Abstract
Sox9 is a member of the gene family of SOX transcription factors, which is highly conserved among vertebrates. It is involved in different developmental processes including gonadogenesis. In all amniote species examined thus far, Sox9 is expressed in the Sertoli cells of the male gonad, suggesting an evolutionarily conserved role in testis development. However, in the anamniotes, fishes and amphibians, it is also expressed in the oocyte but the significance of such an expression remains to be elucidated. Here, we have investigated the nuclear localization of the SOX9 protein in the oocyte of three amphibian species, the urodelan Pleurodeles waltl, and two anurans, Xenopus laevis and Xenopus tropicalis. We demonstrate that SOX9 is associated with ribonucleoprotein (RNP) transcripts of lampbrush chromosomes in an RNA-dependent manner. This association can be visualized by Super-resolution Structured Illumination Microscopy (SIM). Our results suggest that SOX9, known to bind DNA, also carries an additional function in the posttranscriptional processes. We also discuss the significance of the acquisition or loss of Sox9 expression in the oocyte during evolution at the transition between anamniotes and amniotes.
Introduction
The SOX proteins (for “SRY-related HMG box”) are characterized by the presence of the high-mobility-group (HMG) DNA-binding domain. This domain of 79 amino-acids shares at least 50% identity with the HMG box of the mammalian male sex-determining SRY protein1. The SOX proteins constitute a large family of transcription factors (TFs), known to activate or repress gene expression and to be involved in a wide range of biological processes. On the basis of sequence similarity within the HMG box, mammalian SOX proteins have been divided into ten groups (A-J), each including one or several members. Orthologs of these genes have been found in different animal species. SOX TFs influence local chromatin structure through their capacity to bend DNA. In most cases, the HMG box of SOX proteins binds to target DNA sequences and a consensus binding site [(A/T)(A/T)CAA(A/T)G] has been defined2. Regions outside of the HMG domain play also a role in DNA-binding. However, the cell-specific action of SOX proteins is mainly due to their interaction with partners to form stable transcription complexes (for recent reviews see3–9). Besides their activity as TFs, several studies have highlighted a potential role of SOX proteins in RNA processing10–15. In addition, a direct interaction with RNA has been shown for several of them. In Drosophila, the Dichaete Sox protein binds to synthetic target RNAs in a sequence-specific manner16. SOX2 physically interacts with the long noncoding RNA rhabdomyosarcoma 2-associated transcript (RMST) and this interaction is necessary for the activation neural genes17.
The SOX9 protein belongs to the group E that also includes SOX8 and SOX10. SOX9 is highly conserved among vertebrate species and is involved in multiple organogenetic processes, such as chondrogenesis, neurogenesis and gonadogenesis18. The Sox9 gene is the major target of Sry, the master testis-determining gene in mammals, located on the Y chromosome. In mammals, where it has been most extensively studied, Sox9 has been shown to play a critical role in male sex determination and testis differentiation. Its mutation or deletion leads to 46,XY Disorders of Sex Development (formerly known as male-to-female sex reversal) in humans and mice19. During embryogenesis, the SOX9 protein is detected in the cytoplasm of somatic cells in male and female gonads. At the onset of testicular differentiation, SOX9 moves to the nucleus of Sertoli cells precursors where it is detected up to the adult stage. In contrast, the expression of SOX9 is downregulated during ovarian differentiation20–22. Nuclear translocation of SOX9 constitutes a key event in the differentiation of the male gonad and its impairement leads to 46,XY DSD (male-to-female sex-reversed phenotype). In contrast, experimentally-forced nuclear localization of SOX9 in the XX gonad leads to a female-to-male sex reversal23–25. Recently, genetic networks directly regulated by SOX9 in the mammalian foetal testis were identified and a direct or indirect role in the modulation of pre-mRNA splicing of several of its target genes was shown15.
The function of the Sox9 gene during gonadogenesis has been less well investigated in other vertebrate species. In birds26 and reptiles27,28, it is expressed in a male-specific manner, whereas in fishes29–35 and amphibians36–40 it is expressed in both the male and female gonads. Of note, the majority of these investigations focused on the expression of the Sox9 gene only at the RNA level.
We have previously shown that the expression of the SOX9 protein in the amphibian Xenopus tropicalis greatly differs between the male and female gonads38. In the testis, SOX9 is localized in the nuclei of Sertoli-like somatic cells, similarly to what has been observed in mammals. In contrast, it is localized in the germ cells of the female gonad. In the juvenile female gonad, SOX9 is exclusively detected in oocyte cytoplasm. Surprisingly, in the adult gonad, it is detected in both the cytoplasm and the nucleus (also called germinal vesicle or GV). This unexpected observation of a translocation of SOX9 into the GV prompted us to check whether it also takes place in other amphibians and to investigate its functional significance.
We took advantage of the remarkable structure of the giant chromosomes of the amphibian oocyte, the lampbrush chromosomes (LBCs), to analyze the sub-nuclear localization of the SOX9 protein on LBC spreads of three amphibian species, the anurans X. tropicalis and X. laevis, and the urodelan Pleurodeles waltl. Similarly to polytene chromosomes of insects, amphibian LBCs offer the unique possibility to use light microscopy to visualize directly on nuclear spreads the condensed and transcribed uncondensed chromatin regions at the whole genome level and to identify associated proteins41–50 (http://projects.exeter.ac.uk/lampbrush/). In this study, we provide evidence for the association of the SOX9 protein with RNP transcripts in an RNA-dependent manner and we show that this association can be visualized by super-resolution imaging. Our findings suggest a role for SOX9 in post-transcriptional processes.
Results
The SOX9 protein is detected in the nuclear and cytoplasmic extracts from X. tropicalis, X. laevis and P. waltl oocytes
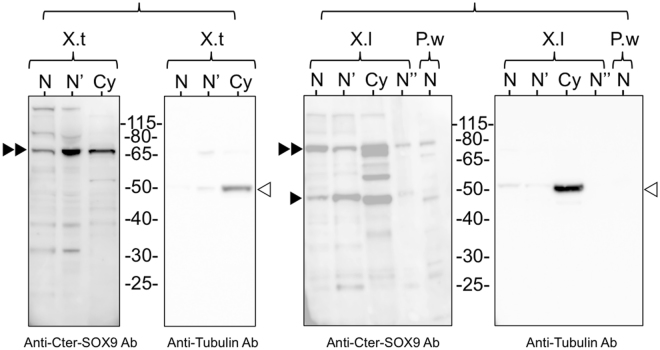
SOX9 nuclear sub-localization was studied using two polyclonal antibodies directed against the N-ter and C-ter regions of the human protein. Although these regions present a high degree of amino acid identity with those of X. tropicalis, X. laevis and P. waltl proteins (Fig. S1), we first confirmed that the antibodies cross-reacted with the exogenous X. tropicalis SOX9 protein expressed in mammalian cells after transfection with a plasmid vector (see Materials and Methods, Fig. S2). We then investigated by western blot the presence of the amphibian SOX9, in both nuclear and cytoplasmic extracts of X. tropicalis, X. laevis and P. waltl oocytes. Envelope-free nuclear extracts were also analyzed to exclude that the detection of SOX9 in the GV was due to a contamination of the nuclear extracts by a small amount of cytoplasm remaining associated with the nuclear envelope. Alpha-tubulin (detected by a monoclonal antibody) was used as a cytoplasmic control. As expected, the two polyclonal anti-SOX9 antibodies recognized one polypeptide of nearly the same apparent molecular weight at around 68-kDa in X. tropicalis, X. laevis and P. waltl nuclear and cytoplasmic extracts (Fig. 1, and Fig. S3), whereas the mouse mAb against alpha-tubulin detected one polypeptide of the expected size (52 kDa) in cytoplasmic extracts only. These results confirmed the presence of SOX9 in the amphibian oocyte and its nuclear localization. They are in contrast to those of Dumond et al.37 who did not detect SOX9 in the GV of P. waltl ovocyte using the same Nter-SOX9 Ab antibody51. One possible explanation for this discrepancy is that they used the anti-SOX9 antibody at a higher dilution (1:1000 instead of 1:400 used here).
Figure 1.
Immunodetection of SOX9 in the oocyte of X. tropicalis, X. laevis and P. walt. Western blot analysis of SOX9 in nuclear and cytoplasmic extracts using the polyclonal anti-Cter-SOX9 antibody and the monoclonal anti-tubulin antibody. (N) corresponds to 25 GVs from X. tropicalis (X.t), 20 from X. laevis (X.l) and 15 from P. waltl (P.w). (N’) nuclear extract without nuclear envelope. (N”) 15 GVs from X. laevis. (Cy) cytoplasms from 10X. tropicalis oocytes and from 5X. laevis oocytes. The anti-Cter- SOX9 antibody recognized a major polypeptide around 68-kDa in the nuclear (N and N’) and cytoplasmic (Cy) extracts of the three species (black double arrowheads) and an additional one at 48 kDa (black arrowhead). The control anti-tubulin antibody recognized a 50-kDa polypeptide only in the cytoplasmic extracts (empty arrowheads).
