Abstract
Genome sequence of Paenibacillus polymyxa ND25 isolated from cow rumen is reported for being a potential candidate in hydrolysis of lignocellulosic plant biomass. Draft genome sequence generated 5.73 Mb data containing 4922 putative protein coding genes, of which 140 are annotated for glycoside hydrolases. P. polymyxa ND25 strain comprises diverse lignocellulolytic components, especially 12 cellulase along with 23 hemicellulases and 11 esterases, signifying its potential for lignocellulose hydrolysis. Subsequent enzyme assay exhibited the potential of strain to produce 0.49, 0.24 and 0.44 U/ml U/ml of endoglucanase, exoglucanase and β-glucosidase, respectively, utilizing sugarcane bagasse as the sole carbon source. This study signifies the efficient application of P. polymyxa ND25 for facilitating plant-biomass utilization.
Electronic supplementary material
The online version of this article (10.1007/s13205-018-1274-3) contains supplementary material, which is available to authorized users.
Keywords: Paenibacillus polymyxa ND25, Genome, Glycoside hydrolases, Cellulase, Hemicellulase, Esterase
Introduction
Lignocellulosic biomass is the most abundant complex biopolymer primarily composed of cellulose, hemicelluloses and lignin. The recalcitrant cellulosic constituent of the lignocelluloses is a rich source of fermentable sugar, obtained by the process known as saccharification (Shirkavand et al. 2016). The enzymatic saccharification process of cellulose is therefore gaining much needed attention worldwide, as it offers a potent renewable energy resource by converting glucose into value-added bioproducts (Aggarwal et al. 2017). In nature, lignocellulosic biomass is degraded by the complex set of synergistically acting enzymes (Sukumaran et al. 2009). Majority of these enzymes belongs to one of the families of glycoside hydrolases (GHs), a subgroup of the carbohydrate-active enzymes (CAZymes) (Lombard et al. 2014). The entire cellulolytic system consists of endoglucanases, exoglucanases and β-glucosidases operating in synergy for complete hydrolysis of cellulose to sugars (He et al. 2017; Sadhu et al. 2013). Cellulases are mainly found in GH1, GH3, GH5, GH6, GH7, GH8 GH9, GH12, GH45, and GH48 families. Similarly hemicellulases hydrolyze bonds within hemicelluloses and are further classified into endoxylanases, xyloglucanases, xylosidases, endomannanases, mannosidases, arabinofuranosidases, fucosidases, glucuronidase, arabinosidase, galactosidase, etc. Hemicellulolytic enzymes are mainly found in GH2, GH10, GH11, GH16, GH26, GH30, GH31, GH36, GH43, GH51, GH74 and GH95 families. Along with GHs, other CAZymes such as carbohydrate esterases (CEs), Polysaccharide lyases (PLs) and carbohydrate-binding modules (CBM) that help in binding enzymes to cellulose or hemicellulose also play crucial role in biomass hydrolysis (López-Mondéjar et al 2016). A broad array of microbes, along with bacteria, fungi, and actinomycetes, are reported to produce cellulases (Bomble et al. 2017; Saini et al. 2015). However, current research is shifted towards bacterial cellulase systems as they have unique cellulolytic mechanisms and high enzyme specificity in their arsenal (Woo et al 2014). P. polymyxa, a member of phyla fermicutes has gained considerable attention primarily for their ability to suppress plant disease or promoting crop growth by nitrogen fixation (Padda et al. 2017). However, its potential for lignocellulosic biomass deconstruction remains largely overlooked as the previous studies are mainly focused on the catalytic efficiency of individual enzymes (Gastelum-Arellanez et al 2014; Weselowski et al 2016). Considering the synergy of the enzymes involved in breakdown of lignocellulosic biomass, the hydrolytic potential of P. polymyxa needs to be systematically studied. This study provides an in-depth analysis of P. polymyxa ND25 genome for cellulolytic and hemicellulolytic potential. The strain was isolated from cow rumen, a naturally adapted and efficient biomass hydrolyzing habitat. Results reveal the presence of a complex multi-component enzymatic system that support the use of P. polymyxa ND25 for effective hydrolysis of cellulosic waste for the large-scale production of value added products.
