Abstract
Unlike common epidermal growth factor receptor gene (EGFR) mutations that confer sensitivity to tyrosine kinase inhibitors (TKIs) in non-small cell lung cancer (NSCLC), mutations in exon 20 of either EGFR or the human EGFR2 gene (HER2) are associated with insensitivity to EGFR-TKIs, with treatment options for patients with such mutations being limited. Clinical characteristics, outcome of EGFR-TKI or nivolumab treatment, and the presence of coexisting mutations were reviewed for NSCLC patients with exon-20 mutations of EGFR or HER2 as detected by routine application of an amplicon-based next-generation sequencing panel. Between July 2013 and June 2017, 206 patients with pathologically confirmed lung cancer were screened for genetic alterations including HER2 and EGFR mutations. Ten patients harbored HER2 exon-20 insertions (one of whom also carried an exon-19 deletion of EGFR), and 12 patients harbored EGFR exon-20 mutations. Five of the 13 patients with EGFR mutations were treated with EGFR-TKIs, two of whom manifested a partial response, two stable disease, and one progressive disease. Among the seven patients treated with nivolumab, one patient manifested a partial response, three stable disease, and three progressive disease, with most (86%) of these patients discontinuing treatment as a result of disease progression within 4 months. The H1047R mutation of PIK3CA detected in one patient was the only actionable mutation coexisting with the exon-20 mutations of EGFR or HER2. Potentially actionable mutations thus rarely coexist with exon-20 mutations of EGFR or HER2, and EGFR-TKIs and nivolumab show limited efficacy in patients with such exon-20 mutations.
Keywords: epidermal growth factor receptor gene (EGFR), human epidermal growth factor receptor 2 gene (HER2), resistance, lung cancer, nivolumab
INTRODUCTION
Lung cancer is the most common cause of cancer-related death worldwide, with non-small cell lung cancer (NSCLC) accounting for ~75% of all lung cancer cases [1]. Recent insight into the molecular basis of lung cancer has led to changes in disease treatment. The identification of driver mutations, such as those affecting the epidermal growth factor receptor (EGFR) [2–4] and anaplastic lymphoma kinase (ALK) [5, 6] genes, has already been successfully translated into clinical practice. Most patients with NSCLC harboring EGFR mutations show an initial marked response to EGFR tyrosine kinase inhibitors (TKIs) [7]. Exon-19 deletions (Ex19del) and the L858R point mutation in exon 21 of EGFR confer high sensitivity to EGFR-TKIs and account for ~85% of all EGFR mutations [8]. The remaining EGFR mutations include point mutations in exon 18 (G719X, ~3% of EGFR mutations) and exon 21 (L861Q, ~2% of EGFR mutations) that confer moderate sensitivity to EGFR-TKIs. On the other hand, exon-20 insertions account for ~5% of all EGFR mutations and define a distinct subset of lung adenocarcinoma characterized by a poor response to the first-generation EGFR-TKIs gefitinib and erlotinib [9–11].
Even patients with TKI-sensitizing EGFR mutations can manifest de novo resistance or only a short-term response to EGFR-TKIs. One mechanism of such de novo resistance is mediated by a T790M mutation in exon 20 of EGFR that impedes drug binding to the ATP pocket of the receptor protein [12, 13]. The S768I mutation in exon 20 of EGFR has similarly been associated with a poor response to EGFR-TKI treatment in a case series [14]. Mutations of the human epidermal growth factor receptor 2 gene (HER2, also known as ERBB2) have also been identified as oncogenic drivers in NSCLC with a frequency of 2% to 3%, with most such mutations consisting of a 12-bp in-frame insertion (encoding YVMA) in exon 20 [15–17]. Preclinical and clinical studies have shown that such mutations in the kinase domain result in constitutive phosphorylation and activation of HER2 [18]. Similar to patients with EGFR exon-20 mutations, treatment with the EGFR- and HER2-selective TKIs afatinib or dacomitinib has met with limited success in patients harboring exon-20 mutations of HER2 [17, 19].
