Abstract
Skeletal myocytes have well-established fast and slow twitch fibers with unique gene and protein specific expression patterns. By immunohistochemical staining, these show a mosaic pattern across myocytes. We hypothesized cardiac myocytes may behave similarly where some proteins are differentially expressed between mature cardiomyocytes. We utilized the tool HPASubC on over 52,000 cardiac images of the Human Protein Atlas to identify differential protein expression patterns by immunohistochemistry across the cardiomyocytes. We matched identified proteins to open chromatin and gene expression data. We identified 143 putative proteins with mosaic patterns of expression across the cardiomyocytes. We validated four of these proteins (MYL3, MYL4, PAM, and MYOM1) and demonstrated unique atrial or ventricular patterns of expression for each. Acetylation of histone H3K27 at the promoters of these four genes were consistent with the atrial/ventricular expression patterns. Despite the generally accepted homogeneity of cardiomyocytes, a small subset of proteins varies between cardiomyocytes in a mosaic pattern. This fundamental process has been previously uncharacterized. These changes may inform on different functional and disease-related activities of proteins in individual cardiomyocytes.
Keywords: myosin light chain, proteomics, myomesin, cardiomyocyte, expression
1. Introduction
It is well-established that skeletal muscle cells can be simplistically divided into slow-twitch (type I) and fast-twitch (type II) fibers [1]. These fibers each have unique and essential activities that are mediated by different patterns of protein expression. Differentially expressed proteins in these fibers include myosin heavy chains (MYH1, MYH2), tropomyosins (TPM1, TPM2), troponins (TNNI1, TNNI2) and others [2]. Individual skeletal myocyte protein expression differences have been noted by ATPase activity, immunohistochemistry (IHC) and immunofluorescence (IF) studies. Depending on the function of a skeletal muscle, the ratio of the fibers will vary [2]. These same proteins are not known to be differentially expressed in human cardiomyocytes, therefore there has been little effort focused on identifying proteins that might vary between cells in the heart. However, there have been some hints in the literature that cardiac myocytes may also vary in protein expression.
In 1984, the Yazaki group reported isozymic changes of human atrial myosin (referred to as HCα, but currently named MYL4) where they showed IF differences across ventricular myocytes [3]. More recently, the work of the Baker laboratory has demonstrated cellular heterogeneity of responses to α1 adrenergic stimulation in isolated myocytes [4]. The Wang laboratory showed that cardiac-specific deletion of Trbp in a mouse model changed the expression of fast and slow-twitch myofiber genes including Tnni2, Tpm2, Myl1, Myl7, Myl9, Tnnc1, Myh7b and Myl3 [5]. The summation of these papers suggests there might be a collection of proteins that differ between cardiac myocytes. However, there has not yet been a way to globally and systematically investigate thousands of proteins for these alternative patterns of expression in cardiac myocytes.
The Human Protein Atlas (HPA) is a web resource of proteomic expression that can be used to investigate this question. The HPA has generated scores of tissue microarrays (TMAs) using 44 normal and 20 cancer tissues. These TMAs have been stained using IHC for >17,000 proteins using >25,000 antibodies, with each antibody generally staining 3 cores of tissue per organ type. This has resulted in ~50,000 heart images (predominately from ventricles) that can be evaluated for any pattern of staining [6].
To investigate interesting staining patterns of HPA tissues as a means to identify similarly expressed proteins, we developed HPASubC [7]. This is a suite of python tools that allows a user to download all of the images of a particular organ, then sequentially review them for a pattern of interest using a PlayStation-style gamepad controller. Images can be rapidly reviewed at ~1/second in an anonymous fashion. We have demonstrated its usefulness in describing the patterns of non-cardiomyocyte expression in the heart and in sinusoidal expression in the liver [7, 8].
We became interested in the concept of cardiac myocyte mosaicism while reviewing HPA heart images for a separate analysis and began noticing occasional images where only a subset of myocytes were stained. We then hypothesized that, like skeletal muscle, there may be different fiber types of the heart denoted by alternative protein expression.
2. Methods
2.1 The Human Protein Atlas and HPASubC
We downloaded 52,737 heart images representing 12,814 proteins from HPA (v16) on 12/9/2016 utilizing the HPASubC script download_images_from_gene_list.py. We then used the HPASubC image_viewer.py to rapidly cycle through the images at ~3,000 per viewing. We captured any image in which the staining intensity varied across the myocytes reasoning it could be the result of a mosaic pattern of protein expression. Staining of other cell types was not interpreted. We then re-evaluated all of the images that corresponded to any protein identified and rescored them as mosaic in a second, stricter screening in which the mosaicism between myocytes was more clear. Although multiple images existed for each protein, the presence of a mosaic pattern on only a single core image was sufficient to call the protein mosaic and be included in this study. Additional scripts were used to download RNA-seq expression data and other protein information from HPA. Skeletal muscle images corresponding to the identified mosaic heart proteins were downloaded along with images corresponding to 1000 random proteins (based on ENSG ID numbers) not identified as mosaic in the heart materials.
