Gerganova and Martin preview work from Allard et al. that describes the Wee1- and Cdr1/2-dependent mechanism by which cells link cell size with mitotic entry.
Abstract
All cells show size homeostasis owing to coordination of division with growth. In this issue, Allard et al. (2018. J. Cell Biol. https://doi.org/10.1083/jcb.201709171) establish that transient inhibitory visits of a negative regulator of Cdk1 to cortical oligomeric platforms increase in number and duration with cell growth, suggesting how Cdk1 activation is coupled to cell size.
To maintain its size over generations, a cell must couple the timing of the cell cycle to its growth. How do cells control division at an optimal size, and what are the mechanisms governing the maintenance of cell size in relation to the cell cycle? Phenomenologically, three classes of size homeostasis behaviors have been described: (a) cells can act as sizers, where cells divide once they reach a given size; (b) as timers, where cell division occurs after a set time; or (c) as adders, where cells grow by a fixed amount at each generation. For instance, bacteria appear to behave as adders (Taheri-Araghi et al., 2015), whereas single-celled yeast eukaryotic cells behave as sizers. For the latter, this means that cells need constant and robust monitoring of their size through effectors that can crosstalk to the regulators of cell cycle progression. Work on these model systems has started revealing the molecular mechanisms by which cells can measure their own size and relay the information to the cell cycle. A study by Allard et al. in this issue of the Journal of Cell Biology now describes how the fission yeast Wee1, a major regulator of the cell cycle, reads the cell size information through burst visits of cortical structures.
The fission yeast Schizosaccharomyces pombe is a well-established system to study cell size regulation. This organism looks like a little rod 14 µm in length that divides in the middle and grows back to its original size before the next division. Previous work had suggested that these cells measure their size geometrically, with two models proposed. In both models, a central component is the protein Cdr2, a kinase that forms a beltlike structure of cortical clusters (also called nodes) around the middle of the cell and signals to the cell cycle machinery. The first model proposes that cell size information is conveyed through the kinase Pom1, a Cdr2 inhibitor that forms invariant concentration gradients from the cell tips (Martin and Berthelot-Grosjean, 2009; Moseley et al., 2009). The central idea is that Pom1 would inhibit Cdr2 more in short cells, allowing Cdr2 activation and thus mitotic entry only in longer cells. However, measurements of Pom1 mean levels at midcell did not detect substantially higher levels in short cells versus long cells, lowering confidence in this model. The second model states that the number of Cdr2 cortical clusters, which increase linearly with cell surface area and therefore double in number during one generation, directly conveys cell size information independently of Pom1 (Pan et al., 2014). By looking at the pathway output, this study by Allard et al. (2018) goes beyond these divergences and proposes a unified model.
The critical point for cell size control in the fission yeast is mitotic entry, which is triggered by the cyclin-dependent kinase Cdk1. During G2 phase, Cdk1 is inhibited by the kinase Wee1, whereas the phosphatase Cdc25 counteracts Wee1 inhibition by dephosphorylating Cdk1 to promote mitotic commitment. Therefore, cell size at division depends strongly on the balance between Wee1 and Cdc25 activity. The study by Allard et al. (2018) focuses on the regulation of Wee1. Previous research has provided genetic evidence for the idea that Cdr2 and the related kinase Cdr1 serve as inhibitors of Wee1 (Breeding et al., 1998). Biochemical work has also shown that Cdr1 phosphorylates Wee1 in vitro (Coleman et al., 1993). In their study, Allard et al. (2018) demonstrate that both Cdr1 and Cdr2 are required for Wee1 phosphorylation in vivo and investigate how these proteins are spatially organized to provide a system of cell size detection. Although Wee1 is known to associate with the spindle pole body and is found in the nucleus, its possible localization to cortical nodes had been debated (Moseley et al., 2009; Masuda et al., 2011). By using total internal reflection fluorescence microscopy, Allard et al. (2018) now unequivocally show that Wee1 transiently associates with the Cdr2 nodes, visiting them in brief bursts of varied duration from subsecond to a minute. In contrast, the Cdr2 nodes, formed by a core of ∼20 Cdr2 molecules and three Cdr1 molecules, are stable over long timescales (at least several minutes). Similar-sized Cdr2 oligomers can also be recovered upon subcellular fractionation on density gradients. Wee1 node visits depend on Wee1 binding Cdr2 and on Cdr2 kinase activity.
