Despite recent progress in treatment, prognostication for T-cell acute lymphoblastic leukemia (T-ALL) patients, including those with early T-cell precursor ALL (ETP-ALL), is rather poor [1]. Therefore, new therapies are needed to extend the complete remission time/cure and/or to treat refractory T-ALL/ETP-ALL patients.
T-ALL accumulate potentially lethal ‘spontaneous’ DNA double-strand breaks (DSBs) caused by RAG1/2 in ATM-deficient cells and/or by reactive oxygen species (ROS) [2,3]. In addition, one of the goals of intensive therapies is to induce DSBs. Thus, T-ALL cells may be ‘addicted’ to DSB repair mechanisms to survive spontaneous/induced DSBs and targeting these pathways should sensitize them to the lethal effect of DSBs.
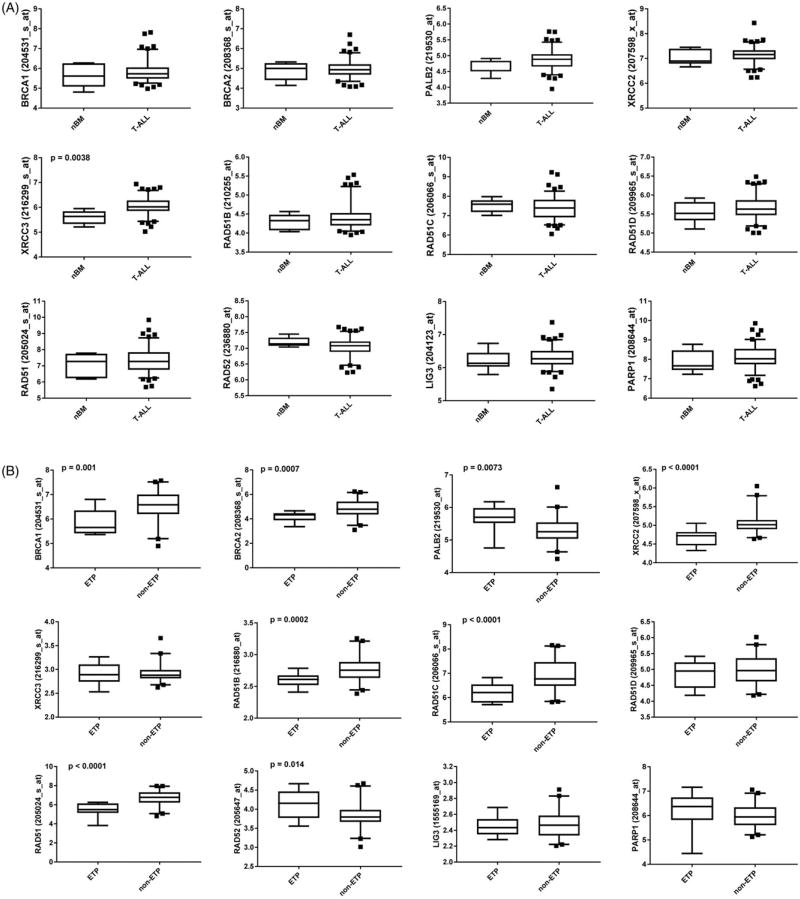
DSBs are usually repaired by two major repair pathways, homologous recombination (HR) repair and non-homologous end-joining (NHEJ) [4]. While NHEJ plays a major role in non-proliferating cells, HR works predominantly on broken replication forks and usually depends on BRCA1, BRCA2, PALB2, RAD51 paralogs (RAD51B, RAD51C, RAD51D, XRCC2, XRCC3), RAD51-mediated pathway [5]. However, in BRCA-deficient cells displaying low expression of at least one of these genes, PARP1-mediated base excision repair (BER) and alternative nonhomologous end-joining (Alt-NHEJ) and/or RAD52-RAD51-dependent alternative HR mechanisms protect cells from the lethal effect of DSBs [6]. Therefore, PARP1 and/or RAD52 are attractive targets to eliminate BRCA-deficient leukemia cells. mRNA microarray gene expression analyses of 117 T-ALLs and 7 normal bone marrow samples revealed wide-range expression levels of HR genes in T-ALLs (Figure 1(A)) suggesting that some leukemias from individual patients were BRCA-deficient. Moreover, the analysis of 12 ETP-ALLs and 40 T-ALLs indicated downregulation of BRCA1, BRCA2, XRCC2, RAD51B, RAD51C, and RAD51 in ETP-ALLs when compared to T-ALLs (Figure 1(B)) suggesting that numerous ETP-ALLs were BRCA-deficient. Although mutations in HR genes are extremely rare in T-ALL [7], these data implicate a malfunctioning of BRCA-RAD51 repair pathway (BRCAness) in a cohort of T-ALLs, especially in ETP-ALLs due to lower levels of at least one protein in BRCA-mediated HR. At the same time, expression of RAD52 seemed upregulated in ETP-ALLs, and PARP1 and LIG3 were unchanged (Figure 1(B)). The mechanisms responsible for relatively high frequency ETP-ALLs displaying “BRCAness’ are currently not known. Genetic aberrations frequently detected in ETP-ALLs, such as RUNX1 mutations may be responsible for this phenomenon [8,9].