Immune-checkpoint inhibitors such as antibodies to programmed cell death–1 (PD-1) have emerged as promising therapeutic agents for NSCLC. Phase III trials of the PD-1 inhibitors nivolumab and pembrolizumab in previously treated patients with NSCLC have thus indicated that immunotherapy strategies will provide new therapeutic options for advanced NSCLC [20–22]. Biomarker studies revealed a significant correlation between the pretreatment expression level of the PD-1 ligand PD-L1 in tumor cells (as determined by immunohistochemical analysis) and the likelihood of a response to PD-1 inhibitors, whereas EGFR mutations appear to be a negative predictive factor for PD-1 inhibitor efficacy. Given that most EGFR mutations are either exon-19 deletions or L858R in exon 21, however, it has been unclear whether such treatment is also without benefit in patients with uncommon EGFR mutations.
Given the limited treatment options for NSCLC patients harboring mutations in exon 20 of either EGFR or HER2, it is important that the clinical features and treatment response of such patients be further explored. We have therefore now evaluated the clinical background of patients with NSCLC positive for EGFR or HER2 exon-20 mutations as well as their response to treatment including that with the immune-checkpoint inhibitor nivolumab. We also examined the patients for coexisting mutations that offer the potential for targeted treatment.
RESULTS
Patient characteristics
Between July 2013 and June 2017, 206 patients with pathologically confirmed lung cancer were screened with a next-generation sequencing (NGS) panel including HER2 and EGFR genes at Kindai University Hospital (Supplementary Figure 1). Nine patients (4%) were found to harbor HER2 exon-20 mutations, 12 patients (6%) to harbor EGFR exon-20 mutations, and one patient (0.5%) to harbor a HER2 exon-20 mutation as well as an EGFR Ex19del mutation. The demographics of these 22 patients included in the present study are shown in Table 1. Thirteen (59%) of the patients were female, and 15 (68%) were never- or light smokers, with the median age of all patients being 70 years (range, 44 to 81). Most (91%) patients had a good Eastern Cooperative Oncology Group performance status (0 or 1) at the onset of initial chemotherapy. Twenty-one patients (95%) had adenocarcinoma, and 18 (82%) had disease of stage IV. With regard to the type of EGFR exon-20 mutations (n = 12), eight patients (36%) had exon-20 insertions, three (14%) had a known exon-20 resistance mutation (T790M or S768I) together with a TKI-sensitizing mutation (Ex19del, G719C, or L858R), and one (5%) had a G779F mutation, whose effect on TKI sensitivity is unknown. All 10 patients with HER2 exon-20 mutations harbored in-frame insertions (A775_G776 ins YVMA in nine patients, and A775_G776 ins V in one).
Table 1. Characteristics of NSCLC patients harboring exon-20 mutations of EGFR or HER2 (n = 22).
| Characteristic | Subset | No. of patients (%) |
|---|---|---|
| [Median (range) age in years | 70 (44–81)] | |
| Sex | Male | 9 (41) |
| Female | 13 (59) | |
| Smoking status | Never | 14 (64) |
| Light smokera | 1 (5) | |
| Smokera | 7 (32) | |
| ECOG PS | 0 | 4 (18) |
| 1 | 16 (73) | |
| 2 | 2 (9) | |
| Tumor histology | Adenocarcinoma | 21 (95) |
| NSCLC, favoring adenocarcinoma | 1 (5) | |
| Disease stage | IIIA | 1 (5) |
| IV | 18 (82) | |
| Postoperative recurrence | 3 (14) | |
| EGFR exon-20 mutation | M766_A767 ins ASV | 2 (9) |
| (n = 12) | A767_S768 ins SVD | 2 (9) |
| A767_S768 ins SVG | 1 (5) | |
| S768I (+ G719C) | 1 (5) | |
| S768I (+ L858R) | 1 (5) | |
| V769_D770 ins ASV | 1 (5) | |
| D770_N771 ins GL | 1 (5) | |
| D770_N771 ins NPG | 1 (5) | |
| G779F | 1 (5) | |
| T790M (+ E746_T751 delins A) | 1 (5) | |
| HER2 exon-20 mutation | A775_G776 ins V | 1 (5) |
| (n = 9) | A775_G776 ins YVMA | 8 (36) |
| EGFR/HER2 double | EGFR E746_A750 del + | 1 (5) |
| mutation (n = 1) | HER2 A775_G776 ins YVMA | |
aLight smoker, ≤10 pack-years; smoker, >10 pack-years. ECOG PS, Eastern Cooperative Oncology Group performance status.
