Abstract
The effect of biofilm formation on bacteraemic pneumonia caused by A. baumannii is unknown. We conducted a 4-year multi-center retrospective study to analyze 71 and 202 patients with A. baumannii bacteraemic pneumonia caused by biofilm-forming and non-biofilm-forming isolates, respectively. The clinical features and outcomes of patients were investigated. Biofilm formation was determined by a microtitre plate assay. The antimicrobial susceptibilities of biofilm-associated cells were assessed using the minimum biofilm eradication concentration (MBEC) assay. Whole-genome sequencing was conducted to identify biofilm-associated genes and their promoters. Quantitative reverse transcription polymerase chain reaction was performed to confirm the expression difference of biofilm-associated genes. There was no significant difference in the clinical characteristics or the outcomes between patients infected with biofilm-forming and non-biofilm-forming strains. Compared with non-biofilm-forming isolates, biofilm-forming isolates exhibited lower resistance to most antimicrobials tested, including imipenem, meropenem, ceftazidime, ciprofloxacin and gentamicin; however, the MBEC assay confirmed the increased antibiotic resistance of the biofilm-embedded bacteria. Biofilm-associated genes and their promoters were detected in most isolates, including the non-biofilm-forming strains. Biofilm-forming isolates showed higher levels of expression of the biofilm-associated genes than non-biofilm-forming isolates. The biofilm-forming ability of A. baumannii isolates might not be associated with worse outcomes in patients with bacteraemic pneumonia.
Introduction
Acinetobacter baumannii is an important pathogen that causes nosocomial bloodstream infections and pneumonia, accounting for high morbidity and mortality rates1,2. A. baumannii is notorious for its multiple antimicrobials resistance. This multidrug resistance, along with its ability to form biofilms3, increases the difficulties in treating infections caused by this microorganism4.
Its prevalence in nosocomial outbreaks and device-related infections has been attributed to its ability to form biofilms in hospital environments and on medical devices attributes, respectively4,5. The ability to produce biofilms is a decisive advantage for colonization of different environments; thus, these bacteria can cause persistent infections6. In most institutions, the majority of A. baumannii isolates are isolated from the respiratory tracts of hospitalized patients7. A previous study reported that biofilm formation was observed in 95% of specimens obtained from patients who were mechanically ventilated for more than 24 hours, and A. baumannii and Pseudomonas aeruginosa were the most frequently isolated bacterial species8.
Biofilm-embedded cells are highly resistant to antimicrobials and disinfectants and therefore are difficult to eradicate9–11. However, whether biofilm-forming ability affects the clinical outcomes of A. baumannii pneumonia is largely unknown. This study aimed to investigate the correlation between the biofilm-forming ability of A. baumannii isolates and clinical outcomes of patients with A. baumannii bacteraemic pneumonia and to evaluate the microbiological features of biofilm-forming A. baumannii isolates.
Results
Clinical characteristics and outcomes of study population
We evaluated 856 patients from the 4 centers (CCH: 135, MMH: 132, TSGH: 186 and TVGH: 403) who had at least one episode of A. calcoaceticus-baumannii complex (Abc) monomicrobial bacteraemia during the 4-year study period. We excluded patients who had bloodstream infections caused by non-baumannii Acinetobacter spp., other concomitant infections at the time of presentation, or bacteraemia from a source other than pneumonia. Finally, 273 patients with documented A. baumannii monomicrobial bacteraemic pneumonia were included in the analysis.
The demographic and clinical characteristics of patients with bacteraemic pneumonia caused by biofilm-forming (n = 71, 26.0%) and non-biofilm-forming (n = 202, 74.0%) isolates of A. baumannii are presented in Table 1. There were no significant differences in the demographic characteristics, comorbidities, invasive procedures, or previous antibiotic use between the two groups. The two groups had similar Charlson comorbidity index (CCI) score, disease severity (Acute Physiology and Chronic Health Evaluation II [APACHE II score]) and appropriateness of antimicrobial therapy. There was also no significant difference in the 14-day and 28-day mortality between the two groups by bivariate analysis (Table 1) or survival analysis (p = 0.092 by log-rank test; Fig. 1). Only two patients with non-biofilm-forming A. baumannii bacteraemic pneumonia had recurrent bacteraemia and no patients in the biofilm-forming group had recurrent bacteraemia.
Table 1.
Demographic and clinical characteristics of patients with bacteraemic pneumonia caused by biofilm-forming and non-biofilm-forming Acinetobacter baumannii.
| Characteristics | Biofilm-forming (n = 71) | Non-biofilm-forming (n = 202) |
p value |