Figure 1.
Expression of HR genes in individual T-ALL/ETP-ALL and normal bone marrow samples. mRNA microarrays genes expression analysis of (A) normal bone marrow samples (nBM, n = 7) and primary T-ALLs (n = 117) from GSE26713 [12] and (B) ETP (n = 12) and non-ETP (n = 40) samples from GSE28703 [7]. Average levels of the probe set signals were obtained from the analysis of Affymetix HG-U133 Plus 2.0 microarrays. Box plots represent the mean ± standard deviation for the selected probes sets (multiple probes sets were tested for each gene). The significance of difference was determined by unpaired two-tailed Student’s t test with Welch’s correction using GraphPad software. Results with a p value <.05 were considered as statistically significant.
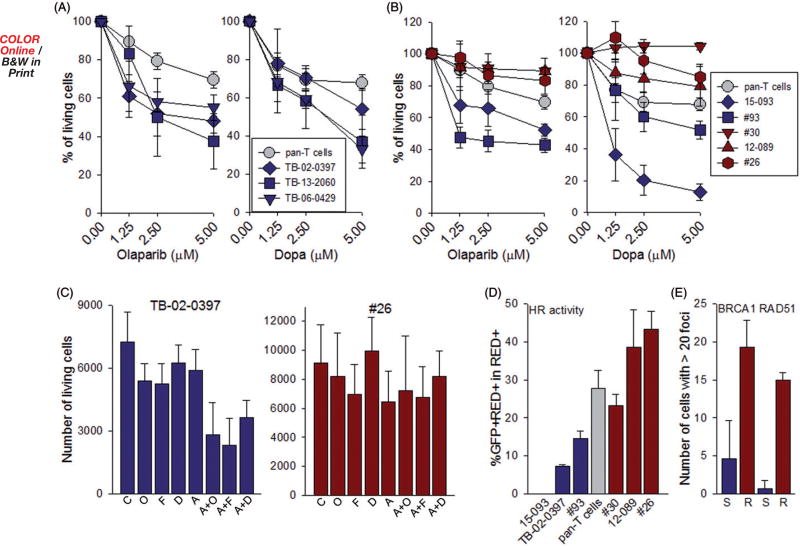
We hypothesized that targeting PARP1 and RAD52 will induce synthetic lethality in T-ALLs/ETP-ALLs displaying ‘BRCAness’ and spare normal cells. Next, we tested the sensitivity of primary leukemia xenograft (PLX) CD3 + cells from three ETP-ALL patients previously identified as BRCA-deficient to DNA repair inhibitors. As controls, we used pan-T cells from healthy donors. All three samples of ETP-ALL cells were more sensitive to PARP1 inhibitor olaparib and RAD52 inhibitor 6(OH)-dl-dopa [10] when compared to normal pan-T cells (Figure 2(A)). However, T-ALL PLXs displayed differences in sensitivity to PARP1 and RAD52 inhibitors; two samples appeared sensitive whereas three seemed resistant when compared to normal pan-T cells (Figure 2(B)). In addition, we tested the combinations of suboptimal concentrations of RAD52 inhibitors 6(OH)-dl-dopa and F79 aptamer [11], PARP1 inhibitor olaparib and/or cytotoxic drug Ara-C. Ara-C combined with PARP1 or RAD52 inhibitors exerted stronger anti-leukemia activity when compared to individual treatments in PARP1/RAD52 inhibitor-sensitive ETP-ALL cells, but not in the resistant T-ALL cells (Figure 2(C)).
