
| KLF4 | GCCACCCACACTTGTGATTA | CCCGTGTGTTTACGGTAGTG |
| HDAC6 | TGGTCCTCAGCTACATCGAC | GCTACCCCTCATCCAAGGTA |
Table 2. Sequences of primers for cloning p62 promoter segments.
| Promoter | Forward primer (5′–3′) | Reverse primer (5′–3′) |
|---|---|---|
| P270 | AACAGATCTCAAGCCTGGGAGGGGCGA | CTCAAGCTTTGTAGCGAACGCGGAGGC |
| P310 | GGGAGATCTGTACCCCCAACTGAGGAT | CTCAAGCTTTGTAGCGAACGCGGAGGC |
| P1068 | GGTAGATCTTTGCCCACTTCGGAGCCC | CTCAAGCTTTGTAGCGAACGCGGAGGC |
Proteasomal peptidase activity
Cell lysates were prepared as for Western blotting, and peptidase activity was assayed in a 100-µl reaction (96-well plate format) containing 1 µg lysate protein, 50 mM Tris-HCl, pH 8.0, 10 mM MgCl2, 1 mM ATP, and 1 mM DTT. To measure the chymotrypsin-like activity, we used 50 µM Suc-LLVY–7-amino-4-methyicoumarin (Suc-LLVY-AMC; 20 mM in DMSO; I-1395.0100; Bachem), and for the caspase-like activity, we used 50 µM Z-Leu-Leu-Glu-AMC (10 mM in DMSO; I-1945.0005; Bachem). Fluorescence of AMC was measured in a SpectraMax M5 microplate reader (0310-5625; VWR International) with an excitation wavelength of 380 nm and emission of 460 nm. Fluorescence was read every 35 s for 1 h at 37°C, and the velocity of peptide hydrolysis was determined by the slope of relative fluorescence units over time.
Measurement of cellular protein degradation
To measure the degradation rate of long-lived proteins, WT or sh-p62 SH-SY5Y cells were first labeled for 24 h with [3H]phenylalanine (Phe l-[3,4,5 3H], ART0614, stock 1 mCi/ml in 0.01 M HCl, final 2 µCi/ml; American Radiolabeled Chemicals) and then chased for 2 h with complete medium containing 2 mM nonradioactive phenylalanine (P5482-25G; Sigma-Aldrich) to allow degradation of short-lived radioactive proteins. Cells were then incubated in chase medium containing 1 µM BTZ. Protein degradation was measured by the conversion of TCA-perceptible [3H]proteins to [3H]phenylalanine in the medium, which was soluble in 10% TCA (BDH3372-2; VWR International). Samples of media were collected after 1-h pretreatment. After TCA precipitation, 200 µl supernatant was mixed with 3 ml Ultima gold scintillation fluid (6013327; PerkinElmer) and measured with a PerkinElmer Tri-Carb 2910TR liquid scintillation analyzer. To calculate the percentage of protein degraded, cells were lysed after the last time point with 0.1 M NaOH, and 100 µl lysate was mixed with 3 ml scintillation fluid to measure total radioactivity incorporated as 100% cellular protein. Six wells of cells were assayed for each condition.
Cell viability and toxicity assays
CellTiter 96 AQueous nonradioactive cell proliferation assay (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium [MTS]; G5421; Promega), which measures the level of functional mitochondria, was used to measure cell viability. To assay adherent cells (M17 and SH-SY5Y), the same numbers of WT, sh-p62, sh-GABARAPL1, or sh-Nbr1 cells were seeded in 96-well plates and treated with BTZ. At the end of the treatment, the medium was replaced by 100 µl prewarmed fresh medium per well. To assay suspension cells (myeloma cells), cells were seeded in six-well plates and treated with BTZ. At the end of the treatment, cells were pelleted gently by centrifugation at 250 g for 5 min and resuspended in prewarmed fresh medium, and 100 µl cells per well were transferred to a 96-well plate. After 20 µl MTS/phenazine methosulfate (20:1) solution was added to each well, absorbance at 490 nM was measured every 30 s for 45 min at 37°C using a SpectraMax M5 microplate reader (0310-5625; VWR International) to determine the rate of MTS reduction into a formazan product by viable cells. Three wells of cells were assayed for each condition. Three wells of empty medium were assayed to determine the background rate of MTS reduction caused by growth medium, which was set at 0. For each cell line, the viability of untreated control cells was set as 100%.
