Abstract
Epidemiological studies suggest a positive association between coronary artery disease (CAD) and late-onset Alzheimer’s disease (LOAD). This large-scale genetic study brings together ‘big data’ resources to examine the causal impact of genetic determinants of CAD on risk of LOAD. A two-sample Mendelian randomization approach was adopted to estimate the causal effect of CAD on risk of LOAD using summary data from 60,801 CAD cases from CARDIoGRAMplusC4D and 17,008 LOAD cases from the IGAP Consortium. Additional analyses assessed the independent relevance of genetic associations at the APOE locus for both CAD and LOAD. Higher genetically determined risk of CAD was associated with a slightly higher risk of LOAD (Odds Ratio (OR) per log-odds unit of CAD [95% CI]: 1.07 [1.01–1.15]; p = 0.027). However, after exclusion of the APOE locus, the estimate of the causal effect of CAD for LOAD was attenuated and no longer significant (OR 0.94 [0.88–1.01]; p = 0.072). This Mendelian randomization study indicates that the APOE locus is the chief determinant of shared genetic architecture between CAD and LOAD, and suggests a lack of causal relevance of CAD for risk of LOAD after exclusion of APOE.
Introduction
Coronary artery disease (CAD), including myocardial infarction and angina, results from atherosclerosis of the underlying coronary arteries that causes obstruction to the blood supply of the heart1,2. Elevated levels of blood pressure and blood lipids, together with cigarette smoking and diabetes are established major risk factors for CAD3. However, CAD is a complex disease involving both environmental and genetic causes, with a heritability of 40–60%4. Genome-wide association studies (GWAS) have identified multiple genetic variants, but each has been associated with only a modest effect on risk of CAD. The CARDIoGRAMplusC4D meta-analysis, involving 60 801 CAD cases and 123 504 controls, identified a total of 57 variants that were associated with CAD5. Few of these variants encoded established risk factors and the effect sizes of these variants were small (OR 1.03–1.37), but together accounted for 13.3% of CAD heritability5.
Dementia is a clinical syndrome characterized by memory loss, difficulties in cognitive function, problem solving or language that affects a high proportion of older people. Alzheimer’s disease (AD) and vascular dementia are the two most common causes of dementia and despite a substantial overlap of their clinical presentation each have distinct neuropathological features6. AD pathology is characterized by extracellular amyloid plaques and intracellular neurofibrillary tangles that precede the onset of clinical symptoms by 1–2 decades7. By contrast, vascular dementia is characterized by micro infarcts and perivascular hyalinosis8. Late onset Alzheimer’s disease (LOAD) is diagnosed by clinical criteria, including insidious onset and progressive impairment of both memory and some other domains of cognitive function in the absence of motor, sensory or coordination deficits in individuals aged over 65 years9.
LOAD has a multifactorial inheritance (with a heritability of 60%)10, the most recent GWAS identified 21 variants that were significantly associated with LOAD11. The APOE locus is the most important locus for LOAD; with individuals with two copies of the ε4 allele (ε4/ε4) having a 15-fold higher risk (OR [95% CI]: 14.9 [10.8–20.6]) and individuals with one copy of the ε4 allele (ε3/ε 4) having a 3-fold higher risk of LOAD (OR [95% CI]: 3.2 [2.8–3.8]) compared with individuals with two copies of the APOE-ε3 allele (ε3/ε3)12. In contrast, carriers of the APOE-ε2 allele had a 40% lower risk (ε2/ε2 OR [95% CI]: 0.6 [0.2–2.0]) and (ε2/ε3 OR [95% CI]: 0.6 [0.5–0.8]) than individuals with two copies of the APOE-ε3 (ε3/ε3)12. Furthermore, the APOE-ε2 (rs7412) has been associated with LDL cholesterol concentrations (effect size [SD units, per C allele] [95% CI]: 0.59 [0.57–0.61]; p = 1.24 × 10−652)13, and rs4420638 (D′ = 1 with rs7412) was associated with a 10% higher risk of CAD (OR [95% CI]: 1.10 [1.07–1.13])5.
Observational studies have implicated atherosclerosis and cardiovascular risk factors in the initiation and progression of dementia, including AD14–18. The Rotterdam study reported a 3-fold higher risk of AD associated with the presence of carotid atherosclerosis (OR [95% CI]: 3.00 [1.50–6.00]; p = 0.001)14. Individuals with both carotid atherosclerosis and the APOE ε4 allele had a 4-fold higher risk of AD (OR [95% CI]: 3.90 [1.60–9.60]) than individuals without either risk factor.
