Abstract
Hox genes can function as key drivers of segment identity, with Hox mutations in Drosophila often resulting in dramatic homeotic transformations. In addition, however, they can serve other essential functions. In mammals, the study of Hox gene roles in development is complicated by the presence of four Hox clusters with a total of 39 genes showing extensive functional overlap. In this study, in order to better understand shared core Hox functions, we examined kidney development in mice with frameshift mutations of multiple Abd-B type Hox genes. The resulting phenotypes included dramatically reduced branching morphogenesis of the ureteric bud, premature depletion of nephron progenitors and abnormal development of the stromal compartment. Most unexpected, however, we also observed a cellular level lineage infidelity in nephron segments. Scattered cells within the proximal tubules, for example, expressed genes normally expressed only in collecting ducts. Multiple combinations of inappropriate nephron segment specific marker expression were found. In some cases, cells within a tubule showed incorrect identity, while in other cases cells showed ambiguous character, with simultaneous expression of genes associated with more than one nephron segment. These results give evidence that Hox genes have an overlapping core function at the cellular level in driving and/or maintaining correct differentiation decisions.
Keywords: Hox genes, kidney development, lineage infidelity, epigenetic
INTRODUCTION
Hox genes gained early notoriety because of their apparent master switch roles during development. Mutation of a single Hox gene in Drosophila can cause an entire developing segment to undergo a homeotic transformation of identity (Lewis, 1978). One particularly dramatic example is the Antennapedia mutation, which results in legs forming on the head in place of antennae (Schneuwly et al., 1987). It was proposed that combinatorial codes of Hox transcription factor expression can determine segment identity (Kessel and Gruss, 1991).
In mammals mutation of a single Hox gene generally results in a much milder phenotype, likely due in part to the presence of four Hox clusters with a total of 39 functionally redundant genes. Hox genes within a paralogous group show the greatest functional similarity, as they are duplicated from a single gene on an ancestral Hox cluster. Of interest, the most 5′ paralog groups (Hox9-13) are particularly closely related and are all thought to be derived from a single ancestral Abd-B type Hox gene. Mutation of multiple Hox genes of a single paralogous group can give homeotic transformations of axial body segments (Horan et al., 1995; McIntyre et al., 2007; Wellik and Capecchi, 2003), hindbrain rhombomeres (Studer et al., 1996; Zhang et al., 1994), and reproductive tract segments (Raines et al., 2013; Small and Potter, 1993). While paralogous Hox genes on different Hox clusters, such as Hoxa11 and Hoxd11, show the greatest functional overlap there is also abundant evidence that adjacent Hox genes, next to each other on a single cluster, are partially redundant. For example, Hoxa10/Hoxa11 trans-heterozygotes show a synergistic phenotype (Branford et al., 2000), and double mutation of the flanking paralog Hoxa10 and Hoxd11 genes shows their functional overlap (Favier et al., 1996). In addition, homeobox swap experiments show partial functional equivalence for adjacent Hox genes (Zhao and Potter, 2001, 2002).
Hox genes have also been proposed to have more evolutionarily primitive functions that are distinct from their involvement in segment identity determination (Hombria and Lovegrove, 2003). Hox genes can regulate cell death (Lohmann et al., 2002), cell proliferation (Bromleigh and Freedman, 2000; Care et al., 1999), and in some cases Hox mutations result in defective cellular differentiation, unrelated to homeotic transformation (Liu and Fire, 2000; Ponzielli et al., 2002). Of particular interest, in the Drosophila larval fat body Hox genes have a shared role in the repression of autophagy (Banreti et al., 2014). The strongly overlapping expression of Hox genes, with shared anterior limits, suggested a function distinct from segment identity determination in the larval fat body. It was shown that repression of all expressed Hox genes was required to initiate normal autophagy during development, while persistent expression of any single Hox gene could curb this process (Banreti et al., 2014).
We have previously shown that there is widespread overlapping Hox gene expression during kidney development (Patterson and Potter, 2004). Indeed, only the most extreme 3′ and 5′ paralog groups, Hox1 and Hox13, are not expressed. Surprisingly, however, kidney developmental defects have only been seen, with rare exception, in mice with multiple mutations in either the Hox10 or Hox11 paralog groups (Davis et al., 1995; Patterson et al., 2001; Wellik et al., 2002; Yallowitz et al., 2011). Mutation of Hoxa11 (Small and Potter, 1993) or Hoxd11 (Davis and Capecchi, 1994) alone gives normal kidneys, while mutation of both results in hypoplastic kidneys (Davis et al., 1995; Patterson et al., 2001) and mutation of all three Hox11 paralogs (Hoxa11, Hoxc11, Hoxd11) completely blocks an initial stage of kidney formation, the outgrowth of the ureteric bud from the nephric duct (Wellik et al., 2002). Hox10 genes are required for proper stromal compartment development (Yallowitz et al., 2011).
It has been observed, in general, that simultaneous mutation of an increased number of closely related Hox genes reveals functions previously concealed by redundancy. One strategy for the mutation of more Hox gene combinations would be to use Cre/Lox to delete blocks of flanking Hox genes from a cluster, but it has been shown that this removes intergenic regional shared enhancers, which results in the misexpression of remaining Hox genes, making interpretation difficult (Di-Poi et al., 2007). Indeed it is possible to use Cre/Lox to remove entire Hox clusters, but unexpectedly this can actually give a phenotype that is milder than the mutation of a single gene within the cluster (Suemori and Noguchi, 2000). This can be due to noncoding RNA crosstalk between Hox clusters that results in compensatory elevated expression of remaining Hox clusters (Rinn et al., 2007). Another approach to the generation of multi-Hox mutants would be to use CRISPR/Cas9 to simultaneously frameshift multiple adjoining Hox genes. This would be challenging, however, since the introduction of nearby double strand breaks by CRISPR/Cas9, when mutating adjacent genes, most often gives deletions.
We previously made mice with mutations of the six Abd-B type Hoxa9,10,11 and the Hoxd9,10,11 genes and described the resulting reproductive tract (Raines et al., 2013) and limb (Raines et al., 2015) malformations. We used a recombineering method, with BAC targeting constructs of over 100 Kb, that allows simultaneous frameshift mutation of multiple flanking genes, leaving shared enhancers intact (Raines et al., 2013). In this report, we show that mice with combined mutation of these six flanking and paralogous Hox genes exhibit a spectrum of kidney development defects, including hypoplasia, dysplasia, and renal agenesis. These results confirm previous work showing the role of Hox genes in determining nephron number by controlling cap mesenchyme (CM) progenitor cell maintenance and differentiation as well as uretic bud (UB) branching (Patterson et al., 2001; Wellik et al., 2002) and better define the redundant relationships of the flanking and paralogous Hox9,10,11 genes in regulating these processes. Of particular interest, however, kidneys with severe loss of Abd-B Hox gene core function showed lineage infidelity. The tubules of the kidney are divided into segments, including the proximal tubules, loop of Henle, distal tubules and collecting duct. The Hoxa9,10,11/Hoxd9,10,11−/− mutant kidneys, with homozygous mutation in six closely related Hox genes, were examined for possible homeotic transformation of nephron segment identity. Unexpectedly, mutant kidney tubules displayed a striking cellular level lineage infidelity. All of the distinct nephron segments were present but they included cells with inappropriate identity, with some simply expressing markers of another tubule segment, while others showed ambiguous differentiation, with simultaneous expression of genes associated with more than one segment. These results give evidence for a novel epigenetic role for Hox genes in helping to drive and/or maintain correct differentiation decisions.
