Medeiros et al. show that mtDNA polymerase POLG controls mtDNA copy number in the context of autophagy-mediated metabolic homeostasis. After prolonged starvation, the mtDNA degradative activity of POLG is activated to adjust the increasing mtDNA copy number in WT cells, whereas in autophagy-deficient cells, POLG’s continued degradative activity causes mtDNA instability and respiratory dysfunction as a result of nucleotide insufficiency.
Abstract
Mitochondria contain tens to thousands of copies of their own genome (mitochondrial DNA [mtDNA]), creating genetic redundancy capable of buffering mutations in mitochondrial genes essential for cellular function. However, the mechanisms regulating mtDNA copy number have been elusive. Here we found that DNA synthesis and degradation by mtDNA polymerase γ (POLG) dynamically controlled mtDNA copy number in starving yeast cells dependent on metabolic homeostasis provided by autophagy. Specifically, the continuous mtDNA synthesis by POLG in starving wild-type cells was inhibited by nucleotide insufficiency and elevated mitochondria-derived reactive oxygen species in the presence of autophagy dysfunction. Moreover, after prolonged starvation, 3′–5′ exonuclease–dependent mtDNA degradation by POLG adjusted the initially increasing mtDNA copy number in wild-type cells, but caused quantitative mtDNA instability and irreversible respiratory dysfunction in autophagy-deficient cells as a result of nucleotide limitations. In summary, our study reveals that mitochondria rely on the homeostatic functions of autophagy to balance synthetic and degradative modes of POLG, which control copy number dynamics and stability of the mitochondrial genome.
Introduction
Derived from endosymbiotic α-proteobacteria, mitochondria evolved as dynamic tubular networks containing highly specialized multicopy genomes with limited coding capacity (Lane and Martin, 2010; Nunnari and Suomalainen, 2012; Garcia et al., 2017). In Saccharomyces cerevisiae, the ∼85-kb mitochondrial DNA (mtDNA) encodes for eight proteins, including seven essential respiratory chain subunits, transfer and ribosomal RNAs, and the RNA subunit of RNase P (Foury et al., 1998; Turk et al., 2013). The 20–100 mtDNA copies per yeast cell are packaged into multiple dynamic nucleoprotein complexes, termed nucleoids, that remodel in number, distribution, protein composition, and mtDNA content (1–10 copies) in response to metabolic cues (MacAlpine et al., 2000; Kucej et al., 2008; Miyakawa, 2017). mtDNA replication occurs independently of the nuclear genome throughout the cell cycle but coordinates with mitochondrial division to distribute nucleoids within the network of mitochondria (Meeusen and Nunnari, 2003; Murley et al., 2013; Lewis et al., 2016). The mitochondrial replisome consists of a DNA helicase, single-strand DNA binding proteins, and a dedicated mtDNA polymerase γ (POLG; Foury, 1989; Young and Copeland, 2016). In addition to its 5′–3′ polymerase, POLG contains an inherent 3′–5′ exonuclease, which provides proofreading activity and inactivation of which causes mtDNA mutator phenotypes (Foury, 1989; Foury and Vanderstraeten, 1992; Trifunovic et al., 2004). The high copy number of mtDNA generates genetic redundancy within mitochondria that can buffer the heteroplasmic presence of mutated or partially deleted mtDNA molecules and maintain cellular function. However, cells have to invest significant resources into synthesis and maintenance of sufficiently high mtDNA copy numbers. In budding yeast, the total mtDNA content of up to 8 million base pairs may rival the 12.5 million base pairs of the haploid nuclear genome. Consistently, available nucleotide pools influence mtDNA copy number, and defects in mitochondrial nucleotide synthesis are associated with mtDNA loss and depletion syndromes in humans (Taylor et al., 2005; Copeland, 2012; Young and Copeland, 2016). Thus, mtDNA content critically determines the susceptibility of cells to develop mitochondrial dysfunction, which is highly relevant for mitochondria-associated diseases (Jiang et al., 2017). How cells control mtDNA copy number in general and during times of nutrient scarcity in particular is still poorly understood.
In response to starvation, cells induce (macro)autophagy, a highly conserved pathway to maintain metabolic homeostasis and cell survival. Core autophagy machinery orchestrates the de novo formation of autophagosomes, double-membrane vesicles that encapsulate portions of cytoplasm for degradation and recycling in lysosomes/vacuoles to refuel essential metabolic processes (Harris and Rubinsztein, 2011; Kraft and Martens, 2012; Feng et al., 2014). Defects in autophagy result in mitochondrial dysfunction during starvation, compromising yeast and cancer cell survival (Guo et al., 2011; Suzuki et al., 2011).
Here, our analyses reveal that mtDNA copy number in yeast cells is dynamically controlled by two opposing functions of POLG, synthesis and degradation of mtDNA, during starvation. Strikingly, both POLG activities crucially depended on metabolic homeostasis provided by autophagy. Our data indicate that, during starvation, sustained mtDNA synthesis required autophagy to provide sufficient nucleotides and prevent the production of elevated reactive oxygen species (ROS) within mitochondria. Moreover, cells adjusted mtDNA content in a manner dependent on the 3′–5′ exonuclease activity of POLG, which, in the absence of autophagy, resulted in mtDNA depletion caused by nucleotide insufficiency.
Results and discussion
mtDNA synthesis and stability require autophagy during starvation
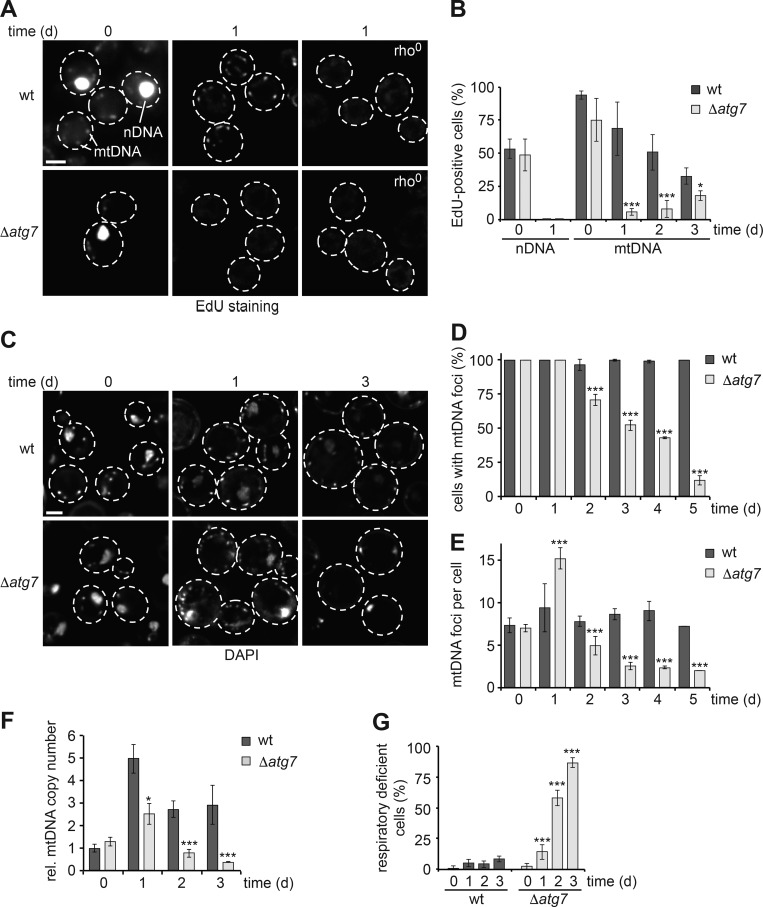
We asked whether asynchronous mtDNA synthesis is maintained during starvation when cells arrest in their cell cycle and, if so, whether it depends on autophagy to provide metabolic homeostasis. We analyzed WT cells and cells deficient for Atg7 (Δatg7), a conserved core autophagy component of the ubiquitin-like system required for autophagy (Tanida et al., 1999; Huang et al., 2000). First, we monitored DNA synthesis by in vivo incorporation of the thymidine analogue 5-ethynyl-2′-deoxyuridine (EdU) during exponential growth (0 d) in minimal medium. EdU incorporation was visualized in fixed cells with Alexa Fluor 647 using copper click chemistry. After 1 h of EdU exposure during growth, we found EdU-positive nuclear DNA in ∼50% and multiple EdU-positive mtDNA foci in ≥70% of WT and Δatg7 cells, respectively, demonstrating sensitive and specific detection of synthesis of both nuclear and mitochondrial genomes and confirming the asynchronous nature of mtDNA replication (Fig. 1 A, B). Next, we exposed cells to starvation medium, lacking external amino acids and ammonium as nitrogen sources. We added EdU after 3, 24, or 48 h of starvation and analyzed its incorporation after an additional 24 h of starvation (time points 1, 2, and 3 d) for sensitive detection of mtDNA synthesis. Nuclear DNA synthesis became undetectable in starved WT or Δatg7 cells, consistent with cell cycle arrest (Fig. 1, A and B). In contrast, we observed punctate EdU staining in 70, 50, and 25% of WT cells in a mtDNA-dependent manner, compared with WT rho0 at 1 d, after 1, 2, and 3 d of starvation, respectively, demonstrating that starving WT cells initially engaged in significant mtDNA synthesis, which gradually diminished during prolonged starvation (Fig. 1, A and B). Notably, only a small fraction of Δatg7 cells showed detectable EdU incorporation in mtDNA, compared with Δatg7 rho0 at 1 d, during starvation (Fig. 1, A and B). Importantly, Δatg7 cells maintained viability during the examined time course (Fig. S1 A). Thus, cells critically depended on autophagy to support mtDNA synthesis during starvation.
