Upon activation, GPCRs must exit cilia for appropriate signal transduction. Using bulk imaging of BBSome and single-molecule imaging of GPCRs, Ye et al. demonstrate that retrograde BBSome trains assemble on demand upon GPCR activation and ferry GPCRs across the transition zone. However, ciliary exit often fails because of a second diffusion barrier.
Abstract
A diffusion barrier at the transition zone enables the compartmentalization of signaling molecules by cilia. The BBSome and the small guanosine triphosphatase Arl6, which triggers BBSome coat polymerization, are required for the exit of activated signaling receptors from cilia, but how diffusion barriers are crossed when membrane proteins exit cilia remains to be determined. In this study, we found that activation of the ciliary G protein–coupled receptors (GPCRs) Smoothened and SSTR3 drove the Arl6-dependent assembly of large, highly processive, and cargo-laden retrograde BBSome trains. Single-molecule imaging revealed that the assembly of BBSome trains enables the lateral transport of ciliary GPCRs across the transition zone. However, the removal of activated GPCRs from cilia was inefficient because a second periciliary diffusion barrier was infrequently crossed. We conclude that exit from cilia is a two-step process in which BBSome/Arl6 trains first move activated GPCRs through the transition zone before a periciliary barrier can be crossed.
Introduction
Diffusion barriers establish the identity of the apical membrane in polarized epithelial cells, of the axon in neurons, of the daughter cell in budding yeast, and of cilia by impeding the lateral movement of membrane proteins (Trimble and Grinstein, 2015). The compartmentalization of cilia enables dynamic changes in ciliary composition through regulated trafficking. Upon Hedgehog pathway activation, the seven-transmembrane-protein Smoothened accumulates in cilia, and ciliary exit of the G protein–coupled receptor (GPCR) GPR161 ensures the appropriate transduction of Hedgehog signals (Bangs and Anderson, 2017; Nager et al., 2017). Although trafficking across the tight junction, the axon initial segment, and the yeast bud neck involves a vesicular carrier intermediate, the mechanisms of ciliary barrier crossing remain undetermined. A ciliary diffusion barrier has been localized to the transition zone, an ultrastructural specialization between the transition fibers of the basal body and the cilium shaft (Garcia-Gonzalo and Reiter, 2012; Gonçalves and Pelletier, 2017). Three hypotheses have been advanced for crossing the transition zone (Nachury et al., 2010; Jensen and Leroux, 2017). First, the detection of vesicles inside the transition zone indicates that a vesicular carrier may transport cargoes across this barrier (Jensen et al., 2004; Chuang et al., 2015). Second, indirect evidence for lateral transport between plasma and ciliary membranes (Hunnicutt et al., 1990; Milenkovic et al., 2009) suggests that membranous cargoes laterally traverse the transition zone by active transport. Third, the regulated opening of a gate inside the transition zone may let selective cargoes move through this membranous barrier (Dyson et al., 2017).
The active transport of proteins inside cilia termed the intraflagellar transport (IFT) is powered by microtubule motors moving along axonemal microtubules. It is now clear that axonemal precursors such as α/β-tubulin are delivered to the tips of cilia by anterograde IFT trains (Lechtreck, 2015; Kubo et al., 2016). In contrast, it is not known where, when, or how membrane proteins are selected for ciliary exit, and prior studies of ciliary signaling receptors dynamics by single-molecule imaging failed to uncover extended IFT movements (Ye et al., 2013; Milenkovic et al., 2015).
Results
Low-level expression recapitulates physiological ciliary trafficking dynamics
To characterize transition zone crossing by membranous cargoes, we sought a system where membrane proteins move across the transition zone in a synchronized manner. GPR161 and the prototypical ciliary GPCR somatostatin receptor 3 (SSTR3) both undergo retrieval from the cilium and back into the cell upon activation (Mukhopadhyay et al., 2013; Green et al., 2015). GPR161, a core component of the Hedgehog pathway that couples to Gαs, exits cilia when Smoothened is activated either indirectly by Hedgehog or directly by Smoothened agonist (SAG; Fig. 1 A; Pal et al., 2016). Meanwhile, SSTR3 is a well-characterized Gαi-coupled receptor that undergoes agonist-dependent retrieval (Fig. 1 A; Green et al., 2015; Nager et al., 2017). Consistent with previous research on SSTR3 in hippocampal neurons (Green et al., 2015), most of SSTR3 immunofluorescence was lost from neuronal cilia after 6 h treatment with the ligand somatostatin-14 (sst) or the SSTR3-specific agonist L796,778 (Figs. 1 B and S1, A–C). Similar to endogenous SSTR3, ciliary exit of endogenous GPR161 proceeds over the course of several hours (Mukhopadhyay et al., 2013). Signal-dependent retrieval is thus a considerably slower process than signal-dependent endocytosis.
Figure 1.
Reconstitution of signal-dependent retrieval of SSTR3 and GPR161. (A) Diagram of the signal-dependent retrieval systems under study. Left: Addition of sst triggers SSTR3 exit from cilia by directly activating SSTR3. Right: Addition of SAG activates the Hedgehog pathway and promotes GPR161 retrieval. SMO, Smoothened. (B) Kinetics of SSTR3 disappearance from cilia of primary hippocampal neurons and of IMCD3 stably expressing APSSTR3NG under the control of the TATA-less EF1α promoter were estimated by quantitation of immunofluorescence signals after addition of sst. The entire dataset for the sst condition is shown in Fig. S1 B. Data were fitted to a single exponential. Error bars indicate 95% confidence interval (CI). n = 280–424 cilia (neurons) and 57–80 cilia (IMCD3). (C) High-level expression of SSTR3 drives elongation of primary cilia. Top: APSSTR3GFP driven by various promoters or APSSTR3NG driven by EF1αΔ promoter was expressed stably at the FlpIn locus of IMCD3 cells, and ciliary fluorescence levels were measured and compared to a GFP calibrator (Breslow et al., 2013) or an NG calibrator (see Materials and methods). Endogenous SSTR3 levels were estimated by comparative immunostaining (see Materials and methods). A Mann-Whitney test was used for pairwise comparisons of the number of SSTR3 molecules per cilia in neurons and in IMCD3 cells expressing APSSTR3NG or APSSTR3GFP under the control of pEF1αΔ. P > 0.05. n = 10–38 cilia. Error bars represent SD. Bottom: Effect of APSSTR3GFP expression on cilium length. Cilia lengths were measured in the GFP channel by live-cell imaging. n = 10–38 cilia. Error bars represent SD. Cilium lengthening upon GPCR overexpression was previously reported by Guadiana et al. (2013). (D) IMCD3-[pCrys-APGPR161NG3] were treated for 2 h with either SAG or vehicle. APGPR161NG3 was visualized by NG fluorescence, and basal bodies of cilia were stained with ninein. All cells were pretreated with the translation inhibitor emetine to eliminate signals from new protein synthesis. Bar, 4 µm. (E) Absolute quantitation of ciliary GPCR abundance. Top: Calibration of single-molecule fluorescence intensity. Bacterially expressed NG3 protein was spotted on glass coverslips (inset), and the fluorescent intensity of each individual NG3 was measured. n = 1,257 particles measured. Bottom: The three-step photobleaching of a representative spot shows that the fluorescence was emitted by a single NG3 molecule. The measured fluorescence intensity of NG3 was used to calibrate NG- and NG3-tagged SSTR3, GPR161, BBS5, and IFT88. Bar, 0.5 μm. (F) IMCD3-[pEF1αΔ-APSSTR3NG] cells were treated with vehicle or sst for 2 h. Stable expression of an ER-localized biotin ligase BirA enables the biotinylation of APSSTR3 with the biotin existing in the DMEM/F-12 cell culture medium. Ciliary APSSTR3 was pulse-labeled by Alexa Fluor 647–conjugated mSA (mSA647) for 5–10 min before imaging (see Materials and methods for details). Bar, 1 μm. The absolute number of APSSTR3NG molecules per cilia at t0 was calculated by measuring the NG signal and using the NG3 calibrator. For all other time points, the ratio in ciliary mSA647 signal compared with t0 was used to calculate the absolute number of molecules (see Materials and methods for details). Data were fitted to a single exponential. Error bars indicate 95% CI. n = 14 cilia. (G) IMCD3-[pCrys-GPR161NG3] cells were treated with SAG or vehicle for 2 h. NG fluorescence was tracked in individual cilia, and the ratio of GPR161NG3 to endogenous GPR161 was used to calculate the total levels of GPR161 as detailed in Materials and methods. Bar, 1 μm. Data were fitted to a single exponential. Error bars indicate 95% CI. n = 12–20 cilia.
To dissect ciliary exit, we expressed GPCRs in mouse inner medullar collecting duct 3 (IMCD3) kidney cells, a widely used cell line for ciliary trafficking studies. GPCRs were tagged on the intracellular C terminus with a fluorescent protein (GFP or NeonGreen [NG]; Shaner et al., 2013), whereas a biotinylation acceptor peptide (AP) on the extracellular N terminus combined with coexpression of the biotin ligase BirA enabled pulse-chase studies with fluorescently labeled monovalent streptavidin (mSA; Howarth and Ting, 2008). When APSSTR3GFP under the control of the EF1α promoter was stably expressed in IMCD3 cells by single integration at the FlpIn locus, agonist-dependent exit of SSTR3 from cilia was undetectable. Molecular counting of GFP and comparison of immunofluorescence intensities revealed that pEF1α-driven expression resulted in SSTR3 levels that were an order of magnitude greater than in neurons (Fig. 1 C, top). Congruently, pEF1α-driven APSSTR3GFP expression resulted in a near doubling of cilia length, likely caused by protein overload driving ciliary membrane expansion and compensatory axoneme growth (Fig. 1 C, bottom; Guadiana et al., 2013). To express SSTR3 at levels closer to those found in neurons, we tested a variety of weak promoters and found that an EF1α promoter lacking the TATA box (pEF1αΔ) produced ciliary amounts of SSTR3 similar to those found in neurons (Fig. 1 C, top). pEF1αΔ-driven expression of SSTR3 did not alter ciliary length (Fig. 1 C, bottom). IMCD3-[pEF1αΔ-APSSTR3NG] cells recapitulated SSTR3 exit from cilia upon sst addition with nearly identical kinetics as in hippocampal neurons (Figs. 1 B and S1 D). Similarly, the Hedgehog signaling–dependent exit of GPR161 was recapitulated by expressing APGPR161NG3 from the δ-crystallin promoter (Fig. 1 D). pEF1αΔ-driven expression of NPY2RNG and NG-tagged melanin-concentrating hormone receptor 1 (MCHR1; MCHR1NG) yielded low ciliary levels (Fig. S1 E) with similar exit kinetics (Nager et al., 2017), thus demonstrating the broad applicability of low-expression promoters for studying the dynamics of ciliary GPCRs.
Calibration of the NG signal with recombinant proteins spotted on glass slides (Fig. 1 E) allowed the measurement of absolute levels of GPCRs per cilia (Fig. S1, F–H) and, together with pulse-chase labeling with mSA647, enabled a specific quantitation of signal-dependent exit rates at close to 500 molecules per hour (Fig. 1, F and G; and Video 1). In support of the precision of our absolute quantitation, using pEF1αΔ-driven SSTR3NG or SSTR3GFP and independent calibrators yielded very similar numbers of SSTR3 molecules per cilium (Fig. 1 C).
Further highlighting the power of AP- and NG-tagged GPCRs, the increased signal/noise ratio afforded by the direct labeling with NG or mSA647 compared with immunofluorescence (Fig. S1 I) made it possible to detect very low-abundance proteins (Fig. 1 C) whose presence in cilia escaped detection by traditional immunostaining techniques (Fig. S1 J). The decreased threshold of detection when pulse-labeling with mSA647 ensured a more faithful visualization of exit kinetics than when exit was monitored by immunostaining (Fig. 1, B and F).
In the absence of an agonist, the NG fluorescence of APSSTR3NG increased over time, whereas the signal from pulse labeling with mSA647 remained constant for 6 h (Fig. S1 K). In the presence of an agonist, the exit kinetics of APSSTR3NG were slower when monitored by direct visualization of the NG tag than by pulse-labeling with mSA647 (Fig. S1 K). Because pulse labeling only reports on SSTR3 exit, whereas the NG signal measures the total ciliary levels, these results indicate that the newly synthesized APSSTR3NG continues to enter cilia during the course of the experiment. We surmise that our previous attempts to assay signal-dependent exit of SSTR3 using strong promoters failed because the entry of newly synthesized GPCRs outpaced the slow exit kinetics.
Sorting complexes for ciliary entry and exit
The low-expression systems made it possible to validate the sorting complexes that carry out ciliary entry and exit. IFT-A is a complex of six proteins with structural elements that suggest a common ancestry with coat complexes (Jékely and Arendt, 2006; van Dam et al., 2013), and IFT-A is recruited to membranes by the PI(4,5)P2-binding protein Tulp3 (Mukhopadhyay et al., 2010). Although IFT-A is often described as the central mediator of retrograde transport (Lechtreck, 2015), IFT-A and TULP3 are required for the import of many GPCRs into cilia (Mukhopadhyay et al., 2010, 2013; Loktev and Jackson, 2013; Fu et al., 2016; Badgandi et al., 2017). We confirmed that Tulp3 is required for ciliary entry of SSTR3, NPY2R, and MCHR1 (Figs. 2 A and S2, A–C) and refined indirect interaction data by showing that the ciliary targeting signal of SSTR3 encoded within the third intracellular loop (i3; Berbari et al., 2008a) was specifically and directly recognized by the purified IFT-A complex (Fig. 2 B) or by the IFT-A subunit IFT140 overexpressed in HEK cells (Fig. S2 D). We conclude that IFT-A/Tulp3 functions as a coat adapter complex that mediates GPCR entry into cilia by directly recognizing sorting signals (Fig. 2 C).
Figure 2.
Roles of IFT-A and BBSome in ciliary entry and exit. (A) Tulp3 is required for ciliary entry of SSTR3. Box plots of ciliary APSSTR3NG intensities measured by NG fluorescence (RFUs) for various IMCD3 lines. Tulp3−/− cells were fixed to identify cilia using antiacetylated tubulin staining, and all other cells were imaged live as cilia were readily identified in the NG channel. NG fluorescence was not affected by fixation (Fig. S2 B). Asterisks indicate ANOVA significance values. ***, P < 0.0001; n.s., P > 0.05. n = 18–59 cilia. (B) IFT-A directly recognizes SSTR3i3. The IFT-A complex was purified from IMCD3-[LAP-IFT43] cells and incubated with beads coated with GST-SSTR3i3 or GST-SSTR5i3. Captured materials were eluted by cleaving off the beads and visualized by silver stain and immunoblotting. Five input equivalents were loaded in the eluate lanes. (C) Model of ciliary entry. (D and E) BBSome subunits were depleted by siRNA; Arl6, Ift27, and β-arrestin 2 (Arrb2) genes were knocked out by genome editing; and SA/TA denotes a phosphomutant of the C tail of SSTR3 that is unable to bind to β-arrestin 2 (Roth et al., 1997). (D) Representative time series of APSSTR3 pulse-labeled with mSA647 under different conditions. Bar, 2 µm. (E) Absolute retrieval rates were calculated by linear fitting of retrieval kinetics measured form SSTR3 pulse-chase labeling as in D (see Materials and methods; Nager et al., 2017). Error bars represent error of the fit. n = 10–35 cilia. (F) BBSome purified to near-homogeneity from bovine retina was incubated with glutathione beads coated with GST, GST-GPR161Ct, and GST-SSTR3i3. Captured materials were cleavage eluted and immunoblotted. Three input equivalents were loaded in the eluate lanes. MW, molecular weight. (G) Signal-dependent retrieval requires the joint activities of Arl6-GTP, BBSome, and β-arrestin 2.
