Abstract
For the 1.7 million patients per year in the U.S. who receive a new cancer diagnosis, treatment decisions are largely made after a histopathology exam. Unfortunately, the gold standard of slide-based microscopic pathology suffers from high inter-observer variability and limited prognostic value due to sampling limitations and the inability to visualize tissue structures and molecular targets in their native 3D context. Here, we show that an open-top light-sheet microscope optimized for non-destructive slide-free pathology of clinical specimens enables the rapid imaging of intact tissues at high resolution over large 2D and 3D fields of view, with the same level of detail as traditional pathology. We demonstrate the utility of this technology for various applications: wide-area surface microscopy to triage surgical specimens (with ~200 μm surface irregularities), rapid intraoperative assessment of tumour-margin surfaces (12.5 sec/cm2), and volumetric assessment of optically cleared core–needle biopsies (1 mm in diameter, 2 cm in length). Light-sheet microscopy can be a versatile tool for both rapid surface microscopy and deep volumetric microscopy of human specimens.
Introduction
Pathologic features provide a gold-standard by which diseases are diagnosed, patient prognoses are determined, and treatment decisions are made [1]. In a pathology lab, the examination of a tissue biopsy or a surgically excised specimen is most reliably performed by the microscopic examination of tissues that have been formalin-fixed and paraffin-embedded (FFPE), thinly sectioned, stained, and mounted on glass slides (see Supplementary Fig. 1a). This is a labor- and time-intensive process in which only a small fraction of a specimen is sampled for microscopy. Frozen sectioning enables tissues to be rapidly sectioned, stained, and mounted on slides. However, this technique can introduce severe artifacts, especially for tissues that do not freeze well (e.g., breast). Furthermore, like FFPE histology, it is destructive of tissue and suffers from the same sampling limitations [2].
Considering the foundational role that pathology plays in patient care, it is concerning to note the large degree of inter-observer and intra-observer variability amongst pathologists in diagnosing and grading malignancies [3–6]. A number of factors limit the reproducibility (precision) and predictive power (accuracy) of conventional pathology, including: (i) sampling errors due to the sparse amounts of tissue that may realistically be prepared on glass slides and visualized [7], (ii) the inability to visualize structures of diagnostic importance within their volumetric context [8], (iii) the destructive nature of routine histology, in which tissues are heavily processed, sectioned, and mounted on slides, thereby compromising their value for downstream molecular and genetic analyses, and (iv) the high overhead of labor, equipment, and consumables (glass slides, reagents, paraffin, cassettes, etc.), all of which increase healthcare costs [9].
The ability to image intact tissues at high resolution over large areas or volumetric fields of view, with the same level of morphological and molecular contrast that is possible through conventional pathology (i.e. H&E staining and immunostaining), promises to advance the field of pathology and thereby to improve patient treatments and outcomes.
Fluorescence microscopy of intact human tissues
While 3D digital pathology using serial sections (on glass slides) is possible, it is not a viable strategy for routine clinical use due to the labor and time involved in preparing and imaging hundreds of glass slides, and aligning the digital images. Nondestructive fluorescence microscopy of human tissues has been attempted using a variety of approaches including confocal microscopy [10, 11], microscopy with UV surface excitation (MUSE) [12], wide-field structured-illumination microscopy (SIM) [13, 14], and nonlinear (e.g., multiphoton and stimulated Raman scattering) microscopy [15–17]. Compared with alternative optical-sectioning approaches, confocal and nonlinear microscopy systems provide superior resolution and contrast. However, these methods are complex and slow, often requiring raster scanning of a tightly focused laser beam in three dimensions to obtain a volumetric dataset (e.g., a recent study required 30 hrs to image a single biopsy specimen [18]). Alternatively, camera-based optical-sectioning microscopy techniques such as MUSE and SIM are advantageous for clinical applications due to their faster imaging speeds and reduced complexity, but are not intended for deep volumetric microscopy. For example, MUSE relies on the limited penetration depth of UV light to obtain a selective image of the tissue surface, thereby precluding subsurface imaging. SIM relies on tissue illumination with spatial patterns that are imaged with high contrast only at the focal plane of a microscope, thereby enabling digital rejection of out-of-focus background light (primarily lower spatial frequencies) through a demodulation algorithm. Imaging depth is therefore limited by the shot noise of the background light that impinges upon the detector array, as well as the finite dynamic range of the detector [19, 20]. Finally, reflectance-based imaging techniques such as optical coherence tomography/microscopy (OCT/OCM) have been successfully translated into clinical settings for label-free imaging of human tissues [21, 22], with a demonstrable impact on patient outcomes [23]. However, these methods are not typically designed to achieve subcellular resolution and are not well-suited for probing multiplexed and molecular targets as desired for standard-of-care clinical pathology.
Light-sheet microscopy, whose technological roots may be traced back over a century [24], has recently seen intense development for biological investigations in which rapid volumetric microscopy is desired [25–28]. The light-sheet microscopy approach achieves “optical sectioning” (rejection of out-of-focus light) by using a thin “selective” illumination plane, which generates fluorescence signal that is imaged in the orthogonal direction. The flexibility of this “dual-axis” configuration, where the illumination and collection beam paths are decoupled and may be individually optimized, contrasts with conventional single-axis microscopes in which the illumination and collection beams travel along an identical path, and where the depth of focus and resolution are coupled by the laws of diffraction (for standard Gaussian beams). For example, by utilizing low-numerical-aperture (NA) Gaussian illumination, the depth of focus of a light-sheet microscope can be extended to hundreds of microns, though at the expense of a moderately thick illumination beam waist (e.g.,7 μm in our current system). Despite this low NA, in which the light sheet is moderately thick, it is still possible to achieve high spatial resolution (sufficient to visualize nuclei) by utilizing a higher NA for the orthogonal collection beam path [29]. This is similar to conventional slide-based histology, where 5-μm thick physically sectioned tissues yield high-quality images when viewed with moderate- to high-NA microscope objectives. Finally, light-sheet microscopy systems can be simple and relatively inexpensive to build, as demonstrated by us and others (e.g., the OpenSPIM project) [30].
In summary, camera-based tissue-microscopy approaches (e.g., SIM and MUSE) are fast, but generally provide poor imaging depth (background suppression), which limits their effectiveness for volumetric imaging applications. On the other hand, 3D laser-scanning microscopy methods can image deeply but are generally complex and slow. Here, we show that light-sheet microscopy is a versatile technology that provides an elegant and simple means of rapidly imaging the irregular surfaces of surgically excised tissues, as well as enabling deep volumetric microscopy of clinical specimens. A qualitative summary of various trades offs for selected thick-tissue microscopy approaches is presented in Supplementary Table 1 for both surface-microscopy and volumetric-microscopy applications.
Open-top light-sheet microscope for clinical pathology
In early light-sheet microscopes, high-resolution volumetric imaging was achieved by slowly rotating or translating a sample embedded in an agarose gel [31]. These configurations were developed for imaging model organisms and tissues for biological investigations (e.g., developmental biology and neuroscience) and were restricted to relatively small transparent objects. A variety of variations of the original light-sheet microscopy architecture have been introduced, each of which has facilitated a growing number of biological imaging applications [32–37]. However, a versatile light-sheet microscope designed to image large centimeter-sized human specimens, for diverse applications in clinical pathology, has not been reported.
Our light-sheet microscope design utilizes an open-top architecture [38] that enables large human tissues to be conveniently and rapidly placed on top of a flat glass-plate sample holder and imaged from underneath (see Fig. 1 and Supplementary Video 1). All optical components in our system are positioned below the glass-plate sample holder, which allows the microscope to accommodate human tissues of arbitrary size and thickness without physically interfering with the illumination and collection optics. However, this geometry also presents significant optical challenges, as the highly off-axis (45-deg) illumination and collection beams are not easily index-matched into the sample, which can result in severe aberrations (e.g., coma, astigmatism, spherical aberrations).