CytoTox 96 nonradioactive cytotoxicity assay (G1780; Promega) was used to measure the percentage of dead cells that have lost membrane integrity and released lactate dehydrogenase (LDH) into the medium. SH-SY5Y cells were seeded in six-well plates and treated with BTZ. At indicated time points, 50 µl medium was collected from each well and stored at 4°C. After the last time points, all medium samples (50 µl each) were mixed in a 96-well plate with 50 µl iodonitro-tetrazolium violet solution, which can be reduced by LDH to a formazan product. Absorbance at 490 nM was measured in a SpectraMax M5 microplate reader (0310-5625; VWR International) every 30 s for 45 min at 37°C to determine the rate of formazan formation by LDH. Four wells of empty medium were assayed to determine the background rate of formazan formation caused by growth medium, which was set as 0. Two wells of untreated cells were lysed at the end of the time course in 0.8% Triton X-100 to measure the total amount of cellular LDH, which was set as 100%.
Dual luciferase reporter assay in M17 cells
Segments of p62 promoters (Fig. 6 A) were cloned into pGL4.20 (E6751; Promega) to drive the expression of firefly luciferase. For normalization purpose, pRL-TK (E2241; Promega) plasmid was used to express renilla luciferase constitutively. To transfect M17 cells, a transfection mixture was prepared that contained 2.5 µg pGL4.20 plasmids bearing different segments of p62 promoters, 2.5 µg pRL-TK, 15 µl Fugene HD (E2311; Promega), and 250 µl Opti-MEM. After mixing at room temperature for 20 min, the mixture was added to each well (of a six-well plate) containing 2.5 ml complete DMEM-F12. After 48 h, transfected cells were treated for 16 h with 0.1 µM BTZ. The activities of firefly luciferase driven by p62 promoter segments and renilla luciferase were measured with the Dual-Luciferase reporter assay system (E1960; Promega). The firefly/renilla ratio was calculated to indicate the transcription activity from the p62 promoter segment, and the fold increase in normalized luciferase activity in BTZ-treated cells was used to indicate the activation of transcription upon proteasome inhibition. Four wells of cells were assayed for each condition.
In vitro deubiquitination by Usp2
Recombinant Usp2 catalytic domain (stock concentration is 40 µM; E-504; R&D Systems) was added to cell lysate to a final concentration of 10 nM and incubated at 4°C for 2 h.
Statistics
Unpaired Student’s t test in Microsoft Excel was used for statistical analysis throughout the study. For mRNA measurements, two separate wells of cells were used for each condition in every figure.
Online supplemental material
Fig. S1 shows that myeloma cells MM1.S upon proteasome inhibition also rapidly induce p62 and GABARAPL1 expression before activation of autophagy or occurrence of widespread cell death. The figure also shows that MM1.S cells ubiquitinate p62 upon BTZ treatment. Fig. S2 shows that other Atg genes are induced only when proteasomes are strongly inhibited and also that prolonged proteasome inhibition activates the expression of several lysosomal genes but not HDAC6. Fig. S3 shows that cells lacking p62 or GABARAPL1 but not Nbr1 release LDH to a greater extent upon proteasome inhibition. Fig. S4 contains data related to Fig. 5 that FoxO3a, p-eIF2α, ATF4, Nrf2, NF-κB, or KLF4 were not playing a major role for the induction of p62 and GABARAPL1 upon proteasome inhibition. Fig. S5 shows that in neuroblastoma cells, where autophagy-mediated degradation of Ub conjugates and p62 is not very robust, p62 mainly forms inclusions of these conjugates rather than promoting their clearance by autophagy.
Supplementary Material
Acknowledgments
We thank Drs. M. Hannink, L. Dick, G. Bianchi, and M. Soustek for kindly providing valuable reagents. Studies on the regulation of p62 expression by Nrf1 benefited from discussions with Drs. R.J. Deshaies and S.K. Radhakrishnan. Microscopy was performed at the Nikon Imaging Center at Harvard Medical School.
Z. Sha is a Novartis Fellow of the Life Sciences Research Foundation and also held a fellowship from the Leukemia and Lymphoma Society. Our research was supported by grants from the National Institute of General Medical Sciences (GM051923-18), Project ALS, and Genentech.