Materials and Methods
Mice
Mice with frame shift mutations in Hoxa9,10,11 and Hoxd9,10,11 were previously described (Raines et al., 2013). The genotypes of adult and embryonic mice were determined by PCR using Direct-PCR lysis reagent (Viagen Biotech) and proteinase K (Sigma-Aldrich) as previously described (Raines et al., 2013). All experiments were carried out with humane protocols in accordance with relevant guidelines and regulations and approved by the Institutional Animal Care and Use Committee (protocol number 2015-0065).
Immunohistochemistry
Wild type and aadd samples for sectioned and whole mount immunofluorescence (IF) were collected and fixed in 4%PFA in PBS at 4°C overnight. Samples to be used for sectioned IF were further processed as previously described (Raines et al 2015). Samples to be used for whole mount IF were dehydrated into methanol and kept at −20°C at least overnight. Whole mount samples were then rehydrated to PBS, incubated with primary antibodies for three days at 4°C, washed three times with PBS, incubated with secondary antibodies for three days at 4°C, and washed three time with PBS. Samples were then cleared for confocal imaging using RIMS (Yang et al., 2014). Primary antibodies against Calbindin (Sigma C9848 1:200), SIX2 (Proteintech 11562-1-AP 1:200), MAFB (Santa Cruz SC-10022 1:50), CRABP1 (Cell Signaling D7F9T 131635 1:200), LTA (Vector Labs LT-1321 1:100), KRT8 (Millipore Troma-1-s 1:50), AlLDH1A2 (Abcam AB96060 1:200), LEF1 (GTX1203 1:200), P57 (Thermo Fisher MS-897-P0 1:200), SLC12A1 (ProteinTech 18970-1-AB 1:200), SLC12A3 (Sigma HPA028748 1:200), and DBA (VectorLabs RL-1032 1:100) were used. Alexa fluor secondary antibodies against Mouse, Rabbit, Goat, and Rat were used as appropriate.
Results and discussion
Hox gene expression in the developing kidney
We have previously described the expression patterns of the thirty nine Hox genes in the developing kidney using radioactive in situ hybridizations (Patterson and Potter, 2004). Of interest, almost all Hox genes, except members of the most terminal Hox1 and Hox13 paralog groups, showed expression during this process. Further, we noted that the Hox genes of a single cluster showed more similar expression patterns than genes of a single paralogous group, suggesting that shared enhancers on clusters drove expression. Finally, no clear compartment or segment specific Hox gene expression codes emerged that might drive distinct differentiation directions.
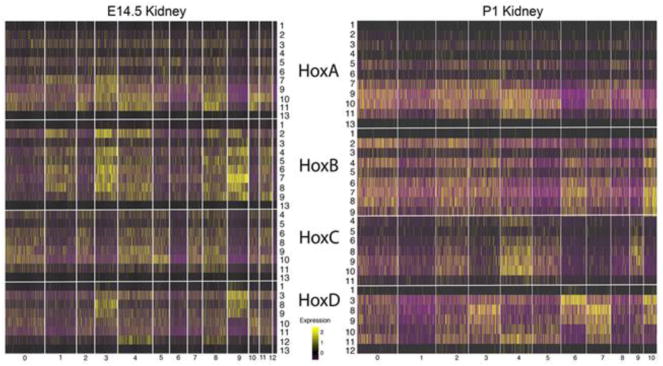
More recently we have used single cell RNA-seq (scRNA-Seq) to examine gene expression patterns in the E14.5 and early postnatal P1 mouse kidney (Adam et al., 2017; Magella et al., 2017). ScRNA-seq offers several advantages over radioactive in situ hybridizations, including single cell resolution and quantitative measure of gene expression levels. These scRNA-Seq datasets can be used to better define Hox gene expression patterns in the developing kidney (Fig. 1). Confirming previous in situ hybridization results, the genes of the Hox1 and Hox13 paralog groups showed very low expression levels. In addition, there was again a strong trend for all Hox genes of a single cluster to show strongly overlapping expression domains. For example, in the E14.5 kidney, almost all of the expressed genes of the HoxB cluster, including members of many different paralog groups (2,3,4,5,6,7,8,), march in lockstep, showing very similar expression patterns (Fig. 1). All HoxB genes show low expression in cell types 0, 2, 5, 6, 7, 11, 13, 14 (stroma, endothelium, podocytes, immune cells), and show higher expression in cell types 1, 3, 4, 8, 9,12 (differentiating nephrons, including renal vesicles, comma and S-shaped bodies, loop of Henle, cap mesenchyme progenitors, collecting duct). HoxA cluster genes also showed widespread and overlapping domains of expression, but in this case with quite different levels of expression for the different paralog genes, with Hoxa7,9,10,11 showing the strongest expression. Almost all cell types showed significant expression of HoxA genes. The most notable exception was type 9 cells (collecting duct), where no HoxA genes were strongly expressed. Similarly, in the E14.5 kidney the HoxC genes showed very strongly overlapping domains of for all expressed paralogs (Hoxc4,5,6,8,9,10,11). In general, if a specific cell type expressed one HoxC gene then it expressed the other HoxC paralog genes as well. Only the HoxD genes showed significant paralog specific expression patterns, in particular within the loop of Henle (type 3), cap mesenchyme (type 4, 8) and collecting duct (type 9). These patterns of Hox expression observed in the E14.5 kidneys were strongly replicated in the later P1 kidneys, providing an important measure of validation (Fig. 1). In summary, expression patterns for the HoxA, B and C genes in the developing kidney appeared more driven by the Hox cluster than paralog group. This result is quite anti-dogmatic, as paralogous Hox genes have generally been observed to show the most similar expression domains during development. These results, therefore, give evidence for Hox functions in the developing kidney that are distinct from their well-known paralog specific Hox Code roles in driving segment identity determination.
Fig. 1. Hox gene expression in the developing kidney shows little evidence for paralog Hox codes.