Figure 2.
Sensitivity of ETP/T-ALL cells from individual patients to PARP1 and RAD52 inhibitors and HR activity. (A) ETP-ALL and (B) T-ALL primary leukemia xenograft cells from individual patients were cultured in RPMI 1640 supplemented with 10% FBS, 10% human AB serum, recombinant human stem cell factor (SCF, 50 ng/ml), FLT3 ligand (20 ng/ml), IL-7 (10 ng/ml) and insulin (116 ng/ml) [13]. Normal human pan-T cells were purchased from Stemcell Technologies and cultured in ImmunoCult™-XF T Cell Expansion Medium supplemented with ImmunoCult™ Human CD3/CD28/CD2 T Cell Activator (Stem Cell Technologies). Cells were treated with olaparib or 6(OH)-DL-dopa (Dopa) at 0 and 48 hours. Cell viability was determined at 96 hours by Trypan blue exclusion. Results represent mean ± SD percentage of living cells when compared to untreated cells from triplicates. (C) Cells were untreated (C) or treated with olaparib (O, 1.25 µM), 6(OH)-dl-dopa (D, 1.25 µM), F79 peptide aptamer (F, 5µM), Ara-C (A, 5nM) and a combination of these drugs at indicated concentrations at 0 and 48 hours. Cell viability was determined at 96 hours by Trypan blue exclusion. Results represent mean ± SD percentage of living cells when compared to untreated cells from triplicates; *p < .025 in comparison to one drug treatment using Student’s t test. (D) HR activity in primary cells was examined as described before [14]. Cells (1–5 × 106) were co-transfected with 2 µg linearized plasmid carrying HR reporter cassette (HR event restores functional GFP expression) and 0.1 µg dsRedMito vector (for transfection efficiency) using a Human CD34 Cell Nucleofector® Kit (Lonza) and Amaxa Nucleofector (Walkersville, MD) as described before [14]. After 72 hours the percentage of GFP+/DsRed + cells in DsRed + cells was analyzed by flow cytometry (FACSCanto, BD Biosciences, San Jose, CA) to assess HR activity. Results represent mean ± SD from triplicates/sample; *p < .02 in comparison to pan-T cells from healthy donors using Student’s t test. (E) BRCA1 and RAD51 nuclear foci were examined as described before with modifications [15]. Briefly, cells were incubated with 3 µg/ml cisplatin for 12 hours followed by staining with anti-BRCA1 (Calbiochem, OP92 1/500) or anti-RAD51 (Thermo scientific, PA5-27195) antibodies. Secondary antibodies conjugated with Alexa Fluor 488 or Alexa Fluor 594 were applied (Molecular Probes, Eugene, OR). DNA was counterstained with 4′,6′diamedino-2-phenylindole (DAPI). Results represent mean ± SD number of cells with >20 foci/nucleus from three samples per sensitive (S) and resistant (R) groups (50 cells examined per sample); *p < .02 using Student’s t test.
To determine if the differences in sensitivity to PARP1 and RAD52 inhibitors were associated with deficiencies in HR activity, sensitive and resistant leukemia cells as well as normal pan-T cells were transfected with the plasmid-containing HR reporter cassette. HR activity was determined by the restoration of a functional GFP gene and detection of GFP + cells. ETP/T-ALL cells which were sensitive to PARP1 and RAD52 inhibitors displayed diminished HR activity in comparison to normal pan-T cells and T-ALL cells resistant to the inhibitors (Figure 2(D)). Moreover, BRCA1 and RAD51 foci formation was inhibited in leukemia cells which displayed reduced HR activity and increased sensitivity to PARP1 and RAD52 inhibitors (Figure 2(E)).
In summary, our results strongly suggest that the majority of ETP-ALLs and a subset of T-ALLs display the ‘BRCAness’ phenotype, which predisposes them to be sensitive to PARP1 and RAD52 inhibitors. While RAD52 inhibitors await clinical development, PARP1 inhibitors such as Lynparza (olaparib), Rubraca (rucaparib), and Zeluja (niraparib) have been FDA approved to treat breast and/or ovarian cancers carrying BRCA1/2 mutations. We postulate that these inhibitors may be used to improve the therapeutic effect of standard cytotoxic drugs especially in patients with ETP-ALLs.