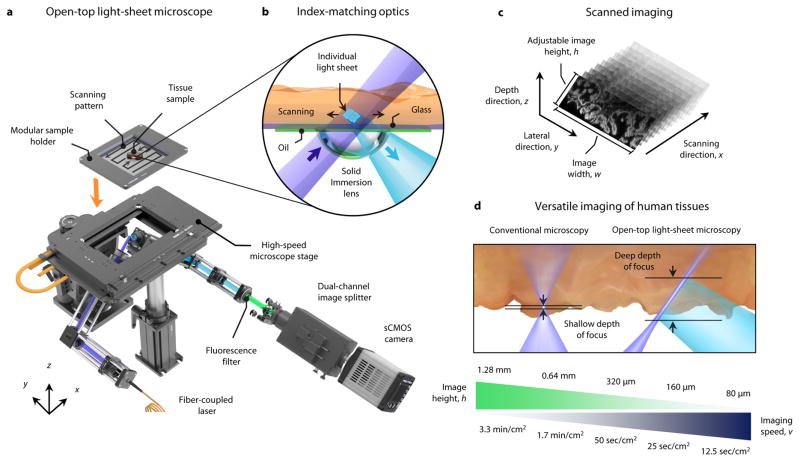
Figure 1. Open-top light-sheet microscope for clinical pathology.
a, An illumination light sheet enters the bottom surface of a tissue sample at an oblique 45-deg angle (purple). The specimen(s) is placed on a modular glass-plate sample holder, which is inserted into a two-axis translation stage and scanned in a serpentine pattern of volumetric image strips to enable 3D imaging over a large lateral extent. Fluorescence emission (cyan), which is generated along the light sheet, is collected in the orthogonal direction by an objective lens. The fluorescence signal is then transmitted through an emission filter (green) and a dual-channel image splitter (for 2 color imaging) before being imaged onto a high-speed sCMOS camera. b, To provide aberration-free imaging, a solid immersion lens (SIL) and oil layer are used for refractive-index matching of both the illumination and collection beams into and out of the glass plate and tissue sample. c, As the sample is translated in the primary scanning direction, x, oblique 2D light-sheet images with a width, w, and adjustable height, h, are captured in succession to form a 3D imaging volume. d, In contrast to conventional microscopy methods that have a shallow fixed depth of focus and slow 3D imaging rates, the deep depth of focus and adjustable vertical field of view of the open-top light-sheet microscope makes it optimal for both rapid microscopy of irregular/tilted tissue surfaces, and deep volumetric microscopy of clinical specimens. The imaging speeds shown correspond to acquiring single-channel images with height, h.
To enable aberration-free imaging, our open-top light-sheet microscope utilizes a hemispherical solid-immersion lens (SIL), which has been utilized in the past in single-axis microscopes and dual-axis confocal microscopes [39, 40], but has not been demonstrated for a light-sheet microscope in which a low-NA cylindrically focused illumination beam, and a higher-NA spherically focused collection beam, must be coupled into a tissue specimen at a large angle of incidence (Fig. 1b). The bottom of the SIL provides a curved surface that approximates the wave-front curvature of the illumination and collection beams as they transition from air into glass (or vice versa), which minimizes the introduction of aberrations at those interfaces (Supplementary Figs. 2 and 3). A thin oil layer at the top surface of the SIL, which does not evaporate or require refilling, provides an index-matched interface between the stationary SIL and a translating glass-plate sample holder (n = 1.464 +/− 0.001). This enables panoramic scanning over theoretically unlimited surface areas with diffraction-limited performance (Supplementary Figs. 3–6). In short, tissues are stage-scanned to allow camera-based imaging of serial oblique light sheets within the tissue sample (Fig. 1c and Supplementary Fig. 3). By translating the sample in one dimension, a 3D trapezoidal volume may be rapidly imaged. Adjacent 3D image strips are acquired in a serpentine pattern, enabling panoramic 3D microscopy over large areas (Supplementary Video 1). The use of a dual-channel image splitter allows simultaneous capture of two independent fluorescent channels for samples stained with multiple dyes.
For the prototype developed in this study, the width of each image is w = 2.0 mm (See Supplementary Fig. 3), which is dictated by the field of view of the collection objective. The image height, h, (i.e. the vertical field of view) is adjustable, by varying the vertical crop size of the camera, which directly affects the imaging speed, v (Fig. 1d). As shown in the results (Figs. 2 – 4), this versatility to trade-off between vertical field of view and imaging speed (Fig. 1d) allows our open-top light-sheet microscope to be optimized for specific clinical applications and tissue types.
Figure 2. Wide-area surface microscopy of fresh human prostate.
a, In preparation for post-operative pathological evaluation, radical prostatectomy specimens are grossly processed by manually cutting the organ into cross-sectional slices (3 to 5 mm in thick). b, A photograph of a fresh prostate slice (3.1 × 3.5 × 0.4 cm) imaged with the open-top light-sheet microscope. c, The tissues are stained with acridine orange for 20 sec and then imaged at a speed of v = 50 sec/cm2, which provides a vertical field of view of h = 320 μm to accommodate for any surface irregularities and tilting errors. A horizontal (en face) 2D “section” from the 3D dataset is shown on the top left, prior to surface extraction, where regions of defocus and incomplete imaging are seen (inset arrows). On the bottom right, the irregular surface of the large specimen has been digitally extracted from the 3D dataset to provide a comprehensive image of the surface. d, A depth profile of the tissue surface, along the dashed pink line, reveals significant surface irregularities (up to 200 μm) as well as a slight tilt in the glass-plate sample holder (2 μm/mm or 0.2% slope). e, A histogram of the tissue surface depth from all N=25 prostate slices imaged in this study (average and 95% confidence intervals, CI). For illustrative purposes, the depth of focus is indicated for both a conventional single-axis microscope and our open-top light-sheet microscope. f, Moderate- and high-magnification images of normal prostate glands, where a layer of both basal and epithelial cells is observed (inset arrows). A corresponding H&E histology image is shown on the right. g, A region with benign prostate (left) transitioning into prostate adenocarcinoma (right), which exhibits a crowding of glands with a single epithelial cell layer (inset arrow). h, High-grade prostate intraepithelial neoplasia, displaying stratified nuclei and tufted projections into the lumen. A corresponding H&E histology image is shown on the right.
