Abstract
Background
Viruses are major etiological agents of childhood gastroenteritis. In recent years, several molecular platforms for the detection of viral enteric pathogens have become available.
Objective/study design
We evaluated the performance of three multiplex platforms including Biofire’s Gastrointestinal Panel (FilmArray), Luminex xTAG® Gastrointestinal Pathogen Panel (GPP), and the TaqMan Array Card (TAC) for the detection of five gastroenteritis viruses using a coded panel of 300 archived stool samples.
Results
The FilmArray detected a virus in 199 (96.1%) and the TAC in 172 (83.1%) of the 207 samples (187 samples positive for a single virus and 20 samples positive for more than one virus) whereas the GPP detected a virus in 100 (78.7%) of the 127 (97 positive for one virus and three positive for more than one virus) samples. Overall the clinical accuracy was highest for the FilmArray (98%) followed by TAC (97.2%) and GPP (96.9%). The sensitivity of the FilmArray, GPP and TAC platforms was highest for rotavirus (100%, 95.8%, and 89.6%, respectively) and lowest for adenovirus type 40/41 (97.4%, 57.9% and 68.4%). The specificity of the three platforms ranged from 95.6% (rotavirus) to 99.6% (norovirus/sapovirus) for the FilmArray, 99.6% (norovirus) to 100% (rotavirus/adenovirus) for GPP, and 98.9% (astrovirus) to 100% (rotavirus/sapovirus) for TAC.
Conclusion
The FilmArray demonstrated the best analytical performance followed by TAC. In recent years, the availability of multi-enteric molecular testing platforms has increased significantly and our data highlight the strengths and weaknesses of these platforms.
Keywords: Gastroenteritis viruses, Rotavirus, Norovirus, Adenovirus type 40/41, Astrovirus, Sapovirus
1. Background
Acute gastroenteritis is an important public health burden causing nearly 2 million cases of diarrheal disease each year globally [1]. The disease has been associated with a diverse group of etiologic agents, which includes bacteria, viruses and parasites [2]. Rotavirus, norovirus, adenovirus types 40 and 41, astrovirus, and sapovirus are the five major viral agents of acute gastroenteritis in humans accounting for nearly 60% of the medically-attended childhood gastroenteritis in the United States [3]. Before the introduction of rotavirus vaccines in the United States, nearly all children were infected with rotavirus before age 5 [4]. After the decline of rotavirus as the results of rotavirus vaccination, noroviruses have become the leading cause of acute gastroenteritis in the United States, especially in children < 5 years of age [5–8].
Different laboratory methods such as viral antigen detection using enzyme immunoassays or immunochromatographic assays or molecular methods such as conventional (RT-)PCR and real-time (RT-)PCR assays have been employed by clinical laboratories to test for gastroenteritis viruses. Some of these methods are time-consuming and most of them usually test for only one virus per assay [9]. In comparison, multiplex molecular assays for the simultaneous detection of known gastrointestinal pathogens, including viruses, reduces turnaround time for accurate results and also identify infections and/or co-infections that remained undiagnosed by routine test methods for single pathogens. In this study, we evaluated the performance characteristics of three multiplex platforms for the detection of gastroenteritis viruses. These included the BioFire gastrointestinal panel (FilmArray), the Luminex xTag GI pathogen panel (GPP) and the TaqMan Array Card (TAC) system. These platform were chosen because they are FDA cleared (FilmArray and GPP) or available for large throughput testing (TAC).
The FilmArray is a cartridge-based integrated system which combines automated sample preparation, nucleic acid extraction and multiplex PCR-based detection using DNA melting curve analysis [10]. Luminex GPP (US-IVD) is based on multiplex RT-PCR for target amplification and detection using Luminex microsphere xMAP and xTAG technologies [10]. The TAC system is custom developed by Life Technologies and contains singleplex or duplex real-time PCR reactions, allowing for multi-target detection through spatial distribution [11].
2. Objective
To evaluate the performance of three multiplex gastrointestinal platforms, a panel of coded stool samples including five different gastroenteritis viruses was compiled and shipped to three study sites each of which tested the panel using one of the three platforms and results were compared to reference methods for each of the individual viruses.
