Abstract
A pea (Pisum sativum) DNA fragment (termed MB3) was isolated by differential display of cDNAs obtained from total leaf RNA of ultraviolet B (UV-B) radiation-treated plants. Longer cDNAs were cloned by rapid amplification of cDNA ends in the 3′ to 5′ direction. Three different, but very similar, cDNAs were cloned, sadA, sadB, and sadC, the major difference between them being a 36-bp deletion in the coding region of sadB. Southern blotting confirmed the occurrence of at least three genes in the pea genome. Database comparisons of the SAD protein sequences revealed high identity (46%) and similarity (77%) with a putative tomato (Lycopersicon esculentum) short-chain alcohol dehydrogenase. Very low levels of UV-B radiation (the biologically effective radiation normalized to 300 nm = 0.08 W m−2) was shown to up-regulate expression, a dose considerably lower than that needed to induce expression of the well-known UV-B defensive chalcone synthase and phenylalanine ammonia lyase genes. RNase protection assay revealed that primarily sadA and sadC mRNA accumulation was enhanced by UV-B. In addition to UV-B irradiation, ozone fumigation, wounding, aluminum stress, and salt stress induced increased transcript levels of the sad genes in pea.
Exposure of plants to UV-B radiation (280–320 nm) results in deleterious effects in plant cells and to the switch-on of defense programs in order to minimize the effects. In particular, the stress responses are directed to protect the genetic content of the nucleus. The adverse effects and the defense responses have been partly characterized in several plant species. For instance, a number of Arabidopsis mutants, hypersensitive to UV-B radiation, have been isolated. These mutants show defects in several types of cellular functions, such as flavonoid biosynthesis (Li et al., 1993; Rao and Ormrod, 1995) and DNA repair (Britt et al., 1993). These mutants indicate what metabolic routes are important for UV-B tolerance in plants.
In pea (Pisum sativum) plants, the effects discovered can roughly be divided into two groups: (a) deleterious effects on chloroplast components, chiefly those involved in photosynthesis (Chow et al., 1992; Jordan et al., 1992; Strid, 1993; Mackerness et al., 1997), and decreases in the expression of genes encoding plastid proteins (Strid et al., 1990, 1996a, 1996b; Jordan et al., 1992; Strid, 1993; Zhang et al., 1994; Mackerness et al., 1997); and (b) increased expression of genes involved in different protective mechanisms, such as the genes for enzymes involved in the scavenging of reactive oxygen species (Mackerness et al., 1998) and those for enzymes of the phenylpropanoid and flavonoid biosynthetic pathways (Strid, 1993; Jordan et al., 1994), giving rise to increased levels of UV-B-absorbing pigments in leaf epidermal cells (Strid and Porra, 1992). It has been shown that the changes in mRNA levels of the above-mentioned genes in most cases are more rapid than the biochemical inhibitory effects of UV-B on the photosynthetic components (Strid et al., 1994), the exception being the degradation of certain plastid-encoded photosynthetic proteins prior to the decrease in mRNA levels (Mackerness et al., 1997). Moreover, the expression of the protective enzyme systems is not solely a UV-B-specific response but seems to also occur under other types of stress, such as oxidative stress (Henkow et al., 1996), ozone fumigation (Willekens et al., 1994), wounding (Thipyapong and Steffens, 1997), salt stress (Zhu et al., 1997), or aluminum stress (Richards et al., 1998).
In our search for other types of protective strategies and to elucidate what components are involved in stress-induced signal transduction within the plant cells, it is of great interest to employ techniques aimed at the identification of genes differentially expressed under stressful conditions. In this study we used the method of differential display (Liang and Pardee, 1992) to search for novel interesting genes regulated by stressful conditions, primarily by UV-B radiation, and we found a gene family encoding putative short-chain alcohol dehydrogenases (ADHs). The transcript levels of these genes were regulated by UV-B radiation at doses lower than for any other UV-B-induced defense genes.
MATERIALS AND METHODS
Plant Material, Lights, Treatments, and RNA Isolation
For the UV-B experiments, pea (Pisum sativum L. cv Greenfeast or cv Kelvedon wonder) was grown in a 12-h light/12-h dark regime at 100 μE m−2 s−1 (fluorescent and incandescent lamps) and at 22°C for 21 d after sowing. On d 21, one-half of the plants were transferred to a UV-B chamber with the same visible light but supplemented with UV-B radiation sources (TL40W/12UV, Philips, Eindhoven, The Netherlands). The biologically effective radiation normalized to 300 nm (UV-BBE,300) was 0.08 (low dose) or 1.6 W m−2 (high dose) according to Caldwell (1971) and Green et al. (1974). Leaves were harvested from the third leaf pair from the base of the plants after 3, 6, or 12 h of exposure and frozen in N2(l) until further analyzed.
For ozone treatments, pea cv Greenfeast plants were grown in open-top chambers supplemented with charcoal-filtered air (approximately 5 nL L−1 ozone). Three weeks after sowing, one-half of the plants were transferred to a chamber with charcoal-filtered air supplemented with approximately 100 nL L−1 ozone for 12 h each 24-h period. Samples were removed after 24 and 48 h after the commencement of the experiment and frozen in N2(l) until further analyzed.
For aluminum or NaCl treatments, pea cv Greenfeast was grown on vermiculite for 7 d. Seedlings were then transferred to hydroponic growth medium at pH 4.1 in the medium described by Hoagland and Arnon (1950): 2 mm KNO3, 3 mm Ca(NO3)2, 2 mm MgSO4, 1 mm KH2PO4, 1 mm NH4Cl, 46 μm H3BO3, 11 μm MnCl2, 1 μm Na2MoO4, 0.7 μm ZnSO4, 0.3 μm CuSO4, 0.2 μm VOSO4, and 0.09 μm FeSO4. On d 21, the nutrient solution was exchanged for fresh medium supplemented with 250 mm NaCl (NaCl treatment) or 100 μm AlK(SO4)2 (aluminum treatment). The control plants were simultaneously moved to fresh medium. Samples were removed after 3, 6, and 24 h.
For wounding experiments, pea cv Greenfeast was grown for 21 d. On d 21, one leaf of the fourth leaf pair was pierced with 10 holes. The wounded leaf and its neighbor were then harvested separately after 3, 6, and 24 h. Controls were taken from nonwounded control plants.
