Abstract
Plants have lived in close association with arbuscular mycorrhizal (AM) fungi for over 400 million years. Today, this endosymbiosis occurs broadly in the plant kingdom where it has a pronounced impact on plant mineral nutrition. The symbiosis develops deep within the root cortex with minimal alterations in the external appearance of the colonized root; however, the absence of macroscopic alterations belies the extensive signaling, cellular remodeling, and metabolic alterations that occur to enable accommodation of the fungal endosymbiont. Recent research has revealed the involvement of a novel N-acetyl glucosamine transporter and an alpha/beta-fold hydrolase receptor at the earliest stages of AM symbiosis. Calcium channels required for symbiosis signaling have been identified, and connections between the symbiosis signaling pathway and key transcriptional regulators that direct AM-specific gene expression have been established. Phylogenomics has revealed the existence of genes conserved for AM symbiosis, providing clues as to how plant cells fine-tune their biology to enable symbiosis, and an exciting coalescence of genome mining, lipid profiling, and tracer studies collectively has led to the conclusion that AM fungi are fatty acid auxotrophs and that plants provide their fungal endosymbionts with fatty acids. Here, we provide an overview of the molecular program for AM symbiosis and discuss these recent advances.
INTRODUCTION
Of the many associations formed between plants and microbes, arbuscular mycorrhizal (AM) symbiosis, in which plants and fungi of the Glomeromycota engage, is one of the most widespread and ancient (Smith and Read, 2008). Fossils of the early land plants (400 million years ago) provide evidence that fungi morphologically similar to the current day Glomales lived within their cells (Remy et al., 1994; Heckman et al., 2001). Phylogenetic analyses indicate that symbiosis signaling genes are present in the genomes of the closest algal relatives to land plants and the function of the encoded proteins is conserved, which suggests that these plant ancestors were preadapted for symbiosis (Delaux et al., 2015). The existence of AM symbiosis in many early diverging land plants including liverworts, hornworts, lycophytes, and ferns reveals that AM symbiosis predates the development of true root systems (Brundrett, 2002; Field et al., 2012), while the broad occurrence of AM in extant plant families is consistent with a single early origin and with retention of the symbiosis over many millions of years. Such a pattern implies that the symbiosis has offered continued advantages and it is speculated that AM fungi helped the early land plants cope with acquisition of nutrients from their new, harsh terrestrial environment (Pirozynski and Malloch, 1975). Nutrient acquisition appears to be their dominant role today as they provide access to phosphorus, which is poorly mobile in the soil and also to a lesser extent, nitrogen and other mineral nutrients. However, additional benefits arise from the symbiosis, including disease and stress resistance (Smith and Read, 2008). Despite the positive attributes of AM symbiosis, several plant species, particularly those with specialized lifestyles such as parasitic, aquatic, and insectivorous plants, have lost the ability to establish AM symbioses (Wang and Qiu, 2006). Noted originally in Arabidopsis thaliana and subsequently in other species, these non-host plants have lost the genes whose functions are required exclusively for symbiosis, and this loss occurred independently in several plant lineages (Delaux et al., 2014; Bravo et al., 2016).
Development of AM symbiosis is initiated by signal exchange between plant roots and germinating fungal spores, which triggers coordinated differentiation of both symbionts to enable their interaction and the development of the symbiotic state. Following physical contact between the symbionts, the fungal hyphae grow through the epidermal cells into the cortex where they differentiate within the cortical cells to form branched hyphae, called arbuscules. Extensive reorganization of the cortical cell, including the deposition of the periarbuscular membrane around the arbuscule, ultimately results in a new membrane-bound apoplastic compartment within the cortical cell, which houses the arbuscule. Both symbionts have access to the common apoplast, and nutrient transfer between the symbionts occurs across this interface. Fueled by a carbon supply from the root, the fungus develops an extraradical mycelium in the surrounding soil. The extraradical and intraradical mycelia are a single continuum and mineral nutrients, particularly phosphorus (as phosphate) captured by the extraradical hyphae, are ultimately transferred to the plant via the arbuscules (for in-depth reviews of symbiotic development, see Gutjahr and Parniske, 2013; Lanfranco et al., 2016).
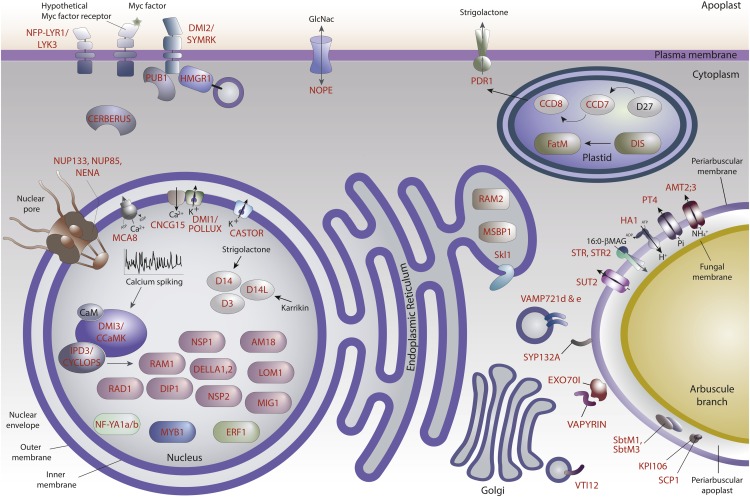
Continual signaling between the symbionts and the expression of an extensive new transcriptional program in the plant host, particularly in the root cortex, are required for symbiosis (reviewed in Hogekamp and Küster, 2013; Bucher et al., 2014). This drives cellular remodeling, including development of new membrane as well as metabolic and physiological alterations necessary for the plant cell to accommodate the fungal endosymbiont and for symbiotic functioning. Labeled many years ago as the accommodation program (Parniske, 2000), key players have gradually been revealed through analyses of plant mutants as summarized in Figure 1 and discussed below.
Figure 1.
An Overview of Plant Proteins That Play Key Roles in AM Symbiosis.
Genetic analyses enabled the identification of genes required for AM symbiosis and the mutants show phenotypes that range from an absence of colonization to a quantitative reduction in plant colonization by AM fungi, and in some cases, an increase in the colonization. For many of these proteins, their subcellular location provides additional clues as to their ultimate function during AM symbiosis. For example, receptors embedded in the plasma membrane perceive external signal molecules that trigger calcium spiking in the nucleus. However, some receptors, such as rice D14 and D14L, could be localized in the nucleus. The downstream transcriptional response, mediated by many transcriptional regulators, results in the production of proteins involved in the cellular remodeling and metabolic regulation necessary for symbiosis. This includes enzymes involved in fatty acid production that are localized in plastids and endoplasmic reticulum and transporters localized in the periarbuscular membrane that are involved in the exchange of nutrients with the AM fungus. This figure shows genes for which there is a knockout/knockdown symbiosis phenotype combined with some direct or indirect information about the location of the encoded protein. The gene names used most frequently are indicated in red and also shown in Table 1.
AM SYMBIOSIS IS INITIATED BY PRE-CONTACT SIGNALING
Development of symbiosis begins with signaling that occurs prior to physical contact between the symbionts, and both symbionts release chemical signals that elicit preparative responses in the other (Buee et al., 2000; Chabaud et al., 2011). Current data suggest that the molecular dialogue is initiated by strigolactones, a group of carotenoid-based phytohormones produced by the plant, that also regulate many aspects of plant development (Akiyama et al., 2005; Gomez-Roldan et al., 2008; Lopez-Obando et al., 2015). In response to phosphate deprivation, strigolactones are secreted into the rhizosphere (Yoneyama et al., 2007a, 2007b; Kretzschmar et al., 2012), where these chemically labile molecules serve as signals by which AM fungi may identify a receptive host in their vicinity. Upon detection of strigolactones, AM fungi activate oxidative metabolism, which drives increases in hyphal growth and branching, enhancing the chance of physical contact with a host root but also committing them to symbiosis (Akiyama and Hayashi, 2006; Besserer et al., 2006, 2008). Strigolactone biosynthesis or export mutants show reduced colonization (Gomez-Roldan et al., 2008; Koltai et al., 2010; Gutjahr et al., 2012; Kretzschmar et al., 2012; Yoshida et al., 2012), indicating the importance of these early signals for establishment of symbiosis.
The recent discovery of a transporter, NO PERCEPTION1 (NOPE1) in maize (Zea mays) and rice (Oryza sativa) that is also required for priming of the fungus (Nadal et al., 2017), suggests that strigolactones may not be the only signal molecules of importance during the precontact phase. NOPE1 encodes a member of the Major Facilitator Superfamily of transport proteins capable of N-acetylglucosamine transport, the first description of such a transport activity in a plant protein. nope1 mutants show almost no interaction with AM fungi and their root exudates fail to elicit transcriptional responses in the fungus leading the authors to hypothesize that NOPE1 transports a plant-derived N-acetylglucosamine-based molecule, that acts to prime signaling in AM fungi to promote symbioses (Nadal et al., 2017). Even in the absence of a full understanding of the interrelationship between these early signals, it is clear that they are important for the establishment of symbiosis.
In rice, the alpha/beta-fold hydrolase and putative receptor protein DWARF 14 LIKE (D14L) is essential for AM symbiosis and is necessary for the establishment of an appropriate transcriptional response in rice roots exposed to germinated spore exudates (Gutjahr et al., 2015). The severity of the symbiotic phenotype and the transcript profiles of the mutant are consistent with D14L-mediated signaling occurring at a very early stage of symbiosis (Gutjahr et al., 2015). D14L is a homolog of the Arabidopsis protein KARRIKIN INSENSITIVE2 (KAI2), a receptor that acts in concert with the F-box protein MORE AXILLIARY GROWTH2 (MAX2) to regulate protein turnover in response to karrikin signaling (Nelson et al., 2011; Waters et al., 2012). Karrikins are butenolide molecules that are related to strigolactones and are generated by the burning of plant tissues during fire; detection of karrikins by dormant seeds triggers germination in fire-chasing species that grow quickly to exploit a lack of competition following a fire (Flematti et al., 2015). The developmental phenotypes exhibited by kai2 mutants that are not related to karrikin perception per se, and broad conservation of KAI2 in basal land plants and species not associated with fire-prone habitats, have led to a hypothesis that the receptor KAI2 recognizes and binds to an as yet unidentified endogenous ligand, presumably a phytohormone that is structurally related to karrikins and strigolactones (Waters et al., 2014, 2015; Conn and Nelson, 2016). The observation that D14L is essential for AM symbiosis in rice (Gutjahr et al., 2015), coupled with an earlier report of a rice d3 mutant (homolog of MAX2) that is likewise unable to support AM symbiosis (Yoshida et al., 2012), suggests this signaling pathway may be involved in AM symbiosis. One possibility is that germinating spore extracts contain a molecule structurally similar to strigolactones and karrikins, which acts via D14L and D3 to enable an appropriate transcriptional response. An alternative scenario is that signaling triggered by the so-called KAI2 ligand (KL), an endogenous ligand that has yet to be identified, is perceived by D14L and triggers a transcriptional response required to establish AM symbiosis. In both scenarios, it is necessary to invoke unique downstream components or modifiers to direct a symbiotic response. As the ability to perceive karrikins and strigolactones may have evolved from an ancestral mechanism involved in the perception of endogenous KL (Conn and Nelson, 2016), it is tempting to speculate the involvement of KL-mediated signaling from the earliest time points of AM symbiosis (Gutjahr et al., 2015).
THE COMMON SYMBIOSIS SIGNALING PATHWAY
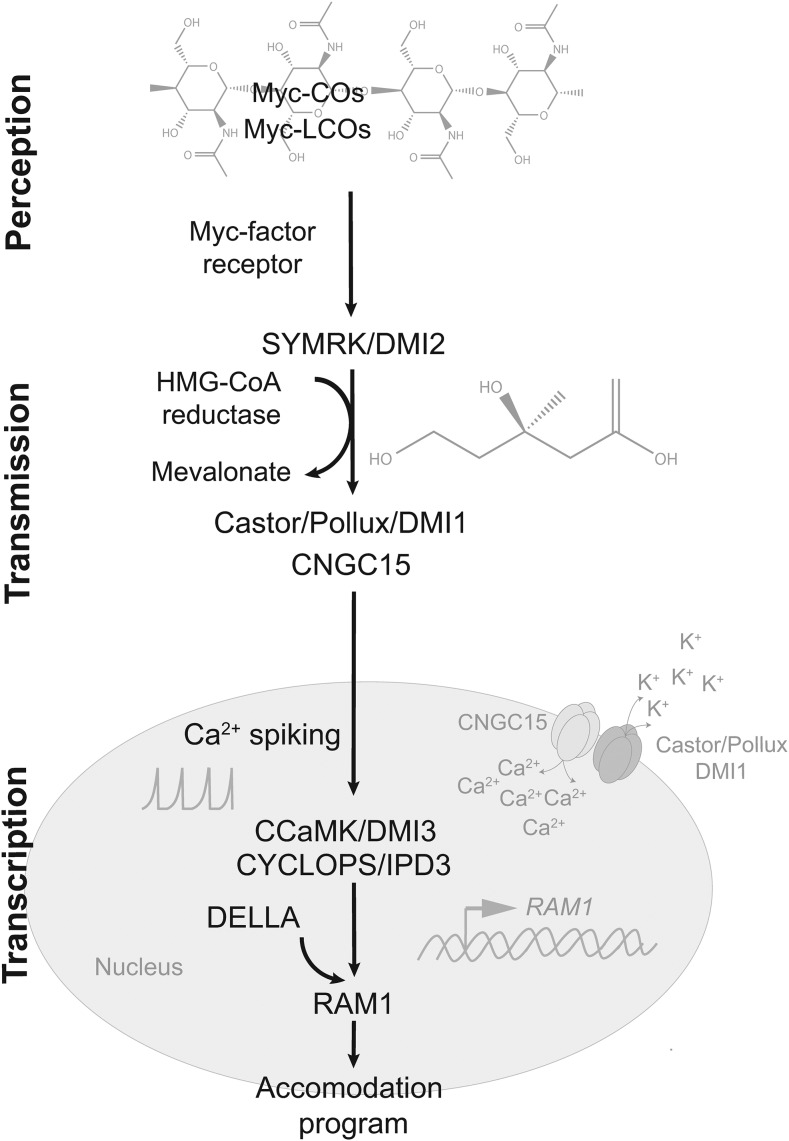
Most of the research over the last few years has focused on a symbiosis signaling pathway, which in legumes is required for symbioses with AM fungi and rhizobia (Oldroyd, 2013). Evolving ∼60 million years ago, the nitrogen-fixing symbiosis with rhizobia recruited components of the already established AM symbiosis signaling pathway resulting in the so-called common symbiosis signaling pathway (Oldroyd, 2013). From a research viewpoint, effort from both symbiosis fields has accelerated our understanding of this pathway and, more recently, of the AM symbiosis-specific downstream responses that it controls. Initiated by AM fungal N-acetylglucosamine based molecules, the events unfolding within the root cells of a host plant may be organized conceptually within a hierarchy of three levels: (1) perception, (2) transmission, and (3) transcription (Figure 2).