The expression patterns for all Hox genes in the compartments of the early developing E14.5 and early postnatal (P1) kidney. The extreme terminal Hox genes from paralog groups 1 and 13 showed very low expression. For the HoxA, HoxB and HoxC clusters all of the genes of a given cluster showed very similar expression patterns, although with varying expression levels. For example, almost all of the HoxB genes, including paralog groups 1,2,3,4,5,6,7,8,9,13, showed stronger expression in cell types 1,3,4,8,9 and 11 and weaker expression in the other compartments. The HoxD cluster was an exception, with paralog Hox gene specific expression patterns in cell types 3,4,5,9 and 10. Cell type identities for E14.5 are 0, Stroma: 1, Differentiating Nephrons: 2, Medullary Stroma: 3, Loop of Henle: 4, Cap Mesenchyme: 5, Stromal Progenitors: 6, Endothelium: 7, Podocytes: 8, Cap Mesenchyme: 9, Collecting Duct: 10, Cortical Stroma: 11, Cap Mesenchyme: 12, Immune Cells. P1 compartments are as follows: 0, Nephron Progenitors: 1, Proximal Tubule: 2, Early Proximal Tubule: 3, Loop of Henle: 4, Cap Mesenchyme: 5, Stroma: 6, Collecting Duct: 7, Distal Tubule: 8, Endothelium: 9, Podocytes: 10, Collecting Duct Tip Cells. Only Hox genes with some measured expression in the developing kidney are included. Genes with no detected expression included at E14.5 Hoxc12 and Hoxd4, and at P1 Hoxb13, Hoxc12,13, and Hoxd4,13.
In order to find possible Hox gene core functions previously concealed by redundancy we examined kidney development in mice with the simultaneous frameshift mutation of six Hox genes, Hoxa9,10,11 and Hoxd9,10,11 (Raines et al., 2015). These are all very closely related Abd-B type Hox genes, they show some of the strongest Hox gene expression levels in the developing kidney, and their expression patterns are overlapping in multiple cell types, all consistent with functional redundancy. In addition, as noted earlier, the Hox10 and Hox11 paralog genes have previously defined kidney development functions, although they have not been heretofore examined with combinations of both paralog and flanking gene mutations. Perhaps surprising, no kidney development function has been reported for the Hox9 genes, even following mutation of all four paralogs (Xu and Wellik, 2011), likely the result of the continued presence of the functionally redundant Hox10 and Hox11 genes.
In sum, the mutation of Hoxa9,10,11 and Hoxd9,10,11 removes functionality for six quite closely related Abd-B type Hox genes yet preserves enough Hox11 paralog function, through the continued presence of Hoxc11, to overcome the early block in kidney development that occurs when all Hox11 paralogs are mutated (Wellik et al., 2002). All of these genes are thought to be derived from a single ancestral Abd-B Hox gene. The goal of this study was to test for possible Abd-B Hox gene core functions that were previously hidden by functional redundancies.
Hoxa9,10,11/Hoxd9,10,11 mutation results in hypoplastic kidneys with reduced ureteric bud branching morphogenesis
Not unexpected, some aspects of the Hoxa9,10,11/Hoxd9,10,11 mutant phenotype were in accord with previously described Hox mutant kidneys. Mutation of only Hoxa11/Hoxd11 results in a hypoplastic kidney with reduced branching morphogenesis of the ureteric bud, likely a result of decreased Gdnf expression by the metanephric mesenchyme/cap mesenchyme (Patterson et al., 2001). In the triple Hoxa11, Hoxc11, Hoxd11 mutant, with all three Hox11 paralog genes mutated, the UB fails to form, with no detectable Gdnf expression by the metanephric mesenchyme (Wellik et al., 2002). In this study, we observed that in Hoxa9,10,11/Hoxd9,10,11 mutants the UB forms but subsequently undergoes severely reduced branching morphogenesis (Fig. S1), similar to Hoxa11/Hoxd11 mutants. Also, similar to the Hoxa11/Hoxd11 mutants, the Hoxa9,10,11/Hoxd9,10,11 mutant kidneys are hypoplastic at birth and unable to support postnatal life.
Hoxa9,10,11/Hoxd9,10,11 mutant kidneys show early depletion of nephron progenitors
During kidney development the progenitors that give rise to all epithelial components of the nephron reside in the cap mesenchyme, overlying the ureteric bud tips (Boyle et al., 2008; Kobayashi et al., 2008). They are marked by the expression of a number of genes, including Six2 and Cited1 (Costantini and Kopan, 2010). These progenitors self renew and normally persist until a few days after birth, when they undergo differentiation in a final round of nephrogenesis (Hartman et al., 2007; Short et al., 2014). Of interest, the Hoxa9,10,11/Hoxd9,10,11 mutant kidney cap mesenchyme progenitors show precocious and severe reduction in number by E14.5 (Fig. 2). There was some variable expressivity, but at E14.5 we typically observed very few nephron progenitors, and by E16.5 we only rarely observed any remaining clusters of these cells (Fig. 2). This further confirms a critical role for Abd-B Hox genes in the regulation of nephron progenitor renewal/maintenance. The continued expression of Six2 is required to prevent premature differentiation of nephron progenitors (Self et al., 2006), and Hox11 paralogs proteins are known to complex with Eya1 and Pax2 to drive expression of both Six2 and Gdnf (Gong et al., 2007; Wellik et al., 2002).
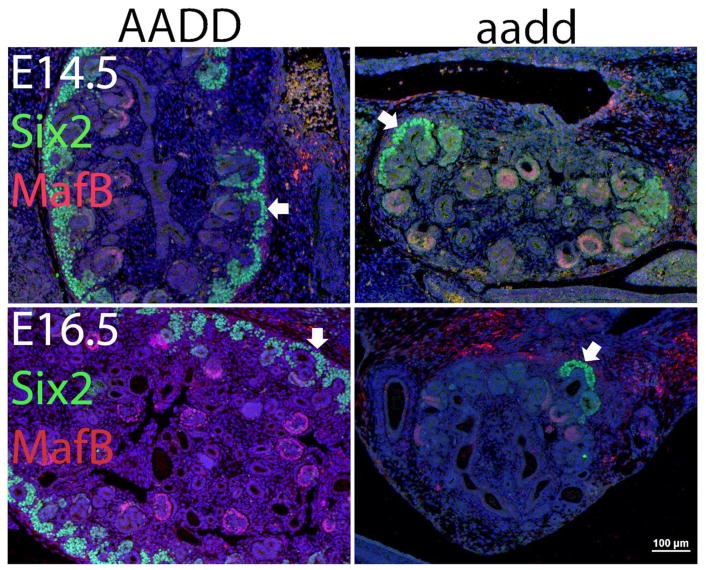
Fig. 2. Nephron progenitors show premature depletion.
At E14.5 and E16.5 the nephron progenitors in wild type kidneys show strong SIX2 (green) labeling. In the Hoxa9,10,11/Hoxd9,10,11 homozygous mutants (aadd) nephron progenitors are rare by E14.5 (top right panel) and with only an occasional cluster of remaining cells at E16.5 (bottom right panel). MAFBP (purple) marks the podocytes in the forming glomeruli (red arrowheads). Glomeruli do form in the aadd mutant kidney but are present at greatly reduced numbers at E16.5 compared to wild type. There was variable expressivity, with some mutant kidneys showing more severe phenotypes than others.