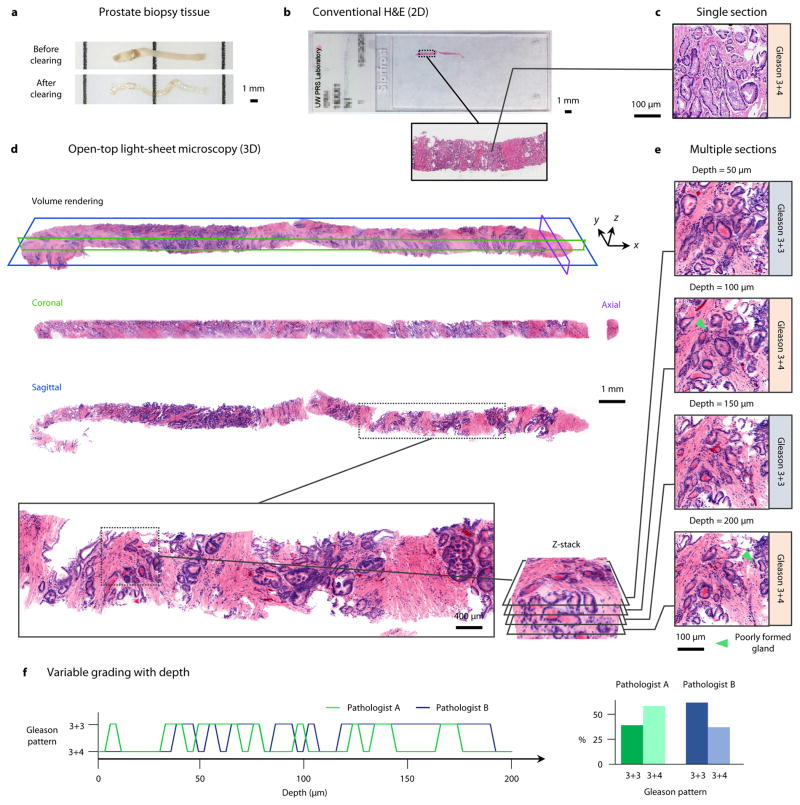
Figure 4. Volumetric dual-channel imaging of a cleared human biopsy.
a, A 2-cm long by 1-mm diameter human prostate core-needle biopsy, before and after optical clearing (overnight procedure). b, A conventional H&E-stained slide of a single 5-μm thick section of the biopsy sample. c, A high-magnification view of a Gleason 3+4 carcinoma region from the histology slide. d, The prostate biopsy was stained with DRAQ5 and eosin, and false-colored to mimic the traditional H&E color palette. A volume rendering of the biopsy is shown, which can be digitally “sectioned” into orthogonal 2D cross sections in the coronal (green), axial (purple), and sagittal (blue) planes. e, A high-magnification view of one sagittal region of interest, visualized at multiple depths (5 μm increments) to a depth of 200 μm. Gleason grading is found to differ between 3+3 and 3+4 due to tangential sectioning artifacts creating the appearance of poorly formed glands (inset arrows). f, Grading of the region of interest as a function of depth (5-μm increments) by two pathologists. When assessing individual depths, the grading varies between 3+3 (40% for Pathologist A, 62.5% for Pathologist B) and 3+4 (60% for Pathologist A, 37.5% for Pathologist B), with inter-observer agreement of 47.5%. However, after comprehensive review of the entire volumetric dataset, both pathologists agreed upon an unambiguous diagnosis of Gleason 3+3 based on the observation that structures which appeared to be “poorly formed” pattern 4 glands were tangential-sectioning artifacts of well-formed pattern 3 glands.
A variety of clinical pathology applications were explored to demonstrate the clinical utility of our open-top light sheet microscope. First, we performed post-operative surface microscopy of fresh radical prostatectomy specimens, with highly irregular surfaces, for triage-inspection of these specimens. Second, we performed rapid intraoperative surface microscopy of fresh breast tissues for guiding surgical oncology. Finally, dual-channel volumetric imaging was achieved of optically cleared core-needle biopsy specimens, demonstrating the potential to improve upon the accuracy of traditional slide-based histopathology for the diagnosis and grading of atypical lesions. Collectively, these examples demonstrate that our open-top light-sheet microscope is optimal and versatile for both rapid microscopy of large irregular tissue surfaces and comprehensive volumetric microscopy. A simplified comparison of a conventional histology protocol and our open-top light-sheet microscopy protocol is provided in Supplementary Fig. 1.
Wide-area surface microscopy of fresh human prostate
For the standard-of-care histological evaluation of a prostatectomy specimen, the organ is first manually cut (“bread loafed”) into thick slices (3 to 5 mm in thickness), Fig. 2a, from which only one histology slide is typically generated per slice. As discussed later, certain institutions will triage up to 50% of the gross slices to conserve resources. In this study, 25 large human prostate slices, approximately 3.1 × 3.5 × 0.4 cm in size (Fig. 2b), were stained with acridine orange for 20 sec, rinsed for 10 sec, and then imaged using the open-top light-sheet microscope (image height, h = 320 μm, imaging speed, v = 50 sec/cm2) within an average imaging time of <10 min per slice (Fig. 2c). A digital surface-extraction algorithm was used to display a comprehensive image of the entire tissue surface from the raw 3D dataset. This process accommodates both the irregularities of the tissue surface (~200 μm in extent for manually sliced prostate tissues), and the minor tilt of the sample plane across the entire tissue surface (typically up to 0.25% slope) (Fig. 2d). An average histogram of the tissue surface depth from all 25 samples demonstrates the typical surface irregularity seen in most unfixed prostate tissues (Fig. 2e). Tissue surface irregularities were found to be less than 200 μm, which is within the depth of focus of the open-top light-sheet microscope (zR > 200 μm, see Supplementary Fig. 4) as well as the vertical field of view (h = 320 μm, see Fig. 1) used in this case.
High-magnification views of relevant prostate pathologies are shown in Figs. 2f – 2h. Benign ducts are observed with clear lumens lined by a bilayer of nuclei (Fig. 2e) and fibromuscular stroma. Carcinoma is characterized by increased cellularity and crowded glands (Fig. 2f) surrounded by fibromuscular stroma. Finally, glands with high-grade prostatic intraepithelial neoplasia are characterized by stratified nuclei and tufted projections into the lumen (Figs. 2g – 2h). Surface-extracted open-top light-sheet microscopy images of all 25 prostate samples are shown in Supplementary Fig. 7 in comparison to slide-based histology. A video of an entire prostate surface, showing the ability to zoom in and out, may be viewed in Supplementary Video 2.
Rapid intra-operative microscopy of fresh human breast
Breast tissue is primarily composed of pliable adipose tissue, which is difficult to freeze for intraoperative assessment [2]. In contrast, our open-top light-sheet microscope can nondestructively and rapidly assess fresh breast tissues with minimal processing. To demonstrate this ability, a large slice of human breast tissue measuring 2.0 × 2.0 × 0.4 cm (Fig. 3a) was stained with acridine orange for 20 sec, rinsed for 10 sec, and rapidly imaged using our open-top light-sheet microscope (Fig. 3c). The tissue was subsequently submitted for post-operative histology of the same tissue surface (Fig. 3b).
Figure 3. Rapid intra-operative microscopy of fresh human breast.
a,b, A freshly excised specimen of human breast tissue (2.0 × 2.0 × 0.4 cm) was stained with acridine orange for 20 sec and then imaged with the open-top light-sheet microscope at a speed of v = 12.5 sec/cm2. The tissue was subsequently submitted for conventional histology. c, A surface-extracted image acquired in <1 min with representative line profiles of the tissue-surface depth, d. In (a–c) the inset arrows denote a region of invasive carcinoma (i.e. a positive margin). e, Moderate- and high-magnification images reveal a transition from benign breast tissue to invasive ductal carcinoma. f, Benign breast lobules are clearly visualized (inset arrows) and correlate with conventional histology. g, Open-top light-sheet microscopy, conventional histology, and frozen-sectioning of fibro-adipose tissue. Open-top light-sheet microscopy images and FFPE histology sections both reveal adipocytes and strands of fibrous tissue with stromal cells, whereas the frozen section contains distorted fibrous tissue and adipocytes that are distorted by the process (inset arrows).
In contrast to firmer prostate tissues [41], the increased adipose content of breast specimens allows them to sit flatter on the glass-plate sample holder with reduced tissue-surface irregularities (Fig. 3d). Thus, the open-top light-sheet microscope, which has the versatility to operate with a reduced vertical field of view, h = 160 μm, can achieve an increased imaging speed (v = 12.5 sec/cm2) that results in a total imaging time of < 1 min for a 4 cm2 tissue surface. This increased imaging speed is particularly advantageous for surgical guidance, where intraoperative pathology consultations must typically be completed within 20 min.