Isolation of total RNA from the plant tissue was carried out as described previously (Strid et al., 1996a).
Differential Display
Differential display was carried out essentially as described previously (Liang and Pardee, 1992). Total RNA was isolated from leaves of UV-B-exposed (3 or 6 h) and control pea plants. A T12GC-primer and one of the following arbitrary primers (d[GCGCCTGCGCGGTGACGCTG], termed MittRp; d[AAGTGGCTGGTGGCCGGTGT], termed MittFp; d[CATTATTTGGATACAGACTA], termed TerminatorRp) were used.
Northern Blotting
Northern blotting was performed according to Strid et al. (1996a). cDNAs excised and reamplified from differential display gels were labeled with [α-32P]dCTP by random labeling. The MB3 (accession no. AF002249), sadA (accession no. AF053638), and polyubiquitin probes were prepared by amplifying the genes from the plasmids pÅS100, pÅS108, and pÅS110, respectively (see below). The resulting PCR products were labeled by random labeling. The northern-blot analyses were repeated with different stress exposure experiments and RNA isolations.
In addition to the sad cDNA clones (see below), the following cDNA clones were used to probe the northern blots for the occurrence of their specific mRNAs: the cab cDNA clone (pAB96), which corresponds to the nuclear gene encoding the chloroplast-localized chlorophyll a/b-binding protein (Coruzzi et al., 1983); the probe for chalcone synthase (the pCC6 cDNA pea clone; Ichinose et al., 1992); the PAL cDNA clone (cPAL-1; Kawamata et al., 1992). An 18S rRNA probe (construct termed pÅS3; Kalbin et al., 1997) was used to check even loading of total RNA on the agarose gel and the transfer efficiency.
Cloning
The cDNA fragment termed MB3 was cloned blunt-ended into a pZErO-2 vector (Invitrogen, Leek, The Netherlands). The construct was termed pÅS100.
Cloning of the sad cDNAs was performed with 5′-RACE by using the Marathon cDNA amplification kit or SMART RACE cDNA amplification kit (CLONTECH Laboratories, Palo Alto, CA) according to the manufacturer's instructions with the MB3 5′-RACE primers d(GCAATGCCTAAAGATAAAAAGATTACA) and d(CATAAATAAGATGCCTGCAGAAGTGTGA). The resulting PCR products were cloned into a T-vector or into pZeRO (Novagen, Madison, WI). The constructs were termed pÅS108 (sadA) and pÅS123 (sadC, accession no. AF097651). sadB (accession no. AF053639) was found with the primers (5′d(TTTCTCGAGCATGGCAGAATCTTCATCA), 5′d(TTTACGCGTTAAATTACAGCGTGACTAGT). This PCR product was cloned into pZeRO (construct pÅS109).
PU1 was cloned by RT-PCR with the primers termed Polyubi1 5′-d(ATGCAAATTTTCGTTAAGACC) and Polyubi25′-d(TTAAAATCCACCACGAAGACG). The PCR product was cloned into a T-vector (Novagen), and the sequence was verified by DNA sequencing as described below. The new construct was named pÅS110.
Slot Blotting
Slot blotting was performed according to the method of Kalbin et al. (1997) using the Bio-Dot equipment from Bio-Rad Laboratories (Hercules, CA). The resulting autoradiographs were scanned in a laser scanner.
RNase Protection Assay
RNA probes were synthesized according the method of Gilman (1993). The probes were purified on denaturing PAGE gels and used together with the RPAII kit (Ambion, Austin, TX) for hybridization and digestion. After RNase T1 digestion and precipitation the samples were separated on a precast TBE-urea PAGE gel (Bio-Rad), which was then autoradiographed. Ambion's century marker template was used to obtain a radiolabeled RNA ladder. To get probes specific for the three sad genes, the following constructs were used: sadA, the construct pÅS108 linearized with NheI and transcribed with T7 RNA polymerase; sadB, the construct pÅS109 restricted with EcoRI removing the 3′ end of the sadB cDNA, after which the vector and the first 126 bp of the cDNA were religated, restricted with XbaI and transcribed with SP6 RNA polymerase; and sadC, the original MB3 clone (pÅS100) linearized with XbaI and transcribed with SP6 RNA polymerase.
DNA Sequencing
Sequencing was carried out by using the ThermoSequenase Fluorescent Sequencing Kit (Amersham-Pharmacia Biotech, Little Chalfont, UK), and the analysis was performed on a Pharmacia ALFred DNA sequencer (Amersham-Pharmacia Biotech, Uppsala). Both strands were sequenced at least three times. Contig assembly and alignments were performed with programs in the Lasergene software package (DNASTAR, Madison, WI).
Southern Blotting
Genomic DNA was isolated according to the method of Doyle and Doyle (1990). Southern blotting was performed according to standard protocols (Brown, 1993). Prehybridization and hybridization were performed in Church buffer (Church and Gilbert, 1984), and labeling of the pÅS108 cDNA probe was as described for northern blotting above.
