Abstract
Obesity is associated with elevated levels of free fatty acids (FFAs) and pro-inflammatory CD11c+ macrophages. However, whether and how FFAs contribute to CD11c+ macrophage differentiation and pro-inflammatory functions remain unclear. Here we report that dietary saturated FAs, but not unsaturated FAs, promoted the differentiation and function of CD11c+ macrophages. Specifically, we demonstrated that stearic acid (SA) significantly induced CD11c expression in monocytes through activation of the nuclear retinoid acid receptor (RAR). More importantly, cytosolic expression of epidermal fatty acid binding protein (E-FABP) in monocytes/macrophages was shown to be critical to the mediation of the SA-induced effect. Depletion of E-FABP not only inhibited SA-induced CD11c upregulation in macrophages in vitro, but also abrogated high saturated fat diet-induced skin lesions in obese mouse models in vivo. Altogether, our data demonstrate a novel mechanism by which saturated FAs promote obesity-associated inflammation through inducing E-FABP/RAR-mediated differentiation of CD11c+ macrophages.
Introduction
In the past several decades, prevalence of obesity has increased dramatically in the U.S. and worldwide. It is estimated that more than half of American adults will be obese by 2030 (1). Accordingly, obesity-related illness, including cardiovascular disease, type 2 diabetes and various cancers, will become serious threats to human health. It is accepted that chronic inflammation is the most common pathogenic signature underlying obesity and its-related disorders. However, the mechanisms of how obesity contributes to chronic inflammation remain largely unknown.
Numerous animal and human studies have shown that macrophages, in particular CD11c+ macrophages, are recruited to adipose tissue during obesity, contributing to obesity-associated chronic inflammation and insulin resistance (2–4). Depletion of CD11c+ cells decreases inflammatory cytokine production and normalizes insulin sensitivity in obese mouse models (5). Our recent studies demonstrated that a high fat diet (HFD) induces the accumulation of CD11c+ macrophages in the skin, promoting obesity-associated skin inflammation through upregulation of inflammasome signaling (6). Thus, it is becoming clear that CD11c+ macrophages are a critical pathogenic cell population mediating obesity-associated chronic inflammation. It is therefore of great interest to determine factors that enhance the differentiation and pro-inflammatory function of CD11c+ macrophages during obesity.
Obesity is generally caused by overnutrition. Excess calories are primarily stored in the form of neutral lipids in tissues and in the circulation. Compared to healthy lean subjects, obese individuals have marked elevated levels of plasma free fatty acids (FFAs), which consist of different types of saturated and unsaturated FAs (7). Palmitic acid (PA, C16:0), stearic acid (SA, 18:0), oleic acid (OA, 18:1) and linoleic acid (LA, 18:2) account for more than 80% of the total plasma FFAs (8). Elevated levels of plasma FFAs are an important contributor to insulin resistance, chronic inflammation, and other obesity-related disorders (9). However, the molecular mechanisms by which individual FAs contribute to obesity-associated inflammation and disorders remain incompletely understood.
Fatty acid binding proteins (FABPs) are a family of cytosolic lipid chaperones which solubilize fatty acids, facilitate their metabolism and coordinate with various nuclear receptors for gene regulation (10,11). FABPs exhibit tightly regulated patterns of tissue distribution. For example, adipose tissues predominantly express adipose FABP (A-FABP) while skin epithelia highly express epidermal FABP (E-FABP), suggesting unique functions of individual FABPs in specific tissues and cells (12). Our recent research focusing on functions of the FABP family in immune cell regulation demonstrate that A-FABP and E-FABP are also highly expressed in macrophages. While E-FABP abundantly expressed in CD11c positive macrophages promotes HFD-induced IL-1β signaling pathways, A-FABP expression in CD11c negative macrophages facilitates fatty acid metabolism for cytotoxic ceramide production and contributes to saturated FA-induced macrophage cell death (6,13). Thus, it appears that individual FABPs play a unique role in linking FA-mediated metabolism and pro-inflammatory functions in different subsets of macrophages.
In the current study, we performed experiments to investigate the role of individual FAs in regulating the differentiation and function of inflammatory CD11c+ macrophages. We demonstrated that saturated FAs (in particular SA), but not unsaturated FAs, greatly promoted CD11c+ macrophage differentiation and pro-inflammatory cytokine production both in vitro and in vivo. Furthermore, E-FABP was identified to be the major FABP member in mediating SA-induced activation of RAR for the differentiation of CD11c+ macrophages. Importantly, deletion of E-FABP significantly reduced accumulation of CD11c+ macrophages in obese mice and abrogated saturated FA-mediated chronic skin lesions in different HFD-induced obese mouse models.
Materials and Methods
Animals
C57BL/6 background E-FABP-deficient mice (E-FABP−/−), A-FABP-deficient mice (A-FABP−/−) and their wild type (WT) littermates were bred and housed in the animal facility of the University of Louisville. All animal manipulations were carried out according to the approval of the Institutional Animal Care and Use Committee of the University of Louisville. Weaned mice were ad libitum fed either with 60% HFD (lard) or 10% LFD (lard) (Research Diets) for 5-9 months before they were sacrificed for analysis of macrophage phenotype in various tissues or organs. To dissect the impact of different fat components in inducing skin inflammation, female mice were fed HFD rich in saturated FAs (45% cocoa butter), unsaturated FAs (45% safflower oil), or control LFD (10% soybean oil), respectively, for 9 months. Skin lesions were measured and CD11c+ macrophages in the skin tissues were analyzed by confocal or flow cytometric staining.
Reagents
M-CSF and GM-CSF were purchased from Cell Signaling Technology (Danvers, MA). All FAs were from Nu-Chek Prep. PA (5 mM), SA (5 mM), OA (5 mM), LA (5 mM), eicosapentaenoic acid (EPA, 5mM) and α-linolenic acid (ALA) (2mM) were prepared with 2 mM of endotoxin-free BSA in PBS (Cat#: BP9705-100, FISHER Scientific), sonicated until dissolved, and filtered through 0.22mm sterile filter as we previously described (14,15). The specific reactive oxygen species (ROS) inhibitors 4-amino-2,4-pyrrolidine-dicarboxylic acid (APDC) and butylated hydroxyanisole (BHA), PPARδ agonist GW0742 and antagonist GSK0660, PPARγ agonist Rosiglitazone and antagonist GW9662, RXR agonist LG100268 and antagonist HX531 were purchased from Sigma-Aldrich. RAR agonist BM753 and antagonist BMS195640 were purchased from TOCRIS. Necrosis inhibitor IM54 and apoptosis inhibitor z-VAD-FMK were purchased from Enzo Life Sciences. TLR4 inhibitor Eritoran, STAT3 inhibitor NSC74859, NFҡB inhibitor CAPE, IKK inhibitor BMS-345541, Cer synthesis inhibitor FB1 (fumonisin B1) and SPT (serine palmitoyltransferase) were purchased from Cayman Chemical.
Cell culture and treatment
For primary macrophage differentiation, bone marrow cells collected from WT, E-FABP−/− or A-FABP−/− mice were differentiated either by M-CSF (20ng/mL) or by GM-CSF (20ng/mL). For some experiments, same as our previous studies (6,13,16), immortalized macrophage cell lines established from WT, A-FABP−/−, or E-FABP−/− mice were also used. All cultured cells were grown in RPMI 1640 supplemented with 5% (v/v) fetal bovine serum (Atlanta Biological) and gentamicin (20μg/ml). Macrophage cell lines (2 ×105 cells/ml/well) or primary BMMs (4 ×105 cells/ml/well) were plated in 24-well plates for all the experiments. After FA treatment for 1-3 days depending on macrophage cell lines or primary BMMs, cells were lifted for flow cytometric analysis and qPCR detection. In the assays including agonists, antagonists or other inhibitors, cells were pre-treated with respective chemicals for 3 hours before addition of different concentrations (50, 100, 200μM) of saturated or unsaturated FAs.
Flow cytometric analyses
For phenotypic analysis of CD11c+ macrophages in obese mice, different tissues including the peripheral blood, lymph nodes, spleen, peritoneum, liver, lung and adipose tissues, bone marrow, and skin, were directly collected from lean and obese mice after PBS perfusion. In order to acquire single cells for flow analysis, liver, lung and adipose tissue were cut into 1 mm3 fragments and digested with collagenase mixture (5% FBS in RMPI 1640, 0.5mg/ml collagenase A (Roche Diagnostic), 0.2 mg/ml hyaluronidase, type V (Sigma-Aldrich), and 0.02mg/ml DNase I (Sigma-Aldrich) on a shaker at 100 RPM at 37°C for 30min (for liver) or 1hr (for other tissues), and then were filtered for use. Skin tissues were digested as our previous studies (5).
Macrophages from different sources were surface stained for 30 min at 4°C in 1% FBS-PBS containing indicated Abs (anti-CD11b, clone M1/70; anti-CD11c, clone HL3; anti-F4/80, clone BM8; anti-MHC class II, clone M5/114.15.2; anti-CD36, clone HM36; anti-CD54,clone YN1/1.7.4; anti-CD80, clone 16-10A1; anti-CD86,clone GL-1). For C16-Bodipy uptake assay, cells were treated with C16-Bodipy (0, 10, 100nM) for 30 min then washed thoroughly with PBS before flow cytometric analysis. All samples were acquired on a BD LSRFortessa flow cytometer, and data analyses were conducted using FlowJo software (Tree Star).