Stromal cells in Hoxa9,10,11/Hoxd9,10,11 mutant kidneys
The cells intertwined between the nephrons and vascular system are referred to as the stroma, or interstitium. There is a population of self renewing multipotent stromal progenitors located within the outer cortex of the developing kidney that give rise to the mesangial cells of the glomerulus, the renin expressing cells of the juxtaglomerular apparatus, the erythropoietin expressing cells flanking the proximal tubules, pericytes and pericyte like myofibroblasts (McMahon, 2016).
The stromal cells also play critical roles in promoting nephrogenesis. Foxd1 is a classic marker gene of stromal progenitor cells. The Foxd1 mutant mouse shows >90% reduction in nephron number, with the cortex having large aggregates of undifferentiated cap mesenchyme cells (Hatini et al., 1996). Stromal cells provide Fat4, required to promote nephron progenitor commitment (Bagherie-Lachidan et al., 2015; Das et al., 2013; Reginensi et al., 2013), and Decorin, which promotes nephron progenitor differentiation by inhibiting Bmp7 (Fetting et al., 2014). Further, it has recently been shown that stromal cells are also a source of GNDF, in addition to the cap mesenchyme (Magella et al., 2017).
In the Hoxa9,10,11/Hoxd9,10,11 mutants the stromal progenitor population persisted, and indeed appeared proportionally over represented at E14.5 (Fig. 3). The layer of stromal progenitors at the outer cortex of the mutant kidney was often thickened (Fig. 3). A Foxd1 positive and therefore presumed stromal cell progenitor population remained even at E18.5, but in a very limited cortical domain (data not shown). It is interesting to note that kidney development appears normal at E11.5, in particular with a normal endowment of nephron progenitors (data not shown), indicating that the disproportionate representation of stromal cells at later times results from disturbed nephrogenesis and not an earlier patterning defect. The simplest explanation is that expansion is more severely impaired for nephron progenitors compared to stromal progenitors. This differential response could be the result of the remaining strong expression of the HoxC cluster genes in the stromal progenitors in the Hoxa9,10,11/Hoxd9,10,11 mutants (Fig. 1). We cannot, however, rule out the possibility that Hox mutant nephron progenitors transdifferentiate into stromal cells, as has been observed for Pax2 mutant kidneys (Naiman et al, 2017).
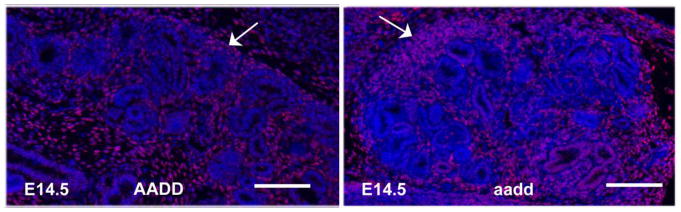
Fig. 3. Stromal progenitors persist.
Stromal cells are marked by expression of MEIS1 (white arrows). At E14.5 the stromal progenitors remain abundant in Hoxa9,10,11/Hoxd9,10,11 homozygous mutants, with a thickened layer of cells compared to the wild type kidneys. White bars are 100 microns. The left panel shows a portion of a wild type kidney, while the entire mutant kidney, which is much smaller, is shown in the right panel.
The stromal cells can be divided into multiple subtypes, including the medullary stroma flanking the forming collecting ducts. P57 is normally expressed in these medullary cells as well as in the podocytes of the glomerulus, while Lef1 is is expressed in these medullary stromal cells as well as multiple cortical cell types. The Hoxa9,10,11/Hoxd9,10,11 mutant medullary stroma maintained Lef1 expression but showed an altered character, with loss of P57 expression (Fig. 4).
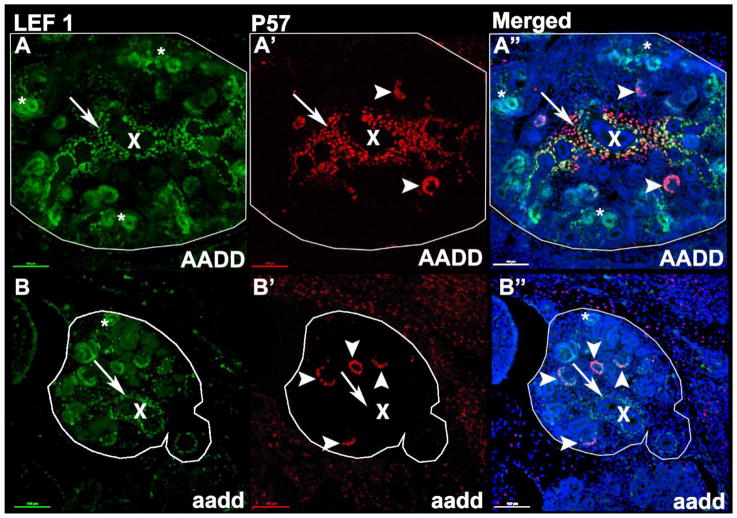
Fig. 4. Altered character of Hox mutant medullary stroma.
LEF1 (green) labels the early nephron segments, podocytes, and medullary stromal cells surrounding the forming collecting ducts, while P57 labels podocytes (arrow heads) and medullary stroma. Asterisks highlight cortical differentiating nephron segments. X labels a central medullary collecting duct. Note that in mutants the medullary stroma is present, and continues to label with LEF1, but does not label with P57 (arrows).
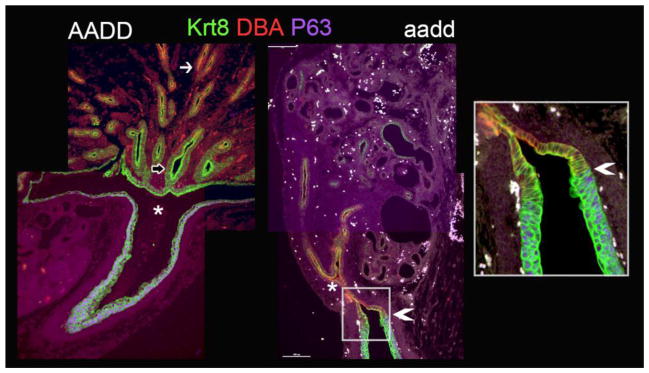
Malformation of the mutant renal pelvis
In the Hoxa9,10,11/Hoxd9,10,11 mutants there is a failure of normal pelvis formation. The ureter is derived from the initial UB stalk and the collecting ducts are derived from the UB branches (Adams and Oxburgh, 2009). In the Hoxa9,10,11/Hoxd9,10,11 mutants the initial UB branch occurs normally at E11.5 (Fig. S1). Later in development, however, at E18.5, the expanded pelvic space, normally located at the site of the initial branch point of the UB (Pietila et al., 2016), does not form (Fig. 5, asterisk). It is also interesting to note that in mutants the DBA staining extends posterior of the UB initial branch point, while it is normally only found well anterior of this branch (Fig. 5). Of the six Hox genes mutated in this study only Hoxd9 shows robust expression in the collecting ducts (Fig. 1).