3. Study design
A panel of 300 stool samples was compiled at CDC and included 187 samples positive for one of the five viruses (rotavirus, norovirus, sapovirus astrovirus and adenovirus 40/41), 20 samples positive for at least two viruses by TaqMan realtime (RT-) PCR and conventional (RT-) PCR followed by Sanger sequencing [12–20], and 93 samples negative for any of these viruses. The stool samples were collected from norovirus outbreaks (children and adults) that occurred between 2008 and 2012 [21] and from sporadic cases (children) of acute gastroenteritis [6,3]. Stool samples had been stored at −70 °C. The panel included 48 samples positive for rotavirus, 41 samples positive for norovirus (GI and/or GII), 41 samples positive for sapovirus, 39 samples positive for astrovirus and 38 samples positive for adenovirus 40/41. The panel included genotypes of each virus circulating in the United States (Table 1). The panel was coded and distributed to three New Vaccine Surveillance Network (NVSN) [22] study sites (Kansas City, MO. Nashville, TN, and Rochester, NY) and one panel was kept at CDC. At CDC, the panel was retested using both reference methods described above by a different laboratorian than who had compiled the panel. At the Children’s Mercy Hospital in Kansas City, MO the panel was tested on the FilmArray, at Vanderbilt University Medical Center, Nashville, TN on Luminex GPP; and at Golisano Children’s Hospital in Rochester, NY on the TAC system. Institutional review approval for the parent NVSN protocols was granted by CDC and each participating site’s Institutional Review Boards.
Table 1.
Virus genotypes included in stool panel to compare performance of three different enteric pathogen platforms.
| Virus | Genotypes |
|---|---|
| Adenovirus | type 41 |
| Astrovirus | type 1–5 and 8 |
| GI Norovirus | GI.1. GI.3, GI.4, GI.5, GI.6, GI.8 and GI.9 |
| GII Norovirus | GII.2, GII.3, GII.4 New Orleans, GII.4 Sydney, GII.6, GII.7, GII.8, GII.12, GII.13, GII,14, GII.16, GII.17 |
| Rotavirus | G2P[4], G3P[8], G9P[8], G12P[8] |
| Sapovirus | GI.1. GI.2, GI.3, GII.1, GII.2, GII.3, GII.5, GII.6, GIV and GV |
The FilmArray and TAC systems are able to detect all five viruses (adenovirus 40/41, astrovirus, norovirus, rotavirus and sapovirus) whereas Luminex GPP detects three viruses (adenovirus 40/41, norovirus and rotavirus) (Table 2). In addition to the viruses, the FilmArray is able to detect 13 bacterial and four parasite organisms; Luminex GPP is able to detect 8 bacterial and 3 parasite organisms; and the TAC system employed in this study could detect 15 enteric bacterial, 3 protozoal, 10 parasite, and 2 fungal organisms as well as enteroviruses (Table 2).
Table 2.
Gastroenteritis pathogens detected by the FilmArray, GPP and TAC multienteric platform systems.
| Target viruses | Other enteric pathogens detected | |
|---|---|---|
| FilmArray | Adenovirus type 40/41, Astrovirus, Norovirus, Rotavirus, Sapovirus | Campylobacter (jejuni, coli and upsaliensis), Clostridium difficile (toxin A/B), Plesiomonas shigelloides, Salmonella, Yersinia enterocolitica, Vibrio (parahaemolyticus, vulnificus and cholerae), Diarrheagenic E. coli/Shigella, Enteroaggregative E. coli, Enteropathogenic E. coli, Enterotoxigenic E. coli lt/st, Shiga-like toxin producing E. coli stx1/stx2, E. coli O157and Shigella/Enteroinvasive E. coli, Cryptosporidium, Cyclospora cayetanensis, Entamoeba histolytica and Giardia lamblia |
| GPP | Adenovirus type 40/41, Norovirus, Rotavirus | Campylobacter, Clostridium difficile Toxin A/B, Escherichia coli O157, Enterotoxigenic E.coli (ETEC) LT/ST, Shiga-like Toxin producing E.coli (STEC) stx1/stx2, Salmonella, Shigella, Vibrio cholerae, Cryptosporidium, Entamoeba histolytica and Giardia |
| TAC system | Adenovirus type 40/41, Astrovirus, Norovirus GI, Norovirus GII, Rotavirus, Rotavirus G1, G3, G8, G9, G10, G11 and P[4], P[8], P[9], P[11], Sapovirus | Campylobacter jejuni/C. coli, Campylobacter spp, Clostridium difficile, Bacteroides fragilis, Salmonella, Vibrio cholerae, diarrheagenic Escherichia coli strains including enteroaggregative E. coli [EAEC], enterotoxigenic E. coli [ETEC], enteropathogenic E. coli [EPEC], and Shiga-toxigenic E. coli [STEC]), Shigella/enteroinvasive E. coli (EIEC), Aeromonas, H. pylori, M. tuberculosis, Cryptosporidium, Giardia lamblia, Entamoeba histolytica, Ascaris lumbricoides, Strongyloides stercoralis, Trichuris trichiura, Ancyclostoma/Necator, Cyclospora/Isospora), E. bieneusi/intestinalis, and Enterovirus |
Testing stool samples on the FilmArray was performed according to the manufacturer’s instructions [10] which requires approximately 5 min of hands-on time and results are available in 1 h (Table 3). Briefly, 200 μL of the stool mixed in sample buffer (provided by the manufacturer) was added to the sample injection port. The FilmArray pouch was rehydrated with hydration solution and then inserted into the instrument (Biofire Diagnostics, Salt Lake City, UT, USA). Each pouch contained an internal nucleic acid extraction control and a PCR control. Runs where the internal controls failed were considered invalid for all panel analytes and those specimens were retested.