A total of 16 nonsynonymous mutations in genes other than EGFR and HER2 was detected in the 22 patients, for a mean of 0.7 such mutations per tumor (Figure 1). The most common such genetic alterations were TP53 mutations, which were detected in 12 patients (55%), whereas SMAD4, PIK3CA, DDR2, and KRAS mutations were each present in one patient (5%). Among these various genetic alterations, only one (H1047R of PIK3CA) was potentially treatable with therapeutic agents.
Figure 1. Somatic gene mutations detected with an NGS panel covering 22 genes in lung cancer specimens positive for exon-20 mutations of EGFR or HER2.
Each column corresponds to one of the 22 patients.
Outcome of EGFR-TKI treatment
Median overall survival (OS) for all patients (n = 22) harboring EGFR or HER2 exon-20 mutations was 23.5 months, with 13 deaths and a median follow-up time of 18.2 months for survivors at last censoring. Most patients (n = 16, 73%) received platinum-based chemotherapy as a first-line drug treatment: pemetrexed plus platinum, with or without bevacizumab, in 13 patients, paclitaxel plus platinum in two patients, and S-1 plus platinum plus bevacizumab in one patient (Supplementary Figure 1). Of the remaining patients (n = 6), two individuals received an EGFR-TKI, two pemetrexed, one docetaxel, and one best supportive care.
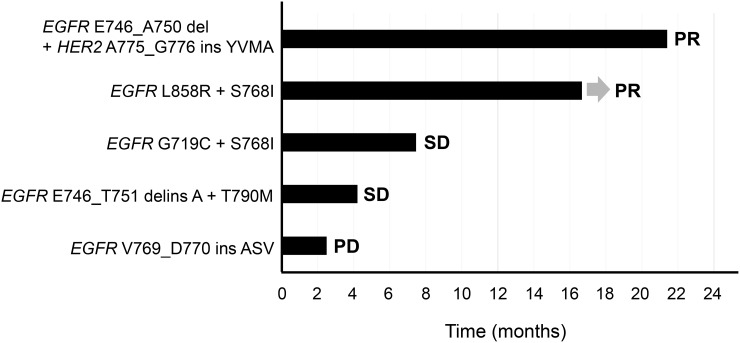
Among the 13 patients positive for EGFR mutations, five individuals received an EGFR-TKI, including three treated with a first-generation agent (gefitinib or erlotinib) and two with a second-generation agent (afatinib). Two of these five patients received the EGFR-TKI as first-line chemotherapy, whereas the remaining three received at least one cytotoxic chemotherapy regimen, including platinum-based combinations, before EGFR-TKI treatment. Two of the five patients manifested a partial response (PR) to EGFR-TKI treatment, two stable disease (SD), and one progressive disease (PD), with the progression-free survival (PFS) of all five individuals during EGFR-TKI therapy being shown in Figure 2. The two patients who achieved a response with long-term remission (>1 year) harbored common TKI-sensitizing EGFR mutations (EGFR E746_A750 del + HER2 A775_G776 ins YVMA in one, and EGFR L858R + S768I in the other).
Figure 2. Swimmer plot for duration of disease stability or response to EGFR-TKI treatment in patients with exon-20 mutations of EGFR or HER2.
Bar length indicates the duration of EGFR-TKI treatment for each patient, with the best response observed before treatment failure indicated on the right. The origin corresponds to treatment start date, and the arrow indicates an ongoing response at the time of data censoring.
