Abstract
Drosophila melanogaster is a genetic model organism that has contributed to the discovery of numerous genes whose human homologues are associated with diseases. The development of sophisticated genetic tools to manipulate its genome accelerates the discovery of the genetic basis of undiagnosed human diseases and the elucidation of molecular pathogenic events of known and novel diseases. Here, we discuss various approaches used in flies to assess the function of the fly homologues of disease-associated genes. We highlight how systematic and combinatorial approaches based on recently established methods provide us with integrated tool sets that can be applied to the study of neurodevelopmental and neurodegenerative disorders.
Keywords: Drosophila, RMCE, MiMIC, T2A-GAL4, flippase recombinase
INTRODUCTION
For over a century, the fruit fly, Drosophila melanogaster, has been widely used as a model organism to study most aspects of biology [1]. Flies have a short life cycle, produce a large number of offspring, and are inexpensive to maintain. Numerous developmental pathways and physiological processes are conserved between flies and mammals and nearly 75% of human disease genes have functional homologue(s) in flies [2]. In addition, genetic manipulations in the fly have no equal in other multicellular model organisms with a complex nervous system. These features provide fly biologists with opportunities to collaborate with human geneticists, discover the genetic basis of new human diseases, unravel pathogenic mechanisms, discover cellular mechanisms and drugs, and establish conserved molecular etiologies underlying numerous neurological diseases.
The goal of this review is to provide examples of techniques used in fly models to gain insight about the pathophysiology of neurological diseases. We discuss recent experimental strategies that build upon ΦC31 recombination-mediated cassette exchange (RMCE) to insert DNA [3]; CRISPR/Cas9 to edit genomes [4,5]; P[acman]/FlyFos genomic libraries to rescue mutants and determine protein localization [6–8]; Minos-mediated integration cassette (MiMIC) to mutate genes and tag proteins [9,10]; and GAL4/UAS approaches [11], including the Trojan-GAL4 (T2A-Gal4) method [12], to express transgenes. We highlight how integrating many of these technologies into new approaches has provided an elegant toolset to unravel molecular mechanisms of neurodevelopmental and neurodegenerative diseases by enabling unsurpassed genetic manipulations in flies.
A. Reverse genetic approaches
Reverse genetic approaches to elucidate disease pathogenesis in flies are based on the discovery of disease-associated genes by human geneticists and the availability of reagents related to the fly homologue(s) of the target gene (Figure 1a). The available information in the literature and databases can be screened through MARRVEL [13] and FlyBase (flybase.org). These include mutations induced by chemical mutagens, deletions, or insertions of transposable elements, which can often be excised to create small deletions. The phenotypes associated with the complete loss of the gene of interest provide an important reference point to compare phenotypes associated with missense mutations identified in patients to gauge if they cause loss-of-function (LOF) (amorph or hypomorph), gain-of-function (GOF) (hypermorph or neomorph) or dominant negative (antimorph) phenotype. Alternatively, spatiotemporally controlled expression of RNAi against the gene of interest, using the GAL4/UAS system [11], allows gene knockdown. However, it often does not provide as rigorous a reference point as the null mutant and the need to address the efficacy and specificity of knock-down with each RNAi increases the workload. On the other hand, a large collection of nervous system-specific GAL4 lines [14,15] is available, permitting knockdown of genes in specific neuronal and glial cell (sub)populations. Finally, CRISPR based strategies can be employed to edit genes [16,17]. In all cases, it is highly recommended that the phenotypes associated with mutations that are being studied are rescued using either a UAS-cDNA approach or better, a genomic fragment that encodes the gene (see Sections C and D). Indeed, second site mutations or RNAi off-target effects may cloud the interpretation of phenotypes associated with the gene of interest.
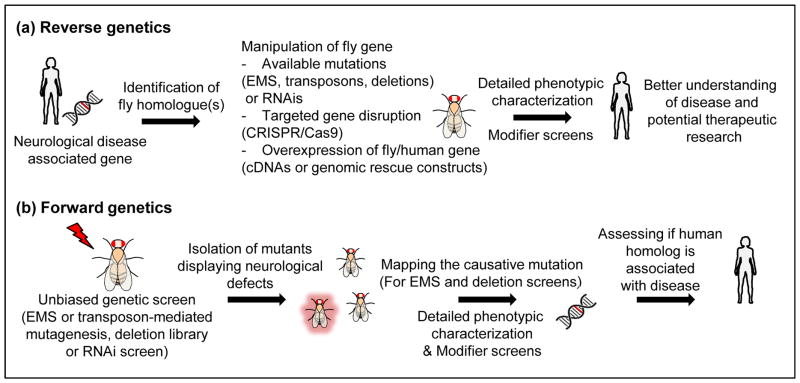
FIGURE 1.
Genetic approaches enabling neurological disease research in Drosophila (a) Reverse genetic approaches in flies are based on the identification of the fly homologue(s) of the disease-associated gene and the assessment of phenotypes caused by genetic manipulations of the gene of interest. Upon examination of the gene’s function, modifier screens can be performed to identify additional genes that act as either enhancers or suppressors of determined phenotypes. (b) Forward genetic approaches identify mutants with a certain phenotype of interest (such as neuronal dysfunction or neurodegeneration) based on unbiased genetic screens. Upon mapping of the causative mutations to genes whose human homologues are associated with disease, detailed phenotypic characterizations can be performed to provide a better understanding of the gene’s function.
The Drosophila community has kept a publicly available database of the majority of fly mutants that were generated. The availability of large collections of LOF alleles from stock centers has been invaluable for many researchers to tackle the pathogenic mechanisms associated with numerous diseases including neurodegenerative and neurodevelopmental disorders. Recent examples include models of Parkinson’s disease (PD) [18], amyotropic lateral sclerosis (ALS) [19], Coffin-Lowry syndrome [20], and lysosomal storage diseases (LSDs) [21]. Similarly, RNAi mediated knockdown has been successfully utilized to assess the in vivo function of disease-associated genes including parkin [22], valosin containing protein (VCP) [23], amyloid precursor protein (APP) [24], and others [25,26].