|---|---|---|---|
| Demographic characteristics | |||
| Age, median (IQR), years | 76 (59–82) | 75 (59–82) | 0.551 |
| Male sex, no. (%) | 55 (77.5) | 151 (74.8) | 0.648 |
| Acquired in the ICU, no. (%) | 32 (45.1) | 105 (52.0) | 0.317 |
| Length of hospitalization before bacteraemia, median (IQR), days | 22 (12–41) | 19 (8.75–42) | 0.572 |
| APACHE II score, median (IQR) | 25 (16–33) | 26 (21–34) | 0.326 |
| Charlson comorbidity index, median (IQR) | 3.75 (2–5) | 3.67 (2–5) | 0.817 |
| Comorbid conditions, no. (%) | |||
| Alcoholism | 7 (9.9) | 9 (4.5) | 0.095 |
| Cerebrovascular disease | 13 (18.3) | 33 (16.3) | 0.702 |
| Coronary artery disease | 15 (21.1) | 33 (16.3) | 0.362 |
| Congestive heart failure | 11 (15.5) | 36 (17.8) | 0.655 |
| Chronic obstructive pulmonary disease | 15 (21.1) | 44 (21.8) | 0.908 |
| Chronic kidney disease | 22 (31.0) | 70 (34.7) | 0.574 |
| Type 2 diabetes mellitus | 26 (36.6) | 66 (32.7) | 0.545 |
| Hypertension | 36 (50.7) | 85 (42.1) | 0.208 |
| Liver cirrhosis | 8 (11.3) | 15 (7.4) | 0.316 |
| Collagen vascular disease | 6 (8.5) | 8 (4.0) | 0.140 |
| Malignancy | 19 (26.8) | 57 (28.2) | 0.814 |
| Neutropenia | 5 (7.0) | 9 (4.5) | 0.367 |
| Immunosuppressive therapy | 17 (23.9) | 49 (24.3) | 0.958 |
| Recent surgery | 13 (18.3) | 54 (26.7) | 0.156 |
| Trauma | 1 (1.4) | 7 (3.5) | 0.685 |
| Septic shock | 25 (35.2) | 79 (39.1) | 0.561 |
| Invasive procedures, no. (%) | |||
| Arterial catheterization | 21 (29.6) | 74 (36.6) | 0.283 |
| Central venous catheter | 40 (56.3) | 129 (63.9) | 0.261 |
| Hemodialysis | 13 (18.3) | 26 (12.9) | 0.260 |
| Nasogastric tube | 21 (29.6) | 84 (41.6) | 0.089 |
| Tracheostomy | 12 (16.9) | 29 (14.4) | 0.606 |
| Thoracic drain | 5 (7.0) | 14 (6.9) | 0.975 |
| Abdominal drainage | 3 (4.2) | 14 (6.9) | 0.599 |
| Mechanical ventilation | 33 (46.5) | 77 (38.1) | 0.217 |
| Previous ICU admission, no. (%) | 44 (62.0) | 145 (71.8) | 0.123 |
| Previous antibiotic exposure, no. (%) | |||
| All | 51 (71.8) | 151 (74.8) | 0.629 |
| All beta-lactam | 44 (62.0) | 128 (63.4) | 0.834 |
| Aminoglycoside | 10 (14.1) | 25 (12.4) | 0.711 |
| Penicillin | 7 (9.9) | 12 (5.9) | 0.264 |
| Beta-lactam/beta-lactamase inhibitor | 9 (12.7) | 38 (18.8) | 0.239 |
| Cephalosporin | 15 (21.1) | 40 (19.8) | 0.811 |
| Anti-pseudomonas cephalosporin | 23 (32.4) | 50 (24.8) | 0.211 |
| Anti-pseudomonas carbapenem | 14 (19.7) | 53 (26.2) | 0.272 |
| Sulbactam | 1 (1.4) | 15 (7.4) | 0.078 |
| Fluoroquinolone | 16 (22.5) | 47 (23.3) | 0.900 |
| Tigecycline | 8 (11.3) | 15 (7.4) | 0.316 |
| Colistin | 5 (7.0) | 8 (4.0) | 0.333 |
| Macrolide | 5 (7.0) | 9 (4.5) | 0.367 |
| Clindamycin | 1 (1.4) | 13 (6.4) | 0.124 |
| Vancomycin | 6 (8.5) | 21 (10.4) | 0.637 |
| Teicoplanin | 20 (28.2) | 51 (25.2) | 0.629 |
| Appropriate antimicrobial therapy, no. (%) | 19 (26.8) | 57 (28.2) | 0.814 |
| Outcome | |||
| 14-day mortality | 26 (36.6) | 93 (46.0) | 0.169 |
| 28-day mortality | 31 (43.7) | 112 (55.4) | 0.087 |
| Recurrent bacteraemia | 0 (0.0) | 2 (1.0) | 1.000 |
*APACHE II = Acute Physiology and Chronic Health Evaluation II, ICU = intensive care unit, IQR = interquartile range.
Figure 1.
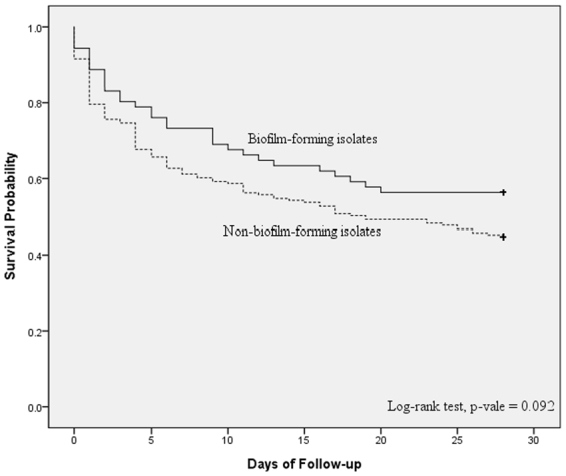
Comparison of Kaplan–Meier survival curves, at 28 days, between patients with Acinetobacter baumannii bacteraemic pneumonia caused by either biofilm-formation isolates or non-biofilm-formation isolates.
Multivariate logistic regression analysis was performed for the overall cohort to delineate the independent risk factors for 28-day mortality in patients with A. baumannii bacteraemic pneumonia (Table 2). A higher APACHE II score and previous intensive care unit (ICU) admission were independently associated with 28-day mortality. Notably, no significant correlation was observed between biofilm formation capability and 28-day mortality (odds ratio [OR], 0.623; 95% confidence interval [CI], 0.361–1.074; p = 0.088).
Table 2.
Logistic regression analysis of predictors for 28-day mortality among patients with bacteraemic pneumonia caused by Acinetobacter baumannii.
| Characteristic | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| Crude OR (95% CI) | p | Adjusted OR (95% CI) | p | |
| Biofilm formation strains infection | 0.623 (0.361–1.074) | 0.088 | ||
| APACHE II score | 1.152 (1.109–1195) | <0.001 | 1.158 (1.113–1.206) | <0.001 |
| Previous ICU admission | 2.022 (1.191–3.436) | 0.009 | 2.723 (1.361–5.450) | 0.005 |
| Hypertension | 0.606 (0.375–0.981) | 0.041 | 0.437 (0.236–0.811) | 0.009 |
| Congestive heart failure | 0.450 (0.235–0.862) | 0.016 | ||
| Cerebrovascular disease | 0.470 (0.244–0.903) | 0.023 | ||
| Previous antibiotic exposure | 1.695 (0.975–2.947) | 0.061 | ||
| Multidrug re sistance strains infectiona | 1.684 (0.751–3.777) | 0.206 | ||
*APACHE II = Acute Physiology and Chronic Health Evaluation II, ICU = intensive care unit; CI = confidence interval, OR = odds ratio.
aDefinition: resistance to three or more of the following classes of antimicrobial agents: anti-pseudomonal cephalosporins, anti-pseudomonal carbapenems, ampicillin/sulbactam, fluoroquinolones, and aminoglycosides.