RESULTS
Differential Display and Cloning of the sad Genes
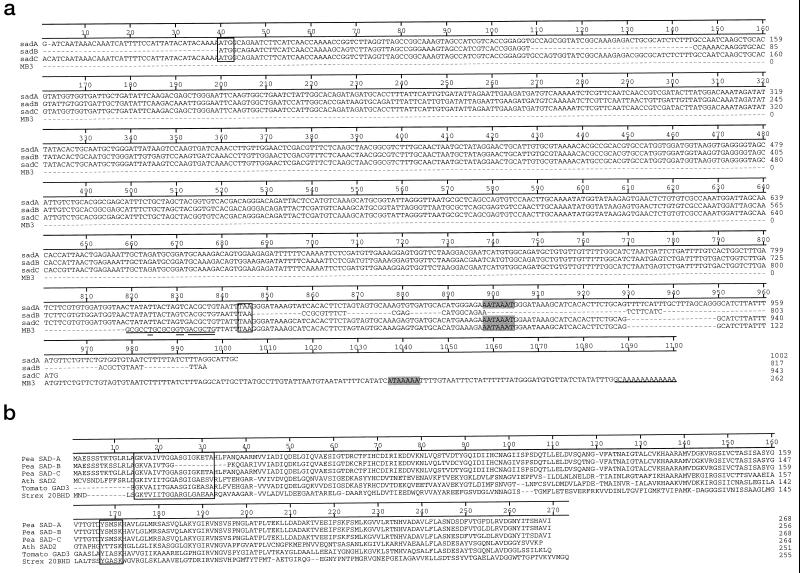
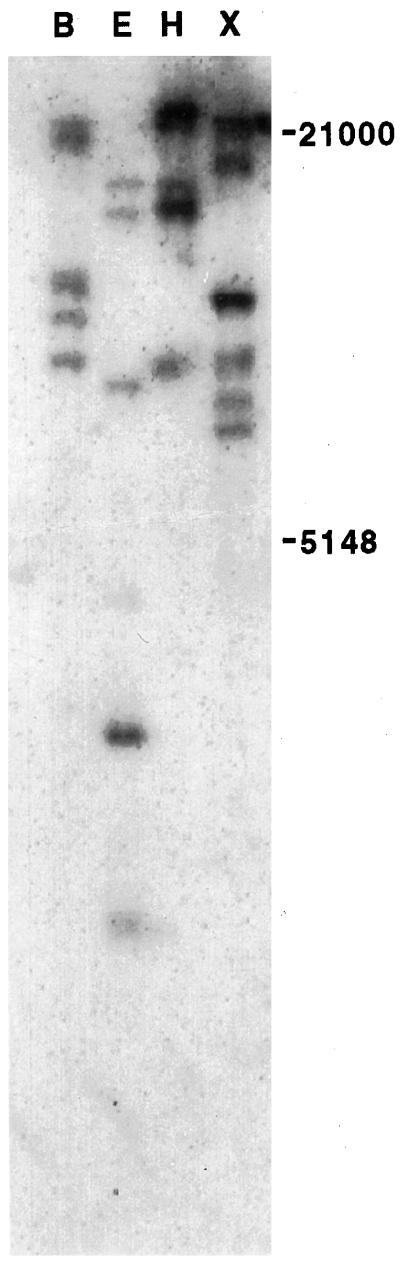
Differential display was carried out using one oligo-dT primer (T12GC) in combination with three different random primers (see “Materials and Methods”). Out of the four cDNAs analyzed, one (termed MB3 since it was the third cDNA fragment isolated by M.B. in the differential display study) was found to be truly differentially regulated by UV-B radiation, as judged by northern blotting (see below). Sequencing of the differentially expressed MB3 cDNA yielded 250 bp with no similarities to any other sequences deposited in GenBank. To determine what gene the MB3 fragment corresponded to, 5′-RACE was performed. In total, three different but homologous sequences (Fig. 1a) were picked up and denoted sadA, sadB, and sadC, respectively, after the order in which they were isolated and since they correspond to stress-induced ADH-like proteins. Of these three, the sadC cDNA corresponded to the original short MB3 differential display fragment. Southern blotting indicated that the sad genes belong to a gene family with at least three members (Fig. 2). sadB showed very strong similarity to sadA, with the exception of 32 single base substitutions and an in-frame deletion of 36 bp (96% identity on the DNA level, 94% identity and 98% similarity on the protein level, the deletion excluded when calculating the similarity score). This deletion was in the protein product situated in the nucleotide binding site of steroid ADHs with similar primary structure (Ghosh et al., 1991). The sadC gene product, on the other hand, contained the same number of amino acids (268) as sadA, and 265 out of these were identical. However, there was considerable difference between both the 3′ (Fig. 1a) and the 5′ (J.G. Gittins and Å. Strid, unpublished results) UTR of the two genes.
Figure 1.
Sequences of the sad genes and the corresponding proteins. a, DNA sequences of the sadA, sadB, sadC, and MB3 cDNAs from pea cv Greenfeast. The start and stop codons are boxed, the MB3 primer sequences are underlined, and the nucleotides of the first primer matching the sad sequences are double underlined. Possible poly(A) signals (Hunt, 1994) are marked with a shaded area. b, Protein sequence alignment of the pea sadA, sadB, and sadC gene products with the translated sequences of the tomato gad3 gene (GenBank accession no. U21801; Jacobsen and Olszewski, 1996), the Arabidopsis Atsad2 gene product (GenBank accession no. AC004411; the second of four sad genes in the same genomic DNA region and by us named AtSad2), and Streptomyces exfoliatus (Marekov et al., 1990; SwissProt accession no. P19992). The catalytic YXXSK region is boxed, as well as the nucleotide binding site (Ghosh et al., 1991).
Figure 2.
Southern blot probed with sadA cDNA, showing the restriction pattern of genomic pea DNA cleaved with the enzymes BamHI (B), EcoRI (E), HindIII (H), and XbaI (X), hybridized with [32P]dCTP-labeled cDNA of the sadA gene. The positions of Mr standards are shown to the right and are expressed in numbers of bases.
In Figure 1a, possible poly(A) signals (shaded area; Hunt, 1994) and start and stop codons (boxed) are also indicated for the three sad cDNAs.
Database searches suggested the sadA cDNA as belonging to a short-chain ADH family of genes. The highest similarity of the corresponding protein (46% identity, 77% similarity; Fig. 1b) was to a tomato enzyme (Jacobsen and Olszewski, 1996; GenBank accession no. U21801) and to the translational product of several genes sequenced (GenBank accession no. AC004411) in the Arabidopsis genome project (49% identity, 84% similarity for the most homologous of these putative gene products; Fig. 1b. This gene product, by us here termed ATH SAD2, corresponds to the second of four SAD genes encoded in the same region of the Arabidopsis genomic DNA). It is possible that the pea SAD proteins correspond to a steroid ADH as judged by the high similarity, 34% identity and 67% similarity (Fig. 1b), of SAD-A to the 3α,20β-hydroxysteroid dehydrogenase (EC 1.1.1.53) from Streptomyces exfoliatus (Marekov et al., 1990; Ghosh et al., 1991; SwissProt accession no. P19992). In addition, considerable similarities were found to the maize TASSELSEED2 gene and its homologs in other plants (not shown; DeLong et al., 1993; GenBank accession no. L20621). The short-chain ADH active site motif YXXSK was found in all three pea translational products, whereas the nucleotide binding motif G(X)6G(X)3G(A)XG(X)3A (Ghosh et al., 1991) was found in SAD-A and SAD-C but not in SAD-B (Fig. 1b).
