Abstract
Radicle protrusion from tomato (Lycopersicon esculentum Mill.) seeds to complete germination requires weakening of the endosperm tissue opposite the radicle tip. In common with other cell wall disassembly processes in plants, polygalacturonases (PGs) may be involved. Only calcium-dependent exo-PG activity was detected in tomato seed protein extracts. Chromatographic profiles of a partially acid-hydrolyzed fraction of polygalacturonic acid further digested with seed extract were consistent with the presence of only calcium-dependent exo-PG activity. In addition, a transcript encoding a previously unknown PG was detected prior to the completion of germination. The mRNA, produced from a gene (LeXPG1) estimated by Southern analysis to be represented once in the genome, was also present in flowers (anthers) and in lower amounts in roots and stems. LeXPG1 mRNA abundance was low during seed development, increased during imbibition, and was even greater in seeds that had completed germination. Expression of LeXPG1 during germination predominates in the endosperm cap and radicle tip, and in the radicle appears as a distinct band possibly associated with vascular tissue differentiation. We suggest that PG is involved in cell wall loosening of the endosperm necessary for radicle protrusion from tomato seeds and in subsequent embryo and seedling growth.
The tomato (Lycopersicon esculentum Mill.) embryo is completely enclosed by the endosperm and testa. Weakening of the micropylar endosperm tissue opposite the radicle tip (the endosperm cap) is the major factor determining whether and when radicle emergence occurs (Groot and Karssen, 1987, 1992; Dahal and Bradford, 1990; Ni and Bradford, 1993). Endosperm cap cell walls are rich in mannans (Groot et al., 1988; Dahal et al., 1997), and weakening of the cap tissue is accompanied by an increase in the activity of endo-β-1,4-mannanase (Bewley, 1997). However, recent results indicate that, at least in tomato, the presence of mannanase activity alone is not sufficient for germination (Toorop et al., 1996; Dahal et al., 1997; Still and Bradford, 1997). It is probable that other hemicellulases, pectinases, and/or cellulases play a role in weakening the endosperm cap permitting radicle protrusion. Sánchez et al. (1986) showed that cellulase activity correlated well with the completion of germination in Datura ferox (like tomato, a member of the Solanaceae), but Leviatov et al. (1995) found that cellulase activity was not closely associated with germination rates of tomato seeds at low temperature.
Based upon microscopic studies of germinating seeds, it has been proposed that the radicle pushes between the cells of the endosperm, which separate rather than break or tear (Karssen et al., 1989). Numerous examples of time- and tissue-dependent loss of cell-to-cell cohesion occur in plant development, including pollen tube growth through transmitting styles (Clarke and Gleeson, 1981; Mu et al., 1994), outgrowth of lateral roots (Peretto et al., 1992), organ abscission (Taylor et al., 1990; Bonghi et al., 1992), dehiscence of seed pods (Jenkins et al., 1996), and intrusive growth of nonarticulated laticifers (Wilson et al., 1976) and fungal hyphae (Hahn et al., 1989). All of these processes are accompanied by partial breakdown in cell wall pectin and are correlated with the presence and activity of polygalacturonase (PG). Furthermore, pollen germination is associated with pectin degradation (Pressey and Reger, 1989; Pressey, 1991). Radicle protrusion from endospermic seeds has much in common with these processes involving loss of cell cohesion or penetration of existing tissues. We therefore tested the hypothesis that a PG may be present in the endosperm cap of tomato seeds. Here we describe the cloning, expression, and activity of a PG during tomato seed germination.
MATERIALS AND METHODS
Plant Material
Tomato (Lycopersicon esculentum Mill. cv Moneymaker) seeds were obtained from immature green, mature green, breaker, and red ripe (mature) fruit. The seeds were cleaned (0.1 m HCl for 1 h) and washed in tap water. Dried seeds (5% moisture content fresh weight basis) were stored at 4°C.
Germination conditions were as described previously (Dahal and Bradford, 1990). One gram of seeds was placed on 15 mL of distilled, deionized water on two, 8.5-cm diameter blotting paper discs (Stults Scientific Eng., Springfield, IL) in a Petri dish. Dishes were placed inside of plastic containers lined with water-saturated paper towels and incubated at 25°C in the dark. Whole tomato seeds were harvested every 12 h for 4 d and separated, after 48 h, into those that had or had not completed germination before being extracted for either RNA or protein (see below). Other seeds were treated as above and dissected into the endosperm cap, radicle tip, and the rest of the seed (includes lateral endosperm, hypocotyl, and cotyledons) and RNA obtained from these seed components.
Endo- and Exo-PG Assays
Enzyme assays for endo-PG activity were conducted using a gel diffusion assay (Buescher and Burgin, 1992), and a viscometric assay (Christensen, 1954; Hadfield et al., 1998). The latter assay is the most sensitive one available for monitoring endo-PG activity (Tagawa and Kaji, 1988). Various commercially prepared PGA and pectic substrates of different degrees of esterification and from a variety of plants were tested.
Exo-PG activity was determined using a reducing sugar assay (Gross, 1982) with PGA from citrus fruit as the substrate (Sigma, St. Louis). One hundred seeds were pulverized in liquid N2 and ground in 1 mL of extraction buffer (1 m NaCl, 2.5 mm PMSF, 10 μm leupeptin, and 50 mm NaOAc buffer, pH 5.0). The homogenate was made 80% (v/v) with respect to ammonium sulfate stepwise while stirring on ice. Subsequently, the suspension was placed in centrifuge tubes and left for 30 min on ice. The suspension was centrifuged (20 min, 10,000g) and the pellet resuspended in 50 mm NaOAc buffer, pH 5.0. The extract was dialyzed overnight in 12- to 14-kD exclusion tubing against 50 mm NaOAc buffer, pH 5.0. After dialysis, some aliquots were made 5 mm with respect to CaCl2, some were boiled for negative controls, substrate was eliminated from other control reactions, and three reactions were spiked with serial dilutions of dialyzed commercial PG (Aspergillus niger; Megazyme, Sidney, Australia). Some experiments included 50 mm EDTA while in others CaCl2 was not added to the extracts. Each time point and replication was assayed at two different dilutions and proportionality determined. All samples were quantified relative to a standard curve of GalUA. Each experiment was performed three times.
The nature of the PG-hydrolyzing activity present in tomato seeds was assayed using HPLC with pulsed amperometric detection (PAD; Townsend et al., 1988). A 1% (w/v) solution of PGA (G12 fraction, Campbell and Labavitch, 1991) was incubated with 1/10th volume of tomato seed extract for 12 h at 37°C. A 25-μL aliquot of each hydrolysate was quantified for monogalacturonic acid and compared with known concentrations from a commercial source (Sigma) using HPLC-PAD as described by Melotto et al. (1994).
PCR Amplification and cDNA Cloning
Degenerate PCR primers were constructed to two flanking conserved sites in known PGs and used according to Hadfield et al. (1998) to obtain putative PG amplicons from diverse plant parts. Amplicons were cloned and used as templates to synthesize DNA probes to screen for expression of their corresponding mRNAs in tomato seeds. Only one of these putative PGs was expressed in seeds prior to radicle protrusion.