The authors declare no competing financial interests.
Author contributions: Z. Sha and A. Goldberg conceived and designed experiments. Z. Sha, H.M. Schnell, and K. Ruoff performed the experiments. Z. Sha and A. Goldberg wrote the manuscript.
References
- Andrews N.C., and Orkin S.H.. 1994. Transcriptional control of erythropoiesis. Curr. Opin. Hematol. 1:119–124. [PubMed] [Google Scholar]
- Arrasate M., Mitra S., Schweitzer E.S., Segal M.R., and Finkbeiner S.. 2004. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 431:805–810. 10.1038/nature02998 [DOI] [PubMed] [Google Scholar]
- Belloni D., Veschini L., Foglieni C., Dell’Antonio G., Caligaris-Cappio F., Ferrarini M., and Ferrero E.. 2010. Bortezomib induces autophagic death in proliferating human endothelial cells. Exp. Cell Res. 316:1010–1018. 10.1016/j.yexcr.2009.11.005 [DOI] [PubMed] [Google Scholar]
- Boehm J.S., Zhao J.J., Yao J., Kim S.Y., Firestein R., Dunn I.F., Sjostrom S.K., Garraway L.A., Weremowicz S., Richardson A.L., et al. 2007. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell. 129:1065–1079. 10.1016/j.cell.2007.03.052 [DOI] [PubMed] [Google Scholar]
- Chen J.J., Tsu C.A., Gavin J.M., Milhollen M.A., Bruzzese F.J., Mallender W.D., Sintchak M.D., Bump N.J., Yang X., Ma J., et al. 2011. Mechanistic studies of substrate-assisted inhibition of ubiquitin-activating enzyme by adenosine sulfamate analogues. J. Biol. Chem. 286:40867–40877. 10.1074/jbc.M111.279984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demishtein A., Fraiberg M., Berko D., Tirosh B., Elazar Z., and Navon A.. 2017. SQSTM1/p62-mediated autophagy compensates for loss of proteasome polyubiquitin recruiting capacity. Autophagy. 13:1697–1708. 10.1080/15548627.2017.1356549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding W.X., Ni H.M., Gao W., Chen X., Kang J.H., Stolz D.B., Liu J., and Yin X.M.. 2009. Oncogenic transformation confers a selective susceptibility to the combined suppression of the proteasome and autophagy. Mol. Cancer Ther. 8:2036–2045. 10.1158/1535-7163.MCT-08-1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran A., Linares J.F., Galvez A.S., Wikenheiser K., Flores J.M., Diaz-Meco M.T., and Moscat J.. 2008. The signaling adaptor p62 is an important NF-kappaB mediator in tumorigenesis. Cancer Cell. 13:343–354. 10.1016/j.ccr.2008.02.001 [DOI] [PubMed] [Google Scholar]
- Duran A., Amanchy R., Linares J.F., Joshi J., Abu-Baker S., Porollo A., Hansen M., Moscat J., and Diaz-Meco M.T.. 2011. p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol. Cell. 44:134–146. 10.1016/j.molcel.2011.06.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fels D.R., and Koumenis C.. 2006. The PERK/eIF2alpha/ATF4 module of the UPR in hypoxia resistance and tumor growth. Cancer Biol. Ther. 5:723–728. 10.4161/cbt.5.7.2967 [DOI] [PubMed] [Google Scholar]
- Fels D.R., Ye J., Segan A.T., Kridel S.J., Spiotto M., Olson M., Koong A.C., and Koumenis C.. 2008. Preferential cytotoxicity of bortezomib toward hypoxic tumor cells via overactivation of endoplasmic reticulum stress pathways. Cancer Res. 68:9323–9330. 10.1158/0008-5472.CAN-08-2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita R., Takayama-Tsujimoto M., Satoh H., Gutiérrez L., Aburatani H., Fujii S., Sarai A., Bresnick E.H., Yamamoto M., and Motohashi H.. 2013. NF-E2 p45 is important for establishing normal function of platelets. Mol. Cell. Biol. 33:2659–2670. 10.1128/MCB.01274-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg A.L. 2012. Development of proteasome inhibitors as research tools and cancer drugs. J. Cell Biol. 199:583–588. 10.1083/jcb.201210077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Giasson B.I., Glavis-Bloom A., Brewer M.D., Shorter J., Gitler A.D., and Yang X.. 2014. A cellular system that degrades misfolded proteins and protects against neurodegeneration. Mol. Cell. 55:15–30. 10.1016/j.molcel.2014.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada M., Hanada S., Toivola D.M., Ghori N., and Omary M.B.. 2008. Autophagy activation by rapamycin eliminates mouse Mallory-Denk bodies and blocks their proteasome inhibitor-mediated formation. Hepatology. 47:2026–2035. 10.1002/hep.22294 [DOI] [PubMed] [Google Scholar]
