SYNOPSIS
Substrate specificity is critically important for enzyme catalysis. In the adrenaline synthesising enzyme phenylethanolamine N-methyltransferase (PNMT), minor changes in substituents can convert substrates into inhibitors. Here we report the crystal structures of 6 human PNMT complexes including the first structure of the enzyme in complex with its physiological ligand R-noradrenaline: determining this structure required rapid soak methods because of the tendency for noradrenaline to oxidize. Comparison of the PNMT:noradrenaline complex with the previously determined PNMT:octopamine complex demonstrates that these two substrates form almost equivalent interactions with the enzyme and show that octopamine is a valid model substrate for PNMT. The crystal structures illustrate the adaptability of the PNMT substrate binding site in accepting multi-fused ring systems such as substituted norbornene as well as noradrenochrome, the oxidation product of noradrenaline. Our data explain why only a subset of ligands recognised by PNMT are methylated by the enzyme: bulky substituents dictate the binding orientation of the ligand and can thereby place the acceptor amine too far from the donor methyl group for methylation to occur. We also show how the critical Glu185 catalytic residue can be replaced by Asp with a loss of only 10-fold catalytic efficiency because protein backbone movements place the Asp185 carboxylate almost coincident with the carboxylate of Glu185. Conversely, replacement of Glu185 by Gln reduces catalytic efficiency almost 300-fold not only because of the loss of charge but also because the variant residue does not adopt the same conformation as Glu185.
Keywords: Noradrenochrome, enzyme catalysis, substrate specificity, PNMT variants, catecholamine synthesis
INTRODUCTION
Small molecule methyltransferase (MT) enzymes catalyse the S-adenosyl-l-methionine (AdoMet) mediated synthesis and degradation of a wide range of molecules including glycine (catalysed by GNMT) [1], histidine (HNMT) [2], creatine (GAMT) [3] and catecholamines (COMT, PNMT) [4–6]. Phenylethanolamine N-methyltransferase, or PNMT, the enzyme that catalyses the biosynthesis of adrenaline (epinephrine) from noradrenaline (norepinephrine) (Figure 1A), is of particular interest because CNS adrenaline is implicated in a wide range of physiological and pathological conditions. These include the central control of blood pressure [7, 8], pituitary hormone secretion [9, 10], ethanol intoxication [11], Parkinson’s disease [12], and the neurodegeneration observed in Alzheimer’s disease [13]. To help elucidate the role of CNS adrenaline, potent selective and CNS-active PNMT inhibitors have been developed [14–22].
Figure 1. Catalytic reaction and chemical structures.
A, Schematic of the reaction catalysed by PNMT involving transfer of a methyl group from AdoMet, 3, to noradrenaline, 1, forming adrenaline, 2, and AdoHcy, 4. B, Chemical structures of substrates and inhibitors described herein.
PNMT can accept substrates other than its physiological substrate noradrenaline (1, Figure 1A), including hydroxyphenylethanolamines and halogen-substituted phenylethanolamines (Figure 1B) [4, 23–25]. Although 1 is the physiological substrate of PNMT, its interaction with PNMT has not yet been characterized structurally due to its rapid air oxidation to noradrenochrome (5, Figure 1B). For kinetic studies, the PNMT model substrate phenylethanolamine is preferred (6, Figure 1B, Table 1) [26], but p-octopamine has been used (7, Figure 1B, Table 1) [26]. A crystal structure of human PNMT (hPNMT) in complex with 7 has been determined previously, and was used to develop a model of the PNMT:1 complex and to propose the enzyme mechanism [27]. In addition, semi-rigid and rigid analogues of substrates and inhibitors of PNMT have been used to develop a structure-activity profile of the enzyme [27–30]. These studies have shown that 3-trifluoromethyl phenylethanolamine (8, Figure 1B) and its rigid analogue anti-9-amino-6-(trifluoromethyl)benzonorbornene (9, Figure 1B) are both substrates for PNMT (Table 1) [31]. However, the position of the aromatic substituent influences recognition and catalysis of PNMT substrates because another rigid analogue of 8, anti-9-amino-5-(trifluoromethyl)benzonorbornene (10, Figure 1B), is not methylated by the enzyme and instead inhibits PNMT [28, 31].
Table 1.
| Enzyme:substrate | Km (µM) | kcat (min−1) | kcat/ Km (M−1min−1) | Ki (µM) |
|---|---|---|---|---|
| wt:6b | 100 ± 4 | 2.8 ± 0.1 | 2.8 × 104 | - |
| E185A:6c | 100 ± 6 | 0.17 ± 0.01 | 1.7 × 103 | - |
| E185D:6 c | 320 ± 32 | 0.85 ± 0.04 | 2.7 × 103 | - |
| E185Q:6c | 100 ± 6 | 0.01 ± 0.001 | 99.0 | - |
| wt:7c | 5.5 ± 0.6 | 1.2 ± 0.1 | 2.2 × 105 | - |
| wt:8b | 0.55 ± 0.08 | 0.50 ± 0.01 | 9.1 × 105 | - |
| wt:9b | 1.5 ± 0.2 | 0.37 ± 0.04 | 2.5 × 105 | - |
| wt:10d | - | - | - | 13 ± 1 |
Our previous work also highlighted a possible role in catalysis for the PNMT residue Glu185 and we had suggested a model whereby, prior to methylation, Glu185 assists in deprotonation of the substrate amine via a water [27, 30]. Mutation of Glu185 supports this model: replacement with Ala (E185A), Gln (E185Q) or Asp (E185D) has little effect on substrate binding (Km is unaffected or increases by 3-fold, Table 1), but by contrast these mutations reduce the catalytic efficiency kcat/ Km by 10–20-fold (E185A and E185D) and up to ~300 fold (E185Q) (Table 1).
We sought to determine the molecular basis for these observations by characterising the crystal structures of PNMT in complex with (i) the physiological substrate noradrenaline 1, (ii) its air-oxidized product noradrenochrome 5, (iii) the 3-trifluoromethyl phenylethanolamine substrate 8 and (iv) the anti-9-amino-5-(trifluoromethyl)benzonorbornene inhibitor 10. In addition, we investigated the structures of (v) the E185D and (vi) the E185Q variants of PNMT in complex with 7. Our results support the previously proposed model of the hPNMT:1 complex, confirm that octopamine is an appropriate model substrate, and reveal that a trifluoromethyl substituent dictates ligand orientation, which determines whether the ligand is a substrate or an inhibitor. Further, analysis of the hPNMT Glu185 variant structures unexpectedly revealed that large conformational changes are induced in the enzyme so that the effects of the mutations cannot be explained simply by direct substitution of charge or size of the side chain.