Outcome of nivolumab treatment
Given the limited efficacy of EGFR-TKIs for the distinct subset of NSCLC patients with EGFR exon-20 mutations as well as the poor response to EGFR-TKI treatment apparent for NSCLC patients with HER2 exon-20 mutations, we examined the efficacy of immune-checkpoint inhibitors in these patients. Five patients with EGFR exon-20 insertions and two with HER2 exon-20 insertions received nivolumab therapy. Five of these seven patients received nivolumab as second-line chemotherapy, whereas the remaining two patients received the drug in the third-line or later setting (Supplementary Figure 1). Among the seven patients treated with nivolumab, one manifested a PR, three SD, and three PD, with the PFS for all these individuals during nivolumab therapy being shown in Figure 3. Most (86%) patients discontinued nivolumab treatment as a result of disease progression within 4 months. The tumors of five patients who received nivolumab therapy were assessed for PD-L1 expression by immunohistochemistry. The tumor proportion score (TPS) for PD-L1 expression [23] was 50% in two patients and 10%, 5%, or 0% in the remaining three (Figure 3).
Figure 3. Swimmer plot for duration of disease stability or response to nivolumab treatment in patients with exon-20 mutations of EGFR or HER2.
Bar length indicates the duration of nivolumab treatment for each patient, with the best response observed before treatment failure indicated on the right. The origin corresponds to treatment start date, and the arrow indicates an ongoing response at the time of data censoring. The TPS for PD-L1 expression in tumor specimens is indicated at the far right. NA, not assessed.
DISCUSSION
Most NSCLC patients harboring TKI-sensitizing EGFR mutations show an initial marked response to EGFR-TKI treatment. The clinical course of EGFR mutation-positive NSCLC during EGFR-TKI treatment shows substantial variability, however [7]. Insertions in exon 20 of EGFR have thus been associated with resistance to EGFR-TKIs in NSCLC patients [9–11]. Case-series studies have found the objective response rate for EGFR-TKI (gefitinib or erlotinib) treatment in patients with EGFR exon-20 mutations to be ~10% to 20%, with a median PFS of ~2 months [9, 11, 14]. The OS of patients with tumors harboring EGFR exon-20 insertions was found to be similar to that for those with tumors wild-type for EGFR [11]. Afatinib was also shown to have limited efficacy in patients with EGFR exon-20 insertions [24]. In addition, a de novo T790M mutation or an S768I mutation in exon 20 of EGFR is associated with moderate efficacy of or resistance to EGFR-TKI treatment in patients whose tumors also harbor TKI-sensitizing EGFR mutations [12–14]. Consistent with these previous observations, our retrospective case series has shown that a patient with an EGFR exon-20 insertion did not respond to EGFR-TKI treatment, although two patients harboring common TKI-sensitizing EGFR mutations together with other resistance mutations achieved a long-term response to such treatment.
The prevalence of HER2 mutations has been found to range from 2% to 3% in NSCLC [15–17]. In the present study, HER2 mutations were detected in 5% of NSCLC patients and were associated with never-smoker status, female sex, and adenocarcinoma histology, consistent with previous observations. A phase II trial of dacomitinib, an irreversible TKI with activity against both EGFR and HER2, revealed that only 12% of NSCLC patients harboring HER2 exon-20 mutations experienced a PR, with none of those harboring a YVMA insertion showing a response [17]. Several next-generation HER2-targeted drugs, such as trastuzumab and trastuzumab emtansine (T-DM1), have been developed to overcome such resistance [25, 26]. Patients with tumors positive for HER2 mutations in the present study were not able to access clinical trials evaluating the efficacy of such HER2-targeted agents for HER2 mutant tumors in Japan (University Medical Hospital Information Network [UMIN] Clinical Trials Registry identifiers UMIN000012551 and UMIN000017709) because of the close of enrollment.
Immunotherapy offers a novel approach to the treatment of NSCLC patients. Immune-checkpoint inhibitors, including antibodies to PD-1 such as nivolumab and pembrolizumab, have been introduced into clinical practice for the treatment of advanced NSCLC on the basis of the results of large phase III studies showing that they confer increased OS compared with standard second-line docetaxel [20–22]. However, the efficacy of such agents tends to be less pronounced in patients with common EGFR mutations, possibly as a result of a low level of mutational heterogeneity [27], with somatic mutation load being thought to be correlated with sensitivity to immune-checkpoint inhibitors. Given the rarity of exon-20 mutations of EGFR and HER2 relative to the major activating mutations of EGFR, clinical information regarding the relation between these exon-20 mutations and the response to nivolumab has not been available. Our study has now shown limited success for nivolumab treatment in patients with exon-20 mutations of EGFR or HER2, similar to the case for those with common EGFR mutations such as Ex19del and L858R.
