Abstract
Objective
The intestinal microbiota plays a central role in the development of many chronic inflammatory diseases including inflammatory bowel disease and metabolic syndrome. Administration of substances that alter microbiota composition, including the synthetic dietary emulsifiers polysorbate 80 (P80) and carboxymethylcellulose (CMC), can promote such inflammatory disorders. However, that inflammation itself impacts microbiota composition has obfuscated defining the extent to which these compounds or other substances act directly upon the microbiota versus acting on host parameters that promote inflammation, which subsequently reshapes the microbiota.
Design
We examined the direct impact of CMC and P80 on the microbiota using the mucosal simulator of the human intestinal microbial ecosystem (M-SHIME) model that maintains a complex stable human microbiota in the absence of a live host.
Results
This approach revealed that both P80 and CMC acted directly upon human microbiota to increase its pro-inflammatory potential, as revealed by increased levels of bioactive flagellin. The CMC-induced increase in flagellin was rapid (1 day) and driven by altered microbiota gene expression. In contrast, the P80-induced flagellin increase occurred more slowly and was closely associated with altered species composition. Transfer of both emulsifier-treated M-SHIME microbiotas to germ-free recipient mice recapitulated many of the host and microbial alterations observed in mice directly treated with emulsifiers.
Conclusion
These results demonstrate a novel paradigm of deconstructing host-microbiota interactions and indicate that the microbiota can be directly impacted by these commonly used food additives, in a manner that subsequently drives intestinal inflammation.
Keywords: Metabolic syndrome, emulsifier, transcriptome, microbiome
INTRODUCTION
A panel of chronic inflammatory diseases are associated with alterations in the species and/or genomic composition of the intestinal microbiota. In animal models, such diseases are typically ameliorated in the absence of a microbiota, achieved via use of antibiotics or germ-free methodologies, indicating that the presence of a microbiota is possibly linked to disease development. Moreover, the observation that transfer of the microbiota from diseased hosts - either mice or humans - to germfree mice can transfer some features of disease suggests that disease-associated alterations in microbiota composition are not purely a consequence of diseases but, rather, play a role in driving disease development. On the other hand, inflammation itself can lead to potent alterations in microbiota composition and does so in a manner that displays similar features that are observed in some inflammatory diseases, for example loss of α-diversity and/or increase in γ-proteobacteria [1, 2, 3, 4, 5, 6, 7]. Together, these results suggest that alterations in gut microbiota that have been associated with inflammatory diseases are likely both a cause and consequence of inflammation. In this respect, it may be very difficult to determine the extent to which a given substance - whose administration results in altered microbiota composition and gut inflammation - acted directly upon the microbiota and/or the host. An example of this concept can be seen in our recent work on the food additives carboxymethylcellulose (CMC) and polysorbate 80 (P80) [8]. CMC and P80 are commonly used synthetic emulsifiers that are added to a variety of processed foods to enhance texture and extend shelf-life. Based on the epidemiology of their usage, and the notion that their detergent properties might promote bacterial translocation across mucosal surfaces, these emulsifiers are hypothesized to have contributed to increased incidence of chronic gut inflammatory diseases [8, 9, 10, 11]. In support of this hypothesis, we observed that administration of CMC and P80 to mice resulted in microbiota encroachment into the mucus, alterations in microbiota composition, including an increase of bacteria that produced pro-inflammatory flagellin and LPS, and development of chronic inflammation [8, 12]. Such inflammation was associated with low-grade inflammation and metabolic disease in WT mice and increased incidence/severity of overt colitis in interleukin-10 (IL-10)−/− mice, which are prone to this disorder. We hypothesized that such events might be initiated by these emulsifiers directly interacting with the mucus, thereby promoting bacterial encroachment that would activate the innate immune system to induce anti-microbial gene expression, thereby more broadly changing microbiota composition. In accordance with this possibility, both CMC and P80 administration resulted in a thinner mucus layer. However, such emulsifier-induced mucus thinning was not observed in germ-free mice, nor did we observe an altered localization of fluorescent beads - of the same size of bacteria - in germ-free animals in presence of CMC or P80 [8]. Such findings argued against the notion that these emulsifiers were directly impacting the mucus layer and, rather, suggests the possibility that CMC and P80 might directly impact the intestinal microbiota.
Hence, the central goal of this study was to investigate the extent to which CMC and P80 might directly alter microbiota using the mucosal simulator of the human intestinal microbial ecosystem (M-SHIME) model, which allows a complex human microbiota to be stably maintained in vitro [13, 14, 15, 16] in the absence of host cells. We observed that P80 and CMC altered the microbiota in the M-SHIME model in a manner reminiscent of, and capable of recapitulating, their effects previously observed in mice, thus suggesting that the microbiota may be a key direct target of CMC and P80.
METHODS
Mice
C57BL/6 mice were maintained in gnotobiotic conditions in a germ-free (GF) or Altered Schaedler flora (ASF) state in a Park Bioservices isolator as previously described [8]. C57 BL/6 Rag−/− mice were maintained in specific pathogen free housing. All mice were bred and housed at Georgia State University (Atlanta, GA, USA) under institutionally-approved protocols (IACUC # A14033). Mice were fed autoclaved Purina rodent chow # 5021 or irradiated (20–40 kGy) purified high-fat diet D12492-1.5V from Research Diet (New Brunswick, NJ, USA) (Table 1). Experiments used both male or female mice (comparisons within a gender) except for figure 7–8 that utilized female mice. ASF mice were established by colonizing WT C57BL/6 GF mice with the complete Altered Schaedler flora (8 strains) using feces purchased from ASF Taconic, Inc. (Hudson, NY, USA) and resuspended in drinking water.
Table 1.
Purified diet used in the study.
| Product # | D12492-1.5V | |
|---|---|---|
| Protein source | Casein | |
| Fiber source | Cellulose | |
| kcal% fat | 60 | |
| Protein (kcal%) | 20 | |
| Carbohydrate (kcal%) | 20 | |
| Fat (kcal%) | 60 | |
| Total | 100 | |
| kcal/gm | 5.2 | |
| Ingredient (g) | kcal/gm | |
| Casein | 4 | 200 |
| L-Cystine | 4 | 3 |
| Corn Starch | 4 | 0 |
| Maltodextrin 10 | 4 | 125 |
| Sucrose | 4 | 63.8 |
| Cellulose | 0 | 50 |
| Inulin | 1 | 0 |
| Soybean Oil | 9 | 25 |
| Lard | 9 | 245 |
| Mineral Mix, S10026 | 0 | 10 |
| DiCalcium Phosphate | 0 | 13 |
| Calcium Carbonate | 0 | 5.5 |
| Potassium Citrate, 1 H2O | 0 | 16.5 |
| Vitamin Mix, V10001 | 4 | 15 |
| Choline Bitartrate | 0 | 2 |
| Total gm | 773.9 | |
| Total kcals | 4057 | |
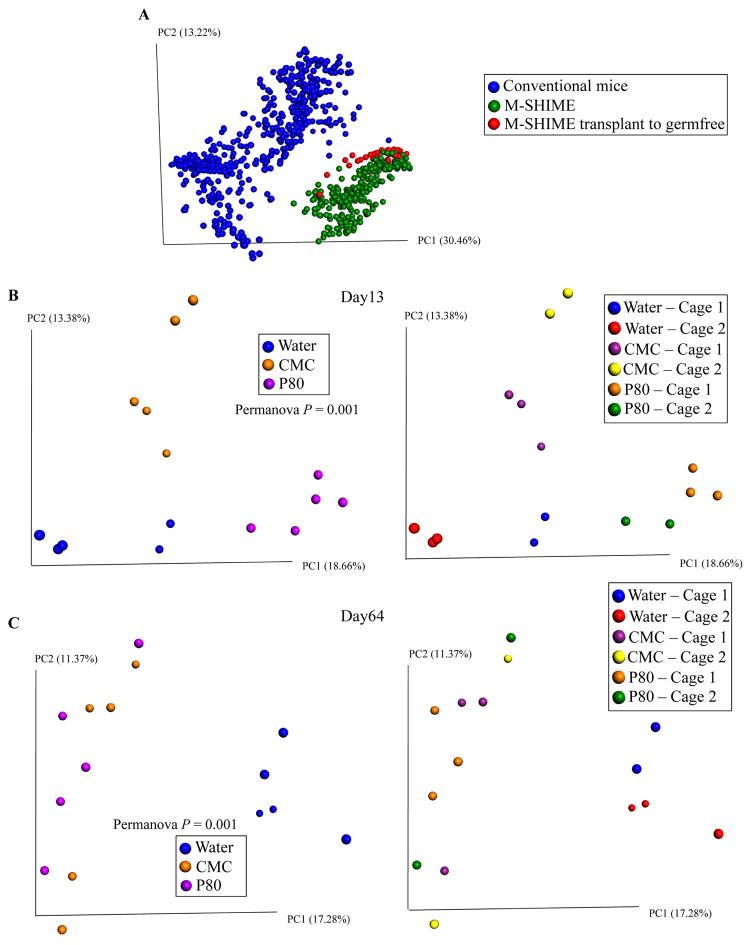
Figure 7. Altered microbiota composition in mice receiving CMC- and P80-treated M-SHIME suspensions.