Expression Pattern of the sad Gene
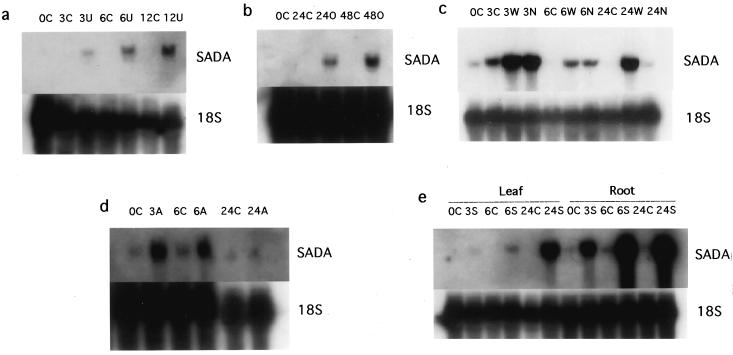
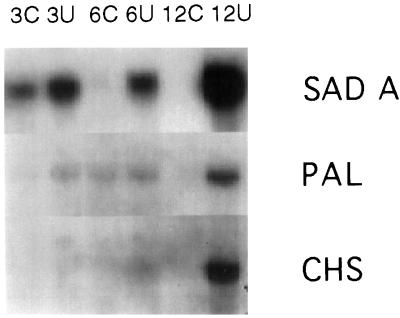
Northern blotting and hybridization of UV-B RNA samples with 32P-labeled sadA cDNA confirmed the same expression pattern (Fig. 3a) as was obtained for the shorter original MB3 fragment (not shown). It is interesting that a very low UV-B dose (0.08 kJ m−2 h−1) also induced accumulation of transcripts of the sad gene in leaves within 3 h (Fig. 4). Other up-regulated genes (pal and chs) were only affected after 12 h of exposure at this dose, whereas the down-regulated cab gene was not affected at all (not shown). To our knowledge, this is the lowest dose of UV-B shown to affect gene expression, and sad is the first gene shown to be regulated at doses lower than those needed to affect pal and chs.
Figure 3.
Northern blot showing the presence of mRNA hybridizing to sadA cDNA under different types of stresses. a, Pea leaves exposed (U) or not exposed (C) to UV-B (UV-BBE,300 = 1.6 W m−2) for 0, 3, 6, or 12 h; b, pea leaves exposed (O) or not exposed (C) to ozone, approximately 100 nL L−1 for 12 h each 24-h experimental period. The experiment was carried out for 0, 24, or 48 h. c, Nonwounded control leaves (C), wounded leaves (W), and neighbors to wounded leaves (N) 0, 3, 6, or 24 h after wounding; d, pea roots exposed (A) or not exposed (C) to aluminum in hydroponic cultures, as described in the “Materials and Methods,” for 0, 3, 6, or 24 h; e, pea roots exposed (S) or not exposed (C) to NaCl in hydroponic cultures, as described in “Materials and Methods,” for 0, 3, 6, or 24 h. The same Northern blots hybridized to cDNA for 18S rRNA are shown below the corresponding sadA-hybridized northern blot to compare the loading of the total RNA samples on the gels and the transfer of the RNA to the membrane.
Figure 4.
Northern blot showing the presence of mRNA hybridizing to [32P]dCTP-labeled cDNA of the sadA, chs, and pal genes after exposure to low dose UV-B (UV-BBE,300 = 0.08 W m−2) for 3, 6, or 12 h. See Figure 3 for abbreviations.
The sadA probe used for the northern blots shown in Figures 3, 4, and 6 was not particularly specific for sadA due to the high similarity between the three sad genes. To address the question whether the different sad genes were regulated differently or in parallel with each other, an RNase protection assay was performed. Probes specific for each sad mRNA were used. For sadA, the probe corresponded to bases 510 to 1,002 of the cDNA, and for sadB the probe was designed toward bases 1 to 126 of this particular mRNA. For sadC, the MB3 differential display fragment, corresponding to the 3′ end of the sadC mRNA, was used. This probe design took advantage of the maximum differences between the nucleotide sequences of the three cDNAs.
Figure 6.
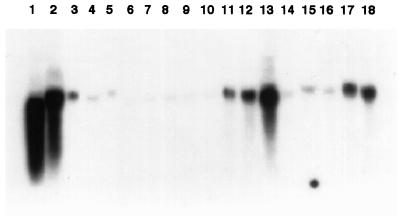
Northern blot showing the presence of mRNA hybridizing to sadA cDNA in different types of tissue. In lanes 1 to 16 the tissue is from cv Greenfeast, whereas in lanes 17 and 18 the tissue is from the mutant argenteum. The tissue types are as follows: 1, Dry seeds; 2, seeds after 16 h of soaking in tap water; 3, shoots 9 d after sowing; 4, cotyledons 14 d after sowing; 5, leaf buds 14 d after sowing; 6, cotyledons 21 d after sowing; 7, second leaf pairs 21 d after sowing; 8, third leaf pairs 21 d after sowing; 9, fourth leaf pairs 21 d after sowing; 10, fifth leaf pairs 21 d after sowing; 11, roots from plants grown in vermiculite 14 d after sowing; 12, roots from plants grown in vermiculite 21 d after sowing; 13, roots from plants grown in vermiculite 42 d after sowing; 14, stems 21 d after sowing; 15, tendrils; 16, flowers (white); 17, flowers (purplish); and 18, pods.