Given the limited therapeutic efficacy of EGFR-TKIs and immune-checkpoint inhibitors in patients with exon-20 mutations of EGFR or HER2, the identification of actionable mutations should be considered for such individuals in order to provide additional therapy options. Exon-20 mutations of EGFR and HER2 were previously found to be mutually exclusive with other driver alterations such as mutations in KRAS and BRAF [28, 29]. To identify additional oncogenic mutations, we prospectively applied an amplicon-based NGS panel that covers mutational hotspots related to lung and colon tumorigenesis. Most coexisting mutations were found to be located in TP53, with only the H1047R mutation of PIK3CA being considered actionable as a result of its association with the therapeutic efficacy of PI3K-AKT-mTOR pathway inhibitors [30]. Other somatic mutations detected in SMAD4, KRAS, and DDR2 are thought to be passenger mutations. A limitation of our study is that detection of other coexisting mutations was limited to those covered by the relatively small NGS panel. Given that EGFR-mutated lung cancer was previously shown in a study based on NGS to have a lower mutation burden compared with lung cancer wild-type for EGFR [27], it may be difficult to identify concurrent actionable driver mutations in patients with EGFR or HER2 mutations. Moreover, the NGS analysis is performed mostly by K.S. (second author of this manuscript). M.T. (first author) is largely responsible for writing an annotation report for each patient and for establishment and maintenance of a database. Therefore, gathering of data by one investigator may produce unbiased results.
Given that the outcome for NSCLC patients with uncommon EGFR or HER2 mutations remains poor, with only a moderate at best benefit conferred by treatment with EGFR-TKIs or immune-checkpoint inhibitors and with the rarity of coexisting actionable mutations, novel therapies that improve outcome in these patients are urgently needed.
PATIENTS AND METHODS
Patients
The study population was selected from the cohort of patients enrolled in an ongoing observational study of the clinical application of NGS to therapeutic decision-making for lung cancer at Kindai University Hospital (UMIN000014782), the design and initial results of which have been presented previously [31]. In brief, the study enrolled patients about to undergo either biopsy for the purpose of diagnosis of or curative surgery for lung cancer, or those with already histologically proven lung cancer. No restrictions on tumor histology, disease stage, subsequent or previous treatment, performance status, or other factors were imposed. Lung cancer staging was performed in all patients according to the 7th TNM classification. All patients provided written informed consent to the performance of genomics analysis, and the study was approved by the Institutional Review Board of Kindai University. Genetic alterations were defined as actionable if they were associated with susceptibility to an approved targeted therapy or if they could serve as a basis for direction of patients toward registered clinical trials (either nationally or at our institution). Tumor response was examined by computed tomography and was evaluated according to the Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1. A best response of SD required the criterion to be met for at least 6 weeks after initiation of treatment.
Molecular testing
Collected formalin-fixed, paraffin-embedded tumor specimens underwent histological review, and only those containing sufficient tumor cells as revealed by hematoxylin-eosin staining were subjected to nucleic acid extraction. DNA was purified with the use of an Allprep DNA FFPE Kit (Qiagen), and 10 ng of the purified material were subjected to multiplex polymerase chain reaction (PCR) amplification and NGS with the use of an Ion AmpliSeq Library Kit 2.0 and Ion AmpliSeq Colon and Lung Cancer Panel (Life Technologies), which targets 504 mutational hotspot regions of 22 cancer-associated genes: AKT1, ALK, BRAF, CTNNB1, DDR2, EGFR, ERBB2, ERBB4, FBXW7, FGFR1, FGFR2, FGFR3, KRAS, MAP2K1, MET, NOTCH1, NRAS, PIK3CA, PTEN, SMAD4, STK11, and TP53 [31, 32]. DNA sequencing data were accessed through the Torrent Suite v.4.0 program (Life Technologies). Reads were aligned with the hg19 human reference genome, and variants were called with the use of Variant Call Format ver. 4.0. Raw variant calls were filtered with the use of the following annotations: homozygous and heterozygous variants, quality score of <100, and depth of coverage of <19. The tumors of five patients who received nivolumab therapy were assessed for PD-L1 expression by immunohistochemistry with the 22C3 antibody as performed as part of clinical practice and in a commercial clinical laboratory (SRL, Tokyo). Scoring for the PD-L1 IHC 22C3 pharmDx assay in NSCLC is based on the percentage of stained tumor cells, or TPS [23].