EXPERIMENTAL
Unless otherwise noted, chemicals were purchased from Sigma-Aldrich (St Louis, Missouri, USA) as the highest purity available. Compound 8 was purchased from Syntech Development Co (New Brunswick, New Jersey, USA). Compounds 9 and 10 were synthesised as hydrochloride salts as described previously [28].
Crystallization of hPNMT
C-terminally His6-tagged wt and mutant (E185Q and E185D) hPNMT enzymes were expressed and purified as described previously [15, 27]. Protein was concentrated to 50 mg/ml and mixed with S-adenosyl-l-homocysteine (AdoHcy) at a final concentration of 2 mM, taking the final protein concentration to 40 mg/mL or 1.25 mM. The protein mixture was crystallized by hanging drop vapour diffusion using drops comprising 1.5 µL of protein and 1.5 µL of precipitant equilibrated over 400 µL of precipitant solution (4–8% polyethylene glycol average molecular weight 6K, 0.25 M LiCl, 0.1 M Na cacodylate pH 5.5–6.0) [32, 33]. All crystal structures reported here include AdoHcy. Crystals grown with no ligand added to occupy the noradrenaline binding pocket were used for compound soaks (see below). Crystals of hPNMT:5 and hPNMT:10 were grown by co-crystallisation whereby 20–30 mM of the appropriate ligand was added to the hPNMT:AdoHcy mix prior to crystallisation. For the complex with 5, we co-crystallised wt hPNMT in the presence of racemic noradrenaline, 1, and this was oxidized to noradrenochrome, 5, during the crystallisation process.
Crystal Soaks
In some cases, ligand soaks were performed to generate the co-crystal structures. For these experiments, crystals of dimensions ~0.25 × 0.25 × 0.25 mm3 were grown in the presence of AdoHcy as described above, and transferred to 1 µL of soaking solution. Crystal soak solutions were prepared by adding 1 µL of 100 mM of the appropriate ligand dissolved in 100% dimethylsulfoxide (DMSO) to 9 µL of stabilizing solution so that the final concentrations of components were 6.6% polyethylene glycol average molecular weight 6K (PEG 6K), 0.3 M LiCl, 0.1 M Na cacodylate pH 5.7, 10 mM ligand and 10% DMSO. In the case of 1, which rapidly oxidizes to 5 in air, the ligand solution was prepared immediately before crystal soaking. For the crystal structures complexed with 1, 7, and 8 we chose to soak with racemic ligands. Crystals in soak solutions were then re-sealed over the original mother liquor solution and incubated at 20°C for 15 min.
X-ray Diffraction Data Measurement
Crystals were cryoprotected by transferring directly from the mother liquor (co-crystals) or soak solution (soaked crystals) into mother liquor supplemented with the cryoprotectant 25% ethylene glycol for 10–15 s. Crystals were then flash-cooled in either a nitrogen gas stream at 100 K or by plunging into liquid nitrogen. X-ray diffraction data for hPNMT E185Q:7 were measured using the UQ ROCX Diffraction Facility Rigaku FR-E Superbright generator with Osmic Vari-Max HF optics and Rigaku Saturn 944 CCD detector. X-ray diffraction data for hPNMT E185D:7, wt PNMT:1 and wt PNMT:8 were measured at the Australian Synchrotron beamline 3BM1 with MAR 165 CCD detector. X-ray diffraction data for wt PNMT:10 were measured using a Rigaku FR-E copper rotating anode generator operating at 45 kV, 45 mA with Osmic Confocal Max-Flux™ optics (HiRes2) with an R-AXIS IV++ imaging plate area detector. Data were processed using Crystal Clear (Rigaku Corporation, (c) 1997–2002) or HKL 2000 [34] and phasing was carried out using CNS v1.1 [35] or PHENIX [36].
Structure Determination
The structures were solved by difference Fourier methods using the structure of PNMT (PDB accession code 1HNN with ligands and waters removed [32]) as the starting model. The crystal structure contains two molecules of hPNMT in the asymmetric unit (PNMTA and PNMTB) [33]. The asymmetric unit was chosen so that the two intermolecular disulfide bonds were formed between molecules in the same asymmetric unit [33]. Model building was performed using COOT [37] and the structures were refined using either CNS v1.1[35] or PHENIX [36]. Coordinates for the ligands were generated using PRODRG [38], restraints files were generated using PHENIX [36] and PRODRG [38]. The procedure used was to model and refine the structure of the protein first, followed by addition of water molecules, then AdoHcy and finally the inhibitor/substrate. The criteria used for including a water molecule were: the presence of 2Fo–Fc density at 1 σ and Fo–Fc density at 3 σ and at least one possible hydrogen bond within 3.2 Å. R-free analysis using 5 – 10% of reflections was used for cross-validation [39]. All of the crystal structure maps revealed density indicative of AdoHcy at the cofactor binding site and density consistent with a ligand at the substrate/inhibitor binding site. Electron density for 1, 7, 8, and 10 was clear and unambiguous in both molecules (hPNMTA and hPNMTB) in the asymmetric unit of hPNMT, and consistent with the R-enantiomers of 1, 7 and 8 and the S-enantiomer of 10. For the complex of hPNMT with 5, electron density corresponding to the ligand was present in hPNMTA and consistent with the S-enantiomer. However, the R-enantiomer also makes a reasonable fit so that binding of both enantiomers cannot be ruled out. In the second molecule in the asymmetric unit, the S-enantiomer of 5 can be modeled into the active site density though the fit is not as good as for hPNMTA and Fo-Fc density remains. It is therefore possible that the hPNMTB density could correspond to (a) varying binding modes of 5, (b) a mix of 1 and 5, and/or (c) a mix of R and S enantiomers. Consequently, our analysis of 5 bound to hPNMT refers only to the hPNMTA molecule. Statistics for the crystal structures are presented in Table 2. A figure showing the OMIT electron density for each of the bound inhibitors or substrates is provided as supplementary information (Figure S1). Modelling of hPNMT:9 was achieved by generating coordinates for 9 using PRODRG [38] and arranging in the hPNMT active site using COOT [37]. The trifluoromethyl group of 9 was placed in the position that the same group occupies in hPNMT:8 and hPNMT:10, and then small adjustments were made in the position of the ring system to minimise unfavourable contacts while maintaining favourable interactions. Modelling of hPNMT:AdoMet structures was performed in COOT [37]. Coordinates for the model of bound AdoMet were generated using PRODRG [38] and the cofactor was then manually positioned in the active site by overlaying equivalent regions over the experimentally observed binding mode of AdoHcy. Previously determined structures of hPNMT bound to AdoMet were used as a comparison [22].