Figure 1.
Autophagy sustains mtDNA synthesis and stability during starvation. (A and B) mtDNA synthesis depends on autophagy during starvation. WT and Δatg7 cells were grown to log-phase (0 d) or shifted to starvation medium, and DNA synthesis was assessed using EdU staining. (A) Single section images after visualization of EdU incorporation in WT, WT rho0, Δatg7, and Δatg7 rho0 cells during log-phase (0 d) or starvation (1 d). (B) Quantification of nDNA and mtDNA synthesis during log-phase (0 d) and starvation (1–3 d) in WT and Δatg7 cells. Data are means ± SD (n ≥ 3; 150 cells). (C–E) Defective autophagy causes mtDNA depletion during starvation. (C) WT and Δatg7 cells were grown to log-phase and shifted to starvation medium. mtDNA foci were visualized by DAPI staining and in vivo fluorescence imaging at indicated time points. Single section images are shown. (D and E) Quantification of cells with mtDNA foci or the number of mtDNA foci in foci-positive cells. Data are means ± SD (n = 3; ≥75 cells). (F) mtDNA copy number dynamics in dependence of autophagy during starvation. Quantitative PCR was performed on isolated DNA from WT and Δatg7 cells at indicated time points after starvation. Data are normalized to mtDNA copy number of WT at 0 d set as 1. Data are means ± SD (n = 6). (G) Respiratory deficiency upon regrowth after starvation of WT and Δatg7 cells at indicated time points. Data are means ± SD (n = 3). Dashed lines indicate cell boundaries. Bars, 2 µm. t tests: *, P < 0.05; ***, P < 0.001. Rel., relative.
Next, we examined whether the lack of autophagy-dependent synthesis affects mtDNA maintenance. To monitor the spatial distribution and stability of mtDNA within nucleoids in vivo, we used DAPI staining of DNA in combination with fluorescent live-cell imaging (Williamson and Fennell, 1975). WT cells showed a mean of approximately eight discrete mtDNA foci per cell during growth, consistent with previous data (Chen and Butow, 2005; Miyakawa, 2017), and maintained this number over 5 d of starvation (Fig. 1, C–E). In contrast, the initial WT-like eight mtDNA foci per cell during growth parsed into ∼15 foci after 1 d of starvation in Δatg7 cells, reminiscent of nucleoid behavior upon general amino acid control pathway activation (MacAlpine et al., 2000). Notably, the initially WT-like tubular mitochondrial network of autophagy-deficient cells quantitatively fragmented within 1 d of starvation, raising the possibility that predominant mitochondrial division redistributed nucleoids (Fig. S1, B and C). After 1 d, the number of mtDNA foci per cell and of cells containing any mtDNA foci gradually decreased, until the vast majority of Δatg7 cells were devoid of detectable mtDNA after 5 d of starvation (Fig. 1, C–E). These data indicate that autophagy deficiency is linked to quantitative depletion of mtDNA during starvation, consistent with a previous study (Suzuki et al., 2011). Blocking autophagy at the stage of initiation (Δatg1) or vacuolar hydrolysis (Δatg15 or Δpep4) caused mtDNA instability indistinguishable from Δatg7 cells after 3 d, whereas absence of selective forms of autophagy (Δatg11), including mitophagy, did not (Fig. S1 D). We concluded that general autophagic turnover is required for mtDNA stability and that compromised metabolic homeostasis drives mtDNA degradation rather than the absence of selective mitochondrial quality control.
The number of cellular mtDNA foci is a rough approximation of mtDNA content, because individual foci may contain 1–10 mtDNA molecules (Chen and Butow, 2005). To accurately assess mtDNA copy number in WT and Δatg7 cells during starvation, we used real-time quantitative PCR (qPCR) measurements and found remarkable mtDNA dynamics. Consistent with ongoing mtDNA replication, we observed a fivefold increase in mtDNA content after starving WT cells for 1 d (Fig. 1 F). The number of mtDNA copies decreased subsequently, suggesting that cells actively adjusted their mtDNA content through degradation in a nondividing state (Fig. 1 F). In Δatg7 cells, the number of mtDNA molecules also increased after 1 d of starvation, however, to a lesser extent than in WT cells, consistent with impaired mtDNA synthesis in the absence of autophagy (Fig. 1 F). Importantly, mtDNA content continuously declined after starving Δatg7 cells past 1 d (Fig. 1 F), providing evidence that mitochondrial genome stability depended on autophagy during nutrient stress. In line with these data, we observed an increasing fraction of Δatg7 cells over time, which was irreversibly respiratory deficient upon regrowth after starvation (Fig. 1 G). Collectively, our data establish that the mitochondrial genome undergoes strikingly dynamic changes in copy number, which are determined by ongoing mtDNA synthesis and subsequent degradation, when cells cease to divide upon starvation. Significantly, mitochondrial genome dynamics and stability require autophagy to support mtDNA synthesis and prevent mtDNA depletion and irreversible loss of respiratory competence.