Materials and methods
Strain isolation, identification and enzymatic activity of P. polymyxa ND25
Paenibacillus polymyxa ND25 was isolated from the cattle rumen sample by plating the sample on Bergs minimal salt (BMS) agar media (Pawar et al. 2015) amended with CMC and incubated at 37 °C. After 48 h, agar plates were stained with 0.1% congo red for the presence of clear halos around bacterial colonies indicating CMC hydrolysis. Cellulase production efficiency of the bacterial isolates was further tested during growth in BMS minimal medium with CMC, avicel, corn starch, rice straw and sugarcane bagasse as the sole carbon source. Cellulose hydrolyzing efficiency was estimated based on endoglucanacse, exogucanase, β-glucosidase activitiy and soluble COD (sCOD) concentration of supernatant (Nitisinpraserta and Temmes 1991; An et al. 2004).
The bacterial 16S rRNA gene of the strain ND25 was sequenced using the primers 27F and 1492R as described by Dafale et al. (2010). For identification sequencing results were compared using the Basic Local Alignment Search Tool (BLAST) program on NCBI.
DNA extraction, genome sequencing, gene assembly prediction
Genome sequencing approach was used for exploring genes responsible for lignocellulose hydrolysis present in the genome of P. polymyxa ND25. Genomic DNA extraction from P. polymyxa ND25 was carried out using Fast DNA SPIN Kit for Soil from MP Biomedicals following manufacturer’s instruction. Nanodrop 8000 and Qubit® 2.0 Fluorometer were used for determination of A260/280 ratio and concentration, respectively. Illumina TruSeq Nano DNA HT Library Preparation Kit was used for generating paired-end sequencing library which was analyzed using High Sensitivity DNA chip in Bioanalyzer 2100. Cluster generation and sequencing was performed using Illumina Miseq platform. The 5.73 Mb high-quality data was assembled into 105 contigs using Velvet on optimized k-mer. The genome was annotated using the NCBI prokaryotic genomes automatic annotation pipeline (PGAAP). Graphical Comparison of P. polymyxa ND25 genome with its closest neighbor, P. polymyxa M1 (accession no. NC_017542.1), P. polymyxa Sb3-1 (accession no NZ_CP010268.1) and P. polymyxa SC2 (accession no. NC_014622.2) was performed using CGView server (http://stothard.afns.ualberta.ca/cgview_server/) using default parameters. Analysis of sequenced genome was achieved using dbCAN carbohydrate-active enzymes (CAZy) annotation algorithm from dbCAN pipelines (http://csbl.bmb.uga.edu/dbCAN/index.php) against the Carbohydrate-Active Enzymes database (http://www.cazy.org/) (Yin et al. 2014; Lombard et al. 2014) for identifying genes responsible in lignocellulose deconstruction. Functional characterization of CDSs annotated by dbCAN as GHs was done against NCBI’s non-redundant (nr) database and protein data bank (pdb) using the BLASTP algorithm (Gujar et al. 2018). For comparing GHs distribution within P. polymyxa strains, genome sequences of 28 available strains were retrieved from NCBI database and were annotated using dbCAN carbohydrate-active enzymes (CAZy) annotation algorithm. Name and accession numbers of the strain retrieved from NCBI database are provided in supplementary table 1.
Results and discussion
Identification and cellulolytic potential of the P. polymyxa ND25
Exploration of natural habitat where lignocelluloses represents an important resource is a promising strategy for isolation of bacterial strains helpful in biomass hydrolysis. In this context, cattle rumen was explored for identifying bacterial strains with novel enzymatic machinery well adapted to harsh conditions in biomass conversion occurring in these ecosystems. Bacterial isolate ND25, initially isolated from cattle rumen exhibited the ability to utilize diverse carbon source—CMC, rice straw, corn powder, avicel and sugarcane bagasse (SB) for efficient production of extracellular cellulase (fig. S1). Maximal enzyme production, 0.49 U/ml endoglucanase, 0.24 U/ml exoglucanase and 0.44 U/ml β-glucosidase activity was observed when 2% SB was used as sole carbon source. SB hydrolysis by the strain ND25 was simultaneously depicted by the significant rise in sCOD from 548 to 1975 mg/l after 48 h with no significant change in sCOD of control variant (fig. S2).The observed rise in sCOD concentration reflects hydrolysis of insoluble organic components of SB into soluble fermentable sugars.