The SOX9 protein binds to LBCs lateral loops and nuclear bodies in the three amphibian species
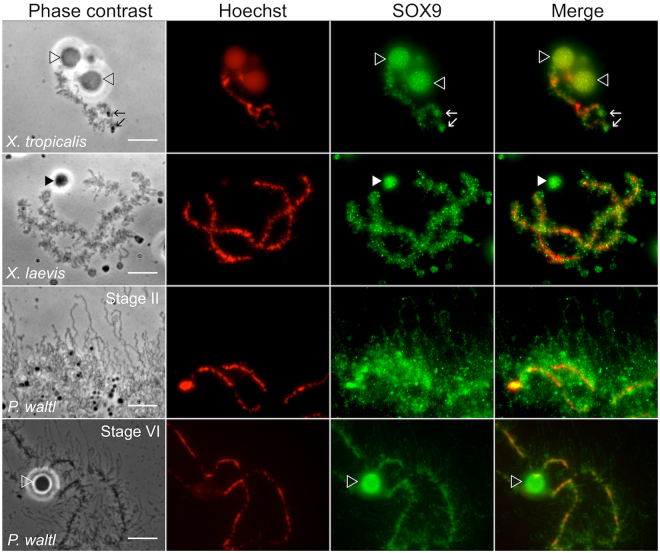
The meiotic lampbrush chromosomes are associated in bivalent pairs. Their axes comprise a succession of highly condensed chromatin regions, the chromomeres, from which lateral loops unfold in opposite directions. The chromomeres constitute the main chromosome axis and can be visualized by DAPI or Hoechst staining. The lateral loops are clearly visible by light microscopy. They serve as template for RNA polymerase II-dependent transcription and comprise one or more transcription units. The nascent RNA transcripts associate with proteins to form the ribonucleoprotein (RNP) matrix whose thickness shows a marked polarity along its length according to the putative direction of transcription. The decondensed transcribed DNA axis of lateral loops is itself neither visible by light microscopy nor after DAPI or Hoechst staining (for review, see:52–54). As shown in Fig. 2, the sizes of the LBCs and their lateral loops differ between the three amphibians with a direct correlation with genome size as reported for many amphibian species55. P. waltl (C value: 19,3 pg) LBCs are larger than those of X. laevis (C value: 3 pg), which are in turn larger than those of X. tropicalis (C value: 1.5)56. The mean size of the lateral loops in P. waltl is 50 μm while it is around 5–10 μm in X. laevis. Furthermore, for a given species, the size of the LBCs and the number and size of their lateral loops, are also directly related to their transcriptional activity. Consistently, the loops are more developed at early oocyte stages, when transcription is intense, than at stage VI when transcription is almost arrested.
Figure 2.
Subnuclear localization of the SOX9 protein in the GV of X. tropicalis, X. laevis and P. waltl. Fluorescent and corresponding phase contrast micrographs of nuclear spreads showing one LBC of X. tropicalis and X. laevis from stage V-VI oocyte, and part of a P. waltl LBC from stages II and V-VI oocyte. Nuclear spreads were immunostained with the anti-Cter-SOX9 antibody (green, Alexa 488 IgG) and counterstained with Hoechst (red). The Hoechst dye stained the chromosome DNA axis and not that of the decondensed lateral loops. The antibody labeled the LBCs, and the nuclear bodies either attached to them (empty arrowheads) or free in the nucleoplasm (full arrowheads). Note that the immunostaining of lateral loops was very clear in P. waltl LBCs from stage II oocyte, because of their extension and density. The intensity of fluorescence in the different experiments was normalized with respect to their relevant controls with the secondary antibody (tagged with Alexa fluor 488) alone. Wide field Leica microscope. Scale bar for all micrographs: 10 μm.
When applied to nuclear spreads of X. tropicalis, X. laevis or P.waltl oocytes, both the anti N-ter and anti C-ter SOX9 antibodies strongly labelled the lateral loops of LBCs and nuclear bodies, which were either free or attached to the LBCs, as it is the case for the Histone Locus Body (HLB) appearing as a sphere bound to chromosome XI of P. waltl (Fig. 2 and Fig. S4-A). Some of the free bodies corresponded to the Cajal bodies (CBs) because they were co-stained for Coilin, one of its major components57,58 (Fig. S4-B). The immunostaining of lateral loops was best displayed in the case P. waltl, particularly at an early oocyte stage, when they were large and well extended from the chromosome axis.
Confirmation of the SOX9 immunostaining pattern
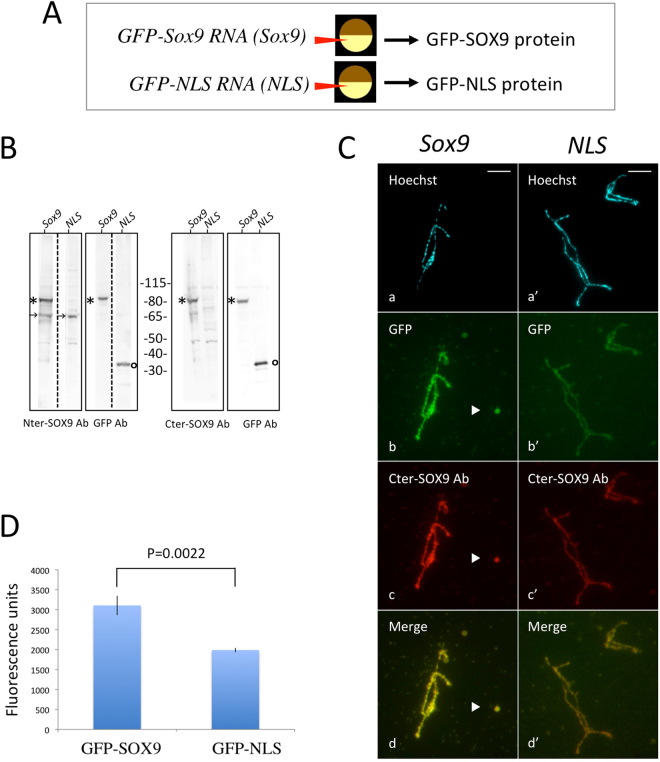
To confirm the immunostaining pattern of the nuclear spreads observed using the anti-SOX9 antibodies, the sub-nuclear localization of an exogenous GFP-tagged SOX9 protein expressed from RNA-injected oocytes was analyzed. In vitro-synthesized capped transcripts coding for either a GFP-tagged Xt- SOX9 (Xt-SOX9-GFP) protein or the NLS-GFP protein as a control, were injected into the cytoplasm of stage V-VI X. laevis oocytes (Fig. 3A). After 24 h of incubation, the Xt-SOX9-GFP and NLS-GFP proteins were detected in the nuclear extracts by western blotting using antibodies against SOX9 and GFP proteins at the expected molecular weights (Fig. 3B and Fig. S5). In parallel, the targeting behavior of these exogenous proteins was analyzed on nuclear spreads by fluorescence microscopy. The Xt-SOX9-GFP protein targets the LBCs and nuclear bodies (Fig. 3C-b). This staining pattern was similar to the one obtained with the anti-SOX9 antibodies (Fig. 3Cc-d). Because of a possible non-specific binding of the NLS-GFP protein to the chromosomes, we quantified the fluorescence intensity using the Otsu’s method by first devising appropriate masks on the Hoechst images to define a region of interest (ROI) (Materials and Methods, Fig. S6). The fluorescence intensity of the Xt-SOX9-GFP protein on the LBCs was much higher than for NLS-GFP (Fig. 3D). Altogether, these and the immunostaning results show that SOX9 binds to the lateral loops and nuclear bodies.
Figure 3.
Targeting of the newly-synthetized Xt-SOX9-GFP protein to LBCs and nuclear bodies. (A) Capped, in vitro-synthetized transcripts from the pcDNA Xt-sox9- CT-GFP (Sox9) vector, or the control pcDNA NLS-CT-GFP (NLS) vector, were injected into the cytoplasm of stage IV-V oocytes of X. laevis. The expressed proteins (Xt-SOX9-GFP or NLS-GFP) were detected 18 hours later, either directly on nuclear spreads by fluorescence microscopy or indirectly on immunoblots using antibodies against the SOX9 or GFP proteins. (B) Immunoblots of nuclear extracts from injected oocytes with the sox9-CT-GFP (Sox9) or NLS-CT-GFP (NLS) transcripts. Each lane corresponds to 15 GVs. kDa: molecular weight markers. A polypeptide (*) around 80 kDa detected with the anti-Nter or Cter- SOX9 antibodies and the anti-GFP antibody in the Sox9-CT-GFP injected oocytes corresponded to Sox9-GFP protein. In the NLS-CT-GFP injected oocytes, the anti-GFP antibody detected a 30 kDa polypeptide (°) corresponding the NLS-GFP protein. The polypeptide of ca. 68 kDa (arrows) corresponds to the endogenous SOX9 protein. The dashed lines separate the lanes which were cut from the original immunoblots shown in the supplementary figure S5. (C) (b, b’) Direct detection of GFP-SOX9 and GFP-NLS proteins on nuclear spreads from the injected oocytes. The GFP-SOX9 protein targeted the LBCs and the free nuclear bodies (arrowhead). A much lower level of GFP-NLS was detected on the LBCs. (c, c’) immunostaining of the nuclear spreads using the anti-Cter-SOX9 antibody. (d, d’) merged images (b, b’ and c, c’) indicating that the staining pattern of the Xt-SOX9-GFP was similar to that using the anti-Cter-SOX9 antibody. Wide field Leica microscope. Scale bar for all micrographs: 10 μm. (D) Quantification of GFP fluorescence density on LBCs with the Otsu’s method. GFP-fluorescence density over the GFP-SOX9 LBCs was significantly higher than that of the control GFP-NLS LBCs (Student’s test, p = 0.002).