A 9-vessel M-SHIME system was set up and inoculated with fresh human feces at day −7. Following a 7-day stabilization period, vessels were treated with either water, CMC 1% or P80 1% for 13 days. Germfree C57BL/6 mice (4 weeks old) were removed from isolator and were orally administered with 200μL of luminal M-SHIME suspension (day 11 sample). Transplanted mice were then housed in isolated ventilated cages. For each condition (water-treated M-SHIME, CMC-treated M-SHIME, P80-treated M-SHIME), 2 cages containing 2–3 animals were used, with each cage receiving suspension from an independent M-SHIME vessel (for each condition, N=2 vessel, N=2 cages, and N=5 animals). Fecal microbiota composition was analyzed using Illumina sequencing of the V4 region of 16S rRNA genes. (A) Microbiota composition was analyzed from GSU conventional mice, luminal M-SHIME suspension and germfree mice colonized with luminal M-SHIME suspension using Illumina sequencing of the V4 region of 16S rRNA genes. (B–C) Principal coordinates analysis (PCoA) of the unweighted UniFrac distance matrix at days 13 (B) and 64 (C). Samples were colored by treatment (left side) or by cage (right side) (N=5). For clustering analyzing on principal coordinate plots, categories were compared and statistical significance of clustering were determined using the Permanova method.
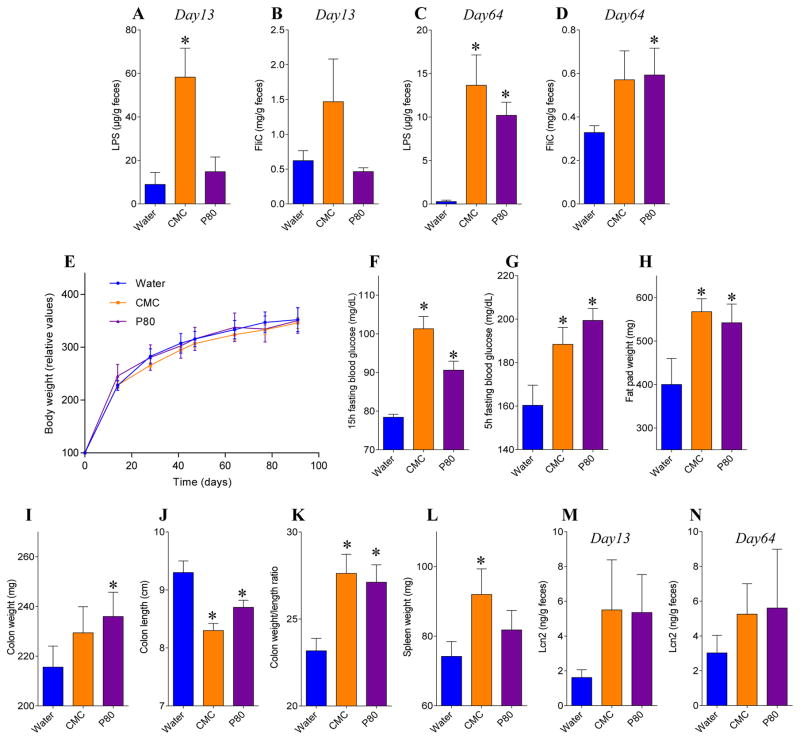
Figure 8. CMC- and P80-treated M-SHIME suspensions promote low-grade inflammation when transplanted to germfree recipient mice.
A 9-vessel mucosal-M-SHIME system was set up and inoculated with fresh human feces at day −7. Following a 7-day stabilization period, vessels were treated with either water, CMC 1% or P80 1% for 13 days. Germfree C57BL/6 mice (4 weeks old) were removed from isolator and were orally administered with 200μL of luminal M-SHIME suspension (day 11 sample). Transplanted mice were then housed in isolated ventilated cages. For each condition (water-, CMC-, and P80-treated M-SHIME), 2 cages containing 2–3 animals were used, with each cage receiving suspension from an independent M-SHIME vessel (for each condition, N=2 vessel, N=2 cages, and N=5 animals). (A, C) LPS and (B, D) FliC levels at days 13 (A, B) and 64 (C, D). (E) Body weight over time, (F) 15hr fasting blood glucose concentration, (G) 5hr fasting blood glucose concentration, (H) fat pad weight, (I) colon weight, (J) colon length, (K) colon weight/length ratio, (L) spleen weight and (M–N) fecal Lcn2 levels at days 13 (M) and 64 (N). Data are the means +/− S.E.M (N=5). Each treatment group was compared with the control group (water-treated) and statistical significance was determined using Student’s t test.
Fasting blood glucose measurement
Mice were placed in a clean cage and fasted for 5h or 15h. Blood glucose concentration was then determined using a Nova Max Plus Glucose Meter and expressed in mg/dL.
Emulsifier treatment of the mucosal simulator of the human intestinal microbial ecosystem (M-SHIME) system
The direct effect of emulsifier on intestinal microbiota was studied in an M-SHIME system, a dynamic in vitro model which simulates the lumen- and mucus-associated human intestinal microbial ecosystem (ProDigest-Ghent University, Ghent, Belgium) [13, 14, 15]. This model consists of consecutive pH controlled, stirred (200 rpm), airtight, double-jacketed glass vessels kept at 37°C and under anaerobic conditions by daily flushing with N2 (15 min). The set-up used in this study consisted of a stomach and a small intestine vessel and three proximal colon vessels in parallel. The system was operated and simulation media were prepared as described earlier [13, 14, 15]. The colon vessels were inoculated at the start with 40 mL human fecal suspension in 500 mL sterile nutritional medium (per L: 3 g yeast extract, 1 g special peptone (Oxoid, Aalst, Belgium), 4 g commercial porcin gastric mucin (Sigma-Aldrich, Bornem, Belgium) and 0.5 g L-cystein). After an initial static incubation of 18 h, 140 mL sterile nutritional medium at pH 2 per colon vessel was supplemented to the stomach vessel: this procedure was conducted three times a day. Stomach digest suspension was pumped into the small intestine vessel together with 60 mL bile and pancreatic juice [per L: 12.5 g NaHCO3, 6 g dehydrated bile extract (Oxgall, Difco) and 0.9 g pancreatin (Sigma)] per colon vessel. Small intestinal digest suspension was distributed over the different proximal colon vessels, which contained simulated colon microbiota. To simulate colon conditions, the residence time in the colon vessels was 20 h and pH was controlled at pH 6.15–6.40 which are both in the range of in vivo observations [17, 18]. Mucus microbiota were simulated by adding carriers coated with agar containing commercially available porcine gastric mucin (Sigma-Aldrich) [13]. Sixty mucin agar-covered microcosms in a polyethylene netting were added to each colon vessel to obtain an appropriate mucus surface area. Every two days, two-third of the mucin agar-covered microcosms were replaced by fresh sterile ones under a flow of N2 to prevent disruption of anaerobic conditions. Seven days after inoculation, colon vessels were treated with either CMC or P80 at a final concentration of 1.00%, 0.50%, 0.25% or 0.10%. Lumen and mucin agar samples were taken every two days. Mucin agar-covered microcosms were washed with sterile PBS to remove lumen bacteria. Mucin agar was removed from microcosms, homogenized and stored immediately at −20°C until further analysis.