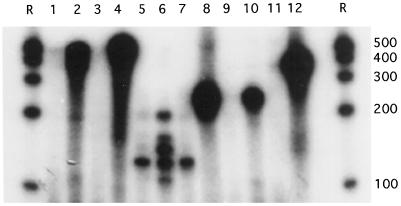
The expected sizes of the full-length probes including vector-derived sequences were 555, 251, and 356 bp for sadA, sadB, and sadC, respectively. As can be seen in the nondigested controls in lanes 4, 8, and 12 of Figure 5, full-length probes were obtained. The sizes of the probes after digestion were expected to be 498, 126, and 240 bp for sadA, sadB, and sadC, respectively, if they had been protected by the correct mRNA species. As expected, the correct sizes could be found for all three samples when the corresponding probes had been hybridized to RNA samples from plants exposed to UV-B (Fig. 5, lanes 2, 6, and 10). In the lanes containing probes hybridized to total RNA from plants kept in the control light-environment before digestion (Fig. 5, lanes 1, 5, and 9), no probe of the right size existed. This indicates that none of the three sad genes were expressed during control conditions.
Figure 5.
RNase protection assay performed with total RNA from UV-B-exposed or control plants. Probes specific for sadA (lanes 1–4), sadB (lanes 5–8), or sadC (lanes 9–12) were used. Equal amounts of 32P-labeled probe (6 × 104 cpm) and total RNA (10 μg) were hybridized in all samples. The samples are as follows: lanes 1, 5, and 9, probe hybridized with total RNA from control plants before digestion; lanes 2, 6, and 10, probe hybridized with total RNA from UV-B-exposed plants (12 h of UV-BBE,300 = 1.6 W m−2) before digestion; lanes 3, 7, and 11, probe hybridized with total RNA from yeast before digestion; lanes 4, 8, and 12, probe hybridized with total RNA from yeast, no digestion (only 5% of the total reaction volume was added in each of these three lanes due to the high specific acivity of these radioactive probes). R denotes the radiolabeled RNA ladder with the distance between each band corresponding to even 100 bases.
However, for the sadB samples, four additional bands were obtained in the lane containing probe hybridized to total RNA from UV-B-irradiated plants (Fig. 5, lane 6). Two of these bands corresponded to larger probe sizes than expected and two corresponded to smaller fragments. One of the lower bands was present not only in lane 6 but also in the lane containing RNA from control plants (lane 5), as well as in the digestion control containing yeast RNA (lane 7). This indicates the formation of a RNase T1-insensitive RNA species by the probe itself in all three lanes. The second smaller band indicates that the radioactive probe could have hybridized to a sad mRNA from yet another gene in the sad gene family. Then, this gene would have a sequence partly identical to sadB. Moreover, for the two larger bands to appear, parts of the vector-derived bases in the probes must have become protected during digestion. Again, this indicates the formation of a stable secondary structure in the probe.
To determine whether the transcript levels of the sad gene were specifically regulated by UV-B radiation, several other stress experiments were performed (Fig. 3). The stresses used were: the air pollutant ozone, mainly thought to act via oxidative damage (Smirnoff, 1996); NaCl, causing osmotic stress (Zhu et al., 1997); aluminum, leading to root damage (Persson and Majdi, 1995); and wounding (Thipyapong and Steffens, 1997).
Under all of the stresses mentioned above, the transcript levels of sad increased. Ozone induced accumulation of sad messages in leaves (Fig. 3b) and salt stress activated sad expression both in roots and in leaves, albeit to a much higher level in the former tissue (Fig. 3e). Aluminum caused a transient increase in sad transcript levels in roots after 3 and 6 h of exposure, whereas after 24 h the sad mRNA levels were the same as in the control (Fig. 3d). As can be seen from the autoradiograph from the aluminum experiment (Fig. 3d), and also in the salt experiment when the autoradiography film had been overexposed (not shown), sad expression appeared to be constitutive at a low level in the roots (see also below).
Wounding gave the most complex expression pattern: sad transcript levels increased after 3 h both in the wounded and in the neighboring leaf. This indicates induction also in undamaged tissue: After 6 h, sad mRNA levels were still elevated in the wounded and neighboring leaves, compared with controls, but to a lesser extent than after 3 h; after 24 h the sad transcript levels were similar in the controls and in the neighboring leaf, while still elevated in the wounded leaf (Fig. 3c).
For a comparison, cDNAs for several other genes, which are likely to be stress regulated (polyubiquitin, PU1; polyubiquitin; Phe ammonia lyase, pal; chalcone synthase, chs; chlorophyll a/b-binding protein, cab), were hybridized to slot blots containing RNA samples. The resulting autoradiographs were quantified with a laser scanner, and the data are presented with the mRNA levels expressed in terms of arbitrary units in Table I. All five genes were more or less regulated by the stresses tested. For instance, cab was down-regulated by ozone and UV-B, with ozone being most effective (signal 5% of the control), pal was up-regulated by UV-B, ozone, and salt stress, with ozone again being most effective (signal 530% of control). PU1 was up-regulated by all stresses (most effectively by NaCl; signal 9- to 10-fold larger than for the control) and so was chs (most efficiently by UV-B; signal 9-fold larger than for the control).
Table I.
Results from the scanning of autoradiographs of hybridized slot blots containing total RNA isolated from pea tissue under a number of stressful conditions
| Treatments | sad | PU1 | cab | pal | chs |
|---|---|---|---|---|---|
| UV-B (leaves; 12 h) | 0.869 | 1.654 | 0.087 | 0.159 | 0.278 |
| Control | 0 | 0.549 | 0.678 | 0.043 | 0.030 |
| % of Control | ∞ | 288 | 13 | 370 | 927 |
| Ozone (leaves; 48 h) | 0.220 | 1.169 | 0.016 | 0.378 | 0.164 |
| Control | 0 | 0.510 | 0.326 | 0.071 | 0.052 |
| % of Control | ∞ | 229 | 5 | 532 | 315 |
| Aluminum (roots; 6 h) | 3.663 | 3.790 | ND | 1.117 | 2.003 |
| Control | 0.594 | 1.546 | ND | 0.842 | 1.517 |
| % of Control | 617 | 245 | — | 133 | 132 |
| NaCl (roots; 24 h) | 3.341 | 1.677 | ND | 0.416 | 0.410 |
| Control | 0.026 | 0.176 | ND | 0.113 | 0.300 |
| % of Control | 12,850 | 953 | — | 368 | 137 |
Leaf tissue from UV-B- or ozone-exposed pea plants was used. Roots from NaCl- and aluminum-exposed plants were also used. The time of exposure is indicated in parentheses. [32P]dCTP-labeled cDNA of the sadA, polyubiquitin (PU1), cab, pal, and chs genes were used for hybridization. cDNA for 18S rRNA was used to normalize the results to the amount of total RNA on the blot. Scanning of the autoradiographs was performed in a laser scanner, and the optical density of the bands is shown in arbitrary units for samples from exposed and control plants. For clarity, the optical density of the exposed samples as a percentage of the corresponding controls is shown. The values are the means of at least three measurements. ND, Not determined.