Statistical analysis
The present observational study was undertaken without prior determination of sample size or power. The primary end point of the study was EGFR-TKI or nivolumab efficacy in patients with NSCLC positive for EGFR or HER2 exon-20 mutations. PFS was calculated from the date of initiation of chemotherapy either to the date of disease progression or to the date of last contact. OS was calculated from the date of diagnosis of metastatic cancer or of recurrence after curative surgery to the date of death from any cause or to the date of last contact. The probability of survival as a function of time was estimated with the Kaplan-Meier method as performed with GraphPad Prism software version 5.0.
SUPPLEMENTARY MATERIALS AND FIGURES
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
FUNDING
The study received no specific funding.
REFERENCES
- 1.Hoffman PC, Mauer AM, Vokes EE. Lung cancer. Lancet. 2000;355:479–85. doi: 10.1016/S0140-6736(00)82038-3. [DOI] [PubMed] [Google Scholar]
- 2.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 3.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 4.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, Mardis E, Kupfer D, Wilson R, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–11. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, Bando M, Ohno S, Ishikawa Y, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–6. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 6.Takeda M, Okamoto I, Sakai K, Kawakami H, Nishio K, Nakagawa K. Clinical outcome for EML4-ALK-positive patients with advanced non-small-cell lung cancer treated with first-line platinum-based chemotherapy. Ann Oncol. 2012;23:2931–6. doi: 10.1093/annonc/mds124. [DOI] [PubMed] [Google Scholar]
- 7.Takeda M, Okamoto I, Nakagawa K. Survival outcome assessed according to tumor response and shrinkage pattern in patients with EGFR mutation-positive non-small-cell lung cancer treated with gefitinib or erlotinib. J Thorac Oncol. 2014;9:200–4. doi: 10.1097/JTO.0000000000000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yasuda H, Kobayashi S, Costa DB. EGFR exon 20 insertion mutations in non-small-cell lung cancer: preclinical data and clinical implications. Lancet Oncol. 2012;13:e23–31. doi: 10.1016/S1470-2045(11)70129-2. [DOI] [PubMed] [Google Scholar]
- 9.Yasuda H, Park E, Yun CH, Sng NJ, Lucena-Araujo AR, Yeo WL, Huberman MS, Cohen DW, Nakayama S, Ishioka K, Yamaguchi N, Hanna M, Oxnard GR, et al. Structural, biochemical, and clinical characterization of epidermal growth factor receptor (EGFR) exon 20 insertion mutations in lung cancer. Sci Transl Med. 2013;5:216ra177. doi: 10.1126/scitranslmed.3007205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kosaka T, Tanizaki J, Paranal RM, Endoh H, Lydon C, Capelletti M, Repellin CE, Choi J, Ogino A, Calles A, Ercan D, Redig AJ, Bahcall M, et al. Response Heterogeneity of EGFR and HER2 Exon 20 Insertions to Covalent EGFR and HER2 Inhibitors. Cancer Res. 2017;77:2712–21. doi: 10.1158/0008-5472.CAN-16-3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oxnard GR, Lo PC, Nishino M, Dahlberg SE, Lindeman NI, Butaney M, Jackman DM, Johnson BE, Janne PA. Natural history and molecular characteristics of lung cancers harboring EGFR exon 20 insertions. J Thorac Oncol. 