Quantitative RT-PCR
For real-time qPCR analysis, RNA was extracted from cells using PureLinkTM RNA Mini Kit (Ambion by Life Technologies). cDNA synthesis was performed with QuantiTect ReverseTranscription Kit (Qiagen). Quantitative realtime PCR using SYBR Green PCR Master Mix was performed on an ABI StepOnePlus Real-Time PCR Systems (Applied Biosystems) to analyze interesting genes, such as FABP family members, common nuclear receptors, FATP1, CD36, ACSL1, inflammatory cytokines, etc (see the detailed gene primer sequence in the supplemental Table 1). Relative mRNA levels were determined using HPRT1 as a reference gene. Gene expression levels were analyzed by the 2−ΔΔCT method.
Small interfering RNA transfection
Duplex small interfering RNAs (siRNAs) targeting the coding region of RXRα were ordered from Integrated DNA Technologies. To knock down RXR gene expression, 2×105 WT macrophage cell line or 5×105 BMMs in 24 well-plates were transfected with designated siRNAs (40nM) using Oligofectamine (Life Technologies) for 4-6 hours. Normal RPMI 1640 medium with 5% FBS was added to the transfected cells for 6 hours before 100μM SA treatment.
Confocal microscopy
The fresh tissue of normal or lesion skin from obese WT or FABP−/− mice were embedded in OCT and stored in −80°C. The embedding blocks were sectioned by freezing microtome (Leica CM1900) at 5 μm thickness. After blocking with anti-mouse CD16/32 (clone: 93, Biolegend cat#:101302, 1:200 in 1% FBS-PBS), anti-mouse CD11c (clone: N418, Biolegend cat#:117310, 1:200 dilution) and DAPI (Cell signaling cat# 4083S, 10mg/mL in H2O, 1:2000 dilution) were applied to the sections for 1h, followed by wash and sealing with anti-fading mounting medium. The sections were analyzed by Nikon A1 laser scanning confocal microscopy. The CD11c fluorescent intensity in the confocal images was analyzed by ImageJ software.
Western blotting
Western blotting was used to determine the levels of A-FABP and E-FABP in primary BMMs, and the levels of RXRα in macrophage transfected with scramble or RXR siRNAs. In brief, 20μg proteins (quantified by BCA assay) in each sample were loaded for SDS-PAGE, transferred to PVDF membrane and blotted with respective antibodies (anti-mFABP5,cat#: AF1476; anti-m/rFABP4 Ab, cat#: AF1443; anti-RXRa, cat#: 3085S). β-actin was probed as a loading control. Image Quant TL system was applied to calculate the relative protein quantification.
Cytokine analyses
Mouse IL-6, TNFα, IL-1β, and IL-10 levels in cell cultural supernatants or serum from obese/lean mice were measured using ELISA MAX™ standard set from BioLegend.
Statistical analyses
Student t test was performed for the comparison of results from different treatments. A p value <0.05 is considered statistically significant.
Results
Consumption of a HFD systemically enhances CD11c+ macrophages in vivo
We previously have shown that mice fed a HFD exhibited increased percentage of CD11c+ macrophages in the skin (6), suggesting a possible systemic increase of pro-inflammatory CD11c+ macrophages during obesity. To address this hypothesis, we put C57BL/6 mice on either a HFD (60% fat) or a low fat diet (LFD) (10% fat) for 20 weeks to induce obesity and then analyzed the phenotype of CD11c+ macrophages in different tissues and organs. As myeloid-derived immune cells express the CD11b marker, we divided CD11c+ macrophages (CD11c+F4/80+) into myeloid-derived CD11b+ and non-myeloid-derived local-residential CD11b- populations (Figure 1A). Interestingly, CD11b+CD11c+ macrophages were significantly increased in immune organs, including bone marrow, draining lymph nodes (LNs), and peripheral blood, in HFD-mice compared to LFD-mice (Figures 1B–1D). In contrast, the percentage of CD11b-CD11c+ macrophages was similar in these immune organs between the HFD and LFD mice, suggesting a specific upregulation of myeloid-derived CD11c+ macrophage differentiation in mice consumption of the HFD. When we further analyzed the phenotype of CD11b+CD11c+ macrophages in non-immune organs, such as adipose tissue, liver and lung, we found that consumption of the HFD also elevated the percentage of CD11b+CD11c+ macrophages, but not CD11b-CD11c+ cells, in these organs (Figure 1E–1G). These data indicate that consumption of the HFD systemically promotes the differentiation of myeloid-derived CD11b+ CD11c+ macrophages in obese mice.
Figure 1. HFD induces CD11c+ macrophage accumulation in major organs in vivo.
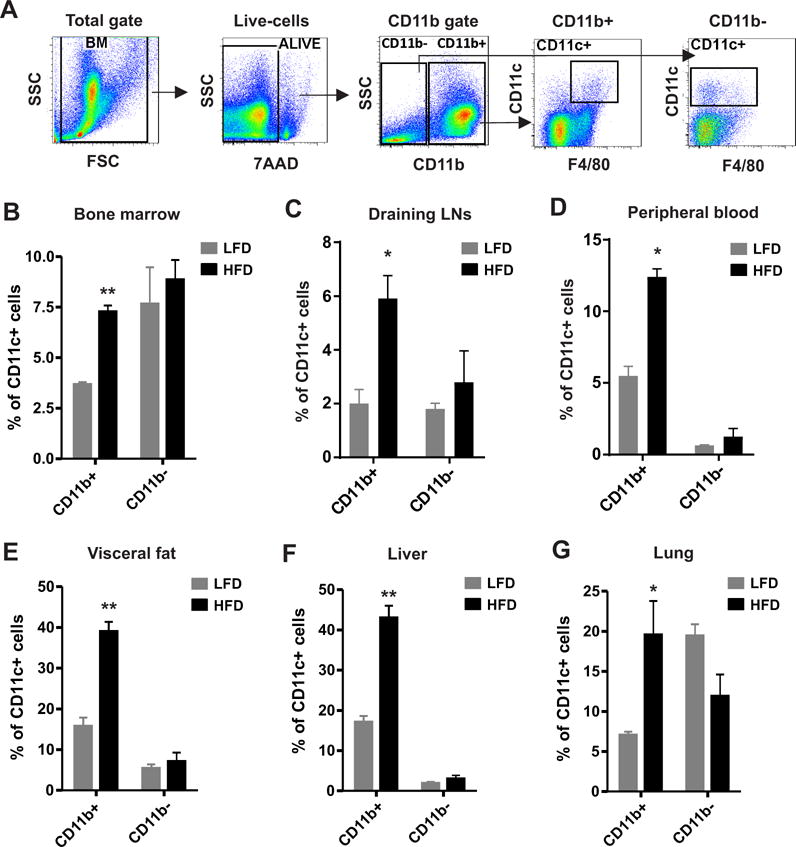
C57BL/6 mice were grouped and fed on HFD (60% fat) and LFD (10% fat), respectively, for 20 weeks. Different tissues and organs were collected from HFD mice and LFD mice (n=4/group) and were phenotypically analyzed for immune cell populations by flow cytometric staining. (A), Gating strategy of flow cytometric surface staining for CD11c+ macrophages. (B-G) Flow cytometric analyses of the percentage of CD11c+ macrophages in bone marrow (B), draining lymph nodes (LNs) (C), peripheral blood (D), visceral adipose tissue (E), liver (F) and lung (G). Data are shown as mean ± SEM from at least three independent experiments (*, p<0.05; **, p<0.01).
Saturated FAs, in particular stearic acid, enhance CD11c+ macrophage differentiation
To investigate why consumption of the HFD induces a systemic increase of myeloid-derived CD11c+ macrophage differentiation in vivo, we noticed that the HFD was rich in both saturated FAs (PA, 19.3% and SA, 10.6%) and unsaturated FAs (OA, 32.9% and LA, 24.4%). We speculated that individual FA components in the HFD might promote the differentiation of myeloid-derived CD11c+ macrophages. To this end, we differentiated bone marrow cells into macrophages under different conditions in the presence or absence of each FA in the diet. When bone marrow cells were differentiated with macrophage colony-stimulating factor (M-CSF), we observed that the percentage of CD11c+ macrophages was elevated in saturated FA-treated groups, in particularly in the SA-treated wells (p<0.01), compared to the BSA-treated control. In contrast, neither OA nor LA treatment increased CD11c expression in macrophages (Figure 2A). Under the differentiating condition with GM-CSF (granulocyte-macrophage colony stimulating factor), we observed the same phenomenon that saturated FAs, but not unsaturated FAs, induced CD11c expression in bone marrow-derived macrophages (Figure 2B). Moreover, when bone marrow cells (BMC) were cultured in vitro without any exogenous colony-stimulating factors, we also observed enhanced expression of CD11c in CD11b+ macrophages in response to the treatment with PA or SA, although to a lesser extent (Figure 2C). These results suggest that saturated FAs, in particular SA, are able to drive bone marrow cell differentiation into CD11c+ macrophages under different conditions.
Figure 2. SA enhances CD11c+ macrophage differentiation in vitro.