As there is no universal standard reference value used for evaluating biofilm formation capacity, strains with optical density of 570 nm (OD570) values at least twice those of the negative controls were classified as biofilm-formation isolates in this study. In order to validate whether there is any bias in assessing the biofilm forming capacity, we conducted the following analysis. Initially, the results were divided into four following categories according to their optic densities as (1) strong biofilm producer (OD570 values ≧ 8 folds those of the negative controls); (2) medium biofilm producer (OD570 values 4–8 folds those of the negative controls); (3) weak biofilm producer (OD570 values 2–4 folds those of the negative controls); and (4) non-biofilm producer (OD570 values ≤2 folds those of the negative controls). We found that there was no significant difference in 14-day and 28-day mortality between the strong biofilm producer, medium biofilm producer, weak biofilm producer and non-biofilm producer (data not shown).
Then we tried to set the cut-off value higher. Specifically, those with the OD570 values at least 4-folds of the negative controls were classified as positive biofilm-formation isolates. Further analysis showed no significant difference in 14-day and 28-day mortality between the biofilm-formation and non-biofilm formation isolates (data not shown).
Next, we used the A. baumannii ATCC 19606 as a reference strain12,13. Strains with OD570 values greater than that of A. baumannii ATCC 19606 were considered positive for biofilm formation. Of the 273 isolates, there were 181 and 92 isolates with biofilm-forming capacity higher than A. baumannii ATCC 19606 and lower than A. baumannii ATCC 19606, respectively. The further analysis showed there was no difference in 14-day and 28-day mortality between the two groups (data not shown).
In the end, we conducted the air-liquid interfaces biofilm formation method to evaluate the ability of biofilm formation3. There were 66 isolates produced air-liquid interfaces biofilm while the others did not. We compared the 14-day and 28-day mortality between the air-liquid interfaces biofilm-formation and non-biofilm formation isolates. The results disclosed there was no significant difference in 14-day and 28-day mortality between the air-liquid interfaces biofilm-formation and non-biofilm formation isolates (data not shown).
Antimicrobial treatment and the outcomes of patients with biofilm-forming A. baumannii bacteraemic pneumonia are listed in Table 3. Patients who were treated with ampicillin/sulbactam or sulbactam had a lower 28-day mortality rate (33.3%) even though they had a higher APACHE II score than those who received other therapeutic regimens. Multivariate analysis revealed that no specific therapeutic regimen was independently associated with higher or lower 28-day morality rates. There was no identifiable link between the antimicrobial treatment regimens and the clinical outcomes in those with non-biofilm-forming A. baumannii bacteraemic pneumonia (see Supplementary Table S1).
Table 3.
Antimicrobials usage and the 28-day survival rates in patients with bacteraemic pneumonia caused by biofilm-forming Acinetobacter baumannii.
| Main agents used | Patients, no. | APACHE II score | Patients, no. (%) | 28-Day survivors | ||
|---|---|---|---|---|---|---|
| Median (interquartile range) | Appropriate antimicrobial therapy | Combination antimicrobial therapy | 28-Day non-survivors | |||
| Anti-pseudomonal penicillinsa | 10 | 22 (16.53–0.75) | 5 (50.0) | 1 (10.0) | 7 (70.0) | 3 (30.0) |
| Anti-pseudomonal cephalosporinsb | 16 | 26 (17.25–30.25) | 4 (25.0) | 1 (7.1) | 6 (37.5) | 10 (62.5) |
| Anti-pseudomonal fluoroquinolonesc | 3 | 39 (20–39) | 0 (0) | 0 (0) | 3 (100.0) | 0 (0) |
| Anti-pseudomonal carbapenemsd | 12 | 29 (21.75–35.75) | 6 (50.0) | 1 (8.3) | 5 (41.7) | 7 (58.3) |
| Ampicillin/sulbactam or sulbactam | 6 | 36.5 (21.75–44) | 1 (16.7) | 3 (50.0) | 2 (33.3) | 4 (66.7) |
| Non-antipseudomonal β-lactamasese | 11 | 19 (13–33) | 0 (0) | 2 (18.2) | 4 (36.4) | 7 (63.6) |
*Data are the median (interquartile range) for continuous variables and number of cases (%) for categorical variables. APACHE II = Acute Physiologic and Chronic Health Evaluation II.
aPiperacillin, piperacillin/tazobactam and ticarcillin/clavulanate.
bCefoperazone, ceftazidime, cefepime and cefpirome.
cCiprofloxacin and levofloxacin.
dImipenem and meropenem.
ePenicillin, amoxicillin/clavulanate, cefazolin, cefuroxime, cefotaxime, cefmetazole and flomoxef.
Clonality and antibiograms of the A. baumannii isolates
To determine the molecular epidemiology of the causative pathogens, 64 biofilm-forming and 126 non-biofilm-forming A. baumannii isolates were randomly selected for pulsed-field gel electrophoresis (PFGE) analysis. There were 68 pulsotypes (P1–P68), based on a threshold of 80% similarity (see Supplementary Fig S1). The biofilm-forming and non-biofilm-forming isolates belonged to 37 and 44 pulsotypes, respectively and 28 biofilm-forming isolates shared the same pulsotype with non-biofilm-forming isolates. P5 (n = 36) and P4 (n = 23) were the most common pulsotypes.
The antimicrobial susceptibilities and the minimum inhibitory concentrations (MICs) of biofilm-forming and non-biofilm-forming A. baumannii isolates against the antimicrobials are presented in Table 4. The proportion of multidrug resistance (MDR) isolates was significantly lower in the biofilm-forming group and biofilm-forming isolates were more susceptible to most commonly used antibiotics, except ampicillin/sulbactam, piperacillin/tazobactam and tigecycline. The MIC90 of ceftazidime, cefepime or cefpirome, piperacillin/tazobactam, ampicillin/sulbactam, ciprofloxacin and meropenem were also lower in the biofilm-forming group. Ten biofilm-forming isolates that belonged to different pulsotypes were selected for the minimum biofilm eradication concentration (MBEC) assay (see Supplementary Table S2). The MBECs of imipenem and meropenem were much higher than the MICs of imipenem and meropenem in biofilm-forming isolates.
Table 4.