Several studies have also reported that clinically manifest cardiovascular disease was associated with a higher risk of LOAD16–18. However, since LOAD has a very long latency period, such studies have been constrained by confounding, reverse causality bias and diagnostic misclassification. Mendelian randomization (MR) studies afford the potential to elucidate the causal relevance of lifelong differences in exposures with disease outcomes that are independent of confounding and reverse causation19,20. Moreover, MR studies have been successful in enhancing our understanding of the causal risk factors for cardiovascular diseases21.
The aim of the present study was to examine the causal relevance of CAD on risk of LOAD, and explore their shared genetic architecture. The objectives of this study were: (i) to assess the impact of genetic determinants of CAD on risk of LOAD; and (ii) to assess the independent relevance of genetic associations at the APOE locus for both CAD and LOAD.
Methods
Participating cohorts
The CARDIoGRAMplusC4D5 summary statistics were derived from a 1000 Genomes-based meta-analysis of 48 studies, involving 60 801 CAD cases and 123 504 controls. Diagnosis of CAD included evidence of myocardial infarction, chronic stable angina with a revascularisation procedure or a coronary stenosis >50% and CAD cases had a mean age of approximately 60 years5. The International Genomics of Alzheimer’s Project (IGAP)11 summary statistics were derived from a 1000 Genomes-based meta-analysis of LOAD cases among European individuals, involving 17 008 LOAD cases and 37 154 controls. Diagnosis of LOAD followed assessment by a neurologist and LOAD cases had a mean age of approximately 74 years11.
This study presents a new analysis of anonymised summary data from previously published meta-analyses in which each of the individual studies had ethics approval by the relevant institutions where participants (or, for those with substantial cognitive impairment, from an appropriate proxy), provided written informed consent5,11.
Selection of variants
Among the 57 genome-wide significant variants identified in the CARDIoGRAMplusC4D meta-analysis, 52 genetic variants were selected for the present study after exclusions5. Variants with recessive associations (n = 2) and for which a suitable proxy was also unavailable (r2 ≥ 0.80) in IGAP (n = 3), were excluded from the analysis11 (eTable 1). Summary estimates (per effect allele) were extracted from the CARDIoGRAMplusC4D5 meta-analysis for CAD and from the IGAP11 meta-analysis for LOAD (eTables 2 and 3).
A sensitivity analysis including 190 genetic variants of the 214 (including 9 with a minor allele frequency <0.05) with a 5% False Discovery Rate (FDR) identified in the CARDIoGRAMplusC4D meta-analysis was performed22. A total of 24 variants were excluded due to the absence of a suitable proxy (i.e. no variant with r2 ≥ 0.80).
Mendelian randomization analysis
The effect of CAD (risk phenotype) on LOAD (outcome phenotype) was analysed by looking at the impact of each genetic marker’s effect size for CAD on its effect size for LOAD. Effects of individual variants are given per copy of the effect allele unless otherwise stated. This was assessed by calculating the ratio of LOAD effect size/CAD effect size for each of the 52 variants, and combining them using a fixed effect meta-analysis model to estimate the causal effect23. The Cochran Q statistic was used to assess heterogeneity in risk estimates between the variants in the fixed effect meta-analysis. An online database (PhenoScanner)24 was used to identify multiple phenotypes associated with individual genetic variants to investigate potential pleiotropy. Sensitivity analyses using Egger regression MR25 were performed, a method that allows for invalid instrumental variables due to pleiotropy, using the MR-BASE R package26.
Cross-trait LD score regression
The genetic correlation of effect statistics for CAD and for LOAD was estimated by cross-trait LD score regression27 using a total of 5,403,795 variants that were studied in both the CARDIoGRAMplusC4D and IGAP meta-analyses. This method estimates the genetic correlation between the two traits using GWAS summary statistics and is unbiased by any overlap of participants in both study populations27.
Scan for shared genetic determinants at the APOE locus
Shared genetic determinants for LOAD and CAD at the APOE locus were investigated using a recently developed approach known as gwas-pw28. This method uses a statistical model to estimate the posterior probability that a genomic region adheres to four separate models. Models 1 and 2 are used to test whether a locus contains a genetic variant for only one of the two phenotypes. Models 3 and 4 are used to test whether a genetic variant exists for both phenotypes within the locus. More specifically, Model 3 assesses whether the same genetic variant influences both phenotypes, whilst Model 4 assesses whether the two phenotypes are influenced by separate genetic variants within a locus.
Data availability
The datasets analysed during the current study are publically available in the CARDIoGRAMplusC4D5 (CAD) repository http://www.cardiogramplusc4d.org and the IGAP11 (LOAD) repository http://web.pasteur-lille.fr/en/recherche/u744/igap/igap_download.php.