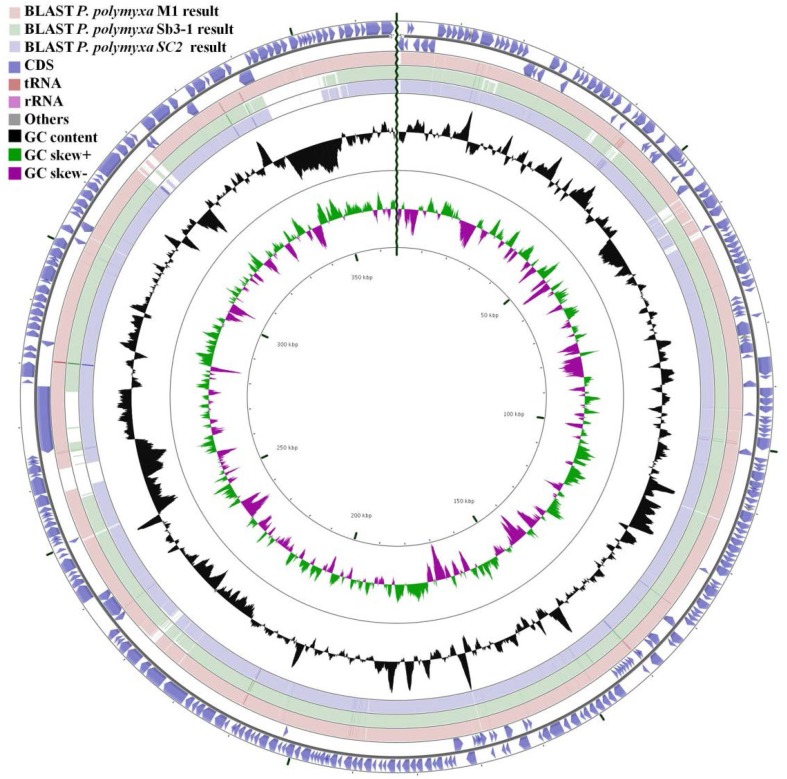
The isolate, referred to ND25, was identified as P. polymyxa by analysis of 16S rRNA gene sequence and showed closest match with P. polymyxa M1, P. polymyxa Sb3-1and P. polymyxa SC2. A circular map comparing P. polymyxa ND25 with the whole genome of above three closest matches can be viewed in Fig. 1. Graphical representation of genome by CGView server shows nearly 90% similarities with the other three strains. The figure also reveals the presence of protein coding genes which are unique to P. polymyxa ND25.
Fig. 1.