Since the PG mRNA expressed in seeds was also present in anthers, 500,000 recombinants in a λgt10 phage anther cDNA library (Twell et al., 1989) were screened and seven hybridizing plaques identified. These were purified, recovered from the vector, and a 1,521-bp cDNA subcloned into pBSIIKS. This cDNA was subsequently digested with ScaI, which eliminated the poly(A) tail along with 17 bp of the 3′-UTR. Sequencing revealed that the cDNA did not contain the entire coding region. To obtain the 5′ portion of the gene, RACE was performed using a 5′RACE kit according to the manufacturer (Life Technologies, Gaithersburg, MD). The nucleotide sequence of the cDNA and expression of this gene in seeds was verified against the exon sequence of a genomic clone (B. Downie and K.J. Bradford, unpublished data) and by cloning and sequencing the PG using RT-PCR with gene-specific primers on seed RNA, respectively.
Sequencing
Sequencing was performed at the Advanced Plant Genetics Facility (University of California, Davis). An ABI Prism 377 DNA Sequencer (ABI; Perkin-Elmer, Foster City, CA) utilizing dye termination chemistry with AmpliTaq DNA polymerase, FS (Taq; FS; Perkin-Elmer/Applied Biosystems Division [PE/ABI], Foster City, CA) was used to read cycle-sequencing reactions using a combination of universal and gene-specific primers (Genset, La Jolla, CA; Operon Technologies, Alameda, CA).
Genomic DNA Isolation and Analysis
Genomic DNA was isolated from lyophilized, expanding tomato leaves using a DNeasy kit (Qiagen, Valencia, CA). Genomic DNA (5 μg per lane) was exhaustively digested with restriction endonucleases, electrophoresed through a 0.8% (w/v) agarose gel in 1× TBE (Sambrook, 1989), and transferred to nylon membrane (Hybond N+, Amersham Life Science, Arlington Heights, IL). The digested DNA was cross-linked to the membrane and hybridized with the 1,504-bp radiolabeled cDNA. Hybridization was performed at 42°C for 12 h in 50% (v/v) formamide, 6× SSC, 5× Denhardt's solution (Denhardt, 1966), 0.5% (w/v) SDS, and 100 μg mL−1 boiled, sheared salmon sperm DNA. Blots were first washed at low stringency (5× SSC and 0.1% [w/v] SDS at 65°C) and exposed to film, prior to being re-hybridized with the PG probe, washed at high stringency (0.2× SSC and 0.1% [w/v] SDS, 65°C) and re-exposed to film.
RNA Isolation and Analysis
Tomato seeds were pulverized in liquid N2 and the RNA extracted and purified by a modification of the method of Ausubel et al. (1987). Extraction buffer consisted of 10 mm Tris-HCl, pH 8.2, 100 mm LiCl, 1 mm EDTA, 1% (w/v) SDS, and 25 mm DTT. Prior to use, aliquots of the RNA were incubated with DNase I for 1 h at 37°C in digestion buffer (40 mm Tris-HCl, pH 7.5, 6 mm MgCl2, 20 units of RNasin, and 10 mm NaCl) followed by extraction with 1 volume of phenol:chloroform:isoamyl alcohol (25:24:1). The RNA was precipitated in ethanol and dissolved in the original aliquot volume of 2 mm EDTA. Poly(A) selection was conducted using Oligotex resin (Qiagen) as directed by the manufacturer. Poly(A) RNA (5 μg) from whole seeds or 5 μg of total RNA from seed pieces were separated on formaldehyde-containing agarose gels and transferred onto positively charged nylon membranes (Amersham Life Science) in 10× SSC overnight and UV cross-linked. After rinsing the membranes for 5 min in 2× SSC, they were placed in pre-hybridization solution (50% [v/v] formamide, 5× Denhardt's solution, 100 μg mL−1 boiled, sheared salmon sperm DNA, 0.2% [w/v] SDS, and 6× SSC, pH 7.0 [Sambrook et al., 1989]) for 4 to 6 h at 42°C (DNA probes) or 62°C (RNA probes). DNA probes to the nearly full-length sequence of the PG (minus the poly[A] tail; see above) were synthesized by random priming (Feinburg and Vogelstein, 1983).
Radiolabeled antisense RNA probes for the PG and for G46, a ubiquitously expressed ribosomal protein transcript used as a loading control, were generated by linearizing the appropriate vector and incubating this template at 37°C with T7 DNA-dependent, RNA polymerase (Pharmacia Biotech, Alameda, CA) in a run-off transcription reaction in the presence of [α-32P]UTP (3,000 Ci mmol−1, New England Nuclear Life Science Products, Boston). The DNA probes were added to the prehybridization solution and the membrane hybridized for at least 12 h at 42°C. The RNA probes were hybridized to membranes at 62°C. Membranes were first hybridized with the PG probe and subsequently were re-exposed to the antisense RNA probe for G46.
Regardless of the probe used, the primary wash was done in 2× SSC and 0.1% (w/v) SDS at room temperature for 5 min, then repeated but at 65°C for 30 min. The two final high stringency washes were at 0.2× SSC and 0.1% (w/v) SDS, 65°C for 30 min each. The hybridized probe was detected by autoradiography or on a phosphorimager (Molecular Dynamics, Sunnyvale, CA).
In Situ Hybridization
PG transcripts were localized in situ using nonradioactive RNA probes labeled with digoxygenin (DIG) using colorometric detection. Labeled sense and antisense strands of RNA were obtained by run-off transcription using the T3 and T7 RNA polymerase promoters of pBSII KS (Invitrogen, Carlsbad, CA), respectively. Template was derived from either XbaI- or XhoI-digested PG cDNA in Bluescript. The transcription reaction was conducted in the presence of digoxygenin-labeled UTP. The nearly full-length transcripts were hydrolyzed in base to generate sheared probes with an average Mr of around 300 bp based on agarose gel electrophoresis.
Seeds germinated on water for 0, 36, and 48 h were sliced longitudinally and asymmetrically under ice-cold primary fixative (0.025 m phosphate buffer, pH 7.5, 2% [v/v] acrolein; 3% [v/v] glutaraldehyde, and 2% [w/v] paraformaldehyde) and fixed twice for 15 min under vacuum and subsequently overnight at 4°C. Fixed seeds were dehydrated in an ethanol series and infiltrated with a 4:1 mixture of nitrogen-sparged n-butyl- and methyl-methacrylate (Ted Pella, Redding, CA) to which benzoin ethyl ether, a UV activated catalyst, had been added. The seeds were arranged in molding trays under methacrylate and embedded by polymerizing the resin with UV light at 4°C for at least 24 h. Embedded seeds were fixed to plastic microtome chucks with glue and 2-μm sections obtained along the median, longitudinal plane on an Ultra-microtome with a diamond knife (LKB, Uppsala).
Sections were affixed to poly-Lys coated slides by heating overnight at 40°C. The plastic was removed from the sections by immersion in acetone for 30 min before rehydration through a graded ethanol series. Sections were treated with 500 μg mL−1 predigested proteinase K (Boehringer Mannheim) for 45 min, rinsed twice in 2 mg mL−1 Gly, and dehydrated through an ethanol series. Prehybridization solution (150 μL per slide; 50% [v/v] formamide, 300 mm NaCl, 10 mm Tris-HCl, pH 7.5, 1 mm EDTA, 5% [w/v] dextran sulfate, 1% [w/v] DIG blocking reagent [Boehringer Mannheim], and 150 μg mL−1 yeast tRNA) was applied for 3 h at 42°C. The sections were hybridized in 100 μL of prehybridization solution at 42°C containing 1 ng μL−1 either sense or antisense sheared probe for 14 to 18 h. The sections were rinsed in 4× SSPE, washed once in 2× SSPE at room temperature, and twice in 0.1× SSPE at 60°C. Sections were washed in 1× PBS and gently agitated in 1:2,500 dilution of anti-DIG-alkaline phosphatase conjugate for 3 h at room temperature.