Table 2.
hPNMT crystallographic data
| Ligand/ Cofactor/ Protein |
1/ ADOHCY/ wt hPNMT |
5/ ADOHCY/ wt hPNMT |
7/ ADOHCY/ E185D hPNMT |
7/ ADOHCY/ E185Q hPNMT |
8/ ADOHCY/ wt hPNMT |
10/ ADOHCY/ wt hPNMT |
|---|---|---|---|---|---|---|
|
| ||||||
| Space Group | P43212 | P43212 | P43212 | P43212 | P43212 | P43212 |
|
| ||||||
| Unit Cell | ||||||
| a,b (Å) | 93.6 | 94.2 | 93.8 | 94.2 | 93.5 | 94.7 |
| c (Å) | 187.3 | 187.9 | 187.6 | 188.1 | 187.5 | 186.2 |
| α,β,γ (°) | 90 | 90 | 90 | 90 | 90 | 90 |
|
| ||||||
| No Observations | 475,660 | 266,070 | 150,614 | 200,537 | 229,180 | 299,134 |
|
| ||||||
| No unique reflections | 33,670 | 33,903 | 20,186 | 30,563 | 23,386 | 38,435 |
|
| ||||||
| Resolution (Å) | 50.00 – 2.40 | 66.50 – 2.40 | 50.0 – 2.85 | 42.11 – 2.40 | 41.84 – 2.70 | 29.95 – 2.30 |
| (top shell) | (2.49 – 2.40) | (2.43 – 2.40) | (2.9 – 2.85) | (2.49 – 2.40) | (2.77 – 2.70) | (2.38 – 2.30) |
|
| ||||||
| Redundancy | 14.1 (14.3) | 7.85 (7.78) | 7.5 (6.6) | 6.56 (3.21) | 9.8 (9.8) | 7.78 (7.84) |
|
| ||||||
| I/σI | 14.8 (3.7) | 14.6 (4.2) | 9.7 (2.1) | 17.0 (1.5) | 10.8 (2.3) | 12.2 (4.1) |
|
| ||||||
| Completenessa (%) | 99.7 (100.0) | 100.0 (100.0) | 99.6 (99.1) | 90.1 (74.4) | 99.3 (100.0) | 100.0 (100.0) |
|
| ||||||
| Rmergeb (%) | 6.9 (78.5) | 7.8 (47.8) | 9.8 (83.3) | 6.9 (55.0) | 6.2 (44.4) | 7.5 (45.2) |
|
| ||||||
| Refinement | ||||||
|
| ||||||
| No. reflections (|F| > 0) | ||||||
| total | 30740 | 33863 | 17371 | 28699 | 20346 | 38227 |
| (test set) | 1828 | 3384 | 1743 | 1423 | 1755 | 3831 |
|
| ||||||
| Rcrystc/Rfreed (%) | 18.4/23.2 | 19.6/24.0 | 18.6/24.6 | 19.4/23.7 | 19.6/23.6 | 20.5/24.0 |
| (top shell) | (23.3/33.0) | (27.0/31.8) | (27.4/33.4) | (31.0/35.5) | (28.9/34.3) | (32.5/33.2) |
|
| ||||||
| No. non-hydrogen atoms | 4422 | 4432 | 4280 | 4499 | 4273 | 4480 |
| protein | 4106 | 4117 | 4104 | 4110 | 4109 | 4127 |
| cofactor | 52 | 52 | 52 | 52 | 52 | 52 |
| inhibitor | 24 | 24 | 22 | 22 | 28 | 32 |
| water | 240 | 239 | 102 | 311 | 80 | 269 |
| other | - | - | - | 4e | 4e | - |
|
| ||||||
| rmsd from ideal geometry | ||||||
| bond length (Å) | 0.008 | 0.007 | 0.009 | 0.003 | 0.014 | 0.011 |
| bond angle (deg) | 1.12 | 1.00 | 1.24 | 0.70 | 1.60 | 1.62 |
|
| ||||||
| Average B-factor (Å2) | ||||||
| all | 51.0 | 48.6 | 52.6 | 44.0 | 63.5 | 58.0 |
| protein | 50.8 | 48.2 | 52.8 | 43.7 | 63.6 | 57.7 |
| cofactor | 46.8 | 48.1 | 52.0 | 41.1 | 61.2 | 49.9 |
| inhibitor | 50.9 | 67.8 | 51.2 | 52.5 | 64.9 | 67.5 |
| water | 55.7 | 53.0 | 42.9 | 48.2 | 56.5 | 62.7 |
| other | - | - | - | 56.2 | 78.6 | - |
|
| ||||||
| Ramachandran plot | ||||||
| % in most favoured region | 93.3 | 94.6 | 90.6 | 91.7 | 90.6 | 92.8 |
| % in disallowed region | 0.0 | 0.0 | 0.0 | 0.2f | 0.0 | 0.0 |
The number of measured unique reflections divided by the number of theoretical reflections, expressed as a percentage.
Rmerge = Σ|Iobs − Iav|/ΣIav, over all symmetry related observations.
Rcryst = Σ|Fobs − Fcalc|/Σ|Fobs|, over all reflections.
Rfree is calculated as for Rcryst from 5–10% of the data excluded from refinement. Values in parentheses are for the top shell of data.
Ethylene glycol (used in the cryoprotectant).
Ala227 of hPNMTB – this residue is poorly ordered (located in a loop with poor electron density).
For structural analysis, all Cα atoms of the hPNMT structures were superimposed using the SSM superimposition tool in COOT. Root mean squared deviations (RMSD) for these superpositions were in the range 0.2–0.3 Å. Molprobity [40] and CCP4 [41]was used to validate the quality of the structures and COOT was used to analyse rotamer probabilities, based on frequency of appearance in the PDB [42]. Hydrogen bond (2.5 – 3.2 Å cutoff) and hydrophobic interactions (3.8 Å) were identified using the COOT environment distance tool. Figures were generated using PyMOL (DeLano, W.L. The PyMOL Molecular Graphics System (2002) DeLano Scientific, San Carlos, CA, USA. http://www.pymol.org), using the hPNMTA molecule.
RESULTS
Binding Mode of the Physiological Substrate
Although noradrenaline, 1, is the physiological substrate of PNMT, determining the crystal structure of PNMT in complex with this substrate has proven challenging because of its tendency to oxidize to noradrenochrome, 5. Thus, our attempts to co-crystallise PNMT with 1 resulted in the structure determination of hPNMT:5.
To overcome this problem, we used crystal soaking methods to minimise the time that 1 is exposed to air prior to cryocooling the crystal and data collection. This approach had not been investigated previously for hPNMT as it had been thought conformational changes might be required for ligands to access the buried binding site [32]. Further, it required that we find crystallisation conditions in the absence of an inhibitor. However, we were able to generate isomorphous crystals of hPNMT:AdoHcy in the absence of substrate/inhibitor and we found that a short soak of 10–15 minutes was sufficient to introduce 1 into the buried hPNMT binding pocket.
