ABSTRACT
Enterococcus faecalis is paradoxically a dangerous nosocomial pathogen and a normal constituent of the human gut microbiome, an environment rich in ethanolamine. E. faecalis carries the eut (ethanolamine utilization) genes, which enable the catabolism of ethanolamine (EA) as a valuable source of carbon and/or nitrogen. EA catabolism was previously shown to contribute to the colonization and growth of enteric pathogens, such as Salmonella enterica serovar Typhimurium and enterohemorrhagic Escherichia coli (EHEC), in the gut environment. We tested the ability of eut mutants of E. faecalis to colonize the gut using a murine model of gastrointestinal (GI) tract competition and report the surprising observation that these mutants outcompete the wild-type strain.
KEYWORDS: Enterococcus, ethanolamine, intestinal colonization
IMPORTANCE
Some bacteria that are normal, harmless colonizers of the human body can cause disease in immunocompromised patients, particularly those that have been heavily treated with antibiotics. Therefore, it is important to understand the factors that promote or negate these organisms’ ability to colonize. Previously, ethanolamine, found in high concentrations in the GI tract, was shown to promote the colonization and growth of bacteria associated with food poisoning. Here, we report the surprising, opposite effect of ethanolamine utilization on the commensal colonizer E. faecalis, namely, that loss of this metabolic capacity made it a better colonizer.
OBSERVATION
Ethanolamine (EA) is a compound found in the gastrointestinal (GI) tract at concentrations of 1 to 2 mM (1, 2). Interestingly, the genes that code for the catabolism of this compound, the eut (ethanolamine utilization) genes, are associated with gut pathogens, including species of Escherichia, Salmonella, Clostridium, and Listeria (3). In species such as Salmonella enterica serovar Typhimurium and enterohemorrhagic Escherichia coli (EHEC), mutants lacking the ability to sense and/or catabolize EA are outcompeted by wild-type strains in the gastrointestinal tract (1, 2, 4) or display neutral colonization efficacy in the case of Clostridium difficile (5).
Enterococcus faecalis also encodes the eut genes and is found in the GI tract, but unlike the above-mentioned examples, it is considered a commensal colonizer rather than a gut pathogen of healthy people. However, the presence of E. faecalis in the GI tract can serve as a source of nosocomial infection (6). Therefore, understanding the factors that promote the colonization and growth of E. faecalis in the gut is important for the development of strategies to mitigate these infections.
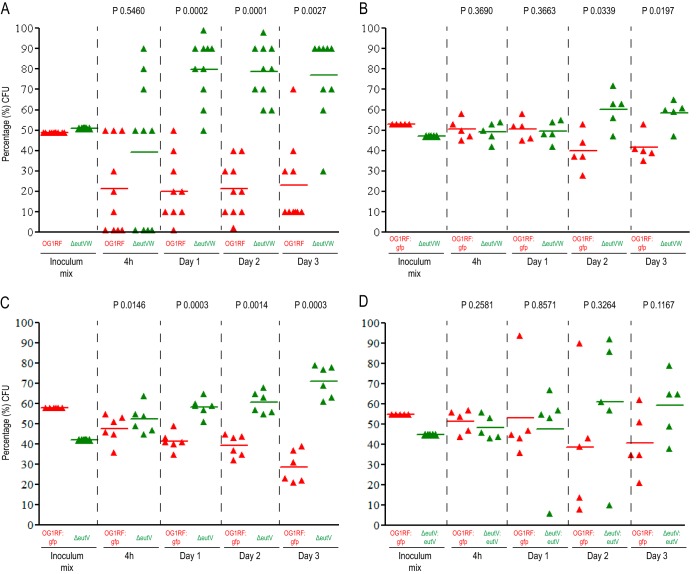
To investigate the role of EA in GI tract colonization, we first tested OG1RF, a wild-type strain of E. faecalis commonly used in animal models (7), and an isogenic ΔeutVW mutant that lacks the two-component system that senses EA (8). EutW is a sensor histidine kinase that, upon binding EA, autophosphorylates and then phosphotransfers to EutV the cognate RNA binding response regulator that activates eut gene expression by an antitermination mechanism (8, 9). The ΔeutVW mutant does not express the eut genes and cannot utilize EA (8, 9). The strains were competed in a murine GI tract model in which the animals were pretreated with an antibiotic cocktail designed to reduce the endogenous flora and facilitate E. faecalis colonization (10). To measure levels of colonization, fecal pellets were collected at 1, 2, and 3 days postinoculation and plated for numbers of CFU on medium selective for enterococci. Fecal pellets were also collected at 4 h to check for a spike in CFU, indicating a failure to colonize. Ten colonies/mouse/time point were screened by PCR for the presence of the deletion that marked the ΔeutVW mutant strain. As shown in Fig. 1A, the ΔeutVW mutant significantly outcompeted the wild type at all colonization time points.