Fig. 5. Hox mutants show malformation of kidney pelvis/ureter junction.
The open pelvic space present in E18.5 wild type kidneys (AADD, left panel, asterisk) is absent in E18.5 Hoxa9,10,11/Hoxd9,10,11 (aadd) mutant kidneys (right two panels). Urothelium is labeled with P63 (purple). The entire collecting duct and urothelium labels with KRT 8 (green, empty arrow), while DBA (red) collecting duct label normally excludes the innermost collecting ducts near the pelvis (left panel, white arrow), but in mutants the DBA label extends to near the ureter (right panel, arrowhead).
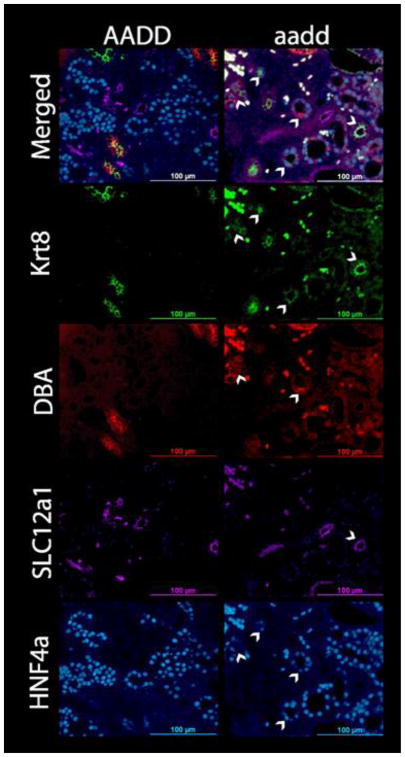
Lineage infidelity
The Hox mutant kidneys showed an unexpected and striking lineage infidelity. The nephron, the functional unit of the kidney, is segmented, with the glomerulus filtration unit followed by the proximal tubule, loop of Henle, and distal tubule, which then connects to the collecting duct. The mutant kidneys were examined for proper formation of nephron segments, given the known role of Hox genes in determining segment identity. We used tubule segment specific markers, with DBA lectin and KRT8 antibody (Aby) for collecting duct, SLC12A1 Aby for loop of Henle, and HNF4A Aby for proximal tubule nuclei. The KRT8 and DBA collecting duct markers normally show overlapping expression patterns, including some inner medullary tubule regions that label with KRT8 but not DBA. In the E18.5 wild type kidney the expected expression patterns for these tubule segment specific markers were observed (Fig. 6). In the mutants, there were also tubules that predominantly labeled for each of these specific markers (Fig. 6). There were also Hox mutant tubules labeling with Aby for SLC12A3, a marker of distal tubules (Fig. S2), further confirming the presence of all tubule segments. There was, therefore, no complete conversion of one segment into another, with resulting loss of a segment. We did, however, observe a surprising differentiation irregularity at the single cell level.
Fig. 6. E18.5 Hox mutant kidneys show lineage infidelity.
In wild type kidneys (AADD, left panels) the cells of each tubule label with a single segment marker, as expected. Surprisingly, however, the tubules of the Hoxa9,10,11/Hoxd9,10,11 (aadd) mutant kidneys include cells with mixed lineage identities. For example, HNF4A labels the nuclei of proximal tubules (bottom panels), but in the mutants proximal tubules include cells that label with DBA and/or KRT8, collecting duct markers (arrowheads). SLC12A1 marks loop of Henle tubules, and for one of the aadd mutant tubules (arrowhead) all of the cells also label with KRT8, suggesting a mixed lineage.
The E18.5 Hoxa9,10,11/Hoxd9,10,11 Hox mutant kidney tubules included individual cells that showed inappropriate cell type. For example, the proximal tubules are the most abundant tubule of the developing and adult kidney, with nuclei that label specifically with Aby to the transcription factor HNF4A (Fig. 6, bottom panels). Surprisingly, within the mutant proximal tubules there were scattered cells that labeled with the collecting duct markers DBA and KRT8 (Fig. 6). This was particularly unexpected, as the proximal tubules and collecting ducts have distinct lineages, with the proximal tubule nephron cells coming from the metanephric mesenchyme while the collecting duct is derived from the ureteric bud. Similarly unanticipated, we also observed tubules with dual identity, for example in some cases all cells in a tubule section showed dual expression of both SLC12A1, a marker of the loop of Henle, and the collecting duct marker KRT8 (Fig. 6). Such cells with apparent incorrect identity, or mixed identity, were observed frequently in the Hox mutant kidneys and absent in the wild type kidneys (Fig. 6, top panels). Fig. S3 shows a higher magnification image of three nearby tubules with each including cells that label for both proximal tubule (HNF4A) and collecting duct (DBA and KRT8) markers.
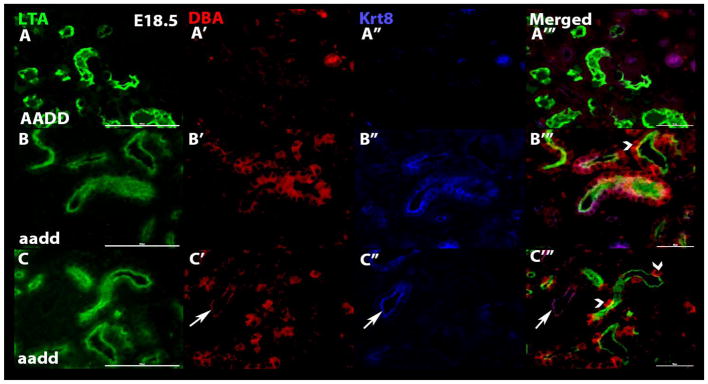
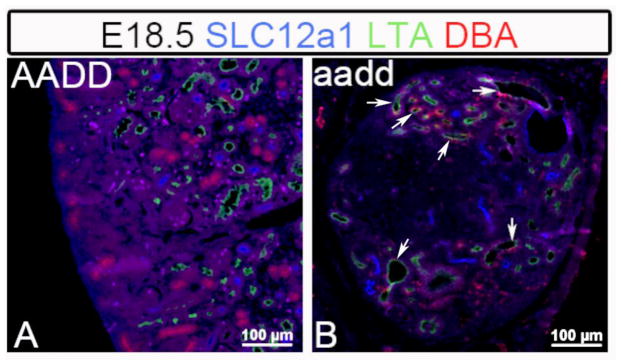
To further confirm and better define the observed lineage infidelity we examined the Hox mutant kidneys with additional combinations of segment specific markers. Again, the E18.5 mutant kidneys showed clear examples of tubules that included cells with incorrect and/or mixed identity. In using the combination of LTA lectin (proximal tubules) with KRT8 and DBA (collecting duct markers) we observed frequent tubules with cells showing lineage infidelity (Fig. 7). As expected, the tubules of wild type kidneys uniformly labeled for only a single segment type (Fig. 7A–A‴). For the Hox mutant E18.5 kidneys, however, most tubules included individual cells showing a mixed identity, simultaneously expressing markers of more than one tubule segment (Fig. 7B–B‴. C–C‴). Such cells with ambiguous identities strongly argue against a simple cell mixing explanation. Instead the cells appear to have differentiated incompletely, or incorrectly, or failed to maintain their correct differentiated state. This was observed once more using the combination of loop of Henle (SLC12A1), proximal tubule (LTA) and collecting duct (DBA) markers, with common occurrence of cells with apparent defects in differentiation (Fig. 8). The inappropriate cells were often observed as single cells, but also often as clusters of cells, suggesting a clonal origin.