High-magnification views of relevant breast pathologies are shown in Figs. 3e – 3g. For example, invasive ductal carcinoma is characterized by nests and cords infiltrating through a desmoplastic stroma with an inflammatory response (Fig. 3e). Benign lobules (Fig. 3f) are visualized clearly at both medium and high magnification. The open-top light-sheet microscope comprehensively images benign fibro-adipose tissue (Fig. 3g) at a quality comparable to conventional archival FFPE histology. In contrast, frozen-section histology is poor, where adipocytes and strands of fibrous tissue with spindled stromal cells are distorted and difficult to visualize. A video of an entire breast surface, showing the ability to zoom in and out, may be viewed in Supplementary Video 3.
Volumetric dual-channel imaging of a cleared human biopsy
A prostate core-needle biopsy, ~2 cm in length by 1 mm in diameter, (Fig. 4a) was optically cleared (overnight procedure) using a protocol based on the CLARITY method [42], fluorescently stained with DRAQ5 and eosin (D&E), and imaged using the open-top light-sheet microscope. Conventional H&E histology was subsequently performed (Fig. 4b). Due to the translucency of the optically cleared biopsy, which allowed for deep imaging through the entire specimen, the image height (vertical field of view) of the microscope was increased to h = 1.28 mm for volumetric dual-channel imaging at a speed of v = 6.6 min/cm2 (total imaging time = 14 min). To minimize disruptions to long-standing clinical practice, and to facilitate rapid clinical adoption, the two fluorescent color channels were false-colored to mimic the color palette of conventional standard-of-care H&E-stained tissue sections (Supplementary Fig. 8). A volume rendering is shown in Fig. 4d, along with orthogonal 2D cross sections (sagittal, coronal, and axial) (Fig. 4d). Inspection of a H&E-stained histology slide reveals a region of carcinoma in a limited 2D view of the biopsy specimen (Fig. 4c). In contrast, the open-top light-sheet microscope enables slide-free comprehensive volumetric imaging of the entire specimen (Figs. 4d – 4e). Analysis of one region at multiple depths demonstrates that the plane of visualization influences whether the carcinoma is classified as Gleason score 3+3=6 (well-formed glands) or if the region contains the “poorly formed gland” variant of Gleason pattern 4, with an overall classification of Gleason score 3+4=7 (Figs. 4e – 4f). When assessed by two pathologists, the grading varies between 3+3 (40% for Pathologist A, 62.5% for Pathologist B) and 3+4 (60% for Pathologist A, 37.5% for Pathologist B), with an inter-observer agreement of 47.5%, which is similar to published studies [6]. An inspection of the entire volumetric dataset by both pathologists (Supplementary Videos 4 & 5) confirms that fully-formed glands exist throughout the specimen (Gleason score 3+3=6), and that any glands that appear to be poorly-formed (Gleason score 3+4 = 7) are an artifact of tangential sectioning.
Discussion
We have developed an open-top light-sheet microscope to image freshly resected human tissues and biopsy specimens for nondestructive slide-free pathology. Unlike previous implementations of light-sheet microscopy, which have focused primarily on investigations in developmental biology and neuroscience, we showcase a highly versatile design that can accommodate specimens of large cross-sectional surface area (limited only by the mechanical travel of the microscope stage, in this case 11 × 7.5 cm) and provide value as an enhancement to “gold-standard” histopathology for a variety of applications (Fig. 1). The scanning area of our open-top light-sheet microscope is larger than any previously reported light-sheet microscope [37] and the rapid 3D imaging capability of our device enables straightforward imaging of irregular and tilted tissues surfaces (Figs. 2 and 3), as well as deep volumetric microscopy of biopsy specimens in toto (Fig. 4). Furthermore, trading off between the vertical field of view and imaging speed allows the microscope to be optimized for specific applications in clinical pathology (Figs. 1 – 4).
Major advantages of our technology over traditional histology include greater sampling of clinical specimens (e.g., comprehensive nondestructive microscopy of tissue surfaces and volumes) and the visualization of tissue microarchitecture in three dimensions, both of which would improve diagnostic accuracy. In addition, slide-free nondestructive microscopic pathology has the potential to conserve healthcare resources and to enable valuable specimens to be used for downstream molecular and genetic assays. Finally, note that our methods are compatible with current pathology standards, including the use of fast-acting fluorescence dyes (acridine orange, DRAQ5, eosin) and optical clearing, which have been shown to not interfere with downstream histology or assays [10, 11, 43–45].
As discussed in greater detail below, the potentially transformative diagnostic paradigm of open-top light-sheet microscopy has been explored for three selected clinical applications: (i) post-operative evaluation and triage of large surgical-resection specimens, (ii) intra-operative imaging of large freshly resected specimens for margin assessment, and (iii) volumetric dual-channel imaging of core-needle biopsies. However, many other applications of this technology are possible, including high-throughput preclinical imaging (Supplementary Fig. 9), imaging of systemically delivered contrast agents for fluorescence-guided surgery (Supplementary Fig. 10), and volumetric immunofluorescence imaging for diagnostic pathology (Supplementary Figs 11 and 12).
Since traditional pathology is relied upon as a clinical “gold standard” for disease diagnosis and patient-management decisions, the translation of open-top light-sheet microscopy into mainstream clinical use would likely benefit from a conservative staged approach. A relatively low-risk but high-value early application of this technology would be for rapid inspection and triage of large surgical excisions such as prostatectomy specimens, breast tumors, and head-and-neck cancers. Currently, these tissues are grossly processed by dividing them into thick 0.3- to 1.0-cm thick slices (“bread loafs”), which are further processed into formalin-fixed paraffin-embedded (FFPE) tissue blocks so that a few thin (~5 μm) tissue sections may be mounted on slides, stained, and viewed by pathologists. Due to the large number of tissues that must be processed from specimens such as a radical prostatectomy, many laboratories utilize partial-sampling methods, which triage the number of inspected specimens by random selection or gross inspection [41, 46, 47]. Our system can be used for rapid nondestructive triage-inspection of fresh specimens to streamline the pathology workflow, where preliminary imaging studies demonstrate the ability to accurately identify prostate tissue slices containing carcinoma (Fig. 2). In particular, we show that our open-top light-sheet microscope is convenient for rapid wide-area imaging of irregular and tilted tissue surfaces (<200 μm for prostate bread loafs), via digital extraction of such surfaces from the 3D datasets, without the need for axial scanning or elaborate tissue-flattening and alignment mechanisms [10].
Current re-excision rates for breast-cancer patients undergoing lumpectomy procedures range from 20 – 50% [48], a statistic that underscores the need for effective intraoperative-guidance techniques. For invasive carcinoma, recent studies and consensus guidelines advocate for lumpectomies to be considered adequate when there is a “negative margin,” i.e. one in which there is no cancer at the surgical margin surface (“no tumor on ink”) [49, 50]. Therefore, a method for accurate intraoperative tumor detection at the surgical margins could significantly reduce the costs, emotional trauma, and potential complications associated with multiple surgeries. Open-top light-sheet microscopy with digital surface extraction (Figs. 2c and 3c) enables rapid wide-area microscopy of large surgical-margin surfaces, unlike traditional histology in which tissues are sectioned in the vertical direction at large intervals (3 to 10 mm) with minimal sampling of the tissue surface. Open-top light-sheet microscopy is nondestructive and does not interfere with conventional post-operative pathology, which can still be relied upon for the identification of close margins (tumor within a few millimeters of the inked margin), if necessary. However, it should be noted that the historical preference to re-excise patients with close margins (e.g., for ductal carcinoma in situ, DCIS) may have arisen because of the sampling limitations of conventional pathology, and that this safety margin may prove to be unnecessary if a comprehensive surface-imaging approach is utilized. We have shown that our technology enables comprehensive surface microscopy of lumpectomy margins at high speed (12.5 sec/cm2, or <1 min for the specimen shown in Fig. 3) with image quality that is superior to frozen-section histology, which requires 20 – 30 min to achieve (Fig. 3f). The imaging times for breast tissues are four times faster than for prostate tissues due to the improved compliance of fatty breast tissues [41], which sit flatter on the microscope stage and require a smaller vertical field of view to reliably capture the tissue surface.