FIG 1 .
Competitive colonization of the gastrointestinal tract by the E. faecalis wild type and mutant strains following combined inoculation. Percentages of CFU of E. faecalis strains from the initial inoculum mix and from stool samples collected 4 h and 1, 2, and 3 days after mixed inoculation of E. faecalis OG1RF and the ΔeutVW mutant (AR2) (A), OG1RF::gfp and the ΔeutVW mutant (AR2) (B), OG1RF::gfp and the ΔeutV mutant (SD54) (C), and OG1RF::gfp and the ΔeutV::PeutS eutV mutant (SD209) (D). In panel A, 10 CFU per mouse per time point were randomly picked and PCR screened to confirm the strains’ identities. For panels B, C, and D, >100 CFU were scored for GFP fluorescence per mouse per time point. An unpaired t test with Welch’s correction for the percentages of bacteria recovered from the stool samples versus the amounts in the initial inoculum mixture was used to calculate the P values.
Because screening the colonies by PCR was labor-intensive, we decided to repeat the experiment using a previously generated marked strain of OG1RF that constitutively expresses gfp present as a chromosomal insertion (11). By exposing the plated colonies from the fecal pellets to a fluorescent stereoscope, the presence of green fluorescent protein (GFP) could easily be discerned, allowing hundreds of colonies from each mouse to be screened. To ensure that GFP expression did not deleteriously affect the fitness of our wild-type strain, it was competed against the parent strain lacking the marker; a significant difference was not observed (see Fig. S1 in the supplemental material). We repeated the competition experiment with the ΔeutVW mutant versus the wild type and again observed the mutant outcompeting the wild-type strain (Fig. 1B). Note that the inoculum of the ΔeutVW mutant was purposely kept lower than that of the wild type so as not to create a bias toward the mutant, since the first experiment indicated that it was more fit.
Colonization of the gastrointestinal tract by OG1RF versus OG1RF::gfp following mixed inoculation. Numbers of CFU (A) and calculated percentages of CFU (B) of E. faecalis strains from stool samples collected at 4 h and 1, 2, and 3 days after combined inoculation. More than 100 CFU were scored for GFP fluorescence per mouse per time point. An unpaired t test with Welch’s correction for the percentages of bacteria recovered from the stool samples versus the amounts in the initial inoculum mixture was used to calculate the P values. Download FIG S1, EPS file, 0.7 MB (734.4KB, eps) .
Copyright © 2018 Kaval et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Considering that EA utilization mutants in other studied bacterial species tended to be less fit in the GI tract, our results showing that an E. faecalis mutant was modestly more fit were surprising. To confirm the finding, we tested a different mutant containing an in-frame deletion of only eutV. Also examined was a complement of this strain, able to induce eut gene expression to levels just slightly lower than those induced by the wild type (11). As shown in Fig. 1C, the ΔeutV mutant outcompeted the wild type at all time points examined, in contrast to the complemented strain (Fig. 1D).
The eut genes in EHEC and S. Typhimuirum are regulated by a mechanism different from that found in E. faecalis. These bacteria encode an EA-sensing transcriptional activator called EutR (12). In addition to the eut genes, EutR was found to bind promoters and directly activate the expression of some virulence factors in these pathogens (13, 14). Because of this additional activity, loss of the regulator resulted in a more severe phenotype than loss of just ethanolamine catabolism, depending on the specifics of the host environment (4). We wondered whether the phenotype observed with the loss of EutV was related solely to its role in regulating genes related to EA catabolism or whether it, like EutR, perhaps had a broader role.