Acknowledgments
This work was supported by the National Health Institutes [R21CA189378] to T.S. Material was provided in part by the Children’s Oncology Group (AALL15B1-Q to KMW).
Footnotes
Potential conflict of interest: Disclosure forms provided by the authors are available with the full text of this article online at https://doi.org.10.1080/10428194.2017.1397662.
References
- 1.Marks DI, Paietta EM, Moorman AV, et al. T-cell acute lymphoblastic leukemia in adults: clinical features, immunophenotype, cytogenetics, and outcome from the large randomized prospective trial (UKALL XII/ECOG 2993) Blood. 2009;114:5136–5145. doi: 10.1182/blood-2009-08-231217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silva A, Girio A, Cebola I, et al. Intracellular reactive oxygen species are essential for PI3K/Akt/mTOR-dependent IL-7-mediated viability of T-cell acute lymphoblastic leukemia cells. Leukemia. 2011;25:960–967. doi: 10.1038/leu.2011.56. [DOI] [PubMed] [Google Scholar]
- 3.Callen E, Jankovic M, Difilippantonio S, et al. ATM prevents the persistence and propagation of chromosome breaks in lymphocytes. Cell. 2007;130:63–75. doi: 10.1016/j.cell.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 4.Chapman JR, Taylor MR, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell. 2012;47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 5.Jasin M, Rothstein R. Repair of strand breaks by homologous recombination. Cold Spring Harb Perspect Biol. 2013;5:a012740. doi: 10.1101/cshperspect.a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tarsounas M, Davies D, West SC. BRCA2-dependent and independent formation of RAD51 nuclear foci. Oncogene. 2003;22:1115–1123. doi: 10.1038/sj.onc.1206263. [DOI] [PubMed] [Google Scholar]
- 7.Liu Y, Easton J, Shao Y, et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet. 2017;49:1211–1218. doi: 10.1038/ng.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J, Ding L, Holmfeldt L, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang CQ, Krishnan V, Tay LS, et al. Disruption of Runx1 and Runx3 leads to bone marrow failure and leukemia predisposition due to transcriptional and DNA repair defects. Cell Rep. 2014;8:767–782. doi: 10.1016/j.celrep.2014.06.046. [DOI] [PubMed] [Google Scholar]
- 10.Chandramouly G, McDevitt S, Sullivan K, et al. Small-molecule disruption of RAD52 rings as a mechanism for precision medicine in BRCA-deficient cancers. Chem Biol. 2015;22:1491–1504. doi: 10.1016/j.chembiol.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cramer-Morales K, Nieborowska-Skorska M, Scheibner K, et al. Personalized synthetic lethality induced by targeting RAD52 in leukemias identified by gene mutation and expression profile. Blood. 2013;122:1293–1304. doi: 10.1182/blood-2013-05-501072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Homminga I, Pieters R, Langerak AW, et al. Integrated transcript and genome analyses reveal NKX2-1 and MEF2C as potential oncogenes in T cell acute lymphoblastic leukemia. Cancer Cell. 2011;19:484–497. doi: 10.1016/j.ccr.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 13.Masiero M, Minuzzo S, Pusceddu I, et al. Notch3-mediated regulation of MKP-1 levels promotes survival of T acute lymphoblastic leukemia cells. Leukemia. 2011;25:588–598. doi: 10.1038/leu.2010.323. [DOI] [PubMed] [Google Scholar]
- 14.Nieborowska-Skorska M, Sullivan K, Dasgupta Y, et al. Gene expression and mutation-guided synthetic lethality eradicates proliferating and quiescent leukemia cells. J Clin Invest. 2017;127:2392–2406. doi: 10.1172/JCI90825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slupianek A, Schmutte C, Tombline G, et al. BCR/ABL regulates mammalian RecA homologs, resulting in drug resistance. Mol Cell. 2001;8:795–806. doi: 10.1016/s1097-2765(01)00357-4. [DOI] [PubMed] [Google Scholar]