Comparison of the antimicrobial susceptibilities of biofilm-forming and non-biofilm-forming Acinetobacter baumannii isolates.
| Antimicrobial agent | Biofilm-forming isolates (n = 71) | Non-biofilm-forming isolates (n = 202) | p value b | ||
|---|---|---|---|---|---|
| Resistance, no. (%) | MIC90 (µg/mL) | Resistance, no. (%) | MIC90 (µg/mL) |
||
| Amikacin | 44 (62.0) | >2048 | 151 (74.8) | >2048 | 0.040 |
| Gentamicin | 57 (80.0) | >2048 | 181 (89.6) | >2048 | 0.043 |
| Ceftazidime | 56 (78.9) | 512 | 186 (92.1) | 1024 | 0.003 |
| Cefepime or cefpirome | 48 (67.6) | 128 | 157 (77.7) | 256 | 0.009 |
| Piperacillin/tazobactam | 52 (73.2) | 4096/4 | 163 (80.7) | >4096/4 | 0.187 |
| Ampicillin/sulbactam | 46 (64.8) | 64/32 | 141 (69.8) | 128/64 | 0.434 |
| Ciprofloxacin | 55 (77.5) | 256 | 187 (92.6) | 512 | 0.001 |
| Imipenem | 39 (54.9) | 64 | 139 (78.1) | 64 | 0.035 |
| Meropenem | 39 (54.9) | 64 | 142 (78.5) | 128 | 0.018 |
| Tigecycline | 19 (26.8) | 4 | 39 (19.3) | 4 | 0.187 |
| Colistin | 0 | 2 | 0 | 2 | — |
| Multidrug resistancea | 58 (81.7) | — | 188 (93.1) | — | 0.006 |
aDefinition: resistance to three or more of the following classes of antimicrobial agents: anti-pseudomonal cephalosporins, anti-pseudomonal carbapenems, ampicillin/sulbactam, fluoroquinolones and aminoglycosides.
bp value: The difference in resistance rate to antibiotics between biofilm-forming isolates and non-biofilm-forming isolates.
The correlation between carbapenemase production and biofilm formation
Carbapenem-resistant A. baumannii strains were less likely to produce biofilms than carbapenem susceptible strains (17.0% vs. 38.6%, p < 0.001). To delineate the impact of carbapenem resistance on biofilm formation capability, we evaluated the biofilm formation capability of A. baumannii transformants harboring different carbapenemase genes. All the transformants had MICs of imipenem and meropenem ≥8 mg/L (data not shown). Carbapenem-resistant transformants containing vectors with various carbapenemase genes produced less biofilms than the parent strain A. baumannii reference strain ATCC 15151 (Ab15151) carrying empty vectors, except those harboring blaOXA-58 (see Supplementary Fig S2). To elucidate if decreased biofilm forming capacity also occurred with the expression of non-carbapenemase genes, a transformant containing a vector with Acinetobacter-derived cephalosporinase (ADC) gene was also generated and showed no significant decrease in biofilm formation capability compared with its parent strain Ab15151 carrying empty vectors (data not shown).
Biofilm-associated genes of the selected isolates and quantitative reverse transcription polymerase chain reaction (qRT-PCR) assay
To investigate the correlation between biofilm-forming capacity and the presence of biofilm-associated genes, 9 biofilm-forming and 30 non-biofilm-forming clonally unrelated isolates were selected for whole genome sequencing. All biofilm-forming strains contained the known biofilm-formation-associated genes except two strains (isolate 156 and 160), which lacked the genes encoding Bap1 and Bap2 (see Supplementary Table S3). Interestingly, most of the non-biofilm-forming isolates also had biofilm-formation-associated genes (see Supplementary Table S3). The promoter sequences of biofilm-associated genes were not different between biofilm-forming and non-biofilm-forming strains (data not shown).
To determine whether the expression of biofilm-associated genes is different between biofilm-forming and non-biofilm-forming strains, we matched two biofilm-forming isolates with two non-biofilm-forming isolates that belonged to the same sequence type (ST) (ST 218 and ST 436). The qRT-PCR assay revealed higher expression levels of the biofilm-formation-associated genes (Bap1, AbaI, Cus A/B) in the biofilm-forming isolates compared with the matched non-biofilm-forming isolates (see Supplementary Fig S3). However, the expression levels of Bap2 and BfmS were not different between biofilm-forming and non-biofilm-forming strains (data not shown).
Discussion
The present study demonstrated no significant differences in the clinical characteristics and 14-day and 28-day mortality between patients with A. baumannii bacteraemic pneumonia caused by biofilm-forming and non-biofilm-forming isolates. Biofilm formation was not independently associated with higher or lower mortality. The biofilm-forming isolates were significantly more susceptible to most antimicrobials tested, including imipenem, meropenem, ceftazidime, ciprofloxacin and gentamicin. No specific regimen exhibited a significant 28-day survival benefit for patients infected with biofilm-forming isolates. Although both biofilm-forming and non-biofilm-forming isolates had biofilm-formation-associated genes and their promoter sequences, the biofilm-forming isolates expressed some of the biofilm-formation-associated genes at a higher level than non-biofilm-forming isolates.
Studies on the relationship between biofilm-formation phenotype and clinical infection outcomes are limited. This is the first study investigating the relationships between biofilm-forming capability and clinical outcomes of A. baumannii bacteraemic pneumonia. A previous study investigated 89 isolates obtained from various infection foci and found no significant difference on the mortality rate between those infected with biofilm-forming and non-biofilm-forming A. baumannii14. The heterogeneity of the infectious diseases and relative low number of cases provided weak evidence to demonstrate correlation between biofilm formation and clinical implication from that study. Another study investigated 221 clinical isolates and assessed the association between biofilm-formation ability and the clinical outcomes15. However, there were only 53 Abc isolates obtained in that study. Recently, Zhang et al.15 studied A. baumannii isolates from 121 patients with hospital-acquired pneumonia and concluded that the previous stay in the ICU, use of antibiotics and less severe disease were likely to lead to infection from biofilm-forming isolates. Although the number of cases was relative large, they did not mention the correlation between biofilm-formation ability and the clinical outcome. Besides, the A. baumannii isolates were obtained from sputum samples in their study. This could not exclude the possibility that these pathogens may just have been the colonizers. Since the clinical conditions and sample sizes were diverse among the above studies, they could not draw robust conclusions on the correlation between biofilm formation ability and the clinical implication.
In our study, all isolates were obtained from the blood samples, indicating true causative pathogens. Patients infected with biofilm-forming and non-biofilm-forming isolates had similar clinical characteristic and outcomes. The finding that biofilm-forming and non-biofilm-forming A. baumannii isolates shared the same pulsotypes ruled out the possibility that biofilm-forming isolates belonged to certain low-virulence pulsotypes. Further investigations are required to determine if biofilm-forming isolates are less virulent than non-biofilm-forming isolates belonging to the same pulsotype.