B. Forward genetic approaches
Even though reverse genetic approaches can generate valuable information about a gene’s function, this research is typically hypothesis-driven. Forward genetic approaches, on the other hand, are unbiased and based on a phenotype of interest. They have led to the discovery of thousands of genes, many of which are relevant to human pathology and disease (Figure 1b). However, in the area of neurobiology there has been a strong bias towards screens that affect neuronal development in flies, and very few high-throughput screens have attempted to isolate mutants that affect neuronal function [27–29] and/or cause the demise of neurons [30,31]. Indeed, most efforts to study genes associated with neurodegeneration have been based on reverse genetic strategies [32,33].
Recently, an unbiased chemical (EMS) mutagenesis screen was performed in flies to isolate essential genes required for neuronal development and maintenance. This screen was designed to isolate lethal mutations on a flippase recognition target (FRT) bearing X-chromosome and to subsequently perform flippase (FLP)-mediated clonal analyses in the eye and thorax/wing. By assessing externally visible eye phenotypes as well as the progressive demise of photoreceptor function based on electroretinograms (ERGs), ~700 mutations were isolated. Unlike other EMS screens, the authors set out to identify the nature of most affected genes by performing duplication mapping [34], whole genome sequencing [35], and systematic transgenic rescue based on BAC clones [36]. This led to the identification of 165 genes associated with neurodevelopmental or neurodegenerative phenotypes [30]. Nearly all isolated mutations were LOF alleles, often providing allelic series of genes whose human homologues were already known to be associated with disease. These alleles have permitted in depth phenotypic characterization of genes associated with Leigh Syndrome (sicily/NDUFAF6, l(1)G0334/PDHA1, and ppr/LRPPRC) [37–39], Charcot-Marie-Tooth Type 2A (CMT2A) (Marf/MFN2) [38,40], PD (Vps35/VPS35 and Vps26/VPS26) [41], and Friedreich’s Ataxia (fh/FXN) [42]. The screen also led to the identification of a novel human gene associated with severe microcephaly (Ankle2/ANKLE2) [30], and two other genes associated with acquired microcephaly and neurodegeneration (Nrd1/NRD1 and dOgdh/OGDH) [43]. In summary, the ability to identify the molecular lesions in genes from a single genetic screen in Drosophila has driven the discovery of genes that are associated with diseases and follow-up studies have revealed a better understanding of the pathology.
C. Expression of fly or human genes using binary systems
Accumulation of toxic proteins in inclusions or aggregates is a key pathological finding associated with many neurological diseases, including AD, PD, ALS, and prion diseases. These may lead to altered protein function or toxic aggregates that can often be mimicked by protein overexpression. The most commonly used strategy to overexpress genes in the flies is based on the GAL4/UAS-mediated binary expression system [11] using tissue and neuronal sub-type specific drivers to model neurodegenerative and neurodevelopmental diseases (Figure 2a). By adding the temperature-sensitive GAL80ts to repress GAL4 activity, fine spatiotemporal control of expression can be obtained [44–46]. Similarly, the GeneSwitch (GS) method utilizes a GAL4-progesterone-receptor chimera protein that can be activated by the hormone progesterone to spatiotemporally control the gene expression [44,47]. Alternative binary overexpression systems such as the LexA/LexA operator (LexAop) system [48] and the Q-system (QF/QUAS/QS) [49] are also used for the analysis of gene function and precise spatiotemporal manipulations of gene expression (Figure 2a). These systems can be used along with the GAL4/UAS/GAL80ts system to perform independent labeling and/or manipulation of separate populations of neurons in the same fly [50–53]. Binary overexpression systems also permit screening for modifiers of a gain of function phenotype and unravel other key players that affect protein toxicity [46,54–56].
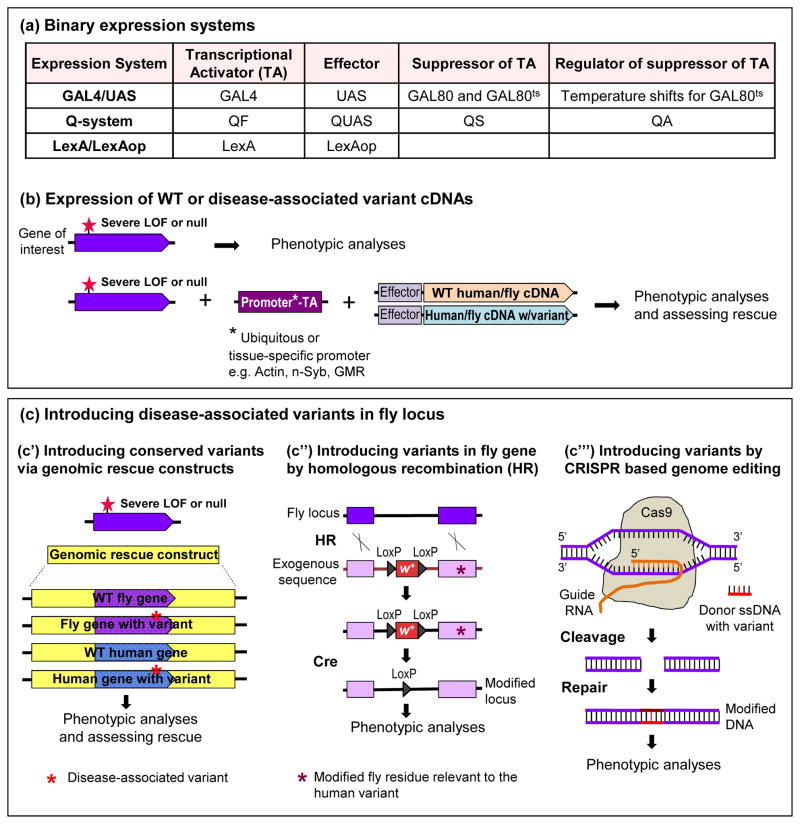
FIGURE 2.
Methods to model Drosophila for assessing disease variants (a) Basic binary expression systems used in Drosophila (b) Upon identification of phenotypes caused by a severe LOF mutation, the effect of overexpression of WT or disease-variant bearing cDNA (of both fly and human) can be tested using binary expression systems: GAL4/UAS, Q-system, and LexA/LexAop. Availability of a wide-range of tissue/developmental stage-specific drivers and conditional use of binary systems allow elegant manipulations on gene expression. (c) Disease-associated variants can be inserted into genomic rescue constructs or into the endogenous fly locus when the variant is conserved in the fly homologue. (c′) If the gene of interest contains a single exon, the fly exon can be replaced by wild type or disease variant-containing human exon. Introducing these genomic rescue constructs into flies that lack the gene of interest and display certain phenotypes allows testing for rescue with different genomic constructs. (c″) Exogenous fly sequence with the disease variant can be replaced by the endogenous fly locus using ends-out homologous recombination (HR). Cre recombinase induces site-specific recombination between LoxP sites that flank mini white marker (w+) and removes w+, leaving a LoxP within the intron. This method can be used to create knock in disease models in flies. (c‴) CRISPR/Cas9 based genome editing can be used to precisely modify nucleotides. RNA-guided Cas9 nuclease induces a double-strand break. Homology directed repair repairs the break by introduction of a donor single-stranded DNA (ssDNA) containing disease-variant.