- Hayes J.D., and McMahon M.. 2001. Molecular basis for the contribution of the antioxidant responsive element to cancer chemoprevention. Cancer Lett. 174:103–113. 10.1016/S0304-3835(01)00695-4 [DOI] [PubMed] [Google Scholar]
- Hoang B., Benavides A., Shi Y., Frost P., and Lichtenstein A.. 2009. Effect of autophagy on multiple myeloma cell viability. Mol. Cancer Ther. 8:1974–1984. 10.1158/1535-7163.MCT-08-1177 [DOI] [PubMed] [Google Scholar]
- Iwata A., Riley B.E., Johnston J.A., and Kopito R.R.. 2005. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J. Biol. Chem. 280:40282–40292. 10.1074/jbc.M508786200 [DOI] [PubMed] [Google Scholar]
- Jain A., Lamark T., Sjøttem E., Larsen K.B., Awuh J.A., Øvervatn A., McMahon M., Hayes J.D., and Johansen T.. 2010. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem. 285:22576–22591. 10.1074/jbc.M110.118976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y., Kovacs J.J., McLaurin A., Vance J.M., Ito A., and Yao T.P.. 2003. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 115:727–738. 10.1016/S0092-8674(03)00939-5 [DOI] [PubMed] [Google Scholar]
- Kirkin V., Lamark T., Sou Y.S., Bjørkøy G., Nunn J.L., Bruun J.A., Shvets E., McEwan D.G., Clausen T.H., Wild P., et al. 2009. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell. 33:505–516. 10.1016/j.molcel.2009.01.020 [DOI] [PubMed] [Google Scholar]
- Kisselev A.F., Callard A., and Goldberg A.L.. 2006. Importance of the different proteolytic sites of the proteasome and the efficacy of inhibitors varies with the protein substrate. J. Biol. Chem. 281:8582–8590. 10.1074/jbc.M509043200 [DOI] [PubMed] [Google Scholar]
- Klionsky D.J., and Ohsumi Y.. 1999. Vacuolar import of proteins and organelles from the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:1–32. 10.1146/annurev.cellbio.15.1.1 [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Tanaka K., Inoue K., and Kakizuka A.. 2002. Functional ATPase activity of p97/valosin-containing protein (VCP) is required for the quality control of endoplasmic reticulum in neuronally differentiated mammalian PC12 cells. J. Biol. Chem. 277:47358–47365. 10.1074/jbc.M207783200 [DOI] [PubMed] [Google Scholar]
- Koizumi S., Irie T., Hirayama S., Sakurai Y., Yashiroda H., Naguro I., Ichijo H., Hamazaki J., and Murata S.. 2016. The aspartyl protease DDI2 activates Nrf1 to compensate for proteasome dysfunction. eLife. 5:e18357 10.7554/eLife.18357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopito R.R. 2000. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 10:524–530. 10.1016/S0962-8924(00)01852-3 [DOI] [PubMed] [Google Scholar]
- Kwak M.K., Wakabayashi N., Greenlaw J.L., Yamamoto M., and Kensler T.W.. 2003. Antioxidants enhance mammalian proteasome expression through the Keap1-Nrf2 signaling pathway. Mol. Cell. Biol. 23:8786–8794. 10.1128/MCB.23.23.8786-8794.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamark T., Kirkin V., Dikic I., and Johansen T.. 2009. NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle. 8:1986–1990. 10.4161/cc.8.13.8892 [DOI] [PubMed] [Google Scholar]