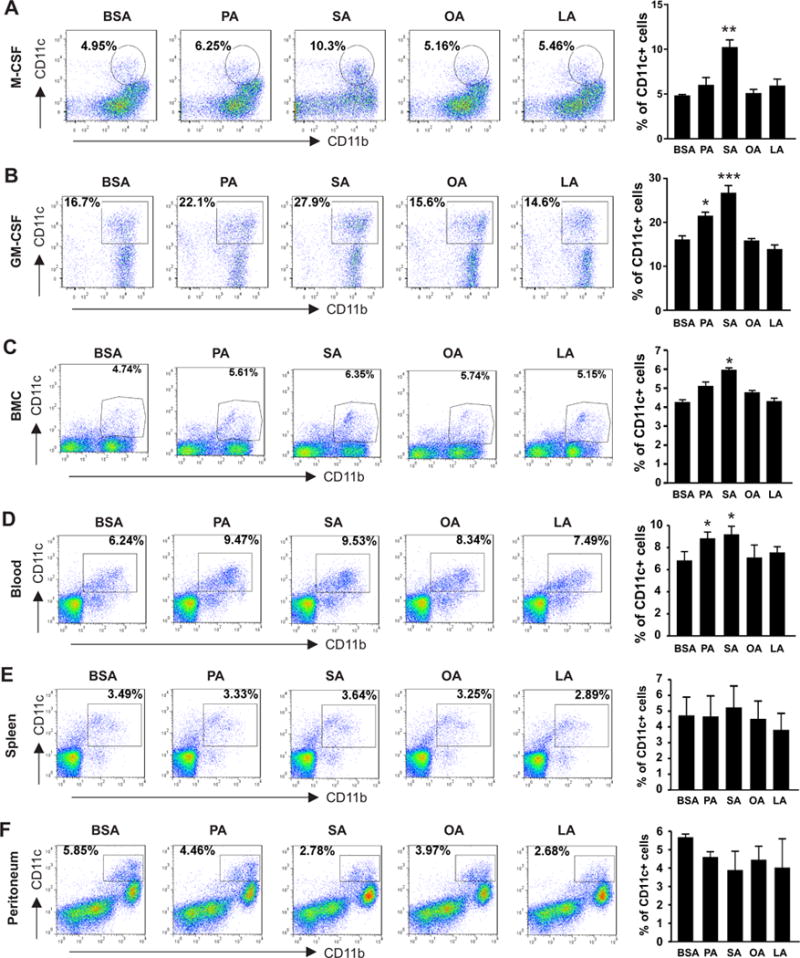
(A-B) Bone marrow cells collected from 6-8 weeks old C57BL/6 mice were differentiated into macrophages under stimulation of M-CSF (20ng/ml) (A) or GM-CSF (20ng/ml) (B) in the presence of individual FAs, including PA (100μM), SA (100μM), OA (100μM) and LA (100μM), and BSA control for 3 days. Flow cytometric staining for analysis of CD11c expression in CD11b+ cells. Average percentage of CD11c+ macrophages is shown in the right panel. (C-F) Cells from bone marrow (BMC) (C), peripheral blood (D), spleen (E) and peritoneum (F) collected from 6-8 weeks old C57BL/6 mice were directly cultured in vitro with PA(100μM), SA(100μM), OA(100μM), LA(100μM) or BSA controls for 36h and flow cytometric analysis of CD11c expression on CD11b+ populations. Average percentage of CD11c+ cells is shown in the right panel. Data are mean value ± SEM of three mice (* p<0.05, ** p<0.01, *** p<0.001 compared to BSA controls), and are representative of three independent experiments.
Due to the distinct origins and functions of tissue macrophages (17), it was unclear whether macrophages in peripheral tissues responded similarly to FA treatment. We collected samples from the peripheral blood, spleen and peritoneum and measured CD11c expression in monocytes/macrophages in response to individual FA treatment. Interestingly, similar to bone marrow cells, blood monocytes responded to the treatment of either PA or SA by enhancing CD11c+ differentiation (Figure 2D). In contrast, FA treatment, regardless of saturated FAs or unsaturated FAs, exhibited marginal impact on CD11c expression in macrophages from the spleen and peritoneum (Figure 2E, 2F). These results suggest that, unlike tissue-resident macrophages, immature monocytes from the bone marrow or the circulation are more susceptible to SA-induced differentiation into CD11c+ macrophages. Altogether, our data indicate that saturated FAs, in particular SA, in the HFD may represent a previously unappreciated factor enhancing differentiation of CD11c+ macrophages from the bone marrow and peripheral blood.
SA promotes inflammatory functions of CD11c+ macrophages
To further confirm the effect of SA in promoting CD11c expression, we treated M-CSF-induced bone marrow cells with different doses of SA ranging from 0-200μM, and found that SA at 100μM was enough to significantly promote CD11c protein expression on the cell surface (Figure 3A, 3B). Moreover, the observation of a significant increase in mRNA levels of CD11c, but not CD11b, by SA treatment validated that SA induced a specific de novo expression of CD11c in bone marrow-derived macrophages (Figure 3C, 3D). As elevated CD11c+ macrophages contribute to chronic inflammation and adaptive immune responses in obesity (4,18), we further examined if SA treatment promoted such functions. First, we assessed whether SA treatment influenced the expression of MHCII and other major co-stimulatory molecules (such as CD86, CD80, CD54), which are critical for antigen presentation in CD11c+ macrophages. As shown in Figure 3E–3H, SA treatment enhanced the expression of all measured co-stimulatory molecules, in particular MHCII (p<0.01), CD80 and CD86 (p<0.05), compared to the BSA control. Next, we measured if SA treatment promoted proinflammatory cytokine production by M-CSF-induced bone marrow macrophages. Using real-time PCR we demonstrated that SA treatment promoted the expression of IL-6, TNFα, IL-1β, IFNα and IFNβ, but had no impact on IL-10, in macrophages (Figure 3J–3O). We further confirmed that pro-inflammatory cytokines (such as IL-6, TNFα and IL-1β) were elevated in the supernatants of SA-treated macrophages by ELISA (Figure 3P–3S). Taken together, our data suggest that SA promotes not only CD11c+ macrophage differentiation, but also their antigen presentation and pro-inflammatory functions.
Figure 3. SA promotes expression of costimulatory molecules and production of inflammatory cytokines in M-CSF-induced bone marrow cells.
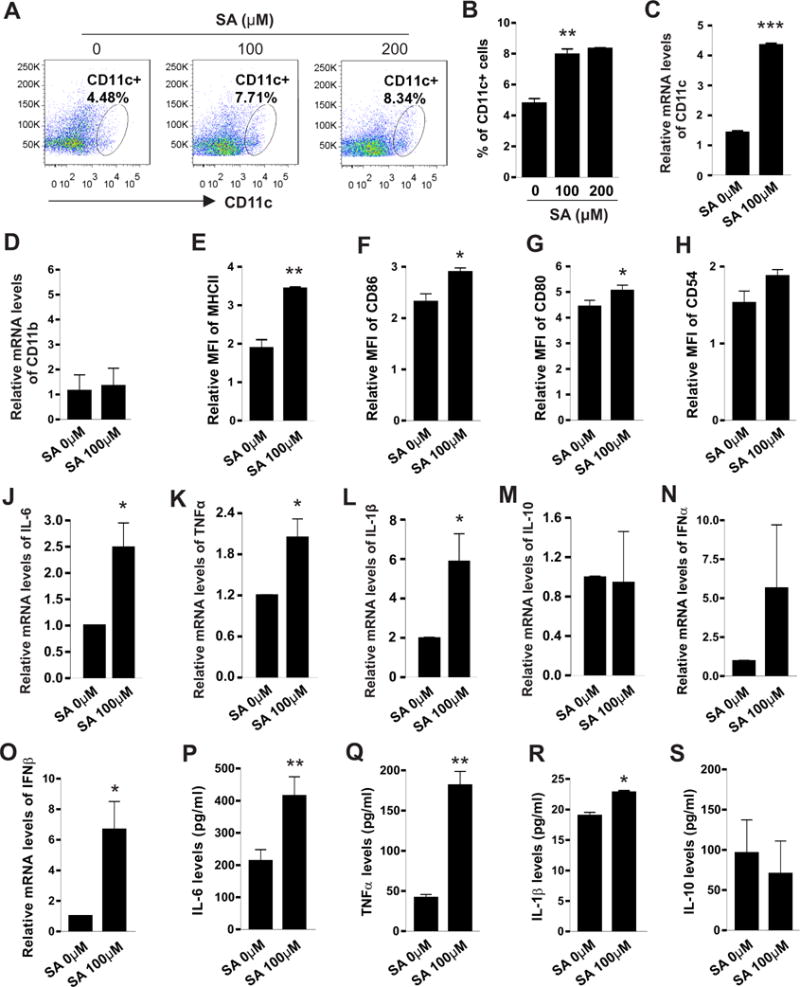
(A-B) Flow cytometric analyses of CD11c expression on bone marrow cells stimulated with M-CSF (20ng/ml) and indicated concentrations of SA for 48h. Average percentage of CD11c+ cells was shown in Panel B. (C-D) Realtime PCR analysis of mRNA levels of CD11c (C) and CD11b (D) in bone marrow cells after stimulation with M-CSF and SA for 48h. (E-H) Flow cytometric analysis of the mean fluorescent intensity (MFI) of MHCII (E), CD86 (F), CD80 (G), and CD54 (H) on bone marrow cells after stimulation with M-CSF and indicated SA for 48h. (J-O) Realtime PCR analysis of mRNA levels of inflammatory cytokines, including IL-6 (J), TNFα (K), IL-1β (L), IL-10 (M), IFNα (N) and IFNβ (O) in M-CSF-stimulated bone marrow cells in the presence or absence SA treatment (100μM) for 48h. (P-S) Measurement of protein levels of IL-6 (P), TNFα (Q) IL-1β (R) and IL-10 (S) in the supernatants of M-CSF-differentiated macrophages in the presence or absence SA treatment (100μM) for 48h. Data are shown as mean ± SEM (* p<0.05, ** p<0.01, ***p<0.001 as compared to controls). Experiments are repeated at least three times.