M-SHIME meta-transcriptomic analysis
Total RNAs were extracted from M-SHIME suspension using Qiagen RNeasy kit, according to the manufacturer’s protocol. After purification, RNA concentration and integrity were determined using Epoch Microplate Spectrophotometer (Bio-Tek, Winooski, VT, USA) and agarose gel electrophoresis, respectively. Total RNA was then prepared for sequencing using Illumina TruSeq RNA kit and according to the manufacturer’s protocol. Briefly, rRNA were depleted using Illumina Ribo-Zero magnetic kit according to the manufacturer’s protocol. rRNA-depleted RNA were fragmented, and converted to cDNA. After end repair and ligation of adapters, mRNA libraries were amplified by PCR and validated using BioAnalyser, according to manufacturer’s recommendations. The purified library was then subjected to sequencing via HiSeq at the Cornell University sequencing core (Ithaca, NY). Sequencing data obtained were analyzed through MG-RAST pipeline [19] and metatranscriptome were determined and summarized using KEGG orthology (KO) [20, 21], Clusters of Orthologous Groups of proteins (COG) [22] and Non-supervised Orthologous Groups (NOG) [23] for each samples and compare using either: principal coordinate analysis of the euclidean distance, volcano plot created using R software, heatmap representation using Gene-E of pathways of interest [24]. Unprocessed sequencing data are deposited at MG-RAST http://metagenomics.anl.gov/linkin.cgi?project=mgp16059, samples mgs411584, mgs411587, mgs411590, mgs411593, mgs411596, mgs411599, mgs411602, mgs411605, mgs411608, mgs411611, mgs411614, mgs411617, mgs411620, mgs411623, mgs411626, mgs411629, mgs411632, mgs411635, mgs411638, mgs411641, mgs411644, mgs411647, mgs411650, mgs411653.
Microbiota transplantation of emulsifier-treated M-SHIME suspension
Germfree C57BL/6 mice (4 weeks old) were removed from isolator and were orally administered with 200μL of luminal M-SHIME suspension (day 11 sample). Transplanted mice were then housed in isolated ventilated cages Isocages (Techniplast, West Chester, PA, USA) [25] and fed autoclaved Purina rodent chow # 5021 or irradiated (20–40 kGy) purified high-fat diet D12492-1.5V from Research Diet (New Brunswick, NJ, USA). For each condition (water-treated M-SHIME, CMC-treated M-SHIME, P80-treated M-SHIME), 2 cages containing 1–3 animals were used, with each cage receiving suspension from an independent M-SHIME vessel (for each condition, N=2 vessels, each one transferred to a cage of 1–3 animals, for a total of 3–5 animals). Body weights were measured every week and are expressed as % compare to the initial body weight (day 0) defined as 100%. Fresh feces were collected every other week for downstream analysis. Eleven weeks post-transplant, mice were fasted for 5-h at which time blood was collected by retrobulbar intraorbital capillary plexus. Hemolysis-free serum was generated by centrifugation of blood using serum separator tubes (Becton Dickinson, Franklin Lakes, NJ). Mice were then euthanized, and colon length, colon weight, spleen weight and adipose weight were measure. Organs were collected for downstream analysis.
Statistical analysis
Normal distribution of the data was assessed using D’Agostino-Pearson omnibus test. Significance was determined using t-test or two-way group ANOVA with Bonferroni’s multiple comparisons test (GraphPad Prism software, version 6.01). Differences were noted as significant *p≤0.05 for t-test and # p≤0.05 for two-way ANOVA. For clustering analysis on principal coordinate plots, categories were compared and statistical significance of clustering was determined via Permanova.
RESULTS
CMC and P80 do not impact intestinal microbiota or host in ASF mice
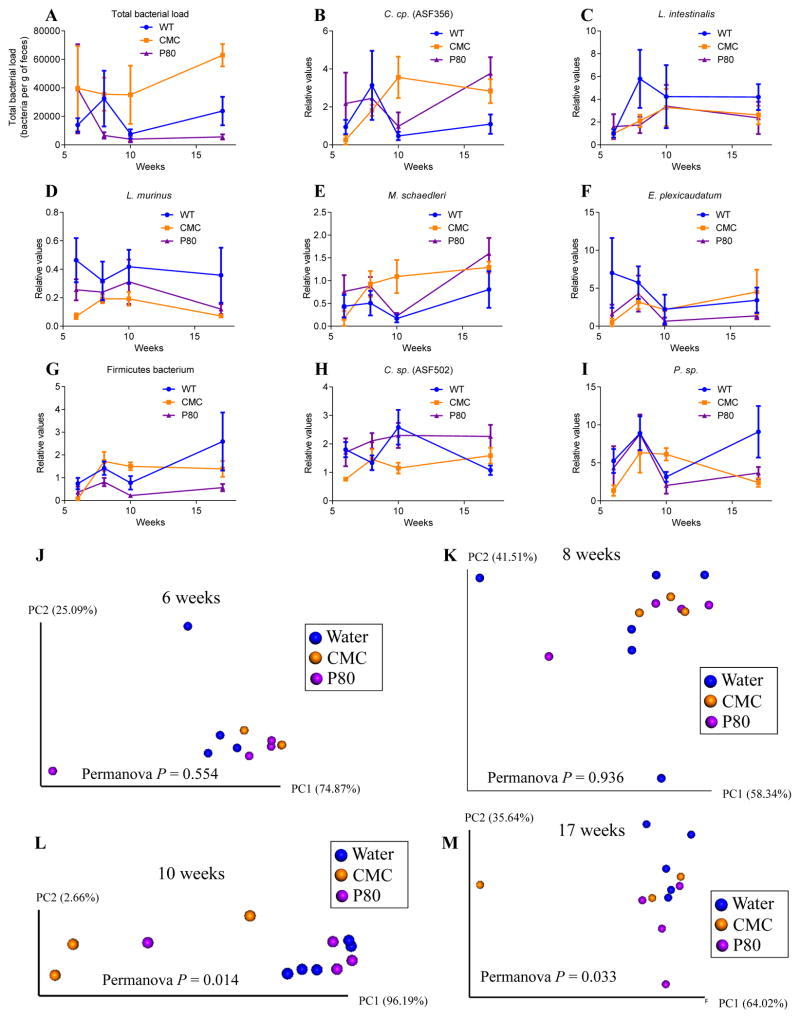
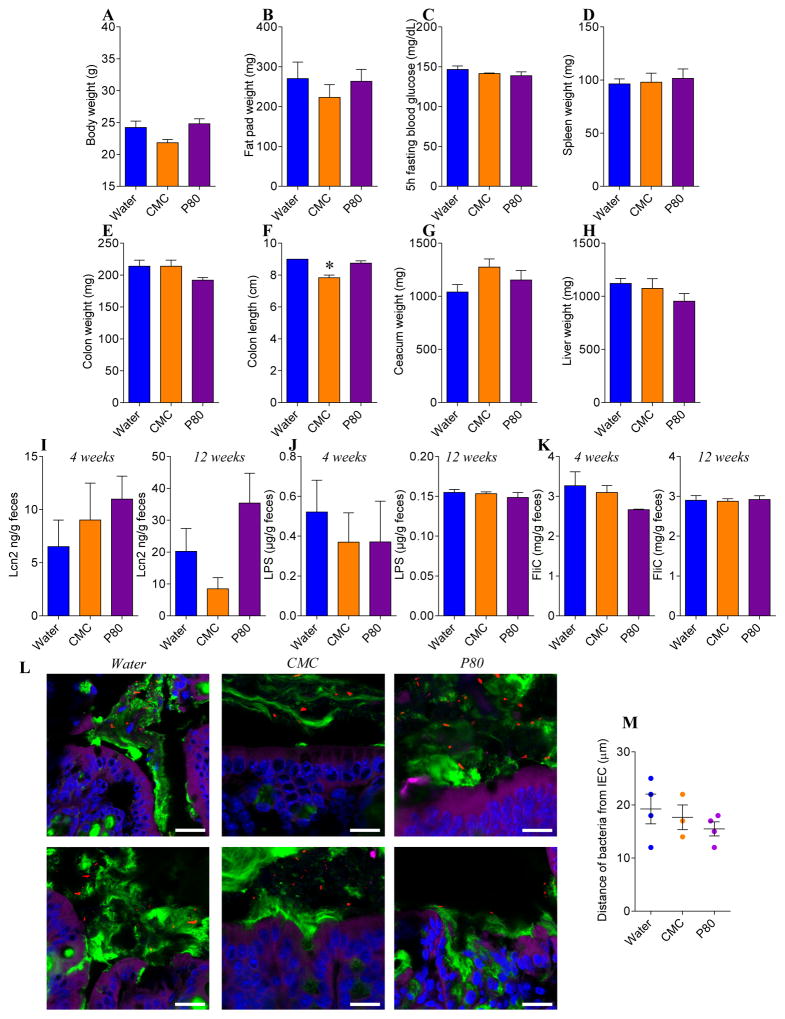
To investigate the extent to which CMC and P80 might impact the host and/or microbiota in mice with a limited defined pathobiont-free microbiota, we administered CMC and P80 to mice colonized exclusively with Altered Schaedler Flora (ASF). As shown in Figure 1 and 2, neither CMC nor P80 had a significant impact on a range of host or microbial parameters in ASF mice. Briefly, whether assessed globally by principal coordinate analysis or comparison of individual species, assay of relative levels of the 8 ASF components by q-PCR indicated that neither CMC nor P80 impacted microbiota composition of ASF mice, nor did they alter fecal levels of TLR-agonists LPS and flagellin (Figure 1 and 2). Moreover, in ASF mice, CMC and P80 administration did not result in microbiota encroachment (Figure 2L–M), mucus thinning, or features of low-grade inflammation/metabolic syndrome that are induced by these compounds in mice with a complex microbiota[8]. Together, these results argue against the notion that these emulsifiers have a strong direct effect on the host and rather suggest the possibility that more subtle effects on a complex microbiota may be proximal mediators of the effects of these compounds.