Table 3.
Features of three enteric pathogens multiplex platforms.
| Parameters | FilmArray | Luminex GPP (IVD)* | TAC system | Reference testing (real-time PCR+ sequencing |
|---|---|---|---|---|
| Number of targets | 22 | 15 | 36 (Max. of 40) | 1 (max 2) |
| Hand-on processing time (minutes) | 5 | 60 | 60 | 60 |
| Total turn-around time (hours) | 1 | 7 | 4 | 48 |
| No. of clinical specimens per assay | 1 | 96 | 6 | 96 |
| Type of system | closed | open | Open extraction, closed system thereafter | open |
| Technology | Nested PCR and melting curve | Fluorescent bead-based detection | fluorescent (TaqMan) detection | fluorescent (TaqMan) detection |
IVD = in vitrodiagnostic device.
Testing on Luminex GPP was performed in accordance with the manufacturer’s instructions [10] and takes approximately 60 min of hands-on time and results are available within 6 h (Table 3). Briefly, 100 μL of 10% clarified stool suspension prepared in phosphate buffered saline was combined with 10 μL xTAG® MS2 and pre-treated by vortexing in a Bertin SK38 bead tube containing 1 mL bioMérieux® NucliSENS® easyMAG® Lysis Buffer. Nucleic acid was extracted by using the NucliSENS easyMAG system (BioMerieux, NC, USA). PCR reactions and subsequent hybridization step were performed according to the manufacturer’s instructions. The data were acquired on the Luminex 200 analyzer, and data analysis was carried out using TDAS data analysis software.
The TAC system can process and analyze up to six samples at a time for up to 40 pathogens tested; and requires approximately 60 min of hand-on time and 3 h to obtain results (Table 3). Nucleic acid was extracted from 200 mg of stool lysed with QiaAmp ASL buffer and homogenized using a bead beater [11] followed by QiaAmp nucleic acid extraction protocol (Qiagen, Valencia, CA). TAC assays were designed as described previously [11] and purchased from Eric R. Houpt (University of Virginia). Ag-Path-ID one step RT-PCR kit (Applied Bio-systems, Carlsbad, CA, USA) was used and the reaction mixture was loaded into each port of the card after which the card was briefly centrifuged twice at 1200 rpm for 1 min. The card was then sealed, the loading ports excised, and run on a ViiA7 real-time PCR (Applied Bio-systems-Thermo Fisher). The reaction conditions consisted of reverse transcription at 45°C for 20 min followed by 10 min at 95°C to activate the Taq polymerase, 45 PCR cycles at 95°C for 15 s and 60°C for 1 min.
Samples were considered true positive for a virus when they tested positive by both reference testing methods (i.e. (RT-)qPCR and sequencing) and samples negative in both methods were considered true negative. We used a cycle threshold (Ct) values of 40 as cut-off for the TaqMan real-time assays. The percent clinical accuracy, sensitivity and specificity for each of the five viruses for each platform were calculated as follows:
% clinical accuracy: 100 × (true positives + true negatives)/(true positives + true negatives + false positive + false negative).
% sensitivity: 100 × (true positives/(true positives + false negatives))
% specificity: 100 × (true negatives/(true negatives + false positives))
4. Results
Among the 207 true virus-positive samples (187 samples positive for a single virus and 20 samples positive for more than one virus), the FilmArray detected 199 (96.1%) samples and the TAC system detected 172 (83.1%) samples (Table 4). The GPP, which is only able to detect rotavirus, adenovirus type 40/41 and norovirus, detected 100 (78.7%) of the 127 true virus-positive samples positive for one or more of the three gastroenteritis viruses. No correlation was found between number of samples missed by each platform and virus genotypes. The overall clinical accuracy for the five viruses was 98% for the FilmArray and 97.2% for the TAC system; for the three target viruses tested by GPP, overall clinical accuracy was 96.9% (Table 3). The sensitivities for rotavirus were 100%, 95.8% and 89.6% for FilmArray, GPP, and TAC system, respectively (Table 4). The sensitivities for norovirus GI and GII were 87.8%, 78.0%, and 87.8% and for adenovirus type 40/41 were 97.4%, 57.9% and 68.4%, respectively (Table 4). The sensitivities of FilmArray and TAC for sapovirus and astrovirus were similar to those for norovirus and rotavirus (Table 4). The specificity ranged from 99.6 to 95.6% (highest for norovirus/sapovirus and lowest for rotavirus) for FilmArray, 100%-99.6% (highest for rotavirus/adenovirus type 40/41 and lowest for norovirus) for GPP and 100%-98.9% (highest for rotavirus/sapovirus and lowest for astrovirus) for the TAC system (Table 4). Among the three platforms, the FilmArray detected most (59/64 (92.10%)) of the samples with a low virus titer Ct value: > 30 followed by the TAC system (35/64 (54.6%)) and the GPP (14/37 (37.8%)) (Fig. 1).