SA-induced CD11c+ macrophage differentiation is dependent on RAR
We next sought to determine the molecular mechanisms by which SA induced CD11c expression in macrophages. M-CSF-induced bone marrow macrophages (M-BMMs) exhibited higher expression of F4/80 and lower expression of CD11c compared to GM-CSF-induced BMMs (GM-BMMs) (supplemental Figure 1A), representing bona fide hematopoietic-derived macrophages in vivo (13,19), we thus focused on primary or immortalized M-BMMs to dissect the molecular mechanisms. Treatment of immortalized macrophages with individual saturated FAs (SA or PA), but not unsaturated FAs (including OA, LA, ALA and EPA), significantly induced a dose-dependent upregulation of CD11c expression (Figure 4A, 4B). To dissect putative mediators or pathways which might be involved in the SA treatment (e.g. SA/TLRs/NFκb pathway or SA/ceramides/cell stress pathway) (13,20–22), we respectively inhibited the activation of TLR4 by Eritoran, STAT3 by NSC74859, and NF-κB by CAPE or BMS-345541, and observed if such factors played a critical role in SA-induced CD11c expression by macrophages. However, none of these treatments showed any significant impact on SA-induced CD11c expression in macrophages (supplemental Figure 1B). We further inhibited ceramide production by myriocin or fumonisin B1, ROS generation by APDC or BHA, apoptosis by z-VAD-FMK, or necrosis by IM-54 in macrophages, and did not have obvious effect on SA-induced CD11c expression either (supplemental Figure 1C).
Figure 4. RAR mediates SA-induced CD11c expression.
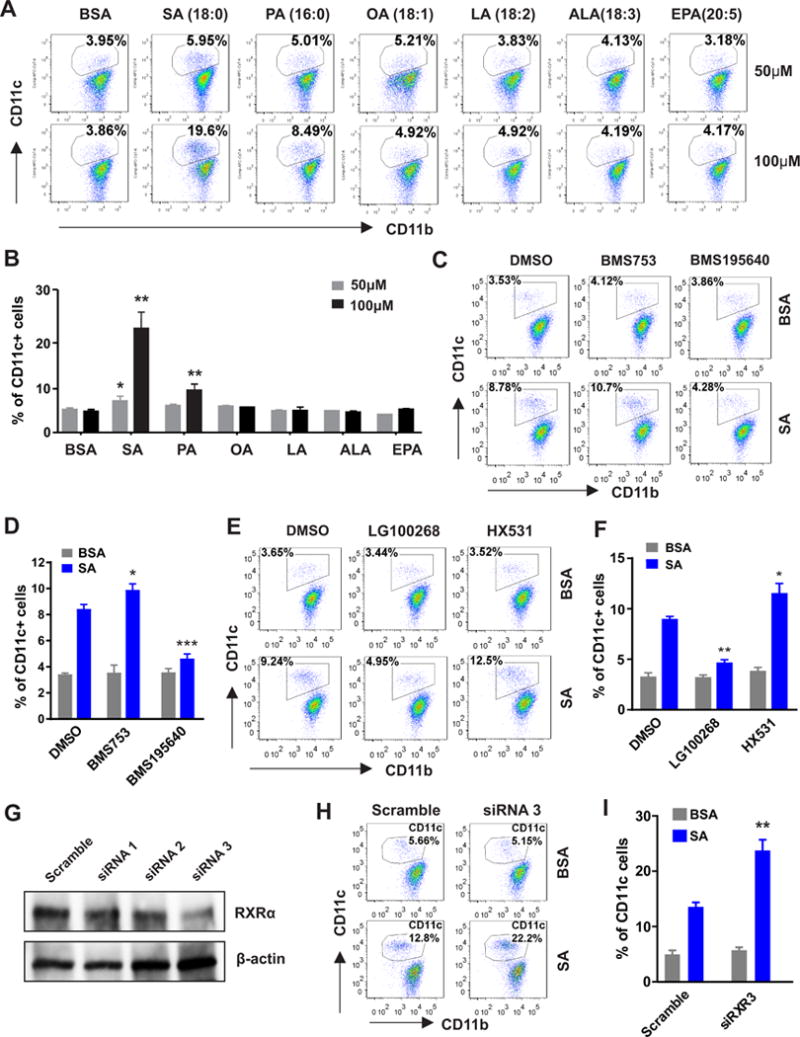
(A-B) Flow cytometric surface staining for CD11c expression in immortalized macrophages stimulated with BSA or indicated concentrations of individual FAs for 48h. Average percentage of CD11c+ macrophages is shown in panel B. (C-D) Analysis of CD11c expression in the immortalized macrophages treated with BMS753 (RAR agonist, 0.3μM) and BMS195640 (RAR antagonist, 0.3μM) in the presence or absence of SA (50μM) for 48h. Average percentage of CD11c+ macrophages is shown in panel D. (E-F) Analysis of CD11c expression in the immortalized macrophages treated with LG100268 (RXR agonist, 0.3μM) and HX531(RXR antagonist, 0.3μM) in the presence or absence of SA (50μM) for 48h. Average percentage of CD11c+ macrophages is shown in panel F. (G) Analyses of RXR protein levels in macrophages transfected with scramble or different sets of RXR siRNAs. (H-I) Immortalized macrophages transfected with RXR siRNA3 or scramble controls were stimulated with BSA or SA (100μM) for 24h. CD11c expression was analyzed by flow cytometry, and average percentage is shown in panel I. Data are shown as mean ± SEM (* p<0.05, ** p<0.01, ***p<0.001 as compared to respective controls). Experiments are repeated at least three times. Also see Figure S1.
As fatty acids and their derivatives are able to regulate gene transcription through various nuclear receptors (23), we measured common nuclear receptor expression profile in bone marrow cells differentiated with M-CSF. PPARβ, RAR and their heterodimer partner RXR were highly expressed as compared to other nuclear receptors at different time points (supplemental Figure 1D). When macrophages were treated with specific agonists or antagonists for PPARβ or PPARγ, none of these treatments demonstrated obvious alterations in CD11c expression (supplemental Figure 1E). Interestingly, treatment with BMS753, a selective RAR agonist, increased while BMS195640, a selective RAR antagonist, decreased SA-induced CD11 expression (Figure 4C, 4D), suggesting that RAR is a major mediator of SA-induced effect in macrophages. As RAR functioned as a heterodimer with RXR to regulate cell differentiation (24), and RXR was able to bind multiple sites on the CD11c promoter region (supplementary Figure 1F), macrophages were further treated with RXR specific agonist LG100268 or antagonist HX531. LG100268 decreased but HX531 increased SA-induced CD11c expression in macrophages (Figure 4E, 4F). Silencing of RXR expression with siRNA also enhanced SA-induced CD11c expression in the immortalized macrophages (Figure 4G–4I). We further used primary M-BMMs to validate these findings. Consistently, RAR antagonist inhibited, but agonist increased, SA-induced CD11c expression (supplemental Figure 1G). Inhibition of RXR with antagonist or siRNA enhanced, but activation of RXR with agonist suppressed, SA-mediated effect in primary BMMs (supplemental Figure 1H, 1I). Altogether, these results suggest that while RXR binds to CD11c promoter serving as a gate keeper to repress CD11c expression in resting macrophages, metabolism of excess SA co-activates RAR/RXR heterodimers leading to the transactivation of CD11c expression in macrophages.
E-FABP expression is critical to SA-mediated CD11c upregulation in macrophages
The importance of nuclear receptor RAR in controlling CD11c gene expression suggested that trafficking of exogenous SA or its derivatives to the nucleus is a key step in SA-induced CD11c+ macrophage differentiation. To this end, we examined the expression of FABPs that mediate fatty acid transport inside macrophages and found that E-FABP and A-FABP were the predominant members expressed in primary BMMs (Figures 5A, 5B). Considering the fact that both A-FABP and E-FABP can bind SA (25), we next investigated which FABP(s) was (were) critical in facilitating SA-induced CD11c expression using WT, A-FABP−/− and E-FABP−/− mice. With real-time PCR and Western blotting, we confirmed the phenotype of macrophages generated from WT, A-FABP−/− and E-FABP−/− mice. As expected, A-FABP−/− and E-FABP−/− macrophages exhibited a compensatory up-expression of E-FABP and A-FABP, respectively (Figures 5C–5E). Importantly, when we measured CD11c expression in BMMs in the presence or absence of SA, we found that deficiency of E-FABP and A-FABP did not impact CD11c expression in macrophages without SA treatment. However, deficiency of E-FABP, but not A-FABP, significantly inhibited SA-induced CD11c expression in macrophages (Figures 5F, 5G). Further analyses demonstrated that deficiency of neither FABP impacted the expression of membrane fatty acid transport proteins including CD36, ASCL1 (long chain acyl-coA synthetase 1), and FATP1 (fatty acid transport protein 1) (Figure 5H–5J), nor the uptake of long-chain fatty acids by these macrophages (Figure 5K). On the basis of these results and our previous study showing that A-FABP facilitated SA-induced ceramide production (13), it seemed clear that deficiency of FABPs did not affect FA uptake by macrophages, but individual FABPs coordinated their unique lipid responses inside macrophages. Unlike A-FABP, E-FABP was critical in trafficking SA or its derivatives for co-activation of RAR-mediated CD11c expression in macrophages. Thus, E-FABP is the main cytosolic chaperone facilitating SA-induced differentiation of CD11c+ macrophages.
