Abstract
Chromatin and associated epigenetic mechanisms stabilize gene expression and cellular states, while also facilitating appropriate responses to developmental or environmental cues. Genetic, environmental or metabolic insults can induce overly restrictive or overly permissive epigenetic landscapes that contribute to pathogenesis of cancer and other diseases. Restrictive chromatin states may prevent appropriate induction of tumor suppressor programs or block differentiation. By contrast, permissive or ‘plastic’ states may allow stochastic oncogene activation or non-physiologic cell fate transitions. While many stochastic events will be inconsequential ‘passengers’, some will confer fitness and be selected as ‘drivers’. We review the broad roles played by epigenetic aberrations in tumor initiation and evolution, and their potential to give rise to all classic hallmarks of cancer.
Introduction
A single human genome gives rise to hundreds of cell types, and adapts to different developmental and environmental conditions with a vast repertoire of gene expression patterns. A mere 2% of its sequence codes for protein, while the remaining 98% is replete with regulatory elements that underlie context-specific gene activity. The 6 billion bases of coding and non-coding DNA are wrapped about ~30 million nucleosomes to form a massive, exquisitely regulated macromolecular complex, termed chromatin. Chromatin is the essential medium through which transcription factors (TFs), signaling pathways and other cues alter gene activity and cellular phenotypes (Fig 1A) (1, 2). Consistent with its broad functions in cellular state and regulation, chromatin aberrations have been associated with a wide range of common diseases, including aging-related diseases, neuropsychiatric disorders, autoimmunity and cancer.
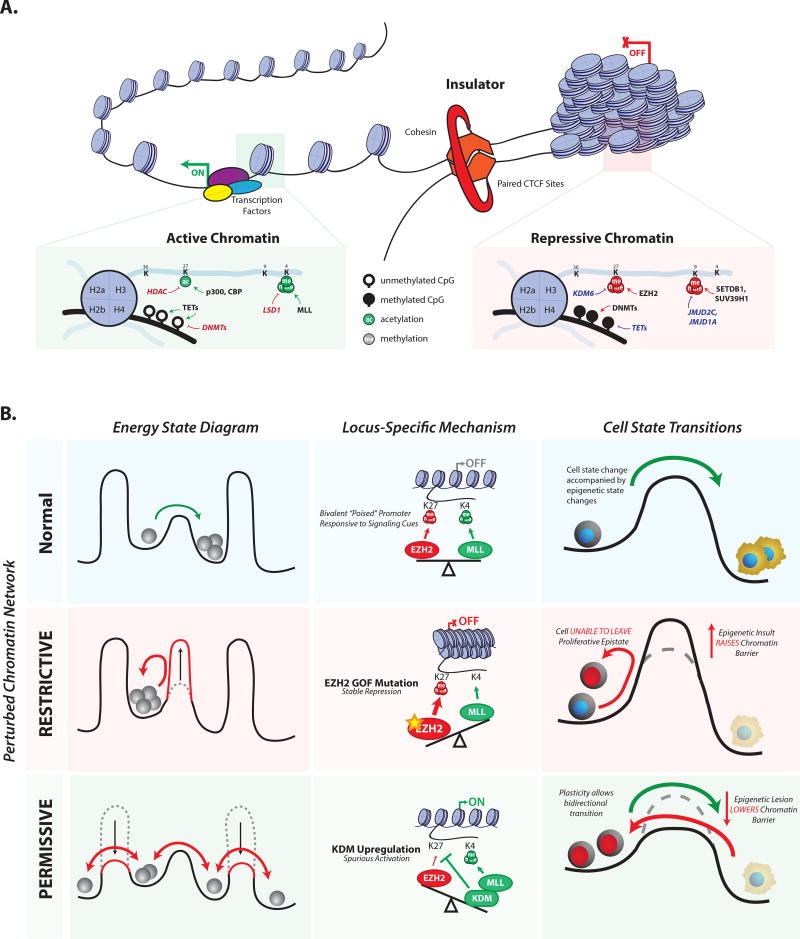
Fig. 1. Chromatin structure affects cellular identity and state transitions.
(A) Chromatin can adopt active and repressive states. Active states are made accessible to TFs and other regulatory factors, and are enriched for histone modifications such as H3K27ac and H3K4me3,. Repressive states are more compact, and characterized by DNA hypermethylation, EZH2-catalyzed H3K27me3 and H3K9me3. CTCF and cohesin partition the genome into discrete regulatory units, termed TADs. (B) Chromatin networks reinforce cell states, and affect responsiveness to intrinsic and extrinsic cues. Cells with perturbed chromatin networks fail to respond appropriately to such cues. Overly restrictive chromatin accentuates epigenetic barriers that prevent cell state transitions. Overly permissive chromatin lowers barriers, allowing promiscuous sampling of alternate cell states. The opposing activities of the H3K27 methyltransferase EZH2 and the H3K4 methyltransferase MLL are given as an example; however, the concept holds for other regulators such as DNMTs and TET enzymes (see main text for details).
Although cancer is typically considered a genetic disease, epigenetic aberrations play profound and ubiquitous roles. In fact, cancers are universally associated with abnormalities in gene expression, cellular identity and responsiveness to internal and external cues (3–6). A major, unanticipated outcome of large-scale cancer genome sequencing projects is that roughly 50% of human cancers harbor mutations in chromatin proteins (7, 8). Malignant cells also exhibit genome-wide alterations in DNA methylation, chromatin structures and regulatory element activities. In addition, many tumors exhibit deranged developmental programs indicative of differentiation block or epigenetic reprogramming (6, 9, 10).
The goal of this review is to synthesize current literature into a general mechanistic model for cancer epigenetics. The over-riding premise is that specific genetic, environmental and metabolic stimuli disrupt the homeostatic balance of chromatin, causing it to become either aberrantly restrictive or aberrantly permissive. Such stimuli may act in a pre-malignant cell to promote tumor initiation and/or in a malignant cell to accelerate tumor evolution and adaptation. This model can explain diverse oncogenic stimuli whose effects are mediated through chromatin aberrations. The ubiquity of such stimuli suggests that epigenetic defects contribute to diverse aspects of cancer biology and may in fact suffice to satisfy every hallmark of cancer (11, 12).
Epigenetic homeostasis in healthy cells
The human genome comprises thousands of expansive genomic loci that contain genes along with proximal (promoters) and distal regulatory elements (enhancers) that control their activity in specific cell types. These loci are organized into topologically associating domains (TADs) and bounded by insulators that ensure their independent and appropriate regulation (13–16). Examples include the β-globin locus that orchestrates developmental stage-specific expression of globin genes, various developmental loci that contain TF genes flanked by enhancers that specify their tissue-specific expression, and gene-rich loci packed with housekeeping genes. The activity of a locus is intimately tied to its chromatin organization. Active genes and elements must be accessible to regulatory factors and transcriptional machinery, while inactive loci are sequestered within compact and inaccessible structures that check their inappropriate activity (1, 2, 17).