2.2 Immunohistochemistry
We obtained multiple human cardiac tissues under an approved Investigational Review Board protocol. This included multiple regions of a human heart explanted for cardiac transplantation and pediatric tissues. Slides were obtained from these formalin-fixed paraffin-embedded blocks and underwent IHC. Formalin-fixed paraffin embedded tissues were sectioned onto plus slides. Paraffin removal and high- temperature antigen retrieval was performed by immersing the slides in Trilogy (Sigma-Aldrich, St. Louis, MO) in a pressure cooker to 126°C and 18-23 psi. Endogenous peroxidase was blocked by incubating the slides in a dual enzyme block (Dako North America Carpinteria, CA). Slides were incubated with primary antibodies to either Myomesin 1 (MYOM1; 1:200); Myosin, light chain 3 (MYL3; 1:500); Myosin, light chain 4 (MYL4; 1:500); or Peptidylglycine alpha-amidating monooxygenase (PAM; 1:500) (Atlas Antibodies, Stockholm, Sweden) followed by an incubation with a polymer HRP IgG (Leica Biosystems, Pleasanton, CA). The antibody complex was detected with ImmPact DAB (Vector Laboratories, Burlingame, CA) and the slides were counterstained with Hematoxylin (Richard-Allen Scientific, Kalamazoo, MI).
2.3 String Analysis, Gene Ontology, and Mitochondrial Localization
The identified mosaic proteins were analyzed using String 10.0 [9] for interactions using the web-based portal at https://string-db.org/. Gene Ontology, a method to find enrichment of functional relationships within gene lists, was ascertained using the Gene Ontology Consortium web-based portal at http://geneontology.org/ [10]. All proteins listed in the Human Mitochondrial Protein Database (http://bioinfo.nist.gov/hmpd/index.html) were obtained and a final list of 813 unique protein names was generated.
2.4 Histone modification ChIP-seq analysis
We downloaded mapped reads, fold change over control, and stable peaks for H3K27ac ChIP-seq experiments for eight heart tissues from the ENCODE Project Consortium [11] website (www.encodeproject.org) (Table 1). For each of the 143 genes that showed mosaic expression patterns, we calculated normalized ChIP-seq read ratio (Ri) in the promoter regions (5,000 basepair of DNA centered at transcription start sites) for each sample using the following formula: Ri = (hi / H) / (ci / C), where hi and ci are the read counts from histone and control ChIP-seq for gene i, and T and C are the total read counts from histone and control ChIP-seq, respectively. We used bedtools (v2.24.0) ‘multicov’ command [12] for counting reads from the bam files. These values were then log2 transformed and averaged for each tissue type (atria vs ventricles). The log ratio between atria and ventricles were calculated lastly. For the gene expression data, GTEx V7 median gene transcripts per million (TPM) values [13] for the heart tissues (atria and ventricles) were used. Median TPMs were further log2 transformed with 0.001 pseudo-count and quantile normalized. We developed in-house scripts for the analysis, which are available upon request. We visualized enhancer and promoter activities around the four genes (MYL4, MYL3, MYOM1, and PAM) using the ChIP-seq signal data (fold change over control) with the Integrative Genomics Viewer (IGV) [14]. Hg19 was used as a reference genome for the analysis. The range of signals for visualization was set between 0 and 30 for all tracks.
Table 1.
H3K27ac ChIP-seq ENCODE data sets for human heart tissues.
| File accession for histon ChIP-seq | File accession for control ChIP-seq | File accession for ChIP-seq signal ratio | File accession for stable peaks | Sample | Gender | Age |
|---|---|---|---|---|---|---|
| ENCFF757FUA | ENCFF783FIH | ENCFF834AET | ENCFF145YCQ | Right ventricle | Male | 34 |
| ENCFF629PMN | ENCFF042HFM | ENCFF951PNB | ENCFF749PTG | Right ventricle | Male | 3 |
| ENCFF024NTE | ENCFF925ZDQ | ENCFF030QXL | ENCFF593HJE | Left ventricle | Male | 34 |
| ENCFF936OFA | ENCFF925ZDQ | ENCFF338CIS | ENCFF942HEX | Left ventricle | Male | 32 |
| ENCFF164FND | ENCFF934RGM | ENCFF890ARB | ENCFF447VLE | Right atrium auricular region | Female | 53 |
| ENCFF072ZDB | ENCFF835OKR | ENCFF630UXZ | ENCFF796AES | Left ventricle | Male | 3 |
| ENCFF995OXQ | ENCFF305OFX | ENCFF006WQL | ENCFF787ATT | Left ventricle | Female | 53 |
| ENCFF287ZVV | ENCFF896AXY | ENCFF456MLR | ENCFF689MQO | Right atrium | Male | 34 |
3. Results
3.1 One hundred and forty-three proteins have possible mosaic patterns of expression
We viewed 52,737 heart images covering 12,814 proteins from the HPA using HPASubC. From these we identified 319 possible mosaic proteins in an initial screen. A second, more stringent screening of the images reduced this number to 152 proteins. We then obtained matched transcripts per million (TPM) RNA-seq data from HPA and removed any protein for which the TPM value was 0. This left 143 putative mosaic proteins (1.1% of all proteins reviewed) (Table 2, Supplemental Table 1). For some proteins (such as FAM3C), the pattern was clear, while for other proteins, the differences were subtler and not always consistent across cores (Fig. 1, Supplemental Fig. 1). Even within a given myocyte, the staining could vary across the cell. The identified proteins included well-known cardiac proteins such as the myosin light chains (MYL3 & MYL4) as well as lesser explored proteins such as zinc finger CCHC-type containing 10 (ZCCHC10) and U-box domain containing 5 (UBOX5). The genes for seven of these proteins reside on the X chromosome. In comparison to the general mitochondrial proteome (813 proteins), three possible mosaic proteins were on that list (CASQ1, PDP1, and RTN4).