Sections were washed three times in BSA washing solution (1% [w/v] phosphatase-free BSA, 0.3% [v/v] Triton X-100, 100 mm Tris-HCl, pH 7.5, and 150 mm NaCl) before being washed in TNM-5 buffer (100 mm Tris-HCl, pH 9.0, 100 mm NaCl, and 5 mm MgCl2). Sections were placed in slide mailers containing 32 mL of TNP buffer (100 mm Tris-HCl, pH 9.0, 100 mm NaCl, 10% [w/v] polyvinyl alcohol, and 5 mm MgCl2) with 16 mg mL−1 5-bromo-4-chloro-3-indolylphosphate and 0.33 mg mL−1 nitroblue tetrazolium. Color development was terminated with 10 mm Tris, pH 8.0, and 1 mm EDTA and the sections coated with Crystal Mount (Biomedia, Foster City, CA), which was hardened at 42°C overnight.
Phylogenetic Analysis
The deduced amino acid sequence of the tomato PG was aligned with 27 full-length deduced amino acid sequences available in the literature and/or database and three melon PGs reported previously (Hadfield et al., 1998) using Clustal V multiple-sequence alignment software (Higgins et al., 1992) and employing a PG from A. niger (2385) as an outgroup. The amino acid sequences and their protein identification (numbers were: apple fruit (1346704, Atkinson, 1994), Arabidopsis gene ADPG1 (2597824, L. Sander, R. Child, P. Uluskov, M. Albrechtsen, B. Joergensen, and B. Borkhardt, unpublished data), gene F5I14.10 (2190556, V.S. Vysotskaia, B.I. Osborne, M. Toriumi, G. Yi, O. Oji, Y.K. Shen, E. Buehler, A.B. Conway, A.R. Conway, K. Dewar, J. Feng, C. Kim, D. Kurtz, Y. Li, P. Shinn, H. Sun, R.W. Davis, J.R. Ecker, N.A. Federspiel, and A. Theologis, unpublished data), gene F6E13.1 (3212875, S.D. Roundsley, S. Kaul, X. Lin, K.A. Ketchum, M.L. Crosby, R.C. Brandon, S.M. Sykes, T.M. Mason, A.R. Kerlavage, M.D. Adams, C.R. Somerville, and J.C. Venter, unpublished data), and PGA2, 3, and 5 (3004442, 3004440, and 2982583, respectively, M. Torki, F. Thomas, R. Mache, P. Mandaron, and D. Falconet, unpublished data), avocado (22631, Dopico et al., 1993; Kutsunai et al., 1993), Japanese cedar (1076242, Komiyama et al., 1994), cotton (3024386, John and Petersen, 1994), kiwifruit (548488, Atkinson and Gardner, 1993), maize (288612 and 288379, Barakate et al., 1993; and 129940, Niogret et al., 1991), melon MPG1, 2, and 3 (3320458, 3320460, and 3320462, respectively, Hadfield et al., 1998), peach (2147957, Lee et al., 1990; 436420, Lester et al., 1994; and 479088, Lee et al., 1990), oilseed rape (304223, Robert et al., 1993; 1212786, Petersen et al., 1996; and 1419408, E.S. Jenkins, W. Paul, S.A. Coupe, S. Bell, L. Davies, and J.A. Roberts, unpublished data), tobacco (22701, Tebbutt et al., 1994), and tomato fruit (129939, Grierson et al., 1986); and tomato abscission zone PGs (TAPG1–5, 2459811, 1575705, 2459809, 2459815, and 2459817, respectively, Hong and Tucker, 1998). The phylogenetic tree was constructed using PAUP software package, version 3.1 (Swofford, 1990). The aligned sequences were analyzed in a heuristic search using simple stepwise addition of taxa. To extend the search beyond local optima on which stepwise addition is prone to converge (Swofford, 1990), the number of trees held for re-evaluation upon adding the next taxon was set to five throughout tree construction (Swofford, 1990). To further guard against defining a phylogenetic tree of local optima, global (tree bisection and reconnection) branch swapping using 100 bootstrapped replications was performed according to the recommendations of the author (Swofford, 1990) once all 32 taxa had been connected. The tree was rooted to an A. niger PG sequence, which was defined as the hypothetical ancestral taxon (outgroup).
RESULTS
A Calcium-Dependent Exo-PG Is Present in Tomato Seeds
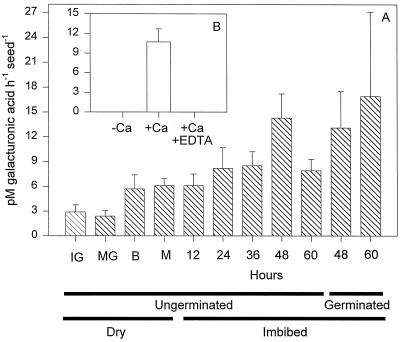
No endo-PG activity was detected in extracts of imbibed tomato seeds regardless of the type of assay used (viscometric, gel diffusion, or HPLC), the substrate employed (PGA or pectin from apple or citrus fruit), or the degree of esterification of citrus fruit pectin (10%, 30%, 60%, or 90%). Crude protein extracts from tomato fruit pericarp and a purified fungal endo-PG consistently exhibited endo-activity in the same assays (data not shown). However, PG activity from tomato seeds was detected using a reducing sugar assay (Fig. 1A), suggesting the presence of exo-activity. This activity was low during seed development but increased during imbibition and germination (Fig. 1A). Like exo-PGs isolated from pollen (Pressey and Reger, 1989; Pressey, 1991), the PG activity from tomato seeds required Ca2+, and adding EDTA to chelate Ca2+ completely inhibited enzyme activity (Fig. 1B).
Figure 1.
PG activity accumulates in conjunction with germination. A, The activity of PG from desiccated developing and mature seeds and mature seeds during germination and subsequent radicle protrusion as assessed using a reducing sugar assay in the presence of 5 mm CaCl2. From 48 h onward seeds were separated into those that had and had not completed germination. IG, Immature green; MG, mature green; B, breaker; M, mature. B, The effect of 5 mm CaCl2 on the activity of PG from whole cv Moneymaker seeds allowed to imbibe for 48 h from which the radicle had protruded. Activity is reported as picomoles of GalUA produced per hour per seed.
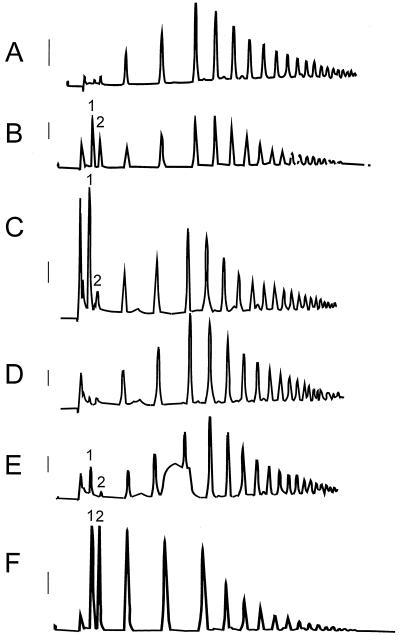
To further characterize the nature of PG activity in tomato seeds, protein extract was incubated with a 1% (w/v) solution of partially hydrolyzed PGA (G12 fraction from Campbell and Labavitch, 1991) and digestion products were analyzed by HPLC. The chromatographic profile of the G12 fraction prior to incubation (Fig. 2A) exhibited little mono- or di-GalUA (Fig. 2B). After incubation with tomato seed extract in the presence of 5 mm CaCl2, a large peak of monogalacturonic acid was present, but the remaining profile was essentially unchanged (Fig. 2C). Little or no monogalacturonic acid was present when the G12 fraction was incubated with boiled tomato seed extract (Fig. 2D) or with extract to which EDTA (Fig. 2E) had been added prior to incubation. In addition, the chromatographic profile upon incubation with tomato seed extract in the presence of CaCl2 (Fig. 2C) was considerably different from that obtained when a commercial source of endo-PG digested the G12 fraction (Fig. 2F). Endo-activity shifted the PGA profile to shorter oligomers (Fig. 2F), which did not occur in the presence of the tomato seed extract (Fig. 2C). These results support the contention that the PG activity present in tomato seeds is calcium-dependent and exo-acting.