Figure 2.
Establishment of AM Symbiosis Involves a Three-Tiered Response of the Host to Accommodate Its Fungal Partner, Comprising Perception, Transmission, and Transcription.
(1) Perception: A receptive plant perceives the presence of AM fungi through the detection of lipochitooligosaccharides (Myc-LCOs) and short-chain chitin oligosaccharides (Myc-COs) by LysM receptor-like kinases such as OsCERK1, SlLYK10, LjNFR1, MtLYK3, and the receptor-like kinase SYMRK/DMI2. (2) Transmission: The enzyme HMG-CoA reductase interacts with DMI2 in M. truncatula and is essential to the generation of Ca2+ spiking that is required to initiate a downstream transcriptional response. These observations have led to a model whereby HMG-CoA reductase generates mevalonate following AM fungal perception by SYMRK/DMI2 (Venkateshwaran et al., 2015). Mevalonate acts as a secondary messenger that transmits fungal perception from the plasma membrane to the nucleus via interaction with the nuclear cation (K+) channel CASTOR and POLLUX/DMI1. The K+ channels CASTOR and POLLUX/DMI1 support symbiotic Ca2+ spiking by enabling K+ efflux to counterbalance Ca2+ influx, which is mediated by the cyclic nucleotide-gated channels CNGC15. (3) Transcription: A calcium and calmodulin-dependent kinase CCaMK is proposed to act as a master decoder and regulatory kinase, deciphering the nuclear Ca2+ oscillations to enact the appropriate transcriptional response. In the presence of calcium, Ca2+-calmodulin associates with CCaMK, promoting a conformational change that induces the phosphorylation of the CCaMK substrate protein CYCLOPS. Phosphorylated CYCLOPS forms a complex with CCaMK, which acts in concert with GRAS transcription factors such as DELLA proteins, to initiate the expression of genes such as RAM1 that are necessary to accommodate the fungal symbiont. For clarity, this diagram shows a subset of the proteins of the symbiosis signaling pathway. The chemical structures underlying Myc-LCOs and Myc-COs are based on Gust et al. (2012).
The discovery of the so-called Myc-factor, a mixture of lipochitooligosaccharides (Myc-LCOs), provided the first clues as to the chemical nature of signaling molecules secreted by AM fungi to communicate with their host plants (Maillet et al., 2011). However, it is important to consider that the discovery of Myc-LCOs was predicated upon a hypothesis that Myc-factor was of a similar chemical composition to rhizobial Nod-factors, and thus additional AM fungal factors that contribute to the perception of AM fungi by plants may have been overlooked. Additionally, the failure of Myc-LCOs to elicit a transcriptional response associated with AM symbiosis in rice raises the possibility that these signaling molecules may not be recognized ubiquitously by all plants (Miyata et al., 2014). The finding that short-chain chitin oligomers elicit symbiotically relevant nuclear calcium oscillations (Ca2+ spiking) within cells derived from AM hosts but not non-AM hosts such as Arabidopsis, and further that the production of these oligomers by Rhizophagus irregularis is strongly stimulated upon exposure to strigolactone, makes these molecules excellent candidates for signaling molecules (Genre et al., 2013).
The lysin motif (LysM) receptor-like kinase OsCERK1 has a clear role in mediating the perception of fungal spore exudates and short-chain chitin oligomers in rice (Carotenuto et al., 2017), with Oscerk1 mutants demonstrating a symbiotic phenotype in which mycorrhizal colonization is reduced (Miyata et al., 2014; Zhang et al., 2015c). OsCERK1 and its homolog in Arabidopsis AtCERK1 were identified originally in the context of their roles in defense-related chitin perception (Miya et al., 2007; Shimizu et al., 2010), revealing a surprising duality of OsCERK1 in mediating both chitin-triggered immunity to restrict the growth of fungal pathogens yet also being necessary to promote the colonization of AM fungi during symbiosis. The LysM membrane protein OsCEBiP acts as a coreceptor to OsCERK1 and is essential for defense-related chitin perception (Kaku et al., 2006; Shimizu et al., 2010), yet the ability of an Oscebip mutant to successfully establish AM symbiosis (Miyata et al., 2014) uncouples OsCERK1-associated chitin-triggered immunity and mycorrhizal symbiotic responses, suggesting the involvement of an additional, as yet unidentified, OsCERK1 coreceptor specific to enabling AM symbiosis. OsCERK1 homologs NFR1 (Lotus japonicus) and LYK3 (Medicago truncatula) have been shown to promote AM symbiosis (Zhang et al., 2015c), as does the LysM receptor-like kinase SILYK10 in the non-legume tomato (Solanum lycopersicum; Buendia et al., 2016); however, genetic redundancy within the LysM-receptor kinase family that is presumed to include the Myc-factor receptor, compounded by the possibility that AM fungi secrete additional chemical signatures to signal symbiosis, has made receptor identification challenging (Zipfel and Oldroyd, 2017). As the recently reported NOPE1 transporter also displays a strong N-acetylglucosamine uptake activity (Nadal et al., 2017), it is interesting to speculate on a potential role of NOPE1 as a means of the plant host detecting a nearby fungal symbiont. Whether there is any relationship between NOPE1 and the chitooligosaccharide signal molecules that activate the common symbiosis signaling pathway remains to be determined.
Perception of Myc-factor by a receptive plant results in transmission of this message from the plasma membrane of the cell to the nucleus and the regulatory proteins located therein. DMI2/SYMRK is an essential protein for signal perception in endosymbiosis (Endre et al., 2002; Stracke et al., 2002) that is thought to act as a coreceptor to the unidentified Myc-factor receptor and has been shown to associate with the LysM receptor-like kinase proteins NFR5 and NFR1 during Rhizobium-legume symbiosis (Antolín-Llovera et al., 2014; Ried et al., 2014). The suggestion that the enzyme 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) reductase of the mevalonate pathway of isoprenoid biosynthesis is the potential link enabling the transmission of Myc- and Nod-factor perception to the generation of symbiotic nuclear Ca2+ oscillations was made via the identification of its interaction with DMI2 (Kevei et al., 2007; Venkateshwaran et al., 2015). Moreover, exogenous application of mevalonate to kidney cells expressing a symbiotically relevant nuclear cation (K+) channel (DMI1) was sufficient to initiate Ca2+ spiking, suggesting a direct interaction between mevalonate and DMI1 (Venkateshwaran et al., 2015). Based on these data, it is hypothesized that mevalonate acts as a secondary messenger to transmit perception of symbiotic factors from the plasma membrane to the cell nucleus (Venkateshwaran et al., 2015); in the absence of a mutant phenotype, what is less clear is whether the pathway is required for symbiosis.
The recent discovery that cyclic nucleotide-gated channels encoded by CNGC15 mediate symbiotic Ca2+ influx into the nucleus represents another important breakthrough (Charpentier et al., 2016), leading to a model whereby Myc-factor perception by a cognate receptor and the coreceptor DMI2 leads to the production of mevalonate by the HMG-CoA reductase that is associated with DMI2 (Kevei et al., 2007). Mevalonate (or its phosphorylated metabolites) then acts as a secondary messenger molecule to transmit signal perception to the nucleus via interaction with DMI1, a K+ permeable channel that itself interacts with CNGC15 cyclic nucleotide-gated channels (Charpentier et al., 2016). The influx of Ca2+ mediated by CNGC15 channels is countered by K+ efflux via DMI1, thereby repolarizing the nuclear membrane to enable sustained Ca2+ oscillations, which complete the transmission of the Myc-factor perception at the level of the plasma membrane to the nucleus and its resident regulatory proteins. Mathematical modeling dictates that sustained Ca2+ oscillations require the simultaneous activation of both DMI1 and CNGC15 channels (Charpentier et al., 2016), which may occur via their co-interactivity; however, it will be interesting to further probe the molecular dynamics of mevalonate association with the DMI1:CNGC15 complex in future research.
A calcium and calmodulin-dependent kinase (CCaMK) is proposed to act as a master decoder and regulatory kinase, deciphering the nuclear Ca2+ oscillations to enact the final stage of this conceptual hierarchy, the transcriptional response (Miller et al., 2013). Primacy of CCaMK at the apex of the regulatory transcriptional cascade is underscored by the observation that expression of an activated gain-of-function CCaMK is sufficient to fully complement the severe symbiotic phenotypes exhibited by mutants of genes upstream in the pathway such as dmi1/pollux, castor, and dmi2/symrk (Hayashi et al., 2010), thereby uncoupling the requirement for Myc-factor perception and the resulting Ca2+ oscillations to elicit the subsequent downstream transcriptional response necessary to support AM symbiosis (see also Tirichine et al., 2006). Nuclear Ca2+ spiking induces association of Ca2+-calmodulin with CCaMK, promoting a conformational change in the kinase that stimulates phosphorylation of a target protein, CYCLOPS (Yano et al., 2008; Miller et al., 2013). CYCLOPS (in L. japonicus) and its ortholog IPD3 (in M. truncatula and rice) (Chen et al., 2008; Horváth et al., 2011) play key roles in AM symbiosis, presumably via interaction with CCaMK to initiate a transcriptional response required to promote symbioses (Pimprikar et al., 2016), as was demonstrated elegantly in a study of CYCLOPS in Rhizobium-legume endosymbiosis (Singh et al., 2014). Nonetheless, differences in the phenotypes exhibited by cyclops/ipd3 mutants indicate these proteins, or their regulatory context within each species, may not be equivalent in all plants. In particular, cyclops mutants in L. japonicus and rice exhibit a severe phenotype during AM symbiosis, in which fungal hyphae show impaired penetration into the outer cortical cells and arbuscules are absent (Chen et al., 2008; Gutjahr et al., 2008; Yano et al., 2008; Floss et al., 2013). Conversely, ipd3 mutants of M. truncatula retain the ability to penetrate the inner cortical cells and establish arbuscules, although colonization levels are reduced (Horváth et al., 2011; Floss et al., 2013). One possibility to account for these different observations is that a degree of functional redundancy in M. truncatula exists because of an IPD3 paralog (Bravo et al., 2016), or alternatively, an additional nonparalogous protein(s) that has yet to be identified (Horváth et al., 2011).
CCaMK and CYCLOPS have been proposed to form a complex that acts in concert with the GRAS (GIBBERELLIC-ACID INSENSITIVE, REPRESSOR of GAI, and SCARECROW) domain regulatory protein DELLA to induce the expression of a downstream regulator(s) (Jin et al., 2016; Pimprikar et al., 2016). DELLA proteins were identified first as repressors of gibberellic acid (GA) signaling (Peng and Harberd, 1993; Alvey and Harberd, 2005), but later emerged as master regulators that interact with, and provide a mechanism for, crosstalk between many hormone and biotic signaling pathways (Gallego-Bartolomé et al., 2012; Davière and Achard, 2013). In M. truncatula and pea (Pisum sativum), two DELLA proteins (DELLA1 and DELLA2 in M. truncatula, and LA and CRY in pea) function redundantly to promote arbuscule development (Floss et al., 2013; Foo et al., 2013; Yu et al., 2014), while in rice, a single DELLA protein, SLENDER RICE1 (SLR1), fulfills this role. The connection between DELLA proteins and the symbiosis signaling pathway provides a mechanism to integrate symbiosis signaling with plant growth and development. For example, during phosphate limitation, DELLA transcripts increase and the protein is stabilized, which serves to restrain growth but to promote arbuscule development (Jiang et al., 2007; Floss et al., 2013)
Direct regulation of GRAS protein Reduced Arbuscular Mycorrhiza 1 (RAM1) gene expression by CCaMK/CYCLOPS in concert with DELLA reflects its central role in enabling arbuscule development (Park et al., 2015; Rich et al., 2015; Xue et al., 2015; Pimprikar et al., 2016). The observation that ectopic RAM1 expression in a cyclops mutant is sufficient to restore arbuscule formation (Pimprikar et al., 2016) implies that the primary, if not sole, function of CYCLOPS relevant to AM symbiosis is to upregulate RAM1 expression. However, both RAM1 expression and arbuscule formation are restored in a cyclops mutant by either overexpression of della1-Δ18 (Floss et al., 2013; Park et al., 2015) or by treatment of roots with an inhibitor of GA biosynthesis (paclobutrazol) (Pimprikar et al., 2016), both of which serve to promote the accumulation of DELLA protein(s) by blocking GA-mediated degradation. Considered as a whole, these data suggest that CYCLOPS is sufficient but may not be necessary for the activation of RAM1 expression in a context where DELLA proteins are no longer subjected to GA-mediated regulation. One possibility is the existence of an additional protein “X” that has some capacity to induce RAM1 expression under conditions in which DELLA proteins are stabilized (i.e., expression of X may be upregulated by DELLA, or alternatively DELLA and X may interact together to promote RAM1 expression). The dynamics of protein-protein interactions between symbiotic GRAS proteins have been studied almost exclusively in heterologous systems, including yeast (yeast two-hybrid), Nicotiana benthamiana, Arabidopsis (protoplasts), and Escherichia coli (pull-down), or in vitro assays, where the biological symbiotic context is lost and analyses of protein-protein interactions in situ represent an important next step to address these limitations.