Table 2.
143 proteins with putative mosaic patterns of expression. The genes of the bolded proteins are located on the X chromosome.
| ABCC8 | CMTR1 | GZF1 | PDILT | TRAM2 |
|---|---|---|---|---|
| ACTA1 | COMMD3- BMI1 | HCN3 | PDLIM5 | TRAPPC4 |
| ACTN2 | CSNK1E | HIGD2A | PDP1* | TRAPPC6B |
| ACVR1 | CTD-2207O23.3 | HSPB6 | PFKM | TRIM14 |
| ADCY6 | CUTA | HTR2B | PKP1 | TRMT2B |
| ADD3 | DACT1 | IFNAR1 | PLBD1 | TRMU |
| ANKRD11 | DDX55 | IRAK4 | PLCG2 | TTN |
| ANKRD2 | DGKH | IRF1 | PPM1F | TXLNB |
| ARHGAP32 | DHRS7B | ITFG1 | PRKG2 | TXNL1 |
| ARMCX4 | DNAJB4 | KATNB1 | PRPF19 | UBAP2 |
| ASB7 | DNAJC19 | KHDRBS3 | PTPRA | UBOX5 |
| ATIC | EFNA4 | LETM1 | PTPRE | UCHL3 |
| ATMIN | ELAC1 | MASTL | RBM25 | UFSP1 |
| ATP13A3 | EPB41L5 | MID1 | RTN4* | UIMC1 |
| ATP5J2-PTCD1 | EPHB3 | MPHOSPH6 | SCAF4 | UNK |
| BBIP1 | FAM188B | MYL3 | SERTAD2 | USP4 |
| BMPR1A | FAM3C | MYL4 | SLC16A10 | WBP5 |
| C1orf116 | FBXW9 | MYL6 | SNAP91 | WDR27 |
| C22orf29 | FHL2 | MYOM1 | SRGAP2 | ZCCHC10 |
| C3orf52 | FITM1 | NBR1 | TAGAP | ZCCHC5 |
| CAMSAP2 | FRAS1 | NCAM1 | TANC2 | ZDHHC9 |
| CAPRIN2 | GABARAPL1 | NEBL | TCP1 | ZKSCAN8 |
| CASQ1* | GALT | NEU3 | TFB2M | ZNF235 |
| CCNDBP1 | GCLM | NFE2 | TMEM173 | ZNF408 |
| CD300C | GFM1 | OBSCN | TNFSF13 | ZNF428 |
| CD53 | GID8 | ODC1 | TPK1 | ZNF81 |
| CERCAM | GSK3A | OSBPL9 | TPM1 | ZSCAN31 |
| CGN | GSTZ1 | PAM | TRAF2 | |
| CHPF2 | GUF1 | PCDHB5 | TRAK2 |
indicates proteins that localize to the mitochondria.
Fig. 1.