Results
Mendelian randomization analyses
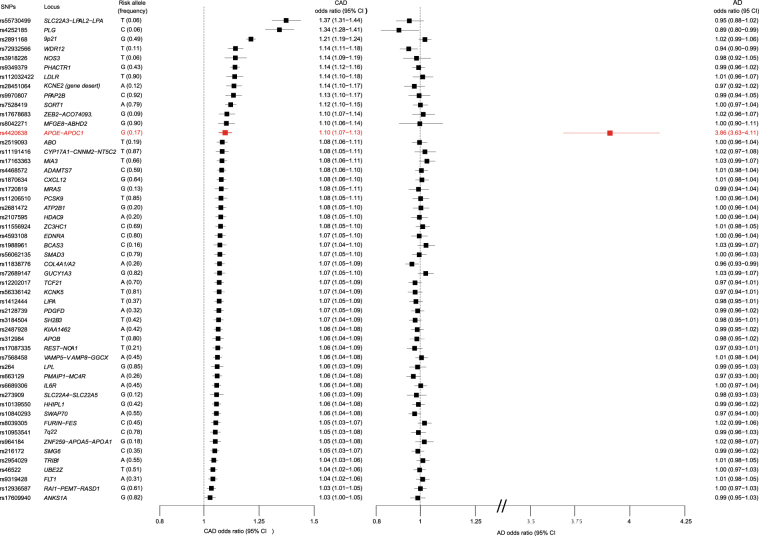
Figure 1 shows the associations of the 52 variants that were used to estimate the causal effect. For each genome-wide significant variant, the odds ratio (OR) and 95% confidence intervals (CI) of the summary statistics in the CARDIoGRAMplusC4D and IGAP meta-analyses are presented. The CAD odds ratios are presented in descending order of strength of their association with CAD and indicate that the APOE rs4420638 genetic variant was the sole variant significantly associated with LOAD.
Figure 1.
Forest plot of variants included in the Mendelian randomization analysis of the causal relevance of CAD for LOAD. Forest plot of odds ratios (OR) for the 52 variants used in the analysis with CAD and LOAD. Variants are reported for the increasing CAD risk allele (i.e. CAD ORs are >1). The APOE locus is marked in red.
Table 1 shows the causal effect estimates on LOAD after combining information across all 52 CAD variants. The results indicate a nominally significant causal association, consistent with a higher risk of CAD being associated with a 7% higher risk of LOAD (OR 1.07 for LOAD per log odds unit of CAD [1.01–1.15]; p = 0.027). However, there was significant heterogeneity between the causal effects for each of the variants included in the analysis (p < 2.2 × 10−308). After removal of the single outlying variant, rs4420638 at the APOE locus, there was no remaining heterogeneity (p = 0.351). Furthermore, after removal of the APOE variant, there was no longer any significant causal association of CAD with LOAD (OR 0.94 for LOAD per log odds unit of CAD [0.88–1.01]; p = 0.072). The causal estimate (based on the 52 CAD variants) from the Egger regression approach was not significant (p = 0.846), suggesting that the APOE variant may not be a valid instrumental variable due to pleiotropy at the APOE locus. Similar results were observed using data for the 190 CAD variants that were significantly associated with CAD at the 5% FDR threshold.
Table 1.
Results of Mendelian randomization analysis for CAD on LOAD.
| Risk Phenotype | Variant count | OR for LOAD[95% CI] | p-value | Cochran’s Q | df | p-value |
|---|---|---|---|---|---|---|
| CAD (GWS) | 52 | 1.07 [1.01–1.15] | 0.027 | 1868.13 | 51 | <2.2 × 10−308 |
| CAD (GWS) (Excluding APOE) | 51 | 0.94 [0.88–1.01] | 0.072 | 53.22 | 50 | 0.351 |
| CAD (GWS) Egger MR | 52 | 1.09 [0.47–2.54] | 0.846 | 1868.13 | 51 | <2.2 × 10−308 |
| CAD (FDR) | 190 | 1.05 [1.01–1,10] | 0.024 | 2059.25 | 189 | <2.2 × 10−308 |
| CAD (FDR) (Excluding APOE) | 189 | 0.99 [0.94–1.03] | 0.553 | 247.40 | 188 | 0.002 |
Results of the analysis of summary statistics from CAD and LOAD. GWS is Genome Wide Significant (p ≥ 5 × 10−8); FDR is a false discovery rate of 5%. APOE refers to the rs4420638 variant within the APOE locus.
Cross-trait LD score regression analysis
A cross-trait regression analysis indicated a non-significant genetic correlation between LOAD and CAD (genetic correlation [95% CI]: −0.02 [−0.20–0.16]; p = 0.84). These results were consistent with the MR analysis after exclusion of the APOE variant.
APOE locus
Figure 2 shows a signal plot of the APOE loci for CAD and LOAD. The peak association signal for CAD was rs4420638 (OR [95% CI]: 1.10 [1.07–1.13]; p = 7.07 × 10−11; effect allele [EA] = G), and the peak signal for LOAD was rs6857 (OR [95% CI]: 3.19 [3.05–3.34]; p = 2.50 × 10−575; EA = G). The variants for the APOE ε2 allele (rs7412) and the APOE ε4 allele (rs429358) were also included in the figure. Table 2 shows the linkage disequilibrium (LD) structure (with D′ and r2) between the variants within the APOE region.