DELLA and RAM1 occupy key positions in the regulation of AM symbiosis, but they are just two of many GRAS domain regulatory proteins with roles in AM symbiosis (Figure 1). In addition to RAM1, REQUIRED FOR ARBUSCULE DEVELOPMENT1 (RAD1), and MYCORRHIZA-INDUCED GRAS1 (MIG1) potentially have specific roles in the regulation of AM symbiosis, while others, including DELLAs, NSP1 and NSP2, show involvement in both mycorrhizal and Rhizobium-legume symbioses as well as other aspects of plant development. For example, NSP1 and NSP2 are essential for regulation of strigolactone biosynthesis (Liu et al., 2011; Lauressergues et al., 2012; Delaux et al., 2013b; Takeda et al., 2013). Analyses of mutant phenotypes, gene expression data, and protein-protein interactions demonstrate the connectivity and importance of GRAS proteins in AM symbiosis (Gobbato et al., 2012; Hohnjec et al., 2015; Park et al., 2015; Rich et al., 2015; Xue et al., 2015; Heck et al., 2016; Pimprikar et al., 2016), but in general it is unclear what genes they regulate and how this occurs. So while the GRAS proteins have emerged as arguably the most important family of regulatory proteins associated with AM symbiosis, there are huge gaps in understanding how they function.
GRAS domain proteins play central roles in regulating a diverse range of processes in plants, including root development (Benfey et al., 1993; Di Laurenzio et al., 1996), phytohormone signaling (Peng et al., 1997; Silverstone et al., 1998; Ogawa et al., 2000; Tong et al., 2009), and phytochrome signaling (Bolle et al., 2000) as well as endosymbiosis (Kaló et al., 2005; Smit et al., 2005; Floss et al., 2013; Gobbato et al., 2013). Interestingly, secondary structure and phylogenetics-based analyses of these proteins first suggested an origin within the Rossman fold methyltransferase superfamily of a bacterial ancestor (Zhang et al., 2012), a prediction that was subsequently supported by crystal structure analyses of the GRAS domains of proteins in rice (Li et al., 2016) and Arabidopsis (Hirano et al., 2017). Sequence comparisons between plant and bacterial GRAS domain proteins reveal conserved residues associated with substrate binding, proposed to have been acquired by the common ancestor of land plants as a single transfer event prior to functional diversification (Zhang et al., 2012). This raises intriguing questions as to whether the plant regulatory factors retain the ability to bind small molecules and, if so, the identity of their substrates.
Despite the significant amount of research that has focused on this family, the primary mechanism by which GRAS domain proteins regulate gene expression remains ambiguous. The crystal structure of the rice GRAS domain protein Os-SCL7 is predicted to form a groove to accommodate DNA binding, which was demonstrated in vitro, albeit with a synthetically designed nucleotide sequence (Li et al., 2016). Furthermore, the symbiosis-associated protein NSP1 has been demonstrated to bind DNA directly (Hirsch et al., 2009) and chromatin immunoprecipitation data support binding of RAM1 to the RAM2 promoter (Gobbato et al., 2013); however, many GRAS domain proteins do not seem to have this ability and instead act within multiprotein complexes in which other regulatory proteins interact with DNA (Gallego-Bartolomé et al., 2012; Yoshida et al., 2014; Hirano et al., 2017). This is exemplified by GRAS proteins SHORT ROOT and SCARECROW, whose crystal structures reveal that these proteins act as transcriptional cofactors that do not bind DNA directly, but form a complex with BIRD/INDETERMINATE DOMAIN (IDD) transcription factors that interact with DNA via zinc finger repeats (Hirano et al., 2017). The multitude of transcription factors induced during symbiosis, including several members of the IDD family, suggests that similar complexes might regulate transcription during symbiosis.
GENES CONSERVED FOR AM SYMBIOSIS AND ACCOMMODATION OF A FUNGAL SYMBIONT
Development of symbiosis is accompanied by transcriptional reprogramming of the root cortex cells, the main site of interaction between the symbionts, with differential expression of many genes associated with transcriptional regulation, transport processes, and lipid metabolism (Gaude et al., 2012). RAM1 is implicated either directly or indirectly in regulating many of these genes (Luginbuehl et al., 2017), but the significant number of symbiosis-induced transcription factors (Hogekamp and Küster, 2013) indicates complex regulation that is not yet fully understood. The transcriptional response drives the cellular changes necessary to accommodate the fungal endosymbiont and through genetic analyses, the roles of the individual genes are gradually being revealed (Table 1, Figure 1). Deposition of the periarbuscular membrane is one of the most prominent alterations to the colonized cell and is achieved via polarized exocytosis, which involves the EXOCYST complex and a unique EXO70 subunit (Zhang et al., 2015a), a symbiosis-specific splice variant of SYP132 (Huisman et al., 2016; Pan et al., 2016), a plant specific protein Vapyrin (Feddermann et al., 2010; Pumplin et al., 2010; Murray et al., 2011), and two symbiosis-specific VAMP721 proteins (Ivanov et al., 2012) (Figure 1). The protein composition of the periarbuscular membrane is distinct relative to that of the plasma membrane and includes unique phosphate, ammonium, and sugar transporters, whose transport activity is energized by the proton gradient generated by a symbiosis-induced periarbuscular membrane-resident proton ATPase (Harrison et al., 2002; Kobae and Hata, 2010; Kobae et al., 2010; Krajinski et al., 2014; Wang et al., 2014; Garcia et al., 2016) as well as ABC transporters (Zhang et al., 2010; Gutjahr et al., 2012), likely involved in export (Figure 1). Surprisingly, trafficking of these transporters to the periarbuscular membrane occurs by default and is achieved as a consequence of gene expression and protein production coincident with deposition of the periarbuscular membrane around the arbuscule branches (Pumplin et al., 2012). Thus, tight transcriptional regulation of the transporter genes is essential not only to ensure expression in the correct cell type but also to ensure their location in the periarbuscular membrane.
Table 1. Genes Involved in AM Symbiosis and Phenotypes Associated with Their Knockout or Knockdown.
| Gene Name | Gene Product | Phenotype | References |
|---|---|---|---|
| Mt-DXS2 | 1-Deoxy-d-xylulose 5-phosphate synthase | RNAi: Increased number of degenerating arbuscules | (Floss et al., 2008a) |
| Mt-CCD1 | Carotenoid cleavage dioxygenase | RNAi: Increased number of degenerating arbuscules | (Floss et al., 2008b) |
| Ps-CCD7 | Carotenoid cleavage dioxygenase | Reduced colonization | (Gomez-Roldan et al., 2008) |
| Ps-CCD8 | Carotenoid cleavage dioxygenase | Reduced colonization | (Gomez-Roldan et al., 2008) |
| Ph-PDR1 | ABC transporter | Reduced colonization | (Kretzschmar et al., 2012) |
| *Mt-DMI2 (Mt-NORK) | Receptor-like kinase | No epidermal penetration | (Endre et al., 2002; Stracke et al., 2002) |
| Lj-SYMRK | |||
| Ps-SYM19 | |||
| * Pa-NFP | LysM receptor kinase | RNAi: Abortion of arbuscule formation in P. andersonii | (Op den Camp et al., 2011; Maillet et al., 2011; Buendia et al., 2016; Madsen et al., 2003) |
| Mt-NFP | Wild-type AMS phenotype in Mt-nfp and Lj-nfr5 but fewer lateral roots in Mt-nfp | ||
| Lj-NRF5 | VIGS: No epidermal penetration in S. lycopersicum | ||
| Sl-LYK10 | |||
| Lj-NFR1 | LysM receptor kinase | Reduced colonization | (Miyata et al., 2014; Zhang et al., 2015a) |
| Mt-LYK3 | |||
| Os-CERK1 | |||
| Os-D14 | Alpha/beta-fold hydrolase | Increased colonization | (Yoshida et al., 2012) |
| Os-D14L | Alpha/beta-fold hydrolase | No colonization | (Gutjahr et al., 2015) |
| Os-D3 | F-box protein | Reduced colonization and arbuscule formation | (Yoshida et al., 2012) |
| Zm-NOPE1 | GlcNAc transporter | No colonization | (Nadal et al., 2017) |
| Os-NOPE1 | |||
| * Lj-Castor | Cation channel | No epidermal penetration | (Imaizumi-Anraku et al., 2005; Gutjahr et al., 2008) |
| Os-Castor | |||
| Mt-DMI1 | Cation channel | No epidermal penetration | (Ané et al., 2004; Imaizumi-Anraku et al., 2005; Banba et al., 2008) |
| Lj-POLLUX | |||
| Ps-SYM8 | |||
| Os-POLLUX | |||
| Lj-NUP133 | Nucleoporin | Delayed colonization | (Kistner et al., 2005; Kanamori et al., 2006) |
| Lj-NUP85 | Nucleoporin | Delayed colonization | (Kistner et al., 2005; Saito et al., 2007) |
| Lj-NENA | Nucleoporin | No epidermal penetration | (Groth et al., 2010) |
| Mt-MCA8 | SERCA-type calcium ATPase | Reduced epidermal penetration | (Capoen et al., 2011) |
| Mt-CNCG15 | Nuclear-localized cyclic nucleotide-gated channel | Reduced colonization | (Charpentier et al., 2016) |
| Mt-DMI3 | Calcium/calmodulin-dependent protein kinase | No epidermal penetration | (Lévy et al., 2004; Tirichine et al., 2006; Chen et al., 2007; Banba et al., 2008) |
| Lj-CCaMK | |||
| Ps-SYM9 | |||
| Os-DMI3 (Os-CCaMK) | |||
| * Lj-CYCLOPS | Coiled-coil domain containing protein | No formation of arbuscules in L. japonicus and O. sativa | (Kistner et al., 2005; Messinese et al., 2007; Yano et al., 2008; Horváth et al., 2011; Ovchinnikova et al., 2011; Larkan et al., 2013) |
| Os-CYCLOPS | Reduced numbers of arbuscules in M. truncatula, P. sativum, and S. lycopersicum | ||
| Mt-IPD3 | |||
| Ps-SYM33 | |||
| Sl-CYCLOPS | |||
| Mt-DELLA1 | GRAS transcription factors | Intraradical colonization but very limited formation of arbuscules | (Floss et al., 2013; Foo et al., 2013; Yu et al., 2014) |
| Mt-DELLA2 | |||
| Ps-LA | |||
| Ps-CRY | |||
| Os-SLR1 | |||
| Ps-NA | Kaurenoic acid oxidase | Increased colonization | (Foo et al., 2013) |
| * Mt-RAM1 | GRAS transcription factor | Cortical cell penetration and trunk formation but almost no hyphal branching | (Gobbato et al., 2012, 2013; Rich et al., 2015; Park et al., 2015) |
| Lj-RAM1 | |||
| Ph-ATA | |||
| * Lj-RAD1 | GRAS transcription factor | Stunted arbuscules in Lj-rad1 | (Xue et al., 2015; Park et al., 2015) |
| Mt-RAD1 | Reduced colonization in Mt-rad1; reduced number of arbuscules and increased arbuscule degeneration | ||
| * Os-DIP1 | GRAS transcription factor | RNAi: Reduced colonization | (Yu et al., 2014) |
| Mt-NSP1 | GRAS transcription factor | Reduced colonization | (Delaux et al., 2013b) |
| Mt-NSP2 | GRAS transcription factor | Reduced colonization | (Maillet et al., 2011) |
| Os-AM18 | GRAS transcription factor | Reduced colonization | (Fiorilli et al., 2015) |
| * Mt-MIG1 | GRAS transcription factor | RNAi: Small and malformed arbuscules | (Heck et al., 2016) |
| Mt-LOM1 | GRAS transcription factor | RNAi: Reduced colonization | (Couzigou et al., 2017) |
| Gm-NF-YA1a/b | CCAAT-binding transcription factor | RNAi: Reduced colonization | (Schaarschmidt et al., 2013) |
| * Mt-ERF1 | AP2 transcription factor | amiR: Collapsed arbuscules | (Devers et al., 2013) |
| * Mt-MYB1 | MYB transcription factor | Suppression of premature arbuscule degeneration in Mt-pt4/myb1 | (Floss et al., 2017) |
| * Mt-VAPYRIN | MSP and ANK repeat-containing protein | No arbuscule formation and reduced epidermal penetration | (Pumplin et al., 2010; Feddermann et al., 2010) |
| Ph-PAM1 | Small protrusions into cortical cells in P. hybrida | ||
| * Mt-VAMP721d/e | R-SNAREs/vesicle-associated membrane proteins | RNAi: Stunted arbuscules | (Ivanov et al., 2012) |
| * Mt-EXO70I | Exocyst complex protein | Stunted arbuscules | (Zhang et al., 2015b) |
| * Mt-SYP132A | Qa-SNARE/syntaxin | RNAi: Collapsed arbuscules | (Pan et al., 2016) |
| Lj-VTI12 | Qb-SNARE | amiR: Increased number of collapsed arbuscules | (Lota et al., 2013) |
| Mt-MSBP1 | Membrane-bound steroid-binding protein | RNAi: Aberrant arbuscule development | (Kuhn et al., 2010) |