Context-specific repression of lineage-specific developmental genes is enforced by Polycomb repressors, such as the histone H3 lysine 27 (H3K27) methyltransferase enhancer of zeste homolog 2 (EZH2)(17, 18). Polycomb repression can be maintained through mitotic cell division by mechanisms that include a conformational switch in the EZH2 complex that is stimulated by H3K27 methylated histones and results in increased enzyme activity (19). Repetitive sequences and gene deserts are silenced by heterochromatin structures, histone H3 lysine 9 (H3K9) methylation and lamin-associated factors (Fig 1A). These repressive states can also be propagated through mitosis via functional interactions between histone modifications, DNA methylation, regulatory proteins and non-coding RNAs (1, 2).
Conversely, active loci may be sustained by TFs and chromatin modifying co-factors that bind promoters and enhancers, engage RNA polymerase, and stimulate transcriptional activity. These regulatory activities present a potent barrier to chromatin repression and compaction, which facilitates robust maintenance of the active state (20, 21)
Because any single locus can assume different transcriptional states in different cellular contexts, chromatin state must be switchable, given appropriate cues and conditions. As discussed below, the likelihood that a locus will respond to a signal for change is dependent on the expression of TFs and their recruitment to the locus, as well as its local chromatin state, and the global chromatin environment in the cell (Fig 1B).
Chromatin homeostasis and Waddington’s landscape
Conrad Waddington famously conceptualized developmental specification as an epigenetic landscape in which differentiating cells proceed downhill along branching canals (22). The canals are separated by walls that constrain lineage and cell identity. Decades of research since Waddington’s prescient description have revealed that TFs are the predominant specifiers of cellular identity, and therefore of the topography of the canals (20, 21). TF networks define and sustain the discrete cellular states represented by the canals. Although chromatin regulators are critical partners, they play a secondary role in the definition of cell fates.
Rather, a primary function of chromatin during development is to reinforce or stabilize these lineages and cell fates. In the context of Waddington’s landscape, chromatin structures and regulators affect the height of the walls that partition canals in that they prevent cells from switching states. This central role for chromatin is strongly supported by genetic, cell biology and biochemistry studies (as reviewed in (17, 23, 24)). Here we highlight just a few key concepts. Drosophila embryo mutants deficient for Polycomb repressors exhibit profound alterations in cell identity while the corresponding mutant cells can trans-differentiate across lineages (25). Polycomb repressors, heterochromatin factors and other histone modifying enzymes act as barriers that hinder cellular reprogramming (26–29). Suppression of these proteins facilitates the conversion of fibroblasts to induced pluripotent stem (iPS) cells. Repressive chromatin structures sequester genomic loci that are unused in a given lineage, including non-lineage TF genes, compacting their DNA and checking their spurious activation. Thus, by restricting changes in gene activity, chromatin increases the heights of energy walls between cell states, and resists changes to their identity.
Further evidence indicates that the magnitude of chromatin restriction changes during development. In embryonic stem (ES) cells, hyper-dynamic nucleosome exchange hinders the establishment of repressive structures, leaving many developmental TF genes in a ‘bivalent’ state with ‘active’ and ‘repressive’ histone marks that ‘poises’ them for alternate fates (1, 15) As developing cells commit along specific lineages, their chromatin becomes more restrictive (23, 24, 26, 30–33). Progressive chromatin restriction correlates with reduction in cell fate potential and is likely to play a causal role in this regard (24, 32). Hence, chromatin structure impedes changes to gene activity (or cellular state more broadly) with a developmentally and contextually appropriate degree of resistance.
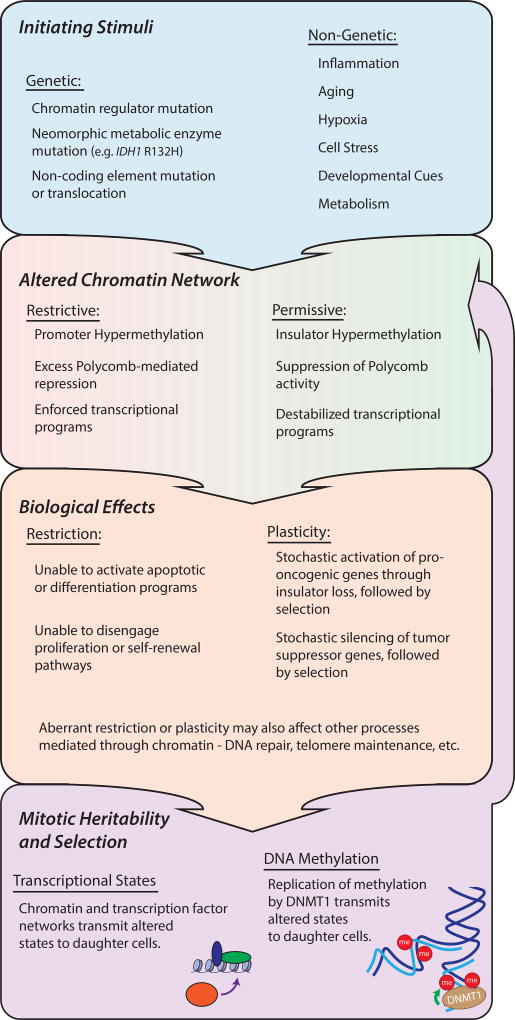
Deviation from a homeostatic chromatin network
Based on a growing body of evidence, we postulate that chromatin resistance must be precisely titrated at each stage of development, and that deviation from the norm is a major factor in tumorigenesis. We discuss genetic, environmental and metabolic ‘stimuli’ that cause such deviations. Certain stimuli may increase chromatin resistance, resulting in a restrictive state that blocks a differentiation program. Others may decrease chromatin resistance, resulting in a permissive state that allows stochastic induction of oncogenes or other adaptive programs.
Epigenetic restriction in tumorigenesis
The homeostatic chromatin network is predicated in large part on interplay between Polycomb-family repressors, trithorax-family activators and nucleosome remodelers (34). Recurrent mutations to these factors represent genetic stimuli likely to disrupt this homeostasis. We begin by considering stimuli that induce chromatin restriction through excessive repressor activity, repressive chromatin marks and/or DNA methylation (Fig 2). Gain-of-function EZH2 mutations are frequent in several lymphoma subtypes and have also been detected in melanoma (35). EZH2 is the catalytic subunit of Polycomb repressive complex 2 (PRC2), which plays broad roles in B-cell development and differentiation. It is highly active in germinal center B cells (GCBs) but rapidly down-regulated upon differentiation, allowing activation of terminal genes. The gain-of-function mutation creates a hyper-active methyltransferase enzyme (36, 37). Genome-wide analyses of EZH2 mutant lymphomas revealed expansive H3K27 tri-methylation and depletion of active chromatin marks over loci encoding terminal genes. The tumorigenic mutant thus appears to induce a restrictive state that checks the induction of differentiation genes and arrests B-cell development in a proliferative state (38, 39).
Fig. 2. Chromatin homeostasis is disrupted in cancer.
Chromatin homeostasis may be disrupted by genetic stimuli (e.g., chromatin regulator mutations or regulatory element translocation) or non-genetic stimuli (e.g., aging, inflammation, hypoxia, etc). Such stimuli can result in an overly permissive or overly restrictive chromatin network. Restrictive or permissive states may create, or allow stochastic adoption of oncogenic epigenetic changes, such as silencing of tumor suppressor genes. Some such events are mitotically heritable and may be selected (Fig 3), giving rise to hallmarks of cancer (Fig 4).