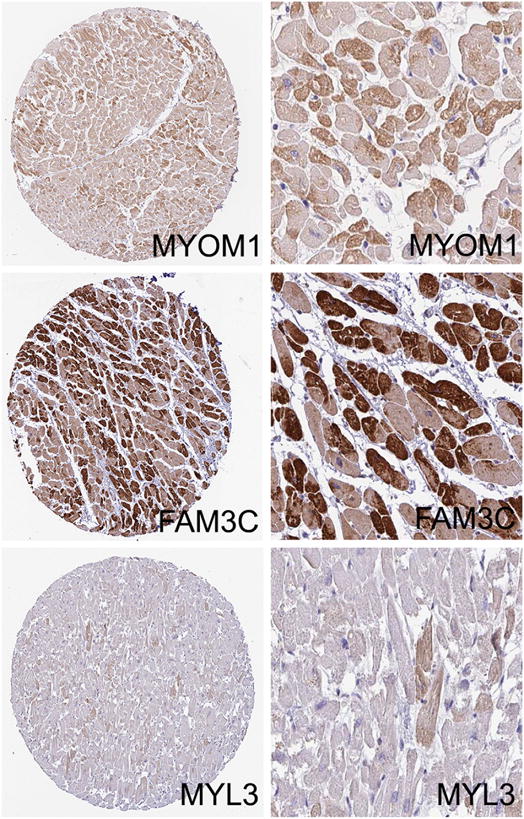
Representative images of mosaic patterns of protein expression as detected by HPASubC. The staining patterns of three proteins MYOM1, FAM3C and MYL3 are shown as the entire TMA core and as a higher power view. Mosaic staining could be subtle as seen in MYL3. All images are from the Human Protein Atlas [19].
3.2 Roughly one-quarter of heart mosaic proteins are also mosaic in skeletal muscle
We then inquired if there was a relationship between cardiac mosaic patterns and skeletal mosaic patterns. To do this, we reviewed all skeletal muscle images at HPA that corresponded to the 143 putative mosaic proteins. Of these, 39 proteins (27%) were mosaic in both cardiac and skeletal muscle (Supplemental Fig. 2). As a comparison group, we evaluated 1,000 random proteins in the skeletal muscle images and found 758 to have some degree of staining. Of these, 216 (28.5%) were mosaic. Thus, there was no enrichment in skeletal muscle.
3.3 Mosaic patterns differ by protein across atria and ventricles
We selected four of the putative mosaic proteins for further review (MYL4, MYL3, MYOM1, PAM). Three of these (MYL4, MYL3, and MYOM1) were abundantly expressed in the heart (>260 TPM) and had mosaic patterns of expression in skeletal muscle. PAM was also abundant (1092 TPM), but was not mosaic in skeletal muscles. We evaluated the atria, ventricles, and septum for evidence of staining mosaicism and interestingly found different patterns for each protein.
MYL3, known as a ventricular light chain myosin with increased expression in slow-twitch skeletal muscle fibers, was robustly and homogeneously present in ventricular cardiomyocytes (Fig. 2A). In the atria, it was present in ~85% of myocytes with some regional variation (Fig. 2B and Supplemental Fig. 3). In contrast to MYL3, MYL4, known as an atrial light chain myosin and an embryonic light chain myosin, was present in a mosaic fashion in ~15% of the ventricular myocytes (Fig. 2C). Curiously, essentially all myocytes in the first layer of the subendocardium were MYL4 positive and this high percent of MYL4 positivity extended through the first few layers of myocytes (Fig. 2D and Supplemental Fig. 4). In the atrium, staining was homogenously positive in all cardiac myocytes (Fig. 2E). MYOM1 was barely mosaic in the ventricles (Fig. 2F) and more strikingly mosaic in the atria with all cells appearing to express some MYOM1 but with ~30% having stronger staining (Fig. 2G and Supplemental Fig. 5). PAM was mosaic in the ventricles with >5% of cells having positive staining (Fig. 2H). In the atria there was a general granular staining pattern that was only faintly variable between cells (Fig. 2I and Supplemental Fig. 6).
Fig. 2.

IHC of MYL3, MYL4, MYOM1, and PAM in the left ventricle and left atrium. A) Ventricular staining of MYL3 showing homogeneous staining. B) Atrial staining of MYL3 showing mosaic staining with ~85% of cells being positive. C) Ventricular staining of MYL4 showing ~15% staining. D) MYL4 staining of the subendocardium of the ventricle showing a marked increase in positive cells just along this edge. E) Atrial staining of MYL4 showing homogeneous staining across all myocytes. F) MYOM1 staining of the ventricle. G) MYOM1 staining of the atrium showing heterogenous staining intensity. H) PAM staining of the ventricle showing rare (~5%) positive cells. I) PAM staining of the atrium showing a granular staining pattern in most cells. Bar = 300 μm.
3.4 Significant interactions are noted across mosaic proteins
Due to the anonymous nature of the images reviewed through HPASubC, there is no bias towards proteins of any given name or function. Therefore, any interactions would suggest a shared functional or regulatory state. We utilized String 10.0 to characterize these interactions. String is a database of known and predicted protein-protein interactions based on genomic context predictions, high throughput laboratory experiments, co-expression studies, automated text mining and information from other databases [9]. The more connecting lines between proteins, the more sources of support for the interaction. Of the 143 proteins, 60 (42%) were found to interact with each other in a large cluster (Fig. 3). Most prominent was a group of 10 proteins anchored to TTN. This group included MYL3, MYL4 and MYOM1. Not surprisingly, Gene Ontology identified this group to be enriched for actin filament organization. A second cluster of six proteins, all attached to PDP1, was noted. Gene Ontology did not identify enrichment in this group for a particular process. PAM was not found to be clustered to other proteins, despite its clear mosaic pattern.