Figure 1. CMC and P80 do not impact intestinal microbiota composition in ASF mice.
Six-week-old ASF C57BL/6 mice were exposed to CMC or P80 diluted in drinking water (1.0%) for 11 weeks. (A–I) Total fecal DNA was extracted and subjected to q-PCR using specific primers for 16sRNA (A), C. sp. (ASF 356) (B), L. intestinalis (C), L. murinus (D), M. schaedleri (E), E. plexicaudatum (F), Firmicutes bacterium (G), C. sp. (ASF 502) (H) and P. sp. (I). Results are expressed as bacteria number per mg of stool (A), using a standard curve, or as relative values (B–I). (J–M) Principal coordinate analysis was performed using euclidean distance with the 8 ASF members at week 6 (J), week 8 (K), week 10 (L) and week 17 (M). Data are the means +/− S.E.M (N=3–6). For clustering analyzing on principal coordinate plots, categories were compared and statistical significance of clustering were determined using Permanova method.
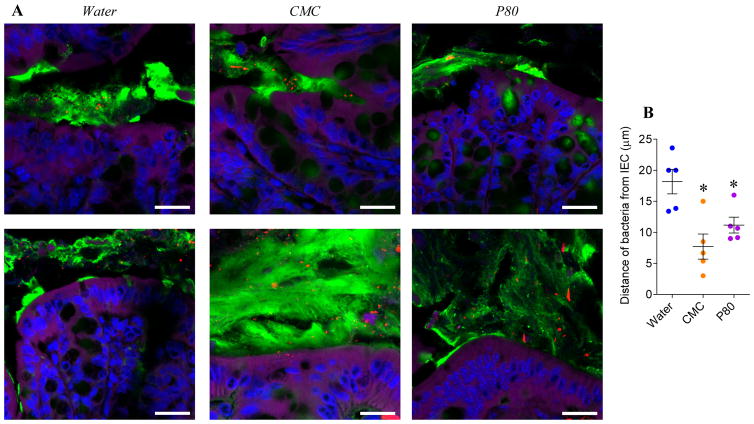
Figure 2. CMC and P80 do not induce intestinal inflammation nor metabolic syndrome nor microbiota encroachment in ASF mice.
Six-week-old ASF C57BL/6 mice were exposed to CMC or P80 diluted in drinking water (1.0%) for 11 weeks. (A) Final body weight, (B) fat pad weight, (C) 5hr fasting blood glucose concentration, (D) spleen weight, (E) colon weight, (F) colon length, (G) ceacum weight, (H) liver weight, (I) fecal Lcn2, (J) LPS and (K) FliC levels at 4 weeks and 12 weeks. (L) Confocal microscopy analysis of microbiota localization; Muc2 (green), actin (purple), bacteria (red), and DNA (Blue). Bar=20μm. (M) Distances of the closest bacteria to intestinal epithelial cells (IEC) per condition over 3 high-powered fields per mouse. Data are the means +/− S.E.M (N=3–6). Each treatment group was compared with the control group (water-treated) and statistical significance was determined using Student’s t test.
P80 alters microbiota composition in the M-SHIME model
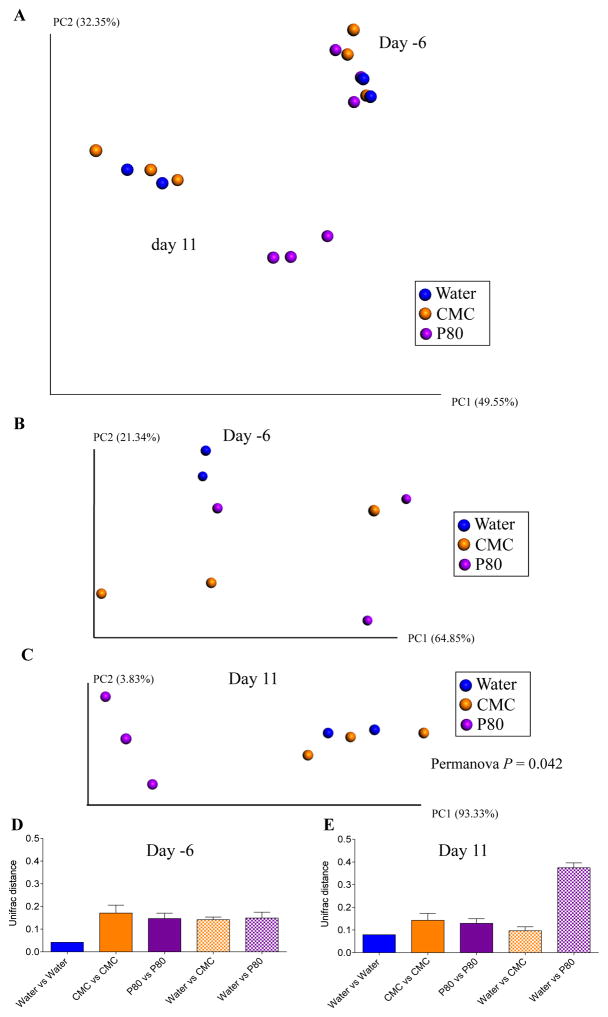
Having not observed an impact of CMC and P80 in a host lacking a complex microbiota, we next utilized the converse approach of examining how these compounds impacted a complex microbiota in the absence of a host by utilizing the M-SHIME model. Application of 16S sequencing to luminal M-SHIME samples indicated that the bacterial composition of this model changed rapidly following initial inoculation with human feces but had largely stabilized 7 days post-inoculation (Figure S1A). Such stabilization was associated with an approximate 50% decline in α-diversity, but nonetheless still afforded a stable complex microbiota for further study (Figure S1B). Hence, we defined this point as time zero and subsequently added CMC, P80, or water (control) to multiple M-SHIME vessels maintained in parallel. As expected, when assayed prior to addition of emulsifiers, M-SHIMEs did not differ significantly from each other (Figure 3A–B). However, in response to P80, microbiota composition of M-SHIMEs gradually shifted such that by day 11, all P80-treated M-SHIMES clearly clustered together quite distinctly from the other M-SHIME vessels (Figure 3C). In contrast, the bacterial compositions of CMC-treated and control M-SHIMEs did not differ significantly from each other at any time point assayed (Figure 3A–C). In accordance with our visual assessment of the PCoA plots, the UniFrac distances between control and P80-treated M-SHIMEs were significantly greater than those of any following (11 days) but not preceding (−6 days) treatment (Figure 3D–E). Such altered composition resulting from P80 treatment was driven by increased α-diversity (Figure S2). All of the above analyses were also applied to M-SHIME mucus samples that yielded a very similar pattern of results (Figure S3). Thus, P80, but not CMC exhibited a clear direct effect on the composition of a complex microbiota in the absence of a live host.
Figure 3. P80 alters M-SHIME microbiota composition.
A 9-vessels mucosal-M-SHIME system was set up and inoculated with fresh human feces at day −7. Following a 7-day stabilization period, vessels were treated with either water (N=2), CMC 1% (w/v, N=3) or P80 1% (v/v, N=3) for 13 days. Luminal microbiota composition was analyzed using Illumina sequencing of the V4 region of 16S rRNA genes. (A–C) Principal coordinates analysis (PCoA) of the weighted UniFrac distance matrix at days −6 and 11 (A), day −6 (B) and day 11 (C). (D–E) Average of the weighted UniFrac distance within group (water, CMC and P80) and between group (Water vs CMC and Water vs P80) has been calculated at days −6 (D) and 11 (E). Data are the means +/− S.E.M (N=2–3). For clustering analyzing on principal coordinate plots, categories were compared and statistical significance of clustering were determined using the Permanova method.