Fig. 7. Hox mutant kidney tubules double label with proximal tubule and collecting duct markers.
To further confirm the observed lineage infidelity additional tubule segment specific marker combinations were used. E18.5 wild type (AADD, top panels) and Hoxa9,10,11/Hoxd9,10,11 mutant (aadd, middle and bottom panels) kidneys were labeled with LTA (proximal tubules, green), DBA (collecting ducts, red) and KRT8 (collecting duct, blue) specific markers. In the wild type kidneys individual tubules uniquely labeled with markers of a single segment type. In the mutant kidneys, however, most tubules included a mix of cells that labeled with collecting duct or proximal tubule markers, and in many cases single cells showed dual identity, labeling for both proximal tubule and collecting duct markers, with arrowheads pointing to a few examples. In the bottom panels one tubule (arrows) uniquely labeled with collecting duct markers, while all others show mixed identity.
Fig. 8. Hox mutant tubules show mixed identities for loop of Henle, proximal tubule and collecting duct.
Hoxa9,10,11/Hoxd9,10,11 (aadd) mutant kidney tubules included cells with mixed linages, with cells expressing proximal tubule (LTA, green), collecting duct (DBA), and loop of Henle (SLC12A1, blue) markers, with some examples marked by arrows.
The most commonly observed lineage infidelity was the presence of cells with collecting duct (DBA or KRT8) complete or partial identity located within proximal tubules (LTA or HNF4A). There was variable expressivity, with some mutants showing a higher frequency than others, and even within a single mutant kidney there was regional variability, with some regions showing more lineage infidelity than others. For example, in one mutant we observed 35% of proximal tubule sections included at least one cell showing collecting duct identity (76/220), while in another mutant the frequency was lower, at 13% (60/454). The next most common observed lineage infidelity was loop of Henle tubules including cells with collecting duct identity. Of interest, we did not observe mutant collecting ducts that included cells with nephron segment identity (LTA, HNF4A, SLC12A1, SLC12A3).
To better define the nature of the observed lineage infidelity we carried out a developmental time course analysis. The observed multilineage state of cells at E18.5 was reminiscent of the multilineage priming that has been shown for kidney cells during development (Brunskill et al., 2014). At the renal vesicle (RV) stage of nephron development single cells can express markers of multiple remaining possible lineage directions. For example, a single RV cell can express genes associated with both differentiated podocytes and proximal tubules. As differentiation proceeds a single lineage is selected, and the genes associated with that developmental direction are more fully expressed while the now inappropriate alternate lineage associated genes are more completely repressed. In this report we show that Hox mutant kidneys exhibit multilineage gene expression patterns in cells within later, more differentiated structures such as proximal tubules, suggesting a possible pronounced delay in the resolution of the earlier multilineage priming state. To our surprise, however, this was not the case.
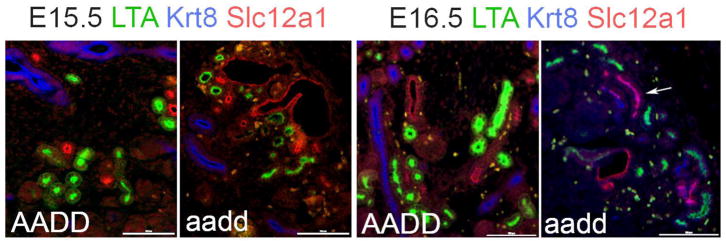
We used a battery of tubule segment specific markers to examine Hoxa9,10,11/Hoxd9,10,11 mutant kidneys for the presence of lineage infidelity at earlier time points. At E15.5 there were very few Hoxa9,10,11/Hoxd9,10,11 mutant cells showing lineage infidelity (Fig. 9). At E16.5 there were more examples of cells with lineage infidelity, but still far fewer than seen at E18.5. These results give evidence for relatively normal early differentiation process, followed by a frequent failure to maintain appropriate differentiation states.
Fig. 9. Time course of lineage infidelity.
At E15.5 the Hoxa9,10,11/Hoxd9,10,11 (aadd) Hox mutant kidney tubules showed few examples of lineage infidelity, as determined using LTA (proximal tubule, KRT8 (collecting duct) and SLC12a1 (loop of Henle) markers. At E16.5 there were some mutant tubules showing expression of markers of more than one tubule segment (arrows mark an example) but compared to E18.5 mutant kidneys there were relatively few. This progressive worsening of lineage infidelity with developmental time gives evidence for a defect in maintaining the proper differentiated state.
Conclusions
Hox genes are multifunctional. While they are best known for their role in segment identity determination they also function in the regulation of cell proliferation, cell death and autophagy. In this report we identify cell identity differentiation defects in the tubules of kidneys with six closely relate Abd-B Hox genes simultaneously mutated. Surprisingly, mutant tubules show the presence of mixed or incorrect identity cell types. These anomalous cells can occur singly or as clusters that suggest a clonal origin.
The Hox code model would predict that changing Hox expression patterns should alter segment identity. We observed, however, differentiation defects that occur at the level of the single cell and not kidney tubule segment. Further, there is not a uniform conversion from one cell fate to another, as would be predicted by an altered Hox code, but rather a seeming stochastic confusion of differentiation, with different cells shifting in diverse developmental directions. Indeed, the Hox expression patterns observed in the developing kidney do not give evidence for distinct Hox codes in different compartments. Nevertheless it remains possible that the combined mutation of six Hox genes produces an indeterminate Hox code that results in a confused differentiation at the cellular level.
In summary, through the mutation of six closely related Abd-B Hox genes with overlapping expression patterns we revealed an unexpected lineage infidelity phenotype. Surprisingly, the results of a developmental time course suggest that the differentiation defect is primarily a memory problem, with lineage infidelity much more common at later time points. This study gives evidence for a novel epigenetic function for Hox genes in maintaining the correct differentiation state.
Supplementary Material
Highlights.