The diagnosis, grading, and subtyping of various cancers, based on core-needle biopsy specimens, provides the basis for patient prognostication and for stratifying patients for various treatment options that are critical for their eventual outcomes and quality of life. Unfortunately, many studies have shown that the evaluation afforded by current histopathology techniques is often neither accurate nor reproducible, with high levels of inter-observer variability between pathologists regarding critical elements such as carcinoma grade and molecular phenotype [6, 51, 52]. For example, a recent study examined the inter-observer agreement within a panel of genitourinary pathologists in distinguishing between the “poorly formed gland” variant of Gleason pattern 4 prostate carcinoma versus tangentially sectioned Gleason pattern 3 glands (i.e., “well-formed glands”), and reported relatively poor inter-observer concordance (κ = 0.34) [53]. The distinction between Gleason pattern 3 and 4 carcinoma can result in dramatically different treatment recommendations: namely, active surveillance (Gleason pattern 3) versus radical prostatectomy (Gleason pattern 4), with its associated iatrogenic risks (incontinence, impotence, etc.). We have shown that the large degree of sampling afforded by open-top light-sheet microscopy in conjunction with optical-clearing, along with the ability to visualize complex 3D structures, has the potential to improve the precision (inter-observer concordance) and accuracy (prognostic value) of the diagnosis and grading of prostate biopsy specimens (Fig. 4), with major implications on patient treatments and outcomes.
While the current system is relatively simple in design and implementation as a versatile general-purpose device for nondestructive pathology, more specialized open-top light-sheet microscopes may be developed to address specific clinical applications. For example, in alternative system designs, the use of propagation-invariant illumination beams or tiling of thin Gaussian light sheets could result in improved resolution while maintaining a large depth of focus [54–56]. In addition, multi-view imaging methods and confocal slit detection could provide enhanced contrast and resolution [57, 58], and the parallelized use of multiple cameras could enable increased imaging speeds [59]. In particular, further improvements to our system aim to improve spatial resolution, which would have value for the definitive diagnosis of diseases.
Outlook
There is institutional inertia that hinders the adoption of incremental changes in the centuries-old technological framework of histopathology, a practice that is foundational to patient-management decisions. For example, attempts to digitize histology with whole-slide scanners have not necessarily simplified the practice of preparing and interpreting histology slides, but have added time and expensive equipment to the pathology workflow without clear diagnostic benefits. In this study, we have developed an open-top light-sheet microscopy system that circumvents the need to prepare histology slides through nondestructive imaging of fresh clinical specimens that are simply placed on a glass plate (Fig. 1). This easy-to-use technology is highly versatile for a diversity of applications in clinical pathology, enabling both rapid surface microscopy and volumetric imaging of clinical specimens, with implications for streamlining the pathology workflow (Fig. 2), guiding surgical oncology (Fig. 3), and improving the diagnosis and grading of biopsied lesions (Fig. 4). Future studies are needed to investigate the capabilities of this technology to improve treatments and patient outcomes. Furthermore, these technologies will lead naturally to efforts in computer-aided diagnosis and automated image interpretation.
Materials and methods
Open-top light-sheet microscope design
An optical schematic of the system is shown in Supplementary Fig. 3, along with ZEMAX (ZEMAX, Kirkland, WA) ray-tracing simulations of the microscope performance. The illumination beam, emitted from a single-mode fiber with numerical aperture (NA) = 0.12, is collimated with a lens, L1 (f = 25 mm), reflected at 45-deg with a mirror, M1, and focused into the sample through a cylindrical lens, C1 (f = 100 mm), resulting in an illumination light sheet of NA = 0.03. These illumination optics generate a light sheet that is approximately 2 mm wide (FWHM), with a full-width half-maximum (FWHM) thickness of 7 μm (wavelength-dependent) at the beam waist, and a depth of focus of ~400 μm (2zR, wavelength-dependent), as shown in Supplementary Fig. 4. The illumination beam is then transmitted through a custom 15-mm diameter fused-silica solid-immersion lens (SIL) (BMW Optics, Ottawa, Canada), a 0.5 mm thick index-matching oil layer (Cargille Labs, Cedar Grove, NJ), and a 1-mm thick glass-plate sample holder (fused silica, 10 × 10 cm). The custom SIL is a hemisphere with a radius of 7.5 mm in which the center thickness has been reduced from 7.5 mm to 7.375 mm such that the illumination and collection beams are index-matched with minimal aberrations when they are positioned at a distance of 125 μm into the sample.
Fluorescence generated from the illumination light sheet within the tissue sample is collected by an objective lens, L2, (f = 43.47 mm, NA = 0.28, Olympus, Tokyo, Japan), reflected at 45-deg from mirror, M2, focused through a tube lens, L3 (f =150 mm), transmitted through an emission filter, F1, and then transmitted through an additional low-power cylindrical lens, C2 (f = 2000 mm, IDEX Optics & Photonics, Albuquerque, New Mexico), to mitigate astigmatism. The total magnification of the collection arm, from sample to detector, is 1.46x(fL3/fL2) = 5, where the SIL provides a magnification factor that is equivalent to refractive index, n = 1.46. A dual-channel image-splitting module (W-VIEW GEMINI, Hamamatsu Photonics, Japan) is used to image two fluorescence imaging channels onto two halves of a high-speed sCMOS detector array with 2048 × 2048 pixels (ORCA Flash v2.0, Hamamatsu Photonics, Japan). Images are transferred at maximum data-transfer rates to a dedicated workstation comprised of a high-speed RAID0 hard disk using a CameraLink interface (Firebird PCI Express, Active Silicon, Severna Park, Maryland). In addition to correcting aberrations, the SIL also increases the illumination and collection NA by a factor that is equivalent to the refractive index of the SIL material (fused silica, n ~ 1.46) [39, 40]. This collection NA provides a field of view of 2 mm, and in-plane resolution (in the plane of the light sheet) of 1.5 μm within the sample. The images are acquired by the sCMOS camera at a sampling pitch of 1.25 μm/pixel along the plane of the light sheets, and a sampling pitch of × = 1.25 μm along the primary scanning direction (see Fig. 1c). Serpentine scanning of serial volumetric imaging strips is achieved with a high-speed motorized stage (MLS-203-1, Thorlabs, Newton, NJ), where the image strips are acquired with a separation of y = 2 mm (secondary scanning direction).