Figure 2.
Profiles of a 1% (w/v) solution of partially hydrolyzed polygalacturonic acid exposed to tomato seed protein extract indicate the activity of an exo-PG. High-pressure liquid chromatography with pulsed amperometric detection (HPLC-PAD) resulted in the fractionation of a 1% (w/v) solution of partially acid hydrolyzed polygalacturonic acid. The solution is comprised of oligomers with a degree of polymerization from 6 to 19 centered on 10 to 12 (G12 fraction; Campbell and Labavitch, 1991). A, Untreated G12; B, G12 spiked with a commercial source of mono- (1) and di-GalUA (2) (made 2.5 nm with respect to both); C, G12 after exposure to tomato seed extract; D, G12 exposed to boiled tomato seed extract; E, G12 exposed to tomato seed extract containing 50 mm EDTA; F, G12 exposed to a commercial source of endo-PG from A. niger. Extracts in chromatograms C to E contained 5 mm CaCl2. Response bars to the left of each chromatogram represent a 100-mV deflection.
Identification and Cloning of a PG Transcript in Tomato Seeds
Six unique approximately 300-bp amplicons were obtained from a variety of tomato tissues using degenerate PCR primers to conserved regions present in all plant PGs and previously used to clone PGs from melon fruit (Hadfield et al., 1998). Of these six amplicons, only one, obtained from tomato leaf mRNA, hybridized to mRNA from tomato seeds (data not shown). The mRNA was also present in flowers, roots, and stems, which enabled a partial-length cDNA to be recovered from a tomato anther library (Twell et al., 1989). The remaining 5′ sequence was obtained from 5′-RACE reactions using cDNA reverse-transcribed from tomato seed RNA. Analysis of the amino acid sequence derived from the full-length cDNA (accession no. AF138858) revealed that the catalytic domain identifying PGs (block BL00502B; CGPGHGISIGSLG, Bussink et al., 1991; Blocks Database Version 9.3, Henikoff and Henikoff, 1991) was conserved in the tomato seed PG (data not shown). The four regions of universal PG homology described by Kester et al. (1996) were also present. An apparent signal peptide with a cleavage site between amino acids 38 and 39 at the amino terminus of the protein included amino acids comprising a potential transmembrane domain (amino acids 17–48 via RAOARGOS; 18–34 via SOAP; PCGene Software, Intelligenetics, Mountain View, CA, data not shown).
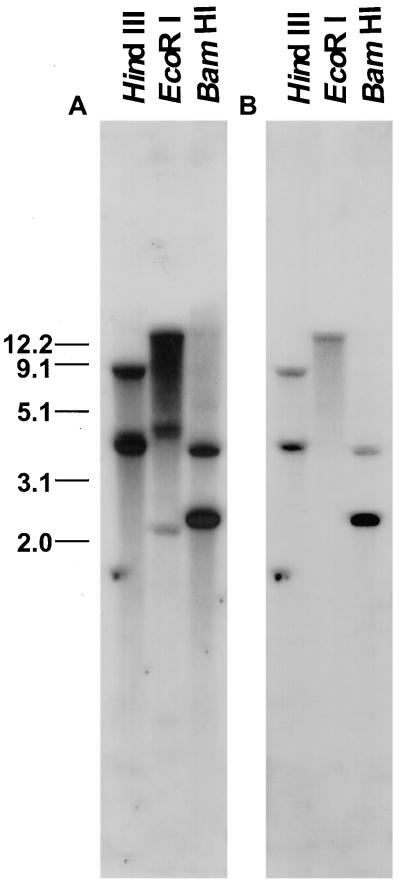
Tomato Seed PG Is Encoded by a Single Gene
A PG cDNA hybridized at high stringency to genomic DNA cleaved with either BamHI or HindIII identified two bands and a single band was detected in DNA cleaved by EcoRI, as predicted from the cDNA sequence (Fig. 3). We conclude that a single gene encodes the PG expressed in tomato seeds. Based on the unique nucleotide sequence and the conservation of catalytic domains characterizing PGs we have termed this gene LeXPG1.
Figure 3.
Southern-blot analysis of tomato genomic DNA probed with 1,504 bp of the poly(A)-truncated PG washed at low (5× SSC and 0.1% SDS at 65°C; A) and high (0.2× SSC and 0.1% SDS, 65°C; B) stringency suggest that there is little homology among the PG transcript and other members of this large gene family in tomato. The number of bands visible on the blot corresponds to the number predicted based on the restriction map of the cDNA (accession no. AF138858) and genomic clone (not shown).
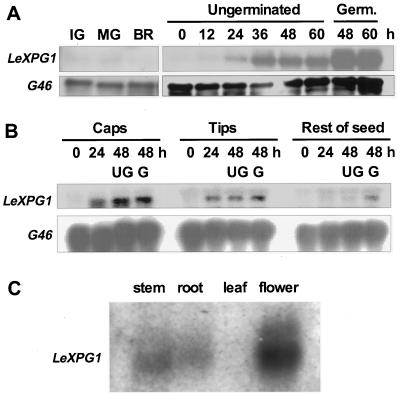
Expression of LeXPG1 Is Associated with Germination
LeXPG1 mRNA was undetectable in developing seeds obtained from fruits at the immature green, mature green, breaker, and ripe stages of development (Fig. 4A). LeXPG1 mRNA began to accumulate within 24 h of imbibition and increased by 36 h of imbibition, just prior to radicle protrusion (Fig. 4A). Abundance of the mRNA increased further in germinated seeds compared with ungerminated seeds at 48 and 60 h (Fig. 4A). To determine the tissue location of expression, imbibed seeds were dissected into the endosperm cap, the radicle tip, and the rest of the seed. LeXPG1 mRNA was present at 24 and 48 h in the endosperm caps and radicle tips, but was present in lesser amounts in the rest of the seed until after radicle emergence had occurred (Fig. 4B). In addition to the germinating seed, LeXPG1 mRNA was also present in tomato stems, roots, and flowers but not in leaves (Fig. 4C). Because the cDNA was recovered from an anther cDNA library, at least part of the expression detected in the flowers is from the anthers, although expression in other floral organs is not ruled out.
Figure 4.