Both CMC and P80 alter microbial gene expression in the M-SHIME model
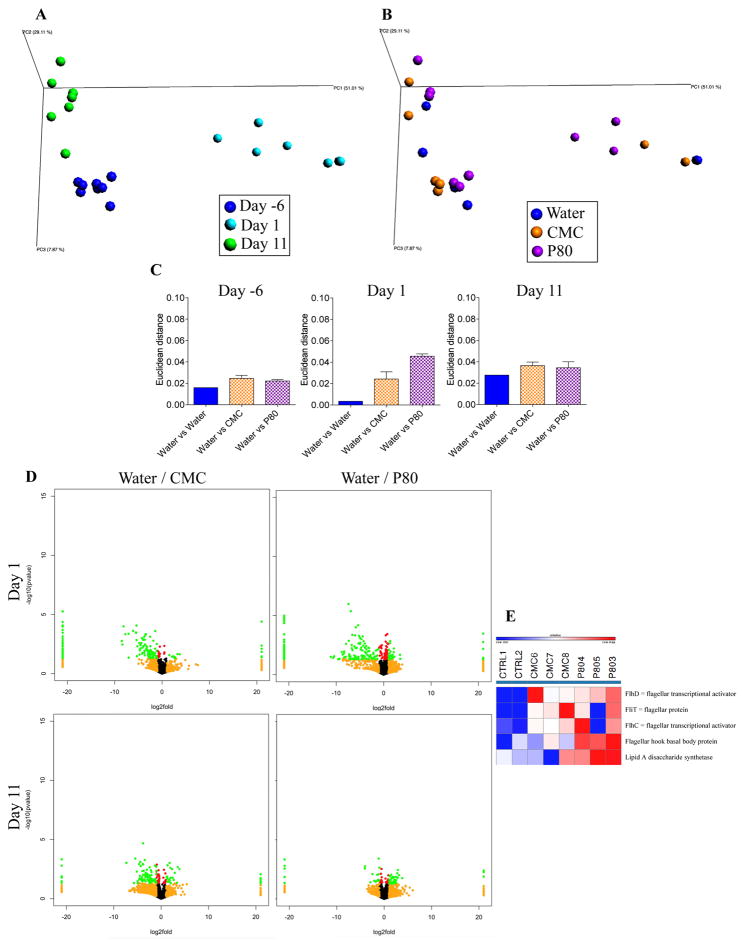
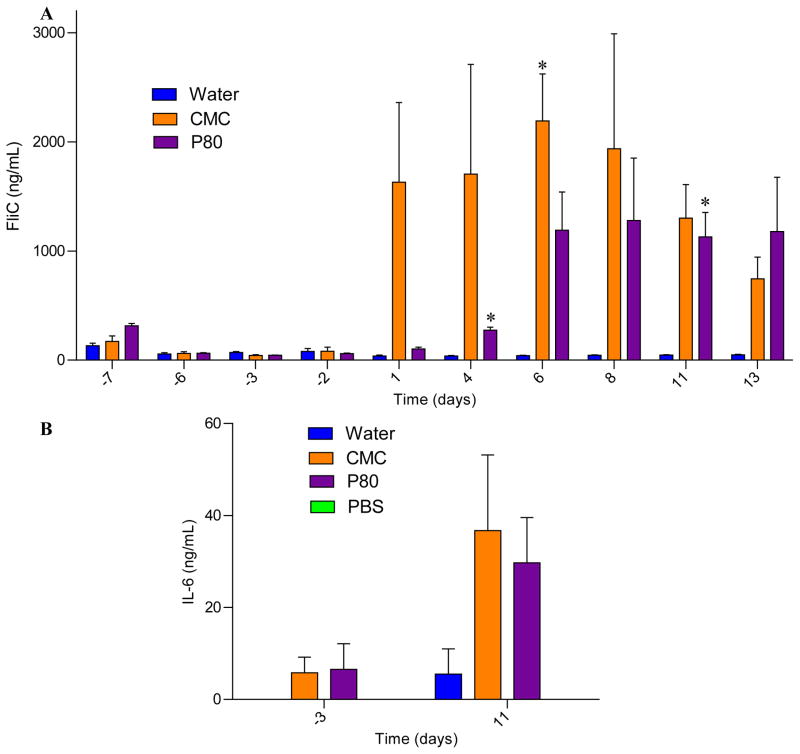
We next examined the extent to which CMC and P80 might influence microbial gene expression in the M-SHIME model. M-SHIME samples were assayed by RNA sequencing at days −6, 6, and 11. PCoA analysis indicated that time in the M-SHIME model had a substantial effect on microbial gene expression irrespective of emulsifier treatment in that all samples uniformly clustered based on the day of isolation (Figure 4A). Nonetheless, M-SHIMEs treated with CMC or P80 following (day 1), but not preceding (day −6) emulsifier treatment exhibited clustering based on treatment as indicated visually and based on multi-dimensional (Euclidian) distance in PCoA coordinates (Figure 4B–C). Display of relative gene expression data in volcano plots (Figure 4D), scatter plots of relative expression between different time points and different conditions (Figure S4) and summarizing sequencing reads at different levels of the KEGG orthology classification (Figure S5) indicated that neither CMC nor P80 broadly influenced gene expression but rather induced changes in a relative limited subset of genes. In accordance with this observation, assay of M-SHIME short-chain fatty acid and branched-chain fatty acid levels, which serve as a general readout for gut microbiota metabolism and fermentation, did not differ appreciably in response to emulsifier treatment (Figure S6 and S7). Importantly, one subset of genes whose transcripts were enriched in both CMC and P80 M-SHIMEs were genes related to flagella expression (Figure 4E). This result was reminiscent of results seen in emulsifier-treated mice, which displayed high level of fecal flagellin, which was hypothesized to promote low-grade inflammation via activation of TLR5 and/or NLRC4 [8]. Hence, we next assayed levels of bioactive flagellin in the M-SHIME model via the use of TLR5 reporter cells. CMC induced a robust increase in bioactive flagellin within 1 day of its administration, that was presumably independent of alterations in microbial composition, which, as shown above, did not appreciably change in this model (Figure 5A). In contrast, P80 induced a slower increase in M-SHIME flagellin levels that was evident 7 days following its administration, which is consistent with it resulting, at least in part, from an alteration in microbial composition (Figure 5A), while no clear pattern was observed for another microbiota-derived pro-inflammatory molecule, lipopolysaccharide (Figure S8). To more broadly examine how pro-inflammatory potential might be altered by CMC and P80 treatment, M-SHIME samples were injected into RAG−/− mice, which can make innate but not adaptive responses to bacteria and their products. As presented figure 5B, suspension from CMC- and P80-treated M-SHIME vessels induced a significantly higher IL-6 expression compared to water-treated M-SHIME vessels, further indicating that these emulsifying agents increased microbiota pro-inflammatory potential.
Figure 4. Both CMC and P80 alter microbial gene expression in the mucosal-M-SHIME model.
A 9-vessel mucosal-M-SHIME system was set up and inoculated with fresh human feces at day −7. Following a 7-day stabilization period, vessels were treated with either water (N=2), CMC 1% (w/v, N=3) or P80 1% (v/v, N=3) for 13 days. M-SHIME-associated luminal metatranscriptome was analyzed by Illumina sequencing. (A–B) Following functional classification of the reads to Kyoto Encyclopedia of genes and genomes (KEGG) orthology classification at level 4 using MG-RAST, principal coordinates analysis (PCoA) of the euclidean distance matrix were ploted at days −6, 1 and 11. (C) Average of the euclidean distance within group (water) and between group (Water vs CMC and Water vs P80) at days −6, 1 and 11. (D) KEGG orthology at level 4 were visualized on a volcano plot. Up/left: water-treated versus CMC-treated at day1; up/right: water-treated versus P80-treated at day1; bottom/left: water-treated versus CMC-treated at day11; bottom/left: water-treated versus P80-treated at day11. For each KEGG identifier level 4, the difference in abundance between the two groups is indicated in log2 fold change on x-axis (with positive values corresponding to an increase in emulsifier-treated group compare to water-treated group, and negative values corresponding to a decrease in emulsifier-treated group compare to water-treated group), and significance between the two groups is indicated by −log10 p-value on the y-axis. Red dots correspond to KEGG identifiers with a p-value <0.05 between emulsifier-treated and water-treated groups. Orange dots correspond to KEGG identifiers with at least a 2-fold decreased or increased abundance in emulsifier-treated group compare to water-treated group. Green dots correspond to KEGG identifiers with at least a 2-fold decreased or increased abundance in emulsifier-treated group compare to water-treated group and with a p-value <0.05. (E) Heatmap representation using Gene-E of pathways related to flagella synthesis identified using KEGG orthology (KO), Clusters of Orthologous Groups of proteins (COG) and Non-supervised Orthologous Groups (NOG). Data are the means +/− S.E.M (N=2–3).
Figure 5. CMC and P80 increase the pro-inflammatory potential of the mucosal-M-SHIME system microbiota.