Nucleotide availability limits mtDNA stability in autophagy mutants
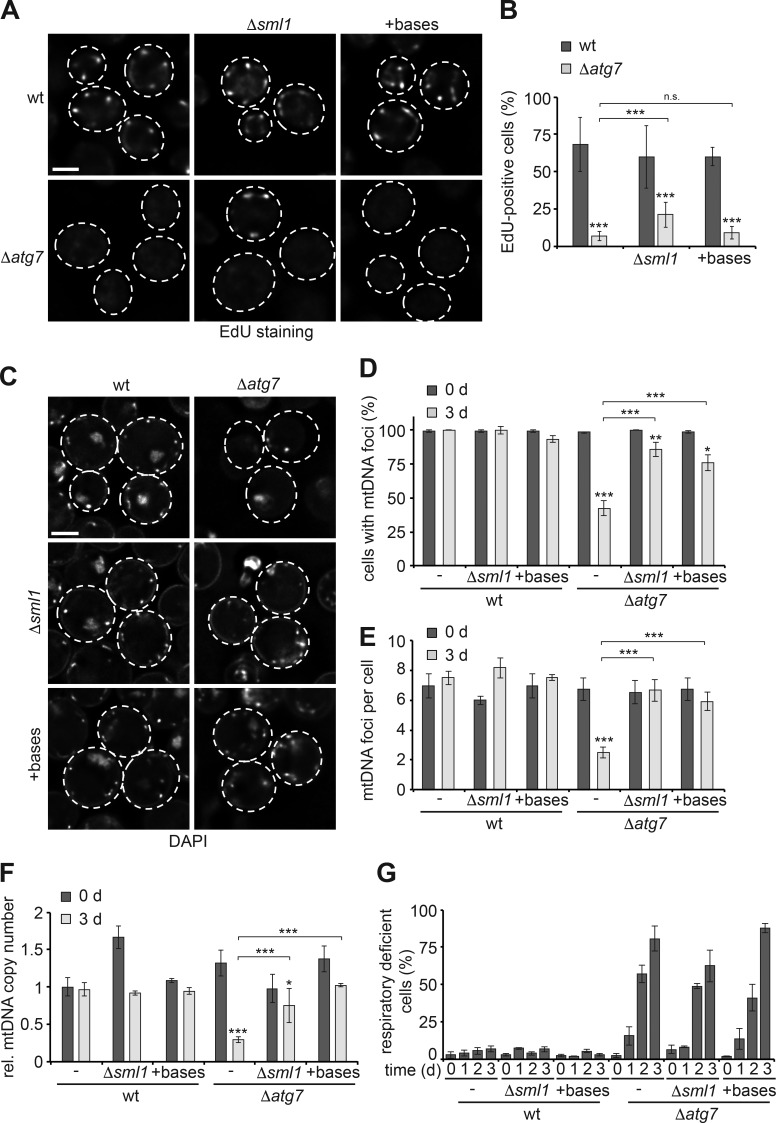
Given size and copy number of mitochondrial genomes, cells likely depend on autophagy to allocate considerable resources to mtDNA replication and maintenance during starvation. Previous work has shown that nucleoside/nucleotide metabolism during starvation relies on autophagy and that defects in nucleotide synthesis are linked to mtDNA depletion syndromes (Copeland, 2012; Huang et al., 2015; Guo et al., 2016; Young and Copeland, 2016). We hypothesized that nucleotide insufficiency might underlie compromised mtDNA synthesis and stability in the absence of autophagy in starving cells. To test this idea, we deleted ribonucleotide reductase inhibitor Sml1 (Δsml1), which has previously been shown to increase deoxynucleotide pools and mtDNA copy number (Zhao et al., 1998; Taylor et al., 2005), or supplemented starvation media with the four nucleobases to metabolically increase nucleotide levels in autophagy-deficient cells. Surprisingly, nucleobase-supplemented Δatg7 or Δatg7Δsml1 cells showed no or only partially improved mtDNA synthesis, respectively, compared with untreated Δatg7 cells after 1 d of starvation (Fig. 2, A and B). Nevertheless, imaging DAPI-stained mtDNA foci after 3 d of starvation revealed that ≥75% of Δatg7Δsml1 and nucleobase-supplemented Δatg7 cells maintained a WT-like number of mtDNA foci in contrast to untreated Δatg7 cells (Fig. 2, C–E), suggesting that increasing nucleotides improved mtDNA maintenance in autophagy-deficient cells. Measuring mtDNA copy number by qPCR analysis demonstrated that nucleobase supplementation and Δsml1 deletion significantly stabilized mtDNA content in Δatg7 cells (Fig. 2 F). These data indicate that mtDNA stability is limited by nucleotide availability in the absence of autophagy. Interestingly, nucleobase-supplemented Δatg7 and Δatg7Δsml1 cells displayed a loss of respiratory competence similar to untreated Δatg7 cells during starvation, indicating that increased nucleotide pools improve structural, but not functional, integrity of mitochondrial genomes (Fig. 3 G).
Figure 2.
Nucleotide availability limits mtDNA stability in autophagy-deficient cells during starvation. (A and B) mtDNA synthesis in dependence of nucleotide levels. WT, Δsml1, Δatg7, and Δsml1Δatg7 cells or WT and Δatg7 cells supplemented with nucleobases were assessed after 1 d of starvation as described in Fig. 1 (A and B). Data are means ± SD (n ≥ 3; 150 cells). (C–E) Increased nucleotide levels stabilize mtDNA in the absence of autophagy. WT, Δsml1, Δatg7, and Δsml1Δatg7 cells or WT and Δatg7 cells supplemented with nucleobases were analyzed by DAPI staining and fluorescence imaging as described in Fig. 1 (C–E). Data are means ± SD (n = 3; ≥75 cells). (F) Increased nucleotide levels stabilize mtDNA in autophagy-deficient cells during starvation. Cells were treated as described in C and analyzed by quantitative PCR as described in Fig. 1 F. Data are means ± SD (n ≥ 3). (G) Increased nucleotide levels do not sustain functional integrity of mtDNA in autophagy mutants. Cells treated as described in C were analyzed as described in Fig. 1 G. Data are means ± SD (n = 3). Dashed lines indicate cell boundaries. Bars, 2 µm. t tests: *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s., not significant. Rel., relative.
Figure 3.
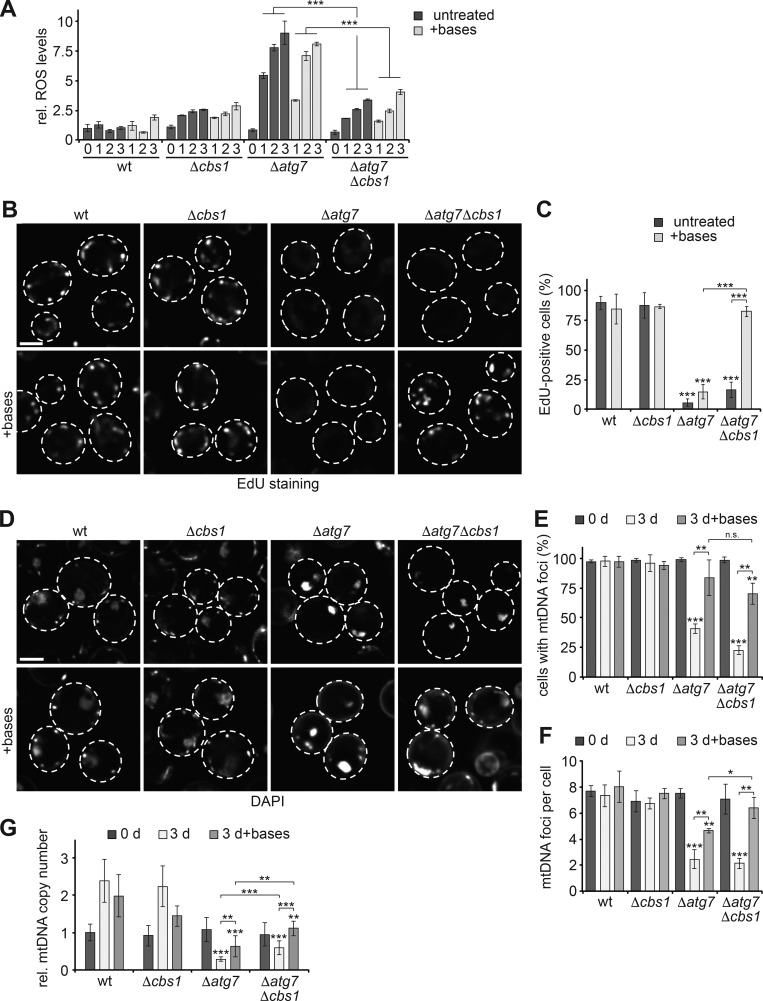
Elevated ROS level and nucleotide insufficiency inhibit mtDNA synthesis and copy number maintenance in autophagy-deficient cells during starvation. (A) Defective autophagy causes complex III–dependent elevated ROS production during starvation. WT, Δcbs1, Δatg7, and Δcbs1Δatg7 cells were grown in log-phase (0 d) or shifted to starvation media ± nucleobases and analyzed for ROS levels using DHE staining and flow cytometry at indicated time points. Data are means ± SD (n = 3). (B and C) mtDNA synthesis in autophagy-deficient cells requires ROS reduction and increased nucleotides. WT, Δcbs1, Δatg7, and Δcbs1Δatg7 cells were treated as described in A and assessed after 1 d of starvation as described in Fig. 1 A. Data are means ± SD (n ≥ 3; 150 cells). (D–F) mtDNA maintenance in autophagy-deficient cells in dependence of ROS production and nucleotide supplementation. WT, Δcbs1, Δatg7, and Δcbs1Δatg7 cells were treated as described in A and analyzed at indicated time points as described in Fig. 1 (C–E). (E and F) Quantification of cells showing mtDNA foci or their number of mtDNA foci. Data are means ± SD (n = 3; ≥75 cells). (G) ROS reduction and nucleotide supplementation synergistically stabilize mtDNA copy number in autophagy-deficient cells. WT, Δcbs1, Δatg7, and Δcbs1Δatg7 cells were treated as described in D and analyzed as described in Fig. 1 F. Data are means ± SD (n = 6). Dashed lines indicate cell boundaries. Bars, 2 µm. t tests: *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s., not significant. Rel., relative.