Consistent with the requirement for the GPCR activation sensor β-arrestin 2 in GPR161 retrieval (Pal et al., 2016), signal-dependent retrieval of SSTR3 required β-arrestin 2 (Fig. 2, D and E; and Fig. S2, E and F; Green et al., 2015). The BBSome, a complex of eight Bardet-Biedl syndrome (BBS) proteins, resembles coat adapters at the structural level and polymerizes into a planar coat upon recruitment to membranes by the GTP-bound form of the small GTPase Arl6/BBS3 (Jin et al., 2010). The function of the BBSome in entry versus exit remains controversial. Although BBSome mutants have decreased ciliary levels of the GPCRs SSTR3, MCHR1, and NPY2R and of the polycystic kidney disease protein PKD1 (Berbari et al., 2008b; Loktev and Jackson, 2013; Su et al., 2014), GPR161, Smoothened, and D1R fail to exit cilia in BBSome or Arl6 mutants (Zhang et al., 2011; Eguether et al., 2014; Liew et al., 2014; Yee et al., 2015; Nager et al., 2017). Finally, systematic studies find that some proteins accumulate whereas others are depleted from Bbs mutant cilia (Lechtreck et al., 2013; Datta et al., 2015; Mick et al., 2015). In our near-endogenous expression systems, deletion of Arl6, of the candidate Arl6 activator Ift27/BBS19, or of β-arrestin 2 did not reduce the steady-state ciliary levels of SSTR3 (Figs. 2 A and S2 F). Instead, the BBSome, Arl6, and Ift27 were required for the signal-dependent retrieval of SSTR3 and GPR161 (Fig. 2, D and E; and Fig. S2, G and H). The carboxy-terminal tail of GPR161 (GPR161Ct) and the i3 of SSTR3 (SSTR3i3) directly interacted with purified BBSome (Fig. 2 F; Jin et al., 2010), suggesting that BBSome coats sort signaling receptors through the direct recognition of cytoplasmic determinants. Because SSTR3 and GPR161 are recognized by β-arrestin 2 in a signal-dependent manner (Roth et al., 1997; Pal et al., 2016), we conclude that the signal-dependent retrieval of GPR161 and SSTR3 is jointly and directly mediated by β-arrestin 2 and the BBSome (Fig. 2 G).
Signal-dependent BBSome redistribution to the tip of cilia triggers GPCR retrieval
To characterize the mechanisms of transition zone crossing by exiting GPCRs, we first sought to determine how BBSome coats facilitate signal-dependent retrieval. The movement of trains consisting of the IFT complex B (IFT-B) can be visualized by imaging foci of the IFT-B subunit IFT88 tagged with NG traveling in the anterograde and retrograde direction inside cilia (Video 2). Imaging of nematode, Chlamydomonas rheinhardtii, and mammalian cilia has shown that BBSome foci frequently comove with IFT-B foci (Ou et al., 2005; Lechtreck et al., 2009; Liew et al., 2014; Williams et al., 2014), suggesting coupling between the two complexes. To follow the dynamics of the BBSome and of IFT-B during SSTR3 and GPR161 signal-dependent retrieval, we expressed NG3BBS5 or NG3IFT88 at near-endogenous levels (Figs. 3 A and S2 I, and Video 3). In the absence of signaling, the BBSome and the IFT-B complex localized in a punctate pattern along the cilium (Fig. 3, B and C; and Fig. S2, J–L). Unexpectedly, SSTR3 activation led to a fourfold enrichment of the BBSome and a twofold enrichment of IFT-B at the tip (Fig. 3, B and C; and Fig. S2, J–L). Likewise, activation of the Hedgehog pathway resulted in BBSome accumulation at the ciliary tip (Fig. 3 C and Video 4). Similar to NG3BBS5, endogenous BBS9 became enriched at the tip upon activation of the Hedgehog pathway or of SSTR3 (Fig. 3 D). The BBSome thus joins a select group of Hedgehog factors that localize to the tip in a signal-dependent manner consisting of Gli2, Gli3, suppressor of fused homolog (SuFu), and Kif7.
Figure 3.
A Gαi–PKA axis promotes BBSome tip accumulation. (A) Near-endogenous expression of NG3BBS5. IMCD3 and IMCD3-[pEF1α-NG3BBS5] cells were subjected to immunoprecipitation (IP) with an anti-BBS5 antibody, and lysates and eluates were immunoblotted (IB) for BBS5. Molecular weights (MWs; kD) are indicated on the left. Measurement of band intensities with Image Lab (Bio-Rad Laboratories) indicates that the molar ratios between NG3BBS5 and endogenous BBS5 is 1.27. (B) IMCD3-[pEF1αΔ-APSSTR3, pEF1α-NG3BBS5] were treated with sst or vehicle for 40 min. Top: Representative images of cilia from live cells. NG3BBS5 is in fire scale, and white crosses mark the location of the basal body (see Fig. S2 J). Bottom: Line scans of NG3BBS5 fluorescence intensities along cilia of live cells. The line marks the mean intensity along length-normalized cilia. The shaded area shows the 95% CI. n = 20–29 cilia. (C) NG3BBS5 tip fluorescence intensities were quantified in live cells after 40 min of incubation with sst, SAG, Rp-cAMPs, or PKI. Cells were preincubated with PTX for 16 h to fully inactivate Gαi. Representative images are shown in the bottom panels with tips marked by yellow arrows. The total number of BBS5 molecules at the tip was calculated using the NG3 calibrator and the measured ratio of NG3BBS5 to total BBS5. The whiskers represent 1.5× the interquartile range. Asterisks indicate Mann-Whitney test significance values. ***, P < 0.0005. n = 20–29 cilia from three independent experiments. (D) IMCD3 or IMCD3-[pEF1αΔ-APSSTR3NG] cells were treated with vehicle, sst, or SAG for 40 min before fixation. The bar graph shows the percentage of BBS9-positive tips detected by immunofluorescence staining of endogenous BBS9. Error bars represent SD. n = 52–99 cilia. (E) The pathways downstream of Smoothened (SMO) and SSTR3 and the site of action of the pharmacological perturbations are shown. (F) Representative immunofluorescence images of ciliary BBS9 (red) in IMCD3-[pEF1α-NPHP3GFP-PKI] and IMCD3-[pEF1α-NPHP3GFP-PKIdead] cells (Mick et al., 2015). Optical sections were deconvolved, and X/Z projections are shown. Arrows mark tips of cilia. Percentages of BBS9-positive ciliary tips are plotted on the right. n = 29–50 cilia. Bars: (B) 1 µm; (C and F) 2 µm.
To determine whether tip enrichment of the BBSome represents a necessary step in GPCR retrieval, we sought to define and manipulate the molecular mechanisms of signal-dependent tip accumulation. Because SSTR3 and Smoothened are known to couple with Gαi and reduce cAMP production through Gαi-mediated inhibition of adenylate cyclases 5 and 6 (AC5/6; Yasuda et al., 1992; Shen et al., 2013), we tested for the role of signaling downstream of Gαi in promoting BBSome tip enrichment (Fig. 3 E). Pharmacological inhibition of Gαi by Pertussis toxin (PTX) blocked SAG- and sst-induced BBSome tip accumulation (Fig. 3 C). Meanwhile, inhibition of AC6 (Fig. S2 M) or inhibition of the cAMP-dependent protein kinase (PKA) by Rp-cAMPs or by a cell-permeable PKA inhibitory peptide (PKI; myr-PKI) led to BBSome tip accumulation in the absence of GPCR activation (Fig. 3 C). Furthermore, cilia-targeted PKI (Mick et al., 2015) was sufficient to redistribute BBSome to the tip (Fig. 3 F). Together with the findings of cilia-localized AC5 and AC6 (Masyuk et al., 2008; Kwon et al., 2010; Mick et al., 2015), these results suggest that activation of Smoothened and SSTR3 reduce the tonically high levels of ciliary cAMP (Moore et al., 2016) through Gαi-mediated inhibition of AC5/6 within cilia. Because PKA was recently shown to reside and function inside cilia (Mick et al., 2015; Moore et al., 2016), we propose that PKA antagonizes the recruitment of BBSome to the tip of cilia in unstimulated cells and that activation of Gαi-coupled GPCRs promotes BBSome tip recruitment by reducing the activity of ciliary PKA (Fig. 3 E).
Importantly, pharmacological alterations of the ciliary Gαi–PKA axis concordantly affected signal-dependent redistribution of BBSome to the tip of cilia and signal-dependent GPCR retrieval as the rates of signal-dependent retrieval of GPR161 and SSTR3 were greatly reduced by PTX (Fig. 4, A and B) and significantly accelerated by myr-PKI (Fig. 4, C and D). We note that PKA inhibition was not sufficient to trigger retrieval of GPR161 or SSTR3 in the absence of receptor stimulation (Fig. 4, C and D), suggesting the existence of mechanisms that act nonredundantly with the Gαi–PKA–BBSome axis (e.g., β-arrestin 2). In support of a Gαi–PKA–BBSome axis promoting GPCR exit, SSTR3 activation was sufficient to elicit the retrieval of GPR161 with identical kinetics to Smoothened activation (Fig. 4 E). In contrast, Smoothened activation was not sufficient to promote SSTR3 retrieval (Fig. 4 F). As the mechanisms that underlie the activation of GPR161 remain unknown, it is conceivable that signaling downstream of SSTR3 (through Gαi or Gβγ) triggers activation of GPR161 and that the subsequent engagement of β-arrestin 2 onto GPR161 cooperates with the Gαi–PKA–BBSome axis to promote GPR161 retrieval. In contrast, SSTR3 can only be activated by specific ligands.
Figure 4.
BBSome tip accumulation is required for the retrieval of SSTR3 and GPR161. (A and B) PTX slows down the exit of SSTR3 (A) and GPR161 (B). IMCD3-[pEF1αΔ-APSSTR3NG] or IMCD3-[pCrys-APGPR161NG3] were pretreated with PTX for 16 h to fully inactivate Gαi. After agonist treatment, ciliary GPCR levels were measured in live cells as described in Fig. 1 (F and G). Error bars represent 95% CI. n = 10–28 cilia. (C and D) PKA inhibition accelerates the exit of SSTR3 (C) and GPR161 (D). IMCD3-[pEF1αΔ-APSSTR3NG] or IMCD3-[pCrys-APGPR161NG3] cells were treated for 3 h with the indicated concentrations of agonist and/or PKI. Ciliary level of GPCRs were measured by NG fluorescence before and after treatment to estimate the rate of exit. Addition of PKI together with subsaturating concentrations of agonist significantly accelerated the GPCR exit rates to the near-maximal values observed with saturating concentrations of agonist. Error bars represent error of the fit. n = 67–112 cilia from three independent experiments. Asterisks indicate multiple regression significance values. *, P < 0.05. (E) GPR161 retrieval can be triggered by sst treatment. IMCD3-[pCrys-APGPR161NG3, pEF1αΔ-APSSTR3] cells were treated with SAG or sst for 2 h. NG fluorescence was tracked in individual cilia. Data were fitted to a single exponential. n = 10–21 cilia. (F) SAG treatment is not sufficient to trigger the retrieval of SSTR3. IMCD3-[pEF1αΔ-APSSTR3NG] cells were treated with SAG for 2 h. APSSTR3 was pulse-labeled with mSA647 and individual cilia were tracked. Data were fitted to a single exponential. Error bars represent 95% CI. n = 12 cilia.
To further establish that BBSome tipping represents a necessary intermediate in GPCR retrieval, we sought to identify molecules that recruit the BBSome to the tip of cilia in a signal-dependent manner. The plus end–directed microtubule motor Kif7 represented a candidate tip recruitment factor because Kif7 accumulates at the tip of cilia upon Hedgehog pathway activation (Endoh-Yamagami et al., 2009; Liem et al., 2009) and is necessary and sufficient to promote tip accumulation of the Hedgehog signaling factors Gli2 and Gli3 (He et al., 2014). Furthermore, KIF7 is a genetic modifier of BBS in human patients (Putoux et al., 2011). To follow the behavior of Kif7 and the BBSome in live IMCD3 cells, we stably coexpressed NG3BBS5 and Kif7 fused to the red fluorescent protein mScarlet. Smoothened activation led to the correlated coaccumulation of BBS5 and Kif7 at the tip of cilia (Fig. 5, A and B). Furthermore, in the rare instances where a second spot of Kif7 was found along cilia, possibly because part of the axoneme terminates before the tip, a similarly intense second spot of BBS5 was observed at the same location (Fig. 5 C). Because Kif7 depletion abolished tip accumulation of the BBSome (Fig. 5, D and E), these data suggest that Kif7 directly mediates the signal-dependent recruitment of BBSomes to the tip of cilia. Alternatively, it is conceivable that structural defects in cilia of Kif7-depeleted cells indirectly affect the recruitment of BBSomes to the tip. Congruent with a Kif7-mediated recruitment of BBSome to tips of cilia, Kif7 was found to coimmunoprecipitate with several BBSome subunits (Fig. S2 N). Given that dephosphorylation of Kif7 leads to the accumulation of Kif7 at the tip of cilia (Liu et al., 2014), we considered that phosphorylation by PKA may directly antagonize Kif7 tip accumulation and the Kif7–BBSome interaction. Concordantly, PKI led to the correlated coaccumulation of BBS5 and Kif7 at the tip of cilia to the same extent as SAG (Fig. 5, A and B), and elevated cAMP levels decreased the Kif7–BBSome interaction (Fig. 5 F). Finally, Kif7 was required for signal-dependent retrieval of SSTR3 (Fig. 5 G). Together, these results suggest that signaling downstream of Smoothened and SSTR3 leads to Kif7 dephosphorylation and that ensuing recruitment of BBSome to the tip initiates GPCR retrieval.
Figure 5.