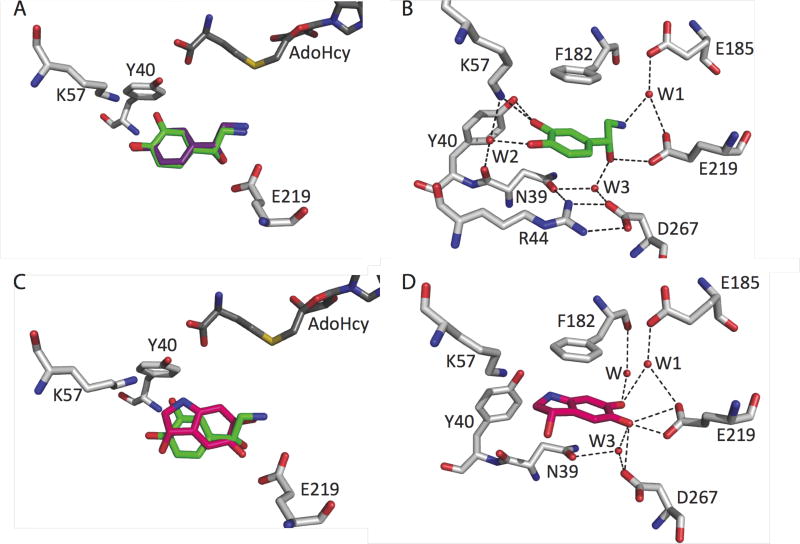
The structure of hPNMT in complex with 1 reveals that the substrate binding pose is largely as predicted from modelling studies [32] and similar to that experimentally determined for octopamine 7 (Figure 2A) [27] confirming that 7 is an appropriate model substrate for hPNMT studies. Thus, the aromatic ring of 1 is wedged between the sidechains of Phe182 and Asn39, and the flexible phenylethylamine chain adopts an extended conformation (Figure 2B). The interactions between the enzyme and the substrates 1 or 7 are conserved (Figure 2B) with the exception that 1 forms additional interactions through its m-hydroxyl group to the side chains of Tyr40 and Lys57. These findings further emphasise the role of Lys57 in orienting substrates for catalysis [27, 32].
Figure 2. Enzyme–ligand interactions and superimpositions of ligand binding positions at the hPNMT active site.
A, Binding pose of 1 (green) overlayed with 7 (purple). B, hPNMT:1 (green). C, binding pose of 5 (pink) overlayed with 1 (green), RMSD 0.3 Å. D, hPNMT:5 (pink).
The overall fold of the enzyme in the hPNMT:5 structure is isomorphous with that of hPNMT:1, with an RMSD of 0.3 Å for all Cα atoms. By comparison with 1, the binding mode of its oxidation product 5 is flipped by ~180° about the short axis of its fused aromatic ring. Additionally, while the R-enantiomer of 1 is observed to bind in the hPNMT crystal structure, it is the S-enantiomer of 5 that is modeled. However, a proportion of bound R-enantiomer of 5 cannot be ruled out. This binding mode places one of the two aromatic carbonyl oxygens (5-oxo) in a position similar to that of the β-hydroxyl of 1 in the PNMT active site (Figure 2C). The β-hydroxyl of 1 interacts directly with the side chain of hPNMT residue Glu219 and—via waters—with the side chains of Asp267 and Asn39 (Figure 2B). Arg44 forms part of a hydrogen bonding network with Asp267, Asn39 and an active site water (W3) (Figure 2B), like that described for the hPNMT:7 structure [27]. The sidechain of Arg44 is within cation-π interaction distance of the benzene ring of 1 (≤ 4.6 Å) [43, 44] and is oriented in a pseudo T-shaped geometry that is favoured for such interactions [45]. However, the Arg44 guanidinium group is offset from the plane of the benzene ring of 1 so that a cation-π interaction is unlikely to contribute significantly to substrate binding.
Surprisingly, the aromatic 5-carbonyl oxygen of 5 (Figure 2D) forms interactions similar to those formed to the β-hydroxyl of 1 supporting the suggestion that hPNMT residue Glu219 may be protonated (unpublished work: P. Georgieva, Q. Wu, M.J. McLeish, F. Himo). The 6-oxo group of 5 forms water-mediated interactions with the side chain of the catalytic residue Glu185 and with the main chain oxygen of Phe182 (Figure 2D). The 3-hydroxyl and the indole amine of 5 do not form hydrogen bonds to hPNMT.
hPNMT Substrate versus hPNMT Inhibitor
To investigate why structurally similar ligands are recognised differently by hPNMT – for example 9 is a substrate whereas 10 is an inhibitor – we determined the crystal structures of hPNMT:8 (Figure 3A, B) and hPNMT:10, and modelled the structure of hPNMT:9. The two crystal structures hPNMT:8 and hPNMT:10 are very similar to each other (RMSD 0.3 Å), and to hPNMT:1 (RMSD values of 0.3 Å and 0.2 Å, respectively). In the crystal structure of hPNMT:8, the substrate 8 has its 3-trifluoromethyl group positioned in a hydrophobic pocket formed by the sidechains of Val53, Met258, Val269 and Val272 and forms a hydrogen bond with Arg44 (Figure 3A). To achieve these interactions, the aromatic ring of 8 is rotated in the active site relative to the aromatic ring of substrate 1 (Figure 3C). However, subtle changes in the flexible β-hydroxyethylamine chain allow the acceptor amine to be placed in a similar position to that of 1 (Figure 3C). These adjustments to the β-hydroxyethylamine chain lead to the displacement of a water molecule that, in the complexes with 1 and 7, mediates an interaction between the β-hydroxyl and the acidic sidechain of Asp267. By contrast, in the hPNMT complex with 8, the β-hydroxyl forms a direct hydrogen bond with Asp267 and a water mediated interaction with the Asn39 sidechain (Figure 3B). The amine of 8, like those of 1 and 7, forms no direct interactions with the enzyme, but does form water-mediated interactions with the sidechains of Glu185 and Glu219 (Figure 3B).
Figure 3. Enzyme–ligand interactions and superimpositions of ligand binding positions at the hPNMT active site.
A, B, Structure of hPNMT:8 (yellow). C, binding pose of 8 (yellow) overlayed with 1 (green), RMSD 0.3 Å. D, binding pose of 10 (orange) overlayed with 8 (yellow), RMSD 0.3 Å. E, F, hPNMT:10 (orange). G, binding pose of 8 (yellow) with modelled AdoMet (dark grey). H, binding pose of 10 (orange) with modelled AdoMet (dark grey).