Another application of the GAL4/UAS approach is to test if the expression of a fly or human cDNA can rescue the phenotypes associated with the loss of a homologous fly gene (Figure 2b). Rescue with the human cDNA indicates functional conservation and permits assessing the consequences of human variant expression in flies. This approach, based on a limited set of roughly 30 genes, is effective for the majority of genes tested (personal communication and [57–59]).
D. Assessing disease variants using genomic rescue constructs
One caveat associated with expression of fly or human cDNAs, especially if the goal is to rescue mutants, is that there are often different isoforms for a single gene, raising the issue as to which isoform should be selected. In addition, when genes are large, full-length fly or human cDNAs are often not available. An alternative strategy to rescue a null mutant phenotype is to use a fly genomic rescue construct that encodes all the transcripts of the gene of interest. Three genomic rescue construct libraries of Drosophila are available: two BAC libraries based on P[acman] [7] (bacpacresources.org) and a fosmid library based on FlyFos clones [8] (sourcebioscience.com). In addition, transgenic flies carrying 80 kb P[acman] clones and FlyFos constructs are available [8,36].
A key requirement to analyze the functional consequences of a patient variant is that the protein sequences that display variants in humans are conserved in the fly homologues. For instance, mutations in the voltage-gated Ca2+ channel CACNA1A cause diverse neurological disorders in humans and two of these mutations with different patient symptoms were recently modeled in flies using a genomic rescue construct [60]. Based on previously identified lethal mutations and neurodegenerative phenotypes caused by mutations in the fly homologue, cac [61], the authors tested the rescuing ability of a wild type 80 kb P[acman] clone and clones derived from this BAC in which the human variants were inserted by recombineering [62]. Whereas the wild type P[acman] clone fully rescued the observed phenotypes, the two variants tested showed very different phenotypes in the flies providing a better understanding of the nature of the patient variants in CACNA1A [60].
In cases where the gene of interest carries a single exon, the fly exon in the genomic rescue constructs can be replaced by wild type and variant-containing human exon to assess/compare the ability of these constructs to rescue mutants (Figure 2c′) [63]. For example, the functional impact of a rare TM2D3 human variant that was predicted to be non-pathogenic was identified in association with late onset AD susceptibility [64]. The loss of the fly homologue of TM2D3, almondex (amx), is known to cause embryonic lethality and severe neurogenic defects. The authors showed that a genomic rescue construct containing the human TM2D3 under the control of endogenous amx regulatory elements can functionally substitute for fly amx, whereas expression of the AD-associated variant under the same conditions causes a severe LOF. These data strongly suggest that the variant in TM2D3 observed in AD patients, which is not evolutionarily conserved in flies, plays a role in AD pathogenesis.
E. Introducing disease variants in conserved residues of fly homologues
An alternative way to assess the functional impact of disease-associated variants that may affect the protein function is to introduce the variants in the endogenous fly gene, rather than in a transgene (Figure 2c″). This requires that the human and fly proteins are likely orthologues and that the residues affected in humans are conserved in flies. Mutations or variants can be integrated into the fly homologue of disease-associated genes by homologous recombination (HR) [65] and this strategy was utilized to introduce ALS-associated SOD1 point mutations in the fly homologue of SOD1 [16]. This study provided valuable insights about the nature of the different mutations that affect SOD1. Currently, the CRISPR/Cas9 technology can be used to precisely modify nucleotides in endogenous genes by injection of single-stranded DNA (ssDNA) [66] (Figure 2c‴). The Cas9 can also be used to perform conditional mutagenesis by ubiquitously expressing the guide RNAs against the gene of interest but restricting the expression of Cas9 with the GAL4/UAS system to specific cells or tissues [17].
F. Integrated approaches to study gene function and disease variants based on MiMIC derived strategies
Some of the basic knowledge required to dissect a gene’s function relates to which cells express the gene and where the protein is localized subcellularly. In addition, a tissue specific knockdown strategy that is conditional and reversible can provide invaluable information. These features, as well as the capability to test the rescuing ability of the fly gene or its human homologues and potential pathogenic variants, were recently integrated using MiMIC technology [9] (Figure 3). MiMIC is a transposable element that incorporates two inverted ΦC31 attP target sites flanking a gene-trap cassette: (Splice Acceptor) SA-three stop codons for each possible reading frame-the polyA tail and a transformation marker, yellow+ (Figure 3a′). Once a MiMIC is in the fly genome, any DNA sequence flanked by attB sites can replace the gene-trap cassette present in MiMIC by ΦC31-mediated RMCE.
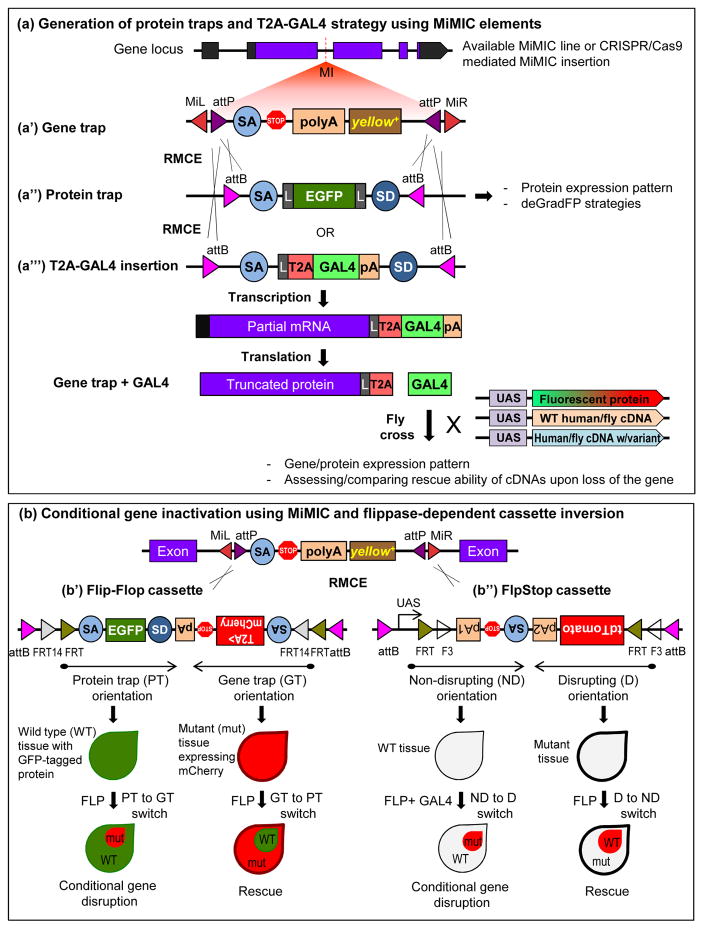
FIGURE 3.