Allard et al. (2018) delve further into the biological relevance of Wee1’s localization at the nodes and present evidence that this localization is cell size dependent. By measuring the number of Wee1 punctae in snapshot images, Allard et al. (2018) find a very strong cell size dependence on the number of punctae, which increase by at least 20-fold through one cell cycle. In these images, the number of Wee1 punctae depends on the number of landing sites but also on the duration of each visit. Interestingly, both of these parameters increase concomitant with cell growth. First, Allard et al. (2018) confirm the previously shown twofold increase in the number of Cdr2 nodes (Pan et al., 2014), which likely increases the number of landing sites for Wee1 during cell growth. Second, burst duration also increases by two- to threefold during one cell cycle. This cell size dependence of burst duration is entirely reliant on Cdr2 activity: when Cdr2 is inactive, Wee1 bursts only last ∼4 s on average; when Cdr2 is hyperactive in the pom1Δ mutant (Deng et al., 2014), Wee1 bursts now last 18 s whatever the size of the cell. To reach a 20-fold increase in Wee1 punctae, it is likely that the frequency of Wee1 visits also increases through the cell cycle. Thus, the massive cell size–dependent increase in Wee1 punctae reflects combinatorial inputs from an increase in Cdr2 nodes and an increase in Cdr2 activity.
These data lend support for the unification of the two existing models for cell size regulation: cell size information is relayed through both an increase in the number of Cdr2 nodes and a progressive loss of its inhibition by Pom1, which together increase the probability of Wee1 kinase visiting a Cdr2 node (Fig. 1). The contribution of the two processes may vary through cell growth, with Pom1 playing a more important role in short cells. Indeed, in mutants lacking Pom1, an increase of Wee1 punctae was observed specifically in short cells (up to 9 µm), indicating that Pom1 prevents Wee1 cortical node visits in short cells, which leads to higher amounts of active Wee1 and a delay of mitotic entry. In contrast, Pom1 plays a more modest role in larger cells, where the system may be dominated by the number of Cdr2 nodes. An open question remains: how does Pom1 confer size-dependent regulation to Cdr2 to modulate Wee1 burst behaviors given its apparent size-independent levels at mid-cell? An intriguing next step in the investigation would be to revisit the relationship between Pom1 and Cdr2 nodes throughout cell cycle progression. Pom1 forms clusters, also apparent at midcell (Hachet et al., 2011; Saunders et al., 2012). Perhaps stochastic visits of these clusters at Cdr2 nodes, masked by previous mean measurements (Bhatia et al., 2014; Pan et al., 2014), are more frequent in short cells. Alternatively, Pom1 inhibition of Cdr2 may take place elsewhere, especially as the role of Pom1 is to inhibit activation of Cdr2 by the cytosolic kinase Ssp1 (Deng et al., 2014). Imaging techniques similar to those used by Allard et al. (2018) may reveal interesting insights about the possible discrete interactions between these regulators.
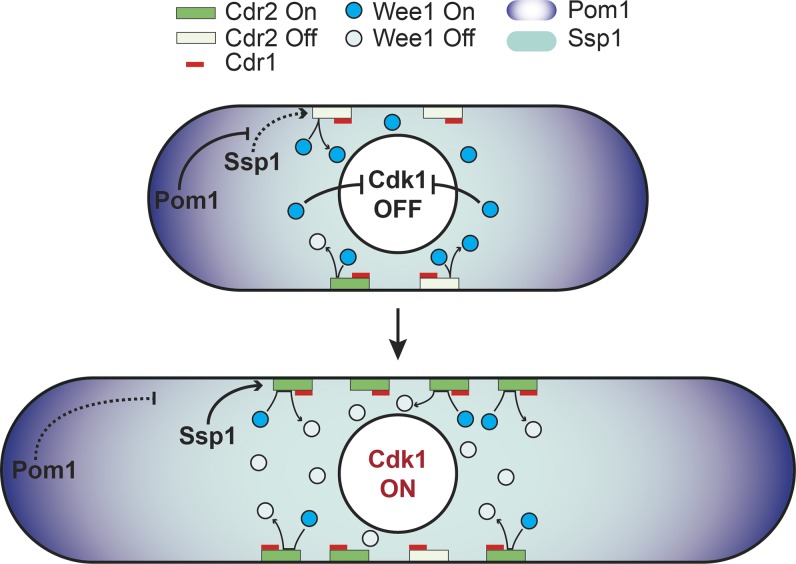
Figure 1.