Simultaneous mutation of six Abd-B Hox genes results in premature loss of nephron progenitors
Hox mutants show disrupted development of renal pelvis
Hox mutant kidneys show lineage infidelity at the single cell level
Hox mutant proximal tubules include interspersed cells with apparent collecting duct identity
Acknowledgments
This work was supported by RO1 DK099995 (SP).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adam M, Potter AS, Potter SS. Psychrophilic proteases dramatically reduce single-cell RNA-seq artifacts: a molecular atlas of kidney development. Development. 2017;144:3625–3632. doi: 10.1242/dev.151142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams DC, Oxburgh L. The long-term label retaining population of the renal papilla arises through divergent regional growth of the kidney. Am J Physiol Renal Physiol. 2009;297:F809–815. doi: 10.1152/ajprenal.90650.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagherie-Lachidan M, Reginensi A, Pan Q, Zaveri HP, Scott DA, Blencowe BJ, Helmbacher F, McNeill H. Stromal Fat4 acts non-autonomously with Dchs1/2 to restrict the nephron progenitor pool. Development. 2015;142:2564–2573. doi: 10.1242/dev.122648. [DOI] [PubMed] [Google Scholar]
- Banreti A, Hudry B, Sass M, Saurin AJ, Graba Y. Hox proteins mediate developmental and environmental control of autophagy. Dev Cell. 2014;28:56–69. doi: 10.1016/j.devcel.2013.11.024. [DOI] [PubMed] [Google Scholar]
- Boyle S, Misfeldt A, Chandler KJ, Deal KK, Southard-Smith EM, Mortlock DP, Baldwin HS, de Caestecker M. Fate mapping using Cited1-CreERT2 mice demonstrates that the cap mesenchyme contains self-renewing progenitor cells and gives rise exclusively to nephronic epithelia. Dev Biol. 2008;313:234–245. doi: 10.1016/j.ydbio.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branford WW, Benson GV, Ma L, Maas RL, Potter SS. Characterization of Hoxa-10/Hoxa-11 transheterozygotes reveals functional redundancy and regulatory interactions. Dev Biol. 2000;224:373–387. doi: 10.1006/dbio.2000.9809. [DOI] [PubMed] [Google Scholar]
- Bromleigh VC, Freedman LP. p21 is a transcriptional target of HOXA10 in differentiating myelomonocytic cells. Genes Dev. 2000;14:2581–2586. doi: 10.1101/gad.817100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunskill EW, Park JS, Chung E, Chen F, Magella B, Potter SS. Single cell dissection of early kidney development: multilineage priming. Development. 2014;141:3093–3101. doi: 10.1242/dev.110601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Care A, Valtieri M, Mattia G, Meccia E, Masella B, Luchetti L, Felicetti F, Colombo MP, Peschle C. Enforced expression of HOXB7 promotes hematopoietic stem cell proliferation and myeloid-restricted progenitor differentiation. Oncogene. 1999;18:1993–2001. doi: 10.1038/sj.onc.1202498. [DOI] [PubMed] [Google Scholar]
- Costantini F, Kopan R. Patterning a complex organ: branching morphogenesis and nephron segmentation in kidney development. Dev Cell. 2010;18:698–712. doi: 10.1016/j.devcel.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Tanigawa S, Karner CM, Xin M, Lum L, Chen C, Olson EN, Perantoni AO, Carroll TJ. Stromal-epithelial crosstalk regulates kidney progenitor cell differentiation. Nat Cell Biol. 2013;15:1035–1044. doi: 10.1038/ncb2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AP, Capecchi MR. Axial homeosis and appendicular skeleton defects in mice with a targeted disruption of hoxd-11. Development. 1994;120:2187–2198. doi: 10.1242/dev.120.8.2187. [DOI] [PubMed] [Google Scholar]
- Davis AP, Witte DP, Hsieh-Li HM, Potter SS, Capecchi MR. Absence of radius and ulna in mice lacking hoxa-11 and hoxd-11. Nature. 1995;375:791–795. doi: 10.1038/375791a0. [DOI] [PubMed] [Google Scholar]
- Di-Poi N, Zakany J, Duboule D. Distinct roles and regulations for HoxD genes in metanephric kidney development. PLoS Genet. 2007;3:e232. doi: 10.1371/journal.pgen.0030232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favier B, Rijli FM, Fromental-Ramain C, Fraulob V, Chambon P, Dolle P. Functional cooperation between the non-paralogous genes Hoxa-10 and Hoxd-11 in the developing forelimb and axial skeleton. Development. 1996;122:449–460. doi: 10.1242/dev.122.2.449. [DOI] [PubMed] [Google Scholar]
- Fetting JL, Guay JA, Karolak MJ, Iozzo RV, Adams DC, Maridas DE, Brown AC, Oxburgh L. FOXD1 promotes nephron progenitor differentiation by repressing decorin in the embryonic kidney. Development. 2014;141:17–27. doi: 10.1242/dev.089078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong KQ, Yallowitz AR, Sun H, Dressler GR, Wellik DM. A Hox-Eya-Pax complex regulates early kidney developmental gene expression. Mol Cell Biol. 2007;27:7661–7668. doi: 10.1128/MCB.00465-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman HA, Lai HL, Patterson LT. Cessation of renal morphogenesis in mice. Dev Biol. 2007;310:379–387. doi: 10.1016/j.ydbio.2007.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatini V, Huh SO, Herzlinger D, Soares VC, Lai E. Essential role of stromal mesenchyme in kidney morphogenesis revealed by targeted disruption of Winged Helix transcription factor BF-2. Genes Dev. 1996;10:1467–1478. doi: 10.1101/gad.10.12.1467. [DOI] [PubMed] [Google Scholar]
- Hombria JC, Lovegrove B. Beyond homeosis--HOX function in morphogenesis and organogenesis. Differentiation. 2003;71:461–476. doi: 10.1046/j.1432-0436.2003.7108004.x. [DOI] [PubMed] [Google Scholar]
- Horan GS, Ramirez-Solis R, Featherstone MS, Wolgemuth DJ, Bradley A, Behringer RR. Compound mutants for the paralogous hoxa-4, hoxb-4, and hoxd-4 genes show more complete homeotic transformations and a dose-dependent increase in the number of vertebrae transformed. Genes Dev. 1995;9:1667–1677. doi: 10.1101/gad.9.13.1667. [DOI] [PubMed] [Google Scholar]
- Kessel M, Gruss P. Homeotic transformations of murine vertebrae and concomitant alteration of Hox codes induced by retinoic acid. Cell. 1991;67:89–104. doi: 10.1016/0092-8674(91)90574-i. [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Valerius MT, Mugford JW, Carroll TJ, Self M, Oliver G, McMahon AP. Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell. 2008;3:169–181. doi: 10.1016/j.stem.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- Liu J, Fire A. Overlapping roles of two Hox genes and the exd ortholog ceh-20 in diversification of the C. elegans postembryonic mesoderm. Development. 2000;127:5179–5190. doi: 10.1242/dev.127.23.5179. [DOI] [PubMed] [Google Scholar]