Imaging speed is limited by the camera frame rate for the acquisition of serial light-sheet images spaced by 1.25 μm. For imaging fresh human tissues, a 256 × 2048 sub-region (crop) of the sCMOS chip is typically utilized (0.32 by 2.56 mm field of view), which enables a frame rate of 800 frames per second. This frame rate results in an overall imaging speed of v = 50 sec/cm2 of tissue. For more-pliable breast tissues, a 64 by 2048 sub-region of the sCMOS chip is utilized (0.80 by 2.56 mm field of view), which enables a frame rate of 3200 frames per second and an overall imaging speed of v = 12.5 sec/cm2 of tissue. Finally, for deeper volumetric imaging of optically cleared samples, a larger 1024 by 2048 sub-region of the sCMOS chip is utilized (1.28 by 2.56 mm field of view) with a frame rate of 200 frames/sec and an overall imaging speed of 3.3 min/cm2 of tissue. For dual-channel imaging, the required imaging sub-region on the sCMOS chip is doubled, resulting in half the frame rate and imaging speed (e.g., 100 frames/sec and an overall imaging speed of 6.6 min/cm2 for the dual-channel imaging shown in Fig. 4).
Image processing
Raw open-top light-sheet microscopy images undergo several image processing steps to render the final images. The processing is written using a combination of MATLAB and ImageJ tools in Miji (a Java package that allows transfer of imaging data between MATLAB and ImageJ).
Each individual image must be flat-field corrected for variations in intensity across the Gaussian light sheet, vignetting of the objective and tube lens collection optics, inter-pixel variations in the sensitivity of the sCMOS camera, and intrinsic aberrations in the open-top light-sheet microscope. To account for all of these effects, prior to imaging, a drop of a homogeneous fluorescence staining solution is imaged to acquire a reference image. Every subsequent raw image is normalized by this reference image to perform flat-field correction.
The captured images correspond to oblique planes oriented at 45-deg with respect to the sample surface. However, when being stored digitally, these images form a data cube in which the images are oriented at 90 deg. Therefore, in post-processing, the images must be sheared by 45-deg in the x-z plane to create a trapezoidal data volume in which the tissue structure is undistorted. The sheared en-face images may exhibit minor striping artifacts due to mechanical vibrations. These artifacts are removed in ImageJ using a stripe filter based on a combined wavelet-Fourier filter [60].
After stripe-filtering, each image strip is registered to adjacent image strips using the ImageJ grid-stitching algorithm [61]. For digital registration of adjacent image strips, an overlap of 500 μm is used between the image strips, and a linear blending of each image strip is used to generate the final image mosaic. This operation is initially performed for a single en-face image mosaic in the x-y plane. The resulting registration and blending operations, determined from this single plane, are then applied to all of the imaging data at each depth, z. The imaging data now represents a 3D volume of multiple registered, blended, and stitched image mosaics. For fresh, scattering tissues, to further reduce the dimensionality of the data, and to extract only the irregular surface of the sample, an extended-depth of field (EDF) algorithm may be used [62]. For images acquired using two fluorophores, the cytoplasmic (eosin) and nuclear (DRAQ5) channels were false-colored to resemble H&E histology as in a recent study [43], using the algorithm described by Giacomelli et al. [63].
Collection and analysis of fresh human prostate tissues
De-identified fresh prostate samples (25 from 19 patients) were obtained prospectively from an IRB-approved genitourinary biorepository, with patient consent, and were imaged using the open-top light-sheet microscope. After imaging, the samples were either snap frozen in a slurry of liquid nitrogen-cooled isopentane (N=24) or formalin-fixed and paraffin-embedded (N=1),. Multiple H&E-stained slides were prepared from each tissue sample (en face sectioning near the imaged tissue surface), and the slide that was most representative of the open-top light-sheet microscope image was scanned to produce a whole-slide image (Aperio, Leica, Germany). Approval was obtained from the University of Washington Institutional Review Board. The descriptive study measuring irregularities in prostate tissue surface topology did not measure an effect size, and thus did not undergo a pre-specified power analysis. We halted the study after collecting 25 samples, when additional samples would have led to minimal changes in the overall standard deviation of the measured tissue surface irregularities.
Collection of fresh human breast tissue
Human breast samples were collected through the University of Washington Northwest BioTrust (NWBT) under an IRB-approved protocol with patient consent. Imaging was performed immediately after surgery, after which H&E slides were obtained. Frozen sectioning was also performed on one human breast sample.
Fresh tissue processing
For fresh tissue imaging, samples were stained with 1 mM of acridine orange for 20 sec, and then rinsed in phosphate-buffered solution (PBS) for 10 sec, blotted dry, and immediately imaged.
Collection of human prostate needle core biopsies
Core-needle biopsy specimens were obtained from fresh ex vivo prostatectomy specimens using an 18-gauge (approximately 1 mm inner diameter) needle biopsy device (Bard Max Core, Bard Biopsy, Tempe, AZ). The prostate core was immediately placed in 10% neutral buffered formalin, where it was maintained at room temperature for 48 hrs. All samples were obtained from an IRB-approved genitourinary biorepository with patient consent.
Human prostate core-needle biopsy processing
The prostate core-needle biopsy was removed from formalin and rinsed for 10 min in 1X PBS. Next, the biopsy was processed using an automated electrophoretic tissue-clearing system (X-CLARITY, Logos, Annandale, VA) for 5 hrs at 33-deg C. Following X-CLARITY processing, the biopsy was rinsed at room temperature in PBST (0.2 % Triton X100 in 1X PBS v/v) for 6 hrs, with agitation. The biopsy was then stained by submerging it in a 1:1000 v/v solution of eosin Y (Surgipath SelecTech Alcoholic Eosin Y 515) in 1X PBS for 20 min. After removal from the eosin solution, the biopsy was rinsed for 6 min in 3 changes of distilled water (2 min per rinse). The biopsy was then incubated in a 1:1000 v/v solution of 5-mM DRAQ5 (Thermo Scientific) in 1X PBS for 20 min. Finally, the biopsy was rinsed in 1X PBS for 1 min before being transferred into dilute 2,2′ thiodiethanol (Sigma Aldrich) with a refractive index of n ~ 1.46 for 30 min (for refractive index-matching). All steps listed in this section were performed at room temperature.
Blinded evaluation of biopsy regions of interest
Forty images (in 5 μm increments to a depth of 200 μm) were obtained from five independent regions of interest for a total of 200 images. The images were randomized and de-identified for a blinded pathology analysis. Pathologists assessed each image for the presence or absence of carcinoma and grade (if applicable). The final analysis consisted of only the single region shown in Fig. 4. Images from the additional four ROIs were included in the randomization to reduce bias during blinded pathologist assessment.
Data availability
All raw and processed imaging data generated in this work, including the representative images provided in the manuscript and supplementary material are available from the authors upon request.
Code availability
Computer code for the results in this study are available in GitHub.
Supplementary Material
Acknowledgments
The authors thank Dr. Nader Sanai of the Barrow Neurological Institute, St. Joseph’s Hospital and Medical Center, Phoenix, AZ for providing the human glioma sample, and Dr. Behzad Najafian for providing the human kidney tissue sample. Human breast specimens were provided by the NorthWest BioTrust, which is supported in part by the NCI of the National Institutes of Health (P30CA015704). Human prostate specimens were provided by the GU Specimen Biorepository, University of Washington, which is supported by resources of the Department of Defense Prostate Cancer Research Program (W81XWH-14-2-0183), the Pacific Northwest Prostate Cancer SPORE (P50CA97186), a PO1 NIH grant (PO1 CA163227), and the Institute for Prostate Cancer Research of the University of Washington. This work was also supported by resources from the NIH/NCI (R01 CA175391 and F32 CA213615-01A1), the NIH/NIDCR (R01 DE023497), the University of Washington Royalty Research Fund, and a UW CoMotion Innovation Award.
Footnotes
Contributions
A.G., N.R., and J.L. designed the studies. A.G. and J.L. designed the open-top light-sheet microscope. A.G., Y.C., C.Y, and P.W fabricated the microscope. A.G., N.R. and Y.C. imaged the tissue samples. E.M. prepared the optically cleared tissue samples. N.R. and L.T. performed the blinded evaluation of prostate samples. N.R. and L.T. provided pathological diagnosis of all samples. A.G., N.R., Y.C., E.M., C.Y., P.W., Y.W., L.T., and J.L. prepared the manuscript.