PKA antagonizes the recruitment of the BBSome to the tip of cilia by Kif7. (A–C) Corecruitment of BBS5 and Kif7 to the tip of cilia in live IMCD3-[pEF1α-NG3BBS5; pCMV-Kif7mScarlet] cells. (A) Bottom: Representative images showing SAG- and PKI-induced accumulation of NG3BBS5 and Kif7mScarlet at ciliary tip. Cells were treated with SAG, PKI, or vehicle for 40 min before imaging. White boxes mark the ciliary tip. b, base; t, tip. Top: Individual channel crops of the tip area. (B) Correlation between the fluorescence signal of NG3BBS5 and Kif7mScarlet at the ciliary tip. Fluorescence signals of NG3BBS5 and Kif7mScarlet were measured at ciliary tip of live cells after 40 min treatment with vehicle, SAG, or PKI. Micrographs of NG3BBS5 and Kif7mScarlet at the ciliary tip for three representative data points are shown. Linear regressions (dotted lines) highlight the positive correlation between ciliary tip levels of NG3BBS5 and Kif7mScarlet in the presence of SAG or PKI. The Pearson correlation coefficient (r) is shown. Student’s t test of the Pearson’s correlation coefficient (r) returned a nonsignificant p-value under control conditions (P > 0.7) but a significant value after SAG or PKI treatment (P < 10−5). n = 40–49 cilia. (C) In SAG-treated cells where a second spot of Kif7 is occasionally found along cilia, a second spot of BBS5 was observed at the same location as Kif7. The yellow arrows mark the location of NG3BBS5 foci that accumulated at the ectopic tip. Bars, 2 µm. B, base; T, tip. (D and E) Kif7 is necessary for the redistribution of BBS5 to the tip of cilia. (D) Line scans of NG3BBS5 fluorescence intensities along cilia of live cells. Cells were transfected with siRNAs for 72 h and treated with sst for 40 min before live imaging of NG3BBS5 fluorescence. The line marks the mean intensity along length-normalized cilia. The shaded areas show the 95% CI (not depicted for the vehicle control). n = 20–27 cilia. (E) The total number of BBS5 molecules at the tip was calculated as in Fig. 3 C. n = 20–28 cilia from three independent experiments. (F) Kif7 interacts with BBSome, and PKA antagonizes this interaction. HEK293 cells cotransfected with Kif7GFP and MycBBS1 were treated with the cAMP phosphodiesterase inhibitor IBMX or vehicle for 30 min before lysis. Complexes were immunoprecipitated (IP) with anti-GFP antibodies. Lysates and eluates were immunoblotted (IB) for Myc. The capture efficiency of MycBBS1 by Kif7GFP was decreased 29 ± 3% upon treatment with IBMX. Molecular weights (MWs; kD) are indicated on the right. n = 3 independent experiments. (G) Kif7 is necessary for SSTR3 exit from cilia. IMCD3-[APSSTR3NG] cells were treated with siRNA targeting Kif7 or luciferase, pulse-labeled with mSA647, and imaged every 10 min after addition of sst. The resulting loss in mSA647 fluorescence was plotted and linearly fitted to determine the rate of SSTR3 retrieval. Asterisks indicate Mann-Whitney test significance values. *, P < 0.05; ***, P < 0.0005. Error bars represent SD. n = 13 cilia.
BBSome tip accumulation drives formation of cargo-laden retrograde trains
Analysis of NG3BBS5 and NG3IFT88 kymographs showed that anterograde BBSome trains, anterograde IFT trains, and retrograde IFT trains moved processively along the length of the cilium regardless of the signaling status (Figs. 6 A and S3, A and B). Yet in unstimulated cells, BBSome trains occasionally detached from retrograde IFT trains before reaching the base (Fig. 6 B), and overall, 90% of BBSome trains failed to reach the base of cilia (Fig. 6, A and C; and Fig. S3 B). Activation of Smoothened or SSTR3 or inhibition of PKA all doubled the number of BBSomes per retrograde train from 10 to 20 (Fig. 6 D) and led to a significant increase in the processivity of retrograde BBSome trains (Figs. 6 C and S3 B). Neither the frequencies nor the velocities of BBSome trains were affected by these treatments (Fig. 6, E and F). Meanwhile, SAG increased the number of IFT-B particles per retrograde train from 62 to 78 (Fig. 6 G). The addition of 106 BBSomes and 206 IFT-Bs to the tip upon SSTR3 activation (Figs. 3 C and S2 L) suggests that the signal-dependent accumulation of BBSomes and IFT-Bs at the tip drives the growth of retrograde trains by increasing the concentration of precursors at the site of assembly. Similar to the BBSome and IFT-B, Arl6 underwent signal-dependent tip accumulation (Fig. 6 H). As Arl6 was required for IFT88 tipping but not BBSome tipping (Fig. S2 L; Nager et al., 2017), Arl6 may recruit IFT-B particles to the tip by increasing the affinity of the BBSome for IFT-B.
Figure 6.
GPCR signaling and Arl6 drive assembly of large processive retrograde BBSome trains. (A) Representative kymographs of BBSome train movement. WT or Arl6−/− IMCD3-[pEF1α-NG3BBS5; pEF1αΔ-APSSTR3] cells were treated with vehicle, sst, SAG, or PKI for 40 min before imaging at 4 Hz for 30 s. Bars, 2 µm. (B) Representative kymograph from an IMCD3-[pEF1α-NG3BBS1, pCMV-tdTomatoIFT88] cell showing comovement and uncoupling between IFT-B and BBSome trains in untreated cells. Nearly 20% of BBSome trains displayed a distinct pause. n = 25 cilia. Bar, 2 μm. (C and D) WT or Arl6−/− IMCD3-[pEF1α-NG3BBS5; pEF1αΔ-APSSTR3] cells were treated with vehicle, sst, SAG, or PKI for 40 min before imaging. (C) The processivity of retrograde BBSome trains was measured by deconvolving kymographs into anterograde and retrograde components (see Materials and methods). The distance traveled by each retrograde train (normalized to the length of cilia) was estimated by manual inspection of the retrograde kymographs. (D) The fluorescence intensity of NG3BBS5 retrograde trains was extracted from deconvolved kymographs, and the total number of BBS5 molecules per train was calculated using the NG calibrator (see Materials and methods). The whiskers represent 1.5× the interquartile range. n = 52–91 cilia from three independent experiments. Asterisks indicate Mann-Whitney U test significance values; ***, P < 0.0001; n.s., P > 0.05. (E) Treatment with sst, SAG, or PKI did not change the frequency of retrograde BBSome trains. (F) Retrograde velocities of IFT trains, BBSome trains, and single SSTR3 molecules. IMCD3-[pEF1α-NG3IFT88], IMCD3-[pEF1α-NG3BBS5; pEF1αΔ-APSSTR3], or IMCD3-[pEF1αΔ-APSSTR3NG] were treated with vehicle, sst, SAG, or PKI for 40 min before imaging. IFT and BBSome train velocities were extracted from kymographs (see Materials and methods). QDSSTR3 velocities were measured from persistent retrograde movements lasting >6 s. Error bars represent SD. n = 9–18 cilia. Pairwise Mann-Whitney tests failed to show significant differences between any two conditions (P > 0.1). (G) The number of IFT88 molecules per retrograde train was measured in IMCD3-[pEF1α-NG3IFT88] cells treated with vehicle or SAG. Counting of molecules is detailed in Materials and methods. n = 11 cilia. (H) Arl6 immunofluorescence of cells treated with vehicle, SAG, or PKI. Optical sections were deconvolved, and X/Z projections are shown. The percentages of Arl6-positive tips are indicated below the micrographs. n = 88–118 cilia from four to five microscopic fields. b, base; t, tip. Bar, 2 μm.
PKI was sufficient to trigger tip accumulation of BBSome and Arl6 as well as the formation of large processive retrograde BBSome trains (Fig. 6, D and H; and Fig. S3 B), suggesting that recruitment of Arl6 and BBSomes to the tip may be sufficient to initiate the assembly of large processive retrograde IFT/BBSome trains. Because Arl6 and its candidate activator Ift27 were required for the signal-dependent formation of large processive BBSome trains (Fig. 6, C and D; and Fig. S3 C), we propose that BBSome coats polymerized upon Arl6-GTP binding become stably coupled to IFT-B trains to generate the large, signal-dependent, and processive retrograde BBSome/IFT trains.
Tip redistribution upon GPCR activation was not limited to the BBSome and IFT-B as GPR161 (Fig. 7, A and B) also underwent tip redistribution. Photobleaching the cilium exclusive of the tip revealed an enrichment of GPR161 at the tip upon Hedgehog pathway activation (Fig. 7, A and B). Recovery kinetics show that GPR161 at the tip became less dynamic after SAG treatment (Fig. 7 C). In support of an association between BBSomes and activated GPCRs at the tip of cilia, we found that WGA-mediated immobilization of membrane proteins increased the amount of BBSomes at the tip, most likely because BBSome–GPCR complexes were unable to leave the tip (Fig. 7 D). Furthermore, WGA treatment increased the amount of BBSomes throughout the length of cilia (Fig. 7 D), indicating that BBSome trains become trapped by immobilized GPCRs.
Figure 7.
Signaling promotes coupling between BBSomes and cargoes. (A) Representative images showing ciliary GPR161NG3 FRAP. IMCD3-[pCrys-APGPR161NG3] cells were treated with vehicle or SAG for 40 min before imaging. White dotted boxes in the first image indicate the photobleaching area that covers >80% of the cilia except the tip. After photobleaching, ciliary GPR161NG3 images were acquired every 5 s. Bar, 2 µm. B, base; T, tip. (B) Line scans of GPR161NG3 fluorescence intensities along cilia at 60 s after photobleaching. The gray and green lines mark mean intensities along length-normalized cilia for control and SAG-treated cells, respectively. Error bars represent SD. n = 10–11 cilia. (C) GPR161NG3 fluorescence at the tip was measured, and the decay of fluorescence signal over time in control and SAG-treated cells was plotted. Data were fitted to a single exponential to calculate the half-life. t1/2(control) = 5.14 ± 0.95 s; t1/2(SAG) = 10.47 ± 1.76 s. Error ranges represent error of fit. n = 10–11 cilia. (D) NG3BBS5 fluorescence was measured in live cells after 40 min of incubation with WGA, sst, or both. The total number of BBS5 at the tip (right) or axoneme (left) was calculated using the NG3 calibrator and the measured ratio of NG3BBS5 to total BBS5. Asterisks indicate Mann-Whitney test significance values. *, P < 0.05; ***, P < 0.0005. n = 20–25 cilia from three independent experiments. (E) Representative kymographs from mSA647-labeled WT or Arl6−/− IMCD3-[pEF1α-NG3BBS5; pEF1αΔ-APSSTR3] cells treated with sst for 1 h before imaging. The cells stably expressed an ER-localized biotin ligase BirA to enable visualization of APSSTR3 by mSA647 labeling. Bar, 2 μm. B, base; T, tip.
The bright and processive retrograde BBSome tracks observed upon sst addition frequently overlapped with faint tracks of SSTR3 (Fig. 7 E and Video 5). No comovement was observed between the faint BBSome trains and SSTR3 in the absence of Arl6 (Figs. 7 E and S3 D). These observations suggest that BBSome/Arl6 coats capture cargoes and move them from the tip to the base of cilia upon coupling to retrograde IFT trains.
Signaling promotes the processive retrograde movement of GPCRs
In bulk fluorescence imaging, the few fluorescent GPCRs that were being trafficked tended to be obscured by the fluorescent GPCRs that remain inside cilia. We measured that the largest retrograde trains contained close to 50 BBSomes. Assuming a 1:1 stoichiometry between BBSomes and cargo, this indicates that the brightest retrograde cargo tracks can carry at most 50/3,129 = 1.6% of the total ciliary SSTR3. It is therefore expected that such tracks will be extremely faint by bulk imaging (Fig. 7 E). To overcome the limitations inherent to ensemble imaging, we set out to visualize the molecular events that underlie GPCR retrieval at single-molecule resolution. Combining site-specific biotinylation of AP-tagged GPCRs (Ye et al., 2013) with Streptavidin (SA)-coupled quantum dots (Qdots [also referred to as QD]; SAQdot) enabled imaging at 2 Hz for >20 min (compared with 30-60 s when using mSA647; Fig. S3 E). By blocking most surface-exposed biotin groups with mSA before labeling with SAQdot, the vast majority of cilia bore no SAQdot, and cilia bearing one SAQdot were expected to possess a single biotinylated APGPCR (Fig. 8 A).
Figure 8.
Activated GPCRs undergo processive retrograde movements and confinements at base and tip. (A) Diagram of the Qdot labeling strategy. IMCD3-[pEF1αΔ-APSSTR3NG] cells stably expressing BirA-ER were first treated with unlabeled mSA to passivate the surface-exposed biotinylated APSSTR3. SAQdots were then added to the medium to label the GPCRs newly arrived at the surface. Finally, biotin was added to the medium to passivate the excess SA on Qdots. (B) The diffusive properties of SSTR3 are not altered by Qdot labeling. The instantaneous velocities of mSA647-labeled APSSTR3NG and Qdot655-labeled APSSTR3NG were measured by single-molecule tracking in the absence of sst. n = 1,223–3,032 instantaneous velocities. (C) Qdot labeling does not alter the exit rate of SSTR3. IMCD3-[pEF1α-APSSTR3GFP] cells were sparsely labeled with Qdot655 as described in Materials and methods and treated with vehicle or sst for 2 h before fixation. The number of Qdots per cilium was counted in both vehicle- and sst-treated conditions. n = 263–303 cilia from three independent experiments. Error bars represent SEM. (D) Representative kymographs showing the movements of SAQdot-labeled ciliary APSSTR3NG (QDSSTR3) in vehicle- or sst-treated cells. Red labels and line coloring highlight four characteristic movement behaviors. Bar, 2 µm. b, base; t, tip. (E) Centroid mapping of QDSSTR3. Left: The contour of the cilium was traced as a dotted line that captures all QDSSTR3 positions. (i) Diffusive movement, (ii) tip confinement followed by retrograde movement, (iii) retrograde movement followed by base confinement and return into the cilium, and (iv) tip confinement followed by fully processive retrograde movement and base confinement. The time dimension is color coded from red to purple. (F and G) Signaling increases tip confinement and processive retrograde movement of SSTR3. Cells were treated with sst (green, sst) or vehicle (gray, control) for 40 min before imaging was initiated for 10–20 min. (F) Durations of persistent movement events for QDSSTR3 in anterograde and retrograde directions. (G) The durations of confinement events for QDSSTR3 at ciliary tip or base were binned into two categories. n = 12 cilia for each condition. (H) Representative kymographs showing the comovement between a single QDSSTR3 and a BBSome retrograde train. IMCD3-[pEF1α-NG3BBS5; pEF1αΔ-APSSTR3] cells were treated with sst for 40 min before imaging. Bar, 2 μm. B, base; T, tip.