In summary, these data suggest that nucleotides are a limiting factor for mtDNA stability in the absence of autophagy and are consistent with a model in which autophagy functions in maintenance of nucleotide homeostasis during starvation to balance mtDNA synthesis and degradation, which determine mtDNA copy number and stability.
ROS and nucleotide levels coregulate mtDNA synthesis and copy number
Intrigued by the seemingly conflicting observations that nucleobase supplementation stabilized mtDNA without promoting detectable mtDNA synthesis, we reasoned that additional factors might stall mtDNA synthesis in starving Δatg7 cells. We assessed the production of ROS in dihydroethidium (DHE)-stained WT and Δatg7 cells by flow cytometry and found that autophagy deficiency enhanced ROS production up to 10-fold compared with WT cells during starvation (Fig. 3 A), consistent with previous work suggesting that elevated ROS might cause mitochondrial dysfunction in cells defective for autophagy (Suzuki et al., 2011). We confirmed the finding of this study that N-acetylcysteine (NAC) treatment specifically lowered ROS levels in Δatg7 cells, which also allowed cells to retain respiratory competence upon regrowth after starvation (Fig. S2, A and B; Suzuki et al., 2011). In addition, we found partially improved mtDNA maintenance after 3 d of starvation when we compared NAC-treated with untreated Δatg7 cells using DAPI staining (Fig. S2, C–E). Moreover, NAC-treated Δatg7 cells engaged in mtDNA synthesis more frequently than untreated Δatg7 cells upon starvation (Fig. S2, F and G). Together with our findings that nucleobase-supplemented Δatg7 and Δatg7Δsml1 cells increased ROS production to a similar extent as untreated Δatg7 cells (Figs. 3 A and S2 H), these data raised the intriguing possibility that elevated ROS inhibited mtDNA synthesis. Because NAC may profoundly affect cellular metabolism aside from enhancing ROS-scavenging capacity, we decided to genetically dissect the potential effects of ROS. For this, we deleted CBS1 (Δcbs1), a nuclear-encoded translation activator of the mitochondria-encoded core subunit of respiratory chain complex III, COB1 (Rödel, 1997). When comparing WT, Δcbs1, Δatg7, and Δcbs1Δatg7 cells, we found that the majority of ROS production in autophagy-deficient cells depended on the integrity of respiratory chain complex III (Fig. 3 A). With this tool in hand, we tested for potential effects of ROS on mtDNA. WT and Δcbs1 cells showed indistinguishable behavior in mtDNA synthesis, maintenance, and copy number during starvation, demonstrating that respiratory deficiency itself did not affect mtDNA homeostasis (Fig. 3, B–G). Lowered ROS in Δcbs1Δatg7 cells barely improved mtDNA synthesis or maintenance but did cause a small, but statistically significant, stabilizing effect on mtDNA copy number compared with Δatg7 cells during starvation (Fig. 3, B–G), suggesting that ROS might affect the rate of mtDNA degradation. Nevertheless, elevated ROS production did not—or at least, not alone—cause impaired mtDNA synthesis or mitochondrial genome instability in the absence of autophagy. We hypothesized that autophagy-deficient cells required not only a reduction in ROS levels, but also sufficient nucleotide resources to fuel mtDNA synthesis, maintenance, and copy number regulation. To test this notion, we supplemented WT, Δcbs1, Δatg7, and Δcbs1Δatg7 cells with nucleobases during starvation. Strikingly, we found that suppressed ROS production of Δcbs1Δatg7 cells in combination with nucleobase supplementation was sufficient to rescue mtDNA synthesis in the absence of autophagy (Fig. 3, B and C), indicating that elevated ROS and nucleotide insufficiency are physiological factors that together suppress mtDNA synthesis in autophagy-deficient cells. Nucleotide supplementation did not cause any further changes in mtDNA maintenance and distribution in Δcbs1Δatg7 cells compared with Δatg7 cells when assessed by mtDNA foci using DAPI staining (Fig. 3, D–F). Notably, when combined with lowered ROS production in Δcbs1Δatg7 cells, nucleotide supplementation synergistically stabilized the number of mtDNA copies measured in cells in log-phase even after 3 d of starvation (Fig. 3 G). In conclusion, our data indicate that autophagy maintains metabolic homeostasis of mitochondria to prevent intramitochondrial ROS production and nucleotide insufficiency, which in combination inhibit mtDNA synthesis and destabilize mitochondrial genomes.
Exonuclease-dependent mtDNA degradation by POLG
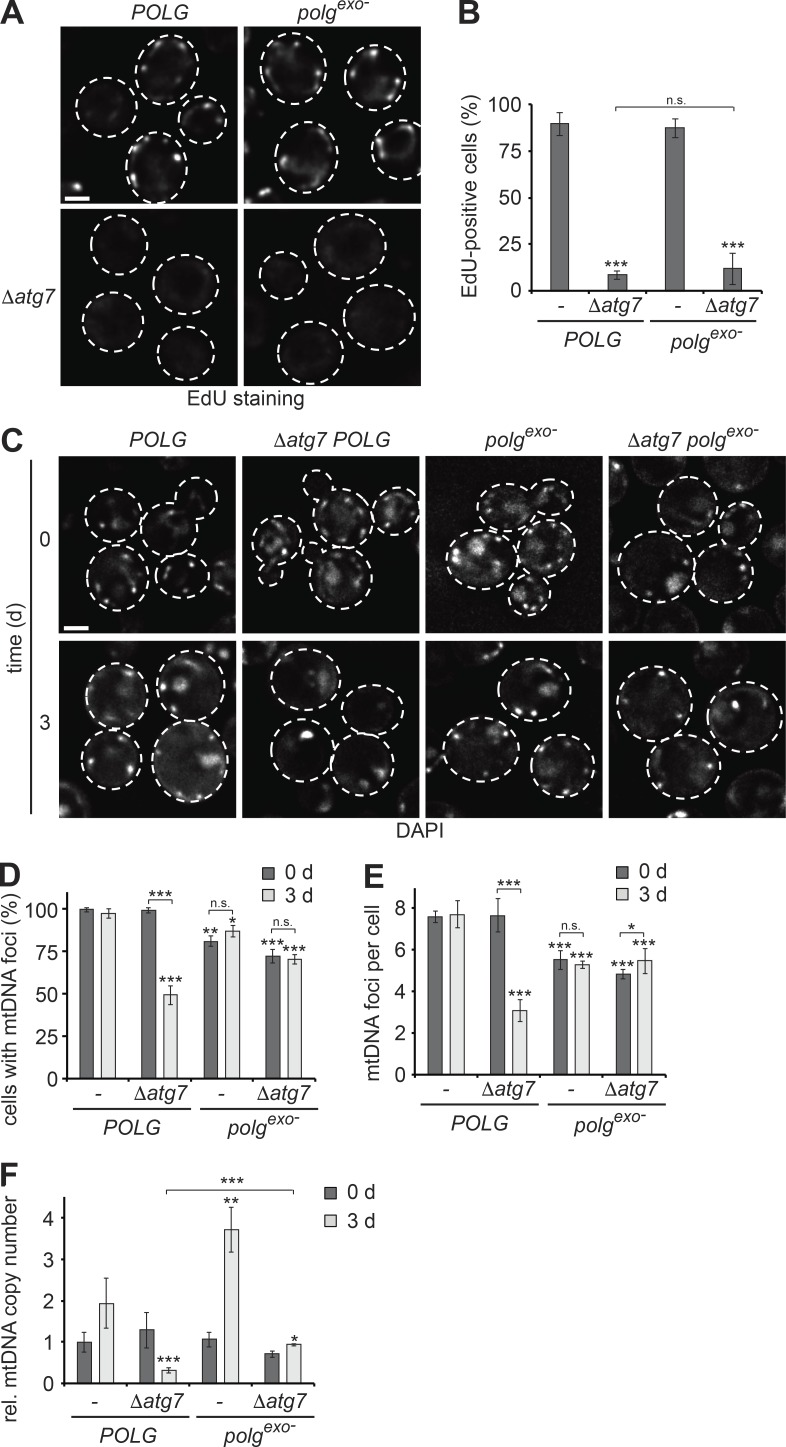