Integrated gene manipulation approaches using Minos-mediated integration cassette (MiMIC) (a′) MiMIC elements contain a gene trap cassette and a yellow+ selection marker flanked by attP sites. Using recombinase mediated cassette exchange (RMCE) an artificial exon nested in attB sites can be integrated into MiMIC. (a″) Protein traps can be generated using cassettes containing SA-EGFP-SD, which allows the assessment of endogenous localization of the GFP-tagged protein as well as deGradFP strategies for conditional manipulations. Flexible linker sequence is abbreviated as L. (a‴) Introduction of an artificial exon that contains a self-cleaving T2A peptide causing ribosomal skipping and a GAL4 sequence creates a truncated protein of interest and a GAL4 protein. GAL4 is expressed in the same spatial and temporal pattern as the fly gene, allowing expression of transgenes including the corresponding UAS-fly or human cDNA as well as UAS-fluorescent protein (e.g. GFP, mCherry). (b) Recently, two flippase-dependent cassettes that can be integrated into MiMIC sites were generated to perform conditional gene inactivation: Flip-Flop and FlpStop. Both cassettes are flanked by two different inverted pairs of FRT sites forming a FLip-Excision (FLEx) switch, thereby making the cassettes invertible in the presence of FLP recombinase. (b′) FlipFlop cassette contains two modules that are placed in opposite orientations: a protein-trap (PT) module and a gene-trap (GT) module. The PT module contains SA-EGFP-SD and thus tags the protein with GFP. The GT module contains SA-T2AmCherry-polyA and thus truncates the gene/protein. The T2AmCherry in the GT module marks the mutant cells with mCherry. Presence of flippase can invert one module to the other, allowing conditional and reversible regulation of gene disruption. (b″) FlpStop cassette is designed to have a non-disrupting (ND) and disrupting (D) mode that are placed in opposite orientations. Flippase activity can invert one orientation to the other providing conditional gene disruption and rescue. Presence of the tdTomato marker in the invertible FLEx switch region allows detection of cells that undergoes inversion.
The MiMIC cassette can be randomly inserted in almost any genomic location and can be used for different applications depending on the insertion location. Among these, the insertions in introns between two coding exons are the most useful. As shown in Figure 3a, they can be used to integrate an artificial exon: SA-EGFP-SD (Splice Donor), creating a GFP-tagged protein trap (Figure 3a″). The internally tagged proteins are most often functional [10] and permit the determination of the cells that express the protein as well as the subcellular localization of the protein. Importantly, the deGradFP system can be used for a reversible temperature-dependent conditional degradation of GFP-tagged proteins [10,67].
Two alternative strategies based on MiMIC and FLP recombinase can be used for conditional gene inactivation in mitotic or post-mitotic cells such as neurons: Flip-Flop [68] and FlpStop [69] (Figure 3b). Both strategies utilize cassettes that are nested in between attB sites and they can easily be integrated into MiMICs using RMCE. Both cassettes are flanked by two different inverted pairs of FRT sites forming a FLip-Excision (FLEx) switch [70]. This allows inversion of the cassette in the presence of FLP. Flip-Flop contains a protein-trap (PT) module that tags the protein with GFP when inserted in PT orientation and a gene-trap (GT) module that truncates the transcript and marks the mutant cells with mCherry when inserted in GT orientation (Figure 3b′). Hence, starting with a PT and inverting the cassette to a GT allows conditional removal of a gene/protein in developing as well as in non-mitotic cells by simultaneously marking wild type and mutant cells. Similarly, the FlpStop cassette can be inserted in two different orientations: non-disrupting (ND) or disrupting (D) (Figure 3b″). Flippase activity can invert one orientation to the other, providing conditional disruption and rescue, similar to Flip-Flop. The presence of the tdTomato marker in the invertible FLEx switch region of the FlpStop cassette allows detection of cells that undergo the inversion. Hence, FlipFlop and FlpStop permit the removal of proteins at any time and in any tissue and provide powerful additions to the Drosophila toolkit.
Another recently developed tool that is also based on intronic MiMIC insertions relies on RMCE-mediated integration of an artificial exon that contains a SA followed by self-cleaving T2A peptide that causes ribosomal skipping, the GAL4 coding sequence, and a polyA signal to create a LOF allele by early transcription termination [12] (Figure 3a‴). Upon translation, the truncated message produces the GAL4 protein that is expressed in the same spatiotemporal expression pattern as the native gene. The GAL4 can then be used to drive a UAS-fly or human cDNA to assess the rescue of mutant phenotype or to express a fluorescent marker like UAS-GFP to assess the cellular pattern of endogenous expression. This strategy was successfully applied to investigate a new neurodevelopmental disorder resulting from de novo variants in EBF3 [71]. The lethality caused by the T2A-GAL4 insertion in the fly EBF3 orthologue, knot, was rescued by expression of human EBF3 cDNA, showing that human EBF3 can functionally replace its Drosophila orthologue. On the other hand, expression of de novo variants observed in patients, abrogated the ability of human EBF3 to rescue the mutant flies, suggesting that these variants are pathogenic. In another example, the T2A-GAL4 strategy was utilized in combination with the forward genetic approach (discussed in section B), revealing a novel role for NRD1 (Nardilysin) as a mitochondrial co-chaperone for OGDH (2-oxoglutarate dehydrogenase) in flies [43]. The authors showed that loss of the fly homologues, Nrd1 or dOgdh cause progressive neurodegeneration and mutations in both human NRD1 and OGDH are linked to rare neurological conditions. They generated a severe LOF allele using the T2A-GAL4 system and showed that the lethality caused by loss of dOgdh can be rescued by expression of wild type fly or human OGDH. Expression of the disease associated variant-containing human or fly OGDH, on the other hand, did not rescue lethality, suggesting that the disease variant in the patient is a severe LOF mutation.