SOX9 does not bind the chromosome axis of the LBCs
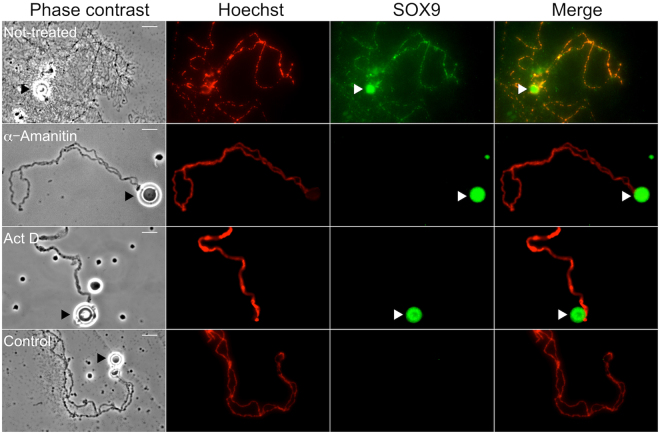
The strong immunostaining which can be observed over the chromosome axis with the SOX9 antibodies as shown in Figs 2 and 3, raised the question as to whether SOX9 actually binds to the condensed chromatin of the chromomeres, which constitute the visible part of the chromosome axis and from which the decondensed chromatin axes of the lateral loops unfold. Three lines of evidence suggest that SOX9 does not bind to the chromomeres. First, in some preparations where the lateral loops were particularly well extended from the chromosome axis and exhibited an obvious polarity as shown in Fig. 4A, a strong SOX9 immunostaining was detected in the close vicinity of the chromomeres over the thickest region of the lateral loops corresponding to the presumed termination of transcription. This pattern may explain why the SOX9 immunostaining appeared to overlap with the chromosome axis in less resolved preparations. Second, SOX9 immunostaining was not detected at the level of the chromomeres when they were devoid of lateral loops as seen with LBCs from oocytes at the end of stage VI (Fig. 4B). A third argument was brought by transcription inhibition experiments using alpha-amanitin or actinomycin D. Although these two drugs act differently, the former being a DNA intercalator blocking the progression of the three RNA polymerases, and the later a highly selective inhibitor of RNA polymerases II and III (for a review see59), their effect on LBCs was the same. As previously described60, after incubation of the oocytes with these drugs, transcription was arrested, the nascent RNP transcripts were released, the lateral loops underwent retraction and the chromosomes being devoid of their lateral loops became extremely condensed. Under these conditions, no labelling of the chromosome axis with the SOX9 antibodies was observed, although it was still detected over free Cajal Bodies or the nuclear bodies associated with them (Fig. 5).
Figure 4.
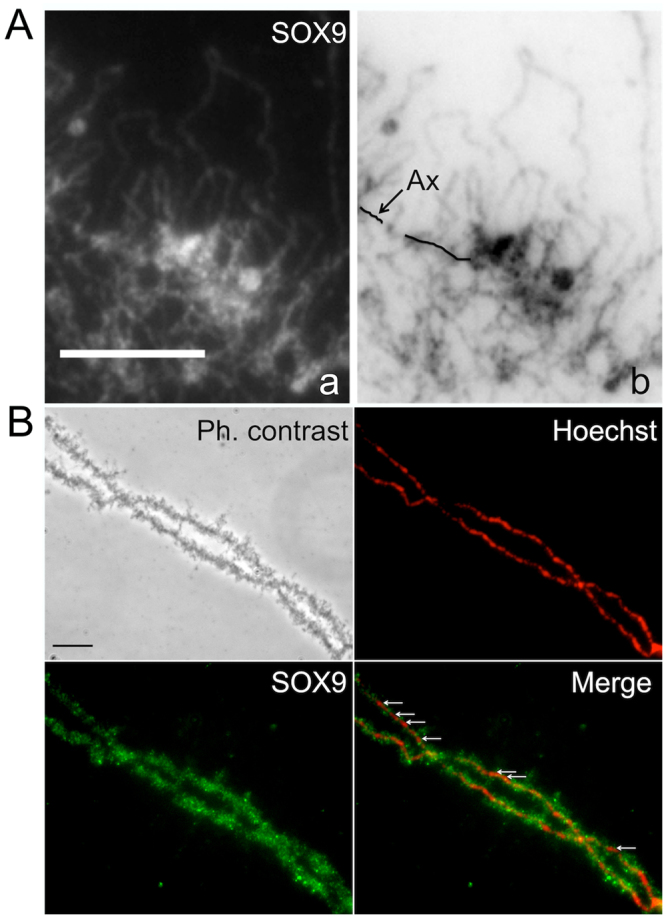
SOX9 protein does not bind the chromosome axis of the LBCs. Immunostaining of P. waltl nuclear spreads using the anti-Cter-SOX9 antibody (green, Alexa 488 IgG) and counterstained with Hoechst (red). (A) Fluorescent micrograph (a) and its corresponding negative (b) showing a strong SOX9 immunostaining detected in the close vicinity of the chromosome axis over the thickest region of the lateral loops (B) Nuclear spreads from an oocyte at the end of stage VI. The SOX9 protein was not detected at the level of the chromosome axis where it was devoid of lateral loops (white arrows, merge panel). Scale bar for all micrographs: 10 μm.
Figure 5.
Immunostaining of SOX9 after inhibition of transcription. Oocytes of P. waltl were treated or not with α-Amanitin or Actinomycin D (Act D). Phase contrast and corresponding fluorescent micrographs of nuclear spreads that were stained with the anti-Cter SOX9 antibody (green, Alexa 488 IgG) and counterstained with Hoechst (red) to show the chromosome axis. In contrast to the LBCs from the not-treated oocytes, those from the oocytes incubated with α-amanitin or Act D were devoid of their lateral loops and and the chromosome axis lacked SOX9 staining. Note that the spheres (arrowheads) known to be storage sites for transcription and post-transcription factors were strongly stained. The intensity of fluorescence in the different experiments was normalized with respect to their relevant controls with the secondary antibodies alone. Wide field Leica microscope. Scale bar for all micrographs: 15 μm.
SOX9 is associated with the RNP matrix of lateral loops in an RNA-dependent manner
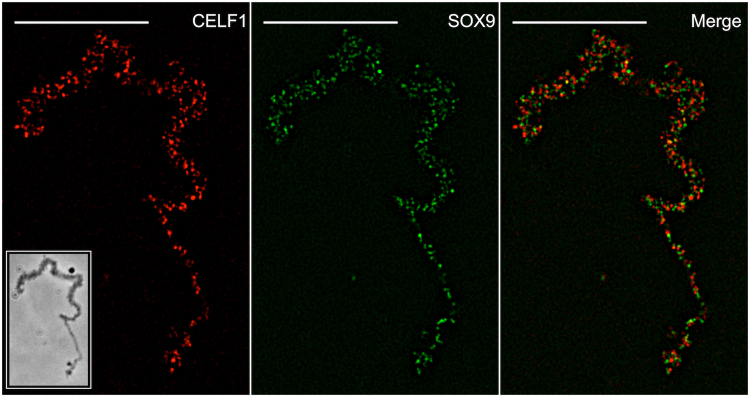
We performed a detailed analysis of the SOX9 immunostaining pattern of lateral loops to determine whether the protein binds to the transcribed DNA axis of the lateral loops, the associated RNPs transcripts or both. Because Hoechst staining was not strong enough to detect the decondensed transcribing chromatin axis of the lateral loops, we carried out co-immunostainings using the SOX9 antibody together with an antibody (mAb H14) directed against phosphorylated RNA Polymerase II (referred to as Pol II), which was previously reported to label the DNA axis of lateral loops undergoing transcription53,54. As expected, both antibodies stained the lateral loops (Fig. 6 and Fig. S7). However, while the Pol II immunostaining appeared continuous over the axis of the lateral loops, the SOX9 immunostaining showed a punctate pattern with granules distributed above the axis of the loops. This suggests that SOX9 is associated with the RNP matrix rather than with the chromatin axis of lateral loops. To confirm this result, co-immunostaining experiments were performed using antibodies directed against CELF1 and SR proteins, previously reported to be associated with the nascent RNA transcripts44,47. CELF1 is a multifunctional RNA-binding protein involved in the regulation of alternative splicing61. Fluorescence microscopy experiments have shown that CELF1 is able to bind to a subset of lateral loops in several amphibian species, Xenopus laevis, Triturus vulgaris and Axolotl44. Its binding to RNP transcripts was confirmed by super-resolution imaging using Spectral Position Determination Microscopy (SPDM) in Notophthalmus viridescens47. In P. waltl we also detected it on a few lateral loops (Fig. S8). The pattern of co-immunostaining performed using antibodies against SOX9 and CELF1 was analyzed using Super-resolution Structured Illumination Microscopy (SIM). The superresolution microscopy images revealed a pattern of granules immunostained with either SOX9 or CELF1 antibodies, which corresponded to packed RNP transcripts (Fig. 7). The average size of these particles as determined using the spot module of the IMARIS software (Bitplane, version 8.4) was similar for those associated with SOX9 (0.192 µm, standard deviation: 0.035) or CELF1 (0. 196 µm, standard deviation: 0.038). The Pearson’s correlation coefficient of the intensities of the fluorescence signal calculated using the Coloc2 plugin of the Image J program, was negative (R = −0.65, p < 0.0001)), excluding a colocalization of SOX9 and CELF1 on the same granules, i.e, on the same RNA-protein complexes.