Figure 2.
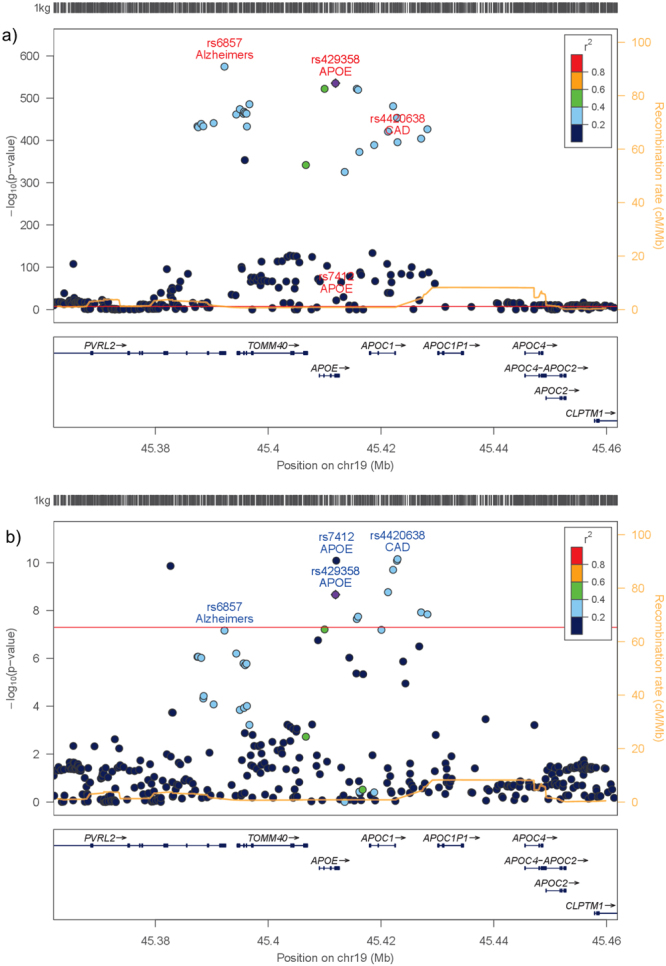
Signal plots of the APOE locus for LOAD and CAD. (a) LocusZoom42 signal plot of the APOE locus from the IGAP cohort. The strongest signal detected was for rs6857. Peak variant for CAD (rs4420638) and the APOE ε2 (rs7412) and ε4 (rs429358) variants are included. (b) Signal plot of the APOE locus from the CARDIoGRAMplusC4D meta-analysis. The strongest association signal detected was for rs4420638. Peak variant for LOAD (rs6857) and the APOE ε2 (rs7412) and ε4 (rs429358) variants are included. LD structure in both plots is in reference to the APOE ε4 (rs429358) variant.
Table 2.
Measures of linkage disequilibrium between variants at the APOE locus.
| rs6857 | rs429358 | rs7412 | rs4420638 | |
|---|---|---|---|---|
| rs6857 | 1.00 (1.00) | 0.64 (0.82) | 0.01 (0.71) | 0.42 (0.74) |
| rs429358 | 1.00 (1.00) | 0.01 (0.68) | 0.66 (0.95) | |
| rs7412 | 1.00 (1.00) | 0.02 (1.00) | ||
| rs4420638 | 1.00 (1.00) |
Linkage disequilibrium estimates taken from European 1000 Genomes for variants within the APOE locus: r2 and D′ (in brackets). Variants shown are as follows: rs6857: Peak variant from the IGAP meta-analysis, rs429358: Variant comprising the APOE ε4 allele, rs7412: Variant comprising the APOE ε2 allele, rs4420638: Peak variant from the CARDIoGRAMplusC4D meta-analysis.
Investigation of the LD structure between the four variants, indicated that rs7412 (the APOE ε2 allele) had a weak r2 with the other variants (rs6857: 0.01, rs429358: 0.01 and rs4420638: 0.02), but moderate to high D′ (rs6857: 0.71, rs429358: 0.68 and rs4420638: 1.00) and with complete LD with rs4420638. Different LD observations for r2 and D′ reflect the differing allele frequencies between rs7412 and the other variants (eTables 4 and 5). The remaining variants were in moderate to high LD with each other for both r2 and D′ (Table 2 and eTable 6) (r2: 0.42–0.65; D′: 0.74–0.95). The peak variant for LOAD (rs6857) was in stronger LD with the APOE-ε4 variant rs429358 (D′: 0.82) than the APOE-ε2 variant rs7412 (D′: 0.71). The peak variant for CAD rs4420638 was in stronger LD with the APOE-ε2 variant rs7412 (D′: 1.00) than the APOE-ε4 variant rs429358 (D′: 0.95).