As WT and Δatg7 cells enter a nondividing state during starvation, a reduction in copy number must be caused by mtDNA degradation independently of autophagy. We considered intramitochondrial nucleases to degrade mitochondrial genomes during starvation. First, we examined the role of the conserved mitochondrial nuclease EndoG (Nuc1; Büttner et al., 2007) and could not detect any significant effect on mtDNA maintenance in the absence of EndoG by itself or in the context of Δatg7 cells (Fig. S3, A and B). Next, we tested POLG (MIP1), as it contains inherent 3′–5′ exonuclease activity that provides proofreading function toward newly synthesized mtDNA (Foury, 1989; Foury and Vanderstraeten, 1992). In vitro, purified human POLG (PolgA/B) has been shown to degrade small linear DNA fragments in an exonuclease-dependent manner in the absence of deoxynucleotides, whereas it polymerizes DNA in their presence (Bratic et al., 2015). We considered the possibility that limiting nucleotide levels in autophagy-deficient cells shift the balance from mtDNA synthesis toward exonuclease-dependent mtDNA degradation by POLG in vivo, resulting in mtDNA instability. To test this hypothesis, we genomically integrated an exonuclease-deficient allele of POLG (polgexo−: mip1D171A,E173A) into the endogenous MIP1 locus in otherwise WT or Δatg7 cells (Longley et al., 1998; Strand et al., 2003). The level of mtDNA synthesis in WT or Δatg7 cells was independent of exonuclease proficiency of POLG (Fig. 4, A and B). The presence of polgexo− resulted in a slightly lower number of cells containing mtDNA foci and a reduction in mtDNA foci per cell independent of autophagy during log-phase (Fig. 4, C–E). Excitingly, when challenged with starvation, Δatg7 cells expressing polgexo− maintained mtDNA foci at the same level as during log-phase, in contrast to POLG-expressing Δatg7 cells, which showed a more than 50% decrease in the number of cells containing mtDNA foci and number of mtDNA foci per cell after 3 d of starvation (Fig. 4, C–E). qPCR-based analysis confirmed that the expression of polgexo− fully prevented starvation-induced mtDNA copy number decline in the presence of autophagy deficiency (Fig. 4 F). Thus, we concluded that the excessive mtDNA degradation in autophagy-deficient cells is driven by the exonuclease activity of POLG depleting mitochondria of mtDNA. Additionally, we found that expression of polgexo− affected mtDNA copy number control in autophagy-proficient cells: in contrast to WT cells, which showed a ∼2-fold increase in relative mtDNA copy number, polgexo− cells maintained an almost fourfold increase in relative mtDNA copy number after 3 d of starvation compared with nonstarving cells (Fig. 4 F). Thus, these data indicate that exonuclease activity of POLG not only affects mtDNA stability in the absence of autophagy, but also generally regulates mtDNA copy number dynamics in nondividing cells.
Figure 4.
Exonuclease activity of POLG regulates mtDNA copy number in WT and is required for mtDNA degradation in autophagy mutants during starvation. (A and B) mtDNA synthesis is unaffected by exonuclease-activity of POLG. POLG, Δatg7 POLG, polgexo−, and Δatg7 polgexo− cells were treated as described in Fig. 1 A. Data are means ± SD (n ≥ 3; 150 cells). (C–E) Exonuclease-deficiency of POLG rescues mtDNA maintenance in autophagy-deficient cells during starvation. POLG, Δatg7 POLG, polgexo−, and Δatg7 polgexo− cells were grown and analyzed as described in Fig. 1 (C–E). Data are means ± SD (n = 3; ≥75 cells). (F) Exonuclease deficiency of POLG prevents decrease in mtDNA copy number during starvation. POLG, Δatg7 POLG, polgexo−, and Δatg7 polgexo− cells were analyzed as described in Fig. 1 F. Data are means ± SD (n = 3). Dashed lines indicate cell boundaries. Bars, 2 µm. t tests: *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s., not significant. Rel., relative.
In summary, our work positions POLG at the center of mtDNA copy number regulation in response to physiological changes by operating in two opposing modes: synthesis and, unexpectedly, degradation of mitochondrial genomes (Fig. 5). We found that yeast cells continued to engage in mtDNA synthesis during starvation-induced cell cycle arrest, resulting in an initial increase in mtDNA content. Our data suggest that prolonged starvation caused POLG to switch from a synthetic to degradative mode, which readjusted mtDNA copy number in a POLG exonuclease-dependent manner. Thus far, the 3′–5′ exonuclease of POLG has been linked to proofreading activity ensuring fidelity (Foury and Vanderstraeten, 1992; Szczepanowska and Trifunovic, 2015; Young and Copeland, 2016). We propose now that it also enables POLG to mediate degradation of mtDNA in vivo. EndoG/Nuc1 has been implicated in degrading mtDNA during spermatogenesis in flies and in participating in paternal mitochondrial elimination (DeLuca and O’Farrell, 2012; Zhou et al., 2016). However, the absence of EndoG/Nuc1 did not affect mtDNA maintenance in our system, supporting the conclusion that POLG is required and likely sufficient for mtDNA degradation. Of note, inducible expression of a restriction enzyme targeted to mitochondria in human cell lines initiates double-strand break–associated mtDNA degradation. However, neither the known nucleases (including ExoG, EndoG, MGME1, and FEN1), individually or in combination, nor mitophagy/autophagy affected mtDNA loss. The role of POLG exonuclease was not assessed in this study (Moretton et al., 2017). These and our data raise the exciting possibility that POLG is sufficient for mtDNA degradation and may constitute an evolutionarily conserved mechanism.
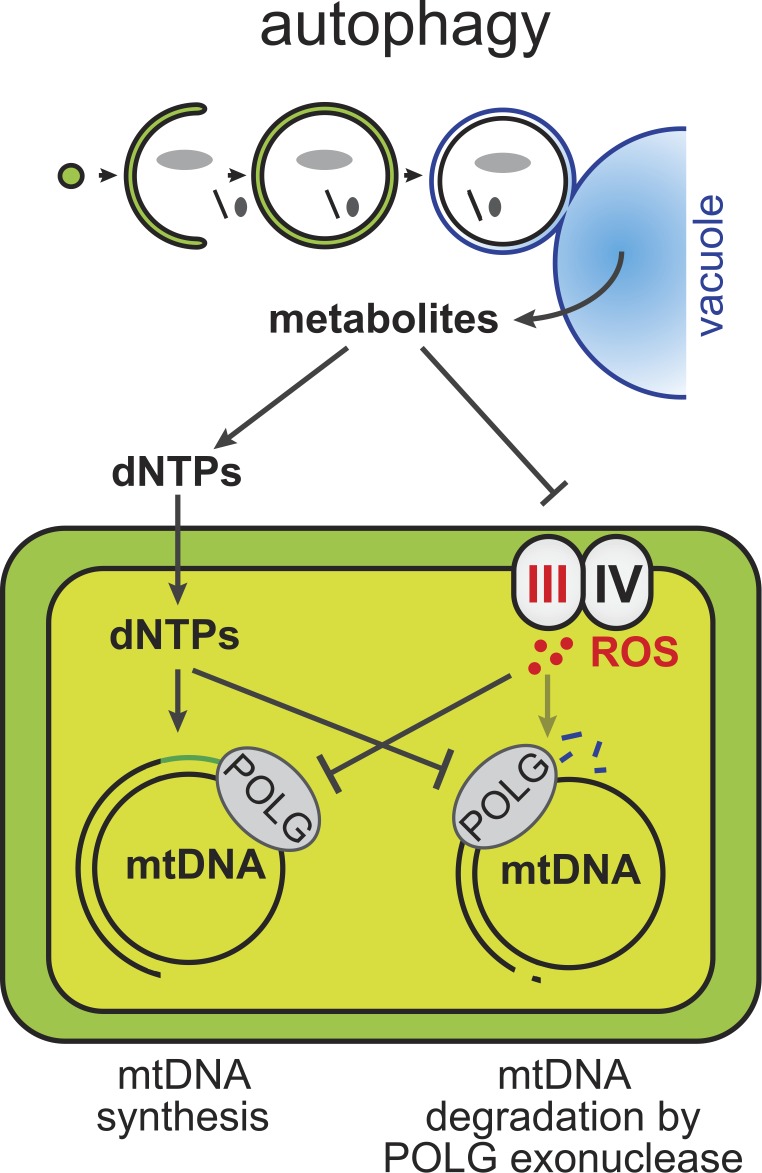
Figure 5.