Figure 6.
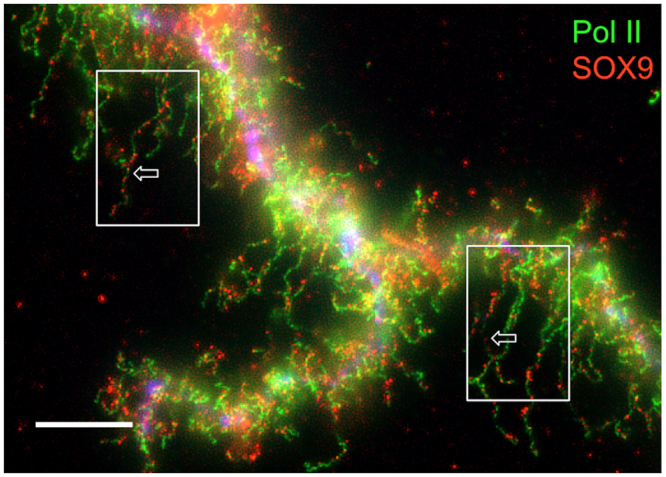
Immunostaining of Pol II and SOX9 in P. waltl LBCs. Nuclear spreads were immunostained for SOX9 with the anti-Cter-SOX9 antibody (red, Alexa 568 IgG) and for Pol II with the mAbH14 (green, Alexa 488 IgM). Pol II immunostaining was continuous over the loop axis while that of SOX9 was concentrated on granules distributed above the loop axis. This double immunostaining pattern was particularly visible on the two lateral loops indicated by arrows in the boxed areas. The intensity of fluorescence in this experiment was normalized with respect to its relevant controls (i.e., secondary antibodies alone). The image was deconvoluted (see Figure S7 for the non deconvoluted image). Wide field Leica microscope. Scale bar: 10 μm.
Figure 7.
Super-resolution images of a P. waltl lateral loop immunostained for CELF1 [mAb3B1 (red, Alexa 568 IgG)] and SOX9 [anti-Cter-SOX9 antibody (green, Alexa 488 IgG)]. The RNP matrix exhibited a pattern of granules immunostained for SOX9 and CELF1. The merged panel showed that the SOX9 and CELF1 granules did not colocalize. The intensity of fluorescence in the different experiments was normalized with respect to their relevant controls with the secondary antibodies alone. Structured Illumination Microscopy (SIM). The image in the boxed area corresponded to the phase contrast image obtained from Wide field Leica microscopy. Scale bar: 10 μm.
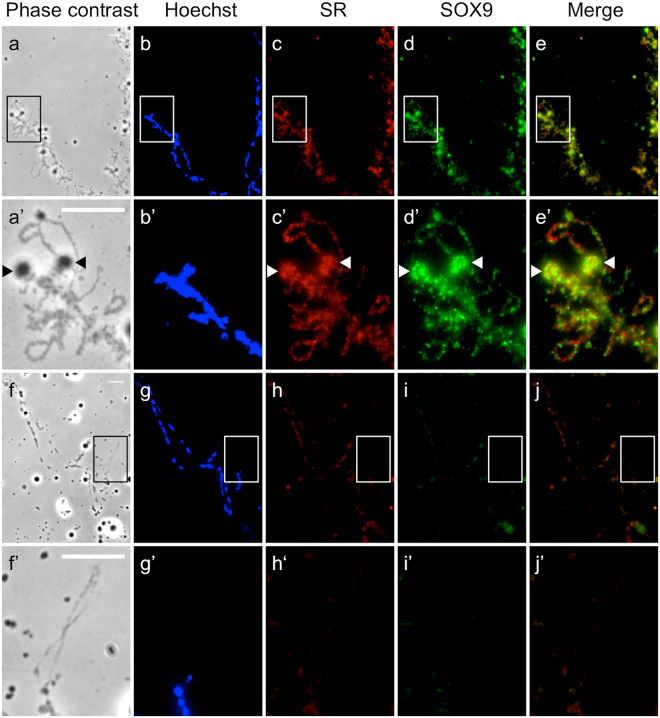
SR proteins belong to a conserved family of pre-mRNA splicing factors that are associated with the RNA transcripts of lateral loops62,63. Co-immunostaining on LBCs of Xenopus laevis using the anti C-ter SOX9 antibody and the monoclonal mAb1H4 antibody directed against SR proteins of X. laevis showed, as expected, that the two antibodies stained almost all the lateral loops (Fig. 8a-e’). In order to determine whether the binding of SOX9 to lateral loops was RNA-dependent, preparations of X. laevis LBCs were treated with RNAse A before co-immunostaining with the SOX9 antibody or the monoclonal mAB1H4. No immunofluorescence was detected over the lateral loops when LBCs preparations were treated with RNAse (Fig. 8f-j’). This clearly indicated that the association of SOX9 to the RNP matrix was mediated directly or indirectly by RNA.
Figure 8.
Localization of SOX9 to the matrix of lateral loops is RNA-dependent. Nuclear spreads of X. laevis were immunostained for SOX9 [anti-Cter-SOX9 antibody (green, Alexa 488 IgG)] and SR proteins [mAb1H4 (red, Alexa 568 IgG)]. (a–e’) LBC from untreated nuclear spreads. (f–j’) LBC from nuclear spread digested with RNase A before immunostaining. (a’–e’) Enlarged images of the regions marked by white boxes (a–e) show several lateral loops immunostained with SOX9 and SR antibodies. The arrowheads point to terminal granules. (f’–j’) enlarged images of the regions marked by white boxes (f–j) show one lateral loop not stained with SOX9 and SR antibodies after digestion with Rnase A. The intensity of fluorescence in the different experiments was normalized with respect to their relevant controls (the secondary antibodies only). Wide field Leica microscopy. Scale bar: 10 μm.
Discussion
The aim of this study was to establish whether the nuclear localization of SOX9, previously reported in the oocyte of X. tropicalis, took also place in other amphibian species such as X. laevis and P.waltl, as a way to get insights into its functional role during oogenesis. Using two different approaches relying either on SOX9 antibodies or on the production of exogenous GFP-tagged SOX9 protein, we have provided evidence for the nuclear localization of SOX9 and its specific association with the nascent RNP matrix of lateral loops in all three amphibian species. Because no SOX9 immunostaining was found on LBCs devoid of lateral loops, either at the late stage VI of oocyte development or after inhibition of transcription by either actinomycin D or alpha-amanitin, it can be excluded that SOX9 binds to the non-transcribed chromatin of the chromosome axis.
As mentioned above, lateral loops are sites of intense RNA synthesis and display a high density of RNA polymerase II complexes. The nascent RNA transcripts associate with proteins that are involved in their packaging, processing, editing, degradation and/or transport. The labelling pattern resulting from the co-immunostaining with antibodies directed against SOX9 and Pol II strongly suggests that the former does not associate with the decondensed DNA axis of lateral loops undergoing transcription but rather with the resulting RNP transcripts. The fact that the association of SOX9 with the RNP matrix is RNA-dependent supports this conclusion. The super resolution microscopy data clearly show that SOX9 exhibits the same granular fluorescence pattern as CELF1 which is involved in the regulation of alternative splicing and known to be associated with the nascent RNP transcripts. SOX9 was also shown to localize to the Cajal and HLBs that are known storage sites for transcriptional factors and those involved in post-transcriptional processes57,58. Altogether these data lead us to propose that the association of SOX9 with the lateral loops results from its binding to the RNA transcripts either directly or indirectly through protein-protein interactions. This raises the question as to whether the amphibian SOX9 protein has a post-transcriptional function. Several studies carried out in different cellular systems have shown that SOX9 proteins could have a role in splicing regulation. Hata et al.12 have demonstrated that SOX9 strongly interacts with the paraspeckle protein p54nrb that regulates both transcription and splicing of the Col2a1 mRNA in the chondrogenic cell line ATDC5. More recently, Rahmoun et al.15 have shown in mammalian fœtal testes that SOX9 not only regulates the transcription of its target genes but also modulates their RNA splicing. A direct role of SOX proteins in pre-mRNA splicing has been already reported in an in vitro assay by Ohe et al.10,11, who showed that the HMG box of either SOX9 or SRY can restore the splicing activity of SOX6-depleted nuclear extracts. They also demonstrated that SRY and SOX6 were co-localized with splicing factors in the nucleus of mammalian cell lines, consistent with their suggestion that SOX6 acts as a general ‘splicing factor’. They proposed that the splicing activity of the SOX HMG domain might be due to its capacity to bend the pre-mRNAs by affecting their structure and therefore supporting or enhancing protein-RNA and RNA-RNA interactions. In the light of these data, it is tempting to propose as suggested by these authors, that in the amphibian oocyte, SOX9 could affect directly or indirectly the structure of the RNP transcripts through its RNA bending activity and promote RNA-protein and protein-protein interactions, thus, contributing to their post-transcriptional processing.
Little data are available as yet, on the interaction between other SOX proteins such as SOX6 and SRY and RNA. The SOX2 protein was shown to interact with rhabdomyosarcoma 2-associated transcript (RMST), a long noncoding RNA essential for neurogenesis17. However, there is mounting evidence from recent investigations64 and hypotheses65 to support RNA binding for TFs which might interact with nascent RNP complexes to exert downstream functions in RNA regulatory events65. A well documented example concerns the transcription factor WT1 (Wilms’ tumor 1)43,66,67.