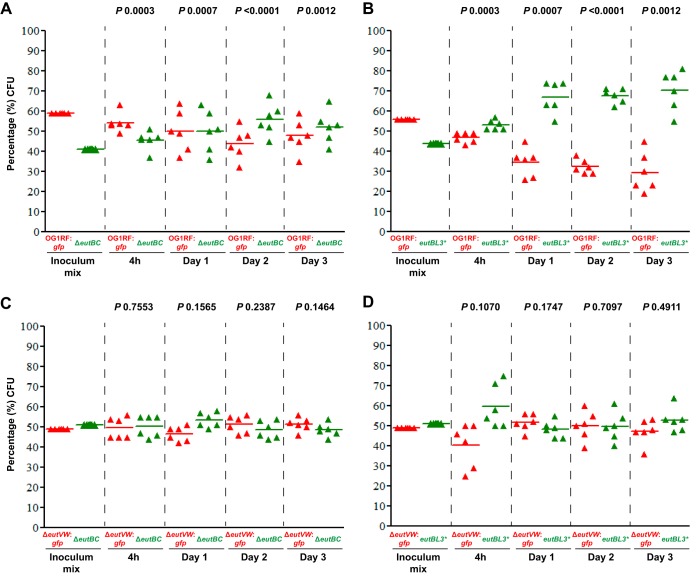
To address this question, we created an in-frame deletion, ΔeutBC, to remove the genes encoding the two subunits of the ethanolamine ammonia lyase, which carries out the first reaction in the breakdown of EA. Many attempts to generate a complement of this mutant failed. Therefore, we created a different, independent mutant by generating a stop codon in eutB, called eutBL3*. Neither the in-frame deletion nor the stop mutant disrupted expression of the downstream eut structural genes, as bacterial microcompartment formation was still observed in both mutants (Fig. S2). Both mutants significantly outcompeted the wild type on days 2 and 3, with the stop codon mutant also displaying significant differences on day 1 (Fig. 2A and B). To test whether loss of EA sensing resulted in a greater fitness increase than loss of EA catabolism, we competed a ΔeutVW strain marked with GFP against the ΔeutBC strains (Fig. 2C and D). A significant difference was not observed for either strain pair at any time point. The data support the conclusion that EA catabolism alone contributes to the phenotype. These data fit former observations that the regulatory sequences recognized by E. faecalis EutV are found only in the eut transcripts (9).
FIG 2 .
Murine gastrointestinal tract colonization by different E. faecalis strains following mixed inoculation. Percentages of CFU of E. faecalis strains from the initial inoculum mix and from stool samples collected 4 h and 1, 2, and 3 days after mixed inoculation of E. faecalis OG1RF::gfp (SD234) with the ΔeutBC strain (EFKK4) (A), OG1RF::gfp with the eutB mutant (eutBL3*) (EFKK12) (B), the ΔeutVW::gfp mutant (EFKK1) with the ΔeutBC mutant (C), and the ΔeutVW::gfp mutant with the eutB mutant (eutBL3*) (D). More than 100 CFU were scored for GFP fluorescence per mouse per time point. P values were calculated using an unpaired t test with Welch’s correction for the percentages of bacteria recovered from the fecal samples versus the amounts in the initial mixed inoculum.
Bacterial microcompartment formation in E. faecalis mutant strains. Transmission electron micrographs showing the formation of bacterial microcompartments (indicated by white arrows) in E. faecalis OG1RF (wild type), the ΔeutBC mutant (EFKK4), and the eutB mutant (eutBL3*) (EFKK12), when grown under inducing conditions. Download FIG S2, EPS file, 3.5 MB (3.6MB, eps) .
Copyright © 2018 Kaval et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplemental materials and methods. Download TEXT S1, DOCX file, 0.03 MB (31.9KB, docx) .
Copyright © 2018 Kaval et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In conclusion, we present the surprising observation that EA catabolism in E. faecalis modestly reduces GI tract colonization efficiency, in contrast to that observed for three gut pathogens (1, 2, 4, 5). The difference might arise from lifestyle, as E. faecalis is a normal gut commensal in mammals; in Caenorhabditis elegans, which E. faecalis kills, a eut mutant was attenuated (15). A deeper understanding awaits further investigation.
ACKNOWLEDGMENTS
This work was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award R01AI110432 to D.A.G. and W.C.W.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Citation Kaval KG, Singh KV, Cruz MR, DebRoy S, Winkler WC, Murray BE, Garsin DA. 2018. Loss of ethanolamine utilization in Enterococcus faecalis increases gastrointestinal tract colonization. mBio 9:e00790-18. https://doi.org/10.1128/mBio.00790-18.