PhenoScanner
The PhenoScanner database24 was searched to detect potential pleiotropy of the 52 variants included in the analysis (eTable 7). The results of the association p-values < 0.0012 (Bonferroni adjusted p < 0.05) are shown in eTable 7. Ten of the 52 variants were significantly associated with plasma LDL cholesterol concentrations, while APOE was the sole variant that was significantly associated with LOAD.
Analysis at the APOE locus (eTables 8–11), indicated that rs6857 (p = 5.12 × 10−110), rs4420638 (p = 1.51 × 10−178) and rs7412 (p = 1.24 × 10−652) were highly significantly associated with plasma LDL-cholesterol concentrations in the Global Lipids Genetics Consortium (GLGC)13. However, the APOE-ε4 variant (rs429358) was not tested in the GLGC study. In another LDL-cholesterol study in which rs429358 was tested29 the APOE-ε2 variant rs7412 (p = 5.54 × 10−30) was more significant than the APOE- ε4 variant rs429358 (p = 4.21 × 10−10). Likewise, rs4420638 (p = 8.80 × 10−139) was also significantly associated with plasma concentrations of C-reactive protein (CRP)30. The variants rs6857 (p = 1.06 × 10−10) and rs429358 (p = 5.45 × 10−14) were also significantly associated with cortical amyloid beta load31.
Shared impact of APOE locus
The gwas-pw method28 was used to detect evidence of shared genetic determinants within the APOE locus (chr19:44,744,147-46,101,600) for CAD and LOAD. There was strong evidence for genetic variants influencing both phenotypes at the locus. Furthermore Model 4 (Posterior Probability: 0.90), which specifies separate genetic variants within the APOE locus influencing CAD and LOAD, had a higher posterior probability than Model 3 (Posterior Probability: 0.10) which specified a shared genetic variant in the APOE locus.
Discussion
Disparate genetic architecture of CAD and LOAD
The present study investigated a shared genetic architecture between CAD and LOAD using large-scale GWAS meta-analyses for both diseases. The initial Mendelian randomization analysis suggested that a higher risk of CAD (per log odds unit) was also associated with a 7% higher risk of LOAD. However, there was significant heterogeneity between the causal effects of individual variants. This heterogeneity was entirely explained by a single variant (rs4420638) at the APOE locus. When the APOE variant was removed from the analysis, the causal effect on LOAD was completely attenuated and no longer significant. Thus, overall, genetic determinants associated with a higher risk of CAD were not significantly associated with LOAD after excluding variants at the APOE locus. In addition, the LD score regression analysis identified little or no genetic correlation between CAD and LOAD.
APOE locus
The APOE locus was investigated in greater detail since the rs4420638 variant was strongly associated with both CAD (p = 7.07 × 10−11) and LOAD (p = 1.67 × 10−396). The gwas-pw analysis28, suggested that the traits were influenced by separate genetic variants within the APOE locus. This indicates that the influence of the APOE locus on both LOAD and CAD may be mediated through different mechanisms.
In the case of the APOE variants (rs7412, rs429358): association with LDL cholesterol, a stronger signal was detected in the APOE-ε2 variant (rs7412, p = 5.54 × 10−30)29 than in the APOE-ε4 variant (rs429358, p = 4.21 × 10−10)29, suggesting that the APOE-ε2 variant is strongly associated with LDL cholesterol pathways. In the case of amyloid beta load, the APOE-ε4 variant has been strongly associated with this phenotype (rs429358; OR not available, p = 5.45 × 10−14)31, suggesting that the APOE-ε4 effect may be mediated by amyloid beta pathways. However the APOE-ε2 variant (rs7412) was not present in the analysis of amyloid beta.
In the case of LOAD, the APOE-ε4 variant (rs429358, OR [95% CI]: 3.86 [3.66–4.07]; p = 6.70 × 10−536; EA = C) had a stronger effect than the APOE-ε2 variant (rs7412, OR [95% CI]: 1.47 [1.36–1.59]; p = 1.23 × 10−22; EA = C)11 suggesting that LOAD may be primarily associated with the APOE-ε4 variant: The APOE-ε4 (rs429358) variant is also the peak signal for cortical amyloid beta load31 suggesting that variants in APOE for LOAD may be primarily associated with amyloid beta pathways. In contrast, CAD had a similar strength of association with both the APOE-ε4 variant (rs429358, OR [95% CI]: 1.10 [1.06–1.13]; p = 2.17 × 10−9; EA = C) and APOE-ε2 variant (rs7412, OR [95% CI]: 1.15 [1.10–1.20]; p = 8.17 × 10−11; EA = C)5. The CAD peak variant (rs4420638) was in complete LD (by measures of D′) with the APOE-ε2 variant (rs7412) suggesting that variants in APOE for CAD may be primarily associated with LDL cholesterol pathways. Future analyses of individual participant data could permit exploration of the ε2/ε3/ε4 APOE haplotypes to further elucidate these relationships.