PRC2 activity is opposed by demethylases that remove H3K27 methylation, by modifying enzymes that catalyze H3K27 acetylation or H3K4 methylation, and by nucleosome remodelers (2, 34). The corresponding enzymes and complex members, including KDM6A/B, p300, MLL components and ARID1A/B, are genetically inactivated in a wide range of cancers (40–42). The various mutations remain poorly understood and are likely to have context-specific and disparate affects. However, in certain cases, they appear to shift the balance of chromatin towards PRC2 repression. For example, inactivating MLL2 and CBP/p300 mutations in lymphoma impede appropriate engagement of promoters or enhancers needed for differentiation, paralleling or potentially cooperating with EZH2 GOF alleles (38, 43, 44).
In pediatric malignant rhabdoid tumors, homozygous inactivation of the remodeling enzyme SNF5 disables enhancers associated with mesenchymal differentiation genes, many of which are PRC2 targets (17, 35) Remarkably, chemical EZH2 inhibitors lead to rhabdoid tumor regression in xenograft models, and are now in clinical trials. Other SWI/SNF complex members, most notably ARID1A/B, are among the most frequently mutated targets in all human cancers (41). Although these factors have been variously implicated in senescence (45) and DNA repair (46), their genetic inactivation may also promote global chromatin restriction.
Chromatin restriction can also arise from alterations to the DNA methylation landscape. DNA methylation plays diverse roles in repetitive element silencing, in parent-of-origin allelic imprinting, as well as in transcriptional elongation and splicing (47). In normal cells, cytosines in CpG islands and other GC-rich loci are largely unmethylated, while GC-poor regions tend to be highly methylated. In many cancers, this pattern is profoundly distorted as CpG islands become hyper-methylated and GC-poor regions become hypo-methylated. The former aberration has been termed CpG island methylator phenotype (CIMP) and is perhaps the most widely studied epigenetic alteration in cancer, having been described in a wide range of phenotypically diverse tumors. CpG island hyper-methylation can silence and/or prevent re-activation of tumor suppressor (p16) (3, 4) and DNA mismatch repair genes (for example MLH1, MSH2 (3, 4, 48)). Although the generality and causality of DNA hyper- and hypo-methylation in cancer remains controversial, these examples are consistent with a role for CpG island hyper-methylation in epigenetic restriction.
Epigenetic plasticity and clonal selection in tumorigenesis
While certain stimuli exert their oncogenic effects by epigenetic restriction, others induce a more permissive state that may be conceptualized as a lowering of the walls between canals in Waddington’s landscape (Fig 2). Permissive or ‘plastic’ chromatin may allow pre-malignant or malignant cells to sample alternative transcriptional states, gene pathways or developmental programs, a subset of which may be pro-oncogenic or otherwise adaptive. Critically, if an adaptive chromatin or transcriptional state change is propagated through mitosis, a new clone will arise and expand due to its increased fitness (Fig 3).
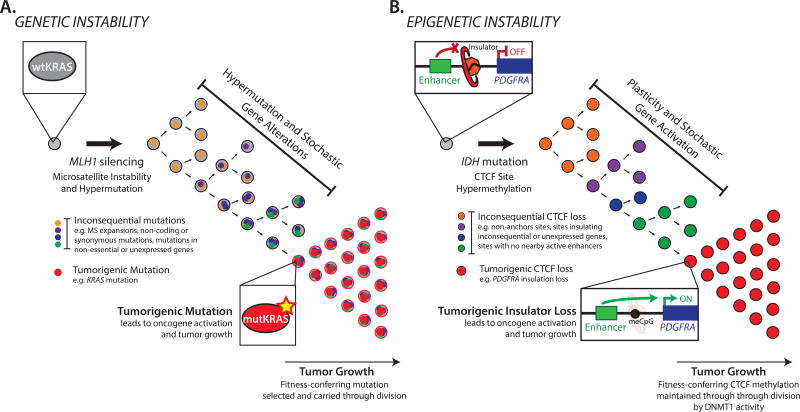
Fig. 3. Genetic and epigenetic evolution in cancer.
(A) Genetic instability in tumor initiation. An initiating event (e.g., MLH1 silencing) causes stochastic hypermutation, leading to inconsequential ‘passenger’ alterations as well as a ‘driver’ mutation (e.g., KRAS) that is selected. (B) Epigenetic instability in tumor initiation. An initiating event (e.g., IDH mutation) causes stochastic hyper-methylation, leading to inconsequential ‘passenger’ CTCF losses, as well as a ‘driver’ event that disrupts insulation of the PDGFRA oncogene and is selected. Selective pressure and mechanisms of epigenetic mitotic heritability may result in persistence of the altered states even if the initiating stimulus is removed.
In considering the epigenetic plasticity model, it is useful to draw an analogy to the more familiar genetic instability induced by carcinogens or repair defects. In that genetic framework, increased mutation frequency leads to ‘driver’ events (e.g., mutations that activate oncogenes) as well as ‘passenger’ events that do not alter fitness. Similarly, in the setting of epigenetic plasticity, we posit that some ensuing chromatin or transcriptional alterations will be drivers (e.g., that induce oncogene expression), while others will be passengers in that they fail to affect the expression of a consequential gene. Epigenetic alterations may occur individually over time or, alternatively, may arise as multiple simultaneous disruptions, analogous to the catastrophic genetic aberrations associated with ‘chromothrypsis’ (49, 50). Thus, our broad hypothesis is that certain stimuli induce epigenetic plasticity that allows pre-malignant or malignant cells to sample alternate chromatin or transcriptional states, a subset of which confer fitness and are maintained through cell division (Fig 3). Indeed, many cancers exhibit marked cell-to-cell variability in gene expression and functional phenotypes (51). The following sections describe potential stimuli for epigenetic plasticity that may promote tumor initiation and/or allow malignant cells to adapt to their environment.
Epigenetic plasticity: DNA methylation and disruption of oncogene insulation
As a first example, we focus on a genetic stimulus that destabilizes chromatin structure and thereby triggers epigenetic instability. Gain-of-function IDH mutations are frequent initiating events in glioma, leukemia and other tumors (52–54). Mutant IDH generates an onco-metabolite that inhibits hydroxylases, including Ten-eleven translocation (TET) enzymes that catalyze DNA demethylation. Consequently, IDH mutant tumors are hyper-methylated (55). Hyper-methylation disrupts binding of the methylation-sensitive DNA binding protein CTCF. CTCF is a critical mediator of chromosomal loops that partition our genome into discrete functional domains and ensure that enhancers regulate their appropriate gene targets. CTCF thus acts as an ‘insulator’ that protects genes from inappropriate activation by overly promiscuous enhancers.