- Lohmann I, McGinnis N, Bodmer M, McGinnis W. The Drosophila Hox gene deformed sculpts head morphology via direct regulation of the apoptosis activator reaper. Cell. 2002;110:457–466. doi: 10.1016/s0092-8674(02)00871-1. [DOI] [PubMed] [Google Scholar]
- Magella B, Adam M, Potter AS, Venkatasubramanian M, Chetal K, Hay SB, Salomonis N, Potter SS. Cross-platform single cell analysis of kidney development shows stromal cells express Gdnf. Dev Biol. 2018;434:36–47. doi: 10.1016/j.ydbio.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre DC, Rakshit S, Yallowitz AR, Loken L, Jeannotte L, Capecchi MR, Wellik DM. Hox patterning of the vertebrate rib cage. Development. 2007;134:2981–2989. doi: 10.1242/dev.007567. [DOI] [PubMed] [Google Scholar]
- McMahon AP. Development of the Mammalian Kidney. Curr Top Dev Biol. 2016;117:31–64. doi: 10.1016/bs.ctdb.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naiman N, Fujioka K, Fujino M, Valerius MT, Potter SS, McMahons AP, Kobayashi A. Repression of interstitial identity in nephron progenitor cells by Pax2 establishes the nephron-interstitum boundary during kidney development. Dev Cell. 22:349–365. doi: 10.1016/j.devcel.2017.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson LT, Pembaur M, Potter SS. Hoxa11 and Hoxd11 regulate branching morphogenesis of the ureteric bud in the developing kidney. Development. 2001;128:2153–2161. doi: 10.1242/dev.128.11.2153. [DOI] [PubMed] [Google Scholar]
- Patterson LT, Potter SS. Atlas of Hox gene expression in the developing kidney. Dev Dyn. 2004;229:771–779. doi: 10.1002/dvdy.10474. [DOI] [PubMed] [Google Scholar]
- Pietila I, Prunskaite-Hyyrylainen R, Kaisto S, Tika E, van Eerde AM, Salo AM, Garma L, Miinalainen I, Feitz WF, Bongers EM, Juffer A, Knoers NV, Renkema KY, Myllyharju J, Vainio SJ. Wnt5a Deficiency Leads to Anomalies in Ureteric Tree Development, Tubular Epithelial Cell Organization and Basement Membrane Integrity Pointing to a Role in Kidney Collecting Duct Patterning. PLoS One. 2016;11:e0147171. doi: 10.1371/journal.pone.0147171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponzielli R, Astier M, Chartier A, Gallet A, Therond P, Semeriva M. Heart tube patterning in Drosophila requires integration of axial and segmental information provided by the Bithorax Complex genes and hedgehog signaling. Development. 2002;129:4509–4521. doi: 10.1242/dev.129.19.4509. [DOI] [PubMed] [Google Scholar]
- Raines AM, Adam M, Magella B, Meyer SE, Grimes HL, Dey SK, Potter SS. Recombineering-based dissection of flanking and paralogous Hox gene functions in mouse reproductive tracts. Development. 2013;140:2942–2952. doi: 10.1242/dev.092569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raines AM, Magella B, Adam M, Potter SS. Key pathways regulated by HoxA9,10,11/HoxD9,10,11 during limb development. BMC Dev Biol. 2015;15:28. doi: 10.1186/s12861-015-0078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reginensi A, Scott RP, Gregorieff A, Bagherie-Lachidan M, Chung C, Lim DS, Pawson T, Wrana J, McNeill H. Yap- and Cdc42-dependent nephrogenesis and morphogenesis during mouse kidney development. PLoS Genet. 2013;9:e1003380. doi: 10.1371/journal.pgen.1003380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneuwly S, Klemenz R, Gehring WJ. Redesigning the body plan of Drosophila by ectopic expression of the homoeotic gene Antennapedia. Nature. 1987;325:816–818. doi: 10.1038/325816a0. [DOI] [PubMed] [Google Scholar]
- Self M, Lagutin OV, Bowling B, Hendrix J, Cai Y, Dressler GR, Oliver G. Six2 is required for suppression of nephrogenesis and progenitor renewal in the developing kidney. EMBO J. 2006;25:5214–5228. doi: 10.1038/sj.emboj.7601381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short KM, Combes AN, Lefevre J, Ju AL, Georgas KM, Lamberton T, Cairncross O, Rumballe BA, McMahon AP, Hamilton NA, Smyth IM, Little MH. Global quantification of tissue dynamics in the developing mouse kidney. Dev Cell. 2014;29:188–202. doi: 10.1016/j.devcel.2014.02.017. [DOI] [PubMed] [Google Scholar]
- Small KM, Potter SS. Homeotic transformations and limb defects in Hox A11 mutant mice. Genes Dev. 1993;7:2318–2328. doi: 10.1101/gad.7.12a.2318. [DOI] [PubMed] [Google Scholar]
- Studer M, Lumsden A, Ariza-McNaughton L, Bradley A, Krumlauf R. Altered segmental identity and abnormal migration of motor neurons in mice lacking Hoxb-1. Nature. 1996;384:630–634. doi: 10.1038/384630a0. [DOI] [PubMed] [Google Scholar]
- Suemori H, Noguchi S. Hox C cluster genes are dispensable for overall body plan of mouse embryonic development. Dev Biol. 2000;220:333–342. doi: 10.1006/dbio.2000.9651. [DOI] [PubMed] [Google Scholar]
- Wellik DM, Capecchi MR. Hox10 and Hox11 genes are required to globally pattern the mammalian skeleton. Science. 2003;301:363–367. doi: 10.1126/science.1085672. [DOI] [PubMed] [Google Scholar]
- Wellik DM, Hawkes PJ, Capecchi MR. Hox11 paralogous genes are essential for metanephric kidney induction. Genes Dev. 2002;16:1423–1432. doi: 10.1101/gad.993302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Wellik DM. Axial Hox9 activity establishes the posterior field in the developing forelimb. Proc Natl Acad Sci U S A. 2011;108:4888–4891. doi: 10.1073/pnas.1018161108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yallowitz AR, Hrycaj SM, Short KM, Smyth IM, Wellik DM. Hox10 genes function in kidney development in the differentiation and integration of the cortical stroma. PLoS One. 2011;6:e23410. doi: 10.1371/journal.pone.0023410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Treweek JB, Kulkarni RP, Deverman BE, Chen CK, Lubeck E, Shah S, Cai L, Gradinaru V. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell. 2014;158:945–958. doi: 10.1016/j.cell.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Kim HJ, Marshall H, Gendron-Maguire M, Lucas DA, Baron A, Gudas LJ, Gridley T, Krumlauf R, Grippo JF. Ectopic Hoxa-1 induces rhombomere transformation in mouse hindbrain. Development. 1994;120:2431–2442. doi: 10.1242/dev.120.9.2431. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Potter SS. Functional specificity of the Hoxa13 homeobox. Development. 2001;128:3197–3207. doi: 10.1242/dev.128.16.3197. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Potter SS. Functional comparison of the Hoxa 4, Hoxa 10, and Hoxa 11 homeoboxes. Dev Biol. 2002;244:21–36. doi: 10.1006/dbio.2002.0595. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.