Expression of LeXPG1 mRNA. A, mRNA (5 μg lane−1) from developing (IG, immature green; MG, mature green; BR, breaker) or mature wild-type cv Moneymaker tomato seeds allowed to imbibe from 0 to 60 h was hybridized with the LeXPG1 cDNA probe. Upon radicle protrusion, mRNA abundance increased as much as 10-fold over that present in mature, desiccated seeds. G46 is a constitutively expressed cDNA coding for a ribosomal protein that was used as an RNA loading control. Germ., Germinated. B, Northern-blot analysis of 5 μg of total RNA from wild-type cv Moneymaker tomato seed parts revealed that LeXPG1 mRNA is present in the endosperm cap and radicle tip by 24 h after imbibition. The mRNA accumulated in the rest of the wild-type seed 48 h after imbibition only if the seed had completed radicle emergence. UG, Ungerminated; G, germinated. C, Expression of LeXPG1 mRNA is not restricted to tomato seeds but also occurs in the stems, roots, and flowers (anthers) of tomato plants. Each lane was loaded with 5 μg of poly(A) RNA.
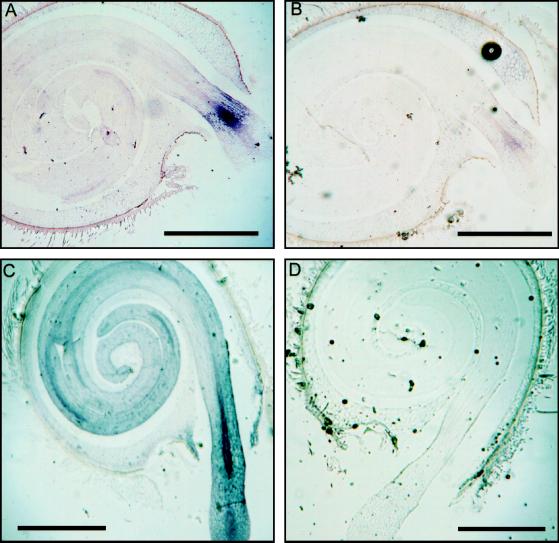
In situ hybridization localized the expression of the LeXPG1 gene in germinated seeds primarily to the embryo and the endosperm cap (Fig. 5, A and C). The expression in the embryo at 36 h was localized to a distinct band of tissue comprised of vascular, cortical, and epidermal cells and, while expression expanded by 48 h to include the whole of the embryo, it was most prominent in the vascular tissues in the region of radicle egress from the endosperm (Fig. 5C). Much weaker expression was also evident in the vascular tissue in the hypocotyl and cotyledons regardless of the time after imbibition (Fig. 5, A and C). Relative to the sense-hybridized control (Fig. 5B), there was also some expression in the peripheral cells of the endosperm cap that remained following radicle protrusion at 36-h imbibition (Fig. 5, compare A and B). The expression in the cap was more evident in the 48-h imbibed seed (Fig. 5C) since there was more cap tissue present in the plane of the section.
Figure 5.
PG mRNA accumulates in the embryo and the endosperm cap. A, In situ analysis detected expression of LeXPG1 mRNA in tomato embryos in a distinct band behind the radicle tip where it first emerged from the surrounding endosperm. Some expression was also detected in the remaining pieces of the endosperm cap. B, Section of the same seed as in A but probed with the sense strand RNA. C, At later stages of radicle protrusion, expression of LeXPG1 occurred throughout the embryo, but was most prominent in the vascular trace around the point of egress from the endosperm. Expression remained high in the endosperm cap. D, Section of the same seed as in C but probed with the sense strand RNA. Message was not observed in situ prior to the completion of germination (data not shown). Bars = 1 mm.
Sequence Homology with Other Known PGs
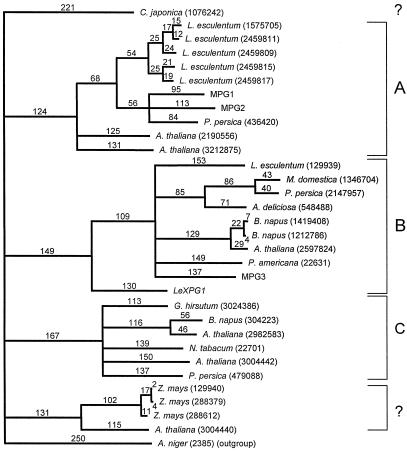
Based on alignment of the deduced amino acid sequences from 30 plant PGs, LeXPG1 is most highly homologous to a PG from avocado fruit (PID 166951), sharing 40% similarity at the amino acid level. LeXPG1 shares 13 highly conserved Cys residues with almost all of the PGs compared in the alignment. However, LeXPG1 does not cluster with PGs associated with angiosperm pollen (Pressey and Reger, 1989; Pressey, 1991). Instead, it is rather distantly related phylogenetically to PGs associated with abscission zones and fruit ripening that generally contain a prosequence (clade B in Hadfield et al., 1998). However, a PG isolated from Japanese cedar pollen also fell outside the clade defining PGs from angiosperm pollen (Fig. 6). In addition, LeXPG1 has been cloned from and detected in tomato anthers and flowers, suggesting that it too might be present in pollen. The assignment of LeXPG1 to clade B (Hadfield and Bennett, 1998; Hadfield et al., 1998) is in accordance with, but not due to, the presence of a predicted prosequence in LeXPG1.
Figure 6.
Homology of LeXPG1 to other reported PGs. Based on alignment of the deduced amino acid sequences from 31 PGs reported in the literature, the tomato seed PG (LeXPG1) is most homologous to a PG from avocado, sharing 40% similarity at the amino acid level. LeXPG1 does not segregate with PGs associated with pollen but is rather distantly related phylogenetically to PGs associated with abscission zones and fruit ripening that are predicted to contain a prosequence (Hadfield et al., 1998). The numbers above the lines on the tree represent theoretical divergence among PGs proportional to the number of amino acid substitutions.
DISCUSSION
This is the first report, to our knowledge, of exo-PG enzyme activity in seeds. The assignment of the tomato seed PG as an exo-enzyme is based on the absence of endo-activity in viscometric and gel diffusion assays and the exclusive detection of calcium-dependent activity in reducing sugar and HPLC assays. In addition, the products of the action of the tomato seed enzyme on oligomeric PGA substrates resolved by HPLC are consistent with exo-activity but not with endo-activity (Fig. 2). Finally, exo-PGs are known to require calcium for activity, as did the enzyme from tomato seeds (Pressey and Reger, 1989; Pressey, 1991). Whereas LeXPG1 was the only PG mRNA detected in tomato seeds, it remains to be confirmed that it codes for the exo-enzyme detected by activity assays. There was, however, a correlation between calcium-dependent exo-PG activity and the accumulation of LeXPG1 mRNA (Figs. 1 and 4, A and B).
An exo-PG present in liverwort cell cultures increased in parallel with cell growth until the cessation of the exponential growth phase, when the activity declined (Konno et al., 1983), and an exo-PG extracted from cultured carrot cells degrades pectic polymers from these cultures (Konno et al., 1984). Based on this developmental correlation, Konno et al. (1983) suggested that exo-PGs act coordinately with other cell wall-modifying enzymes to promote remodeling and/or loosening of the plant cell wall, permitting growth. The expression of LeXPG1 in a conspicuous band of cells in the radicle tip and later throughout the expanding embryo, is consistent with a role for PGs in potentiating the cell elongation necessary for growth. The detection of LeXPG1 in stems and roots undergoing secondary thickening and its presence in young expanding (initial PCR amplicon) but not mature expanded leaves (Fig. 4C) also correlates well with a role in mediating cell elongation. Pectin is deposited in a highly esterified form abundant in cell walls prior to the onset of elongation (Moore and Staehelin, 1988; Carpita and Gibeaut, 1993; Catesson, 1994). Pectin methylesterase activity is necessary to modify this esterified pectin to permit PG-mediated hydrolysis (Jona, 1989; Konno et al., 1989). PME is abundant from the earliest stage of tomato seed germination in both the endosperm cap and radicle tip (Downie et al., 1998). In protruded radicles, PME activity occurs in the tip of the radicle and in a distinct band of cells (Downie et al., 1998) reminiscent of the PG gene expression observed here. It is possible that this localized region of expression is associated with the transition between root and hypocotyl tissue.