In the crystal structure of the hPNMT:10 complex, the 5-trifluoromethyl group occupies a position similar to that of the 3-trifluoromethyl group of 8 (Figure 3D) forming hydrophobic interactions with side chains of Val53, Met258, Val269 and Val272 and a hydrogen bond with Arg44 (Figure 3E). The fused ring adopts a binding mode that is rotated and skewed compared with 8 (Figure 3D) and the positions of the amines of 8 and 10 are thus separated by almost 1 Å. The amine of 10 forms direct rather than water-mediated hydrogen bonds with the side chain of Glu219, in addition to the usual water-mediated hydrogen bond with Glu185 (Figure 3F). A water-mediated interaction is also formed between the amine of 10 and the main chain oxygen of Phe182, similar to that observed between the 6-oxo substituent in the noradrenochrome-bound structure hPNMT:5 (Figure 2D, 3F).
We investigated why the binding of 10 does not result in amine methylation by hPNMT (10 is an inhibitor not a substrate of the enzyme). We modelled the cofactor AdoMet into the binding site by adding the donor methyl group to AdoHcy. Previously, we had proposed that the angle of attack between the AdoMet donor methyl group and the substrate C–N bond influences whether the amine is methylated [27]: substrates had angles of 89–93° and an inhibitor had angles of 129–132°. Of the substrates reported in the present work, 1 has angles of 100–114° and 8 has angles of 73–84° (Figure 3G). However, the inhibitor 10 has values for this angle of 85–89° (Figure 3H), suggesting that it is not the angle of attack that prevents hPNMT from methylating 10. We also investigated the distance between the amine and the cofactor donor methyl group. In the present work, this distance ranges between 3.8 and 3.9 Å for the substrates 1 and 8 (Figure 3G), in agreement with our previous work (3.7–3.9 Å for substrates [27]) and within the expected distance for catalysed methyl transfer (3.0–4.0 Å) [46]. By contrast, the distance between the amine of 10 and the AdoMet methyl group is 4.4 Å (Figure 3H). We therefore conclude that 10 is an inhibitor and not a substrate because its amine is too distant from the cofactor for methylation to occur.
Despite extensive trials, we were unable to measure crystallographic data for the complex of hPNMT:9 (a rigid analogue of 8): soaking crystals in solutions containing 9 induced crystal disorder or cracking and co-crystallisation resulted in poorly diffracting crystals that gave low resolution or streaky diffraction patterns that could not be processed. Instead, the structure of hPNMT:9 was modeled from the structures of hPNMT:8 and hPNMT:10 (Figure S2, supplementary information). We took the refined molecular structure of 10, and moved the trifluoromethyl group from the 5- to the 6-position. The model of 9 was then placed into the hPNMT binding site, by positioning the 6-trifluoromethyl group into the same hydrophobic pocket occupied by the trifluoromethyl groups of 10 and 8 (Figure S2 panels A–C). In this modelled structure, the fit of 9 within the active site is tight: steric clashes are formed between 9 and both Tyr35 and Tyr222 (Figure S2 panel B), perhaps explaining why crystals crack and diffraction is poor in the presence of this ligand. Presumably, binding of 9 destabilises the hPNMT crystalline lattice or requires conformational changes in the enzyme that disrupt crystal contacts. To minimize unfavourable clashes, we modelled different rotameric positions of Tyr35 and Tyr222 sidechains, but this introduced clashes with other residues. Nonetheless, conformational changes in Tyr35 and/or Tyr222 may still be possible if accompanied by cooperative changes in these other residues, as we have observed for other inhibitors [22]. If similar changes occur on binding of the substrate 9, this could explain why it remains an excellent hPNMT substrate (Table 1). Notwithstanding, the model of hPNMT:9 gave angle and distance measurements for the amine to AdoMet donor methyl group of 3.3 Å and 106°, respectively, consistent with values obtained for other hPNMT substrates (Figure S2D).
Glu185 Variants
Our previous studies identified Glu185 as a key catalytic residue of hPNMT: mutation of Glu185 significantly reduces the rate of catalysis but has little effect on the affinity of the enzyme for the substrate [27] (Table 1). To investigate the molecular basis for these effects, we determined the crystal structures of the hPNMT variants E185D and E185Q in complex with the model substrate 7. These two point mutations do not affect the overall protein fold, in that the crystal structures of the two variants are isomorphous with each other and with wthPNMT:7 (RMSD values 0.3 Å in each case). Mutation of the large acidic residue Glu185 to the smaller acidic residue Asp (E185D) leads to a ~1Å movement of the backbone atoms of residue 185 and of nearby residues Thr230 and Val231 compared with the wt structure (Figure 4A). Surprisingly, these backbone changes in E185D place the carboxylate group of Asp185 almost coincident with the carboxylate of Glu185, and interactions formed to the acidic sidechain are conserved between the wt and E185D structures. The backbone changes occur with little or no disruption to bound water molecules in the active site or to intramolecular interactions of the protein (Figure 4B). However, an interaction between the main chain oxygen of Asp185 and the mainchain amine of Val231 is lost and an unfavourable interaction is formed between the Asp185 mainchain oxygen and the sidechain of Val231, suggesting that binding of substrate to the E185D variant will be less favourable than binding to the wt enzyme.
Figure 4. Effect of hPNMT Glu185 variants on residue position and binding pose of 7.
A, Structure of wt hPNMT:7 (dark gray) superimposed with E185D hPNMT:7 (light gray), RMSD 0.3 Å. B, E185D hPNMT:7 (light gray). C Structure of wt hPNMT:7 (dark gray) superimposed with E185Q hPNMT:7 (light gray), RMSD 0.3 Å. D, E185Q hPNMT:7 (light gray). Residues 185 and Val231 which exhibit conformational movement are displayed in stick representation. Distances are measured between Cα atoms and shown as black dashed lines.