Model for mtDNA copy number regulation by POLG in dependence of autophagy. POLG regulates mtDNA copy number by balanced DNA synthesis and degradation. During starvation, autophagy provides metabolites to fuel nucleotide pools (dNTPs) and prevent elevated ROS production in mitochondria, which is required for sustained mtDNA synthesis and suppressed mtDNA degradation by POLG.
We found that the striking dynamics of mtDNA copy number controlled by POLG strictly depended on autophagy during starvation. As our work indicates, autophagy affected two physiological factors, nucleotide availability and mitochondrial ROS production, which together determined the balance of the two POLG activities. Our manipulations to improve nucleotide availability genetically and biochemically were sufficient to stall the degradation of mtDNA in the absence of autophagy, suggesting that nucleotide binding to POLG might directly interfere with exonuclease-driven mtDNA degradation (Fig. 5). In this context, it will be interesting to assess nucleotide pools within mitochondria, as insufficient general nucleotide pools are linked to mtDNA depletion syndromes, and although impaired replication likely constitutes a major factor during cell division, POLG-mediated mtDNA degradation might contribute to mtDNA loss in nonproliferative tissues (Suomalainen and Isohanni, 2010; Copeland, 2012). In addition, similar to our system, cell lines derived from Kras-driven lung cancer tumors defective for autophagy display mitochondrial dysfunction, elevated ROS production, and starvation sensitivity caused by impaired maintenance of nucleotide pools (Guo et al., 2011, 2016), indicating that nucleotide homeostasis is a conserved key function of autophagy to sustain mitochondrial function. It is noteworthy that, based on genetic evidence, we found that elevated mitochondria-derived ROS inhibited mtDNA synthesis in autophagy-deficient cells even in the presence of improved nucleotide pools. Interestingly, nuclear genome replication occurs during the nonrespiratory phase of the cell cycle in yeast to preserve genome integrity, and the mammalian nuclear replisome undergoes architectural alterations to mitigate oxidative stress and fluctuations in nucleotide levels (Chen et al., 2007; Somyajit et al., 2017). Thus, regulatory mechanisms integrating oxidative stress with DNA replication may be universally conserved and, in mitochondria, might affect POLG to repress mtDNA replication within nucleoids exposed to elevated ROS levels and limited nucleotide pools.
Materials and methods
Yeast strains
Strains W303 (ade2-1; leu2-3; his3-11, 15; trp1-1; ura3-1; can1-100) and W303 Δatg7::kanMX6 (Graef and Nunnari, 2011) were described previously. W303 URA3::GDP-Tk(5x) AUR1c::ADH-hENT1 (E3087; Talarek et al., 2015) was a gift from E. Schwob (University of Montpellier, Montpellier, France). The following gene deletions were generated in the W303 or E3087 background by replacing complete ORFs by indicated marker cassettes using PCR-based targeted homologous recombination as previously described (Longtine et al., 1998): Δcbs1::natMX6, Δatg7::kanMX6 Δcbs1::natMX6, Δnuc1::natMX6, Δatg7::kanMX6 Δnuc1::natMX6, Δsml1::HIS3MX6, and Δatg7::kanMX6 Δsml1::HIS3MX6. To introduce POLG alleles into the genome, we generated the deletion Δmip1::URA3 in W303 or E3087 backgrounds and replaced the URA3 selection marker with the ORF for MIP1 or mip1D171A,E173A in conjunction with a 3′-natMX6 selection cassette. These strains were backcrossed to W303 Δatg7::kanMX6 to reintroduce mitochondrial genomes and generate Δatg7::kanMX6 deletion variants. Diploid strains were sporulated, and desired haploid strains were identified by appropriate marker selection and confirmed by colony PCR to give rise to W303 or E3087 Δmip1::MIP1-natMX6 (POLG), Δmip1::MIP1-natMX6 (POLG) Δatg7::kanMX6, Δmip1::mip1D171A,E173A-natMX6 (polgexo−; bases A512C, A518C), and Δmip1::mip1D171A,E173A-natMX6 (polgexo−) Δatg7::kanMX6. Indicated rho0 strains were generated by growth in YPD medium supplemented with ethidium bromide (25 µg/ml).
Media
Strains were grown in synthetic complete medium containing 0.7% (wt/vol) yeast nitrogen base (BD Difco) and 2% (wt/vol) α-d-glucose (Sigma) at 30°C. Cells were washed three to five times and starved in nitrogen starvation medium with 0.17% (wt/vol) yeast nitrogen base without amino acids and ammonium sulfate (BD Difco) and 2% (wt/vol) α-d-glucose buffered at pH 6.2 using 50 mM MES-KOH as described in Suzuki et al. (2011), supplemented with the nucleobases adenine, guanine, cytosine, and thymidine at a final concentration of 0.4 mM each or NAC (Sigma) at a final concentration of 10 mM when indicated.
To analyze respiratory competence, cells were plated onto YPD plates containing 1% (wt/vol) yeast extract (Serva), 2% (wt/vol) bacto-peptone (Merck), and 2% (wt/vol) α-d-glucose, and colony color was assessed and quantified. Because all strains contained the ade2 mutation, on YPD, white and sectored colonies were classified as respiratory deficient and red colonies were classified as respiratory competent.
Staining of mitochondrial morphology and DNA
To monitor mtDNA foci or mitochondrial morphology in vivo, 2.5 DAPI (Sigma) or 100 nM MitoTracker green FM (Invitrogen), respectively, was added to the medium for 30 min at 30°C. Cells were examined by fluorescence microscopy. The number of mtDNA foci was assessed conservatively and counted only when clearly discrete foci were visible.
In vivo detection of mtDNA synthesis
EdU incorporation was detected as described in Talarek et al. (2015) and optimized for mtDNA detection as follows. All strains are derivatives of yeast strain E3087 (gift from E. Schwob; URA3::GPD-TK5x, AUR1c::ADH-hENT1, and W303 RAD5) expressing herpes simplex virus thymidine kinase and human equilibrative nucleoside transporter to allow for incorporation of EdU in yeast. Cells were exposed to 100 µM EdU for 1 h in growth medium (0 d) or for 24 h in starvation medium after 3, 24, or 48 h of starvation, resulting in time points 1, 2, and 3 d, respectively. Cells were harvested; fixed in 2% (vol/vol) PFA for 20 min; permeabilized in 70% (vol/vol) ethanol for 3 h; incubated in 1× PBS, pH 7.4, and 1% (wt/vol) BSA for 30 min; washed in 1× PBS; resuspended in 150 µl Click-iT EdU Alexa Fluor 674 reaction mix (Invitrogen); and incubated for 30 min protected from light. Cells then were washed twice with 1× PBS and analyzed by fluorescence microscopy.
Fluorescence microscopy
Cells were imaged in 96-well glass-bottom microplates (Greiner Bio-One) containing indicated media with an inverted microscope (Ti-E; Nikon) using a Plan Apochromat IR 60× 1.27 NA objective (Nikon) and Spectra X LED light source (Lumencor) at room temperature. 3D light microscopy data were collected using the triggered Z-Piezo system (Nikon) and orca flash 4.0 camera (Hamamatsu). 3D data were processed using NIS-elements AR (Nikon), Huygens Professional 16.10 (Scientific Volume Imaging), ImageJ v.2.0.0., and Photoshop (Adobe Systems) software.