The prominent expression of LeXPG1 in the vascular tissue of the radicle tip and its less prominent expression in the vascular tissue of the rest of the embryo (Fig. 5, A and C) implies a role for LeXPG1 in vascular tissue differentiation. Differentiation of the vasculature is one of the few changes discernable microscopically during seed germination prior to radicle protrusion (Sundås et al., 1992). The localization of a PG to the vascular cylinder in maize anther filaments (Dubald et al., 1993) may indicate a role for PGs in vascular tissue differentiation. Esterified pectin has been shown to be rapidly de-esterified in the stele of growing roots, particularly the vessel elements (Dolan and Roberts, 1995), which would potentiate hydrolysis by PGs. Not surprisingly, differentiation of Zinnia cell cultures into tracheary elements is accompanied by a marked decrease in pectin (Ingold et al., 1988) potentially brought about by PG activity.
The expression of LeXPG1 in the endosperm cap is correlated with the well-characterized weakening of this region necessary for radicle protrusion. Our understanding of the biochemical action of PGs on the plant cell wall is limited (Hadfield and Bennett, 1998), so a role for a single PG in both endosperm weakening and embryo growth (and/or vascular tissue differentiation) is plausible.
Alternatively, the PG in the endosperm cap could function as part of a plant defense mechanism that is up-regulated prior to radicle protrusion through the endosperm. Some PGs are thought to play a role in trimming large pectic oligomers cleaved by exogenous (pathogen produced) PGs to sizes effective in eliciting plant protective responses and/or in the removal of these signals, enabling the plant to stand down upon the cessation of pathogen attack (Aldington and Fry, 1993; García-Romera and Fry, 1995). PGs are capable of stimulating ethylene evolution (Baldwin and Pressey, 1990) leading to pathogenesis-related protein induction (Boller, 1988). Conceivably, LeXPG1 transcription and protein accumulation in the endosperm cap may be initiated in response to imminent endosperm rupture by the radicle, which would expose the endosperm to the external biotic environment. Pathogenesis-related proteins, such as a vacuolar targeted, class I β-1,3-glucanase, have been shown to accumulate exclusively in the cells of the endosperm cap of germinating tobacco seeds (Vögeli-Lange et al., 1994; Leubner-Metzger et al., 1996). In conjunction with endosperm weakening in tomato, both β-1,3-glucanase and chitinase are expressed in the endosperm caps of tomato seeds prior to the completion of germination (C.-T. Wu, G. Leubner-Metzger, F. Meins, Jr., and K.J. Bradford, unpublished results). Whether the PG in tomato seeds is involved in cell wall disassembly or functions in a manner to increase resistance to pathogens remains unresolved.
Phylogenetic analysis of LeXPG1 puts it into a clade of PGs associated with abscission zones and fruit softening, although the clade distinctions are by no means absolute. Hence, a PG from cedar pollen, like the PG from seeds and anthers, falls outside the clade containing most pollen PGs (Fig. 6). In addition, a Cys residue thought to be conserved in pollen PGs (Kester et al., 1996) is present in neither LeXPG1 (amino acid 306) nor in the PG from cedar pollen. Conversely, a PG cloned from melon fruit (MPG2) does contain this Cys (Hadfield et al., 1998). The presence of a potential prosequence in LeXPG1 is also consistent with its placement with clade B, as other PGs identified with clade C do not contain prosequences (Hadfield and Bennett, 1998).
The identification of LeXPG1 in germinating tomato seeds provides another candidate for enzymes that may be involved in endosperm weakening and/or embryo expansion related to the mechanism of radicle protrusion. Hydrolysis of PG-susceptible, acidic pectin in the middle lamella of the walls of endosperm cap cells would likely reduce cell-to-cell cohesion and facilitate penetration by the radicle. Alternatively, the PG in the cap could be performing a purely protective function. The localization of the message for the enzyme in the vascular tissues corroborates previous evidence of PG expression in the vascular tissue of maize (Dubald et al., 1993) and implicates PG activity in vascular tissue differentiation (Ingold et al., 1988). The enzyme could also potentially be involved in cell wall modification permitting turgor-driven elongation of the radicle culminating in radicle protrusion through the endosperm.
ACKNOWLEDGMENTS
We are grateful to Dr. Sheila McCormick (Plant Gene Expression Center, U.S. Department of Agriculture-Agricultural Research Service, Albany, CA) for providing us with the tomato anther cDNA library, to Dr. John Labavitch (Department of Pomology, University of California, Davis) for the G12 PGA, and to Carl Greve (Department of Pomology, University of California, Davis) for the HPLC elution protocol for separating the same. Dr. Donald Nevins permitted us the use of his Dionex HPLC-PAD.
Footnotes
This work was supported by the National Science Foundation (grant nos. IBN 9407264 and 9722978 to K.J.B.) and by the National Research Initiative Competitive Grants Program/U.S. Department of Agriculture (grant no. 9701534 to A.B.B.). Y.S. was supported by the United States-Israel Binational Agricultural Research and Development Fund Post-Doctoral Fellowship no. FI–0169–93. A.B.D. was supported by Natural Sciences and Engineering Research Council of Canada Post-Doctoral Scholarships.