The above strategies show the power and versatility of the MiMIC system, yet the MiMIC collection contains intronic insertions for only 1,800 genes [10]. Hence, the Gene Disruption Project is creating an additional collection of intronic MiMIC-like elements using a CRISPR based technology termed CRIMIC (CRISPR-mediated integration of MiMIC-like elements). CRIMIC allows the efficient targeting of MiMIC-like elements in introns of fly homologues of human genes [12,72] and should therefore expedite our understanding of neurological disease pathologies in fly models.
In summary, integrating numerous technologies in a pipeline based on MiMIC allows us to assess the rescue ability of wild type and variants of human genes, to determine the cells expressing the gene of interest, to probe the subcellular localization of wild type and mutant proteins, and enables elegant conditional manipulations in vivo at various developmental stages as well as in adults.
CONCLUSION
Drosophila has been a powerful model organism to discover and study human genes. Recently developed techniques further facilitate the study of disease-associated genes and permit assessing the pathogenicity of identified human genetic variants in flies. Integrating these techniques in seamless approaches will continue to drive gene discovery and assessment of pathogenic mechanisms.
Highlights.
Expression of reference and variant human genes in flies permit functional testing
Innovative genetic strategies in flies drive discovery of disease associated genes
Integration of different approaches accelerates discovery of disease mechanisms
Acknowledgments
We would like to thank Nele Haelterman, Sonal Nagarkar-Jaiswal, Oguz Kanca and Megan Campbell for helpful comments and suggestions. We apologize to our colleagues whose work we were unable to cite due to space limitations. This study was supported by National Institutes of Health (NIH) grants U54NS093793, R24OD022005, R01GM067858 to H.J.B. H.J.B. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Conflict of Interest Statement
Nothing declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wangler MF, Yamamoto S, Bellen HJ. Fruit flies in biomedical research. Genetics. 2015;199:639–653. doi: 10.1534/genetics.114.171785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bier E. Drosophila, the golden bug, emerges as a tool for human genetics. Nat Rev Genet. 2005;6:9–23. doi: 10.1038/nrg1503. [DOI] [PubMed] [Google Scholar]
- 3.Bateman JR, Lee AM, Wu CT. Site-specific transformation of Drosophila via phiC31 integrase-mediated cassette exchange. Genetics. 2006;173:769–777. doi: 10.1534/genetics.106.056945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bier E, Harrison MM, O’Connor-Giles KM, Wildonger J. Advances in Engineering the Fly Genome with the CRISPR-Cas system. Flybook, Genetics. 2017 doi: 10.1534/genetics.117.1113. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanca O, Bellen H, Schnorrer F. Gene tagging strategies to assess protein localization, protein inactivation, and conditionally express other genes in Drosophila. Flybook, Genetics. 2017 doi: 10.1534/genetics.117.199968. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Venken KJ, Carlson JW, Schulze KL, Pan H, He Y, Spokony R, Wan KH, Koriabine M, de Jong PJ, White KP, et al. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat Methods. 2009;6:431–434. doi: 10.1038/nmeth.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sarov M, Barz C, Jambor H, Hein MY, Schmied C, Suchold D, Stender B, Janosch S, KJV, Krishnan RT, et al. A genome-wide resource for the analysis of protein localisation in Drosophila. Elife. 2016;5:e12068. doi: 10.7554/eLife.12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Venken KJ, Schulze KL, Haelterman NA, Pan H, He Y, Evans-Holm M, Carlson JW, Levis RW, Spradling AC, Hoskins RA, et al. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat Methods. 2011;8:737–743. doi: 10.1038/nmeth.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagarkar-Jaiswal S, Lee PT, Campbell ME, Chen K, Anguiano-Zarate S, Gutierrez MC, Busby T, Lin WW, He Y, Schulze KL, et al. A library of MiMICs allows tagging of genes and reversible, spatial and temporal knockdown of proteins in Drosophila. Elife. 2015:4. doi: 10.7554/eLife.05338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 12.Diao F, Ironfield H, Luan H, Shropshire WC, Ewer J, Marr E, Potter CJ, Landgraf M, White BH. Plug-and-play genetic access to drosophila cell types using exchangeable exon cassettes. Cell Rep. 2015;10:1410–1421. doi: 10.1016/j.celrep.2015.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J, Al-Ouran R, Hu Y, Kim SY, Wan YW, Wangler MF, Yamamoto S, Chao HT, Comjean A, Mohr SE, et al. MARRVEL: Integration of Human and Model Organism Genetic Resources to Facilitate Functional Annotation of the Human Genome. Am J Hum Genet. 2017;100:843–853. doi: 10.1016/j.ajhg.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jenett A, Rubin GM, Ngo TT, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, et al. A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2012;2:991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Panser K, Tirian L, Schulze F, Villalba S, Jefferis GS, Buhler K, Straw AD. Automatic Segmentation of Drosophila Neural Compartments Using GAL4 Expression Data Reveals Novel Visual Pathways. Curr Biol. 2016;26:1943–1954. doi: 10.1016/j.cub.2016.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16**.Sahin A, Held A, Bredvik K, Major P, Achilli TM, Kerson AG, Wharton K, Stilwell G, Reenan R. Human SOD1 ALS Mutations in a Drosophila Knock-In Model Cause Severe Phenotypes and Reveal Dosage-Sensitive Gain- and Loss-of-Function Components. Genetics. 2017;205:707–723. doi: 10.1534/genetics.116.190850. Using homologous recombination, the authors genetically engineered four human ALS-causing SOD1 point mutations into the endogenous fly SOD1 locus and showed that only enzymatically inactive mutants cause neurodegeneration. This is the first study that created fly knock-in models of ALS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xue Z, Wu M, Wen K, Ren M, Long L, Zhang X, Gao G. CRISPR/Cas9 mediates efficient conditional mutagenesis in Drosophila. G3 (Bethesda) 2014;4:2167–2173. doi: 10.1534/g3.114.014159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar A, Christian PK, Panchal K, Guruprasad BR, Tiwari AK. Supplementation of Spirulina (Arthrospira platensis) Improves Lifespan and Locomotor Activity in Paraquat-Sensitive DJ-1betaDelta93 Flies, a Parkinson’s Disease Model in Drosophila melanogaster. J Diet Suppl. 2017;14:573–588. doi: 10.1080/19390211.2016.1275917. [DOI] [PubMed] [Google Scholar]