To address the question whether the sad gene expression was tissue dependent or developmental stage dependent, RNA was isolated from different tissues and different developmental stages. As is seen in Figure 6, the sad gene was constitutively expressed in seeds, in roots, and also in purplish flowers and pods of the pea mutant argenteum, and to a certain extent in young shoots. No or only very minute amounts of sad mRNA were found in cotyledons, leaves of different ages, stems, tendrils, or white flowers (cv Greenfeast).
DISCUSSION
sad cDNAs and Genes
The differential display fragment termed MB3 labeled an mRNA species on a northern blot containing total RNA from UV-B-exposed pea leaves. In contrast, no hybridization with total RNA from control leaves was seen. Database searches with MB3 revealed no similarities to other genes present in the database. To assign a function to the MB3 cDNA, the sadA, sadB, and sadC cDNAs were then isolated by using 5′-RACE. The SAD-A protein had the largest numbers of identical amino acid residues in common with the short-chain ADH family of proteins (Ghosh et al., 1991), which might infer a similar role for the SAD proteins in pea. The largest similarity score between the pea SAD polypeptides and other proteins (46% identity, 77% homology) was with a tomato short-chain ADH. According to recent results, the tomato short-chain ADH gene (termed gad3) was constitutively expressed but down-regulated by GA and ABA (Jacobsen and Olszewski, 1996).
When comparing the primary structures of the three pea sad gene translational products, it was found that the difference between the coding regions of sadA and sadC was minute (265 identical amino acids out of 268). Larger differences were seen between these two gene products and the SAD-B polypeptide. The major, a deletion of 36 bases at the beginning of sadB (Fig. 1b) removes one-half of the putative nucleotide binding site in the N-terminal region of the protein. These amino acids in the related and previously crystallized 3α,20β-hydroxysteroid dehydrogenase protein (Ghosh et al., 1991) form one complete α-helix within the nucleotide binding site. The role of the sadB protein with this truncated nucleotide binding site is unknown. We do not consider it likely that the sadB is a pseudo-gene, at least not as judged by the information contained in the cDNA: All codons including the 36-bp deletion are in-frame, there are no internal stop codons, and start and stop codons match with the sadA gene. In addition, the presence of specific sadB mRNA, shown in RNase protection assay (Fig. 5), supports this. It is possible that SAD-B is a member of an oligomeric structure together with SAD-A and/or SAD-C. In fact, one of the proteins homologous to the pea SAD proteins forms active tetramers (Ghosh et al., 1991), and in an MS analysis, we found that SAD-C, purified after overexpression in Escherichia coli, formed stable homodimers (A. Olsson, M. Brosché, and Å. Strid, unpublished data). Another possibility is that the sadB mRNA emanates from a splicing error of the message for a sad gene other than sadA and sadC. However, we consider this very unlikely since the genomic sequences of sadA and sadC do not contain any introns close to the position where the deletion occurs in the sadB mRNA (J.G. Gittins and Å. Strid, unpublished results).
Regarding the function of the SAD proteins, several genes with high similarity to the sad genes are considered to encode proteins involved in steroid metabolism. Thus, it is possible that the SAD proteins are involved in the metabolism of phytosteroids. The proposed pathway for brassinolide biosynthesis includes several steps that could be catalyzed by an ADH-type of protein (Li and Chory, 1999). However, it is also clear that short-chain ADHs are involved in oxidation of hydroxyl groups of diverse substrates such as sugars, acetoacetyl-CoA, mammalian prostaglandins, and diols, in addition to steroids (Persson et al., 1991). Thus, these compounds, in addition to other unidentified hydroxyl-containing chemical species, are possible substrates for the SAD proteins.
In addition to the similarities between the pea SAD proteins and short-chain ADHs from tomato and prokaryotes, considerable similarities were found between the SAD amino acid sequences and the gene products of the male sex-specific genes of the TASSELSEED2 class from maize (DeLong et al., 1993), and homologs from other species.
sad Expression Pattern
Judged by the results of the RNase protection assay (Fig. 5), both sadA and sadC are strongly up-regulated in plants exposed to UV-B radiation. sadB transcript levels were also increased but to a considerably smaller extent. The fact that sad was regulated by a very low dose rate of UV-B radiation (0.08 kJ m−2) is very interesting. None of the other genes tested (chs, pal, and cab) had altered transcript levels by short exposures at the same low UV-B dose (Fig. 4). The expression of these three genes has previously been shown to be affected by the lowest doses of UV-B (Strid et al., 1994). This implies that the sad gene products might regulate or participate in an early event after the onset of stress.
So far, none of the genes known to be regulated by UV-B radiation have been shown to be exclusively so. The sad gene is no exception. All types of stresses used in this study (ozone fumigation, NaCl stress, aluminum stress, and wounding) lead to elevated levels of sad transcript levels, although the temporal and spatial expression patterns differed among the stresses (Fig. 3).
An interesting finding is that the wounding of one leaflet of a pea leaf pair leads to accumulation of sad mRNA transcripts not only in the wounded leaflet but also in its neighbor (Fig. 3c). Wounding is considered to be a biotic stress factor, whereas the other types of stresses used in this study are abiotic. This means that wounding a plant is reminiscent of an attack by microbial organisms, such as fungal infection (Thipyapong and Steffens, 1997). It is well known that pathogen attack induces a massive stress response known as systemic acquired resistance (SAR). A result of SAR is the expression of defense genes distal from the site of infection, in order to fend off infectious organisms throughout the plant (Ryals et al., 1996). Our results imply that the increase in expression of a sad gene could be a component of the SAR development.