Reduced CTCF binding in IDH mutant gliomas is associated with a global transcriptional signature indicative of insulator dysfunction (56). Whereas gene pairs that are separated by a CTCF boundary exhibit relatively lower correlation in normal cell types (57), many such cross-boundary pairs become correlated in IDH mutant tumors. This suggests that IDH oncogenicity might be mediated by loss of gene insulation. Indeed, one consistently deregulated CTCF boundary is near PDGFRA, a prominent glioma oncogene. Loss of this boundary allows a potent enhancer in a neighboring domain to aberrantly activate PDGFRA and drive proliferation of hyper-methylated gliomas.
While the PDGFRA insulator may be preferentially sensitive to disruption, the totality of findings is consistent with an epigenetic plasticity model. The human genome contains thousands of chromosomal loops, hundreds of which appear to be disrupted in IDH mutant tumors (56). This suggests that hyper-methylation causes stochastic CTCF insulator disruption in a pre-malignant IDH mutant cell. Loss of any specific insulator may then be preserved through cell division due to the epigenetic stability of DNA methylation (Fig 3). Thus, a new ‘epigenetic clone’ will arise with altered chromosomal topology and proximal gene activity. In most cases, transcriptional changes will be inconsequential ‘passengers’ and the new clone will be maintained at low frequency or lost entirely. However, in a subset of cases, a ‘driver’ transcriptional change will activate an oncogene or otherwise confer fitness. This adaptive clone will expand and, given appropriate conditions and subsequent hits, give rise to tumor.
The proposed model of insulator loss is of general importance because many oncogenes are sequestered within insulated neighborhoods, presumably owing to their tumorigenic potential (58). Indeed, frequent mutations of CTCF motifs in the vicinity of oncogenes have been reported in colorectal, liver and esophageal cancer (58, 59). Furthermore, the CTCF protein and its associated boundary factor cohesion are both subject to recurrent mutation in multiple tumor types (60–63). CTCF haploinsufficiency has also been shown to promote tumor formation in mice (64). Importantly, CTCF haploinsufficiency in this setting also destabilizes DNA methylation, providing further support of interplay between DNA methylation and CTCF function. Thus, multiple genetic and epigenetic mechanisms can compromise CTCF-mediated genome topology, each with the potential to drive stochastic insulator dysfunction, epigenetic plasticity and oncogene activation.
Epigenetic plasticity: permissive chromatin states
In considering chromatin aberrations likely to confer plasticity, EZH2 and its substrate histone H3K27 are of particular interest. EZH2 can repress a wide range of genes, but does so in a highly context-dependent manner (17). Thus, while EZH2 gain-of-function mutations may be oncogenic in B-cell lineages, EZH2 loss-of-function mutations are tumorigenic in other settings. EZH2 is genetically inactivated in myelodysplastic syndromes (MDS), T cell acute lymphoblastic leukemia (T-ALL), and other cancers (35). In addition, somatic mutation of the histone substrate (e.g., the H3.3 Lys27→Met ‘oncohistone’) in pediatric brain tumors dominantly suppress EZH2 function (65–68). While the underlying mechanisms remain unclear, suppression of Polycomb activity by EZH2 inactivation or histone mutation may create an overly permissive chromatin state that allows spurious gene activation, prevents differentiation-associated silencing, or destabilizes other processes, such as telomere regulation.
Histone lysine demethylases (KDMs) have also been widely implicated in cancer. There are more than 25 human KDMs that target different lysine positions in the histone tails, and thus differ in their regulatory functions (69). Families of related KDMs are up-regulated under stress conditions and are responsive to tumor microenvironments. Model organism studies have established roles for KDMs in erasing epigenetic memory, raising the possibility that cancer-associated deregulation of these enzymes may confer plasticity and facilitate reprogramming or adaptation (70, 71). H3K4 demethylases (KDM5) enable lung and melanoma cell lines to evade anti-proliferative therapies by adopting a slow-cycling persister state (72, 73). H3K27 demethylases (KDM6) enable glioblastoma stem cells to tolerate similar drug pressures by regressing to a more ‘primitive’ developmental state (74). KDM4 family enzymes, which demethylate H3K9 and H3K36, are upregulated in many cancer types, where they deregulate heterochromatin, affect replication timing and prime chromosomal copy number alterations (75, 76).
Finally, compromised fidelity of DNA methylation patterning may also contribute to stochastic activation of stem cell or proliferation genes. DNA methyltransferase mutations can lead to hypo-methylation and aberrant activation of enhancers that drive leukemogenic gene patterns (77, 78). Fidelity may also be perturbed by altered cellular contexts, including the increased demands of a rapidly replicating cancer genome. Accordingly, tumor cell methylomes can exhibit increased heterogeneity and variability of large numbers of CpG sites genome-wide (79). DNA methylation instability has also been linked to stochastic activation of cancer-associated genes that are stably repressed in non-neoplastic tissue, including cell cycle and epithelial-mesenchymal transition (EMT)-related genes (80).
Non-genetic stimuli of plasticity or restriction
The preceding sections are biased towards chromatin modifier mutations and other genetic stimuli whose relatively clearer effects on chromatin provide strongest support for our model. However, an essential element of our thesis is that oncogenic chromatin aberrations can also be induced by non-genetic (purely epigenetic) stimuli. In support, we note that chromatin state and stability can differ markedly between cells with identical genetic backgrounds, as a function of development (see above, “Chromatin Homeostasis”), metabolic conditions, aging and environment. We also note that some pediatric tumors arise with very few or no detectable genetic mutations (e.g., ependymoma) (81, 82). We review below non-genetic stimuli that alter chromatin state, followed by specific examples of pro-oncogenic epigenetic changes that arise in the absence of any genetic stimulus (Fig. 2).
Chromatin state is intimately tied to metabolic conditions. DNA and histone modifying enzymes utilize many metabolites as donors and cofactors, including alpha-ketoglutarate (alpha-KG), methyl donors in the folate pathway, and acetyl-CoA (83). Because the dissociation constants for these cofactors are close to their physiologic cellular concentrations, chromatin enzymes are exquisitely sensitive to shifts in metabolite levels. Physiologic methylation of repetitive DNA is dependent on folate (84), while maintenance of unmethylated CpG islands requires vitamin C, which is critical for demethylase activity (85). In stem cells, the maintenance of pluripotency is dependent on finely tuned levels of the methyl donor, S-adenosylmethionine, and the demethylase co-factor, alpha-KG (86, 87). In the adipocyte lineage, the AMPK/alpha-KG axis regulates a master determinant of brown fat differentiation and maintenance, PRDM16, by controlling demethylase activity at its promoter (88).
Chromatin and methylation states also change during aging. Methylation signatures associated with aging have been identified in epigenomic studies, and parallel some changes seen in cancer, such as increased CpG island methylation and global hypo-methylation (89). Model organism aging studies have documented changes in global histone methylation patterns and, moreover, have established causal roles for the corresponding modifying enzymes in longevity (45, 89). Finally, a recent single-cell transcriptomic study found that T-cells from aging mice show a highly heterogeneous response to stimulation, consistent with increased epigenetic state variability or plasticity (90).