Tracking of SAQdot-labeled GPCRs (QDGPCRs) demonstrated that their dynamics were unperturbed by SAQdot labeling, consistent with a single valence of SAQdot per GPCR. First, consistent with prior single-molecule tracking studies (Ye et al., 2013; Milenkovic et al., 2015), QDSSTR3 displayed a diffusive behavior in the absence of agonist (Figs. 8 D and S4 A; Video 6), and instant velocities of diffusing GPCRs were nearly identical when labeled by mSA647 or SAQD655 (Fig. 8 B). Second, congruency of exit rates between bulk imaging and QDSSTR3 imaging indicated that SAQdot labeling did not impair exit (Fig. 8 C). Mapping the centroid of the Qdot enabled a 7-nm measurement precision (Fig. S3 F) that resolved the lateral displacement of QDSSTR3 around the 250-nm diameter of the ciliary membrane during diffusive events (Fig. 8 E, i). Addition of sst led to QDSSTR3 undergoing frequent confinements at the base and tip as well as processive retrograde movements (Fig. 8, D and E, ii–iv; Fig. S4 B; and Video 7). Although we occasionally observed apparent directional movement during brief periods of time for QDSSTR3 in control-treated cells (Figs. 8 D and S4 A), persistent retrograde movements of QDSSTR3 lasting ≥6 s were strictly dependent on SSTR3 activation (Figs. 8 F and S3 G), confirming that signaling drives the formation of cargo-laden IFT/BBSome retrograde trains. This signal-dependent increase in long processive movements was not observed for anterograde movements (Fig. 8 F). Processive transport events did not exhibit lateral displacement (Fig. 8 E, ii–iv), strongly suggesting that each cargo-laden IFT/BBSome train moves along a single axonemal microtubule. GPCRs occasionally resided for several seconds at either the tip or the base, and the frequency of confinement events, defined as residence >15 s, significantly increased in the presence of sst (Figs. 8 G and S3 H). Because processive retrograde transport events were frequently preceded by tip confinement (Fig. 8 E, ii and iv), it is likely that tip confinement of GPCRs reflects a step of cargo capture by BBSome coats assembling at the tip. Nevertheless, QDSSTR3 exhibited retrograde movements starting at any point along the cilium (Fig. 8 E, iii; and Fig. S4 B), indicative of activated GPCRs hopping onto retrograde BBSome trains.
Similar to QDSSTR3, QDGPR161 underwent mostly diffusive behavior in unstimulated cells, and addition of SAG led to frequent retrograde processive transport events (Fig. S4, C and D). Consistent with IFT/BBSome trains powering the retrograde transport of activated GPCRs, long processive retrograde movements of QDGPR161 were absent when Arl6 was deleted (Fig. S4 E). The mean velocity of the processive retrograde movements of QDSSTR3 was similar to the velocities of IFT and BBSome trains (Fig. 6 F), suggesting that signaling promotes coupling between cargoes and retrograde IFT trains. In support of this hypothesis, coimaging of NG3BBS5 and QDSSTR3 uncovered instances of comovement between the retrograde BBSome train and QDSSTR3 (Fig. 8 H). We conclude that activated GPR161 and SSTR3 are transported in the retrograde direction by the large processive BBSome trains that couple to retrograde IFT trains upon GPCR activation.
Arl6 and signaling enable transition zone crossing by GPR161
The expression of APGPCRs at near-endogenous levels enabled the SAQdot-mediated visualization of exit from the ciliary compartment at single-molecule resolution. In combined 21 h of single-molecule imaging, three exit events of QDGPR161 were observed. In all three events, QDGPR161 exit followed a stereotypical sequence of processive retrograde transport, base confinement, and diffusion away from the ciliary compartment (Fig. 9, A and B, iii). Surprisingly, many QDGPCRs that underwent retrograde transport and base confinement returned into the shaft of the cilium seemingly by processive anterograde transport (Fig. 9, A and B, i and ii; and Fig. 8, D and E, iii). During two out of the three preexit confinement events we observed, QDGPR161 first diffused within an area of <360-nm diameter before near-complete immobilization before exit (Figs. 9 C and S5 A; Videos 8 and 9). We did not observe a clear correlation between successful exit and either base residence time (Fig. 9 D) or area explored before exit (Fig. 9 E), suggesting that completion of exit is a stochastic process. Because QDGPR161 diffused within the 0.4-µm-deep focal plane after exit from the ciliary compartment, it is most likely that the GPCR moved into the plasma membrane after exit (Figs. 9 C and S5 A; Videos 8 and 9). Exit events were observed at the expected frequency based on measurements of bulk exit rates (Fig. S5 B), and this confirms that SAQdot labeling did not interfere with exit.
Figure 9.
GPCR base confinement results in either exit or reentry. (A) Cilium kymograph of QDGPR161. Cells were treated with SAG for 40 min before the start of imaging. Green, red, and purple labels and line coloring indicate retrograde movements, confinement and anterograde transport (events i and ii), or exit from cilia (iii). (B) Time series of the three confinement events. A reference image of bulk NG fluorescence from APGPR161NG3 was overlaid with images of QDGPR161. Time stamp (s) is in the top left corner. Bars, 2 µm. b, base; t, tip. (C) Centroid mapping of QDGPR161 during event iii. The QDGPR161 locations captured during the entire imaging session are shown as gray dots. The green track represents a processive retrograde transport event that precedes confinement of QDGPR161 (shown in red dots) at the base. Immediately before diffusion away from the cilium (purple track), QDGPR161 was nearly immobile for 4.5 s (purple dots). (D) Dot plot showing the base residence time of QDGPR161 during unproductive events (gray circles; n = 82) or before exit (green boxes; n = 3). IMCD3-[pCrys-APGPR161NG3] cells were treated with SAG for 40 min before imaging. (E) Scatter plot of areas explored by QDGPR161 during base residence events versus time. Gray circles represent unproductive events, and green circles represent preexit base residence events. Linear regression shows no obvious correlation between these two variables (Pearson correlation coefficient; r = 0.3). n = 17.
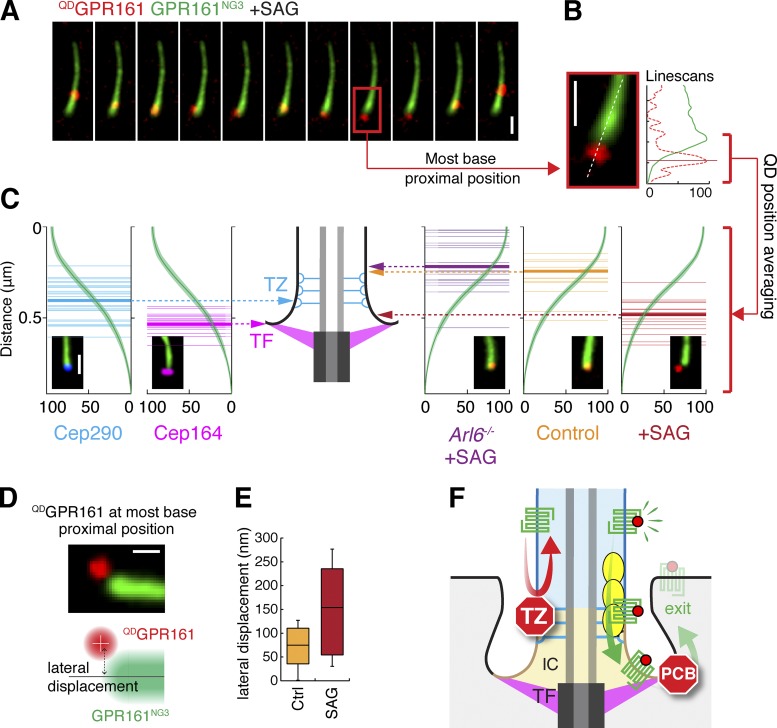
Unexpectedly, the position of QDGPR161 during signal-dependent base confinement events was often separate from the bulk fluorescence of GPR161NG3 (Fig. 10 A). Using the profile of the NG channel as a common reference (Fig. 10, A–C; and Fig. S5 C), we mapped the location of the most base-proximal position explored by QDGPR161 relative to the transition zone marker Cep290 and the transition fiber marker Cep164 and found that activated QDGPR161 moved into the 100-nm space between these two markers (Fig. 10, A–C). We termed the region between the 10th and 50th percentile of GPR161NG3 fluorescence the intermediate compartment as it defined the area visited by QDGPR161 before exit was completed. Systematic analysis of QDGPR161 position demonstrated that GPR161 enters the intermediate compartment in a signal- and Arl6-dependent manner (Fig. 10 C), thereby demonstrating that large, processive BBSome trains ferry GPR161 through the transition zone and deliver it to the intermediate compartment.
Figure 10.
GPR161 traverses the transition zone in an Arl6- and signaling-dependent manner. (A) Representative images of a base residence event for QDGPR161 after 40 min of SAG treatment. The most base-proximal position of QDGPR161 (red) relative to bulk APGPR161NG3 fluorescence (green) is highlighted by a red box, and the enlarged image is shown in B. (B) Line scans of the fluorescence intensities of QDGPR161 (red dotted line) and APGPR161NG3 (green line) along the length of cilia. (C) Because the longitudinal profile of APGPR161NG3 fluorescence is highly reproducible and unchanged by fixation (Fig. S5 C), the profile of APGPR161NG3 can be used as a common reference to align the positions of QDGPR161, Cep290, and Cep164 with respect to one another. WT or Arl6−/− IMCD3-[pCrys-APGPR161NG3] cells were treated with SAG or vehicle for 40 min before imaging. The most base-proximal positions of the centroid of QDGPR161 relative to the profile of APGPR161NG3 were plotted as thin lines, and the means were plotted as thick lines. The same method was used to plot the positions of immunofluorescence-stained Cep290 and Cep164 relative to APGPR161NG3 fluorescence. n = 15–26. (D and E) The lateral displacement between the centroid of QDGPR161 at its most base-proximal position and the center axis of APGPR161NG3 fluorescence was box plotted. The lateral displacement informs the half-width of the cilium base (control) or the intermediate compartment (SAG). IMCD3-[pCrys-APGPR161NG3] cells were treated with SAG or vehicle for 40 min before imaging. Bars: (A–C) 1 µm; (D) 0.5 µm. n = 11–16 cilia. (F) Two-barrier model for exit from cilia. The intermediate compartment is displayed in tan, the cilium shaft is blue, and the cell is gray. GPCR is green, agonist is red, and BBSome/Arl6 coats are yellow. IC, intermediate compartment; PCB, periciliary barrier; TF, transition fiber; TZ, transition zone. The diagram is not drawn to scale.
Tracking QDGPR161 over a combined 70-min period of imaging in the presence of SAG revealed that the probability of QDGPR161 entering the intermediate compartment is 0.66 during a 1-min interval (Fig. S5 D). Meanwhile, the absolute exit rate (Fig. 1 G) of 0.0043 molecules/min is equivalent to the probability of a single molecule of GPR161 experiencing exit during a 1-min interval. Thus, a comparison of intermediate compartment entry frequency and ciliary exit rates revealed that <1% of intermediate compartment visits productively lead to exit. Meanwhile, 99.3% of intermediate compartment visits are resolved by the GPCR returning into the ciliary compartment (Fig. 9, A and B, i and ii). Thus, although crossing the first diffusion barrier at the transition zone appears to be a prerequisite for ciliary exit, it is not sufficient to commit GPCRs for exit because a periciliary barrier blocks lateral diffusion between the intermediate compartment and the plasma membrane (Fig. 10 F).
Examining the lateral displacement of QDGPR161 to the center of the axoneme when QDGPR161 reached its most base-proximal position in unstimulated cells confirmed that GPR161 stayed within the expected 250-nm diameter of the ciliary membrane during rapid turnaround events in unstimulated cells (Fig. 10, D and E). The mean and maximal lateral displacements of QDGPR161 doubled when QDGPR161 reached its most base-proximal position in SAG-treated cells (Fig. 10, D and E). Because QDGPR161 visits its most base-proximal location in SAG-treated cells while residing within the intermediate compartment, these data indicate that the diameter of the intermediate compartment is close to 550 nm, similar to the 450-nm diameter defined by the tip of the transition fibers (Lau et al., 2012; Yang et al., 2015, 2017; Kanie et al., 2017). The periciliary barrier thus appears to be located near the point where the transition fibers attach to the ciliary membrane.
Discussion
Our pharmacological and live-cell imaging manipulations indicate that downstream of SSTR3 and Smoothened, a Gαi-mediated decrease in PKA activity promotes BBSome tip redistribution and subsequent retrieval of SSTR3 and GPR161. Intriguingly, the Gαs-coupled GPCR dopamine receptor 1 (DRD1) becomes enriched in cilia of amygdala neurons when BBSome function is compromised (Domire et al., 2011). It will be important for future studies to determine how Gα-coupled GPCRs are retrieved from cilia and whether BBSome tipping is induced by DRD1 activation.
We note that in olfactory receptor neurons, BBS4 is present in all IFT88-marked trains (Williams et al., 2014). In contrast, IFT tracks in unstimulated IMCD3 cells are often devoid of BBSomes, and we observed frequent uncoupling of BBSomes from IFT trains (Fig. 6 B). This suggests that olfactory receptor neurons resemble IMCD3 cells under signaling conditions with respect to BBSome transport. In this context, it is notable that BBS2 and BBS4 accumulate at the tip of olfactory cilia (Williams et al., 2014), and we propose that BBSome transport is highly active in olfactory receptor neurons. According to our absolute quantitation, an average retrograde train contains 10 BBSomes and 62 IFT-B complexes in untreated cells (Fig. 6, D and G). The BBSome/IFT stoichiometry (6.2:1) we measured by quantitative imaging is in close concordance with that measured using quantitative mass spectrometry in Chlamydomonas (6.5:1; Lechtreck et al., 2009), thus suggesting a low basal rate of BBSome-mediated transport in Chlamydomonas under vegetative conditions.
We found that base confinement events are prerequisites to exit. In contrast to GPR161 and SSTR3 whose activation increased their base confinement frequency, Smoothened activation decreases its base confinement frequency (Milenkovic et al., 2015). As Smoothened accumulates in cilia of unstimulated Bbs mutant cells (Zhang et al., 2011; Eguether et al., 2014; Goetz et al., 2017), this suggests that Smoothened is constitutively retrieved from cilia by the BBSome and that a reduction in Smoothened retrieval underlies the signal-dependent accumulation of Smoothened in cilia.
Although the idea of a periciliary diffusion barrier was initially considered (Nachury et al., 2010), the transition zone has come to be viewed as the sole diffusion barrier of the cilium in recent years (Garcia-Gonzalo and Reiter, 2012; Gonçalves and Pelletier, 2017; Jensen and Leroux, 2017). Our finding that the ciliary membrane is individualized from the plasma membrane by two successive diffusion barriers suggests the existence of an intermediate compartment located between the transition zone and a nearly impassable periciliary barrier. Crossing of the second barrier is extremely infrequent and is often preceded by a near-complete immobilization before exit. In all the exit events we imaged, QDGPR161 diffused within the plane of imaging after exit, suggesting that the QDGPR161 stayed in the plasma membrane. However, we cannot rule out that this mobility reflects that of an endosome or that endocytosis mediates crossing of the second barrier.
The periciliary barrier is likely to correspond with the recently described distal appendage matrix (DAM) because depletion of the DAM component FBF1 results in the leakage of GPCRs from cilia (Yang et al., 2017). Remarkably, when the receptors for insulin-like growth factor 1 (IGF1) or for transforming growth factor β (TGF-β) undergo signal-dependent exit from cilia, they transiently localize to a zone at the base of cilia that does not overlap with axonemal markers and may correspond to the intermediate compartment. Furthermore, residence of activated IGF1R and TGF-βR at the base of cilia appears to organize downstream signaling for these two pathways (Clement et al., 2013; Yeh et al., 2013). The intermediate compartment may harbor specific lipids as both PI(4,5)P2 and PI(3,4,5)P3 are dynamically enriched at a zone at the base of cilia that is clearly nonoverlapping from axonemal markers in mammalian cells (Dyson et al., 2017). The intermediate compartment may therefore constitute a privileged signaling locale. Finally, the existence of a second barrier explains why transition zone mutants have only mild mislocalization phenotypes and can still assemble cilia.