Exploration of potentially pleiotropic effects in each of the 52 variants for CAD (eTable 7) suggest that LDL cholesterol is unlikely to be involved in any shared biological pathways between LOAD and CAD, other than via the effects of APOE. Among the 10 CAD loci that were associated with significant differences in LDL-cholesterol concentrations, only one of these (APOE) was also associated with LOAD.
Other MR analyses
Recently, several studies have conducted MR analyses of vascular risk factors with LOAD. Østergaard and colleagues32 reported that a genetically predicted 1-SD (15.4 mmHg) higher systolic blood pressure was associated with lower risk of AD (OR [95% CI]: 0.75 [0.62–0.91]; p = 3.4 × 10−3); and that a 1-SD (0.91 mmol/l) higher LDL cholesterol was associated with a higher risk of AD (OR [95% CI]: 2.31 [2.12–2.50]; p = 3.0 × 10−87); and that a 1-SD (0.41 mmol/l) higher HDL cholesterol levels was associated with lower risk of AD (OR [95% CI]: 0.75 [0.69–0.82]; p = 1.0 × 10−11). However, after removing the APOE locus (rs6857), the associations were no longer significant for LDL-cholesterol (OR [95% CI]: 1.07 [0.98–1.17]; p = 0.14) or HDL-cholesterol (OR [95% CI]: 1.01 [0.93–1.09]; p = 0.87).
Another study reported null associations between genetically-predicted body mass index with LOAD33. Likewise, variants encoding Type-2 diabetes were also unrelated to LOAD34.
The METASTROKE consortium35 examined the genetic association between ischemic stroke (IS) and LOAD using GREML36, and reported evidence of a shared genetic contribution of LOAD with small vessel stroke (genetic correlation [95% CI]: 0.37 [0.04–0.70]; p = 0.01). However, variants encoding large vessel stroke, which has a shared genetic architecture with CAD37, were unrelated to LOAD (genetic correlation [95% CI]: 0.00 [−0.22–0.22]; p = 0.49). Variants encoding cardioembolic stroke were also unrelated to LOAD (genetic correlation [95% CI]: 0.08 [−0.16–0.32]; p = 0.25)
Limitations of the study
The present analysis was performed on 52 variants selected from the CARDIoGRAMplusC4D meta-analysis, at which a genome-wide significant signal (p ≤ 5 × 10−8) had been identified5,38–41. The results of this analysis were not materially altered when including 190 variants based on an FDR 5% threshold. Furthermore, results from LD score regression, examining information across the genome, also provide further support.
Results of an post-hoc power analysis are presented in eTable 12, which suggest that the analyses with genome wide significant and FDR 5% instrumental variables had ~80% power to identify ~10% effect on LOAD. Thus, later studies are unlikely to discover a material shared genetic architecture between CAD and LOAD.
The available evidence on the APOE locus indicated separate mechanisms by which this locus acts upon LOAD and CAD, which raises questions about the assumptions for MR analysis involving this locus. Further fine mapping studies of the APOE locus are needed to assess the genetic associations at these highly correlated variants. These studies could include populations with greater haplotype diversity to resolve tightly linked genetic signals that appear intractably interwoven in Europeans. However, we encountered difficulties with exploratory fine mapping studies of the APOE locus for LOAD, due to collinearity induced by the strong linkage disequilibrium between the variants in this locus.
The present study also had several limitations. Firstly, the study maybe constrained by selection bias due to differences in age. The average age of onset of CAD was approximately 60 years, while the average age of onset of LOAD cases was 74 years. Individuals predisposed to developing both LOAD and CAD may not have survived to old age, which may have underestimated any association with LOAD. Moreover, the diagnosis of probable LOAD excludes a prior history of cerebrovascular disease, so studies of LOAD may have reduced risk of overlap between CAD and LOAD. However, neuroimaging studies show that pathological changes in the brain precede the development of mild cognitive impairment and LOAD by 1–2 decades7.
Selection bias may also have influenced summary statistics between CAD and LOAD due to overlapping participants for each disease in some studies. However, the number of overlapping cases in AGES, Rotterdam and Framingham Heart studies is very limited (up to 1000 LOAD cases in IGAP could potentially overlap with CAD cases included in the CARDIoGRAMplusC4D study).
Another possible limitation is that since the CARDIoGRAMplusC4D data included some non-European samples, these could increase heterogeneity between the samples. However, the CARDIoGRAMplusC4D study reported that no heterogeneity between studies was observed at any of the genome wide significant variants apart from the 9p21 locus5.