LITERATURE CITED
- Aldington S, Fry SC. Oligosaccharins. Adv Bot Res. 1993;19:1–101. [Google Scholar]
- Atkinson RG. A cDNA clone for endopolygalacturonase from apple. Plant Physiol. 1994;105:1437–1438. doi: 10.1104/pp.105.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson RG, Gardner RC. A polygalacturonase gene from kiwifruit (Actinidia deliciosa) Plant Physiol. 1993;103:669–670. doi: 10.1104/pp.103.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kinston RE, Moore DD, Smith JA, Seidman JG, Struhl K. Current Protocols in Molecular Biology. New York: Wiley-Interscience; 1987. [Google Scholar]
- Baldwin EA, Pressey R. Exopolygalacturonase elicits ethylene production in tomato. HortScience. 1990;25:779–780. [Google Scholar]
- Barakate A, Martin W, Quigley F, Mache R. Characterization of a multigene family encoding an exopolygalacturonase in maize. J Mol Biol. 1993;229:797–801. doi: 10.1006/jmbi.1993.1084. [DOI] [PubMed] [Google Scholar]
- Bewley JD. Seed germination and dormancy. Plant Cell. 1997;9:1055–1066. doi: 10.1105/tpc.9.7.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T. Ethylene and the regulation of antifungal hydrolases in plants. Oxf Surv Plant Mol Cell Biol. 1988;5:145–174. [Google Scholar]
- Bonghi C, Rascio N, Ramina A, Casadoro G. Cellulase and polygalacturonase involvement in the abscission of leaf and fruit explants of peach. Plant Mol Biol. 1992;20:839–848. doi: 10.1007/BF00027155. [DOI] [PubMed] [Google Scholar]
- Buescher RW, Burgin C. Diffusion plate assay for measurement of polygalacturonase activity in pickle brines. J Food Biochem. 1992;16:59–68. [Google Scholar]
- Bussink HJD, Buxton FP, Visser J. Expression and sequence comparison of the Aspergillus niger and Aspergillus tubigensis genes encoding polygalacturonase II. Curr Genet. 1991;19:467–474. doi: 10.1007/BF00312738. [DOI] [PubMed] [Google Scholar]
- Campbell AD, Labavitch JM. Induction and regulation of ethylene biosynthesis by pectic oligomers in cultured pear cells. Plant Physiol. 1991;97:699–705. doi: 10.1104/pp.97.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut M. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993;3:1–30. doi: 10.1111/j.1365-313x.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- Catesson A-M. Cambial ultrastructure and biochemistry: changes in relation to vascular tissue differentiation and the seasonal cycle. Int J Plant Sci. 1994;155:251–261. [Google Scholar]
- Christensen PE. Methods of grading pectin in relation to the molecular weight (intrinsic viscosity) of pectin. Food Res. 1954;19:163–172. [Google Scholar]
- Clarke AE, Gleeson PA. Molecular aspects of recognition and response in pollen-stigma interactions. Rec Adv Phytochem. 1981;15:161–211. [Google Scholar]
- Dahal P, Bradford KJ. Effects of priming and endosperm integrity on seed germination rates of tomato genotypes. II. Germination at reduced water potential. J Exp Bot. 1990;41:1441–1453. [Google Scholar]
- Dahal P, Nevins DJ, Bradford KJ. Relationship of endo-β-d-mannanase activity and cell wall hydrolysis in tomato endosperm to germination rates. Plant Physiol. 1997;113:1243–1252. doi: 10.1104/pp.113.4.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denhardt DT. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966;23:641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Dolan L, Roberts K. Secondary thickening in roots of Arabidopsis thaliana: anatomy and cell surface changes. New Phytol. 1995;131:121–128. doi: 10.1111/j.1469-8137.1995.tb03061.x. [DOI] [PubMed] [Google Scholar]
- Dopico B, Lowe AL, Wilson ID, Merodio C, Grierson D. Cloning and characterization of avocado fruit mRNAs and their expression during ripening and low-temperature storage. Plant Mol Biol. 1993;21:437–449. doi: 10.1007/BF00028802. [DOI] [PubMed] [Google Scholar]
- Downie B, Dirk LMA, Hadfield KA, Wilkins TA, Bennett AB, Bradford KJ. A gel diffusion assay for quantification of pectin methylesterase activity. Anal Biochem. 1998;264:149–157. doi: 10.1006/abio.1998.2847. [DOI] [PubMed] [Google Scholar]
- Dubald M, Barakate A, Mandaron P, Mache R. The ubiquitous presence of exopolygalacturonase in maize suggests a fundamental cellular function of this enzyme. Plant J. 1993;4:781–791. doi: 10.1046/j.1365-313x.1993.04050781.x. [DOI] [PubMed] [Google Scholar]
- Feinburg AP, Vogelstein B. A technique for radiolabelling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- García-Romera I, Fry SC. The longevity of biologically-active oligogalacturonides in rose cell cultures: degradation by exo-polygalacturonase. J Exp Bot. 1995;46:1853–1857. [Google Scholar]
- Grierson D, Tucker GA, Keen J, Ray J, Bird CR, Schuch W. Sequencing and identification of a cDNA clone for tomato polygalacturonase. Nucleic Acids Res. 1986;14:8595–8603. doi: 10.1093/nar/14.21.8595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groot SPC, Karssen CM. Gibberellins regulate seed germination in tomato by endosperm weakening: a study with gibberellin-deficient mutants. Planta. 1987;171:525–531. doi: 10.1007/BF00392302. [DOI] [PubMed] [Google Scholar]
- Groot SPC, Karssen CM. Dormancy and germination of abscisic acid-deficient tomato seeds: studies with the sitiens mutant. Plant Physiol. 1992;99:952–958. doi: 10.1104/pp.99.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groot SPC, Kieliszewska-Rokicka B, Vermeer E, Karssen CM. Gibberellin-induced hydrolysis of endosperm cell walls in gibberellin-deficient tomato seeds prior to radicle protrusion. Planta. 1988;174:500–504. doi: 10.1007/BF00634479. [DOI] [PubMed] [Google Scholar]
- Gross KC. A rapid and sensitive spectrophotometric method for assaying polygalacturonase using 2-cyanoacetamide. HortScience. 1982;17:933–934. [Google Scholar]
- Hadfield KA, Bennett AB. Polygalacturonases: many genes in search of a function. Plant Physiol. 1998;117:337–343. doi: 10.1104/pp.117.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield KA, Rose JKC, Yaver DS, Berka RM, Bennett AB. Polygalacturonase gene expression in ripe melon fruit supports a role for polygalacturonase in ripening associated pectin disassembly. Plant Physiol. 1998;117:363–373. doi: 10.1104/pp.117.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MG, Bucheli P, Cervone F, Doares SH, O'Neill RA, Darvill A, Albersheim P. The role of the cell wall constituents in plant-pathogen interactions. In: Nester E, Kosuge T, editors. Plant-Microbe Interactions. Vol. 3. New York: McGraw-Hill; 1989. pp. 1–38. [Google Scholar]
- Henikoff S, Henikoff JG. Automated assembly of protein blocks for database searching. Nucleic Acids Res. 1991;19:6565–6572. doi: 10.1093/nar/19.23.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins DG, Bleasby AJ, Fuchs R. Clustal V: improved software for multiple sequence alignment. Comput Appl Biosci. 1992;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- Hong SB, Tucker ML. Genomic organization of six tomato polygalacturonases and 5′ upstream sequence identity with tap1 and win2 genes. Mol Gen Genet. 1998;258:479–487. doi: 10.1007/s004380050758. [DOI] [PubMed] [Google Scholar]
- Ingold E, Sugiyama M, Komamine A. Secondary cell wall formation: changes in cell wall constituents during the differentiation of isolated mesophyll cells of Zinnia elegans to tracheary elements. Plant Cell Physiol. 1988;29:295–303. [Google Scholar]
- Jenkins ES, Paul W, Coupe SA, Bell SJ, Davies EC, Roberts JA. Characterization of an mRNA encoding a polygalacturonase expressed during pod development in oilseed rape (Brassica napus L.) J Exp Bot. 1996;47:111–115. [Google Scholar]
- John ME, Petersen MW. Cotton (Gossypium hirsutum L.) pollen-specific polygalacturonase mRNA: tissue and temporal specificity of its promoter in transgenic tobacco. Plant Mol Biol. 1994;26:1989–1993. doi: 10.1007/BF00019509. [DOI] [PubMed] [Google Scholar]