Graphical comparision of P. polymyxa ND25 genome with P. polymyxa M1 (accession no. NC_017542.1), P. polymyxa Sb3-1(accession no NZ_CP010268.1) and P. polymyxa SC2 (accession no. NC_014622.2) genome retrireved from NCBI database using CGView server using default parameters
Genomic features of P. polymyxa ND25
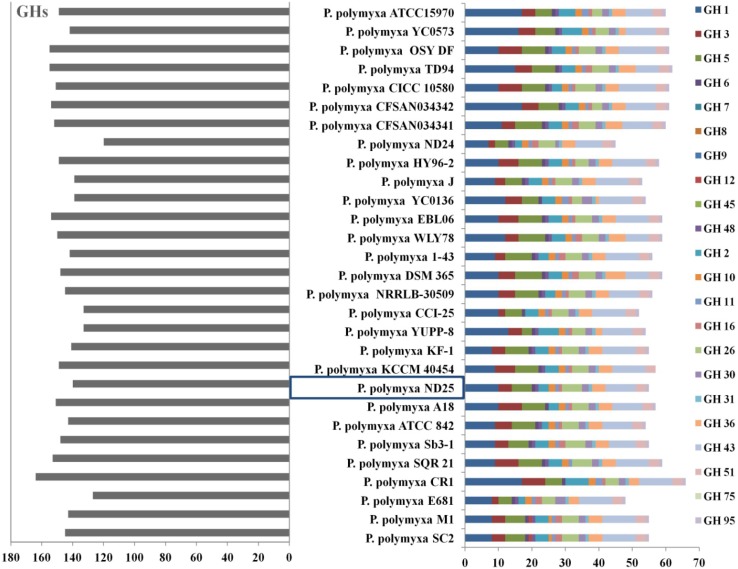
Genome assembly generated 105 scaffolds containing 5.73 Mb of DNA. Gene prediction revealed 4922 protein-coding genes, 100 tRNA genes and 14 rRNA genes (Table 1). dbCAN carbohydrate-active enzymes (CAZy) annotation algorithm identified 328 (6.66%) genes among the predicted protein belong to CAZy family, comprising 140 glycoside hydrolases (GHs), 52 carbohydrate esterases (CEs), 13 polysaccharide lyases (PLs), 72 Glycosyl transferases (GTs) and 5 proteins with auxiliary activities (AAs). Cazymes often display associated modular structure which helps in adhesion to the carbohydrates, additionally covering 66 carbohydrate binding modules (CBMs) and 38 surface layer homology domains (SLH). The predicted GHs belonged to 46 different families and several genes were assigned to GH families engaged in cellulolytic and hemicellulolytic deconstruction. Genes belonging to GH6, GH48 (cellulolytic) and GH11, GH16, GH31, GH95 (hemicelulolytic) families were found in single copy number in the genome. Genes from families GH1 (10), GH2 (3), GH3(4), GH5 (6), GH510 (2), GH26 (6), GH30 (2) GH36 (4), GH43 (9) and GH51 (3) encoding putative β-glucosidases, β-xylanases and α-glucuronidase, α-N-arabinofuranosidase, α-arabinosidase, α-galactosidase, β-mannanase, β-mannosidase β-xylosidase, 1,3-β-glucanase, xyloglucanase, and other hemicellulases were more abundant. The number of genes belonging to these families in P. polymyxa ND25 were higher (55) in comparison with other P. polymyxa strains such as E681, ATCC 842, YUPP-8, CCI-25, YC0136, J and ND25, but were similar in strains SC2, M1, SQR 21, Sb3-1, A18, KCCM 40454, KF-1, NRRLB-30509, DSM 365, 1–43, WLY78, EBL06 and HY96-2. Although the strains SC2, M1, SQR 21, CR1 and A18 have comparatively higher number of GHs, the total count of genes responsible for cellulolytic and hemicellulolytic activity remains same in SC2, M1, SQR 21 whereas the CR1 lacks hydrolytic enzyme from GH48 family, responsible for exoglucanase activity (Fig. 2). Further analysis of P. polymyxa ND25 genome revealed the presence of 12 cellulase genes comprising five endoglucanases gene, two exoglucanases and five β-glucosidase genes. Additionally 42 GH’s responsible for hydrolysis of hemicelluloses, four 1,4-β-xylanase, one α-glucuronidase, three β-xylosidase, three α-N-arabinofuranosidase, three α-N-arabinofuranosidase, two α-arabinosidase, six α-galactosidase, one β-mannanase, two β-mannosidase, one 1,3-β-glucanase, one xyloglucanase were also annotated. Moreover, 11 esterase genes, including two acetyl esterase, two acyl-CoA thioesterase, two carboxylesterase and five esterases responsible for complex carbohydrate degradation have also been annotated in P. polymyxa ND25 genome (Table 2). The presence of both types of plant cell wall-degrading enzyme is crucial for hydrolyzing lignocellulosic biomass. The presence of wide ranges of cellulase and hemicellulase in the genome depicts the great potential of P. polymyxa ND25 for hydrolysis of various types of hemicellulosic biomass. Therefore, the enzyme system of P. polymyxa ND25 exhibits itself as potentially more efficient in degrading lignocellulosic biomass.