We also investigated the effect of removing the negative charge of Glu185 by determining the crystal structure of hPNMT E185Q in complex with the model substrate 7. This mutation has been characterised kinetically, and reduces the catalytic activity of the enzyme by ~300 fold (Table 1) [27]. The structure of hPNMT E185Q:7 (Figure 4C, D) revealed that Gln185 undergoes a conformational change compared with Glu185. In the wt hPNMT:7 complex, the Glu185 sidechain adopts an unusual conformation involving a hydrogen bond to its own mainchain nitrogen. This Glu rotamer has a 2% probability of occurrence based on frequency of appearance in the pdb. The other carboxylate oxygen forms a hydrogen bond with the main chain nitrogen of Ala216, and water mediated interactions with the amine of 7 and the main chain oxygen of Ile214. These same set of interactions are unfavourable when the acidic sidechain is replaced by the amide of a Gln sidechain: the amide nitrogen cannot form a hydrogen bond with amides, so the interactions with its own backbone amide or with the backbone amide of Ala216 cannot occur. We assume that this is the reason Gln185 adopts a conformation with a rotamer probability of 1% in the E185Q hPNMT:7 structure [42]. This position places its sidechain nitrogen within hydrogen bond distance of the mainchain oxygen of Val232, and is not favourable in that it is in close contact with the Val231 sidechain, which induces static disorder in Val231 despite a shift of 0.8–1.0 Å in the backbone (Figure 4C). Gln185 does form an indirect interaction with the amine of 7, via an ordered water located in a position equivalent to that of W1 in the wt hPNMT:7 structure (Figure 4D) [27]. Interestingly, the binding position of 7 changes subtly in the structure of E185Q hPNMT:7 in comparison to wt hPNMT:7 (Figure 4A). This change in the binding position results in the acceptor amine of 7 moving further than 4 Å from the modeled cofactor methyl group, and may also contribute to the reduced catalytic activity of the E185Q hPNMT variant.
DISCUSSION
We have reported for the first time the structure of hPNMT in complex with its physiological ligand noradrenaline, 1, obtained by using rapid crystal soaking methods. We used the same methods to determine the structures of hPNMT:8, hPNMT E185D:7 and hPNMT E185Q:7. The results show that ligands can be diffused into hPNMT crystals over 10–15 minutes, without generating stresses that lead to crystal cracking or disorder in the diffraction pattern. These same soaking methods should therefore be suitable for fragment-screening approaches to identify novel inhibitor scaffolds for hPNMT. During efforts to determine the structure of hPNMT:1 the structure of hPNMT:5 from co-crystals grown in the presence of noradrenaline was also determined. This result confirms that noradrenaline 1 is converted to noradrenochrome 5 during the crystallisation experiment and unexpectedly shows that 5 binds to hPNMT in a reverse orientation: the amine of 5 is located in the same relative position of the active site as the catechol hydroxyls of 1, and 6.6 Å from the methylating cofactor. Our findings predict that noradrenochrome is not methylated by hPNMT. Oxidation of 1 limits its usefulness in kinetic studies of hPNMT. The crystal structure of hPNMT:7 has been determined previously by co-crystallisation [27], and the new structure of hPNMT:1 allowed a direct comparison of the physiological and model substrates. We found that the binding of 7 to hPNMT is comparable to that of 1, confirming that 7 is a valid model substrate.
Analysis of crystal structures of two trifluoromethyl-substituted ligands of hPNMT, and of the modeled structure of a third, showed that this substituent is the driving force in orienting these ligands within the active site and thereby determining whether they are methylated by hPNMT. These results further confirm the importance of geometry in methylation of ligands by hPNMT indicating that the amine should be within 4Å of the donor methyl group and have an orientation close to tetrahedral. The structures also explain why Asp267 is important for catalysis of 8 but does not affect 9, a rigid analogue of 8 [30]: 8 forms a direct interaction with Asp267 through its β-hydroxyl group whereas the modeled structure of the hPNMT:9 complex suggests that there is no interaction with Asp267.
This research further emphasises previous findings that the binding pocket of hPNMT is extraordinarily plastic. Previous studies have shown that the enzyme is capable of movement of active site helix α4 and large conformational changes of sidechains (Lys57, Tyr126, Met258) to accommodate the binding of some ligands [14, 22]. Our current research shows that mutations in active site residues can also lead to structural rearrangements, involving both sidechain and mainchain atoms. In the E185D variant, enzyme backbone movements are observed that together position the acidic carboxyl group of Asp185 in the same relative location as the acidic carboxyl group of Glu185 in the native enzyme. Furthermore, many of the same intramolecular interactions involving residue Glu185 are observed in the E185D variant, suggesting that these may be important for catalytic activity. The observed reduction in catalytic efficiency of the E185D variant enzyme is likely due to the energetic cost required to reposition Asp185 so that it can deprotonate the substrate.
Mutation of Glu185 to Gln causes the most dramatic reduction in catalytic activity of hPNMT [30] and this was thought to be due to the loss of the acidic sidechain. However, the crystal structure of hPNMT E185Q reveals that the Gln185 sidechain adopts a different conformation to that of Glu185 suggesting that the specific conformation of the sidechain might also be important for catalytic activity. We were unable to produce hPNMT E185A in sufficient quantities for structural studies due to low expression levels [30]. However, the fact that the E185A variant retains reasonable catalytic activity compared with E185Q suggests that additional factors may be involved. In the conversion of noradrenaline to adrenaline, these factors are likely to be that proton transfer is not rate limiting and that in the absence of Glu185, the nearby Glu219 residue may be able to accept the proton. Mutation of Glu185 to Gln (E185Q) disrupts the hydrogen bonding network between residue 185, Glu219, W1 and the substrate, leading to a more dramatic decrease in catalytic activity compared with E185A. Overall, our results support an important role for residue 185 in catalysis and suggest that evolution of hPNMT has given rise to interactions with the E185 sidechain, including an internal hydrogen bond, that position it optimally for catalysis. These results have wider implications for the interpretation of kinetic data and emphasise the value of obtaining structural evidence in combination with kinetic data to provide a comprehensive understanding of enzyme activity.
Supplementary Material
Acknowledgments
We acknowledge use of the UQ ROCX Diffraction Facility and thank Karl Byriel for his help and assistance. We are also grateful for access to beamline 3BM1 at the Australian Synchrotron and thank staff at the beamline for their support and advice in measuring data.
FUNDING:
ND is a recipient of an Australian Postgraduate Award; JLM is funded by an Australian National Health and Medical Research Fellowship. This work was supported by Australian Research Council Discovery Project Grant [DP0664564] and by the US National Institutes of Health [R01 HL034193].