Table 4.
Performance characteristics of the FilmArray, GPP and TAC system compared with reference test methods for five established gastroenteritis viruses.
| (a)
| |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Target Virus | Reference testing methods
|
FilmArray Results
|
FilmArray performance
|
||||||
| True Positive samples (single infections + mixed infections) |
True Negative samples |
True Positive samples (single infections + mixed infections) |
True Negative samples |
False Positive samples |
False Negative samples |
Percent clinical accuracy |
Percent Sensitivity |
Percent Specificity |
|
| Adenovirus 40/41 | 38 (27 + 11) | 262 | 37 (27 + 10) | 256 | 6 | 1 | 97.7 | 97.4 | 97.7 |
| Astrovirus | 39 (29 + 10) | 261 | 38 (28 + 10) | 258 | 3 | 1 | 98.7 | 97.4 | 98.9 |
| Norovirus | 41 (33 + 8) | 259 | 36 (28 + 8) | 258 | 1 | 5 | 98.0 | 87.8 | 99.6 |
| Rotavirus | 48 (43 + 5) | 252 | 48 (43 + 5) | 241 | 11 | 0 | 96.3 | 100.0 | 95.6 |
| Sapovirus | 41 (34 + 7) | 259 | 40 (33 + 7) | 258 | 1 | 1 | 99.3 | 97.6 | 99.6 |
|
| |||||||||
| (b)
| |||||||||
| Target Virus | Reference testing methods
|
GPP Results
|
GPP Performance
|
||||||
| True Positive samples (single infections + mixed infections) |
True Negative samples |
True Positive samples (single infections + mixed infections) |
True Negative samples |
False Positive samples |
False Negative samples |
Percent clinical accuracy |
Percent Sensitivity |
Percent Specificity |
|
|
| |||||||||
| Adenovirus 40/41 | 38 (27 + 11) | 262 | 22 (18 + 4) | 262 | 0 | 16 | 94.7 | 57.9 | 100.0 |
| Norovirus | 41 (33 + 8) | 259 | 32 (25 + 7) | 258 | 1 | 9 | 96.7 | 78.0 | 99.6 |
| Rotavirus | 48 (43 + 5) | 252 | 46 (41 + 5) | 252 | 0 | 2 | 99.3 | 95.8 | 100.0 |
|
| |||||||||
| (c)
| |||||||||
| Target Virus | Reference testing methods
|
TAC system Results
|
TAC system Performance
|
||||||
| True Positive samples (single infections + mixed infections) |
True Negative samples |
True Positive samples (single infections + mixed infections) |
True Negative samples |
False Positive samples |
False Negative samples |
Percent clinical accuracy |
Percent Sensitivity |
Percent Specificity |
|
|
| |||||||||
| Adenovirus 40/41 | 38 (27 + 11) | 262 | 26 (20 + 6) | 260 | 2 | 12 | 95.3 | 68.4 | 99.2 |
| Astrovirus | 39 (29 + 10) | 261 | 36 (26 + 10) | 258 | 3 | 3 | 98.0 | 92.3 | 98.9 |
| Norovirus | 41 (33 + 8) | 259 | 36 (28 + 8) | 257 | 2 | 5 | 97.7 | 87.8 | 99.2 |
| Rotavirus | 48 (43 + 5) | 252 | 43 (39 + 4) | 252 | 0 | 5 | 98.3 | 89.6 | 100.0 |
| Sapovirus | 41 (34 + 7) | 259 | 31 (28 + 3) | 259 | 0 | 10 | 69.7 | 75.6 | 100.0 |
Fig. 1.
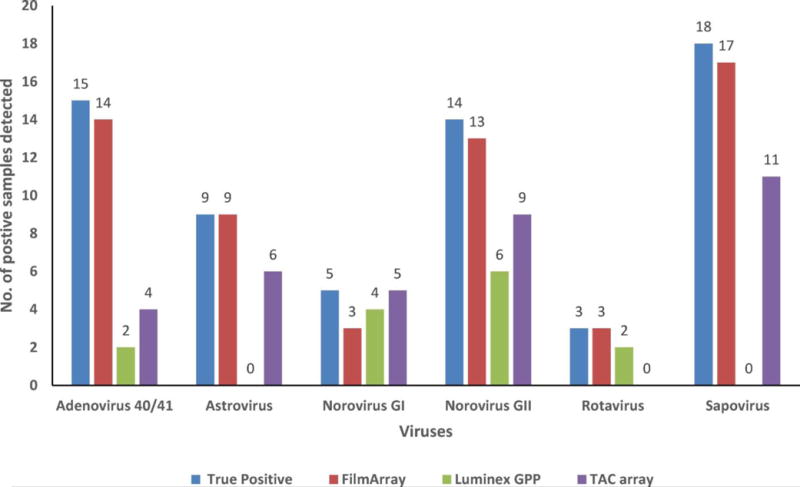
Detection of virus-positive samples with low copy number (Ct value > 30) by the FilmArray, GPP and TAC system.