Our results raise the question of whether the binding of SOX9 to RNPs also takes place in the oocyte of other vertebrates. It is noteworthy that oocyte expression of the Sox9 gene is restricted to the anamniotes clade (fishes and amphibians). No expression has been reported to date in ovarian germ cells of amniotes (reptiles, birds and mammals). In fishes, two sox9 genes (sox9a and sox99b/a2) are present in several teleosts [Medaka33,34,68; Zebrafish29,30,69; Rice field eel70]. One paralog is expressed in somatic Sertoli like-cells of the male gonad, similarly to other vertebrates. The second duplicate is expressed in the oocyte. However, almost all of the studies carried out on fishes were performed at the level of mRNA expression only and the presence of the SOX9 protein in the nucleus was not explored. In this context, it would be interesting to investigate the association of SOX9 with the RNP transcripts in the zebrafish oocyte where LBCs have been described71. In amphibians, the expression of a single sox9 gene in the testis and ovary was reported in X. tropicalis38, Pleurodeles waltl37 and Bufo marinus36. In Rana Rugosa, a sox9 alpha gene and a truncated beta isoform have been reported, and both are expressed during gonad development in males and females40. In the allotetraploid frog Xenopus laevis72, two separate sox9A and B genes are present, sox9B being expressed in the gonads of both sexes. It therefore appears that among vertebrates, only the amphibians exhibit a sox9 gene that displays the dual expression pattern found in teleost fish gonads, i.e., in the somatic cells of the testis and the germ cells of the ovary.
It is widely accepted that the Sox9 gene has been conserved as a Sertoli-expressed gene in all vertebrate species. However, few hypotheses have been proposed to explain its expression in the oocyte. Until recently, the expression of Sox9 in the ovary was considered as a fish-specific feature31. The teleost fishes underwent an additional round of whole-genome duplication (WGD) as a fish-specific whole-genome duplication event 225 to 333 million years ago73,74. It has been suggested that, as a result of this duplication, the zebrafish sox9a gene has retained a testis-specific function while sox9b acquired a new function in the ovary29. Alternatively, the ancestral Sox9 gene in vertebrates may have been expressed in both the male and female gonads. This pattern would have been conserved until the divergence between the anamniotes, which kept the dual pattern, and the amniotes, which retained a testicular expression only. In this context, it would be interesting to investigate the expression of soxE3, the ortholog of the gnathostome sox9, in the gonads of lamprey, one of the jawless vertebrate lineages, which diverged from the jawed vertebrates 500 million years ago.
In conclusion, our study demonstrates that the LBCs of the amphibian oocyte and that of P. waltl in particular (whose genome has been recently sequenced75 provide a powerful in vivo system for the further investigation of the RNP-binding capacity of other TFs and a characterization of the protein domains involved because they offer the possibility of a direct and simultaneous visualization of active and inactive chromatin compartments.
Methods
Ethical statement
Animal studies were approved (Agreement: N° 01612.01) by the Animal Experimentation Ethical Committee Buffon (CEEA-40) supervised by the French Ministry of Education and Research, and conducted in accordance with the guidelines of the EU directive 2010/63/EU for the housing and care of laboratory animals used for scientific purposes.
Oocyte collection
Xenopus laevis and Pleurodeles waltl were raised at 20° and Xenopus tropicalis at 25° in separate Techniplast aquariums, illuminated with a photoperiod of 12 h, in the animal housing facility at the Jacques Monod Institute (licence N° B-75-13-17). Ovarian biopsies were performed on adult females that were anesthetized in 0.15% MS222 (Amino-benzoic Acid Ethyl, Fluka). Stage II-VI oocytes were selected and maintained in MBS buffer (Modified Barth’s solution) at 18°, as detailed in76. The total number of different females involved was: 14 P. waltl; 8X. laevis; 6X. tropicalis.
X. tropicalis sox9 gene amplification and cloning
Total ovary RNA from two Xenopus tropicalis females was extracted using Trizol Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. RNA extract was treated with DNase I (Invitrogen, Carlsbad, CA, USA). RNA was reverse transcribed to obtain cDNA using Transcriptor Reverse Transcriptase following manufacturer’s protocols (Lifescience. Roche.com). The open reading frame (ORF) of X. tropicalis sox9 was amplified from total ovaries cDNA using the following primers FW: ATGATGAATCTCTTGGATCCCTTCATG; Rev:TGGGGCCTGGTGAGCTGTGTATA and PFX Taq polymerase (Invitrogen, Carlsbad, CA, USA) under PCR Touch Down conditions: 30 s denaturation at 94°, 30 s annealing at 65° to 58°, 1.30 mn elongation at 68° (10 cycles) followed by 24 cycles with a 30 s annealing step at 58°. The sox9 PCR product containing the ORF and the sequence (GATCCAAAAAAGAAGAGAAAGGTAGATCCAAAAAAGAAGAGAAAGGTGGAT) encoding an NLS as control were cloned in the pcDNA3.1/CT-GFP-TOPO® vector (Invitrogen, Carlsbad, CA, USA) following manufacturer’s protocols.
Cell culture and transient transfections
Cos-7 cells were grown in DMEM F-12 medium supplemented with 10% FBS and 1% penicillin/streptomycin. Cells were seeded 16 h prior to transfection at a density of 2.5 × 104 cells.cm−2 and transfected using the calcium-phosphate method with the X. tropicalis pcDNA sox9-CT-GFP or the control pcDNA NLS-CT-GFP vector. For subcellular localization analysis, cells were fixed 48 hr after transfection in 2% PFA for 30 min at 4°, DNA was stained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) (Merck) (1:1000 for 2 min). Coverslips were mounted on glass slides with DakoCytomaton (Dako). Cells were visualized by epifluorescence microscopy using a Leitz Aristoplan microscope with a Leitz objective lens 6100/1.37 Oil with a numerical aperture of 0.17. Image acquisition was carried out 48 hr after transfection in the 2X buffer (Tris 100 mM, pH 6,5, SDS4%, Bromophenol blue 0.2%, glycerol 20%). For western blot analyses, cells were lysed 48 h after transfection in lysis buffer supplemented with 50 mM Tris, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, pH 7.6, supplemented with protease inhibitors (Complete mini EDTA-free cocktail, Roche) and phosphatase inhibitors (PhosSTOP, Roche). The homogenate was sonicated 1 × 10 min at 4°. After centrifugation (12,000xg for 20 min at 4°), the supernatant was mixed with 5% Beta-mercaptoethanol and heated at 95 °C for 5 min.
Control of the specifity of the anti-human SOX9 antibodies against the Xenopus tropicalis SOX9 protein
Cos-7 cells were transfected with the pcDNA Xt-sox9-CT-GFP vector encoding theX. tropicalis SOX9 protein (XtSOX9-GFP). As a negative control, cells were transfected with the pcDNA NLS CT-GFP vector encoding the NLS-GFP protein, which contains a nuclear localization signal (NLS) that drives GFP translocation to the nucleus. As shown in Figure S2A–B, the newly synthesized XtGFP-tagged SOX9 and NLS-GFP proteins were expressed in the nuclei of mammalian cells. Western blot analyses showed that the Xt GFP-SOX9 protein synthesized in the human cells was recognized by the two polyclonal anti-SOX9 antibodies (Fig. S2C–D).
RNA synthesis and oocyte microinjection
The X. tropicalis sox9-CT-GFP Topo TA and the control NLS-CT-GFP topo TA vectors were linearized and used as templates for in vitro synthesis of capped and tailed sense-strand RNA using the mMESSAGE mMACHINE® Kit procedure and the Poly (A) Tailing kit (AmbionTM). Oocytes were defolliculated for 2–3 hours in saline buffer OR2 containing 0.15% collagenase type II (Sigma Chemical, St. Louis, MO). Fully grown oocytes (stage V-VI) were selected and maintained overnight in MBS buffer (Modified Barth’s solution) at 18°. Ten nanoliters of the RNA solution (1 µg/µl) was injected into the cytoplasm of oocytes at stage V-VI. All injections were performed under a dissection microscope using the nanoject II (Drummond scientific Company) as previously described46. After injection, the oocytes were incubated for 24 h at 18° and the newly synthesized GFP-tagged translation products were analyzed by fluorescence microscopy on nuclear spreads or on immunoblots of extracted proteins.
Oocyte proteins extracts
Two methods were used for the preparation of nuclear extracts49,77. In the first one, the GVs were placed in Tris-ethanol solution (6 mmol/L Tris, 45% ethanol, 45% glycerol), right after their extraction from the oocytes to precipitate the nuclear content. A few minutes later, the precipitated nuclear extract became retracted and well detached from the nuclear envelope. The nuclear envelope was then removed and the envelope-free nuclear extracts were stored at −20° in ethanol-glycerol (6 mmol/L Tris, 3 mmol/L MgCl2, 70% ethanol). In the second, oocytes were incubated in MBS containing 2% TCA (W/V) for 20 min at 4° to precipitate the proteins, then washed several times in MBS. GVs were isolated, transferred to microtubes, washed in water, and air-dried. Before electrophoresis, the precipitate was recovered by 5 min centrifugation at 12,000 × g at 4°. Nuclear extracts were homogenized in Laemmli buffer, heated for 10 min at 95°, centrifuged for 5 min at 12,000 × g.