- Lee J.Y., Koga H., Kawaguchi Y., Tang W., Wong E., Gao Y.S., Pandey U.B., Kaushik S., Tresse E., Lu J., et al. 2010. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 29:969–980. 10.1038/emboj.2009.405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Chou T.F., Pittman S.K., Keith A.L., Razani B., and Weihl C.C.. 2017. Keap1/Cullin3 modulates p62/SQSTM1 activity via UBA domain ubiquitination. Cell Rep. 19:188–202. 10.1016/j.celrep.2017.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrbach N.J., and Ruvkun G.. 2016. Proteasome dysfunction triggers activation of SKN-1A/Nrf1 by the aspartic protease DDI-1. eLife. 5:e17721 10.7554/eLife.17721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares J.F., Duran A., Yajima T., Pasparakis M., Moscat J., and Diaz-Meco M.T.. 2013. K63 polyubiquitination and activation of mTOR by the p62-TRAF6 complex in nutrient-activated cells. Mol. Cell. 51:283–296. 10.1016/j.molcel.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling J., Kang Y., Zhao R., Xia Q., Lee D.F., Chang Z., Li J., Peng B., Fleming J.B., Wang H., et al. 2012. KrasG12D-induced IKK2/β/NF-κB activation by IL-1α and p62 feedforward loops is required for development of pancreatic ductal adenocarcinoma. Cancer Cell. 21:105–120. 10.1016/j.ccr.2011.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K., den Brave F., and Jentsch S.. 2017. Receptor oligomerization guides pathway choice between proteasomal and autophagic degradation. Nat. Cell Biol. 19:732–739. 10.1038/ncb3531 [DOI] [PubMed] [Google Scholar]
- Manasanch E.E., and Orlowski R.Z.. 2017. Proteasome inhibitors in cancer therapy. Nat. Rev. Clin. Oncol. 14:417–433. 10.1038/nrclinonc.2016.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S., Wilkinson K.A., Nishimune A., and Henley J.M.. 2007. Emerging extranuclear roles of protein SUMOylation in neuronal function and dysfunction. Nat. Rev. Neurosci. 8:948–959. 10.1038/nrn2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto G., Wada K., Okuno M., Kurosawa M., and Nukina N.. 2011. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol. Cell. 44:279–289. 10.1016/j.molcel.2011.07.039 [DOI] [PubMed] [Google Scholar]
- Milan E., Perini T., Resnati M., Orfanelli U., Oliva L., Raimondi A., Cascio P., Bachi A., Marcatti M., Ciceri F., and Cenci S.. 2015. A plastic SQSTM1/p62-dependent autophagic reserve maintains proteostasis and determines proteasome inhibitor susceptibility in multiple myeloma cells. Autophagy. 11:1161–1178. 10.1080/15548627.2015.1052928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani M., Rzymski T., Mellor H.R., Pike L., Bottini A., Generali D., and Harris A.L.. 2009. The role of ATF4 stabilization and autophagy in resistance of breast cancer cells treated with Bortezomib. Cancer Res. 69:4415–4423. 10.1158/0008-5472.CAN-08-2839 [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yamamoto A., Hatano M., Kobayashi Y., Kabeya Y., Suzuki K., Tokuhisa T., Ohsumi Y., and Yoshimori T.. 2001. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 152:657–668. 10.1083/jcb.152.4.657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myeku N., Clelland C.L., Emrani S., Kukushkin N.V., Yu W.H., Goldberg A.L., and Duff K.E.. 2016. Tau-driven 26S proteasome impairment and cognitive dysfunction can be prevented early in disease by activating cAMP-PKA signaling. Nat. Med. 22:46–53. 10.1038/nm.4011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaso K., Yoshimoto Y., Nakano T., Takeshima T., Fukuhara Y., Yasui K., Araga S., Yanagawa T., Ishii T., and Nakashima K.. 2004. Transcriptional activation of p62/A170/ZIP during the formation of the aggregates: Possible mechanisms and the role in Lewy body formation in Parkinson’s disease. Brain Res. 1012:42–51. 10.1016/j.brainres.2004.03.029 [DOI] [PubMed] [Google Scholar]
- Palombella V.J., Rando O.J., Goldberg A.L., and Maniatis T.. 1994. The ubiquitin-proteasome pathway is required for processing the NF-kappa B1 precursor protein and the activation of NF-kappa B. Cell. 78:773–785. 10.1016/S0092-8674(94)90482-0 [DOI] [PubMed] [Google Scholar]