Note: Samples were considered true positive for a virus when they tested positive by TaqMan realtime (RT-) PCR and conventional (RT-)PCR followed by Sanger sequencing reference methods.
Twenty (9.6%) of 207 true positive samples were co-infected with more than one gastroenteritis virus, results that were confirmed by reference testing (Table 5). Overall among true positive samples, adenovirus type 40/41 positive samples had the highest co-infection rate (11/38 (28.9)%) followed by astrovirus (10/39 (25.6%)), norovirus (8/41 (19.5%)), sapovirus (7/41 (17.0%)) and rotavirus (5/48 (10.4%)) (Table 5). The FilmArray was able to detect 19 (95%) of the 20 co-infections in true positive samples whereas the TAC system detected 10 (50%) of the 20 co-infections and GPP detected two (28.5%) of the seven co-infections (Table 5).
Table 5.
Viral co-infections detected in 20 stool samples by FilmArray, Luminex GPP and TAC platforms.
| True co-infections (number of samples) | Co-infection detected by FilmArray (number of samples) | Co-infection detected by Luminex GPPa (number of samples) | Co-infection detected by TAC Systems (number of samples) |
|---|---|---|---|
| Adenovirus + Astrovirus (3) | Adenovirus (3); Astrovirus (3) | Adenovirus (1); Astrovirus (n/a)
|
Adenovirus (1); Astrovirus (3) |
| Adenovirus + Norovirus (4) | Adenovirus (3); Norovirus (4) | Adenovirus (1); Norovirus (4) | Adenovirus (2); Norovirus (4) |
| Adenovirus + Rotavirus (2) | Adenovirus (2); Rotavirus (2) | Adenovirus (1); Rotavirus (2) | Adenovirus (1); Rotavirus (2) |
| Adenovirus + Sapovirus (1) | Adenovirus (1); Sapovirus (1) | Adenovirus (1); Sapovirus (n/a)) | Adenovirus (1); Sapovirus (1) |
| Adenovirus + Norovirus + Astrovirus (1) | Adenovirus (1); Norovirus (1); Astrovirus (1) | Adenovirus (0); Norovirus (0); Astrovirus (n/a) | Adenovirus (1); Norovirus (1); Astrovirus (1) |
| Astrovirus + Norovirus (1) | Astrovirus (1); Norovirus (1) | Astrovirus (N/A); Norovirus (1) | Astrovirus (1); Norovirus (1) |
| Astrovirus + Rotavirus (2) | Astrovirus (2); Rotavirus (2) | Astrovirus (N/A); Rotavirus (2) | Astrovirus (2); Rotavirus (1) |
| Astrovirus + Sapovirus (3) | Astrovirus (3); Sapovirus (3) | Astrovirus (n/a); Sapovirus (n/a) | Astrovirus (3); Sapovirus (2) |
| Norovirus + Sapovirus (2) | Norovirus (2); Sapovirus (2) | Norovirus (2); Sapovirus (n/a) | Norovirus (2); Sapovirus (0) |
| Rotavirus + Sapovirus (1) | Rotavirus (1); Sapovirus (1) | Rotavirus (1); Sapovirus (n/a) | Rotavirus (1); Sapovirus (0) |
= GPP platform only detects adenovirus 40/41, norovirus and rotavirus.
5. Discussion
We compared the performance characteristics of three multi-enteric diagnostic molecular platforms for the detection of five gastroenteritis viruses to the reference methods for each individual virus using a coded panel of 300 stool specimens. Overall, the FilmArray had the highest clinical accuracy rate followed by the TAC system. The FilmArray was able to detect most samples with high Ct values for all five viruses.
Most of the previous studies that compared the performance of one or more of these platforms were focused on the detection of bacterial enteric pathogens [23–27]. Given that acute gastroenteritis caused by the five established viruses accounts for nearly 60% of the medically-attended childhood cases in the United States [3], we focused primarily on the evaluation of the performance of the detection of viral gastroenteritis pathogens in these multiplex platforms. In contrast to most of the previous studies that used only one confirmation method to confirm true viral positive samples [23,25,2,27], we used both RT-qPCR as well as conventional RT-PCR followed by bidirectional sequencing as reference methods which helped us to address discrepancies among the platforms and more accurately assess the individual performance characteristics.