ABBREVIATIONS
- PNMT
phenylethanolamine N-methyltransferase
- hPNMT
human PNMT
- wt
wild type
- hPNMTA
molecule A in the asymmetric unit of hPNMT crystals
- hPNMTB
molecule B in the asymmetric unit of hPNMT crystals
- AdoMet
S-adenosyl-L-methionine
- AdoHcy
S-adenosyl-L-homocysteine
- CNS
Central Nervous System
- CNS v1.1
Crystallography and NMR System version 1.1
- SEM
standard error of the mean
Footnotes
STRUCTURAL DATA:
Coordinates and structure factors for all six crystal structures reported in this manuscript have been deposited with the Protein Data Bank at the Research Collaboratory for Structural Bioinformatics and will be released upon publication. (hPNMT:1 3HCD; hPNMT:5 3HCB; hPNMT E185D:7 3HCE; hPNMT E185Q:7 3HCA; hPNMT:8 3HCF; hPNMT:10 3HCC)
References
- 1.Fu ZJ, Hu YB, Konishi K, Takata Y, Ogawa H, Gomi T, Fujioka M, Takusagawa F. Crystal structure of glycine N-methyltransferase from rat liver. Biochemistry. 1996;35:11985–11993. doi: 10.1021/bi961068n. [DOI] [PubMed] [Google Scholar]
- 2.Horton JR, Sawada K, Nishibori M, Zhang X, Cheng XD. Two polymorphic forms of human histamine methyltransferase: Structural, thermal and kinetic comparisons. Structure. 2001;9:837–849. doi: 10.1016/s0969-2126(01)00643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Komoto J, Yamada T, Takata Y, Konishi K, Ogawa H, Gomi T, Fujioka M, Takusagawa F. Catalytic mechanism of guanidinoacetate methyltransferase: crystal structures of guanidinoacetate methyltransferase ternary complexes. Biochemistry. 2004;43:14385–14394. doi: 10.1021/bi0486785. [DOI] [PubMed] [Google Scholar]
- 4.Axelrod J. Purification and properties of phenylethanolamine-N-methyl transferase. J. Biol. Chem. 1962;237:1657–1660. [PubMed] [Google Scholar]
- 5.Kirshner N, Goodall M. The formation of adrenaline from noradrenaline. Biochimica. Et. Biophysica. Acta. 1957;24:658–659. doi: 10.1016/0006-3002(57)90271-8. [DOI] [PubMed] [Google Scholar]
- 6.Vidgren J, Svensson LA, Liljas A. Crystal-structure of catechol Omethyltransferase. Nature. 1994;368:354–358. doi: 10.1038/368354a0. [DOI] [PubMed] [Google Scholar]
- 7.Fuller RW. Pharmacology of brain epinephrine neurons. Ann. Rev. Pharmacol. Toxicol. 1982;22:31–55. doi: 10.1146/annurev.pa.22.040182.000335. [DOI] [PubMed] [Google Scholar]
- 8.Rothballer AB. The effects of catecholamines on the central nervous system. Pharmacol. Rev. 1959;11:494–547. [PubMed] [Google Scholar]
- 9.Crowley WR, Terry LC, Johnson MD. Evidence for the involvement of central epinephrine systems in the regulation of luteinizing-hormone, prolactin, and growth-hormone release in female rats. Endocrinology. 1982;110:1102–1107. doi: 10.1210/endo-110-4-1102. [DOI] [PubMed] [Google Scholar]
- 10.Hokfelt T, Fuxe K, Goldstei M, Johansso O. Immunohistochemical evidence for existence of adrenalin neurons in rat-brain. Brain Res. 1974;66:235–251. [Google Scholar]
- 11.Mefford IN, Lister RG, Ota M, Linnoila M. Antagonism of ethanol intoxication in rats by inhibitors of phenylethanolamine N-methyltransferase. Alcoholism Clin. Exp. Res. 1990;14:53–57. doi: 10.1111/j.1530-0277.1990.tb00446.x. [DOI] [PubMed] [Google Scholar]
- 12.Gearhart DA, Neafsey EJ, Collins MA. Phenylethanolarnine N-methyltransferase has beta-carboline 2N-methyltransferase activity: hypothetical relevance to Parkinson's disease. Neurochemistry Int. 2002;40:611–620. doi: 10.1016/s0197-0186(01)00115-2. [DOI] [PubMed] [Google Scholar]
- 13.Kennedy BP, Bottiglieri T, Arning E, Ziegler MG, Hansen LA, Masliah E. Elevated S-adenosylhomocysteine in Alzheimer brain: influence on methyltransferases and cognitive function. J. Neural Transm. 2004;111:547–567. doi: 10.1007/s00702-003-0096-5. [DOI] [PubMed] [Google Scholar]
- 14.McMillan FM, Archbold J, McLeish MJ, Caine JM, Criscione KR, Grunewald GL, Martin JL. Molecular recognition of sub-micromolar inhibitors by the epinephrine-synthesizing enzyme phenylethanolamine N-methyltransferase. J. Med. Chem. 2004;47:37–44. doi: 10.1021/jm0205752. [DOI] [PubMed] [Google Scholar]
- 15.Romero FA, Vodonick SM, Criscione KR, McLeish MJ, Grunewald GL. Inhibitors of phenylethanolamine N-methyltransferase that are predicted to penetrate the blood-brain barrier: Design, synthesis and evaluation of 3-fluoromethyl-7-(N-substituted aminosulfonyl)-1,2,3,4-tetrahydroisoquinolines that possess low affinity toward the α2-adrenoreceptor. J. Med. Chem. 2004;47:4483–4493. doi: 10.1021/jm0400653. [DOI] [PubMed] [Google Scholar]
- 16.Grunewald GL, Lu J, Criscione KR, Okoro CO. Inhibitors of phenylethanolamine N-methyltransferase devoid of alpha2-adrenoceptor affinity. Bioorg Med Chem Lett. 2005;15:5319–5323. doi: 10.1016/j.bmcl.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 17.Grunewald GL, Romero FA, Chieu AD, Fincham KJ, Bhat SR, Criscione KR. Exploring the active site of phenylethanolamine N-methyltransferase, 3-alkyl-7-substituted-1,2,3,4-tetrahydroisoquinoline inhibitors. Bioorganic & Medicinal Chemistry. 2005;13:1261–1273. doi: 10.1016/j.bmc.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 18.Grunewald GL, Romero FA, Criscione KR. 3-hydroxymethyl-7-(N-substituted aminosulfonyl)-1,2,3,4-tetrahydroisoquinoline inhibitors of phenylethanolamine N-methyltransferase that display remarkable potency and selectivity. J. Med. Chem. 2005;48:134–140. doi: 10.1021/jm049368n. [DOI] [PubMed] [Google Scholar]
- 19.Grunewald GL, Romero FA, Criscione KR. Nanomolar inhibitors of CNS epinephrine biosynthesis: (R)-(+)-3-fluoromethyl-7-(N-substituted aminosulfonyl)-1,2,3,4-tetrahydroisoquinolines as potent and highly selective inhibitors of phenylethanolamine N-methyltransferase. Journal of Medicinal Chemistry. 2005;48:1806–1812. doi: 10.1021/jm049594x. [DOI] [PubMed] [Google Scholar]