- Pandey U.B., Nie Z., Batlevi Y., McCray B.A., Ritson G.P., Nedelsky N.B., Schwartz S.L., DiProspero N.A., Knight M.A., Schuldiner O., et al. 2007. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 447:859–863. 10.1038/nature05853 [DOI] [PubMed] [Google Scholar]
- Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H., Øvervatn A., Bjørkøy G., and Johansen T.. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282:24131–24145. 10.1074/jbc.M702824200 [DOI] [PubMed] [Google Scholar]
- Pankiv S., Lamark T., Bruun J.A., Øvervatn A., Bjørkøy G., and Johansen T.. 2010. Nucleocytoplasmic shuttling of p62/SQSTM1 and its role in recruitment of nuclear polyubiquitinated proteins to promyelocytic leukemia bodies. J. Biol. Chem. 285:5941–5953. 10.1074/jbc.M109.039925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Hahm E.R., Lee D.K., and Yang C.H.. 2004. Inhibition of AP-1 transcription activator induces myc-dependent apoptosis in HL60 cells. J. Cell. Biochem. 91:973–986. 10.1002/jcb.10768 [DOI] [PubMed] [Google Scholar]
- Peng H., Yang J., Li G., You Q., Han W., Li T., Gao D., Xie X., Lee B.H., Du J., et al. 2017. Ubiquitylation of p62/sequestosome1 activates its autophagy receptor function and controls selective autophagy upon ubiquitin stress. Cell Res. 27:657–674. 10.1038/cr.2017.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilli M., Arko-Mensah J., Ponpuak M., Roberts E., Master S., Mandell M.A., Dupont N., Ornatowski W., Jiang S., Bradfute S.B., et al. 2012. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity. 37:223–234. 10.1016/j.immuni.2012.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan S.K., Lee C.S., Young P., Beskow A., Chan J.Y., and Deshaies R.J.. 2010. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol. Cell. 38:17–28. 10.1016/j.molcel.2010.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter-Landsberg C., and Leyk J.. 2013. Inclusion body formation, macroautophagy, and the role of HDAC6 in neurodegeneration. Acta Neuropathol. 126:793–807. 10.1007/s00401-013-1158-x [DOI] [PubMed] [Google Scholar]
- Riz I., Hawley T.S., and Hawley R.G.. 2015. KLF4-SQSTM1/p62-associated prosurvival autophagy contributes to carfilzomib resistance in multiple myeloma models. Oncotarget. 6:14814–14831. 10.18632/oncotarget.4530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockel T.D., Stuhlmann D., and von Mikecz A.. 2005. Proteasomes degrade proteins in focal subdomains of the human cell nucleus. J. Cell Sci. 118:5231–5242. 10.1242/jcs.02642 [DOI] [PubMed] [Google Scholar]
- Rogov V., Dötsch V., Johansen T., and Kirkin V.. 2014. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol. Cell. 53:167–178. 10.1016/j.molcel.2013.12.014 [DOI] [PubMed] [Google Scholar]
- Rozenknop A., Rogov V.V., Rogova N.Y., Löhr F., Güntert P., Dikic I., and Dötsch V.. 2011. Characterization of the interaction of GABARAPL-1 with the LIR motif of NBR1. J. Mol. Biol. 410:477–487. 10.1016/j.jmb.2011.05.003 [DOI] [PubMed] [Google Scholar]
- Sanz L., Diaz-Meco M.T., Nakano H., and Moscat J.. 2000. The atypical PKC-interacting protein p62 channels NF-kappaB activation by the IL-1-TRAF6 pathway. EMBO J. 19:1576–1586. 10.1093/emboj/19.7.1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel J., Larsen K.M., Matic I., van Hagen M., Cox J., Mann M., Andersen J.S., and Vertegaal A.C.. 2008. The ubiquitin-proteasome system is a key component of the SUMO-2/3 cycle. Mol. Cell. Proteomics. 7:2107–2122. 10.1074/mcp.M800025-MCP200 [DOI] [PubMed] [Google Scholar]