Figure 5. Expression of E-FABP, but not A-FABP, is critical to SA-induced CD11c expression in macrophages.
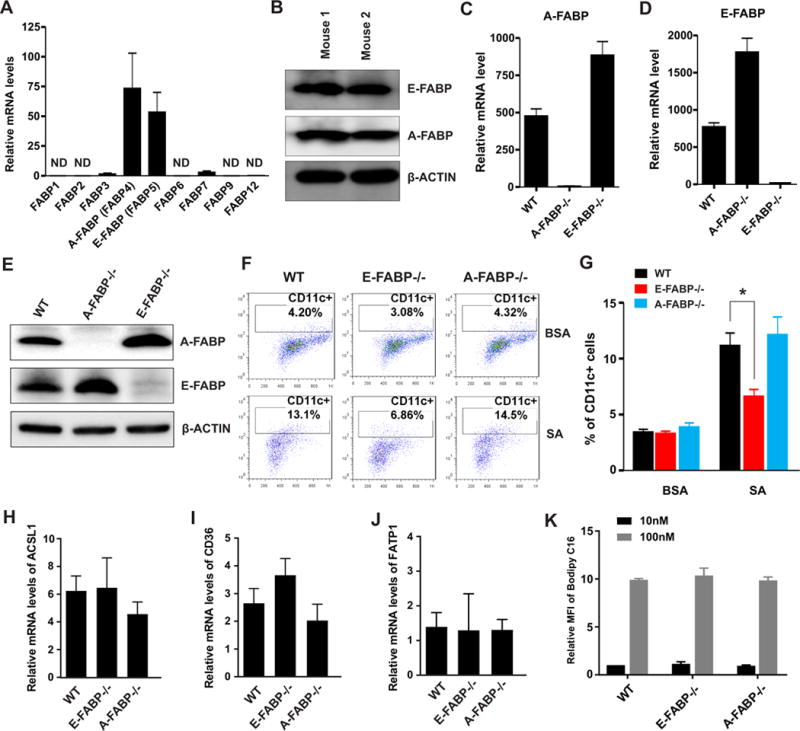
(A-B) Analysis of expression of FABP family members by realtime PCR (A) and western blotting (B) in bone marrow-derived primary macrophages (BMMs) differentiated by M-CFS for 7 days. (C-E) Analyses of A-FABP (C) and E-FABP (D) expression by realtime PCR and Western blotting (E) in differentiated BMMs from WT, A-FABP−/− and E-FABP−/− mice. (F-G) Bone marrow cells collected from WT, A-FABP−/− and E-FABP−/− mice were stimulated with M-CSF in the presence or absence of SA treatment (100μM) for 3 days. CD11c expression on these primary macrophages was analyzed by flow surface staining. Average percentage of CD11c+ macrophages is shown in panel G. (H-J) Analysis of expression of major membrane fatty acid transport proteins, including ACSL1 (H), CD36 (I) and FATP1 (J), in differentiated BMMs by realtime PCR. (K) Flow cytometric analysis of C16-Bodipy uptake by WT, A-FABP−/− and E-FABP−/− BMMs. Data are shown as mean ± SEM (*p<0.05 as compared to the WT macrophages). Experiments are repeated at least three times.
E-FABP deficiency inhibits saturated FA-induced CD11c+ macrophage differentiation in obese mice
Given our in vitro cellular studies that E-FABP expression was critical for SA-induced differentiation of CD11c+ macrophages, we reasoned that HFD-induced CD11c+ macrophage differentiation in different tissues would be compromised in the absence of E-FABP in obese mice in vivo. To this end, WT and E-FABP−/− mice were fed a HFD for 20 weeks to induce obesity. E-FABP deficiency did not impact body weight increase in E-FABP−/− mice compared to WT mice, but when we measured CD11b+CD11c+ macrophages in major organs of these seemingly normal mice, we found that obese E-FABP−/− mice exhibited reduced levels of CD11c+ macrophages in all analyzed tissues compared to obese WT mice, including the bone marrow, liver, lung and skin (Figure 6A–6D). Moreover, serum levels of the pro-inflammatory cytokines IL-6 and IL-1β (but not TNFα) were significantly lower in E-FABP−/− mice than in WT mice (Figure 6E–6G). Interestingly, we also analyzed the impact of A-FABP deficiency on CD11c+ macrophage differentiation and pro-inflammatory cytokine production in obese mice. Consistent with the in vitro studies, A-FABP deficiency did not affect CD11c+ macrophage differentiation (supplemental Figure 2A-2D), nor influence cytokine levels (supplemental Figure 2E-2G). Thus, E-FABP, but A-FABP, plays a critical role in promoting HFD-induced CD11c+ macrophage differentiation and pro-inflammatory cytokine production in obese mice.
Figure 6. E-FABP deficiency reduces CD11c+ macrophages in obese mice.
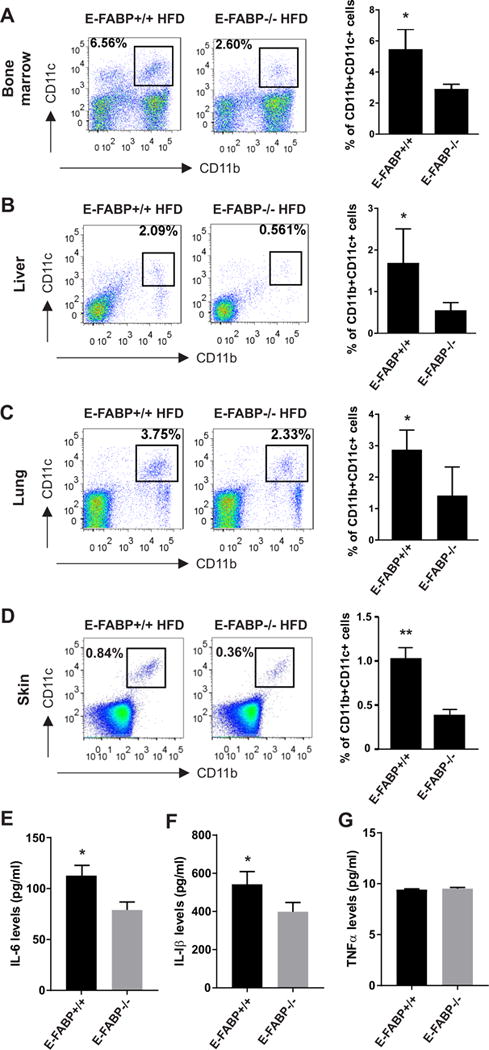
WT and E-FABP−/− mice were fed on HFD (60% fat) for 20 weeks. Different tissues or organs were collected, respectively from the obese WT and E-FABP−/− mice (n=5) for analysis of the presence of CD11b+CD11c+ macrophages. (A-D) Flow cytometric surface staining for analysis of CD11b+CD11c+ macrophages in bone marrow (A), liver (B), lung (C) and skin (D). Average percentage of CD11c+ cells is shown in the right panel. (E-G) Measurement of cytokine levels of IL-6 (E), IL-1β (F) and TNFα (G) in the serum collected from obese WT and E-FABP−/− mice by ELISA. Data shown as mean ± SEM (*p<0.05, **p<0.01), and are representative of at least two independent experiments. Also see Figure S2.