Determination of relative mtDNA copy number
To isolate total DNA at indicated time points, five OD600 units of cells were harvested, washed in dH2O, resuspended, and incubated for 30 min at 37°C in 50 mM Tris/HCl, pH 7.4, 25 mM EDTA, and 0.01% (wt/vol) Lyticase (Sigma). Cells were resuspended in 200 µl lysis solution (0.2 M NaOH, 1% [wt/vol] SDS, 10 mM Tris/HCl, pH 7.4, and 1 mM EDTA) and incubated at 65°C for 10 min. DNA was precipitated by addition of 150 µl of 5 M potassium acetate, washed in ice-cold ethanol, dried, and resuspended in 10 mM Tris/HCl, pH 7.4. Total DNA concentrations were determined using a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific) and adjusted to 100 ng/ml.
Total DNA was subjected to qPCR using LightCycler 480 SYBR Green (Roche) according to the manufacturer’s instructions. In brief, 1 µl total DNA (100 ng/ml) was mixed with 4 µl dH2O and 5 µl PCR mix (LightCycler 480 SYBR Green and 0.5 µM of each primer). mtDNA or nuclear DNA was amplified using primers against COX3 (forward, 5′-TTGAAGCTGTACAACCTACC-3′; reverse, 5′-CCTGCGATTAAGGCATGATG-3′) or primers against ACT1 (forward, 5′-CACCCTGTTCTTTTGACTGA-3′; reverse, 5′-CGTAGAAGGCTGGAACG TTG-3′), respectively. Reactions were performed in 96-well Lightcycler plates (Roche) sealed with adhesive qPCR seal (Roche) with the following cycler program: 95°C for 7 min followed by 35 cycles of 95°C for 10 s, 60°C for 30 s. Dissociation curves confirmed single PCR products, and signals were analyzed within the linear amplification range. Each sample was run in triplicate. qPCR data were analyzed using LightCycler 96 Application v.1.01.00. Relative mtDNA copy number changes were calculated using the comparative ΔΔCt method (Livak and Schmittgen, 2001) by determining Ct threshold values and using equations ΔΔCt = (Ct_COX2 − Ct_ACT1) at t(n) − (Ct_COX2 − Ct_ACT1) at t(0) and 2−ΔΔCt.
ROS level and cell viability assays
Cellular ROS levels were measured using superoxide anion–sensitive DHE (Molecular Probes). 107 cells were resuspended in medium containing 5 µM DHE and incubated at 30°C for 15 min. Mean fluorescence intensities (MFIs) of 10,000 events per sample were measured using the PE-Cy5 channel of a BD FACS Canto II flow cytometer with 488-nm excitation and ≥670-nm emission settings.
Cell viability was determined by phloxine B (Sigma) staining. 107 cells were resuspended in medium containing 5 µg/ml phloxine B and incubated at 30°C for 15 min. MFIs of 10,000 events per sample were measured using the PE channel of a Miltenyi MACSQuant VYB flow cytometer with 488-nm excitation and 586-nm emission settings. Data analysis was performed with FlowJo v.10 software.
Statistical analysis
Error bars represent SD as indicated in the figure legends. Data were processed in Excel. Statistical analysis of differences between two groups was performed using a two-tailed, unpaired t test. Only statistically significant comparisons are indicated in graphs.
Online supplemental material
Fig. S1 provides data analyzing cell viability and mitochondrial morphology of WT and autophagy-deficient cells during starvation. Furthermore, evidence is shown that mtDNA stability depends on autophagy flux and is independent of selective forms of autophagy. Fig. S2 analyzes the effects of NAC treatment on ROS production, respiratory competence, mtDNA maintenance, and mtDNA synthesis in WT and autophagy-deficient cells. Fig. S3 provides data showing that loss of mtDNA in autophagy-deficient cells occurs in an EndoG/Nuc1-independent manner.
Supplementary Material
Acknowledgments
We thank members of the Graef laboratory as well as Professor Adam Antebi, Professor Rudolf Wiesner, and Dr. James Stewart for discussion and Professor Etienne Schwob for providing materials. We thank the FACS and Imaging Core Facility at the Max Planck Institute for Biology of Ageing for support.
This work was supported by the Max Planck Society and the Deutsche Forschungsgemeinschaft (SFB 1218/TP A04 to M. Graef).
The authors declare no competing financial interests.
Author contributions: T.C. Medeiros and M. Graef were the lead contributors to the conception and design of the experiments and the analysis and interpretation of data. T.C. Medeiros was the lead contributor to the acquisition of data. R.L. Thomas and R. Ghillebert supported data acquisition and analysis. M. Graef wrote the manuscript.
References
- Bratic A., Kauppila T.E., Macao B., Grönke S., Siibak T., Stewart J.B., Baggio F., Dols J., Partridge L., Falkenberg M., et al. 2015. Complementation between polymerase- and exonuclease-deficient mitochondrial DNA polymerase mutants in genomically engineered flies. Nat. Commun. 6:8808 10.1038/ncomms9808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner S., Eisenberg T., Carmona-Gutierrez D., Ruli D., Knauer H., Ruckenstuhl C., Sigrist C., Wissing S., Kollroser M., Fröhlich K.U., et al. 2007. Endonuclease G regulates budding yeast life and death. Mol. Cell. 25:233–246. 10.1016/j.molcel.2006.12.021 [DOI] [PubMed] [Google Scholar]
- Chen X.J., and Butow R.A.. 2005. The organization and inheritance of the mitochondrial genome. Nat. Rev. Genet. 6:815–825. 10.1038/nrg1708 [DOI] [PubMed] [Google Scholar]
- Chen Z., Odstrcil E.A., Tu B.P., and McKnight S.L.. 2007. Restriction of DNA replication to the reductive phase of the metabolic cycle protects genome integrity. Science. 316:1916–1919. 10.1126/science.1140958 [DOI] [PubMed] [Google Scholar]
- Copeland W.C. 2012. Defects in mitochondrial DNA replication and human disease. Crit. Rev. Biochem. Mol. Biol. 47:64–74. 10.3109/10409238.2011.632763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca S.Z., and O’Farrell P.H.. 2012. Barriers to male transmission of mitochondrial DNA in sperm development. Dev. Cell. 22:660–668. 10.1016/j.devcel.2011.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., He D., Yao Z., and Klionsky D.J.. 2014. The machinery of macroautophagy. Cell Res. 24:24–41. 10.1038/cr.2013.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foury F. 1989. Cloning and sequencing of the nuclear gene MIP1 encoding the catalytic subunit of the yeast mitochondrial DNA polymerase. J. Biol. Chem. 264:20552–20560. [PubMed] [Google Scholar]
- Foury F., and Vanderstraeten S.. 1992. Yeast mitochondrial DNA mutators with deficient proofreading exonucleolytic activity. EMBO J. 11:2717–2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foury F., Roganti T., Lecrenier N., and Purnelle B.. 1998. The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett. 440:325–331. 10.1016/S0014-5793(98)01467-7 [DOI] [PubMed] [Google Scholar]
- Garcia I., Jones E., Ramos M., Innis-Whitehouse W., and Gilkerson R.. 2017. The little big genome: The organization of mitochondrial DNA. Front. Biosci. 22:710–721. 10.2741/4511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graef M., and Nunnari J.. 2011. Mitochondria regulate autophagy by conserved signalling pathways. EMBO J. 30:2101–2114. 10.1038/emboj.2011.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J.Y., Chen H.Y., Mathew R., Fan J., Strohecker A.M., Karsli-Uzunbas G., Kamphorst J.J., Chen G., Lemons J.M., Karantza V., et al. 2011. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 25:460–470. 10.1101/gad.2016311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J.Y., Teng X., Laddha S.V., Ma S., Van Nostrand S.C., Yang Y., Khor S., Chan C.S., Rabinowitz J.D., and White E.. 2016. Autophagy provides metabolic substrates to maintain energy charge and nucleotide pools in Ras-driven lung cancer cells. Genes Dev. 30:1704–1717. 10.1101/gad.283416.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris H., and Rubinsztein D.C.. 2011. Control of autophagy as a therapy for neurodegenerative disease. Nat. Rev. Neurol. 8:108–117. 10.1038/nrneurol.2011.200 [DOI] [PubMed] [Google Scholar]