| * Mt-STR | Half-ABC transporter | Stunted arbuscules | (Zhang et al., 2010; Gutjahr et al., 2012; Kojima et al., 2014) |
| Os-STR | |||
| Lj-STR | |||
| * Mt-STR2 | Half-ABC transporter | Stunted arbuscules | (Zhang et al., 2010; Gutjahr et al., 2012) |
| Os-STR2 | |||
| * Mt-RAM2 | Glycerol-3-phosphate acyl transferase | Collapsed arbuscules | (Wang et al., 2012; Keymer et al., 2017) |
| Lj-RAM2 | |||
| * Mt-FatM | Acyl-(ACP) thioesterase | Collapsed arbuscules | (Bravo et al., 2016) |
| * Lj-DIS | Ketoacyl-ACP synthase | Collapsed arbuscules | (Keymer et al., 2017) |
| * Lj-CERBERUS | E3 ubiquitin ligase | Reduced intercellular hyphal elongation | (Takeda et al., 2013) |
| Mt-PUB1 | E3 ubiquitin ligase | Increased colonization | (Vernié et al., 2016) |
| Mt-Skl1 | NRAMP-like integral membrane protein | Increased number of infections | (Penmetsa et al., 2008) |
| Mt-SUNN | Leucine-rich repeat (LRR) receptor kinase | Increased arbuscule formation in P. sativum | (Morandi et al., 2000; Krusell et al., 2002; Nishimura et al., 2002; Searle et al., 2003; Schnabel et al., 2005) |
| Lj-HAR1 | |||
| Ps-SYM29 | |||
| Gm-NARK | |||
| Sl-SUT2 | Sucrose transporter | RNAi: increased mycorrhizal colonization | (Bitterlich et al., 2014) |
| Lj-SbtM1 | Subtilisin-like protease | RNAi: reduced colonization and decrease in arbuscule formation | (Takeda et al., 2009) |
| Lj-SbtM3 | Subtilisin-like protease | RNAi: reduced colonization and decrease in arbuscule formation | (Takeda et al., 2009) |
| Mt-SCP1 a | Serine carboxypeptidase | RNAi: malformed arbuscules | (Rech et al., 2013) |
| * Mt-PT4 | Phosphate transporter | Premature arbuscule degeneration | (Javot et al., 2007; Yang et al., 2012) |
| Os-PT11 | |||
| * Os-PT13 | Phosphate transporter | Small arbuscules | (Yang et al., 2012) |
| Mt-HA1 | ATPase | Degenerating arbuscules | (Krajinski et al., 2014; Wang et al., 2014) |
| * Mt-AMT2;3 | Ammonium transporter | Suppression of premature arbuscule degeneration in Mt-pt4/amt2;3 | (Breuillin-Sessoms et al., 2015) |
| * Mt-KIN2 | Protein kinase | Reduced colonization | (Bravo et al., 2016) |
| * Mt-KIN3 | Protein kinase | Reduced colonization | (Bravo et al., 2016) |
| * Mt-KIN5 | Serine-threonine protein kinase | Reduced colonization | (Bravo et al., 2016) |
| * Mt-RFCb | Replication factor C | Reduced colonization | (Bravo et al., 2016) |
| * Mt-CYT733A1 | P450 enzyme | Reduced colonization | (Bravo et al., 2016) |
| * Mt-PP2AB’1 | Protein phosphatase 2A | Reduced colonization | (Charpentier et al., 2014) |
The genes marked with an asterisk are present exclusively in plants that form AM symbiosis and are not present in non-hosts as described by Bravo et al. (2016). DIS, MIG1, MYB1, VAMP721d/e, PT13, AMT2;3, and CERBERUS are AM symbiosis-conserved genes but were not listed by Bravo et al. (2016) because the criteria used by these authors to define AM symbiosis conserved genes were exceptionally stringent and required broad conservation in monocot hosts including grass and non-grass monocot hosts. DIS, MIG1, MYB1, VAMP721d/e, PT13, and CERBERUS are present in either grass or non-grass monocot hosts but not in both of these groups. When a phenotype was reported from a knockdown of the gene, the method used is reported as RNAi, RNA interference; VIGS, virus-induced gene silencing; or amiR, artificial microRNA. Species names: Gm, Glycine max; Lj, Lotus japonicus; Mt, Medicago truncatula; Os, Oryza sativa; Ph, Petunia hybrida; Ps, Pisum sativum; Pa, Parasponia andersonii; Sl, Solanum lycopersicum; Zm, Zea mays.
Silencing of many homologs.
Accommodation of the arbuscule involves not only the development of the periarbuscular membrane and apoplast, but later, the active disassembly and removal of the membrane, arbuscule, and interface during a senescence phase known as arbuscule degeneration (Bonfante-Fasolo, 1984). The degeneration phase is accompanied by the expression of secreted hydrolase genes, regulated by a Myb transcription factor, as well as GRAS factors NSP1 and DELLA (Floss et al., 2017). The latter two proteins are also required for arbuscule development, which suggests that changes in composition of a transcription factor complex may regulate the transition between the development and degeneration phases of the accommodation program.
With several thousand plant genes showing differential expression during AM symbiosis, genetic dissection of the symbiotic program is a daunting task. However, the early single origin of AM symbiosis, the broad taxonomic distribution within the vascular plant lineage, and the observation that all mycorrhizal plants contain the same set of genes for AM symbiosis provided a unique opportunity to use phylogenomics to identify genes conserved for AM symbiosis, which provides a point of focus for reverse genetics analyses.
The observation that some genes essential for symbiosis were absent from the Arabidopsis genome provided the first hints that non-host plants had lost the genes whose functions are required exclusively for symbiosis (Harrison et al., 2002; Yano et al., 2008; Zhang et al., 2010; Gobbato et al., 2012). With the availability of sequenced genomes, it was noted that this multiple gene loss occurred independently in several plant lineages (Delaux et al., 2014; Bravo et al., 2016). This evolutionary pattern of conservation in hosts and loss in non-host plants was visualized by constructing phylogenies and exploited to identify genes conserved for AM symbiosis (Delaux et al., 2014; Favre et al., 2014; Bravo et al., 2016). The most stringent analysis identified 138 AM symbiosis-conserved genes, of which 15 had known roles in AM symbiosis and mutants in an additional six also revealed their involvement (Bravo et al., 2016). The 138 AM symbiosis-conserved genes show a variety of molecular functions, but in several cases, they were found to interact or to function at different points of a cellular process or single metabolic pathway, leading to a proposal that the AM conserved genes function in small modules to fine-tune cellular processes for symbiosis (Bravo et al., 2016). For example, EXO70I, Vapyrin, and SYP132 are AM symbiosis-conserved proteins that modulate exocytosis to enable deposition of the periarbuscular membrane (Feddermann et al., 2010; Pumplin et al., 2010; Murray et al., 2011; Zhang et al., 2015b; Huisman et al., 2016; Pan et al., 2016). The conserved category provides a useful filter when selecting candidates for functional analyses; however, by no means are these the only genes required for AM symbiosis. Genes that also have functions outside of the symbiotic context (Delaux et al., 2013a), for example, M. truncatula DELLA1 and DELLA2 (Floss et al., 2013), D14L (Gutjahr et al., 2015), and NOPE1 (Nadal et al., 2017), as well as several others indicated in Table 1, are present in AM non-host and host plants and are required for AM symbiosis.
NUTRIENT EXCHANGE DURING SYMBIOSIS
The exchange of nutrients between the symbionts is central to the AM symbiosis, and this occurs mostly across the extensive interface between the arbuscule and cortical cell. Phosphorus (as phosphate) is the major mineral nutrient contributed by the fungal symbiont and its acquisition and delivery is a remarkable process. Transport from the soil into the extraradical hyphae is followed by synthesis of polyphosphate and long distance transfer through the coenocytic hyphae to the arbuscules where polyphosphate catabolism releases Pi that is exported out of the arbuscule to the common apoplast (Ezawa et al., 2002; Tani et al., 2009; Hijikata et al., 2010). Plant phosphate transporters in the periarbuscular membrane then import phosphate into the cortical cell. While the complement of phosphate transporters in the periarbuscular membrane varies with the plant species, the AM symbiosis conserved transporter MtPT4/OsPT11 plays a significant role and is essential to maintain the symbiosis (Javot et al., 2007; Yang et al., 2012). Loss of MtPT4 function leads to premature degeneration of the arbuscule and loss of symbiosis, which indicates a regulatory role for this nutrient. Although all arbuscules will degenerate eventually, acceleration of this process in the mtpt4 mutant indicates a causal link between symbiotic phosphate delivery and arbuscule lifespan (Javot et al., 2007). This provides a mechanism to maintain balance in the symbiosis as ineffective endosymbionts would not be maintained. RNAi-mediated suppression of MYB1, the regulator of the transcriptional program associated with arbuscule degeneration, relieves premature arbuscule degeneration in the mtpt4 mutant, suggesting that MYB1 could be the degeneration program trigger. However, constitutive overexpression of MYB1 did not totally abolish full arbuscule development, suggesting that MYB1 is only part of the story (Floss et al., 2017). Based on the fact that the fungi are obligate symbionts, it is tempting to speculate that regulation of arbuscule lifespan and its reduction in mtpt4 mutants might involve the withholding of carbon.
AM fungi obtain their entire carbon supply from the plant, and it is estimated that they acquire up to 20% of the carbon fixed during photosynthesis (Bago et al., 2000). Labeling studies coupled with NMR provided evidence for hexose transfer to the fungus (Shachar-Hill et al., 1995; Pfeffer et al., 1999), with recent studies pinpointing specific fungal hexose transporters involved (Helber et al., 2011). The finding that the plant sucrose transporter/sensor SUT2 negatively regulates colonization levels indicates that the host plant likely regulates sugar fluxes to the apoplast spaces around the intraradical hyphae and arbuscules (Bitterlich et al., 2014; Roth and Paszkowski, 2017). Thus, perhaps as anticipated given their status as obligate symbionts, it is clear that the fungus obtains sugars from its host. The recent discoveries that the fungus also obtains fatty acids from the plant, and that they are actually fatty acid auxotrophs, is perhaps more surprising, particularly given the tremendous amounts of triacylglycerols that they synthesize and move through their mycelia to support metabolism, growth, and sporulation (Bago et al., 2002).
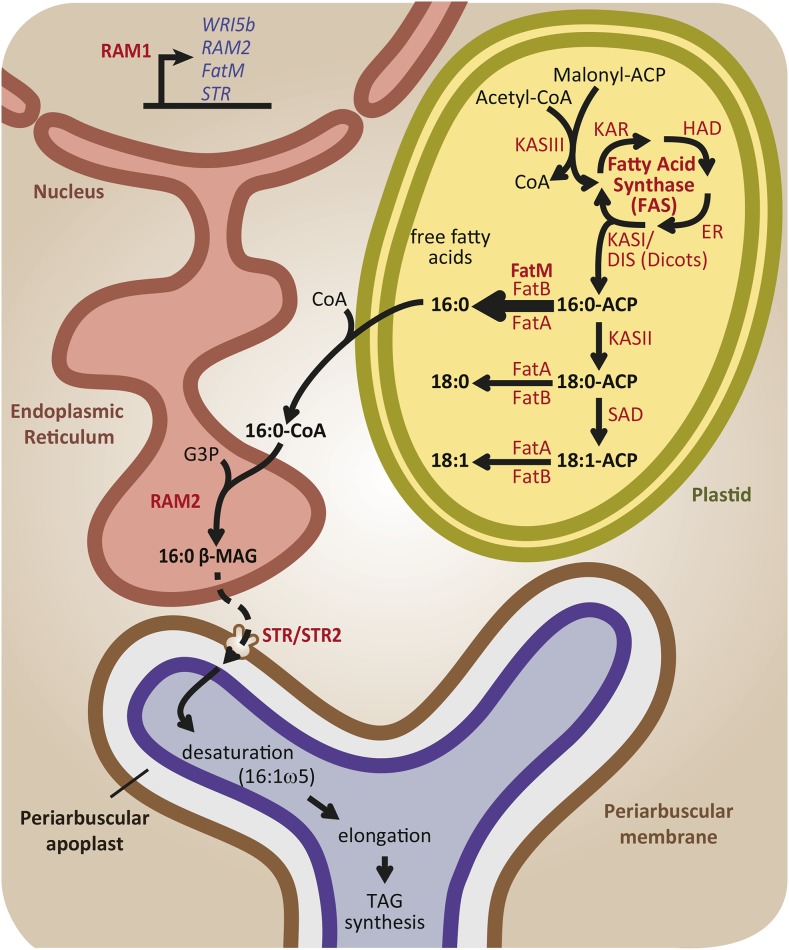
The first suggestion of fatty acid auxotrophy was based on the astute observation that genes encoding the multidomain FAS complex, which is required for de novo fatty acid biosynthesis and is highly conserved in eukaryotes, were missing from the R. irregularis genome. By contrast, genes encoding enzymes for the elongation and desaturation of fatty acids beyond chain length C16, as well as enzymes for the generation of complex lipids were all present (Wewer et al., 2014). This raised the possibility that the fungus relied on its host for a source of fatty acids that it would then modify to generate the necessary array of membrane and storage lipids (Wewer et al., 2014). Previous labeling experiments were consistent with this idea as they had shown that de novo fatty acid biosynthesis could be detected only in mycorrhizal roots and not in extraradical hyphae or spores. However, at that time, the authors had favored the idea of specific in planta expression of fungal fatty acid biosynthetic capacity, rather than fatty acid auxotrophy (Trépanier et al., 2005). Addressing the topic from different angles and with different approaches, four groups recently provided complementary lines of evidence that collectively demonstrate that the plant provides fatty acids, most likely 16:0 β-monoacylglycerol (16:0 β-MAG) but possibly a derivative, to the fungus and that transfer occurs at the interface with the arbuscule (Figure 3) (Bravo et al., 2017; Jiang et al., 2017; Keymer et al., 2017; Luginbuehl et al., 2017).