Expression of Other Defense Genes
Ubiquitin is a protein found in all eukaryotes analyzed so far and is encoded by a family of polyubiquitin genes. The ubiquitin protein is used by the cell to label proteins for degradation (Belknap and Garbarino, 1996). Wounding, heat stress, and dehydration have all been shown to increase expression of polyubiquitin genes. The results of our present study indicate that the PU1 (polyubiquitin) gene in pea is generally up-regulated in response to stress. As can be seen in Table I, all types of stresses lead to increased levels of the PU1 transcript.
During aluminum stress, the mRNA levels for two of the genes tested (sad and PU1) are strongly but transiently increased, returning to the control levels after 24 h from commencement of the exposure (not shown). Richards et al. (1998) showed that some of the genes regulated by aluminum stress were also regulated by ozone fumigation. Our results confirm this notion. In fact, it appears that genes uniquely regulated by only one stress factor are scarce. The explanation for this could be that most stress factors, at least as one mode of action, cause oxidative stress, which in turn triggers a general defense response.
CONCLUSION
We have shown the expression of three members of a gene family, called sad, which possibly encode short-chain ADHs, and which are induced by several stressful conditions: ozone fumigation, wounding, NaCl and aluminum stress, and UV-B irradiation, the latter factor causing accumulation of mRNA even at very low doses. This implies a role for these proteins at an early stage in stress responses in plants. Of the three sad cDNAs, sadB differed from sadA and sadC by lacking 36 bp encoding an α-helix of the nucleotide binding site of short-chain ADHs.
ACKNOWLEDGMENTS
We would like to thank Prof. Gun Selldén for help with the ozone fumigation experiments and Dr. John Gittins for discussions. Karl Axel Strid corrected our English.
Footnotes
This work was supported by grants to Å.S. from the Swedish Natural Science Research Council, the Crafoord Foundation, the Carl Trygger Foundation, and the Swedish Research Council for Engineering Sciences and to M.B. from the Lawski Foundation, Hierta-Retzius fond för vetenskaplig forskning, and from Kungliga och Hvitfeldtska stipendiestiftelsen.
LITERATURE CITED
- Belknap WR, Garbarino JE. The role of ubiquitin in plant senescence and stress response. Trends Plant Sci. 1996;1:331–335. [Google Scholar]
- Britt AB, Chen JJ, Wykoff D, Mitchell D. A UV-sensitive mutant of Arabidopsis defective in the repair of pyrimidine-pyrimidinone (6–4) dimers. Science. 1993;264:1571–1574. doi: 10.1126/science.8372351. [DOI] [PubMed] [Google Scholar]
- Brown T. Southern blotting. In: Ausubel FM, Brent R, Kingston RE, Moore D, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. New York: John Wiley & Son; 1993. pp. 2.9.1–2.9.15. [Google Scholar]
- Caldwell MM. Solar UV irradiation and the growth and development of higher plants. In: Giese AC, editor. Current Topics in Photobiology and Photochemistry. 4: Photophysiology. New York: Academic Press; 1971. pp. 131–177. [Google Scholar]
- Chow WS, Strid Å, Anderson JM. Short-term treatment of pea plants with supplementary ultraviolet-B radiation: recovery time-courses of some photosynthetic functions and components. In: Murata N, editor. Research in Photosynthesis. IV. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1992. pp. 361–364. [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coruzzi G, Broglie R, Cashmore A, Chua N-H. Nucleotide sequences of two pea cDNA clones encoding the small subunit of ribulose 1,5-bisphosphate carboxylase and the major chlorophyll a/b-binding thylakoid polypeptide. J Biol Chem. 1983;258:1399–1402. [PubMed] [Google Scholar]
- DeLong A, Caldero-Urrea A, Dellaporta S. Sex determination gene TASSELSEED2 of maize encodes a short-chain alcohol dehydrogenase required for stage specific floral organ abortion. Cell. 1993;74:757–768. doi: 10.1016/0092-8674(93)90522-r. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- Ghosh D, Weeks CM, Grochulski P, Duax WL, Erman M, Rimsay RL, Orr JC. Three-dimensional structure of holo 3α,20β-hydroxysteroid dehydrogenase: a member of a short-chain dehydrogenase family. Proc Natl Acad Sci USA. 1991;88:10064–10068. doi: 10.1073/pnas.88.22.10064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman M. Ribonuclease protection assay. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1993. pp. 4.7.1–4.7.2. [Google Scholar]
- Green AES, Sawada T, Shettle EP. The middle ultraviolet reaching the ground. Photochem Photobiol. 1974;19:251–259. [Google Scholar]
- Henkow L, Strid Å, Berglund T, Rydström J, Ohlsson AB. Alteration of gene expression in Pisum sativum tissue cultures caused by the free radical-generating agent 2, 2′-azobis (2-amidinopropane) dihydrochloride. Physiol Plant. 1996;96:6–12. [Google Scholar]
- Hoagland DR, Arnon DI. California Agricultural Experiment Station Circular 347. The College of Agriculture, University of California, Berkeley; 1950. The water-culture method for growing plants without soil. [Google Scholar]
- Hunt AG. Messenger RNA 3′ end formation in plants. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:47–60. [Google Scholar]
- Ichinose Y, Kawamata S, Yamada T, An CC, Kajiwara T, Shiraishi T, Oku H. Molecular cloning of chalcone synthase cDNAs from Pisum sativum. Plant Mol Biol. 1992;18:1009–1012. doi: 10.1007/BF00019221. [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Olszewski NE. Gibberellins regulate the abundance of RNAs with sequence similarity to proteinase inhibitors, dioxygenases and dehydrogenases. Planta. 1996;198:78–86. doi: 10.1007/BF00197589. [DOI] [PubMed] [Google Scholar]