- Sebens S., Bauer I., Geismann C., Grage-Griebenow E., Ehlers S., Kruse M.L., Arlt A., and Schäfer H.. 2011. Inflammatory macrophages induce Nrf2 transcription factor-dependent proteasome activity in colonic NCM460 cells and thereby confer anti-apoptotic protection. J. Biol. Chem. 286:40911–40921. 10.1074/jbc.M111.274902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C., Di Malta C., Polito V.A., Garcia Arencibia M., Vetrini F., Erdin S., Erdin S.U., Huynh T., Medina D., Colella P., et al. 2011. TFEB links autophagy to lysosomal biogenesis. Science. 332:1429–1433. 10.1126/science.1204592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha Z., and Goldberg A.L.. 2014. Proteasome-mediated processing of Nrf1 is essential for coordinate induction of all proteasome subunits and p97. Curr. Biol. 24:1573–1583. 10.1016/j.cub.2014.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J. 1998. P62 and the sequestosome, a novel mechanism for protein metabolism. Arch. Pharm. Res. 21:629–633. 10.1007/BF02976748 [DOI] [PubMed] [Google Scholar]
- Sidrauski C., McGeachy A.M., Ingolia N.T., and Walter P.. 2015. The small molecule ISRIB reverses the effects of eIF2α phosphorylation on translation and stress granule assembly. eLife. 4:e05033 10.7554/eLife.05033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen J., Seeger M., Koch A., and Krüger E.. 2010. Proteasomal degradation is transcriptionally controlled by TCF11 via an ERAD-dependent feedback loop. Mol. Cell. 40:147–158. 10.1016/j.molcel.2010.09.012 [DOI] [PubMed] [Google Scholar]
- Strnad P., Zatloukal K., Stumptner C., Kulaksiz H., and Denk H.. 2008. Mallory-Denk-bodies: Lessons from keratin-containing hepatic inclusion bodies. Biochim. Biophys. Acta. 1782:764–774. 10.1016/j.bbadis.2008.08.008 [DOI] [PubMed] [Google Scholar]
- Tsvetkov P., Mendillo M.L., Zhao J., Carette J.E., Merrill P.H., Cikes D., Varadarajan M., van Diemen F.R., Penninger J.M., Goldberg A.L., et al. 2015. Compromising the 19S proteasome complex protects cells from reduced flux through the proteasome. eLife. 4:e08467 10.7554/eLife.08467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemura A., He F., Taniguchi K., Nakagawa H., Yamachika S., Font-Burgada J., Zhong Z., Subramaniam S., Raghunandan S., Duran A., et al. 2016. p62, upregulated during preneoplasia, induces hepatocellular carcinogenesis by maintaining survival of stressed HCC-initiating cells. Cancer Cell. 29:935–948. 10.1016/j.ccell.2016.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadlamudi R.K., and Shin J.. 1998. Genomic structure and promoter analysis of the p62 gene encoding a non-proteasomal multiubiquitin chain binding protein. FEBS Lett. 435:138–142. 10.1016/S0014-5793(98)01021-7 [DOI] [PubMed] [Google Scholar]
- Vaeteewoottacharn K., Kariya R., Matsuda K., Taura M., Wongkham C., Wongkham S., and Okada S.. 2013. Perturbation of proteasome function by bortezomib leading to ER stress-induced apoptotic cell death in cholangiocarcinoma. J. Cancer Res. Clin. Oncol. 139:1551–1562. 10.1007/s00432-013-1473-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.W., and Klionsky D.J.. 2003. The molecular mechanism of autophagy. Mol. Med. 9:65–76. [PMC free article] [PubMed] [Google Scholar]
- Zhang D.D., and Hannink M.. 2003. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol. Cell. Biol. 23:8137–8151. 10.1128/MCB.23.22.8137-8151.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Brault J.J., Schild A., Cao P., Sandri M., Schiaffino S., Lecker S.H., and Goldberg A.L.. 2007. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 6:472–483. 10.1016/j.cmet.2007.11.004 [DOI] [PubMed] [Google Scholar]
- Zhao J., Zhai B., Gygi S.P., and Goldberg A.L.. 2015. mTOR inhibition activates overall protein degradation by the ubiquitin proteasome system as well as by autophagy. Proc. Natl. Acad. Sci. USA. 112:15790–15797. 10.1073/pnas.1521919112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu K., Dunner K. Jr., and McConkey D.J.. 2010. Proteasome inhibitors activate autophagy as a cytoprotective response in human prostate cancer cells. Oncogene. 29:451–462. 10.1038/onc.2009.343 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.