The biofilm-forming isolates in this study were significantly more susceptible to amikacin, gentamicin, ceftazidime, cefepime or cefpirome, ciprofloxacin, imipenem and meropenem. The results suggested a possible inverse association between antibiotic resistance and biofilm-forming ability. Previous reports showed that biofilm-forming A. baumannii were more frequently susceptible to imipenem and ciprofloxacin13,14. One recent study also demonstrated that strong biofilm-forming A. baumannii isolates exhibited low-level resistance to gentamicin, minocycline and ceftazidime16. Our results are consistent with these previous findings. While several attributes can confer antimicrobial resistance to bacteria within a biofilm, even less-resistant biofilm-forming bacteria can survive6,17. Based on the aforementioned results, it is likely that the biofilm-forming strains rely less frequently on intrinsic antimicrobial resistance for survival.
The biofilm-forming isolates in our study exhibited lower rates of carbapenem resistance than non-biofilm-forming isolates. Since the correlation between biofilm-forming capacity and carbapenem resistance in previous studies was questionable18,19, we compared the biofilm-forming ability of A. baumannii transformants harboring various carbapenemase genes to that of the parent strain with an empty vector to clarify this correlation. We found that most carbapenem-resistant transformants had decreased biofilm-forming capacity. Nevertheless, we could not find a similar trend in transformants carrying a vector with non-carbapenemase gene (ADC-type β-lactamase). The energy necessary for expressing the carbapenemase genes may decrease the biofilm formation, but not in the case of ADC genes. Research efforts should be directed to obtain a deeper understanding of this issue. Among A. baumannii expressed different carbapenemase genes, those harvesting blaOXA-58 produce more biofilm than other carbapenem-resistant transformants. It seems that the influence of carbapenemase genes on biofilm formation varied between A. baumannii expressed different carbapenemase genes. Further experiments are needed to elucidate its mechanism.
According to the results of the MBEC assay, the biofilm-forming isolates showed extremely high antibiotic resistance within the biofilm. Although biofilm-forming isolates were more susceptible than non-biofilm-forming isolates, the increased resistance of bacteria embedded in the biofilm might result in treatment failure and recurrent infection in patients infected with biofilm-forming isolates. Because of the high drug resistance of bacteria within biofilms, clinicians should manage infections caused by biofilm-forming isolates very carefully. Currently, there are no standard therapeutic guidelines for the management of A. baumannii biofilm-associated infections. Our previous in vitro study demonstrated that meropenem plus sulbactam showed synergism against biofilm-embedded carbapenem-resistant A. baumannii20. We analyzed therapeutic regimens and outcomes and found that patients who received ampicillin/sulbactam or sulbactam treatment had a more favorable 28-day survival rate in this study. However, we could not draw any conclusion due to the small number of cases. Additional large-scale clinical studies are needed to determine the optimal therapeutic regimens for A. baumannii biofilm-associated infections.
The whole genome sequencing analysis demonstrated the presence of biofilm-formation-associated genes and their promoter sequences in biofilm-forming isolates as well as non-biofilm-forming isolates. Further qRT-PCR experiments showed that the biofilm-forming isolates expressed higher levels of some biofilm-formation-associated genes. There may be several explanations for these findings. First, since the development of biofilms is regulated by a variety of factors, such as the environment, nutrition, stress and the pathogen itself, pathogens that contain biofilm-associated genes may only produce biofilms under certain conditions21,22. Second, some biofilm-associated proteins function in processes other than biofilm formation5,23. The presence of biofilm-associated genes in non-biofilm-forming isolates might indicate involvement in other physiological processes rather than biofilm formation. For example, pgaABCD encodes genes involved in the synthesis of poly-β-(1–6)-N-acetylglucosamine, which is an essential component of bacterial cell walls24. Third, the expression of biofilm-associated genes may be upregulated by factors other than promoters, such as transcriptional activators.
The limitations of this study included several confounding factors, such as variations in patient care and different patient backgrounds, which are inherent to a retrospective study. In addition, the biofilms established in vitro may be quite different from those in human bodies. It should be of concern that the results of our study may not be applied to the clinical settings. Yet, there is no standard laboratory protocol to assess in vivo biofilm formation. To overcome the inadequacy, we conducted a crystal violet assay, MBEC assay and genetic analysis to validate the biofilm-forming phenotype. We also conducted the qRT-PCR to confirm the expression of biofilm-associated genes. The inclusion of a large number of patients from multiple medical centers located in representative regions of Taiwan is another major strength of this study.
In conclusion, there might be no significant difference in the clinical characteristics and infectious outcomes between patients with bacteraemic pneumonia caused by biofilm-forming and non-biofilm-forming A. baumannii isolates. Although the biofilm-forming isolates were more susceptible to most antibiotics than non-biofilm-forming isolates, the biofilm-embedded cells indeed exhibited much higher antibiotic resistance. The antibiotic susceptibilities of biofilm-forming A. baumannii isolates should be interpreted cautiously. Further studies are required to determine the optimal treatment for bacteraemic pneumonia caused by biofilm-forming A. baumannii.
Methods
Study population
This retrospective study was performed during a 4-year period from January 2012 to December 2015 at four medical centers in Taiwan: Changhua Christian Hospital (CCH, 1676 beds) in Central Taiwan and Mackay Memorial Hospital (MMH, 2055 beds), Tri-Service General Hospital (TSGH, 1712 beds) and Taipei Veterans General Hospital (TVGH, 2900 beds) in Northern Taiwan. Patients who had at least one positive blood culture for A. baumannii, accompanied with the signs and symptoms of infection, were enrolled. Only the first blood culture from patients with two or more positive blood cultures was included. Patients under 20 years of age and those with incomplete medical records were excluded. The study protocol was approved by the institutional review boards (IRBs) of each site (CCH: IRB No. 140514, MMH: IRB No. 14MMHIS125, TSGH: IRB No. 1-103-05-100, TVGH: IRB No. 2015-04-001AC). All methods were performed in accordance with the relevant guidelines and regulations. Written informed consent was waived by the IRBs due to the retrospective nature of the analysis using information contained in medical charts and records, which were anonymized and de-identified prior to analysis.