Understanding how epigenetic changes associated with metabolism, environment and aging drive tumorigenesis remains a formidable challenge. However, concrete examples are emerging. DNA methylation is of particular interest as a mediator of non-genetic stimuli given its stability and mitotic heritability. Only a minority of cancers with aberrant methylation can be explained by an underlying genetic event. Methylation changes are particularly prevalent in colorectal cancer and may also be detected in pre-malignant hyperplastic polyps that have yet to acquire characteristic genetic mutations (91). Indeed, the epigenome of gastrointestinal cells appears particularly sensitive to environmental stimuli, such as inflammation or butyrate levels associated with diet and microbiome (92). Cancer-specific rewiring of glucose metabolism (“Warburg effect”) can further butyrate accumulation, resulting in histone deacetylase inhibition and increased cancer cell proliferation (93).
Promoter methylation events are the best-studied epigenetic mediator of oncogenic effects. Methylation and silencing of the DNA repair factor O6-methylguanine-DNA methyltransferase (MGMT) promoter is a key example. MGMT loss drives a hypermutator phenotype that generates many genetic subclones in the tumor (94). This methylation event underlies a field defect wherein multiple sporadic colorectal tumors arise within a larger region of cells. This strongly suggests that the epigenetic aberration precedes the genetic change, and likely arises in response to an environmental insult. Another notable example is methylation of the succinate dehydrogenase (SDH) gene promoter in gastrointestinal stromal tumors. SDH loss increases succinate levels and inhibits DNA demethylation, potentially reinforcing the initial methylation event and causing global hyper-methylation (95). MLH1, MSH2 and PTEN methylation are also commonly observed without any obvious genetic trigger, and are associated with poor prognosis (3, 4, 48, 96).
Tumor microenvironment is widely implicated as influencing cancer epigenomes. As discussed above, stress induced by microenvironment and/or therapeutic intervention up-regulates histone demethylases that reshape the chromatin landscape, potentially inducing plasticity and adaptation to therapy (72–74). Microenvironmental hypoxia has been shown to suppress DNA demethylase activity in breast cancer (97). Altered cellular contexts can also increase the rate of DNA methylation changes that affect enhancer activity and increase transcriptional plasticity in melanoma (98). Finally, the observation that non-malignant cells in the tumor ecosystem can also undergo striking phenotypic changes suggests that microenvironmental effects on epigenetic states and plasticity may extend beyond the malignant compartment (99). These collective examples likely portend a far broader role for oncogenic epigenetic alterations that arise from non-genetic stimuli in tumorigenesis.
Relating genetic and epigenetic models of cancer
An expanded view of epigenetics raises important questions regarding interplay between genetic and epigenetic oncogenic lesions (8). Such lesions may arise concurrently or step-wise, potentially in defined order. In some cases, epigenetic changes precede characteristic oncogenic mutations (e.g., hyper-methylation in pre-malignant polyps) (Fig 3). Moreover, certain epigenetic lesions prime genetic lesions. For example, KDM4 over-expression is causally associated with chromosomal copy number aberrations, while promoter silencing of MGMT causes increased mutational rates. It remains to be seen whether a genetically unstable cancer cell still requires such initiating epigenetic lesions once it has acquired downstream oncogenic mutations.
The converse case wherein a genetic lesion disrupts epigenetic regulation also occurs. Consider for example the epigenetic plasticity associated with IDH mutation (see “Epigenetic plasticity”, above). The initiating genetic stimulus (IDH mutation) may become irrelevant once a downstream epigenetic event has occurred (stable oncogene induction). Such a ‘hit and run’ mechanism has important therapeutic implications as targeting the initiator of plasticity would become futile (Fig 3). New diagnostic strategies that integrate genetic and epigenetic biomarkers might thus provide critical insight into cancer subtypes, prognosis and therapeutic susceptibility.
The hallmarks of cancer
In two influential essays, Hanahan and Weinberg distilled a set of biological capabilities or ‘hallmarks’ that must be acquired for development of a human cancer (11, 12). It framed efforts to define the mechanisms by which cells and tumor ecosystems progress through subsequent malignant stages. In large part, it was assumed that these mechanisms are fundamentally rooted in genetic alterations. However, pervasive alterations in chromatin state, methylation and gene expression suggest that the contributions of epigenetic alterations must also be carefully considered (100, 101). Indeed, cancer’s hallmarks may also be achieved through tumor suppressor silencing, oncogene activation by repurposed enhancers, or cell fate transitions.
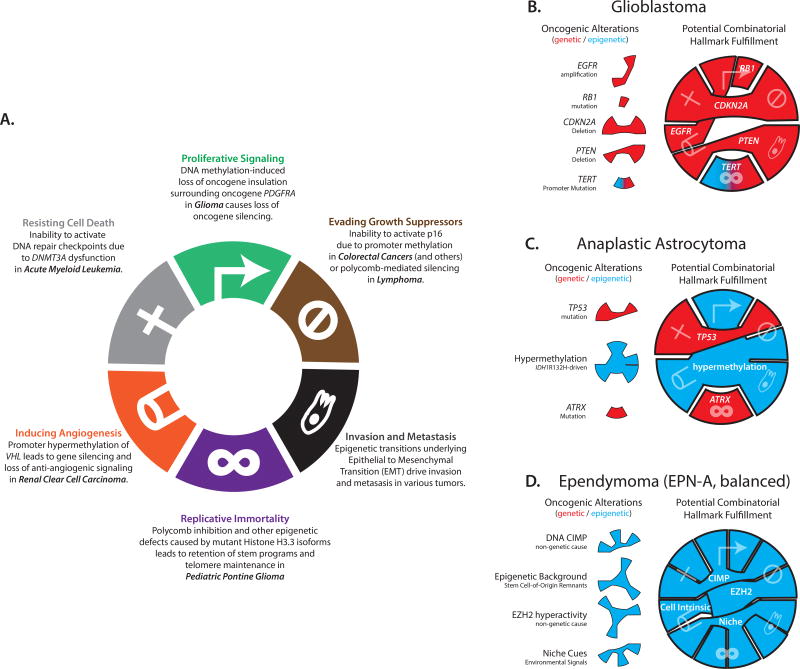
It is worthwhile to consider how epigenetic mechanisms might contribute to each hallmark (Fig 4A). Proliferative signaling can be achieved by PDGFRA activation due to insulator disruption (56) (Fig 4). Evasion of growth suppressors, such as p16/INK4a, can be mediated by promoter hyper-methylation or EZH2 hyperactivity (3, 35). Invasion and metastasis depend on cell fate transitions, such as epithelial-mesenchymal transition (EMT), with epigenetic etiology (98, 102–104). Replicative immortality may be driven by mutations to histone H3.3 or its chaperone proteins that promote alternative telomere lengthening (65, 105), or by non-genetic mechanisms that simulate self-renewing stem cell states (9, 106). Angiogenesis may be rooted in hyper-methylation of the von Hippel-Lindau (VHL) tumor suppressor promoter (107, 108). Finally, resisting cell death may be achieved by DNMT3A or IDH mutations that alter DNA damage responses (109, 110) or epigenetic mechanisms that alter expression of apoptosis or pro-survival genes (111, 112).
Fig. 4. Genetic and epigenetic mechanisms underlie the hallmarks of cancer.