A 9-vessel mucosal-M-SHIME system was set up and inoculated with fresh human feces at day −7. Following a 7-day stabilization period, vessels were treated with either water (N=2), CMC 1% (w/v, N=3) or P80 1% (v/v, N=3) for 13 days. (A) Luminal M-SHIME suspension FliC levels at days −7, −6, −3, −2, 1, 4, 6, 8, 11 and 13 days. (B) Luminal M-SHIME suspension was injected intraperitoneally to Rag−/− mice, and IL-6 were measured in the serum 2 h post-injection. Data are the means +/− S.E.M (N=3). Each treatment group was compared with the control group (water-treated) and statistical significance was determined using Student’s t test.
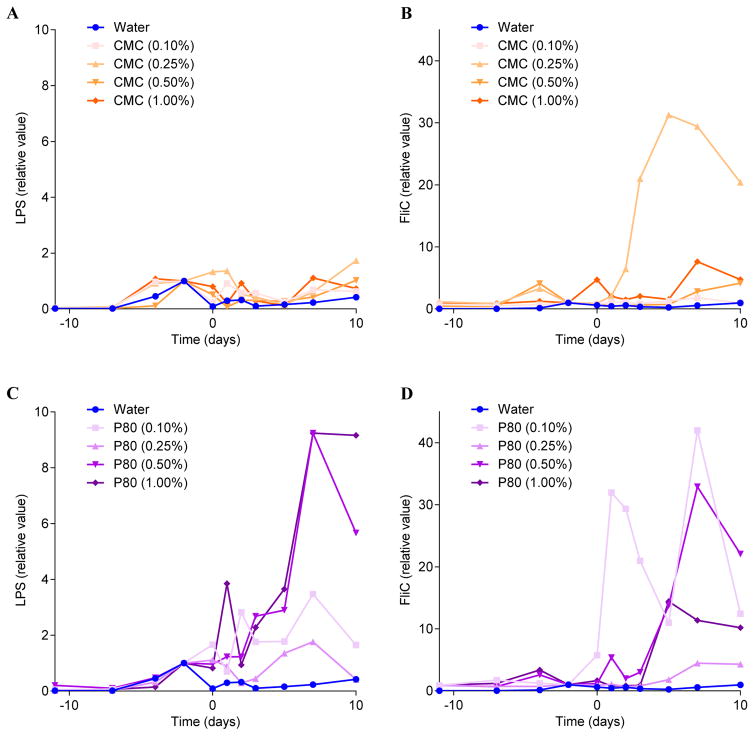
We next sought to investigate the concentration dependence of the effects of CMC and P80 in the M-SHIME model. M-SHIMEs were treated with 0.00, 0.10, 0.25, 0.50, or 1.00 % CMC or P80 (v/v and w/v, respectively). The relatively complex apparatus needed to maintain each M-SHIME unit as an independent ecosystem precluded us from running parallel replicates. Moreover, although these experiments used the same human donor used above to inoculate the M-SHIME vessels, the possibility that this donor’s microbiome may have differed on the days the samples were provided for this and the above-described experiments cannot be eliminated. Such caveats notwithstanding, we observed that all tested doses of P80 and all but the lowest dose of CMC elicited an increase in levels of flagellin at multiple time points, albeit not displaying a clear dose-dependent linear relationship (Figure 6A and 6C). P80 treatment also resulted in a clear increase in levels of the TLR4-agonist LPS especially at higher doses (Figure 6B and 6D). Together, these results indicated that CMC and P80 can act directly on a complex microbiota in a manner that might increase its potential to promote inflammation in the host.
Figure 6. CMC and P80 increase the pro-inflammatory potential of the mucosal-M-SHIME system microbiota.
A 9-vessel mucosal-M-SHIME system was set up and inoculated with fresh human feces at day −11. Following an 11-day stabilization period, vessels were treated with either water, CMC (0.10%, 0.25%, 0.50% or 1.00%) or P80 (0.10%, 0.25%, 0.50% or 1.00%) for 10 days. (A–B) Luminal M-SHIME suspension LPS (A) and FliC (B) levels of CMC-treated M-SHIME at days −10, −7, −4, −2, 0, 1, 2, 3, 5, 7 and 10 days. (C–D) Luminal M-SHIME LPS (C) and FliC (D) levels of CMC- and P-80-treated M-SHIME at days −10, −7, −4, −2, 0, 1, 2, 3, 5, 7 and 10 days. Data are expressed as relative values with day −1 defined as 1 (N=1).
CMC and P80 treated M-SHIME suspensions promote low-grade inflammation associated phenotypes in mice
We next examined the extent to which emulsifier-induced alterations of M-SHIME microbiotas might have potential to impact a host by transferring them to germfree mice. 16S sequencing analysis of feces from M-SHIME recipient mice revealed that, relative to a range of actual M-SHIME samples and fecal samples from conventional mice (from ongoing-microbiome-related studies), M-SHIME-recipient mice had microbiomes that, although distinct from both aforementioned groups, were much closer in overall composition to M-SHIME microbiomes, indicating that our approach preserved much of the basic nature of the M-SHIME microbiota many weeks following transplant (Figure 7A). Moreover, at both times assayed, microbiota composition of M-SHIME recipient mice clearly clustered based on M-SHIME treatment, irrespective of cage clustering (Figure 7B–C), including alteration of α-diversity in mice receiving emulsifier-treated M-SHIME suspensions compared to those receiving water-treated M-SHIMEs (Figure S9A–B). Such altered microbiota composition was characterized by an increase in inflammation-associated bacteria, such as Proteobacteria and Enterobacteriacae, and decreased levels of health-associated bacteria, such as Bacteroidaceae, in mice receiving emulsifier-treated M-SHIME microbiotas (Figure S9C–F). Predicted metagenomes based on this 16S composition, using PICRUSt [26], indicated that the mice receiving emulsifier-treated M-SHIME-derived microbiotas harbored an altered metagenome compare to those receiving water-treated M-SHIMEs, characterized by an enrichment of genes related to flagella/motility (Figure S10 and S11). Accordingly, at both time points assayed, recipients of CMC- and P80-treated M-SHIMEs displayed higher levels of bioactive flagellin and LPS (Figure 8A–D). Thus, some aspects of CMC- and P80-induced M-SHIME microbiota alterations persisted for a prolonged period in vivo following transfer to germfree mice.
We next analyzed the extent to which such conventionalization of germfree mice with emulsifier-altered microbiotas might correlate with altered metabolic and or intestinal phenotypes. Conventionalization of germfree mice is known to restore a more normal gut morphology. Accordingly, relative to germfree mice, those colonized with M-SHIME water-treated suspension exhibited an increase in intestinal mass, which was not associated with any indications of metabolic syndrome (Figure S12). Conventionalization of germfree mice is also known to be followed by weight gain. Accordingly, although we did not track weight of germfree mice (as it risks breaching the germfree state), we observed that all groups of germfree mice receiving M-SHIMEs, more than doubled their body mass with in 2 weeks of receiving M-SHIMEs. The extent of such weight gain did not differ between mice receiving water-treated and emulsifier-treated M-SHIMEs (Figure 8E), but, recipients of both CMC- and P80-treated M-SHIMEs exhibited higher levels of fasting blood glucose following a 15 h or 5 h fasting period (Figure 8F–G), often utilized for human and mouse studies respectively, thus suggesting that emulsifier-treated microbiotas might promote this central feature of metabolic syndrome. This notion was further supported by post-euthanasia assay of these animals, which indicated that mice receiving emulsifier-treated M-SHIMES displayed a range of features that are associated with low-grade inflammation and metabolic syndrome. Specifically, relative to mice receiving water-treated M-SHIME suspensions, recipients of CMC- and P80-treated M-SHIMEs displayed shortened thickened colons, mild splenomegaly, elevated fecal Lcn2 levels, and increased adiposity, as indicated by epididymal fat pad mass (Figure 8H–N). Importantly, such low-grade inflammatory phenotypes correlated with microbiota encroachment by the emulisifer-treated M-SHIME microbiotas (Figure 9).
Figure 9. CMC- and P80-treated M-SHIME suspensions promote microbiota encroachment when transplanted to germfree recipient mice.
A 9-vessel mucosal-M-SHIME system was set up and inoculated with fresh human feces at day −7. Following a 7-day stabilization period, vessels were treated with either water, CMC 1% or P80 1% for 13 days. Germfree C57BL/6 mice (4 weeks old) were removed from isolator and were orally administered with 200μL of luminal M-SHIME suspension (day 11 sample). Transplanted mice were then housed in isolated ventilated cages Isocages (Techniplast). (A) Confocal microscopy analysis of microbiota localization; Muc2 (green), actin (purple), bacteria (red), and DNA (Blue). Bar=20μm. (B) Distances of the closest bacteria to intestinal epithelial cells (IEC) per condition over 3 high-powered fields per mouse. Data are the means +/− S.E.M (N=5). Each treatment group was compared with the control group (water-treated) and statistical significance was determined using Student’s t test.