The ultrastructural location of the diffusion barrier within the transition zone is beyond the resolution of our imaging study. However, considering that QDGPR161 explores a compartment that is ∼220 nm long during base confinement events (Fig. 9 C), and given that the distance between the tip of the distal appendages (marked by Cep164) and the proximal end of the transition zone (marked by Cep290) is ∼100 nm (Yang et al., 2015), it is likely that the intermediate compartment encompasses part of the transition zone and that the diffusion barrier is located within the most distal part of the transition zone (Fig. 10 F).
Furthermore, because activated GPR161 only crosses the transition zone when BBSome/Arl6 coat assembly is permitted, the hypothesis that Hedgehog signaling loosens the diffusion barrier of the transition zone (Dyson et al., 2017) cannot account for GPR161 exit. Instead, our data suggest that BBSome/Arl6 coats bound to retrograde IFT trains on the axoneme-facing side and to cargoes on the membrane-facing side facilitate lateral transport through the transition zone. Thus, in contrast to all other known diffusion barriers, the transition zone is a porous barrier that allows the selective permeation of GPCRs bound to BBSome/Arl6 coats. Physical and genetic interactions between the BBSome and transition zone proteins such as Cep290 (Zhang et al., 2014; Barbelanne et al., 2015; Yee et al., 2015; Goetz et al., 2017) are in agreement with a general model where BBSome/Arl6 coats contact the transition zone during crossing. Taking our data into account, we propose that Arl6-GTP increases the coupling between cargoes, BBSomes, and IFT trains to facilitate lateral transport through the transition zone. However, the intimate details of transition zone crossing remain to be determined. In particular, the lack of precedent for selective permeation though a membrane diffusion barrier points to distinguishing biophysical features of the transition zone whose definition promises to enrich the concepts underlying diffusion barriers.
Materials and methods
Cell line construction
For all experiments, a mouse IMCD3-FlpIn cell line was used (gift from P.K. Jackson, Stanford University, Stanford, CA). IMCD3-FlpIn cells were cultured in DMEM/F-12 (11330-057; Gibco) supplemented with 5% FBS, 100 U/ml penicillin-streptomycin, and 2 mM l-glutamine.
Cell lines expressing SSTR3, GPR161, BBS1, BBS5, and IFT88 were generated using the FlpIn System (Thermo Fisher Scientific). Plasmids with multiple expression cassettes driven by low-expression promoters were built by modification of the pEF5B/FRT plasmid. The second cassette was inserted into an NsiI site that had been introduced in the pEF5B/FRT plasmid between the ampicillin cassette and an EF1α promoter. Subsequent cloning into the NsiI site was used to incorporate NsiI-pPGK-BirA-ER or NsiI-pEF1αΔ-APSSTR3-IRES-BirA-ER followed by a herpes simplex virus polyadenylation signal (Nager et al., 2017). Coding sequences were amplified from plasmids encoding human BBS1 and BBS5 (gifts from V. Sheffield, University of Iowa, Iowa City, IA), BirA-ER (a gift from A. Ting, Stanford University, Stanford, CA; 20856; Addgene; Howarth and Ting, 2008), mouse GPR161 (BC028163; Mammalian Gene Collection [MGC]; GE Healthcare), human NPY2R (BC075052; MGC; GE Healthcare), mouse IFT88 (IOM20300; UltimateORF; Invitrogen), human MCHR1 (BC001736; MGC; GE Healthcare), PACT (pericentrin and AKAP450 centrosome-targeting domain; gift from S. Munro, Medical Research Council, Cambridge, UK; Gillingham and Munro, 2000), mouse Kif7 (gift from K. Anderson, Memorial Sloan Kettering Cancer Center, New York, NY; He et al., 2014), mouse SSTR3 (gift from K. Mykytyn, Ohio State University, Columbus, OH), and IFT43. BBS1, BBS5, and IFT88 were expressed by the EF1α promoter, SSTR3, NPY2R, and MCHR1 were expressed by the EF1αΔ promoter, and GPR161 was expressed by the Crys promoter. GFP, NG (Shaner et al., 2013), mScarlet (gift from D. Gadella, University of Amsterdam, Amsterdam, Netherlands; 85042; Addgene; Bindels et al., 2017), tdTomato (gift from M. Davidson, Florida State University, Tallahassee, FL; 54653; Addgene; Shaner et al., 2004), and an acceptor peptide (GLNDIFEAQKIEWHE) for the biotin ligase BirA (AP) were used in fusion proteins. Kif7 cDNA was subcloned into a modified pCMV-based plasmid (pmScarlet-C; 85042; Addgene) that was transfected into IMCD3 cells using Lipofectamine 2000, and clones were selected using neomycin resistance. The expression level of NG3BBS5 and NG3IFT88 relative to endogenous BBS5 and IFT88 were determined by Western blotting (Figs. 3 A and S2 I), and the expression levels of exogenous SSTR3 or GPR161 compared with endogenous GPCRs were determined by measuring the intensity of ciliary fluorescence after immunostaining (Figs. 1 C and S1 B).
CRISPR-based genome editing was conducted by transiently expressing Cas9 and guide RNA from plasmids derivative of pX330 (gift from F. Zhang, Massachusetts Institute of Technology, Cambridge, MA; 42230; Addgene; Cong et al., 2013). The guide sequences were: 5′-AAGCCGCGATATGGGCTTGC-3′ for Arl6 (Liew et al., 2014), 5′-GGAAATGGGTCCCGTCGCTG-3′ for Ift27 (Liew et al., 2014), 5′-ACTCACCCACGGGGTCCACG-3′ for Arrb1 (Nager et al., 2017), 5′-TCTAGGCAAACTTACCCACA-3′ for Arrb2 (Nager et al., 2017), and 5′-ACGTCGCTGCGAGGCATCTG-3′ for Tulp3 (this study).
CRISPR-modified clones were isolated by limited dilution and verified by Western blotting for the disrupted gene product. To confirm gene editing, the modified genomic locus was isolated using the CloneJET PCR cloning kit (K1231; Thermo Fisher Scientific). The genotypes were Arl6, NM_001347244.1:c.-10_25del; c.3_6del; Ift27, the genotype was not characterized; Tulp3, NM_011657.2:c.142_143del; c.142_143insA; Arrb1, NM_177231.2:c.108del;c.108del; and Arrb2, NM_001271358.1:c.112del;c112_113del.
Low-expression promoters
To build constructs with low-expression promoters, an NsiI restriction cloning site was inserted by site-directed mutagenesis before the EF1α promoter in pEF5B/FRT (Nager et al., 2017). The EF1α promoter was then excised by NsiI and SpeI and replaced by either the ubiquitin C promoter (UbC), thymidine kinase (TK), EF1αΔ, CMVΔ, or a minimal chicken lens δ-crystallin promoter (Kamachi and Kondoh, 1993; Ferreira et al., 2011; Morita et al., 2012). The UbC promoter was cloned from pLenti6/UbC/V5-Dest (V49910; Thermo Fisher Scientific). The TK promoter was from pRL TK (E2241; Promega). EF1αΔ consists of a TATA-less EF1α promoter from pEF5/FRT/V5-Dest (V602020; Thermo Fisher Scientific), wherein the TATA box sequence TATAA was mutated to TCCCC. CMVΔ was designed after the CMV(Δ6) promoter (Morita et al., 2012) and was cloned from pcDNA3.1 (V79020; Thermo Fisher Scientific). The chicken lens δ-crystallin promoter was cloned from a pGL3-8×Gli–firefly luciferase plasmid (gift from P. Beachy, Stanford University, Stanford, CA).
Hippocampal neurons
Rat hippocampal neurons were dissected from P0 or P1 rat pups (gifts from M. Lin, Stanford University, Stanford, CA) and plated on poly-d-lysine–coated 12-mm #0 cover glass (633009; Carolina Biological Supply). Neurons were cultured in neurobasal medium with serum-free B27 (21103049; Thermo Fisher Scientific) and Gibco GlutaMAX (35050061; Thermo Fisher Scientific). Neurons were identified by nuclear NeuN staining in immunofluorescence experiments.
Transfection
Plasmids were reverse-transfected by Lipofectamine 2000 (11668027; Thermo Fisher Scientific). In brief, detached cells were plated with the transfection reagent and plasmid in OptiMEM (31985070; Thermo Fisher Scientific). The transfection reagent was replaced by fresh DMEM/F-12 medium after 4-h siRNAs were reverse-transfected by Lipofectamine RNAiMAX (13778030; Thermo Fisher Scientific). In brief, detached cells were plated with the transfection reagent and the siRNA duplex in OptiMEM. Cells were then grown for 48 h before 24 h starvation and subsequent imaging. Control (D-001210-04-05), BBS1 (D-019180-03), BBS2 (D-010080-02), and BBS4 (D-054691-03) were from GE Healthcare, and the Kif7 siRNA (GS16576) and matched control (1027280) were from QIAGEN. siRNA targeting Arl6 (5′-CTTTAGGACTTGAGACATT-3′) was previously validated (Jin et al., 2010).
Pharmacology
Small molecules were added at the following concentrations and for the indicated pretreatment times unless otherwise indicated: ACQ090 (20 µM; Novartis; Bänziger et al., 2003), 3-isobutyl-1-methylxanthine (IBMX; 500 µM; 30 min preincubation; I5879; Sigma-Aldrich), L796,778 (10 µM; Merck; Rohrer et al., 1998), PKI (50 µM; 40 min preincubation; BML-P210-0500; Enzo Life Sciences), PTX (10 ng/ml; 16 h preincubation; 180; List Biological Laboratories), Rp-cAMPS (10 µM; 2 h preincubation; sc-24010; Santa Cruz Biotechnology, Inc.), SAG (200 nM; ALX-270-426-M001; Enzo Life Sciences), sst (10 µM; ASR-003; Alomone Labs), and SQ22536 (500 µM; 40 min preincubation; S153; Sigma-Aldrich).
Antibodies and affinity reagents
The following antibodies were used: AC3 (rabbit; sc-588; Santa Cruz Biotechnology, Inc.), acetylated tubulin (mouse; clone 6-11B-1; Sigma-Aldrich), Arl6 (rabbit; Jin et al., 2010), β-arrestin 1 (rabbit; 15361-1-AP; ProteinTech), β-arrestin 2 (rabbit; 10171-1-AP; ProteinTech), BBS5 (rabbit; 14569-1-AP; ProteinTech; used for immunoblotting), BBS5 (mouse monoclonal; used for immunoprecipitation; gift from C. Smith, University of Florida, Gainesville, FL; Smith et al., 2013), BBS9 (rabbit; HPA021289; Sigma-Aldrich), centrin (mouse; clone 20H5; EMD Millipore), Cep164 (rabbit; gift from T. Stearns, Stanford University, Stanford, CA), Cep290 (rabbit; gift from S. Saunier, Institut Imagine, Paris, France), c-Myc (mouse; sc-40; Santa Cruz Biotechnology, Inc.), GFP (rabbit; A11122; Invitrogen), GPR161 (rabbit; gift from S. Mukhopadhyay, University of Texas Southwestern, Dallas, TX), IFT139 (rabbit; gift from P. Tran, University of Kansas, Kansas City, KS; Tran et al., 2008), IFT140 (rabbit; 17460-1-AP; ProteinTech), NeuN (mouse; MAB377; EMD Millipore), ninein (rabbit; gift from M. Bornens, Institut Curie, Paris, France; Mogensen et al., 2000), phospho-p44/42 MAPK Erk1/2 Thr202/Tyr204 (rabbit; 4370; Cell Signaling Technologies), SSTR3 (goat; sc-11617; Santa Cruz Biotechnology, Inc.), and TULP3 (rabbit; gift from J. Eggenschwiler, University of Georgia, Athens, GA; Norman et al., 2009). Biotinylated SSTR3 and GPR161 were detected using Alexa Fluor 647–labeled mSA (Ye et al., 2013).
Protein–protein interaction assays
The IFT-A complex was purified from IMCD3 cells stably expressing IFT43 with an N-terminal LAP tag. 3 × 108 cells were harvested and lysed in 6 ml LAP200N buffer (50 mM Hepes, pH 7.4, 200 mM KCl, 1 mM EGTA, 1 mM MgCl2, 10% glycerol, and 0.05% NP-40) with protease inhibitors. The IFT-A complex was captured on 100 µl Affiprep protein A beads (Bio-Rad Laboratories) coupled to 100 µg GFP antibody, and complexes were eluted in 250 µl LAP200N containing 3 µg TEV protease (Nachury, 2008). The BBSome was purified to near-homogeneity from bovine retinal extract by affinity chromatography on GST-Arl6GTP followed by MonoS (Jin et al., 2010). GST and GST-SSTRi3 fusion proteins were purified from Escherichia coli and used for capture of pure BBSome or pure IFT-A samples (Jin et al., 2010).
The interaction of GFPKif7 with MycBBS1 was assayed by cotransfection/coimmunoprecipitation. HEK293 cells were forward transfected in a 6-cm plate with plasmids expressing GFPKif7 and either MycRFP or a Myc-tagged BBSome subunit. After 2 d, cells were washed and treated with media containing either IBMX or DMSO for 30 min. Cells were then trypsinized, pelleted, and cleaned in a flacon tube and then lysed for 10 min with cold coimmunoprecipitation buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% Triton X-100, 1 mM DTT, 1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride, 800 nM aprotonin, 15 µM E-64, 10 mg/ml leupeptin, 10 mg/ml pepstatin A, and 10 mg/ml bestatin). The resulting lysates were then centrifuged for 15 min, concentration matched, and then added to anti-GFP antibody–coupled beads in coimmunoprecipitation buffer. The beads were rotated for 20 min at 4°C, pelleted, washed with coimmunoprecipitation buffer four times, and eluted by boiling in SDS-PAGE loading buffer.
Microscopy
All imaging was conducted on an DeltaVision Elite (GE Healthcare) equipped with a Plan Apochromat 60× 1.40 NA oil objective lens and a Plan Apochromat 60× 1.49 NA total internal reflection microscopy (TIRF) oil objective lens (Olympus) and a 488-nm laser from DeltaVision Quantifiable Laser Module, and images were captured with a pco.edge sCMOS camera with near-perfect linearity across its 15-bit dynamic range. The pixel size of the sCMOS camera was 0.1077 µm. The entire microscope was housed within a temperature-controlled environmental chamber. All image acquisition was done in SoftWoRx (6.0; GE Healthcare). Files were imported into ImageJ (National Institutes of Health) and analyzed as described in the following sections. No gamma adjustment was applied during figure preparation.