Data collection
Medical records were reviewed to extract patient information, including demographic characteristics, comorbidities, admission to ICU, duration of hospitalization and the schedule and doses of antimicrobials administered. The presence of arterial cauterization, a central venous catheter, nasogastric tube, tracheostomy, ventilator, thoracic drain, or abdominal drain for more than 48 hours prior to bacteraemia onset was also recorded. The inclusion criteria25 for A. baumannii bacteraemic pneumonia consisted of: (a) at least one positive respiratory sample (sputum, endotracheal aspirate, or bronchoalveolar lavage) for A. baumannii obtained within 48 hours before or after the first positive blood culture; (b) a clinical course compatible with the diagnosis of pneumonia as defined by Centers for Disease Control and Prevention guidelines26 and (c) the positive blood culture could not be attributed to another source of infection. The onset of bacteraemia was defined as the day when then blood culture that yielded A. baumannii was obtained. Episodes of bloodstream infection were considered acquired in the ICU if they appeared 48 hours after ICU admission. Chronic kidney disease was defined as an estimated glomerular filtration rate <60 mL/min/1.73 m2. Neutropenia was defined as an absolute neutrophil count <0.5 × 109 cells/L. Immunosuppressive therapy was defined as use of immunosuppressive agents within 2 weeks or use of corticosteroids (≥15 mg of prednisolone daily for 1 week) within 4 weeks before bacteraemia onset. Recent surgery was defined as surgery within 4 weeks before bacteraemia onset. CCI was also calculated for further analysis. The severity of illness was evaluated using the APACHE II score within 24 h before bacteraemia onset. Previous ICU admission was defined as admission to ICU within 30 days before bacteraemia onset. Previous use of antimicrobials was defined as the use of antimicrobials within 30 days before bacteraemia onset. Appropriate antimicrobial therapy was defined as administration of at least one antimicrobial agent to which the causative pathogen was susceptible within 24 h after bacteraemia onset, via the approved route and at the recommended dosage for the affected organ(s). Antimicrobial therapy that did not meet this definition was considered inappropriate. Monotherapy with an aminoglycoside was considered an inappropriate therapy. The 28-day mortality rate was used as an endpoint and it was defined as death occurring within 28 days after bacteraemia onset. For patients who were discharged before the 28-day limit, the status was determined by review of outpatient records or by contacting the patient. No patient was lost to follow-up. Recurrent bacteraemia was defined as the redetection of A. baumannii 14 days after the index day.
Species identification and clonal study
Bacterial isolates were phenotypically identified as members of the Abc using the Vitek 2 system (bioMérieux, Marcy l’Étoile, France). A multiplex-polymerase chain reaction (PCR) method was used to identify A. baumannii to the species level27. The clonal relationship of clinical isolates was determined by PFGE as previously described28. Isolates were considered to be different pulsotypes if they had more than three DNA fragment differences and a similarity of <80% in dendrogram analysis.
Antimicrobial susceptibility testing
The MICs of antimicrobial agents were determined by agar dilution according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI)29. Antimicrobial susceptibility was interpreted according to the standards of the CLSI30. MDR was defined as resistance to ≥3 antimicrobial classes: aminoglycosides, antipseudomonal carbapenems, fluoroquinolones, antipseudomonal cephalosporins and β-lactam/β-lactamase inhibitor combinations. The MIC90 indicated the concentration of each antimicrobial agent required to inhibit 90% of the strains29.
Biofilm formation and measurement
Biofilm formation experiments were conducted using a microtitre plate assay as previously described31 with slight modifications. A. baumannii strains were cultured in 5 mL of Luria-Bertani (LB) broth supplemented with 1% D-glucose (LBglu) at 37 °C for 1 day. Thereafter, the cultures were diluted in LBglu to 0.03 at an OD570 and 200-μL aliquots were added to each well of a 96-well polystyrene tissue culture plate. The plates were subsequently incubated at 37 °C with shaking (180 rpm) for 48 hours. The suspensions were removed and the wells were washed with phosphate buffered saline (PBS). Thereafter, 200 μL of 0.1% crystal violet in H2O was added to stain the cells. The plates were incubated for 20 minutes with gentle agitation, thoroughly washed with PBS and then the stained biofilms were solubilized with 200 μL of 95% ethanol for 10 minutes with gentle agitation. The amount of biofilm formed was quantified by measuring the OD570. The OD570 values of un-inoculated wells were used as a negative control. We classified isolates as biofilm-forming if the OD570 values were at least twice those of the negative controls14. All experiments were performed in triplicate and repeated on three separate occasions. When an isolate was positive for biofilm formation on at least two occasions, the isolate was considered to be biofilm-positive.
Biofilm susceptibility testing
The antimicrobial susceptibility of A. baumannii biofilm cells was determined using the MBEC assay as previously described with some modification32,33. In brief, the tested isolates were cultivated using a 96-well microtitre plate (with a sterile peg lid) containing fresh Mueller-Hinton broth (MHB) for 16 hours at 37 °C to allow for biofilm formation. Thereafter, the biofilms were treated with serial dilutions of imipenem and meropenem for 20 hours at 37 °C. After incubation, the peg lids were rinsed three times in PBS and placed into antibiotic-free MHB in a flat-bottom microtitre plate. To transfer the biofilms from the pegs to the wells, each plate was centrifuged at 800 × g for 20 min and 20 µL was removed from each well and placed in the corresponding well of a fresh, sterile 96-well microtitre plate containing 180 µL of fresh MHB and incubated overnight to allow for recovery. The minimum antibiotic concentration at which no viable cell counts were recovered (OD570 <0.07) was considered to be the MBEC. All the experiments were performed in triplicate and repeated on three separate occasions.