Competing interests
The authors declare no competing financial interests.
References
- 1.Surveillance Research Program, N.C.I. [12-13-2016];Fast Stats: An interactive tool for access to SEER cancer statistics. 2016 Available from: https://seer.cancer.gov/faststats/
- 2.Barakat FH, Sulaiman I, Sughayer MA. Reliability of frozen section in breast sentinel lymph node examination. Breast Cancer. 2014;21(5):576–82. doi: 10.1007/s12282-012-0431-5. [DOI] [PubMed] [Google Scholar]
- 3.McKenney JK, et al. The potential impact of reproducibility of Gleason grading in men with early stage prostate cancer managed by active surveillance: a multi-institutional study. J Urol. 2011;186(2):465–9. doi: 10.1016/j.juro.2011.03.115. [DOI] [PubMed] [Google Scholar]
- 4.Shah RB, et al. Diagnosis of Gleason pattern 5 prostate adenocarcinoma on core needle biopsy: an interobserver reproducibility study among urologic pathologists. Am J Surg Pathol. 2015;39(9):1242–9. doi: 10.1097/PAS.0000000000000442. [DOI] [PubMed] [Google Scholar]
- 5.Meyer JS, et al. Breast carcinoma malignancy grading by Bloom-Richardson system vs proliferation index: reproducibility of grade and advantages of proliferation index. Mod Pathol. 2005;18(8):1067–78. doi: 10.1038/modpathol.3800388. [DOI] [PubMed] [Google Scholar]
- 6.Tozbikian G, et al. Atypical Ductal Hyperplasia Bordering on Ductal Carcinoma In Situ: Interobserver Variability and Outcomes in 105 Cases. Int J Surg Pathol. 2016 doi: 10.1177/1066896916662154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bedossa P, Dargere D, Paradis V. Sampling variability of liver fibrosis in chronic hepatitis C. Hepatology. 2003;38(6):1449–57. doi: 10.1016/j.hep.2003.09.022. [DOI] [PubMed] [Google Scholar]
- 8.Roberts N, et al. Toward Routine Use of 3D Histopathology as a Research Tool. Am J Pathol. 2012;180(5):1835–42. doi: 10.1016/j.ajpath.2012.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlson RO, Amirahmadi F, Hernandez JS. A primer on the cost of quality for improvement of laboratory and pathology specimen processes. Am J Clin Pathol. 2012;138(3):347–54. doi: 10.1309/AJCPSMQYAF6X1HUT. [DOI] [PubMed] [Google Scholar]
- 10.Gareau DS, et al. Confocal mosaicing microscopy in Mohs skin excisions: feasibility of rapid surgical pathology. J Biomed Opt. 2008;13(5):054001. doi: 10.1117/1.2981828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Royen ME, et al. Three-dimensional microscopic analysis of clinical prostate specimens. Histopathology. 2016;69(6):985–992. doi: 10.1111/his.13022. [DOI] [PubMed] [Google Scholar]
- 12.Fereidouni F, et al. SPIE BiOS. International Society for Optics and Photonics; 2016. Microscopy with UV Surface Excitation (MUSE) for slide-free histology and pathology imaging. [Google Scholar]
- 13.Wang M, et al. High-Resolution Rapid Diagnostic Imaging of Whole Prostate Biopsies Using Video-Rate Fluorescence Structured Illumination Microscopy. Cancer Res. 2015;75(19):4032–41. doi: 10.1158/0008-5472.CAN-14-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang M, et al. Gigapixel surface imaging of radical prostatectomy specimens for comprehensive detection of cancer-positive surgical margins using structured illumination microscopy. Sci Rep. 2016;6:27419. doi: 10.1038/srep27419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tao YK, et al. Assessment of breast pathologies using nonlinear microscopy. Proc Natl Acad Sci U S A. 2014;111(43):15304–9. doi: 10.1073/pnas.1416955111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orringer DA, et al. Rapid intraoperative histology of unprocessed surgical specimens via fibre-laser-based stimulated Raman scattering microscopy. Nature Biomedical Engineering. 2017;1(2):0027. doi: 10.1038/s41551-016-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tu H, et al. Stain-free histopathology by programmable supercontinuum pulses. Nat Photonics. 2016;10(8):534–540. doi: 10.1038/nphoton.2016.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olson E, Levene MJ, Torres R. Multiphoton microscopy with clearing for three dimensional histology of kidney biopsies. Biomed Opt Express. 2016;7(8):3089–96. doi: 10.1364/BOE.7.003089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jonkman J, Brown CM. Any Way You Slice It-A Comparison of Confocal Microscopy Techniques. J Biomol Tech. 2015;26(2):54–65. doi: 10.7171/jbt.15-2602-003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mertz J. Optical sectioning microscopy with planar or structured illumination. Nat Methods. 2011;8(10):811–9. doi: 10.1038/nmeth.1709. [DOI] [PubMed] [Google Scholar]
- 21.Vakoc BJ, et al. Three-dimensional microscopy of the tumor microenvironment in vivo using optical frequency domain imaging. Nat Med. 2009;15(10):1219–23. doi: 10.1038/nm.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Assayag O, et al. Large field, high resolution full-field optical coherence tomography: a pre-clinical study of human breast tissue and cancer assessment. Technol Cancer Res Treat. 2014;13(5):455–68. doi: 10.7785/tcrtexpress.2013.600254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zysk AM, et al. Optical coherence tomography: a review of clinical development from bench to bedside. J Biomed Opt. 2007;12(5):051403. doi: 10.1117/1.2793736. [DOI] [PubMed] [Google Scholar]
- 24.Siedentopf H, Zsigmondy R. Uber Sichtbarmachung und Gröβenbestimmung ultramikoskopischer Teilchen, mit besonderer Anwendung auf Goldrubingläser. Annalen der Physik. 1902;315(1):1–39. [Google Scholar]
- 25.Dodt HU, et al. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat Methods. 2007;4(4):331–6. doi: 10.1038/nmeth1036. [DOI] [PubMed] [Google Scholar]
- 26.Keller PJ, et al. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science. 2008;322(5904):1065–9. doi: 10.1126/science.1162493. [DOI] [PubMed] [Google Scholar]
- 27.Cella Zanacchi F, et al. Live-cell 3D super-resolution imaging in thick biological samples. Nat Methods. 2011;8(12):1047–9. doi: 10.1038/nmeth.1744. [DOI] [PubMed] [Google Scholar]
- 28.Huisken J, et al. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science. 2004;305(5686):1007–9. doi: 10.1126/science.1100035. [DOI] [PubMed] [Google Scholar]
- 29.Glaser AK, Wang Y, Liu JT. Assessing the imaging performance of light sheet microscopies in highly scattering tissues. Biomed Opt Express. 2016;7(2):454–66. doi: 10.1364/BOE.7.000454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pitrone PG, et al. OpenSPIM: an open-access light-sheet microscopy platform. Nat Methods. 2013;10(7):598–9. doi: 10.1038/nmeth.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reynaud EG, et al. Guide to light-sheet microscopy for adventurous biologists. Nat Methods. 2015;12(1):30–4. doi: 10.1038/nmeth.3222. [DOI] [PubMed] [Google Scholar]
- 32.Kumar A, et al. Dual-view plane illumination microscopy for rapid and spatially isotropic imaging. Nat Protoc. 2014;9(11):2555–73. doi: 10.1038/nprot.2014.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strnad P, et al. Inverted light-sheet microscope for imaging mouse pre-implantation development. Nat Methods. 2016;13(2):139–42. doi: 10.1038/nmeth.3690. [DOI] [PubMed] [Google Scholar]