Figure 3.
Lipid Metabolism in the Colonized Cell and Transfer to the AM Fungus
Lipid biosynthesis increases in cortical cells containing arbuscules to produce lipids that will be transferred to the AM fungus. Regulation of lipid biosynthesis begins with the transcriptional induction of several genes encoding enzymes involved in lipid biosynthesis by a transcriptional regulator, RAM1. The downstream targets of RAM1 identified so far are present exclusively in plant species that form AM symbiosis, as is RAM1 itself, and the encoded enzymes regulate key points in fatty acid and monoacylglycerol biosynthesis. Plant de novo fatty acid synthesis begins in plastids with the elongation of the two-carbon acyl moiety of malonyl-ACP. The elongation is performed by the multimeric enzyme fatty acid synthase whose product is a 16-carbon acyl molecule attached to an acyl carrier protein. In dicots and monocots but not in grasses, an additional KASI protein named DIS is responsible for increasing the biosynthesis of fatty acids in plastids of colonized cells. During AM symbiosis, the thioesterase FatM boosts the release of 16:0 fatty acids (palmitic acid) that, when attached to CoA, are used as a substrate by RAM2 to produce 16:0 β-MAG. 16:0 β-MAG or a derivative of this molecule is subsequently exported across the PAM by the half ABC transporters STR and STR2 where it is accessed by the AM fungus. The fungus modifies the 16:0 acyl moiety through desaturation and elongation reactions to produce a variety of fungal lipids. The main proteins (red) and lipid products (black) involved in redirecting lipid metabolism in the colonized cell are shown. Genes regulated by RAM1 are shown in blue.
Several profiling studies had documented increases in lipid biosynthesis during symbiosis (Schliemann et al., 2008; Wewer et al., 2014), but evidence for increased de novo fatty acid production in the colonized cells and its significance for symbiosis was provided by the L. japonicus dis mutant. DIS encodes a Keto-acyl ACP synthase 1 (KAS1), part of the fatty acid biosynthetic machinery responsible for the elongation of fatty acid chains specifically between C4:0 and C16:0 carbon chain length. In dis mutants, arbuscule development is impaired and fungal fatty acid levels are low (Groth et al., 2013; Keymer et al., 2017). Thus, DIS initiates increased fatty acid biosynthesis in the colonized cell. Evidence to support the direction of lipid flux toward 16:0 β-MAG and potentially transfer to the periarbuscular space was provided by analyses of M. truncatula loss-of-function mutants of three AM symbiosis-conserved proteins, FatM, an acyl ACP-thioesterase, RAM2, a glycerol-3-phosphate acyl transferase (GPAT), and STR, a periarbuscular membrane-resident ABC transporter. The genes encoding these three proteins are highly induced in colonized cells, and in all three mutants arbuscule development is impaired, fungal lipid levels are low, and symbiosis is not maintained (Zhang et al., 2010; Gobbato et al., 2012; Wang et al., 2012; Bravo et al., 2016, 2017).
Building on the concept of modular functions for AM symbiosis-conserved genes and a high degree of similarity between the fatm, ram2, and str phenotypes, Bravo et al. (2017) used complementation analyses and comparative lipid profiling to determine whether these proteins might function in a single pathway. The data support the model that FatM directs the generation of high levels of C16:0 fatty acids in the plastid by releasing the C16:0 acyl chains from their carrier protein, therefore terminating chain elongation and initiating export from the plastid. Following transfer to the endoplasmic reticulum, these C16:0 molecules serve as a substrate for the glycerol-3-phosphate acyl transferase, RAM2. RAM2 is an sn-2 GPAT that generates monoacylglycerols (Gobbato et al., 2012), in this case 16:0 β-MAG, which are members of a class of lipids typically exported from the cell. Based on several lipid profiles, which show the accumulation of 16:0 β-MAG in str to higher levels than fatm and ram2, coupled with the identity of STR/STR2 as an ABCG transporter (a subfamily of ABC transporters whose members include lipid transporters), it was proposed that 16:0 β-MAG are exported across the periarbuscular membrane by the STR/STR2 transporter (Bravo et al., 2017). The same model was proposed through isotopolog and lipid profiles of L. japonicus dis, ram2, and str mutants (Keymer et al., 2017). The lipid profiles fully support this model, but the evidence is indirect with roles inferred based on the accumulation of lipids or lack thereof and on the appearance of new lipid products arising from the redirection of flux through the biosynthetic pathway (Bravo et al., 2017).
Direct evidence for lipid transfer from the plant to the fungus was provided via creative experiments in which M. truncatula roots were engineered to express a FatB gene from Umbellularia californica, which encodes a C12:0 acyl ACP thioesterase (Jiang et al., 2017; Luginbuehl et al., 2017). This results in release of C12:0 fatty acids, a chain length not usually observed in M. truncatula roots or AM fungi. Lipid profiles of the colonized transgenic roots revealed that the fungal triacylglycerols now contained C12:0 fatty acids, indicating that they originated in the plant. As a second line of evidence, Luginbuehl et al. (2017) obtained a M. truncatula plastid acetyl-CoA synthetase mutant that is unable to incorporate acetate into fatty acid production and comparative analyses with radiolabeled acetate or sucrose enabled them to demonstrate that the fungal fatty acids originated in the plant (Luginbuehl et al., 2017). Finally, Keymer et al. (2017) used comparative isotopolog profiling to provide evidence for direct transfer of a C16:0 containing lipid to the fungus (Keymer et al., 2017). Coupled with the native lipid profiles (Bravo et al., 2017), the data collectively provide strong evidence that the colonized cell increases fatty acid biosynthesis and redirects flux through lipid metabolism to generate 16:0 β-monoacylglycerols (16:0 β-MAG) and these, or a derivative thereof, are transferred to the periarbuscular apoplast and subsequently accessed by the fungus (Figure 3). The key proteins that increase and direct lipid flux within the colonized cell are DIS, FatM, and RAM2 (Gobbato et al., 2012; Bravo et al., 2017; Jiang et al., 2017; Keymer et al., 2017; Luginbuehl et al., 2017). Furthermore, RAM1 is a direct regulator of RAM2 (Gobbato et al., 2012), and likely of FatM as well as an AP2 domain transcription factor, which potentially regulates other fatty acid biosynthesis genes (Luginbuehl et al., 2017). Thus, these data reveal that during AM symbiosis, signaling through the common symbiosis signaling pathway triggers the reprogramming of lipid metabolism in the colonized cells to enable production and export of essential fatty acids for the fungus. Furthermore, output from the signaling pathway is directed by genes that are conserved for AM symbiosis, and so while the current analyses are focused on legumes, it is likely that this scenario occurs broadly within the AM host plants.
SUMMARY AND CONCLUSIONS
In summary, research over the past few years has enhanced our understanding of the common symbiosis signaling pathway and established direct connections between signaling and downstream events in the colonized cells. Yet, despite these advances, a full understanding of the receptor complexes and signaling molecules that generate input to the pathway, and the transcriptional regulatory networks that control downstream gene expression, remains to be achieved. Currently, GRAS factors dominate the regulatory landscape, yet spatial and temporal details are sparse, and their regulatory roles within multiprotein complexes remain to be established in a symbiotically relevant context. While the importance of the common symbiosis signaling pathway cannot be denied, new data from rice and maize indicate that additional signaling pathways play significant roles. Whether signaling through the D14L/D3 pathway connects directly to the symbiosis signaling pathway or is necessary to establish an appropriate molecular environment to enable symbiosis remains to be determined. Additionally, the rice and maize data emphasize the importance of studies in a diversity of plant species and provide opportunities for comparisons between hosts with different evolutionary trajectories.
While this review has focused on events taking place within a plant host, a true understanding of AM symbiosis requires comprehensive knowledge of both symbiotic partners. Like the host plant, the fungal hyphae must undergo physiological and cellular reprogramming during the transitions from spore germination, to the colonization of the root and terminal differentiation to form arbuscules, yet we know little of how this occurs. The first genome and transcriptome data afforded valuable insight into the biology of AM fungi (Tisserant et al., 2013; Lin et al., 2014; Kamel et al., 2017), providing a strong foundation for future experimentation. The power of integrating information about both partners has been illustrated recently, where the prediction of fatty acid auxotrophy, gleaned through analyses of the genome and transcriptome data (Wewer et al., 2014), contributed to the recent discoveries that the plant specifically redirects lipid metabolism in the colonized cell for the purpose of provisioning the fungus. Future analyses that focus on integrating available resources and data relevant to both the plant and fungi will undoubtedly reveal that we have many more surprises in store.
Acknowledgments
This work was supported by a National Research Initiative Competitive grant from the USDA National Institute of Food and Agriculture (2014-67013-21571), by U.S. National Science Foundation Grant IOS-1353367, and the Office of Science (BER), U.S. Department of Energy, Grant DE-SC0014260. We apologize to the many authors whose work we could not include because of space constraints.
AUTHOR CONTRIBUTIONS
A.M.M., A.B., and M.J.H. conceived the review and contributed text. A.M.M. contributed Figure 2. A.B. contributed Figures 1 and 3 and Table 1. M.J.H. edited the review.