Transient visits of Wee1 kinase to stable cortical Cdr2 nodes monitor cell size. In short cells, Wee1 visits to nodes are rare and transient because of the small number of Cdr2 nodes and their inactivation by Pom1. Wee1 thus remains active and inhibits Cdk1 in the nucleus. In long cells, the increased number of Cdr2 nodes and the relief of Cdr2 inhibition combine to increase the frequency of productive Wee1 node visits in which Wee1 is inhibited by Cdr1. Inactive Wee1 no longer inhibits Cdk1, which promotes mitotic entry.
The net output of the Wee1 visits to Cdr2 nodes is the inhibition of Wee1. The details of the reaction still need to be worked out, but the data presented show that Cdr2 binds Wee1 and recruits it to the nodes. When Wee1 lacks its Cdr2-binding region, it is not able to localize to nodes and appears hyperactive, delaying mitotic entry, suggesting that node visit is essential for inhibition. At the nodes, Cdr2 activity extends the duration of Wee1 visits, but it is neither required to form nodes nor to recruit Cdr1. Cdr1 phosphorylates Wee1, but it is required neither for the formation of Cdr2 nodes nor for their visit by Wee1. Thus, a likely scenario is that Cdr2 binds Wee1 directly to recruit it to nodes. If Cdr2 is inactive because of inhibition by Pom1, binding is short-lived and unproductive. If Cdr2 is active, phosphorylation of Wee1 promotes its retention at the node. In turn, the less abundant Cdr1 may now modify Wee1 with inhibitory phosphorylation. This makes the Wee1 node visit productive by decreasing Wee1 activity.
One intriguing question is how the number and duration of Wee1 visits are computed. An important side observation of the study is that even though the mean duration of Wee1 visits scales with cell size and is statistically longer in pom1Δ cells, individual measures of burst duration are highly variable, with SD similar to the mean. This raises the question of precision and robustness of the sizing system. Perhaps the stochastic nature of the Wee1 visits to nodes offers a way to overcome the noisiness of the system, for instance if most visits are nonproductive, with only the longest-lasting ones leading to inhibition. Alternatively, the cell may compute a time-averaged signal from Wee1 visits.
Finally, an open question is how the cell size information encoded into Wee1 phosphorylation status is transmitted to Cdk1 in the cell nucleus. It is interesting to note that the Saccharomyces cerevisiae homologue of Wee1, the protein kinase Swe1, performs an analogous function in relaying geometric information to the cell cycle. In that case, Swe1 does not monitor cell size but blocks cell cycle progression until formation of a bud in the so-called bud morphogenesis checkpoint (Keaton and Lew, 2006). Similar to the fission yeast situation, Swe1 shuttles out of the nucleus and transiently visits the bud neck, where it gets phosphorylated. This modification primes it for degradation. As Wee1 phosphorylation increases during the cell cycle and its levels drop at mitosis (Lucena et al., 2017), this raises the possibility that although the specific geometric sensed input is distinct, its output on Wee1/Swe1 and ultimately on Cdk1 may be conserved.
In summary, the data presented by Allard et al. (2018) suggest a revised model for cell size regulation in which cell growth promotes both an increase in the number of Cdr2 nodes and a loss of their inhibition by Pom1. In consequence, transient node visits by Wee1 are stabilized, leading to its inhibition. In turn, this shifts the balance in favor of Cdc25, which promotes Cdk1 activation and mitotic entry. This mechanism may therefore contribute to the size-dependent decision to divide. However, it is likely to only be a part of the answer as at least in some conditions, cells lacking the Wee1/Cdc25 target site on Cdk1 retain homeostatic behaviors (Wood and Nurse, 2013). One major challenge for the future is determining the extent to which this beautiful size sensing system contributes to size homeostasis.
Acknowledgments
This work was funded by a Swiss National Science Foundation Sinergia grant CRSII3 160728.
The authors declare no competing financial interests.