- 19.Xia Q, Wang H, Hao Z, Fu C, Hu Q, Gao F, Ren H, Chen D, Han J, Ying Z, et al. TDP-43 loss of function increases TFEB activity and blocks autophagosome-lysosome fusion. Embo J. 2016;35:121–142. doi: 10.15252/embj.201591998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beck K, Ehmann N, Andlauer TF, Ljaschenko D, Strecker K, Fischer M, Kittel RJ, Raabe T. Loss of the Coffin-Lowry syndrome-associated gene RSK2 alters ERK activity, synaptic function and axonal transport in Drosophila motoneurons. Dis Model Mech. 2015;8:1389–1400. doi: 10.1242/dmm.021246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong CO, Palmieri M, Li J, Akhmedov D, Chao Y, Broadhead GT, Zhu MX, Berdeaux R, Collins CA, Sardiello M, et al. Diminished MTORC1-Dependent JNK Activation Underlies the Neurodevelopmental Defects Associated with Lysosomal Dysfunction. Cell Rep. 2015;12:2009–2020. doi: 10.1016/j.celrep.2015.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lehmann S, Costa AC, Celardo I, Loh SH, Martins LM. Parp mutations protect against mitochondrial dysfunction and neurodegeneration in a PARKIN model of Parkinson’s disease. Cell Death Dis. 2016;7:e2166. doi: 10.1038/cddis.2016.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson AE, Shu H, Hauswirth AG, Tong A, Davis GW. VCP-dependent muscle degeneration is linked to defects in a dynamic tubular lysosomal network in vivo. Elife. 2015:4. doi: 10.7554/eLife.07366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farca Luna AJ, Perier M, Seugnet L. Amyloid Precursor Protein in Drosophila Glia Regulates Sleep and Genes Involved in Glutamate Recycling. J Neurosci. 2017;37:4289–4300. doi: 10.1523/JNEUROSCI.2826-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li A, Hooli B, Mullin K, Tate RE, Bubnys A, Kirchner R, Chapman B, Hofmann O, Hide W, Tanzi RE. Silencing of the Drosophila ortholog of SOX5 leads to abnormal neuronal development and behavioral impairment. Hum Mol Genet. 2017;26:1472–1482. doi: 10.1093/hmg/ddx051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dubos A, Castells-Nobau A, Meziane H, Oortveld MA, Houbaert X, Iacono G, Martin C, Mittelhaeuser C, Lalanne V, Kramer JM, et al. Conditional depletion of intellectual disability and Parkinsonism candidate gene ATP6AP2 in fly and mouse induces cognitive impairment and neurodegeneration. Hum Mol Genet. 2015;24:6736–6755. doi: 10.1093/hmg/ddv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neely GG, Hess A, Costigan M, Keene AC, Goulas S, Langeslag M, Griffin RS, Belfer I, Dai F, Smith SB, et al. A genome-wide Drosophila screen for heat nociception identifies alpha2delta3 as an evolutionarily conserved pain gene. Cell. 2010;143:628–638. doi: 10.1016/j.cell.2010.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stowers RS, Megeath LJ, Gorska-Andrzejak J, Meinertzhagen IA, Schwarz TL. Axonal transport of mitochondria to synapses depends on milton, a novel Drosophila protein. Neuron. 2002;36:1063–1077. doi: 10.1016/s0896-6273(02)01094-2. [DOI] [PubMed] [Google Scholar]
- 29.Bellen HJ, Tong C, Tsuda H. 100 years of Drosophila research and its impact on vertebrate neuroscience: a history lesson for the future. Nat Rev Neurosci. 2010;11:514–522. doi: 10.1038/nrn2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30**.Yamamoto S, Jaiswal M, Charng WL, Gambin T, Karaca E, Mirzaa G, Wiszniewski W, Sandoval H, Haelterman NA, Xiong B, et al. A drosophila genetic resource of mutants to study mechanisms underlying human genetic diseases. Cell. 2014;159:200–214. doi: 10.1016/j.cell.2014.09.002. The authors generated a large collection of X-chromosome mutants through an unbiased forward genetic screen performed to isolate essential genes involved in neuronal function and maintenance. This screen allowed identification and detailed characterization of numerous genes whose human homologues are associated with neurological diseases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osterloh JM, Yang J, Rooney TM, Fox AN, Adalbert R, Powell EH, Sheehan AE, Avery MA, Hackett R, Logan MA, et al. dSarm/Sarm1 is required for activation of an injury-induced axon death pathway. Science. 2012;337:481–484. doi: 10.1126/science.1223899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGurk L, Berson A, Bonini NM. Drosophila as an In Vivo Model for Human Neurodegenerative Disease. Genetics. 2015;201:377–402. doi: 10.1534/genetics.115.179457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chouhan AK, Guo C, Hsieh YC, Ye H, Senturk M, Zuo Z, Li Y, Chatterjee S, Botas J, Jackson GR, et al. Uncoupling neuronal death and dysfunction in Drosophila models of neurodegenerative disease. Acta Neuropathol Commun. 2016;4:62. doi: 10.1186/s40478-016-0333-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cook RK, Deal ME, Deal JA, Garton RD, Brown CA, Ward ME, Andrade RS, Spana EP, Kaufman TC, Cook KR. A new resource for characterizing X-linked genes in Drosophila melanogaster: systematic coverage and subdivision of the X chromosome with nested, Y-linked duplications. Genetics. 2010;186:1095–1109. doi: 10.1534/genetics.110.123265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haelterman NA, Jiang L, Li Y, Bayat V, Sandoval H, Ugur B, Tan KL, Zhang K, Bei D, Xiong B, et al. Large-scale identification of chemically induced mutations in Drosophila melanogaster. Genome Res. 2014;24:1707–1718. doi: 10.1101/gr.174615.