Western blots
Protein extracts were loaded on 10% or 4–12% NuPage gels (Invitrogen). After transfer, nitrocellulose membranes (Hybond-ECL, GE Healthcare) were incubated overnight in PBS with 0.1% Tween 20 (PBST) and 5% fat-free dry milk at 4°. Membranes were then incubated 1 hr with the primary rabbit polyclonal anti-SOX9 antibodies raised against either the N-terminal (amino acids 1 to 62)51 or the C-terminal (amino acids 408 to 504) region of the human SOX9 protein21 diluted to 1:400, or with an anti-GFP monoclonal antibody (Lifescience.Roche.com) diluted to 1:1000 or with the monoclonal anti-alpha-tubulin (Sigma) diluted to 1:2000 Membranes were then washed in PBST, incubated with either an HRP conjugated goat anti-rabbit or an HRP conjugated rabbit anti-mouse (Jackson ImmunoResearch) diluted to 1:10,000 in PBS with 0.5% BSA for 1 hr and washed in PBST. Detection was performed using the ECL “SuperSignal West Femto” (Thermo Scientific). Membranes were scanned with the LAS-3000 imaging system (FUJI).
Nuclear spreads of amphibian oocytes
Stage II-VI oocytes from P. waltl were selected and maintained in MBS buffer (Modified Barth’s solution) at 18 °C, as detailed in76. Stages V-VI GVs, were manually isolated from X. tropicalis and X. laevis females in 83 mM KCl, 17 mM NaCl, 6.5 mM NA2HPO4, 3.5 mM KH2PO4, 1.0 mM MgCl2, 1.0 mM Dithiothreitol (DTT)) at pH 7.0 and dissected in a centrifugation chamber filled with dispersal medium (20.7 mM KCl, 4.3 mM NaCl, 1.6 mM NA2HPO4, 0.9 mM KH2PO4, 1.0 mM MgCl2, 1.0 mM Dithiothreitol (DTT), 0.1% w/v paraformaldehyde) at pH 6.6–6.8 for X. laevis or pH 7.0 for X. tropicalis76,78,79. Germinal vesicles were manually isolated from stage II to VI oocytes of P. Waltl, and dissected in 75 mM KCl, 25 mM NaCl, 0.01 mM MgCl2 and 0.01 mM CaCl2, pH7.2 and LBCs were prepared as previously described60. Nuclear spread preparations were centrifugated at 300×g for 10 min, and at 3,000×g for 30 min at 4° before processing for immunostaining.
Transcription inhibition
P. waltl oocytes were incubated for 24 h in MBS Buffer supplemented or not (controls) with either Actinomycin D (5μg/ml, Sigma) or alpha-amanitin (20μg/ml, a gift of O. Bensaude). LBCs spreads were prepared as previously described.
RNase treatment
Nuclear spreads of X. laevis were prepared as previously described. They were then digested 30 min at 37° with RNase A (Roche Life Science, France) (20μg/ml). After washing 3 × 10 min in PBS, preparations were processed for immunostaining experiments as described below.
Immunostaining of nuclear spreads
Nuclear spread preparations were fixed for 30 min at 4° in phosphate buffer saline (PBS) containing 2% paraformaldehyde, washed 3 × 10 mn in PBS and blocked for 10 mn with Horse serum at 10% dilution. They were then incubated with the relevant primary antibody (see below) for 1 hr at room temperature, washed 3 × 10 mn with PBS, incubated with the secondary antibody for 1 hr at room temperature, washed again in PBS for 1 hr. Preparations were post-stained with Hoechst 33342 (1:1,000; Invitrogen) and mounted using Citifluor (Biovalley). The two rabbit polyclonal antibodies raised against N-terminal (amino acids 1 to 62) (Nter-SOX9 Ab)51 or C-terminal (amino acids 408 to 504) regions of the human SOX9 protein(Cter-SOX9 Ab)21 were used at 1:100 and 1:400 dilution, respectively.The anti-coilin antibody (H1) (Santa Cruz Biotechnology) was used at 1:200 dilution. The mouse IgM anti-RNA pol II CTD phospho-epitope (mAb H14) and the mouse IgG anti-CELF1 (mAb 3B1) (kindly supplied by Dr. J. Gall) were used at 1:5000 and 1 μg/ml dilution, respectively. The mouse anti-SR antibody (1H4) raised against X. laevis SR proteins (Santa Cruz Biotechnology) was used at 1:200 dilution. The secondary antibodies Alexa fluor 488, Alexa fluor 568 IgG, goat anti-rabbit, Alexa fluor 568 IgG goat anti-mouse (Invitrogen Corp., Carlsbad, CA) or Alexa 488 goat anti-mouse IgM (Life technlogies) were used at 1:1000 dilution. Standard transmitted light and fluorescence microscopy were performed using a wide field Leica microscope with a plan Apo 100X oil objective (NA = 1.4). Images were captured using a CoolSnap HQ, Photometrics camera driven by the software Metamorph6 (Universal imaging). In order to probe accurately the fluorescence, series of confocal Z-planes (0.7 µm distance) were collected. Pixel size of the image was 64.5 nm. Structured illumination microscopy was using an ELYRA PS.1 Zeiss microscope. Structured illumination was achieved using appropriate grids for the different excitation wavelengths (405 nm, 488 nm and 561 nm).For each plane three grid rotations and 5 different phases were taken. A Zeiss PLAN-APO 63x NA 1.40 objective was used. The microscope was coupled to an EMCCD camera (Andor iXon 885). For each series of experiments, image acquisition was conducted using the same settings. The intensity of fluorescence in the different experiments was normalized with respect to their relevant controls with the secondary antibodies Alexa fluor alone.
Quantification of fluorescence density and colocalization
A specific algorithm for image processing was developed to quantify fluorescence density of each chromosome using the ImageJ software. Chromatin areas were first isolated on the Hoechst images using an automatic thresholding with the Otsu’s method after removing noise with a filter and background correction. The resulting region of interest (R.O.I) designated as MASK was then applied to the GFP images. Fluorescence density was quantified by measuring the average intensity of the GFP images inside masks previously built (Fig. S6). Colocalization analyses were carried out using the ImageJ plugin “Coloc2”.
Statistics
Statistical significance for mean values was estimated by Student t-Tests using Microsoft’s Excell. Error bars represent the standard deviations. The degree of colocalization between SOX9 and CELF1 granules was estimated using a Pearson correlation coefficient over a large number of pixels.
Electronic supplementary material
Acknowledgements
This work was supported by the Centre National de la Recherche Scientifique (CNRS), and the University Paris Diderot-Paris7. We thank Dr Olivier Bensaude (IBENS) for the gift of alpha-amanitin, Dr Jacques Moreau (Institut Jacques Monod) for his help with oocyte injection experiments, Dr Orestis Falkaris and France Lam from the ImagoSeine core facility for their help with the Structured Illumination microscopy analyses. The ImagoSeine core facility is a member of the IBiSA and the France-BioImaging (ANR-10-INBS-04) infrastructure. We thank Ludovic Maingault and Daniel Quintas at the animal housing facility of the Jacques Monod Institute for their care of the amphibians. We thank Pr Michel Vervoort (Institut Jacques Monod) for helpful discussions on the origin and evolution of vertebrates.