(A) Epigenetic mechanisms involving aberrant chromatin restriction or plasticity can give rise to each classic hallmark of cancer (see main text for citations). Figure adapted from (11). Human cancers are underpinned by varying degrees of epigenetic and genetic contributions, as conceptualized for three central nervous system tumors (B–D). While most hallmarks can be traced to genetic drivers in glioblastoma, epigenetic factors predominate in pediatric tumors such as ependymoma, which exhibits DNA hyper-methylation but lacks recurrent mutations.
Clearly, our understanding of how epigenetic plasticity and alterations contribute to cancer’s hallmarks lags that of the genetic culprits. Yet new epigenetic mechanisms are rapidly being uncovered and, as with genetic lesions, will be powerfully selected for their tumorigenicity (101). Adaptive epigenetic states may then be maintained as cancer cells divide by the stability of DNA methylation, repressive chromatin or transcriptional regulatory programs. Thus, epigenetic plasticity and restriction should be considered alongside the more familiar genetic events that underlie each hallmark of cancer (Fig 4B, C & D).
Outlook for cancer biology and therapeutics
The altered epigenetics of cancer cells suggests that epigenetic therapies could have a major clinical impact (113). However, realizing their promise will require a far deeper understanding of how epigenetic lesions drive cancers. Towards this end, the field must develop, test, validate and refute conceptual and mechanistic models for cancer epigenetics, and place them in context with prevailing genetic models.
Investigating the specific hypotheses presented here will require a new generation of assays and experimental models. First, new single-cell technologies are urgently needed to evaluate the state and variability of insulators, enhancers, methylation and expression in situ within human tumors. We predict that such approaches will detect heterogeneous ‘passenger’ changes at low frequency as well as higher frequency ‘driver’ events that are recurrent across tumors. Such changes will likely be accompanied by increased cell-to-cell variability in gene expression (e.g., in single-cell RNA-seq) and other phenotypes. These technologies might also be applied to pre-malignant lesions, such as benign polyps, with the goal to ascertain whether stochastic epigenetic changes in individual cells represent early indicators of tumorigenic potential. Second, in vitro and in vivo tumor models that recapitulate the nature, dynamics and heterogeneity of successive tumorigenic epigenetic alterations are needed. Current cancer models are biased towards genetic lesions (e.g., genetically engineered mouse models) and may not recapitulate aspects of the tumor microenvironment that may profoundly influence the epigenetic state of malignant cells. Such experimental systems will need to be complemented by new technologies capable of tracking epigenetic alterations, such as insulator loss, with temporal resolution. Here again we predict that plasticity stimuli (e.g., a metabolic insult that disable DNA demethylation) will increase the rate at which epigenetic changes arise, yielding some epiclones with advantageous alterations that will overtake the population over the course of tumor development.
We are optimistic that fuller understanding of epigenetic plasticity and restriction in cancer could advance diagnostic tools for detecting early epigenetic lesions and evaluating tumor stage and heterogeneity (114, 115). It could also yield new therapeutic strategies for correcting epigenetic lesions or exploiting vulnerabilities of epigenetically altered cells. These new diagnostics and therapies would powerfully complement those rooted in cancer genetics. The road ahead is long, but vital to capture this major component of cancer biology and human disease in general.
Acknowledgments
We would like to thank members of the Bernstein lab for thoughtful discussion, and Rhiannon Macrae, Yotam Drier and Peter van Galen for helpful comments on the manuscript. Research in the Bernstein laboratory is supported by funds from the National Cancer Institute (DP1CA216873), the National Human Genome Research Institute, the American Cancer Society, the Leukemia and Lymphoma Society and the Starr Foundation. W.A.F. is supported by an F32 from the National Cancer Institute.
References
- 1.Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nat. Rev. Genet. 2010;11:285–296. doi: 10.1038/nrg2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016;17:487–500. doi: 10.1038/nrg.2016.59. [DOI] [PubMed] [Google Scholar]
- 3.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat. Rev. Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esteller M. Epigenetics in cancer. N. Engl. J. Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 5.Feinberg AP, Koldobskiy MA, Göndör A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat. Rev. Genet. 2016;17:284–299. doi: 10.1038/nrg.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laugesen A, Helin K. Chromatin repressive complexes in stem cells, development, and cancer. Cell Stem Cell. 2014;14:735–751. doi: 10.1016/j.stem.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 7.You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell. 2012;22:9–20. doi: 10.1016/j.ccr.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen H, Laird PW. Interplay between the cancer genome and epigenome. Cell. 2013;153:38–55. doi: 10.1016/j.cell.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suvà ML, Riggi N, Bernstein BE. Epigenetic reprogramming in cancer. Science. 2013;339:1567–1570. doi: 10.1126/science.1230184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim J, Orkin SH. Embryonic stem cell-specific signatures in cancer: insights into genomic regulatory networks and implications for medicine. Genome Med. 2011;3:75. doi: 10.1186/gm291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 12.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 13.Dixon JR, Gorkin DU, Ren B. Chromatin Domains: The Unit of Chromosome Organization. Mol. Cell. 2016;62:668–680. doi: 10.1016/j.molcel.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hnisz D, Day DS, Young RA. Insulated Neighborhoods: Structural and Functional Units of Mammalian Gene Control. Cell. 2016;167:1188–1200. doi: 10.1016/j.cell.2016.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dekker J, Misteli T. Long-Range Chromatin Interactions. Cold Spring Harb Perspect Biol. 2015;7:a019356. doi: 10.1101/cshperspect.a019356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rao SSP, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Comet I, Riising EM, Leblanc B, Helin K. Maintaining cell identity: PRC2-mediated regulation of transcription and cancer. Nat. Rev. Cancer. 2016;16:803–810. doi: 10.1038/nrc.2016.83. [DOI] [PubMed] [Google Scholar]
- 18.Simon JA, Kingston RE. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell. 2013;49:808–824. doi: 10.1016/j.molcel.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Margueron R, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zaret KS, Mango SE. Pioneer transcription factors, chromatin dynamics, and cell fate control. Curr. Opin. Genet. Dev. 2016;37:76–81. doi: 10.1016/j.gde.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell. 2013;152:1237–1251. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waddington CH. The Strategy of the Genes, a Discussion of Some Aspects of Theoretical Biology, by C. H. Waddington,… With an Appendix [Some Physico-chemical Aspects of Biological Organisation] by H. Kacser,…. 1957. [Google Scholar]
- 23.Perino M, Veenstra GJC. Chromatin Control of Developmental Dynamics and Plasticity. Developmental Cell. 2016;38:610–620. doi: 10.1016/j.devcel.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 24.Becker JS, Nicetto D, Zaret KS. H3K9me3-Dependent Heterochromatin: Barrier to Cell Fate Changes. Trends Genet. 2016;32:29–41. doi: 10.1016/j.tig.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee N, Maurange C, Ringrose L, Paro R. Suppression of Polycomb group proteins by JNK signalling induces transdetermination in Drosophila imaginal discs. Nature. 2005;438:234–237. doi: 10.1038/nature04120. [DOI] [PubMed] [Google Scholar]