E-FABP deficiency protects mice from development of high saturated FA diet-induced skin lesions
We previously reported that mice fed a HFD (lard) developed skin lesions (6). As the HFD was rich in both saturated (31.8%) and unsaturated FAs (61.9%), it was unclear of which dietary FA components contributed to skin lesion formation. To this end, we fed mice a HFD rich in either saturated FAs (cocoa butter) or unsaturated FA (safflower oil) (supplemental Figure 3A), and observed mouse body weight and skin lesion development. After 6 months, both saturated and unsaturated HFDs significantly increased mouse body weight (from 18.32±0.73g at the beginning to 34.05±2.54g for the cocoa butter diet and from 19±0.72g to 29.01±1.25g for the safflower diet). Although average weight of the cocoa butter group appeared higher than safflower oil group, the weight change between the groups was not statistically different (p=0.167). Of note, both saturated and unsaturated HFDs led to similar weight gain among E-FABP−/− and WT mice (Figure 7A). After WT mice were fed either the cocoa butter or safflower oil HFD for 9 months, 6/16 mice (37.5%) in the cocoa butter group vs. 1/10 (10%) in the safflower oil group developed skin lesions (p<0.01), suggesting that HFD rich in saturated FAs, but not unsaturated FAs, is more pro-inflammatory. By contrast, none of the E-FABP−/− mice in either group developed skin lesions (n=15/group) (Figure 7B). As CD11c+ macrophages are the major pathogenic cells in HFD-induced skin lesions (6), we analyzed this population in skin tissue specimens from these mice. In the cocoa butter fed group, there was massive infiltration of CD11c+ macrophages in the skin of WT, but not E-FABP−/−, mice (Figure 7C, 7D). Accordingly, the expression of MHCII, CD80 and CD86 on CD11c+ macrophages was significantly downregulated in E-FABP−/− mice (Figure 7E–7G). Levels of circulating IL-6 and IL-1β were increased in WT but not E-FABP−/− mice (Figure 7H–7I). Notably, TNFα levels were very low and not increased, suggesting that TNFα does not drive this inflammatory process (Figure 7J). As neither WT nor E-FABP−/− mice that consumed the safflower oil developed significant skin inflammation, it was not surprising that we did not observe obvious accumulation of CD11c+ macrophages in the skin of these mice (supplemental Figure 3B, 3C). In addition, there were no differences in the expression of co-stimulatory molecules on CD11c+ macrophages isolated from the skin of safflower oil-fed WT and E-FABP−/− mice (supplemental Figure 3D-3F), suggesting that unsaturated FAs in the safflower oil diet are not the main factor in the induction of CD11c+ macrophage differentiation and function.
Figure 7. E-FABP deficiency protects high saturated fat-induced skin lesions.
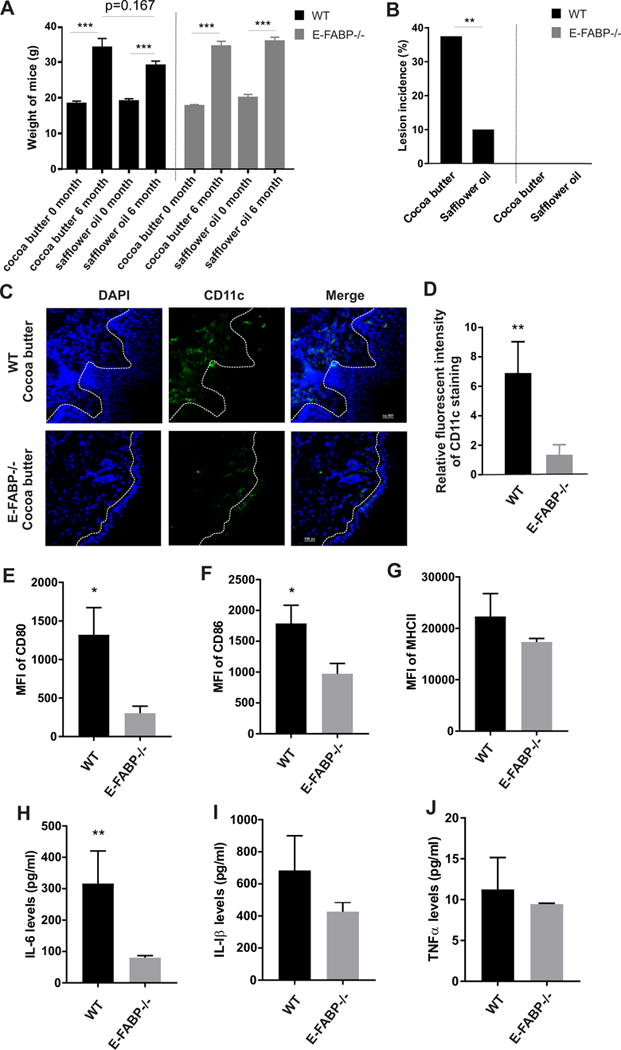
(A) Weight of WT mice and E-FABP−/− mice before and after cocoa butter diet (45% fat) (n=16 for WT mice, and n=15/E-FABP−/− mice) and safflower diet (45% fat) (n=10/WT mice and n=15/E-FABP−/−) for 6 months. (B) The incidence of skin lesions in WT and E-FABP−/− mice fed with cocoa butter diet or safflower oil diet for 9 months. (C-D) Confocal analysis of CD11c+ macrophage infiltration (green color) in the skin tissue of WT and E-FABP−/− mice fed with cocoa butter for 9 months. Relative CD11c fluorescent intensity is shown in panel D. (E-G) Flow cytometric analysis of CD80 (E), CD86 (F) and MHCII (G) expression on CD11c+ macrophages in the skin of WT and E-FABP−/− mice fed with the cocoa butter diet for 9 months. (H-I) Measurement of serum levels of IL-6 (H), IL-1β (I) and TNFα (J) collected from WT and E-FABP−/− mice fed with the cocoa butter diet for 9 months. Data are shown as mean ± SEM (*** p<0.001, ** p<0.01, and * p<0.05). Also see Figure S3.
In another independent experiment comparing WT and A-FABP−/− mice on the same cocoa butter or safflower oil diets, we found that A-FABP deficiency did not affect body weight increases of mice on either diets (supplemental Figure 3G). On the other hand, 40% (6/15) of WT mice on the saturated FA diet compared to 0% on the unsaturated FA diet developed a skin lesion. Interestingly, A-FABP deficiency failed to protect mice against cocoa butter-induced skin lesions (7/14 mice exhibiting skin lesions in the cocoa butter diet). No mice on the unsaturated FA diet development a skin lesion (supplemental Figure 3H). Taken together, our data clearly showed that diets rich in saturated FAs promoted E-FABP-mediated differentiation of CD11c+ macrophages and skin inflammation.
Discussion
Studies in both mice and humans have documented CD11c as a hallmark for pro-inflammatory macrophages in obesity (2,4,5,26). However, the etiology of how CD11c+ macrophages are developed and activated during obesity remains unclear. Although new evidence suggests that lipid uptake and metabolism, rather than inflammatory cytokines, may determine the activation of inflammatory macrophages (27,28), little is known about which species of lipids are critical and how they mechanistically induce pro-inflammatory macrophage activation. Here, we reported that dietary fatty acids, in particular saturated FAs, induced CD11c+ macrophage differentiation in vitro, which contributed to the systematic increase of CD11c+ macrophages in HFD-induced obese mice. Mechanistically, SA metabolized by macrophages was channeled by the cytosolic lipid sensor E-FABP to activate nuclear receptor RAR, thus inducing CD11c transcription. Importantly, deletion of E-FABP reduced the differentiation of CD11c+ macrophages and led to complete abrogation of saturated FA diet-induced skin inflammation in E-FABP−/− mice.
It has long been recognized that obesity is associated with elevated levels of circulating FFAs, which contribute to chronic inflammation, insulin resistance and other metabolic disturbances (7,29,30). Monocytes/macrophages function as professional phagocytes that sense the increase of FFAs through surface lipid receptors, such as CD36 and scavenger receptors, and attempt to clear them via enhanced uptake (31). Questions remain as to how macrophages respond to these overloaded lipids. Macrophages can be polarized in vitro into inflammatory and anti-inflammatory (tissue reparative) phenotypes historically designated as M1 and M2, respectively. However, it is now recognized that macrophages are exposed to a wide variety of environmental triggers in vivo, these strictly polarized phenotypes are rare (32). In the context of obesity, excess FAs or abnormal lipid metabolism play a role in the differentiation and activation of monocytes. For example, it has been shown that obesity induces lysosomal-dependent metabolism of lipids by CD11c+ macrophages independent of inflammatory function typical for “classical” activation (33). Likewise, studies of human adipose tissue macrophages reveal a profile of gene expression distinct from a M1 phenotype, and mimicking the influence of obesity-associated metabolic syndrome by treating macrophages with glucose, insulin, and palmitate resulted in a “metabolically activated” distinct from classical activation (34). Clearly, the influences of various FAs on macrophage differentiation and function are likely to be diverse. In our previous studies, we have demonstrated that high levels of saturated FAs can induce different levels of macrophage cell death depending on FA concentrations and macrophage origins (13). We were curious as to what happened to these FA-exposed but surviving populations. By gating of the live cells exposed to various FA stimulations, we found that myeloid-derived (CD11b+) monocytes could be driven to differentiate into CD11c+ macrophages, in particularly following SA treatment in vitro. The number of CD11b+CD11c+ macrophages is enhanced in various tissues, including lipid-enriched (e.g. visceral fat) and unenriched tissues (e.g. lung), in HFD-induced obese mice in vivo. Thus, our studies suggest a new concept that excess circulating FFAs or abnormal lipid metabolism determine differentiation and activation of monocytes/macrophages during obesity-associated sterile inflammation.
Inside cells water-insoluble fatty acids often form complexes with members of FABP family (35,36). FABPs exhibit tissue specific distribution, whereby macrophages mainly express A-FABP and E-FABP (37,38). Our recent studies further identified that A-FABP and E-FABP are not expressed equally among different subsets of macrophages. While E-FABP is widely expressed in macrophages with the highest levels in the CD11c+ subset, A-FABP is preferentially expressed in the CD11c-CD36+ subset (13,39), suggesting unique roles of individual FABPs in macrophages. In dissecting which FABP members are critical to SA-induced CD11c transcription, we have revealed that E-FABP is responsible for facilitating SA-induced transcriptional activity. Given our previous study showing that A-FABP facilitates saturated FA-induced ceramide production, it is becoming clear that individual FABP members exhibit unique functions in determining the phenotype and differentiation of macrophages in response to environmental lipids.