- 20.Grunewald GL, Romero FA, Seim MR, Criscione KR, Deupree JD, Spackman CC, Bylund DB. Exploring the active site of phenylethanolamine N-methyltransferase with 3-hydroxyethyl- and 3-hydroxypropyl-7-substituted-1,2,3,4-tetrahydroisoquinolines. Bioorg Med Chem Lett. 2005;15:1143–1147. doi: 10.1016/j.bmcl.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 21.Grunewald GL, Seim MR, Regier RC, Martin JL, Gee CL, Drinkwater N, Criscione KR. Comparison of the binding of 3-fluoromethyl-7-sulfonyl-1,2,3,4-tetrahydroisoquinolines with their isosteric sulfonamides to the active site of phenylethanolamine N-methyltransferase. J. Med. Chem. 2006;49:5424–5433. doi: 10.1021/jm060466d. [DOI] [PubMed] [Google Scholar]
- 22.Gee CL, Drinkwater N, Tyndall JDA, Grunewald GL, Wu Q, McLeish MJ, Martin JL. Enzyme adaptation to inhibitor binding: A cryptic binding site in phenylethanolamine N-methyltransferase. J. Med. Chem. 2007;50:4845–4853. doi: 10.1021/jm0703385. [DOI] [PubMed] [Google Scholar]
- 23.Fuller RW, Hemrick-Luecke SK, Midgley JM. Comparison of o-octopamine and related phenylethanolamines as substrates for norepinephrine N-methyltransferase. Res. Commun. Chem. Pathol. Pharmacol. 1981;33:207–213. [PubMed] [Google Scholar]
- 24.Fuller RW, Hunt JM. Substrate specificity of phenethanolamine N-methyl transferase. Biochem. Pharmacol. 1965;14:1896–1897. doi: 10.1016/0006-2952(65)90281-9. [DOI] [PubMed] [Google Scholar]
- 25.Fuller RW, Roush BW, Snoddy HD, Day WA, Molloy BB. Norepinephrine N-methyltransferase inhibition by benzamidines, phenylacetamidines, benzylguanidines, and phenylethylguanidines. J. Med. Chem. 1975;18:304–307. doi: 10.1021/jm00237a019. [DOI] [PubMed] [Google Scholar]
- 26.Wu Q, Criscione KR, Grunewald GL, McLeish MJ. Phenylethanolamine N-methyltransferase inhibition: Re-evaluation of kinetic data. Bioorg. Med. Chem. Lett. 2004;14:4217–4220. doi: 10.1016/j.bmcl.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 27.Gee CL, Tyndall JD, Grunewald GL, Wu Q, McLeish MJ, Martin JL. Mode of binding of methyl acceptor substrates to the adrenaline-synthesizing enzyme phenylethanolamine N-methyltransferase: Implications for catalysis. Biochemistry. 2005;44:16875–16885. doi: 10.1021/bi051636b. [DOI] [PubMed] [Google Scholar]
- 28.Grunewald GL, Markovich KM, Sall DJ. Binding orientation of amphetamine and norfenfluramine analogues in the benzonorbornene and benzobicyclo[3.2.1]octane ring systems at the active site of phenylethanolamine N-methyltransferase (PNMT) J. Med. Chem. 1987;30:2191–2208. doi: 10.1021/jm00395a005. [DOI] [PubMed] [Google Scholar]
- 29.Rafferty MF, Grunewald GL. The remarkable substrate activity for phenylethanolamine N-methyltransferase of some conformationally defined phenylethylamines lacking a side-chain hydroxyl group. Conformationally defined adrenergic agents. Mol. Pharmacol. 1982;22:127–132. [PubMed] [Google Scholar]
- 30.Wu Q, Gee CL, Lin F, Tyndall JD, Martin JL, Grunewald GL, McLeish MJ. Structural, mutagenic, and kinetic analysis of the binding of substrates and inhibitors of human phenylethanolamine N-methyltransferase. J. Med. Chem. 2005;48:7243–7252. doi: 10.1021/jm050568o. [DOI] [PubMed] [Google Scholar]
- 31.Grunewald GL, McLeish MJ, Criscione KR. Phenylethanolamine N-methyltransferase kinetics: bovine versus recombinant human enzyme. Bioorg Med Chem Lett. 2001;11:1579–1582. doi: 10.1016/s0960-894x(01)00245-1. [DOI] [PubMed] [Google Scholar]
- 32.Martin JL, Begun J, McLeish MJ, Caine JM, Grunewald GL. Getting the adrenaline going: Crystal structure of the adrenaline-synthesizing enzyme PNMT. Structure. 2001;9:977–985. doi: 10.1016/s0969-2126(01)00662-1. [DOI] [PubMed] [Google Scholar]
- 33.Gee CL, Nourse A, Hsin AY, Wu Q, Tyndall JD, Grunewald GL, McLeish MJ, Martin JL. Disulfide-linked dimers of human adrenaline synthesizing enzyme PNMT are catalytically active. BBA-Proteins Proteom. 2005;1750:82–92. doi: 10.1016/j.bbapap.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 34.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method. Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 35.Brünger AT, Adams PD, Clore GM, Delano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges N, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography and NMR system (CNS): A new software system for macromolecular structure determination. Acta. Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 36.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. 2002;D58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 37.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta. Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 38.Schuettelkopf AW, van Aalten DMF. PRODRG - a tool for high-throughput crystallography of protein-ligand complexes. Acta. Crystallogr. 2004;D60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 39.Brünger AT. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:672–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 40.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bailey S. The CCP4 Suite - Programs For Protein Crystallography Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 42.Dunbrack RL, Jr, Cohen FE. Bayesian statistical analysis of protein side-chain rotamer preferences. Protein Sci. 1997;6:1661–1681. doi: 10.1002/pro.5560060807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Flocco MM, Mowbray SL. Planar stacking interactions of arginine and aromatic side-chains in proteins. J. Mol. Biol. 1994;235:709–717. doi: 10.1006/jmbi.1994.1022. [DOI] [PubMed] [Google Scholar]
- 44.Momany FA, Carruthe Lm, McGuire RF, Scheraga HA. Intermolecular potentials from crystal data. III. Determination of empirical potentials and application to packing configurations and lattice energies in crystals of hydrocarbons, carboxylic-acids, amines, and amides. J. Phys. Chem. 1974;78:1595–1620. [Google Scholar]
- 45.Gallivan JP, Dougherty DA. Cation-pi interactions in structural biology. Proc. Natl. Adad. Sci. U S A. 1999;96:9459–9464. doi: 10.1073/pnas.96.17.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Svetlitchnaia T, Svetlitchnyi V, Meyer O, Dobbek H. Structural insights into methyltransfer reactions of a corrinoid iron-sulfur protein involved in acetyl-CoA synthesis. Proc. Natl. Adad. Sci. U S A. 2006;103:14331–14336. doi: 10.1073/pnas.0601420103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.