- Jordan BR, Chow WS, Strid Å, Anderson JM. Reduction in cab and psbA RNA transcripts in response to supplementary ultraviolet-B radiation. FEBS Lett. 1991;284:5–8. doi: 10.1016/0014-5793(91)80748-r. [DOI] [PubMed] [Google Scholar]
- Jordan BR, He J, Chow WS, Anderson JM. Changes in mRNA levels and polypeptide subunits of ribulose 1,5-bisphosphate carboxylase in response to supplementary ultraviolet-B radiation. Plant Cell Environ. 1992;15:91–98. [Google Scholar]
- Jordan BR, James PE, Strid Å, Anthony RG. The effect of ultraviolet-B radiation on gene expression and pigment composition in etiolated and green pea leaf tissue: UV-B-induced changes are gene-specific and dependent upon the developmental stage. Plant Cell Environ. 1994;17:45–54. [Google Scholar]
- Kalbin G, Ohlsson AB, Berglund T, Rydström J, Strid Å. Ultraviolet-B-radiation-induced changes in nicotinamide and glutathione metabolism and gene expression in plants. Eur J Biochem. 1997;249:465–472. doi: 10.1111/j.1432-1033.1997.00465.x. [DOI] [PubMed] [Google Scholar]
- Kawamata S, Yamada T, Tanaka Y, Sriprasertsak P, Kato H, Ichinose Y, Kato H, Shiraishi T, Oku H. Molecular cloning of phenylalanine ammonia-lyase cDNA from Pisum sativum. Plant Mol Biol. 1992;20:167–170. doi: 10.1007/BF00029163. [DOI] [PubMed] [Google Scholar]
- Li J, Chory J. Brassinosteroid action in plants. J Exp Bot. 1999;50:275–282. [Google Scholar]
- Li J, Ou-Lee TM, Raba R, Amundson RG, Last RL. Arabidopsis flavonoid mutants are hypersensitive to UV-B irradiation. Plant Cell. 1993;5:171–179. doi: 10.1105/tpc.5.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- Mackerness SAH, Surplus SL, Jordan BR, Thomas B. Effects of supplementary ultraviolet-B radiation on photosynthetic transcripts at different stages of leaf development and light levels in pea (Pisum sativum L.): role of active oxygen species and antioxidant enzymes. Photochem Photobiol. 1998;68:88–96. [Google Scholar]
- Mackerness SAH, Thomas B, Jordan BR. The effect of supplementary ultraviolet-B radiation on mRNA transcripts, translation and stability of chloroplast proteins and pigment formation in Pisum sativum L. J Exp Bot. 1997;48:729–738. [Google Scholar]
- Marekov L, Krook M, Jörnvall H. Prokaryotic 20-β-hydroxysteroid dehydrogenase is an enzyme of the 'short-chain, non-metalloenzyme' alcohol dehydrogenase type. FEBS Lett. 1990;266:51–54. doi: 10.1016/0014-5793(90)81504-h. [DOI] [PubMed] [Google Scholar]
- Persson B, Krook M, Jörnvall H. Characteristics of short-chain alcohol dehydrogenases and related enzymes. Eur J Biochem. 1991;200:537–543. doi: 10.1111/j.1432-1033.1991.tb16215.x. [DOI] [PubMed] [Google Scholar]
- Persson H, Majdi H. Effects of acid deposition on tree roots in Swedish forest stands. Water Air Soil Pollut. 1995;85:1287–1292. [Google Scholar]
- Rao MV, Ormrod DP. Ozone exposure decreases UV-B sensitivity in a UV-B-sensitive flavonoid mutant of Arabidopsis. Photochem Photobiol. 1995;61:71–78. doi: 10.1111/j.1751-1097.1995.tb09245.x. [DOI] [PubMed] [Google Scholar]
- Richards KD, Schott EJ, Sharma YK, Davis KR, Gardner RC. Aluminum induces oxidative stress genes in Arabidopsis thaliana. Plant Physiol. 1998;116:409–418. doi: 10.1104/pp.116.1.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals JA, Neuenschwander UH, Willits MG, Molina A, Steiner HY, Hunt MD. Systemic acquired resistance. Plant Cell. 1996;8:1809–1819. doi: 10.1105/tpc.8.10.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnoff N. The function and metabolism of ascorbic acid in plants. Ann Bot. 1996;78:661–669. [Google Scholar]
- Strid Å. Increased expression of defence genes in Pisum sativum after exposure to supplementary ultraviolet-B radiation. Plant Cell Physiol. 1993;34:949–953. [Google Scholar]
- Strid Å, Chow WS, Anderson JM. Effects of supplementary ultraviolet-B radiation on photosynthesis in Pisum sativum. Biochim Biophys Acta. 1990;1020:260–268. [Google Scholar]
- Strid Å, Chow WS, Anderson JM. UV-B damage and protection at the molecular level in plants. Photosynth Res. 1994;39:475–489. doi: 10.1007/BF00014600. [DOI] [PubMed] [Google Scholar]
- Strid Å, Chow WS, Anderson JM. Changes in the relaxation of electrochromic shifts of photosynthetic pigments and in the levels of mRNA transcripts in leaves of Pisum sativum as a result of exposure to supplementary UV-B radiation. The dependency on the intensity of the photosynthetically active radiation. Plant Cell Physiol. 1996a;37:61–67. [Google Scholar]
- Strid Å, Chow WS, Anderson JM. Temperature-dependency of changes in the relaxation of electrochromic shifts, of chlorophyll fluorescence, and in the levels of mRNA transcripts in detached leaves from Pisum sativum exposed to supplementary UV-B radiation. Plant Sci. 1996b;115:199–206. [Google Scholar]
- Strid Å, Porra RJ. Alterations in pigment content in leaves of Pisum sativum after exposure to supplementary UV-B. Plant Cell Physiol. 1992;32:1015–1023. [Google Scholar]
- Thipyapong P, Steffens JC. Tomato polyphenol oxidase: differential response of the polyphenol oxidase F promoter to injuries and wound signals. Plant Physiol. 1997;115:409–418. doi: 10.1104/pp.115.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willekens H, Van Camp W, Van Montagu M, Inzé D, Langebartels C, Sandermann H. Ozone, sulfur dioxide, and ultraviolet B have similar effects on mRNA accumulation of antioxidant genes in Nicotiana plumbaginifolia L. Plant Physiol. 1994;106:1007–1014. doi: 10.1104/pp.106.3.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Hu X, Henkow L, Jordan BR, Strid Å. The effects of ultraviolet-B radiation on the CFoF1-ATPase. Biochim Biophys Acta. 1994;1185:295–302. [Google Scholar]
- Zhu J-K, Hasegawa PM, Bressan RA. Molecular aspects of osmotic stress in plants. Crit Rev Plant Sci. 1997;16:253–277. [Google Scholar]