Fixed imaging
In a 24-well plate, 50,000–100,000 cells were seeded on acid-washed cover glass (12 mm #1.5; 12-545-81; Thermo Fisher Scientific). Cells were grown for 24 h and then starved for 16–24 h in 0.2% FBS media before experimental treatment. After treatment, cells were fixed with room-temperature 4% paraformaldehyde in PBS for 5 min, extracted in −20°C methanol for 5 min, permeabilized in PBS containing 0.1% Triton X-100, 5% normal donkey serum (017-000-121; Jackson ImmunoResearch Laboratories, Inc.), and 3% bovine serum albumin (BP1605-100; Thermo Fisher Scientific) for 30 min, and subsequently immunolabeled for imaging. In brief, cells were incubated with primary antibodies for 1 h, washed three times with PBS, incubated with dye-coupled secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.) for 30 min, washed two times with PBS, stained with Hoechst DNA dye, washed twice more with PBS, and mounted on slides using fluoromount-G (17984-25; Electron Microscopy Sciences). For nearly every experiment, cilia closest to the coverslip were imaged (ventral cilia) as these cilia often lay perpendicular to the objective and within a single focal plane. In a few select cases (Figs. 3 F and 6 H), cilia pointing away from the coverslip (dorsal cilia) were imaged to reduce background fluorescence. To do so, a z stack of images with 0.3-µm separation was collected and deconvolved using SoftWoRx 6.0 based on iterative-constrained algorithms and subsequently projected by maximum-intensity projections.
Live-cell imaging
400,000 cells were seeded on acid-washed 25-mm cover glass (72223; Electron Microscopy Sciences) in a 35-mm dish. After 24 h of growth, cells were starved for 16 h and transferred to the DeltaVision stage for imaging at 37°C. Cells were imaged in DMEM/F-12 media with Hepes, no phenol red, and 0.2% FBS (11039-021; Gibco). For all >1-h imaging experiments, the imaging chamber was overlaid with a Petri dish containing a moist towel to maintain the imaging volume and pH.
Absolute quantitation of cilia-localized proteins
In Fig. 1 C, the number of APSSTR3GFP molecules per cilium resulting from expression driven by the EF1α, UbC, TK, EF1αΔ, and CMVΔ promoters was estimated by comparison with a viral particle containing exactly 120 GFP molecules (Breslow et al., 2013). As ciliary GFP was not detectable by direct imaging of the IMCD3–pCrys-APSSTR3GFP line, immunostaining for GFP was used to amplify the fluorescent signal and compare with immunostained GFP in IMCD3-[pEF1αΔ-SSTR3GFP] cells. To estimate the number of SSTR3 molecules per hippocampal neuron, hippocampal neurons and IMCD3-[APSSTR3GFP] cells using the aforementioned promoters were compared by SSTR3 immunostaining. The epitope recognized by the SSTR3 antibody (sc-11617; Santa Cruz Biotechnology, Inc.) is identical between rat and mouse (Santa Cruz Biotechnology, Inc.; personal communication with K. Griffin).
For quantitation of NG-tagged proteins including APSSTR3NG, APGPR161NG3, NG3BBS5, and NG3IFT88, ciliary molecules were quantified using single NG trimers (NG3) as a calibrator (Prevo et al., 2015). NG3 protein was recombinantly expressed in E. coli and purified by nickel affinity. The purified protein was then sparsely immobilized on a 18 × 18–mm coverslip for imaging on a DeltaVision microscope. Imaging was done in live-cell imaging solution (A14291DJ; Invitrogen). To confirm that fluorescent foci originate from a single NG3, we confirmed that fluorescence was lost by photobleaching in three discrete steps (Figs. 1 E and S1 H). The mean fluorescent intensity from >1,257 molecules was then used to estimate the number of NG-tagged molecules in cilia.
To quantify the absolute number of ciliary proteins, serum-starved live IMCD3 cells were imaged in live-cell imaging solution with the same exposure setting as used in single NG3 fluorescent quantitation. Therefore, Ncilia = (Fcilia_NG − Fbackground)/((FNG3/n) * Rexpression) where Ncilia is the absolute number of ciliary protein, Fcilia_NG is the total ciliary fluorescence detected from NG- or NG3-labeled proteins, Fbackground is the background fluorescence measured in the adjacent area, FNG3 is the fluorescent intensity of a single NG3 protein, and n determined by the single or triple NG tag was used. For example, n = 3 for APSSTR3NG, and n = 1 for GPR161NG3. Rexpression is the abundance ratio between the NG-tagged form of a given protein and the total amount of that protein in cilia. For pEF1α-NG3BBS5 and pEF1α-NG3IFT88, Rexpression = 0.55 and 0.51, respectively, which were measured by Western blotting (Figs. 3 A and S2 I). For pEF1αΔ-APSSTR3NG, Rexpression = 1 because IMCD3 cells do not express SSTR3. For APGPR161NG3, Rexpression could not be directly measured because tagging of GPR161 at the C terminus (e.g., in GPR161NG3) interferes with recognition by the anti-GPR161 antibody developed by Mukhopadhyay et al. (2013). We thus used APGPR161 as an intermediate calibrator between APGPR161NG3 and endogenous GPR161. A plasmid encoding pCrys-APGPR161 was transfected into IMCD3 cells, and the relative expression levels of transiently expressed APGPR161 and stably expressed APGPR161NG3 were measured by mSA647 pulse-labeling. We used the equation RAP.GPR161.NG3:AP.GPR161 = (FmSA.Gpr161.NG3)/(FmSA.GPR161) where RAP.GPR161.NG3:AP.GPR161 is the ratio between stably expressed APGPR161NG3 and transiently expressed APGPR161 in cilia, FmSA.GPR161 is the ciliary fluorescence signal measured by mSA647 labeling of transiently expressed APGPR161, and FmSA.Gpr161.NG3 is the ciliary fluorescence signal measured by mSA647 labeling of stably expressed APGPR161.
The relative ciliary expression levels of endogenous GPR161 and APGPR161 were measured by anti-GPR161 antibody. We used the equation RendoGPR161:AP.GPR161 = FAb/(FAb + APGPR161 − FAb) where RendoGPR161:AP.GPR161 is the ratio between endogenous GPR161 and transiently expressed APGPR161 in cilia, FAb is the ciliary fluorescence signal measured by immunofluorescence of untransfected cells with the anti-GPR161 antibody, and FAb + APGPR161 is the ciliary fluorescence signal measured by immunofluorescence of cells transiently expressing with APGPR161 with the anti-GPR161 antibody.
Therefore, NAP.GPR161.NG3 = (Fcilia.NG − Fbackground)/FNG3; RendoGPR161:AP.GPR161.NG3 = RendoGPR161:AP.GPR161/RAP.GPR161.NG3:AP.GPR161; NendoGPR161 = NAP.GPR161.NG3/RendoGPR161:AP.GPR161.NG3; and Rexpression = NAP.GPR161.NG3/(NAP.GPR161.NG3 + NendoGPR161), where RendoGPR161:AP.GPR161.NG3 is the ciliary abundance ratio between endogenous GPR161 and stably expressed APGPR161NG3; NendoGPR161 and NAP.GPR161.NG3 are the absolute numbers of endogenous GPR161 and stably expressed APGPR161NG3 in cilia; Fcilia_NG is the total ciliary fluorescence detected in the green channel from APGPR161NG3; Fbackground is the background fluorescence measured in the adjacent area; FNG3 is the fluorescent intensity of a single NG3 protein; and Rexpression is the ciliary abundance ratio between stably expressed APGPR161NG3 and total GPR161. Using this strategy, we determined that NAP.GPR161.NG3 = 1,226, NendoGPR161 = 949, and Rexpression = 0.56.
Bulk GPCR exit assays
To measure SSTR3 exit, cells expressing APSSTR3NG were first washed three times with PBS containing 5 mM MgCl2 (PBS-Mg) and then pulse-labeled with Alexa Fluor 647–labeled mSA (mSA647) for 5–10 min. To remove the unbound mSA647, cells were washed three times with PBS containing 5 mM MgCl2. Cells were then imaged after addition of sst (Nager et al., 2017). For each time point, the integrated Alexa Fluor 647 fluorescence density was measured using ImageJ. The cilia-adjacent fluorescence was subtracted as the background, and a mathematical photobleaching correction was applied: Fcilia = (FmSA647_measured/FmSA647_1) + ((1 – e-λ) * (n − 1)), where λ is the photobleaching decay constant, n is the number of images taken, FmSA647_measured is the integrated mSA647 fluorescence measured for image n, FmSA657_1 is the measurement for the first time point, and Fcilia is the reported fluorescence. In this equation, Fcilia is reported in relative fluorescence units (RFUs).
APGPR161NG3 was assayed similarly except that quantitations were done by NG fluorescence intensity using the following equation: GPR161cilia = (FNG3_n/FNG3_1) * (µF_NG3_xilia * µF_NG3), where GPR161cilia is the number of GPR161 molecules in the cilium, FNG3_n is the photobleaching-corrected integrated NG fluorescence measured for image n, FNG3_1 is the measurement for the first time point, µF_NG3_cilia is the mean integrated NG fluorescence from a population of APGPR161NG3 cilia, and µF_NG3 is the mean integrated green fluorescence from individual NG3 molecules.
To measure the exit rate of SSTR3 and GPR161 in the first 2 h after adding agonist (Fig. 1, F and G), Rexit/h = Ncilia * (Fcilia_1 − Fcilia_2h)/(2 * Fcilia_1), where Rexit/h is the number of SSTR3 or GPR161 that exited cilia per hour, Fcilia_1 is the background-corrected ciliary fluorescent intensity before adding agonist, Fcilia_2h is the background-corrected ciliary fluorescent intensity after 2 h of agonist treatment, and Ncilia is the absolute number of SSTR3 or GPR161 molecules in cilia as described in previous section.
To assess significant differences in GPCR removal, we used multiple regression. First, raw data were linearly fitted (Fcilia = m * time + c). Conditions were then compared using a z statistic: z = (m1 – m2)/(se1,2) where m1 and m2 are the fitted slopes for two experiments, and se1,2 is the propagated standard error of the slopes: se1,2 = √(sm12 + sm22) where sm1 and sm2 are the SDs for m1 and m2, respectively. Z statistics were converted to p-values for statistical interpretation.
Kymograph analysis and processivity
To analyze intraciliary trafficking, IFT88 and BBS5 proteins were genetically labeled with NG3 and rapidly imaged with TIRF (4 Hz) for short time periods (30–60 s). TIRF illumination reduced nonciliary (background) fluorescence, permitting visualization of dim trains. The resulting videos were analyzed by ImageJ for generating kymographs. KymographClear and KymographDirect (Mangeol et al., 2016) were used to deconvolve anterograde from retrograde trains for measuring relative intensities for BBSome trains, whereas Multi Kymograph was used for unaltered presentation (e.g., Fig. 6 A).
The processivity of IFT-B or BBSome trains along axonemal microtubules was defined as the duration for which a fluorescent focus of IFT88 or BBS5 unidirectionally moved along the cilium at a rate expected for kinesin- or dynein-mediated transport (0.3–0.6 µm/s). Only events occurring for >3 s were considered. Measurements were made from kymographs of NG3IFT88, NG3BBS5, or tdTomatoIFT88 and NG3BBS5. In brief, kymographs were visualized in ImageJ, and lines were drawn along the long axis of the cilium (“the leg”) and the processive movement (“the hypotenuse”). The angle between the leg and the hypotenuse was measured (“the included angle”). Comparison of the leg and the hypotenuse was used to quantify the processivity of an IFT- or BBS-containing train. Comparison of the hypotenuse and the included angle was used to measure the velocity of IFT and BBS trains.
Quantitation of IFT/BBS trains
To estimate the number of IFT88 or BBS5 molecules per train, the total NG3 fluorescence along the ciliary axoneme was first divided by the number of trains: TRFU_mean = µRFU_NG3_axoneme/Ntrain, where TRFU_mean is the estimated NG3IFT88 or NG3BBS5 fluorescence of a single train, µRFU_NG3_axoneme is the mean integrated fluorescence along the axonemes of several cilia (excluding the base and the tip), and Ntrain is the number of trains counted by kymograph analysis in the cilia when µRFU_NG3_axoneme was measured. To increase measurement precision for calculating µRFU_NG3_axoneme, cilia were imaged by epifluorescence rather than TIRF illumination as the TIRF field did not reproducibly illuminate cilia from different cells. TRFU_mean was then used to calculate the absolute number of IFT88 or BBS5 molecules per train by using the NG3 standard described in the Absolute quantitation of cilia-localized markers section: Tlabeled_abs = TRFU_average/µF_NG3, where Tlabeled_abs is the mean number of labeled molecules per train, and µF_NG3 is the previously measured mean fluorescence of a single NG3. As Tlabeled_abs does not account for unlabeled IFT88 or BBS5 within a train, a correction must be applied that relates the ratio of unlabeled to labeled molecules in the cell: Tabs = Tlabeled_abs * ((WNG3 + Wendogenous)/WNG3), where Tabs is the number of IFT88 or BBS5 molecules per train, and WNG3 and Wendogenous are the integrated intensities of Western blot bands for NG3-tagged and endogenous molecules using antibodies against mouse IFT88 or BBS5 (Figs. 3 A and S2 I). Tabs was used to calculate the number of molecules in an anterograde (Tabs_anterograde) versus retrograde (Tabs_anterograde) train: Tabs = (fanterograde * Tabs_anterograde + fretrograde * Tabs_retrograde)/(fanterograde + fretrograde), where fanterograde and fretrograde are the frequency of either anterograde or retrograde trains. Using KymographClear analysis on NG3IFT88, we measured fanterograde = 21.4 trains/min and fretrograde = 18.5 trains/min. Tabs_anterograde and Tabs_retrograde can now be solved using the size ratio (RT) of anterograde versus retrograde trains: RT = µRFU_T_anterograde/µRFU_T_retrograde, where µRFU_T_anterograde and µRFU_T_retrograde are the mean NG3 intensity of anterograde or retrograde trains. By KymographClear analyses, RT = 1.69 for NG3IFT88, and RT = 1.78 for NG3BBS5. Relating Tabs_anterograde to Tabs_retrograde: Tabs_anterograde = Tabs_retrograde * RT.
One can calculate the number of IFT88/BBS5 particles in anterograde and retrograde trains: Tabs_anterograde = (fanterograde + fretrograde) * Tabs/(fanterograde + fretrograde/RT); Tabs_retrograde = (fanterograde + fretrograde) * Tabs/(fanterograde * RT + fretrograde).
Line scan and tipping quantitation
The longitudinal fluorescence intensities of BBS5, IFT88, SSTR3, and GPR161 were measured in ImageJ by a plot profile of a 5-pixel-wide line along the long axis of the cilium. To average data from multiple cilia, pixel intensities were assigned a length percentage, with 0% referring to the base and 100% referring to the tip. Values were then grouped into 5% bins and averaged. Bin means were then averaged across multiple cilia and plotted (e.g., Fig. 3 B).
To quantify NG3BBS5 tipping, a 5 × 7–pixel box was centered at the cilium tip, and the integrated fluorescence intensity within the box was measured by ImageJ. The resulting values were background corrected by subtracting the fluorescence immediately adjacent to the cilium. The values from multiple ciliary tips were averaged and then converted to absolute numbers of BBS5 particles using the NG3 calibration.