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Venken KJ, Popodi E, Holtzman SL, Schulze KL, Park S, Carlson JW, Hoskins RA, Bellen HJ, Kaufman TC. A molecularly defined duplication set for the X chromosome of Drosophila melanogaster. Genetics. 2010;186:1111–1125. doi: 10.1534/genetics.110.121285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang K, Li Z, Jaiswal M, Bayat V, Xiong B, Sandoval H, Charng WL, David G, Haueter C, Yamamoto S, et al. The C8ORF38 homologue Sicily is a cytosolic chaperone for a mitochondrial complex I subunit. J Cell Biol. 2013;200:807–820. doi: 10.1083/jcb.201208033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu L, Zhang K, Sandoval H, Yamamoto S, Jaiswal M, Sanz E, Li Z, Hui J, Graham BH, Quintana A, et al. Glial lipid droplets and ROS induced by mitochondrial defects promote neurodegeneration. Cell. 2015;160:177–190. doi: 10.1016/j.cell.2014.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jaiswal M, Haelterman NA, Sandoval H, Xiong B, Donti T, Kalsotra A, Yamamoto S, Cooper TA, Graham BH, Bellen HJ. Impaired Mitochondrial Energy Production Causes Light-Induced Photoreceptor Degeneration Independent of Oxidative Stress. PLoS Biol. 2015;13:e1002197. doi: 10.1371/journal.pbio.1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sandoval H, Yao CK, Chen K, Jaiswal M, Donti T, Lin YQ, Bayat V, Xiong B, Zhang K, David G, et al. Mitochondrial fusion but not fission regulates larval growth and synaptic development through steroid hormone production. Elife. 2014:3. doi: 10.7554/eLife.03558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang S, Tan KL, Agosto MA, Xiong B, Yamamoto S, Sandoval H, Jaiswal M, Bayat V, Zhang K, Charng WL, et al. The retromer complex is required for rhodopsin recycling and its loss leads to photoreceptor degeneration. PLoS Biol. 2014;12:e1001847. doi: 10.1371/journal.pbio.1001847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen K, Lin G, Haelterman NA, Ho TS, Li T, Li Z, Duraine L, Graham BH, Jaiswal M, Yamamoto S, et al. Loss of Frataxin induces iron toxicity, sphingolipid synthesis, and Pdk1/Mef2 activation, leading to neurodegeneration. Elife. 2016:5. doi: 10.7554/eLife.16043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43*.Yoon WH, Sandoval H, Nagarkar-Jaiswal S, Jaiswal M, Yamamoto S, Haelterman NA, Putluri N, Putluri V, Sreekumar A, Tos T, et al. Loss of Nardilysin, a Mitochondrial Co-chaperone for alpha-Ketoglutarate Dehydrogenase, Promotes mTORC1 Activation and Neurodegeneration. Neuron. 2017;93:115–131. doi: 10.1016/j.neuron.2016.11.038. By combining forward genetic approaches and the MiMIC-derived T2A-GAL4 strategy, the authors identified a novel role for Nardilysin as a mitochondrial co-chaperone for 2-oxoglutarate dehydrogenase and identified patients with recessive mutations exhibiting an acquired microcephaly for both genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McGuire SE, Mao Z, Davis RL. Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci STKE. 2004:pl6. doi: 10.1126/stke.2202004pl6. [DOI] [PubMed] [Google Scholar]
- 45.Niehues S, Bussmann J, Steffes G, Erdmann I, Kohrer C, Sun L, Wagner M, Schafer K, Wang G, Koerdt SN, et al. Impaired protein translation in Drosophila models for Charcot-Marie-Tooth neuropathy caused by mutant tRNA synthetases. Nat Commun. 2015;6:7520. doi: 10.1038/ncomms8520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sreedharan J, Neukomm LJ, Brown RH, Jr, Freeman MR. Age-Dependent TDP-43-Mediated Motor Neuron Degeneration Requires GSK3, hat-trick, and xmas-2. Curr Biol. 2015;25:2130–2136. doi: 10.1016/j.cub.2015.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shaw JL, Zhang S, Chang KT. Bidirectional Regulation of Amyloid Precursor Protein-Induced Memory Defects by Nebula/DSCR1: A Protein Upregulated in Alzheimer’s Disease and Down Syndrome. J Neurosci. 2015;35:11374–11383. doi: 10.1523/JNEUROSCI.1163-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lai SL, Lee T. Genetic mosaic with dual binary transcriptional systems in Drosophila. Nat Neurosci. 2006;9:703–709. doi: 10.1038/nn1681. [DOI] [PubMed] [Google Scholar]
- 49.Potter CJ, Tasic B, Russler EV, Liang L, Luo L. The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell. 2010;141:536–548. doi: 10.1016/j.cell.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50**.Pearce MM, Spartz EJ, Hong W, Luo L, Kopito RR. Prion-like transmission of neuronal huntingtin aggregates to phagocytic glia in the Drosophila brain. Nat Commun. 2015;6:6768. doi: 10.1038/ncomms7768. Using the GAL4 and Q- system, the authors identified an essential role for glia in clearance of Huntingtin aggregates from axons undergoing Wallerian degeneration. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51*.Babcock DT, Ganetzky B. Transcellular spreading of huntingtin aggregates in the Drosophila brain. Proc Natl Acad Sci U S A. 2015;112:E5427–5433. doi: 10.1073/pnas.1516217112. By combining the GAL4/UAS and LexA/LexAop binary systems, the authors monitored the spreading of fluorescently tagged Huntingtin aggregates in Drosophila brain. This study contributed to our understanding of the mechanisms responsible for transcellular spreading of huntingtin aggregates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang L, Hagemann TL, Kalwa H, Michel T, Messing A, Feany MB. Nitric oxide mediates glial-induced neurodegeneration in Alexander disease. Nat Commun. 2015;6:8966. doi: 10.1038/ncomms9966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53**.Aguilar JI, Dunn M, Mingote S, Karam CS, Farino ZJ, Sonders MS, Choi SJ, Grygoruk A, Zhang Y, Cela C, et al. Neuronal Depolarization Drives Increased Dopamine Synaptic Vesicle Loading via VGLUT. Neuron. 