Table 1.
Genome feature of P. polymyxa ND25
| Features | P. polymyxa ND25 |
|---|---|
| NCBI accession no. | LZEL00000000 |
| Genome size (Mb) | 5.73 |
| G + C content (%) | 45.2 |
| Number of contigs | 105 |
| Total number of genes | 5265 |
| Protein coding genes | 4922 |
| tRNA genes | 100 |
| rRNA | 14 |
| Pseudo gene | 225 |
Fig. 2.
Predicted numbers of glycosyl hydrolases (GH) in the genome of P. polymyxa ND25 and other strains of P. polymyxa. On the left total number of GHs annotated in the genome; on the right gene content in GH families containing enzymes involved in the hydrolysis of cellulose and hemicelluloses
Table 2.
Predicted CAZymes and other potential lignocellulolytic enzymes in the P. polymyxa ND25 genome
Conclusion
Genomic approach for analyzing the hydrolytic arsenal for new cellulolytic isolates reveals a promising strategy for ultimately enhancing the biomass conversion process. Preliminary genomic analysis of P. polymyxa ND 25 revealed diverse lignocellulolytic enzyme system covering five endoglucanases (GH5), two exoglucanases (GH6 and GH48) and five β-glucosidase genes (GH1, GH3). Additionally 42 GHs responsible for hydrolysis of hemicelluloses along with 11 esterase genes were observed which support in effective utilization of plant biomass. Genome information will be useful for exploring the potential of P. polymyxa ND 25 and substantiate its application for the production of biogas and/or other value-added products.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Miss Varsha Bohra acknowledges Department of Science and technology (DST) of India for awarding Junior Research Fellowship (JRF). The funding from DBT project G-1-2282 is gratefully acknowledged for carrying out the work. The manuscript has been checked for plagiarism by Knowledge Resource Centre, CSIR-NEERI, Nagpur, India and assigned KRC No.: CSIR-NEERI/KRC/2018/April/EBGD/2.
Compliance with ethical standards
Conflict of interest
The authors declare no financial or commercial conflict of interest.
Footnotes
Accession numbers: The 16S rDNA gene sequence of P. polymyxa ND25 was deposited in the GenBank database under accession number KX404921. The whole genome shotgun sequence (WGS) of the strain ND25 was deposited in the NCBI-WGS database under accession number LZEL00000000. The accession numbers and locus tag of cellulase, hemicellulase and esterases are listed in Table 2.
Electronic supplementary material
The online version of this article (10.1007/s13205-018-1274-3) contains supplementary material, which is available to authorized users.
References
- Aggarwal NK, Goyal V, Saini A, Yadav A, Gupta R. Enzymatic saccharification of pretreated rice straw by cellulases from Aspergillus niger BK01. 3 Biotech. 2017;7(3):158. doi: 10.1007/s13205-017-0755-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An CL, Lim WJ, Hong SY, Kim EJ, Shin EC, Kim MK, Lee JR, Park SR, Woo JG, Lim YP, Yun HD. Analysis of bgl operon structure and characterization of β-glucosidase from Pectobacterium carotovorum subsp. carotovorum LY34. Biosci Biotechnol Biochem. 2004;68:2270–2278. doi: 10.1271/bbb.68.2270. [DOI] [PubMed] [Google Scholar]
- Bomble YJ, Lin CY, Amore A, Wei H, Holwerda EK, Ciesielski PN, Donohoe BS, Decker SR, Lynd LR, Himmel ME. Lignocellulose deconstruction in the biosphere. Curr Opin Chem Biol. 2017;41:61–70. doi: 10.1016/j.cbpa.2017.10.013. [DOI] [PubMed] [Google Scholar]
- Dafale N, Agrawal L, Kapley A, Meshram S, Purohit H, Wate S. Selection of indicator bacteria based on screening of 16S rDNA metagenomic library from a two-stage anoxic–oxic bioreactor system degrading azo dyes. Bioresour Technol. 2010;101:476–484. doi: 10.1016/j.biortech.2009.08.006. [DOI] [PubMed] [Google Scholar]