REFERENCES
- 1.Bertin Y, Girardeau JP, Chaucheyras-Durand F, Lyan B, Pujos-Guillot E, Harel J, Martin C. 2011. Enterohaemorrhagic Escherichia coli gains a competitive advantage by using ethanolamine as a nitrogen source in the bovine intestinal content. Environ Microbiol 13:365–377. doi: 10.1111/j.1462-2920.2010.02334.x. [DOI] [PubMed] [Google Scholar]
- 2.Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, Sterzenbach T, Tsolis RM, Roth JR, Bäumler AJ. 2011. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci U S A 108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsoy O, Ravcheev D, Mushegian A. 2009. Comparative genomics of ethanolamine utilization. J Bacteriol 191:7157–7164. doi: 10.1128/JB.00838-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson CJ, Clark DE, Adli M, Kendall MM. 2015. Ethanolamine signaling promotes Salmonella Niche recognition and adaptation during infection. PLoS Pathog 11:e1005278. doi: 10.1371/journal.ppat.1005278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nawrocki KL, Wetzel D, Jones JB, Woods EC, McBride SM. 2018. Ethanolamine is a valuable nutrient source that impacts Clostridium difficile pathogenesis. Environ Microbiol 20:1419–1435. doi: 10.1111/1462-2920.14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lebreton F, Willems RJL, Gilmore MS. 2014. Enterococcus diversity, origins in nature, and gut colonization. In Gilmore MS, Clewell DB, Ike Y, Shankar N (ed), Enterococci: from commensals to leading causes of drug resistant infection. Massachusetts Eye and Ear Infirmary, Boston, MA. [PubMed] [Google Scholar]
- 7.Dunny GM, Brown BL, Clewell DB. 1978. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc Natl Acad Sci U S A 75:3479–3483. doi: 10.1073/pnas.75.7.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fox KA, Ramesh A, Stearns JE, Bourgogne A, Reyes-Jara A, Winkler WC, Garsin DA. 2009. Multiple posttranscriptional regulatory mechanisms partner to control ethanolamine utilization in Enterococcus faecalis. Proc Natl Acad Sci U S A 106:4435–4440. doi: 10.1073/pnas.0812194106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramesh A, DebRoy S, Goodson JR, Fox KA, Faz H, Garsin DA, Winkler WC. 2012. The mechanism for RNA recognition by ANTAR regulators of gene expression. PLoS Genet 8:e1002666. doi: 10.1371/journal.pgen.1002666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montealegre MC, Singh KV, Murray BE. 2016. Gastrointestinal tract colonization dynamics by different Enterococcus faecium clades. J Infect Dis 213:1914–1922. doi: 10.1093/infdis/jiv597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Debroy S, van der Hoeven R, Singh KV, Gao P, Harvey BR, Murray BE, Garsin DA. 2012. Development of a genomic site for gene integration and expression in Enterococcus faecalis. J Microbiol Methods 90:1–8. doi: 10.1016/j.mimet.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaval KG, Garsin DA. 2018. Ethanolamine utilization in bacteria. mBio 9:e00066-18. doi: 10.1128/mBio.00066-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kendall MM, Gruber CC, Parker CT, Sperandio V. 2012. Ethanolamine controls expression of genes encoding components involved in interkingdom signaling and virulence in enterohemorrhagic Escherichia coli O157:H7. mBio 3:e00050-12. doi: 10.1128/mBio.00050-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luzader DH, Clark DE, Gonyar LA, Kendall MM. 2013. EutR is a direct regulator of genes that contribute to metabolism and virulence in enterohemorrhagic Escherichia coli O157:H7. J Bacteriol 195:4947–4953. doi: 10.1128/JB.00937-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maadani A, Fox KA, Mylonakis E, Garsin DA. 2007. Enterococcus faecalis mutations affecting virulence in the Caenorhabditis elegans model host. Infect Immun 75:2634–2637. doi: 10.1128/IAI.01372-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Colonization of the gastrointestinal tract by OG1RF versus OG1RF::gfp following mixed inoculation. Numbers of CFU (A) and calculated percentages of CFU (B) of E. faecalis strains from stool samples collected at 4 h and 1, 2, and 3 days after combined inoculation. More than 100 CFU were scored for GFP fluorescence per mouse per time point. An unpaired t test with Welch’s correction for the percentages of bacteria recovered from the stool samples versus the amounts in the initial inoculum mixture was used to calculate the P values. Download FIG S1, EPS file, 0.7 MB (734.4KB, eps) .
Copyright © 2018 Kaval et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bacterial microcompartment formation in E. faecalis mutant strains. Transmission electron micrographs showing the formation of bacterial microcompartments (indicated by white arrows) in E. faecalis OG1RF (wild type), the ΔeutBC mutant (EFKK4), and the eutB mutant (eutBL3*) (EFKK12), when grown under inducing conditions. Download FIG S2, EPS file, 3.5 MB (3.6MB, eps) .
Copyright © 2018 Kaval et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplemental materials and methods. Download TEXT S1, DOCX file, 0.03 MB (31.9KB, docx) .
Copyright © 2018 Kaval et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.