Fig. 3.

Interactome map of putative mosaic proteins. A string 10.0 analysis of 143 proteins identified 60 proteins in a single large interactome. (smaller interactions and non-interacting proteins were removed for clarity). Two subclusters, one around TTN and one containing PDP1 are noted.
3.5 Age-related patterns
MYL3 and MYL4 staining were evaluated in a range of young specimens to determine the earliest instance of mosaic staining. The youngest specimen was from a 24-week fetus. It displayed mosaic pattern atrial MYL3 staining but consistent, non-mosaic pattern MYL4 ventricular staining. We then looked across a broader range of ages for MYL4 and noted consistent ventricular staining through 6 months of age and the earliest appearance of mosaic staining at 14 months of age. The percent of MYL4+ cells remained elevated compared to adult specimens through specimens taken from the first 3 years of life and seemed similar to adult samples by 5-10 years of age (Fig. 4).
Fig. 4.
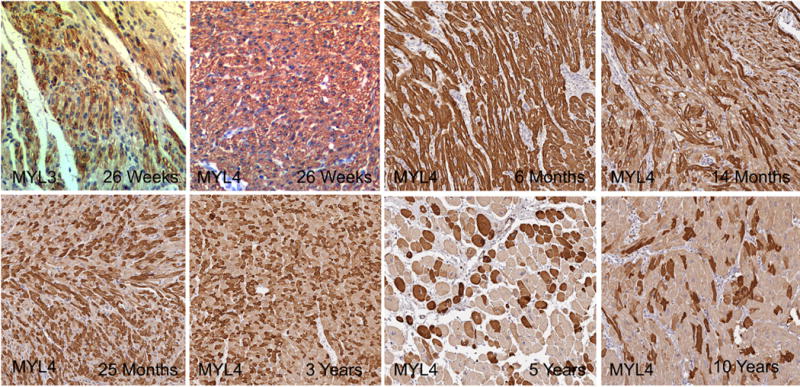
Age related changes in mosaicism. MYL3 was mosaic at 26 weeks in the atria, while MYL4 remained consistently expressed in the ventricle through 6 months and then decreased the percent of MYL4+ cells to 10 years of age (Original Magnification 20× for all images).
3.6 Classic fast and slow muscle fiber myosin heavy chain genes are not mosaic
The discovery that MYL3, MYL4 and MYL6 (myosin light chain 6, another myosin noted in Table 2) were all mosaic suggested a potential wider role for myosin proteins in this pattern. Myosin heavy chains, particularly MYH1, MYH2, and MYH4 are known as fast myosin heavy chain isoforms, while MYH7 is known as a slow isoform [15]. Although no myosin heavy chain proteins were found in our HPASubC scan, we specifically investigated 9 MYH proteins in the HPA that had mosaic skeletal muscle expression in the HPA. All but one (MYH9), had cardiac staining, but none of these proteins were mosaic in the heart (Supplemental Fig. 7).
3.7 Histone modification pattern is consistent with gene expression
We were intrigued by the differences in expression between atria and ventricles for these four proteins: MYL3, MYL4, PAM and MYOM1. Therefore, we sought additional methods to identify other mosaic proteins that may vary between these two regions of the heart. To do this, we systematically evaluated the 143 proteins/genes using H3K27ac histone modification and gene expression to determine how these genes are differentially regulated between atria and ventricles.
We quantified promoter activities of these genes in the heart tissues using the H3K27ac histone modification signal, a well-known marker for active cis-regulatory elements (CREs) [16] and compared this signal at each gene locus to the gene’s mRNA expression from GTEx RNA-seq data. The H3K27ac data was obtained from eight publicly available data sets from the ENCODE project [11]; two of them being from right atria, four from left ventricles, and the remaining two from right ventricles (Table 1).
First we established a high correlation of H3K27ac signal and TPM expression signal ratios between atria and ventricles (Pearson correlation coefficient=0.55) (Supplementary Table 2, Supplementary Fig. 8). In this dataset, we found MYL4 to be the gene most skewed toward atrial expression and MYL3 to be the second most skewed gene toward ventricular expression (Fig. 5A and 5B). Both MYOM1 and PAM were unremarkably different by H3K27ac signals and gene expression between atria and ventricles (Fig. 5C and 5D). The entire locus of the MYL3 gene including its promoter region shows significantly elevated CRE activities only in the ventricles (Fig. 5A). Conversely, the MYL4 promoter as well as its putative enhancers in the introns and the upstream region of the gene are highly active only in the two atria samples, compared to the left and right ventricles (Fig. 5B).
Fig. 5.