References
- Akiyama K., Hayashi H. (2006). Strigolactones: chemical signals for fungal symbionts and parasitic weeds in plant roots. Ann. Bot. (Lond.) 97: 925–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama K., Matsuzaki K., Hayashi H. (2005). Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435: 824–827. [DOI] [PubMed] [Google Scholar]
- Alvey L., Harberd N.P. (2005). DELLA proteins: integrators of multiple plant growth regulatory inputs? Physiol. Plant. 123: 153–160. [Google Scholar]
- Ané J.-M., et al. (2004). Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303: 1364–1367. [DOI] [PubMed] [Google Scholar]
- Antolín-Llovera M., Ried M.K., Parniske M. (2014). Cleavage of the SYMBIOSIS RECEPTOR-LIKE KINASE ectodomain promotes complex formation with Nod factor receptor 5. Curr. Biol. 24: 422–427. [DOI] [PubMed] [Google Scholar]
- Bago B., Pfeffer P.E., Shachar-Hill Y. (2000). Carbon metabolism and transport in arbuscular mycorrhizas. Plant Physiol. 124: 949–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bago B., Pfeffer P.E., Zipfel W., Lammers P., Shachar-Hill Y. (2002). Tracking metabolism and imaging transport in arbuscular mycorrhizal fungi. Metabolism and transport in AM fungi. Plant Soil 244: 189–197. [Google Scholar]
- Banba M., Gutjahr C., Miyao A., Hirochika H., Paszkowski U., Kouchi H., Imaizumi-Anraku H. (2008). Divergence of evolutionary ways among common sym genes: CASTOR and CCaMK show functional conservation between two symbiosis systems and constitute the root of a common signaling pathway. Plant Cell Physiol. 49: 1659–1671. [DOI] [PubMed] [Google Scholar]
- Benfey P.N., Linstead P.J., Roberts K., Schiefelbein J.W., Hauser M.T., Aeschbacher R.A. (1993). Root development in Arabidopsis: four mutants with dramatically altered root morphogenesis. Development 119: 57–70. [DOI] [PubMed] [Google Scholar]
- Besserer A., Bécard G., Jauneau A., Roux C., Séjalon-Delmas N. (2008). GR24, a synthetic analog of strigolactones, stimulates the mitosis and growth of the arbuscular mycorrhizal fungus Gigaspora rosea by boosting its energy metabolism. Plant Physiol. 148: 402–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besserer A., Puech-Pagès V., Kiefer P., Gomez-Roldan V., Jauneau A., Roy S., Portais J.C., Roux C., Bécard G., Séjalon-Delmas N. (2006). Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol. 4: e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitterlich M., Krügel U., Boldt-Burisch K., Franken P., Kühn C. (2014). The sucrose transporter SlSUT2 from tomato interacts with brassinosteroid functioning and affects arbuscular mycorrhiza formation. Plant J. 78: 877–889. [DOI] [PubMed] [Google Scholar]
- Bolle C., Koncz C., Chua N.H. (2000). PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes Dev. 14: 1269–1278. [PMC free article] [PubMed] [Google Scholar]
- Bonfante-Fasolo P. (1984). Anatomy and morphology of VA mycorrhizae. Mycorrhizae V.A., Powell C.L. Bagyaraj D.J., eds (Boca Raton, FL: CRC Press; ), pp. 5–33. [Google Scholar]
- Bravo A., York T., Pumplin N., Mueller L.A., Harrison M.J. (2016). Genes conserved for arbuscular mycorrhizal symbiosis identified through phylogenomics. Nat. Plants 2: 15208. [DOI] [PubMed] [Google Scholar]
- Bravo A., Brands M., Wewer V., Dörmann P., Harrison M.J. (2017). Arbuscular mycorrhiza-specific enzymes FatM and RAM2 fine-tune lipid biosynthesis to promote development of arbuscular mycorrhiza. New Phytol. 214: 1631–1645. [DOI] [PubMed] [Google Scholar]
- Breuillin-Sessoms F., et al. (2015). Suppression of arbuscule degeneration in Medicago truncatula phosphate transporter4 mutants is dependent on the Ammonium Transporter 2 family protein AMT2;3. Plant Cell 27: 1352–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brundrett M.C. (2002). Coevolution of roots and mycorrhiza of land plants. New Phytol. 154: 275–304. [DOI] [PubMed] [Google Scholar]
- Bucher M., Hause B., Krajinski F., Küster H. (2014). Through the doors of perception to function in arbuscular mycorrhizal symbioses. New Phytol. 204: 833–840. [DOI] [PubMed] [Google Scholar]
- Buee M., Rossignol M., Jauneau A., Ranjeva R., Bécard G. (2000). The pre-symbiotic growth of arbuscular mycorrhizal fungi is induced by a branching factor partially purified from plant root exudates. Mol. Plant Microbe Interact. 13: 693–698. [DOI] [PubMed] [Google Scholar]
- Buendia L., Wang T., Girardin A., Lefebvre B. (2016). The LysM receptor-like kinase SlLYK10 regulates the arbuscular mycorrhizal symbiosis in tomato. New Phytol. 210: 184–195. [DOI] [PubMed] [Google Scholar]
- Capoen W., Sun J., Wysham D., Otegui M.S., Venkateshwaran M., Hirsch S., Miwa H., Downie J.A., Morris R.J., Ané J.M., Oldroyd G.E.D. (2011). Nuclear membranes control symbiotic calcium signaling of legumes. Proc. Natl. Acad. Sci. USA 108: 14348–14353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carotenuto G., Chabaud M., Miyata K., Capozzi M., Takeda N., Kaku H., Shibuya N., Nakagawa T., Barker D.G., Genre A. (2017). The rice LysM receptor-like kinase OsCERK1 is required for the perception of short-chain chitin oligomers in arbuscular mycorrhizal signaling. New Phytol. 214: 1440–1446. [DOI] [PubMed] [Google Scholar]
- Chabaud M., Genre A., Sieberer B.J., Faccio A., Fournier J., Novero M., Barker D.G., Bonfante P. (2011). Arbuscular mycorrhizal hyphopodia and germinated spore exudates trigger Ca2+ spiking in the legume and nonlegume root epidermis. New Phytol. 189: 347–355. [DOI] [PubMed] [Google Scholar]
- Charpentier M., Sun J., Wen J., Mysore K.S., Oldroyd G.E.D. (2014). Abscisic acid promotion of arbuscular mycorrhizal colonization requires a component of the PROTEIN PHOSPHATASE 2A complex. Plant Physiol. 166: 2077–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier M., Sun J., Vaz Martins T., Radhakrishnan G.V., Findlay K., Soumpourou E., Thouin J., Véry A.A., Sanders D., Morris R.J., Oldroyd G.E.D. (2016). Nuclear-localized cyclic nucleotide-gated channels mediate symbiotic calcium oscillations. Science 352: 1102–1105. [DOI] [PubMed] [Google Scholar]
- Chen C., Gao M., Liu J., Zhu H. (2007). Fungal symbiosis in rice requires an ortholog of a legume common symbiosis gene encoding a Ca2+/calmodulin-dependent protein kinase. Plant Physiol. 145: 1619–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Ané J.M., Zhu H. (2008). OsIPD3, an ortholog of the Medicago truncatula DMI3 interacting protein IPD3, is required for mycorrhizal symbiosis in rice. New Phytol. 180: 311–315. [DOI] [PubMed] [Google Scholar]
- Conn C.E., Nelson D.C. (2016). Evidence that KARRIKIN-INSENSITIVE2 (KAI2) receptors may perceive an unknown signal that is not karrikin or strigolactone. Front. Plant Sci. 6: 1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couzigou J.M., Lauressergues D., André O., Gutjahr C., Guillotin B., Bécard G., Combier J.P. (2017). Positive gene regulation by a natural protective miRNA enables arbuscular mycorrhizal symbiosis. Cell Host Microbe 21: 106–112. [DOI] [PubMed] [Google Scholar]
- Davière J.M., Achard P. (2013). Gibberellin signaling in plants. Development 140: 1147–1151. [DOI] [PubMed] [Google Scholar]
- Delaux P.-M., Séjalon-Delmas N., Bécard G., Ané J.-M. (2013a). Evolution of the plant-microbe symbiotic ‘toolkit’. Trends Plant Sci. 18: 298–304. [DOI] [PubMed] [Google Scholar]
- Delaux P.-M., Varala K., Edger P.P., Coruzzi G.M., Pires J.C., Ané J.-M. (2014). Comparative phylogenomics uncovers the impact of symbiotic associations on host genome evolution. PLoS Genet. 10: e1004487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaux P.M., Bécard G., Combier J.P. (2013b). NSP1 is a component of the Myc signaling pathway. New Phytol. 199: 59–65. [DOI] [PubMed] [Google Scholar]
- Delaux P.M., et al. (2015). Algal ancestor of land plants was preadapted for symbiosis. Proc. Natl. Acad. Sci. USA 112: 13390–13395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devers E.A., Teply J., Reinert A., Gaude N., Krajinski F. (2013). An endogenous artificial microRNA system for unraveling the function of root endosymbioses related genes in Medicago truncatula. BMC Plant Biol. 13: 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Laurenzio L., Wysocka-Diller J., Malamy J.E., Pysh L., Helariutta Y., Freshour G., Hahn M.G., Feldmann K.A., Benfey P.N. (1996). The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86: 423–433. [DOI] [PubMed] [Google Scholar]
- Endre G., Kereszt A., Kevei Z., Mihacea S., Kaló P., Kiss G.B. (2002). A receptor kinase gene regulating symbiotic nodule development. Nature 417: 962–966. [DOI] [PubMed] [Google Scholar]
- Ezawa T., Smith S.E., Smith F.A. (2002). P metabolism and transport in AM fungi. Plant Soil 244: 221–230. [Google Scholar]
- Favre P., Bapaume L., Bossolini E., Delorenzi M., Falquet L., Reinhardt D. (2014). A novel bioinformatics pipeline to discover genes related to arbuscular mycorrhizal symbiosis based on their evolutionary conservation pattern among higher plants. BMC Plant Biol. 14: 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feddermann N., Muni R.R.D., Zeier T., Stuurman J., Ercolin F., Schorderet M., Reinhardt D. (2010). The PAM1 gene of petunia, required for intracellular accommodation and morphogenesis of arbuscular mycorrhizal fungi, encodes a homologue of VAPYRIN. Plant J. 64: 470–481. [DOI] [PubMed] [Google Scholar]
- Field K.J., Cameron D.D., Leake J.R., Tille S., Bidartondo M.I., Beerling D.J. (2012). Contrasting arbuscular mycorrhizal responses of vascular and non-vascular plants to a simulated Palaeozoic CO2 decline. Nat. Commun. 3: 835. [DOI] [PubMed] [Google Scholar]
- Fiorilli V., Volpe V., Zanini S., Vallino M., Abbà S., Bonfante P. (2015). A rice GRAS gene has an impact on the success of arbuscular mycorrhizal colonization. Am. J. Plant Sci. 6: 1905–1915. [Google Scholar]
- Flematti G.R., Dixon K.W., Smith S.M. (2015). What are karrikins and how were they ‘discovered’ by plants? BMC Biol. 13: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floss D.S., Schliemann W., Schmidt J., Strack D., Walter M.H. (2008a). RNA interference-mediated repression of MtCCD1 in mycorrhizal roots of Medicago truncatula causes accumulation of C27 apocarotenoids, shedding light on the functional role of CCD1. Plant Physiol. 148: 1267–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floss D.S., Levy J.G., Levesque-Tremblay V., Pumplin N., Harrison M.J. (2013). DELLA proteins regulate arbuscule formation in arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. USA 110: E5025–E5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floss D.S., Hause B., Lange P.R., Küster H., Strack D., Walter M.H. (2008b). Knock-down of the MEP pathway isogene 1-deoxy-D-xylulose 5-phosphate synthase 2 inhibits formation of arbuscular mycorrhiza-induced apocarotenoids, and abolishes normal expression of mycorrhiza-specific plant marker genes. Plant J. 56: 86–100. [DOI] [PubMed] [Google Scholar]
- Floss D.S., Gomez S.K., Park H.J., MacLean A.M., Müller L.M., Bhattarai K.K., Lévesque-Tremblay V., Maldonado-Mendoza I.E., Harrison M.J. (2017). A transcriptional program for arbuscule degeneration during AM symbiosisis regulated by MYB1. Curr. Biol. 27: 1206–1212. [DOI] [PubMed] [Google Scholar]
- Foo E., Ross J.J., Jones W.T., Reid J.B. (2013). Plant hormones in arbuscular mycorrhizal symbioses: an emerging role for gibberellins. Ann. Bot. (Lond.) 111: 769–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé J., Minguet E.G., Grau-Enguix F., Abbas M., Locascio A., Thomas S.G., Alabadí D., Blázquez M.A. (2012). Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 13446–13451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia K., Doidy J., Zimmermann S.D., Wipf D., Courty P.E. (2016). Take a trip through the plant and fungal transportome of mycorrhiza. Trends Plant Sci. 21: 937–950. [DOI] [PubMed] [Google Scholar]
- Gaude N., Bortfeld S., Duensing N., Lohse M., Krajinski F. (2012). Arbuscule-containing and non-colonized cortical cells of mycorrhizal roots undergo extensive and specific reprogramming during arbuscular mycorrhizal development. Plant J. 69: 510–528. [DOI] [PubMed] [Google Scholar]
- Genre A., Chabaud M., Balzergue C., Puech-Pagès V., Novero M., Rey T., Fournier J., Rochange S., Bécard G., Bonfante P., Barker D.G. (2013). Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol. 198: 190–202. [DOI] [PubMed] [Google Scholar]
- Gobbato E., Wang E., Higgins G., Bano S.A., Henry C., Schultze M., Oldroyd G.E.D. (2013). RAM1 and RAM2 function and expression during arbuscular mycorrhizal symbiosis and Aphanomyces euteiches colonization. Plant Signal. Behav. 8: e26049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobbato E., et al. (2012). A GRAS-type transcription factor with a specific function in mycorrhizal signaling. Curr. Biol. 22: 2236–2241. [DOI] [PubMed] [Google Scholar]
- Gomez-Roldan V., et al. (2008). Strigolactone inhibition of shoot branching. Nature 455: 189–194. [DOI] [PubMed] [Google Scholar]
- Groth M., Takeda N., Perry J., Uchida H., Dräxl S., Brachmann A., Sato S., Tabata S., Kawaguchi M., Wang T.L., Parniske M. (2010). NENA, a Lotus japonicus homolog of Sec13, is required for rhizodermal infection by arbuscular mycorrhiza fungi and rhizobia but dispensable for cortical endosymbiotic development. Plant Cell 22: 2509–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth M., Kosuta S., Gutjahr C., Haage K., Hardel S.L., Schaub M., Brachmann A., Sato S., Tabata S., Findlay K., Wang T.L., Parniske M. (2013). Two Lotus japonicus symbiosis mutants impaired at distinct steps of arbuscule development. Plant J. 75: 117–129. [DOI] [PubMed] [Google Scholar]
- Gust A.A., Willmann R., Desaki Y., Grabherr H.M., Nürnberger T. (2012). Plant LysM proteins: modules mediating symbiosis and immunity. Trends Plant Sci. 17: 495–502. [DOI] [PubMed] [Google Scholar]