Although the overall performance for the three platforms was > 90% in terms of sensitivity and specificity, the sensitivity for certain viral targets was relatively low. For example, the sensitivity for adenovirus type 40/41 was 57% and for norovirus was 78% on the GPP whereas on the TAC system the sensitivity for adenovirus type 40/41 was 68.4% and for sapovirus was 75.6%. The low sensitivity for each of these viruses may be directly linked to the difficulty of each platform to detect low viral load samples and/or related to mismatches with the viral templates and the oligonucleotide primers and probes used in these systems. The GPP and TAC systems were able to detect 37.8% and 54.6% of the samples with low viral load whereas the FilmArray performed best for the detection of these samples except for noroviruses (84.2%). The high sensitivity for the detection of noroviruses on the GPP compared to the FilmArray (100% vs 91.7%) that was recently reported [23], could not be confirmed in our study (78% vs 87.8%).
The FilmArray demonstrated the highest number of false positive samples including eleven rotavirus and six adenovirus type 40/41 samples. High rates of false positivity for norovirus detection using this platform was recently reported by others [2]. Although the FilmArray is a closed system and the chances of cross contamination are minimum, we retested all rotavirus positive samples on the FilmArray and confirmed previous results. The number of false positive samples reduced the percent specificity of the FilmArray for rotavirus (95.6%) and adenovirus 40/41 (97.7%) as compared to what has been reported in the other comparative studies (> 99%) [23,2]. Because true positives were defined by the reference methods, we cannot rule out the possibility that these methods for may have missed the detection of rotavirus or adenovirus 40/41 in these samples. Another explanation could be that the FilmArray is more sensitive than the reference methods.
The most advanced feature of multiplex molecular diagnostic platforms is the simultaneous detection of more than one gastroenteritis pathogen in a single reaction. Co-infections with more than one virus were identified in 9.6% of the true positive samples with adenovirus 40/41 as the most frequently detected virus and co-infections with non-viral pathogens have been reported previously [23,11]. However, in this study we only focused on the performance of detection of gastroenteritis viruses and did not evaluate the detection of the bacterial and parasitic enteric pathogens.
Aspects such as workflow, turnaround time, cost and number of samples to test per run may impact the choice of platform which may be different for a clinical laboratory, large hospital laboratory, or reference laboratory. Since the turnaround time of a single sample on a Biofire is about 90 min which includes nucleic acid extraction which is incorporated in the completely automated system, this platform may be highly attractive for laboratories that receive fewer than 8 samples per 8 h work shift. To increase testing capacity in a regular shift, more than one instrument or round the clock testing is needed. The GPP and TAC systems involve a unidirectional work flow which requires a higher level of training for staff to avoid amplicon contamination and both platforms require nucleic acid extraction prior to testing [23]. For a high throughput of samples in a single run, GPP has the capacity to process 96 samples in approximately 7 h. The TAC system is not FDA approved but can process 6 samples in a single run in 4 h. The large number of targets simultaneously tested, however, could make the GPP and TAC system better suited for outbreak investigations in which the etiologic agents are unknown.
A limitation of this study was that we did not evaluate the detection of the bacterial and parasitic enteric pathogens and therefore it was difficult to assess the true co-infections in the panel. Additionally, the clinical meaning of samples with a high Ct is uncertain as viruses with low copy number could be passing through the gut, be the result of an earlier infection that resulted in long-term shedding or not be associated with clinical symptoms [28].
In summary, each multi-enteric molecular testing platform has strengths and weaknesses in detecting the five most important gastroenteritis viruses. The fact that to date more than 15% of medical facilities and diagnostic laboratories are implementing multiplex enteric pathogens platforms [29,30] requires careful evaluation of the performance characteristics and limitations of each platform as well as the interpretation of the results (e.g., co-infections). Therefore, our data may help in choosing the appropriate platform for laboratories to determine the etiology of the most important causes of epidemic and endemic acute viral gastroenteritis and thus aid in making informed decisions for their control and prevention.
Acknowledgments
We thank Eric R Houpt for performing QA/QC of TAC cards and Lynne Shelley, Jennifer Carnahan, Jenelle Putzig, and Ken Schnabel for excellent technical assistance.
Funding
This study was supported by a grant from the intramural food safety program of CDC.
Abbreviations
- GPP
Gastrointestinal Pathogen Panel
- TAC
TaqMan Array Card
Footnotes
Disclaimer
The findings and conclusions in this article are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention (CDC). Names of specific vendors, manufacturers, or products are included for public health and informational purposes; inclusion does not imply endorsement of the vendors, manufacturers, or products by the CDC or the US Department of Health and Human Services.
Conflict of interest
James Chappell has served as a consultant to Luminex Molecular Diagnostics on the xTAG Gastrointestinal Pathogen Panel (GPP) and received research support from Luminex to perform GPP analysis of specimens collected through the CDC New Vaccine Surveillance Network. Additionally, he collaborates with Luminex evaluating new technology for the diagnosis of C. difficile infection. Rangaraj Selvarangan serves on the scientific advisory board for BioFire Diagnostics and has received research funding from BioFire Diagnostics and Luminex Corporation for other unrelated studies.