- Gastelum-Arellanez A, Paredes-Lopez O, Olalde-Portugal V. Extracellular endoglucanase activity from Paenibacillus polymyxa BEb-40: production, optimization and enzymatic characterization. World J Microbiol Biotechnol. 2014;30:2953–2965. doi: 10.1007/s11274-014-1723-z. [DOI] [PubMed] [Google Scholar]
- Gujar VV, Fuke P, Khardenavis AA, Purohit HJ. Draft genome sequence of Penicilliumchrysogenum strain HKF2, a fungus with potential for production of prebiotic synthesizing enzymes. 3 Biotech. 2018;8(2):106. doi: 10.1007/s13205-018-1132-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R, Bai X, Cai P, Sun C, Zhang D, Chen S. Genome sequence of Talaromycespiceus 9-3 provides insights into lignocellulose degradation. 3 Biotech. 2017;7(6):368. doi: 10.1007/s13205-017-1001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard V, GolacondaRamulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:490–495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Mondéjar R, Zühlke D, Větrovský T, Becher D, Riedel K, Baldrian P. Decoding the complete arsenal for cellulose and hemicellulose deconstruction in the highly efficient cellulose decomposer Paenibacillus O199. Biotechnol Biofuels. 2016;9(1):104. doi: 10.1186/s13068-016-0518-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitisinprasert S, Temmes A. The characteristics of a new non-spore-forming cellulolytic mesophilic anaerobe strain CM126 isolated from municipal sewage sludge. J Appl Bacteriol. 1991;71:154–161. doi: 10.1111/j.1365-2672.1991.tb02972.x. [DOI] [PubMed] [Google Scholar]
- Padda KP, Puri A, Chanway CP. Agriculturally important microbes for sustainable agriculture. Singapore: Springer; 2017. Paenibacillus polymyxa: a prominent biofertilizer and biocontrol agent for sustainable agriculture; pp. 165–191. [Google Scholar]
- Pawar KD, Dar MA, Rajput BP, Kulkarni GJ. Enrichment and identification of cellulolytic bacteria from the gastrointestinal tract of giant African snail, Achatinafulica. Biotechnol Appl Biochem. 2015;175:1971–1980. doi: 10.1007/s12010-014-1379-z. [DOI] [PubMed] [Google Scholar]
- Sadhu S, Saha P, Sen SK, Mayilraj S, Maiti TK (2013) Production, purification and characterization of a novel thermotolerant endoglucanase (CMCase) from Bacillus strain isolated from cow dung. Springer Plus 2(1):10. http://www.springerplus.com/content/2/1/1 [DOI] [PMC free article] [PubMed]
- Saini JK, Saini R, Tewari L. 3 Biotech. 2015;5(4):337–353. doi: 10.1007/s13205-014-0246-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirkavand E, Baroutian S, Gapes DJ, Brent R, Young Combination of fungal and physicochemical processes for lignocellulosic biomass pretreatment—a review. Renew Sust Energy Re. 2016;54:217–234. doi: 10.1016/j.rser.2015.10.003. [DOI] [Google Scholar]
- Sukumaran RK, Singhania RR, Mathew GM, Pandey A. Cellulase production using biomass feed stock and its application in lignocellulose saccharification for bio-ethanol production. Renew Energy. 2009;34(2):421–424. doi: 10.1016/j.renene.2008.05.008. [DOI] [Google Scholar]
- Weselowski B, Nathoo N, Eastman AW, MacDonald J, Yuan ZC. Isolation, identification and characterization of Paenibacillus polymyxa CR1 with potentials for biopesticide, biofertilization, biomass degradation and biofuel production. BMC Microbiol. 2016;16(1):244. doi: 10.1186/s12866-016-0860-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HL, Hazen TC, Simmons BA, DeAngelis KM. Enzyme activities of aerobic lignocellulolytic bacteria isolated from wet tropical forest soils. Syst Appl Microbiol. 2014;37:60–67. doi: 10.1016/j.syapm.2013.10.001. [DOI] [PubMed] [Google Scholar]
- Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2014;40(W1):W445–W451. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.