2013;8:179–84. doi: 10.1097/JTO.0b013e3182779d18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu HA, Arcila ME, Hellmann MD, Kris MG, Ladanyi M, Riely GJ. Poor response to erlotinib in patients with tumors containing baseline EGFR T790M mutations found by routine clinical molecular testing. Ann Oncol. 2014;25:423–8. doi: 10.1093/annonc/mdt573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu JY, Yu CJ, Chang YC, Yang CH, Shih JY, Yang PC. Effectiveness of tyrosine kinase inhibitors on “uncommon” epidermal growth factor receptor mutations of unknown clinical significance in non-small cell lung cancer. Clin Cancer Res. 2011;17:3812–21. doi: 10.1158/1078-0432.CCR-10-3408. [DOI] [PubMed] [Google Scholar]
- 14.Wu JY, Wu SG, Yang CH, Gow CH, Chang YL, Yu CJ, Shih JY, Yang PC. Lung cancer with epidermal growth factor receptor exon 20 mutations is associated with poor gefitinib treatment response. Clin Cancer Res. 2008;14:4877–82. doi: 10.1158/1078-0432.CCR-07-5123. [DOI] [PubMed] [Google Scholar]
- 15.Shigematsu H, Takahashi T, Nomura M, Majmudar K, Suzuki M, Lee H, Wistuba II, Fong KM, Toyooka S, Shimizu N, Fujisawa T, Minna JD, Gazdar AF. Somatic mutations of the HER2 kinase domain in lung adenocarcinomas. Cancer Res. 2005;65:1642–6. doi: 10.1158/0008-5472.CAN-04-4235. [DOI] [PubMed] [Google Scholar]
- 16.Stephens P, Hunter C, Bignell G, Edkins S, Davies H, Teague J, Stevens C, O’Meara S, Smith R, Parker A, Barthorpe A, Blow M, Brackenbury L, et al. Lung cancer: intragenic ERBB2 kinase mutations in tumours. Nature. 2004;431:525–6. doi: 10.1038/431525b. [DOI] [PubMed] [Google Scholar]
- 17.Kris MG, Camidge DR, Giaccone G, Hida T, Li BT, O’Connell J, Taylor I, Zhang H, Arcila ME, Goldberg Z, Janne PA. Targeting HER2 aberrations as actionable drivers in lung cancers: phase II trial of the pan-HER tyrosine kinase inhibitor dacomitinib in patients with HER2-mutant or amplified tumors. Ann Oncol. 2015;26:1421–7. doi: 10.1093/annonc/mdv186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang SE, Narasanna A, Perez-Torres M, Xiang B, Wu FY, Yang S, Carpenter G, Gazdar AF, Muthuswamy SK, Arteaga CL. HER2 kinase domain mutation results in constitutive phosphorylation and activation of HER2 and EGFR and resistance to EGFR tyrosine kinase inhibitors. Cancer Cell. 2006;10:25–38. doi: 10.1016/j.ccr.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 19.Herter-Sprie GS, Greulich H, Wong KK. Activating Mutations in ERBB2 and Their Impact on Diagnostics and Treatment. Front Oncol. 2013;3:86. doi: 10.3389/fonc.2013.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brahmer J, Reckamp KL, Baas P, Crino L, Eberhardt WE, Poddubskaya E, Antonia S, Pluzanski A, Vokes EE, Holgado E, Waterhouse D, Ready N, Gainor J, et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. N Engl J Med. 2015;373:123–35. doi: 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, Chow LQ, Vokes EE, Felip E, Holgado E, Barlesi F, Kohlhaufl M, Arrieta O, et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N Engl J Med. 2015;373:1627–39. doi: 10.1056/NEJMoa1507643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herbst RS, Baas P, Kim DW, Felip E, Perez-Gracia JL, Han JY, Molina J, Kim JH, Arvis CD, Ahn MJ, Majem M, Fidler MJ, de Castro G, Jr, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet. 2016;387:1540–50. doi: 10.1016/S0140-6736(15)01281-7. [DOI] [PubMed] [Google Scholar]