Once bound to a specific ligand, FABPs are activated to deliver the ligand to the nucleus for assistance of transcriptional regulation. For example, L-FABP can deliver fibrate for transactivation of PPARα (40,41), and A-FABP delivers troglitazone to enhance transcriptional activity of PPARγ (42). Similarly, E-FABP has been shown to transfer retinoic acids and saturated FAs for activation of RAR and PPARβ/δ in mammary tumor cells (43,44). As bone marrow-derived macrophages highly expressed PPARβ/δ, RAR and RXR, we reasoned that they might be involved in the SA-mediated effect in macrophages. When macrophages were treated with specific agonists or antagonists to PPARβ or PPARγ, none of interventions appeared to be critical in saturated FA-induced CD11c upregulation. However, RAR activation by agonist and inhibition by antagonist significantly enhanced and inhibited SA-induced CD11c expression, respectively. Given the evidence that other SA-associated inflammatory and metabolic pathways, including TLR4, NFκB, STAT3, ROS and ceramide production, were not involved in regulation of CD11c expression, it is very likely that E-FABP/RAR axis mediates SA-induced CD11c activation in macrophages. Of note, in resting macrophages RXR may function as a check point to repress CD11c expression, but when monocytes/macrophages are activated by excess SA (e.g. in the setting of obesity), E-FABP-facilitated SA metabolism will co-activate RAR/RXR for CD11c transactivation.
In general, unsaturated FAs are believed to be healthier to consume than saturated FAs since consumption of excess saturated fatty acids increases the risk of cardiovascular disease, type 2 diabetes and some types of cancer (45,46). However, the underlying mechanisms by which saturated FAs increase disease risk remain largely unknown. Our studies provide insight into how saturated FAs negatively affect macrophage biology in several ways. When saturated FAs are taken up by macrophages, they are transported by the cytoplasmic lipid chaperones A-FABP or E-FABP. On one hand, excess saturated FAs delivered by A-FABP can be metabolized for de novo production of ceramides, triggering intrinsic macrophage toxicity. On the other hand, E-FABP is able to mediate saturated FAs-induced CD11c upregulation in macrophages. Unlike saturated FAs, unsaturated FAs are taken up and stored in the form of lipid droplets in macrophages, averting their metabolism for ceramide production and CD11c transactivation (13). The reasons why unsaturated FAs impose less toxicity to macrophages may be multifaceted: 1) uptake of unsaturated FAs increases macrophage membrane fluidity and neutral-lipid formation, which promotes lipid droplet formation; 2) binding of unsaturated FAs alters A-FABP structure for coordinating other lipid-mediated responses (47); and 3) double bonds in unsaturated FAs pose extra requirements for efficient utilization and metabolism of unsaturated FAs in macrophages. Therefore, distinct responses induced by saturated FAs and unsaturated FAs in macrophages may determine their different effects in vivo.
The pathogenic role of CD11c+ macrophages in obesity is well established (2,5). Our previous studies demonstrated that consumption of diets rich in both saturated and unsaturated FAs induced CD11c+ macrophage-mediated skin inflammation (6). To further validate the different effects of saturated vs unsaturated FAs in vivo, we fed mice with diets rich in either saturated (cocoa butter) or unsaturated (safflower oil) FAs and measured CD11c+ macrophage differentiation and skin inflammation. Consistent with our in vitro cellular studies, mice that consumed diets rich in saturated FAs exhibited a significantly higher percentage of CD11c+ macrophages and skin lesions as compared to mice fed the unsaturated FA diet. More strikingly, deletion of E-FABP, but not A-FABP, dramatically reduced the development of high saturated FA diet-induced skin lesions. Thus, the present study demonstrates that consumption of HFD, in particularly high saturated fat diet, promotes skin lesions through inducing differentiation of pro-inflammatory function of CD11c+ macrophages. Recently, we identified specific E-FABP inhibitors through virtual screening and showed the therapeutic efficacy of this inhibitor in suppression of inflammation in an autoimmune disease model (48). It will be of great interest to test if E-FABP inhibition reduces obesity-associated skin inflammation.
In summary, the current study identified a novel mechanism by which saturated FAs promote the differentiation of CD11c+ macrophages through the E-FABP/RAR axis. This finding not only provides a scientific explanation for harmful effects of consuming excess saturated FAs, but suggests E-FABP as a new target for the treatment of obesity-related chronic inflammation and other diseases.
Supplementary Material
Acknowledgments
Grant Support
We thank Dr. Hyeran Jang for assistance with design of the custom rodent diets made by Research Diets, Inc. This work was supported partially by the University of Louisville start-up funds and grants R01CA177679, R01CA180986 funded by the National Cancer Institute (Bethesda, MD).
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors’ Contributions
B.L. conceived ideas and designed the experiments. J.Z., Y.Z., J.H., and Y.W. performed the experiments; J.Z., Y.Z., J.H. and B.L. analyzed the data. Y.S., and Y.W helped with obese mouse models. S.J., D.A.B., E.R.S., M.P.C., J.S. and B.L. wrote the manuscript with inputs from all authors. B.L. oversaw the entire research project.
References
- 1.Finkelstein EA, Khavjou OA, Thompson H, Trogdon JG, Pan L, Sherry B, Dietz W. Obesity and severe obesity forecasts through 2030. Am J Prev Med. 2012;42:563–570. doi: 10.1016/j.amepre.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 2.Lumeng CN, Bodzin JL, Saltiel AR. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J Clin Invest. 2007;117:175–184. doi: 10.1172/JCI29881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakajima S, Koh V, Kua LF, So J, Davide L, Lim KS, Petersen SH, Yong WP, Shabbir A, Kono K. Accumulation of CD11c+CD163+ Adipose Tissue Macrophages through Upregulation of Intracellular 11beta-HSD1 in Human Obesity. J Immunol. 2016 doi: 10.4049/jimmunol.1600895. [DOI] [PubMed] [Google Scholar]
- 4.Wentworth JM, Naselli G, Brown WA, Doyle L, Phipson B, Smyth GK, Wabitsch M, O’Brien PE, Harrison LC. Pro-inflammatory CD11c+CD206+ adipose tissue macrophages are associated with insulin resistance in human obesity. Diabetes. 2010;59:1648–1656. doi: 10.2337/db09-0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patsouris D, Li PP, Thapar D, Chapman J, Olefsky JM, Neels JG. Ablation of CD11c-positive cells normalizes insulin sensitivity in obese insulin resistant animals. Cell Metab. 2008;8:301–309. doi: 10.1016/j.cmet.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y, Li Q, Rao E, Sun Y, Grossmann ME, Morris RJ, Cleary MP, Li B. Epidermal Fatty Acid binding protein promotes skin inflammation induced by high-fat diet. Immunity. 2015;42:953–964. doi: 10.1016/j.immuni.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boden G. Obesity and free fatty acids. Endocrinol Metab Clin North Am. 2008;37:635–6ix. doi: 10.1016/j.ecl.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Almeida IT, Cortez-Pinto H, Fidalgo G, Rodrigues D, Camilo ME. Plasma total and free fatty acids composition in human non-alcoholic steatohepatitis. Clin Nutr. 2002;21:219–223. doi: 10.1054/clnu.2001.0529. [DOI] [PubMed] [Google Scholar]
- 9.Boden G. Obesity, insulin resistance and free fatty acids. Curr Opin Endocrinol Diabetes Obes. 2011;18:139–143. doi: 10.1097/MED.0b013e3283444b09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furuhashi M, Hotamisligil GS. Fatty acid-binding proteins: role in metabolic diseases and potential as drug targets. Nat Rev Drug Discov. 2008;7:489–503. doi: 10.1038/nrd2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hertzel AV, Bernlohr DA. The mammalian fatty acid-binding protein multigene family: molecular and genetic insights into function. Trends Endocrinol Metab. 2000;11:175–180. doi: 10.1016/s1043-2760(00)00257-5. [DOI] [PubMed] [Google Scholar]