Some alterations in microbiota predispose to weight gain upon exposure to an obesogenic high-fat diet (HFD) [27, 28, 29]. Hence, we analyzed the extent to which emulsifier-treated M-SHIME suspension inoculation would impact germfree recipient mice upon feeding the colonized animals HFD. We observed that upon high fat diet-feeding mice receiving emulsifier-treated M-SHIME suspensions gained more weight than those receiving water-treated M-SHIMEs. Such increased weight gain correlated with elevated levels of fasting blood glucose (Figure S13B) and inflammation, such as a colon shortening (Figure S13E) and splenomegaly (Figure S13G). Together, these results suggest that direct effects of CMC and P80 on the microbiota play a role in driving the inflammo-metabolic phenotypes induced by these compounds in vivo.
DISCUSSION
The gut microbiota is a complex ecosystem that wields great influence over the health of its host [30]. Conversely, mammalian hosts utilize a broad array of mechanisms to shape the microbiota in a way that maximizes its benefit and minimizes its potential to harm [31]. That, at least amongst healthy persons, intra-individual variance in microbiota composition (i.e. repeat sampling of individual subjects) is much smaller than that seen when comparing different individuals [31, 32, 33], indicates that the ability of host and microbiota to influence each other typically results in a relatively stable equilibrium within specific hosts. Yet, some exogenous factors, e.g. pathogens or antibiotics, can dramatically and lastingly disturb this equilibrium in a manner that promotes persistent disease [34]. Moreover, even factors that cause relatively modest disturbances of the host-microbiota relationship have been associated with chronic low-grade inflammatory diseases, including metabolic syndrome [34]. The genetics of the host is surely a pivotal determinant of the resiliency of host-microbiota equilibrium following exogenous to perturbants. For example, pathobionts that are readily cleared by WT mice without evidence of an associated phenotype can trigger lasting colitis in mice with an array of genetic abnormalities [35]. Such colitis is associated with, and promoted by, the altered microbiota, which can remain lastingly disturbed long after the pathobiont has been cleared. Analogously, the dietary emulsifiers CMC and P80 induce a moderate disturbance of microbiota in WT mice, that associates with low-grade inflammation and metabolic disease, but promotes robust colitis, associated with a more severe alteration of gut microbiota composition, in mice lacking IL-10 or TLR5 [8].
On the one hand, the ability of the host and microbiota to influence each other precludes the use of microbiota-free hosts (i.e. germfree mice) or host-free microbiotas (e.g. M-SHIMEs) in fully defining how CMC or P80 (or other substances) ultimately impacts the host or microbiota. But on the other hand, such bi-directional host-microbiota interactions dictates that use of these methodologies may be the only means to define direct mechanisms by which an exogenous agent perturbs the host-microbiota relationship. Indeed, we reasoned that the detergent-like properties of CMC and P80 might allow them to alter physiochemical properties of the mucus layer that protects the gut from the microbiota [8, 9, 12]. However, that we have not observed such effects by these compounds in germfree mice prompted us to consider that the microbiota itself may be a direct target of CMC or P80. That CMC and P80 altered gene expression and composition in the M-SHIME model in a manner somewhat reminiscent of their effects in mice supports the notion that, in vivo, the bioactions of CMC and P80 effects involve direct effects on the microbiota. Conversely, we speculate that CMC not inducing a clear change in microbial composition in the M-SHIME model, but doing so in mice, may reflect that the altered composition in vivo is driven, in part, by the host response to the CMC-induced changes microbiota gene expression. Indeed, an increase in flagella-related gene expression, which CMC induced in the M-SHIME model, can be readily envisaged, in vivo, to increase activation of TLR5 and/or NLRC4 by both providing higher intestinal concentrations of flagellin and greater ability of motile bacteria to penetrate the mucus layer, increasing the likelihood that flagellin released by bacteria would reach host receptors capable of recognizing it. Activation of these receptors would increase host expression of a range of antimicrobial peptides that would have the potential to shift microbiota composition.
That transfer of CMC- and P80-treated M-SHIMEs to germfree mice transferred features of low-grade inflammation/metabolic syndrome, supports the notion that the direct actions of these compounds observed in the M-SHIME model play a central role in mediating their detrimental effects on the host. We hypothesize that the key feature of emulsifier-treated M-SHIME microbiotas that drove their ability to promote inflammation following transplant was their greater ability to encroach upon host cells whereupon their products (e.g. LPS and flagellin) would induce pro-inflammatory gene expression, including production of cytokines that can cross-desensitize metabolic receptor signaling [12, 36].
Differences in microbiota and host parameters between germfree mice receiving water-treated and emulsifier-treated M-SHIMEs persisted many weeks following M-SHIMEs transplant. Yet, given that any residual CMC or P80 present during the transplant would be quickly washed out upon transfer to germfree mice, it might have been reasonable to presume that the microbiota would quickly revert to its untreated state. We speculate such persistence reflects that the initial host response to the altered M-SHIMEs maintains and/or enhances alterations in microbiota gene expression, and leads to further alterations in composition. The lack of beneficial microbiota reversion in mice receiving CMC and P80-treated M-SHIMEs might also be a consequence of our approach not permitting a normal mouse microbiota to outcompete a disturbed human microbiome, which may be more difficult for the mouse gut to corral. Regardless, these observations highlight the importance of early host-microbiota interactions in having a lasting impact on the host-microbiota relationship and, consequently, host health.
In conclusion, while we do not believe the M-SHIME or any host-free microbiota can fully define how an agent would impact the microbiota within its host, our results demonstrate that this system can be a very useful tool to explore underlying mechanisms. The M-SHIME model has enabled us to conclude that our observation that the emulsifiers CMC and P80, which have been previously observed to impact the mouse microbiome, also impact human microbiomes, at least in this ex vivo model. This observation should enable us to better design human clinical trial on dietary emulsifiers, which we hope to initiate soon. Furthermore, our results described herein strongly suggest that the microbiota is one direct important target of the food additives CMC and P80. We envision that this model may allow further mechanistic investigation into how these compounds directly alter the microbiota. Moreover, our results suggest that such models may be a useful means to develop food additives and medicine vehicles that maintain their intended functions without negatively impacting the microbiome.
Supplementary Material
Table 2.
Primers used in the study.
| ASF Number | Forward (5′ > 3′) | Reverse (5′ > 3′)) | |
|---|---|---|---|
| 16S | N.A. | 515F GTGCCAGCMGCCGCGGTAA | 806R GGACTACHVGGGTWTCTAAT |
| Clostridium sp. | 356 | AAAATAATTAGGAGCTTGCTTTTAA | TTAGAAGATGCCTCCTAAGAACC |
| Lactobacillus intestinalis | 360 | GGTGATGACGCTGGGAAC | AAGCAATAGCCATGCAGC |
| Lactobacillus murinus | 361 | GAACGAAACTTCTTTATCACC | TAGCATAGCCACCTTTTACA |
| Mucispirillum shaedleri | 457 | TCTCTTCGGGGATGATTAAAC | AACTTTTCCTATATAAACATGCAC |
| Eubacterium plexicaudatum | 492 | AATTCCTTCGGGGAGGAAGC | TAAAACCATGCGGTTTTAAAAAC |
| Firmicutes bacterium | 500 | ACGGAGGACCCCTGAAGG | AGCGATAAATCTTTGATGTCC |
| Clostridium sp. | 502 | GAGCGAAGCACTTTTTTAGAAC | TTACACCACCTCAGTTTTTACC |
| Parabacteroides sp. | 519 | GCAGCACGATGTAGCAATACA | TTAACAAATATTTCCATGTGGAAC |
SUMMARY BOX.
What is already known about this subject
Chronic inflammatory diseases are associated with alterations in the species and genomic composition of the intestinal microbiota.
Dietary emulsifiers polysorbate 80 (P80) and carboxymethylcellulose (CMC) consumption result in microbiota encroachment into the mucus, alterations in microbiota composition, and development of chronic inflammation.
Inflammation itself can impact microbiota composition, making very difficult to discern the extent to which these compounds act directly upon the microbiota versus promoting host inflammation, which subsequently reshapes the microbiota.
What are the new findings
CMC and P80 do not impact intestinal microbiota or host in mice harboring a pathobiont-free microbiota.
Use of the mucosal simulator of the human intestinal microbial ecosystem (M-SHIME), reveals that both P80 and CMC directly alter the microbiota, increasing its pro-inflammatory potential.