- Jona R. Cell wall pectic content as an early signal for cell separation. NATO ASI Adv Sci Inst Ser Ser H Cell Biol. 1989;35:421–437. [Google Scholar]
- Karssen CM, Haigh A, van der Toorn P, Weges R. Physiological mechanisms involved in seed priming. In: Taylorson RB, editor. Recent Advances in the Development and Germination of Seeds. New York: Plenum Press; 1989. pp. 269–280. [Google Scholar]
- Kester HCM, Kusters-van Someren MA, Müller Y, Visser J. Primary structure and characterization of an exopolygalacturonase from Aspergillus tubingensis. Eur J Biochem. 1996;240:738–746. doi: 10.1111/j.1432-1033.1996.0738h.x. [DOI] [PubMed] [Google Scholar]
- Komiyama N, Sone T, Shimizu K, Morikubo K, Kino K. cDNA cloning and expression of Cry j II the second major allergen of Japanese cedar pollen. Biochem Biophys Res Commun. 1994;201:1021–1028. doi: 10.1006/bbrc.1994.1804. [DOI] [PubMed] [Google Scholar]
- Konno H, Yamasaki Y, Katoh K. Exopolygalacturonase from suspension cultures of Marchantia polymorpha: its presence and involvement in pectic polysaccharide degradation. Plant Physiol. 1983;73:216–222. doi: 10.1104/pp.73.2.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno H, Yamasaki Y, Katoh K. Degradation of pectic polysaccharides extracted from suspension cultures of carrot by purified exo-polygalacturonase. Physiol Plant. 1984;61:20–26. [Google Scholar]
- Konno H, Yamasaki Y, Katoh K. Extracellular exo-polygalacturonase secreted from carrot cell cultures: its purification and involvement in pectic polymer degradation. Physiol Plant. 1989;76:514–520. [Google Scholar]
- Kutsunai SY, Lin AC, Percival FW, Laties GG, Christoffersen RE. Ripening-related polygalacturonase cDNA from avocado. Plant Physiol. 1993;103:289–290. doi: 10.1104/pp.103.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Speirs J, Gray J, Brady CJ. Homologies to the tomato endopolygalacturonase gene in the peach genome. Plant Cell Environ. 1990;13:513–521. [Google Scholar]
- Lester DR, Speirs J, Orr G, Brady CJ. Peach (Prunus persica) endopolygalacturonase cDNA isolation and mRNA analysis in melting and nonmelting peach cultivars. Plant Physiol. 1994;105:225–231. doi: 10.1104/pp.105.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leubner-Metzger G, Fründt C, Meins F., Jr Effects of gibberellins, darkness and osmotica on endosperm rupture and class I β-1,3-glucanase induction in tobacco seed germination. Planta. 1996;199:282–288. [Google Scholar]
- Leviatov S, Shoseyov O, Wolf S. Involvement of endomannanase in the control of tomato seed germination under low temperature conditions. Ann Bot. 1995;76:1–6. [Google Scholar]
- Melotto E, Greve LC, Labavitch JM. Cell wall metabolism in ripening fruit. VII. Biologically active pectin oligomers in ripening tomato (Lycopersicon esculentum Mill.) fruits. Plant Physiol. 1994;106:575–581. doi: 10.1104/pp.106.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PJ, Staehelin LA. Immunogold localization of the cell-wall-matrix polysaccharides rhamnogalacturonan I and xyloglucan during cell expansion and cytokinesis in Trifolium pratense L.: implications for secretory pathway. Planta. 1988;174:433–445. doi: 10.1007/BF00634471. [DOI] [PubMed] [Google Scholar]
- Mu J-H, Stains JP, Kao T. Characterization of a pollen-expressed gene encoding a putative pectin esterase of Petunia inflata. Plant Mol Biol. 1994;25:539–544. doi: 10.1007/BF00043881. [DOI] [PubMed] [Google Scholar]
- Ni BR, Bradford KJ. Germination and dormancy of abscisic acid and gibberellin-deficient mutant tomato (Lycopersicon esculentum) seeds: sensitivity of germination to abscisic acid, gibberellin and water potential. Plant Physiol. 1993;101:607–617. doi: 10.1104/pp.101.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niogret MF, Dubald M, Mandaron P, Mache R. Characterization of pollen polygalacturonase encoded by several cDNA clones in maize. Plant Mol Biol. 1991;17:1155–1164. doi: 10.1007/BF00028732. [DOI] [PubMed] [Google Scholar]
- Peretto R, Favaron F, Bettini V, DeLornezo G, Marini S, Alghisi P, Cervone F, Bonfante P. Expression and localization of polygalacturonase during the outgrowth of lateral roots in Allium porrum L. Planta. 1992;188:164–172. doi: 10.1007/BF00216810. [DOI] [PubMed] [Google Scholar]
- Petersen M, Sander L, Child R, van Onckelen H, Ulvskov P, Borkhardt B. Isolation and characterization of a pod dehiscence zone-specific polygalacturonase from Brassica napus. Plant Mol Biol. 1996;31:517–527. doi: 10.1007/BF00042225. [DOI] [PubMed] [Google Scholar]
- Pressey R. Polygalacturonase in tree pollens. Phytochemistry. 1991;30:1753–1755. [Google Scholar]
- Pressey R, Reger BJ. Polygalacturonase in pollen from corn and other grasses. Plant Sci. 1989;59:57–62. [Google Scholar]
- Robert LS, Allard S, Gerster JL, Cass L, Simmonds J. Isolation and characterization of a polygalacturonase gene highly expressed in Brassica napus pollen. Plant Mol Biol. 1993;23:1273–1278. doi: 10.1007/BF00042360. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. A Laboratory Manual. Ed 2. I–III. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sánchez RA, de Miguel L, Mercuri O. Phytochrome control of cellulase activity in Datura ferox L. seeds and its relationship with germination. J Exp Bot. 1986;37:1574–1580. [Google Scholar]
- Still DW, Bradford KJ. Endo-β-mannanase activity from individual tomato endosperm caps and radicle tips in relation to germination rates. Plant Physiol. 1997;113:21–29. doi: 10.1104/pp.113.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundås A, Tandre K, Holmstedt E, Engström P. Differential gene expression during germination and after the induction of adventitious bud formation in Norway spruce embryos. Plant Mol Biol. 1992;18:713–724. doi: 10.1007/BF00020013. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP: Phylogenetic Analysis Using Parsimony, Version 3.0. Champaign, IL: Illinois Natural History Survey; 1990. [Google Scholar]
- Tagawa K, Kaji A. Polygalacturonase from Corticium rolfsii. Methods Enzymol. 1988;161:361–366. [Google Scholar]
- Taylor JE, Tucker GA, Lasslett Y, Smith CJS, Arnold CM, Watson CF, Schuch W, Grierson D, Roberts JA. Polygalacturonase expression during leaf abscission of normal and transgenic tomato plants. Planta. 1990;183:133–138. doi: 10.1007/BF00197577. [DOI] [PubMed] [Google Scholar]
- Tebbutt SJ, Rogers HJ, Lonsdale DM. Characterization of a tobacco gene encoding a pollen-specific polygalacturonase. Plant Mol Biol. 1994;23:397–400. doi: 10.1007/BF00023244. [DOI] [PubMed] [Google Scholar]
- Toorop PE, Bewley JD, Hilhorst HWM. Endo-β-mannanase isoforms are present in the endosperm and embryo of tomato seeds, but are not essentially linked to the completion of germination. Planta. 1996;200:153–158. [Google Scholar]
- Townsend RR, Hardy MR, Hindsgaul O, Lee YC. High-performance anion-exchange chromatography of oligosaccharides using pellicular resins and pulsed amperometric detection. Anal Biochem. 1988;174:459–470. doi: 10.1016/0003-2697(88)90044-9. [DOI] [PubMed] [Google Scholar]
- Twell D, Wing R, Yamaguchi J, McCormick S. Isolation and expression of an anther-specific gene from tomato. Mol Gen Genet. 1989;217:240–245. doi: 10.1007/BF02464887. [DOI] [PubMed] [Google Scholar]
- Vögeli-Lange R, Fründt C, Hart CM, Beffa R, Nagy F, Meins F., Jr Evidence for a role of β-1,3-glucanase in dicot seed germination. Plant J. 1994;5:273–278. [Google Scholar]
- Wilson KJ, Nessler CL, Mahlberg PG. Pectinase in Asclepias latex and its possible role in laticifer growth and development. Am J Bot. 1976;63:1140–1144. [Google Scholar]