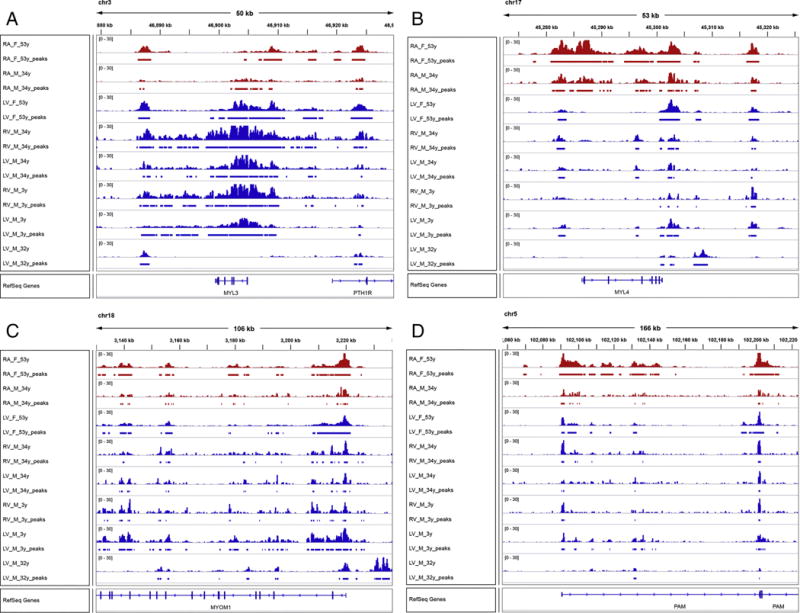
Histone modification data. Colored peaks represent regions of enhanced H3K27ac signal signifying open chromatin. The top two red rows are right atria and the bottom 6 rows are ventricle samples. Peak heights are normalized to 30.
If other genes behaved similarly to MYL3 and MYL4, with consistent expression in the atria or ventricle and mosaic expression in the opposite, we surmised this dataset would be useful. Other genes with a similar skew as MYL3 included C3orf52, SRGAP2 and SNAP91, although all of these genes had overall low (TPM) expression. Other genes with lower expression in the ventricle compared to the atria, consistent with MYL4 were PRKG2, TANC2, TAGAP and CASQ1. Again, the overall TPM expression of these four genes was low. This genetic expression data validated our protein expression results in MYL3 and MYL4 and suggested a small number of putative mosaic proteins may also vary between the atria and ventricles.
3.8 Relationship to disease expression changes
In general, gene or protein expression studies are performed on a tissue sample containing thousands of cells. Therefore, no appreciation of the mosaic patterns described here have been made in these studies. It is possible that, rather than expression changes of a given protein in a myocyte, simply more or fewer myocytes are recruited to express the protein at a steady state. We inquired as to whether or not the mosaic proteins described herein are altered in disease states. We found 10 examples in the literature of genes, corresponding to these proteins, as being altered in diseases of the cardiac myocardium (Table 3). This included 7 genes upregulated in dilated cardiomyopathy and MYL4 being dysregulated in a variety of diseases. Other genes in this group, such as nebulette (NEBL), have mutations associated with dilated and hypertrophic cardiomyopathy [17, 18].
Table 3.
Mosaic protei006Es with known dysregulation in cardiac disease.
| Protein/Gene | Disease | Expression Change | Samples Evaluated | Citation(s) |
|---|---|---|---|---|
| MYL4, ACTN2, ACTA1, RTN4, MYOM1, ODC1, PAM | Dilated Cardiomyopathy | ↑ | 181 | [20], [21], [22], [23], [24], [25] |
| MYL4, ACTN2, ACTA1, RTN4 | Ischemic Cardiomyopathy | ↑ | 57 | [20], [21], [25] |
| FHL2 | Dilated Cardiomyopathy | ↓ | 73 | [24] |
| FHL2 | Ischemic Cardiomyopathy | ↓ | 73 | [24] |
| MYL4 | Congenital heart disease | ↑ | 27 | [26] |
| MYL4, MYL6 | End stage LV failure | ↓ | 8a | [27] |
| PFKM | Right heart failure | ↑ | 24b | [28] |
Samples are human unless otherwise denoted.
Dog samples;
Rat samples
4. Discussion
This is the first wide-scale description of mosaic patterns of expression of the heart. We identified 143 proteins from HPA staining of over 12,800 proteins that have some degree of mosaic staining. The method that we used, HPASubC, presents the images in a fully anonymous fashion, thus our selections were not biased by what is known about mosaic patterns in skeletal muscle. Indeed, many of the classically described proteins with mosaic patterns in skeletal muscle such as MYH2 and MYH4 are not mosaic in the heart. Interestingly, the four proteins that we chose to further explore had diverse patterns of expression across the atria and ventricles that were inconsistent amongst each other. Therefore, a concept as simple as fast-twitch and slow-twitch seen in skeletal muscles is unlikely to be useful in the heart. Of course, skeletal muscle research has shown these phenotypes are more complex and have identified at least 4 major fiber types and numerous protein isoforms of myosin heavy and light chains that vary in muscles [2].