When transplanted to germfree mice, CMC- and P80-treated M-SHIME suspensions promote low-grade inflammation associated phenotypes indicating that the direct effects of these emulsifiers on the microbiota are sufficient to drive low-grade inflammation and metabolic disease.
How might it impact on clinical practice in the foreseeable future?
The observation that emulsifiers CMC and P80 directly impact human microbiomes in a manner reminiscent of their effects in mice supports the use of this model as a practical means to test food additives for safety.
Our results demonstrate the utility of the M-SHIME system as a tool to explore underlying mechanisms of any host/microbiota alterations.
Acknowledgments
Funding: This work was supported by NIH grant DK099071 (A.T.G.) and DK083890 (A.T.G.). B.C. is a recipient of the Career Development Award from the Crohn’s and Colitis Foundation of America (CCFA).
We thank Dr. Amanda Ramer-Tait for the sequences of ASF members specifics primers [37].
Abbreviations
- ASF
altered Schaedler flora
- CMC
carboxymethylcellulose
- IL-10
interleukin-10
- M-SHIME
mucosal simulator of the human intestinal microbial ecosystem
- P80
polysorbate-80
- IBD
inflammatory bowel disease
- LEfSe
LDA Effect Size
- PICRUSt
phylogenetic investigation of communities by reconstruction of unobserved states
- RAG
recombination activating gene
- TLR
Toll-like receptor
Footnotes
Contributorship
Conception and design: Chassaing B; Van de Wiele T; Gewirtz AT
Development of methodology: Chassaing B; Van de Wiele T; Gewirtz AT
Acquisition of data: Chassaing B and De Bodt J
Analysis and interpretation of data: Chassaing B; Van de Wiele T; De Bodt J; Marzorati M; Gewirtz AT
Writing, review, and/or revision of the manuscript: Chassaing B and Gewirtz AT
Study supervision: Gewirtz AT
Competing interests: none.
References
- 1.Dalal SR, Chang EB. The microbial basis of inflammatory bowel diseases. J Clin Invest. 2014;124:4190–6. doi: 10.1172/JCI72330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:204. doi: 10.1016/j.chom.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Spees AM, Wangdi T, Lopez CA, Kingsbury DD, Xavier MN, Winter SE, et al. Streptomycin-induced inflammation enhances Escherichia coli gut colonization through nitrate respiration. mBio. 2013:4. doi: 10.1128/mBio.00430-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faber F, Baumler AJ. The impact of intestinal inflammation on the nutritional environment of the gut microbiota. Immunol Lett. 2014;162:48–53. doi: 10.1016/j.imlet.2014.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–9. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature. 2012;487:104–8. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devkota S, Chang EB. Diet-induced expansion of pathobionts in experimental colitis: implications for tailored therapies. Gut Microbes. 2013;4:172–4. doi: 10.4161/gmic.23589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chassaing B, Koren O, Goodrich JK, Poole AC, Srinivasan S, Ley RE, et al. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature. 2015;519:92–6. doi: 10.1038/nature14232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roberts CL, Keita AV, Duncan SH, O’Kennedy N, Soderholm JD, Rhodes JM, et al. Translocation of Crohn’s disease Escherichia coli across M-cells: contrasting effects of soluble plant fibres and emulsifiers. Gut. 2010;59:1331–9. doi: 10.1136/gut.2009.195370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberts CL, Rushworth SL, Richman E, Rhodes JM. Hypothesis: Increased consumption of emulsifiers as an explanation for the rising incidence of Crohn’s disease. J Crohns Colitis. 2013;7:338–41. doi: 10.1016/j.crohns.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 11.Swidsinski A, Ung V, Sydora BC, Loening-Baucke V, Doerffel Y, Verstraelen H, et al. Bacterial overgrowth and inflammation of small intestine after carboxymethylcellulose ingestion in genetically susceptible mice. Inflamm Bowel Dis. 2009;15:359–64. doi: 10.1002/ibd.20763. [DOI] [PubMed] [Google Scholar]
- 12.Chassaing B, Gewirtz AT. Has provoking microbiota aggression driven the obesity epidemic? Bioessays. 2016;38:122–8. doi: 10.1002/bies.201500116. [DOI] [PubMed] [Google Scholar]
- 13.Van den Abbeele P, Roos S, Eeckhaut V, MacKenzie DA, Derde M, Verstraete W, et al. Incorporating a mucosal environment in a dynamic gut model results in a more representative colonization by lactobacilli. Microb Biotechnol. 2012;5:106–15. doi: 10.1111/j.1751-7915.2011.00308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van den Abbeele P, Belzer C, Goossens M, Kleerebezem M, De Vos WM, Thas O, et al. Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. ISME J. 2013;7:949–61. doi: 10.1038/ismej.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geirnaert A, Wang J, Tinck M, Steyaert A, Van den Abbeele P, Eeckhaut V, et al. Interindividual differences in response to treatment with butyrate-producing Butyricicoccus pullicaecorum 25-3T studied in an in vitro gut model. FEMS Microbiol Ecol. 2015:91. doi: 10.1093/femsec/fiv054. [DOI] [PubMed] [Google Scholar]
- 16.Marzorati M, Vilchez-Vargas R, Bussche JV, Truchado P, Jauregui R, El Hage RA, et al. High-fiber and high-protein diets shape different gut microbial communities, which ecologically behave similarly under stress conditions, as shown in a gastrointestinal simulator. Mol Nutr Food Res. 2016 doi: 10.1002/mnfr.201600150. [DOI] [PubMed] [Google Scholar]
- 17.Metcalf AM, Phillips SF, Zinsmeister AR, MacCarty RL, Beart RW, Wolff BG. Simplified assessment of segmental colonic transit. Gastroenterology. 1987;92:40–7. doi: 10.1016/0016-5085(87)90837-7. [DOI] [PubMed] [Google Scholar]
- 18.Nugent SG, Kumar D, Rampton DS, Evans DF. Intestinal luminal pH in inflammatory bowel disease: possible determinants and implications for therapy with aminosalicylates and other drugs. Gut. 2001;48:571–7. doi: 10.1136/gut.48.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meyer F, Paarmann D, D’Souza M, Olson R, Glass EM, Kubal M, et al. The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–62. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–6. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huerta-Cepas J, Szklarczyk D, Forslund K, Cook H, Heller D, Walter MC, et al. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016;44:D286–93. doi: 10.1093/nar/gkv1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.http://www.broadinstitute.org/cancer/software/GENE-E/index.html.
- 25.Hecht G, Bar-Nathan C, Milite G, Alon I, Moshe Y, Greenfeld L, et al. A simple cage-autonomous method for the maintenance of the barrier status of germ-free mice during experimentation. Laboratory animals. 2014;48:292–7. doi: 10.1177/0023677214544728. [DOI] [PubMed] [Google Scholar]
- 26.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–21. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujisaka S, Ussar S, Clish C, Devkota S, Dreyfuss JM, Sakaguchi M, et al. Antibiotic effects on gut microbiota and metabolism are host dependent. J Clin Invest. 2016;126:4430–43. doi: 10.1172/JCI86674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Backhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci U S A. 2007;104:979–84. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–31. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 30.Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. 2016;16:341–52. doi: 10.1038/nri.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lloyd-Price J, Abu-Ali G, Huttenhower C. The healthy human microbiome. Genome Med. 2016;8:51. doi: 10.1186/s13073-016-0307-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hisada T, Endoh K, Kuriki K. Inter- and intra-individual variations in seasonal and daily stabilities of the human gut microbiota in Japanese. Archives of microbiology. 2015;197:919–34. doi: 10.1007/s00203-015-1125-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lazarevic V, Whiteson K, Hernandez D, Francois P, Schrenzel J. Study of inter- and intra-individual variations in the salivary microbiota. BMC Genomics. 2010;11:523. doi: 10.1186/1471-2164-11-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cox LM, Yamanishi S, Sohn J, Alekseyenko AV, Leung JM, Cho I, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014;158:705–21. doi: 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chassaing B, Koren O, Carvalho FA, Ley RE, Gewirtz AT. AIEC pathobiont instigates chronic colitis in susceptible hosts by altering microbiota composition. Gut. 2014;63:1069–80. doi: 10.1136/gutjnl-2013-304909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gregor MF, Hotamisligil GS. Inflammatory mechanisms in obesity. Annu Rev Immunol. 2011;29:415–45. doi: 10.1146/annurev-immunol-031210-101322. [DOI] [PubMed] [Google Scholar]
- 37.Gomes-Neta JC, Mantz S, Held K, Sinha R, Benson AK, Walter J, et al. Evaluation of a new real-time qPCR assay for quantification of the Altered Schaedler Flora microbiota in gnotobiotic mice. 2016 doi: 10.1016/j.mimet.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.