Competing interests
None declared.
Ethical approval
Institutional review approval for the parent NVSN protocols was granted by CDC and each participating site’s Institutional Review Boards.
References
- 1.Farthing M, Salam MA, Lindberg G, Dite P, Khalif I, Salazar-Lindo E, et al. Acute diarrhea in adults and children: a global perspective. J Clin Gastroenterol. 2013;47:12–20. doi: 10.1097/MCG.0b013e31826df662. [DOI] [PubMed] [Google Scholar]
- 2.Buss SN, Leber A, Chapin K, Fey PD, Bankowski MJ, Jones MK, et al. Multicenter evaluation of the BioFire FilmArray gastrointestinal panel for etiologic diagnosis of infectious gastroenteritis. J Clin Microbiol. 2015;53:915–925. doi: 10.1128/JCM.02674-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chhabra P, Payne DC, Szilagyi PG, Edwards KM, Staat MA, Shirley SH, et al. Etiology of viral gastroenteritis in children < 5 years of age in the United States, 2008–2009. J Infect Dis. 2013;208:790–800. doi: 10.1093/infdis/jit254. [DOI] [PubMed] [Google Scholar]
- 4.Staat MA, Payne DC, Donauer S, Weinberg GA, Edwards KM, Szilagyi PG, et al. Effectiveness of pentavalent rotavirus vaccine against severe disease. Pediatrics. 2011;128:e267–75. doi: 10.1542/peds.2010-3722. [DOI] [PubMed] [Google Scholar]
- 5.Wikswo ME, Desai R, Edwards KM, Staat MA, Szilagyi PG, Weinberg GA, et al. Clinical profile of children with norovirus disease in rotavirus vaccine era. Emerg Infect Dis. 2013;19:1691–1693. doi: 10.3201/eid1910.130448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Payne DC, Vinje J, Szilagyi PG, Edwards KM, Staat MA, Weinberg GA, et al. Norovirus and medically attended gastroenteritis in U.S. children. N Engl J Med. 2013;368:1121–1130. doi: 10.1056/NEJMsa1206589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tate JE, Cortese MM, Payne DC, Curns AT, Yen C, Esposito DH, et al. Uptake, impact, and effectiveness of rotavirus vaccination in the United States: review of the first 3 years of postlicensure data. Pediatr Infect Dis J. 2011;30:S56–60. doi: 10.1097/INF.0b013e3181fefdc0. [DOI] [PubMed] [Google Scholar]
- 8.Koo HL, Neill FH, Estes MK, Munoz FM, Cameron A, DuPont HL, et al. Noroviruses: the most common pediatric viral enteric pathogen at a large university hospital after introduction of rotavirus vaccination. J Pediatric Infect Dis Soc. 2013;2:57–60. doi: 10.1093/jpids/pis070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunbar SA. Molecular revolution entering GI diagnostic testing. Med Lab Obs. 2013;45:28. [PubMed] [Google Scholar]
- 10.Zhang H, Morrison S, Tang YW. Multiplex polymerase chain reaction tests for detection of pathogens associated with gastroenteritis. Clin Lab Med. 2015;35:461–486. doi: 10.1016/j.cll.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J, Gratz J, Amour C, Kibiki G, Becker S, Janaki L, et al. A laboratory-developed TaqMan Array Card for simultaneous detection of 19 enteropathogens. J Clin Microbiol. 2013;51:472–480. doi: 10.1128/JCM.02658-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mijatovic-Rustempasic S, Tam KI, Kerin TK, Lewis JM, Gautam R, Quaye O, et al. Sensitive and specific quantitative detection of rotavirus A by one-step real-time reverse transcription-PCR assay without antecedent double-stranded-RNA denaturation. J Clin Microbiol. 2013;51:3047–3054. doi: 10.1128/JCM.01192-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oka T, Yamamoto M, Miyashita K, Ogawa S, Katayama K, Wakita T, et al. Self-assembly of sapovirus recombinant virus-like particles from polyprotein in mammalian cells. Microbiol Immunol. 2009;53:49–52. doi: 10.1111/j.1348-0421.2008.00086.x. [DOI] [PubMed] [Google Scholar]
- 14.Okada M, Yamashita Y, Oseto M, Ogawa T, Kaiho I, Shinozaki K. Genetic variability in the sapovirus capsid protein. Virus Genes. 2006;33:157–161. doi: 10.1007/s11262-005-0051-7. [DOI] [PubMed] [Google Scholar]
- 15.Grant L, Vinje J, Parashar U, Watt J, Reid R, Weatherholtz R, et al. Epidemiologic and clinical features of other enteric viruses associated with acute gastroenteritis in american Indian infants. J Pediatr. 2012;161:110–115 e1. doi: 10.1016/j.jpeds.2011.12.046. [DOI] [PubMed] [Google Scholar]