This expression complexity in cardiac myocytes is further supported and confirmed by open chromatin information. The histone modification data was consistent for all 4 proteins (MYL3, MYL4, MYOM1, and PAM) with their general levels of expression in the atria and ventricle. This partially validated the antibody as having specificity to each protein and suggested that the changes were at the gene expression level and not the result of protein isotypes that may change the epitope of the antibody.
It is provocative to think that rather than a change of expression of a protein within a cardiomyocyte, global expression changes may simply reflect more myocytes turning on a gene to make the protein. It is clear from this work, that an evaluation of these mosaic proteins should be done to determine if that is indeed the case. It may be that the expression of many of these proteins are at an optimal state in a cell, and that within the cell the expression does not change. What causes a protein to be expressed in one cardiomyocyte but not in an adjacent myocyte is a second needed area of inquiry. One possibility could be random X inactivation. However, only seven proteins were derived from genes on the X chromosome and that percentage (~5% of all mosaic proteins) is in line with the percentage of all human genes on the X chromosome (~5%). That would suggest no obvious enrichment of random X inactivation as a mechanism. Mosaicism of the mitochondria, unlike for fast/slow twitch skeletal muscle fibers, also does not appear to play a significant role as only 3 mitochondrial proteins were amongst our list of 143 proteins. As we noted, some myocytes had internal heterogenous expression. This may be technical or biological. At least for some proteins, we believe this pattern may be the result of the staining of intracellular structures that may be cut through in such a way as to render the cell only partly stained. This may also explain some false positives that are surely present in the dataset.
There are important limitations to this study. As we have stated repeatedly, HPA IHC contains numerous false positives (and false negatives) [7, 8]. The staining was unlikely to have been optimized to identify this particular mosaic pattern of staining. Therefore, many of the proteins we have identified are certain to be false positives and they are (with the exception of the four we validated) best thought of as putative mosaic proteins. The majority of cardiac images in HPA are listed as “heart” but are thought to be ventricle tissue. Thus, proteins that are exclusively mosaic in the atria (such as MYL3) would have likely been missed in our survey and await discovery. We note that MYL3 was only marginally mosaic (only 2 of 12 images from 4 antibodies had weak, variable staining) in the HPA data, yet was clearly mosaic in the atria when we evaluated it in our validation study. Thus it is not readily apparent which proteins are mosaic from our list purely from their behavior amongst the HPA images.
In conclusion, we present the first large scale demonstration of mosaic patterns of expression among human cardiomyocytes. Many of these 143 proteins are known to change their expression levels in disease. The extent to which more or fewer cardiomyocytes express any given protein, rather than an upregulation in a cell is a potentially exciting and fundamentally new field of inquiry as we learn more about cardiovascular disease.
Supplementary Material
Highlights.
There are 143 proteins with a putative mosaic pattern of expression in the heart.
Four proteins, MYL3, MYL4, MYOM1 and PAM are all mosaic, but each has a different pattern of mosaicism.
MYL3 and MYL4 are variably mosaic relative to patient age.
cis Regulatory elements and gene expression confirm the regional differences in protein mosaicism.
Acknowledgments
MKH is supported by grant 1R01HL137811 from the NIH and an American Heart Association Grant-in-Aid 17GRNT33670405. DEA is supported by grant R01HL111267 from the NIH. The authors thank Loyal Goff for his help with data analysis.
Sources of support: None
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare they have no conflict of interest.