- 26.Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors' initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Apostolou E, Hochedlinger K. Chromatin dynamics during cellular reprogramming. Nature. 2013;502:462–471. doi: 10.1038/nature12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hörmanseder E, et al. H3K4 Methylation-Dependent Memory of Somatic Cell Identity Inhibits Reprogramming and Development of Nuclear Transfer Embryos. Cell Stem Cell. 2017 doi: 10.1016/j.stem.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Onder TT, et al. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483:598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stergachis AB, et al. Developmental fate and cellular maturity encoded in human regulatory DNA landscapes. Cell. 2013;154:888–903. doi: 10.1016/j.cell.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harikumar A, Meshorer E. Chromatin remodeling and bivalent histone modifications in embryonic stem cells. EMBO Rep. 2015;16:1609–1619. doi: 10.15252/embr.201541011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fisher CL, Fisher AG. Chromatin states in pluripotent, differentiated, and reprogrammed cells. Curr. Opin. Genet. Dev. 2011;21:140–146. doi: 10.1016/j.gde.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 33.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piunti A, Shilatifard A. Epigenetic balance of gene expression by Polycomb and COMPASS families. Science. 2016;352:aad9780–aad9780. doi: 10.1126/science.aad9780. [DOI] [PubMed] [Google Scholar]
- 35.Kim KH, Roberts CWM. Targeting EZH2 in cancer. Nat. Med. 2016;22:128–134. doi: 10.1038/nm.4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sneeringer CJ, et al. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc. Natl. Acad. Sci. U.S.A. 2010;107:20980–20985. doi: 10.1073/pnas.1012525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yap DB, et al. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood. 2011;117:2451–2459. doi: 10.1182/blood-2010-11-321208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Béguelin W, et al. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell. 2013;23:677–692. doi: 10.1016/j.ccr.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCabe MT, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 40.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 41.Hodges C, Kirkland JG, Crabtree GR. The Many Roles of BAF (mSWI/SNF) and PBAF Complexes in Cancer. Cold Spring Harb Perspect Med. 2016;6:a026930. doi: 10.1101/cshperspect.a026930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ryan RJH, Bernstein BE. Molecular biology. Genetic events that shape the cancer epigenome. Science. 2012;336:1513–1514. doi: 10.1126/science.1223730. [DOI] [PubMed] [Google Scholar]
- 43.Zhang J, et al. Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagenesis. Nat. Med. 2015;21:1190–1198. doi: 10.1038/nm.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ryan RJH, et al. Detection of Enhancer-Associated Rearrangements Reveals Mechanisms of Oncogene Dysregulation in B-cell Lymphoma. Cancer Discov. 2015;5:1058–1071. doi: 10.1158/2159-8290.CD-15-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sen P, Shah PP, Nativio R, Berger SL. Epigenetic Mechanisms of Longevity and Aging. Cell. 2016;166:822–839. doi: 10.1016/j.cell.2016.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brownlee PM, Meisenberg C, Downs JA. The SWI/SNF chromatin remodelling complex: Its role in maintaining genome stability and preventing tumourigenesis. DNA Repair (Amst.) 2015;32:127–133. doi: 10.1016/j.dnarep.2015.04.023. [DOI] [PubMed] [Google Scholar]
- 47.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 48.Hesson LB, Hitchins MP, Ward RL. Epimutations and cancer predisposition: importance and mechanisms. Curr. Opin. Genet. Dev. 2010;20:290–298. doi: 10.1016/j.gde.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 49.Leibowitz ML, Zhang C-Z, Pellman D. Chromothripsis: A New Mechanism for Rapid Karyotype Evolution. Annu. Rev. Genet. 2015;49:183–211. doi: 10.1146/annurev-genet-120213-092228. [DOI] [PubMed] [Google Scholar]
- 50.Davis A, Gao R, Navin N. Tumor evolution: Linear, branching, neutral or punctuated? Biochim. Biophys. Acta. 2017 doi: 10.1016/j.bbcan.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brock A, Chang H, Huang S. Non-genetic heterogeneity--a mutation-independent driving force for the somatic evolution of tumours. Nat. Rev. Genet. 2009;10:336–342. doi: 10.1038/nrg2556. [DOI] [PubMed] [Google Scholar]
- 52.Shlush LI, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506:328–333. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yan H, et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cairns RA, Mak TW. Oncogenic isocitrate dehydrogenase mutations: mechanisms, models, and clinical opportunities. Cancer Discov. 2013;3:730–741. doi: 10.1158/2159-8290.CD-13-0083. [DOI] [PubMed] [Google Scholar]
- 56.Flavahan WA, et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016;529:110–114. doi: 10.1038/nature16490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nora EP, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hnisz D, et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science. 2016;351:1454–1458. doi: 10.1126/science.aad9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Katainen R, et al. CTCF/cohesin-binding sites are frequently mutated in cancer. Nat. Genet. 2015;47:818–821. doi: 10.1038/ng.3335. [DOI] [PubMed] [Google Scholar]
- 60.Filippova GN, et al. Tumor-associated zinc finger mutations in the CTCF transcription factor selectively alter tts DNA-binding specificity. Cancer Res. 2002;62:48–52. [PubMed] [Google Scholar]
- 61.Kon A, et al. Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat. Genet. 2013;45:1232–1237. doi: 10.1038/ng.2731. [DOI] [PubMed] [Google Scholar]
- 62.Gibson WJ, et al. The genomic landscape and evolution of endometrial carcinoma progression and abdominopelvic metastasis. Nat. Genet. 2016;48:848–855. doi: 10.1038/ng.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marshall AD, et al. CTCF genetic alterations in endometrial carcinoma are pro-tumorigenic. Oncogene. 2017 doi: 10.1038/onc.2017.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kemp CJ, et al. CTCF haploinsufficiency destabilizes DNA methylation and predisposes to cancer. Cell Rep. 2014;7:1020–1029. doi: 10.1016/j.celrep.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schwartzentruber J, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 66.Lewis PW, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340:857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bender S, et al. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell. 2013;24:660–672. doi: 10.1016/j.ccr.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 68.Wu G, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat. Genet. 2012;44:251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol. Cell. 2012;48:491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ragunathan K, Jih G, Moazed D. Epigenetics. Epigenetic inheritance uncoupled from sequence-specific recruitment. Science. 2015;348:1258699–1258699. doi: 10.1126/science.1258699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Audergon PNCB, et al. Epigenetics. Restricted epigenetic inheritance of H3K9 methylation. Science. 2015;348:132–135. doi: 10.1126/science.1260638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sharma SV, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Roesch A, et al. Overcoming intrinsic multidrug resistance in melanoma by blocking the mitochondrial respiratory chain of slow-cycling JARID1B(high) cells. Cancer Cell. 2013;23:811–825. doi: 10.1016/j.ccr.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liau BB, et al. Adaptive Chromatin Remodeling Drives Glioblastoma Stem Cell Plasticity and Drug Tolerance. Cell Stem Cell. 2016 doi: 10.1016/j.stem.