References
- Allard C.A.H., Opalko H.E., Liu K.-W., Medoh U., and Moseley J.B.. 2018. Cell size–dependent regulation of Wee1 localization by Cdr2 cortical nodes. J. Cell Biol. 10.1083/jcb.201709171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia P., Hachet O., Hersch M., Rincon S.A., Berthelot-Grosjean M., Dalessi S., Basterra L., Bergmann S., Paoletti A., and Martin S.G.. 2014. Distinct levels in Pom1 gradients limit Cdr2 activity and localization to time and position division. Cell Cycle. 13:538–552. 10.4161/cc.27411 [DOI] [PubMed] [Google Scholar]
- Breeding C.S., Hudson J., Balasubramanian M.K., Hemmingsen S.M., Young P.G., and Gould K.L.. 1998. The cdr2(+) gene encodes a regulator of G2/M progression and cytokinesis in Schizosaccharomyces pombe. Mol. Biol. Cell. 9:3399–3415. 10.1091/mbc.9.12.3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman T.R., Tang Z., and Dunphy W.G.. 1993. Negative regulation of the wee1 protein kinase by direct action of the nim1/cdr1 mitotic inducer. Cell. 72:919–929. 10.1016/0092-8674(93)90580-J [DOI] [PubMed] [Google Scholar]
- Deng L., Baldissard S., Kettenbach A.N., Gerber S.A., and Moseley J.B.. 2014. Dueling kinases regulate cell size at division through the SAD kinase Cdr2. Curr. Biol. 24:428–433. 10.1016/j.cub.2014.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachet O., Berthelot-Grosjean M., Kokkoris K., Vincenzetti V., Moosbrugger J., and Martin S.G.. 2011. A phosphorylation cycle shapes gradients of the DYRK family kinase Pom1 at the plasma membrane. Cell. 145:1116–1128. 10.1016/j.cell.2011.05.014 [DOI] [PubMed] [Google Scholar]
- Keaton M.A., and Lew D.J.. 2006. Eavesdropping on the cytoskeleton: progress and controversy in the yeast morphogenesis checkpoint. Curr. Opin. Microbiol. 9:540–546. 10.1016/j.mib.2006.10.004 [DOI] [PubMed] [Google Scholar]
- Lucena R., Alcaide-Gavilán M., Anastasia S.D., and Kellogg D.R.. 2017. Wee1 and Cdc25 are controlled by conserved PP2A-dependent mechanisms in fission yeast. Cell Cycle. 16:428–435. 10.1080/15384101.2017.1281476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S.G., and Berthelot-Grosjean M.. 2009. Polar gradients of the DYRK-family kinase Pom1 couple cell length with the cell cycle. Nature. 459:852–856. 10.1038/nature08054 [DOI] [PubMed] [Google Scholar]
- Masuda H., Fong C.S., Ohtsuki C., Haraguchi T., and Hiraoka Y.. 2011. Spatiotemporal regulations of Wee1 at the G2/M transition. Mol. Biol. Cell. 22:555–569. 10.1091/mbc.E10-07-0644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley J.B., Mayeux A., Paoletti A., and Nurse P.. 2009. A spatial gradient coordinates cell size and mitotic entry in fission yeast. Nature. 459:857–860. 10.1038/nature08074 [DOI] [PubMed] [Google Scholar]
- Pan K.Z., Saunders T.E., Flor-Parra I., Howard M., and Chang F.. 2014. Cortical regulation of cell size by a sizer cdr2p. eLife. 3:e02040 10.7554/eLife.02040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders T.E., Pan K.Z., Angel A., Guan Y., Shah J.V., Howard M., and Chang F.. 2012. Noise reduction in the intracellular pom1p gradient by a dynamic clustering mechanism. Dev. Cell. 22:558–572. 10.1016/j.devcel.2012.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taheri-Araghi S., Bradde S., Sauls J.T., Hill N.S., Levin P.A., Paulsson J., Vergassola M., and Jun S.. 2015. Cell-size control and homeostasis in bacteria. Curr. Biol. 25:385–391. 10.1016/j.cub.2014.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood E., and Nurse P.. 2013. Pom1 and cell size homeostasis in fission yeast. Cell Cycle. 12:3417–3425. 10.4161/cc.26462 [DOI] [PMC free article] [PubMed] [Google Scholar]