Whole genome sequencing
Biofilm-associated genes and their promoters in biofilm-forming and non-biofilm-forming strains were identified by whole genome sequencing. Bacterial genomic DNA was purified from a single bacterial colony sub-cultured on Muller-Hinton agar and incubated for 16–24 hours using the fully automated LabTurbo 48 Compact System DNA extraction system. Genomic DNA isolated from a well-characterized A. baumannii strain was used to prepare a sequencing library in 90 minutes using Illumina’s Nextera DNA Sample Preparation Kit. For sequencing on the MiSeq instrument, prepared samples were placed in the reagent cartridge and loaded on the instrument along with the flow cell. All subsequent steps were performed on the instrument, including cluster generation, single or paired-end sequencing and primary data analysis. For de novo sequencing, the A. baumannii library prepared using Nextera sample preparation reagents was sequenced using a 2 × 150 read length on the MiSeq. Sequence read data were analyzed and assembled using a pipeline developed specifically for bacterial sequencing. The National Center for Biotechnology Information nucleotide database was searched for A. baumannii. Sequences were filtered by position to keep only the genomic regions corresponding to biofilm-associated genes, including BfmS, AbaI, Bap1, Bap2, PgaABCD and CsuAB/ABCDE. These regions were then organized as a sequence list in the Workbench and converted to a reference track. Post-adaptor-clipped and quality-trimmed reads per sample were then mapped in pairs against this reference track under default parameters. The number of mapped reads and the type of variants per gene falling within each biofilm-associated gene were then enumerated. In addition, the promoter sequences of these genes were also analyzed.
Multilocus sequence typing (MLST)
MLST scheme of selected isolated were tested as previous described34. This scheme involves PCR amplification and sequencing of seven housekeeping genes (gltA, gyrB, gdhB, recA, cpn60, gpi and rpoD).
Quantification of expression of biofilm-associated genes
A qRT-PCR assay was conducted for detection and quantification of mRNA of biofilm-associated genes. Total RNA was isolated using RNAprotect bacterial reagent and an RNeasy Mini kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. Genomic DNA contamination was eliminated by RNase-free DNase I treatment. RNA was reverse transcribed using random hexamer primers (MBI Fermentas, Vilnius, Lithuania) with Moloney Murine Leukemia Virus reverse transcriptase (Epicenter, Madison, WI, USA). PCR of total RNA without reverse transcription was used to examine the DNA contamination of RNA samples. Real-time PCR was carried out in final reaction volumes of 20 μL with 10 μL of Fast SYBR® Green Master Mix (2×) (Thermo Fisher Scientific Taiwan Co., Ltd. Taipei, Taiwan), 1 μL of each primer set (2 μM), 1 μL of template DNA and 3 μL of ddH2O. Thermal cycling was performed on the 7500 Fast (7500 Fast Real-Time PCR System) using the cycling conditions: pre-denaturation at 95 °C for 20s followed by 50 cycles of denaturation at 95 °C for 3s, annealing and extension at 60 °C for 30s. The amplification program was then followed by a melting cycle of 95 °C for 15s, 60 °C for 1 min, 95 °C for 15s and 60 °C for 15s. The qPCR amplification of the targeted DNA was monitored by the increase in the fluorescence in real time. The positive amplification of 16S rRNA gene was determined when the PCR cycle crossed the threshold cycle (CT) value and confirmed by the DNA melt curve analysis. All experiments were performed in duplicate.
Construction of carbapenem-resistant transformants of A. baumannii and Acinetobacter-derived cephalosporinase transformant of A. baumannii
To elucidate the impact of carbapenem resistance on biofilm formation, the vectors containing different carbapenemase genes with or without the upstream insertion sequence as a strong promoter were transformed into a biofilm-forming A. baumannii reference strain Ab15151 as previously described35 and the biofilm-forming capabilities of carbapenem-resistant transformants and Ab15151 with empty vectors were measured. Transformation of the vectors containing ADC genes with the upstream insertion sequence was also performed and the biofilm-forming capabilities of ADC transformant and Ab15151 with empty vectors were compared.
Statistical analysis
The PASW statistical package for Windows (version 20; SPSS, Chicago, IL, USA) was used for all data analyses. The χ2 test with Yates correction or Fisher’s exact test was used to compare categorical variables. Continuous variables were analyzed using the two-sample t-test. The time to mortality, defined as the interval between bacteraemia onset and death, was analyzed using the Kaplan–Meier survival analysis and the log-rank test was used to compare univariate survival distribution between different groups of patients. Univariate analyses were performed separately for each risk factor to ascertain the OR and 95% CI. All biologically plausible variables with p values less than 0.10 in the univariate analysis were considered for inclusion in the logistic regression model in the multivariate analysis. A backward selection process was utilized. A p value less than 0.05 was considered statistically significant.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Electronic supplementary material
Acknowledgements
This work was supported by grants from Taipei Veterans General Hospital [V105B-005, V106B-002 and VTA106-T-5-3], Tri-Service General Hospital [TSGH-C105-112, TSGH-C107-099 and VTA106-T-5-1], Academia Sinica [VTA106-T-5-2], National Defense Medical Center [MAB-106-076 and MAB-107-095] and the Ministry of Science and Technology [MOST 104-2314-B-075-043-MY3, MOST 105-2314-B-016-039-MY3 and MOST 105-2628-B-016-003-MY2]. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Contributions
Y.C.W., T.L.C. and Y.T.L. conceived the project and wrote the manuscript; F.Y.C., S.H.W. and C.K.H. designed the experiments and manuscript revisions; T.W.H., Y.S.Y. and S.C.K. performed experiments. C.T.C., C.P.L. and Y.M.L. conducted statistical analysis. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-25661-9.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cisneros JM, et al. Bacteremia due to Acinetobacter baumannii: epidemiology, clinical findings and prognostic features. Clin. Infec. Dis. 1996;22:1026–1032. doi: 10.1093/clinids/22.6.1026. [DOI] [PubMed] [Google Scholar]
- 2.Dijkshoorn L, Nemec A, Seifert H. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 2007;5:939–951. doi: 10.1038/nrmicro1789. [DOI] [PubMed] [Google Scholar]