- Huang H., Kawamata T., Horie T., Tsugawa H., Nakayama Y., Ohsumi Y., and Fukusaki E.. 2015. Bulk RNA degradation by nitrogen starvation-induced autophagy in yeast. EMBO J. 34:154–168. 10.15252/embj.201489083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W.P., Scott S.V., Kim J., and Klionsky D.J.. 2000. The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J. Biol. Chem. 275:5845–5851. 10.1074/jbc.275.8.5845 [DOI] [PubMed] [Google Scholar]
- Jiang M., Kauppila T.E.S., Motori E., Li X., Atanassov I., Folz-Donahue K., Bonekamp N.A., Albarran-Gutierrez S., Stewart J.B., and Larsson N.G.. 2017. Increased total mtDNA copy number cures male infertility despite unaltered mtDNA mutation load. Cell Metab. 26:429–436. [DOI] [PubMed] [Google Scholar]
- Kraft C., and Martens S.. 2012. Mechanisms and regulation of autophagosome formation. Curr. Opin. Cell Biol. 24:496–501. 10.1016/j.ceb.2012.05.001 [DOI] [PubMed] [Google Scholar]
- Kucej M., Kucejova B., Subramanian R., Chen X.J., and Butow R.A.. 2008. Mitochondrial nucleoids undergo remodeling in response to metabolic cues. J. Cell Sci. 121:1861–1868. 10.1242/jcs.028605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N., and Martin W.. 2010. The energetics of genome complexity. Nature. 467:929–934. 10.1038/nature09486 [DOI] [PubMed] [Google Scholar]
- Lewis S.C., Uchiyama L.F., and Nunnari J.. 2016. ER-mitochondria contacts couple mtDNA synthesis with mitochondrial division in human cells. Science. 353:aaf5549 10.1126/science.aaf5549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., and Schmittgen T.D.. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Longley M.J., Ropp P.A., Lim S.E., and Copeland W.C.. 1998. Characterization of the native and recombinant catalytic subunit of human DNA polymerase gamma: Identification of residues critical for exonuclease activity and dideoxynucleotide sensitivity. Biochemistry. 37:10529–10539. 10.1021/bi980772w [DOI] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A. III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., and Pringle J.R.. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- MacAlpine D.M., Perlman P.S., and Butow R.A.. 2000. The numbers of individual mitochondrial DNA molecules and mitochondrial DNA nucleoids in yeast are co-regulated by the general amino acid control pathway. EMBO J. 19:767–775. 10.1093/emboj/19.4.767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeusen S., and Nunnari J.. 2003. Evidence for a two membrane–spanning autonomous mitochondrial DNA replisome. J. Cell Biol. 163:503–510. 10.1083/jcb.200304040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakawa I. 2017. Organization and dynamics of yeast mitochondrial nucleoids. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 93:339–359. 10.2183/pjab.93.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretton A., Morel F., Macao B., Lachaume P., Ishak L., Lefebvre M., Garreau-Balandier I., Vernet P., Falkenberg M., and Farge G.. 2017. Selective mitochondrial DNA degradation following double-strand breaks. PLoS One. 12:e0176795 10.1371/journal.pone.0176795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murley A., Lackner L.L., Osman C., West M., Voeltz G.K., Walter P., and Nunnari J.. 2013. ER-associated mitochondrial division links the distribution of mitochondria and mitochondrial DNA in yeast. eLife. 2:e00422 10.7554/eLife.00422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J., and Suomalainen A.. 2012. Mitochondria: In sickness and in health. Cell. 148:1145–1159. 10.1016/j.cell.2012.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rödel G. 1997. Translational activator proteins required for cytochrome b synthesis in Saccharomyces cerevisiae. Curr. Genet. 31:375–379. 10.1007/s002940050219 [DOI] [PubMed] [Google Scholar]
- Somyajit K., Gupta R., Sedlackova H., Neelsen K.J., Ochs F., Rask M.B., Choudhary C., and Lukas J.. 2017. Redox-sensitive alteration of replisome architecture safeguards genome integrity. Science. 358:797–802. 10.1126/science.aao3172 [DOI] [PubMed] [Google Scholar]
- Strand M.K., Stuart G.R., Longley M.J., Graziewicz M.A., Dominick O.C., and Copeland W.C.. 2003. POS5 gene of Saccharomyces cerevisiae encodes a mitochondrial NADH kinase required for stability of mitochondrial DNA. Eukaryot. Cell. 2:809–820. 10.1128/EC.2.4.809-820.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomalainen A., and Isohanni P.. 2010. Mitochondrial DNA depletion syndromes—Many genes, common mechanisms. Neuromuscul. Disord. 20:429–437. 10.1016/j.nmd.2010.03.017 [DOI] [PubMed] [Google Scholar]
- Suzuki S.W., Onodera J., and Ohsumi Y.. 2011. Starvation induced cell death in autophagy-defective yeast mutants is caused by mitochondria dysfunction. PLoS One. 6:e17412 10.1371/journal.pone.0017412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczepanowska K., and Trifunovic A.. 2015. Different faces of mitochondrial DNA mutators. Biochim. Biophys. Acta. 1847:1362–1372. 10.1016/j.bbabio.2015.05.016 [DOI] [PubMed] [Google Scholar]
- Talarek N., Petit J., Gueydon E., and Schwob E.. 2015. EdU incorporation for FACS and microscopy analysis of DNA replication in budding yeast. Methods Mol. Biol. 1300:105–112. 10.1007/978-1-4939-2596-4_7 [DOI] [PubMed] [Google Scholar]
- Tanida I., Mizushima N., Kiyooka M., Ohsumi M., Ueno T., Ohsumi Y., and Kominami E.. 1999. Apg7p/Cvt2p: A novel protein-activating enzyme essential for autophagy. Mol. Biol. Cell. 10:1367–1379. 10.1091/mbc.10.5.1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S.D., Zhang H., Eaton J.S., Rodeheffer M.S., Lebedeva M.A., O’Rourke T.W., Siede W., and Shadel G.S.. 2005. The conserved Mec1/Rad53 nuclear checkpoint pathway regulates mitochondrial DNA copy number in Saccharomyces cerevisiae. Mol. Biol. Cell. 16:3010–3018. 10.1091/mbc.E05-01-0053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifunovic A., Wredenberg A., Falkenberg M., Spelbrink J.N., Rovio A.T., Bruder C.E., Bohlooly-Y M., Gidlöf S., Oldfors A., Wibom R., et al. 2004. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 429:417–423. 10.1038/nature02517 [DOI] [PubMed] [Google Scholar]
- Turk E.M., Das V., Seibert R.D., and Andrulis E.D.. 2013. The mitochondrial RNA landscape of Saccharomyces cerevisiae. PLoS One. 8:e78105 10.1371/journal.pone.0078105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson D.H., and Fennell D.J.. 1975. The use of fluorescent DNA-binding agent for detecting and separating yeast mitochondrial DNA. Methods Cell Biol. 12:335–351. 10.1016/S0091-679X(08)60963-2 [DOI] [PubMed] [Google Scholar]
- Young M.J., and Copeland W.C.. 2016. Human mitochondrial DNA replication machinery and disease. Curr. Opin. Genet. Dev. 38:52–62. 10.1016/j.gde.2016.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Muller E.G., and Rothstein R.. 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell. 2:329–340. 10.1016/S1097-2765(00)80277-4 [DOI] [PubMed] [Google Scholar]
- Zhou Q., Li H., Li H., Nakagawa A., Lin J.L., Lee E.S., Harry B.L., Skeen-Gaar R.R., Suehiro Y., William D., et al. 2016. Mitochondrial endonuclease G mediates breakdown of paternal mitochondria upon fertilization. Science. 353:394–399. 10.1126/science.aaf4777 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.