- Gutjahr C., Parniske M. (2013). Cell and developmental biology of arbuscular mycorrhiza symbiosis. In Annual Review of Cell and Developmental Biology, Vol. 29, Schekman R., ed (Palo Alto, CA: Annual Reviews), pp. 593–617. [DOI] [PubMed] [Google Scholar]
- Gutjahr C., Banba M., Croset V., An K., Miyao A., An G., Hirochika H., Imaizumi-Anraku H., Paszkowski U. (2008). Arbuscular mycorrhiza-specific signaling in rice transcends the common symbiosis signaling pathway. Plant Cell 20: 2989–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutjahr C., et al. (2012). The half-size ABC transporters STR1 and STR2 are indispensable for mycorrhizal arbuscule formation in rice. Plant J. 69: 906–920. [DOI] [PubMed] [Google Scholar]
- Gutjahr C., et al. (2015). Rice perception of symbiotic arbuscular mycorrhizal fungi requires the karrikin receptor complex. Science 350: 1521–1524. [DOI] [PubMed] [Google Scholar]
- Harrison M.J., Dewbre G.R., Liu J. (2002). A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 14: 2413–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T., Banba M., Shimoda Y., Kouchi H., Hayashi M., Imaizumi-Anraku H. (2010). A dominant function of CCaMK in intracellular accommodation of bacterial and fungal endosymbionts. Plant J. 63: 141–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck C., Kuhn H., Heidt S., Walter S., Rieger N., Requena N. (2016). Symbiotic fungi control plant root cortex development through the novel GRAS transcription factor MIG1. Curr. Biol. 26: 2770–2778. [DOI] [PubMed] [Google Scholar]
- Heckman D.S., Geiser D.M., Eidell B.R., Stauffer R.L., Kardos N.L., Hedges S.B. (2001). Molecular evidence for the early colonization of land by fungi and plants. Science 293: 1129–1133. [DOI] [PubMed] [Google Scholar]
- Helber N., Wippel K., Sauer N., Schaarschmidt S., Hause B., Requena N. (2011). A versatile monosaccharide transporter that operates in the arbuscular mycorrhizal fungus Glomus sp is crucial for the symbiotic relationship with plants. Plant Cell 23: 3812–3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijikata N., Murase M., Tani C., Ohtomo R., Osaki M., Ezawa T. (2010). Polyphosphate has a central role in the rapid and massive accumulation of phosphorus in extraradical mycelium of an arbuscular mycorrhizal fungus. New Phytol. 186: 285–289. [DOI] [PubMed] [Google Scholar]
- Hirano Y., Nakagawa M., Suyama T., Murase K., Shirakawa M., Takayama S., Sun T.P., Hakoshima T. (2017). Structure of the SHR-SCR heterodimer bound to the BIRD/IDD transcriptional factor JKD. Nat. Plants 3: 17010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch S., Kim J., Muñoz A., Heckmann A.B., Downie J.A., Oldroyd G.E.D. (2009). GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 21: 545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogekamp C., Küster H. (2013). A roadmap of cell-type specific gene expression during sequential stages of the arbuscular mycorrhiza symbiosis. BMC Genomics 14: 306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohnjec N., Czaja-Hasse L.F., Hogekamp C., Küster H. (2015). Pre-announcement of symbiotic guests: transcriptional reprogramming by mycorrhizal lipochitooligosaccharides shows a strict co-dependency on the GRAS transcription factors NSP1 and RAM1. BMC Genomics 16: 994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horváth B., et al. (2011). Medicago truncatula IPD3 is a member of the common symbiotic signaling pathway required for rhizobial and mycorrhizal symbioses. Mol. Plant Microbe Interact. 24: 1345–1358. [DOI] [PubMed] [Google Scholar]
- Huisman R., Hontelez J., Mysore K.S., Wen J., Bisseling T., Limpens E. (2016). A symbiosis-dedicated SYNTAXIN OF PLANTS 13II isoform controls the formation of a stable host-microbe interface in symbiosis. New Phytol. 211: 1338–1351. [DOI] [PubMed] [Google Scholar]
- Imaizumi-Anraku H., et al. (2005). Plastid proteins crucial for symbiotic fungal and bacterial entry into plant roots. Nature 433: 527–531. [DOI] [PubMed] [Google Scholar]
- Ivanov S., Fedorova E.E., Limpens E., De Mita S., Genre A., Bonfante P., Bisseling T. (2012). Rhizobium-legume symbiosis shares an exocytotic pathway required for arbuscule formation. Proc. Natl. Acad. Sci. USA 109: 8316–8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javot H., Penmetsa R.V., Terzaghi N., Cook D.R., Harrison M.J. (2007). A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. USA 104: 1720–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C., Gao X., Liao L., Harberd N.P., Fu X. (2007). Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the gibberellin-DELLA signaling pathway in Arabidopsis. Plant Physiol. 145: 1460–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Wang W., Xie Q., Liu N., Liu L., Wang D., Zhang X., Yang C., Chen X., Tang D., Wang E. (2017). Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science 356: 1172–1175. [DOI] [PubMed] [Google Scholar]
- Jin Y., Liu H., Luo D., Yu N., Dong W., Wang C., Zhang X., Dai H., Yang J., Wang E. (2016). DELLA proteins are common components of symbiotic rhizobial and mycorrhizal signalling pathways. Nat. Commun. 7: 12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaku H., Nishizawa Y., Ishii-Minami N., Akimoto-Tomiyama C., Dohmae N., Takio K., Minami E., Shibuya N. (2006). Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. USA 103: 11086–11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaló P., et al. (2005). Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308: 1786–1789. [DOI] [PubMed] [Google Scholar]
- Kamel L., Keller-Pearson M., Roux C., Ané J.M. (2017). Biology and evolution of arbuscular mycorrhizal symbiosis in the light of genomics. New Phytol. 213: 531–536. [DOI] [PubMed] [Google Scholar]
- Kanamori N., et al. (2006). A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc. Natl. Acad. Sci. USA 103: 359–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevei Z., et al. (2007). 3-Hydroxy-3-Methylglutaryl Coenzyme A Reductase1 interacts with NORK and is crucial for nodulation in Medicago truncatula. Plant Cell 19: 3974–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keymer A., et al. (2017). Lipid transfer from plants to arbuscular mycorrhiza fungi. eLife 6: e29107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistner C., Winzer T., Pitzschke A., Mulder L., Sato S., Kaneko T., Tabata S., Sandal N., Stougaard J., Webb K.J., Szczyglowski K., Parniske M. (2005). Seven Lotus japonicus genes required for transcriptional reprogramming of the root during fungal and bacterial symbiosis. Plant Cell 17: 2217–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobae Y., Hata S. (2010). Dynamics of periarbuscular membranes visualized with a fluorescent phosphate transporter in arbuscular mycorrhizal roots of rice. Plant Cell Physiol. 51: 341–353. [DOI] [PubMed] [Google Scholar]
- Kobae Y., Tamura Y., Takai S., Banba M., Hata S. (2010). Localized expression of arbuscular mycorrhiza-inducible ammonium transporters in soybean. Plant Cell Physiol. 51: 1411–1415. [DOI] [PubMed] [Google Scholar]
- Kojima T., Saito K., Oba H., Yoshida Y., Terasawa J., Umehara Y., Suganuma N., Kawaguchi M., Ohtomo R. (2014). Isolation and phenotypic characterization of Lotus japonicus mutants specifically defective in arbuscular mycorrhizal formation. Plant Cell Physiol. 55: 928–941. [DOI] [PubMed] [Google Scholar]
- Koltai H., LekKala S.P., Bhattacharya C., Mayzlish-Gati E., Resnick N., Wininger S., Dor E., Yoneyama K., Yoneyama K., Hershenhorn J., Joel D.M., Kapulnik Y. (2010). A tomato strigolactone-impaired mutant displays aberrant shoot morphology and plant interactions. J. Exp. Bot. 61: 1739–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajinski F., Courty P.E., Sieh D., Franken P., Zhang H., Bucher M., Gerlach N., Kryvoruchko I., Zoeller D., Udvardi M., Hause B. (2014). The H+-ATPase HA1 of Medicago truncatula is essential for phosphate transport and plant growth during arbuscular mycorrhizal symbiosis. Plant Cell 26: 1808–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar T., Kohlen W., Sasse J., Borghi L., Schlegel M., Bachelier J.B., Reinhardt D., Bours R., Bouwmeester H.J., Martinoia E. (2012). A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching. Nature 483: 341–344. [DOI] [PubMed] [Google Scholar]
- Krusell L., et al. (2002). Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature 420: 422–426. [DOI] [PubMed] [Google Scholar]
- Kuhn H., Küster H., Requena N. (2010). Membrane steroid-binding protein 1 induced by a diffusible fungal signal is critical for mycorrhization in Medicago truncatula. New Phytol. 185: 716–733. [DOI] [PubMed] [Google Scholar]
- Lanfranco L., Bonfante P., Genre A. (2016). The mutualistic interaction between plants and arbuscular mycorrhizal fungi. Microbiol. Spectr. 4: 4. [DOI] [PubMed] [Google Scholar]
- Larkan N.J., Ruzicka D.R., Edmonds-Tibbett T., Durkin J.M.H., Jackson L.E., Smith F.A., Schachtman D.P., Smith S.E., Barker S.J. (2013). The reduced mycorrhizal colonisation (rmc) mutation of tomato disrupts five gene sequences including the CYCLOPS/IPD3 homologue. Mycorrhiza 23: 573–584. [DOI] [PubMed] [Google Scholar]
- Lauressergues D., Delaux P.M., Formey D., Lelandais-Brière C., Fort S., Cottaz S., Bécard G., Niebel A., Roux C., Combier J.P. (2012). The microRNA miR171h modulates arbuscular mycorrhizal colonization of Medicago truncatula by targeting NSP2. Plant J. 72: 512–522. [DOI] [PubMed] [Google Scholar]
- Lévy J., et al. (2004). A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303: 1361–1364. [DOI] [PubMed] [Google Scholar]
- Li S., Zhao Y., Zhao Z., Wu X., Sun L., Liu Q., Wu Y. (2016). Crystal structure of the GRAS domain of SCARECROW-LIKE7 in Oryza sativa. Plant Cell 28: 1025–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K., et al. (2014). Single nucleus genome sequencing reveals high similarity among nuclei of an endomycorrhizal fungus. PLoS Genet. 10: e1004078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., et al. (2011). Strigolactone biosynthesis in Medicago truncatula and rice requires the symbiotic GRAS-type transcription factors NSP1 and NSP2. Plant Cell 23: 3853–3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Obando M., Ligerot Y., Bonhomme S., Boyer F.D., Rameau C. (2015). Strigolactone biosynthesis and signaling in plant development. Development 142: 3615–3619. [DOI] [PubMed] [Google Scholar]
- Lota F., Wegmüller S., Buer B., Sato S., Bräutigam A., Hanf B., Bucher M. (2013). The cis-acting CTTC-P1BS module is indicative for gene function of LjVTI12, a Qb-SNARE protein gene that is required for arbuscule formation in Lotus japonicus. Plant J. 74: 280–293. [DOI] [PubMed] [Google Scholar]
- Luginbuehl L.H., Menard G.N., Kurup S., Van Erp H., Radhakrishnan G.V., Breakspear A., Oldroyd G.E.D., Eastmond P.J. (2017). Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plant. Science 356: 1175–1178. [DOI] [PubMed] [Google Scholar]
- Madsen E.B., Madsen L.H., Radutoiu S., Olbryt M., Rakwalska M., Szczyglowski K., Sato S., Kaneko T., Tabata S., Sandal N., Stougaard J. (2003). A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425: 637–640. [DOI] [PubMed] [Google Scholar]
- Maillet F., et al. (2011). Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469: 58–63. [DOI] [PubMed] [Google Scholar]
- Messinese E., Mun J.H., Yeun L.H., Jayaraman D., Rougé P., Barre A., Lougnon G., Schornack S., Bono J.J., Cook D.R., Ané J.M. (2007). A novel nuclear protein interacts with the symbiotic DMI3 calcium- and calmodulin-dependent protein kinase of Medicago truncatula. Mol. Plant Microbe Interact. 20: 912–921. [DOI] [PubMed] [Google Scholar]
- Miller J.B., Pratap A., Miyahara A., Zhou L., Bornemann S., Morris R.J., Oldroyd G.E.D. (2013). Calcium/calmodulin-dependent protein kinase is negatively and positively regulated by calcium, providing a mechanism for decoding calcium responses during symbiosis signaling. Plant Cell 25: 5053–5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miya A., Albert P., Shinya T., Desaki Y., Ichimura K., Shirasu K., Narusaka Y., Kawakami N., Kaku H., Shibuya N. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 19613–19618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata K., et al. (2014). The bifunctional plant receptor, OsCERK1, regulates both chitin-triggered immunity and arbuscular mycorrhizal symbiosis in rice. Plant Cell Physiol. 55: 1864–1872. [DOI] [PubMed] [Google Scholar]
- Morandi D., Sagan M., Prado-Vivant E., Duc G. (2000). Influence of genes determining supernodulation on root colonization by the mycorrhizal fungus Glomus mosseae in Pisum sativum and Medicago truncatula mutants. Mycorrhiza 10: 37–42. [Google Scholar]
- Murray J.D., et al. (2011). Vapyrin, a gene essential for intracellular progression of arbuscular mycorrhizal symbiosis, is also essential for infection by rhizobia in the nodule symbiosis of Medicago truncatula. Plant J. 65: 244–252. [DOI] [PubMed] [Google Scholar]
- Nadal M., et al. (2017). An N-acetylglucosamine transporter required for arbuscular mycorrhizal symbioses in rice and maize. Nat. Plants 3: 17073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D.C., Scaffidi A., Dun E.A., Waters M.T., Flematti G.R., Dixon K.W., Beveridge C.A., Ghisalberti E.L., Smith S.M. (2011). F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 108: 8897–8902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura R., Hayashi M., Wu G.J., Kouchi H., Imaizumi-Anraku H., Murakami Y., Kawasaki S., Akao S., Ohmori M., Nagasawa M., Harada K., Kawaguchi M. (2002). HAR1 mediates systemic regulation of symbiotic organ development. Nature 420: 426–429. [DOI] [PubMed] [Google Scholar]