- 12.Storch J, Corsico B. The emerging functions and mechanisms of mammalian fatty acid-binding proteins. Annu Rev Nutr. 2008;28:73–95. doi: 10.1146/annurev.nutr.27.061406.093710. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, Rao E, Zeng J, Hao J, Sun Y, Liu S, Sauter ER, Bernlohr DA, Cleary MP, Suttles J, Li B. Adipose Fatty Acid Binding Protein Promotes Saturated Fatty Acid-Induced Macrophage Cell Death through Enhancing Ceramide Production. J Immunol. 2017;198:798–807. doi: 10.4049/jimmunol.1601403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Hao J, Sun Y, Li B. Saturated Fatty Acids Induce Ceramide-associated Macrophage Cell Death. J Vis Exp. 2017 doi: 10.3791/56535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Rao E, Zeng J, Hao J, Sun Y, Liu S, Sauter ER, Bernlohr DA, Cleary MP, Suttles J, Li B. Adipose Fatty Acid Binding Protein Promotes Saturated Fatty Acid-Induced Macrophage Cell Death through Enhancing Ceramide Production. J Immunol. 2017;198:798–807. doi: 10.4049/jimmunol.1601403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Sun Y, Rao E, Yan F, Li Q, Zhang Y, Silverstein KA, Liu S, Sauter E, Cleary MP, Li B. Fatty acid-binding protein E-FABP restricts tumor growth by promoting IFN-beta responses in tumor-associated macrophages. Cancer Res. 2014;74:2986–2998. doi: 10.1158/0008-5472.CAN-13-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Epelman S, Lavine KJ, Randolph GJ. Origin and functions of tissue macrophages. Immunity. 2014;41:21–35. doi: 10.1016/j.immuni.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li P, Lu M, Nguyen MT, Bae EJ, Chapman J, Feng D, Hawkins M, Pessin JE, Sears DD, Nguyen AK, Amidi A, Watkins SM, Nguyen U, Olefsky JM. Functional heterogeneity of CD11c-positive adipose tissue macrophages in diet-induced obese mice. J Biol Chem. 2010;285:15333–15345. doi: 10.1074/jbc.M110.100263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomez PE, Klapproth K, Schulz C, Busch K, Azzoni E, Crozet L, Garner H, Trouillet C, de Bruijn MF, Geissmann F, Rodewald HR. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature. 2015;518:547–551. doi: 10.1038/nature13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong SW, Kwon MJ, Choi AM, Kim HP, Nakahira K, Hwang DH. Fatty acids modulate Toll-like receptor 4 activation through regulation of receptor dimerization and recruitment into lipid rafts in a reactive oxygen species-dependent manner. J Biol Chem. 2009;284:27384–27392. doi: 10.1074/jbc.M109.044065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leamy AK, Egnatchik RA, Young JD. Molecular mechanisms and the role of saturated fatty acids in the progression of non-alcoholic fatty liver disease. Prog Lipid Res. 2013;52:165–174. doi: 10.1016/j.plipres.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weigert C, Brodbeck K, Staiger H, Kausch C, Machicao F, Haring HU, Schleicher ED. Palmitate, but not unsaturated fatty acids, induces the expression of interleukin-6 in human myotubes through proteasome-dependent activation of nuclear factor-kappaB. J Biol Chem. 2004;279:23942–23952. doi: 10.1074/jbc.M312692200. [DOI] [PubMed] [Google Scholar]
- 23.Georgiadi A, Kersten S. Mechanisms of gene regulation by fatty acids. Adv Nutr. 2012;3:127–134. doi: 10.3945/an.111.001602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le MA, Alvarez S, Shankaranarayanan P, Lera AR, Bourguet W, Gronemeyer H. Retinoid receptors and therapeutic applications of RAR/RXR modulators. Curr Top Med Chem. 2012;12:505–527. doi: 10.2174/156802612799436687. [DOI] [PubMed] [Google Scholar]
- 25.Lee CW, Kim JE, Do H, Kim RO, Lee SG, Park HH, Chang JH, Yim JH, Park H, Kim IC, Lee JH. Structural basis for the ligand-binding specificity of fatty acid-binding proteins (pFABP4 and pFABP5) in gentoo penguin. Biochem Biophys Res Commun. 2015;465:12–18. doi: 10.1016/j.bbrc.2015.07.087. [DOI] [PubMed] [Google Scholar]
- 26.Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW., Jr Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112:1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu X, Grijalva A, Skowronski A, van E M, Serlie MJ, Ferrante AW., Jr Obesity activates a program of lysosomal-dependent lipid metabolism in adipose tissue macrophages independently of classic activation. Cell Metab. 2013;18:816–830. doi: 10.1016/j.cmet.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kratz M, Coats BR, Hisert KB, Hagman D, Mutskov V, Peris E, Schoenfelt KQ, Kuzma JN, Larson I, Billing PS, Landerholm RW, Crouthamel M, Gozal D, Hwang S, Singh PK, Becker L. Metabolic dysfunction drives a mechanistically distinct proinflammatory phenotype in adipose tissue macrophages. Cell Metab. 2014;20:614–625. doi: 10.1016/j.cmet.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bjorntorp P, Bergman H, Varnauskas E. Plasma free fatty acid turnover rate in obesity. Acta Med Scand. 1969;185:351–356. doi: 10.1111/j.0954-6820.1969.tb07347.x. [DOI] [PubMed] [Google Scholar]
- 30.Jensen MD, Haymond MW, Rizza RA, Cryer PE, Miles JM. Influence of body fat distribution on free fatty acid metabolism in obesity. J Clin Invest. 1989;83:1168–1173. doi: 10.1172/JCI113997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore KJ, Sheedy FJ, Fisher EA. Macrophages in atherosclerosis: a dynamic balance. Nat Rev Immunol. 2013;13:709–721. doi: 10.1038/nri3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, Gordon S, Hamilton JA, Ivashkiv LB, Lawrence T, Locati M, Mantovani A, Martinez FO, Mege JL, Mosser DM, Natoli G, Saeij JP, Schultze JL, Shirey KA, Sica A, Suttles J, Udalova I, van Ginderachter JA, Vogel SN, Wynn TA. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu X, Grijalva A, Skowronski A, van E M, Serlie MJ, Ferrante AW., Jr Obesity activates a program of lysosomal-dependent lipid metabolism in adipose tissue macrophages independently of classic activation. Cell Metab. 2013;18:816–830. doi: 10.1016/j.cmet.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kratz M, Coats BR, Hisert KB, Hagman D, Mutskov V, Peris E, Schoenfelt KQ, Kuzma JN, Larson I, Billing PS, Landerholm RW, Crouthamel M, Gozal D, Hwang S, Singh PK, Becker L. Metabolic dysfunction drives a mechanistically distinct proinflammatory phenotype in adipose tissue macrophages. Cell Metab. 2014;20:614–625. doi: 10.1016/j.cmet.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smathers RL, Petersen DR. The human fatty acid-binding protein family: evolutionary divergences and functions. Hum Genomics. 2011;5:170–191. doi: 10.1186/1479-7364-5-3-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Storch J, McDermott L. Structural and functional analysis of fatty acid-binding proteins. J Lipid Res. 2009;50(Suppl):S126–S131. doi: 10.1194/jlr.R800084-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li B, Reynolds JM, Stout RD, Bernlohr DA, Suttles J. Regulation of Th17 differentiation by epidermal fatty acid-binding protein. J Immunol. 2009;182:7625–7633. doi: 10.4049/jimmunol.0804192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reynolds JM, Liu Q, Brittingham KC, Liu Y, Gruenthal M, Gorgun CZ, Hotamisligil GS, Stout RD, Suttles J. Deficiency of fatty acid-binding proteins in mice confers protection from development of experimental autoimmune encephalomyelitis. J Immunol. 2007;179:313–321. doi: 10.4049/jimmunol.179.1.313. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y, Li B. E-FABP: regulator of immune function. Oncoscience. 2014;1:398–399. doi: 10.18632/oncoscience.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petrescu AD, Huang H, Martin GG, McIntosh AL, Storey SM, Landrock D, Kier AB, Schroeder F. Impact of L-FABP and glucose on polyunsaturated fatty acid induction of PPARalpha-regulated beta-oxidative enzymes. Am J Physiol Gastrointest Liver Physiol. 2013;304:G241–G256. doi: 10.1152/ajpgi.00334.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hostetler HA, McIntosh AL, Atshaves BP, Storey SM, Payne HR, Kier AB, Schroeder F. L-FABP directly interacts with PPARalpha in cultured primary hepatocytes. J Lipid Res. 2009;50:1663–1675. doi: 10.1194/jlr.M900058-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ayers SD, Nedrow KL, Gillilan RE, Noy N. Continuous nucleocytoplasmic shuttling underlies transcriptional activation of PPARgamma by FABP4. Biochemistry. 2007;46:6744–6752. doi: 10.1021/bi700047a. [DOI] [PubMed] [Google Scholar]
- 43.Schug TT, Berry DC, Shaw NS, Travis SN, Noy N. Opposing effects of retinoic acid on cell growth result from alternate activation of two different nuclear receptors. Cell. 2007;129:723–733. doi: 10.1016/j.cell.2007.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levi L, Wang Z, Doud MK, Hazen SL, Noy N. Saturated fatty acids regulate retinoic acid signalling and suppress tumorigenesis by targeting fatty acid-binding protein 5. Nat Commun. 2015;6:8794. doi: 10.1038/ncomms9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hooper L, Summerbell CD, Thompson R, Sills D, Roberts FG, Moore H, Davey SG. Reduced or modified dietary fat for preventing cardiovascular disease. Cochrane Database Syst Rev. 2011:CD002137. doi: 10.1002/14651858.CD002137.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Makarem N, Chandran U, Bandera EV, Parekh N. Dietary fat in breast cancer survival. Annu Rev Nutr. 2013;33:319–348. doi: 10.1146/annurev-nutr-112912-095300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gillilan RE, Ayers SD, Noy N. Structural basis for activation of fatty acid-binding protein 4. J Mol Biol. 2007;372:1246–1260. doi: 10.1016/j.jmb.2007.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rao E, Singh P, Li Y, Zhang Y, Chi YI, Suttles J, Li B. Targeting epidermal fatty acid binding protein for treatment of experimental autoimmune encephalomyelitis. BMC Immunol. 2015;16:28. doi: 10.1186/s12865-015-0091-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