References
- 1.Close RI. Dynamic properties of mammalian skeletal muscles. Physiological reviews. 1972;52:129–97. doi: 10.1152/physrev.1972.52.1.129. [DOI] [PubMed] [Google Scholar]
- 2.Bottinelli R, Reggiani C. Human skeletal muscle fibres: molecular and functional diversity. Progress in biophysics and molecular biology. 2000;73:195–262. doi: 10.1016/s0079-6107(00)00006-7. [DOI] [PubMed] [Google Scholar]
- 3.Tsuchimochi H, Sugi M, Kuro-o M, Ueda S, Takaku F, Furuta S, et al. Isozymic changes in myosin of human atrial myocardium induced by overload. Immunohistochemical study using monoclonal antibodies. The Journal of clinical investigation. 1984;74:662–5. doi: 10.1172/JCI111466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chu C, Thai K, Park KW, Wang P, Makwana O, Lovett DH, et al. Intraventricular and interventricular cellular heterogeneity of inotropic responses to alpha(1)-adrenergic stimulation. Am J Physiol Heart Circ Physiol. 2013;304:H946–53. doi: 10.1152/ajpheart.00822.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding J, Chen J, Wang Y, Kataoka M, Ma L, Zhou P, et al. Trbp regulates heart function through microRNA-mediated Sox6 repression. Nat Genet. 2015;47:776–83. doi: 10.1038/ng.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 7.Cornish TC, Chakravarti A, Kapoor A, Halushka MK. HPASubC: A suite of tools for user subclassification of human protein atlas tissue images. Journal of pathology informatics. 2015;6:36. doi: 10.4103/2153-3539.159213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anene DF, Rosenberg AZ, Kleiner DE, Cornish TC, Halushka MK. Utilization of HPASubC for the identification of sinusoid-specific proteins in the liver. Journal of proteome research. 2016 doi: 10.1021/acs.jproteome.6b00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D8. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–61. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dale RK, Pedersen BS, Quinlan AR. Pybedtools: a flexible Python library for manipulating genomic datasets and annotations. Bioinformatics. 2011;27:3423–4. doi: 10.1093/bioinformatics/btr539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Consortium GT, Laboratory DA, Coordinating Center -Analysis Working G, Statistical Methods groups-Analysis Working G, Enhancing Gg, Fund NIHC et al. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–13. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nature biotechnology. 2011;29:24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quiat D, Voelker KA, Pei J, Grishin NV, Grange RW, Bassel-Duby R, et al. Concerted regulation of myofiber-specific gene expression and muscle performance by the transcriptional repressor Sox6. Proc Natl Acad Sci U S A. 2011;108:10196–201. doi: 10.1073/pnas.1107413108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–6. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purevjav E, Varela J, Morgado M, Kearney DL, Li H, Taylor MD, et al. Nebulette mutations are associated with dilated cardiomyopathy and endocardial fibroelastosis. Journal of the American College of Cardiology. 2010;56:1493–502. doi: 10.1016/j.jacc.2010.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perrot A, Tomasov P, Villard E, Faludi R, Melacini P, Lossie J, et al. Mutations in NEBL encoding the cardiac Z-disk protein nebulette are associated with various cardiomyopathies. Archives of medical science : AMS. 2016;12:263–78. doi: 10.5114/aoms.2016.59250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, et al. Towards a knowledge-based Human Protein Atlas. Nature biotechnology. 2010;28:1248–50. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 20.Steenman M, Chen YW, Le Cunff M, Lamirault G, Varro A, Hoffman E, et al. Transcriptomal analysis of failing and nonfailing human hearts. Physiological genomics. 2003;12:97–112. doi: 10.1152/physiolgenomics.00148.2002. [DOI] [PubMed] [Google Scholar]
- 21.Grzeskowiak R, Witt H, Drungowski M, Thermann R, Hennig S, Perrot A, et al. Expression profiling of human idiopathic dilated cardiomyopathy. Cardiovasc Res. 2003;59:400–11. doi: 10.1016/s0008-6363(03)00426-7. [DOI] [PubMed] [Google Scholar]
- 22.Asakura M, Kitakaze M. Global gene expression profiling in the failing myocardium. Circulation journal : official journal of the Japanese Circulation Society. 2009;73:1568–76. doi: 10.1253/circj.cj-09-0465. [DOI] [PubMed] [Google Scholar]
- 23.Yung CK, Halperin VL, Tomaselli GF, Winslow RL. Gene expression profiles in end-stage human idiopathic dilated cardiomyopathy: altered expression of apoptotic and cytoskeletal genes. Genomics. 2004;83:281–97. doi: 10.1016/j.ygeno.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 24.Lal S, Nguyen L, Tezone R, Ponten F, Odeberg J, Li A, et al. Tissue microarray profiling in human heart failure. Proteomics. 2016;16:2319–26. doi: 10.1002/pmic.201600135. [DOI] [PubMed] [Google Scholar]
- 25.Morano I, Hadicke K, Haase H, Bohm M, Erdmann E, Schaub MC. Changes in essential myosin light chain isoform expression provide a molecular basis for isometric force regulation in the failing human heart. Journal of molecular and cellular cardiology. 1997;29:1177–87. doi: 10.1006/jmcc.1996.0353. [DOI] [PubMed] [Google Scholar]
- 26.Morano M, Zacharzowski U, Maier M, Lange PE, Alexi-Meskishvili V, Haase H, et al. Regulation of human heart contractility by essential myosin light chain isoforms. The Journal of clinical investigation. 1996;98:467–73. doi: 10.1172/JCI118813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao Z, Xu H, DiSilvestre D, Halperin VL, Tunin R, Tian Y, et al. Transcriptomic profiling of the canine tachycardia-induced heart failure model: global comparison to human and murine heart failure. Journal of molecular and cellular cardiology. 2006;40:76–86. doi: 10.1016/j.yjmcc.2005.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drake JI, Bogaard HJ, Mizuno S, Clifton B, Xie B, Gao Y, et al. Molecular signature of a right heart failure program in chronic severe pulmonary hypertension. American journal of respiratory cell and molecular biology. 2011;45:1239–47. doi: 10.1165/rcmb.2010-0412OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