The present report demonstrated a different genetic architecture of CAD and LOAD. While further studies are required to further elucidate links between cardiovascular disease risk factors and LOAD, additional MR studies of CAD are unlikely to be informative about the causes of LOAD.
Conclusions
Analyses were performed to investigate whether CAD and LOAD have a shared genetic architecture and whether CAD is a causal risk factor for LOAD, given the findings of observational studies. However, the present study demonstrated that although genetic predisposition to CAD was significantly associated with LOAD, this association was entirely mediated through the APOE locus. After exclusion of the APOE locus, CAD variants were no longer significantly associated with LOAD. Additional fine mapping studies are needed to dissect the independent relevance of APOE for both CAD and LOAD.
Electronic supplementary material
Acknowledgements
We would like to thank the IGAP and GERAD consortiums for access to GWAS data on LOAD and the CARDIoGRAMplusC4D Consortium for access to GWAS data on CAD. This work was supported by the British Heart Foundation [FS/14/55/30806 to JCH], [RE/13/1/30181 to MF], and the Wellcome Trust [090532/Z/09/Z to MF], British Heart Foundation [grant number: CH/1996001/9454 to Clinical Trial Service Unit], Medical Research Council [grant number: Clinical Trial Service Unit A310], and British Heart Foundation Centre for Research Excellence. AG acknowledge European Community Sixth Framework Program (LSHM-CT- 2007-037273), European Union Seventh Framework Programme FP7/2007-2013 under grant agreement no. HEALTH-F2-2013-601456 (CVGenes@Target), TriPartite Immunometabolism Consortium [TrIC]- Novo Nordisk Foundation’s Grant number NNF15CC0018486.
Author Contributions
R.C., M.F., H.W. and J.C.H. conceived and designed the study. C.G. prepared the analyses and figures. C.G., R.C., M.F. and J.C.H. wrote the initial draft of the manuscript. All authors provided critical revision of the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-25460-2.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lusis AJ. Atherosclerosis. Nature. 2000;407:233–41. doi: 10.1038/35025203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weber C, Noels H. Atherosclerosis: current pathogenesis and therapeutic options. Nat Med. 2011;17:1410–22. doi: 10.1038/nm.2538. [DOI] [PubMed] [Google Scholar]
- 3.Yusuf S, et al. Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): case-control study. Lancet. 2004;364:937–52. doi: 10.1016/S0140-6736(04)17018-9. [DOI] [PubMed] [Google Scholar]
- 4.Zdravkovic S, et al. Heritability of death from coronary heart disease: a 36-year follow-up of 20 966 Swedish twins. J Intern Med. 2002;252:247–54. doi: 10.1046/j.1365-2796.2002.01029.x. [DOI] [PubMed] [Google Scholar]
- 5.CARDIoGRAMplusC4D Consortium. A comprehensive 1000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 47, 1121–30 (2015). [DOI] [PMC free article] [PubMed]
- 6.Masters CL, et al. Alzheimer’s disease. Nat Rev Dis Primers. 2015;1:1–18. doi: 10.1038/nrdp.2015.56. [DOI] [PubMed] [Google Scholar]
- 7.Jack CR, et al. Tracking pathophysiological processes in Alzheimer’s disease: an updated hypothetical model of dynamic biomarkers. Lancet Neurol. 2013;12:207–16. doi: 10.1016/S1474-4422(12)70291-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matthews FE, et al. A two-decade comparison of prevalence of dementia in individuals aged 65 years and older from three geographical areas of England: results of the Cognitive Function and Ageing Study I and II. Lancet. 2013;382:1405–12. doi: 10.1016/S0140-6736(13)61570-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKhann GM, et al. The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:263–9. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gatz M, et al. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry. 2006;63:168–74. doi: 10.1001/archpsyc.63.2.168. [DOI] [PubMed] [Google Scholar]
- 11.Lambert JC, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–8. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu CC, Kanekiyo T, Xu H, Bu G. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol. 2013;9:106–18. doi: 10.1038/nrneurol.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Global Lipids Genetics Consortium. Discovery and refinement of loci associated with lipid levels. Nat Genet. 45, 1274–83 (2013). [DOI] [PMC free article] [PubMed]
- 14.Hofman A, et al. Atherosclerosis, apolipoprotein E, and prevalence of dementia and Alzheimer’s disease in the Rotterdam Study. Lancet. 1997;349:151–4. doi: 10.1016/S0140-6736(96)09328-2. [DOI] [PubMed] [Google Scholar]
- 15.Van Oijen M, et al. Atherosclerosis and risk for dementia. Ann Neurol. 2007;61:403–10. doi: 10.1002/ana.21073. [DOI] [PubMed] [Google Scholar]