Single-molecule labeling with SAQdot
Molecules of SSTR3 or GPR161 present on the surface of IMCD3-[pEF1αΔ-APSSTR3NG] and IMCD3-[pCrys-APGPR161NG3] cells were labeled with SAQdot through the affinity of SA for the extracellular biotinylated AP tag. Both cell lines express an ER-targeted BirA ligase that biotinylates the N-terminal AP tag of each GPCR (Howarth and Ting, 2008; Nager et al., 2017). Single GPCR labeling was achieved through a blocking strategy. First, to block surface-exposed biotinylated receptors, cells were washed three times with PBS and then incubated with 10 nM mSA for 15 min. Cells were washed three times with PBS and incubated with 1 nM SAQdot (Q10123MP; Thermo Fisher Scientific) solution for 15 min. As the SAQdot only binds to receptors newly arrived at the surface during the 15 min of labeling, this approach labels very few receptors. Lastly, cells were washed three times with PBS and transferred to imaging media (DMEM/F-12, Hepes, and no phenol red media; 11039-021; Gibco) with 0.2% FBS and 1 µM biotin to block unbound SA on SAQdot.
Qdot-labeled SSTR3 and GPR161 molecules exited the cilium at rates congruent with bulk imaging (Fig. 1, F and G). By counting the number of QDSSTR3 per cilia after 2 h treatment with vehicle or sst (Fig. 8 C), we determined that sst treatment resulted in the loss of 25% of cilia-localized Qdots, consistent with the SSTR3 exit rate measured by bulk fluorescence (Fig. 1 F). As a second test, the frequencies of QDGPR161 exit events captured by live imaging matched the frequencies predicted from imaging the decrease in bulk fluorescence of GPR161NG3 (Fig. S5 B).
Qdot localization and analysis
To monitor single receptor trafficking, cilia bearing one Qdot were imaged on the DeltaVision microscope at 2 Hz for 5–20 min. Cilia were identified by the NG signal from the C-terminal NG and NG3 tags. The centroid of SAQdot-labeled single SSTR3 and GPR161 were tracked with dynamic programming method. To achieve subpixel accuracy, each detected spot position was finally replaced by the center of gravity of a small neighborhood (3 * 3) in a postprocessing step (Sage et al., 2005). The localization precision of Qdot was measured by mapping the centroid of an immobile Qdot, where localization precision σxy = (σx2 + σy2)1/2 (Deschout et al., 2014).
Processivity, residence, and confinement frequencies were based on kymograph analyses (examples shown in Fig. S4). Processive QDGPCR movements were defined as consecutive frames where a Qdot moved longitudinally along the ciliary axoneme. To distinguish motor-driven from diffusive events, only processive motions that extend for at least six consecutive frames (Fig. S3 G) or 10 consecutive frames (Fig. 8 F) were plotted. Residence at the base and tip was operationally defined as residing at the basal or distal 10% length of the cilium for >2.5 s (Fig. 8 G). As freely diffusing receptors occasionally remain at the cilia base or tip for >2.5 s, confinement was defined by a more stringent criterion of events lasting >15 s (Fig. 8 G).
The most base-proximal position of QDGPR161 was mapped on a coordinate system established by the profile of ciliary GPR161NG3 (Fig. 10, A–C). The same approach was applied to map the immunofluorescence intensity center of Cep290 and Cep164 (Fig. 10, A–C). Intermediate compartment visits were defined as events where the Qdot centroid crossed the 50th percentile of NG intensity in the GPR161NG longitudinal scan profile (Fig. 10, C and F).
Box blots
All box plots display the second, third, and fourth quartiles along with whiskers that represent values within the 1.5× the interquartile range. Outliers exceeding the whiskers are plotted as points.
Online supplemental material
Fig. S1 shows that low-expression promoters recapitulate the physiological exit kinetics of SSTR3. Fig. S2 identifies IFT-A/Tulp3 as the importer of GPCRs into cilia and BBSome/Arl6 as the exporter. Fig. S3 presents BBSome and IFT train processivity and single-molecule quantitation. Fig. S4 shows kymographs of single molecules of QDSSTR3 and QDGPR161. Fig. S5 shows visualization of QDGPR161 exit at single-molecule resolution. Video 1 shows sst-dependent removal of SSTR3 from cilia. Video 2 shows IFT-B foci movements in cilia. Video 3 shows comovement of BBSome and IFT-B. Video 4 shows BBSome foci movement in cilia. Video 5 shows dynamics of BBSome-mediated retrieval. Video 6 shows behavior of Qdot-labeled SSTR3 in control-treated cells. Video 7 shows behavior of Qdot-labeled SSTR3 in sst-treated cells. Videos 8 and 9 show direct observations of ciliary exit of Qdot-labeled GPR161.
Supplementary Material
Acknowledgments
We thank A. Kopp and J. Goldstein for technical assistance, and we thank K. Hill and A. Benmerah as well as the Nachury laboratory for comments on the manuscript.
This work was supported by National Institutes of Health funding (GM089933) to M.V. Nachury and a Damon Runyon Cancer Research Foundation Fayez Sarofim Fellowship to A.R. Nager (DRG 2160-13).
The authors declare no competing financial interests.
Author contributions: M.V. Nachury conceived and coordinated the project and wrote the paper with contributions from all authors. F. Ye and A.R. Nager jointly developed the cell lines used for imaging of GPCRs and BBSomes. F. Ye conducted all single-molecule imaging and the quantitative analysis of BBSome trains and exit.
References
- Badgandi H.B., Hwang S., Shimada I.S., Loriot E., and Mukhopadhyay S.. 2017. Tubby family proteins are adapters for ciliary trafficking of integral membrane proteins. J. Cell Biol. 216:743–760. 10.1083/jcb.201607095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangs F., and Anderson K.V.. 2017. Primary Cilia and Mammalian Hedgehog Signaling. Cold Spring Harb. Perspect. Biol. 9:a028175 10.1101/cshperspect.a028175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bänziger M., Cercus J., Hirt H., Laumen K., Malan C., Spindler F., Struber F., and Troxler T.. 2003. The development of a practical synthesis of the potent and selective somatostatin sst3 receptor antagonist [4-(3,4-difluoro-phenyl)-piperazine-1-yl]-{(4S,4aS,8aR)-2[(S)-3-(6-methoxy-pyridin-3-yl)-2-methyl-propyl]-decahydroisoquinoline-4-yl}-methanone (NVP-ACQ090). Tetrahedron Asymmetry. 14:3469–3477. 10.1016/j.tetasy.2003.07.024 [DOI] [Google Scholar]
- Barbelanne M., Hossain D., Chan D.P., Peränen J., and Tsang W.Y.. 2015. Nephrocystin proteins NPHP5 and Cep290 regulate BBSome integrity, ciliary trafficking and cargo delivery. Hum. Mol. Genet. 24:2185–2200. 10.1093/hmg/ddu738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbari N.F., Johnson A.D., Lewis J.S., Askwith C.C., and Mykytyn K.. 2008a Identification of ciliary localization sequences within the third intracellular loop of G protein-coupled receptors. Mol. Biol. Cell. 19:1540–1547. 10.1091/mbc.E07-09-0942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbari N.F., Lewis J.S., Bishop G.A., Askwith C.C., and Mykytyn K.. 2008b Bardet-Biedl syndrome proteins are required for the localization of G protein-coupled receptors to primary cilia. Proc. Natl. Acad. Sci. USA. 105:4242–4246. 10.1073/pnas.0711027105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindels D.S., Haarbosch L., van Weeren L., Postma M., Wiese K.E., Mastop M., Aumonier S., Gotthard G., Royant A., Hink M.A., and Gadella T.W. Jr. 2017. mScarlet: a bright monomeric red fluorescent protein for cellular imaging. Nat. Methods. 14:53–56. 10.1038/nmeth.4074 [DOI] [PubMed] [Google Scholar]
- Breslow D.K., Koslover E.F., Seydel F., Spakowitz A.J., and Nachury M.V.. 2013. An in vitro assay for entry into cilia reveals unique properties of the soluble diffusion barrier. J. Cell Biol. 203:129–147. 10.1083/jcb.201212024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang J.-Z., Hsu Y.-C., and Sung C.-H.. 2015. Ultrastructural visualization of trans-ciliary rhodopsin cargoes in mammalian rods. Cilia. 4:4 10.1186/s13630-015-0013-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement C.A., Ajbro K.D., Koefoed K., Vestergaard M.L., Veland I.R., Henriques de Jesus M.P.R., Pedersen L.B., Benmerah A., Andersen C.Y., Larsen L.A., and Christensen S.T.. 2013. TGF-β signaling is associated with endocytosis at the pocket region of the primary cilium. Cell Reports. 3:1806–1814. 10.1016/j.celrep.2013.05.020 [DOI] [PubMed] [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., and Zhang F.. 2013. Multiplex genome engineering using CRISPR/Cas systems. Sci. N. Y. NY. 339:819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta P., Allamargot C., Hudson J.S., Andersen E.K., Bhattarai S., Drack A.V., Sheffield V.C., and Seo S.. 2015. Accumulation of non-outer segment proteins in the outer segment underlies photoreceptor degeneration in Bardet-Biedl syndrome. Proc. Natl. Acad. Sci. USA. 112:E4400–E4409. 10.1073/pnas.1510111112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschout H., Cella Zanacchi F., Mlodzianoski M., Diaspro A., Bewersdorf J., Hess S.T., and Braeckmans K.. 2014. Precisely and accurately localizing single emitters in fluorescence microscopy. Nat. Methods. 11:253–266. 10.1038/nmeth.2843 [DOI] [PubMed] [Google Scholar]
- Domire J.S., Green J.A., Lee K.G., Johnson A.D., Askwith C.C., and Mykytyn K.. 2011. Dopamine receptor 1 localizes to neuronal cilia in a dynamic process that requires the Bardet-Biedl syndrome proteins. Cell. Mol. Life Sci. 68:2951–2960. 10.1007/s00018-010-0603-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson J.M., Conduit S.E., Feeney S.J., Hakim S., DiTommaso T., Fulcher A.J., Sriratana A., Ramm G., Horan K.A., Gurung R., et al. 2017. INPP5E regulates phosphoinositide-dependent cilia transition zone function. J. Cell Biol. 216:247–263. 10.1083/jcb.201511055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguether T., San Agustin J.T., Keady B.T., Jonassen J.A., Liang Y., Francis R., Tobita K., Johnson C.A., Abdelhamed Z.A., Lo C.W., and Pazour G.J.. 2014. IFT27 links the BBSome to IFT for maintenance of the ciliary signaling compartment. Dev. Cell. 31:279–290. 10.1016/j.devcel.2014.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh-Yamagami S., Evangelista M., Wilson D., Wen X., Theunissen J.-W., Phamluong K., Davis M., Scales S.J., Solloway M.J., de Sauvage F.J., and Peterson A.S.. 2009. The mammalian Cos2 homolog Kif7 plays an essential role in modulating Hh signal transduction during development. Curr. Biol. 19:1320–1326. 10.1016/j.cub.2009.06.046 [DOI] [PubMed] [Google Scholar]
- Ferreira J.P., Peacock R.W.S., Lawhorn I.E.B., and Wang C.L.. 2011. Modulating ectopic gene expression levels by using retroviral vectors equipped with synthetic promoters. Syst. Synth. Biol. 5:131–138. 10.1007/s11693-011-9089-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W., Wang L., Kim S., Li J., and Dynlacht B.D.. 2016. Role for the IFT-A Complex in Selective Transport to the Primary Cilium. Cell Reports. 17:1505–1517. 10.1016/j.celrep.2016.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo F.R., and Reiter J.F.. 2012. Scoring a backstage pass: mechanisms of ciliogenesis and ciliary access. J. Cell Biol. 197:697–709. 10.1083/jcb.201111146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham A.K., and Munro S.. 2000. The PACT domain, a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep. 1:524–529. 10.1093/embo-reports/kvd105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz S.C., Bangs F., Barrington C.L., Katsanis N., and Anderson K.V.. 2017. The Meckel syndrome- associated protein MKS1 functionally interacts with components of the BBSome and IFT complexes to mediate ciliary trafficking and hedgehog signaling. PLoS One. 12:e0173399 10.1371/journal.pone.0173399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves J., and Pelletier L.. 2017. The Ciliary Transition Zone: Finding the Pieces and Assembling the Gate. Mol. Cells. 40:243–253. 10.14348/molcells.2017.0054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J.A., Schmid C.L., Bley E., Monsma P.C., Brown A., Bohn L.M., and Mykytyn K.. 2015. Recruitment of β-Arrestin into Neuronal Cilia Modulates Somatostatin Receptor Subtype 3 Ciliary Localization. Mol. Cell. Biol. 36:223–235. 10.1128/MCB.00765-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guadiana S.M., Semple-Rowland S., Daroszewski D., Madorsky I., Breunig J.J., Mykytyn K., and Sarkisian M.R.. 2013. Arborization of dendrites by developing neocortical neurons is dependent on primary cilia and type 3 adenylyl cyclase. J. Neurosci. 33:2626–2638. 10.1523/JNEUROSCI.2906-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M., Subramanian R., Bangs F., Omelchenko T., Liem K.F. Jr., Kapoor T.M., and Anderson K.V.. 2014. The kinesin-4 protein Kif7 regulates mammalian Hedgehog signalling by organizing the cilium tip compartment. Nat. Cell Biol. 16:663–672. 10.1038/ncb2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth M., and Ting A.Y.. 2008. Imaging proteins in live mammalian cells with biotin ligase and monovalent streptavidin. Nat. Protoc. 3:534–545. 10.1038/nprot.2008.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunnicutt G.R., Kosfiszer M.G., and Snell W.J.. 1990. Cell body and flagellar agglutinins in Chlamydomonas reinhardtii: the cell body plasma membrane is a reservoir for agglutinins whose migration to the flagella is regulated by a functional barrier. J. Cell Biol. 111:1605–1616. 10.1083/jcb.111.4.1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jékely G., and Arendt D.. 2006. Evolution of intraflagellar transport from coated vesicles and autogenous origin of the eukaryotic cilium. BioEssays. 28:191–198. 10.1002/bies.20369 [DOI] [PubMed] [Google Scholar]
- Jensen V.L., and Leroux M.R.. 2017. Gates for soluble and membrane proteins, and two trafficking systems (IFT and LIFT), establish a dynamic ciliary signaling compartment. Curr. Opin. Cell Biol. 47:83–91. 10.1016/j.ceb.2017.03.012 [DOI] [PubMed] [Google Scholar]
- Jensen C.G., Poole C.A., McGlashan S.R., Marko M., Issa Z.I., Vujcich K.V., and Bowser S.S.. 2004. Ultrastructural, tomographic and confocal imaging of the chondrocyte primary cilium in situ. Cell Biol. Int. 28:101–110. 10.1016/j.cellbi.2003.11.007 [DOI] [PubMed] [Google Scholar]