- Ogawa M., Kusano T., Katsumi M., Sano H. (2000). Rice gibberellin-insensitive gene homolog, OsGAI, encodes a nuclear-localized protein capable of gene activation at transcriptional level. Gene 245: 21–29. [DOI] [PubMed] [Google Scholar]
- Oldroyd G.E.D. (2013). Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 11: 252–263. [DOI] [PubMed] [Google Scholar]
- Op den Camp R., Streng A., De Mita S., Cao Q., Polone E., Liu W., Ammiraju J.S.S., Kudrna D., Wing R., Untergasser A., Bisseling T., Geurts R. (2011). LysM-type mycorrhizal receptor recruited for rhizobium symbiosis in nonlegume Parasponia. Science 331: 909–912. [DOI] [PubMed] [Google Scholar]
- Ovchinnikova E., et al. (2011). IPD3 controls the formation of nitrogen-fixing symbiosomes in pea and Medicago spp. Mol. Plant Microbe Interact. 24: 1333–1344. [DOI] [PubMed] [Google Scholar]
- Pan H., Oztas O., Zhang X., Wu X., Stonoha C., Wang E., Wang B., Wang D. (2016). A symbiotic SNARE protein generated by alternative termination of transcription. Nat. Plants 2: 15197. [DOI] [PubMed] [Google Scholar]
- Park H.J., Floss D.S., Levesque-Tremblay V., Bravo A., Harrison M.J. (2015). Hyphal branching during arbuscule development requires Reduced Arbuscular Mycorrhiza 1. Plant Physiol. 169: 2774–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parniske M. (2000). Intracellular accommodation of microbes by plants: a common developmental program for symbiosis and disease? Curr. Opin. Plant Biol. 3: 320–328. [DOI] [PubMed] [Google Scholar]
- Peng J., Harberd N.P. (1993). Derivative alleles of the Arabidopsis gibberellin-insensitive (GAI) mutation confer a wild-type phenotype. Plant Cell 5: 351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., Carol P., Richards D.E., King K.E., Cowling R.J., Murphy G.P., Harberd N.P. (1997). The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 11: 3194–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penmetsa R.V., et al. (2008). The Medicago truncatula ortholog of Arabidopsis EIN2, sickle, is a negative regulator of symbiotic and pathogenic microbial associations. Plant J. 55: 580–595. [DOI] [PubMed] [Google Scholar]
- Pfeffer P.E., Douds D.D., Becard G., Shachar-Hill Y. (1999). Carbon uptake and the metabolism and transport of lipids in an arbuscular mycorrhiza. Plant Physiol. 120: 587–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimprikar P., Carbonnel S., Paries M., Katzer K., Klingl V., Bohmer M.J., Karl L., Floss D.S., Harrison M.J., Parniske M., Gutjahr C. (2016). A CCaMK-CYCLOPS-DELLA complex activates transcription of RAM1 to regulate arbuscule branching. Curr. Biol. 26: 987–998. [DOI] [PubMed] [Google Scholar]
- Pirozynski K.A., Malloch D.W. (1975). The origin of land plants: a matter of mycotrophism. Biosystems 6: 153–164. [DOI] [PubMed] [Google Scholar]
- Pumplin N., Zhang X., Noar R.D., Harrison M.J. (2012). Polar localization of a symbiosis-specific phosphate transporter is mediated by a transient reorientation of secretion. Proc. Natl. Acad. Sci. USA 109: E665–E672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumplin N., Mondo S.J., Topp S., Starker C.G., Gantt J.S., Harrison M.J. (2010). Medicago truncatula Vapyrin is a novel protein required for arbuscular mycorrhizal symbiosis. Plant J. 61: 482–494. [DOI] [PubMed] [Google Scholar]
- Rech S.S., Heidt S., Requena N. (2013). A tandem Kunitz protease inhibitor (KPI106)-serine carboxypeptidase (SCP1) controls mycorrhiza establishment and arbuscule development in Medicago truncatula. Plant J. 75: 711–725. [DOI] [PubMed] [Google Scholar]
- Remy W., Taylor T.N., Hass H., Kerp H. (1994). Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc. Natl. Acad. Sci. USA 91: 11841–11843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich M.K., Schorderet M., Bapaume L., Falquet L., Morel P., Vandenbussche M., Reinhardt D. (2015). The petunia GRAS transcription factor ATA/RAM1 regulates symbiotic gene expression and fungal morphogenesis in arbuscular mycorrhiza. Plant Physiol. 168: 788–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ried M.K., Antolín-Llovera M., Parniske M. (2014). Spontaneous symbiotic reprogramming of plant roots triggered by receptor-like kinases. eLife 3: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth R., Paszkowski U. (2017). Plant carbon nourishment of arbuscular mycorrhizal fungi. Curr. Opin. Plant Biol. 39: 50–56. [DOI] [PubMed] [Google Scholar]
- Saito K., et al. (2007). NUCLEOPORIN85 is required for calcium spiking, fungal and bacterial symbioses, and seed production in Lotus japonicus. Plant Cell 19: 610–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaarschmidt S., Gresshoff P.M., Hause B. (2013). Analyzing the soybean transcriptome during autoregulation of mycorrhization identifies the transcription factors GmNF-YA1a/b as positive regulators of arbuscular mycorrhization. Genome Biol. 14: R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliemann W., Ammer C., Strack D. (2008). Metabolite profiling of mycorrhizal roots of Medicago truncatula. Phytochemistry 69: 112–146. [DOI] [PubMed] [Google Scholar]
- Schnabel E., Journet E.P., de Carvalho-Niebel F., Duc G., Frugoli J. (2005). The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Mol. Biol. 58: 809–822. [DOI] [PubMed] [Google Scholar]
- Searle I.R., Men A.E., Laniya T.S., Buzas D.M., Iturbe-Ormaetxe I., Carroll B.J., Gresshoff P.M. (2003). Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science 299: 109–112. [DOI] [PubMed] [Google Scholar]
- Shachar-Hill Y., Pfeffer P.E., Douds D., Osman S.F., Doner L.W., Ratcliffe R.G. (1995). Partitioning of intermediary carbon metabolism in vesicular-arbuscular mycorrhizal leek. Plant Physiol. 108: 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T., Nakano T., Takamizawa D., Desaki Y., Ishii-Minami N., Nishizawa Y., Minami E., Okada K., Yamane H., Kaku H., Shibuya N. (2010). Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 64: 204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone A.L., Ciampaglio C.N., Sun T. (1998). The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10: 155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S., Katzer K., Lambert J., Cerri M., Parniske M. (2014). CYCLOPS, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe 15: 139–152. [DOI] [PubMed] [Google Scholar]
- Smit P., Raedts J., Portyanko V., Debellé F., Gough C., Bisseling T., Geurts R. (2005). NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308: 1789–1791. [DOI] [PubMed] [Google Scholar]
- Smith S.E., Read D.J. (2008). Mycorrhizal Symbiosis. (San Diego, CA: Academic Press; ). [Google Scholar]
- Stracke S., Kistner C., Yoshida S., Mulder L., Sato S., Kaneko T., Tabata S., Sandal N., Stougaard J., Szczyglowski K., Parniske M. (2002). A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417: 959–962. [DOI] [PubMed] [Google Scholar]
- Takeda N., Sato S., Asamizu E., Tabata S., Parniske M. (2009). Apoplastic plant subtilases support arbuscular mycorrhiza development in Lotus japonicus. Plant J. 58: 766–777. [DOI] [PubMed] [Google Scholar]
- Takeda N., Tsuzuki S., Suzaki T., Parniske M., Kawaguchi M. (2013). CERBERUS and NSP1 of Lotus japonicus are common symbiosis genes that modulate arbuscular mycorrhiza development. Plant Cell Physiol. 54: 1711–1723. [DOI] [PubMed] [Google Scholar]
- Tani C., Ohtomo R., Osaki M., Kuga Y., Ezawa T. (2009). ATP-dependent but proton gradient-independent polyphosphate-synthesizing activity in extraradical hyphae of an arbuscular mycorrhizal fungus. Appl. Environ. Microbiol. 75: 7044–7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirichine L., et al. (2006). Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature 441: 1153–1156. [DOI] [PubMed] [Google Scholar]
- Tisserant E., et al. (2013). Genome of an arbuscular mycorrhizal fungus provides insight into the oldest plant symbiosis. Proc. Natl. Acad. Sci. USA 110: 20117–20122. Erratum. Proc. Natl. Acad. Sci. USA 111: 563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong H., Jin Y., Liu W., Li F., Fang J., Yin Y., Qian Q., Zhu L., Chu C. (2009). DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J. 58: 803–816. [DOI] [PubMed] [Google Scholar]
- Trépanier M., Bécard G., Moutoglis P., Willemot C., Gagné S., Avis T.J., Rioux J.A. (2005). Dependence of arbuscular-mycorrhizal fungi on their plant host for palmitic acid synthesis. Appl. Environ. Microbiol. 71: 5341–5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkateshwaran M., Jayaraman D., Chabaud M., Genre A., Balloon A.J., Maeda J., Forshey K., den Os D., Kwiecien N.W., Coon J.J., Barker D.G., Ané J.M. (2015). A role for the mevalonate pathway in early plant symbiotic signaling. Proc. Natl. Acad. Sci. USA 112: 9781–9786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernié T., et al. (2016). PUB1 interacts with the receptor kinase DMI2 and negatively regulates rhizobial and arbuscular mycorrhizal symbioses through its ubiquitination activity in Medicago truncatula. Plant Physiol. 170: 2312–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Qiu Y.L. (2006). Phylogenetic distribution and evolution of mycorrhizas in land plants. Mycorrhiza 16: 299–363. [DOI] [PubMed] [Google Scholar]
- Wang E., Schornack S., Marsh J.F., Gobbato E., Schwessinger B., Eastmond P., Schultze M., Kamoun S., Oldroyd G.E.D. (2012). A common signaling process that promotes mycorrhizal and oomycete colonization of plants. Curr. Biol. 22: 2242–2246. [DOI] [PubMed] [Google Scholar]
- Wang E., et al. (2014). A H+-ATPase that energizes nutrient uptake during mycorrhizal symbioses in rice and Medicago truncatula. Plant Cell 26: 1818–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters M.T., Scaffidi A., Flematti G., Smith S.M. (2015). Substrate-induced degradation of the α/β-fold hydrolase KARRIKIN INSENSITIVE2 requires a functional catalytic triad but is independent of MAX2. Mol. Plant 8: 814–817. [DOI] [PubMed] [Google Scholar]
- Waters M.T., Scaffidi A., Sun Y.K., Flematti G.R., Smith S.M. (2014). The karrikin response system of Arabidopsis. Plant J. 79: 623–631. [DOI] [PubMed] [Google Scholar]
- Waters M.T., Nelson D.C., Scaffidi A., Flematti G.R., Sun Y.K.M., Dixon K.W., Smith S.M. (2012). Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 139: 1285–1295. [DOI] [PubMed] [Google Scholar]
- Wewer V., Brands M., Dörmann P. (2014). Fatty acid synthesis and lipid metabolism in the obligate biotrophic fungus Rhizophagus irregularis during mycorrhization of Lotus japonicus. Plant J. 79: 398–412. [DOI] [PubMed] [Google Scholar]
- Xue L., Cui H., Buer B., Vijayakumar V., Delaux P.-M., Junkermann S., Bucher M. (2015). Network of GRAS transcription factors involved in the control of arbuscule development in Lotus japonicus. Plant Physiol. 167: 854–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.Y., et al. (2012). Nonredundant regulation of rice arbuscular mycorrhizal symbiosis by two members of the phosphate transporter1 gene family. Plant Cell 24: 4236–4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K., et al. (2008). CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc. Natl. Acad. Sci. USA 105: 20540–20545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama K., Yoneyama K., Takeuchi Y., Sekimoto H. (2007a). Phosphorus deficiency in red clover promotes exudation of orobanchol, the signal for mycorrhizal symbionts and germination stimulant for root parasites. Planta 225: 1031–1038. [DOI] [PubMed] [Google Scholar]
- Yoneyama K., Xie X., Kusumoto D., Sekimoto H., Sugimoto Y., Takeuchi Y., Yoneyama K. (2007b). Nitrogen deficiency as well as phosphorus deficiency in sorghum promotes the production and exudation of 5-deoxystrigol, the host recognition signal for arbuscular mycorrhizal fungi and root parasites. Planta 227: 125–132. [DOI] [PubMed] [Google Scholar]
- Yoshida H., et al. (2014). DELLA protein functions as a transcriptional activator through the DNA binding of the indeterminate domain family proteins. Proc. Natl. Acad. Sci. USA 111: 7861–7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S., Kameoka H., Tempo M., Akiyama K., Umehara M., Yamaguchi S., Hayashi H., Kyozuka J., Shirasu K. (2012). The D3 F-box protein is a key component in host strigolactone responses essential for arbuscular mycorrhizal symbiosis. New Phytol. 196: 1208–1216. [DOI] [PubMed] [Google Scholar]
- Yu N., et al. (2014). A DELLA protein complex controls the arbuscular mycorrhizal symbiosis in plants. Cell Res. 24: 130–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Iyer L.M., Aravind L. (2012). Bacterial GRAS domain proteins throw new light on gibberellic acid response mechanisms. Bioinformatics 28: 2407–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Blaylock L.A., Harrison M.J. (2010). Two Medicago truncatula half-ABC transporters are essential for arbuscule development in arbuscular mycorrhizal symbiosis. Plant Cell 22: 1483–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Pumplin N., Ivanov S., Harrison M.J. (2015a). EXO70I is essential for development of a sub-domain of the periarbuscular membrane during arbuscular mycorrhizal symbiosis. Curr. Biol. 25: 2189–2195. [DOI] [PubMed] [Google Scholar]
- Zhang X., Pumplin N., Ivanov S., Harrison M.J. (2015b). EXO70I is required for development of a sub-domain of the periarbuscular membrane during arbuscular mycorrhizal symbiosis. Curr. Biol. 25: 2189–2195. [DOI] [PubMed] [Google Scholar]
- Zhang X., Dong W., Sun J., Feng F., Deng Y., He Z., Oldroyd G.E.D., Wang E. (2015c). The receptor kinase CERK1 has dual functions in symbiosis and immunity signalling. Plant J. 81: 258–267. [DOI] [PubMed] [Google Scholar]
- Zipfel C., Oldroyd G.E.D. (2017). Plant signalling in symbiosis and immunity. Nature 543: 328–336. [DOI] [PubMed] [Google Scholar]