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Black JC, et al. Hypoxia drives transient site-specific copy gain and drug-resistant gene expression. Genes Dev. 2015;29:1018–1031. doi: 10.1101/gad.259796.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Black JC, et al. KDM4A lysine demethylase induces site-specific copy gain and rereplication of regions amplified in tumors. Cell. 2013;154:541–555. doi: 10.1016/j.cell.2013.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang L, et al. DNMT3A Loss Drives Enhancer Hypomethylation in FLT3-ITD-Associated Leukemias. Cancer Cell. 2016;29:922–934. doi: 10.1016/j.ccell.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lu R, et al. Epigenetic Perturbations by Arg882-Mutated DNMT3A Potentiate Aberrant Stem Cell Gene-Expression Program and Acute Leukemia Development. Cancer Cell. 2016;30:92–107. doi: 10.1016/j.ccell.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Landau DA, et al. Locally disordered methylation forms the basis of intratumor methylome variation in chronic lymphocytic leukemia. Cancer Cell. 2014;26:813–825. doi: 10.1016/j.ccell.2014.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hansen KD, et al. Increased methylation variation in epigenetic domains across cancer types. Nat. Genet. 2011;43:768–775. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bayliss J, et al. Lowered H3K27me3 and DNA hypomethylation define poorly prognostic pediatric posterior fossa ependymomas. Sci Transl Med. 2016;8:366ra161–366ra161. doi: 10.1126/scitranslmed.aah6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mack SC, et al. Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature. 2014;506:445–450. doi: 10.1038/nature13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sharma U, Rando OJ. Metabolic Inputs into the Epigenome. Cell Metab. 2017;25:544–558. doi: 10.1016/j.cmet.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 84.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol. Cell. Biol. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hore TA, et al. Retinol and ascorbate drive erasure of epigenetic memory and enhance reprogramming to naïve pluripotency by complementary mechanisms. Proc. Natl. Acad. Sci. U.S.A. 2016;113:12202–12207. doi: 10.1073/pnas.1608679113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shyh-Chang N, et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2013;339:222–226. doi: 10.1126/science.1226603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Carey BW, Finley LWS, Cross JR, Allis CD, Thompson CB. Intracellular α-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature. 2015;518:413–416. doi: 10.1038/nature13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang Q, et al. AMPK/a-Ketoglutarate Axis Dynamically Mediates DNA Demethylation in the Prdm16 Promoter and Brown Adipogenesis. Cell Metab. 2016;24:542–554. doi: 10.1016/j.cmet.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Booth LN, Brunet A. The Aging Epigenome. Mol. Cell. 2016;62:728–744. doi: 10.1016/j.molcel.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Martinez-Jimenez CP, et al. Aging increases cell-to-cell transcriptional variability upon immune stimulation. Science. 2017;355:1433–1436. doi: 10.1126/science.aah4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Curtin K, Slattery ML, Samowitz WS. CpG island methylation in colorectal cancer: past, present and future. Patholog Res Int. 2011;2011:902674–8. doi: 10.4061/2011/902674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.O'Keefe SJD. Diet, microorganisms and their metabolites, and colon cancer. Nat Rev Gastroenterol Hepatol. 2016;13:691–706. doi: 10.1038/nrgastro.2016.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Donohoe DR, et al. The Warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation. Mol. Cell. 2012;48:612–626. doi: 10.1016/j.molcel.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shen L, et al. MGMT promoter methylation and field defect in sporadic colorectal cancer. J. Natl. Cancer Inst. 2005;97:1330–1338. doi: 10.1093/jnci/dji275. [DOI] [PubMed] [Google Scholar]
- 95.Killian JK, et al. Recurrent epimutation of SDHC in gastrointestinal stromal tumors. Sci Transl Med. 2014;6:268ra177–268ra177. doi: 10.1126/scitranslmed.3009961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumor suppression. Cell. 2008;133:403–414. doi: 10.1016/j.cell.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 97.Thienpont B, et al. Tumour hypoxia causes DNA hypermethylation by reducing TET activity. Nature. 2016;537:63–68. doi: 10.1038/nature19081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bell RE, et al. Enhancer methylation dynamics contribute to cancer plasticity and patient mortality. Genome Res. 2016;26:601–611. doi: 10.1101/gr.197194.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Madar S, Goldstein I, Rotter V. “Cancer associated fibroblasts--”more than meets the eye. Trends Mol Med. 2013;19:447–453. doi: 10.1016/j.molmed.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 100.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Timp W, Feinberg AP. Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat. Rev. Cancer. 2013;13:497–510. doi: 10.1038/nrc3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Andrieu G, Tran AH, Strissel KJ, Denis GV. BRD4 Regulates Breast Cancer Dissemination through Jagged1/Notch1 Signaling. Cancer Res. 2016;76:6555–6567. doi: 10.1158/0008-5472.CAN-16-0559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nieto MA, Huang RY-J, Jackson RA, Thiery JP. EMT: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 104.Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat. Med. 2013;19:1438–1449. doi: 10.1038/nm.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Heaphy CM, et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333:425–425. doi: 10.1126/science.1207313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CLL, Rich JN. Cancer stem cells in glioblastoma. Genes Dev. 2015;29:1203–1217. doi: 10.1101/gad.261982.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cowey CL, Rathmell WK. VHL gene mutations in renal cell carcinoma: role as a biomarker of disease outcome and drug efficacy. Curr Oncol Rep. 2009;11:94–101. doi: 10.1007/s11912-009-0015-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Carmeliet P. VEGF as a key mediator of angiogenesis in cancer. Oncology. 2005;69(Suppl 3):4–10. doi: 10.1159/000088478. [DOI] [PubMed] [Google Scholar]
- 109.Inoue S, et al. Mutant IDH1 Downregulates ATM and Alters DNA Repair and Sensitivity to DNA Damage Independent of TET2. Cancer Cell. 2016;30:337–348. doi: 10.1016/j.ccell.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Guryanova OA, et al. DNMT3A mutations promote anthracycline resistance in acute myeloid leukemia via impaired nucleosome remodeling. Nat. Med. 2016 doi: 10.1038/nm.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tessema M, et al. Re-expression of CXCL14, a common target for epigenetic silencing in lung cancer, induces tumor necrosis. Oncogene. 2010;29:5159–5170. doi: 10.1038/onc.2010.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Elias A, et al. Epigenetic silencing of death receptor 4 mediates tumor necrosis factor-related apoptosis-inducing ligand resistance in gliomas. Clin. Cancer Res. 2009;15:5457–5465. doi: 10.1158/1078-0432.CCR-09-1125. [DOI] [PubMed] [Google Scholar]
- 113.Jones PA, Issa J-PJ, Baylin S. Targeting the cancer epigenome for therapy. Nat. Rev. Genet. 2016;17:630–641. doi: 10.1038/nrg.2016.93. [DOI] [PubMed] [Google Scholar]
- 114.Easwaran H, Tsai H-C, Baylin SB. Cancer epigenetics: tumor heterogeneity, plasticity of stem-like states, and drug resistance. Mol. Cell. 2014;54:716–727. doi: 10.1016/j.molcel.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mazor T, Pankov A, Song JS, Costello JF. Intratumoral Heterogeneity of the Epigenome. Cancer Cell. 2016;29:440–451. doi: 10.1016/j.ccell.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]