2017;95:1074–1088. e1077. doi: 10.1016/j.neuron.2017.07.038. By combining genetic, pharmacological, and imaging approaches, the authors studied mechanisms underlying regulation of the presynaptic machinery for neurotransmission. This is the first study to show the regulation of dopamine content in response to depolarization in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boeynaems S, Bogaert E, Michiels E, Gijselinck I, Sieben A, Jovicic A, De Baets G, Scheveneels W, Steyaert J, Cuijt I, et al. Drosophila screen connects nuclear transport genes to DPR pathology in c9ALS/FTD. Sci Rep. 2016;6:20877. doi: 10.1038/srep20877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55*.Freibaum BD, Lu Y, Lopez-Gonzalez R, Kim NC, Almeida S, Lee KH, Badders N, Valentine M, Miller BL, Wong PC, et al. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature. 2015;525:129–133. doi: 10.1038/nature14974. Using the GAL4/UAS system, these authors created transgenic flies expressing 8, 28 or 58 G4C2-repeat-containing transcripts and showed that a G4C2-repeat expansion compromises nucleocytoplasmic transport. This study revealed a novel mechanism of neurodegeneration in c9FTD/ALS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Coyne AN, Yamada SB, Siddegowda BB, Estes PS, Zaepfel BL, Johannesmeyer JS, Lockwood DB, Pham LT, Hart MP, Cassel JA, et al. Fragile X protein mitigates TDP-43 toxicity by remodeling RNA granules and restoring translation. Hum Mol Genet. 2015;24:6886–6898. doi: 10.1093/hmg/ddv389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chihara T, Luginbuhl D, Luo L. Cytoplasmic and mitochondrial protein translation in axonal and dendritic terminal arborization. Nat Neurosci. 2007;10:828–837. doi: 10.1038/nn1910. [DOI] [PubMed] [Google Scholar]
- 58.Wangler MF, Yamamoto S, Chao HT, Posey JE, Westerfield M, Postlethwait J, Hieter P, Boycott KM, Campeau PM, Bellen HJ. Model Organisms Facilitate Rare Disease Diagnosis and Therapeutic Research. Genetics. 2017;207:9–27. doi: 10.1534/genetics.117.203067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sujkowski A, Rainier S, Fink JK, Wessells RJ. Delayed Induction of Human NTE (PNPLA6) Rescues Neurodegeneration and Mobility Defects of Drosophila swiss cheese (sws) Mutants. PLoS One. 2015;10:e0145356. doi: 10.1371/journal.pone.0145356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luo X, Rosenfeld JA, Yamamoto S, Harel T, Zuo Z, Hall M, Wierenga K, Pastore MT, Bartholomew D, Delgado MR, et al. Clinically severe CACNA1A alleles affect synaptic function and neurodegeneration differentially. PLoS Genet. 2017;13:e1006905. doi: 10.1371/journal.pgen.1006905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tian X, Gala U, Zhang Y, Shang W, Nagarkar Jaiswal S, di Ronza A, Jaiswal M, Yamamoto S, Sandoval H, Duraine L, et al. A voltage-gated calcium channel regulates lysosomal fusion with endosomes and autophagosomes and is required for neuronal homeostasis. PLoS Biol. 2015;13:e1002103. doi: 10.1371/journal.pbio.1002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Venken KJ, He Y, Hoskins RA, Bellen HJ. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 2006;314:1747–1751. doi: 10.1126/science.1134426. [DOI] [PubMed] [Google Scholar]
- 63**.Weinberger S, Topping MP, Yan J, Claeys A, Geest N, Ozbay D, Hassan T, He X, Albert JT, Hassan BA, et al. Evolutionary changes in transcription factor coding sequence quantitatively alter sensory organ development and function. Elife. 2017:6. doi: 10.7554/eLife.26402. By knocking in different atonal orthologs and a fly paralog in fly locus, the authors explored the contribution of Atonal family of proneural transcription factor coding sequence variation in the evolution of sense organs. This is the first direct functional comparison of closely related developmental gene orthologs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jakobsdottir J, van der Lee SJ, Bis JC, Chouraki V, Li-Kroeger D, Yamamoto S, Grove ML, Naj A, Vronskaya M, Salazar JL, et al. Rare Functional Variant in TM2D3 is Associated with Late-Onset Alzheimer’s Disease. PLoS Genet. 2016;12:e1006327. doi: 10.1371/journal.pgen.1006327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rong YS, Golic KG. Gene targeting by homologous recombination in Drosophila. Science. 2000;288:2013–2018. doi: 10.1126/science.288.5473.2013. [DOI] [PubMed] [Google Scholar]
- 66.Housden BE, Lin S, Perrimon N. Cas9-based genome editing in Drosophila. Methods Enzymol. 2014;546:415–439. doi: 10.1016/B978-0-12-801185-0.00019-2. [DOI] [PubMed] [Google Scholar]
- 67.Caussinus E, Kanca O, Affolter M. Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nat Struct Mol Biol. 2011;19:117–121. doi: 10.1038/nsmb.2180. [DOI] [PubMed] [Google Scholar]
- 68*.Nagarkar-Jaiswal S, Manivannan SN, Zuo Z, Bellen HJ. A cell cycle-independent, conditional gene inactivation strategy for differentially tagging wild-type and mutant cells. Elife. 2017;6 doi: 10.7554/eLife.26420. The authors developed a cell cycle independent FRT mediated conditional gene inactivation strategy that marks wild-type and mutant cells with different fluorescent marker. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69*.Fisher YE, Yang HH, Isaacman-Beck J, Xie M, Gohl DM, Clandinin TR. FlpStop, a tool for conditional gene control in Drosophila. Elife. 2017:6. doi: 10.7554/eLife.22279. The authors describe a conditional gene disruption method based on FRT in post-mitotic cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schnutgen F, Doerflinger N, Calleja C, Wendling O, Chambon P, Ghyselinck NB. A directional strategy for monitoring Cre-mediated recombination at the cellular level in the mouse. Nat Biotechnol. 2003;21:562–565. doi: 10.1038/nbt811. [DOI] [PubMed] [Google Scholar]
- 71.Chao HT, Davids M, Burke E, Pappas JG, Rosenfeld JA, McCarty AJ, Davis T, Wolfe L, Toro C, Tifft C, et al. A Syndromic Neurodevelopmental Disorder Caused by De Novo Variants in EBF3. Am J Hum Genet. 2017;100:128–137. doi: 10.1016/j.ajhg.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang X, Koolhaas WH, Schnorrer F. A versatile two-step CRISPR- and RMCE-based strategy for efficient genome engineering in Drosophila. G3 (Bethesda) 2014;4:2409–2418. doi: 10.1534/g3.114.013979. [DOI] [PMC free article] [PubMed] [Google Scholar]