- 16.Noel JS, Lee TW, Kurtz JB, Glass RI, Monroe SS. Typing of human astroviruses from clinical isolates by enzyme immunoassay and nucleotide sequencing. J Clin Microbiol. 1995;33:797–801. doi: 10.1128/jcm.33.4.797-801.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyman WH, Walsh JF, Kotch JB, Weber DJ, Gunn E, Vinje J. Prospective study of etiologic agents of acute gastroenteritis outbreaks in child care centers. J Pediatr. 2009;154:253–257. doi: 10.1016/j.jpeds.2008.07.057. [DOI] [PubMed] [Google Scholar]
- 18.Allard A, Albinsson B, Wadell G. Detection of adenoviruses in stools from healthy persons and patients with diarrhea by two-step polymerase chain reaction. J Med Virol. 1992;37:149–157. doi: 10.1002/jmv.1890370214. [DOI] [PubMed] [Google Scholar]
- 19.Park GW, Chhabra P, Vinje J. Swab sampling method for the detection of human norovirus on surfaces. J Vis Exp. 2017;120(6) doi: 10.3791/55205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cannon JL, Barclay L, Collins NR, Wikswo ME, Castro CJ, Magana LC, et al. Genetic and epidemiologic trends of norovirus outbreaks in the United States from 2013 to 2016 demonstrated emergence of novel GII.4 recombinant viruses. J Clin Microbiol. 2017;55:2208–2221. doi: 10.1128/JCM.00455-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vega E, Barclay L, Gregoricus N, Shirley SH, Lee D, Vinje J. Genotypic and epidemiologic trends of norovirus outbreaks in the United States, 2009–2013. J Clin Microbiol. 2014;52:147–155. doi: 10.1128/JCM.02680-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Payne DC, Staat MA, Edwards KM, Szilagyi PG, Weinberg GA, Hall CB, et al. Direct and indirect effects of rotavirus vaccination upon childhood hospitalizations in 3 US counties, 2006–2009. Clin Infect Dis. 2011;53:245–253. doi: 10.1093/cid/cir307. [DOI] [PubMed] [Google Scholar]
- 23.Khare R, Espy MJ, Cebelinski E, Boxrud D, Sloan LM, Cunningham SA, et al. Comparative evaluation of two commercial multiplex panels for detection of gastrointestinal pathogens by use of clinical stool specimens. J Clin Microbiol. 2014;52:3667–3673. doi: 10.1128/JCM.01637-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deng J, Luo X, Wang R, Jiang L, Ding X, Hao W, et al. A comparison of Luminex xTAG(R) Gastrointestinal Pathogen Panel (xTAG GPP) and routine tests for the detection of enteropathogens circulating in Southern China. Diagn Microbiol Infect Dis. 2015;83:325–330. doi: 10.1016/j.diagmicrobio.2015.07.024. [DOI] [PubMed] [Google Scholar]
- 25.Patel A, Navidad J, Bhattacharyya S. Site-specific clinical evaluation of the Luminex xTAG gastrointestinal pathogen panel for detection of infectious gastroenteritis in fecal specimens. J Clin Microbiol. 2014;52:3068–3071. doi: 10.1128/JCM.01393-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rand KH, Tremblay EE, Hoidal M, Fisher LB, Grau KR, Karst SM. Multiplex gastrointestinal pathogen panels: implications for infection control. Diagn Microbiol Infect Dis. 2015;82:154–157. doi: 10.1016/j.diagmicrobio.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 27.Murphy CN, Fowler RC, Iwen PC, Fey PD. Evaluation of the BioFire FilmArray (R) Gastrointestinal Panel in a midwestern academic hospital. Eur J Clin Microbiol Infect Dis. 2016;36(4):747–754. doi: 10.1007/s10096-016-2858-7. [DOI] [PubMed] [Google Scholar]
- 28.Tate JE, Mijatovic-Rustempasic S, Tam KI, Lyde FC, Payne DC, Szilagyi P, et al. Comparison of 2 assays for diagnosing rotavirus and evaluating vaccine effectiveness in children with gastroenteritis. Emerg Infect Dis. 2013;19:1245–1252. doi: 10.3201/eid1908.130461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hurd S, Patrick M, Hatch J, Clogher P, Wymore K, Cronquist AB, et al. Clinical laboratory practices for the isolation and identification of Campylobacter in Foodborne Diseases Active Surveillance Network (FoodNet) sites: baseline information for understanding changes in surveillance data. Clin Infect Dis. 2012;54(Suppl. 5):S440–5. doi: 10.1093/cid/cis245. [DOI] [PubMed] [Google Scholar]
- 30.Iwamoto M, Huang JY, Cronquist AB, Medus C, Hurd S, Zansky S, et al. Bacterial enteric infections detected by culture-independent diagnostic tests-FoodNet, United States, 2012–2014. Morb Mortal Wkly Rep. 2015;64:252–257. [PMC free article] [PubMed] [Google Scholar]