- 34.Yang Z, et al. An inverted light sheet microscope optimized for studies in neuroscience. Conference on Lasers and Electro-Optics (2016), paper ATu3O.5; 2016. p. ATu3O.5. [Google Scholar]
- 35.Strnad P, et al. Inverted light-sheet microscope for imaging mouse pre-implantation development. Nature Methods. 2016;13(2):139–42. doi: 10.1038/nmeth.3690. [DOI] [PubMed] [Google Scholar]
- 36.Wu Y, et al. Inverted selective plane illumination microscopy (iSPIM) enables coupled cell identity lineaging and neurodevelopmental imaging in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2011;108(43):17708–13. doi: 10.1073/pnas.1108494108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Power RM, Huisken J. A guide to light-sheet fluorescence microscopy for multiscale imaging. Nature Methods. 2017;14(4):360–373. doi: 10.1038/nmeth.4224. [DOI] [PubMed] [Google Scholar]
- 38.McGorty R, et al. Open-top selective plane illumination microscope for conventionally mounted specimens. Opt Express. 2015;23(12):16142–53. doi: 10.1364/OE.23.016142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kino GS S.U. (USA) Optoelectronics ‘99 - Integrated Optoelectronic Devices. International Society for Optics and Photonics; 1999. Applications and theory of the solid immersion lens. [Google Scholar]
- 40.Liu JT, et al. Efficient rejection of scattered light enables deep optical sectioning in turbid media with low-numerical-aperture optics in a dual-axis confocal architecture. J Biomed Opt. 2008;13(3):034020. doi: 10.1117/1.2939428. [DOI] [PubMed] [Google Scholar]
- 41.Hall GS, Kramer CE, Epstein JI. Evaluation of radical prostatectomy specimens. A comparative analysis of sampling methods. Am J Surg Pathol. 1992;16(4):315–24. doi: 10.1097/00000478-199204000-00001. [DOI] [PubMed] [Google Scholar]
- 42.Chung K, et al. Structural and molecular interrogation of intact biological systems. Nature. 2013;497(7449):332–7. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elfer KN, et al. DRAQ5 and Eosin (‘D&E’) as an Analog to Hematoxylin and Eosin for Rapid Fluorescence Histology of Fresh Tissues. PLoS One. 2016;11(10):e0165530. doi: 10.1371/journal.pone.0165530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moffitt JR, et al. High-performance multiplexed fluorescence in situ hybridization in culture and tissue with matrix imprinting and clearing. Proc Natl Acad Sci U S A. 2016;113(50):14456–14461. doi: 10.1073/pnas.1617699113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen F, et al. Nanoscale imaging of RNA with expansion microscopy. Nat Methods. 2016;13(8):679–84. doi: 10.1038/nmeth.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmid HP, McNeal JE. An abbreviated standard procedure for accurate tumor volume estimation in prostate cancer. Am J Surg Pathol. 1992;16(2):184–91. doi: 10.1097/00000478-199202000-00012. [DOI] [PubMed] [Google Scholar]
- 47.Sehdev AE, Pan CC, Epstein JI. Comparative analysis of sampling methods for grossing radical prostatectomy specimens performed for nonpalpable (stage T1c) prostatic adenocarcinoma. Hum Pathol. 2001;32(5):494–9. doi: 10.1053/hupa.2001.24322. [DOI] [PubMed] [Google Scholar]
- 48.Jacobs L. Positive margins: the challenge continues for breast surgeons. Ann Surg Oncol. 2008;15(5):1271–2. doi: 10.1245/s10434-007-9766-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moran MS, et al. Society of Surgical Oncology-American Society for Radiation Oncology consensus guideline on margins for breast-conserving surgery with whole-breast irradiation in stages I and II invasive breast cancer. J Clin Oncol. 2014;32(14):1507–15. doi: 10.1200/JCO.2013.53.3935. [DOI] [PubMed] [Google Scholar]
- 50.Adams BJ, et al. The role of margin status and reexcision in local recurrence following breast conservation surgery. Ann Surg Oncol. 2013;20(7):2250–5. doi: 10.1245/s10434-012-2831-3. [DOI] [PubMed] [Google Scholar]
- 51.Meyer JS, et al. Breast carcinoma malignancy grading by Bloom-Richardson system vs proliferation index: reproducibility of grade and advantages of proliferation index. Mod Pathol. 2005;18(8):1067–78. doi: 10.1038/modpathol.3800388. [DOI] [PubMed] [Google Scholar]
- 52.Singletary SE, et al. Revision of the American Joint Committee on Cancer staging system for breast cancer. J Clin Oncol. 2002;20(17):3628–36. doi: 10.1200/JCO.2002.02.026. [DOI] [PubMed] [Google Scholar]
- 53.Zhou M, et al. Diagnosis of “Poorly Formed Glands” Gleason Pattern 4 Prostatic Adenocarcinoma on Needle Biopsy: An Interobserver Reproducibility Study Among Urologic Pathologists With Recommendations. Am J Surg Pathol. 2015;39(10):1331–9. doi: 10.1097/PAS.0000000000000457. [DOI] [PubMed] [Google Scholar]
- 54.Vettenburg T, et al. Light-sheet microscopy using an Airy beam. Nat Methods. 2014;11(5):541–4. doi: 10.1038/nmeth.2922. [DOI] [PubMed] [Google Scholar]
- 55.Fahrbach FO, Rohrbach A. Propagation stability of self-reconstructing Bessel beams enables contrast-enhanced imaging in thick media. Nat Commun. 2012;3:632. doi: 10.1038/ncomms1646. [DOI] [PubMed] [Google Scholar]
- 56.Fu Q, et al. Imaging multicellular specimens with real-time optimized tiling light-sheet selective plane illumination microscopy. Nat Commun. 2016;7:11088. doi: 10.1038/ncomms11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Medeiros G, et al. Confocal multiview light-sheet microscopy. Nat Commun. 2015;6:8881. doi: 10.1038/ncomms9881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tomer R, et al. Quantitative high-speed imaging of entire developing embryos with simultaneous multiview light-sheet microscopy. Nat Methods. 2012;9(7):755–63. doi: 10.1038/nmeth.2062. [DOI] [PubMed] [Google Scholar]
- 59.Dean KM, et al. Imaging subcellular dynamics with fast and light-efficient volumetrically parallelized microscopy. Optica. 2017;4(2):263–271. doi: 10.1364/OPTICA.4.000263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Munch B, et al. Stripe and ring artifact removal with combined wavelet--Fourier filtering. Opt Express. 2009;17(10):8567–91. doi: 10.1364/oe.17.008567. [DOI] [PubMed] [Google Scholar]
- 61.Preibisch S, Saalfeld S, Tomancak P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics. 2009;25(11):1463–5. doi: 10.1093/bioinformatics/btp184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aguet F, Van De Ville D, Unser M. Model-based 2.5-d deconvolution for extended depth of field in brightfield microscopy. IEEE Trans Image Process. 2008;17(7):1144–53. doi: 10.1109/TIP.2008.924393. [DOI] [PubMed] [Google Scholar]
- 63.Giacomelli MG, et al. Virtual Hematoxylin and Eosin Transillumination Microscopy Using Epi-Fluorescence Imaging. PLoS One. 2016;11(8):e0159337. doi: 10.1371/journal.pone.0159337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw and processed imaging data generated in this work, including the representative images provided in the manuscript and supplementary material are available from the authors upon request.