- 3.Marti S, et al. Biofilm formation at the solid-liquid and air-liquid interfaces by Acinetobacter species. BMC Res. Notes. 2011;4:5. doi: 10.1186/1756-0500-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Villegas MV, Hartstein AI. Acinetobacter outbreaks, 1977–2000. Infect. Control Hosp. Epidemiol. 2003;24:284–295. doi: 10.1086/502205. [DOI] [PubMed] [Google Scholar]
- 5.Tomaras AP, Dorsey CW, Edelmann RE, Actis LA. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology. 2003;149:3473–3484. doi: 10.1099/mic.0.26541-0. [DOI] [PubMed] [Google Scholar]
- 6.Lewis K. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 2001;45:999–1007. doi: 10.1128/AAC.45.4.999-1007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 2008;21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gil-Perotin S, et al. Implications of endotracheal tube biofilm in ventilator-associated pneumonia response: a state of concept. Crit. Care. 2012;16:R93. doi: 10.1186/cc11357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents. 2010;35:322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 10.Olsen, I. Biofilm-specific antibiotic tolerance and resistance. Eur J Clin Microbiol Infect Dis., 10.1007/s10096-015-2323-z (2015). [DOI] [PubMed]
- 11.Mah TF. Biofilm-specific antibiotic resistance. Future Microbiol. 2012;7:1061–1072. doi: 10.2217/fmb.12.76. [DOI] [PubMed] [Google Scholar]
- 12.Lee HW, et al. Capacity of multidrug-resistant clinical isolates of Acinetobacter baumannii to form biofilm and adhere to epithelial cell surfaces. Clin. Microbiol. Infect. 2008;14:49–54. doi: 10.1111/j.1469-0691.2007.01842.x. [DOI] [PubMed] [Google Scholar]
- 13.Qi L, et al. Relationship between Antibiotic Resistance, Biofilm Formation and Biofilm-Specific Resistance in Acinetobacter baumannii. Front. microbiol. 2016;7:483. doi: 10.3389/fmicb.2016.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodriguez-Bano J, et al. Biofilm formation in Acinetobacter baumannii: associated features and clinical implications. Clin. Microbiol. Infect. 2008;14:276–278. doi: 10.1111/j.1469-0691.2007.01916.x. [DOI] [PubMed] [Google Scholar]
- 15.Barsoumian AE, et al. Clinical infectious outcomes associated with biofilm-related bacterial infections: a retrospective chart review. BMC Infect. Dis. 2015;15:223. doi: 10.1186/s12879-015-0972-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang D, et al. Biological features of biofilm-forming ability of Acinetobacter baumannii strains derived from 121 elderly patients with hospital-acquired pneumonia. Clin. Exp. Med. 2016;16:73–80. doi: 10.1007/s10238-014-0333-2. [DOI] [PubMed] [Google Scholar]
- 17.Mah TF, O’Toole GA. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001;9:34–39. doi: 10.1016/S0966-842X(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 18.Gallant CV, et al. Common beta-lactamases inhibit bacterial biofilm formation. Mol. Microbiol. 2005;58:1012–1024. doi: 10.1111/j.1365-2958.2005.04892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nucleo E, et al. Differences in biofilm formation and aggregative adherence between beta-lactam susceptible and beta-lactamases producing P. mirabilis clinical isolates. New Microbiol. 2010;33:37–45. [PubMed] [Google Scholar]
- 20.Wang YC, et al. Individual or Combined Effects of Meropenem, Imipenem, Sulbactam, Colistin and Tigecycline on Biofilm-Embedded Acinetobacter baumannii and Biofilm Architecture. Antimicrob. Agents Chemother. 2016;60:4670–4676. doi: 10.1128/AAC.00551-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 22.Jefferson KK. What drives bacteria to produce a biofilm? FEMS Microbiol. Lett. 2004;236:163–173. doi: 10.1111/j.1574-6968.2004.tb09643.x. [DOI] [PubMed] [Google Scholar]
- 23.Mihara K, et al. Identification and transcriptional organization of a gene cluster involved in biosynthesis and transport of acinetobactin, a siderophore produced by Acinetobacter baumannii ATCC 19606T. Microbiology. 2004;150:2587–2597. doi: 10.1099/mic.0.27141-0. [DOI] [PubMed] [Google Scholar]
- 24.Choi AH, Slamti L, Avci FY, Pier GB, Maira-Litran T. The pgaABCD locus of Acinetobacter baumannii encodes the production of poly-beta-1-6-N-acetylglucosamine, which is critical for biofilm formation. J. Bacteriol. 2009;191:5953–5963. doi: 10.1128/JB.00647-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee YT, et al. Bacteremic nosocomial pneumonia caused by Acinetobacter baumannii and Acinetobacter nosocomialis: a single or two distinct clinical entities? Clin. Microbiol. Infect. 2013;19:640–645. doi: 10.1111/j.1469-0691.2012.03988.x. [DOI] [PubMed] [Google Scholar]
- 26.Garner JS, Jarvis WR, Emori TG, Horan TC, Hughes JM. CDC definitions for nosocomial infections, 1988. Am. J. Infect. Control. 1988;16:128–140. doi: 10.1016/0196-6553(88)90053-3. [DOI] [PubMed] [Google Scholar]
- 27.Chen TL, et al. Comparison of one-tube multiplex PCR, automated ribotyping and intergenic spacer (ITS) sequencing for rapid identification of Acinetobacter baumannii. Clin. Microbiol. Infect. 2007;13:801–806. doi: 10.1111/j.1469-0691.2007.01744.x. [DOI] [PubMed] [Google Scholar]
- 28.Huang LY, et al. Dissemination of multidrug-resistant, class 1 integron-carrying Acinetobacter baumannii isolates in Taiwan. Clin. Microbiol. Infect. 2008;14:1010–1019. doi: 10.1111/j.1469-0691.2008.02077.x. [DOI] [PubMed] [Google Scholar]
- 29.Wayne, P. CLSI. Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically: Approved Standard M7–A7. (2006).
- 30.Wayne, P. CLSI. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: twenty-fifth informational supplement. M100–S25. (2015).
- 31.O’Toole GA, et al. Genetic approaches to study of biofilms. Methods Enzymol. 1999;310:91–109. doi: 10.1016/S0076-6879(99)10008-9. [DOI] [PubMed] [Google Scholar]
- 32.Ceri H, et al. The Calgary Biofilm Device: New Technology for Rapid Determination of Antibiotic Susceptibilities of Bacterial Biofilms. J. Clin. Microbiol. 1999;37:1771–1776. doi: 10.1128/jcm.37.6.1771-1776.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moskowitz SM, Foster JM, Emerson J, Burns JL. Clinically feasible biofilm susceptibility assay for isolates of Pseudomonas aeruginosa from patients with cystic fibrosis. J. Clin. Microbiol. 2004;42:1915–1922. doi: 10.1128/JCM.42.5.1915-1922.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jolley KA, Maiden MC. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen TL, et al. Contribution of a plasmid-borne blaOXA-58 gene with its hybrid promoter provided by IS1006 and an ISAba3-like element to beta-lactam resistance in Acinetobacter genomic species 13TU. Antimicrob. Agents Chemother. 2010;54:3107–3112. doi: 10.1128/AAC.00128-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.