- 23.Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, Patnaik A, Aggarwal C, Gubens M, Horn L, Carcereny E, Ahn MJ, Felip E, et al. KEYNOTE-001 Investigators Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372:2018–28. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 24.Yang JC, Sequist LV, Geater SL, Tsai CM, Mok TS, Schuler M, Yamamoto N, Yu CJ, Ou SH, Zhou C, Massey D, Zazulina V, Wu YL. Clinical activity of afatinib in patients with advanced non-small-cell lung cancer harbouring uncommon EGFR mutations: a combined post-hoc analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung 6. Lancet Oncol. 2015;16:830–8. doi: 10.1016/S1470-2045(15)00026-1. [DOI] [PubMed] [Google Scholar]
- 25.Mazieres J, Barlesi F, Filleron T, Besse B, Monnet I, Beau-Faller M, Peters S, Dansin E, Fruh M, Pless M, Rosell R, Wislez M, Fournel P, et al. Lung cancer patients with HER2 mutations treated with chemotherapy and HER2-targeted drugs: results from the European EUHER2 cohort. Ann Oncol. 2016;27:281–6. doi: 10.1093/annonc/mdv573. [DOI] [PubMed] [Google Scholar]
- 26.Ohashi K, Hotta K, Hirata T, Aoe K, Kozuki T, Ninomiya K, Kayatani H, Yanai H, Toyooka S, Hinotsu S, Takata M, Kiura K. Trastuzumab Emtansine in HER2+ Recurrent Metastatic Non-Small-Cell Lung Cancer: Study Protocol. Clin Lung Cancer. 2017;18:92–5. doi: 10.1016/j.cllc.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 27.Dong ZY, Zhong WZ, Zhang XC, Su J, Xie Z, Liu SY, Tu HY, Chen HJ, Sun YL, Zhou Q, Yang JJ, Yang XN, Lin JX, et al. Potential Predictive Value of TP53 and KRAS Mutation Status for Response to PD-1 Blockade Immunotherapy in Lung Adenocarcinoma. Clin Cancer Res. 2017;23:3012–24. doi: 10.1158/1078-0432.CCR-16-2554. [DOI] [PubMed] [Google Scholar]
- 28.Arcila ME, Nafa K, Chaft JE, Rekhtman N, Lau C, Reva BA, Zakowski MF, Kris MG, Ladanyi M. EGFR exon 20 insertion mutations in lung adenocarcinomas: prevalence, molecular heterogeneity, and clinicopathologic characteristics. Mol Cancer Ther. 2013;12:220–9. doi: 10.1158/1535-7163.MCT-12-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki M, Shiraishi K, Yoshida A, Shimada Y, Suzuki K, Asamura H, Furuta K, Kohno T, Tsuta K. HER2 gene mutations in non-small cell lung carcinomas: concurrence with Her2 gene amplification and Her2 protein expression and phosphorylation. Lung Cancer. 2015;87:14–22. doi: 10.1016/j.lungcan.2014.10.014. [DOI] [PubMed] [Google Scholar]
- 30.Janku F, Wheler JJ, Naing A, Falchook GS, Hong DS, Stepanek VM, Fu S, Piha-Paul SA, Lee JJ, Luthra R, Tsimberidou AM, Kurzrock R. PIK3CA mutation H1047R is associated with response to PI3K/AKT/mTOR signaling pathway inhibitors in early-phase clinical trials. Cancer Res. 2013;73:276–84. doi: 10.1158/0008-5472.CAN-12-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takeda M, Sakai K, Terashima M, Kaneda H, Hayashi H, Tanaka K, Okamoto K, Takahama T, Yoshida T, Iwasa T, Shimizu T, Nonagase Y, Kudo K, et al. Clinical application of amplicon-based next-generation sequencing to therapeutic decision making in lung cancer. Ann Oncol. 2015;26:2477–82. doi: 10.1093/annonc/mdv475. [DOI] [PubMed] [Google Scholar]
- 32.Scarpa A, Sikora K, Fassan M, Rachiglio AM, Cappellesso R, Antonello D, Amato E, Mafficini A, Lambiase M, Esposito C, Bria E, Simonato F, Scardoni M, et al. Molecular typing of lung adenocarcinoma on cytological samples using a multigene next generation sequencing panel. PLoS One. 2013;8:e80478. doi: 10.1371/journal.pone.0080478. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.