Author Contributions
Conceived, designed, performed the experiments, and drafted MS: M.P.M. Performed the experiments: C.P. Contributed reagents/materials/analysis tools: D.L., V.C., B.B.B., F.P. and X.B., Discussed data and drafted MS: J.A.L. and R.A.V.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-25356-1.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Stevanović M, Lovell-Badge R, Collignon J, Goodfellow PN. SOX3 is an X-linked gene related to SRY. Hum. Mol. Genet. 1993;2:2013–2018. doi: 10.1093/hmg/2.12.2013. [DOI] [PubMed] [Google Scholar]
- 2.Harley VR, Lovell-Badge R, Goodfellow PN. Definition of a consensus DNA binding site for SRY. Nucleic Acids Res. 1994;22:1500–1501. doi: 10.1093/nar/22.8.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilson M, Koopman P. Matching SOX: partner proteins and co-factors of the SOX family of transcriptional regulators. Curr. Opin. Genet. Dev. 2002;12:441–446. doi: 10.1016/S0959-437X(02)00323-4. [DOI] [PubMed] [Google Scholar]
- 4.Lefebvre V, Dumitriu B, Penzo-Méndez A, Han Y, Pallavi B. Control of cell fate and differentiation by Sry-related high-mobility-group box (Sox) transcription factors. Int. J. Biochem. Cell Biol. 2007;39:2195–2214. doi: 10.1016/j.biocel.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kiefer JC. Back to basics: Sox genes. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2007;236:2356–2366. doi: 10.1002/dvdy.21218. [DOI] [PubMed] [Google Scholar]
- 6.Lovell-Badge R. The early history of the Sox genes. Int. J. Biochem. Cell Biol. 2010;42:378–380. doi: 10.1016/j.biocel.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Wegner M. All purpose Sox: The many roles of Sox proteins in gene expression. Int. J. Biochem. Cell Biol. 2010;42:381–390. doi: 10.1016/j.biocel.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 8.Kamachi Y, Kondoh H. Sox proteins: regulators of cell fate specification and differentiation. Dev. Camb. Engl. 2013;140:4129–4144. doi: 10.1242/dev.091793. [DOI] [PubMed] [Google Scholar]
- 9.Hou L, Srivastava Y, Jauch R. Molecular basis for the genome engagement by Sox proteins. Semin. Cell Dev. Biol. 2016 doi: 10.1016/j.semcdb.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 10.Ohe K, Lalli E, Sassone-Corsi P. A direct role of SRY and SOX proteins in pre-mRNA splicing. Proc. Natl. Acad. Sci. USA. 2002;99:1146–1151. doi: 10.1073/pnas.022645899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohe K, Tamai KT, Parvinen M, Sassone-Corsi P. DAX-1 and SOX6 molecular interplay results in an antagonistic effect in pre-mRNA splicing. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2009;238:1595–1604. doi: 10.1002/dvdy.21957. [DOI] [PubMed] [Google Scholar]
- 12.Hata K, et al. Paraspeckle protein p54nrb links Sox9-mediated transcription with RNA processing during chondrogenesis in mice. J. Clin. Invest. 2008;118:3098–3108. doi: 10.1172/JCI31373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mallanna SK, et al. Proteomic analysis of Sox2-associated proteins during early stages of mouse embryonic stem cell differentiation identifies Sox21 as a novel regulator of stem cell fate. Stem Cells Dayt. Ohio. 2010;28:1715–1727. doi: 10.1002/stem.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cox JL, Mallanna SK, Luo X, Rizzino A. Sox2 uses multiple domains to associate with proteins present in Sox2-protein complexes. PloS One. 2010;5:e15486. doi: 10.1371/journal.pone.0015486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rahmoun M, et al. In mammalian foetal testes, SOX9 regulates expression of its target genes by binding to genomic regions with conserved signatures. Nucleic Acids Res. 2017;45:7191–7211. doi: 10.1093/nar/gkx328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mukherjee A, Melnattur KV, Zhang M, Nambu JR. Maternal expression and function of the Drosophila sox gene Dichaete during oogenesis. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2006;235:2828–2835. doi: 10.1002/dvdy.20904. [DOI] [PubMed] [Google Scholar]
- 17.Ng S-Y, Bogu GK, Soh BS, Stanton LW. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol. Cell. 2013;51:349–359. doi: 10.1016/j.molcel.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 18.Weider M, Wegner M. SoxE factors: Transcriptional regulators of neural differentiation and nervous system development. Semin. Cell Dev. Biol. 2017;63:35–42. doi: 10.1016/j.semcdb.2016.08.013. [DOI] [PubMed] [Google Scholar]
- 19.Wagner T, et al. Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell. 1994;79:1111–1120. doi: 10.1016/0092-8674(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 20.Morais da Silva S, et al. Sox9 expression during gonadal development implies a conserved role for the gene in testis differentiation in mammals and birds. Nat. Genet. 1996;14:62–68. doi: 10.1038/ng0996-62. [DOI] [PubMed] [Google Scholar]
- 21.De Santa Barbara, et al. Direct interaction of SRY-related protein SOX9 and steroidogenic factor 1 regulates transcription of the human anti-Müllerian hormone gene. Mol. Cell. Biol. 1998;18:6653–6665. doi: 10.1128/MCB.18.11.6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanley NA, et al. SRY, SOX9, and DAX1 expression patterns during human sex determination and gonadal development. Mech. Dev. 2000;91:403–407. doi: 10.1016/S0925-4773(99)00307-X. [DOI] [PubMed] [Google Scholar]
- 23.Gasca S, et al. A nuclear export signal within the high mobility group domain regulates the nucleocytoplasmic translocation of SOX9 during sexual determination. Proc. Natl. Acad. Sci. USA. 2002;99:11199–11204. doi: 10.1073/pnas.172383099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sim H, Argentaro A, Harley VR. Boys, girls and shuttling of SRY and SOX9. Trends Endocrinol. Metab. TEM. 2008;19:213–222. doi: 10.1016/j.tem.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 25.Malki S, Boizet-Bonhoure B, Poulat F. Shuttling of SOX proteins. Int. J. Biochem. Cell Biol. 2010;42:411–416. doi: 10.1016/j.biocel.2009.09.020. [DOI] [PubMed] [Google Scholar]
- 26.Kent J, Wheatley SC, Andrews JE, Sinclair AH, Koopman PA. Male-specific role for SOX9 in vertebrate sex determination. Dev. Camb. Engl. 1996;122:2813–2822. doi: 10.1242/dev.122.9.2813. [DOI] [PubMed] [Google Scholar]
- 27.Spotila LD, Spotila JR, Hall SE. Sequence and expression analysis of WT1 and Sox9 in the red-eared slider turtle, Trachemys scripta. J. Exp. Zool. 1998;281:417–427. doi: 10.1002/(SICI)1097-010X(19980801)281:5<417::AID-JEZ7>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 28.Western PS, Harry JL, Graves JA, Sinclair AH. Temperature-dependent sex determination in the American alligator: AMH precedes SOX9 expression. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 1999;216:411–419. doi: 10.1002/(SICI)1097-0177(199912)216:4/5<411::AID-DVDY9>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 29.Chiang, E. F. et al. Two sox9 genes on duplicated zebrafish chromosomes: expression of similar transcription activators in distinct sites. Dev. Biol. 231, 149–163 (2001). [DOI] [PubMed]
- 30.Yokoi H, et al. Sox9 in a teleost fish, medaka (Oryzias latipes): evidence for diversified function of Sox9 in gonad differentiation. Mol. Reprod. Dev. 2002;63:5–16. doi: 10.1002/mrd.10169. [DOI] [PubMed] [Google Scholar]
- 31.Klüver N, Kondo M, Herpin A, Mitani H, Schartl M. Divergent expression patterns of Sox9 duplicates in teleosts indicate a lineage specific subfunctionalization. Dev. Genes Evol. 2005;215:297–305. doi: 10.1007/s00427-005-0477-x. [DOI] [PubMed] [Google Scholar]
- 32.Rodríguez-Marí A, et al. Characterization and expression pattern of zebrafish Anti-Müllerian hormone (Amh) relative to sox9a, sox9b, and cyp19a1a, during gonad development. Gene Expr. Patterns GEP. 2005;5:655–667. doi: 10.1016/j.modgep.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 33.Nakamoto M, Suzuki A, Matsuda M, Nagahama Y, Shibata N. Testicular type Sox9 is not involved in sex determination but might be in the development of testicular structures in the medaka, Oryzias latipes. Biochem. Biophys. Res. Commun. 2005;333:729–736. doi: 10.1016/j.bbrc.2005.05.158. [DOI] [PubMed] [Google Scholar]
- 34.Liu J, Liu S, Tao M, Li W, Liu Y. Isolation and expression analysis of testicular type Sox9b in allotetraploid fish. Mar. Biotechnol. N. Y. N. 2007;9:329–334. doi: 10.1007/s10126-006-6123-4. [DOI] [PubMed] [Google Scholar]
- 35.Nakamura S, et al. Analysis of medaka sox9 orthologue reveals a conserved role in germ cell maintenance. PloS One. 2012;7:e29982. doi: 10.1371/journal.pone.0029982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abramyan J, Feng C-W, Koopman P. Cloning and expression of candidate sexual development genes in the cane toad (Bufo marinus) Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2009;238:2430–2441. doi: 10.1002/dvdy.22055. [DOI] [PubMed] [Google Scholar]
- 37.Dumond H, et al. Temporal and spatial SOX9 expression patterns in the course of gonad development of the caudate amphibian Pleurodeles waltl. J. Exp. Zoolog. B Mol. Dev. Evol. 2011;316B:199–211. doi: 10.1002/jez.b.21390. [DOI] [PubMed] [Google Scholar]
- 38.El Jamil A, Kanhoush R, Magre S, Boizet-Bonhoure B, Penrad-Mobayed M. Sex-specific expression of SOX9 during gonadogenesis in the amphibian Xenopus tropicalis. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2008;237:2996–3005. doi: 10.1002/dvdy.21692. [DOI] [PubMed] [Google Scholar]
- 39.Osawa N, Oshima Y, Nakamura M. Molecular cloning of Dmrt1 and its expression in the gonad of Xenopus. Zoolog. Sci. 2005;22:681–687. doi: 10.2108/zsj.22.681. [DOI] [PubMed] [Google Scholar]