- 16.Newman AB, et al. Dementia and Alzheimer’s disease incidence in relationship to cardiovascular disease in the Cardiovascular Health Study cohort. J Am Geriatr Soc. 2005;53:1101–7. doi: 10.1111/j.1532-5415.2005.53360.x. [DOI] [PubMed] [Google Scholar]
- 17.Laurin D, Masaki KH, White LR, Launer LJ. Ankle-to-brachial index and dementia: the Honolulu-Asia Aging Study. Circulation. 2007;116:2269–74. doi: 10.1161/CIRCULATIONAHA.106.686477. [DOI] [PubMed] [Google Scholar]
- 18.Qiu C, et al. Heart failure and risk of dementia and Alzheimer disease: a population-based cohort study. Arch Intern Med. 2006;166:1003–8. doi: 10.1001/archinte.166.9.1003. [DOI] [PubMed] [Google Scholar]
- 19.Smith GD, Ebrahim S. Mendelian randomization: prospects, potentials, and limitations. Int J Epidemiol. 2004;33:30–42. doi: 10.1093/ije/dyh132. [DOI] [PubMed] [Google Scholar]
- 20.Smith GD, Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 21.Jansen H, Samani NJ, Schunkert H. Mendelian randomization studies in coronary artery disease. Eur Heart J. 2014;35:1917–24. doi: 10.1093/eurheartj/ehu208. [DOI] [PubMed] [Google Scholar]
- 22.Magosi LE, Goel A, Hopewell JC, Farrall M. & CARDIoGRAMplusC4D Consortium. Identifying systematic heterogeneity patterns in genetic association meta-analysis studies. PLoS Genet. 2017;13:e1006755. doi: 10.1371/journal.pgen.1006755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37:658–65. doi: 10.1002/gepi.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Staley JR, et al. PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics. 2016;32:3207–9. doi: 10.1093/bioinformatics/btw373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512–25. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hemani, G. et al. MR-Base: a platform for systematic causal inference across the phenome using billions of genetic associations. bioRxiv. 10.1101/078972 (2016).
- 27.Bulik-Sullivan B, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–41. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pickrell JK, et al. Detection and interpretation of shared genetic influences on 42 human traits. Nat Genet. 2016;48:709–17. doi: 10.1038/ng.3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson JF, et al. Comprehensive whole-genome and candidate gene analysis for response to statin therapy in the Treating to New Targets (TNT) cohort. Circ Cardiovasc Genet. 2009;2:173–81. doi: 10.1161/CIRCGENETICS.108.818062. [DOI] [PubMed] [Google Scholar]
- 30.Dehghan A, et al. Meta-analysis of genome-wide association studies in 80 000 subjects identifies multiple loci for C-reactive protein levels. Circulation. 2011;123:731–8. doi: 10.1161/CIRCULATIONAHA.110.948570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramanan VK, et al. APOE and BCHE as modulators of cerebral amyloid deposition: a florbetapir PET genome-wide association study. Mol Psychiatry. 2014;19:351–7. doi: 10.1038/mp.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Østergaard SD, et al. Associations between Potentially Modifiable Risk Factors and Alzheimer Disease: A Mendelian Randomization Study. PLoS Med. 2015;12:e1001841. doi: 10.1371/journal.pmed.1001841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mukherjee S, et al. Genetically predicted body mass index and Alzheimer’s disease-related phenotypes in three large samples: Mendelian randomization analyses. Alzheimers Dement. 2015;11:1439–51. doi: 10.1016/j.jalz.2015.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walter S, et al. Diabetic Phenotypes and Late-Life Dementia Risk: A Mechanism-specific Mendelian Randomization Study. Alzheimer Dis Assoc Disord. 2016;30:15–20. doi: 10.1097/WAD.0000000000000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Traylor M, et al. Shared genetic contribution to Ischaemic Stroke and Alzheimer’s Disease. Ann Neurol. 2016;79:739–747. doi: 10.1002/ana.24621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bellenguez C, et al. Genome-wide association study identifies a variant in HDAC9 associated with large vessel ischemic stroke. Nat Genet. 2012;44:328–33. doi: 10.1038/ng.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Coronary Artery Disease (C4D) Genetics Consortium. A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat Genet. 43, 339–44 (2011). [DOI] [PubMed]
- 39.Deloukas P, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schunkert H, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–8. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dichgans M, et al. Shared genetic susceptibility to ischemic stroke and coronary artery disease: a genome-wide analysis of common variants. Stroke. 2014;45:24–36. doi: 10.1161/STROKEAHA.113.002707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pruim RJ, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analysed during the current study are publically available in the CARDIoGRAMplusC4D5 (CAD) repository http://www.cardiogramplusc4d.org and the IGAP11 (LOAD) repository http://web.pasteur-lille.fr/en/recherche/u744/igap/igap_download.php.