- Jin H., White S.R., Shida T., Schulz S., Aguiar M., Gygi S.P., Bazan J.F., and Nachury M.V.. 2010. The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell. 141:1208–1219. 10.1016/j.cell.2010.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamachi Y., and Kondoh H.. 1993. Overlapping positive and negative regulatory elements determine lens-specific activity of the delta 1-crystallin enhancer. Mol. Cell. Biol. 13:5206–5215. 10.1128/MCB.13.9.5206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanie T., Abbott K.L., Mooney N.A., Plowey E.D., Demeter J., and Jackson P.K.. 2017. The CEP19-RABL2 GTPase Complex Binds IFT-B to Initiate Intraflagellar Transport at the Ciliary Base. Dev. Cell. 42:22–36. 10.1016/j.devcel.2017.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T., Brown J.M., Bellve K., Craige B., Craft J.M., Fogarty K., Lechtreck K.F., and Witman G.B.. 2016. Together, the IFT81 and IFT74 N-termini form the main module for intraflagellar transport of tubulin. J. Cell Sci. 129:2106–2119. 10.1242/jcs.187120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon R.Y., Temiyasathit S., Tummala P., Quah C.C., and Jacobs C.R.. 2010. Primary cilium-dependent mechanosensing is mediated by adenylyl cyclase 6 and cyclic AMP in bone cells. FASEB J. 24:2859–2868. 10.1096/fj.09-148007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau L., Lee Y.L., Sahl S.J., Stearns T., and Moerner W.E.. 2012. STED microscopy with optimized labeling density reveals 9-fold arrangement of a centriole protein. Biophys. J. 102:2926–2935. 10.1016/j.bpj.2012.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck K.F. 2015. IFT-Cargo Interactions and Protein Transport in Cilia. Trends Biochem. Sci. 40:765–778. 10.1016/j.tibs.2015.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck K.-F., Johnson E.C., Sakai T., Cochran D., Ballif B.A., Rush J., Pazour G.J., Ikebe M., and Witman G.B.. 2009. The Chlamydomonas reinhardtii BBSome is an IFT cargo required for export of specific signaling proteins from flagella. J. Cell Biol. 187:1117–1132. 10.1083/jcb.200909183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck K.F., Brown J.M., Sampaio J.L., Craft J.M., Shevchenko A., Evans J.E., and Witman G.B.. 2013. Cycling of the signaling protein phospholipase D through cilia requires the BBSome only for the export phase. J. Cell Biol. 201:249–261. 10.1083/jcb.201207139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liem K.F. Jr., He M., Ocbina P.J.R., and Anderson K.V.. 2009. Mouse Kif7/Costal2 is a cilia-associated protein that regulates Sonic hedgehog signaling. Proc. Natl. Acad. Sci. USA. 106:13377–13382. 10.1073/pnas.0906944106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew G.M., Ye F., Nager A.R., Murphy J.P., Lee J.S., Aguiar M., Breslow D.K., Gygi S.P., and Nachury M.V.. 2014. The intraflagellar transport protein IFT27 promotes BBSome exit from cilia through the GTPase ARL6/BBS3. Dev. Cell. 31:265–278. 10.1016/j.devcel.2014.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.C., Couzens A.L., Deshwar A.R., McBroom-Cerajewski L.D.B., Zhang X., Puviindran V., Scott I.C., Gingras A.-C., Hui C.-c., and Angers S.. 2014. The PPFIA1-PP2A protein complex promotes trafficking of Kif7 to the ciliary tip and Hedgehog signaling. Sci. Signal. 7:ra117 10.1126/scisignal.2005608 [DOI] [PubMed] [Google Scholar]
- Loktev A.V., and Jackson P.K.. 2013. Neuropeptide Y family receptors traffic via the Bardet-Biedl syndrome pathway to signal in neuronal primary cilia. Cell Reports. 5:1316–1329. 10.1016/j.celrep.2013.11.011 [DOI] [PubMed] [Google Scholar]
- Mangeol P., Prevo B., and Peterman E.J.G.. 2016. KymographClear and KymographDirect: two tools for the automated quantitative analysis of molecular and cellular dynamics using kymographs. Mol. Biol. Cell. 27:1948–1957. 10.1091/mbc.E15-06-0404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masyuk A.I., Gradilone S.A., Banales J.M., Huang B.Q., Masyuk T.V., Lee S.-O., Splinter P.L., Stroope A.J., and Larusso N.F.. 2008. Cholangiocyte primary cilia are chemosensory organelles that detect biliary nucleotides via P2Y12 purinergic receptors. Am. J. Physiol. Gastrointest. Liver Physiol. 295:G725–G734. 10.1152/ajpgi.90265.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mick D.U., Rodrigues R.B., Leib R.D., Adams C.M., Chien A.S., Gygi S.P., and Nachury M.V.. 2015. Proteomics of Primary Cilia by Proximity Labeling. Dev. Cell. 35:497–512. 10.1016/j.devcel.2015.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milenkovic L., Scott M.P., and Rohatgi R.. 2009. Lateral transport of Smoothened from the plasma membrane to the membrane of the cilium. J. Cell Biol. 187:365–374. 10.1083/jcb.200907126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milenkovic L., Weiss L.E., Yoon J., Roth T.L., Su Y.S., Sahl S.J., Scott M.P., and Moerner W.E.. 2015. Single-molecule imaging of Hedgehog pathway protein Smoothened in primary cilia reveals binding events regulated by Patched1. Proc. Natl. Acad. Sci. USA. 112:8320–8325. 10.1073/pnas.1510094112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen M.M., Malik A., Piel M., Bouckson-Castaing V., and Bornens M.. 2000. Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: the role of ninein. J. Cell Sci. 113:3013–3023. [DOI] [PubMed] [Google Scholar]
- Moore B.S., Stepanchick A.N., Tewson P.H., Hartle C.M., Zhang J., Quinn A.M., Hughes T.E., and Mirshahi T.. 2016. Cilia have high cAMP levels that are inhibited by Sonic Hedgehog-regulated calcium dynamics. Proc. Natl. Acad. Sci. USA. 113:13069–13074. 10.1073/pnas.1602393113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E., Arii J., Christensen D., Votteler J., and Sundquist W.I.. 2012. Attenuated protein expression vectors for use in siRNA rescue experiments. Biotechniques. 0:1–5. 10.2144/000113909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S., Wen X., Chih B., Nelson C.D., Lane W.S., Scales S.J., and Jackson P.K.. 2010. TULP3 bridges the IFT-A complex and membrane phosphoinositides to promote trafficking of G protein-coupled receptors into primary cilia. Genes Dev. 24:2180–2193. 10.1101/gad.1966210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S., Wen X., Ratti N., Loktev A., Rangell L., Scales S.J., and Jackson P.K.. 2013. The ciliary G-protein-coupled receptor Gpr161 negatively regulates the Sonic hedgehog pathway via cAMP signaling. Cell. 152:210–223. 10.1016/j.cell.2012.12.026 [DOI] [PubMed] [Google Scholar]
- Nachury M.V. 2008. Tandem Affinity Purification of the BBSome, a Critical Regulator of Rab8 in Ciliogenesis. In Methods in Enzymology. Academic Press; pp. 501–513. 10.1016/S0076-6879(07)00434-X [DOI] [PubMed] [Google Scholar]
- Nachury M.V., Seeley E.S., and Jin H.. 2010. Trafficking to the ciliary membrane: how to get across the periciliary diffusion barrier? Annu. Rev. Cell Dev. Biol. 26:59–87. 10.1146/annurev.cellbio.042308.113337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nager A.R., Goldstein J.S., Herranz-Pérez V., Portran D., Ye F., Garcia-Verdugo J.M., and Nachury M.V.. 2017. An Actin Network Dispatches Ciliary GPCRs into Extracellular Vesicles to Modulate Signaling. Cell. 168:252–263. 10.1016/j.cell.2016.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman R., Norman R.X., Ko H., Ko H.W., Huang V., Huang V., Eun C.M., Eun C., Abler L.L., Abler L., et al. 2009. Tubby-like protein 3 (TULP3) regulates patterning in the mouse embryo through inhibition of Hedgehog signaling. Hum. Mol. Genet. 18:1740–1754. 10.1093/hmg/ddp113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou G., Blacque O.E., Snow J.J., Leroux M.R., and Scholey J.M.. 2005. Functional coordination of intraflagellar transport motors. Nature. 436:583–587. 10.1038/nature03818 [DOI] [PubMed] [Google Scholar]
- Pal K., Hwang S.H., Somatilaka B., Badgandi H., Jackson P.K., DeFea K., and Mukhopadhyay S.. 2016. Smoothened determines β-arrestin-mediated removal of the G protein-coupled receptor Gpr161 from the primary cilium. J. Cell Biol. 212:861–875. 10.1083/jcb.201506132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevo B., Mangeol P., Oswald F., Scholey J.M., and Peterman E.J.G.. 2015. Functional differentiation of cooperating kinesin-2 motors orchestrates cargo import and transport in C. elegans cilia. Nat. Cell Biol. 17:1536–1545. 10.1038/ncb3263 [DOI] [PubMed] [Google Scholar]
- Putoux A., Thomas S., Coene K.L.M., Davis E.E., Alanay Y., Ogur G., Uz E., Buzas D., Gomes C., Patrier S., et al. 2011. KIF7 mutations cause fetal hydrolethalus and acrocallosal syndromes. Nat. Genet. 43:601–606. 10.1038/ng.826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrer S.P., Birzin E.T., Mosley R.T., Berk S.C., Hutchins S.M., Shen D.-M., Xiong Y., Hayes E.C., Parmar R.M., Foor F., et al. 1998. Rapid identification of subtype-selective agonists of the somatostatin receptor through combinatorial chemistry. Science. 282:737–740. 10.1126/science.282.5389.737 [DOI] [PubMed] [Google Scholar]
- Roth A., Kreienkamp H.J., Meyerhof W., and Richter D.. 1997. Phosphorylation of four amino acid residues in the carboxyl terminus of the rat somatostatin receptor subtype 3 is crucial for its desensitization and internalization. J. Biol. Chem. 272:23769–23774. 10.1074/jbc.272.38.23769 [DOI] [PubMed] [Google Scholar]
- Sage D., Neumann F.R., Hediger F., Gasser S.M., and Unser M.. 2005. Automatic tracking of individual fluorescence particles: application to the study of chromosome dynamics. IEEE Trans. Image Process. 14:1372–1383. 10.1109/TIP.2005.852787 [DOI] [PubMed] [Google Scholar]
- Shaner N.C., Campbell R.E., Steinbach P.A., Giepmans B.N.G., Palmer A.E., and Tsien R.Y.. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22:1567–1572. 10.1038/nbt1037 [DOI] [PubMed] [Google Scholar]
- Shaner N.C., Lambert G.G., Chammas A., Ni Y., Cranfill P.J., Baird M.A., Sell B.R., Allen J.R., Day R.N., Israelsson M., et al. 2013. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods. 10:407–409. 10.1038/nmeth.2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen F., Cheng L., Douglas A.E., Riobo N.A., and Manning D.R.. 2013. Smoothened is a fully competent activator of the heterotrimeric G protein G(i). Mol. Pharmacol. 83:691–697. 10.1124/mol.112.082511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T.S., Spitzbarth B., Li J., Dugger D.R., Stern-Schneider G., Sehn E., Bolch S.N., McDowell J.H., Tipton J., Wolfrum U., and Smith W.C.. 2013. Light-dependent phosphorylation of Bardet-Biedl syndrome 5 in photoreceptor cells modulates its interaction with arrestin1. Cell. Mol. Life Sci. 70:4603–4616. 10.1007/s00018-013-1403-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X., Driscoll K., Yao G., Raed A., Wu M., Beales P.L., and Zhou J.. 2014. Bardet-Biedl syndrome proteins 1 and 3 regulate the ciliary trafficking of polycystic kidney disease 1 protein. Hum. Mol. Genet. 23:5441–5451. 10.1093/hmg/ddu267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran P.V., Haycraft C.J., Besschetnova T.Y., Turbe-Doan A., Stottmann R.W., Herron B.J., Chesebro A.L., Qiu H., Scherz P.J., Shah J.V., et al. 2008. THM1 negatively modulates mouse sonic hedgehog signal transduction and affects retrograde intraflagellar transport in cilia. Nat. Genet. 40:403–410. 10.1038/ng.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimble W.S., and Grinstein S.. 2015. Barriers to the free diffusion of proteins and lipids in the plasma membrane. J. Cell Biol. 208:259–271. 10.1083/jcb.201410071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam T.J.P., Townsend M.J., Turk M., Schlessinger A., Sali A., Field M.C., and Huynen M.A.. 2013. Evolution of modular intraflagellar transport from a coatomer-like progenitor. Proc. Natl. Acad. Sci. USA. 110:6943–6948. 10.1073/pnas.1221011110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C.L., McIntyre J.C., Norris S.R., Jenkins P.M., Zhang L., Pei Q., Verhey K., and Martens J.R.. 2014. Direct evidence for BBSome-associated intraflagellar transport reveals distinct properties of native mammalian cilia. Nat. Commun. 5:5813 10.1038/ncomms6813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T.T., Su J., Wang W.-J., Craige B., Witman G.B., Tsou M.-F.B., and Liao J.-C.. 2015. Superresolution Pattern Recognition Reveals the Architectural Map of the Ciliary Transition Zone. Sci. Rep. 5:14096 10.1038/srep14096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T.T., Chong W.M., Wang W.-J., Mazo G., Tanos B., Chen Z., Tran M.N.T., Chen Y.-D., Weng R.R., Huang C.-E., et al. 2017. Architecture of mammalian centriole distal appendages accommodates distinct blade and matrix functional elements. bioRxiv. accessed September 25, 2017. 10.1101/193474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda K., Rens-Domiano S., Breder C.D., Law S.F., Saper C.B., Reisine T., and Bell G.I.. 1992. Cloning of a novel somatostatin receptor, SSTR3, coupled to adenylylcyclase. J. Biol. Chem. 267:20422–20428. [PubMed] [Google Scholar]
- Ye F., Breslow D.K., Koslover E.F., Spakowitz A.J., Nelson W.J., and Nachury M.V.. 2013. Single molecule imaging reveals a major role for diffusion in the exploration of ciliary space by signaling receptors. eLife. 2:e00654 10.7554/eLife.00654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee L.E., Garcia-Gonzalo F.R., Bowie R.V., Li C., Kennedy J.K., Ashrafi K., Blacque O.E., Leroux M.R., and Reiter J.F.. 2015. Conserved Genetic Interactions between Ciliopathy Complexes Cooperatively Support Ciliogenesis and Ciliary Signaling. PLoS Genet. 11:e1005627 10.1371/journal.pgen.1005627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh C., Li A., Chuang J.-Z., Saito M., Cáceres A., and Sung C.-H.. 2013. IGF-1 activates a cilium-localized noncanonical Gβγ signaling pathway that regulates cell-cycle progression. Dev. Cell. 26:358–368. 10.1016/j.devcel.2013.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Nishimura D., Seo S., Vogel T., Morgan D.A., Searby C., Bugge K., Stone E.M., Rahmouni K., and Sheffield V.C.. 2011. Bardet-Biedl syndrome 3 (Bbs3) knockout mouse model reveals common BBS-associated phenotypes and Bbs3 unique phenotypes. Proc. Natl. Acad. Sci. USA. 108:20678–20683. 10.1073/pnas.1113220108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Seo S., Bhattarai S., Bugge K., Searby C.C., Zhang Q., Drack A.V., Stone E.M., and Sheffield V.C.. 2014. BBS mutations modify phenotypic expression of CEP290-related ciliopathies. Hum. Mol. Genet. 23:40–51. 10.1093/hmg/ddt394 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.