- 40.Takase M, Noguchi S, Nakamura M. Two Sox9 messenger RNA isoforms: isolation of cDNAs and their expression during gonadal development in the frog Rana rugosa. FEBS Lett. 2000;466:249–254. doi: 10.1016/S0014-5793(00)01078-4. [DOI] [PubMed] [Google Scholar]
- 41.Lacroix JC, et al. Monoclonal antibodies to lampbrush chromosome antigens of Pleurodeles waltlii. Chromosoma. 1985;92:69–80. doi: 10.1007/BF00327246. [DOI] [PubMed] [Google Scholar]
- 42.Bellini M, Lacroix JC, Gall JG. A putative zinc-binding protein on lampbrush chromosome loops. EMBO J. 1993;12:107–114. doi: 10.1002/j.1460-2075.1993.tb05636.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ladomery M, Sommerville J, Woolner S, Slight J, Hastie N. Expression in Xenopus oocytes shows that WT1 binds transcripts in vivo, with a central role for zinc finger one. J. Cell Sci. 2003;116:1539–1549. doi: 10.1242/jcs.00324. [DOI] [PubMed] [Google Scholar]
- 44.Morgan GT. Localized co-transcriptional recruitment of the multifunctional RNA-binding protein CELF1 by lampbrush chromosome transcription units. Chromosome Res. Int. J. Mol. Supramol. Evol. Asp. Chromosome Biol. 2007;15:985–1000. doi: 10.1007/s10577-007-1179-1. [DOI] [PubMed] [Google Scholar]
- 45.Austin C, Novikova N, Guacci V, Bellini M. Lampbrush chromosomes enable study of cohesin dynamics. Chromosome Res. Int. J. Mol. Supramol. Evol. Asp. Chromosome Biol. 2009;17:165–184. doi: 10.1007/s10577-008-9015-9. [DOI] [PubMed] [Google Scholar]
- 46.Kanhoush R, et al. Novel domains in the hnRNP G/RBMX protein with distinct roles in RNA binding and targeting nascent transcripts. Nucl. Austin. Tex. 2010;1:109–122. doi: 10.4161/nucl.1.1.10857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaufmann R, Cremer C, Gall JG. Superresolution imaging of transcription units on newt lampbrush chromosomes. Chromosome Res. Int. J. Mol. Supramol. Evol. Asp. Chromosome Biol. 2012;20:1009–1015. doi: 10.1007/s10577-012-9306-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morgan GT, Jones P, Bellini M. Association of modified cytosines and the methylated DNA-binding protein MeCP2 with distinctive structural domains of lampbrush chromatin. Chromosome Res. Int. J. Mol. Supramol. Evol. Asp. Chromosome Biol. 2012;20:925–942. doi: 10.1007/s10577-012-9324-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Penrad-Mobayed M, Perrin C, Lepesant J-A. Precocious detection on amphibian oocyte lampbrush chromosomes of subtle changes in the cellular localisation of the Ro52 protein induced by in vitro culture. Chromosome Res. Int. J. Mol. Supramol. Evol. Asp. Chromosome Biol. 2012;20:1033–1044. doi: 10.1007/s10577-012-9325-9. [DOI] [PubMed] [Google Scholar]
- 50.Tang Z, et al. CTCF-Mediated Human 3D Genome Architecture Reveals Chromatin Topology for Transcription. Cell. 2015;163:1611–1627. doi: 10.1016/j.cell.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Notarnicola C, Malki S, Berta P, Poulat F, Boizet-Bonhoure B. Transient expression of SOX9 protein during follicular development in the adult mouse ovary. Gene Expr. Patterns GEP. 2006;6:695–702. doi: 10.1016/j.modgep.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 52.Angelier N, Penrad-Mobayed M, Billoud B, Bonnanfant-Jaïs ML, Coumailleau P. What role might lampbrush chromosomes play in maternal gene expression? Int. J. Dev. Biol. 1996;40:645–652. [PubMed] [Google Scholar]
- 53.Morgan GT. Lampbrush chromosomes and associated bodies: new insights into principles of nuclear structure and function. Chromosome Res. Int. J. Mol. Supramol. Evol. Asp. Chromosome Biol. 2002;10:177–200. doi: 10.1023/A:1015227020652. [DOI] [PubMed] [Google Scholar]
- 54.Gall, J. G. Xenopus Development. (M. Kloc and J. Z. Kubiak, 2014).
- 55.Scheer U, Sommerville J. Sizes of chromosome loops and hnRNA molecules in oocytes of amphibia of different genome sizes. Exp. Cell Res. 1982;139:410–416. doi: 10.1016/0014-4827(82)90268-3. [DOI] [PubMed] [Google Scholar]
- 56.Horner HA, Macgregor HC. C value and cell volume: their significance in the evolution and development of amphibians. J. Cell Sci. 1983;63:135–146. doi: 10.1242/jcs.63.1.135. [DOI] [PubMed] [Google Scholar]
- 57.Gall JG, Bellini M, Wu Z, Murphy C. Assembly of the nuclear transcription and processing machinery: Cajal bodies (coiled bodies) and transcriptosomes. Mol. Biol. Cell. 1999;10:4385–4402. doi: 10.1091/mbc.10.12.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nizami ZF, Deryusheva S, Gall JG. Cajal bodies and histone locus bodies in Drosophila and Xenopus. Cold Spring Harb. Symp. Quant. Biol. 2010;75:313–320. doi: 10.1101/sqb.2010.75.005. [DOI] [PubMed] [Google Scholar]
- 59.Bensaude O. Inhibiting eukaryotic transcription: Which compound to choose? How to evaluate its activity? Transcription. 2011;2:103–108. doi: 10.4161/trns.2.3.16172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Penrad-Mobayed M, Bonnanfant-Jaïs ML, N’Da E, Angelier N. Evidence for a particular mode of transcription in globular loops of lampbrush chromosomes of the newt Pleurodeles waltlii. Chromosoma. 1986;94:319–328. doi: 10.1007/BF00328631. [DOI] [PubMed] [Google Scholar]
- 61.Barreau C, Paillard L, Méreau A, Osborne HB. Mammalian CELF/Bruno-like RNA-binding proteins: molecular characteristics and biological functions. Biochimie. 2006;88:515–525. doi: 10.1016/j.biochi.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 62.Roth MB, Murphy C, Gall JG. A monoclonal antibody that recognizes a phosphorylated epitope stains lampbrush chromosome loops and small granules in the amphibian germinal vesicle. J. Cell Biol. 1990;111:2217–2223. doi: 10.1083/jcb.111.6.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu ZA, Murphy C, Callan HG, Gall JG. Small nuclear ribonucleoproteins and heterogeneous nuclear ribonucleoproteins in the amphibian germinal vesicle: loops, spheres, and snurposomes. J. Cell Biol. 1991;113:465–483. doi: 10.1083/jcb.113.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Han H, et al. Multilayered Control of Alternative Splicing Regulatory Networks by Transcription Factors. Mol. Cell. 2017;65:539–553.e7. doi: 10.1016/j.molcel.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 65.Scherrer K. Regulation of gene expression and the transcription factor cycle hypothesis. Biochimie. 2012;94:1057–1068. doi: 10.1016/j.biochi.2011.12.010. [DOI] [PubMed] [Google Scholar]
- 66.Ladomery MR, Slight J, Mc Ghee S, Hastie ND. Presence of WT1, the Wilm’s tumor suppressor gene product, in nuclear poly(A)(+) ribonucleoprotein. J. Biol. Chem. 1999;274:36520–36526. doi: 10.1074/jbc.274.51.36520. [DOI] [PubMed] [Google Scholar]
- 67.Bharathavikru R, et al. Transcription factor Wilms’ tumor 1 regulates developmental RNAs through 3’ UTR interaction. Genes Dev. 2017;31:347–352. doi: 10.1101/gad.291500.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nakamura S, et al. Sox9b/sox9a2-EGFP transgenic medaka reveals the morphological reorganization of the gonads and a common precursor of both the female and male supporting cells. Mol. Reprod. Dev. 2008;75:472–476. doi: 10.1002/mrd.20764. [DOI] [PubMed] [Google Scholar]
- 69.Plavicki JS, et al. Construction and characterization of a sox9b transgenic reporter line. Int. J. Dev. Biol. 2014;58:693–699. doi: 10.1387/ijdb.140288jp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou R, et al. Similar gene structure of two Sox9a genes and their expression patterns during gonadal differentiation in a teleost fish, rice field eel (Monopterus albus) Mol. Reprod. Dev. 2003;66:211–217. doi: 10.1002/mrd.10271. [DOI] [PubMed] [Google Scholar]
- 71.Baumeister H. Lampbrush chromosomes and RNA-synthesis during early oogenesis of Brachydanio rerio (Cyprinidae, Teleostei) Z Zellforsh. 1973;145:145–150. doi: 10.1007/BF00307195. [DOI] [PubMed] [Google Scholar]
- 72.Session AM, et al. Genome evolution in the allotetraploid frog Xenopus laevis. Nature. 2016;538:336–343. doi: 10.1038/nature19840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Berthelot C, et al. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat. Commun. 2014;5:3657. doi: 10.1038/ncomms4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Santini F, Harmon LJ, Carnevale G, Alfaro ME. Did genome duplication drive the origin of teleosts? A comparative study of diversification in ray-finned fishes. BMC Evol. Biol. 2009;9:194. doi: 10.1186/1471-2148-9-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Elewa A, et al. Reading and editing the Pleurodeles waltl genome reveals novel features of tetrapod regeneration. Nat. Commun. 2017;8:2286. doi: 10.1038/s41467-017-01964-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Penrad-Mobayed M, Kanhoush R, Perrin C. Tips and tricks for preparing lampbrush chromosome spreads from Xenopus tropicalis oocytes. Methods San Diego Calif. 2010;51:37–44. doi: 10.1016/j.ymeth.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 77.Penrad-Mobayed M, Moreau N, Angelier N. Evidence for specific RNA/protein interactions in the differential segment of the W sex chromosome in the amphibian Pleurodeles waltl. Dev. Growth Differ. 1998;40:147–156. doi: 10.1046/j.1440-169X.1998.00004.x. [DOI] [PubMed] [Google Scholar]
- 78.Gall JG, Murphy C, Callan HG, Wu ZA. Lampbrush chromosomes. Methods Cell Biol. 1991;36:149–166. doi: 10.1016/S0091-679X(08)60276-9. [DOI] [PubMed] [Google Scholar]
- 79.Gall JG, Wu Z. Examining the contents of isolated Xenopus germinal vesicles. Methods San Diego Calif. 2010;51:45–51. doi: 10.1016/j.ymeth.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.